Summary
Monoclonal IgG antibodies are the fastest growing class of biologics, but large differences exist in their plasma half-life in humans. Thus, to design IgG antibodies with favorable pharmacokinetics, it is crucial to identify the determinants of such differences. Here, we demonstrate that the variable region sequences of IgG antibodies greatly affect cellular uptake and subsequent recycling and rescue from intracellular degradation by endothelial cells. When the variable sequences are masked by the cognate antigen, it influences both their transport behavior and binding to the neonatal Fc receptor (FcRn), a key regulator of IgG plasma half-life. Furthermore, we show how charge patch differences in the variable domains modulate both binding and transport properties and that a short plasma half-life, due to unfavorable charge patches, may partly be overcome by Fc-engineering for improved FcRn binding.
Subject areas: Biological sciences, Immunology, Biophysics
Graphical abstract
Highlights
-
•
IgG variable region sequences greatly affect cellular uptake and recycling
-
•
Variable region charge patches affect FcRn binding and transport
-
•
The presence of cognate antigen modulates cellular transport and FcRn binding
-
•
Fc-engineering for improved FcRn binding can overcome unfavorable charge patches
Biological sciences; Immunology; Biophysics
Introduction
IgG recognizes target antigen by its two antigen binding fragments (Fab) that are linked via a hinge to a constant crystallizable fragment (Fc), which engages Fc receptors and induce effector functions (Nimmerjahn and Ravetch, 2008). The combination of specific antigen binding and potent effector functions have made IgG and IgG Fc-fusions successful therapeutics (Jefferis, 2007; Reichert, 2017; Kang and Jung, 2019). Important in this regard is the long serum half-life of IgG. Humans have four IgG subclasses (IgG1–4), three of which have a serum half-life of roughly 3 weeks (IgG1,2,4), whereas it is 1 week for IgG3 (Morell et al., 1970). Both IgG1 and IgG3 are efficient triggers of effector functions (Vidarsson et al., 2014), but because IgG1 has a longer half-life, it is the subclass of choice for therapeutics if potent effector functions are required (Reichert, 2017). Despite this, the plasma half-life of therapeutic IgG1 molecules has been reported to vary greatly in humans, from 6–32 days (Carter and Lazar, 2018).
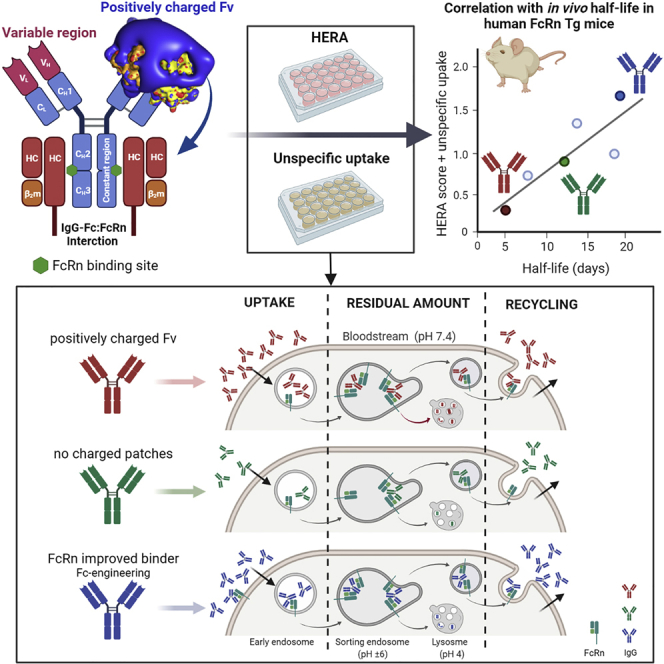
A key player in plasma half-life of IgG antibodies is a cellular receptor named the neonatal Fc receptor (FcRn) (Ghetie et al., 1996; Junghans and Anderson, 1996; Israel et al., 1996). The short half-life of IgG3 is due to R435 in the Fc that modulates pH-dependent binding to the receptor (Stapleton et al., 2011). FcRn is a heterodimer consisting of an N-glycosylated transmembrane MHC class I-like heavy chain (HC) that is noncovalently associated with soluble β2-microglobulin (Simister and Mostov, 1989). Two FcRn molecules can engage IgG by binding to each side of the symmetric CH2-CH3 interface (Abdiche et al., 2015; Burmeister et al., 1994; Kim et al., 1994; Martin et al., 2001). Binding is strictly pH dependent; strong binding occurs at acidic pH < 6.5 and no binding or release at neutral pH (Ghetie et al., 1997; Kim et al., 1999; Raghavan et al., 1995; Vaughn and Bjorkman, 1998). In general, the cellular model for half-life regulation relies on uptake of IgG, likely via fluid-phase pinocytosis, followed by binding to FcRn in acidified endosomes, where the receptor predominately resides. The FcRn-IgG complex is then routed away from lysosomal degradation and recycled to the cell surface where exposure to a near neutral blood pH results in release of IgG into the extracellular environment (Goebl et al., 2008; Grevys et al., 2018; Lencer and Blumberg, 2005; Ober et al., 2004a, 2004b; Prabhat et al., 2007; Vaughn and Bjorkman, 1998).
Modulation of pH-dependent FcRn binding by Fc-engineering has a major influence on rescue from degradation, where enhanced binding at neutral pH results in inefficient release at the outer cell membrane and shorter plasma half-life (Dall'Acqua et al., 2002; Ghetie et al., 1997; Vaccaro et al., 2005). In contrast, Fc-engineering for improved binding at acidic pH without affecting binding at neutral pH translates into extended half-life beyond that of the parental antibody (Dall'Acqua et al., 2006; Ghetie et al., 1997; Lee et al., 2019; Zalevsky et al., 2010). One such engineered antibody has three Fc substitutions (M252Y/S254T/T256E; YTE) that results in approximately 10-fold improved binding to human FcRn at pH 6.0 and almost 4-fold extended plasma half-life in nonhuman primates (Dall'Acqua et al., 2006). When the YTE substitutions were introduced into IgG1 with specificity for respiratory syncytial virus, it extended the terminal half-life to more than 100 days in humans (Booth et al., 2018; Griffin et al., 2017; Robbie et al., 2013). Design of IgG antibodies with extended half-life is of great interest, as it enables reduced frequency of dosing and thus increases patient compliance. In particular, this is an attractive strategy for prophylaxis and treatment of infectious diseases, as recently exemplified for broadly neutralizing IgG antibodies targeting HIV (Gautam et al., 2016; Gaudinski et al., 2018; Ko et al., 2014).
However, some monoclonal IgG1 antibodies have surprisingly poor pharmacokinetics (Carter and Lazar, 2018; Jain et al., 2017). Because such antibodies are of the same subclass and have identical Fc sequence, the differences in half-life must necessarily be dictated by the fragment variable (Fv) domains. Some reports suggest that the Fab arms may directly engage FcRn (Jensen et al., 2015; Suzuki et al., 2010; Wang et al., 2011), but this is in disagreement with other observations (Neuber et al., 2014). Rather, recent reports have highlighted isoelectric point (pI), charge patch differences, and off-target reactivity as factors that affect half-life (Datta-Mannan et al., 2015; Igawa et al., 2010b; Kelly et al., 2017). For instance, the surface of cells is negatively charged, and high pI may facilitate cellular uptake and transport to lysosomes for degradation (Igawa et al., 2010b). In addition, substitutions of amino acids in the Fv complementarity-determining regions (CDRs) as well as the Fv framework regions, or change of the light chain (LC), have been shown to greatly modulate plasma half-life (Datta-Mannan et al., 2015; Li et al., 2014; Piche-Nicholas et al., 2018; Schoch et al., 2015). Interestingly, it was recently found that IgG molecules with only one to five amino acid differences in their CDRs could modulate FcRn binding affinity by as much as nearly 80-fold and that affinity correlated with the pI values of the Fv and CDR3 loop of the LC (Piche-Nicholas et al., 2018). As such, contribution of the Fab arms to FcRn binding must be a consequence of the amino acid composition of the CDRs and/or Fv framework of each antibody.
As there seems to be a complex relationship between how both the Fab arms and the Fc affect pharmacokinetics, we aimed to gain insights into how Fv sequence differences and engineering are modulating FcRn binding, cellular transport, and in vivo properties. Therefore, we conducted a thorough biochemical, cellular, and in vivo analysis of two monoclonal IgG1 antibodies, briakinumab and ustekinumab, that target the same antigen, the p40-subunit of interleukin (IL)-12/23, but that have very distinct plasma half-lives in humans, 9 versus 23 days, respectively (Gandhi et al., 2010; Zhu et al., 2009). Our findings provide insights into how charged patches in the Fv affect FcRn binding and cellular handling in an FcRn-independent and -dependent manner, in both the absence and presence of cognate antigen. We also demonstrate that unfavorable Fv charge patches that cause limited plasma half-life can be partly counteracted by Fc-engineering for improved FcRn binding.
Results
Charge differences in the Fv domains of ustekinumab and briakinumab
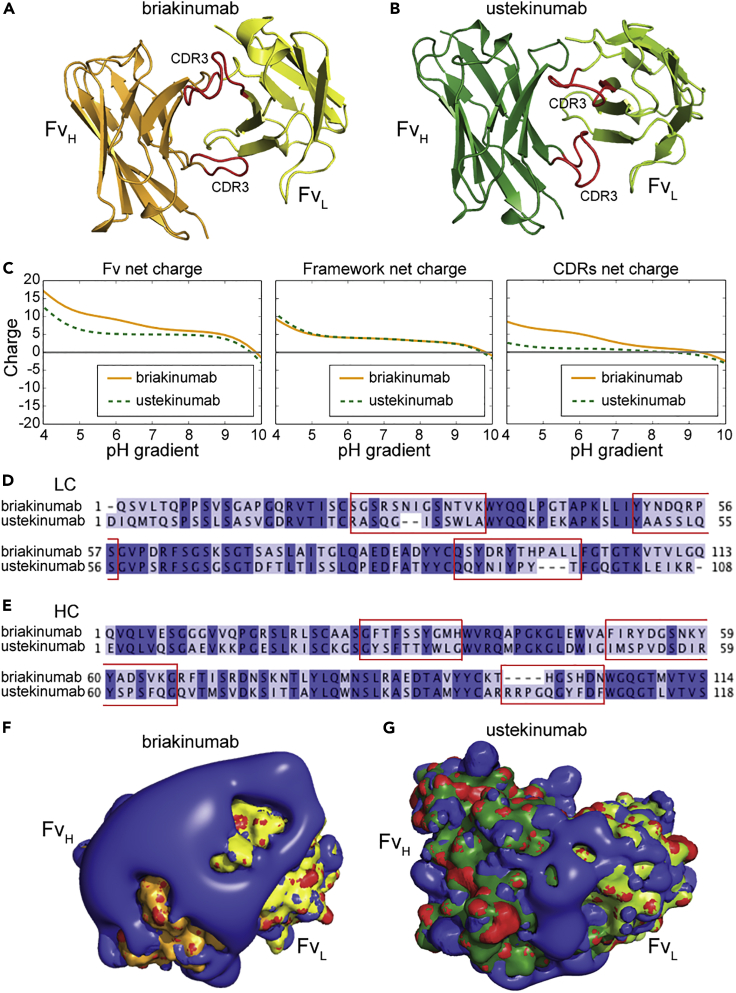
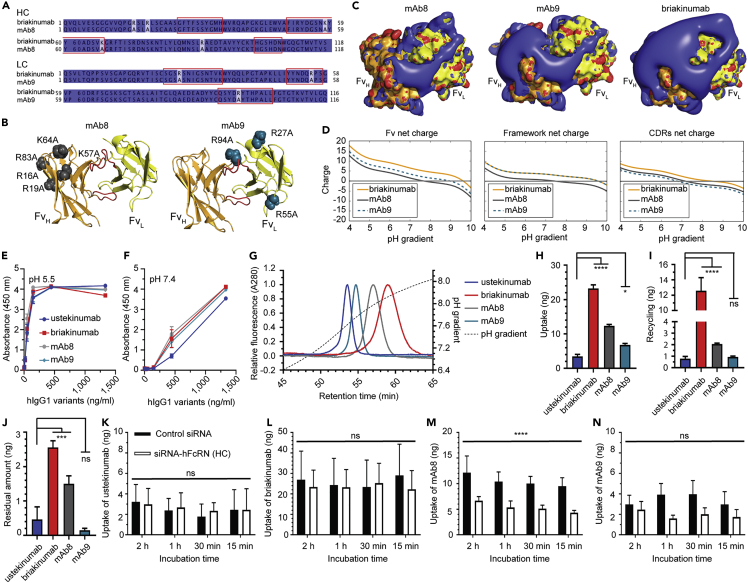
To gain insight into charge distribution of Fv as a function of pH, we investigated the crystal structures of Fab fragments derived from ustekinumab and briakinumab (Figures 1A and 1B). The net charge of the Fv domain of briakinumab was more positive than that of ustekinumab throughout the entire pH range, and the effect was most pronounced at acidic pH (Figures 1C and S1), in agreement with previous observations (Schoch et al., 2015). This was solely due to CDR sequence variations and not the framework, where briakinumab and ustekinumab have 10 and 4 positively charged residues, respectively (Figures 1D and 1E, Table S1). The results revealed a large difference in charge distribution, where briakinumab showed a positively charged area localized in the CDRs (Figures 1F and 1G). Despite these charge differences, briakinumab and ustekinumab have a similar pI of 9.6 and 9.3 for the full-length antibodies, respectively.
Figure 1.
Charge distribution of the Fv domains of ustekinumab and briakinumab
(A and B) (A) Crystal structures of top orientated briakinumab and (B) ustekinumab variable domains. CDR loop three of LC and HC are highlighted in red color. The HC of ustekinumab and briakinumab are shown in green and orange, and the LC of ustekinumab and briakinumab are colored in light green and yellow, respectively.
(C–G) (C) Sequence-based calculation of net charge through pH range of ustekinumab and briakinumab variable domains, framework (without CDRs) and CDRs, respectively. CDRs were defined as indicated in the material and methods. Sequence alignments of ustekinumab and briakinumab (D) LC and (E) HC, conserved residues are marked with dark blue, distinct residues with light blue, dash indicate missing amino acid residues, and CDR sequences are highlighted by red square. Sequence alignments have been made by Jalview. The charge distribution of top oriented (F) briakinumab and (G) ustekinumab Fv domains at pH 7.4. Blue color indicates positive charge, red negative charge, the HC of ustekinumab and briakinumab are shown in green and orange, and the LC of ustekinumab and briakinumab are colored in light green and yellow, respectively. The figures were visualized using PyMOL (www.pymol.org) with the crystallography data of human IgG1 (Luo et al., 2010; Bloch et al., 2017) (PDB ID codes 3HMW [ustekinumab] and 5N2K [briakinumab]), and net charges were calculated with Emboss iep (www.bioinformatics.nl).
Distinct FcRn binding properties
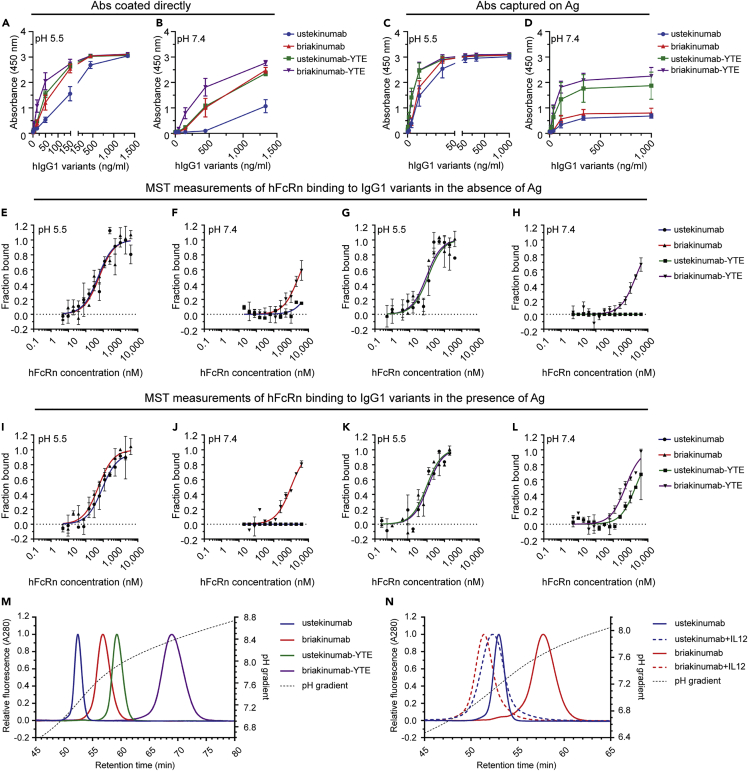
Recombinant versions of the two antibodies were produced in HEK293E cells, which gave similar amounts, and they migrated with expected molecular weights in SDS-PAGE (Figures 2A–2C). FcRn binding was measured by ELISA, where titrated amounts of antibodies were coated or captured on the antigen IL-12, before GST-tagged human FcRn (hFcRn) was added; this was done at either pH 5.5 or 7.4 in both cases, followed by detection using an HRP-conjugated anti-GST antibody. When the antibodies were coated directly, hFcRn bound in a concentration- and pH-dependent manner, but binding was stronger to briakinumab than to ustekinumab at both pH conditions (Figures 2A and 2B). In contrast, when the antibodies were captured on IL-12, no difference in binding was measured at pH 5.5, whereas a distinct difference was observed at pH 7.4 (Figures 2C and 2D), where briakinumab bound more strongly than ustekinumab.
Figure 2.
Influence of IL-12 on IgG binding to hFcRn
(A–N) ELISA binding of titrated amounts (1400.0–0.07 ng/mL) of WT IgG1 and Fc-engineered variants to hFcRn at pH (A) 5.5 and (B) 7.4, when the antibodies were directly coated to ELISA wells. ELISA binding of titrated amounts (1000.0–0.02 ng/mL) of WT IgG1 and Fc-engineered variants to hFcRn at pH (C) 5.5 and (D) 7.4, when the antibodies were captured on IL-12 coated in wells. Data are mean ± SD of three independent experiments performed in duplicates. Microscale Thermophoresis analysis where constant amount (20 nM) of ustekinumab and briakinumab at pH (E) 5.5 or (F) 7.4, ustekinumab-YTE and briakinumab-YTE at pH (G) 5.5 or (H) 7.4, ustekinumab and briakinumab were preincubated with IL-12 at pH (I) 5.5 or (J) 7.4, and further ustekinumab-YTE and briakinumab-YTE were preincubated with IL12 at pH (K) 5.5 or (L) 7.4 and added to titrated (40,000.0–0.3 nM) amounts of hFnRn. Binding data are derived from the specific change in the thermophoresis mobility and the ratio of normalized time averaged (1s) fluorescence intensities at T-jump points of the Microscale Thermophoresis traces (−1 and 1.5s). To form IgG-IL-12 complexes, the IgGs variants were mixed with IL-12 at ratio 1 to 2, and incubated for 10 min at room temperature. The results are mean ± SD from two independent experiments performed in triplicates. Analytical hFcRn affinity chromatography of (M) ustekinumab, briakinumab, and YTE variants, and (N) ustekinumab and briakinumab as monomeric fractions as well as in a complex with the IL-12. The elution profiles are shown as relative fluorescence intensity as a function of the pH gradient. Fluorescence intensity was normalized and set to one for the clarity. Data are shown as one representative experiment out of three independent experiments.
Next, we asked how Fc-engineering for strong hFcRn interaction affected binding by introducing the YTE substitutions (Dall'Acqua et al., 2006; Robbie et al., 2013). The Fc-engineered variants were expressed in slightly higher amounts than the WT counterparts (Figure S2A). When tested in ELISA, the YTE substitutions increased receptor binding for both antibodies at pH 5.5 and pH 7.4, but a clear difference in binding between the antibodies was still detected when they were coated directly in wells (Figures 2A and 2B). Briakinumab-YTE showed the strongest binding at both pH conditions (Figures 2A and 2B). When the two antibodies were captured on IL-12, binding was equally strong to the receptor at pH 5.5, whereas briakinumab-YTE binding was stronger than ustekinumab-YTE binding at pH 7.4 (Figures 2C and 2D).
In addition, we measured hFcRn binding to the antibodies by surface plasmon resonance (SPR). Titrated amounts of monomeric His-tagged receptor were injected over immobilized antibodies (Figures S3A–S3F, Table S2), and the obtained sensorgrams were fitted to the 1:1 Langmuir binding model. The results showed no difference in on-rates between the WT antibodies, whereas about 1.5-fold faster on-rates were measured for the Fc-engineered antibodies. In regard to dissociation, ustekinumab showed a slightly faster off-rate than briakinumab, both for the WT IgG1 Fc versions (1.2-fold) and the Fc-engineered versions (1.5-fold); this resulted in small but distinct differences in KD values, where briakinumab-YTE bound hFcRn with the strongest affinity (KD 64.0 nM), followed by ustekinumab-YTE (KD 95.5 nM), briakinumab (KD 545.2 nM), and ustekinumab (KD 653.0 nM). These data are in agreement with the hFcRn binding hierarchy observed in ELISA.
To further dissect the binding differences, we measured hFcRn binding to the antibodies in solution using Microscale Thermophoresis. Titrated amounts of monomeric His-tagged receptor were added to a constant amount of labeled antibody at pH 5.5 or pH 7.4. At acidic pH, hFcRn bound briakinumab and ustekinumab equally well (0.5 μM) (Figure 2E), whereas briakinumab (>25 μM) bound more strongly than ustekinumab at neutral pH (Figure 2F). The YTE substitutions improved binding by 5- to 7-fold at acidic pH (Figure 2G). The experiments were then repeated in the presence of IL-12, by preincubating antibodies with the antigen at a 1:2 ratio, as ustekinumab has been shown to bind two IL-12 molecules per antibody (Luo et al., 2010). The results revealed that the presence of IL-12 does not affect hFcRn binding at pH 5.5 (Figures 2I and 2K). At neutral pH, briakinumab and briakinumab-YTE bound more strongly than in the absence of the antigen (Figures 2F, 2H, 2J, and 2L, Table 1), which was also the case for ustekinumab-YTE (Figures 2H and 2L). The derived KD values are summarized in Table 1. Thus, Fv charge differences of the antibodies studied were shown to affect pH-dependent hFcRn binding.
Table 1.
KD values derived from microscale thermophoresis analysis
| hIgG1 variants | KD (nM) pH 5.5 | KD (μM) pH 7.4 |
|---|---|---|
| ustekinumab | 497 ± 214 | NA |
| briakinumab | 599 ± 99 | >25 |
| ustekinumab-YTE | 100 ± 56 | NA |
| briakinumab-YTE | 79 ± 45 | >20 |
| ustekinumab/IL12 | 1000 ± 700 | NA |
| briakinumab/IL12 | 533 ± 65 | >10 |
| ustekinumab-YTE/IL12 | 78 ± 45 | >20 |
| briakinumab-YTE/IL12 | 98 ± 50 | >4 |
NA – not acquired due to negligible binding affinity.
The results are mean ± SD from two independent experiments performed in duplicates.
Release from FcRn varies between antibodies and the presence of antigen
To investigate how the antibodies bind hFcRn across a pH gradient, equal amounts of the antibodies were applied to a column immobilized with a recombinant form of hFcRn. Injections were done at pH 5.5, and the pH of the buffer was gradually increased toward 8.8 (Figures 2M and 2N, Table S3). The obtained elution profiles were distinct showing that ustekinumab eluted first (pH 7.2) followed by briakinumab (pH 7.7). The YTE substitutions delayed dissociation to pH 7.9 for ustekinumab-YTE and 8.3 for briakinumab-YTE (Figure 2M, Table S2). In the presence of IL-12, the greatest effect on dissociation was measured for briakinumab, as it was released at pH 7.0. Ustekinumab eluted slightly earlier (pH 7.1) than the uncomplexed ustekinumab (pH 7.2) (Figure 2N), and the same effect was observed for the YTE variants (Figure S4). As such, the presence of IL-12 clearly affected dissociation from the receptor.
Fc-engineering extends the plasma half-life of briakinumab
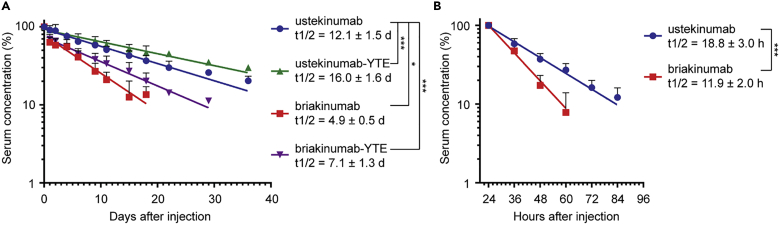
As YTE-containing briakinumab was shown to elute as late as at pH 8.3 from the receptor coupled column, we hypothesized that this would compromise efficient release at the cell surface during recycling, and as such, result in a shorter half-life than that of WT briakinumab, as previously postulated (Grevys et al., 2018; Schoch et al., 2015). To test this, we measured the half-life of the WT and YTE-containing antibodies in hFcRn transgenic mice. First, we confirmed that briakinumab was cleared at a faster rate than ustekinumab, as the half-life values were 4.9 versus 12.1 days, respectively. Although the YTE substitutions increased the half-life of ustekinumab by 4 days, we unexpectedly found that the half-life of briakinumab was extended by almost 2-fold (Figure 3A).
Figure 3.
Half-life of ustekinumab and briakinumab variants in FcRn KO and hFcRn transgenic mice
(A and B) Log-linear changes in the serum concentration of hIgG1 ustekinumab (blue), briakinumab (red), ustekinumab-YTE (green), and briakinumab-YTE (purple) in (A) hFcRn transgenic mice (n = 5, per group) and (B) FcRn KO mice (n = 6, per group). The antibodies were administrated as a single i.v. injection at 2 mg/kg (hFcRn transgenic mice) or i.p. injection at 4 mg/kg (FcRn deficient mice), followed by collection of serum samples and determination of IgG concentrations by ELISA. The results are shown as mean ± SD from one representative experiment. (A) ∗p = 0.0491 and ∗∗∗p < 0.0001 by Mixed effect model (Dunnett multiple comparison test), and (B) ∗∗∗p = 0.0009 by unpaired t test. The results were derived from a single experiment with five or six mice per group.
In addition, the half-lives of the WT antibodies were determined in FcRn KO mice, which confirmed the importance of FcRn for rescue of both antibodies, as the half-life decreased by 9- and almost 15-fold for briakinumab and ustekinumab, respectively, compared with the data from the hFcRn transgenic mice (Figures 3A and 3B). In addition, briakinumab was cleared at a faster rate than ustekinumab in KO mice, with a half-life of 11.9 compared to 18.8 h, respectively (Figure 3B). Thus, both FcRn-dependent and -independent mechanisms regulate the half-life of the antibodies with a dominant role for the receptor, and the shorter half-life of briakinumab could be prolonged by Fc-engineering.
HERA screening reveals differences in cellular properties
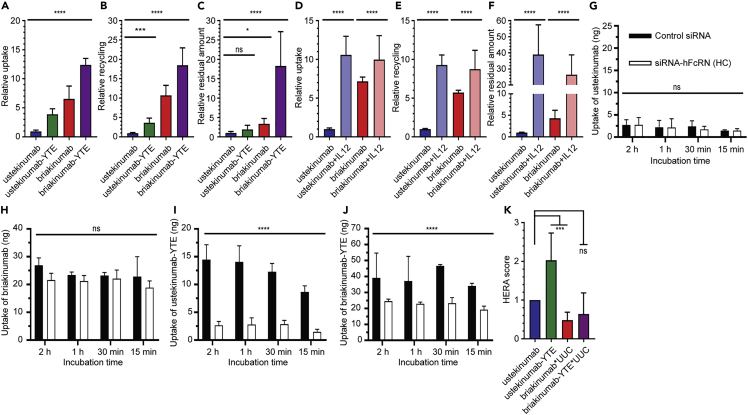
To gain insight into how the antibodies are taken up and sorted in FcRn-expressing cells, we took advantage of a human endothelial-cell-based recycling assay (HERA) that is based on a human microvascular endothelial cell line (HMEC1) overexpressing hFcRn, which can be used to measure cellular uptake and FcRn-dependent rescue from degradation that correlates with plasma half-life in hFcRn transgenic mice (Grevys et al., 2018). Adherent cells were incubated with equal amounts of the antibodies for 4 h, before the cells were gently washed, and then lysed. Lysates were analyzed in ELISA to quantify the amounts of antibodies present inside the cells, which is a result of uptake and recycling taking place during the incubation step. The results revealed 6-fold more briakinumab than ustekinumab (Figure 4A). Instead of lysis, cells were also incubated for additional 4 h in fresh medium, before the medium was collected and analyzed. The amounts measured reflect recycling of antibodies from inside the cells to the cell surface followed by release into the medium, which showed that 10-fold more of briakinumab than ustekinumab was detected (Figure 4B). In addition, we quantified the amounts that remained inside the cells at termination of the assay, which demonstrated 3.5-fold more accumulation of briakinumab (Figure 4C).
Figure 4.
Cellular transport properties of ustekinumab and briakinumab
(A) Relative uptake of WT and Fc-engineered human IgG1 variants when 400 nM of each variant was added to the cells followed by 4 h incubation, washing, and lysis of the cells.
(B) Relative recycling of the Fc-engineered human IgG1 variants when 400 nM of each variant was added to the cells and incubated for 4 h followed by extensive washing and additional 4 h incubation before sample collection.
(C) Relative residual amount of WT and Fc-engineered human IgG1 variants. The same procedure as in (B) followed by lysis of the cells.
(D–F) Relative (D) uptake, (E) recycling, and (F) residual amount of ustekinumab and briakinumab in the absence or presence of IL-12. The amounts of IgG variants in all samples were quantified by ELISA, and obtained data are shown as mean ± SD of (A–C) five or (D–F) three independent experiments performed in (A–C) triplicates or (D–F) duplicates. ns, not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001, by one-way ANOVA (Dunnett’s multiple comparison test for A–C and for D–F Sidak’s multiple comparisons test). (A–F) Values for ustekinumab were set to 1, and the other antibody variants relative to this.
(G–J) Uptake of (G) ustekinumab, (H) briakinumab, (I) ustekinumab-YTE, and (J) briakinumab-YTE at pH 7.4 when 400 nM of each was added to HMEC1-hFcRn cells treated with a mixture of control siRNA or siRNA targeting the hFcRn HC followed by incubation for 15 min, 30 min, 1 h, and 2 h. After extensive washing, the cells were lysed and added to ELISA for quantification of the amounts of antibody variants. Obtained data are presented as mean ± SD from one representative experiment performed in duplicates out of five independent experiments. ns, not significant and ∗∗∗∗p < 0.0001, by two-way ANOVA (Sidak’s multiple comparisons test).
(K) HERA score for the ustekinumab and briakinumab and the respective YTE variants were calculated from the data shown in A, C, H, and J. ns, not significant and ∗∗∗p < 0.001, by one-way ANOVA (Dunnett’s multiple comparison test).
Next, introduction of the YTE substitutions resulted in enhanced uptake and accumulation inside the cells of both antibodies with the more pronounced effect detected for briakinumab-YTE (Figures 4A–4C). In addition, the release step was run overnight without affecting the relative differences between the antibodies, except for briakinumab that showed 2-fold reduced recycling (Figures 4B and S5A–S5C). Thus, the results revealed a remarkable influence of Fv differences and Fc-engineering on cellular transport properties.
To address how the presence of IL-12 affected the cellular transport, WT antibodies were mixed with the antigen in a 1:2 ratio, followed by incubation for 30 min to allow for immune complex formation. Repeating HERA revealed that similar amounts of ustekinumab and briakinumab were detected inside the cells, 7-fold more than ustekinumab alone (Figure 4D). Further, about 10-fold more of both antigen-antibody complexes were detected in the medium compared with ustekinumab (Figure 4E). Quantification of the amounts inside the cells at termination showed roughly 30- to 40-fold more of both IL-12-bound briakinumab and ustekinumab compared to ustekinumab alone (Figures 4F). Hence, ustekinumab and briakinumab in complex with IL-12 behaved similarly in HERA, and the presence of cognate antigen greatly increased cellular uptake, recycling, and accumulation inside the cells.
Distinct differences in FcRn-dependent uptake
To address whether FcRn is responsible for the difference in cellular uptake, we treated HMEC1-hFcRn cells with a mixture of control siRNA or siRNA targeting the HC of hFcRn, as previously established (Grevys et al., 2018), before the antibodies were added. The result showed that uptake of the antibodies was not dependent on the receptor (Figures 4G and 4H). When the Fc-engineering variants were tested, 3-fold lower levels were measured inside the cells lacking hFcRn (Figures 4I and 4J). Although the uptake of briakinumab was partly dependent on hFcRn, the higher levels of the antibody compared with ustekinumab were not a direct result of more active receptor engagement at or near the cell surface.
Recently, we described a so-called HERA score, which can be used to predict the half-life of IgG1 Fc-engineered variants in hFcRn transgenic mice (Grevys et al., 2018). However, for antibodies with features that greatly affect cellular uptake, the HERA score will not give a reliable measurement. As such, we modified the HERA score to include an enhanced unspecific uptake coefficient (UUC), which is based on dividing the difference between total relative uptake and relative FcRn-independent uptake by relative FcRn-independent uptake. The HERA score was then multiplied with the UUC and plotted against the half-life values obtained from the hFcRn transgenic mice (Figures 4K and S6). By taking the differences in uptake into account, we found a correlation (r2 = 0.946, p = 0.054) between the cellular data and the half-life values, however, to finally conclude, the number of antibody variants should be increased in future studies.
Pronounced effect of charged LC residues
Interestingly, briakinumab has one histidine in the LC CDR3 (H97) and three in the HC CDR1 (H52) and CDR3 (H98 and H101), whereas ustekinumab does not have any histidines (Figures 1D, 1E, S7A, and S7B; Table S1). These histidines will protonate as a function of pH during cellular recycling and can potentially affect cellular sorting. To test this, we engineered briakinumab variants where the histidines were replaced with alanines in the LC or HC or both. Although no major influence on hFcRn binding was detected, and the antibodies were equally well taken up by cells, recycling was reduced by 3-fold for the histidine-engineered variants (Figures S7C–S7H).
To further address the influence of charged amino acids of the Fv domains, we took advantage of two mutated versions of briakinumab, named mAb8 and mAb9, which contain five substitutions in the HC (framework: R16A, R19A, R83A, CDR1: K57A and CDR2: K64A) and three substitutions in the LC (CDR1: R27A, CDR2: R55A and CDR3: R94A), respectively (Schoch et al., 2015) (Figures 5A–5D). Although the engineered variants were shown to bind hFcRn pH-dependently, the LC substitutions (mAb9) reduced binding at both pH conditions. In contrast, the variant with the HC substitutions (mAb8) did not differ from the parental antibody (Figures S7E and S7F). Injection of the antibodies on the hFcRn coupled column (Figure 5G, Table S3) showed that elution of mAb8 shifted toward ustekinumab by 0.1 in pH, whereas mAb9 dissociated earlier by 0.3 in pH value compared with briakinumab.
Figure 5.
Fv-engineering of briakinumab modulates binding to FcRn and cellular uptake
(A) Sequence alignments of HC and LC of briakinumab, mAb8, and mAb9. Conserved and nonconserved amino acid residues are marked in dark and light blue, respectively, whereas CDR sequences are highlighted by red squares.
(B) Crystal structure of top orientated briakinumab Fv showing the substitutions within mAb8 and mAb9, respectively. CDR loop three of LC and HC are highlighted in red. The HC of briakinumab is shown in orange, and the LC is colored in yellow.
(C) The charge distribution of top oriented briakinumab, mAb8, and mAb9 Fv at pH 7.4. The blue and red colors indicate positive and negative charges. The coloring of the HC and LC of Fvs is kept the same as in (B).
(D) Sequence-based calculation of net charge through pH range of briakinumab, mAb8, and mAb9 Fv, frameworks (without CDRs) and CDRs, respectively. Sequence alignments have been made by Jalview. The figures were designed using PyMOL (www.pymol.org) with the crystallography data (PDB ID code 5N2K [briakinumab]) of human IgG1 (Bloch et al., 2017), and Emboss iep was used for net charge calculation (www.bioinformatics.nl).
(E and F) ELISA binding of titrated amounts (1400.0–0.07 ng/mL) of ustekinumab, briakinumab, mAb8, and mAb9 to hFcRn at pH (E) 5.5 and (F) 7.4. Data are mean ± SD of three independent experiments performed in duplicates.
(G) Analytical hFcRn affinity chromatography of ustekinumab, briakinumab, mAb8, and mAb9 as monomeric fractions. The elution profiles are shown as relative fluorescence intensity and as a function of pH gradient. Fluorescence intensity was normalized and set to one for the clarity. Data are shown as one representative experiment out of three independent experiments.
(H) Uptake of ustekinumab, briakinumab, mAb8, and mAb9 when 400 nM of each variant was added to the cells followed by 4 h incubation, washing, and lysis of the cells.
(I) Recycling of the WTs and Fv-engineered variants when 400 nM of each variant was added to the cells and incubated for 4 h followed by extensive washing and additional 4 h incubation before sample collection.
(J) Residual amount of WTs and Fv-engineered variants. The amounts of IgG variants in all samples were quantified by ELISA, and obtained data are shown as mean ± SD of three independent experiments performed in triplicates. ns, not significant, ∗p = 0.0472 and ∗∗∗∗p < 0.0001, by one-way ANOVA (Dunnett’s multiple comparison test).
(K–N) Uptake of (K) ustekinumab, (L) briakinumab, (M) mAb8, and (N) mAb9 at pH 7.4 when 400 nM of each was added to HMEC1-hFcRn cells treated with a mixture of control siRNA or siRNA targeting the hFcRh HC followed by incubation for 15 min, 30 min, 1 h, and 2 h. After extensive washing, the cells were lysed and added to ELISA for quantification of the amounts of antibody variants. Obtained data are presented as mean ± SD of five experiments performed in duplicates. ns, not significant and ∗∗∗∗p < 0.0001, by two-way ANOVA (Sidak’s multiple comparisons test).
The antibodies were then compared in HERA, which revealed that 2- and 4-fold less of mAb8 and mAb9, respectively, were taken up by the cells compared with briakinumab, and 6- and 12-fold less of mAb8 and mAb9, respectively, were detected in the medium (Figures 5H–5J). Quantification of the remaining amounts in the cells showed 2-fold less of mAb8 than briakinumab, whereas similar amounts of ustekinumab and mAb9 were detected, 6- to 8-fold less than briakinumab (Figure 5J). Cells were then treated with hFcRn HC-specific siRNA, which reduced uptake of mAb8, yet interestingly, uptake was shown to be partly dependent on hFcRn (Figures 5L and 5M). On the other hand, uptake of mAb9 was independent of hFcRn but reduced to a level similar to that of ustekinumab (Figures 5K and 5N). As such, the results illustrate how alteration of the amino acid composition of the Fv may affect cellular uptake and recycling in both FcRn-dependent and -independent manners.
Discussion
Although the FcRn-IgG Fc interaction has been dissected in great detail (Burmeister et al., 1994; Kim et al., 1999; Oganesyan et al., 2014; Medesan et al., 1996; West and Bjorkman, 2000), recent reports have challenged our current view and provided evidence that not only the Fc but also the Fv regions of the Fab arms may modulate receptor binding (Jensen et al., 2015; Piche-Nicholas et al., 2018; Rossini et al., 2020; Schoch et al., 2015; Wang et al., 2011). In this study, we aimed to gain a better understanding on how sequence differences in the Fv domains affect cellular transport and FcRn binding properties. To do so, we chose two monoclonal antibodies, ustekinumab and briakinumab, that bind the same antigen and have the same IgG1 Fc sequence. These antibodies show a remarkable 13 days difference in plasma half-life in humans (Gandhi et al., 2010; Lima et al., 2009; Zhu et al., 2009; Weger, 2010). When we determined the half-life of the recombinant antibody versions in hFcRn expressing mice, a 2.5-fold difference between the antibodies was measured.
Despite this difference in half-life, we show that both antibodies are rescued in an FcRn-dependent manner, as the half-life of both was dramatically reduced in mice lacking expression of the receptor. But the half-life of briakinumab was roughly 7 h shorter than that of ustekinumab in these mice, which confirms a previous report (Kelly et al., 2016). A greater drop in the α-phase of briakinumab indicates an FcRn-independent factor that negatively affects the half-life. The reason for this is very likely related to positively charged patches in the Fv that facilitate increased tissue retention and clearance due to accumulation in highly vascularized organs, such as the liver and spleen (Boswell et al., 2010; Li et al., 2014). Differences in Fv charge patches may also modulate FcRn binding and how efficient antibodies are rescued from intracellular degradation. For instance, it has been postulated that positively charged patches may strengthen the Fab contribution by making contacts with negative charge patches on FcRn, which then will compromise dissociation at neutral pH (Schoch et al., 2015). However, it was very recently suggested that the Fv domains modulate the Fc by allosteric effects (Rossini et al., 2020).
Here, we show that recombinant forms of both briakinumab and ustekinumab bind pH-dependently to hFcRn with minor but distinct differences between the antibodies. By direct coating of the antibodies in ELISA, we detected slightly stronger binding of briakinumab than ustekinumab at both neutral and acidic pH. When antibodies were captured on the antigen, this difference disappeared at acidic pH but not at neutral pH. Using Microscale Thermophoresis technology to measure binding in solution, we detected a clear difference at neutral pH, both in the absence and presence of IL-12. When running through a pH gradient on the hFcRn coupled column, briakinumab dissociated at a more basic pH than ustekinumab, which was not the case in the presence of antigen. These data demonstrate that differences in Fv sequences and binding to antigen can modulate FcRn binding and release, as in agreement with previous reports (Jensen et al., 2017; Rossini et al., 2020).
FcRn resides predominately within endosomal compartments, and as such, only a minor fraction is displayed at the cell surface (Dickinson et al., 1999; D'Hooghe et al., 2017; Ober et al., 2004a; Prabhat et al., 2007). Uptake of IgG has been shown to be mediated by fluid-phase pinocytosis due to no or very weak affinity for the receptor at or near neutral pH (Ober et al., 2004b). However, some data also support that the receptor may be involved (Goebl et al., 2008). Here, we studied the antibodies using HERA (Grevys et al., 2018), which revealed that considerably more of briakinumab was taken up by the cells and then also released at the cell surface following recycling. However, more of briakinumab also accumulated inside the cells compared with ustekinumab. When expression of hFcRn was downregulated by siRNA, we found that uptake of briakinumab and ustekinumab was receptor independent. We also demonstrated that uptake of the antibodies differs greatly, which was a direct result of differences in charge patches of the Fvs. In particular, positive charges of briakinumab were shown to be responsible for enhanced uptake, as replacement of positively charged amino acids by alanines resulted in decreased uptake with the most pronounced effect seen for the LC substitutions. The reason for enhanced uptake is likely due to increased interaction with the negatively charged cell surface that promotes fluid-phase pinocytosis and intracellular accumulation of briakinumab. When five positive charged residues in the HC were replaced, uptake was reduced by 2-fold, whereas a more remarkable effect was seen when three residues in the LC were targeted, which gave a similar HERA profile as that of ustekinumab. Interestingly, the substitutions also shifted the trend of receptor binding toward that of ustekinumab, both by ELISA and analytical hFcRn chromatography, where the latter is in accordance with a previous study (Schoch et al., 2015). Again, the effect on receptor binding was most pronounced for the LC substitutions; this is in line with hydrogen/deuterium exchange mass spectrometry analysis of an IgG1-hFcRn complex, supporting that the LC, but not the HC, of the Fab is modulating FcRn binding (Jensen et al., 2015). Of note, one of the targeted residues (R55A) of the LC of mAb9 is located to the area identified to be required for receptor binding. When we targeted histidines in the LC or HC of briakinumab, none of the engineered variants affected cellular uptake, but less of the variants were recycled back to the medium. Thus, the presence of the histidines seems to favor rescue from degradation, which may be due to a direct or indirect effect on receptor engagement or biophysical properties as a result of engineering.
Previously, it has been demonstrated that FcRn-expressing endothelial cells recycle IgG in complex with antigen but that the efficacy depends on the size of the complex, as large complexes were sorted to lysosomes to a larger degree than small complexes (Weflen et al., 2013). In our study, we did not detect any significant difference in FcRn binding affinity in the absence and presence of antigen, whereas a recent study observed that binding of the antigen may cause changes in the binding kinetics (Rossini et al., 2020). These incongruences may be due to different methods used to study the FcRn-IgG interaction.
Further, we found that briakinumab dissociated from the hFcRn column at lower pH in the presence of IL-12, which gave a similar profile as ustekinumab, as in agreement with previous observations (Jensen et al., 2015). Thus, masking of the Fv sequences altered binding to the receptor through a pH gradient. In line with this, almost equal amounts of IL-12-bound ustekinumab and briakinumab were detected inside the cells following uptake. However, drastic effects on cellular transport were observed when IL-12 was present, as an increase in the uptake range of 2- to 40-fold was measured. Thus, higher amounts of both antibodies returned to the medium than in the absence of antigen, but 10-fold more also accumulated inside the cells, which means that complexes are recycled, but because of increased uptake and accumulation, a larger fraction of complexes may undergo lysosomal degradation than that of monomeric antibodies. Hence, endothelial cells handle antibodies and antibodies in complex with monovalent antigen very differently. As such, Fv charge patches may be masked in the presence of antigen, affecting cellular uptake. The reason for the enhanced cellular uptake in the presence of antigen is not fully understood, and there is therefore a need for further studies that include a larger panel of antibody-antigen pairs. In addition, when we inspected a crystal structure of the monovalent antigen, the surface charge was found to be mostly negative and efficiently shielding the positive charge patch on the briakinumab paratope (Figures S8A–S8H).
Moreover, positive Fv charge patches may affect release from the receptor following recycling and exocytosis, as supported by the distinct differences in dissociation measured on the hFcRn coupled column; this may explain the shorter in vivo half-life of briakinumab. To test this experimentally, we added the YTE substitutions to ustekinumab and briakinumab and determined their half-life in hFcRn transgenic mice. Although YTE extended the half-life of ustekinumab by 1.3-fold, the substitutions also extended the half-life of briakinumab by nearly 1.5-fold. Thus, in this case, Fc-engineering counteracted unfavorable effects of Fv charge patches; this finding may be of value in design of IgG therapeutics with tailored half-life.
When we studied the effect of the YTE substitutions in vitro, enhanced binding to hFcRn was measured, with the same differences observed between the two antibodies as that of the parental versions. In agreement with this, elution was delayed for both antibodies on the hFcRn coupled column where YTE-briakinumab eluted latest with a peak at pH 8.3; this may suggest that release of the antibody is hampered following exocytosis. Using HERA, YTE-briakinumab was taken up more efficiently, which was due to FcRn engagement. Although increased accumulation inside the cells occurred, recycling was also efficient. These findings are interesting in light of previous experiments where we studied an IgG1 variant with five Fc substitutions (M252Y/S254T/T256E/H433K/N434F) that considerably improve binding to hFcRn at both pH conditions, but still with a 100-fold difference in KD at acidic and neutral pH (Grevys et al., 2018). This variant eluted from the hFcRn column at pH 8.6. In HERA, the antibody was very efficiently taken up, accumulated to a larger degree than the WT counterpart but a fraction of the antibodies returned to the medium. This cellular behavior reflects a measured half-life of 3.3 days in hFcRn transgenic mice, which is more than 3-fold longer than the half-life of a WT antibody measured in mice lacking hFcRn. Furthermore, when we aimed to correlate the in vitro cellular data with in vivo half-life values by calculation of the HERA score, it failed for briakinumab and YTE-briakinumab. This was solely due to their enhanced ability to be taken up by cells. When the UUC was taken into consideration, we obtained a reasonable correlation with half-life values in hFcRn transgenic mice. However, to finally conclude, a larger panel of antibodies should be included in follow-up studies.
Taken together, our study demonstrates that the FcRn-IgG interaction is more complex than previously anticipated. We show how charged patch differences in the Fv of monoclonal antibodies can affect FcRn binding, cellular uptake, and transport in both an FcRn-independent and -dependent manner. We also show how the presence of monomeric antigen, which may partly or fully mask Fv features, can drastically alter how bound antibodies are handled by cells. At last, we demonstrate that Fc-engineering for improved FcRn binding may compensate for unfavorable Fv features and provide prolonged half-life. As such, design of antibodies should take into consideration both how the Fab arms and the constant Fc are modulating FcRn-dependent and -independent properties as to secure optimal antigen engagement combined with preferred biodistribution and pharmacokinetics. Our results encourage investigation of larger panels of IgG antibodies with distinct Fab and Fc differences, as well as alternative antibody formats, to gain a more complete understanding of the complex relationship between the structure and functions of antibodies and antibody-mimicking molecules. In this regard, HERA combined with the other in vitro and in vivo systems reported study should be good tools.
Furthermore, antibodies typically bind one or two soluble antigens, which may prolong their persistence, but it may also increase clearance and result in lower effective antibody doses (Chaparro-Riggers et al., 2012; Fukuzawa et al., 2017). To circumvent these drawbacks, so-called acid-switched antibodies have been developed that bind the antigens more strongly at neutral pH than at acidic pH. As such, the antibodies dissociate from their antigen in acidic endosomes following cellular uptake (Igawa et al., 2010a; Devanaboyina et al., 2013; Yang et al., 2017; Bonvin et al., 2015; Kang et al., 2019), which results in rescue from degradation via FcRn while the antigen is degraded. The antibodies can then be reused multiple times. However, when acid-switching has been combined with Fc-engineering for half-life extension, conflicting data have been obtained regarding antigen clearance (Fukuzawa et al., 2017; Igawa et al., 2010a; Henne et al., 2015; Yang et al., 2017). In addition, antibodies that are both acid-switched and engineered for stronger FcRn binding at neutral pH, so-called “sweeping” antibodies, have been developed (Igawa et al., 2013, 2016; Yang et al., 2017). Consequently, less pH-dependent FcRn binding results in rapid accumulation within cells (Vaccaro et al., 2005) and increased clearance of bound antigen (Igawa et al., 2013, 2016; Yang et al., 2017), but also shorter antibody half-life (Dall'Acqua et al., 2002; Gan et al., 2009). Hence, there is a trade-off between increasing cellular accumulation and decreasing half-life of the antibody. Insight into such scenarios may be studies by the use of HERA combined with imaging to track the behavior such of engineered antibody formats as well as their target antigens.
Limitation of the study
A limitation of the study is the number of monoclonal IgG1 antibodies included with defined variable domain biophysical properties, such as positive charged patches. To make firm conclusions, this study should be expanded to assess how different properties of both the light and heavy chains of a large panel of antibodies are affecting cellular transport, in an FcRn-dependent and -independent manner, as well as plasma half-life in human FcRn-expressing mice.
STAR★Methods
Key resource table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human Fc specific antibody | Sigma-Aldrich | I1886; RRID:AB_260125 |
| Polyclonal alkaline phosphatase-conjugated anti-human Fc antibody produced in goat | Sigma-Aldrich | A9544; RRID:AB_258459 |
| monoclonal horseradish peroxidase-conjugated anti-GST antibody produced in goat | Rockland | 600-103-200; RRID:AB_218192 |
| Chemicals, peptides, and recombinant proteins | ||
| Human FcRn | In-house | NA |
| Recombinant human IL-12 | PeproTech | 200-12 |
| Mouse epidermal growth factor (BD | ThermoFisher | PMG8043 |
| MES | Sigma-Aldrich | M3671 |
| NaCl | Sigma-Aldrich | S9888 |
| Tris/HCl | Sigma-Aldrich | T3253 |
| Na2HPO4x2H2O, | Sigma-Aldrich | 71645 |
| NaH2PO4xH2O | Sigma-Aldrich | S9638 |
| 3,3′5,5′-tetramethylbenzine substrate | Merck | CL07 |
| Tween-20 | Sigma-Aldrich | P1379 |
| Skimmed milk | ITW reagents | A0830 |
| phosphate-buffered saline | Sigma-Aldrich | D8537 |
| Hydrocortisone | Sigma-Aldrich | H0888 |
| G418 | ThermoFisher | 10131027 |
| Blasticidin S | ThermoFisher | A1113903 |
| Alkaline phosphatase substrate | Sigma-Aldrich | S0942 |
| K3-EDTA | Sigma-Aldrich | 03664 |
| cOmplete protease inhibitor cocktail | Roche | 11836145001 |
| NT-647-HNS fluorescent dye | Nanotemper technologies | MO-L001 |
| Phosphate-buffered saline | Sigma-Aldrich | D8537 |
| MCDB 131 medium | Gibco | 10372019 |
| RPMI medium | Sigma-Aldrich | R2405 |
| Heat-inactivated fetal calf serum | Sigma-Aldrich | F7524 |
| L-glutamine | ThermoFisher | 25030123 |
| Streptomycin and penicillin | Sigma-Aldrich | P4458 |
| Hank’s Balanced salt solution | ThermoFisher | 14025100 |
| MEM non-essential amino acids | ThermoFisher | 11140050 |
| RIPA buffer | ThermoFisher | 89900 |
| Hank’s Balanced salt solution | ThermoFisher | 14025100 |
| Experimental models: Cell lines | ||
| HEK293E | ATCC | CRL-1573 |
| HMEC1 stably expressing HA-hFcRn-EGFP (HMEC1-hFcRn | Weflen et al., 2013 | Harvard Medical School and Harvard Digestive Diseases Center |
| Experimental models: Organisms/strains | ||
| Hemizygous hFcRn transgenic Tg32 mice, B6.Cg-Fcgrttm1DcrTg(FCGRT)32Dcr/DcrJ | The Jackson Laboratory | NA |
| FcRn-deficient B6-Fcgrttm1Dcr mice | The Jackson Laboratory | NA |
| Oligonucleotides | ||
| Control siRNA | Santa Cruz Biotechnology Inc. | sc-37007 |
| Mixture of three hFcRn HC-specific siRNAs | Santa Cruz Biotechnology Inc | sc-45632 |
| Other | ||
| ustekinumab Fab | RCSB PDB (www.rcsb.org) | 3HMW |
| briakinumab Fab | RCSB PDB | 5N2K |
| ustekinuamb-IL-12 complex | RCSB PDB | 3HMX |
| briakinumab-IL23 complex | RCSB PDB | 5NJD |
| Software and algorithms | ||
| GraphPad Prism 7 | GraphPad Software Inc | https://www.graphpad.com/scientific-software/prism/ |
| Jalview 2 | Waterhouse et al., 2009 | Jalview.org |
| PyMOL | PyMOL by Schrödinger | Pymol.org |
| Emboss iep | Emboss | http://www.bioinformatics.nl/cgi-bin/emboss/iep?_pref_hide_optional=0 |
| pdb2pqr sever | Dolinsky et al., 2004 | http://nbcr-222.ucsd.edu/pdb2pqr_2.1.1/ |
| Other | ||
| 24 well plates | Costar | 10732552 |
| 96-well ELISA plates | Costar | 10544522 |
| CaptureSelecet™ pre-packed anti-human IgG-CH1 column | Life technologies | 494346201 |
| Amicon Ultra-15 mL 50K columns | Millipore | C7715 |
| Superdex 200 increase 10/300 GL column | GE Healthcare | 28–9909-44 |
| Amicon Ultra-0.5 mL 100K columns | Millipore | UFC510096 |
| SDS-PAGE | ThermoFisher | NW00125BOX |
| MycoAlertTM PLUS Mycoplasma detection kit | Lonza | LT07-703 |
| analytical FcRn column | Roche | 08128057001 |
| GSTrap HP column | GE Healthcare | 17–5282-02 |
| HisTrap HP column supplied with Ni2+ ions | GE Healthcare | 17–5027-68 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact: Jan Terje Andersen (j.t.andersen@medisin.uio.no)
Material availability
This study did not generated new unique reagents
Experimental model and subject details
Mice
FcRn-deficient B6-Fcgrttm1Dcr mice of different ages and gender were obtained from The Jackson Laboratory (Bar Harbor, ME). All experiments were performed with 6-12 weeks old male and female mice. Mice were housed in microisolator cages and fed autoclaved food. Institutional Care and Use Committee approved all mouse protocols. Hemizygous hFcRn transgenic Tg32 mice (B6.Cg-Fcgrttm1DcrTg(FCGRT)32Dcr/DcrJ) were used directly at The Jackson Laboratory (by JAX service, Bar Harbor, ME) in accordance with the approved guidelines and regulations.
Method details
Cell culture
Human embryonic kidney (HEK) 293E cells (#CRL-1573, ATCC) were cultured in RPMI 1640 (#R2405, Sigma-Aldrich) supplemented with 10% heat-inactivated fetal calf serum (FCS) (#F7524, Sigma-Aldrich), 2 mM L-glutamine (#25030123, ThermoFisher), 25 μg/mL streptomycin, and 25 U/mL penicillin (#P4458, Sigma-Aldrich). HMEC1 stably expressing HA-hFcRn-EGFP (HMEC1-hFcRn) (Weflen et al., 2013) were grown in MCDB 131 medium (#10372019, Gibco) supplemented with 10% heat-inactivated FCS, 2 mM L-glutamine, 25 μg/mL streptomycin, and 25 U/mL penicillin, 10 ng/mL mouse epidermal growth factor (#PMG8043, ThermoFisher) and 1 μg/mL hydrocortisone (#H0888, Sigma-Aldrich). In addition, 100 μg/mL G418 (#10131027, ThermoFisher) and 50 μg/mL Blasticidine (#A1113903, ThermoFisher) were added to keep the stable expression of hFcRn in HMEC-1-hFcRn cells. All cells were negative for mycoplasma contamination (MycoAlertTM PLUS Mycoplasma detection kit, # LT07-703, Lonza).
Visualization of surface charge distribution
We used the available crystal structures of ustekinumab (3HMW), ustekinuamb-IL-12 complex (3HMX), briakinumab (5N2K) and briakinumab-IL23 complex (5NJD) to calculate and visualize charge distributions on the Fv surfaces. Pqr files were generated using the pdb2pqr sever (http://nbcr-222.ucsd.edu/pdb2pqr_2.1.1/) with pdb2pqr v. 2.1.1 (Dolinsky et al., 2004) and default settings. Then APBS v. 1.3 (Baker et al., 2001) was used to calculate electrostatics with different settings for the pH, using the PARSE force field. Isopotential surfaces were visualized at ± 3 kT/e using PyMOL.
Protein sequence alignment
Jalview 2 (Waterhouse et al., 2009) was used to align ustekinumab and briakinumab protein sequences of Fv.
Calculation of net charges
Emboss iep (http://www.bioinformatics.nl/cgi-bin/emboss/iep?_pref_hide_optional=0) was used to calculate sequence dependent net charges. All occurring cysteines were assumed to form disulfide bridges. Modified Kabat CDR definitions (Jeliazkov et al., 2021) were used: CDR L1 24-34, CDR L2 50-56, CDR L3 89-97, CDR H1 26-35, CDR H2 50-65 and CDR H3 95-102 in Chothia numbering.
Antibody production
cDNA sequences encoding the HC and LC of ustekinumab and briakinumab were designed based on protein sequences (Schoch et al., 2015) and synthesized in the vectors pLNOH2-OriP for HC (pLNOH2-OriP-Ustekinumab-HC and pLNOH2-OriP-Briakinumab-HC) and puC57 with restriction sites BsmI/BamHI for LC (GenScript). The gene fragment of ustekinumab and briakinumab LC were sub-cloned into the vector pLNOH2-OriP (pLNOH2-OriP-Ustekinumab-LC and pLNOH2-OriP-Briakinumab-LC) using restriction endonucleases BsmI and BamHI. Vectors encoding briakinumab CDR variants with substitutions in the CDR3 of LC (H97A) and in the CDR1 and CDR3 of HC (H52A, H98A and H101A) and ustekinumab and briakinumab variants with YTE (M252Y/S2546T/T256E) substitutions were generated by site directed mutagenesis (GenScript). The vectors containing HC and LC of antibodies were transiently transfected at a molar ratio of 1:1.5 into HEK 293E cells using Lipofectamine 2000 as described by the manufacture (#11668500, ThermoFisher), respectively. Growth medium was harvested and replaced every day for the first week and every second day for the second week prior to purification using a CaptureSelecetTM pre-packed anti-human IgG-CH1 column (#494346201, Life technologies) as described by the manufacturer. The collected proteins were up-concentrated and buffer-changed to phosphate-buffered saline (PBS, #D8537, Sigma-Aldrich) using Amicon Ultra-15 mL 50K columns (#C7715, Millipore) prior to size exclusion chromatography using Superdex 200 increase 10/300 GL column (#28–9909-44, GE Healthcare) and an ÄKTA Avant25 (GE Healthcare). The monomeric fractions were up-concentrated by Amicon Ultra-0.5 mL 100K columns (#UFC510096, Millipore). Purified fractions were tested for integrity by reduced and non-reduced 12% SDS-PAGE (#NW00125BOX, ThermoFisher). CDR engineered antibodies mAb8 and mAb9 have been described (Schoch et al., 2015).
Production of recombinant hFcRn
A combi-vector encoding human β2-microglobulin and a truncated version of hFcRn fused in frame with a GST-tag, based on a pcDNA3.1 vector (#V79020, Invitrogen) (Andersen et al., 2008) was transiently transfected into HEK293E cells, and secreted receptor was purified using a GSTrap HP column (#17–5282-02, GE Healthcare) (Andersen et al., 2008).
Production of truncated monomeric His-tagged hFcRn was done in a Baculovirus production system (Firan et al., 2001) with a viral stock obtained from Dr. Sally Ward (University of Southampton, UK). The receptor was purified using a HisTrap HP column supplied with Ni2+ ions (#17–5027-68, GE Healthcare) (Grevys et al., 2015).
FcRn ELISA
Titrated amounts of the antibodies (1400.0-4.5 ng/mL) diluted in PBS were coated directly on 96-well ELISA plates (#10544522, Costar) and incubated overnight followed by washing with PBS containing 0.05% Tween-20 (#P1379, Sigma-Aldrich) (PBS/T) (pH 7.4) or 67 mM phosphate buffer with 0.05% Tween-20 (pH 5.5). 0.5 μg/mL hFcRn-GST was diluted into PBS/T containing 4% of skimmed milk powder (#P1379, ITW reagents) (PBS/T/S) pH 7.4 or pH 5.5 added to the wells and incubated for 1.5 hours. Bound receptor was detected using a monoclonal horseradish peroxidase-conjugated anti-GST antibody produced in goat (#600-103-200; 1:8000, Rockland). Binding was visualised by adding 100 μL of 3,3′5,5′-tetramethylbenzine substrate (#CL07, Merck) and reaction was stopped by adding 50 μL of 1 M HCl. Absorbance was measured at 450 nm using a TECAN spectrophotometer (Sunrise). The same procedure was repeated, when titrated amounts of the antibodies were captured on recombinant human IL-12 (#200-12, PeproTech; 0.5 μg/mL) coated 96-well ELISA.
SPR
SPR was performed using Biacore T200 (GE Healthcare) and CM5 Series S sensor chips following the manufacturer’s protocol. Antibodies (5 μg/mL in 10 mM sodium acetate pH 4.5) were immobilized using amine-coupling chemistry to ∼100 RU, and unreacted moieties on the chips were blocked with 1 M ethanolamine. Phosphate buffer (155 mM phosphate buffer, 85 mM NaCl, 0.05% Tween 20) pH 5.5 and phosphate buffer (195 mM phosphate, 85 mM NaCl, 0.05% Tween 20) pH 7.4 were used as running-buffer and regeneration buffer, respectively. Binding measurements were performed by injecting titrated amounts of monomeric his-tagged hFcRn (32.06–10000.0 nM for ustekinumab, briakinumab, mAb8 and mAb9, and 3.9–1000 nM for ustekinumab-YTE and briakinumab-YTE) with a flow rate of 30 μL/min at 25 °C. The sensorgrams were zero adjusted, and double referenced by subtracting the response from the reference cell and a blank sample. The kinetic rate constants were estimated using the Langmuir 1:1 binding model available in the Biacore T200 Evaluation Software, version 3.0.
Analytical hFcRn affinity chromatography
Analytical hFcRn affinity chromatography was performed using an ÄKTA Avant25 instrument (GE Healthcare). Briefly, 77 μL monomeric or IL12 bound (2 to 1 molar ratio) WT IgG1 and Fc-engineered variants (1 mg/mL) were injected to analytical FcRn column (#08128057001, Roche) and eluted by a linear pH gradient from pH 5.5 to 8.8 within 110 min using 20 mM MES (#M3671, Sigma-Aldrich), 140 mM NaCl (#S9888, Sigma-Aldrich), pH 5.5 and 20 mM Tris/HCl (#T3253, Sigma-Aldrich), 140 NaCl, pH 8.8 as eluent. To determine the elution pH at particular retention time, the pH was monitored by pH detector (GE Healthcare).
Microscale thermophoresis assay
A Monolith NT.115 instrument (Nano Temper Technologies GmbH, Munich, Germany) (Wienken et al., 2010) was used where ustekinumab, ustekinumab-YTE, briakinumab and briakinumab-YTE were labelled with NT-647-HNS fluorescent dye (#MO-L001, Nanotemper Technologies). Further constant amount of each Ab (20 nM) was added to titrated amounts of his-tagged hFcRn (produced in-house) (40000.0-0.3 nM) and incubated for 10 min at RT in PBS pH 7.4 or 100 mM phosphate buffer (6 mM Na2HPO4x2H2O (#71645, Sigma-Aldrich), 94 mM NaH2PO4xH2O (#S9638, Sigma-Aldrich), 150 nM NaCl) pH 5.5. Samples were loaded onto premium-coated capillaries (Nano Temper Technologies). Measurements were performed at 25°C using 50% LED and 50% microscale thermophoresis power for all human IgG1 variants. To determine the affinities of IL-12 bound IgG1 variants to hFcRn, the antibodies and recombinant human IL-12 (PeproTech) were pre-incubated with ustekinumab and briakinumab variants at a 2 to1 molar ratio for 10 min at RT, respectively. All experiments were repeated 2 times in duplicates for each measurement. Data analyses were done using Nano Temper analysis software.
HERA
7.5 × 104 HMEC1 cells stably expressing HA-FcRn-EGFP were seeded into 24 well plates (#10732552, Costar) and cultured for two days in growth medium. The cells were washed twice and starved for 1 hour in Hank’s Balanced salt solution (HBSS; #14025100, ThermoFisher). Next, 400 nM of the monomeric or IL12 bound (2 to 1 molar ratio) antibodies were diluted into 250 μL HBSS (pH 7.4) and added to the cells followed by 4 hours incubation. The medium was removed and the cells were washed 4 times with ice cold HBSS (pH 7.4), before fresh warm HBSS (pH 7.4) was added. Samples were collected at 0 and 4 hours or after overnight incubation. During overnight incubation the cells were provided with the fresh growth medium without FCS and supplemented with non-essential amino acids. Total protein lysates were obtained using the RIPA buffer (#89900, ThermoFisher) supplied with cOmplete protease inhibitor cocktail (#11697498001, Roche). The mixture was incubated on a shaker for 10 min on ice followed by centrifugation for 10 min at 10,000×g to remove cellular debris. Quantification of the amounts of IgG1 was done by ELISA as described below.
The derived values for recycling and residual amount for each antibody were used to calculate the HERA score by the formula: (RX/RWT)/RAX/RWT). The obtained values for uptake by cells treated with control siRNA or siRNA targeting hFcRn HC for each antibody were used to calculate UUC. The following formula was used: ((Control-siRNAUX/Control-siRNAUWT)-(siRNA-hFcRnUX/siRNA-hFcRnUWT))/(siRNA-hFcRnUX/siRNA-hFcRnUWT). If UUC value was between 0 and 1 (0 < UUC <1), it indicates enhanced uptake dominated by FcRn-independent fluid-phase endocytosis. If UUC was more than 1 (UUC >1), it indicates FcRn-dependent uptake. The HERA score was multiplied by UUC when 0 < UUC<1, but not when UUC>1. The parameters R (in ng) is recycling at given time, RA (in ng) is residual amount, siRNA-hFcRnU (in ng) is uptake by cells treated with siRNA targeting hFcRn HC and Control-siRNAU (in ng) is total uptake or uptake by cells treated with control siRNA. X is the protein of interest and WT it the parental variant used to standardize results.
siRNA knockdown of FcRn expression
HMEC1-hFcRn cells were transfected with a control siRNA (sc-37007, Santa Cruz Biotechnology Inc) or mixture of three hFcRn HC-specific siRNAs (sc-45632, Santa Cruz Biotechnology Inc). A siRNA mixture was diluted in siRNA transfection medium and transfection regent for each transfection as described by the manufacturer (Santa Cruz Biotechnology Inc). Transfection mixture was added to cells and incubated for 5-7 h at 37°C in a CO2 incubator followed by adding MCDB 131medium with 20% FCS and 200 U/mL PS. Cells were then incubated for additional 24 h before medium was replaced with MCDB 131 medium.
Quantification of antibodies by ELISA
96-well ELISA plates (Costar) were coated with recombinant human IL-12 (0.5 μg/mL; PeproTech), or goat polyclonal anti-human Fc specific antibody (produced in-house) or anti-human IgG (#I1886, 1:3000, Sigma-Aldrich) diluted to 1 μg/mL in PBS. The plates were blocked with PBS/S for 1 hour at RT followed by washing 4 times with PBS/T. Samples collected from HERA experiments were added directly to wells and titrated 4 times by diluting 1:1 in PBS/S/T and incubated for 2 hours before washing as described above. Captured antibodies were detected with a polyclonal alkaline phosphatase-conjugated anti-human Fc antibody produced in goat (#A9544, 1:4000, Sigma-Aldrich). Binding was visualised by adding of 100 μL alkaline phosphatase substrate (#S0942, Sigma-Aldrich) solved in diethanolamine buffer (made in-house, pH 9.8). Absorbance was measured at 405 nm using a TECAN spectrophotometer (Sunrise).
In vivo studies
The animal experiments and protocols used were reviewed and approved by The Jackson Laboratory Animal Care and Use Committee. The mouse strains used in this study were obtained from The Jackson Laboratory (Bar Harbor, ME). FcRn-deficient B6-Fcgrttm1Dcr mice (aged 9 weeks, weight between 19 and 32 g, 6 mice/group) received 4 mg/kg ustekinumab or briakinumab in 20 mL/kg 1× PBS by intraperitoneal injection. Blood samples (25 μL) were drawn from the retro-orbital sinus at 24, 36, 48, 60, 72, 84, 96, 120 and 144 hours after injection. Hemizygous hFcRn transgenic Tg32 mice (B6.Cg-Fcgrttm1DcrTg(FCGRT)32Dcr/DcrJ, aged 7-8 weeks, weight between 20 and 25 g, 5 mice/group) received 2 mg/kg of IgG antibody in 20 mL/kg 1× PBS by intraperitoneal injection. Blood samples (25 μL) were drawn from the retro-orbital sinus at 1, 3, 5, 7, 10, 12, 16, 19, 23 and 37 days after injection. Following sample collection, the blood was immediately mixed with 1 μL 1% K3-EDTA (#03664, Sigma-Aldrich) to prevent coagulation and then centrifuged at 17,000 × g for 5 min at 4°C. Plasma was isolated, diluted 1:10 in 50% glycerol/PBS solution and then stored at −20°C until analysis. The studies were performed at The Jackson Laboratory (Tg32 study by JAX service, Bar Harbor, ME) in accordance with the approved guidelines and regulations.
Half-life calculation
The plasma concentration of the human IgG1 variants is presented as percentage remaining in circulation at different time points after injection compared to the concentration on day 1 in the Tg32 and FcRn-deficient mice. Nonlinear regression analysis was performed to fit a straight line through the data using the Prism 7 software, and the β-phase half-life was calculated using the formula: t1/2 = log 0.5/(log Ae/A0) × t, where t1/2 is the half-life of the human IgG1 evaluated, Ae is the amount of human IgG1 remaining, A0 is the amount of human IgG1 on day 1 and t is the elapsed time.
Quantification and statistical analysis
Statistical analysis
Data were analysed using GraphPad Prism 7 for Windows (Version 7.02; GraphPad Software Inc.) and Microsoft Excel 2016 (Microsoft). The specific statistical analysis used is given in the figure legends.
Acknowledgments
We are grateful to Sathiaruby Sivaganesh for excellent technical assistance, and Dr. Wayne I. Lencer (Boston Children’|'s Hospital, Harvard Medical School and Harvard Digestive Diseases Center) for the HMEC1 cell line stably expressing HA-hFcRn-EGFP. In part, this work was supported by the Research Council of Norway through its Center of Excellence funding scheme (project no. 179573), while JTA, KFK, and JN were supported by the Research Council of Norway (Grant no. 230526, 274993 and 287927) and JTA and SM by South-Eastern Norway Regional Health Authority (Grant no. 2018052). AG was funded by the Roche postdoctoral fellowship program. The visual abstract was made by BioRender.com.
Author contributions
A.G. and J.T.A. designed research; A.G., J.N., R.F., S.M., K.F.K., K.M.K.S., T.E., and S.F. performed research; A.G., J.N., R.F., S.M., K.F.K., F.M.S., S.F., V.G., K.M.K.S., T.E., J.A.A.F., T.S., I.S., and J.T.A. analyzed data; A.G., S.M., R.F., V.G., K.F.K., T.E., J.A.A.F., T.S., I.S., and J.T.A. revised the paper; A.G. and J.T.A. wrote the paper.
Declaration of interests
T.S., T.E., F.A.A.J., and A.G. are under paid employment by the company F. Hoffmann-La Roche. Other authors declare no conflict of interest.
Published: February 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103746.
Contributor Information
Algirdas Grevys, Email: algirdas.grevys@roche.com.
Jan Terje Andersen, Email: j.t.andersen@medisin.uio.no.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. The protein crystal structures used in this paper have been already published and are available at RCSB PDB (www.rcsb.org). PDB ID codes of each crystal has been added in the REAGENT or RESOURCE table, and in the figure legends. Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- Abdiche Y.N., Yeung Y.A., Chaparro-Riggers J., Barman I., Strop P., Chin S.M., Pham A., Bolton G., Mcdonough D., Lindquist K., et al. The neonatal Fc receptor (FcRn) binds independently to both sites of the IgG homodimer with identical affinity. MAbs. 2015;7:331–343. doi: 10.1080/19420862.2015.1008353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J.T., Justesen S., Fleckenstein B., Michaelsen T.E., Berntzen G., Kenanova V.E., Daba M.B., Lauvrak V., Buus S., Sandlie I. Ligand binding and antigenic properties of a human neonatal Fc receptor with mutation of two unpaired cysteine residues. FEBS J. 2008;275:4097–4110. doi: 10.1111/j.1742-4658.2008.06551.x. [DOI] [PubMed] [Google Scholar]
- Baker N.A., Sept D., Joseph S., Holst M.J., Mccammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch Y., Bouchareychas L., Merceron R., Adanowska K., Bossche L., Detry S., Govindarajan S., Elewaut D., Haerynck F., Dullaers M. Structural activation of pro-inflammatory human cytokine IL-23 by cognate IL-23 receptor enables recruitment of the shared receptor IL-12r 1. Immunity. 2017;48:45–58. doi: 10.1016/j.immuni.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonvin P., Venet S., Fontaine G., Ravn U., Gueneau F., Kosco-Vilbois M., Proudfoot A.E., Fischer N. De novo isolation of antibodies with pH-dependent binding properties. MAbs. 2015;7:294–302. doi: 10.1080/19420862.2015.1006993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth B.J., Ramakrishnan B., Narayan K., Wollacott A.M., Babcock G.J., Shriver Z., Viswanathan K. Extending human IgG half-life using structure-guided design. MAbs. 2018;10:1098–1110. doi: 10.1080/19420862.2018.1490119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boswell C.A., Tesar D.B., Mukhyala K., Theil F.P., Fielder P.J., Khawli L.A. Effects of charge on antibody tissue distribution and pharmacokinetics. Bioconjug. Chem. 2010;21:2153–2163. doi: 10.1021/bc100261d. [DOI] [PubMed] [Google Scholar]
- Burmeister W.P., Huber A.H., Bjorkman P.J. Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature. 1994;372:379–383. doi: 10.1038/372379a0. [DOI] [PubMed] [Google Scholar]
- Carter P.J., Lazar G.A. Next generation antibody drugs: pursuit of the 'high-hanging fruit. Nat. Rev. Drug Discov. 2018;17:197–223. doi: 10.1038/nrd.2017.227. [DOI] [PubMed] [Google Scholar]
- Chaparro-Riggers J., Liang H., Devay R.M., Bai L., Sutton J.E., Chen W., Geng T., Lindquist K., Casas M.G., Boustany L.M., et al. Increasing serum half-life and extending cholesterol lowering in vivo by engineering antibody with pH-sensitive binding to PCSK9. J. Biol. Chem. 2012;287:11090–11097. doi: 10.1074/jbc.M111.319764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Hooghe L., Chalmers A.D., Heywood S., Whitley P. Cell surface dynamics and cellular distribution of endogenous FcRn. PLoS One. 2017;12:e0182695. doi: 10.1371/journal.pone.0182695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall'Acqua W.F., Kiener P.A., Wu H. Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn) J. Biol. Chem. 2006;281:23514–23524. doi: 10.1074/jbc.M604292200. [DOI] [PubMed] [Google Scholar]
- Dall'Acqua W.F., Woods R.M., Ward E.S., Palaszynski S.R., Patel N.K., Brewah Y.A., Wu H., Kiener P.A., Langermann S. Increasing the affinity of a human IgG1 for the neonatal Fc receptor: biological consequences. J. Immunol. 2002;169:5171–5180. doi: 10.4049/jimmunol.169.9.5171. [DOI] [PubMed] [Google Scholar]
- Datta-Mannan A., Thangaraju A., Leung D., Tang Y., Witcher D.R., Lu J., Wroblewski V.J. Balancing charge in the complementarity-determining regions of humanized mAbs without affecting pI reduces non-specific binding and improves the pharmacokinetics. MAbs. 2015;7:483–493. doi: 10.1080/19420862.2015.1016696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanaboyina S.C., Lynch S.M., Ober R.J., Ram S., Kim D., Puig-Canto A., Breen S., Kasturirangan S., Fowler S., Peng L., et al. The effect of pH dependence of antibody-antigen interactions on subcellular trafficking dynamics. MAbs. 2013;5:851–859. doi: 10.4161/mabs.26389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson B.L., Badizadegan K., Wu Z., Ahouse J.C., Zhu X., Simister N.E., Blumberg R.S., Lencer W.I. Bidirectional FcRn-dependent IgG transport in a polarized human intestinal epithelial cell line. J. Clin. Invest. 1999;104:903–911. doi: 10.1172/JCI6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky T.J., Nielsen J.E., Mccammon J.A., Baker N.A. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firan M., Bawdon R., Radu C., Ober R.J., Eaken D., Antohe F., Ghetie V., Ward E.S. The MHC class I-related receptor, FcRn, plays an essential role in the maternofetal transfer of gamma-globulin in humans. Int. Immunol. 2001;13:993–1002. doi: 10.1093/intimm/13.8.993. [DOI] [PubMed] [Google Scholar]
- Fukuzawa T., Sampei Z., Haraya K., Ruike Y., Shida-Kawazoe M., Shimizu Y., Gan S.W., Irie M., Tsuboi Y., Tai H., et al. Long lasting neutralization of C5 by SKY59, a novel recycling antibody, is a potential therapy for complement-mediated diseases. Sci. Rep. 2017;7:1080. doi: 10.1038/s41598-017-01087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Z., Ram S., Vaccaro C., Ober R.J., Ward E.S. Analyses of the recycling receptor, FcRn, in live cells reveal novel pathways for lysosomal delivery. Traffic. 2009;10:600–614. doi: 10.1111/j.1600-0854.2009.00887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi M., Alwawi E., Gordon K.B. Anti-p40 antibodies ustekinumab and briakinumab: blockade of interleukin-12 and interleukin-23 in the treatment of psoriasis. Semin. Cutan. Med. Surg. 2010;29:48–52. doi: 10.1016/j.sder.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Gaudinski M.R., Coates E.E., Houser K.V., Chen G.L., Yamshchikov G., Saunders J.G., Holman L.A., Gordon I., Plummer S., Hendel C.S., et al. Safety and pharmacokinetics of the Fc-modified HIV-1 human monoclonal antibody VRC01LS: a Phase 1 open-label clinical trial in healthy adults. PLoS Med. 2018;15:e1002493. doi: 10.1371/journal.pmed.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam R., Nishimura Y., Pegu A., Nason M.C., Klein F., Gazumyan A., Golijanin J., Buckler-White A., Sadjadpour R., Wang K., et al. A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature. 2016;533:105–109. doi: 10.1038/nature17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetie V., Hubbard J.G., Kim J.K., Tsen M.F., Lee Y., Ward E.S. Abnormally short serum half-lives of IgG in beta 2-microglobulin-deficient mice. Eur. J. Immunol. 1996;26:690–696. doi: 10.1002/eji.1830260327. [DOI] [PubMed] [Google Scholar]
- Ghetie V., Popov S., Borvak J., Radu C., Matesoi D., Medesan C., Ober R.J., Ward E.S. Increasing the serum persistence of an IgG fragment by random mutagenesis. Nat. Biotechnol. 1997;15:637–640. doi: 10.1038/nbt0797-637. [DOI] [PubMed] [Google Scholar]
- Goebl N.A., Babbey C.M., Datta-Mannan A., Witcher D.R., Wroblewski V.J., Dunn K.W. Neonatal Fc receptor mediates internalization of Fc in transfected human endothelial cells. Mol. Biol. Cell. 2008;19:5490–5505. doi: 10.1091/mbc.E07-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevys A., Bern M., Foss S., Bratlie D.B., Moen A., Gunnarsen K.S., Aase A., Michaelsen T.E., Sandlie I., Andersen J.T. Fc engineering of human IgG1 for altered binding to the neonatal Fc receptor affects Fc effector functions. J. Immunol. 2015;194:5497–5508. doi: 10.4049/jimmunol.1401218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevys A., Nilsen J., Sand K.M.K., Daba M.B., Oynebraten I., Bern M., Mcadam M.B., Foss S., Schlothauer T., Michaelsen T.E., et al. A human endothelial cell-based recycling assay for screening of FcRn targeted molecules. Nat. Commun. 2018;9:621. doi: 10.1038/s41467-018-03061-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin M.P., Khan A.A., Esser M.T., Jensen K., Takas T., Kankam M.K., Villafana T., Dubovsky F. Safety, tolerability, and pharmacokinetics of MEDI8897, the respiratory syncytial virus prefusion F-targeting monoclonal antibody with an extended half-life, in healthy adults. Antimicrob. Agents Chemother. 2017;61:e01714. doi: 10.1128/AAC.01714-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne K.R., Ason B., Howard M., Wang W., Sun J., Higbee J., Tang J., Matsuda K.C., Xu R., Zhou L., et al. Anti-PCSK9 antibody pharmacokinetics and low-density lipoprotein-cholesterol pharmacodynamics in nonhuman primates are antigen affinity-dependent and exhibit limited sensitivity to neonatal Fc receptor-binding enhancement. J. Pharmacol. Exp. Ther. 2015;353:119–131. doi: 10.1124/jpet.114.221242. [DOI] [PubMed] [Google Scholar]
- Igawa T., Haraya K., Hattori K. Sweeping antibody as a novel therapeutic antibody modality capable of eliminating soluble antigens from circulation. Immunol. Rev. 2016;270:132–151. doi: 10.1111/imr.12392. [DOI] [PubMed] [Google Scholar]
- Igawa T., Ishii S., Tachibana T., Maeda A., Higuchi Y., Shimaoka S., Moriyama C., Watanabe T., Takubo R., Doi Y., et al. Antibody recycling by engineered pH-dependent antigen binding improves the duration of antigen neutralization. Nat. Biotechnol. 2010;28:1203–1207. doi: 10.1038/nbt.1691. [DOI] [PubMed] [Google Scholar]
- Igawa T., Maeda A., Haraya K., Tachibana T., Iwayanagi Y., Mimoto F., Higuchi Y., Ishii S., Tamba S., Hironiwa N., et al. Engineered monoclonal antibody with novel antigen-sweeping activity in vivo. PLoS One. 2013;8:e63236. doi: 10.1371/journal.pone.0063236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa T., Tsunoda H., Tachibana T., Maeda A., Mimoto F., Moriyama C., Nanami M., Sekimori Y., Nabuchi Y., Aso Y., Hattori K. Reduced elimination of IgG antibodies by engineering the variable region. Protein Eng. Des. Sel. 2010;23:385–392. doi: 10.1093/protein/gzq009. [DOI] [PubMed] [Google Scholar]
- Israel E.J., Wilsker D.F., Hayes K.C., Schoenfeld D., Simister N.E. Increased clearance of IgG in mice that lack beta(2)-microglobulin: possible protective role of FcRn. Immunology. 1996;89:573–578. doi: 10.1046/j.1365-2567.1996.d01-775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain T., Sun T., Durand S., Hall A., Houston N.R., Nett J.H., Sharkey B., Bobrowicz B., Caffry I., Yu Y., et al. Biophysical properties of the clinical-stage antibody landscape. Proc. Natl. Acad. Sci. U S A. 2017;114:944–949. doi: 10.1073/pnas.1616408114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis R. Antibody therapeutics: isotype and glycoform selection. Expert Opin. Biol. Ther. 2007;7:1401–1413. doi: 10.1517/14712598.7.9.1401. [DOI] [PubMed] [Google Scholar]
- Jeliazkov J.R., Frick R., Zhou J., Gray J.J. Robustification of RosettaAntibody and rosetta SnugDock. PLoS One. 2021;16:e0234282. doi: 10.1371/journal.pone.0234282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P.F., Larraillet V., Schlothauer T., Kettenberger H., Hilger M., Rand K.D. Investigating the interaction between the neonatal Fc receptor and monoclonal antibody variants by hydrogen/deuterium exchange mass spectrometry. Mol. Cell Proteomics. 2015;14:148–161. doi: 10.1074/mcp.M114.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P.F., Schoch A., Larraillet V., Hilger M., Schlothauer T., Emrich T., Rand K.D. A two-pronged binding mechanism of IgG to the neonatal Fc receptor controls complex Stability and IgG serum half-life. Mol. Cell Proteomics. 2017;16:451–456. doi: 10.1074/mcp.M116.064675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junghans R.P., Anderson C.L. The protection receptor for IgG catabolism is the beta2-microglobulin-containing neonatal intestinal transport receptor. Proc. Natl. Acad. Sci. U S A. 1996;93:5512–5516. doi: 10.1073/pnas.93.11.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.C., Sun W., Khare P., Karimi M., Wang X., Shen Y., Ober R.J., Ward E.S. Engineering a HER2-specific antibody-drug conjugate to increase lysosomal delivery and therapeutic efficacy. Nat. Biotechnol. 2019;37:523–526. doi: 10.1038/s41587-019-0073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T.H., Jung S.T. Boosting therapeutic potency of antibodies by taming Fc domain functions. Exp. Mol. Med. 2019;51:1–9. doi: 10.1038/s12276-019-0345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R.L., Geoghegan J.C., Feldman J., Jain T., Kauke M., LE D., Zhao J., Wittrup K.D. Chaperone proteins as single component reagents to assess antibody nonspecificity. MAbs. 2017;9:1036–1040. doi: 10.1080/19420862.2017.1356529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R.L., Yu Y., Sun T., Caffry I., Lynaugh H., Brown M., Jain T., Xu Y., Wittrup K.D. Target-independent variable region mediated effects on antibody clearance can be FcRn independent. MAbs. 2016;8:1269–1275. doi: 10.1080/19420862.2016.1208330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.K., Firan M., RADU C.G., Kim C.H., Ghetie V., Ward E.S. Mapping the site on human IgG for binding of the MHC class I-related receptor, FcRn. Eur. J. Immunol. 1999;29:2819–2825. doi: 10.1002/(SICI)1521-4141(199909)29:09<2819::AID-IMMU2819>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Kim J.K., Tsen M.F., Ghetie V., Ward E.S. Localization of the site of the murine IgG1 molecule that is involved in binding to the murine intestinal Fc receptor. Eur. J. Immunol. 1994;24:2429–2434. doi: 10.1002/eji.1830241025. [DOI] [PubMed] [Google Scholar]
- Ko S.Y., Pegu A., Rudicell R.S., Yang Z.Y., Joyce M.G., CHEN X., Wang K., Bao S., Kraemer T.D., Rath T., et al. Enhanced neonatal Fc receptor function improves protection against primate SHIV infection. Nature. 2014;514:642–645. doi: 10.1038/nature13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.H., Kang T.H., Godon O., Watanabe M., Delidakis G., Gillis C.M., Sterlin D., Hardy D., Cogne M., Macdonald L.E., et al. An engineered human Fc domain that behaves like a pH-toggle switch for ultra-long circulation persistence. Nat. Commun. 2019;10:5031. doi: 10.1038/s41467-019-13108-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencer W.I., Blumberg R.S. A passionate kiss, then run: exocytosis and recycling of IgG by FcRn. Trends Cell Biol. 2005;15:5–9. doi: 10.1016/j.tcb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Li B., Tesar D., Boswell C.A., Cahaya H.S., Wong A., Zhang J., Meng Y.G., Eigenbrot C., Pantua H., Diao J., et al. Framework selection can influence pharmacokinetics of a humanized therapeutic antibody through differences in molecule charge. MAbs. 2014;6:1255–1264. doi: 10.4161/mabs.29809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima X.T., Abuabara K., Kimball A.B., Lima H.C. Briakinumab. Expert Opin. Biol. Ther. 2009;9:1107–1113. doi: 10.1517/14712590903092188. [DOI] [PubMed] [Google Scholar]
- Luo J., Wu S.J., Lacy E.R., Orlovsky Y., Baker A., Teplyakov A., Obmolova G., Heavner G.A., Richter H.T., Benson J. Structural basis for the dual recognition of IL-12 and IL-23 by ustekinumab. J. Mol. Biol. 2010;402:797–812. doi: 10.1016/j.jmb.2010.07.046. [DOI] [PubMed] [Google Scholar]
- Martin W.L., West A.P., Jr., Gan L., Bjorkman P.J. Crystal structure at 2.8 A of an FcRn/heterodimeric Fc complex: mechanism of pH-dependent binding. Mol. Cell. 2001;7:867–877. doi: 10.1016/s1097-2765(01)00230-1. [DOI] [PubMed] [Google Scholar]
- Medesan C., Radu C., Kim J.K., Ghetie V., Ward E.S. Localization of the site of the IgG molecule that regulates maternofetal transmission in mice. Eur. J. Immunol. 1996;26:2533–2536. doi: 10.1002/eji.1830261038. [DOI] [PubMed] [Google Scholar]
- Morell A., Terry W.D., Waldmann T.A. Metabolic properties of IgG subclasses in man. J. Clin. Invest. 1970;49:673–680. doi: 10.1172/JCI106279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuber T., Frese K., Jaehrling J., Jager S., Daubert D., Felderer K., Linnemann M., Hohne A., Kaden S., Kolln J., et al. Characterization and screening of IgG binding to the neonatal Fc receptor. MAbs. 2014;6:928–942. doi: 10.4161/mabs.28744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F., Ravetch J.V. Fcgamma receptors as regulators of immune responses. Nat. Rev. Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- Ober R.J., Martinez C., Lai X., Zhou J., Ward E.S. Exocytosis of IgG as mediated by the receptor, FcRn: an analysis at the single-molecule level. Proc. Natl. Acad. Sci. U S A. 2004;101:11076–11081. doi: 10.1073/pnas.0402970101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober R.J., Martinez C., Vaccaro C., Zhou J., Ward E.S. Visualizing the site and dynamics of IgG salvage by the MHC class I-related receptor, FcRn. J. Immunol. 2004;172:2021–2029. doi: 10.4049/jimmunol.172.4.2021. [DOI] [PubMed] [Google Scholar]
- Oganesyan V., Damschroder M.M., Cook K.E., Li Q., Gao C., Wu H., Dall'Acqua W.F. Structural insights into neonatal Fc receptor-based recycling mechanisms. J. Biol. Chem. 2014;289:7812–7824. doi: 10.1074/jbc.M113.537563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piche-Nicholas N.M., Avery L.B., King A.C., Kavosi M., Wang M., O'hara D.M., Tchistiakova L., Katragadda M. Changes in complementarity-determining regions significantly alter IgG binding to the neonatal Fc receptor (FcRn) and pharmacokinetics. MAbs. 2018;10:81–94. doi: 10.1080/19420862.2017.1389355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhat P., Gan Z., Chao J., Ram S., Vaccaro C., Gibbons S., Ober R.J., Ward E.S. Elucidation of intracellular recycling pathways leading to exocytosis of the Fc receptor, FcRn, by using multifocal plane microscopy. Proc. Natl. Acad. Sci. U S A. 2007;104:5889–5894. doi: 10.1073/pnas.0700337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan M., Bonagura V.R., Morrison S.L., Bjorkman P.J. Analysis of the ph-dependence of the neonatal Fc receptor immunoglobulin-G interaction using antibody and receptor variant. Biochemistry. 1995;34:14649–14657. doi: 10.1021/bi00045a005. [DOI] [PubMed] [Google Scholar]
- Reichert J.M. Antibodies to watch in 2017. MAbs. 2017;9:167–181. doi: 10.1080/19420862.2016.1269580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbie G.J., Criste R., Dall'Acqua W.F., Jensen K., Patel N.K., Losonsky G.A., Griffin M.P. A novel investigational Fc-modified humanized monoclonal antibody, motavizumab-YTE, has an extended half-life in healthy adults. Antimicrob. Agents Chemother. 2013;57:6147–6153. doi: 10.1128/AAC.01285-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossini S., Noe R., Daventure V., Lecerf M., Justesen S., Dimitrov J.D. V region of IgG controls the molecular properties of the binding site for neonatal Fc receptor. J. Immunol. 2020;205:2850–2860. doi: 10.4049/jimmunol.2000732. [DOI] [PubMed] [Google Scholar]
- Schoch A., Kettenberger H., Mundigl O., Winter G., Engert J., Heinrich J., Emrich T. Charge-mediated influence of the antibody variable domain on FcRn-dependent pharmacokinetics. Proc. Natl. Acad. Sci. U S A. 2015;112:5997–6002. doi: 10.1073/pnas.1408766112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simister N.E., Mostov K.E. An Fc receptor structurally related to MHC class I antigens. Nature. 1989;337:184–187. doi: 10.1038/337184a0. [DOI] [PubMed] [Google Scholar]
- Stapleton N.M., Andersen J.T., Stemerding A.M., Bjarnarson S.P., Verheul R.C., Gerritsen J., Zhao Y., Kleijer M., Sandlie I., De Haas M., et al. Competition for FcRn-mediated transport gives rise to short half-life of human IgG3 and offers therapeutic potential. Nat. Commun. 2011;2:599. doi: 10.1038/ncomms1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Ishii-Watabe A., Tada M., Kobayashi T., Kanayasu-Toyoda T., Kawanishi T., Yamaguchi T. Importance of neonatal FcR in regulating the serum half-life of therapeutic proteins containing the Fc domain of human IgG1: a comparative study of the affinity of monoclonal antibodies and Fc-fusion proteins to human neonatal FcR. J. Immunol. 2010;184:1968–1976. doi: 10.4049/jimmunol.0903296. [DOI] [PubMed] [Google Scholar]
- Vaccaro C., Zhou J., Ober R.J., Ward E.S. Engineering the Fc region of immunoglobulin G to modulate in vivo antibody levels. Nat. Biotechnol. 2005;23:1283–1288. doi: 10.1038/nbt1143. [DOI] [PubMed] [Google Scholar]
- Vaughn D.E., Bjorkman P.J. Structural basis of pH-dependent antibody binding by the neonatal Fc receptor. Structure. 1998;6:63–73. doi: 10.1016/s0969-2126(98)00008-2. [DOI] [PubMed] [Google Scholar]
- Vidarsson G., Dekkers G., Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front Immunol. 2014;5:520. doi: 10.3389/fimmu.2014.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lu P., Fang Y., Hamuro L., Pittman T., Carr B., Hochman J., Prueksaritanont T. Monoclonal antibodies with identical Fc sequences can bind to FcRn differentially with pharmacokinetic consequences. Drug Metab. Dispos. 2011;39:1469–1477. doi: 10.1124/dmd.111.039453. [DOI] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weflen A.W., Baier N., Tang Q.J., Van den Hof M., Blumberg R.S., Lencer W.I., Massol R.H. Multivalent immune complexes divert FcRn to lysosomes by exclusion from recycling sorting tubules. Mol. Biol. Cell. 2013;24:2398–2405. doi: 10.1091/mbc.E13-04-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger W. Current status and new developments in the treatment of psoriasis and psoriatic arthritis with biological agents. Br. J. Pharmacol. 2010;160:810–820. doi: 10.1111/j.1476-5381.2010.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.P., Jr., Bjorkman P.J. Crystal structure and immunoglobulin G binding properties of the human major histocompatibility complex-related Fc receptor(,) Biochemistry. 2000;39:9698–9708. doi: 10.1021/bi000749m. [DOI] [PubMed] [Google Scholar]
- Wienken C.J., Baaske P., Rothbauer U., Braun D., Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- Yang D., Giragossian C., Castellano S., Lasaro M., Xiao H., Saraf H., Hess Kenny C., Rybina I., Huang Z.F., Ahlberg J., et al. Maximizing in vivo target clearance by design of pH-dependent target binding antibodies with altered affinity to FcRn. MAbs. 2017;9:1105–1117. doi: 10.1080/19420862.2017.1359455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J., Chamberlain A.K., Horton H.M., Karki S., Leung I.W., Sproule T.J., Lazar G.A., Roopenian D.C., Desjarlais J.R. Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 2010;28:157–159. doi: 10.1038/nbt.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Hu C., Lu M., Liao S., Marini J.C., Yohrling J., Yeilding N., Davis H.M., Zhou H. Population pharmacokinetic modeling of ustekinumab, a human monoclonal antibody targeting IL-12/23p40, in patients with moderate to severe plaque psoriasis. J. Clin. Pharmacol. 2009;49:162–175. doi: 10.1177/0091270008329556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. The protein crystal structures used in this paper have been already published and are available at RCSB PDB (www.rcsb.org). PDB ID codes of each crystal has been added in the REAGENT or RESOURCE table, and in the figure legends. Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.