Abstract
Phosphorylation of viral proteins serves as a regulatory mechanism during the intracellular life cycle of infected viruses. There is therefore a pressing need to develop a method to efficiently purify and enrich phosphopeptides derived from viral particles in biological samples. In this study, we utilized Phos-tag technology to analyze the functional phosphorylation of the nucleocapsid protein (N protein; NP) of severe respiratory syndrome coronavirus 2 (SARS-CoV-2). Viral particles were collected from culture supernatants of SARS-CoV-2-infected VeroE6/TMPRSS2 cells by ultracentrifugation, and phosphopeptides were purified by Phos-tag magnetic beads for LC-MS/MS analysis. Analysis revealed that NP was reproducibly phosphorylated at serine 79 (Ser79). Multiple sequence alignment and phylogenetic analysis showed that the Ser79 was a distinct phospho-acceptor site in SARS-CoV-2 but not in other beta-coronaviruses. We also found that the prolyl-isomerase Pin1 bound to the phosphorylated Ser79 in NP and positively regulated the production of viral particles. These results suggest that SARS-CoV-2 may have acquired the potent virus-host interaction during its evolution mediated by viral protein phosphorylation. Moreover, Phos-tag technology can provide a useful means for analyzing the functional phosphorylation of viral proteins.
Significance
In this study, we aimed to investigate the functional phosphorylation of SARS-CoV-2 NP. For this purpose, we used Phos-tag technology to purify and enrich virus-derived phosphopeptides with high selectivity and reproducibility. This method can be particularly useful in analyzing viral phosphopeptides from cell culture supernatants that often contain high concentrations of fetal bovine serum and supplements. We newly identified an NP phosphorylation site at Ser79, which is important for Pin1 binding. Furthermore, we showed that the interaction between Pin1 and phosphorylated NP could enhance viral replication in a cell culture model.
Keywords: Phos-tag, Phosphorylation, SARS-CoV-2, Pin1
Graphical abstract
1. Introduction
The molecular link between virus and host cells is vital to the efficient and persistent replication of all types of viruses [[1], [2], [3]]. The phosphorylation of viral components by host protein kinases can serve as a dynamic way to regulate their activity, subcellular localization, and stability as part of the process of elaborate virus-host interactions. Moreover, the phosphorylation of viral proteins may alter their interactions with cellular components and thereby affect cellular functions and pathogenicity associated with viral infection [[1], [2], [3]]. Phosphorylation has been shown to be one of the most effective post-translational modifications in terms of impacting protein function [4], and can affect the ability of infected cells to generate progeny viruses.
Dramatic advances in LC-MS/MS technology have enabled high-throughput, comprehensive analysis of proteins and peptides. On the other hand, the peptide enrichment step is prerequisite for efficient measurement of phosphoproteins (phosphopeptides), especially those with low abundance. These conditions often result in inefficient peptide ionization on LC-MS/MS analysis [5,6]. In phospho-proteomics, the most common method for phosphopeptide enrichment is based on the affinity purification of phosphate groups with metal ions such as Fe (III), Ga (III), Ni (II), or Zn (II) [7,8]. Recently developed Phos-tag ligands are alkoxide-bridged dinuclear metal complexes that can selectively capture phosphate groups in aqueous solutions at neutral pH [9,10]. Phos-tag beads conjugated with zinc ions have been used for enrichment of phosphopeptides as a pretreatment method for LC-MS/MS analysis [11,12]. Hence, Phos-tag-based methods enable rapid, highly selective, and reproducible phosphopeptide enrichment [13,14].
In the present study, we utilized Phos-tag technology to analyze the functional phosphorylation of SARS-CoV-2 NP. We demonstrated that phosphorylation at Ser79 of NP was crucial for viral interaction with host protein Pin1 and for efficient viral replication. Interestingly, Ser79 was a distinct phosphorylation site in SARS-CoV-2 but not in other coronaviruses.
2. Materials and methods
2.1. Cell culture
VeroE6/TMPRSS2 cells were cultured at 37 °C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). HEK 293A cells were cultured at 37 °C in 5% CO2 in DMEM supplemented with 10% FBS.
2.2. Viral infection
SARS-CoV-2 strain (JPN/TY/WK-521) was provided by the National Institute of Infectious Diseases (Japan), and virus titers were determined by a standard plaque assay. VeroE6/TMPRSS2 cells were infected with SARS-CoV-2 for 72 h at a multiplicity of infection (MOI) of 0.05 at room temperature. Infected cells were washed with PBS and then lysed. Lysates were rotated for 30 min and centrifuged at 15,000 rpm at 4 °C. The supernatant was treated or not treated with calf intestine alkaline phosphatase (CIAP), and then used for Phos-tag SDS-PAGE and SDS-PAGE. The culture medium was collected in a conical tube and centrifuged at 17,700 xg for isolation of viral particles. Viral particles were washed with PBS and dissolved in urea buffer containing 8 M urea, 50 mM NH4HCO3, and 10 mM DTT, followed by heating at 60 °C for 30 min. The resulting viral protein solution was used for phosphopeptide enrichment using Phos-tag magnetic beads and Pin1 detection by immunoblotting analysis with anti-Pin1 antibody (R&D Systems, Minneapolis, Minnesota, USA).
2.3. Phos-tag SDS-PAGE
A homemade gel for Phos-tag SDS-PAGE was composed of 7.5%T (total acrylamide), 1%C (bis-acrylamide for cross-linking), 25 μM Phos-tag acrylamide (Wako Pure Chemical Industries, Ltd., Osaka, Japan), and 50 μM MnCl2. SDS-PAGE was performed using SuperSep Ace 5–20% gradient gel (Wako Pure Chemical Industries, Ltd.). The running buffer for electrophoresis was composed of 25 mM Tris, 192 mM glycine, and 0.1% SDS. Whole-cell lysates were diluted with SDS sample buffer and heated for 5 min at 95 °C. After electrophoresis, the gels were soaked in running buffer containing 1 mM EDTA for 20 min to chelate Mn2+ coordinated to Phos-tag in the gel, and washed twice with running buffer for 10 min. The separated proteins in the gel were electroblotted onto a polyvinylidene difluoride membrane using a Trans-Blot Turbo Transfer system (Bio Rad, Hercules, CA, USA), and detected by immunoblotting analysis with anti-NP antibody. Proteins derived from host cells within the viral particles were subjected to trypsin digestion by SDS-PAGE followed by in-gel digestion and identification by LC-MS/MS analysis. We performed a parallel experiment using culture supernatants of uninfected cells to exclude cellular contaminants that were subtracted from suspected viral proteins.
2.4. Phosphopeptide enrichment using Phos-tag magnetic beads
Tryptic digestion and peptide desalting of viral particle lysates were performed according to our previous report [15], and desalted peptides were used for phosphopeptide enrichment. The experimental procedure and solution composition for phosphopeptide enrichment are depicted in Supplemental Fig. S1. In brief, Phos-tag magnetic beads were provided by NARD Institute, Ltd., and the beads were activated with a solution containing zinc acetate and then washed. Unbound peptides (non-phosphopeptides) were removed after incubation of the beads with desalted peptides dissolved in wash solution. After washing the beads, phosphopeptides were eluted with sodium phosphate solution. Enriched phosphopeptides were desalted and subjected to LC-MS/MS analysis.
2.5. LC-MS/MS analysis and protein identification
Phosphopeptides were analyzed using an Orbitrap Q-Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) coupled with a Dionex Ultimate 3000 RSLC nano system (U3000, Thermo Fisher Scientific). The analytical conditions were as follows: Nano HPLC capillary column (75 μm × 180 mm, C18; Nikkyo Technos Co., Ltd., Tokyo, Japan); temperature = room temperature (22 ± 1.5 °C); mobile phase = (A) 0.1% formic acid and 2% ACN, (B) 0.1% formic acid and 95% ACN; gradient = 2–41% mobile phase B for 45 min; flow rate = 300 nL/min; full-mass scan range = m/z 300–1500; full-mass resolution = 70,000; normalized collision energy = 27; isolation windows = 1.6 m/z. The top 20 most intense ions in the full-mass scan were selected for MSMS analysis. MS raw files were converted to mgf files by Proteome Discoverer software (version 2.2; Thermo Fisher Scientific). Protein identifications were performed using the MASCOT search engine (version 2.7.0; Matrix Science, London, United Kingdom) with the following parameters: database, SARS-CoV-2 amino acid sequences downloaded from the UniProtKD (2021.04); enzyme, trypsin; peptide mass tolerance, ± 5 ppm; fragment mass tolerance, ± 0.05 Da; maximum missed cleavages, 2; variable modifications, acetyl (Protein N-term), carbamidomethyl (C), carbamyl (N-term), oxidation (M), phospho (ST), phospho (Y). For the phosphopeptide identification, a significance threshold of p < 0.05 and a peptide score ≥ 25 were adopted as the acceptance criteria. Peptides from in-gel digestion samples were analyzed using an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) coupled with a U3000. Protein identification was performed with the following parameters; database, Macaca fascicularis amino acid sequences downloaded from the UniProtKD (2020.01); enzyme, trypsin; peptide mass tolerance, ± 5 ppm; fragment mass tolerance, ± 0.5 Da; maximum missed cleavages, 2; variable modification, propionamide (C), oxidation (M). For the protein identification, peptides with a false discovery rate of less than 1% were used. Proteomics data were deposited in the ProteomeXchange Consortium (PXD027939, http://www.proteomexchange.org/) via the jPOST partner repository (JPST001292, https://jpostdb.org/). Amino acid sequence alignment and phylogenetic tree construction were conducted to confirm homology by GENETYX software (GENETYX CO., Tokyo, Japan). The BioGRID website tool (https://thebiogrid.org/) [16] was used to search for previously reported NP-interacting proteins.
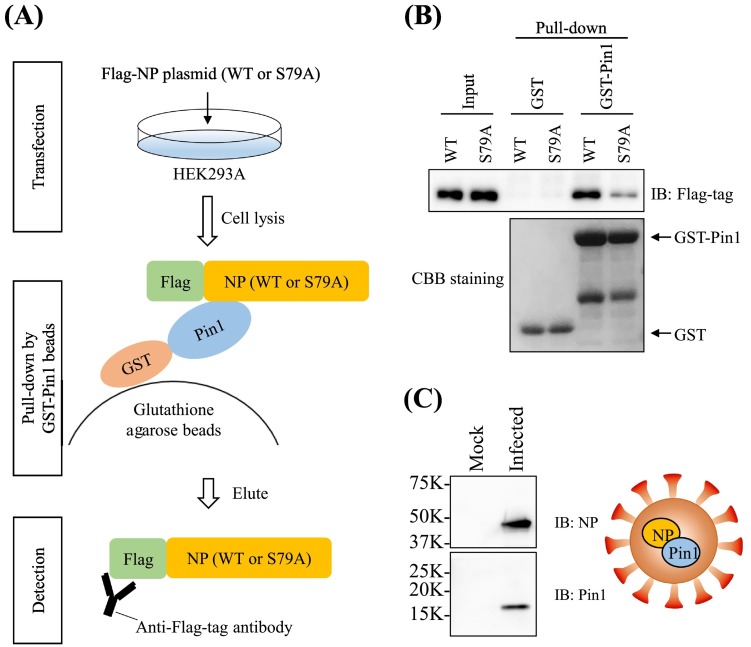
2.6. Pull-down using GST-tagged Pin1
The open reading frame of SARS-CoV-2 NP was cloned into pcDNA 3.1 plasmids with an N-terminal Flag-tag. The codon for Ser79 was substituted with GCC (alanine) by inverse PCR to generate a plasmid that expressed a non-phosphorylatable mutant of NP (S79A). The construct was verified by restriction enzyme digestion and DNA sequencing. Plasmids were transfected into HEK 293A cells using Effectene Transfection Reagent (QIAGEN, Hilden, Germany). Pull-down using a GST-tagged Pin1 assay was performed according to a previous report [17]. In brief, harvested cells were lysed in pull-down buffer containing 50 mM Hepes (pH 7.4), 200 mM NaCl, 10% glycerol, 1% TritonX-100, 1 mM EDTA, and 1.5 mM MgCl2. Cell lysates were added to glutathione agarose beads containing GST or GST-Pin1 and incubated at 4 °C for 4 h. Proteins bound to beads were washed with pull-down buffer twice, eluted with SDS sample buffer, and then used for NP detection by immunoblotting analysis.
2.7. Lentiviral shRNA knockdown of Pin1
Plasmids encoding shRNAs targeting Pin1 (TRCN0000001035 and TRCN0000010577) were obtained from Sigma-Aldrich (St Louis, MO, USA) and co-transfected with the packaging plasmid mix into 293 T-lenti-X cells using Lipofectamine 3000 (Thermo Fisher Scientific). The supernatants were filtered using 0.45-μm filters and added to VeroE6/TMPRSS2 cells, followed by selection using 2 μg/mL puromycin. Knockdowns of Pin1 were confirmed by immunoblotting analysis with anti-Pin1 antibody.
2.8. Quantification RT-PCR and virus titration
Pin1 knockdown cells and control cells were infected with SARS-CoV-2 at an MOI of 0.05, and supernatants were collected after 48 h. Total viral RNA was extracted from the supernatant using a QIAamp Viral RNA Mini Kit (QIAGEN). Quantification RT-PCR (qRT-PCR) was performed using TaqMan Fast 1-step master mix reagent (Thermo Fisher Scientific) with SARS-CoV-2-specific primers (5’-AAATTTTGGGGACCAGGAAC-3′, 5’-TGGCAGCTGTGTAGGTCAAC-3′, and 5’-FAM-ATGTCGCGCATTGGCATGGA-BHQ-3′) [18,19] and a CFX 96 Real-Time PCR system (Bio Rad). The amount of viral RNA was calculated by plotting the Ct value on a standard curve constructed based on the N2 standard product (Nihon Gene Research Laboratories, catalog number JP-NN2-PC). Viral titers were measured using VeroE6/TMPRSS2 cells in 96-well plates with four wells per viral dilution. Cells were inoculated with 100 μL of diluted virus (1:100–1:100,000,000 dilution). Three days after inoculation, the Cell-Titer Glo assay (Promega, Madison, WI, USA) was used to determine the cytopathic effects induced by virus infection. The infection percentage was calculated as follows:
The 50% tissue culture infectious dose (TCID50) value was calculated by Prism 8 software (GraphPad).
3. Results
3.1. Confirmation of SARS-CoV-2 NP phosphorylation using Phos-tag gel electrophoresis
SARS-CoV-2 NP is a most abundant viral structural protein that play an important role in viral particle formation. To investigate if SARS-CoV-2 NP underwent phosphorylation in cells, cell lysates from virus-infected VeroE6/TMPRSS2 cells were subjected to electrophoresis using a Phos-tag SDS-PAGE gel, and the band patterns were compared between cell lysates treated with and without calf intestinal alkaline phosphatase (CIAP) to dephosphorylate proteins. Several upshifted bands were observed, and they disappeared after CIAP treatment, indicating NP phosphorylation. Some upshifted bands remained even after CIAP treatment, suggesting incomplete dephosphorylation of NP. On the other hand, conventional SDS-PAGE exhibited no such difference between the samples (Fig. 1 ). These results indicate that NP is subjected to phosphorylation in infected cells, and that this phosphorylation can be detected by Phos-tag ligand.
Fig. 1.
Confirmation of the phosphorylation of NP in SARS-CoV-2. VeroE6/TMPRSS2 cells were infected with SARS-CoV-2 and the cells and culture medium were collected separately. Infected whole cells were used for Phos-tag SDS-PAGE, followed by immunoblotting with anti-NP antibody. Phosphorylated forms are observed as upshifted bands. Viral particles in culture medium were collected by centrifugation and used for phosphopeptide enrichment with Phos-tag magnetic beads (Supplemental Fig. S2).
Confirmation of the phosphorylation of NP in SARS-CoV-2. VeroE6/TMPRSS2 cells were infected with SARS-CoV-2 and the cells and culture medium were collected separately. Infected whole cells were used for Phos-tag SDS-PAGE, followed by immunoblotting with anti-NP antibody. Phosphorylated forms are observed as upshifted bands. Viral particles in culture medium were collected by centrifugation and used for phosphopeptide enrichment with Phos-tag magnetic beads (Supplemental Fig. S2).
3.2. Identification of phosphorylated sites in NP
To investigate the viral protein phosphorylation more precisely, viral particles were collected by centrifugation of the culture supernatant derived from SARS-CoV-2-infected VeroE6/TMPRSS2 cells. The phosphorylation of proteins within viral particles was confirmed by Phos-tag SDS-PAGE analysis, which showed several upshifted bands indicative of protein phosphorylation (Supplemental Fig. S2). We next attempted to identify the phosphorylated viral protein(s). The viral particles were extracted, digested by trypsin, and enriched using Phos-tag magnetic beads (Supplemental Fig. S1). Purified peptides were processed for LC-MS/MS analysis (Fig. 1). The MSMS data were searched against a SARS-CoV-2 amino acid sequence database downloaded from UniProtKD. We identified several peptide fragments derived from NP. Among them, one phosphorylated peptide was reproducibly identified. This phosphopeptide, GQGVPINTNSpSPDDQIGYYR (#66-#89), was phosphorylated at Ser79 (Fig. 2A and Supplemental Fig. S3).
Fig. 2.
Identification of phosphorylated sites in NP. (A) SARS-CoV-2 viral particles were used for phosphopeptide enrichment with Phos-tag magnetic beads, followed by LC-MS/MS analysis. The phosphorylation of Ser79 in NP was identified. (B) Phylogenetic tree analysis of NP confirmed that WIV1-CoV and SARS-CoV had high similarity to SARS-CoV-2. Subsequent multi-alignment analysis showed that Ser79 was a unique phosphor-acceptor Ser in SARS-CoV-2 only. The phosphorylated Ser79 preceded Pro80, and the two amino acids constitute a recognition/binding motif for the prolyl-isomerase Pin1 (pSer/Thr-Pro).
Phylogenetic tree analysis of NP confirmed that WIV1-CoV and SARS-CoV viruses had high similarity to SARS-CoV-2 as reported by previous study [20,21] (Fig. 2B). Subsequent multi-alignment analysis revealed that Ser79 was a unique phosphor-acceptor serine residue observed only in SARS-CoV-2 but not in other coronaviruses (Fig. 2B). Additionally, we performed multiple alignments of amino acid sequences around the Ser79-Pro80 of NP in variants of interest (VOIs) and variants of concern (VOCs) of SARS-CoV-2. Our results showed that the Pin1 target motif (Ser79-Pro80) was largely conserved among the variants, including the delta and omicron strains currently spreading worldwide. Interestingly, the gamma variant demonstrated a substitution of Pro80 to Arg (Supplemental Fig. S4), implying that Pin1 may not interact with NP from this variant.
3.3. Pin1 binds to the phosphorylated Ser79-Pro80 motif in NP
We found that the phosphorylated Ser79 preceded Pro80, constituting a binding motif for the prolyl-isomerase Pin1 (pSer/Thr-Pro). Although there are five other potential Pin1 recognition/binding motifs in the entire amino acid sequence of NP, only Ser79 is a distinct phosphor-acceptor site for SARS-CoV-2 (Supplemental Fig. S5). To confirm this hypothesis, we next used a GST pull-down assay to determine if Pin1 could directly interact with NP. Our results showed that NP did bind to GST-Pin1, but not to GST only (Fig. 3A). Furthermore, the binding of NP to GST-Pin1 was markedly reduced by substituting Ser79 with Ala (S79A), suggesting that Pin1 binds to this site (Fig. 3B).
Fig. 3.
Interaction of phosphorylated NP and host Pin1. (A) 293A cell lysates expressing Flag-tagged wild-type NP (WT) or non-phosphorylatable mutant NP (S79A) were subjected to pull-down by GST or GST-Pin1 conjugated beads. (B) NP bound to Pin1 was detected by immunoblotting with anti-Flag-tag antibody. It was observed that NP bound to Pin1 and that the amount of binding was reduced by substitution of Ser79 to Ala in NP. The amounts of GST and GST-Pin1 bound to beads were verified by SDS-PAGE and CBB staining. (C) Detection of host cell-derived Pin1 in SARS-CoV-2 particles by immunoblot analysis. The presence of Pin1 in viral particles was confirmed.
NP is a structural protein of the viral nucleocapsid and is known to transport the binding host protein into the viral particle during viral packaging. Therefore, the next step was to examine whether Pin1 was incorporated into viral particles via interaction with NP. We confirmed the presence of Pin1 in viral particles by immunoblot analysis (Fig. 3C).
A separate analysis was conducted in parallel to comprehensively analyze host proteins present in the viral particles. LC-MS/MS identified 145 proteins in the culture supernatant of infected cells. Excluding 26 proteins that were also identified in the culture supernatant of uninfected cells, 119 proteins were considered to be host cell-derived proteins (Supplemental Fig. S6). However, the probability of the inclusion of cellular components released from dead cells due to viral infection remains unclear. Many of these proteins were involved in transcription and translation, and 13 have already been reported to bind to NP (Supplemental Table S1 and Supplemental Fig. S6) (BioGRID; https://thebiogrid.org/). Peptide spectrum matches (PSMs), protein scores, and coverages are shown in Table S1. Of note, Pin1 was not detected in this analysis, presumably because the protein concentration was below the detection limit.
3.4. Pin1 is involved in viral replication
To investigate the functional significance of Pin1 interaction to NP, we used two Pin1-specific shRNAs (sh_Pin1#1 and sh_Pin1#2) to establish VeroE6/TMPRSS2 cells in which Pin1 was stably depleted. Immunoblot analysis confirmed that Pin1 expression was sufficiently reduced by both shRNAs (Fig. 4A). These cells were infected with live SARS-CoV-2 (MOI = 0.5), and the amount of viral RNA in the culture supernatant was measured by qRT-PCR after 48 h. In Pin1 knockdown cells, the amount of viral RNA was significantly decreased compared to control cells (33% in sh_Pin1#1 and 38% in sh_Pin1#2) (Fig. 4B). Similarly, the viral titers were prominently decreased in Pin1 knockdown cells as compared with control cells (26% in sh_Pin1 #1 and #2) (Fig. 4C). These results suggest that Pin1 may positively regulate viral replication.
Fig. 4.
Functions of Pin1 in viral replication. (A) Immunoblot analysis of Pin1 expression in control Pin1 knockdown VeroE6/TMPRSS2 cells. (B) The amounts of viral RNA in control and Pin1 knockdown cells were detected by qRT-PCR with specific primers. (C) Viral titers of control and Pin1 knockdown cells were determined as TCID50.
4. Discussion
The analysis of post-translational modifications of viral proteins is an important means of understanding the molecular mechanisms of viral replication and pathogenesis. Protein phosphorylation in particular is essential for many viruses to achieve productive infection. In this study, we utilized Phos-tag technology to reveal the functional phosphorylation of SARS-CoV-2 NP and delineate the molecular link for underlying pathogen-host interaction. We found that the Ser79 residue in NP is constitutively phosphorylated in infected cells and that it initiates the interaction with Pin1. Furthermore, we showed that targeted knock-down of Pin1 in virus-infected cells resulted in a reduction of viral RNA in cell supernatant, suggesting that Pin1 might positively regulate viral replication.
Phosphorylation is one of the most common post-translational protein modifications and is regulated by intracellular kinases and phosphatases. The total amounts of phosphoproteins in cells are usually lower than those of non-phosphorylated proteins, and therefore pretreatments such as enrichment and/or separation of phosphorylated proteins/peptides are essential for proteomic analysis [22]. The most commonly used method is affinity purification based on the coordination between an immobilized metal ion and phosphate groups on the protein surface, followed by identification of the protein and phosphorylation site by LC-MS/MS analysis. For phosphopeptide enrichment, immobilized-metal affinity chromatography [[23], [24], [25]] and metal-oxide affinity chromatography [[26], [27], [28], [29]] are widely used as pretreatment measures in LC-MS/MS analysis. The recently developed Phos-tag ligand is an alkoxide-bridged dinuclear metal complex that can selectively capture phosphate groups in aqueous solution at neutral pH [9,10]. Phos-tag ligands can be combined with a variety of compounds for targeted phosphorylation analysis. Indeed, Phos-tag acrylamide can be apply for the polyacrylamide gel electrophoresis (Phos-tag SDS-PAGE) in which different phosphorylated forms of the same protein can be detected as separate bands. The Phos-tag SDS-PAGE has recently been used to analyze phosphorylation patterns of proteins in different external and/or internal conditions [30]. For example, the phosphorylation of HBc, one of the component proteins of hepatitis B virus, was found to be enhanced by co-expression of serine-arginine-rich protein kinase 1 (SRPK1), which was discovered by Phos-tag SDS PAGE [31]. In addition, the phosphorylation of Tyr79 in the NP of influenza A virus has been confirmed by Phos-tag SDS PAGE [32]. In this study, we used Phos-tag SDS PAGE to demonstrate phosphorylation of the SARS-CoV-2 NP. Phosphorylation of NP is theoretically rendered by host cell proline-directed Ser/Thr kinases (CDK, GSK3, MAPK, and CLK) after viral infection as this virus does not encode protein kinase(s). Since NP is involved in various steps of the viral life cycle [33], functional phosphorylation analysis might be useful for identifying its functions during virus replication and virus-induced cytopathogenesis.
In order to identify viral phosphoproteins and their phosphorylation sites, we purified viral particles collected from the supernatants of infected cell cultures. Although we were unable to perform density gradient ultracentrifugation due to biosafety limitations, we successfully purified viral particles using a simple centrifugation procedure and performed protein extraction and trypsin digestion to enrich phosphopeptides with Phos-tag magnetic beads before LC-MS/MS analysis. In fact, high concentrations of fetal bovine serum and multiple supplements in cell culture supernatants often cause troublesome in LC-MS/MS analysis. Phos-tag magnetic beads conjugated to zinc ion can be useful for the efficient purification of phosphopeptides from cell culture supernatant with high selectivity and reproducibility [13]. In fact, our recent study demonstrated that the Phos-tag magnetic bead enrichment method is more suitable than other methods for analyzing serum containing high-abundant proteins [34].
Several studies have demonstrated that SARS-CoV-2 NP is heavily phosphorylated on multiple phosphorylation sites in infected cells [[35], [36], [37], [38], [39]]. On the other hand, the current study identifies Ser79 as the prominent and consistent phosphorylation site of NP in viral particles. A possible explanation for this is that protein dephosphorylation may alter the phosphorylation profile of NP during and after viral particle production. Consistent with this hypothesis, Phos-tag SDS-PAGE in this study showed that NP had fewer phosphorylated bands in viral particles than in cell lysates (Fig. 1 and Supplemental Fig. S2). In addition, it has been well documented that viral core protein is subjected to dephosphorylation during virus particle formation as observed in hepatitis virus B infection [40,41]. These results indicate that dynamic phosphorylation/dephosphorylation of viral core protein regulates multiple steps of viral replication. Additionally, several phosphorylation bands were detected in viral particles, indicating that phosphorylation occurs at other sites besides S79. However, phosphorylation at all other examined sites either had low scores or were not reproducible. Only the phosphorylation at S79 was reproducible with a prominent high score, suggesting it as the major phosphorylated site in NP. Further studies are needed to characterize the phosphorylation dynamics of the NP protein.
In the current study, we found that Pin1 binds to Ser79 of NP and positively regulates viral replication. Pin1 is the only known peptidyl-prolyl cis-trans isomerase (PPIase) that specifically recognizes and isomerizes the phosphorylated Ser/Thr-Pro motif. Pin1 has been shown to catalyze conformational changes through cis-trans isomerization of peptide bonds, thus altering the catalytic activity, localization, and stability of target proteins [42,43]. As an example, interaction between host cell Pin1 and viral proteins has been reported for HBc, the core protein of hepatitis B. It has been shown that functional regulation of Pin1 binding to the phosphorylation site of HBc contributes to HBc stability [44]. The functional regulation of phosphoproteins by Pin1 also affects virus-host cell interactions [45]. The role of intra-particle Pin1 has been extensively studied in RNA virus infection. Misumi et al. showed that the uncoating process of HIV required the interaction of the capsid (CA) protein with Pin1. Suppression of Pin1 expression by RNA interference in a target cell attenuated HIV-1 replication and increased the number of particulate CA cores in the cytosol of target cells [46]. Furthermore, Manganaro et al. reported that Pin1 catalyzed the conformational modification of HIV-1 integrase and increased its stability related to efficient HIV-1 integration and infection [47]. Interestingly, Yamamotoya et al. recently reported that Pin1-targeted small-molecule inhibitors could serve as potential therapeutic agents for COVID-19 [48]. Together, these results suggest that Pin1 plays an important role in the molecular crosstalk between virus and host cells and could be a unique target for antiviral therapy.
Our multiple sequence alignment of the NPs of several beta-coronaviruses revealed that Ser79 is a distinct phosphor-acceptor site specifically located in SARS-CoV-2 but not in other coronaviruses. We consider that SARS-CoV-2 has evolved to achieve more efficient proliferation and/or greater adaptation to host cells. Thereby, a novel form of virus-host cell interaction may have been acquired as a consequence of viral evolution. Further detailed analyses are needed to delineate this intriguing hypothesis in terms of viral protein phosphorylation.
One limitation of our study was that we used only a single-cell culture model of Vero E6-TMPRSS2 and therefore the observed phenomenon may not reflect in vivo events in terms of NP phosphorylation or Pin1 interaction. Also, we did not evaluate the efficacy of our phosphoprotein purification strategy using related technologies. Furthermore, the performance and sensitivity of MS system to detect the phosphorylation sites depends on sample pretreatment and instrument used. Although the MS system utilized in the current study performs well to detect phosphorylation sites, use of a higher performance MS system might possibly reveal other phosphorylation sites [49,50]. Hence, further analysis is needed to clarify the feasibility of our method for virus phosphoproteomics using Phos-tag technology.
In summary, we utilized Phos-tag technology to reveal physiologically relevant phosphorylation of SARS-CoV-2 NP. Subsequent functional studies revealed that the interaction of NP with host factor Pin1 played a role in viral replication. Furthermore, it was suggested that the interaction between NP and Pin1 may contribute to the stability and infectivity of viral particles. This study highlights a previously undescribed virus-host interaction mediated by Pin1-dependent prolyl-isomerization and shows that Pin1 is a potential therapeutic target for COVID-19.
The following are the supplementary data related to this article.
Phosphopeptide enrichment using Phos-tag magnetic beads. Enriched phosphopeptides were desalted and subjected to LC-MS/MS analysis.
Confirmation of NP phosphorylation in SARS-CoV-2 viral particles. Viral particle lysates were subjected to Phos-tag SDS-PAGE followed by immunoblotting with anti-NP antibody. Phosphorylated forms were observed as upshifted bands.
Matched spectra ions showing that Ser79 was phosphorylated.
Multiple alignment of amino acid sequences, including Ser 79 in the NP amino acid sequence of SARS-CoV-2 and its variants of interest (VOIs) and variants of concern (VOCs). Residues that are conserved in all sequences are highlighted in grey.
Sequences of NP Pin1 target motifs (pSer/pThr-Pro) involving amino acids other than Ser79.
LC-MS/MS analysis of host cell-derived proteins contained in SARS-CoV-2 particles. Proteins identified in uninfected cells were considered as cellular contaminants and were subtracted from the proteins identified within the virus particles. *Proteins already reported to bind to NP in BioGRID (https://thebiogrid.org/).
Proteins identified by in-gel digestion and LC-MS/MS analysis of SARS-CoV-2 particles. For protein identification, peptides with a false discovery rate of less than 1% were used. Proteins identified in uninfected cells were considered as cellular contaminants and were subtracted from the proteins identified within the virus particles. *Count of PSMs (PSMs that have significant scores). **Proteins that have already been reported to bind to NP in BioGRID (https://thebiogrid.org/).
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
We thank Natsumi Takaira, Kenji Yoshihara, and Kazuo Horikawa for their technical assistance. This work was in part supported by Japan Agency for Medical Research and Development (AMED) (JP20he0522001, JP21fk0108104) and Kanagawa Institute of Industrial Science and Technology (KISTEC) grant to AR.
Data availability
Data will be made available on request.
References
- 1.Appel N., Pietschmann T., Bartenschlager R. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J. Virol. 2005;79(5):3187–3194. doi: 10.1128/JVI.79.5.3187-3194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goonawardane N., Gebhardt A., Bartlett C., Pichlmair A., Harris M. Phosphorylation of serine 225 in hepatitis C virus NS5A regulates protein-protein interactions. J. Virol. 2017;91(17) doi: 10.1128/JVI.00805-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teppor M., Zusinaite E., Merits A. Phosphorylation sites in the Hypervariable domain in Chikungunya virus nsP3 are crucial for viral replication. J. Virol. 2021;95(9) doi: 10.1128/JVI.02276-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen P. The regulation of protein function by multisite phosphorylation--a 25 year update. Trends Biochem. Sci. 2000;25(12):596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 5.Ma Y., Lu Y., Zeng H., Ron D., Mo W., Neubert T.A. Characterization of phosphopeptides from protein digests using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and nanoelectrospray quadrupole time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2001;15(18):1693–1700. doi: 10.1002/rcm.426. [DOI] [PubMed] [Google Scholar]
- 6.Asara J.M., Allison J. Enhanced detection of phosphopeptides in matrix-assisted laser desorption/ionization mass spectrometry using ammonium salts. J. Am. Soc. Mass Spectrom. 1999;10(1):35–44. doi: 10.1016/S1044-0305(98)00129-9. [DOI] [PubMed] [Google Scholar]
- 7.Liu S., Zhang C., Campbell J.L., Zhang H., Yeung K.K., Han V.K., Lajoie G.A. Formation of phosphopeptide-metal ion complexes in liquid chromatography/electrospray mass spectrometry and their influence on phosphopeptide detection. Rapid Commun. Mass Spectrom. 2005;19(19):2747–2756. doi: 10.1002/rcm.2105. [DOI] [PubMed] [Google Scholar]
- 8.Cheung R.C., Wong J.H., Ng T.B. Immobilized metal ion affinity chromatography: a review on its applications. Appl. Microbiol. Biotechnol. 2012;96(6):1411–1420. doi: 10.1007/s00253-012-4507-0. [DOI] [PubMed] [Google Scholar]
- 9.Kinoshita E., Kinoshita-Kikuta E., Koike T. Advances in Phos-tag-based methodologies for separation and detection of the phosphoproteome. Biochim. Biophys. Acta. 2015;1854(6):601–608. doi: 10.1016/j.bbapap.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Kinoshita E., Takahashi M., Takeda H., Shiro M., Koike T. Recognition of phosphate monoester dianion by an alkoxide-bridged dinuclear zinc(II) complex. Dalton Trans. 2004;(8):1189–1193. doi: 10.1039/b400269e. [DOI] [PubMed] [Google Scholar]
- 11.Kinoshita E., Yamada A., Takeda H., Kinoshita-Kikuta E., Koike T. Novel immobilized zinc(II) affinity chromatography for phosphopeptides and phosphorylated proteins. J. Sep. Sci. 2005;28(2):155–162. doi: 10.1002/jssc.200401833. [DOI] [PubMed] [Google Scholar]
- 12.Nabetani T., Kim Y.J., Watanabe M., Ohashi Y., Kamiguchi H., Hirabayashi Y. Improved method of phosphopeptides enrichment using biphasic phosphate-binding tag/C18 tip for versatile analysis of phosphorylation dynamics. Proteomics. 2009;9(24):5525–5533. doi: 10.1002/pmic.200900341. [DOI] [PubMed] [Google Scholar]
- 13.Tsunehiro M., Meki Y., Matsuoka K., Kinoshita-Kikuta E., Kinoshita E., Koike T. A Phos-tag-based magnetic-bead method for rapid and selective separation of phosphorylated biomolecules. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;925:86–94. doi: 10.1016/j.jchromb.2013.02.039. [DOI] [PubMed] [Google Scholar]
- 14.Yuan E.T., Ino Y., Kawaguchi M., Kimura Y., Hirano H., Kinoshita-Kikuta E., Kinoshita E., Koike T. A Phos-tag-based micropipette-tip method for rapid and selective enrichment of phosphopeptides. Electrophoresis. 2017;38(19):2447–2455. doi: 10.1002/elps.201700175. [DOI] [PubMed] [Google Scholar]
- 15.Ino Y., Arakawa N., Ishiguro H., Uemura H., Kubota Y., Hirano H., Toda T. Phosphoproteome analysis demonstrates the potential role of THRAP3 phosphorylation in androgen-independent prostate cancer cell growth. Proteomics. 2016;16(7):1069–1078. doi: 10.1002/pmic.201500365. [DOI] [PubMed] [Google Scholar]
- 16.Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database issue):D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishi M., Akutsu H., Masui S., Kondo A., Nagashima Y., Kimura H., Perrem K., Shigeri Y., Toyoda M., Okayama A., Hirano H., Umezawa A., Yamamoto N., Lee S.W., Ryo A. A distinct role for Pin1 in the induction and maintenance of pluripotency. J. Biol. Chem. 2011;286(13):11593–11603. doi: 10.1074/jbc.M110.187989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., Katoh H., Sakata M., Nakatsu Y., Mori Y., Kageyama T., Matsuyama S., Takeda M. Development of genetic diagnostic methods for detection for novel coronavirus 2019(nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020;73(4):304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- 19.Tsukagoshi H., Shinoda D., Saito M., Okayama K., Sada M., Kimura H., Saruki N. Relationships between viral load and the clinical course of COVID-19. Viruses. 2021;13(2) doi: 10.3390/v13020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khazeei Tabari M.A., Khoshhal H., Tafazoli A., Khandan M., Bagheri A. Applying computer simulations in battling with COVID-19, using pre-analyzed molecular and chemical data to face the pandemic. Inform Med Unlocked. 2020;21 doi: 10.1016/j.imu.2020.100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., Sigal A., Schmidt A.G., Iafrate A.J., Naranbhai V., Balazs A.B. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184(9):2523. doi: 10.1016/j.cell.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beltran L., Cutillas P.R. Advances in phosphopeptide enrichment techniques for phosphoproteomics. Amino Acids. 2012;43(3):1009–1024. doi: 10.1007/s00726-012-1288-9. [DOI] [PubMed] [Google Scholar]
- 23.Feng S., Pan C., Jiang X., Xu S., Zhou H., Ye M., Zou H. Fe3+ immobilized metal affinity chromatography with silica monolithic capillary column for phosphoproteome analysis. Proteomics. 2007;7(3):351–360. doi: 10.1002/pmic.200600045. [DOI] [PubMed] [Google Scholar]
- 24.Stensballe A., Andersen S., Jensen O.N. Characterization of phosphoproteins from electrophoretic gels by nanoscale Fe(III) affinity chromatography with off-line mass spectrometry analysis. Proteomics. 2001;1(2):207–222. doi: 10.1002/1615-9861(200102)1:2<207::AID-PROT207>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Thingholm T.E., Larsen M.R. Phosphopeptide enrichment by immobilized metal affinity chromatography. Methods Mol. Biol. 2016;1355:123–133. doi: 10.1007/978-1-4939-3049-4_8. [DOI] [PubMed] [Google Scholar]
- 26.Imami K., Sugiyama N., Kyono Y., Tomita M., Ishihama Y. Automated phosphoproteome analysis for cultured cancer cells by two-dimensional nanoLC-MS using a calcined titania/C18 biphasic column. Anal. Sci. 2008;24(1):161–166. doi: 10.2116/analsci.24.161. [DOI] [PubMed] [Google Scholar]
- 27.Wolschin F., Wienkoop S., Weckwerth W. Enrichment of phosphorylated proteins and peptides from complex mixtures using metal oxide/hydroxide affinity chromatography (MOAC) Proteomics. 2005;5(17):4389–4397. doi: 10.1002/pmic.200402049. [DOI] [PubMed] [Google Scholar]
- 28.Sugiyama N., Masuda T., Shinoda K., Nakamura A., Tomita M., Ishihama Y. Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol. Cell. Proteomics. 2007;6(6):1103–1109. doi: 10.1074/mcp.T600060-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Lo C.Y., Chen W.Y., Chen C.T., Chen Y.C. Rapid enrichment of phosphopeptides from tryptic digests of proteins using iron oxide nanocomposites of magnetic particles coated with zirconia as the concentrating probes. J. Proteome Res. 2007;6(2):887–893. doi: 10.1021/pr060333g. [DOI] [PubMed] [Google Scholar]
- 30.Kimura Y., Nagata K., Suzuki N., Yokoyama R., Yamanaka Y., Kitamura H., Hirano H., Ohara O. Characterization of multiple alternative forms of heterogeneous nuclear ribonucleoprotein K by phosphate-affinity electrophoresis. Proteomics. 2010;10(21):3884–3895. doi: 10.1002/pmic.201000349. [DOI] [PubMed] [Google Scholar]
- 31.Heger-Stevic J., Zimmermann P., Lecoq L., Bottcher B., Nassal M. Hepatitis B virus core protein phosphorylation: identification of the SRPK1 target sites and impact of their occupancy on RNA binding and capsid structure. PLoS Pathog. 2018;14(12) doi: 10.1371/journal.ppat.1007488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui L., Zheng W., Li M., Bai X., Yang W., Li J., Fan W., Gao G.F., Sun L., Liu W. Phosphorylation status of tyrosine 78 residue regulates the nuclear export and Ubiquitination of influenza a virus nucleoprotein. Front. Microbiol. 2019;10:1816. doi: 10.3389/fmicb.2019.01816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cubuk J., Alston J.J., Incicco J.J., Singh S., Stuchell-Brereton M.D., Ward M.D., Zimmerman M.I., Vithani N., Griffith D., Wagoner J.A., Bowman G.R., Hall K.B., Soranno A., Holehouse A.S. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021;12(1):1936. doi: 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ino Y., Kinoshita E., Kinoshita-Kikuta E., Akiyama T., Nakai Y., Nishino K., Osada M., Ryo A., Hirano H., Koike T., Kimura Y. Evaluation of four phosphopeptide enrichment strategies for mass spectrometry-based proteomic analysis. Proteomics. 2021 doi: 10.1002/pmic.202100216. [DOI] [PubMed] [Google Scholar]
- 35.Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Marrero M. Correa, Polacco B.J., Melnyk J.E., Ulferts S., Kaake R.M., Batra J., Richards A.L., Stevenson E., Gordon D.E., Rojc A., Obernier K., Fabius J.M., Soucheray M., Miorin L., Moreno E., Koh C., Tran Q.D., Hardy A., Robinot R., Vallet T., Nilsson-Payant B.E., Hernandez-Armenta C., Dunham A., Weigang S., Knerr J., Modak M., Quintero D., Zhou Y., Dugourd A., Valdeolivas A., Patil T., Li Q., Huttenhain R., Cakir M., Muralidharan M., Kim M., Jang G., Tutuncuoglu B., Hiatt J., Guo J.Z., Xu J., Bouhaddou S., Mathy C.J.P., Gaulton A., Manners E.J., Felix E., Shi Y., Goff M., Lim J.K., McBride T., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., De Wit E., Leach A.R., Kortemme T., Shoichet B., Ott M., Saez-Rodriguez J., Ten Oever B.R., Mullins R.D., Fischer E.R., Kochs G., Grosse R., Garcia-Sastre A., Vignuzzi M., Johnson J.R., Shokat K.M., Swaney D.L., Beltrao P., Krogan N.J. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell. 2020;182(3) doi: 10.1016/j.cell.2020.06.034. (685–712 e19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlson C.R., Asfaha J.B., Ghent C.M., Howard C.J., Hartooni N., Safari M., Frankel A.D., Morgan D.O. Phosphoregulation of Phase Separation by the SARS-CoV-2 N Protein Suggests a Biophysical Basis for its Dual Functions. Mol Cell. 2020;80(6) doi: 10.1016/j.molcel.2020.11.025. (1092–1103 e4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davidson A.D., Williamson M.K., Lewis S., Shoemark D., Carroll M.W., Heesom K.J., Zambon M., Ellis J., Lewis P.A., Hiscox J.A., Matthews D.A. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12(1):68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klann K., Bojkova D., Tascher G., Ciesek S., Munch C., Cinatl J. Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication. Mol Cell. 2020;80(1) doi: 10.1016/j.molcel.2020.08.006. (164–174 e4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stukalov A., Girault V., Grass V., Karayel O., Bergant V., Urban C., Haas D.A., Huang Y., Oubraham L., Wang A., Hamad M.S., Piras A., Hansen F.M., Tanzer M.C., Paron I., Zinzula L., Engleitner T., Reinecke M., Lavacca T.M., Ehmann R., Wolfel R., Jores J., Kuster B., Protzer U., Rad R., Ziebuhr J., Thiel V., Scaturro P., Mann M., Pichlmair A. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature. 2021;594(7862):246–252. doi: 10.1038/s41586-021-03493-4. [DOI] [PubMed] [Google Scholar]
- 40.Xi J., Luckenbaugh L., Hu J. Multiple roles of PP2A binding motif in hepatitis B virus core linker and PP2A in regulating core phosphorylation state and viral replication. PLoS Pathog. 2021;17(1) doi: 10.1371/journal.ppat.1009230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Q., Hu Z., Cheng J., Wu S., Luo Y., Chang J., Hu J., Guo J.T. Hepatitis B virus Core protein Dephosphorylation occurs during Pregenomic RNA Encapsidation. J. Virol. 2018;92(13) doi: 10.1128/JVI.02139-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu K.P., Zhou X.Z. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8(11):904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 43.Ryo A., Liou Y.C., Lu K.P., Wulf G. Prolyl isomerase Pin1: a catalyst for oncogenesis and a potential therapeutic target in cancer. J. Cell Sci. 2003;116(Pt 5):773–783. doi: 10.1242/jcs.00276. [DOI] [PubMed] [Google Scholar]
- 44.Nishi M., Miyakawa K., Matsunaga S., Khatun H., Yamaoka Y., Watashi K., Sugiyama M., Kimura H., Wakita T., Ryo A. Prolyl Isomerase Pin1 regulates the stability of hepatitis B virus Core protein. Front Cell Dev Biol. 2020;8:26. doi: 10.3389/fcell.2020.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kojima Y., Ryo A. Pinning down viral proteins: a new prototype for virus-host cell interaction. Front. Microbiol. 2010;1:107. doi: 10.3389/fmicb.2010.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Misumi S., Inoue M., Dochi T., Kishimoto N., Hasegawa N., Takamune N., Shoji S. Uncoating of human immunodeficiency virus type 1 requires prolyl isomerase Pin1. J. Biol. Chem. 2010;285(33):25185–25195. doi: 10.1074/jbc.M110.114256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manganaro L., Lusic M., Gutierrez M.I., Cereseto A., Del Sal G., Giacca M. Concerted action of cellular JNK and Pin1 restricts HIV-1 genome integration to activated CD4+ T lymphocytes. Nat. Med. 2010;16(3):329–333. doi: 10.1038/nm.2102. [DOI] [PubMed] [Google Scholar]
- 48.Yamamotoya T., Nakatsu Y., Kanna M., Hasei S., Ohata Y., Encinas J., Ito H., Okabe T., Asano T., Sakaguchi T. Prolyl isomerase Pin1 plays an essential role in SARS-CoV-2 proliferation, indicating its possibility as a novel therapeutic target. Sci. Rep. 2021;11(1):18581. doi: 10.1038/s41598-021-97972-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leitner A., Sturm M., Lindner W. Tools for analyzing the phosphoproteome and other phosphorylated biomolecules: a review. Anal. Chim. Acta. 2011;703(1):19–30. doi: 10.1016/j.aca.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 50.Steen H., Jebanathirajah J.A., Rush J., Morrice N., Kirschner M.W. Phosphorylation analysis by mass spectrometry: myths, facts, and the consequences for qualitative and quantitative measurements. Mol. Cell. Proteomics. 2006;5(1):172–181. doi: 10.1074/mcp.M500135-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phosphopeptide enrichment using Phos-tag magnetic beads. Enriched phosphopeptides were desalted and subjected to LC-MS/MS analysis.
Confirmation of NP phosphorylation in SARS-CoV-2 viral particles. Viral particle lysates were subjected to Phos-tag SDS-PAGE followed by immunoblotting with anti-NP antibody. Phosphorylated forms were observed as upshifted bands.
Matched spectra ions showing that Ser79 was phosphorylated.
Multiple alignment of amino acid sequences, including Ser 79 in the NP amino acid sequence of SARS-CoV-2 and its variants of interest (VOIs) and variants of concern (VOCs). Residues that are conserved in all sequences are highlighted in grey.
Sequences of NP Pin1 target motifs (pSer/pThr-Pro) involving amino acids other than Ser79.
LC-MS/MS analysis of host cell-derived proteins contained in SARS-CoV-2 particles. Proteins identified in uninfected cells were considered as cellular contaminants and were subtracted from the proteins identified within the virus particles. *Proteins already reported to bind to NP in BioGRID (https://thebiogrid.org/).
Proteins identified by in-gel digestion and LC-MS/MS analysis of SARS-CoV-2 particles. For protein identification, peptides with a false discovery rate of less than 1% were used. Proteins identified in uninfected cells were considered as cellular contaminants and were subtracted from the proteins identified within the virus particles. *Count of PSMs (PSMs that have significant scores). **Proteins that have already been reported to bind to NP in BioGRID (https://thebiogrid.org/).
Data Availability Statement
Data will be made available on request.