Highlights
-
•
In this study we characterized in vivo large-scale propagation pathways of α-synuclein pathology and identified different patterns of functional connectivity disruptions in PD patients at different stages of the disease.
-
•
Our results identified key genetic signatures of large-scale PD pathology, highlighting their contribution to focal neuronal vulnerability to disease progression.
-
•
Our results pave the way toward better accounting for the brain networks complexity and mechanisms underlying distinct spatial vulnerability to PD pathology, thus informing early diagnosis and future novel therapeutic strategies.
Abbreviations: AHBA, Allen Human Brain Atlas; AZSAND, Arizona Study of Aging and Neurodegenerative Disorders; BBDP, Brain and Body Donation Program; CAT12, Computational Anatomy Toolbox 12; DK, Desikan-Killiany; FDR, False Discovery Rate; GO, Gene Ontology; HY, Hoehn and Yahr; LEDD, levodopa equivalent daily-dose; PANTHER, Protein Analysis Through Evolutionary Relationships; PALS, Population-Average Landmark and Surface-based; PD, Parkinson's disease; RS-fMRI, resting-state functional MRI;; SD, standard deviations; SFC, stepwise functional connectivity; UPDRS, Unified Parkinson’s Disease Rating Scale
Keywords: Parkinson’s disease, fMRI, Connectomics, Cortical Gene Expression
Abstract
The genetic traits that underlie vulnerability to neuronal damage across specific brain circuits in Parkinson’s disease (PD) remain to be elucidated. In this study, we characterized the brain topological intersection between propagating connectivity networks in controls and PD participants and gene expression patterns across the human cortex – such as the SNCA gene. We observed that brain connectivity originated from PD-related pathology epicenters in the brainstem recapitulated the anatomical distribution of alpha-synuclein histopathology in postmortem data. We also discovered that the gene set most related to cortical propagation patterns of PD-related pathology was primarily involved in microtubule cellular components. Thus, this study sheds light on new avenues for enhancing detection of PD neuronal vulnerability via an evaluation of in vivo connectivity trajectories across the human brain and successful integration of neuroimaging-genetic strategies.
1. Introduction
Parkinson's disease (PD) is a frequently diagnosed neurodegenerative disorder that is characterized by an accumulation of Lewy bodies and Lewy neurites in the brain, including α-synuclein aggregates (Poewe et al., 2017). According to the presence of this pathologically misfolded protein, Braak and colleagues hypothesized that the pathologic process progresses in a stereotyped fashion along specific brain systems, from one susceptible brain region to the next, sometimes postulated as a trans-synaptic spreading mechanism (Braak et al., 2003a, Kalia and Lang, 2015, Soto and Pritzkow, 2018). The observed pattern of PD pathology distribution at postmortem led Braak et al. to suggest a six-stage system of disease progression, beginning from the brainstem and progressively affecting subcortical areas and cerebral hemispheres along neuronal pathways, following a caudal-rostral axis within the brain (Braak et al., 2003b, Braak et al., 2006b, Goedert et al., 2013). While histological postmortem studies have helped establish patterns of α-synuclein distribution in PD brains, it is less understood how α-synuclein, as well as other PD-related pathologic factors, relate to in vivo disruptions of large-scale neuronal circuits across the entire cerebral topology. Furthermore, the genetic background that underlies the vulnerability of specific neuronal circuits related to PD pathology remains to be elucidated.
Recent in vivo neuroimaging studies have shown that deposition and aggregation of misfolded proteins in neurodegenerative diseases, such as Alzheimer’s disease and PD, are not randomly distributed but follow connectivity networks, supporting the assumed trans-neuronal spread model of progression (Goedert et al., 2017, Greicius, 2008, Sepulcre et al., 2018, Zheng et al., 2019, Zhou et al., 2012). In the last few years, the graph-theory neuroimaging approach has widely aided the study of complex human brain network architectures (Crossley et al., 2014, Fornito et al., 2015, Rubinov and Sporns, 2010). The implementation of such a framework in the study of PD pathology networks would help toward a deeper understanding of the spatial distribution and potential propagation of PD (Yau et al., 2018). Using graph theory-based methods, previous studies have investigated models of PD propagation through brain networks using resting-state functional MRI (RS-fMRI). For example, functional and anatomical connectivity of a supposed epicenter of the disease in the substantia nigra has been shown to be correlated with degree of regional atrophy (Zeighami et al., 2015). Such findings have been corroborated in more recent studies, suggesting that cortical areas with greater functional or structural connectivity to subcortical pathology reservoirs may present greater cortical impairment (Yau et al., 2018). Finally, graph theory-based approaches may help in understanding the neural correlates of clinical evolution in PD (Filippi et al., 2020b, Zarkali et al., 2021). A recent study has examined changes in structural–functional connectivity coupling in PD patients in relation to underlying different neurotransmitter expression, identifying the organisational patterns that possibly drive cognitive impairment (Zarkali et al., 2021).
In this study, we applied a novel graph theory metric based on stepwise functional connectivity (SFC) analysis (Bueichekú et al., 2020, Gao et al., 2019, Qian et al., 2018, Sepulcre et al., 2012). This method allows for the mapping of connectivity patterns of brain ‘seed’ regions and considers direct and indirect connectivity routes. In this way, we aimed to reveal how disease might alter functional brain network organization and how pathology may spread across such networks. Moreover, a detailed account of the relationship between brain connectivity networks and spatial gene expression patterns would aid in the understanding of the pathogenic mechanisms underlying progression of pathology and its interaction with genes in neurodegenerative diseases (Sepulcre et al., 2018). In the emerging field of neuroimaging-genetics, the advent of a comprehensive brain-wide gene expression atlas (Allen Human Brain Atlas (Jones et al., 2009, Shen et al., 2012); AHBA) helped position the field toward innovation by combining RS-fMRI with gene expression data, opening up an exciting avenue for discovery in neurobiology and brain network architectures (Diez et al., 2021, Freeze et al., 2018, Ortiz-Teran et al., 2017). To date, the vulnerability that the SNCA gene (encoding α-synuclein protein) and other gene expression profiles confer to functional connectivity networks in PD patients is not well understood. In keeping with the hypothesis that α-synuclein deposits follow specific propagation connectivity pathways in the human brain, our study first aimed to investigate functional brain network organization and putative pathways of α-synuclein deposits in a sample of PD participants at different stages of the disease using functional connectivity MRI and a new graph theory metrics. Secondly, we examined the topological similarity between large-scale functional connectivity networks emanating from postulated brain-stem epicenters of PD pathology and gene expression patterns of the SNCA gene. We then evaluated if functional connectivity disruptions were associated with the abnormal deposition of α-synuclein in PD participants by using postmortem density scores of α-synuclein-immunoreactive Lewy bodies. Finally, we investigated the biological functional meaning of our connectomic-genetic findings.
2. Materials and methods
The main cohort of participants of this study were recruited by University of Belgrade, Belgrade, Serbia, where they performed clinical and cognitive evaluations, and RS-fMRI scans (Filippi et al., 2020b). The replication dataset of PD participants for neuroimaging purposes was recruited at the Hospital Universitario Donostia, San Sebastian, Spain. Both studies were approved by the local ethics committees at University of Belgrade and Gipuzkoa Clinical Research Ethics Committee respectively and written informed consent to participate in the study was obtained from all participants. All the acquired data were sent to Neuroimaging Research Unit, Division of Neuroscience of San Raffaele Scientific Institute and Vita-Salute San Raffaele University of Milan, Italy, and analyzed at Gordon Center for Medical Imaging, Department of Radiology, Massachusetts General Hospital and Harvard Medical School, Boston, Massachusetts, USA.
2.1. Participants and cluster/subtype definition
154 PD patients were recruited and underwent comprehensive evaluation including clinical, cognitive/behavioral and MRI assessments (Supplemental Table 1) as previously described (Filippi et al., 2020b, Filippi et al., 2020c). Sixty age- and sex-matched healthy controls performed the same study protocol (Supplemental Table 1). Eight PD patients were excluded from analysis due to excessive movement. Supplementary materials report details on (i) inclusion/exclusion criteria; (ii) cognitive/behavioral evaluations; and (iii) cluster analysis for data partitioning (Filippi et al., 2020b). According to cluster analysis, two main PD subgroups were identified: 86 mild and 60 moderate-to-severe PD patients (Supplemental Tables 1 and 4) (Filippi et al., 2020b, Filippi et al., 2020c).
A replication cohort of PD participants (N = 58, mean age 70.80 ± 5.76 years, 20/38F/M) was recruited at the Hospital Universitario Donostia, and scanned at the Basque Center on Cognition, Brain and Language, San Sebastian, Spain. Replication data were used for neuroimaging purposes only, namely to assess the spatial similarity between SFC maps with our PD study sample for three alternative seeds of the brainstem (medulla, pons and midbrain) (see MRI analysis for further details).
2.2. MRI analysis
Supplementary materials report the MRI protocol, RS-fMRI pre-processing details and functional connectivity brain networks construction (Fig. 1-A).
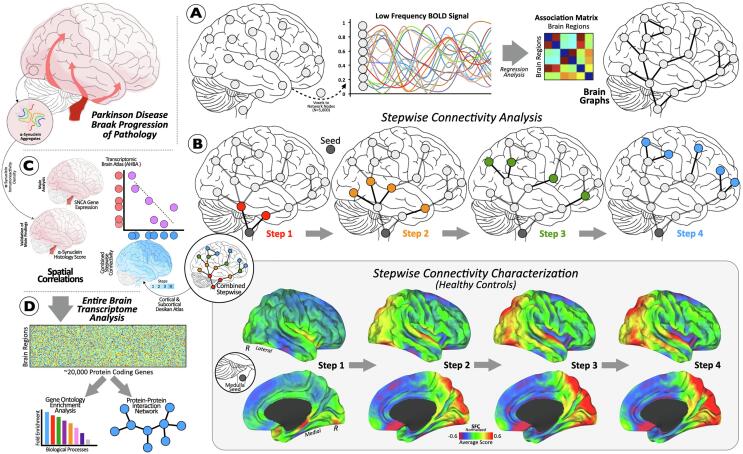
Fig. 1.
Progression of Parkinson’s disease pathology based on Braak stages. (A) Pipeline of intrinsic connectivity analysis. (B) Cortical connectivity diagram of an a priori selected area in the brainstem using stepwise connectivity analysis. Cortical maps show characterization of stepwise connectivity analysis from medulla oblongata in healthy controls. (C) Small diagram showing the cortical maps and scatterplot of our spatial similarity approach. (D) Connectomic-genetic analyses. Abbreviations: SFC = stepwise functional connectivity; N = number; AHBA = Allen Human Brain Atlas; R = right.
2.2.1. Stepwise functional connectivity analysis
SFC analysis is a novel graph theory metric that detects both direct and indirect functional couplings between a predefined seed region and other brain regions (Fig. 1-B) (Costumero et al., 2020, Gao et al., 2019, Qian et al., 2018, Sepulcre et al., 2012). SFC analysis aims to characterize regions connected to specific seed brain areas at different levels of link-step distances (Costumero et al., 2020, Gao et al., 2019, Qian et al., 2018, Sepulcre et al., 2012) (see Supplementary materials for details).
Given the goal of the study was to investigate a model of PD-related pathology propagation, we used seeds from brainstem regions described in the first stage of Braak’s staging scheme, namely medulla oblongata, pons, and midbrain. Binary masks were created for these three seeds using FreeSurfer version 6.0.0 software package (Fig. 1-B) (http://surfer.nmr.mgh.harvard.edu/) and its extension of brainstem substructure segmentation (https://surfer.nmr.mgh.harvard.edu/fswiki/BrainstemSubstructures). As our replication analysis showed a high reproducibility of the medulla oblongata connectivity distribution compared to pons and midbrain (Supplemental Fig. 1), and since the pathology occurs in the medulla in the first stage and only in the pontine tegmentum in the second stage (see Supplementary Table 5 for details), we used the medulla oblongata mask in our main analyses. To test if the obtained correlations with the replication dataset were due to spatial dependencies introduced by comparison of autocorrelated brain maps (Markello and Misic, 2021), the likelihood of the obtained similarity with randomly generated autocorrelated maps were evaluated. 1,000 surrogate maps were generated with a spatial autocorrelation matching target brain map using BrainSMASH software (Burt et al., 2020) and compared with the similarity obtained with replication dataset. All maps across different link-step distances from one to four (i.e., SFC maps 1 to 4) were used in the characterization of connectivity alterations between healthy controls and PD participants. Considering only healthy subjects, combined version of all SFC 1 to 4 maps into one single map from non-disrupted connectivity pathways (combined SFC map; Fig. 1-B) was employed to investigate the relationships between standard neuroimaging patterns and the AHBA transcriptome data. To build the SFC combined map, the functional connectivity between the seed region and all the other brain voxels has been identified in the four different steps. Subsequently, the highest functional connectivity per each voxel of the four previous considered values were selected. Finally, the values of each voxel in the SFC combined map were set to the number of step (from 1 to 4) in which the functional connectivity was maximized.
Thus, we obtained a SFC combined map for each subject whose values ranged from 1 to 4 steps (1 = closer to the epicenter; 4 = far from the epicenter), as a proxy for the spreading of pathology (1 denoting more pathology; 4 referring to less pathology). A mean combined SFC map of controls was obtained averaging all the healthy subject maps. Consequently, SFC maps were projected onto the cerebral hemispheres of the Population-Average Landmark and Surface-based (PALS) surface (PALS-B12) provided with Caret software (Van Essen and Dierker, 2007) using the “enclosing voxel algorithm” and “multifiducial mapping” settings (Fig. 2).
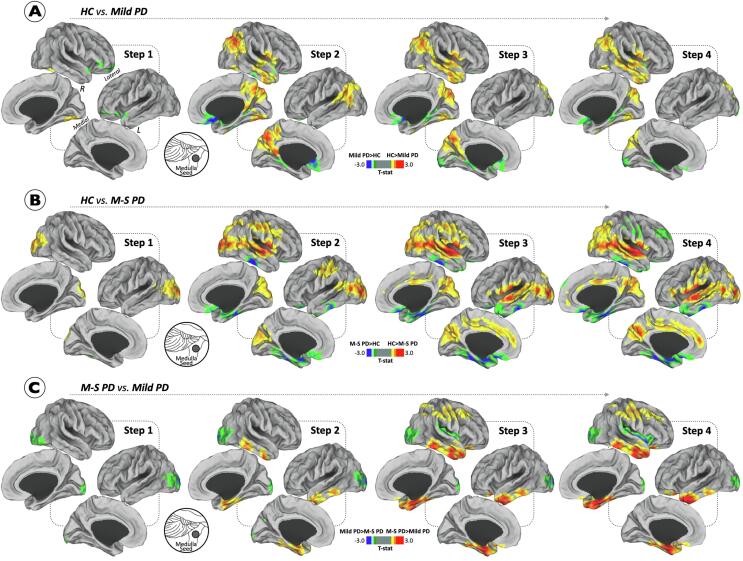
Fig. 2.
Differences between Parkinson’s disease participants and healthy controls in stepwise functional connectivity of the medulla oblongata. Cortical maps represent the significant differences in stepwise functional connectivity values between PD severity subtypes and healthy controls (A-B) and between PD groups (C). Insets show the approximate location of the seed. Statistical analysis was adjusted for age, gender and levodopa equivalent daily-dose (only in the comparison between patient groups). Results were corrected for multiple comparisons using a threshold-free cluster enhancement method combined with nonparametric permutation testing at p < 0.05 FWE-corrected. Color bars show the t-statistic applicable to the image. Abbreviations: HC = healthy controls; M−S = moderate-to-severe; PD = Parkinson’s disease.
In order to verify whether the null hypothesis is uniform, a network randomization analysis was performed by considering each individual functional connectivity matrix in the healthy controls and randomly shuffling all the connections. All data were normally distributed, and the randomization created also normally distributed data, including the degree distribution. Then, we re-run SFC maps across different link-step distances from one to four starting from the medulla oblongata seed (Supplemental Fig. 3). With this method, we used the resulting maps of healthy controls obtained after network randomisation to confirm disease progression in PD patients .
2.2.2. Statistical analyses
Voxel-wise analyses were performed using general linear models in SPM12 (Wellcome Department of Imaging Neuroscience, London, England; www.fil.ion.ucl.ac.uk/spm). Whole-brain two-sample t-test comparisons between groups were performed, including age, gender and LEDD (only in the comparison between patients’ groups) as covariates. A threshold-free cluster enhancement method (Smith and Nichols, 2009) combined with nonparametric permutation testing (5000 permutations) as implemented in the Computational Anatomy Toolbox 12 (CAT12, http://www.neuro.uni-jena.de/cat/) was used to detect statistically significant differences at p < 0.05, family-wise error (FWE) corrected. These analyses allowed for the identification of specific regions that demonstrated between-group differences in SFC.
2.2.3. Spatial associations between SFC and human transcriptome (AHBA)
Aprioristic knowledge of PD risk genes and data-driven approaches based on the full genome-wide (protein-coding) transcriptome of the AHBA (Hawrylycz et al., 2012) was used to investigate cortical and subcortical gene expression profiles associated with PD pathology (Fig. 1-C and 1-D). The AHBA provides whole-brain genome-wide expression of 20,737 protein-coding genes patterns extracted from 3,702 brain samples covering the whole brain (58,692 measurements) in six human subjects. To reduce noise and variability in the genetic measures a brain atlas was used and all the samples falling into each region averaged. We generated 105 regions to account for both cortical and subcortical structures. 68 cortical regions of the Desikan-killiany (DK) atlas were used, 16 subcortical from Freesurfer segmentation, and 21 Parkinson related nuclei. These nuclei included each hemisphere medullary reticular group, substantia nigra, subthalamic nucleus, lateral hypothalamic area, ventromedial hypothalamic nucleus, reticular nucleus, paraventricular nucleus, locus coeruleus, dorsal motor nucleus vagus and medial raphe nuclei, pontine raphe and midbrain raphe. Three processing stages were followed to derive projected transcriptome data to our 105 regions atlas: (i) expression values from multiple probes were mean averaged for each gene; (ii) each sample was mapped to a region of our atlas. All samples falling inside the cortical or subcortical regions of the atlas were assigned to the corresponding region and for samples belonging to each nuclei we used the classification provided by AHBA. The distance of the samples not assigned to any region was evaluated and the samples with a distance < 2.5 mm to any cortical or subcortical region was assigned to it; (iii) the average genetic expression across all samples mapped into a specific atlas region was computed for each individual brain. A group expression map was derived computing the median values between the 6 donors.
To investigate spatial similarities between regional gene expression profiles and brain areas involved in PD pathology, the mean combined SFC map of healthy subjects was converted from the voxel-level spatial resolution to a cortical atlas based on DK parcellation and subcortical FreeSurfer regions (using the average intensity) to allow for comparison with the AHBA data. Subsequently, we correlated SFC value of each cortical or subcortical region with the expression levels of genes of that region from AHBA using the Pearson’s correlation (MATLAB v8.0 software; Mathworks Inc.). As a data-driven strategy, we calculated the spatial similarity between the combined SFC map and gene expression level for each of the 20,737 genes (the entire transcriptome of AHBA data; histogram in Fig. 1-D). Then, as SFC spreading patterns display an inverse spatial relationship with potential PD-related pathological biomarkers (please see Fig. 1), we identified all genes for which expression levels presented negative spatial similarities with the SFC map (considering a significant cutoff of −1.96 standard deviations [SD] in the lower bound of the tail of the null hypothesis distribution; red area in Fig. 3-A). To test that the obtained spatial similarity of SFC map with the genes set in the lower tail are not due to spurious similarities due to spatial autocorrelation of the data (Markello and Misic, 2021), we computed the likelihood of obtaining this spatial similarity values in a randomly generated maps preserving the autocorrelation of the input map. BrainSMASH software (Burt et al., 2020) was used to generate 1000 surrogate maps and compute a distribution of random association between surrogate maps and 20,737 genes. Finally, the significance of the association of SFC map with SNCA expression was evaluated computing a p-value based on the likelihood of obtaining a higher association between SNCA expression and the generated 1000 random maps.
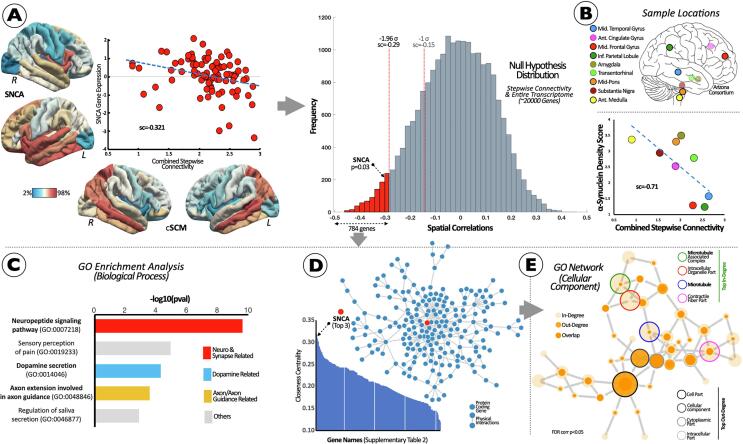
Fig. 3.
Brain Co-localization of In Vivo SFC Patterns and Allen Gene Expression Data to detect PD Vulnerable Pathways and Genetic/Pathological Signatures. (A) Scatterplot between in vivo spreading connectivity pattern (bottom cortical maps) and SNCA (left cortical maps). Null hypothesis distributions of similarity scores between in vivo SFC patterns and the entire protein-coding transcriptome. Similarity scores were converted to z-scores, corresponding to one-tailed p-values for the SNCA z-score. Red area shows similarity scores below −1.96σ. (B) Relationship between SFC and α-synuclein-immunoreactive density scores. (C) Interactome and Gene Ontology Analyses of the genetic set obtained in II (red bars in histogram). Interactome network showing significant Gene Ontology (GO) overrepresented functionality (FDR-corrected q < 0.05). (D) Protein-protein interactome network with closeness centrality, and (E) GO network for Cellular Components assessment. Abbreviations: Ant = anterior; cSCM = combined stepwise connectivity map; Inf = inferior; GO = gene ontology; L = left; Mid = middle; R = right; sc = spatial correlations; SFC = stepwise functional connectivity. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.2.4. Relationship between post-mortem neuropathological data and imaging metrics
We used the combined SFC maps and postmortem density scores of α-synuclein-immunoreactive Lewy bodies to gain knowledge concerning how the cortical spreading from PD-related epicenters related to abnormal cortical deposition of PD pathology. Density scores of α-synuclein-immunoreactive Lewy bodies were obtained postmortem from 186 PD volunteer donors from the Arizona Study of Aging and Neurodegenerative Disorders (AZSAND)/Banner Sun Health Research Institute Brain and Body Donation Program (BBDP; www.brainandbodydonationprogram.org). Particularly, α-synuclein-immunoreactive density scores were evaluated in the following brain regions: olfactory bulb and tract, anterior medulla (two levels anterior to the obex), anterior and mid-pons, mid-amygdala with adjacent trans-entorhinal area, anterior cingulate gyrus (1–3 cm posterior to the coronal slice containing the genu of the corpus callosum), middle temporal gyrus (at the level of the lateral geniculate nucleus), middle frontal gyrus (4–5 cm posterior to the frontal pole), and inferior parietal lobule (Beach et al., 2009). As we spatially compared these regions to functional connectivity-derived values, we excluded olfactory bulb and tract areas due to the low reliability of the BOLD signals in those areas. Subsequently, the α-synuclein-immunoreactive density scores of the eight leftover brain regions were averaged over all 186 PD subjects. Similarly as the previous section, the Pearson’s correlation between the combined SFC map and synucleinopathy density scores from postmortem brain tissue was used to evaluate the spatial similarity between corresponding regions (Fig. 1-C).
2.2.5. Gene Ontology and interactome analysis
Using the entire profile of AHBA genes with significant negative spatial similarity with the SFC map based on the full protein-coding transcriptome distribution, we performed a gene set enrichment analysis and Interactome analyses to gain insights about their genetic functionalities (Diez and Sepulcre, 2021). First, we investigated whether the data-driven genetic imaging profile displayed any specific biological process. To overcome false-positive bias in gene set enrichment of spatially resolved transcriptomics (Fulcher et al., 2021) we adapted over-representation analysis to evaluate the likelihood of obtaining each functional annotation by surrogate maps with same autocorrelation as the input map (Bueicheku et al., 2021). First, BrainSMASH software (Burt et al., 2020) was used to generate 1000 surrogate maps with matching spatial autocorrelation properties to SFC map. For each of the surrogate maps the list of genes in the tail was used to perform an overrepresentation analysis using a Fisher test. For each Biological process annotation, a p-value and fold enrichment were computed. Then the overrepresentation of the genes in the original lower tail was computed for Biological Process annotations. For each annotation a p-value was computed using the Fisher test and compared with the distribution of p-values obtained for the surrogate maps. A new p-value with the likelihood of the annotation appearing by chance was computed. A false discovery rate of the neuro related annotations was computed to correct for multiple comparisons with a q-level < 0.01. The neuro related annotations were defined as those annotations with at least 50% of their genes been classify as neuro genes (3,157 genes derived from Amigo). The resulting annotation were clustered into groups based on a similarity approach to reduce the information and help int the interpretability. The following steps were done for clustering the annotations: (a) a binary gene-term matrix was generated. Genes belonging to a particular term had a value of 1 and 0 otherwise; (b) a term-term kappa score matrix was generated. Each entry measured the similarity of 2 terms with kappa score based on observed occurrence of genes and was compared to chance occurrence; (c) hierarchical clustering was applied were terms with a kappa threshold > 0.3 were merged; (d) for each cluster the term with the most significant p value was used as the representative term. The enrichment analysis has been carried out for Biological Process terms from Gene Ontology (the reference genome and Gene Ontology annotations were retrieved from http://www.webgestalt.org/ on 01/14/2019). Additional analysis details were as follow: (a) genes that were not annotated as Biological Process were not used for the analyses; (b) terms with less than 5 or more than 2,000 genes were removed; (c) annotations with less than 3 genes overlapping with the candidate gene list or with less than an uncorrected p value of 0.01 (Fisher test) were discarded. The spatial similarity analysis was done by means of in-house Matlab code. We used Cytoscape (www.cytoscape.org (Lopes et al., 2010)) and the Genemania toolbox (http://www.genemania.org; default settings (Mostafavi et al., 2008)) to perform a network interactome analysis based on protein–protein interactions in order to identify key metabolic players within our AHBA-derived gene list. Using this interactome approach and node-level closeness centrality, we identified the most central interacting genes at the protein–protein level, as well as their particular role in cellular components (FDR-corrected at q-level < 0.05).
3. Results
3.1. Connectivity reorganization in PD subjects
In the early step maps of healthy controls, medulla displayed a direct functional connection to the midbrain, thalamus, insula, parahippocampus, superior temporal lateral occipital and lingual gyri (Fig. 1; please see Supplemental Fig. 1 for a complete left and right display, as well as alternative seeds in pons and midbrain). After the initial steps of connectivity, medullary pathways then reached the parietal lobe (particularly its superior and inferior parts and precuneus cortex), temporal lobe (superior temporal and fusiform gyri) and occipital lobe (including lateral occipital gyrus and lingual gyri and cuneus). At the final steps 3 and 4, other temporal regions (middle and inferior temporal and isthmus cingulate gyri) were related to the medullary connectivity pathways (Fig. 1; Supplemental Fig. 1). Medullary connectivity pathways were spatially similar in healthy controls and the two PD subgroups in all the four different step maps (Supplemental Fig. 2-A and 2-B). The correspondence of SFC maps across different link-step distances (from one to four) with PD pathological stages and clinical evolution according to the Braak system is reported in Supplemental Table 5.
Analogous results were obtained with the replication PD dataset: indeed, spatial similarity analysis among the SFC maps in the two PD cohorts (PD study sample and PD replication sample) revealed significant similarity starting from the three alternative seeds (medulla, pons and midbrain; Supplemental Fig. 1-D, 1-E and 1-F).
When comparing PD subgroups with healthy controls, we found significant differences in the stepwise connectivity of the medullary epicenter (Fig. 2). Mild PD patients presented lower connectivity in fusiform and lingual gyri compared to controls (Fig. 2-A). At intermediate link-step distances, mild PD patients showed a lower functional connectivity in sensorimotor cortex (precentral and postcentral gyri), parietal (superior and inferior parietal gyri and precuneus), temporal (superior, middle, and inferior temporal gyri) regions and cuneus (Fig. 2-A). Referring to the opposite contrast, mild PD patients exhibited enhanced connectivity across 2–4 link-steps from medulla seed to medial orbitofrontal, parahippocampal and lateral occipital gyri (Fig. 2-A). The analysis also revealed that at one-link step distance from medulla seed, moderate-to-severe PD patients showed decreased connectivity in the inferior parietal gyrus and occipital lobe (lateral occipital gyrus and cuneus) relative to controls (Fig. 2-B). Across 2–4 link-steps, reduced functional connectivity was found in sensorimotor cortex (precentral and postcentral gyri), frontal (superior frontal gyrus and caudal/rostral anterior cingulate), parietal (superior and inferior parietal gyri, posterior cingulate cortex and precuneus), temporal (superior and middle temporal gyri) regions and cuneus (Fig. 2-B). With regard to the opposite contrast, moderate-to-severe PD patients exhibited significantly increased functional connectivity at intermediate link-step distances in frontal (medial orbitofrontal gyri) and temporal (inferior temporal, parahippocampal, fusiform gyri and entorhinal cortex) regions (Fig. 2-B).
When comparing PD patient subgroups, at one-link step distance from the medulla seed, moderate-to-severe PD patients showed decreased connectivity only in the lateral occipital gyrus relative to mild PD cases (Fig. 2-C). Across 2–4 link-steps, moderate-to-severe PD patients were characterized by additional decreased functional connectivity within inferior parietal gyrus and insula relative to the mild PD group (Fig. 2-C). Furthermore, moderate-to-severe PD patients showed increased connectivity relative to mild PD cases only at intermediate link-step distances within sensorimotor cortex (precentral and postcentral gyri), caudal middle frontal gyrus, superior parietal gyrus, and temporal areas (superior, middle, and inferior temporal, parahippocampal gyri and entorhinal cortex).
3.2. Connectomic-Genetic relationships
The analysis of spatial similarity between SNCA gene expression levels and functional brain organization is reported in Fig. 3-A. Our analysis showed that SNCA gene expression levels are spatially -and inversely- related to the normal pattern of stepwise connectivity from medulla (Fig. 3-A). In the data-driven approach taking into account the entire human transcriptome as null hypothesis distribution, 784 genes (Supplemental Table 2) were similarly distributed along the cortical mantle as in the medulla stepwise connectivity maps in healthy controls (<-1.96 SD; see histogram in Fig. 3-A). These genes were significantly associated when testing with 1,000 surrogate maps with same autocorrelation as the SFC map. Importantly, SNCA gene expression was found in the lower bound of the tail of the overall distribution of similarities between the medulla connectivity map and the expression of 20,737 protein-coding genes (Fig. 3-A). The cortico-subcortical similarity analysis among the combined SFC map and postmortem α-synuclein-immunoreactive density scores from 186 PD brains revealed statistically significant similarities. In particular, synucleinopathy density in the aforementioned brain regions showed a notable negative correlation with the spreading connectivity map (Fig. 3-B), that is, the greater synucleinopathy density score the closer step or connectivity proximity to the medulla epicenter.
Using FDR-corrected GO annotation analysis (q-level < 0.05) targeting biological processes, we found several overrepresented gene sets engaged in the regulation of dopamine secretion, neuropeptide transmission, and axon guidance assemblies (Fig. 3-C). Moreover, we found that SNCA was a top central gene in the protein–protein interactome (top 3; Fig. 3-D; Supplemental Table 3). Finally, the main gene set recollecting the PD-related spreading pattern was predominantly involved in microtubule cellular components (Fig. 3-E).
4. Discussion
Recent evidence supports the hypothesis postulated by Braak, according to which pathogenic forms of α-synuclein protein in PD spread throughout the nervous system in a predefined pattern (Braak et al., 2003b, Braak et al., 2006b, Goedert et al., 2013). In this study, we identified a model of PD propagation, starting from a supposed epicenter of the disease (according to Braak staging), by using novel graph theory metrics (i.e., SFC analysis). The findings are strongly consistent with the staging of brain pathology in PD patients presumed by Braak et al (Braak et al., 2003b). SFC also revealed patterns of functional network reorganization for early- or late-stage PD patients. Moreover, AHBA data of gene expression levels supported us in the characterization of the α-synuclein propagation using functional connectivity patterns from the epicenter region in healthy controls. We found that the spatial topology of a rich set of genes devoted to the regulation of dopamine secretion, in which SNCA played a central role, recapitulated the identified pathways of connectivity emanating from the medullary epicenter. Collectively, our findings demonstrate a parsimonious approach for prediction of α-synuclein pathology along the disease trajectory using an SFC approach to map aberrations in streams of connectivity from a priori epicenters of disease.
With the medulla as a candidate epicenter region for PD in mind, we first found in healthy controls that direct functional connections from a medulla seed (one step-link distance) included functional connections to the insular cortex, pericalcarine, lingual and parahippocampal gyri. With subsequent steps in the connectivity network, the seed region was connected to parietal, occipital, temporal lobes and the isthmus cingulate gyrus. Considering those functional connections form ‘highways’ through which a disease-causing agent might spread, our findings are in line with the idea that pathological proteins propagate firstly in regions directly connected to the epicenter (one step-link distance) and then progress to areas at further link-step distances. These findings are strongly consistent with the ascending staging system of brain pathology in PD patients assumed by Braak (Braak et al., 2003a) (Supplemental Table 5). This observation is also supported by the randomization analysis. Indeed, the average maps of healthy controls obtained after network randomization (Supplemental Fig. 3) did not confirm the ascending staging system (Braak et al., 2003a). Importantly, the medullary connectivity patterns were also found to be spatially consistent in healthy and diseased populations as the average maps of mild and moderate-to-severe PD patients displayed similar pathways of healthy controls in all the four different step maps, albeit with altered functional connectivity values. The stepwise connectivity characterization in healthy controls lays a foundation for studying the emerging concept that misfolded proteins follow connectivity-based models of spreading, highlighting the essential role of connectivity in susceptible brain regions (Filippi et al., 2020a, Frost et al., 2009, Raj et al., 2012, Vogel et al., 2020, Zhou et al., 2012).
PD has been characterized by atypical brain network organization. Conventional strategies as well as more-sophisticated graph theory analyses have shown that PD patients are characterized by hypo- and hyper-connectivity patterns (Filippi et al., 2019). Our results highlight the potential for studying functional network reorganization at different stages of disease through SFC analysis: early- and late-stages of PD. In particular, SFC analysis showed different patterns of disruption among PD subgroups. For example, in mild PD patients, the involvement of direct and indirect connections from the medulla to the sensorimotor network (precentral and postcentral gyri), cuneus, lateral and medial parietal regions and lateral temporal gyri suggests an initial disruption of functional connectivity, in accordance with mild motor and clinical symptoms in addition to mild cognitive impairment (Filippi et al., 2019, Wolters et al., 2019). RS-fMRI techniques have been applied in recent studies in order to disentangle the role of sensorimotor network in PD patients (Agosta et al., 2014a, Baudrexel et al., 2011, Helmich et al., 2010, Kwak et al., 2010, Seibert et al., 2012). Considering the characteristic motor manifestations found in PD, dysfunction of somatomotor networks is not surprising and is even expected as confirmed in many studies (Campbell et al., 2015, Canu et al., 2015, Gorges et al., 2015, Göttlich et al., 2013, Guimaraes et al., 2016, Olde Dubbelink et al., 2014, Peraza et al., 2017, Tan et al., 2015).
At a one-link step distance, moderate-to-severe PD patients showed decreased connectivity in the lateral occipital and inferior parietal gyri, and cuneus. Across 2–4 link-steps, we detected extended reductions in functional connectivity within parietal (superior and inferior parietal gyrus and precuneus), temporal (superior and middle temporal gyrus), frontal (superior frontal gyrus and caudal/rostral anterior cingulate) and limbic (posterior cingulate) lobes. These findings are in support of non-motor symptom presentation in moderate-to-severe PD patients, including cognitive deficits and other non-motor manifestations (Agosta et al., 2014a, Baudrexel et al., 2011, Helmich et al., 2010, Kwak et al., 2010, Seibert et al., 2012). Our results are also consistent with previous studies highlighting that cognitive functions result from alterations in RS-fMRI networks (default mode, dorsal-attention, fronto-parietal, salience or associative visual networks) (Filippi et al., 2019).
Interestingly, SFC analysis highlighted a greater extent of damage in moderate-to-severe PD than in mild PD patients, suggesting that functional organization of brain networks worsens with progression of motor and non-motor symptom severity. Consistently with previous studies (Agosta et al., 2014b, de Schipper et al., 2018, Fang et al., 2017, Filippi et al., 2020a, Filippi et al., 2019, Tuovinen et al., 2018), both mild and moderate-to-severe PD patients exhibited significantly increased functional connectivity at intermediate link-step distances in frontal and temporal regions, with a greater extent of functional alterations in moderate-to-severe PD than in mild PD cases. The explanation of this finding is that increased connectivity may be a compensation or might also be caused by loss of cortical inhibitory influence.
Importantly, the SFC approach was able to provide information regarding not only the strength or number of connections, but also nodal properties of network distance of voxels relative to the rest of the voxels in the entire brain, providing essential information about putative flows across brain networks. Owing to this, the functional connectivity results found at different stages of PD patients resembles assumed PD pathology progression (Braak et al., 2006a), in such a way that it demonstrates greater impairment along the suggested propagation patterns of α-synuclein deposits in the human brain.
Understanding ‘if’ and ‘how’ genetic factors are related to the identified PD propagation pathways is challenging. In this study, we investigated whether genetic biomarkers confer functional vulnerability to PD-related pathology in brain regions directly and indirectly connected to the epicenter. Expression of the SNCA gene (encoding α-synuclein protein) in healthy controls was associated with the proximity of a region to the epicenter of pathology, likely shaping functional architectures of the human brain. In our study, we investigated gene expression profiles and functional connectomes as possible vulnerability factors, using a new stepwise connectivity technique that might improve the possibility to reveal vulnerable networks. In light of this, our findings support the idea that brain regions closer to the epicenter and with greater SNCA gene expression may have higher probability of accumulating pathological proteins before other brain regions do, and, thus, confer greater vulnerability to disease spread (Achard and Bullmore, 2007). These results are in line with findings demonstrating that SNCA has a dose–response relationship with PD severity, posited to arise from an increasing presence of pathological α-synuclein aggregates (Devine et al., 2011). Notably, while previous imaging studies evaluating functional connectivity at the whole-brain level have found weak associations between SNCA gene expression and loss of functional connectivity (Rittman et al., 2016), the present stochastic SFC method demonstrated greater spatial sensitivity for aberrant flow across streams of functional connectivity than whole-brain level assays.
While SNCA played a central role in regional propagation, it has been demonstrated that the coordination of multiple genes may relate to progression of PD pathology (Hibar et al., 2015). Indeed, here we further performed an exploratory analysis that identified 784 genes (including SNCA gene) from the 20,737 protein-coding genes of the human transcriptome. Importantly, these data-driven ascertained genes shared similar spatial distributions with the maps of medulla connectivity characterized in healthy controls. We also observed that these genes exhibited an overrepresented biological functionality in key domains for PD pathology, such as dopamine neurotransmission. Moreover, we observed that genes displaying a high spatial overlap with the PD-related spreading pattern were predominantly implicated in microtubule dynamics. Lewy bodies have been found to contain tubulin and neurofilaments, key elements of the neuronal cytoskeleton (Goldman et al., 1983). Neurons overexpressing α-synuclein show microtubule dysfunction, including decreased microtubule networks, impaired microtubule-dependent trafficking, Golgi fragmentation and neuritic degeneration (Lee et al., 2006). In addition, a multitude of other proteins and pathways of varying functional relationships with microtubules are suggested to be implicated in PD, including parkin, PINK1, and the Leucine-rich repeat kinase 2 (Pellegrini et al., 2017). These proteins interact with microtubules, either directly or via adaptor and motor proteins, modulating their assembly and stability. Their aberrant activity may thus ultimately result in disruption of cytoskeletal architecture and loss of functional localization of any protein or organelle whose transport is secondarily delayed/affected. Notably, stabilization of microtubules has emerged as a beneficial therapeutic strategy for PD (Ballatore et al., 2012).
Few years ago, knowledge on PD progression was limited to inferences about underlying pathology ascertained from histopathological examination (Beach et al., 2009). In this study, we investigated the relation between functional connectivity (SFC maps) and density scores of α-synuclein-immunoreactive Lewy bodies in PD subjects. In line with previous genetic findings, we demonstrate that regions closer to the epicenter present higher density of α-synuclein. The combination of Lewy-type synucleinopathy density on postmortem brain tissue information with modern neuroimaging tools bolster our hypothesis of a convergence between spatially stereotyped accumulation of protein aggregates and putative contributions from functional connectivity network organization.
The present study is not without limitations. The most important one is that a confirmed relationship between Braak staging, and the clinical evolution of PD patients has not been provided yet. The validity of Braak staging has been suggested by several studies demonstrating that between 51 and 83% of PD followed this pathological system (Beach et al., 2009, Dickson et al., 2010, Halliday et al., 2008, Halliday et al., 2006, Jellinger, 2009, Kingsbury et al., 2010, Muller et al., 2005). On the other hand, the relationship between neuropathological staging and clinical symptoms has been only partially confirmed by other (Braak et al., 2005, Jellinger, 2003, Mori et al., 2008, Muller et al., 2005). Secondly, the data was cross-sectional in nature, and by clustering participants into different stages of the disease (mild PD and moderate-to-severe PD) does not entirely represent the progression of the disease. Another limitation of this study is that we considered only gene expression profiles and functional connectome as possible vulnerability factors to a specific pathophysiological process. However, it has been demonstrated that different factors are able to confer regional vulnerability to pathological process in neurodegenerative disorders (Bischof et al., 2019, Raj et al., 2012, Raj et al., 2015, Warren et al., 2013). These models include protein strains, susceptible cell features, gene expression level, structural and functional connectome and activity-demands (Bischof et al., 2019). Finally, different approaches can be used to detect spatial spread of pathology in vivo possibly leading to different findings. A group of studies tested the hypothesis of Braak staging using standard structural and functional MRI techniques supporting or not the assumed spreading (Guimaraes et al., 2016, Kamagata et al., 2013, Li et al., 2017, Potgieser et al., 2014, Tinaz et al., 2011). On the other hand, recent studies analysed the utility of connectomics approach applied to structural and functional data to understand the temporo-spatial pattern of accumulation and spreading of misfolded proteins in neurodegenerative diseases (Yau et al., 2018, Zarkali et al., 2021, Zeighami et al., 2015). However, to date, there is no agreement about the best method to apply.
In conclusion, in this study we characterized in vivo large-scale propagation pathways of α-synuclein pathology and identified different patterns of functional connectivity disruptions in PD patients at different stages of the disease. Our results identified the genetic underpinnings of PD pathology, highlighting their contribution to focal neuronal vulnerability to progression of PD. Our results pave the way toward better accounting for the brain networks complexity and mechanisms underlying distinct spatial vulnerability to PD pathology, thus informing early diagnosis and future novel therapeutic strategies.
5. Disclosure statement
S.B. reports no disclosures. F.A. is Section Editor of NeuroImage: Clinical; received honoraria for consulting services and/or speaking activities from Philips, Novartis, Biogen Idec and Roche; and receives or has received research supports from the Italian Ministry of Health, AriSLA (Fondazione Italiana di Ricerca per la SLA), and the European Research Council. I.D. reports no disclosures. E.B. reports no disclosures. F.d.O.U. reports no disclosures. M.D-A. report honoraria for lectures, travel, and accommodation to attend scientific meetings from Alter, Bial, Merk, Novartis, Sanofi Genzyme, UCB, and Zambon. C.C-G. reports no disclosures. M.R-O. reports honoraria for lectures, travel and accommodation to attend scientific meetings from Boston Scientific and Bial. T.S. has received speaker honoraria from Actavis and Alzheimer's Association International Research Grant. V.S.K. has received speaker honoraria from Roche and Alkaloid and receives research supports from the Swiss Pharm and Serbian Ministry of Education, Science, and Development and Serbian Academy of Sciences and Art. M.F. is Editor-in-Chief of the Journal of Neurology and Associate Editor of Human Brain Mapping; received compensation for consulting services and/or speaking activities from Almiral, Alexion, Bayer, Biogen, Celgene, Eli Lilly, Genzyme, Merck-Serono, Novartis, Roche, Sanofi, Takeda, and Teva Pharmaceutical Industries; and receives research support from Biogen Idec, Merck-Serono, Novartis, Roche, Teva Pharmaceutical Industries, Italian Ministry of Health, Fondazione Italiana Sclerosi Multipla, and ARiSLA (Fondazione Italiana di Ricerca per la SLA). J.S. reports no disclosures.
6. Data availability
The codes that support the findings of this study are openly available in GitHub public repository at https://doi.org/10.5281/zenodo.4672562.
CRediT authorship contribution statement
Silvia Basaia: Conceptualization, Formal analysis, Data curation, Writing – original draft, Writing – review & editing. Federica Agosta: Conceptualization, Data curation, Funding acquisition, Writing – original draft, Writing – review & editing. Ibai Diez: Conceptualization, Formal analysis, Data curation, Writing – review & editing. Elisenda Bueichekú: Conceptualization, Formal analysis, Data curation, Writing – review & editing. Federico d'Oleire Uquillas: Conceptualization, Data curation, Writing – review & editing. Manuel Delgado-Alvarado: Conceptualization, Data curation, Writing – review & editing. César Caballero-Gaudes: Conceptualization, Data curation, Writing – review & editing. MariCruz Rodriguez-Oroz: Conceptualization, Data curation, Writing – review & editing. Tanja Stojkovic: Conceptualization, Data curation, Writing – review & editing. Vladimir S. Kostic: Conceptualization, Formal analysis, Data curation, Writing – review & editing. Massimo Filippi: Conceptualization, Data curation, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. Jorge Sepulcre: Conceptualization, Formal analysis, Data curation, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors thank the patients and their families for the time and effort they dedicated to the research. Funding This research was supported by grants from the National Institutes of Health (NIH; R01AG061811, and R01AG061445 to J.S.), the Ministry of Education, Science, and Technological Development of the Republic of Serbia (grant number 175090), the Italian Ministry of Health (grant number RF-2018-12366746), the Carlos III Institute of Health (PI11/02109) Spain, the Basque Government through the BERC 2018-2021 program and the Spanish State Research Agency through BCBL Severo Ochoa excellence accreditation SEV-2015-0490.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2022.102941.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Achard S., Bullmore E. Efficiency and cost of economical brain functional networks. PLoS Comput. Biol. 2007;3(2):e17. doi: 10.1371/journal.pcbi.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosta F., Canu E., Inuggi A., Chio A., Riva N., Silani V., Calvo A., Messina S., Falini A., Comi G., Filippi M. Resting state functional connectivity alterations in primary lateral sclerosis. Neurobiol. Aging. 2014;35(4):916–925. doi: 10.1016/j.neurobiolaging.2013.09.041. [DOI] [PubMed] [Google Scholar]
- Agosta F., Caso F., Stankovic I., Inuggi A., Petrovic I., Svetel M., Kostic V.S., Filippi M. Cortico-striatal-thalamic network functional connectivity in hemiparkinsonism. Neurobiol. Aging. 2014;35(11):2592–2602. doi: 10.1016/j.neurobiolaging.2014.05.032. [DOI] [PubMed] [Google Scholar]
- Ballatore C., Brunden K.R., Huryn D.M., Trojanowski J.Q., Lee V.M., Smith A.B., 3rd Microtubule stabilizing agents as potential treatment for Alzheimer's disease and related neurodegenerative tauopathies. J. Med. Chem. 2012;55(21):8979–8996. doi: 10.1021/jm301079z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudrexel S., Witte T., Seifried C., von Wegner F., Beissner F., Klein J.C., Steinmetz H., Deichmann R., Roeper J., Hilker R. Resting state fMRI reveals increased subthalamic nucleus-motor cortex connectivity in Parkinson's disease. Neuroimage. 2011;55(4):1728–1738. doi: 10.1016/j.neuroimage.2011.01.017. [DOI] [PubMed] [Google Scholar]
- Beach T.G., Adler C.H., Lue L., Sue L.I., Bachalakuri J., Henry-Watson J., Sasse J., Boyer S., Shirohi S., Brooks R., Eschbacher J., White C.L., 3rd, Akiyama H., Caviness J., Shill H.A., Connor D.J., Sabbagh M.N., Walker D.G., Arizona Parkinson's Disease, C. Unified staging system for Lewy body disorders: correlation with nigrostriatal degeneration, cognitive impairment and motor dysfunction. Acta Neuropathol. 2009;117:613–634. doi: 10.1007/s00401-009-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof G.N., Ewers M., Franzmeier N., Grothe M.J., Hoenig M., Kocagoncu E., Neitzel J., Rowe J.B., Strafella A., Drzezga A., van Eimeren T., faculty, M., Connectomics and molecular imaging in neurodegeneration. Eur. J. Nucl. Med. Mol. Imaging. 2019;46:2819–2830. doi: 10.1007/s00259-019-04394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H., Bohl J.R., Muller C.M., Rub U., de Vos R.A., Del Tredici K. Stanley Fahn Lecture 2005: The staging procedure for the inclusion body pathology associated with sporadic Parkinson's disease reconsidered. Mov. Disord. 2006;21(12):2042–2051. doi: 10.1002/mds.21065. [DOI] [PubMed] [Google Scholar]
- Braak H., Tredici K.D., Rüb U., de Vos R.A., Jansen Steur E.N., Braak E. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol. Aging. 2003;24(2):197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- Braak H., Rb U., Gai W.P., Del Tredici K. Idiopathic Parkinson's disease: possible routes by which vulnerable neuronal types may be subject to neuroinvasion by an unknown pathogen. J. Neural Transm. (Vienna) 2003;110(5):517–536. doi: 10.1007/s00702-002-0808-2. [DOI] [PubMed] [Google Scholar]
- Braak H., Rub U., Jansen Steur E.N.H., Del Tredici K., de Vos R.A.I. Cognitive status correlates with neuropathologic stage in Parkinson disease. Neurology. 2005;64(8):1404–1410. doi: 10.1212/01.WNL.0000158422.41380.82. [DOI] [PubMed] [Google Scholar]
- Braak H., Rub U., Schultz C., Del Tredici K. Vulnerability of cortical neurons to Alzheimer's and Parkinson's diseases. J. Alzheimers Dis. 2006;9(s3):35–44. doi: 10.3233/jad-2006-9s305. [DOI] [PubMed] [Google Scholar]
- Bueicheku E., Aznarez-Sanado M., Diez I., d'Oleire Uquillas F., Ortiz-Teran L., Qureshi A.Y., Sunol M., Basaia S., Ortiz-Teran E., Pastor M.A., Sepulcre J. Central neurogenetic signatures of the visuomotor integration system. Proc. Natl. Acad. Sci. U. S. A. 2020;117(12):6836–6843. doi: 10.1073/pnas.1912429117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueicheku E., Gonzalez-de-Echavarri J.M., Ortiz-Teran L., Montal V., d'Oleire Uquillas F., De Marcos L., Orwig W., Kim C.M., Ortiz-Teran E., Basaia S., Diez I., Sepulcre J. Divergent connectomic organization delineates genetic evolutionary traits in the human brain. Sci. Rep. 2021;11:19692. doi: 10.1038/s41598-021-99082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt J.B., Helmer M., Shinn M., Anticevic A., Murray J.D. Generative modeling of brain maps with spatial autocorrelation. Neuroimage. 2020;220:117038. doi: 10.1016/j.neuroimage.2020.117038. [DOI] [PubMed] [Google Scholar]
- Campbell M.C., Koller J.M., Snyder A.Z., Buddhala C., Kotzbauer P.T., Perlmutter J.S. CSF proteins and resting-state functional connectivity in Parkinson disease. Neurology. 2015;84(24):2413–2421. doi: 10.1212/WNL.0000000000001681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canu E., Agosta F., Sarasso E., Volonte M.A., Basaia S., Stojkovic T., Stefanova E., Comi G., Falini A., Kostic V.S., Gatti R., Filippi M. Brain structural and functional connectivity in Parkinson's disease with freezing of gait. Hum. Brain Mapp. 2015;36:5064–5078. doi: 10.1002/hbm.22994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costumero V., d'Oleire Uquillas F., Diez I., Andorra M., Basaia S., Bueicheku E., Ortiz-Teran L., Belloch V., Escudero J., Avila C., Sepulcre J. Distance disintegration delineates the brain connectivity failure of Alzheimer's disease. Neurobiol. Aging. 2020;88:51–60. doi: 10.1016/j.neurobiolaging.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley N.A., Mechelli A., Scott J., Carletti F., Fox P.T., McGuire P., Bullmore E.T. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 2014;137(8):2382–2395. doi: 10.1093/brain/awu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Schipper L.J., Hafkemeijer A., van der Grond J., Marinus J., Henselmans J.M.L., van Hilten J.J. Altered Whole-Brain and Network-Based Functional Connectivity in Parkinson's Disease. Front. Neurol. 2018;9:419. doi: 10.3389/fneur.2018.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine M.J., Gwinn K., Singleton A., Hardy J. Parkinson's disease and alpha-synuclein expression. Mov. Disord. 2011;26:2160–2168. doi: 10.1002/mds.23948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson D.W., Uchikado H., Fujishiro H., Tsuboi Y. Evidence in favor of Braak staging of Parkinson's disease. Mov. Disord. 2010;25(Suppl 1):S78–82. doi: 10.1002/mds.22637. [DOI] [PubMed] [Google Scholar]
- Diez I., Larson A.G., Nakhate V., Dunn E.C., Fricchione G.L., Nicholson T.R., Sepulcre J., Perez D.L. Early-life trauma endophenotypes and brain circuit-gene expression relationships in functional neurological (conversion) disorder. Mol. Psychiatry. 2021;26(8):3817–3828. doi: 10.1038/s41380-020-0665-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez I., Sepulcre J. Unveiling the neuroimaging-genetic intersections in the human brain. Curr. Opin. Neurol. 2021;34:480–487. doi: 10.1097/WCO.0000000000000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Chen H., Cao Z., Jiang Y., Ma L., Ma H., Feng T. Impaired brain network architecture in newly diagnosed Parkinson's disease based on graph theoretical analysis. Neurosci. Lett. 2017;657:151–158. doi: 10.1016/j.neulet.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Filippi M., Basaia S., Canu E., Imperiale F., Magnani G., Falautano M., Comi G., Falini A., Agosta F. Changes in functional and structural brain connectome along the Alzheimer's disease continuum. Mol. Psychiatry. 2020;25(1):230–239. doi: 10.1038/s41380-018-0067-8. [DOI] [PubMed] [Google Scholar]
- Filippi M., Basaia S., Sarasso E., Stojkovic T., Stankovic I., Fontana A., Tomic A., Piramide N., Stefanova E., Markovic V., Kostic V.S., Agosta F. Longitudinal brain connectivity changes and clinical evolution in Parkinson's disease. Mol. Psychiatry. 2020 doi: 10.1038/s41380-020-0770-0. [DOI] [PubMed] [Google Scholar]
- Filippi M., Sarasso E., Agosta F. Resting-state functional MRI in Parkinsonian syndromes. Mov Disord Clin Pract. 2019;6(2):104–117. doi: 10.1002/mdc3.12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi M., Sarasso E., Piramide N., Stojkovic T., Stankovic I., Basaia S., Fontana A., Tomic A., Markovic V., Stefanova E., Kostic V.S., Agosta F. Progressive brain atrophy and clinical evolution in Parkinson's disease. Neuroimage Clin. 2020;28:102374. doi: 10.1016/j.nicl.2020.102374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A., Zalesky A., Breakspear M. The connectomics of brain disorders. Nat. Rev. Neurosci. 2015;16(3):159–172. doi: 10.1038/nrn3901. [DOI] [PubMed] [Google Scholar]
- Freeze B., Acosta D., Pandya S., Zhao Y., Raj A. Regional expression of genes mediating trans-synaptic alpha-synuclein transfer predicts regional atrophy in Parkinson disease. Neuroimage Clin. 2018;18:456–466. doi: 10.1016/j.nicl.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B., Ollesch J., Wille H., Diamond M.I. Conformational diversity of wild-type Tau fibrils specified by templated conformation change. J. Biol. Chem. 2009;284(6):3546–3551. doi: 10.1074/jbc.M805627200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher B.D., Arnatkeviciute A., Fornito A. Overcoming false-positive gene-category enrichment in the analysis of spatially resolved transcriptomic brain atlas data. Nat. Commun. 2021;12:2669. doi: 10.1038/s41467-021-22862-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Yu Y., Su X., Tao Z., Zhang M., Wang Y., Leng J., Sepulcre J., Chen H. Adaptation of brain functional stream architecture in athletes with fast demands of sensorimotor integration. Hum. Brain Mapp. 2019;40(2):420–431. doi: 10.1002/hbm.24382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M., Masuda-Suzukake M., Falcon B. Like prions: the propagation of aggregated tau and alpha-synuclein in neurodegeneration. Brain. 2017;140:266–278. doi: 10.1093/brain/aww230. [DOI] [PubMed] [Google Scholar]
- Goedert M., Spillantini M.G., Del Tredici K., Braak H. 100 years of Lewy pathology. Nat. Rev. Neurol. 2013;9(1):13–24. doi: 10.1038/nrneurol.2012.242. [DOI] [PubMed] [Google Scholar]
- Goldman J.E., Yen S.H., Chiu F.C., Peress N.S. Lewy bodies of Parkinson's disease contain neurofilament antigens. Science. 1983;221(4615):1082–1084. doi: 10.1126/science.6308771. [DOI] [PubMed] [Google Scholar]
- Gorges M., Muller H.P., Lule D., Consortium L., Pinkhardt E.H., Ludolph A.C., Kassubek J. To rise and to fall: functional connectivity in cognitively normal and cognitively impaired patients with Parkinson's disease. Neurobiol. Aging. 2015;36(4):1727–1735. doi: 10.1016/j.neurobiolaging.2014.12.026. [DOI] [PubMed] [Google Scholar]
- Gottlich M., Munte T.F., Heldmann M., Kasten M., Hagenah J., Kramer U.M. Altered resting state brain networks in Parkinson's disease. PLoS ONE. 2013;8(10):e77336. doi: 10.1371/journal.pone.0077336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius M. Resting-state functional connectivity in neuropsychiatric disorders. Curr. Opin. Neurol. 2008;21:424–430. doi: 10.1097/WCO.0b013e328306f2c5. [DOI] [PubMed] [Google Scholar]
- Guimaraes R.P., Arci Santos M.C., Dagher A., Campos L.S., Azevedo P., Piovesana L.G., De Campos B.M., Larcher K., Zeighami Y., Scarparo Amato-Filho A.C., Cendes F., D'Abreu A.C. Pattern of reduced functional connectivity and structural abnormalities in Parkinson's disease: an exploratory study. Front. Neurol. 2016;7:243. doi: 10.3389/fneur.2016.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday G., Hely M., Reid W., Morris J. The progression of pathology in longitudinally followed patients with Parkinson's disease. Acta Neuropathol. 2008;115(4):409–415. doi: 10.1007/s00401-008-0344-8. [DOI] [PubMed] [Google Scholar]
- Halliday G.M., Del Tredici K., Braak H. Critical appraisal of brain pathology staging related to presymptomatic and symptomatic cases of sporadic Parkinson's disease. J. Neural Transm. Suppl. 2006:99–103. doi: 10.1007/978-3-211-45295-0_16. [DOI] [PubMed] [Google Scholar]
- Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L., Abajian C., Beckmann C.F., Bernard A., Bertagnolli D., Boe A.F., Cartagena P.M., Chakravarty M.M., Chapin M., Chong J., Dalley R.A., David Daly B., Dang C., Datta S., Dee N., Dolbeare T.A., Faber V., Feng D., Fowler D.R., Goldy J., Gregor B.W., Haradon Z., Haynor D.R., Hohmann J.G., Horvath S., Howard R.E., Jeromin A., Jochim J.M., Kinnunen M., Lau C., Lazarz E.T., Lee C., Lemon T.A., Li L., Li Y., Morris J.A., Overly C.C., Parker P.D., Parry S.E., Reding M., Royall J.J., Schulkin J., Sequeira P.A., Slaughterbeck C.R., Smith S.C., Sodt A.J., Sunkin S.M., Swanson B.E., Vawter M.P., Williams D., Wohnoutka P., Zielke H.R., Geschwind D.H., Hof P.R., Smith S.M., Koch C., Grant S.G.N., Jones A.R. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489(7416):391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmich R.C., Derikx L.C., Bakker M., Scheeringa R., Bloem B.R., Toni I. Spatial remapping of cortico-striatal connectivity in Parkinson's disease. Cereb. Cortex. 2010;20(5):1175–1186. doi: 10.1093/cercor/bhp178. [DOI] [PubMed] [Google Scholar]
- Hibar, D.P., Stein, J.L., Renteria, M.E., Arias-Vasquez, A., Desrivieres, S., Jahanshad, N., Toro, R., Wittfeld, K., Abramovic, L., Andersson, M., Aribisala, B.S., Armstrong, N.J., Bernard, M., Bohlken, M.M., Boks, M.P., Bralten, J., Brown, A.A., Chakravarty, M.M., Chen, Q., Ching, C.R., Cuellar-Partida, G., den Braber, A., Giddaluru, S., Goldman, A.L., Grimm, O., Guadalupe, T., Hass, J., Woldehawariat, G., Holmes, A.J., Hoogman, M., Janowitz, D., Jia, T., Kim, S., Klein, M., Kraemer, B., Lee, P.H., Olde Loohuis, L.M., Luciano, M., Macare, C., Mather, K.A., Mattheisen, M., Milaneschi, Y., Nho, K., Papmeyer, M., Ramasamy, A., Risacher, S.L., Roiz-Santianez, R., Rose, E.J., Salami, A., Samann, P.G., Schmaal, L., Schork, A.J., Shin, J., Strike, L.T., Teumer, A., van Donkelaar, M.M., van Eijk, K.R., Walters, R.K., Westlye, L.T., Whelan, C.D., Winkler, A.M., Zwiers, M.P., Alhusaini, S., Athanasiu, L., Ehrlich, S., Hakobjan, M.M., Hartberg, C.B., Haukvik, U.K., Heister, A.J., Hoehn, D., Kasperaviciute, D., Liewald, D.C., Lopez, L.M., Makkinje, R.R., Matarin, M., Naber, M.A., McKay, D.R., Needham, M., Nugent, A.C., Putz, B., Royle, N.A., Shen, L., Sprooten, E., Trabzuni, D., van der Marel, S.S., van Hulzen, K.J., Walton, E., Wolf, C., Almasy, L., Ames, D., Arepalli, S., Assareh, A.A., Bastin, M.E., Brodaty, H., Bulayeva, K.B., Carless, M.A., Cichon, S., Corvin, A., Curran, J.E., Czisch, M., de Zubicaray, G.I., Dillman, A., Duggirala, R., Dyer, T.D., Erk, S., Fedko, I.O., Ferrucci, L., Foroud, T.M., Fox, P.T., Fukunaga, M., Gibbs, J.R., Goring, H.H., Green, R.C., Guelfi, S., Hansell, N.K., Hartman, C.A., Hegenscheid, K., Heinz, A., Hernandez, D.G., Heslenfeld, D.J., Hoekstra, P.J., Holsboer, F., Homuth, G., Hottenga, J.J., Ikeda, M., Jack, C.R., Jr., Jenkinson, M., Johnson, R., Kanai, R., Keil, M., Kent, J.W., Jr., Kochunov, P., Kwok, J.B., Lawrie, S.M., Liu, X., Longo, D.L., McMahon, K.L., Meisenzahl, E., Melle, I., Mohnke, S., Montgomery, G.W., Mostert, J.C., Muhleisen, T.W., Nalls, M.A., Nichols, T.E., Nilsson, L.G., Nothen, M.M., Ohi, K., Olvera, R.L., Perez-Iglesias, R., Pike, G.B., Potkin, S.G., Reinvang, I., Reppermund, S., Rietschel, M., Romanczuk-Seiferth, N., Rosen, G.D., Rujescu, D., Schnell, K., Schofield, P.R., Smith, C., Steen, V.M., Sussmann, J.E., Thalamuthu, A., Toga, A.W., Traynor, B.J., Troncoso, J., Turner, J.A., Valdes Hernandez, M.C., van 't Ent, D., van der Brug, M., van der Wee, N.J., van Tol, M.J., Veltman, D.J., Wassink, T.H., Westman, E., Zielke, R.H., Zonderman, A.B., Ashbrook, D.G., Hager, R., Lu, L., McMahon, F.J., Morris, D.W., Williams, R.W., Brunner, H.G., Buckner, R.L., Buitelaar, J.K., Cahn, W., Calhoun, V.D., Cavalleri, G.L., Crespo-Facorro, B., Dale, A.M., Davies, G.E., Delanty, N., Depondt, C., Djurovic, S., Drevets, W.C., Espeseth, T., Gollub, R.L., Ho, B.C., Hoffmann, W., Hosten, N., Kahn, R.S., Le Hellard, S., Meyer-Lindenberg, A., Muller-Myhsok, B., Nauck, M., Nyberg, L., Pandolfo, M., Penninx, B.W., Roffman, J.L., Sisodiya, S.M., Smoller, J.W., van Bokhoven, H., van Haren, N.E., Volzke, H., Walter, H., Weiner, M.W., Wen, W., White, T., Agartz, I., Andreassen, O.A., Blangero, J., Boomsma, D.I., Brouwer, R.M., Cannon, D.M., Cookson, M.R., de Geus, E.J., Deary, I.J., Donohoe, G., Fernandez, G., Fisher, S.E., Francks, C., Glahn, D.C., Grabe, H.J., Gruber, O., Hardy, J., Hashimoto, R., Hulshoff Pol, H.E., Jonsson, E.G., Kloszewska, I., Lovestone, S., Mattay, V.S., Mecocci, P., McDonald, C., McIntosh, A.M., Ophoff, R.A., Paus, T., Pausova, Z., Ryten, M., Sachdev, P.S., Saykin, A.J., Simmons, A., Singleton, A., Soininen, H., Wardlaw, J.M., Weale, M.E., Weinberger, D.R., Adams, H.H., Launer, L.J., Seiler, S., Schmidt, R., Chauhan, G., Satizabal, C.L., Becker, J.T., Yanek, L., van der Lee, S.J., Ebling, M., Fischl, B., Longstreth, W.T., Jr., Greve, D., Schmidt, H., Nyquist, P., Vinke, L.N., van Duijn, C.M., Xue, L., Mazoyer, B., Bis, J.C., Gudnason, V., Seshadri, S., Ikram, M.A., Alzheimer's Disease Neuroimaging, I., Consortium, C., Epigen, Imagen, Sys, Martin, N.G., Wright, M.J., Schumann, G., Franke, B., Thompson, P.M., Medland, S.E., 2015. Common genetic variants influence human subcortical brain structures. Nature 520, 224-229. [DOI] [PMC free article] [PubMed]
- Jellinger K.A. Alpha-synuclein pathology in Parkinson's and Alzheimer's disease brain: incidence and topographic distribution–a pilot study. Acta Neuropathol. 2003;106:191–201. doi: 10.1007/s00401-003-0725-y. [DOI] [PubMed] [Google Scholar]
- Jellinger K.A. A critical evaluation of current staging of alpha-synuclein pathology in Lewy body disorders. BBA. 2009;1792:730–740. doi: 10.1016/j.bbadis.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Jones Allan R., Overly Caroline C., Sunkin Susan M. The Allen Brain Atlas: 5 years and beyond. Nat. Rev. Neurosci. 2009;10(11):821–828. doi: 10.1038/nrn2722. [DOI] [PubMed] [Google Scholar]
- Kalia Lorraine V, Lang Anthony E. Parkinson's disease. Lancet. 2015;386(9996):896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]
- Kamagata K., Tomiyama H., Motoi Y., Kano M., Abe O., Ito K., Shimoji K., Suzuki M., Hori M., Nakanishi A., Kuwatsuru R., Sasai K., Aoki S., Hattori N. Diffusional kurtosis imaging of cingulate fibers in Parkinson disease: comparison with conventional diffusion tensor imaging. Magn. Reson. Imaging. 2013;31:1501–1506. doi: 10.1016/j.mri.2013.06.009. [DOI] [PubMed] [Google Scholar]
- Kingsbury A.E., Bandopadhyay R., Silveira-Moriyama L., Ayling H., Kallis C., Sterlacci W., Maeir H., Poewe W., Lees A.J. Brain stem pathology in Parkinson's disease: an evaluation of the Braak staging model. Mov. Disord. 2010;25:2508–2515. doi: 10.1002/mds.23305. [DOI] [PubMed] [Google Scholar]
- Kwak Y., Peltier S., Bohnen N.I., Muller M.L., Dayalu P., Seidler R.D. Altered resting state cortico-striatal connectivity in mild to moderate stage Parkinson's disease. Front. Syst. Neurosci. 2010;4:143. doi: 10.3389/fnsys.2010.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.J., Khoshaghideh F., Lee S., Lee S.J. Impairment of microtubule-dependent trafficking by overexpression of alpha-synuclein. Eur. J. Neurosci. 2006;24:3153–3162. doi: 10.1111/j.1460-9568.2006.05210.x. [DOI] [PubMed] [Google Scholar]
- Li X., Xing Y., Schwarz S.T., Auer D.P. Limbic grey matter changes in early Parkinson's disease. Hum. Brain Mapp. 2017;38:3566–3578. doi: 10.1002/hbm.23610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes C.T., Franz M., Kazi F., Donaldson S.L., Morris Q., Bader G.D. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markello R.D., Misic B. Comparing spatial null models for brain maps. Neuroimage. 2021;236 doi: 10.1016/j.neuroimage.2021.118052. [DOI] [PubMed] [Google Scholar]
- Mori F., Tanji K., Zhang H., Kakita A., Takahashi H., Wakabayashi K. alpha-Synuclein pathology in the neostriatum in Parkinson's disease. Acta Neuropathol. 2008;115:453–459. doi: 10.1007/s00401-007-0316-4. [DOI] [PubMed] [Google Scholar]
- Mostafavi S., Ray D., Warde-Farley D., Grouios C., Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C.M., de Vos R.A., Maurage C.A., Thal D.R., Tolnay M., Braak H. Staging of sporadic Parkinson disease-related alpha-synuclein pathology: inter- and intra-rater reliability. J. Neuropathol. Exp. Neurol. 2005;64:623–628. doi: 10.1097/01.jnen.0000171652.40083.15. [DOI] [PubMed] [Google Scholar]
- Olde Dubbelink K.T., Schoonheim M.M., Deijen J.B., Twisk J.W., Barkhof F., Berendse H.W. Functional connectivity and cognitive decline over 3 years in Parkinson disease. Neurology. 2014;83:2046–2053. doi: 10.1212/WNL.0000000000001020. [DOI] [PubMed] [Google Scholar]
- Ortiz-Teran L., Diez I., Ortiz T., Perez D.L., Aragon J.I., Costumero V., Pascual-Leone A., El Fakhri G., Sepulcre J. Brain circuit-gene expression relationships and neuroplasticity of multisensory cortices in blind children. Proc Natl Acad Sci U S A. 2017;114:6830–6835. doi: 10.1073/pnas.1619121114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L., Wetzel A., Granno S., Heaton G., Harvey K. Back to the tubule: microtubule dynamics in Parkinson's disease. Cell. Mol. Life Sci. 2017;74:409–434. doi: 10.1007/s00018-016-2351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peraza L.R., Nesbitt D., Lawson R.A., Duncan G.W., Yarnall A.J., Khoo T.K., Kaiser M., Firbank M.J., O'Brien J.T., Barker R.A., Brooks D.J., Burn D.J., Taylor J.P. Intra- and inter-network functional alterations in Parkinson's disease with mild cognitive impairment. Hum. Brain Mapp. 2017;38:1702–1715. doi: 10.1002/hbm.23499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poewe W., Seppi K., Tanner C.M., Halliday G.M., Brundin P., Volkmann J., Schrag A.E., Lang A.E. Parkinson disease. Nat. Rev. Dis. Primers. 2017;3:17013. doi: 10.1038/nrdp.2017.13. [DOI] [PubMed] [Google Scholar]
- Potgieser A.R., van der Hoorn A., Meppelink A.M., Teune L.K., Koerts J., de Jong B.M. Anterior temporal atrophy and posterior progression in patients with Parkinson's disease. Neurodegener. Dis. 2014;14:125–132. doi: 10.1159/000363245. [DOI] [PubMed] [Google Scholar]
- Qian J., Diez I., Ortiz-Teran L., Bonadio C., Liddell T., Goni J., Sepulcre J. Positive connectivity predicts the dynamic intrinsic topology of the human brain network. Front. Syst. Neurosci. 2018;12:38. doi: 10.3389/fnsys.2018.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., Kuceyeski A., Weiner M. A network diffusion model of disease progression in dementia. Neuron. 2012;73:1204–1215. doi: 10.1016/j.neuron.2011.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., LoCastro E., Kuceyeski A., Tosun D., Relkin N., Weiner M., Alzheimer's Disease Neuroimaging, I. Network diffusion model of progression predicts longitudinal patterns of atrophy and metabolism in Alzheimer's disease. Cell Rep. 2015;10:359–369. doi: 10.1016/j.celrep.2014.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittman T., Rubinov M., Vertes P.E., Patel A.X., Ginestet C.E., Ghosh B.C.P., Barker R.A., Spillantini M.G., Bullmore E.T., Rowe J.B. Regional expression of the MAPT gene is associated with loss of hubs in brain networks and cognitive impairment in Parkinson disease and progressive supranuclear palsy. Neurobiol. Aging. 2016;48:153–160. doi: 10.1016/j.neurobiolaging.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M., Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Seibert T.M., Murphy E.A., Kaestner E.J., Brewer J.B. Interregional correlations in Parkinson disease and Parkinson-related dementia with resting functional MR imaging. Radiology. 2012;263:226–234. doi: 10.1148/radiol.12111280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre J., Grothe M.J., d'Oleire Uquillas F., Ortiz-Teran L., Diez I., Yang H.S., Jacobs H.I.L., Hanseeuw B.J., Li Q., El-Fakhri G., Sperling R.A., Johnson K.A. Neurogenetic contributions to amyloid beta and tau spreading in the human cortex. Nat. Med. 2018;24:1910–1918. doi: 10.1038/s41591-018-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre J., Sabuncu M.R., Yeo T.B., Liu H., Johnson K.A. Stepwise connectivity of the modal cortex reveals the multimodal organization of the human brain. J. Neurosci. 2012;32:10649–10661. doi: 10.1523/JNEUROSCI.0759-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen E.H., Overly C.C., Jones A.R. The Allen Human Brain Atlas: comprehensive gene expression mapping of the human brain. Trends Neurosci. 2012;35:711–714. doi: 10.1016/j.tins.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Smith S.M., Nichols T.E. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage. 2009;44:83–98. doi: 10.1016/j.neuroimage.2008.03.061. [DOI] [PubMed] [Google Scholar]
- Soto C., Pritzkow S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 2018;21:1332–1340. doi: 10.1038/s41593-018-0235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Tan J., Luo C., Cui W., He H., Bin Y., Deng J., Tan R., Tan W., Liu T., Zeng N., Xiao R., Yao D., Wang X. Altered brain activation in early drug-naive Parkinson's disease during heat pain stimuli: an fMRI study. Parkinsons Dis. 2015;2015 doi: 10.1155/2015/273019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinaz S., Courtney M.G., Stern C.E. Focal cortical and subcortical atrophy in early Parkinson's disease. Mov. Disord. 2011;26:436–441. doi: 10.1002/mds.23453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuovinen N., Seppi K., de Pasquale F., Muller C., Nocker M., Schocke M., Gizewski E.R., Kremser C., Wenning G.K., Poewe W., Djamshidian A., Scherfler C., Seki M. The reorganization of functional architecture in the early-stages of Parkinson's disease. Parkinsonism Relat Disord. 2018;50:61–68. doi: 10.1016/j.parkreldis.2018.02.013. [DOI] [PubMed] [Google Scholar]
- Van Essen D.C., Dierker D.L. Surface-based and probabilistic atlases of primate cerebral cortex. Neuron. 2007;56:209–225. doi: 10.1016/j.neuron.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Vogel J.W., Iturria-Medina Y., Strandberg O.T., Smith R., Levitis E., Evans A.C., Hansson O., Alzheimer's Disease Neuroimaging, I., Swedish BioFinder, S., Spread of pathological tau proteins through communicating neurons in human Alzheimer's disease. Nat. Commun. 2020;11:2612. doi: 10.1038/s41467-020-15701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren J.D., Rohrer J.D., Schott J.M., Fox N.C., Hardy J., Rossor M.N. Molecular nexopathies: a new paradigm of neurodegenerative disease. Trends Neurosci. 2013;36:561–569. doi: 10.1016/j.tins.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters A.F., van de Weijer S.C.F., Leentjens A.F.G., Duits A.A., Jacobs H.I.L., Kuijf M.L. Resting-state fMRI in Parkinson's disease patients with cognitive impairment: a meta-analysis. Parkinsonism Relat Disord. 2019;62:16–27. doi: 10.1016/j.parkreldis.2018.12.016. [DOI] [PubMed] [Google Scholar]
- Yau Y., Zeighami Y., Baker T.E., Larcher K., Vainik U., Dadar M., Fonov V.S., Hagmann P., Griffa A., Misic B., Collins D.L., Dagher A. Network connectivity determines cortical thinning in early Parkinson's disease progression. Nat. Commun. 2018;9:12. doi: 10.1038/s41467-017-02416-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkali A., McColgan P., Leyland L.A., Lees A.J., Rees G., Weil R.S. Organisational and neuromodulatory underpinnings of structural-functional connectivity decoupling in patients with Parkinson's disease. Commun Biol. 2021;4:86. doi: 10.1038/s42003-020-01622-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeighami Y., Ulla M., Iturria-Medina Y., Dadar M., Zhang Y., Larcher K.M., Fonov V., Evans A.C., Collins D.L., Dagher A. Network structure of brain atrophy in de novo Parkinson's disease. Elife. 2015;4 doi: 10.7554/eLife.08440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.Q., Zhang Y., Yau Y., Zeighami Y., Larcher K., Misic B., Dagher A. Local vulnerability and global connectivity jointly shape neurodegenerative disease propagation. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.3000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Gennatas E.D., Kramer J.H., Miller B.L., Seeley W.W. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron. 2012;73:1216–1227. doi: 10.1016/j.neuron.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The codes that support the findings of this study are openly available in GitHub public repository at https://doi.org/10.5281/zenodo.4672562.
CRediT authorship contribution statement
Silvia Basaia: Conceptualization, Formal analysis, Data curation, Writing – original draft, Writing – review & editing. Federica Agosta: Conceptualization, Data curation, Funding acquisition, Writing – original draft, Writing – review & editing. Ibai Diez: Conceptualization, Formal analysis, Data curation, Writing – review & editing. Elisenda Bueichekú: Conceptualization, Formal analysis, Data curation, Writing – review & editing. Federico d'Oleire Uquillas: Conceptualization, Data curation, Writing – review & editing. Manuel Delgado-Alvarado: Conceptualization, Data curation, Writing – review & editing. César Caballero-Gaudes: Conceptualization, Data curation, Writing – review & editing. MariCruz Rodriguez-Oroz: Conceptualization, Data curation, Writing – review & editing. Tanja Stojkovic: Conceptualization, Data curation, Writing – review & editing. Vladimir S. Kostic: Conceptualization, Formal analysis, Data curation, Writing – review & editing. Massimo Filippi: Conceptualization, Data curation, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. Jorge Sepulcre: Conceptualization, Formal analysis, Data curation, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.