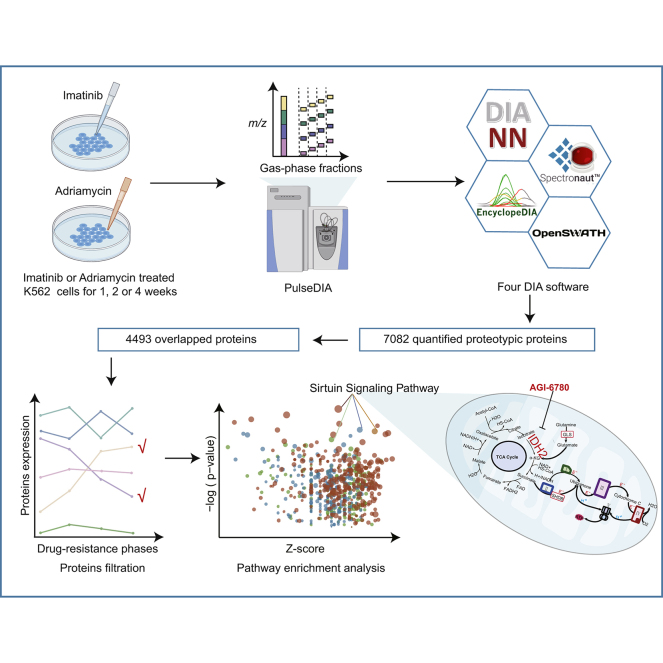
Abstract
Drug resistance is a critical obstacle to effective treatment in patients with chronic myeloid leukemia. To understand the underlying resistance mechanisms in response to imatinib mesylate (IMA) and adriamycin (ADR), the parental K562 cells were treated with low doses of IMA or ADR for 2 months to generate derivative cells with mild, intermediate, and severe resistance to the drugs as defined by their increasing resistance index. PulseDIA-based (DIA [data-independent acquisition]) quantitative proteomics was then employed to reveal the proteome changes in these resistant cells. In total, 7082 proteins from 98,232 peptides were identified and quantified from the dataset using four DIA software tools including OpenSWATH, Spectronaut, DIA-NN, and EncyclopeDIA. Sirtuin signaling pathway was found to be significantly enriched in both ADR-resistant and IMA-resistant K562 cells. In particular, isocitrate dehydrogenase (NADP(+)) 2 was identified as a potential drug target correlated with the drug resistance phenotype, and its inhibition by the antagonist AGI-6780 reversed the acquired resistance in K562 cells to either ADR or IMA. Together, our study has implicated isocitrate dehydrogenase (NADP(+)) 2 as a potential target that can be therapeutically leveraged to alleviate the drug resistance in K562 cells when treated with IMA and ADR.
Keywords: drug resistance, DIA, imatinib, adriamycin, IDH2, Chronic Myeloid Leukemia
Abbreviations: α-KG, α-ketoglutarate; ADR, adriamycin; CiRT, common index retention time standards; CML, chronic myeloid leukemia; CV, coefficient of variation; DDA, data-dependent acquisition; DIA, data-independent acquisition; FA, formic acid; FDR, false discovery rate; GLS, glutaminase; IDH2, isocitrate dehydrogenase (NADP(+)) 2; IMA, imatinib mesylate; IPA, ingenuity pathway analysis; MS, mass spectrometry; PCT, pressure cycling technology; RT, retention time; TKI, tyrosine kinase inhibitor; TOMM, translocase of the outer mitochondrial membrane; VDAC, voltage-dependent anion channel
Graphical Abstract
Highlights
-
•
Temporal proteomic dynamics in the imatinib or adriamycin-induced drug resistance.
-
•
Comparison of four DIA software tools (OpenSWATH, Spectronaut, DIA-NN, and EncyclopeDIA).
-
•
Sirtuin signaling pathway was significantly regulated in resistant K562 cells.
-
•
IDH2 was identified as a potential drug target correlated for resistant K562 cells.
In Brief
To understand the underlying resistance mechanisms in response to imatinib (IMA) and adriamycin (ADR), we explored two unique drug resistance models of K562 cells. We applied an optimized DIA–MS method to quantify 98,232 peptides from 7082 proteotypic proteins from these samples using four DIA software tools including OpenSWATH, Spectronaut, DIA-NN, and EncyclopeDIA. The sirtuin signaling pathway was found significantly regulated in both models, and IDH2 was identified as a druggable regulator of acquired drug resistance.
The treatment of chronic myeloid leukemia (CML) patients includes targeted therapy (tyrosine kinase inhibitors [TKIs]), chemotherapy, biological therapy, hematopoietic cell transplant, and donor lymphocyte infusion (1, 2, 3). Chemotherapy inhibits the rapidly proliferating tumor cells by interfering with cell replication. However, drug resistance leads to failed chemotherapy treatment in 90% patients (4). Adriamycin (ADR) is a traditional chemotherapeutic drug that disturbs the DNA replication process, which can be therapeutically leveraged against certain hematologic tumors. Using a K562 cell model, various mechanisms have been uncovered to explain ADR-induced drug resistance, such as transporter-mediated drug efflux (5, 6), altered mitochondrial function (7, 8), and changes in survival-related signaling pathways including epidermal growth factor receptor, extracellular-regulated kinase, NF-κB, phosphatase and tensin homolog, and AKT pathways (9, 10, 11).
As one of the most effective clinical regimens in the chronic phase, TKIs have dramatically improved survival rate in CML patients (12). Indeed, imatinib mesylate (IMA) is among the first generation of TKIs and the first-line drug for CML treatment, which targets BCR–ABL1 and inhibits tumor growth (13). Notably, the 5-year survival rate of CML patients with IMA treatment has increased to 89% (14). However, about 20 to 25% of CML patients showed a suboptimal response to IMA, who have likely developed drug resistance (15). Several mechanisms have been proposed to explain the failed IMA treatment in CML patients, including, for example, altered conformation of the BCR–ABL1 kinase domain by mutations that reduce its binding affinity to IMA (12). Other resistance mechanisms independent of BCR–ABL1 have also been reported, including P-glycoprotein upregulation, activation of alternative PI3K/AKT, Janus kinase-2, or mitogen-activated protein kinase signaling (16, 17), and changes in the intracellular environment such as endoplasmic reticulum stress–induced autophagy (18).
Drug resistance remains a clinical hurdle to traditional chemotherapy and targeted therapy (19). Given its complex nature, both genetic mutations and nongenetic changes (such as epigenetics) may contribute to a drug-resistant phenotype (20). Mass spectrometry (MS)-based proteomics could quantify thousands of proteins and provide unique insights into the dysregulated pathways (21); thus, it can be used to explore the mechanisms of drug resistance. A deep proteome profiling is essential for characterizing relevant signaling proteins responsible for drug resistance. Some studies of CML drug resistance reported relatively small protein numbers, that is, 2059 proteins (22), 1344 proteins (23), and 477 proteins (24). Some studies tried to characterize the changes of proteomes related to either drug-resistant or drug-sensitive phenotypes of CML cells, bone marrow extracellular fluids (24, 25, 26), or IMA treatment of CML cells in just 24 h (23). However, the difference in phenotypes may only be partially attributed to IMA. Furthermore, these are all proteomic profiling of CML cells under a certain physiological state (27). Here, we focused on the dynamic and temporal changes of proteomes responsive to the IMA-induced drug resistance of CML cells at different stages.
Data-independent acquisition (DIA) is an effective proteomic method with rigorous quantitative accuracy and reproducibility (28, 29). Based on DIA–MS, our group has recently developed PulseDIA–MS as an improved approach that utilizes gas phase fractionation to achieve a greater proteome depth (30). In this work, we employed the pressure cycling technology (PCT)-based peptide preparation (31, 32) and the PulseDIA–MS strategy to quantify the dynamic proteome changes upon drug treatment, using time-series K562 drug-resistant cell line models treated by ADR and IMA, respectively. Because of the lack of correspondence information between precursor ions and fragment ions in DIA data, DIA data analysis has become a huge challenge. Different DIA tools have different scoring methods and core algorithms for prediction of peptides, which may lead to certain technical deviations in the analytical results of the same DIA data (33). To reduce the technical deviations of the quantitative proteome by DIA software tools, we quantified the proteome with four commonly used independent tools, Spectronaut (34), DIA-NN (35), EncyclopeDIA (36), and OpenSWATH (37). With this cell model and quantitative approaches, we were able to pinpoint and characterize pathways that were significantly altered upon ADR or IMA treatment. Notably, we identified isocitrate dehydrogenase (NADP(+)) 2 (IDH2), a previously unknown potential target that can be therapeutically leveraged to reverse drug resistance in K562 cells.
Experimental Procedures
Experimental Design and Statistical Rationale
In this study, a total of 21 samples from seven different drug sensitivities of K562 cell lines were subjected to MS analysis. For each cell line, we harvested three individually cultured cell samples as biological replicates and performed sample preparation independently. We also selected five samples for duplicate injection in MS acquisition as technical replicates. All 26 samples were subjected to four-part PulseDIA analysis as described previously (30) and generated a total of 104 DIA–MS raw data. To ensure the stability and reliability of the MS data, a mouse liver digest was used for instrument performance evaluation, and blank samples (buffer A) were also run for every four injections to minimize carryover. We carried out library-based method analysis of the 104 PulseDIA–MS data using four DIA tools. About 37 data-dependent acquisition (DDA) acquired samples from 10 in-solution digestion files, seven PCT-assisted digestion files, and 20 high-PH fractionations were used to generate the spectral library. Because of the presence of a gap in the MS2 window of each PulseDIA data, the standard peptides or proteins were not suitable for retention time (RT) correction (30). Therefore, we used the software's internal default algorithm for RT correction or the common index retention time standards (CiRT) method (38). We obtained quantitative results of peptide levels by DIA software with peptide precursors of 0.01 Q value (false discovery rate [FDR]) cutoff. At the protein level, the linear regression of top three precursor intensities was used to peptide to protein inference (39).
Establishment of Drug-Resistant K562 Cell Models
The parental-sensitive K562 cells were purchased from Nanjing KeyGen Biotech Co, Ltd and authenticated via short tandem repeat profiling by Shanghai Biowing Applied Biotechnology Co, Ltd on February 28, 2019. The parental K562 cells were cultured in RPMI medium (Cromwell) with 10% fetal bovine serum (Waltham) and 1% penicillin–streptomycin (HyClone) at 37 °C with 5% CO2 and 95% humidity.
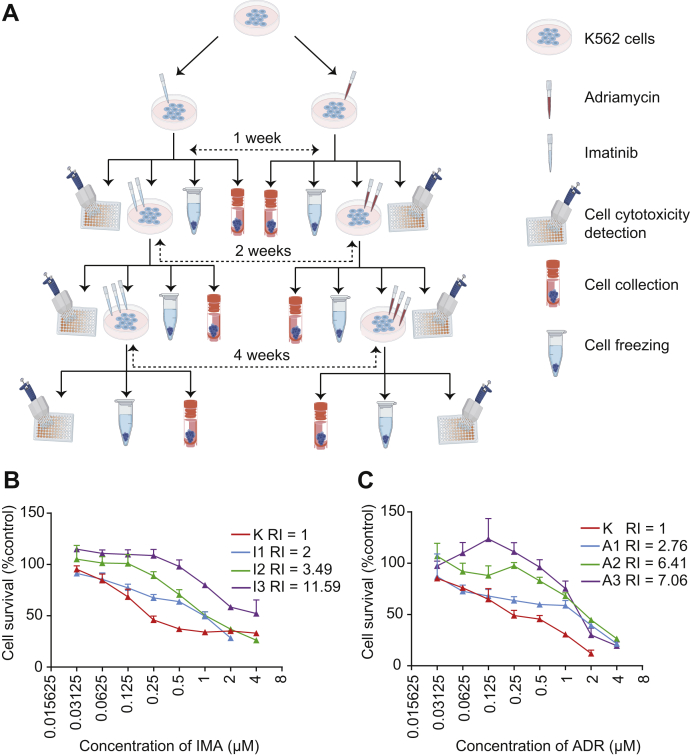
The building of drug-resistant K562 cell models includes three phases (Fig. 1A). In the first phase, 1 × 105 K562 cells were treated with 0.1 μM ADR (CAS: 25316-40-9; meilunbio) (A1) or IMA (CAS: 220127-57-1; meilunbio) (I1). After 1 week, K562 cells were cultured to about 10 million cells. Then drugs were removed by centrifugation, and cells were divided into four aliquots for cell cytotoxicity determination, subsequent drug-resistant induction, cell collection, and cell freezing. In the second phase, the concentration of ADR and IMA was increased to 0.4 μM (A2) and 0.8 μM (I2), respectively. The treatment lasted for 2 weeks. In the third phase, the concentration of ADR and IMA increased to 0.8 μM (A3) and 1.6 μM (I3), respectively. The treatment lasted for 4 weeks. In this way, we established two drug-resistant K562 models for ADR and IMA, separately.
Fig. 1.
Establish derivative K562 cells with mild, intermediate, and severe resistance to ADR and IMA.A, overview of the drug resistance model. B, native K562 cells and IMA-resistant K562 cells from each of the three phases were treated with a series of cytotoxicity concentrations (4, 2, 1, 0.5, … 0 μM) for IMA for 48 h. C, native K562 cells and ADR-resistant K562 cells from each of the three phases were treated with a series of concentrations (4, 2, 1, 0.5, … 0 μM) for ADR for 48 h. The cell survival rate was calculated and plotted in each group. The downward shift of the survival curves by increasing treating concentration (I1, I2, I3 and A1, A2, A3) indicated suppressed proliferation. ADR, adriamycin; IMA, imatinib.
Cytotoxicity Assay
The cytotoxicity of ADR, IMA, or AGI-6780 (CAS: 1432660-47-3; MedChemExpress) to native K562 cells and the model cells was detected by Cell Counting Kit-8 (Bimake). Cells were planted into 96-well plates in a density of 5000 cells/100 μl medium/well. After 24 h, cells were treated with drugs for 48 h. After 48 h, 10 μl of Cell Counting Kit-8 was added to each well and incubated for about 2 h in dark. The absorbance value of each well was determined by a Synergy H1 (BioTek) at 450 nm. The IC50 value was calculated by SPSS (IBM, version 22.0).
PCT-Based Peptide Extraction
The workflow of PCT-based peptide extraction is described by Shao et al. (32). For each sample, 500,000 cells were harvested and cleaned by PBS three times to remove all traces of fetal bovine serum.
Cells were transferred into PCT-MicroTube (Pressure Biosciences, Inc) with 30 μl of lysis buffer, 5 μl of 1 μg/μl DNAase (STEMCELL), and 15 μl of 0.1 M ammonium bicarbonate (General Reagent). The lysis buffer includes 6 M urea (Sigma–Aldrich) and 2 M thiourea (Sigma–Aldrich). Then the cells were lysed by Barocycler NEP2320-45k (PressureBioSciences, Inc) with 90 cycles containing 25 s of 45,000 psi high pressure plus 10 s at ambient pressure, at 30 °C. The lysate solution was added to 5 μl of 100 mM Tris(2-carboxyethyl)phosphine (Aldrich) and 2.5 μl of 800 mM iodoacetamide (Sigma) in PCT-MicroTube to dilute into a final concentration of 10 and 40 mM, followed by a 30 min of incubation in the dark with gentle vortexing (800 rpm) at room temperature in a shaker.
After reduction and alkylation, the protein solution was added with 57.5 μl of 0.1 M ammonium bicarbonate and 25 μl of 0.1 mg/ml Lys-C (Hualishi) and digested using Barocycler NEP2320-45k (Pressure Biosciences, Inc) with 45 cycles containing 50 s of 20,000 psi high pressure plus 10 s at ambient pressure, at 30 °C.
After Lys-C digestion, the solution was added with 10 μl of 0.2 mg/ml trypsin (Hualishi) and was tryptic digested by Barocycler NEP2320-45k with 90 cycles containing 50 s of 20,000 psi high pressure plus 10 s at ambient pressure, at 30. Then, 15 μl of 10% trifluoroacetic acid (Fisher Scientific) was added to the lysate solution at a final concentration of 1% to stop digestion.
Then the digested peptides were cleaned in microspin columns (The Nest Group, Inc) and dried in CentriVap DNA Vacuum Concentrators (Labconco). Peptides were redissolved in MS buffer (0.1% formic acid [FA] and 2% acetonitrile in HPLC water). The peptide concentration was measured using ScanDrop2 (Analytik Jena).
PulseDIA–MS
The PulseDIA–MS method was performed as previously described (30). The redissolved peptides of each sample were analyzed by EASY-nLC 1200 System (Thermo Fisher Scientific) coupled to a QE HF-X mass spectrometer (Thermo Fisher Scientific). The MS1 was acquired in an m/z range of 390 to 1210 with the resolution at 60,000, automatic gain control target of 3e6, and the maximum ion injection time of 80 ms. The MS2 was performed with the resolution at 30,000, automatic gain control target of 1e6, and the maximum ion injection time of 50 ms. Different from the conventional DIA method (the 400–1200 m/z mass range is divided into 24 windows) (28), the PulseDIA symmetrically divided each window into four parts and acquired the data of each part independently. For one sample, we acquired four injections with a different MS2 range. For each pulse acquisition, 0.2 μg of peptides was injected and separated across a linear 45 min LC gradient (from 8% to 40% buffer B) at a flow rate of 300 nl/min (precolumn, 3 μm, 100 Å, 20 mm ∗ 75 μm i.d.; analytical column, 1.9 μm, 120 Å, 150 mm ∗ 75 μm i.d.). Buffer A was HPLC-grade water containing 0.1% FA, and buffer B was 80% acetonitrile and 20% water containing 0.1% FA.
Quality Control Samples
Cells with mild, intermediate, and severe resistance to ADR and IMA were analyzed in three duplicates for peptide extraction and acquisition as biological replicates. Five samples were repeatedly injected by PulseDIA as the technical replicates to evaluate the data quality.
Generating Spectral Library for DIA–MS
To build the spectral library, we acquired 37 DDA files including 10 in-solution digestion files, seven PCT-assisted digestion files, and 20 high-PH fraction files on a QE-HFX mass spectrometer in DDA mode. A library was built by Spectronaut (version 13.5.190902.43655) for Spectronaut and DIA-NN analysis. Another library was built by OpenSWATH (version 2.0) for OpenSWATH and EncyclopeDIA analysis. In the two libraries, data were searched against the SwissProt Human database (20,269 entries). Trypsin and Lys-C were used to generate peptides in silico.
For Spectronaut library building, carbamidomethyl (C) was set as the fixed modification, and acetyl (protein N-term) and oxidation (M) were set as the variable modifications. Two missed trypsin cleavages were allowed. The precursor and fragment tolerance were set as dynamic. Two calibration searches were performed: based on the first-pass calibration (rough calibration), the ideal tolerance for the second-pass calibration was defined; based on the second-pass calibration (finer calibration), the ideal tolerance for the main search was defined (Spectronaut Manual, https://biognosys.com/resources/spectronaut-manual/). The FDR cutoff for precursor and protein identification was 0.01. Finally, a K562 library containing 191,008 precursors, 133,025 peptides, 8226 protein groups, and 8313 proteins was built.
For OpenSWATH library building, the pFind (40) (version 3.1.5) was used as a search engine with the parameters including carbamidomethyl (C) as a fixed modification and oxidation (O) as a variable modification. DDA library was built according to the workflow (41) (http://openswath.org/en/latest/docs/pqp.html#id3). No missed trypsin cleavage was allowed by default. The precursor peptide mass tolerance was 20 ppm, and fragment ion mass tolerance was 0.05 Da. The FDR cutoff for precursor and protein identification was 0.01, and other parameters were set as default. Finally, a K562 library containing 110,583 transition groups, 84,548 target peptides, 84,910 decoy peptides, 9511 target protein groups, 9575 decoy protein groups, and 7935 target proteotypic proteins was built.
Based on the analysis of K562 spectral library, we finally identified a total of 8524 proteins based on proteotypic peptides and up to 10,732 protein groups.
PulseDIA Data Analysis
Library-based PulseDIA data analysis was performed by Spectronaut, OpenSWATH, DIA-NN, and EncyclopeDIA.
For Spectronaut (version 13.5.190902.43655) analysis, the default setting of library-based DIA analysis was used for PulseDIA analysis. PulseDIA analysis was performed according to the standard workflow in Spectronaut (Spectronaut Manual, https://biognosys.com/resources/spectronaut-manual/). RT prediction type was set to dynamic iRT (correction factor for window 1). The MS mass tolerance was set as dynamic, which means Spectronaut calculated the ideal mass tolerances for data extraction and scoring based on its extensive mass calibration. At least three fragment ions were used per peptide identification and major and minor group quantities were set to mean peptide and mean precursor quantity. The FDR was set to 1% at the peptide precursor level.
For DIA-NN (version 1.6.0) analysis, we used the same library with Spectronaut analysis. Library search was performed according to the DIA-NN manual (https://github.com/vdemichev/DiaNN/blob/master/DIA-NN%20GUI%20manual.pdf). For RT prediction and extraction mass accuracy, we used the default parameter 0.0, which means DIA-NN performed automatic mass and RT correction. Top six fragments (ranked by their library intensities) were used for peptide identification and quantification. The FDR was set to 1% at the peptide precursor level.
For OpenSWATH (version 2.4) analysis, the RT extraction window was 300 s, and m/z extraction for MS2 was performed with 30 ppm tolerance, whereas m/z extraction for MS1 was performed with 20 ppm tolerance. RT was then calibrated using CiRT peptides. The m/z extraction for CiRT peptides was performed with 50 ppm tolerance. Peak groups were used for peptide identification if they contained peaks in three of five transitions, and the most intense peaks were prioritized. Peptide precursors were identified by OpenSWATH and Pyprophet with an FDR of 0.01.
For EncyclopeDIA (version 0.9.0) analysis, the library was converted from OpenSWATH library. The precursor, fragment, and library mass tolerance were set as 10 ppm for the PulseDIA data. The RT model was generated from peptides detected at 1% FDR using a nonparametric kernel density estimation algorithm that follows the density mode over time. Peptide quantities were set to the sum of the top five transitions that pass EncyclopeDIA criteria, and peptides with at least three quantitative transitions were considered to be trustworthy. The search results were filtered at a 1% peptide-level FDR.
The peptide matrices from four DIA software tools were converted to protein matrices by an in-house R code (https://github.com/Allen188/PulseDIA/blob/master/Pulsedia_DIANNresult_combine.R) and the ProteomeExpert server (38).
Calculation of IC50 Values
The IC50 values were calculated by a nonlinear least-squares regression model to fit the data log (inhibitor) versus normalized response in GraphPad Prism 6 (GraphPad Software, Inc).
Bliss Independence Model Analysis
Bliss independence model analysis was performed using Combenefit software (Cancer Research UK Cambridge Institute, version 2.021) (42). With the cytotoxicity assay results as input, we calculated the cell survival rate after jointly treated with AGI-6780 and IMA or ADR.
Statistical Analysis
The statistical significance of protein intensity in different types of resistant cells and parental K562 cells was determined by one-way ANOVA, and p values were adjusted using Benjamini and Hochberg correction. Proteins with p values less than 0.05 were considered as statistically significant. Soft clustering analysis of statistically significant proteins was performed by the R/Bioconductor package Mfuzz (43). The average protein intensity of the parental K562 cells and each type of resistant cells was used as the input data for clustering. The time series were separated according to the cell sensitivity to ADR or IMA, with the initial being the parental K562 cells. Ingenuity pathway analysis (IPA; QIAGEN) was performed to outline the significant canonical pathways (44). In IPA, the p value was calculated using the right-tailed Fisher's exact test, and a p value less than 0.05 is considered as significant (45).
Results
Establishment of Drug-Resistant Cell Models
Drug-resistant K562 cell models were developed as shown in the workflow (Fig. 1A). The parental K562 cells were sensitive to ADR and IMA treatment and then treated with increasing concentrations of these drugs, respectively, for 1, 2, and 4 weeks to obtain derivative cell lines with differential drug sensitivities. Each model contained three time points during the drug resistance acquisition, with each time point containing cells showing a different degree of drug sensitivity (Fig. 1A, method).
At this point, we have generated derivative K562 cells with different degrees of IMA resistance: mild, intermediate, and severe, which were defined by resistance indexes of 2.76, 6.41, and 7.06 (Fig. 1B), respectively, as shown in Table 1. Similarly, we generated mild, intermediate, and severe ADR-resistant K562 cells with resistance indexes at 2.00, 3.49, and 11.59, respectively (Fig. 1C and Table 1). These K562 cells with different degrees of IMA and ADR resistance were all collected for PulseDIA-based proteomic analysis (Fig. 2A).
Table 1.
IC50 values and resistance index of the derivative K562 cells
| Model | IC50 (μM, 95% confidence interval)a |
Resistance indexb | |
|---|---|---|---|
| IMA | ADR | ||
| Native K562 cells | 0.37 (0.26–0.54) | 0.29 (0.24–0.35) | 1 |
| Model IMA phase 1 | 0.74 (0.61–0.90) | — | 2 |
| Model IMA phase 2 | 1.29 (1.06–1.58) | — | 3.49 |
| Model IMA phase 3 | 4.29 (3.03–6.36) | — | 11.59 |
| Model ADR phase 1 | — | 0.80 (0.51–1.24) | 2.76 |
| Model ADR phase 2 | — | 1.86 (1.5–2.30) | 6.41 |
| Model ADR phase 3 | — | 2.05 (1.32–3.32) | 7.06 |
IC50 values represent the best-fit values and 95% confidence interval of three independent experiments performed.
Resistance index was calculated by dividing the best-fit IC50 values of native K562 cells in response to ADR and IMA to the best-fit IC50 values of model cells.
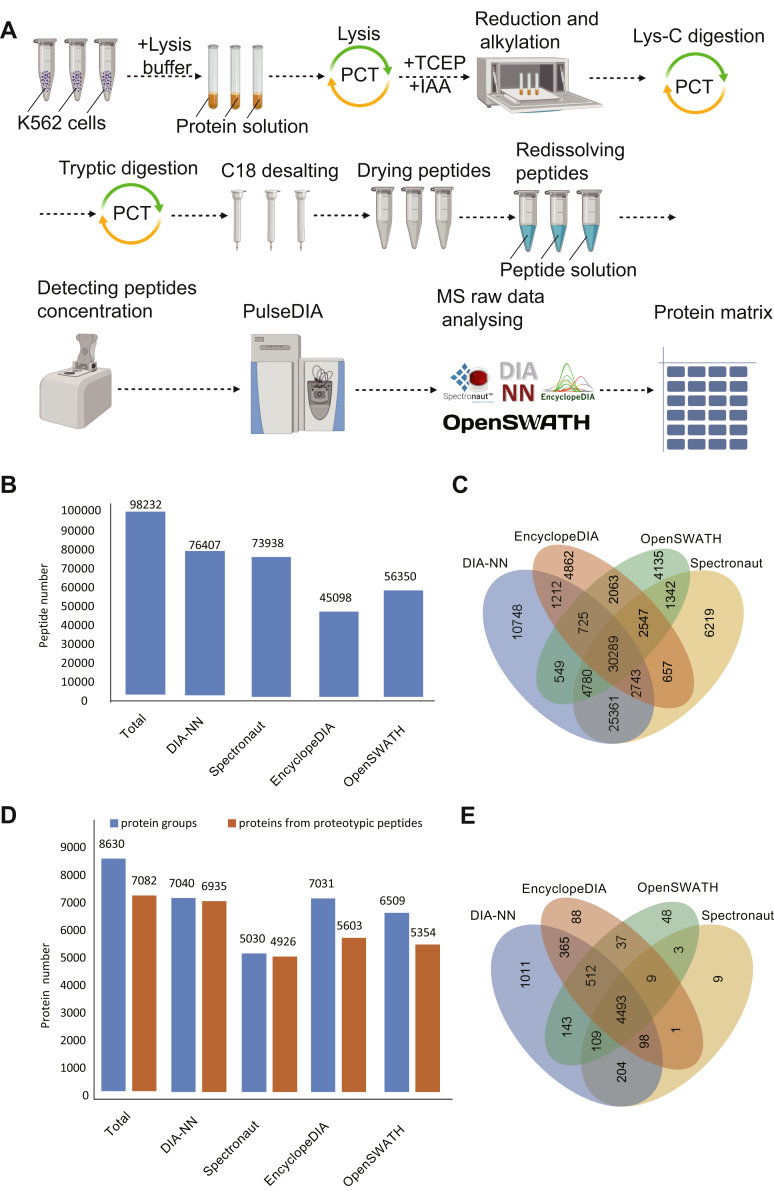
Fig. 2.
PulseDIA raw data acquisition and analysis.A, the workflow of peptide extraction from samples and analysis of mass spectrometry raw data. B, the numbers of identified peptides by four DIA software tools and their Venn diagram (C). D, the numbers of identified proteins by four DIA software and their Venn diagram (E). DIA, data-independent acquisition.
Peptide and Protein Identification
PCT-assisted peptide preparation and PulseDIA–MS were then carried out to analyze the parental K562 cells and the derivative resistant cells in biological triplicates (Fig. 2A). In total, 98,232 peptides, 8630 protein groups, and 7082 proteins from proteotypic peptides were quantified from 26 independent PulseDIA–MS runs (Fig. 2, B and D and supplemental Table S1). About 30,289 peptides and 4493 proteins from proteotypic peptides were consistently quantified by four DIA software (Fig. 2, C and E). We used these commonly identified proteins for the subsequent quantitative analysis of proteome changes during the development of drug resistance in K562 cells.
Quality Control of PulseDIA Proteome Dataset
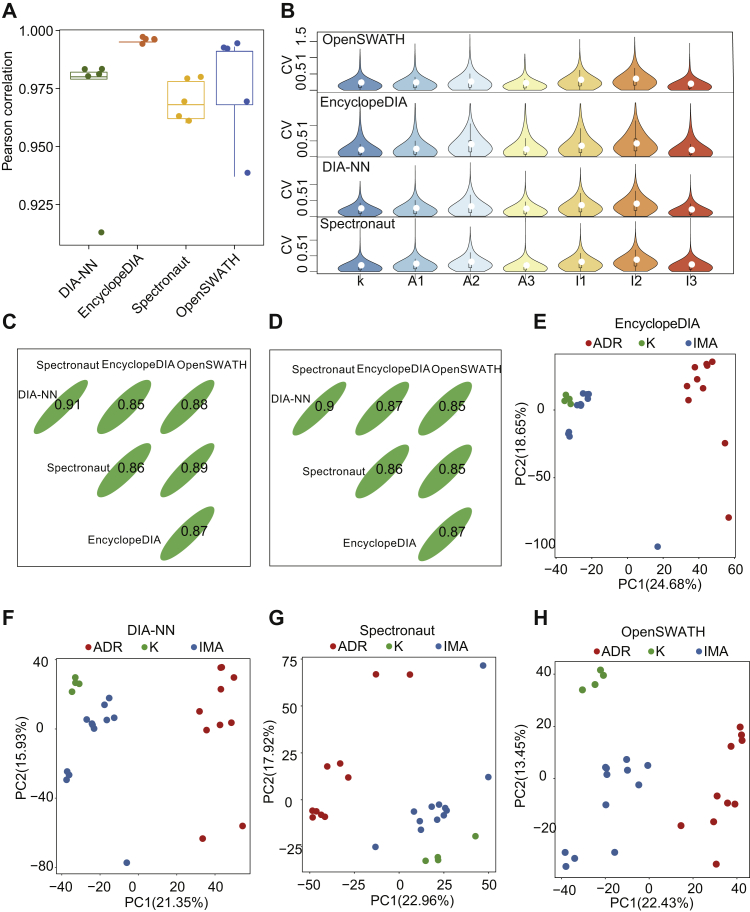
We examined the reproducibility of PulseDIA data by calculating the Pearson correlation coefficient between technical duplicates and the coefficient of variation (CV) of the protein intensity among the biological triplicates. Using four different DIA analysis tools as described previously, the five pairs of technical duplicates showed a strong correlation (r > 0.9) (Fig. 3A), and the median CV of protein intensity among biological triplicates was around 20% (Fig. 3B). These results confirmed the high degree of reproducibility of MS data acquired by the PulseDIA method. To compare the quantified results of the same peptides and proteins in the four DIA software, we calculated the Spearman correlation of the overlapped 30,289 peptides and 4493 proteins quantified in the four DIA software. At the peptide and protein levels, DIA-NN and Spectronaut exhibited the strongest correlation (r > 0.9), with the correlation of any two DIA software as no less than 0.85 (Fig. 3, C and D). To check whether the quantified proteome thus acquired can classify different drug resistance models, we performed principal component analysis between the cells in the ADR-resistant and IMA-resistant cells as well as the parental K562 cells. The results showed that the two resistant cell lines and parental K562 cells were separated into three clusters (Fig. 3, E–H).
Fig. 3.
PulseDIA proteome data quality control (QC) analysis.A, the box plot shows the Pearson correlation coefficient of five technical replicates in four DIA software quantification. B, the violin plot shows the distribution of the coefficient of variation (CV) of each protein's quantitative values in the three biological replicates. Three lines of the black box inside the violin represented lower quartile, median, and higher quartile, respectively. C, Spearman correlation coefficient of overlapped 30,289 peptide quantitative values in the four DIA software. D, Spearman correlation coefficients of overlapped 4493 protein in the four DIA software. E–H, PCA plot shows the distribution of the samples in the first two principal component levels. The red, blue, and green dots indicate samples from model ADR, model IMA, native K562 cells, respectively. ADR, adriamycin; DIA, data-independent acquisition; IMA, imatinib; PCA, principal component analysis.
Dynamic Proteomic Changes During Acquisition of Drug Resistance
To minimize the statistical variation for each analytic step, these 4493 overlapped proteins were selected, and those with less than 25% missing ratio in each derivative-resistant cell line were subjected to the ANOVA analysis. Proteins with adjusted p value less than 0.05 were selected for further downstream analysis.
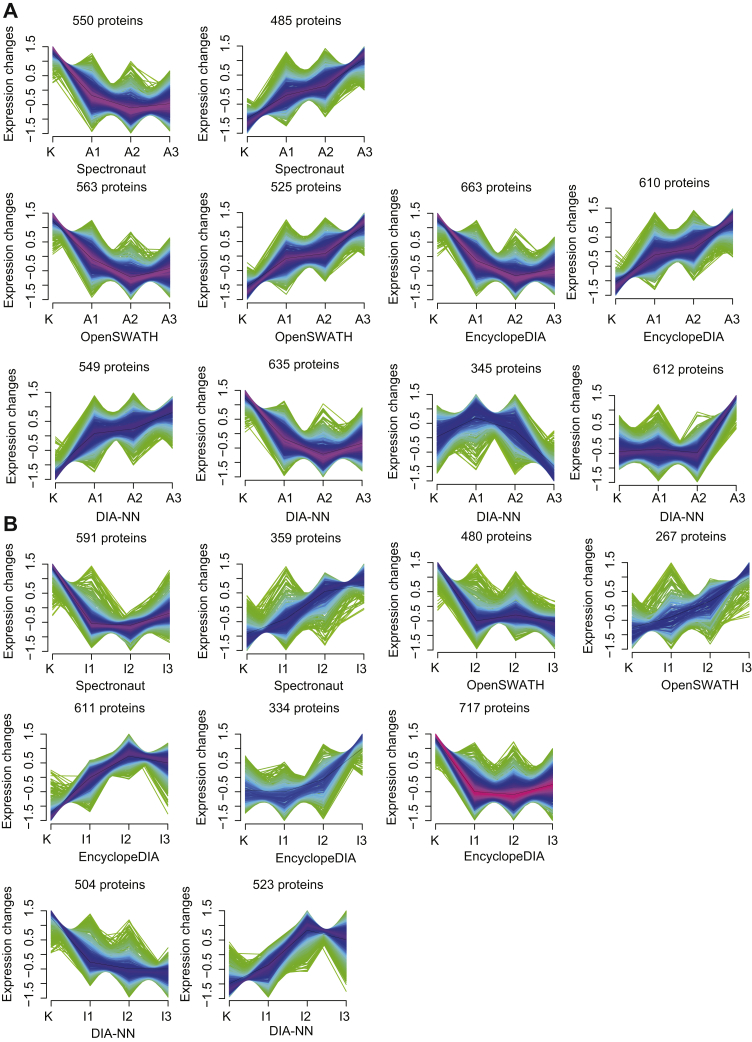
By fuzzy c-means clustering (43, 46), we identified four clusters related to the resistance to ADR and IMA (supplemental Fig. S1 and Fig. 4), among which only those that were continuously upregulated and downregulated were further selected and studied (Fig. 4). In addition, we selected 1035 (by Spectronaut), 1273 (by EncyclopeDIA), 1088 (by OpenSWATH), and 2161 (by DIA-NN) proteins related to resistance to ADR (supplemental Table S2 and Fig. 4A), and 1662 (EncyclopeDIA), 747 (OpenSWATH), 950 (Spectronaut), and 1027 (DIA-NN) proteins related to resistance to IMA (supplemental Table S3 and Fig. 4B).
Fig. 4.
Protein cluster analysis. The cluster for proteins that were continuously upregulated and downregulated from K562 cells resistant to ADR (A) and IMA (B).The horizontal axis represents the progress of the model (K-native K562 cells, A1/I1—first phase of model ADR/IMA, A2/I2—second phase of model ADR/IMA, and A3/I3—third phase of model ADR/IMA). The vertical axis represents protein expression changes in each cluster. ADR, adriamycin; IMA, imatinib.
Activated Sirtuin Signaling Pathway and Abnormal Mitochondrial Function in Drug-Resistant K562 Cells
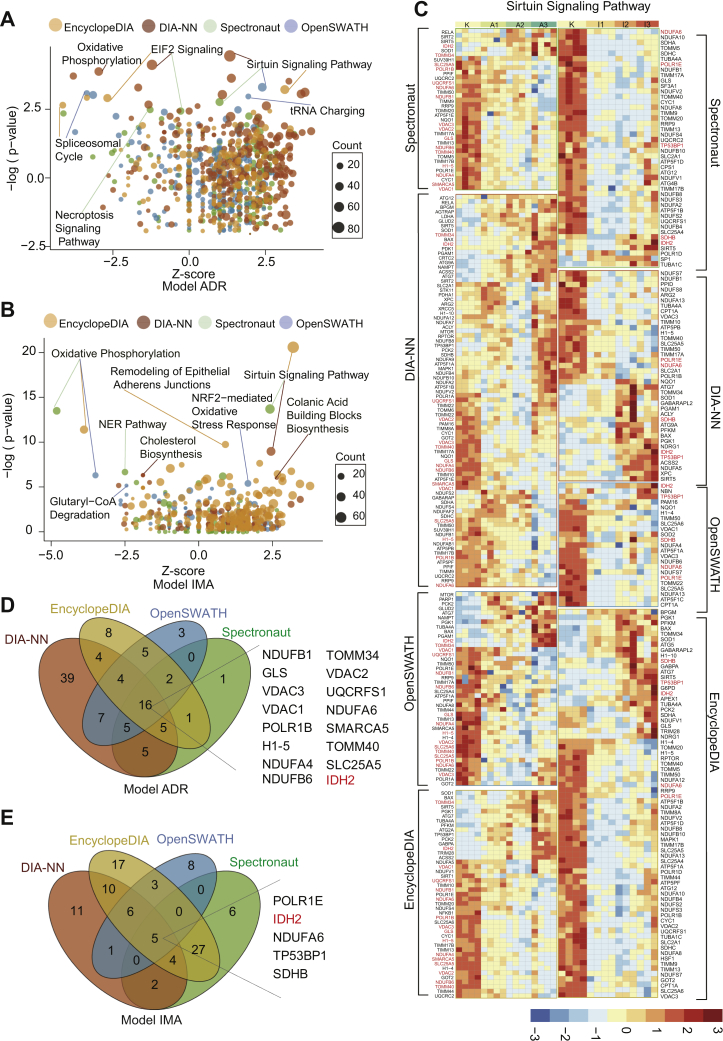
The selected proteins from the four DIA analytic tools as described previously in resistance to ADR and IMA were further analyzed by IPA (supplemental Tables S4 and S5), with the top three pathways (sorted by p value) as listed in Figure 5, A and B. Notably, eukaryotic initiation factor 2 signaling and sirtuin signaling pathway were enriched from at least three DIA tools related to ADR resistance. In contrast, sirtuin signaling pathway and oxidative phosphorylation were frequently enriched related to IMA resitance (Fig. 5, A and B). These data indicate that the sirtuin signaling pathway was significantly enriched in both types of drug resistance with a positive activation mode (Z-score > 0; Table 2).
Fig. 5.
Enriched proteins that exhibit continuous changes as identified by IPA in ADR-resistant and IMA-resistant cells.A, in the ADR-resistant cells, the three most significantly changed pathways were enriched by IPA from four DIA software. B, in the IMA-resistant cells, the three most significantly changed pathways were enriched by IPA from four DIA software. C, the heatmap shows the proteins (adjusted p value < 0.05 calculated by ANOVA) involved in sirtuin signaling pathway from four DIA software expression in the IMA-resistant and ADR-resistant cells, with the overlapped quantified proteins being highlighted in red. Each row indicats a protein, and each column indicats a sample. The protein intensity matrix was normalized by Z-score and colored in the heatmap. D, Venn diagram shows the 16 overlapped proteins involved in the sirtuin signaling pathway in the ADR-resistant cells. E, Venn diagram shows the five overlapped proteins involved in the sirtuin signaling pathway in the IMA-resistant cells. ADR, adriamycin; DIA, data-independent acquisition; IMA, imatinib; IPA, ingenuity pathway analysis.
Table 2.
The enrichment statistics of sirtuin signaling pathway in four DIA software
| Model | Software | pa | Ratiob | Z-scorec |
|---|---|---|---|---|
| Model IMA | OpenSWATH | 4.48 | 0.0825 | 1.155 |
| Model IMA | Spectronaut | 13.7 | 0.155 | 2.414 |
| Model IMA | DIA-NN | 8.97 | 0.134 | 2.449 |
| Model IMA | EncyclopeDIA | 20.6 | 0.251 | 3.202 |
| Model ADR | OpenSWATH | 9.87 | 0.144 | 2.4 |
| Model ADR | Spectronaut | 6.8 | 0.12 | 1 |
| Model ADR | DIA-NN | 21.8 | 0.296 | 1.336 |
| Model ADR | EncyclopeDIA | 9.31 | 0.155 | 3.138 |
p Value is the result of Fisher's exact test.
Ratio is the number of proteins from different DIA software that map to the pathway divided by the total number of proteins that map to the same pathway.
Z-score is calculated by the IPA z-score algorithm to predict the direction of change for the function. An absolute z-score of ≥2 is considered as significant.
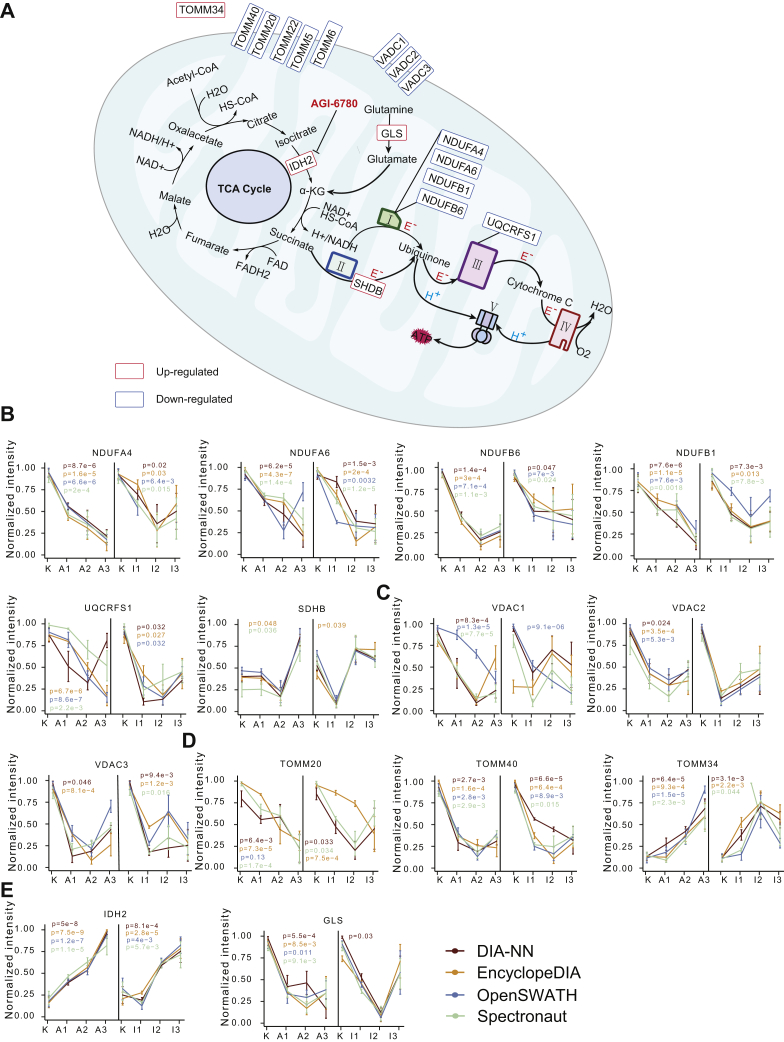
We then focused on the proteins involved in the sirtuin signaling pathway, whose expression levels were shown in a heatmap (Fig. 5C). As a result, 16 proteins, which were involved in the sirtuin signaling pathway, were quantified between all four DIA analytic tools in the model ADR (Fig. 5D), and five proteins in the model IMA (Fig. 5E). The overlapped proteins between all four DIA analytic tools showed the same expression pattern (Fig. 5C). Among the overlapped proteins, 13 of the 16 related to ADR resistance were involved in maintaining mitochondrial function, and three of the five related to IMA resistance participated in the formation of the mitochondrial respiratory chain (Fig. 6A).
Fig. 6.
Proteins involved in mitochondrial function.A, schematic diagram of TCA cycle and electron respiratory chain in mitochondria. B, C, D, and E, normalized protein expression in ADR-resistant and IMA-resistant cells from four DIA software. ADR, adriamycin; DIA, data-independent acquisition; IMA, imatinib; TCA, tricarboxylic acid.
NDUFB1, NDUFB6, NDUFA4, and NDUFA6 are the subunits of NADH–ubiquinone oxidoreductase (complex I) of the mitochondrial respiratory chain (Fig. 6A) (47). UQCRFS1 is a subunit of ubiquinol–cytochrome c oxidoreductase (complex III) (Fig. 6A). The main physiological function of complex I and complex III is to oxidize NADH, transfer electrons to ubiquinone and from ubiquinone to cytochrome C, and carry out the next electron transfer in the mitochondrial respiratory chain (Fig. 5A) (48). In addition, complex I and complex III also regulate mitochondrial production of reactive oxygen species (49). Our results showed decreased expression levels of complex I and III components (Fig. 6, A and B), suggesting low oxidative stress levels in the drug-resistant cells.
We also identified upregulation of succinate dehydrogenase complex iron sulfur subunit B, which participates in the electron transfer process of succinate dehydrogenase (complex II) from succinate to ubiquinone (Fig. 6, A and B), and when mutated, is closely related to pheochromocytoma (50).
Voltage-dependent anion channel (VDAC) proteins are the most abundant proteins (51) on the outer mitochondrial membrane (Fig. 6A) and function to maintain free diffusion of small molecules across the mitochondrial membrane (52). In tumor cells, the interaction between VDAC and hexokinase inhibits apoptosis (53), therefore targeting both molecules could potentially offer improved antitumor benefits. Our results showed that the three VDAC isoforms, namely VDAC1, VDAC2, and VDAC3, were significantly downregulated upon acquisition of ADR resistance (Fig. 6C), suggesting that targeting VDAC may not be an effective strategy in the drug-resistant tumor cells.
The translocase of the outer mitochondrial membrane (TOMM) complex proteins regulates entry of mitochondrial protein precursors into the mitochondria cytoplasm (Fig. 6A) (54). Our data showed that the subunits of TOMMs, TOMM40, TOMM5, TOMM6, TOMM22, and TOMM20 were significantly downregulated in cells resistant to ADR or IMA (Figs. 5C and 6, A and D). In contrast, we found increased expression of TOMM34 upon acquisition of drug resistance (Fig. 5D). Located in the cytoplasm, TOMM34 is known to interact with HSP70 and HSP90 and regulate the activity of ATPase (55). High expression of TOMM34 has been found in colon cancer (56), breast cancer (57), and ovarian cancer (58).
Glutaminase (GLS) can convert glutamine into glutamic acid, which constitutes the major source for α-ketoglutarate (α-KG) production (Fig. 6A) (59, 60), which promotes cell differentiation through dioxygenases (61). Our results showed significantly downregulated expression of GLS when K562 cells became resistant to ADR (Fig. 6E, right panel).
IDH2 catalyzes the oxidative decarboxylation of isocitrate into α-KG in tricarboxylic acid cycle, and NADPH was synchronously produced at the biochemical process (62) (Fig. 6A). NADPH is essential in protecting cells from oxidative damage (63). We found that the IDH2 abundance increased upon resistance to both ADR and IMA (Fig. 6E, left panel), indicating that IDH2 overexpression may promote cellular resistance against high doses of both therapeutic drugs.
Taken together, our results showed significantly changed abundance of proteins involved in mitochondrial functions that may sustain the survival of drug-resistant cells, and might be therapeutically targeted to enhance drug sensitivity and response in tumor cells.
IDH2 Is a Potential Target for Reversing Drug Resistance in K562 Cells
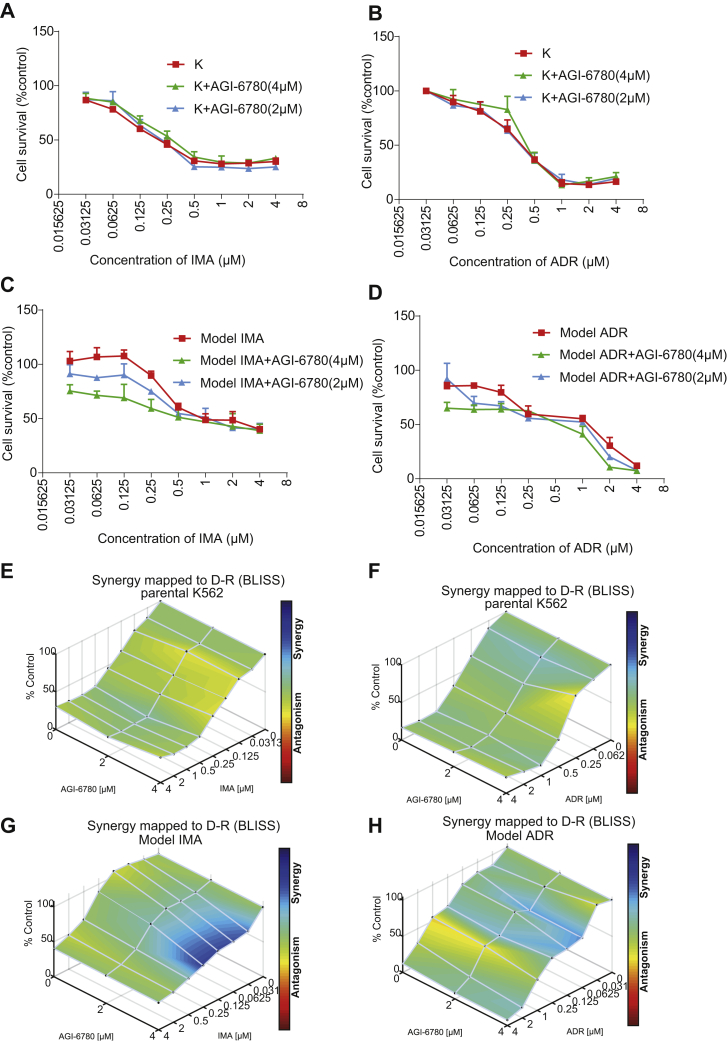
As discussed previously, among the dysregulated proteins, IDH2 was upregulated in K562 cells resistant to both ADR and IMA. To validate its biological function, we utilized a selective inhibitor of IDH2 (64), AGI-6780, and treated sensitive or resistant K562 cells with ADR and IMA alone or combined with AGI-6780, followed by monitoring cell survival with a cytotoxicity assay. Our results showed that, compared with ADR and IMA treatment alone, combination with AGI-6780 did not affect the survival in the parental-sensitive K562 cells (Fig. 7, A and B and Table 3). However, in resistant K562 cells, the sensitivity to ADR and IMA significantly increased (IMA + AGI-6780, IC50 = 0.53 μM; ADR + AGI-6780, IC50 = 0.29 μM), when combined with AGI-6780. We further showed that in IMA-resistant cells the reversal indexes of 2 and 4 μM AGI-6780 were 1.92 and 2.94, respectively (Fig. 7C and Table 3), whereas in ADR-resistant cells, the reversal indexes of 2 and 4 μM AGI-6780 were 1.60 and 2.74, respectively (Fig. 7D and Table 3). To further confirm the synergistic sensitization effect of AGI-6780 in combination with IMA or ADR on drug resistance models, we performed Bliss independence model using Combenefit software (version 2.021) (42). The surface plots showed the dose–response results of AGI-6780 in combination with IMA or ADR (Fig. 7, E–H). As shown in the surface plot, for the parental K562 cell line, the combination of AGI-6780 with IMA (Fig. 7E) or ADR (Fig. 7F) did not produce a significant synergistic effect. In the IMA resistance model, the combination of 0.03 to 0.25 μM IMA and 4 μM AGI-6780 produced a significant synergistic effect (Fig. 7G). In the ADR resistance model (Fig. 7H), the combination of 0.03 to 0.125 μM ADR and 2 to 4 μM AGI-6780 produced a synergistic effect, although this synergy is relatively weak.
Fig. 7.
IDH2 enhanced the sensitivity to ADR and IMA in K562 cells.A and C, the parental-sensitive and IMA-resistant K562 cells were treated with various concentrations (4, 2, 1, 0.5, … 0 μM) of IMA alone or combined with 4 μM or 2 μM AGI-6780 for 48 h. B and D, the parental-sensitive and ADR-resistant K562 cells were treated with various concentrations (4, 2, 1, 0.5, … 0 μM) of ADR alone or combined with 4 μM or 2 μM AGI-6780 for 48 h. The cell survival rate was calculated and plotted in each group. The downward shift of the survival curves indicated suppressed proliferation. Columns are expressed as mean ± SD. E–H, surface plots show the synergistic or antagonistic effects from the combination of two drugs. The parental K562 treated with AGI-6780 (0, 2, and 4 μM) combined with IMA (0–4 μM) (E) or ADR (0–4 μM) (F). G, the IMA resistance model cell treated with AGI-6780 (0, 2, and 4 μM) combined with IMA (0–4 μM). H, the ADR resistance model cell treated with AGI-6780 (0, 2, and 4 μM) combined with ADR (0–4 μM). Each point represents the mean of two independent cytotoxicity assay results. Plots were generated using Combenefit by applying the Bliss independence model. ADR, adriamycin; IDH2, isocitrate dehydrogenase (NADP(+)) 2; IMA, imatinib.
Table 3.
IC50 values and reverse index of the derivative resistant K562 cells
| Drugs | IC50 (μM, 95% confidence interval)a |
Resistance indexb | Reversal indexc | ||
|---|---|---|---|---|---|
| Native K562 | Model IMA | Model ADR | |||
| IMA | 0.26 (0.19–0.36) | 1.55 (1.12–2.19) | — | 5.96 | 1 |
| IMA + AGI-6780 (2 μM) | 0.35 (0.26–0.48) | 1.09 (0.80–1.49) | — | 3.11 | 1.92 |
| IMA + AGI-6780 (4 μM) | 0.26 (0.20–0.33) | 0.53 (0.27–1.16) | — | 2.03 | 2.94 |
| ADR | 0.37 (0.31–0.45) | — | 0.67 (0.49–0.95) | 1.81 | 1 |
| ADR + AGI-6780 (2 μM) | 0.38 (0.31–0.46) | — | 0.43 (0.27–0.68) | 1.13 | 1.60 |
| ADR + AGI-6780 (4 μM) | 0.44 (0.31–0.61) | — | 0.29 (0.18–0.47) | 0.66 | 2.74 |
IC50 values represent the best-fit values, and 95% confidence interval of three independent experiments were performed.
Resistance index was calculated by dividing the best-fit IC50 values of parental K562 cells to ADR or IMA by the best-fit IC50 values of the model cells.
Reversal index was calculated by dividing the resistance index in the absence of ACI-6780 by the resistance index in the presence of AGI-6780.
Taken together, the cytotoxicity assay confirmed the ability of AGI-6780 to reverse drug resistance in vitro, implicating IDH2 as a potential target to improve drug sensitivity and clinical responses.
Discussion
Two major paradigms have been widely proposed to elucidate the underlying mechanism of drug resistance. One is based on the principle of Darwinian selection, which posits that a fraction of tumor cells is inherently more tolerant and thus becomes enriched after drug treatment, thereby gradually developing and exhibiting a drug-resistant phenotype. The other mechanism, which is called Lamarckian induction, proposes that drug treatment induces a resistance phenotype in tumor cells (65). In our resistant cell line model, we focused on the proteins that showed continuous and consistent changes upon acquisition of drug resistance. Here, we uncovered potential association between multiple proteins and drug responsiveness in K562 cells. These proteins serve as potential therapeutic targets for combating drug resistance.
While establishing the resistance cell lines, we also constantly monitored drug sensitivity in the parental-sensitive K562 cells (Fig. 1, B and C). Different from other similar studies (66), we did not consider morphology changes as a parameter, primarily because K562 cells are suspension cells in the medium in a spherical shape when the drug-resistant phenotype is established. After K562 cells with mild, intermediate, and severe resistance to ADR and IMA were developed and collected, we performed PCT-based peptide extraction. The efficient and prompt sample preparation under high pressure (lysis in 45,000 psi; digest in 20,000 psi) by PCT ensures that the process of peptide extraction is reproducible and stable (32).
The resistant model K562 cells and parental K526 cells were acquired by PulseDIA (30). Because of the inherent complexity of DIA data, multiple computational software tools have been developed to analyze DIA data, among which Spectronaut (34), DIA-NN (35), EncyclopeDIA (36), and OpenSWATH (37) are all widely used. These software tools identify and quantify peptides and proteins by various different algorithms with some commonality and difference. The exact algorithm of commercial software Spectronaut is not available to users. However, all software tools meet the rigorous statistical criteria of FDR. Navarro et al. (67) reported the quantified proteins of several DIA analysis software (OpenSWATH, SWATH2.0, Skyline, Spectronaut, and DIA Umpire) for the same SWATH–MS proteomics data, with the number of proteins quantified in the library-based parsing algorithm ranging from 3673 to 4692 and with 3064 proteins shared by all. In our study, the number of proteins quantified from the four DIA tools varied from 4926 to 6935 for the same set of proteomics raw data, with 4493 shared by all (Fig. 2D). Therefore, we focused on the 4493 shared proteins for a more unbiased discovery of potential candidates. This approach is a useful way to further narrow down the candidates for functional validation. While most DIA studies used only one software tool for data interpretation and prioritization of proteins of interest, our study presents a more rigorous approach for protein selection. Nevertheless, this approach may miss certain useful candidates and may not be the best option for all DIA studies.
Our results showed that the CV of protein intensity in three biological replicates was around 20% (Fig. 3B). For PulseDIA-based MS raw data acquisition, we identified and quantified 83.1% (7082/8524) proteotypic proteins of the DDA library from 26 samples by the four DIA datasets, and 63.4% (4493/7082) proteotypic proteins were detected by all four tools (supplemental Table S1 and Fig. 2, D and E). The PulseDIA data stability was assessed by the Pearson correlation of two technical replicates, which was no less than 0.9 (Fig. 3A). The principal component analysis results demonstrated that the resistant K562 cells can be discriminated from the parental cells at the whole proteome level (Fig. 2, E–H). These high-quality MS data provide a key basis for our subsequent data mining analysis.
We independently analyzed 4493 proteins identified by all four tools. Based on cluster analysis and ANOVA (p ≤ 0.05), we selected proteins that exhibited continuous changes during development of IMA and ADR resistance (supplemental Table S2 and S3; Fig. 4), followed by IPA analysis. Pathway analysis revealed common characteristics associated with drug resistance. Significantly changed stress signaling pathways including oxidative phosphorylation and the sirtuin signaling pathway (Fig. 5, A and B) are known to enhance the resistance and plasticity of tumor cells (68, 69, 70). We then decided to focus on the identified proteins involved in the sirtuin signaling pathway as identified by all four DIA tools, including IDH2, NDUFB1, NDUFB6, NDUFA4, NDUFA6, SHDB, and GLS, which are involved in mitochondrial ATP generation (Figs. 5, D and E and 6A) (47, 71). These results suggest that inhibiting ATP production and blocking P-glycoprotein energy supply could potentially enable reversal of drug resistance (5, 72).
IDH2 mutation catalyzes D-2-hydroxyglutarate production, which leads to competitive inhibition of α-KG-dependent DNA demethylases and consequently promotes tumorigenesis (61). Preclinical experiments have shown that IDH2 inhibitors promote leukemic cell differentiation in 40% of the patients with relapsed/refractory AML (64). Here, we analyzed the therapeutic potential of IDH2 using a the selective inhibitor AGI-6780. Our data nominate IDH2 as a potential target for reversing drug resistance in tumor cells. Recent research has shown that 5 μM of AGI-6780 selectively impaired wildtype IDH2 enzymatic activity in multiple myeloma cells in vitro (59). In contrast, our data showed no effect on cell proliferation even 48 h after 4 μM of AGI-6780 treatment in either sensitive or resistant K562 cells (supplemental Fig. S2). As a potential therapeutic avenue for reversing drug resistance, IDH2 inhibitors are specifically efficacious in resistant tumor cells.
Derivative cell lines that have developed drug resistance could provide an important and informative model to help elucidate the underlying mechanism that can be strategically leveraged to manipulate and improve the sensitivity of tumor cells to therapeutic treatment. Single-cell proteomics studies of melanoma-derived cells with different levels of drug sensitivity revealed changes in intracellular signals before drug resistance has developed (73). By constructing a cisplatin-resistant neuroblastoma cell line, Piskareva et al. (66) characterized the epithelial to mesenchymal transition during the development of drug resistance. Herein, we have developed multiple K562 cell lines with different degrees of resistance to ADR and IMA treatment, which could be used for preclinical investigation of the molecular and cellular mechanisms that underlie the therapeutic resistance in CML patients. We further identified and characterized IDH2 as a potentially useful target that can be utilized to enhance tumor cell sensitivity to targeted treatment.
Data Availability
All data are available in the article or the supporting information. The proteomics data are deposited in ProteomeXchange Consortium (Project ID: PXD030553) via the iProX repository partner with the dataset identifier IPX0002882000. The project data analysis codes are deposited in GitHub (https://github.com/guomics-lab/KMDR).
Supplemental data
This article contains supplemental data.
Conflict of interest
T. G. and Y. Z. are shareholders of Westlake Omics, Inc. W. G. is an employee of Westlake Omics, Inc. The remaining authors declare no competing interests.
Acknowledgments
This work is supported by grants from National Key R&D Program of China (grant no.: 2020YFE0202200), the National Natural Science Foundation of China (81874324, 81972492, 21904107, 81473280, and U1608283), Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholars (LR19C050001), Hangzhou Agriculture and Society Advancement Program (20190101A04), and Westlake Education Foundation. We thank Westlake University Supercomputer Center for assistance in data storage and computation.
Author contributions
T. G., W. L., Y. S., and K. L.: methodology; W. L. and W. G.: experiments; W. L., Y. S., W. G., F. Z., L. G., Y. Z., and K. L.: data curation; W. L., Y. Z., and T. G.: manuscript writing; T. G. and K. L.: supervision.
Contributor Information
Tiannan Guo, Email: guotiannan@westlake.edu.cn.
Kexin Liu, Email: liukexin89@163.com.
Supplemental Data
Protein cluster analysis. The protein cluster from model ADR (A) and IMA (B).The horizontal axis represented the progress of the model (K-native K562 cells, A1/I1-first phase of model ADR/IMA, A2/I2-second phase of model ADR/IMA, A3/I3-third phase of model ADR/IMA). The vertical axis represented changes in proteins expression in each cluster. ADR, adriamycin; IMA, imatinib.
Cell survival analysis with treatment of various concentrations of AGI-6780 to IMA-resistant (A), ADR-resistant (B), and parental K562 cell (C) for 48 h. ADR, adriamycin; IMA, imatinib.
Protein and peptide matrix of quantified proteins in 26 samples by DIA-NN, Spectronaut, EncyclopeDIA, and OpenSWATH. The quantitative protein abundance was log2 converted.
In ADR-resistant cells, the list of continuously changed proteins (adjusted p<0.05) with associated p values. ADR, adriamycin.
In IMA-resistant cells, the list of continuously changed proteins (adjusted p<0.05) with associated p values. IMA, imatinib.
IPA analysis of continuously changed proteins from ADR-resistant cells. ADR, adriamycin; IPA, ingenuity pathway analysis.
IPA analysis of continuously changed proteins from IMA-resistant cells. IMA, imatinib; IPA, ingenuity pathway analysis.
References
- 1.Radujkovic A., Guglielmi C., Bergantini S., Iacobelli S., van Biezen A., Milojkovic D., Gratwohl A., Schattenberg A.V.M.B., Verdonck L.F., Niederwieser D.W., de Witte T., Kröger N., Olavarria E. Donor lymphocyte infusions for chronic myeloid leukemia relapsing after allogeneic stem cell transplantation: May we predict graft-versus-leukemia without graft-versus-host disease? Biol. Blood Marrow Transpl. 2015;21:1230–1236. doi: 10.1016/j.bbmt.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Radich J.P., Deininger M., Abboud C.N., Altman J.K., Berman E., Bhatia R., Bhatnagar B., Curtin P., DeAngelo D.J., Gotlib J., Hobbs G., Jagasia M., Kantarjian H.M., Maness L., Metheny L., et al. Chronic myeloid leukemia, version 1.2019, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc. Netw. 2018;16:1108–1135. doi: 10.6004/jnccn.2018.0071. [DOI] [PubMed] [Google Scholar]
- 3.Pal S.K., Miller M.J., Agarwal N., Chang S.M., Chavez-MacGregor M., Cohen E., Cole S., Dale W., Magid Diefenbach C.S., Disis M.L., Dreicer R., Graham D.L., Henry N.L., Jones J., Keedy V., et al. Clinical cancer advances 2019: Annual report on progress against cancer from the American Society of Clinical Oncology. J. Clin. Oncol. 2019;37:834–849. doi: 10.1200/JCO.18.02037. [DOI] [PubMed] [Google Scholar]
- 4.Mansoori B., Mohammadi A., Davudian S., Shirjang S., Baradaran B. The different mechanisms of cancer drug resistance: A brief review. Adv. Pharm. Bull. 2017;7:339–348. doi: 10.15171/apb.2017.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu W., Meng Q., Sun Y., Wang C., Huo X., Liu Z., Sun P., Sun H., Ma X., Liu K. Targeting P-glycoprotein: Nelfinavir reverses adriamycin resistance in K562/ADR cells. Cell Physiol. Biochem. 2018;51:1616–1631. doi: 10.1159/000495650. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y., Wang C., Meng Q., Liu Z., Huo X., Sun P., Sun H., Ma X., Peng J., Liu K. Targeting P-glycoprotein and SORCIN: Dihydromyricetin strengthens anti-proliferative efficiency of adriamycin via MAPK/ERK and Ca(2+) -mediated apoptosis pathways in MCF-7/ADR and K562/ADR. J. Cell Physiol. 2018;233:3066–3079. doi: 10.1002/jcp.26087. [DOI] [PubMed] [Google Scholar]
- 7.De Oliveira F., Chauvin C., Ronot X., Mousseau M., Leverve X., Fontaine E. Effects of permeability transition inhibition and decrease in cytochrome c content on doxorubicin toxicity in K562 cells. Oncogene. 2006;25:2646–2655. doi: 10.1038/sj.onc.1209293. [DOI] [PubMed] [Google Scholar]
- 8.Li R.J., Zhang G.S., Chen Y.H., Zhu J.F., Lu Q.J., Gong F.J., Kuang W.Y. Down-regulation of mitochondrial ATPase by hypermethylation mechanism in chronic myeloid leukemia is associated with multidrug resistance. Ann. Oncol. 2010;21:1506–1514. doi: 10.1093/annonc/mdp569. [DOI] [PubMed] [Google Scholar]
- 9.Zhao L., Shan Y., Liu B., Li Y., Jia L. Functional screen analysis reveals miR-3142 as central regulator in chemoresistance and proliferation through activation of the PTEN-AKT pathway in CML. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.223. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Jiang L., Wang P., Sun Y.J., Wu Y.J. Ivermectin reverses the drug resistance in cancer cells through EGFR/ERK/Akt/NF-κB pathway. J. Exp. Clin. Cancer Res. 2019;38:265. doi: 10.1186/s13046-019-1251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong J., Qin Z., Zhang W.D., Cheng G., Yehuda A.G., Ashby C.R., Chen Z.S., Cheng X.D., Qin J.J. Medicinal chemistry strategies to discover P-glycoprotein inhibitors: An update. Drug Resist. Updat. 2020;49:100681. doi: 10.1016/j.drup.2020.100681. [DOI] [PubMed] [Google Scholar]
- 12.Holyoake T.L., Helgason G.V. Do we need more drugs for chronic myeloid leukemia? Immunol. Rev. 2015;263:106–123. doi: 10.1111/imr.12234. [DOI] [PubMed] [Google Scholar]
- 13.Buchdunger E., O'Reilly T., Wood J. Pharmacology of imatinib (STI571) Eur. J. Cancer. 2002;38 Suppl 5:S28–S36. doi: 10.1016/s0959-8049(02)80600-1. [DOI] [PubMed] [Google Scholar]
- 14.Druker B.J., Guilhot F., O'Brien S.G., Gathmann I., Kantarjian H., Gattermann N., Deininger M.W., Silver R.T., Goldman J.M., Stone R.M., Cervantes F., Hochhaus A., Powell B.L., Gabrilove J.L., Rousselot P., et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 15.Milojkovic D., Apperley J. Mechanisms of resistance to imatinib and second-generation tyrosine inhibitors in chronic myeloid leukemia. Clin. Cancer Res. 2009;15:7519–7527. doi: 10.1158/1078-0432.CCR-09-1068. [DOI] [PubMed] [Google Scholar]
- 16.Burchert A. Roots of imatinib resistance: A question of self-renewal? Drug Resist. Updat. 2007;10:152–161. doi: 10.1016/j.drup.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Weisberg E., Griffin J.D. Resistance to imatinib (glivec): Update on clinical mechanisms. Drug Resist. Updat. 2003;6:231–238. doi: 10.1016/s1368-7646(03)00062-1. [DOI] [PubMed] [Google Scholar]
- 18.Bellodi C., Lidonnici M.R., Hamilton A., Helgason G.V., Soliera A.R., Ronchetti M., Galavotti S., Young K.W., Selmi T., Yacobi R., Van Etten R.A., Donato N., Hunter A., Dinsdale D., Tirrò E., et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J. Clin. Invest. 2009;119:1109–1123. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cree I.A., Charlton P. Molecular chess? Hallmarks of anti-cancer drug resistance. BMC Cancer. 2017;17:10. doi: 10.1186/s12885-016-2999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aleksakhina S.N., Kashyap A., Imyanitov E.N. Mechanisms of acquired tumor drug resistance. Biochim. Biophys. Acta Rev. Cancer. 2019;1872:188310. doi: 10.1016/j.bbcan.2019.188310. [DOI] [PubMed] [Google Scholar]
- 21.Aebersold R., Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 22.Monteleone F., Taverna S., Alessandro R., Fontana S. SWATH-MS based quantitative proteomics analysis reveals that curcumin alters the metabolic enzyme profile of CML cells by affecting the activity of miR-22/IPO7/HIF-1α axis. J. Exp. Clin. Cancer Res. 2018;37:170. doi: 10.1186/s13046-018-0843-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong L., Zhang J., Yuan B., Dong X., Jiang X., Wang Y. Global proteome quantification for discovering imatinib-induced perturbation of multiple biological pathways in K562 human chronic myeloid leukemia cells. J. Proteome Res. 2010;9:6007–6015. doi: 10.1021/pr100814y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corrêa S., Pizzatti L., Du Rocher B., Mencalha A., Pinto D., Abdelhay E. A comparative proteomic study identified LRPPRC and MCM7 as putative actors in imatinib mesylate cross-resistance in Lucena cell line. Proteome Sci. 2012;10:23. doi: 10.1186/1477-5956-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrari G., Pastorelli R., Buchi F., Spinelli E., Gozzini A., Bosi A., Santini V. Comparative proteomic analysis of chronic myelogenous leukemia cells: Inside the mechanism of imatinib resistance. J. Proteome Res. 2007;6:367–375. doi: 10.1021/pr0603708. [DOI] [PubMed] [Google Scholar]
- 26.Gjertsen B.T., Wiig H. Investigation of therapy resistance mechanisms in myeloid leukemia by protein profiling of bone marrow extracellular fluid. Expert Rev. Proteomics. 2012;9:595–598. doi: 10.1586/epr.12.55. [DOI] [PubMed] [Google Scholar]
- 27.Hrdinova T., Toman O., Dresler J., Klimentova J., Salovska B., Pajer P., Bartos O., Polivkova V., Linhartova J., Machova Polakova K., Kabickova H., Brodska B., Krijt M., Zivny J., Vyoral D., et al. Exosomes released by imatinib-resistant K562 cells contain specific membrane markers, IFITM3, CD146 and CD36 and increase the survival of imatinib-sensitive cells in the presence of imatinib. Int. J. Oncol. 2021;58:238–250. doi: 10.3892/ijo.2020.5163. [DOI] [PubMed] [Google Scholar]
- 28.Gillet L.C., Navarro P., Tate S., Röst H., Selevsek N., Reiter L., Bonner R., Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.O111.016717. O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins B.C., Hunter C.L., Liu Y., Schilling B., Rosenberger G., Bader S.L., Chan D.W., Gibson B.W., Gingras A.C., Held J.M., Hirayama-Kurogi M., Hou G., Krisp C., Larsen B., Lin L., et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat. Commun. 2017;8:291–312. doi: 10.1038/s41467-017-00249-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai X., Ge W., Yi X., Sun R., Zhu J., Lu C., Sun P., Zhu T., Ruan G., Yuan C., Liang S., Lyu M., Huang S., Zhu Y., Guo T. PulseDIA: Data-independent acquisition mass spectrometry using multi-injection pulsed gas-phase fractionation. J. Proteome Res. 2021;20:279–288. doi: 10.1021/acs.jproteome.0c00381. [DOI] [PubMed] [Google Scholar]
- 31.Guo T., Kouvonen P., Koh C.C., Gillet L.C., Wolski W.E., Röst H.L., Rosenberger G., Collins B.C., Blum L.C., Gillessen S., Joerger M., Jochum W., Aebersold R. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nat. Med. 2015;21:407–413. doi: 10.1038/nm.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao S., Guo T., Koh C.C., Gillessen S., Joerger M., Jochum W., Aebersold R. Minimal sample requirement for highly multiplexed protein quantification in cell lines and tissues by PCT-SWATH mass spectrometry. Proteomics. 2015;15:3711–3721. doi: 10.1002/pmic.201500161. [DOI] [PubMed] [Google Scholar]
- 33.Zhang F., Ge W., Ruan G., Cai X., Guo T. Data-independent acquisition mass spectrometry-based proteomics and software tools: A glimpse in 2020. Proteomics. 2020;20:1900276. doi: 10.1002/pmic.201900276. [DOI] [PubMed] [Google Scholar]
- 34.Bruderer R., Bernhardt O.M., Gandhi T., Miladinović S.M., Cheng L.Y., Messner S., Ehrenberger T., Zanotelli V., Butscheid Y., Escher C., Vitek O., Rinner O., Reiter L. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteomics. 2015;14:1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Demichev V., Messner C.B., Vernardis S.I., Lilley K.S., Ralser M. DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods. 2020;17:41–44. doi: 10.1038/s41592-019-0638-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Searle B.C., Pino L.K., Egertson J.D., Ting Y.S., Lawrence R.T., MacLean B.X., Villén J., MacCoss M.J. Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat. Commun. 2018;9:5128. doi: 10.1038/s41467-018-07454-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rost H.L., Rosenberger G., Navarro P., Gillet L., Miladinović S.M., Schubert O.T., Wolski W., Collins B.C., Malmström J., Malmström L., Aebersold R. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 38.Zhu T., Zhu Y., Xuan Y., Gao H., Cai X., Piersma S.R., Pham T.V., Schelfhorst T., Haas R.R.G.D., Bijnsdorp I.V., Sun R., Yue L., Ruan G., Zhang Q., Hu M., et al. Dphl: A DIA pan-human protein mass spectrometry library for robust biomarker discovery. Genomics Proteomics Bioinformatics. 2020;18:104–119. doi: 10.1016/j.gpb.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu T., Chen H., Yan X., Wu Z., Zhou X., Xiao Q., Ge W., Zhang Q., Xu C., Xu L., Ruan G., Xue Z., Yuan C., Chen G.-B., Guo T. ProteomeExpert: A docker image-based web server for exploring, modeling, visualizing and mining quantitative proteomic datasets. Bioinformatics. 2021;37:273–275. doi: 10.1093/bioinformatics/btaa1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chi H., Liu C., Yang H., Zeng W.F., Wu L., Zhou W.J., Wang R.M., Niu X.N., Ding Y.H., Zhang Y., Wang Z.W., Chen Z.L., Sun R.X., Liu T., Tan G.M., et al. Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat. Biotechnol. 2018;36:1059–1061. doi: 10.1038/nbt.4236. [DOI] [PubMed] [Google Scholar]
- 41.Schubert O.T., Gillet L.C., Collins B.C., Navarro P., Rosenberger G., Wolski W.E., Lam H., Amodei D., Mallick P., MacLean B., Aebersold R. Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat. Protoc. 2015;10:426–441. doi: 10.1038/nprot.2015.015. [DOI] [PubMed] [Google Scholar]
- 42.Di Veroli G.Y., Fornari C., Wang D., Mollard S., Bramhall J.L., Richards F.M., Jodrell D.I. Combenefit: An interactive platform for the analysis and visualization of drug combinations. Bioinformatics. 2016;32:2866–2868. doi: 10.1093/bioinformatics/btw230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar L., Futschik M.E. Mfuzz: A software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jimenez-Marin Á., Collado-Romero M., Ramirez-Boo M., Arce C., Garrido J.J. Biological pathway analysis by ArrayUnlock and ingenuity pathway analysis. BMC Proc. 2009;3 Suppl 4 doi: 10.1186/1753-6561-3-S4-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kramer A., Green J., Pollard J., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Futschik M.E., Carlisle B. Noise-robust soft clustering of gene expression time-course data. J. Bioinform. Comput. Biol. 2005;3:965–988. doi: 10.1142/s0219720005001375. [DOI] [PubMed] [Google Scholar]
- 47.Brandt U. Energy converting NADH:quinone oxidoreductase (complex I) Annu. Rev. Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 48.Sarewicz M., Osyczka A. Electronic connection between the quinone and cytochrome C redox pools and its role in regulation of mitochondrial electron transport and redox signaling. Physiol. Rev. 2015;95:219–243. doi: 10.1152/physrev.00006.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma L.K., Lu J., Bai Y. Mitochondrial respiratory complex I: Structure, function and implication in human diseases. Curr. Med. Chem. 2009;16:1266–1277. doi: 10.2174/092986709787846578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Astuti D., Latif F., Dallol A., Dahia P.L., Douglas F., George E., Sköldberg F., Husebye E.S., Eng C., Maher E.R. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am. J. Hum. Genet. 2001;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Camara A.K.S., Zhou Y., Wen P.C., Tajkhorshid E., Kwok W.M. Mitochondrial VDAC1: A key gatekeeper as potential therapeutic target. Front. Physiol. 2017;8:460. doi: 10.3389/fphys.2017.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathupala S.P., Pedersen P.L. Voltage dependent anion channel-1 (VDAC-1) as an anti-cancer target. Cancer Biol. Ther. 2010;9:1053–1056. doi: 10.4161/cbt.9.12.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pastorino J.G., Hoek J.B. Regulation of hexokinase binding to VDAC. J. Bioenerg. Biomembr. 2008;40:171–182. doi: 10.1007/s10863-008-9148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sokol A.M., Sztolsztener M.E., Wasilewski M., Heinz E., Chacinska A. Mitochondrial protein translocases for survival and wellbeing. FEBS Lett. 2014;588:2484–2495. doi: 10.1016/j.febslet.2014.05.028. [DOI] [PubMed] [Google Scholar]
- 55.Durech M., Trcka F., Man P., Blackburn E.A., Hernychova L., Dvorakova P., Coufalova D., Kavan D., Vojtesek B., Muller P. Novel entropically driven conformation-specific interactions with Tomm34 protein modulate Hsp70 protein folding and ATPase activities. Mol. Cell. Proteomics. 2016;15:1710–1727. doi: 10.1074/mcp.M116.058131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimokawa T., Matsushima S., Tsunoda T., Tahara H., Nakamura Y., Furukawa Y. Identification of TOMM34, which shows elevated expression in the majority of human colon cancers, as a novel drug target. Int. J. Oncol. 2006;29:381–386. [PubMed] [Google Scholar]
- 57.Muller P., Coates P.J., Nenutil R., Trcka F., Hrstka R., Chovanec J., Brychtova V., Vojtesek B. Tomm34 is commonly expressed in epithelial ovarian cancer and associates with tumour type and high FIGO stage. J. Ovarian Res. 2019;12:30. doi: 10.1186/s13048-019-0498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aleskandarany M.A., Negm O.H., Rakha E.A., Ahmed M.A., Nolan C.C., Ball G.R., Caldas C., Green A.R., Tighe P.J., Ellis I.O. TOMM34 expression in early invasive breast cancer: A biomarker associated with poor outcome. Breast Cancer Res. Treat. 2012;136:419–427. doi: 10.1007/s10549-012-2249-4. [DOI] [PubMed] [Google Scholar]
- 59.Matés J.M., Segura J.A., Martín-Rufián M., Campos-Sandoval J.A., Alonso F.J., Márquez J. Glutaminase isoenzymes as key regulators in metabolic and oxidative stress against cancer. Curr. Mol. Med. 2013;13:514–534. doi: 10.2174/1566524011313040005. [DOI] [PubMed] [Google Scholar]
- 60.Wu N., Yang M., Gaur U., Xu H., Yao Y., Li D. alpha-Ketoglutarate: Physiological functions and applications. Biomol. Ther. (Seoul) 2016;24:1–8. doi: 10.4062/biomolther.2015.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dang L., Yen K., Attar E.C. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann. Oncol. 2016;27:599–608. doi: 10.1093/annonc/mdw013. [DOI] [PubMed] [Google Scholar]
- 62.Reitman Z.J., Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: Alterations at a crossroads of cellular metabolism. J. Natl. Cancer Inst. 2010;102:932–941. doi: 10.1093/jnci/djq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Biaglow J.E., Miller R.A. The thioredoxin reductase/thioredoxin system: Novel redox targets for cancer therapy. Cancer Biol. Ther. 2005;4:6–13. doi: 10.4161/cbt.4.1.1434. [DOI] [PubMed] [Google Scholar]
- 64.Quek L., David M.D., Kennedy A., Metzner M., Amatangelo M., Shih A., Stoilova B., Quivoron C., Heiblig M., Willekens C., Saada V., Alsafadi S., Vijayabaskar M.S., Peniket A., Bernard O.A., et al. Clonal heterogeneity of acute myeloid leukemia treated with the IDH2 inhibitor enasidenib. Nat. Med. 2018;24:1167–1177. doi: 10.1038/s41591-018-0115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pisco A.O., Huang S. Non-genetic cancer cell plasticity and therapy-induced stemness in tumour relapse: 'What does not kill me strengthens me'. Br. J. Cancer. 2015;112:1725–1732. doi: 10.1038/bjc.2015.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Piskareva O., Harvey H., Nolan J., Conlon R., Alcock L., Buckley P., Dowling P., Henry M., O'Sullivan F., Bray I., Stallings R.L. The development of cisplatin resistance in neuroblastoma is accompanied by epithelial to mesenchymal transition in vitro. Cancer Lett. 2015;364:142–155. doi: 10.1016/j.canlet.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 67.Navarro P., Kuharev J., Gillet L.C., Bernhardt O.M., MacLean B., Röst H.L., Tate S.A., Tsou C.C., Reiter L., Distler U., Rosenberger G., Perez-Riverol Y., Nesvizhskii A.I., Aebersold R., Tenzer S. A multicenter study benchmarks software tools for label-free proteome quantification. Nat. Biotechnol. 2016;34:1130–1136. doi: 10.1038/nbt.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vellinga T.T., Borovski T., de Boer V.C., Fatrai S., van Schelven S., Trumpi K., Verheem A., Snoeren N., Emmink B.L., Koster J., Rinkes I.H., Kranenburg O. SIRT1/PGC1α-dependent increase in oxidative phosphorylation supports chemotherapy resistance of colon cancer. Clin. Cancer Res. 2015;21:2870–2879. doi: 10.1158/1078-0432.CCR-14-2290. [DOI] [PubMed] [Google Scholar]
- 69.Verdin E., Hirschey M.D., Finley L.W., Haigis M.C. Sirtuin regulation of mitochondria: Energy production, apoptosis, and signaling. Trends Biochem. Sci. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alexa-Stratulat T., Pešić M., Gašparović A.Č., Trougakos I.P., Riganti C. What sustains the multidrug resistance phenotype beyond ABC efflux transporters? Looking beyond the tip of the iceberg. Drug Resist. Updat. 2019;46:100643. doi: 10.1016/j.drup.2019.100643. [DOI] [PubMed] [Google Scholar]
- 71.Parker S.J., Metallo C.M. Metabolic consequences of oncogenic IDH mutations. Pharmacol. Ther. 2015;152:54–62. doi: 10.1016/j.pharmthera.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hedley D.W., Xie S.X., Minden M.D., Choi C.H., Chen H., Ling V. A novel energy dependent mechanism reducing daunorubicin accumulation in acute myeloid leukemia. Leukemia. 1997;11:48–53. doi: 10.1038/sj.leu.2400538. [DOI] [PubMed] [Google Scholar]
- 73.Su Y., Wei W., Robert L., Xue M., Tsoi J., Garcia-Diaz A., Homet Moreno B., Kim J., Ng R.H., Lee J.W., Koya R.C., Comin-Anduix B., Graeber T.G., Ribas A., Heath J.R. Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc. Natl. Acad. Sci. U. S. A. 2017;114:13679–13684. doi: 10.1073/pnas.1712064115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein cluster analysis. The protein cluster from model ADR (A) and IMA (B).The horizontal axis represented the progress of the model (K-native K562 cells, A1/I1-first phase of model ADR/IMA, A2/I2-second phase of model ADR/IMA, A3/I3-third phase of model ADR/IMA). The vertical axis represented changes in proteins expression in each cluster. ADR, adriamycin; IMA, imatinib.
Cell survival analysis with treatment of various concentrations of AGI-6780 to IMA-resistant (A), ADR-resistant (B), and parental K562 cell (C) for 48 h. ADR, adriamycin; IMA, imatinib.
Protein and peptide matrix of quantified proteins in 26 samples by DIA-NN, Spectronaut, EncyclopeDIA, and OpenSWATH. The quantitative protein abundance was log2 converted.
In ADR-resistant cells, the list of continuously changed proteins (adjusted p<0.05) with associated p values. ADR, adriamycin.
In IMA-resistant cells, the list of continuously changed proteins (adjusted p<0.05) with associated p values. IMA, imatinib.
IPA analysis of continuously changed proteins from ADR-resistant cells. ADR, adriamycin; IPA, ingenuity pathway analysis.
IPA analysis of continuously changed proteins from IMA-resistant cells. IMA, imatinib; IPA, ingenuity pathway analysis.
Data Availability Statement
All data are available in the article or the supporting information. The proteomics data are deposited in ProteomeXchange Consortium (Project ID: PXD030553) via the iProX repository partner with the dataset identifier IPX0002882000. The project data analysis codes are deposited in GitHub (https://github.com/guomics-lab/KMDR).