Abstract
The mitochondrial enzyme glutaminase C (GAC) is upregulated in many cancer cells to catalyze the first step in glutamine metabolism, the hydrolysis of glutamine to glutamate. The dependence of cancer cells on this transformed metabolic pathway highlights GAC as a potentially important therapeutic target. GAC acquires maximal catalytic activity upon binding to anionic activators such as inorganic phosphate. To delineate the mechanism of GAC activation, we used the tryptophan substitution of tyrosine 466 in the catalytic site of the enzyme as a fluorescent reporter for glutamine binding in the presence and absence of phosphate. We show that in the absence of phosphate, glutamine binding to the Y466W GAC tetramer exhibits positive cooperativity. A high-resolution X-ray structure of tetrameric Y466W GAC bound to glutamine suggests that cooperativity in substrate binding is coupled to tyrosine 249, located at the edge of the catalytic site (i.e., the “lid”), adopting two distinct conformations. In one dimer within the GAC tetramer, the lids are open and glutamine binds weakly, whereas, in the adjoining dimer, the lids are closed over the substrates, resulting in higher affinity interactions. When crystallized in the presence of glutamine and phosphate, all four subunits of the Y466W GAC tetramer exhibited bound glutamine with closed lids. Glutamine can bind with high affinity to each subunit, which subsequently undergo simultaneous catalysis. These findings explain how the regulated transitioning of GAC between different conformational states ensures that maximal catalytic activity is reached in cancer cells only when an allosteric activator is available.
Keywords: glutaminase, enzyme mechanism, structure-function, cancer biology, mitochondrial metabolism
Abbreviations: GAC, glutaminase C; GLS, glutaminase gene; GLS2, glutaminase gene variant 2; PDB, Protein Data Bank; TCA, tricarboxylic acid
Targeting the metabolic changes that tumor cells undergo has offered new opportunities for the development of cancer therapeutics with less detrimental effects on normal and healthy cells compared with traditional radiation and chemotherapy treatments (1, 2, 3, 4). The metabolic reprogramming of cancer cells involves the uncoupling of the glycolytic pathway from the tricarboxylic acid (TCA) cycle, such that pyruvate is almost entirely converted to lactic acid, rather than being oxidized to acetyl-CoA as is the case in normal and healthy cells. Thus, cancer cells use aerobic glycolysis for the rapid production of ATP (5, 6), as well as to generate building blocks for fatty acid and nucleotide synthesis, and other biosynthetic processes necessary for malignant transformation. These metabolic changes are commonly referred to as the “Warburg effect.” In order to meet their metabolic needs and compensate for the Warburg effect and the uncoupling of glycolysis from the TCA cycle, cancer cells show a significant increase in their ability to metabolize glutamine, resulting in the production of α-ketoglutarate that can feed into and sustain the TCA cycle (7, 8, 9, 10). The dependence of cancer cells on glutamine metabolism, known as “glutamine addiction,” has led to strategies that attempt to inhibit malignant growth by targeting glutaminolysis by starving the cells of glutamine or inhibiting the catalytic activity of members of the family of enzymes collectively known as glutaminase (9, 11, 12, 13, 14). The mitochondrial glutaminase enzymes catalyze the first step in glutamine metabolism, specifically, the hydrolysis of glutamine to glutamate with the accompanying production of ammonia.
In humans, two genes encode the glutaminase enzymes, designated as glutaminase gene (GLS) (kidney type) and glutaminase gene variant 2 (GLS2) (liver type), which are expressed in a wide range of tissues under different conditions. A number of cancer cells are known to preferentially upregulate the activity and expression of GLS (15, 16), whereas others appear to specifically express and to be dependent upon GLS2 (9). GLS encodes for the expression of two active isoforms, kidney-type glutaminase isoform (the full-length isoform) and glutaminase C (GAC) (a carboxyl-terminal splice variant). The GAC isoform is more active and has frequently been found to promote tumorigenesis in response to upstream oncogenic signals (17, 18, 19). This has led to GAC being examined as a therapeutic target for cancer treatments, and in fact, an allosteric inhibitor of its enzymatic activity, CB-839, is in clinical trials (12) (also see “clinicaltrials.gov”). Additional small molecules targeting GAC are also being developed as potential anticancer drugs (20). Therefore, gaining further insights into the mechanistic basis by which this enzyme is activated should benefit the development of new therapeutic strategies.
GAC exists predominantly as either dimers or tetramers in solution, with the tetrameric form of the enzyme being essential for catalytic activity (21). The enzyme has been shown to be activated by multiple anionic activators (22), of which inorganic phosphate induces tetramer formation and gives rise to maximal specific activity. However, the underlying mechanism by which anionic activators markedly increase GAC activity is still not completely understood. Thus, we set out to address this important question. We found that in the absence of inorganic phosphate, GAC adopts an asymmetric arrangement between the two dimers that comprise the tetrameric enzyme, with substrate binding being highly cooperative and with the enzyme showing only weak catalytic activity. Upon the addition of the activator phosphate, a conformational transition occurs that results in a symmetrical arrangement of the four subunits comprising the GAC tetramer, such that glutamine binds independently and with high affinity at each substrate site, enabling all four subunits to simultaneously undergo hydrolysis at maximal rates of catalytic turnover. Overall, these findings provide new mechanistic insights into the regulation of this key enzyme in cancer cell metabolism by its anionic activators.
Results
Substrate binding to GAC reveals communication across its subunits
In order to monitor substrate (glutamine) binding to GAC without the complications of its conversion to product (glutamate) because of enzyme catalysis, we generated a recombinant human GAC mutant in which the active site residue Tyr466 was changed to tryptophan. The GAC (Y466W) mutant is unable to catalyze the hydrolysis of glutamine to glutamate; however, the fluorescence of Trp466 is quenched specifically upon the binding of substrate (glutamine) but not the product (glutamate) (23), thereby providing a direct readout for the interactions of glutamine with the enzyme.
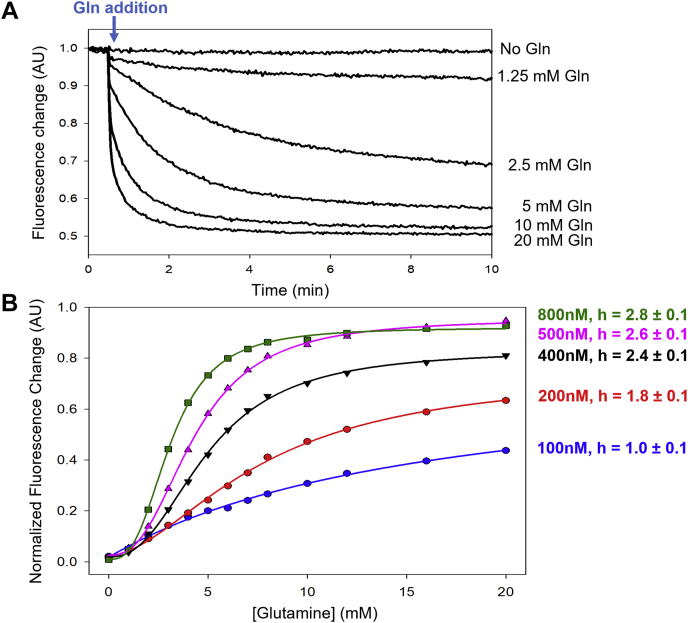
The activation of GAC has been shown to require its transition from an inactive dimer to an active tetramer (21, 24, 25, 26, 27). Higher concentrations of GAC (i.e., [GAC] ≥ 400 nM) favor the formation of tetramers (28), whereas the enzyme exists predominantly in its dimeric state at lower concentrations, with the KD value for tetramer formation being ∼160 nM (29). We therefore examined the binding profiles for glutamine over a range of enzyme concentrations, (i.e., from 100 to 800 nM). For each concentration of GAC (Y466W), we measured real-time fluorescence traces prior to and after the addition of increasing amounts of glutamine (Fig. 1A). The equilibrium fluorescence (Ff) measured at each concentration of glutamine was normalized against the intrinsic fluorescence of the GAC (Y466W) mutant (Fo) and plotted as a function of the substrate concentration (Fig. 1B). The data were then fit to the Hill equation (see “Experimental procedures” section, Equation 1). At GAC concentrations greater than 200 nM (i.e., above the KD value for tetramer formation), the substrate-binding profiles exhibited positive cooperativity with Hill coefficients ranging between 1.8 and 2.8. However, at the lowest GAC concentration examined (100 nM), a single class of glutamine-binding sites was observed with a Hill coefficient of 1.0. The titration curve in this case can be fitted to Equation 1 to yield a single KD value of 16.0 ± 4.5 mM.
Figure 1.
Glutamine binding to GAC (Y466W) shows increasing positive cooperativity as the concentration of enzyme increases.A, the fluorescence emission (λex = 285 nm and λem = 340 nm) of 800 nM GAC (Y466W) was monitored in real time. At the indicated time (30 s, arrow), glutamine was added to the indicated final concentrations, resulting in a quench in fluorescence. The signal was normalized to the initial fluorescence value (F0). B, equilibrium fluorescence from A for different concentrations of GAC (Y466W) was plotted as a function of glutamine concentration. A nonleast square fit of the data at each concentration to Equation 1 (solid lines) yielded the Hill coefficients (h) as listed in the figure (±SE of the fit). The data points are representative of three independent experiments. GAC, glutaminase C.
The anionic activator inorganic phosphate synchronizes glutamine-binding events at each of the four subunits of GAC
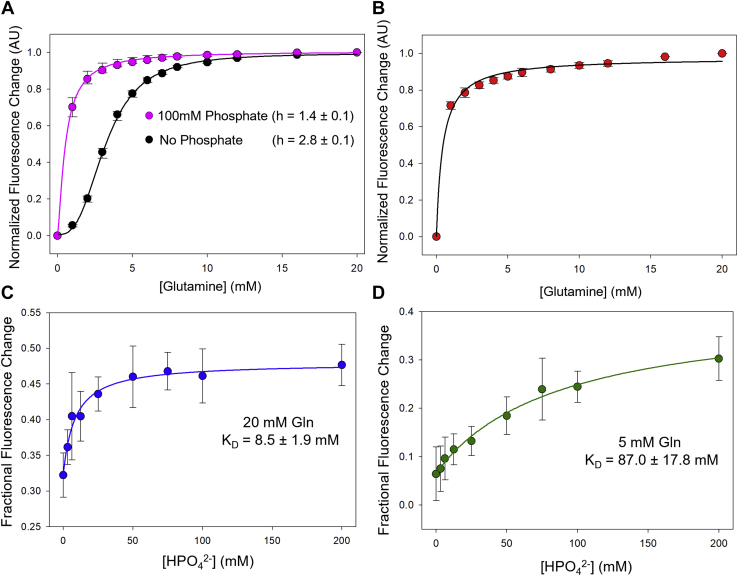
The binding of the allosteric anionic activator, inorganic phosphate, to GAC has been shown to promote its transition from an inactive dimer to an activated tetramer (21, 24, 25, 26, 27). Thus, we were interested in seeing how inorganic phosphate influenced the binding profiles for substrate. As shown in Figure 2A, the binding of glutamine to 800 nM GAC (Y466W) in the absence of inorganic phosphate again yielded a profile indicative of positive cooperativity with a Hill coefficient of 2.8 ± 0.1. However, when glutamine binding was examined in the presence of 100 mM inorganic phosphate, a concentration of the anionic activator that maximally activates GAC, the substrate-binding profile was shifted to lower glutamine concentrations, indicative of an increase in the affinity of the substrate for the enzyme. Moreover, in the presence of phosphate, the glutamine-binding profile showed no sigmoidal behavior indicative of positive cooperativity and instead yielded a Hill coefficient of 1.4 ± 0.1.
Figure 2.
The interactions of the anionic activator inorganic phosphate and glutamine with GAC.A, the equilibrium fluorescence (λex = 285 nm and λem = 340 nm) for 800 nM of GAC (Y466W) was monitored in real time, and different glutamine concentrations were added at 30 s. The equilibrium fluorescence in the presence and absence of 100 mM K2HPO4 (inorganic phosphate) was normalized with the initial fluorescence and plotted as a function of glutamine concentration in magenta and black, respectively. A nonleast square fit of the data in each case to Equation 1 (solid lines) yielded the Hill coefficients (h) of h = 1.4 ± 0.1 in the presence of phosphate and h = 2.8 ± 0.1 in the absence of phosphate. Each data point shown is the mean ± SE from three independent experiments. B, the equilibrium fluorescence (λex = 285 nm and λem = 340 nm) from real-time fluorescence emission for 800 nM of GAC (K320A and Y466W) in the absence of phosphate was normalized with the initial fluoresence and plotted as a function of glutamine concentration. A nonleast square fit (solid black line) of the data in each case to Equation 1, fixing the Hill coefficient h = 1, was shown. C and D, in the presence of different concentrations of inorganic phosphate (0–100 mM K2HPO4), the equilibrium fluorescence (λex = 285 nm and λem = 340 nm) was monitored in real time, and glutamine was added at 30 s to the indicated concentrations of 20 mM (C) and 5 mM (D). The equilibrium fluorescence emission for each concentration of phosphate was normalized to the initial fluoresence of the enzyme and plotted as a function of phosphate concentration. A nonleast square fit (solid black line) of the data in each case to Equation 2 yields KD values of 8.5 ± 1.9 mM and 87.0 ± 17.8 mM for A and B, respectively. Each data point shown is the mean ± SE from three independent experiments. GAC, glutaminase C.
The presence of inorganic phosphate induces all four GAC subunits to adopt a single class of high-affinity binding sites for glutamine. The titration curve in the presence of phosphate (magenta trace, Fig. 2A) in this case can be fitted to Equation 1 yielding an apparent KD value of 0.5 ± 0.1 mM. In contrast, in the absence of inorganic phosphate, the Hill coefficients larger than 1 (seen previously in Fig. 1) suggest the existence of multiple conformations of the enzyme with varying affinity for the substrate. We have now modeled this sigmoidal behavior with the Monod–Wyman–Changeux model (30) (or the symmetry model), which assumes that there are two different conformations of the enzyme: the T state that binds glutamine weakly and the R state that binds glutamine with higher affinity. For example, in the case of 800 nM GAC (Y466W), the sigmoidal titration curve that gives a Hill coefficient of 2.8 (black trace, Fig. 2A) was modeled with Equation 2, yielding two different dissociation constants: KT = 21 ± 0.1 mM and KR = 0.3 ± 0.1 mM. These two constants suggest that glutamine initially binds to GAC with low affinity, similar to the apparent KD value determined for the binding of glutamine to 100 nM GAC (Y466W), that is, when the enzyme exists predominantly as a dimer. This initial binding of glutamine causes conformational changes within GAC such that subsequent substrate binding occurs with a higher affinity similar to what we observe when substrate binding is measured in the presence of the allosteric activator inorganic phosphate.
We and others have previously shown that changing the lysine residue at position 320 within the activation loop of GAC to alanine yielded an enzyme that was constitutively active (21, 31), with its catalytic activity matching that measured when the WT enzyme was activated by inorganic phosphate. We therefore generated a double-mutant GAC (K320A and Y466W) and examined glutamine binding by monitoring changes in Trp466 emission. The glutamine-binding profile obtained with the GAC (K320A and Y466W) double mutant mirrored what we observed when assaying substrate binding in the presence of inorganic phosphate (Fig. 2B). Specifically, glutamine bound to a single class of high-affinity sites on the GAC (K320A and Y466W) double mutant, again with no detectable cooperativity.
We then examined whether glutamine can exert a reciprocal effect and increase the affinity of phosphate for the enzyme. The same fluorescence readout was used to monitor glutamine-induced changes in Trp466 fluorescence; however, lower enzyme concentrations were examined to ensure that fewer GAC tetramers were present at the outset of the assay, in order to better observe GAC tetramer formation induced by the binding of phosphate and/or glutamine. These assays were performed at two different glutamine concentrations, which represented saturating (20 mM) and nonsaturating (5 mM) doses of the substrate. In the presence of saturating (20 mM) glutamine, we measured an apparent KD value of 8.5 ± 1.9 mM for inorganic phosphate (Fig. 2C), compared with a value of 87.0 ± 17.8 mM when the assays were performed under conditions of subsaturating levels of substrate (i.e., 5 mM glutamine) (Fig. 2D). These studies indicate that the presence of glutamine in the active sites increases the affinity of the enzyme for phosphate, establishing the reciprocal communication between the binding of the anionic activator and substrate.
X-ray structure of glutamine bound to GAC (Y466W) in the absence of an anionic activator
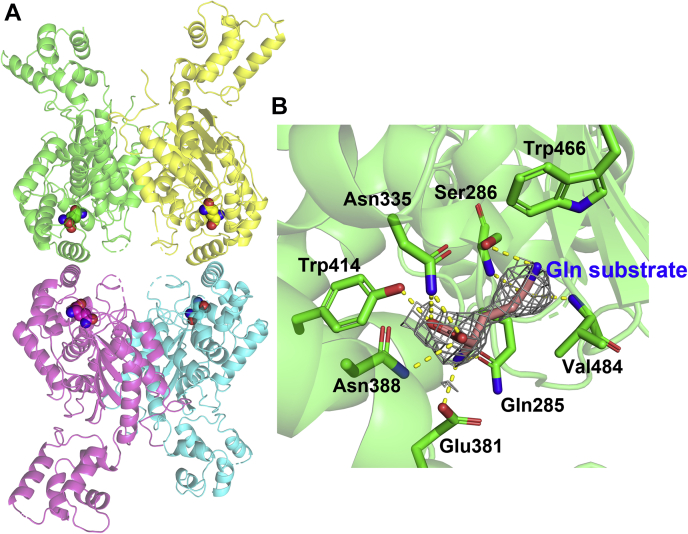
To better understand the structural basis of glutamine binding to GAC, we determined a 2.8 Å resolution X-ray crystal structure for the GAC (Y466W)–substrate complex (see Table 1 for the crystallographic parameters). Four GAC monomers were observed in the asymmetric unit, with electron density for glutamine being present in each enzyme subunit making up the tetramer (Fig. 3A). In the X-ray crystal structures (28, 32, 33) determined thus far for GAC bound to either the product of the catalytic reaction, glutamate, or the glutamine analog, DON, most of the key catalytic residues, including Ser286, Tyr466, Lys289, and Tyr414, appear to be essentially in the same positions as observed in the structure for the apo enzyme. The same is true for the crystal structure that we determined for the GAC (Y466W) mutant bound to glutamine. Figure 3B shows the electron density calculated with the 2Fo–Fc map at the contour level of 0.7 rmsd for glutamine and the side chains engaging in hydrogen bond interactions with the substrate, together with the catalytically active residues. However, one structural element of GAC that we suspected to be more dynamic during the catalytic reaction is the region that serves as a “lid” to cover the active site pocket.
Table 1.
X-ray crystal data collection and refinement statistics
| Structure | Human glutaminase C (Y466W) with l-Gln without phosphate (PDB: 7SBM) | Human glutaminase C (Y466W) with l-Gln plus phosphate (PDB: 7SBN) |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P1211 |
| Cell dimensions | ||
| a, b, c (Å) | 98.8, 139.1, 177.4 | 51.1, 140.1, 177.2 |
| α, β, γ (°) | 90, 90, 90 | 90, 95.4, 90 |
| Resolution (Å) | 2.8 | 2.1 |
| Completeness (%) | 99.40 (98.52) | 98.46 (88.80) |
| Redundancy | 6.3 (6.3) | 3.3 (2.8) |
| Refinement | ||
| Resolution (Å) | 37.9–2.8 | 47.8–2.1 |
| No. of reflections | 60,927 (5953) | 133,794 (12,085) |
| Rwork/Rfree | 0.1993/0.2435 | 0.1879/0.2301 |
| Avg B-factors (Å2) | 38.04 | 40.33 |
| Rmsd | ||
| Bond lengths (Å) | 0.010 | 0.007 |
| Bond angles (°) | 1.38 | 0.87 |
Values in parentheses are for highest-resolution shell.
Figure 3.
Structure of the GAC (Y466W) tetramer, with glutamine bound to all four subunits.A, glutamine-binding sites in the GAC (Y466W) tetramer are shown. B, the electron density for glutamine and the side chains engaging in hydrogen bonding with the substrate, together with the catalytically active residues. The electron density shown was calculated with the 2Fo–Fc map at the contour level of 0.7 rmsd, which had been crossverified with the overlapping green density of the Fo–Fc map at the contour level of 2.0 rmsd. GAC, glutaminase C.
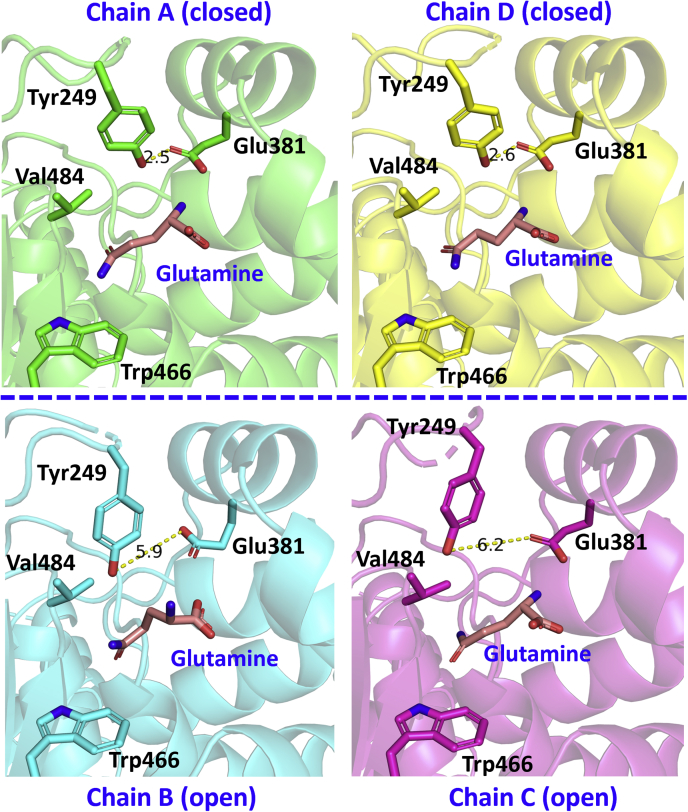
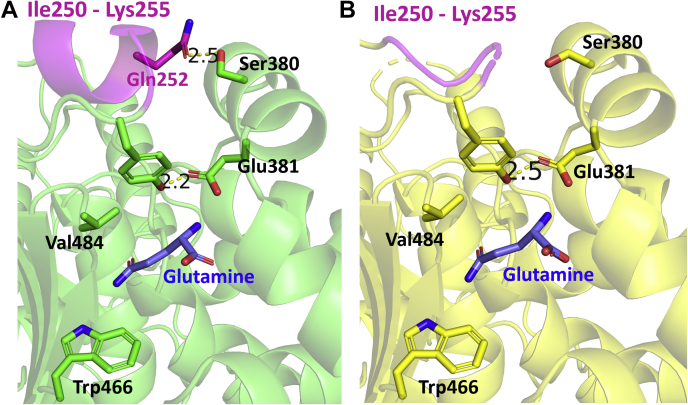
The lid is made up of the “YIP” motif, which consists of Tyr249, Ile250, and Pro251, and is highly conserved in glutaminase enzymes across many species (21). In analyzing the crystal structure for the glutamine-bound GAC (Y466W) tetrameric complex, we found that each GAC dimer within the tetramer shows Tyr249 in a different conformation, resulting in the lid adopting two distinct states (Fig. 4). Within one of the GAC dimers, Tyr249 in chains A and D form a hydrogen bond with Glu381 (with a measured distance of 2.5 Å), such that the lid completely covers the active site (Fig. 4A, top dimer). This is referred to as the “closed” lid conformation of GAC and presumably represents a high-affinity state for substrate binding as glutamine would effectively be “locked” into its binding site. Closing the lid through hydrogen bonding between Tyr249 and Glu381 is critical both for high-affinity glutamine binding at the active site and for enzymatic catalysis to occur. In the adjacent GAC dimer comprising the tetrameric enzyme (i.e., chains B and chain C), this hydrogen bond is broken, and Tyr249 moves away from Glu381 in each of the two subunits (with measured distances of 6.1 and 5.9 Å) (Fig. 4, bottom dimer). Instead, the phenyl ring in Tyr249 appears to engage in a hydrophobic interaction with the alkyl chain of Val484. Therefore, we refer to this as the “open” lid conformation of GAC.
Figure 4.
Two distinct conformations of the “lid”, Tyr249, are observed in each dimer of GAC (Y466W) in the presence of glutamine. The GAC (Y466W) tetramer (made up of four chains) is shown, with the blue dashed line showing the dimer–dimer interface. The dimer at the top (chains A and D) shows the “closed” conformation: Tyr249 is forming hydrogen bonds with Glu381 (measured distance of 2.5 Å). The dimer at the bottom (chains B and C) shows Tyr249 moving away from Glu381 (measured distance of 6.1 and 5.9 Å). The hydrophobic residue Val484 is also shown to be in close proximity with Tyr249 in chains B and C. GAC, glutaminase C.
Because glutamine can enter and leave the active site freely when the lids assume an open conformation, this should represent a lower affinity substrate-binding state as the rate of substrate dissociation from the enzyme would be significantly increased compared with when the lids are closed. We tested this idea by examining the GAC (Y249F) mutant, since by changing the tyrosine to phenylalanine, the hydrogen bond interaction between Tyr249 and Glu381 is weakened, allowing the lids to rapidly switch between their open and closed conformations. Indeed, we observed that in the absence of inorganic phosphate, glutamine has a significantly weaker affinity for the GAC (Y249F and Y466W) double mutant compared with the GAC (466W) mutant (Fig. S1). In the presence of phosphate, the dose response for glutamine is still shifted to much lower concentrations; however, interestingly, positive cooperativity for substrate binding was observed, unlike the case for the WT enzyme. Thus, the Y249F mutation, which allows the lids to transition more easily between their closed and open states, apparently makes it more difficult for phosphate to synchronize substrate binding to each of the subunits comprising the GAC tetramer.
X-ray structure of glutamine bound to GAC (Y466W) in the presence of inorganic phosphate
To investigate how the presence of inorganic phosphate influences the lids and affects glutamine binding to GAC, we determined a 2.1 Å resolution crystal structure for GAC (Y466W) bound to glutamine in the presence of H2PO42− (see Table 1 for the crystallographic parameters). The structure shows that there are four GAC monomers in the asymmetric unit, with electron density for glutamine being present in all four active sites. Inorganic phosphate appears to induce a symmetrical arrangement of the GAC tetramer, such that all four subunits bind glutamine with high affinity and closed lids, as evident by Tyr249 engaging in hydrogen bonding with Glu381 (Fig. S2). Moreover, the density for the entire YIP motif and the neighboring residues are well resolved in this structure (Fig. 5A), unlike in the structures of GAC (Y466W) and WT GAC (21) obtained in the absence of phosphate, where we were not able to observe electron density in this region (Fig. 5B). The residues from Pro251 to Lys255 now form an α-helix, positioning Gln252 to engage in a hydrogen bond with Ser380, further closing the glutamine-binding pocket and stabilizing the “closed” conformation. However, clear density for phosphate was not observed. This may be due to inorganic phosphate binding preferentially to and stabilizing the open lid conformation of the enzyme, since we found that the GAC (Y249F and Y466W) double mutant exhibits a greatly increased affinity for the anionic activator (21). Therefore, as considered further in the “Discussion” section later, once phosphate has helped increase substrate access to the active site by stabilizing the open conformation, the lids then close over the bound substrates at each active site, weakening the affinity of the anionic activator for the enzyme. This likely makes it difficult to trap a complex between phosphate with GAC tetramers that have all four subunits in the closed state.
Figure 5.
A more stable “closed” conformation for GAC crystallized in the presence of phosphate.A, the “closed” conformation was observed in all four chains in the structure for GAC (Y466W) with bound glutamine when protein crystallization was performed in the presence of both substrate and inorganic phosphate (figure shows the residues in chain A). The residues following Tyr249 now form an α-helix, further closing off the active site. B, comparison to the conformation observed in the absence of phosphate, where the density for Ile250 to Lys255 is not well resolved and results in a partially disordered loop. GAC, glutaminase C.
Examining the interplay between glutamine and inorganic phosphate during enzyme catalysis
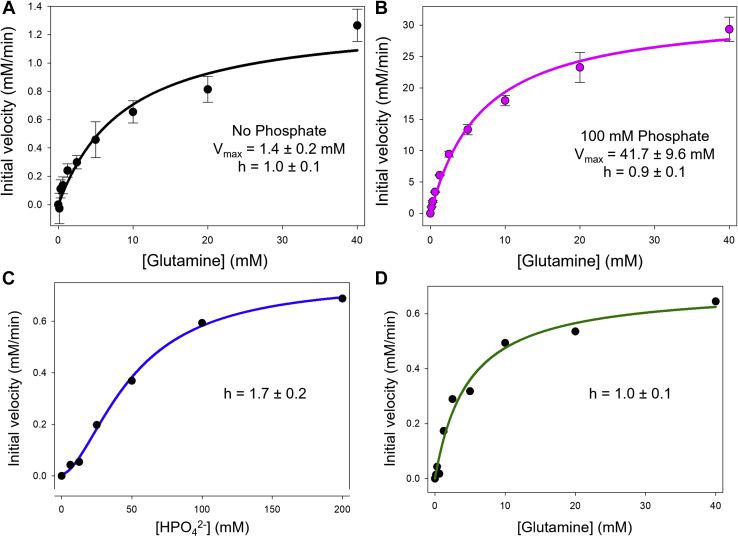
We next compared the initial velocity for glutamine hydrolysis measured for 800 nM WT GAC in the presence of glutamine alone, versus the catalytic rate determined in the presence of substrate and inorganic phosphate. As expected, the anionic activator gave rise to a significant increase in the Vmax of the reaction (Fig. 6, A and B). However, especially interesting was the fact that the dose curve for the substrate glutamine in the absence of phosphate did not show any detectable cooperativity, contrary to what we observed when we measured glutamine binding in the absence of the anionic activator. Moreover, the KM for glutamine is very similar in the presence and absence of phosphate (KM = 7.7 ± 0.7 mM and 8.6 ± 1.3 mM, respectively). This would suggest that without the anionic activator, only those enzyme subunits that bind substrate with high affinity (i.e., with their lids closed) are capable of catalytic turnover at any given time, thus yielding a similar KM value for substrate as observed in the presence of phosphate. However, in the absence of the anionic activator, a very low kcat is measured. The presence of the anionic activator then induces a symmetrical GAC tetramer, allowing each of the enzyme subunits to undergo catalysis simultaneously and with a maximum kcat.
Figure 6.
Catalysis of glutamine by WT GAC shows no cooperativity.A and B, initial velocities of glutamine hydrolysis by 800 nM WT GAC was measured at the indicated glutamine concentrations in the absence (black) or the presence (magenta) of 100 mM K2HPO4 using the two-step assay method. Each data point shown is the mean ± SE from three independent experiments. A simultaneous nonleast square fit of the data from three independent experiments to Equation 4 (solid line) yielded Hill coefficients, h = 1.0 ± 0.1 and h = 0.9 ± 0.1 (±SE of the fit) in the absence and presence of phosphate, respectively, indicative of no cooperativity in both cases. C, initial velocities of glutamine hydrolysis by GAC (D386A) was measured at the indicated phosphate concentrations and 20 mM glutamine using the two-step assay. Each data point shown is the mean from three independent experiments. A simultaneous nonleast square fit of the data from three independent experiments to Equation 4 (solid line) yielded a Hill coefficient, h = 1.7 ± 0.2 (±SD of there independent fits). D, initial velocities of glutamine hydrolysis by GAC (D386A) was measured at the indicated glutamine concentrations and 100 mM K2HPO4 using the two-step assay. Each data point shown is the mean from three independent experiments. A simultaneous nonleast square fit of the data from three independent experiments to Equation 4 (solid line) yielded a Hill coefficient, h = 1.0 ± 0.1 (±SD of there independent fits). GAC, glutaminase C.
The two dimers comprising the GAC tetramer are held together by four salt bridge interactions between Asp386 and Lys396 that are present on the opposing subunit pairs. Weakening the electrostatic interactions at the GAC dimer–dimer interface and reducing the ability of dimers to engage to form a tetramer would allow us to further examine how glutamine and phosphate are influencing the dimer to tetramer transition during catalysis. Therefore, we changed the aspartic acid at position 386 to an alanine residue to create GAC (D386A), which is less effective at forming tetramers as indicated by analytical size-exclusion chromatography (Fig. S3). When the enzymatic activity of the GAC (D386A) mutant was assayed as a function of inorganic phosphate concentration, at a fixed saturating level of glutamine, the dose–response profile for the anionic activator appeared to be cooperative with a Hill coefficient of 1.7 ± 0.2 (Fig. 6C). This differs from what we observed with the WT enzyme, where no cooperativity was observed for the phosphate dose response when measuring catalytic activity. The hindered ability of the GAC (D386A) mutant to form tetramers, even at saturating substrate concentrations, apparently requires the initial binding of phosphate (at low doses) to fully drive tetramer formation, which then enables the activator to bind with higher affinity to the tetrameric enzyme, thereby accounting for the cooperativity observed. However, when the enzymatic activity of the GAC (D386A) mutant was assayed as a function of glutamine concentration, at a fixed saturating level of inorganic phosphate, a strict hyperbolic dependence on substrate concentration was observed with a Hill coefficient of 1.0 ± 0.1 (Fig. 6D), similar to what we observed when assaying WT GAC. Thus, saturating levels of inorganic phosphate are able to fully drive the GAC (D386A) mutant to form tetramers and synchronize glutamine binding to all four enzyme subunits with high affinity, thereby giving rise to maximal catalytic activity.
Discussion
Members of the glutaminase family of enzymes play a key role in cancer progression by catalyzing the first step in glutamine metabolism, the hydrolysis of glutamine to glutamate with the production of ammonia, generating α-ketoglutarate as a carbon source that enters the TCA cycle (7, 8, 9, 10). This provides a mechanism to compensate for the uncoupling of the glycolytic pathway from the mitochondrial TCA cycle, characteristic of the Warburg effect. GAC, a carboxyl-terminal splice variant of the glutaminase family member GLS, has been shown to be highly upregulated in a number of cancers and consequently has received a good deal of attention as a potential therapeutic target (9, 11, 12, 13). Therefore, much effort has been directed toward understanding how the catalytic activity of GAC is regulated.
In order to gain additional insights into the mechanisms responsible for GAC activation, we have developed fluorescence spectroscopic assays to directly monitor the binding of the substrate glutamine as well as the anionic activator inorganic phosphate and allosteric inhibitors to the enzyme (23, 34). Here, we have used one of these readouts in which a tryptophan was substituted for a tyrosine residue at position 466 within human GAC, immediately proximal to the substrate-binding site. This substitution prevented catalysis and thereby made it possible to directly monitor the binding of glutamine to GAC, both in the presence and absence of inorganic phosphate, without complications arising from hydrolysis of the substrate to the product glutamate. When using this spectroscopic readout to assay glutamine binding in the absence of the activator, at low GAC concentrations where the enzyme would exist predominantly in its dimeric state, we observed a single class of substrate interactions with relatively weak affinity. However, when these assays were performed at progressively higher GAC concentrations, which increase the potential for tetramers to form, substrate binding showed positive cooperativity. Thus, under conditions where GAC exists in a dimeric state, glutamine binds to the two subunits of the dimer equally and independently but relatively weakly. At higher GAC concentrations, the initial low-affinity binding of glutamine to the enzyme dimer helps promote tetramer formation, albeit less effectively than inorganic phosphate (Fig. S4), thus enabling substrates to bind to the tetramers that form with higher affinity.
Structural analysis of the glutamine-bound GAC (Y466W) tetramer in the absence of the anionic activator phosphate shed additional light on how substrate binds to enzyme. The X-ray structure showed that each of the four enzyme subunits were occupied with glutamine, although there were differences in the orientation of their lids that open and close over the active site. In particular, the two subunits comprising one of the dimers making up the GAC tetramer showed the lids in an open conformation, whereas the two subunits of the adjacent dimer had closed lids. In the case of the GAC tetramer in the apo state (i.e., without bound substrate or anionic activators), the lids likely undergo transitions between the open and closed conformations, as X-ray crystal structures of the apo enzyme show that their lids are poorly defined. Glutamine binding to a GAC dimer with one or both subunits having an open lid conformation is likely to be relatively weak, consistent with our finding that substitutions within the lid that enable it to more easily transition to the open-state conformation result in the enzyme having a significantly weaker affinity for substrate. However, the initial weak binding of glutamine to a GAC dimer with open lids, by promoting tetramer formation and transmitting conformational changes to the adjoining enzyme dimer within the tetramer, would enable the lids to close over the bound substrates, resulting in higher affinity interactions that account for the positive cooperativity observed in the glutamine-binding profiles.
When the same fluorescence readout was used to assay glutamine binding to GAC (Y466W) in the presence of the anionic activator, inorganic phosphate, a different profile was obtained. Instead of observing positive cooperativity for the interactions of glutamine with GAC at enzyme concentrations where tetramers are able to form, the presence of the activator made it possible for the substrate to bind equally and with high affinity to each of the four enzyme subunits. The same was true when we examined glutamine binding to a double mutant, GAC (K320A and Y466W), which mimics the effects of an anionic activator. Moreover, when GAC (Y466W) was crystallized in the presence of both glutamine and phosphate, the resulting X-ray structure for the enzyme tetramer bound to glutamine did not show the same asymmetry regarding the lid orientations that we observed in the absence of the anionic activator. Instead, all four enzyme subunits contained glutamine with closed lid conformations. While anionic activators such as inorganic phosphate, as well as GAC mutants that mimic the actions of activators, significantly increase substrate-binding affinity, we found that the reverse is also true, that is the interactions of inorganic phosphate with GAC are enhanced at increasing substrate concentrations. This is likely because of the ability of glutamine to promote the formation of GAC tetramers, although again less effectively than inorganic phosphate (Fig. S4).
An analysis of the initial rates of enzyme catalysis observed under different conditions can then be interpreted in light of the glutamine-binding profiles that we have obtained. Although the activity of GAC in the absence of inorganic phosphate was markedly decreased compared with that measured in the presence of the anionic activator, a hyperbolic response rather than a sigmoidal profile indicative of positive cooperativity was obtained when GAC activity was measured as a function of glutamine concentration, even at enzyme levels where it exists as a tetramer. This finding indicates that in the absence of the anionic activator, catalysis is only occurring at those subunits in which the lids have closed over the bound substrates but with a low turnover rate (24, 29, 30). The binding of the anionic activator then causes the situation to change, by triggering conformational changes that immediately induce GAC tetramer formation (24, 28, 31) and enabling substrates to have immediate access to all four subunits of the enzyme tetramer by stabilizing their open lid conformations. Upon glutamine binding, the lids then close over the active sites, increasing substrate affinity by impairing substrate dissociation from the enzyme, thus enabling equal and simultaneous maximal catalytic turnover at each active site.
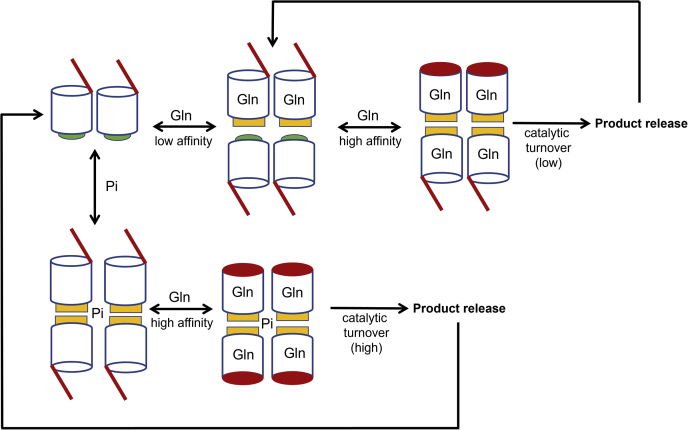
These findings, together with those from earlier studies (21, 23, 28), lead to a working model for the interactions of substrates and anionic activators with GAC. As depicted in Figure 7, under conditions where apo GAC initially exists as a dimer, glutamine binds with relatively low affinity only to those subunits that have an open lid. Substrate binding can help promote tetramer formation, thereby enabling the subsequent binding of substrates to occur with higher affinity as the lids close over their binding sites, accounting for the positive cooperativity observed in the glutamine-binding profiles. In the absence of allosteric activators, such as inorganic phosphate, catalytic turnover is only occurring at those subunits where the lids have closed but is still relatively low. The binding of allosteric anionic activators to GAC more strongly induces tetramer formation and stabilizes the open lid conformation, enabling glutamine to immediately have access to all four active sites of the enzyme tetramer. However, upon glutamine binding, the lids close over the substrates, locking them into the active sites and enabling maximum catalytic turnover to occur at all four subunits simultaneously, whereas weakening the affinity for the anionic activators. The formation of products (glutamate) would then prompt the opening of the lids to enable their release and allow the rebinding of activators to start another catalytic cycle. Such a mechanism would provide for a highly tuned regulation of GAC catalytic activity, which would ensure that even at relatively high glutamine concentrations, while the enzyme is fully loaded with substrate, it would only be capable of very low catalytic activity. When an anionic activator is made available, GAC is then immediately converted from an asymmetric tetrameric state with relatively weak enzymatic activity, to a symmetrical tetramer that can undergo high rates of catalysis occurring at all four subunits. Future studies will be directed toward further testing different aspects of this working model and identifying those anionic activators produced by cancer cells that drive GAC to an optimal catalytic state, thus ensuring that optimal levels of glutamine metabolism occur to appropriately compensate for the Warburg effect.
Figure 7.
Schematic diagram depicting the interactions of the substrate glutamine and the anionic activator phosphate with GAC. Shown is the initial weak binding of glutamine to dimeric GAC in which the lids are in an open conformation. However, this leads to the formation of GAC tetramers, as a result of conformational changes through the activation loops at the dimer–dimer interface, and the subsequent high-affinity binding of glutamine to the remaining substrate sites, caused by the closing of the lids, resulting in (low) catalytic turnover. The production of glutamate causes the lids to reopen, enabling the reaction product to be released. In the presence of inorganic phosphate, GAC dimers are driven to form tetramers, which then enable all four subunits to bind glutamine with high affinity and to have their lids closed. The closing of the lids results in the dissociation of the activator phosphate and maximal catalytic turnover, at which point, the production of glutamate opens the lids so that it can dissociate from the enzyme. The activator phosphate would then rebind to the enzyme with open lids and start another catalytic cycle. GAC, glutaminase C.
Experimental procedures
Recombinant protein expression and purification
An N-terminal His-tagged form of the full-length human kidney-type glutaminase isoform GAC (NP_001106854.1) gene without the mitochondrial localization sequence (residue Leu72–Ser598) was cloned into pQE80L plasmid. The equivalent construct encoding the mouse GAC gene was cloned into pET28a plasmid. Site-directed mutagenesis was performed for desired mutations using Phusion DNA polymerase (NEB). To express each construct, plasmids were transformed into Escherichia coli BL21(DE3) competent cells. Overnight cultures were grown in LB medium with 100 mg/ml ampicillin (for pQE80L) or 50 mg/ml kanamycin (for pET28a) and subsequently inoculated at ∼1:100 ratio in 6 l cultures with the same antibiotic concentrations. The culture was left shaking at 37 °C and 180 rpm for 3 to 4 h until an absorbance reached between 0.6 and 0.8 at 600 nm. The flasks were then chilled at 4 °C for 1 to 2 h before being induced by the addition of 30 μM IPTG and shaking at room temperature for 16 to 18 h. Cells were collected by rapid centrifugation and resuspended in 150 ml lysis buffer (50 mM Tris–HCl, pH 8.5, 500 mM NaCl, and 10% glycerol) with ∼2 mg of DNase I for 20 to 30 min. The cells were disrupted mechanically using sonication, and the soluble fractions were separated from the debris by ultracentrifugation (40,000g). The lysate was then loaded onto Co2+-charged TALON resin, which had been equilibrated previously with the wash buffer (50 mM Tris–HCl, pH 8.5, 10 mM NaCl, and 10 mM imidazole). The protein that bound to the column was washed with the wash buffer and eluted with 320 mM imidazole. Further purification was performed by anion exchange chromatography using MonoQ column (GE Healthcare) and size-exclusion chromatography using Superdex 200 pg 16/600 column (GE Healthcare). Proteins were kept in 20 mM Tris–HCl (pH 8.5), 500 mM NaCl for crystallization, and 20 mM Tris–HCl (pH 8.5), 150 mM NaCl for assay purposes and otherwise snap-frozen in liquid nitrogen and stored at −80 °C. Protein concentrations were determined by absorbance at 280 nm using extinction coefficients calculated using Expasy ProtParam tool. The residue numbers for GAC were used throughout the text to avoid confusion.
Fluorescence assays
Measurements were carried out using a Varian Cary Eclipse fluorimeter in the “Kinetics” mode. Stock solutions of glutamine or glutamate (200 mM) and K2HPO4 (1 M) were prepared in the assay buffer (50 mM Tris acetate, pH 8.5, 0.125 mM EDTA, and 100 mM NaCl). One hundred microliter of recombinant GAC was added to 900 μl of assay buffer, supplemented with 100 mM NaCl in a 1 ml quartz cuvette to the appropriate final concentration. Samples were stirred constantly while being held at 25 °C. Subsequently, 100 μl of glutamine at the indicated concentrations were added to the protein solution at 30 s. The signal was recorded at an excitation wavelength of 285 nm (5 nm bandpass) and an emission wavelength of 340 nm (20 nm bandpass). K2HPO4 was supplemented as part of the 900 μl of initial buffer when needed.
Glutaminase activity assays
Glutaminase activity was measured by coupling NAD+ reduction to glutamine hydrolysis in a two-step reaction. In the first reaction, glutaminase catalyzes the hydrolysis of glutamine to glutamate, and in the second reaction, glutamate dehydrogenase catalyzes the oxidative deamination of glutamate to form α-ketoglutarate and NADH, which can be measured through its absorbance at 340 nm. Assay buffer for the first reaction contains 50 mM Tris acetate (pH 8.5) and 0.125 mM EDTA. Stock solutions of glutamine or glutamate (200 mM) and K2HPO4 (1 M) were prepared in the assay buffer. The first reaction was initiated upon the addition of 10 μl of WT GAC (0.5 μM stock) to 90 μl of a reaction mix (20 μl glutamine, 10 μl K2HPO4, and 60 μl assay buffer). The reaction was quenched after 2 min (in the absence of K2HPO4) or 15 s (in the presence of K2HPO4) by the addition of 10 μl HCl (3 M), and the samples were subsequently stored on ice. In the second reaction, the reaction mixture containing 130 mM Tris–HCl (pH 9.4), 7 units/ml of glutamate dehydrogenase, and 2 mM NAD+. Two hundred microliter of the reaction mix was added to a single well of a 96-well Costar clear plate (Corning), and 10 μl of each sample from the first step was added, and the reaction was incubated with gentle rocking at room temperature for 40 to 60 min. The absorbance at 340 nm was measured on a plate reader. A standard curve was obtained with glutamate solutions in place of the first reaction. The slope of the linear part of the standard curve was used to convert the absorbance values to the concentration of glutamine hydrolyzed. The initial velocities of glutamine hydrolysis were calculated by dividing the concentration of glutamine hydrolyzed by the time of the first reaction.
Equations for data analysis
Dose–response curves of fluorescence as a function of glutamine concentration were fit to Equations 1 and 2,
| (1) |
where F is the equilibrium fluorescence attained upon the addition of glutamine at a concentration LT, F0 is the initial fluorescence before the addition of glutamine, Ff is the fluorescence attained in the presence of saturating concentrations of glutamine, K0.5 is the concentration of glutamine at which 50% of the fluorescence change occurs, and h is the Hill coefficient.
| (2) |
where Y is the fractional fluorescence change, α is the normalized ligand concentration describing how many times of KR the ligand concentration is (α = S/[KR], where S is the concentration of glutamine and KR is the dissociation constant for the tight binding state), n is the number of binding sites (n = 4 for GAC), L is the equilibrium constant between R and T states, and c = (KR)/(KT), where KT is the dissociation constant for the weak binding state.
| (3) |
where F is the equilibrium fluorescence attained upon the addition of glutamine at a concentration LT, F0 is the initial fluorescence before the addition of glutamine, Ff is the fluorescence attained in the presence of saturating concentrations of glutamine, RT is the concentration of glutaminase used, and KD is the dissociation constant.
Initial velocities of glutamine hydrolysis as a function of glutamine concentration were fit to the Michaelis–Menten equation,
| (4) |
where V is the initial velocity at any glutamine concentration, S is the concentration of glutamine, Vmax is the maximal velocity at saturating concentrations of glutamine, KM is the Michaelis constant, and h is the Hill coefficient.
Crystallization, data collection, and structure determination
Human GAC (Y466W) was concentrated to 5 mg/ml using an Amicon ultrafiltration device (10 KDa cutoff; Millipore). Stock solutions of glutamine (200 mM) and K2HPO4 (1 M) were made using the buffer from the size-exclusion purification step (20 mM Tris–HCl [pH 8.5] and 150 mM NaCl). GAC (Y466W) was mixed with glutamine so that the final concentration was 3 mg/ml for the protein and 40 mM glutamine and kept on ice for ∼1 h. Crystallization experiments were performed at 18 °C using the conventional hanging drop vapor diffusion technique (2 μl of protein solution and 2 μl of reservoir solution), with the reservoir containing 11.5% PEG 6000, 0.95 M LiCl, and 0.1 M Tris (pH 8.5). Crystals formed after 5 days and grew to full size in 14 days and did not require cryoprotectant for plunge freezing. For crystallization in the presence of inorganic phosphate, GAC (Y466W) was mixed with the ligand solution so that the final concentrations were 3 mg/ml for the protein, 40 mM glutamine, and 40 mM K2HPO4. Crystals formed with 9% PEG 3350, 0.9 M LiCl, and 0.1 M Tris (pH 8.5) after 5 days and grew to full size in 14 days. The harvested crystals were soaked for 15 min with a solution of mother liquor supplemented with 40 mM Gln, 200 mM K2HPO4, and 10% ethylene glycol immediately prior to freezing.
The diffraction data were collected at cryogenic temperature (100 K) at the 23-ID-D and 23-ID-B-GM/CA beamline at the GM/CA beamline of the Advanced Photon Source (APS) with 0.2° oscillation/image. The monomer extracted from the apo hGAC (WT) structure (Protein Data Bank [PDB]: 5D3O) was used as the search model for molecular replacement for both structures. The data reduction was performed with HKL2000 (35), prior to phasing and refinement by Phenix (36) and Coot (37).
Analytical size-exclusion chromatography
Different running buffers were made to obtain elution profiles of apo GAC (20 mM Tris [pH 8.5] and 100 mM NaCl), GAC with saturating glutamine (20 mM Tris [pH 8.5], 100 mM NaCl, and 20 mM glutamine), and GAC with saturating inorganic phosphate (20 mM Tris [pH 8.5], 100 mM NaCl, and 100 mM K2HPO4). A superdex 200 5/150 GL (Cytiva) column was pre-equilibrated with the running buffers, and 50 μl of protein (10 μM) was injected in the column.
Data availability
The structures of GAC (Y466W) co-crystallized with l-glutamine in the absence and presence of K2HPO4 can be found in the PDB (PDB IDs: 7SBM and 7SBN, respectively).
Supporting information
This article contains supporting information.
Conflict of interest
R. A. C. reports financial support provided by the National Institutes of Health. All other authors declare no conflicts of interest with the contents of this article.
Acknowledgments
We thank Dr Marc Antonyak, Dr Shawn Milano, and Cody Aplin for valuable discussions. This study was supported by the National Institutes of Health grants GM122575 and CA201402. This study is dedicated to the memory of our dear colleague Jon Erickson. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
T.-T. T. N. and S.R. conceptualization; T.-T. T. N. and S. R. methodology; S. R. validation; T.-T. T. N. and M. J. H. investigation; T.-T. T. N. formal analysis; T.-T. T. N. writing–original draft; T.-T. T. N. and R. A. C. writing–review & editing; T.-T. T. N. visualization; R. A. C. supervision; R. A. C. project administration; R. A. C. funding acquisition.
Edited by Phyllis Hanson
Supporting information
References
- 1.Vander Heiden M.G. Targeting cancer metabolism: A therapeutic window opens. Nat. Rev. Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 2.Luengo A., Gui D.Y., Vander Heiden M.G. Targeting metabolism for cancer therapy. Cell Chem. Biol. 2017;24:1161–1180. doi: 10.1016/j.chembiol.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kroemer G., Pouyssegur J. Tumor cell metabolism: Cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Teicher B.A., Linehan W.M., Helman L.J. Targeting cancer metabolism. Clin. Cancer Res. 2012;18:5537–5545. doi: 10.1158/1078-0432.CCR-12-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D., Weinberg R.A. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Wise D.R., Thompson C.B. Glutamine addiction: A new therapeutic target in cancer. Trends Biochem. Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lukey M.J., Katt W.P., Cerione R.A. Targeting amino acid metabolism for cancer therapy. Drug Discov. Today. 2017;22:796–804. doi: 10.1016/j.drudis.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Méndez-Lucas A., Lin W., Driscoll P.C., Legrave N., Novellasdemunt L., Xie C., Charles M., Wilson Z., Jones N.P., Rayport S., Rodríguez-Justo M., Li V., MacRae J.I., Hay N., Chen X., et al. Identifying strategies to target the metabolic flexibility of tumours. Nat. Metab. 2020;2:335–350. doi: 10.1038/s42255-020-0195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang C., Ko B., Hensley C.T., Jiang L., Wasti A.T., Kim J., Sudderth J., Calvaruso M.A., Lumata L., Mitsche M., Rutter J., Merritt M.E., DeBerardinis R.J. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol. Cell. 2014;56:414–424. doi: 10.1016/j.molcel.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J.B., Erickson J.W., Fuji R., Ramachandran S., Gao P., Dinavahi R., Wilson K.F., Ambrosio A.L., Dias S.M., Dang C.V., Cerione R.A. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross M.I., Demo S.D., Dennison J.B., Chen L., Chernov-Rogan T., Goyal B., Janes J.R., Laidig G.J., Lewis E.R., Li J., MacKinnon A.L., Parlati F., Rodriguez M.L., Shwonek P.J., Sjogren E.B., et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther. 2014;13:890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- 13.DeBerardinis R.J., Cheng T. Q’s next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukey M.J., Cluntun A.A., Katt W.P., Lin M.J., Druso J.E., Ramachandran S., Erickson J.W., Le H.H., Wang Z.E., Blank B., Greene K.S., Cerione R.A. Liver-type glutaminase GLS2 is a druggable metabolic node in luminal-subtype breast cancer. Cell Rep. 2019;29:76–88.e7. doi: 10.1016/j.celrep.2019.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Den Heuvel A.P., Jing J., Wooster R.F., Bachman K.E. Analysis of glutamine dependency in non-small cell lung cancer: GLS1 splice variant GAC is essential for cancer cell growth. Cancer Biol. Ther. 2012;13:1185–1194. doi: 10.4161/cbt.21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang L., Mou J., Shao B., Wei Y., Liang H., Takano N., Semenza G.L., Xie G. Glutaminase 1 expression in colorectal cancer cells is induced by hypoxia and required for tumor growth, invasion, and metastatic colonization. Cell Death Dis. 2019;10:40. doi: 10.1038/s41419-018-1291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao P., Tchernyshyov I., Chang T., Lee Y., Kita K., Ochi T., Zeller K.I., De Marzo A.M., Van Eyk J.E., Mendell J.T., Dang C.V. C-myc suppression of MiR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukey M.J., Greene K.S., Erickson J.W., Wilson K.F., Cerione R.A. The oncogenic transcription factor c-Jun regulates glutaminase expression and sensitizes cells to glutaminase-targeted therapy. Nat. Commun. 2016;7:11321. doi: 10.1038/ncomms11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu L., Yin Y., Li Y., Chen X., Chang Y., Zhang H., Liu J., Beasley J., McCaw P., Zhang H., Young S., Groth J., Wang Q., Locasale J.W., Gao X., et al. A glutaminase isoform switch drives therapeutic resistance and disease progression of prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2012748118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leone R.D., Zhao L., Englert J.M., Sun I.M., Oh M.H., Sun I.H., Arwood M.L., Bettencourt I.A., Patel C.H., Wen J., Tam A., Blosser R.L., Prchalova E., Alt J., Rais R., et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science. 2019;366:1013–1021. doi: 10.1126/science.aav2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y., Erickson J.W., Stalnecker C.A., Katt W.P., Huang Q., Cerione R.A., Ramachandran S. Mechanistic basis of glutaminase activation: A key enzyme that promotes glutamine metabolism in cancer cells. J. Biol. Chem. 2016;291:20900–20910. doi: 10.1074/jbc.M116.720268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weil-Malherbe H. Activators and inhibitors of brain glutaminase. J. Neurochem. 1969;16:855–864. doi: 10.1111/j.1471-4159.1969.tb08973.x. [DOI] [PubMed] [Google Scholar]
- 23.Li Y., Ramachandran S., Nguyen T.-T.T., Stalnecker C.A., Cerione R.A., Erickson J.W. The activation loop and substrate-binding cleft of glutaminase C are allosterically coupled. J. Biol. Chem. 2020;295:1328–1337. doi: 10.1074/jbc.RA119.010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godfrey S., Kuhlenschmidt T., Curthoys N.P. Correlation between activation and dimer formation of rat renal phosphate dependent glutaminase. J. Biol. Chem. 1977;252:1927–1931. [PubMed] [Google Scholar]
- 25.Morehouse R.F., Curthoys N.P. Properties of rat renal phosphate-dependent glutaminase coupled to sepharose. Evidence that dimerization is essential for activation. Biochem. J. 1981;193:709–716. doi: 10.1042/bj1930709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson M.M., McBryant S.J., Tsukamoto T., Rojas C., Ferraris D.V., Hamilton S.K., Hansen J.C., Curthoys N.P. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-Yl)ethyl sulfide (BPTES) Biochem. J. 2007;406:407–414. doi: 10.1042/BJ20070039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenny J., Bao Y., Hamm B., Taylor L., Toth A., Wagers B., Curthoys N.P. Bacterial expression, purification, and characterization of rat kidney-type mitochondrial glutaminase. Protein Expr. Purif. 2003;31:140–148. doi: 10.1016/s1046-5928(03)00161-x. [DOI] [PubMed] [Google Scholar]
- 28.Cassago A., Ferreira A.P., Ferreira I.M., Fornezari C., Gomes E.R., Greene K.S., Pereira H.M., Garratt R.C., Dias S.M., Ambrosio A.L. Mitochondrial localization and structure-based phosphate activation mechanism of glutaminase C with implications for cancer metabolism. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1092–1097. doi: 10.1073/pnas.1112495109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stalnecker C.A., Ulrich S.M., Li Y., Ramachandran S., McBrayer M.K., De Berardinis R.J., Cerione R.A., Erickson J.W. Mechanism by which a recently discovered allosteric inhibitor blocks glutamine metabolism in transformed cells. Proc. Natl. Acad. Sci. U. S. A. 2015;112:394–399. doi: 10.1073/pnas.1414056112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monod J., Wyman J., Changeux J.P. On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 31.Ferreira A.P., Cassago A., De Almeida Gonçalves K., Dias M.M., Adamoski D., Ascenção C.F., Honorato R.V., de Oliveira J.F., Ferreira I.M., Fornezari C., Bettini J., Oliveira P.S., Leme A.F., Portugal R.V., Ambrosio A.L., et al. Active glutaminase C self-assembles into a supratetrameric oligomer that can be disrupted by an allosteric inhibitor. J. Biol. Chem. 2013;288:28009–28020. doi: 10.1074/jbc.M113.501346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thangavelu K., Pan C.Q., Karlberg T., Balaji G., Uttamchandani M., Suresh V., Schüler H., Low B.C., Sivaraman J. Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by raf-mek-erk signaling in cancer cell metabolism. Proc. Natl. Acad. Sci. U. S. A. 2012;109:7705–7710. doi: 10.1073/pnas.1116573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thangavelu K., Chong Q.Y., Low B.C., Sivaraman J. Structural basis for the active site inhibition mechanism of human kidney-type glutaminase (KGA) Sci. Rep. 2014;4:3827. doi: 10.1038/srep03827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stalnecker C.A., Erickson J.W., Cerione R.A. Conformational changes in the activation loop of mitochondrial glutaminase C: A direct fluorescence readout that distinguishes the binding of allosteric inhibitors from activators. J. Biol. Chem. 2017;292:6095–6107. doi: 10.1074/jbc.M116.758219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., Moriarty N.W., Oeffner R.D., Poon B.K., Prisant M.G., Read R.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta. Crystallogr. D. Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta. Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structures of GAC (Y466W) co-crystallized with l-glutamine in the absence and presence of K2HPO4 can be found in the PDB (PDB IDs: 7SBM and 7SBN, respectively).