Abstract
The aberrant self-assembly of intrinsically disordered proteins (IDPs) into soluble oligomers and their interactions with biological membranes underlie the pathogenesis of numerous neurodegenerative diseases, including Alzheimer’s disease. Catechins have emerged as useful tools to reduce the toxicity of IDP oligomers by modulating their interactions with membranes. However, the structural determinants of catechin binding to IDP oligomers and membranes remain largely elusive. Here, we assemble a catechin library by combining several naturally occurring chemical modifications and, using a coupled NMR-statistical approach, we map at atomic resolution the interactions of such library with the Alzheimer’s-associated amyloid-beta (Aβ) oligomers and model membranes. Our results reveal multiple catechin affinity drivers and show that the combination of affinity-reducing covalent changes may lead to unexpected net gains in affinity. Interestingly, we find that the positive cooperativity is more prevalent for Aβ oligomers than membrane binding, and that the determinants underlying catechin recognition by membranes are markedly different from those dissected for Aβ oligomers. Notably, we find that the unanticipated positive cooperativity arises from the critical regulatory role of the gallate catechin moiety, which recruits previously disengaged substituents into the binding interface and leads to an overall greater compaction of the receptor-bound conformation. Overall, the previously elusive structural attributes mapped here provide an unprecedented foundation to establish structure-activity relationships of catechins.
Keywords: Alzheimer disease, amyloid-beta (Aβ), catechins, membrane, protein aggregation, nuclear magnetic resonance (NMR), small molecule, structure-function
Abbreviations: Aβ, amyloid-beta; CG, (-)-catechin-3-gallate; EC, (-)-epicatechin; ECG, (-)-epicatechin-3-gallate; EGC, (-)-epigallocatechin; EGCG, (-)-epigallocatechin-3-gallate; GCG, (-)-gallocatechin-3-gallate; IDPs, intrinsically disordered proteins; MG, methyl 3,4,5-trihydroxybenzoate; NOE, Nuclear Overhauser effects; ROE, Rotational Overhauser effect; ROESY, Rotational Overhauser effect spectroscopy; STD, Saturation Transfer Difference; STDaf, STD amplification factors; STR, Saturation Transfer Reference; SUVs, small unilamellar vesicles; TrNOESY, transfer Nuclear Overhauser effect spectroscopy
The self-association of intrinsically disordered proteins (IDPs) into toxic oligomers underlies the pathogenesis of several neurodegenerative and non-neuropathic systemic disorders (1, 2). One of the key mechanisms by which oligomers confer toxicity is through disrupting the integrity of biological membranes (3). Thus, it is critical to identify compounds that modulate the deleterious interactions of IDP oligomers with membranes. In this regard, several classes of compounds have been identified to date, including small molecules (4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17), peptides/peptidomimetics (18, 19), molecular chaperones (20, 21, 22, 23, 24, 25), and antibodies (22, 26). One notable ligand that has garnered significant attention in recent years is (-)-epigallocatechin-3-gallate (EGCG). Catechins, such as EGCG, remodel toxic oligomers of IDPs into nontoxic species by modulating the solvent exposure of such assemblies and their interactions with lipid membranes (4, 6, 7, 27, 28, 29).
EGCG and other catechins bind oligomeric species of amyloid-beta (Aβ) and several other IDPs such as ⍺-synuclein, tau, IAPP, p53, and MSP2 (28, 30, 31, 32, 33, 34). Molecular dynamics (MD) simulations have shown that the central core region of Aβ aggregates interacts strongly with EGCG, due to the presence of hydrophobic residues (27). In addition, we previously revealed by NMR that EGCG can bind Aβ oligomers at multiple equivalent and independent sites with a sub-mM affinity per-site (6). In contrast, EGCG binds monomeric Aβ more weakly with mM affinities (6, 30, 35). Furthermore, Wang et al. (35) found that the binding of EGCG to Aβ monomers spans a wide range of stoichiometries consistent with multiple, largely aspecific-binding modes. In addition, Fusco et al. (30) determined through NMR that EGCG only marginally affects the conformational properties of monomeric Aβ, in agreement with the weak binding between the catechin and Aβ monomers. Taken together, these results suggest that the binding of catechins to Aβ oligomeric species largely accounts for their inhibitory effect on amyloid formation.
Despite significant progress in understanding the interactions between catechins and the IDP oligomers they detoxify, the underlying mechanism of action remains still largely elusive. This is primarily because most prior studies used catechins simply as chemical modulators of IDP-oligomer neurotoxicity and focused primarily on the analysis of IDPs as opposed to the actual catechins (4, 6, 7, 27, 36, 37). Furthermore, it remains unclear to what extent the reduction in neurotoxicity arises from the binding of catechins to IDP oligomers versus membranes. Taken together, the current understanding of how catechin structure relates to IDP oligomer versus membrane binding is at best scant. Addressing this gap and finding the drivers of catechins-oligomeric Aβ/membrane interactions is essential to inform the rational design of new amyloid inhibitors with enhanced potency, pharmacokinetic properties, and bioavailability (9, 37, 38, 39).
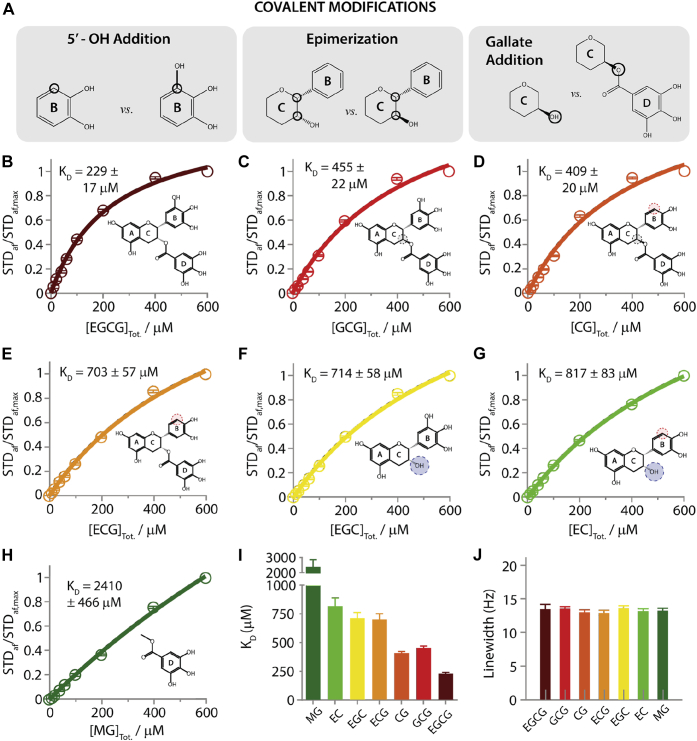
In an effort to establish structure-activity relationships (SARs) for catechins, we assembled a tripartite system composed of a prototypical IDP prone to form toxic oligomers, that is, amyloid beta 1 to 40 (Aβ40), biological membrane mimetics, that is, DOPE:DOPS:DOPC small unilamellar vesicles (SUVs), and a targeted catechin library. The catechin library was generated starting from the parent EGCG compound (Fig. 1B) and combining three modifications that preserve the catechin flavan-3-ol scaffold spanning rings A to C: (i) the elimination of the ‘gallo’ 5′-hydroxyl in ring B (denoted by the first G in EGCG; Fig. 1A, left), (ii) the epimerization of ring C (denoted by the first E in EGCG; Fig. 1A, center), and (iii) the removal of the ‘gallate’ ring D (denoted by the second G in EGCG; Fig. 1A, right). The resulting library includes seven catechins: EGCG, (-)-gallocatechin-3-gallate (GCG), (-)-catechin-3-gallate (CG), (-)-epicatechin-3-gallate (ECG), (-)-epigallocatechin (EGC), (-)-epicatechin (EC) (Fig. 1, B–G), and the separate gallate moiety methyl 3,4,5-trihydroxybenzoate (MG) (Fig. 1H). For each member of the library we measured affinities for Aβ40 oligomers and SUVs, the corresponding binding epitope maps and the inter-ring interactions either free or bound to oligomers or membranes.
Figure 1.
Binding affinities of catechins to Aβ40 oligomers.A, schematic indicating the three different covalent modifications of the catechin backbone used to build the catechin library. B–H, STD-derived binding isotherms for the interactions of Aβ40 oligomers with (B) EGCG, (C) GCG, (D) CG, (E) ECG, (F) EGC, (G) EC, and (H) MG. I, the binding affinities of catechins for Aβ40 oligomers derived from fitting the normalized STD amplification factors shown in (B–H) using a Scatchard-like binding model. J, linewidths of Aβ40 aromatic resonances, which are sensitive to monomer – oligomer exchange, for the Aβ40 samples to which catechins were titrated. Aβ, amyloid-beta; CG, (-)-catechin-3-gallate; EC, (-)-epicatechin; ECG, (-)-epicatechin-3-gallate; EGC, (-)-epigallocatechin; EGCG, (-)-epigallocatechin-3-gallate; GCG, (-)-gallocatechin-3-gallate; MG, methyl 3,4,5-trihydroxybenzoate; STD, Saturation Transfer Difference; STDaf, STD amplification factors.
The resulting comprehensive datasets were systematically analyzed through agglomerative clustering and other statistical approaches to distill out the key drivers of catechin interactions with Aβ oligomers and membranes. The comparative analysis of the affinities measured for our targeted catechin library revealed the contribution of distinct catechin moieties to the binding affinity for Aβ oligomers versus membranes. In addition, the comparison of binding epitope maps and inter-ring interactions revealed how a given structural component modulates the interactions of other components with the target receptor. Through this integrated experimental approach, we unexpectedly found unique oligomer versus membrane differences in terms of catechin interactions and underlying drivers, including the degree to which covalent modifications contribute additively to the overall binding. Moreover, we show that although our catechin library interacts with both Aβ oligomers and membranes, only one set of interactions correlates with Aβ oligomer toxicity under our conditions. Thus, the structural information emerging from this work provides a foundation to develop catechin SARs.
Results
Dissecting the drivers of catechin-binding affinities for Aβ40 oligomers
To evaluate the contribution of distinct catechin moieties to Aβ40 oligomer binding, we measured oligomer-binding affinities for the catechins in our library through Saturation Transfer Difference (STD) NMR. Comparisons of such affinities (Fig. 1, B–H) reveals that our library spans KD values varying by approximately one-order of magnitude in the mM to sub-mM range (Fig. 1I). The most significant contributions to the binding affinity of EGCG arise from the 5′-OH of ring B and the gallate (ring D). In going from either ECG or EGC to EGCG, the KD values decreases by ∼three-fold (Fig. 1, B, E and F), whereas the epimerization of ring C results in only a ∼two-fold enhancement of affinity for Aβ40 oligomers (Fig. 1, B and C). These variations in affinities within our catechin library are unlikely to arise from shifts in the Aβ40 monomer-oligomer equilibria, as indicated by the similarity in Aβ40 linewidths observed for different catechin samples (Fig. 1J) (20, 40, 41, 42), suggesting similar fractions of oligomers. Hence, the affinity gradient observed within the catechin library (Fig. 1I) is expected to genuinely reflect differences in substituent-specific contributions to Aβ40 oligomer-catechin binding and in first approximation, these rank in the order: 5′-OH ring B (gallo) ∼ gallate (ring D) > epimerization (ring C). However, it is notable that such contributions are not fully independent of each other. For example, the 5′-OH or gallate modification of EC (Fig. 1G) to EGC (Fig. 1F) and ECG (Fig. 1E), respectively, only marginally enhances the binding affinity, whereas the simultaneous addition of both modifications, as in EGCG (Fig. 1B), enhances the binding affinity ∼four-fold.
To quantitatively evaluate the coupling between the 5′-OH and gallate contributions to Aβ40 oligomer binding, we relied on a double-substituent cycle that includes the unmodified parent catechin EC, the two singly modified EC-analogs (ECG and EGC), and the doubly modified compound (EGCG), as shown in Figure 2A. The Aβ40 oligomer binding nonadditivity is then quantified by the gamma parameter defined as:
| (1) |
where implies full additivity, whereas (γ > 1) implies nonadditivity with positive (negative) cooperativity. Based on the data of Figure 1, is 0.37 ± 0.06 (Fig. 2C), suggesting that the 5′-OH and gallate catechin contributions to Aβ40 oligomer binding exhibit positive cooperativity.
Figure 2.
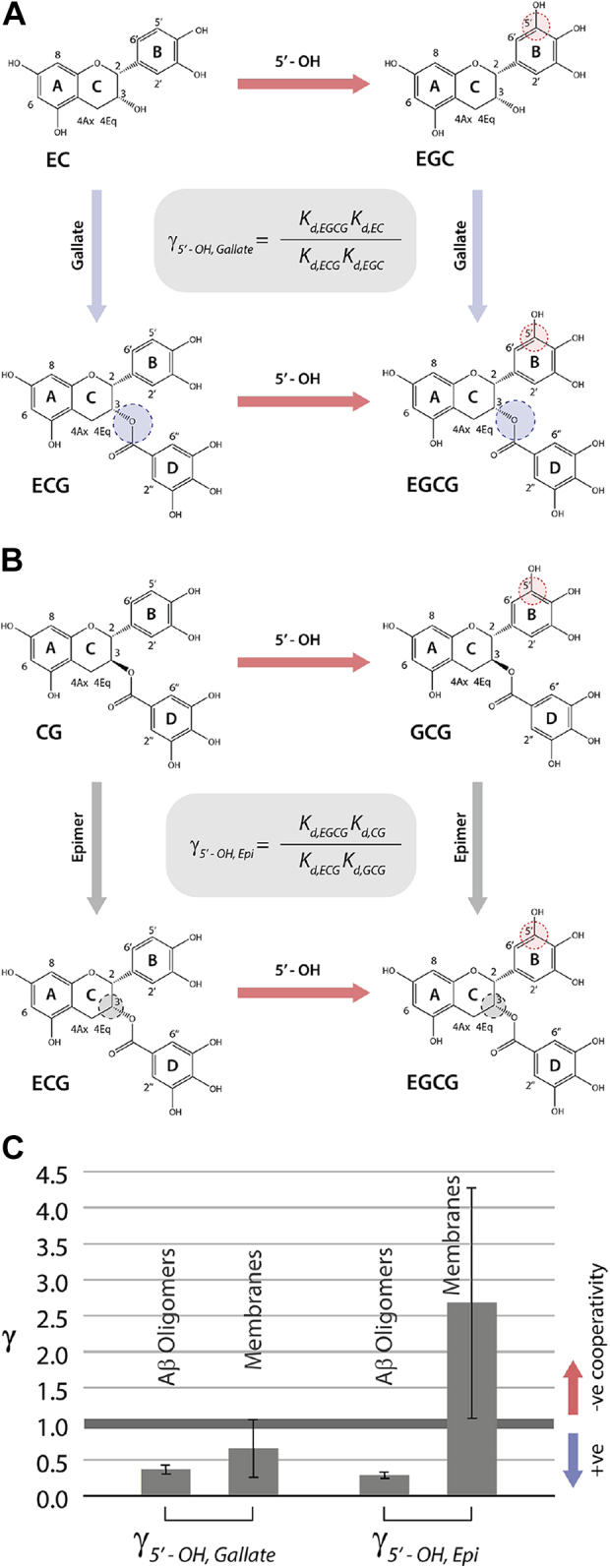
Binding nonadditivity of catechin structural modifications for Aβ oligomers versus membranes.A, double-substituent cycle for the 5′-OH (ring B) and gallate (ring D) substituents of the catechin scaffold. B, as (A) but for the 5′-OH and epimerization modification (ring C). C, corresponding nonadditivity factors (γ) for the cycles in (A and B) computed using Equations 1 and 2, respectively, as applied to the binding to Aβ oligomers and membranes. Aβ, amyloid-beta; CG, (-)-catechin-3-gallate; EC, (-)-epicatechin; ECG, (-)-epicatechin-3-gallate; EGC, (-)-epigallocatechin; EGCG, (-)-epigallocatechin-3-gallate; GCG, (-)-gallocatechin-3-gallate.
We also evaluated the coupling between the 5′-OH and epimerization contributions to Aβ40 oligomer binding through a similar double-substituent cycle that combines CG, the two singly modified CG-analogs (ECG and GCG), and the doubly modified compound (EGCG), as shown in Figure 2B. In this case, the Aβ40 oligomer binding nonadditivity quantified by the gamma parameter is defined as:
| (2) |
Using the data of Figure 1, is 0.29 ± 0.04 (Fig. 2C), which is significantly <1 and points to the presence of positive cooperativity also between the 5′-OH and epimerization contributions to Aβ40 oligomer binding. Hence, our data reveal that catechin binding to Aβ40 oligomers is enhanced by the synergy of the 5′-OH in ring B with both the stereochemistry of ring C and the gallate (ring D). Yet, the gallate moiety alone is insufficient to bind Aβ40 oligomers with sub-mM affinity, as shown by the mM Kd value observed for MG (Fig. 1H). To assess to what extent these binding patterns are conserved in the interactions between catechins and membranes, we extended similar affinity measurements to DOPE:DOPS:DOPC SUVs (Fig. 3).
Figure 3.
Binding affinities of catechins to DOPE:DOPS:DOPC Small Unilamellar Vesicles (SUVs). STD-derived binding isotherms for the interactions of SUVs with (A) EGCG, (B) GCG, (C) CG, (D) ECG, (E) EGC, (F) EC, and (G) MG. H, the binding affinities of catechins for SUVs derived from fitting the normalized STD amplification factors shown in (A–H) using a Scatchard-like binding model. I, correlation between catechin-binding affinities for Aβ oligomers versus membranes. The affinities are reported as log scales. Aβ, amyloid-beta; CG, (-)-catechin-3-gallate; EC, (-)-epicatechin; ECG, (-)-epicatechin-3-gallate; EGC, (-)-epigallocatechin; EGCG, (-)-epigallocatechin-3-gallate; GCG, (-)-gallocatechin-3-gallate; MG, methyl 3,4,5-trihydroxybenzoate; STD, Saturation Transfer Difference; STDaf, STD amplification factors.
Dissecting the drivers of catechin-binding affinities for membranes
We also measured the membrane-binding affinities for the same catechin library used to probe the catechin-Aβ oligomer interactions (Fig. 1). To this end and to allow for an unbiased membrane versus Aβ oligomers binding comparison, we relied again on STD NMR, the same technique used for the Aβ oligomers. The resulting membrane-binding isotherms and corresponding Kd values are summarized in Figure 3, A–H. The catechins bind membranes with affinities consistently falling in the sub-mM range (Fig. 3H). No correlations were detected between the Kd values measured for Aβ40 oligomers and membranes (Fig. 3I), and the Kd gradient observed for Aβ oligomers (Fig. 1I) is to a large extent lost in the case of membrane binding, for which a more uniform distribution of affinities is observed (Fig. 3H). The two major drivers of Aβ40 oligomer binding, that is, the 5′-OH ring B (gallo) and the gallate (ring D), now appear to contribute only marginally to membrane binding (Fig. 3, A and D–F), whereas the epimerization of ring C enhances the affinity for membranes (Fig. 3B) rather than decreasing it (Fig. 1C). Overall, our data reveal markedly different patterns in the SARs for catechin binding to Aβ40 oligomers versus membranes (Figure 1, Figure 2, Figure 3).
Interestingly, the cooperativity between the epimerization and 5′-OH contributions switches from positive to negative in the case of membrane binding (Fig. 2C). In addition, the synergy between the 5′-OH and gallate substituents is also diminished in going from Aβ40 oligomers to membranes (Fig. 2C), and the gallate alone without the catechin scaffold still exhibits significantly lower affinities compared to the other members of the library (Fig. 3, G–I). Taken together, our data show distinct differences between Aβ40 oligomers and membranes in terms of the catechin-binding determinants and their cooperativity (Figure 1, Figure 2, Figure 3). To obtain further insight into such Aβ40 oligomer versus membrane dissimilarities, and to understand how the various modifications work cooperatively to enhance (reduce) binding, we mapped the catechin-binding epitopes (Fig. 4) and the changes in free versus bound catechin conformations (Fig. 5).
Figure 4.
Catechins exhibit unique binding epitopes for Aβ40 oligomers versus membranes.A, violin plots indicating the normalized STD:STR ratios of protons in rings A to D for catechins from our library either containing or devoid of ring D, both in the presence of Aβ40 oligomers. One-way ANOVA and subsequent Tukey's post-hoc test was used to determine statistical significance between rings, with ∗, ∗∗, ∗∗∗, and ∗∗∗∗ representing p-values of <0.05, <0.01, <0.001, and <0.0001, respectively. Additional details are provided in the methods. B, violin plots indicating the normalized STD:STR ratios of protons in rings A to D for catechins from our library with the ring C stereocenters in cis or trans configuration, both in the presence of Aβ40 oligomers. C, schematic diagram displaying the coupling between catechin rings in the presence of Aβ40 oligomers, as indicated by the dashed lines. The sizes of the colored circles qualitatively represent the relative extent of saturation transfer among the various catechin rings A to D. D, dendrogram displaying the clusters obtained through single linkage agglomerative clustering of the normalized STD:STR ratios in the presence of Aβ40 oligomers using Spearman rank correlation. E–H, as (A–D) except in the presence of DOPE:DOPS:DOPC membranes. Aβ, amyloid-beta; CG, (-)-catechin-3-gallate; EC, (-)-epicatechin; ECG, (-)-epicatechin-3-gallate; EGC, (-)-epigallocatechin; EGCG, (-)-epigallocatechin-3-gallate; GCG, (-)-gallocatechin-3-gallate; n.s., not significant; STD, Saturation Transfer Difference; STR, Saturation Transfer Reference.
Figure 5.
Conformation of catechins free and bound to Aβ40 oligomers and membranes. Off-resonance ROESY and NOESY intensity heat maps for free, Aβ40 oligomer-, and membrane-bound (A) EGCG, (B) GCG, (C) CG, (D) ECG, (E) EGC and (F) EC, respectively. Stronger normalized NOE (ROE) intensities are reported in red, whereas weaker intensities are shown in yellow. Gray bars indicate protons that overlap with the H2O resonance. G, bar graph indicating the normalized intensities of NOE (ROE) cross-peaks between ring D and B protons in the free (purple), oligomer bound (red), and membrane bound (blue) states, with ∗, ∗∗, ∗∗∗, and ∗∗∗∗ representing p-values of <0.05, <0.01, <0.001, and <0.0001, respectively. H, as (G) except between ring B and C protons. I, as (G) except between ring C and D protons. CG, (-)-catechin-3-gallate; EC, (-)-epicatechin; ECG, (-)-epicatechin-3-gallate; EGC, (-)-epigallocatechin; EGCG, (-)-epigallocatechin-3-gallate; GCG, (-)-gallocatechin-3-gallate; NOESY, Nuclear Overhauser effect spectroscopy; ns, not significant; ROESY, Rotational Overhauser effect spectroscopy.
Aβ40 oligomer versus membrane-binding epitopes in catechins
Using saturation transfer NMR, we mapped for each of the catechins in our library the epitopes for binding Aβ40 oligomers (Figs. 4, A–D and S1) and membranes (Figs. 4, E–H and S2). Focusing first on the catechin epitope maps measured for Aβ40 oligomers, the highest STD/Saturation Transfer Reference (STR) ratios consistently observed for all catechins are those of protons in ring A (Figs. 4, A and B and S1). Whenever present, ring D receives saturation to levels approaching ring A (Figs. 4, A and B and S1), explaining why ring D increases Aβ40 oligomer–binding affinities (Fig. 1). Hence, rings A and D emerge as the primary anchoring points to Aβ40 oligomers, whereas rings B and C are generally subject to a lower degree of saturation transfer relative to rings A and D (Fig. 4C).
Another difference between the saturation patterns observed for rings A,D versus B,C is that the STD/STR ratios of the latter are quite variable across our catechin library and are dependent on the presence of the former (Fig. 4, A and B). For example, in the absence of ring D, we observe marginal saturation transfer from Aβ40 oligomers to rings B and C (Fig. 4A, left). In contrast, catechins containing ring D further recruit rings B and C into the binding interface, as evidenced by the significantly higher STD/STR ratios (Fig. 4A, right versus left; Fig. S1D versus Fig. S1F). These observations offer a viable explanation for the positive-binding cooperativity observed between the 5′-OH (ring B) and gallate (ring D) modifications (Fig. 2C). Unlike the inclusion of ring D, epimerization of ring C does not significantly affect the STD/STR ratios observed for other rings, but it reduces the saturation transfer differential between rings B and C (Fig. 4B left versus right). Furthermore, when comparing the various catechins in our library based on their Aβ40 oligomer binding epitopes, we observe that catechins containing ring D are effectively partitioned away from those without ring D (Fig. 4D). Taken together, our STD data reveal that the ring-specific engagement in Aβ oligomer binding ranks as A ≥ D > B > C (Fig. 4C), with ring D serving not only as a critical anchoring point that enhances affinities but also as a recruiter of rings B and C.
To assess to what degree similar patterns are transferable to the interactions with membranes, we extended our statistical STD analyses to catechin-SUV interactions (Figs. 4, E–H and S2). Similar to the case of Aβ oligomer binding, also for membrane binding, ring A remains the primary anchoring point with the highest STD/STR ratios consistently observed for all catechins (Figs. 4, E and F and S2). However, in the case of membranes, no clear ranking emerges across the catechin library for the extent of saturation transferred to the other rings. This observation is consistent with the more uniform distribution of binding affinities observed across our catechin library in the case of membranes (Fig. 3H). Furthermore, inclusion of ring D does not result in significant enhancement of saturation transfer from membranes to rings B and C (Fig. 4E left versus right), pointing to a loss of the couplings between rings D and B,C detected for Aβ oligomers (Fig. 4A left versus right). Such losses are consistent with the reduction in positive cooperativity between the binding contributions of the 5′-OH gallo modification in ring B and the gallate (ring D; Fig. 2C). In addition, ring D-containing catechins cannot be fully partitioned away from catechins lacking ring D based on the binding epitopes (Fig. 4H). Overall, the STD-based analyses of binding epitopes across our catechin library reveal that the distinct ring rankings and couplings observed for Aβ oligomer binding (Fig. 4, A–D) are mostly lost in the case of membrane binding (Fig. 4, E–H). To better understand these Aβ oligomer versus membrane differences in catechin recognition, we then proceeded to probe the inter-ring interactions through Rotational Overhauser effects (ROEs) and Nuclear Overhauser effects (NOEs).
Inter-ring interactions in free, Aβ40 oligomer-, and membrane-bound catechins
To probe the interactions between the different catechin rings in the free and bound forms, we acquired for each member of our catechin family off-resonance Rotational Overhauser effect spectroscopy (ROESY) (Figs. S3, A–D and S4, A–F) and transfer-Nuclear Overhauser effect spectroscopy data in the presence of Aβ oligomers (Figs. S3, E–H and S4, G–L) or membranes (Figs. S3, I–L and S4, M–R). The resulting ROE and NOE datasets are summarized in the heat maps of Figure 5, A–F and bar plots of Figure 5, G–I. In general, we observe more inter-ring cross-relaxation in the presence of receptors than in the free forms of the catechins (Fig. 5), suggesting a more compact catechin conformation upon binding to either Aβ oligomers or membranes. However, the extent of compaction upon binding differs for Aβ oligomers versus membranes.
Figure 5, G–I indicates that the free versus bound differences in the distribution of inter-ring cross-relaxation across the catechin library are consistently more significant in the case of Aβ oligomers than membranes. These observations point to the presence of more pronounced interactions between catechin rings D/B/C for Aβ40 oligomers versus membrane. The enhanced catechin compaction detected for Aβ oligomers versus membranes agrees with the increased positive cooperativity between ring-specific contributions to binding observed for Aβ40 oligomers versus membranes (Fig. 2C). The more compact conformation of catechins bound to Aβ40 oligomers versus membranes is also consistent with the inter-ring couplings revealed by the binding epitope analyses for Aβ oligomers, but not for membranes (Fig. 4, C and G).
Discussion
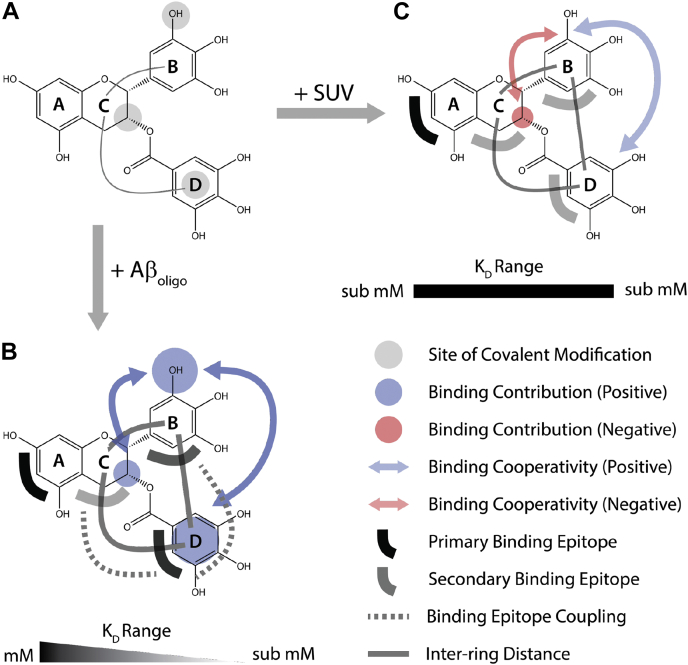
Our catechin library based on the combination of the 5′-OH gallo, epimerization, and gallate modifications of rings B, C, and D, respectively (Fig. 6A), has revealed key attributes of catechin recognition by Aβ oligomers and membranes, which are summarized in Figure 6, B and C. The primary sites to anchor catechins to Aβ oligomers are rings A and D (Fig. 6B, black, dark gray semi circles). This is likely a general feature as rings A and D consistently emerge as preferred Aβ oligomer–binding epitopes for all gallate containing catechins in our library. In addition, similar conclusions on the A and D rings have been reported for catechin binding to assemblies of different IDPs, including Aβ 1 to 42 and amyloidogenic peptides from the prion and ataxin-3 proteins (9).
Figure 6.
Schematic model for the recognition of catechins by Aβ oligomers and membranes.A, sites of covalent modifications (gray circles) used to build our catechin library starting from the conserved catechin scaffold spanning rings A to C. B, summary of observables across the catechin library pertaining to Aβ oligomer binding, as supported by the data in Figures 1, 2, 4, A–D and 5. The legend is shown in the figure. Larger, thicker, or darker symbols denote enhancement of the observable encoded by the symbols. C, as (B) but for membrane binding, as supported by the data in Figures 2, 3, 4, E–H and 5. Aβ, amyloid-beta.
Ring D not only serves as one of the primary epitopes for anchoring Aβ oligomers but also recruits both rings B and C (Fig. 6B, dashed gray lines), which act as secondary anchoring points (Fig. 6B, light gray semi circle). Upon Aβ oligomer binding, rings B and C form a compact aromatic cluster with ring D (Fig. 6B, thick solid gray lines). Within this cluster, the contributions of the B and D moieties to Aβ oligomer–binding affinities are more significant than that of ring C (Fig. 6B, blue circles). In addition, the binding contribution of ring B exhibits positive cooperativity with those of the other two rings (Fig. 6B, dark blue arrows), resulting in a gradient of Kd values across the catechin library spanning the mM – sub-mM range (Fig. 6B). The significant contribution of the gallate (D) to Aβ oligomer–binding affinity is likely another general attribute, as suggested by previous experiments on competitive binding of catechins with and without the gallate to assemblies of other amyloidogenic peptides (9) as well as MD simulations (43, 44). The latter have revealed how removal of the gallate moiety from EGCG significantly remodels the interaction patterns with Aβ 1 to 42 protofibrils with the losses of cation-π interactions in favor of more hydrophobic contacts in the case of EGC (43). Furthermore, the tight coupling of rings B and D may explain the concerted rotations previously reported for these aromatic moieties upon EGCG binding to Aβ40 oligomers (6).
Markedly, different recognition patterns are observed for binding of our catechin library to membranes (Fig. 6C). Only a single primary anchor epitope could be identified (i.e., ring A; Fig. 6C, black semi circle), and the cluster formed by rings B, C and D upon membrane binding is less compact than in the case of Aβ oligomer binding (Fig. 6C, thin solid gray lines). Such reduced compactness leads to losses in the positive cooperativity between rings B and C, D (Fig. 6C, light blue arrow) and to an overall more uniform distribution of binding affinities across the catechin library (Fig. 6C). In addition, in the case of SUVs, rings B and D are not major affinity drivers, and the epimerization of ring C contributes negatively to binding affinities (Fig. 6C light red arrow; Fig. 3H). Such patterns are distinctly different from the more synergistic contributions of rings B-D to Aβ oligomer binding (Fig. 6B).
It is important to emphasize that the catechin affinities reported here reflect the window of Kd values accessible to STD NMR, which is ideally suited to probe weak binding. Hence, our results complement previous studies in which catechin–lipid interactions were probed with other techniques more sensitive to tighter binding, such as centrifugation and quartz-crystal microbalance analyses, which revealed catechin affinities for membranes in the sub-μM range (45, 46). Notably, these studies were conducted under experimental conditions different from ours, for example, higher temperatures (25 °C versus 10 °C, used here) that may possibly cause catechin partial degradation and self-oxidation, and different lipid mimetics were used (45, 46). Although such differences may at least partially account for some of the different affinities observed by STD versus other methods, it is also possible that catechins bind lipid membranes at multiple sites with a distribution of affinities spanning both tight and weak binding. STD experiments report primarily on the latter, whereas other methods on the former.
Interestingly, in the case of tight catechin-binding sites within membranes, the key affinity determinants appear to be more similar to those identified here for Aβ oligomers. For example, both the 5′-OH gallo substituent in ring B and the gallate (ring D) are drivers of tight membrane binding, in agreement with the notion that the key determinants of catechin-lipid bilayer interactions are hydrogen bonds between the catechin hydroxyls and surface lipid headgroups (47), together with cation-pi and hydrophobic interactions (48). The epimerization of ring C was also reported to reduce affinity for the tight binding sites of membranes (45, 46), again confirming the similarity between the determinants of Aβ oligomer binding and those of tight membrane binding.
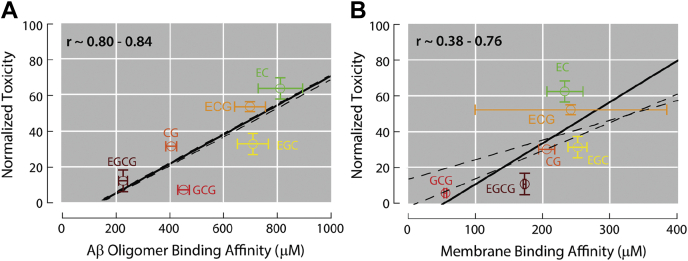
Notably, the weak binding affinities of catechins to membranes do not appear to correlate with the relative cellular toxicities of catechin-stabilized Aβ oligomers as strongly as the catechin affinities for the Aβ oligomers (Fig. 7). Although correlation is not causation, the data in Figure 7 suggests that the catechin binding to the Aβ assemblies is more relevant to the toxicities of the latter than the sub-mM interactions of catechins with membranes. However, membrane binding is likely to still play an important role in understanding the mechanism of action of catechins as inhibitors of neurodegeneration, because lipid bilayers may function as catechin sinks that compete with Aβ oligomers for binding to these polyphenols. In addition, lipid–catechin interactions are also relevant for how catechins disrupt the membranes of viruses and bacteria, thus shedding light on the mechanism of action of catechins not only as modulators of neurodegeneration but also as antiviral and antibacterial agents (49, 50, 51).
Figure 7.
Relationship between cellular toxicities of catechin-stabilized Aβ oligomer species and binding affinities of catechins to Aβ oligomers or membranes.A and B, correlations between cellular toxicities of Aβ oligomers stabilized by the presence of various catechins and catechin-binding affinities to (A) Aβ oligomers (4) and (B) DOPE:DOPS:DOPC small unilamellar vesicles. The lines of best fit for the data points are shown as solid lines. Dashed lines indicate the lines of best fit computed when replacing the ECG data point with the data point plus or minus the error. The range of Pearson’s correlation coefficients (r) for the relationships are annotated. Aβ, amyloid-beta; ECG, (-)-epicatechin-3-gallate.
In conclusion, the data obtained here provide unprecedented atomic-resolution insight on the SAR of catechins. Specifically, we highlight the role of the gallate (ring D) as a master regulator of catechin interactions with nanoparticles, wherein ring D is poised to recruit other catechin rings to the binding interface. Notably, this cooperative effect is receptor dependent, as the synergy between the various catechin rings is present when binding to Aβ40 oligomers but not membranes. Such results also highlight the potential of the integrated solution NMR and statistical approaches used here to dissect the drivers of small molecule binding to nanoparticles, as exemplified by the tripartite system of catechins, Aβ40 oligomers, and SUVs.
Experimental procedures
Catechin stock preparation
(-)-epigallocatechin-3-gallate, (-)-gallocatechin-3-gallate, (-)-catechin-3-gallate, (-)-epicatechin-3-gallate (ECG), (-)-epigallocatechin (EGC), (-)-epicatechin (EC) and methyl 3,4,5-trihydroxybenzoate (MG) were purchased from Sigma Aldrich, with a purity greater than 95%. A 2.8 mM stock solution was prepared in 20 mM sodium phosphate buffer pH 6.8 in 100% D2O with 0.05% NaN3.
Preparation of amyloid-beta oligomers and DOPE:DOPS:DOPC SUVs
Aβ (1–40) was purchased as a lyophilized powder from EZBiolab Inc, with purity greater than 95% and treated as described previously (4). Briefly, 1 mg of lyophilized peptide was resuspended in 80 μl of 1% NH4OH/ddH2O and subsequently diluted to a concentration of 1 mg/ml with ddH2O. The solution was then lyophilized and subsequently resuspended in 10 mM NaOH to a concentration of 1 mg/ml. The peptide solution in NaOH was then divided into aliquots, lyophilized, and frozen at −20 °C until use. The oligomers were prepared through a variation of a previously established protocol (4, 6). Specifically, lyophilized Aβ40 aliquots were resuspended in 20 mM sodium phosphate buffer pH 6.8, 100% D2O, 0.05% NaN3 to a final concentration of 30 μM and subsequently incubated at 10 °C for 24 h. In addition, SUVs composed of a 5:3:2 lipid molar ratio mixture of DOPE:DOPS:DOPC lipids were prepared, as described previously (4).
General NMR spectroscopy
All NMR spectra were recorded at 10 °C on a Bruker 700 MHz spectrometer equipped with a TCI cryoprobe and analyzed with Topspin 4.0.6. Catechin 1H NMR assignments were based on those previously published by Sironi et al. (9). Additional details are discussed below.
Saturation Transfer Difference NMR experiments to probe-binding affinities of catechins to Aβ40 oligomers and membranes, and determine binding epitopes
STD spectra (5, 52, 53, 54) were recorded with either 1024, 512 or 256 scans, as necessary to gain sufficient sensitivity, 32K complex points, and a spectral width of 11.98 ppm. Selective saturation of Aβ40 oligomers or membranes was implemented through methyl irradiation (∼0.61 and 1.01 ppm for Aβ40 oligomers and membranes, respectively), using a train of 40 Gaussian-shaped pulses of 50 ms, separated by a 1 ms interpulse delay. A 30 ms spin lock was also used to suppress peptide signals that overlap with catechin resonances. Subtraction of on-resonance versus off-resonance intensities was completed through phase cycling. Saturation transfer reference spectra were recorded with 256 or 512 scans, 32K complex points, and a spectral width of 11.98 ppm. Off-resonance saturation was introduced at 30 ppm. The intensities in the STD and STR spectra were scaled to account for differences in the number of scans acquired for each experiment.
Binding isotherms were created by measuring the STD amplification factors (STDaf) at each catechin concentration using the most intense of the Ring B catechin proton peaks (2′, 5′ or 6′) in the STD spectra. Notably, the proton peak used for creating the binding isotherms was held constant for each catechin, but differed between catechins given that the 5′-OH removal from ring B results in additional nonchemically equivalent protons. The STDaf was calculated as:
where I0 – Isat represents the signal intensity in the STD spectrum, I0 is the intensity in the STR spectrum, and Ltot. is the total concentration of catechin. The binding isotherm obtained using the normalized STDaf was fitted to a Scatchard-like model assuming the presence of multiple catechin-binding sites that are in first approximation equivalent and independent:
where [catechin] is the concentration of free catechin and Kd is an effective site-specific Kd value averaged over the heterogenous distribution of Aβ40 oligomers or SUVs. The catechin epitopes for binding to the Aβ40 oligomers or membranes were determined through the STD:STR ratio of each catechin proton resonance at ligand concentrations where the protein or membrane is nearly saturated ([catechin] ∼ 600 μM). For the purpose of epitope mapping (55), the STD:STR ratios for each proton were normalized to the proton with the highest ratio, that is, proton 6 or 8 of ring A, which was set to 100%.
Off-resonance ROESY and transfer NOESY NMR experiments to probe the conformations of catechins in the free and Aβ40 oligomer- or membrane-bound states
The transfer Nuclear Overhauser effect spectroscopy (TrNOESY) NMR spectra were collected at the end of each STD titration, that is, using catechin concentrations of 600 μM and Aβ40 oligomer and DOPE:DOPS:DOPC membrane concentrations of 30 and 50 μM, respectively. The TrNOESY spectra were recorded with 64 scans, a recycle delay of 1.2 s, 2048 (t2) and 400 (t1) complex points, and spectral widths of 11.98 ppm for each 1H dimension. A mixing time of 60 ms was used to measure the NOEs. Similarly, Off-resonance ROESY NMR spectra were acquired in the same buffer conditions used for the TrNOESY NMR with catechin concentrations of 180 μM. The spectra were recorded with 64 scans, a recycle delay of 1.5 s, 2048 (t2) and 512 (t1) complex points, and spectral widths of 11.98 ppm for each 1H dimension. The off-resonance ROESY mixing time was 158 ms for the ROEs reported, including two 4 ms adiabatic ramps, and the effective field angle was 54.7°.
Statistical analyses of catechin STD-based epitope maps to determine structural features required for binding to Aβ40 oligomers and membranes
To determine changes in binding epitopes upon covalent modification or alterations in catechin stereochemistry, the normalized STD:STR ratios of protons originating from catechins without and with the modification were compared. The protons were further subdivided into groups corresponding to their location in the various catechin rings (A, B, C, or D) to gauge to what extent the modification influences the engagement of the various rings. For instance, to determine how the presence of the gallate ester (ring D) influences the extent of saturation transfer to the other catechin protons, the normalized proton STD:STR ratios of the gallate-containing catechins in our library (i.e., EGCG, GCG, CG, and ECG) were compared to those measured for catechins devoid of gallate (i.e., EC and EGC). One-way ANOVA and subsequent post-hoc Tukey test with 95% confidence interval were used to determine the significance of the binding epitope changes due to the catechin modifications.
Violin plots were used to visualize the binding epitopes in the absence and presence of the structural modifications. The median is indicated by the thick solid black line, and the interquartile range is indicated by the space between the two thin solid black lines. The probability density of the data is shown through the rotated kernel density plot, which was lightly smoothened using a kernel density estimator, as implemented in GraphPad Prism 8.4.2. Multimodal distributions of the STD-based binding epitopes are emphasized through the violin plots based on the position and relative amplitude of the peaks in the kernel density map.
To identify structural features that explains the variations in binding epitopes of the catechins either in the presence of Aβ40 oligomers or membranes, single-linkage agglomerative clustering analyses of the normalized STD:STR ratios were performed. Briefly, the normalized STD:STR ratios for each catechin proton (columns) measured for the six different catechins (rows) were used to create a data matrix (D). The Spearman rank correlations (r) were computed between each pair of catechins in D using the normalized STD:STR ratios for the various protons to build the correlation. The resulting correlation matrix was analyzed by single-linkage agglomerative clustering analysis through the Cluster 3.0 program (http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm), and the dendrogram trees were generated using Java Treeview (http://jtreeview.sourceforge.net/). All catechin protons were used for the analysis, whenever available.
Statistical analyses of catechin ROEs and NOEs to identify significant changes in the conformations of free and Aβ40 oligomer- and membrane-bound catechins
The cross-peak and diagonal intensities in the trNOESY and Off-resonance ROESY spectra were measured on TopSpin 4.0.6 using the peak picking tool. The ROE and NOE intensity heat maps were generated in Microsoft Excel by color coding each individual intensity matrix according to the following three-color gradient scheme: lowest value is indicated in yellow, 50th percentile is indicated in orange, and the highest value is indicated in red. All values in between are linear combinations of the three specified colors.
Bar-plots of the normalized NOE (ROE) intensities for catechins in the free, oligomer-, and membrane-bound states were generated by normalizing all the NOE (ROE) cross-peak intensities for a given catechin to the highest intensity proton in the diagonal. The normalized cross-peak intensities for all the protons in a given ring pair, including those above and below the diagonal, were plotted and the mean was shown as a bar plot. One-way ANOVA and subsequent post-hoc Tukey test with 95% confidence interval were used to determine the significance of the NOE (ROE) differences between the free, oligomer-, and membrane-bound states.
Data availability
All data are contained within the article and the supporting information or available upon request from the corresponding author: melacin@mcmaster.ca.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank J.C. Bozelli Jr and R. Epand (McMaster University) for helpful discussions and providing the DOPE:DOPS:DOPC small unilamellar vesicles. This project was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC RGPIN-2019-05990).
Author contributions
R. A. and G. M. conceptualization; R. A. and G. M. methodology; R. A. validation; R. A., R. L., and G. M., formal analysis; R. A. and J. H. investigation; R. A., K. M. P., and G. M. writing–original draft; R. A., J. H., R. L., K. M. P., and G. M. writing–review and editing; R. A. and G. M. visualization; G. M. supervision; G. M. project administration; G. M. funding acquisition.
Funding and additional information
This project was funded by the Canadian Institutes of Health Research scholarship (201710GSD-402345-288638 to R. A.).
Edited by Karen Fleming
Supporting information
References
- 1.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F., Dobson C.M. Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- 3.Lee S.J.C., Nam E., Lee H.J., Savelieff M.G., Lim M.H. Towards an understanding of amyloid-β oligomers: Characterization, toxicity mechanisms, and inhibitors. Chem. Soc. Rev. 2017;46:310–323. doi: 10.1039/c6cs00731g. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed R., Akcan M., Khondker A., Rheinstädter M.C., Bozelli J.C., Epand R.M., Huynh V., Wylie R.G., Boulton S., Huang J., Verschoor C.P., Melacini G. Atomic resolution map of the soluble amyloid beta assembly toxic surfaces. Chem. Sci. 2019;10:6072–6082. doi: 10.1039/c9sc01331h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed R., Melacini G. A solution NMR toolset to probe the molecular mechanisms of amyloid inhibitors. Chem. Commun. 2018;54:4644–4652. doi: 10.1039/c8cc01380b. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed R., VanSchouwen B., Jafari N., Ni X., Ortega J., Melacini G. Molecular mechanism for the (−)-epigallocatechin gallate-induced toxic to nontoxic remodeling of Aβ oligomers. J. Am. Chem. Soc. 2017;139:13720–13734. doi: 10.1021/jacs.7b05012. [DOI] [PubMed] [Google Scholar]
- 7.Fusco G., Chen S.W., Williamson P.T.F., Cascella R., Perni M., Jarvis J.A., Cecchi C., Vendruscolo M., Chiti F., Cremades N., Ying L., Dobson C.M., de Simone A. Structural basis of membrane disruption and cellular toxicity by α-synuclein oligomers. Science. 2017;358:1440–1443. doi: 10.1126/science.aan6160. [DOI] [PubMed] [Google Scholar]
- 8.García-Viñuales S., Ahmed R., Sciacca M.F.M., Lanza V., Giuffrida M.L., Zimbone S., Romanucci V., Zarrelli A., Bongiorno C., Spinella N., Galati C., Di Fabio G., Melacini G., Milardi D. Trehalose conjugates of silybin as prodrugs for targeting toxic Aβ aggregates. ACS Chem. Neurosci. 2020;11:2566–2576. doi: 10.1021/acschemneuro.0c00232. [DOI] [PubMed] [Google Scholar]
- 9.Sironi E., Colombo L., Lompo A., Messa M., Bonanomi M., Regonesi M.E., Salmona M., Airoldi C. Natural compounds against neurodegenerative diseases: Molecular characterization of the interaction of catechins from green tea with Aβ1–42, PrP106–126, and ataxin-3 oligomers. Chem. Eur. J. 2014;20:13793–13800. doi: 10.1002/chem.201403188. [DOI] [PubMed] [Google Scholar]
- 10.Ciaramelli C., Palmioli A., de Luigi A., Colombo L., Sala G., Riva C., Zoia C.P., Salmona M., Airoldi C. NMR-driven identification of anti-amyloidogenic compounds in green and roasted coffee extracts. Food Chem. 2018;252:171–180. doi: 10.1016/j.foodchem.2018.01.075. [DOI] [PubMed] [Google Scholar]
- 11.Cox S.J., Lam B., Prasad A., Marietta H.A., Stander N.V., Joel J.G., Sahoo B.R., Guo F., Stoddard A.K., Ivanova M.I., Ramamoorthy A. High-throughput screening at the membrane interface reveals inhibitors of amyloid-β. Biochemistry. 2020;59:2249–2258. doi: 10.1021/acs.biochem.0c00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox S.J., Rodriguez Camargo D.C., Lee Y.-H., Dubini R.C.A., Rovó P., Ivanova M.I., Padmini V., Reif B., Ramamoorthy A. Small molecule induced toxic human-IAPP species characterized by NMR. Chem. Commun. 2020;56:13129–13132. doi: 10.1039/d0cc04803h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perni M., Galvagnion C., Maltsev A., Meisl G., Müller M.B.D., Challa P.K., Kirkegaard J.B., Flagmeier P., Cohen S.I.A., Cascella R., Chen S.W., Limbocker R., Sormanni P., Heller G.T., Aprile F.A., et al. A natural product inhibits the initiation of α-synuclein aggregation and suppresses its toxicity. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E1009–E1017. doi: 10.1073/pnas.1610586114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinha S., Du Z., Maiti P., Klärner F.-G., Schrader T., Wang C., Bitan G. Comparison of three amyloid assembly inhibitors: The sugar scyllo- inositol, the polyphenol epigallocatechin gallate, and the molecular tweezer CLR01. ACS Chem. Neurosci. 2012;3:451–458. doi: 10.1021/cn200133x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorenzen N., Nielsen S.B., Yoshimura Y., Vad B.S., Andersen C.B., Betzer C., Kaspersen J.D., Christiansen G., Pedersen J.S., Jensen P.H., Mulder F.A.A., Otzen D.E. How epigallocatechin gallate can inhibit α-synuclein oligomer toxicity in vitro. J. Biol. Chem. 2014;289:21299–21310. doi: 10.1074/jbc.M114.554667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen C.B., Yoshimura Y., Nielsen J., Otzen D.E., Mulder F.A.A. How epigallocatechin gallate binds and assembles oligomeric forms of human alpha-synuclein. J. Biol. Chem. 2021;296:100788. doi: 10.1016/j.jbc.2021.100788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Errico S., Lucchesi G., Odino D., Muscat S., Capitini C., Bugelli C., Canale C., Ferrando R., Grasso G., Barbut D., Calamai M., Danani A., Zasloff M., Relini A., Caminati G., et al. Making biological membrane resistant to the toxicity of misfolded protein oligomers: A lesson from trodusquemine. Nanoscale. 2020;12:22596–22614. doi: 10.1039/d0nr05285j. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh A., Pradhan N., Bera S., Datta A., Krishnamoorthy J., Jana N.R., Bhunia A. Inhibition and degradation of amyloid beta (Aβ40) fibrillation by designed small peptide: A combined spectroscopy, microscopy, and cell toxicity study. ACS Chem. Neurosci. 2017;8:718–722. doi: 10.1021/acschemneuro.6b00349. [DOI] [PubMed] [Google Scholar]
- 19.Pariary R., Ghosh B., Bednarikova Z., Varnava K.G., Ratha B.N., Raha S., Bhattacharyya D., Gazova Z., Sarojini V., Mandal A.K., Bhunia A. Targeted inhibition of amyloidogenesis using a non-toxic, serum stable strategically designed cyclic peptide with therapeutic implications. Biochim. Biophys. Acta. Proteins Proteom. 2020;1868:140378. doi: 10.1016/j.bbapap.2020.140378. [DOI] [PubMed] [Google Scholar]
- 20.Ahmed R., Huang J., Akimoto M., Shi T., Melacini G. Atomic resolution map of hierarchical self-assembly for an amyloidogenic protein probed through thermal 15 N–R 2 correlation matrices. J. Am. Chem. Soc. 2021;143:4668–4679. doi: 10.1021/jacs.0c13289. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed R., Huang J., Weber D.K., Gopinath T., Veglia G., Akimoto M., Khondker A., Rheinstädter M.C., Huynh V., Wylie R.G., Bozelli J.C., Epand R.M., Melacini G. Molecular mechanism for the suppression of alpha synuclein membrane toxicity by an unconventional extracellular chaperone. J. Am. Chem. Soc. 2020;142:9686–9699. doi: 10.1021/jacs.0c01894. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed R., Melacini G. A biophysical toolset to probe the microscopic processes underlying protein aggregation and its inhibition by molecular chaperones. Biophys. Chem. 2021;269:106508. doi: 10.1016/j.bpc.2020.106508. [DOI] [PubMed] [Google Scholar]
- 23.Algamal M., Ahmed R., Jafari N., Ahsan B., Ortega J., Melacini G. Atomic-resolution map of the interactions between an amyloid inhibitor protein and amyloid β (Aβ) peptides in the monomer and protofibril states. J. Biol. Chem. 2017;292:17158–17168. doi: 10.1074/jbc.M117.792853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burmann B.M., Gerez J.A., Matečko-Burmann I., Campioni S., Kumari P., Ghosh D., Mazur A., Aspholm E.E., Šulskis D., Wawrzyniuk M., Bock T., Schmidt A., Rüdiger S.G.D., Riek R., Hiller S. Regulation of α-synuclein by chaperones in mammalian cells. Nature. 2020;577:127–132. doi: 10.1038/s41586-019-1808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheidt T., Łapińska U., Kumita J.R., Whiten D.R., Klenerman D., Wilson M.R., Cohen S.I.A., Linse S., Vendruscolo M., Dobson C.M., Knowles T.P.J., Arosio P. Secondary nucleation and elongation occur at different sites on Alzheimer’s amyloid-β aggregates. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aau3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aprile F.A., Sormanni P., Podpolny M., Chhangur S., Needham L.-M., Ruggeri F.S., Perni M., Limbocker R., Heller G.T., Sneideris T., Scheidt T., Mannini B., Habchi J., Lee S.F., Salinas P.C., et al. Rational design of a conformation-specific antibody for the quantification of Aβ oligomers. Proc. Natl. Acad. Sci. U. S. A. 2020;117:13509–13518. doi: 10.1073/pnas.1919464117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tavanti F., Pedone A., Menziani M.C. Insights into the effect of curcumin and (–)-epigallocatechin-3-gallate on the aggregation of Aβ(1–40) monomers by means of molecular dynamics. Int. J. Mol. Sci. 2020;21:5462. doi: 10.3390/ijms21155462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehrnhoefer D.E., Bieschke J., Boeddrich A., Herbst M., Masino L., Lurz R., Engemann S., Pastore A., Wanker E.E. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 2008;15:558–566. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- 29.Bieschke J., Russ J., Friedrich R.P., Ehrnhoefer D.E., Wobst H., Neugebauer K., Wanker E.E. EGCG remodels mature α-synuclein and amyloid-β fibrils and reduces cellular toxicity. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7710–7715. doi: 10.1073/pnas.0910723107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fusco G., Sanz-Hernandez M., Ruggeri F.S., Vendruscolo M., Dobson C.M., de Simone A. Molecular determinants of the interaction of EGCG with ordered and disordered proteins. Biopolymers. 2018;109 doi: 10.1002/bip.23117. [DOI] [PubMed] [Google Scholar]
- 31.Zhao J., Blayney A., Liu X., Gandy L., Jin W., Yan L., Ha J.-H., Canning A.J., Connelly M., Yang C., Liu X., Xiao Y., Cosgrove M.S., Solmaz S.R., Zhang Y., et al. EGCG binds intrinsically disordered N-terminal domain of p53 and disrupts p53-MDM2 interaction. Nat. Commun. 2021;12:986. doi: 10.1038/s41467-021-21258-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wobst H.J., Sharma A., Diamond M.I., Wanker E.E., Bieschke J. The green tea polyphenol (−)-epigallocatechin gallate prevents the aggregation of tau protein into toxic oligomers at substoichiometric ratios. FEBS Lett. 2015;589:77–83. doi: 10.1016/j.febslet.2014.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young L.M., Cao P., Raleigh D.P., Ashcroft A.E., Radford S.E. Ion mobility spectrometry–mass spectrometry defines the oligomeric intermediates in amylin amyloid formation and the mode of action of inhibitors. J. Am. Chem. Soc. 2014;136:660–670. doi: 10.1021/ja406831n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandrashekaran I.R., Adda C.G., MacRaild C.A., Anders R.F., Norton R.S. Inhibition by flavonoids of amyloid-like fibril formation by Plasmodium falciparum merozoite surface protein 2. Biochemistry. 2010;49:5899–5908. doi: 10.1021/bi902197x. [DOI] [PubMed] [Google Scholar]
- 35.Wang S.-H., Liu F.-F., Dong X.-Y., Sun Y. Thermodynamic analysis of the molecular interactions between amyloid β-peptide 42 and (−)-epigallocatechin-3-gallate. J. Phys. Chem. B. 2010;114:11576–11583. doi: 10.1021/jp1001435. [DOI] [PubMed] [Google Scholar]
- 36.Lopez del Amo J.M., Fink U., Dasari M., Grelle G., Wanker E.E., Bieschke J., Reif B. Structural properties of EGCG-induced, nontoxic Alzheimer's disease Aβ oligomers. J. Mol. Biol. 2012;421:517–524. doi: 10.1016/j.jmb.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Palhano F.L., Lee J., Grimster N.P., Kelly J.W. Toward the molecular mechanism(s) by which EGCG treatment remodels mature amyloid fibrils. J. Am. Chem. Soc. 2013;135:7503–7510. doi: 10.1021/ja3115696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez Pomier K., Ahmed R., Melacini G. Catechins as tools to understand the molecular basis of neurodegeneration. Molecules. 2020;25:3571. doi: 10.3390/molecules25163571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomaselli S., La Vitola P., Pagano K., Brandi E., Santamaria G., Galante D., D'Arrigo C., Moni L., Lambruschini C., Banfi L., Lucchetti J., Fracasso C., Molinari H., Forloni G., Balducci C., et al. Biophysical and in vivo studies identify a new natural-based polyphenol, counteracting Aβ oligomerization in vitro and Aβ oligomer-mediated memory impairment and neuroinflammation in an acute mouse model of Alzheimer’s disease. ACS Chem. Neurosci. 2019;10:4462–4475. doi: 10.1021/acschemneuro.9b00241. [DOI] [PubMed] [Google Scholar]
- 40.Esposito V., Das R., Melacini G. Mapping polypeptide self-recognition through 1 H off-resonance relaxation. J. Am. Chem. Soc. 2005;127:9358–9359. doi: 10.1021/ja051714i. [DOI] [PubMed] [Google Scholar]
- 41.Milojevic J., Esposito V., Das R., Melacini G. Understanding the molecular basis for the inhibition of the Alzheimer's Abeta-peptide oligomerization by human serum albumin using saturation transfer difference and off-resonance relaxation NMR spectroscopy. J. Am. Chem. Soc. 2007;129:4282–4290. doi: 10.1021/ja067367+. [DOI] [PubMed] [Google Scholar]
- 42.Fawzi N.L., Ying J., Ghirlando R., Torchia D.A., Clore G.M. Atomic-resolution dynamics on the surface of amyloid-β protofibrils probed by solution NMR. Nature. 2011;480:268–272. doi: 10.1038/nature10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhan C., Chen Y., Tang Y., Wei G. Green tea extracts EGCG and EGC display distinct mechanisms in disrupting Aβ42 protofibril. ACS Chem. Neurosci. 2020;11:1841–1851. doi: 10.1021/acschemneuro.0c00277. [DOI] [PubMed] [Google Scholar]
- 44.Minh Hung H., Nguyen M.T., Tran P.-T., Truong V.K., Chapman J., Le Quynh Anh H., Derreumaux P., van Vu V., Ngo S.T. Impact of the astaxanthin, betanin, and EGCG compounds on small oligomers of amyloid Aβ 40 peptide. J. Chem. Inf. Model. 2020;60:1399–1408. doi: 10.1021/acs.jcim.9b01074. [DOI] [PubMed] [Google Scholar]
- 45.Hashimoto T., Kumazawa S., Nanjo F., Hara Y., Nakayama T. Interaction of tea catechins with lipid bilayers investigated with liposome systems. Biosci. Biotechnol. Biochem. 1999;63:2252–2255. doi: 10.1271/bbb.63.2252. [DOI] [PubMed] [Google Scholar]
- 46.Kajiya K., Kumazawa S., Nakayama T. Effects of external factors on the interaction of tea catechins with lipid bilayers. Biosci. Biotechnol. Biochem. 2002;66:2330–2335. doi: 10.1271/bbb.66.2330. [DOI] [PubMed] [Google Scholar]
- 47.Sirk T.W., Brown E.F., Sum A.K., Friedman M. Molecular dynamics study on the biophysical interactions of seven green tea catechins with lipid bilayers of cell membranes. J. Agric. Food Chem. 2008;56:7750–7758. doi: 10.1021/jf8013298. [DOI] [PubMed] [Google Scholar]
- 48.Uekusa Y., Kamihira-Ishijima M., Sugimoto O., Ishii T., Kumazawa S., Nakamura K., Tanji K.-i., Naito A., Nakayama T. Interaction of epicatechin gallate with phospholipid membranes as revealed by solid-state NMR spectroscopy. Biochim. Biophys. Acta. 2011;1808:1654–1660. doi: 10.1016/j.bbamem.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 49.Steinmann J., Buer J., Pietschmann T., Steinmann E. Anti-infective properties of epigallocatechin-3-gallate (EGCG), a component of green tea. Br. J. Pharmacol. 2013;168:1059–1073. doi: 10.1111/bph.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ikigai H., Nakae T., Hara Y., Shimamura T. Bactericidal catechins damage the lipid bilayer. Biochim. Biophys. Acta. 1993;1147:132–136. doi: 10.1016/0005-2736(93)90323-r. [DOI] [PubMed] [Google Scholar]
- 51.Yamaguchi K., Honda M., Ikigai H., Hara Y., Shimamura T. Inhibitory effects of (−)-epigallocatechin gallate on the life cycle of human immunodeficiency virus type 1 (HIV-1) Antiviral Res. 2002;53:19–34. doi: 10.1016/s0166-3542(01)00189-9. [DOI] [PubMed] [Google Scholar]
- 52.Meyer B., Peters T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew. Chem. Int. Ed. Engl. 2003;42:864–890. doi: 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- 53.Mayer M., Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 54.Dalvit C., Flocco M., Knapp S., Mostardini M., Perego R., Stockman B.J., Veronesi M., Varasi M. High-throughput NMR-based screening with competition binding experiments. J. Am. Chem. Soc. 2002;124:7702–7709. doi: 10.1021/ja020174b. [DOI] [PubMed] [Google Scholar]
- 55.Mayer M., Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and the supporting information or available upon request from the corresponding author: melacin@mcmaster.ca.