Abstract
Since the beginning of the COVID-19 pandemic, accumulating mutations have led to marked changes in the genetic sequence of SARS-CoV-2. Of these, mutations in the spike (S) protein can alter the properties of the virus, particularly transmissibility and antigenicity. However, it is difficult to detect antigenic variants of the SARS-CoV-2 S protein by immunoassay. Here, we developed an ACE2-based biosensor designed to detect both SARS-CoV-2 S1 mutations and neutralizing antibodies. In “binding mode”, the biosensor works by detecting binding of the S protein to an immobilized ACE2 receptor. The ACE2-based biosensor was able to detect S1 proteins of the alpha (500 pg/mL) and beta variants (10 ng/mL), as well as wild-type S1 (10 ng/mL), of SARS-CoV-2. The biosensor distinguished wild-type SARS-CoV-2 S1 from the S1 alpha and beta variants via color differences. In addition, a slight modification to the protocol enabled the ACE2-based biosensor to operate in “blocking mode” to detect neutralizing antibodies in serum samples from COVID-19 patients. Therefore, the ACE2-based biosensor is a versatile test for detecting wild-type S1, S1 mutants, and neutralizing antibodies against SARS-CoV-2. This approach to targeting both the mechanism by which SARS-CoV-2 enters host cells and the subsequent adaptive immune response will facilitate the development of various biosensors against SARS-CoV-2.
Keywords: COVID-19, SARS-CoV-2, ACE2, Spike mutations, Neutralizing antibodies
Abbreviations: COVID-19, Coronavirus disease; ACE2, Angiotensin-converting enzyme 2; S1, Spike 1
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) first appeared in Wuhan city, China, in 2019 and has since caused a global pandemic of coronavirus disease 2019 (COVID-19) (Wu et al., 2020; Zhou et al., 2020). COVID-19 causes respiratory syndromes ranging from mild to severe, with symptoms such as fever, fatigue, sore throat, diarrhea, and cough (Huang et al., 2020; Liu et al., 2020). Although new RNA, DNA, and recombinant antigen vaccines against COVID-19 have been developed, transmission remains high due to virus mutations (Kyriakidis et al., 2021). Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) is the standard technique for the diagnosis of SARS-CoV-2 (Jung et al., 2020). Various immunoassays, including lateral flow assays and enzyme-linked immunosorbent assays, have been developed to detect viral antigens or antibodies, respectively, specific for SARS-CoV-2 (Kim et al., 2021). However, immunoassays have low sensitivity and do not discriminate different SARS-CoV-2 mutants, or wild-type virus from mutants. Furthermore, gold-standard molecular diagnosis by RT-qPCR is labor-intensive and expensive, and requires specific laboratory equipment (Kilic et al., 2020).
The SARS-CoV-2 genome comprises a single positive-strand RNA molecule that encodes four structural proteins: spike (S), envelope, matrix, and nucleocapsid. Angiotensin-converting enzyme 2 (ACE2), a receptor required for entry of SARS-CoV-2 into host cells, binds to the receptor-binding domain (RBD) of the S protein (Lan et al., 2020; Wang et al., 2020). Therefore, receptor recognition is essential for viral infectivity, pathogenesis, and host cell tropism (Perlman and Netland, 2009; Shang et al., 2020). Previous reports show that the binding affinity of SARS-CoV S protein for ACE2 correlates with virus transmissibility, disease severity, and viral replication rates. Interestingly, several studies show that SARS-CoV-2 has a greater affinity for ACE2 than SARS-CoV, which could (partially) explain the more efficient and frequent transmission of SARS-CoV-2 between humans.
Mutation of SARS-CoV-2 alters its genetic sequence when compared with the reference sequence, Wuhan-Hu1 (Harvey et al., 2021). In particular, mutations in the S protein of SARS-CoV-2 circulating in the population result in amino acid substitutions within certain epitopes, thereby increasing receptor-binding avidity and changing the glycosylation pattern (Walls et al., 2020). Some of these mutations are associated with increased transmissibility because they increase S protein density and enhance the affinity of the new S variants for the ACE2 receptor (Ozono et al., 2021).
A biosensor based on a matched receptor-antibody pair (ACE2 and an anti-SARS-CoV-2 S1 antibody) was previously developed for rapid detection of the SARS-CoV-2 S1 antigen (Lee et al., 2021). The affinity of ACE2 for the wild-type S1 protein of SARS-CoV-2 has been reported (Shang et al., 2020). The affinity of ACE2 for the S1 proteins of the alpha and beta variants has also been measured (Gan et al., 2021; Ozono et al., 2021; Ramanathan et al., 2021), although the results are variable. In addition, other studies used ACE2 to detect neutralizing antibodies specific for SARS-CoV-2 (Fulford et al., 2021; Tan et al., 2020). Taken together, the data suggest that ACE2 and anti-SARS-CoV-2 antibodies could form the basis of a biosensor that can detect both variant S1 proteins and neutralizing antibodies present in serum after vaccination or recovery from COVID-19.
In this study, we developed an ACE2-based biosensor designed to detect both mutations in the S protein and neutralizing antibodies utilizing the binding and blocking mode of ACE2. We examined the interaction between ACE2 and the S1 protein (wild-type S1 and S1 proteins of the alpha and beta variants), as well as the interactions between these S1 proteins and commercially available antibodies specific for wild-type S1. In the binding mode, the data show that the biosensor detects the S1 proteins of both variants, and can distinguish them from the S1 protein of wild-type SARS-CoV-2. In addition, the ACE2-based biosensor to operate in the blocking mode can be used for the detection of neutralizing antibodies in patients with COVID-19.
2. Materials and methods
2.1. Biolayer interferometry (BLI)
Fc-tag tagged human ACE2 (ACE2, Cat. No. 10108-H05H), anti-SARS-CoV-2 spike antibodies [(Commercial Ab No.1, S1-mAb, Cat. No. 40150-R007; and Commercial Ab No.2, S1-pAb, Cat. No. 40591-T62)] were purchased from Sino Biological (Beijing, China). Recombinant SARS-CoV-2 spike antigens (S1 and RBD) and their variants were also purchased from Sino Biological. The list of purchased recombinant antigens is as follows: SARS-CoV-2 spike S1 (Cat. No. 40591-V08H), SARS-CoV-2 RBD (Cat. No. 40592-V08B), SARS-CoV-2 spike S1 alpha variant (B.1.1.7, Cat No. 40591-V08H12), SARS-CoV-2 RBD alpha variant (B.1.1.7, Cat. No. 40592-V08H82), SARS-CoV-2 spike S1 beta variant (B.1.351, Cat. No. 40591-V08H10), and SARS-CoV-2 RBD beta variant (B.1.351, Cat. No. 40592-V08H85). In addition, an anti-SARS-CoV-2 NTD (N-terminal domain) antibody (180E11, Cat. No. ab-P0032) was purchased from Abclon (Seoul, Korea). The affinity/avidity of the target antigens (wild-type and variants) for the ACE2 receptor (or S1-mAb and S1-pAb) was analyzed by BLI using a BLITz instrument (ForteBio, CA, USA). An NTA biosensor (Part No. 18-5102) was purchased from ForteBio. The BLI assay began by hydrating the NTA biosensor tip for 10 min, followed by five steps: initial baseline (30 s), antigen immobilization (300 s), second baseline (120 s), antigen-receptor association (300 s), and dissociation (300 s). After the initial baseline step, each histidine-tagged SARS-CoV-2 antigen (100 μg/mL) was loaded onto the NTA biosensor. The nickel attached to the NTA biosensor tip captures the histidine-tagged SARS-CoV-2. After eliminating unbound antigen through the second baseline step, four different concentrations (100 μg/mL, 50 μg/mL, 25 μg/mL, and 10 μg/mL) of ACE2 (or antibody) were incubated with the antigen coated-biosensor tip, and the strength of the association between antigen and the receptor was measured. All BLI analysis steps, including baseline, association, dissociation, and sample dilution, were performed in sample diluent buffer (0.02% Tween 20, 150 mM NaCl, and 1 mg/mL BSA in 10 mM PBS with 0.05% sodium azide, pH 7.4). Binding constants were calculated from four association-dissociation curves obtained at each ACE2 (or antibody) concentration (based on a 1:1 binding model).
2.2. Preparation of antibody-conjugated cellulose nanobeads (CNB)
Red and blue CNB were purchased from Asahi Kasei Fibers Corporation (NanoAct, Cat. No. RE2AA(Red)/BL1AA(Blue); Miyazaki, Japan); the red and blue CNBs have an average diameter of 340 nm and 325 nm, respectively. A CNB conjugation kit containing conjugation, blocking, and wash buffer was purchased from DCN Diagnostics (Cat. No. CKNB-010, CA, USA). Conjugation of SARS-CoV-2-specific antibodies to the CNBs was performed as recommended by the manufacturer. Briefly, 0.12 mL of antibody (0.5 mg/mL) was mixed with 0.12 mL of conjugation buffer and 0.06 mL of 1% (wt) stock CNB. This mixture was incubated for 2 h at 37 °C. Thereafter, 7.2 mL of a blocking solution was added to the mixture and further incubated for 1 h at 37 °C to block the surface of the CNBs. The residual blocking agent was removed by centrifugation (14,400×g, 20 min, 4 °C), and the pellet was resuspended in 7.2 mL of wash buffer and re-centrifugated (14,400×g, 20 min, 4 °C). The washed CNB pellet was then suspended in 0.1 mL of wash buffer. The concentration of the suspended antibody-conjugated CNB was approximately 0.2%. To minimize signal interference between the test and the control lines, two different colored CNBs were used: red CNB for immobilization of the SARS-CoV-2-specific antibody and blue CNB for immobilization of a chicken IgY antibody (Cat. No. 40200, Bore Da Biotech, Seongnam, Korea). The exact concentrations of red and blue CNBs were calculated by measuring the absorbance maxima at 554 nm and 632 nm, respectively, using UV-vis spectrophotometry (Synergy H1; BioTek).
2.3. Preparation of ACE2-based biosensor
The ACE2-based biosensor, which is based on a lateral flow immunoassay (LFIA), comprises paper-based pads and a membrane, which provides the capillary force that drives sample flow along the LFIA strip. As shown in supplementary figure 1, the ACE2-based biosensor strip comprises a sample pad (Ahlstrom, Helsinki, Finland), a conjugate pad (Ahlstrom, Helsinki, Finland), a nitrocellulose membrane (Advanced Microdevices, Haryana, India), and an absorbent pad (Ahlstrom, Helsinki, Finland). The conjugate pad was pre-treated with 0.1% Triton X-100 (Cat. No. T8787; Sigma-Aldrich, MO, USA) and dried in a 37 °C oven for 4 h. After complete drying, a mixture comprising 0.05% SARS-CoV-2 specific antibody-conjugated CNBs solution (red color) and 0.01% Chicken IgY antibody-conjugated CNB solution (blue color) in stabilizing buffer (10 mM 2-amino-2-methyl-1-propanol (pH 9.0), 0.5% BSA, 0.5% β-Lactose, 0.05% Triton X-100, and 0.05% sodium azide) was sprayed onto the conjugate pad and incubated for 1 h at 37 °C in a vacuum oven (FDU-1200, EYELA, Tokyo, Japan; JSVO-30T, JSR, Gongju, Korea). A line dispenser (BTM Inc., Uiwang, Korea) was used to dispense the test and control lines onto the nitrocellulose membrane under the following conditions: dispensing speed, 50 mm/s; dispensing rate, test line 1 μL/cm; control line 0.5 μL/cm. Next, ACE2 receptor (1 mg/mL; Cat. No. 10108-H05H) in 10 mM sodium carbonate buffer, and a goat anti-chicken IgY antibody (0.5 mg/mL; Cat. No. 23000, Bore Da Biotech, Seongnam, Korea) in 10 mM sodium carbonate buffer were dispensed to form the test and control lines, respectively. Immediately after the lines were dispensed, the nitrocellulose membrane was placed in the vacuum oven and dried for 1 h at 37 °C. Each component of the ACE2-based biosensor was assembled, cut into strips (38 mm wide), and incorporated into a housing prior to testing.
2.4. Sensitivity of the ACE2-based biosensor for detection of recombinant SARS-CoV-2 wild-type and variant S1 proteins
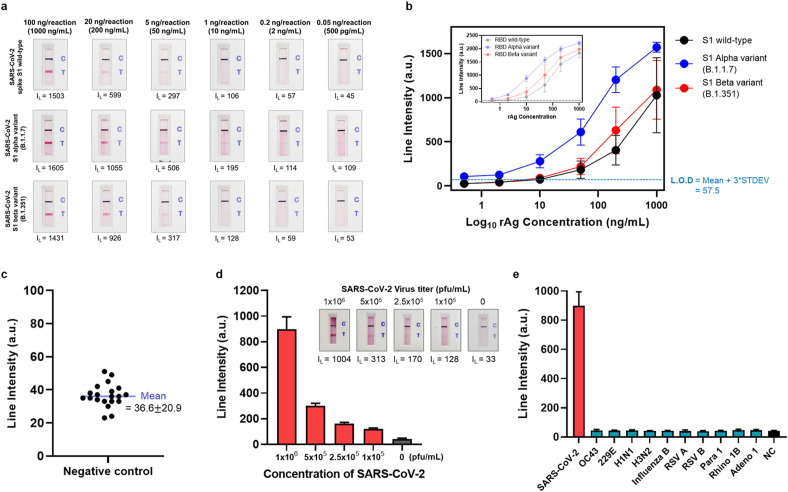
To evaluate the ability of the ACE2-based biosensor to detect recombinant wild-type SARS-CoV-2 S1, and RBD, recombinant SARS-CoV-2 alpha (B.1.1.7) S1 and RBD, and recombinant SARS-CoV-2 beta (B.1.351) S1 and RBD, the antigens were serially diluted and mixed with running buffer [Borex, 5 mM EDTA, 200 mM urea, 1% Triton X-100, 0.5% Tween 20, 500 mM NaCl, 1% PEG (MW 200)]. The final concentration of the serially diluted samples ranged from 1 mg/mL to 500 pg/mL. When 100 μL of running buffer containing the target antigen is loaded into the LFIA device, the target antigen in the sample first encounters the antibody-conjugated CNB, which is pre-immobilized on the conjugate pad. Next, the antigen-antibody-CNB complex is captured by a capture probe (the ACE2 receptor) pre-dispensed onto the nitrocellulose membrane. After 20 min, the appearance of red and blue test and control lines is confirmed with the naked eye. In addition, the line intensity was semi-quantified by a portable line analyzer (Light-G; WellsBio, Seoul, Korea). The mean values and standard deviations from three replicates were obtained; the limit of detection (L.O.D; see Fig. 3) was obtained by adding three times the standard deviation to the mean value of the negative control.
Fig. 3.
Sensitivity and specificity of the ACE2-based biosensor a) Sensitivity of the ACE2-based biosensor for the wild-type SARS-CoV-2 S1 antigen and the alpha and beta variants. Serially diluted samples containing S1 wild-type, S1 alpha variant, and S1 beta variant (concentration: 1000 ng/mL–500 pg/mL) were tested. Twenty minutes after sample loading, the test and control lines were photographed with a smartphone, and the line intensities were measured by a portable line analyzer. b) Sensitivity analysis results for the wild-type SARS-CoV-2 S1 antigen (black line) and the alpha (blue line) and beta variants (red line). Inset) Sensitivity analysis results for the SARS-CoV-2 RBD. The limit of detection (L.O.D) is determined as the mean value of the negative control plus three times the standard deviation. c) The line intensity of 20 negative control samples was also measured. The negative control samples do not contain target antigen (i.e., buffer only). d) Results of sensitivity analysis of live SARS-CoV-2. The live virus was serially diluted (106 pfu/mL–105 pfu/mL) and tested. e) Specificity analysis using human coronaviruses (OC32 and 229E), and other respiratory viruses [human influenza A virus H1N1 (H1N1), human influenza A virus H3N2 (H3N2), human influenza B virus (Influenza B), human respiratory syncytial virus A (RSV A), human respiratory syncytial virus B (RSV B), human parainfluenza virus 1 (Para 1), human rhinovirus 1B (Rhino 1B), and human adenovirus 1 (Adeno 1)].
2.5. SARS-CoV-2 culture and quantification
Vero (African green monkey, kidney) cells were incubated at 37 °C/5% CO2 in Dulbecco's modified Eagle's medium (Hyclone) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin (P/S; ThermoFisher Scientific). The SARS-CoV-2 (Korea/KCDC-03/2020) used for the infection experiments was provided by the Korea Centers for Disease Control and Prevention. SARS-CoV-2 was propagated and amplified in Vero cells for 3 days (three to five passages). The titer of the purified SARS-CoV-2 was determined in a plaque-forming assay. All experiments were performed in a Biosafety level 3 facility.
2.6. Specificity of the ACE2-based biosensor
For the specificity tests, human coronaviruses OC43 (KBPV-VR-8) and 229E (KBPV-VR-9), and eight respiratory viruses that cause clinical symptoms similar to those of SARS-CoV-2 [i.e., human influenza A virus H1N1 (KBPV-VR-33), human influenza A virus H3N2 (KBPV-VR-32), human influenza B virus (KBPV-VR-34), human respiratory syncytial virus A (KBPV-VR-41), human respiratory syncytial virus B (KBPV-VR-42), human parainfluenza virus 1 (KBPV-VR-44), human rhinovirus 1B (KBPV-VR-81), and human adenovirus 1 (KBPV-VR-1)] were purchased from the Korea Bank for Pathogenic Viruses (KBPV, Seoul, Korea). The virus samples were diluted to 106 pfu/mL in running buffer (except human influenza A viruses H1N1 and H3N2 and human respiratory syncytial virus B; the concentration of these samples were adjusted to 105 pfu/mL because the initial concentration of the samples was lower than 106 pfu/mL)). Similar to the sensitivity analysis, a 100 μL sample was loaded onto the biosensor. Twenty minutes after initiation of sample flow, line intensities were confirmed by the naked eye and by the portable line analyzer.
2.7. Detection of SARS-CoV-2 wild-type and variant S1 antigens
The anti-SARS-CoV-2 NTD antibody 180E11 (180E11) recognizes the N-terminal region of SARS-CoV-2. The affinity of 180E11 for the S1 wild-type was significantly higher than that of the S1 beta variant. To take advantage of these characteristics, we designed the new ACE2-based biosensor such that it can distinguish SARS-CoV-2 S1 variants from the wild-type S1 antigen. To detect both the wild-type and variant antigens, an anti-S1-mAb was conjugated to red CNB. The 180E11 antibody was conjugated to blue CNB to detect only the wild-type S1 antigen. The two CNBs were then co-sprayed onto the conjugate pad at a 5:1 (red:blue) (v/v) ratio. The wild-type and each SARS-CoV-2 S1 variant (alpha and beta) were serially diluted from 1000 ng/mL to 20 ng/mL and loaded onto the cartridge. The line intensity and color were evaluated after 20 min. The line intensity was measured by the portable line analyzer, and the RGB composition of the test lines was analyzed using software (ImageJ, NIH, MD, United States) and an image analyzer (Sapphire Biomolecular Imager, Azure Biosystems, CA, USA).
2.8. Competitive assay for detecting neutralizing antibodies
Addition of SARS-CoV-2 RBD to the running buffer alters the detection characteristics of the ACE2-based biosensor from S1 detection (binding mode) to neutralizing antibody detection (blocking mode). Four different RBD concentrations (100 ng, 50 ng, 20 ng, and 10 ng) were tested to identify the optimum concentration of the best L.O.D. A SARS-CoV-2 spike neutralizing antibody (Cat. No. 40591-MM43, Sino Biological) was used as a standard neutralizing antibody; three concentrations of this neutralizing antibody (10 μg/mL, 2 μg/mL, and 500 ng/mL) were tested. After the sample flow, the intensity of each line was normalized against that in the absence of the neutralizing antibody. Based on the optimization results, 20 ng of RBD was chosen for the detection of neutralizing antibodies. Neutralizing antibodies (serially diluted from 5 μg/mL to 100 ng/mL) in RBD-containing running buffer were loaded onto the biosensor. Twenty minutes after loading, the line intensities were measured by a portable line analyzer. To evaluate the ability of the biosensor to detect neutralizing antibodies in patient serum, serum samples from COVID-19 patients (n = 10) and healthy individuals (n = 5) were applied. The clinical specimens from COVID-19 patients and healthy donors were kindly provided by Chungbuk National University Hospital (Korea). Detailed information about the COVID-19 patients and healthy donors is presented in Supplementary Table 1. Ten microliters of serum from COVID-19 patients or healthy donors were mixed in running buffer at a 1:9 (v/v) ratio and then loaded onto the biosensor. After 20 min, the reduction in line intensity for COVID-19 samples compared with that for healthy samples was confirmed by both the naked eye and by a portable line analyzer.
3. Results and discussion
3.1. Binding and blocking modes of the biosensor
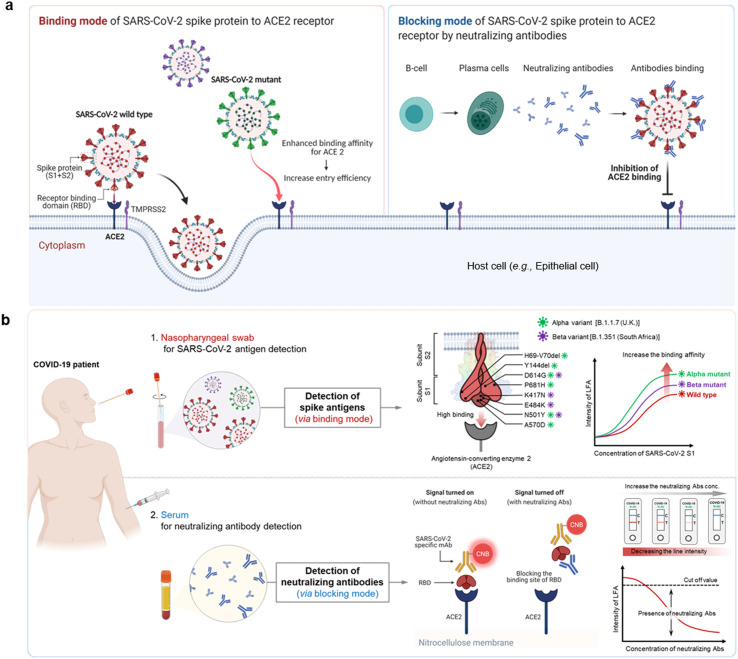
ACE2, a type I membrane protein expressed in the lungs, heart, kidneys, and intestine, is a functional receptor for SARS-CoV-2 (Yan et al., 2020). During the initial phase of viral infection, the transmembrane S glycoprotein is cleaved into S1 and S2 subunits; the RBD of the S1 subunit binds directly to ACE2 (Walls et al., 2020; Wang et al., 2020). In addition, because the binding of SARS-CoV-2 to ACE2 is a major target for neutralizing antibodies during infection, it is attracting attention as a significant target for vaccination and antiviral strategies (Walls et al., 2020).
Previous studies examined the interactions between ACE2 and the SARS-CoV-2 RBD to better understand the entry process and to develop antivirals, including therapeutic antibodies and small molecules, against SARS-CoV-2 (Izda et al., 2021; Taylor et al., 2021). ACE2 plays an essential role in the entry of SARS-CoV-2 into host cells. During infection, ACE2 binds viruses prior to cell entry (binding mode); however, during the subsequent immune response, binding of the virus to ACE2 is blocked by neutralizing antibodies produced by B cells (blocking mode) (Fig. 1 ). The S protein of SARS-CoV-2 binds to ACE2 and TMPRSS2 to initiate viral infection of cells lining the host upper respiratory tract. Some S proteins of SARS-CoV-2 variants show enhanced binding to ACE2, which is associated with increased transmissibility (Fig. 1a). However, infection activates the adaptive immune response so that B cells produce neutralizing antibodies targeting the RBD of the SARS-CoV-2 S protein; these antibodies block the binding of S proteins to ACE2 on the surface of the host cells (Fig. 1b). These two modes of action mean that we can design a versatile biosensor that not only detects SARS-CoV-2 wild-type and variant S1, but also neutralizing antibodies (Fig. 1b).
Fig. 1.
The binding and blocking modes of the SARS-CoV-2 the ACE receptor. a) Schematic illustration showing the mechanism by which SARS-CoV-2 enters cells via the ACE2 receptor (left), and the adaptive immune response generated in response (right). The receptor-binding domain (RBD) of the SARS-CoV-2 spike protein binds to ACE2 and RMPRSS2, thereby allowing host cell entry and infection. Mutations in the SARS-Cov-2 spike protein increase its affinity for ACE2, thereby increasing the efficiency with which the virus enters cells (binding mode). Infection with COVID-19 activates the adaptive immune response, during which B cells generate neutralizing antibodies that bind to the spike protein, inhibit its interaction with ACE2, and block entry to the cell (blocking mode). b) The ACE2-based biosensor can detect both viral antigens (upper) and neutralizing antibodies (lower). Nasopharyngeal swabs from COVID-19 patients contain SARS-CoV-2. Mutations in the SARS-CoV-2 spike antigen increase affinity for ACE2; therefore, ACE2-based biosensors could be an effective diagnostic platform in situations where SARS-CoV-2 variants are proliferating. The ACE2-based biosensor can also detect neutralizing antibodies in serum from COVID-19 patients. By simply adding SARS-CoV-2 RBD to the running buffer, the detection mode of the ACE2-based biosensor changes from binding to blocking. Neutralizing antibodies block the interaction between RBD and ACE2, resulting in a dose-dependent decrease in test line intensity.
3.2. Effects of SARS-CoV-2 mutations on binding affinity for the ACE2 receptor (or antibodies)
Since the beginning of the COVID-19 pandemic, SARS-CoV-2 has accumulated mutations. Of these, mutations in the S protein are of particular importance because they affect virus transmission and vaccine efficacy. Recent studies show an increase in the efficiency with which the virus enters cells; mutations in the S protein of SARS-Cov-2 have increased its affinity for the ACE2 receptor (Cheng et al., 2021; Li et al., 2021; Luan et al., 2021; Ozono et al., 2021), particularly the D614G and N501Y mutations are hotspots in both the alpha and beta variants.
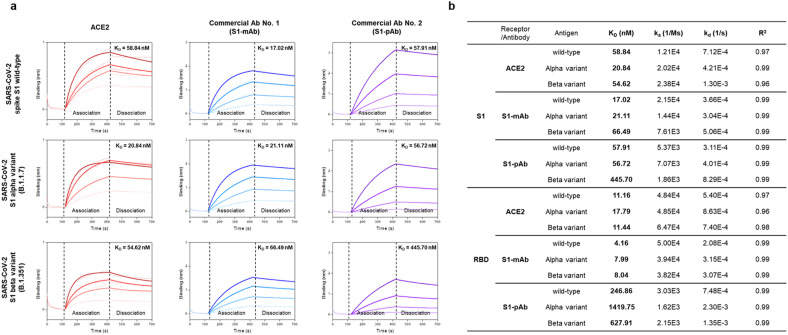
The BLI measures biomolecular interactions based on differences in the interference pattern resulting from binding events. To evaluate the binding affinity between ACE2 and wild-type and variant SARS-CoV-2 S antigens, we measured the affinity of ACE2 for six different recombinant SARS-CoV-2 S antigens: wild-type SARS-CoV-2 S1 and its RBD, the alpha variant (B.1.1.7) of SARS-CoV-2 S1 and its RBD, and the beta variant (B.1.351) of SARS-CoV-2 S1 and its RBD. The recombinant SARS-CoV-2 S1 alpha variant (B.1.1.7) harbors three deletion and four point mutations (deletions: H69del, V70del, and Y144del; point mutations: N501Y, A570D, D614G, and P681H), whereas the RBD alpha variant harbors a single point mutation (N501Y). Moreover, the recombinant SARS-CoV-2 S1 beta variant (B.1.351) harbors four point mutations (K417N, E484K, N501Y, and D614G), and the RBD beta variant harbors three point mutations (K417N, E484K, and N501Y). In addition, the affinity/aviditiy of two commercial antibodies (S1-mAb and S1-pAb) for the SARS-CoV-2 S antigens was measured by BLI.
Each recombinant SARS-CoV-2 S1 antigen was immobilized onto the NTA biosensor via specific interaction between nickel and the histidine-tag. Next, four different concentrations of ACE2 receptor (or S1-mAb and S1-pAb antibodies) were added, and the binding affinity/avidity was evaluated. Representative real-time binding sensorgrams (dotted lines) and their fitted curves (solid line) for the receptor (or antibody)-antigen interaction are shown in Fig. 2 a and in Supplementary Figure 2. The kinetic constants were calculated from these four curves based on a 1:1 binding model (Fig. 2b). The KD values of ACE2 for the wild-type S1 and RBD were 58.84 and 11.16 nM, respectively. Due to the fact that the RBD is the key functional component within the S1 subunit and is responsible for the binding of SARS-CoV-2 to ACE2 (Wang et al., 2020), the KD value of ACE2 for the RBD was lower than that for S1.
Fig. 2.
Affinity/avidity of ACE2 and commercial antibodies (S1-mAb and S1-pAb) for the wild-type SARS-CoV-2 antigens and its variants, as assessed by biolayer interferometry (BLI). a) Association and dissociation curves for the interaction between SARS-CoV-2 S1 antigens (wild-type and variants) and the ACE2 receptor (or commercial antibodies). Dotted lines represent the response curves. Solid lines represent the fitted curves. b) The kinetic constants for the interactions between ACE2 (or commercial antibodies) and SARS-CoV-2 spike antigens. Binding constants were calculated from the fitted curves using a 1:1 binding model.
On the other hand, the affinity of ACE2 for the S1 subunit and RBD of the alpha and beta variants was similar or slightly higher than that of ACE2 and wild-type S1 and RBD (Fig. 2b). Notably, the KD value for ACE2 binding to the alpha variant of the S1 antigen was 20.84 nM, which is significantly lower than that for the wild-type S1 antigen (58.84 nM). These results are consistent with recently reported findings showing that mutations in the S antigen increase the strength of ACE2 binding (Cheng et al., 2021; Li et al., 2021; Luan et al., 2021; Ozono et al., 2021). For example, the D614G mutation confers the S protein with structural flexibility (Ozono et al., 2021), while N501Y forms two hydrogen bonds with ACE2, making the S1 antigen more stable when bound to ACE2 (Cheng et al., 2021). Lower KD values indicate a higher binding affinity for ACE2, which reflects better cell entry by SARS-CoV-2, as well as increased viral transmission rates.
The binding affinity/avidity of two different commercial antibodies (S1-mAb and S1-pAb) for SARS-CoV-2 wild-type antigens and variants were also evaluated. The KD values of the S1-mAb for S1 wild-type, the S1 alpha variant, and the S1 beta variant were 17.02 nM, 21.11 nM, and 66.49 nM, respectively. In addition, the KD values of S1-pAb for S1 wild-type, the S1 alpha variant, and the S1 beta variant were 57.91 nM, 56.72 nM, and 445.70 nM, respectively. The KD values of the commercial antibodies for the alpha variant S1 were similar to those for the wild-type S1; however, the binding affinity/avidity for the beta variant S1 were significantly lower (approximately 3.9- and 7.7-fold lower for S1-mAb and S1-pAb, respectively) than for wild-type S1.
Furthermore, we performed additional binding affinity/avidity analysis between various antigens (S1 wild-type, S1 alpha variant, S1 beta variant) and receptors such as ACE2 and antibodies using SPR utilizing EDC/NHS coupling chemistry for protein immobilization. Biolayer interferometry (BLI) Blitz is one of the powerful instruments in a small laboratory. It, however, has several disadvantages, including analyte rebinding (no flow-through system) during dissociation phases. In particular, this problem may occur imprecise measurement of affinity between protein and protein interaction with a low concentration of ligand protein. On the other hand, SPR is more precise than BLI on these issues due to the flow-through system. Most of the affinities/avidities (KD) tendency (Supplemental Figure 3) between each receptor (ACE2, S1-mAb, and S1-pAb) and each antigen (S1 wild-type, S1 alpha variant, S1 beta variant) were similar to previous results using BLI. The KD values of ACE2 for the S1 wild-type, S1 alpha variant, and S1 beta variant were 0.56 nM, 0.07 nM, and 0.05 nM, respectively. The affinity of ACE2 for the alpha and beta variants was clearly higher than that of the S1 wild-type, confirming once again that it was caused by the mutation of the S antigen that increases the strength of ACE2 binding. On the other hand, the binding affinity/avidity of two different commercial antibodies (S1-mAb and S1-pAb) to S1 alpha and beta variants were lower than that of the S1 wild-type. These results were also similar to those of previous BLI analyses. Detailed materials and methods for SPR experiments were described in Supplementary information.
The ability of each antibody to bind to its target antigen depends on the antigenic determinant that first triggers antibody production. Unfortunately, most SARS-CoV-2 virus has accumulated mutations. Thus, the antigenic determinants in the mutant that trigger antibody production inevitably differ from the wild-type, and these differences have made the virus more transmissible and, in some cases, better able to evade vaccines (at least partially). As shown in Fig. 2, all the alpha and beta variants showed a reduced affinity/avidity for commercial antibodies, even though there were some differences. Since mutations accumulate rapidly, and information about the paratope, also known as the antigen-binding site, is insufficient, it is difficult to speculate how the binding affinity of the antibody for the mutant antigen will change. However, an important fact that does not change is that the antibody's ability to detect a mutant antigen decreases as the number of mutations increases. SARS-CoV-2 mutants show increased affinity for the ACE2 receptor, which has increased their infectivity and transmissibility. Therefore, a strategy based on using the ACE2 receptor as a detection probe, rather than antibodies, may be more effective for diagnosing COVID-19 in the future.
3.3. Detection of SARS-CoV-2 S1 antigens
For highly sensitive and selective detection of disease-specific antigens, two types of probes (capture and detection) that recognize different regions of the target antigen are essential. Using ACE2 as a diagnostic probe has significant benefits with respect to preventing the spread of the SARS-CoV-2, especially SARS-CoV-2 variants. In addition, another diagnostic antibody that forms a sandwich pair with ACE2, and is less affected by antigen mutations, is also critical for the diagnostic performance of the biosensor. Previously, we described an LFIA device based on a matched sandwich pair comprising ACE2 and a commercial antibody (S1-mAb) specific for SARS-CoV-2; this pairing allowed detection of SARS-CoV-2 S1 antigen in clinical samples from COVID-19 patients (Lee et al., 2021). To improve the diagnostic performance of the biosensor for detection of SARS-CoV-2 variants (alpha and beta), we optimized it with respect to buffer composition and membrane treatment conditions (see Materials and Methods). In addition, the signal molecules for test and control lines were separated into different colors to minimize the effect on the line intensity of the control line by that of the test line. The detection antibody (S1-mAb) used for the test line was conjugated to red CNB, and the chicken IgY antibody used for the control line was conjugated to blue CNB. As with the previously reported LFIA device, the ACE2 receptor was immobilized onto a nitrocellulose membrane to form the test line; however, the new biosensor reported herein used the anti-chicken IgY antibody to generate the control line.
The sensitivity of the ACE2-based biosensor was evaluated using serially diluted samples of recombinant SARS-CoV-2 S antigens (S1 and RBD) and their alpha and beta variants (concentration range: 1000 ng/mL to 500 pg/mL), as shown in Fig. 3 a and b, and in Supplementary Figure 4. Twenty minutes after sample loading, the results were photographed using a smartphone, and the line intensities were measured by a portable line analyzer. Both the S1 and RBD of the SARS-CoV-2 alpha variant showed markedly stronger line intensities than the wild-type antigens at the same concentration (an approximately 2-fold increase). By contrast, the line intensities generated by the beta variant were slightly stronger than those of the wild-type. Considering the BLI results for the binding affinity between ACE2 and several SARS-CoV-2 antigens, the increased line intensities in the detection of alpha variants are attributed to the enhanced binding affinity of ACE2 to the SARS-CoV-2 variants. Thus, as we mentioned previously, the ACE2-based biosensor would be more effective in the circumstance of spreading the mutant SARS-CoV-2. The L.O.D was determined as the mean value of 20 negative controls (buffer only, 0 ng/mL of antigen) plus three times the standard deviation (Fig. 3c). Based on this, the L.O.D for detecting S1 wild-type, the S1 alpha variant, and the S1 beta variant was 10 ng/mL, 500 pg/mL, and 10 ng/mL, respectively. In addition, for all SARS-CoV-2 antigens, the detection signal was stronger for the RBD than for S1, and the L.O.D was slightly better (RBD wild-type, 2 ng/mL; RBD alpha mutant, 500 pg/mL; and RBD beta variant, 2 ng/mL). The increased sensitivity for the RBD might be associated with the lower KD value of the ACE2 S1-mAb for the RBD.
Cultured virus and clinical samples were used to assess the clinical performance of the ACE2-based biosensor. SARS-CoV-2 was propagated and amplified in Vero cells for 3 days (three to five passages), and titers were determined in a plaque-forming assay. The L.O.D. of the ACE2-based biosensor was investigated using serially diluted live SARS-CoV-2 at concentrations ranging from 1 × 106 pfu/mL to 1 × 105 pfu/mL. Twenty minutes after sample loading, the test and control lines were photographed with a smartphone, and the line intensities were measured using a portable line analyzer (Fig. 3d). The ACE2-based biosensor successfully detected live SARS-CoV-2 at 1 × 105 pfu/mL. Furthermore, we tested three nasopharyngeal (NP) swabs from COVID-19 patients and seven NP swabs from healthy donors to evaluate clinical performance (Supplementary Figure 5). The ACE2-based biosensor successfully detected SARS-CoV-2 S1 antigens in clinical specimens of all three COVID-19 patients; there were no positive signals for the NP swabs from healthy patients. Although the number of clinical samples was small, the results confirm that the ACE2-based biosensor is capable of detecting SARS-CoV-2 S antigens in COVID-19 clinical specimens, as demonstrated in our previous study.
We also performed specificity tests to examine cross-reactivity with other human coronaviruses (OC43 and 229E), as wells as other respiratory viruses that cause clinical symptoms similar to those of SARS-CoV-2. We tested human influenza A virus H1N1 (H1N1), human influenza A virus H3N2 (H3N2), human influenza B virus (influenza B), human respiratory syncytial virus A (RSV A), human respiratory syncytial virus B (RSV B), human parainfluenza virus 1 (Para 1), human rhinovirus 1B (Rhino 1B), and human adenovirus 1 (Adeno 1). Despite testing these viruses at high concentrations (106 pfu/mL or 105 pfu/mL), there was no cross-reactivity (Fig. 3e and Supplementary Figure 6). These results indicate that the ACE2-based biosensor is specific for the SARS-CoV-2 S antigen.
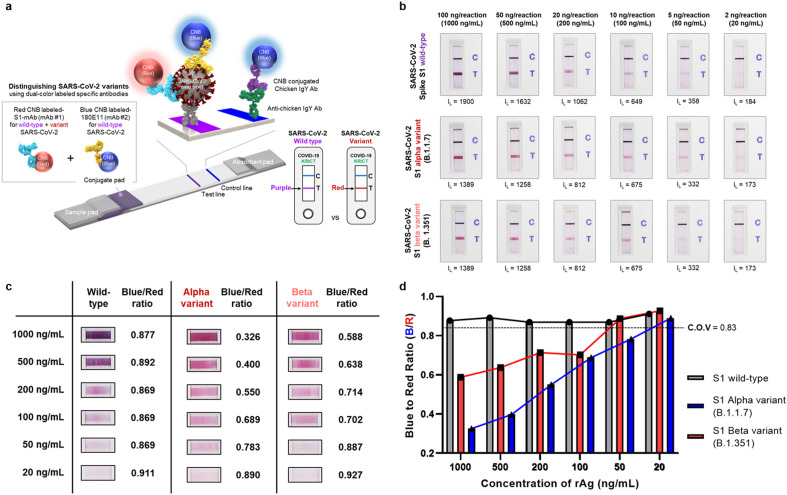
3.4. The ACE2-based biosensor can distinguish SARS-CoV-2 S1 variant antigens from wild-type antigens
Distinguishing between wild-type SARS-CoV-2 and its variants could provide crucial information for vaccine development and help to establish strategies aimed at preventing the spread of COVID-19. Fig. 4 a illustrates the construction and principles underlying the advanced ACE2 biosensor. Previously, we used a single specific antibody as a probe (i.e., S1-mAb) to detect SARS-CoV-2 S antigen (both wild-type and variant); however, in this study, we introduced a second S-specific antibody as a detection probe. This antibody (180E11) was generated using the N-terminal domain of wild-type SARS-CoV-2 as the antigenic determinant. As shown in Supplementary Figure 7a, it has a high affinity (KD = 18.17 nM) for the wild-type SARS-CoV-2 S1 antigen. Interestingly, it has no affinity for the SARS-CoV-2 S1 variants (Supplementary Figure 7b). SPR analysis was further performed to clarify the binding affinity of 180E11 for the S1 variant. SPR results showed that the 180E11 antibody had an affinity for both S1 wild-type (KD = 0.66 nM) and S1 beta variants (KD = 21.27 nM) (Supplementary Figure 8). However, the affinity of 180E11 for S1 wild-type was significantly higher than that of beta spike S1 (approximately 32-fold). We speculate that mutations in the S1 protein caused structural and sequence changes that weakened binding by 180E11. In addition, the reason 180E11 did not show an affinity for the SARS-CoV-2 S1 beta variant in BLI experiments might be caused by an imprecise reaction of His-tagged protein on the nickel-charged tri-nitriloacetic acid (Tris-NTA) biosensor.
Fig. 4.
Distinguishing SARS-CoV-2 wild-type antigens from variant antigens. a) Schematic illustration showing the use of the ACE2-based biosensor to distinguish SARS-CoV-2 wild-type antigens from the variants. Two color-labeled SARS-CoV2-specific antibodies were co-deposited onto the conjugate pad. A specific antibody labeled with red CNB detects both wild-type S1 and the S1 variants; however, a second specific antibody labeled with blue CNB detects only wild-type S1. Therefore, the color of the test line depends on whether wild-type S1 (purple) or an S1 variant (red) is detected. b) The results of the tests. Performance was evaluated using SARS-CoV-2 S1 antigens (wild-type, alpha variant, and beta variant) serially diluted from 1000 ng/mL to 20 ng/mL c) Representative images of test lines and the blue-to-red ratios. At the reaction end-point (after 20 min), the test lines were analyzed using commercial software (ImageJ) to examine the RGB composition. d) Bar graph showing the blue-to-red ratio obtained from the serially diluted sample tests. The cut-off-value (C.O.V) was determined as the mean value of the blue-to-red ratio for the wild-type S1 minus three times the standard deviation.
As mentioned above, each detection antibody was immobilized to CNB (either red or blue) and co-deposited onto the conjugate pad. Detection of wild-type SARS-CoV-2 or its variant S antigens yields a different colored line. The test line is purple when a sample contains the wild-type S1 because both the red CNB-labeled S1-mAb and the blue CNB-labeled 180E11 antibody bind to the target antigen. However, if the variant SARS-CoV-2 S antigen is present, the test line is red because the antigen is recognized only by the S1-mAB labeled with red CNB. It could be concluded that the reason why the blue CNB-labeled 180E11 antibody could not recognize the S1 variant was due to the difference in affinity between S1-mAb (red CNB-labeled) and 180E11 (blue CNB-labeled) for the S1 variant (approximately 18-fold). We assumed that this affinity difference induces S1-mAb to preferentially bind to the variant spike S1. Furthermore, appropriate combinations of sandwich pairs, such as ACE2-180E11 for wild-type antigens and ACE2-(S1-mAb) for both wild-type and variant antigens, contributed to distinguishing SARS-CoV-2 variant antigens from wild-type antigen.
We evaluated the ability of the advanced biosensor to distinguish between wild-type and variant S antigens by serially diluting the antigens (wild-type, alpha variant, and beta variant) from 1000 ng/mL to 20 ng/mL. These samples were loaded onto the biosensor, and the color and intensity of the lines were evaluated after 20 min (Fig. 4b). Again, the line intensity was measured by a portable line analyzer (Supplementary Figure 9), and the RGB composition of the test lines was analyzed by an image analyzer and commercial software (ImageJ). At high antigen concentrations (up to 200 ng/mL), the color of the line denoting S1 wild-type and S1 mutants could be distinguished even with the naked eye. In addition, the blue-to-red ratio of the test lines was examined (Fig. 4c and d). When red and blue CNB co-existed (i.e., detection of wild-type S1), the blue-to-red ratio was approximately 0.9. However, when only variant S1 was present, the blue-to-red ratio was lower. The cut-off-value (C.O.V) was determined as the mean value of the blue-to-red ratio for wild-type S1 detection minus three times the standard deviation. Based on this, it was possible to reliably distinguish SARS-CoV-2 S1 variants from wild-type S1, even at low concentrations (C.O.V for the alpha variant = 50 ng/mL; C.O.V for the beta variant, 100 ng/mL).
3.5. Detection of neutralizing antibodies
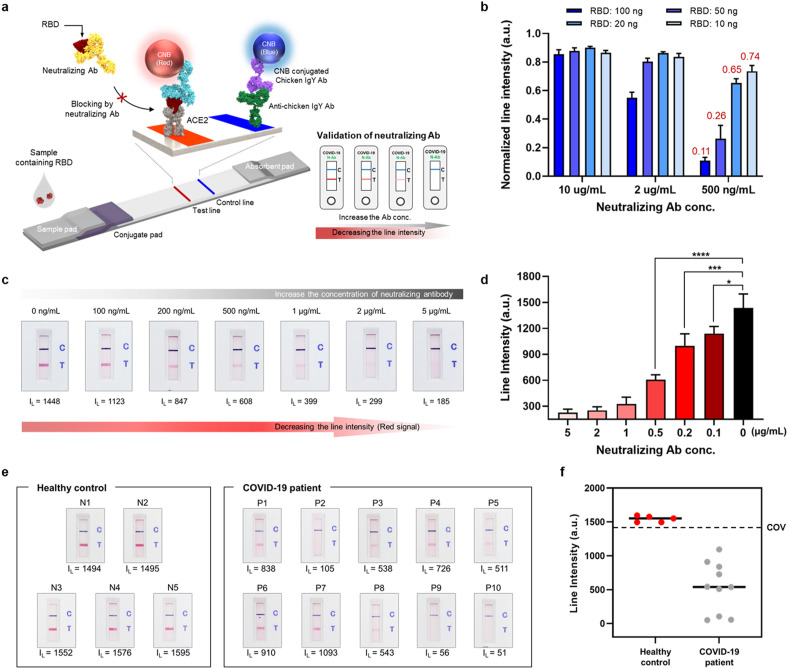
Fig. 5a illustrates how the ACE2-based biosensor detects neutralizing antibodies. By simply adding SARS-CoV-2 S RBD to the running buffer, the mode of detection used by the ACE2-based biosensor changes from binding to blocking. Pre-loaded RBD is responsible for the target antigen of the competitive reaction between neutralizing antibodies and ACE2 (or S1-mAb). In the absence of neutralizing antibodies, the pre-loaded RBD was detected by ACE2 and CNB-labeled S1-mAb, resulting in a clear red test line. By contrast, when neutralizing antibodies are present, the intensity of the test line decreases in a dose-dependent manner by blocking the interaction between RBD and the ACE2.
Fig. 5.
Use of the ACE2-based biosensor to detect neutralizing antibodies a) Schematic illustration showing detection of neutralizing antibodies by the ACE2-based biosensor. By adding the SARS-CoV-2 RBD antigen into the running buffer, the ACE2-based biosensor changes from detecting SARS-Cov-2 spike antigens (binding mode) to detecting neutralizing antibodies (blocking mode). When neutralizing antibodies are present in a sample, the line intensity decreases in a dose-dependent manner. b) Detection of neutralizing antibodies depends on the amount of loaded RBD. c) Sensitivity of the ACE2-based biosensor for neutralizing antibodies. Serially diluted neutralizing antibodies (5 μg/mL to 100 ng/mL) were tested. d) Bar graph showing the intensity of the test lines. (P-values: ns > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001). e) Ability of the ACE2-based biosensor to detect neutralizing antibodies. Clinical samples from COVID-19 patients (n = 10) and healthy controls (n = 5) were tested. All samples from COVID-19 patients showed a clear reduction in line intensity; no decrease was observed for healthy controls. f) Results presented as a dot graph. The cut-off-value (C.O.V) was determined as the mean value of the line intensity for healthy controls (n = 5) minus three times the standard deviation.
At first, the amount of pre-loaded RBD was optimized to increase the sensitivity and dynamic range of the biosensor (Fig. 5b, and Supplementary Figures 10 and 11). A large amount of RBD (50 ng) resulted in a clear test line, but limited sensitivity. By contrast, a small amount of RBD (10 ng) improved the sensitivity, but the intensity of the test line was too weak for assessment with the naked eye. Fig. 5b shows the normalized line intensity after the amount of RBD had been optimized. The closer the line intensity value is to 1, the better the inhibitory effect of the neutralizing antibody on the ACE2-RBD interaction. The optimal amount of RBD was 20 ng. The sensitivity of the biosensor was tested using serially diluted samples of commercially available neutralizing antibodies. These neutralizing antibodies (serially diluted from 5 μg/mL to 100 ng/mL) in RBD (20 ng) containing running buffer were loaded onto the ACE2-based biosensor, and the test and control lines were photographed with a smartphone 20 min later. In addition, line intensities were measured by a portable line analyzer. The ACE2-based biosensor detected neutralizing antibodies, even at a low concentration (approximately 100 ng/mL) (Fig. 5c and d).
Finally, the biosensor was tested using clinical serum samples from COVID-19 patients (n = 10) and healthy controls (n = 5). Detailed information about the patients and healthy controls is provided in Supplementary Table 1. Titers of virus-specific IgG and IgM antibodies increase during the first 3 weeks after symptom onset; in particular, the proportion of patients with virus-specific IgG reaches 100% approximately 17–19 days after symptom onset (Long et al., 2020). In this experiment, serum samples were collected 1–3 weeks after hospitalization. In all tests, the line intensity generated from sera of COVID-19 patients was significantly lower than in the absence of neutralizing antibodies (i.e., buffer only). Moreover, serum from healthy controls did not reduce the test line intensity (Fig. 5e and f). These results indicate that the ACE2-based biosensor can detect neutralizing antibodies in clinical specimens without any cross-reactivity. Production of neutralizing antibodies specific for SARS-CoV-2 is induced either by natural infection or vaccination. Therefore, we expect the ACE2-based biosensor will detect neutralizing antibodies in both naturally infected COVID-19 patients as well as in vaccinated individuals.
4. Conclusions
SARS-CoV-2 has continued to evolve via mutation of its gene and protein sequences. These mutations alter the properties of the virus, particularly transmissibility and antigenicity. Detection of S antigen variants is important, particularly with respect to vaccination status and quarantine. Here, we developed an ACE2-based biosensor to detect S1 mutants and neutralizing antibodies specific for SARS-CoV-2. The dual binding modes of the ACE2-based biosensor mean that it can distinguish S1 antigen variants from wild-type S1, as well detecting neutralizing antibodies in clinical samples from COVID-19 patients. The approach used herein to design this new biosensor may facilitate the development of other biosensors specific for SARS-CoV-2 variants.
CRediT authorship contribution statement
Jong-Hwan Lee: Conceptualization, Methodology, Investigation, Data curation, Writing – original draft. Yungmin Lee: Methodology, Investigation, Data curation. Sung Kyun Lee: Methodology, Investigation. Jung Kim: Investigation. Chang-Seop Lee: Methodology, Investigation. Nam Hoon Kim: Investigation. Hong Gi Kim: Project administration, Funding acquisition, Supervision, Conceptualization, Investigation, Writing – original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the National Culture Collection for Pathogens of the Korean CDC for providing clinical SARS-CoV-2 isolates. This work was supported by the National Research Council of Science and Technology (NST) and National Research Foundation (NRF), funded by the Ministry of Science and ICT, Republic of Korea (grant numbers CRC-16-01-KRICT, NRF-2020M3E9A1043749, and NRF-2018R1D1A3B07049557), and by the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number HI20C0363). This Research has also been performed as a project No. (KK21- SI2161-119) and supported by the Korea Research Institute of Chemical Technology (KRICT), Republic of Korea.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bios.2022.114034.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Cheng M.H., Krieger J.M., Kaynak B., Arditi M., Bahar I. Impact of South African 501.V2 variant on SARS-CoV-2 spike infectivity and neutralization: a structure-based computational assessment. bioRxiv. 2021 2021.2001.2010. [Google Scholar]
- Fulford T.S., Van H., Gherardin N.A., Zheng S., Ciula M., Drummer H.E., Redmond S., Tan H.-X., Center R.J., Li F., Grimley S.L., Wines B.D., Nguyen T.H.O., Mordant F.L., Rowntree L.C., Cheng A.C., Doolan D.L., Bond K., Hogarth P.M., McQuilten Z., Subbarao K., Kedzierska K., Juno J.A., Wheatley A.K., Kent S.J., Williamson D.A., Purcell D.F.J., Anderson D.A., Godfrey D.I. medRxiv; 2021. A Point-Of-Care Lateral Flow Assay for Neutralising Antibodies against SARS-CoV-2; p. 21255368. 2021.2004.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H.H., Twaddle A., Marchand B., Gunsalus K.C. Structural modeling of the SARS-CoV-2 spike/human ACE2 complex interface can identify high-affinity variants associated with increased transmissibility. J. Mol. Biol. 2021;433(15) doi: 10.1016/j.jmb.2021.167051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L., Consortium C.-G.U. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. 10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izda V., Jeffries M.A., Sawalha A.H. COVID-19: a review of therapeutic strategies and vaccine candidates. Clin. Immunol. 2021;222 doi: 10.1016/j.clim.2020.108634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y., Park G.-S., Moon J.H., Ku K., Beak S.-H., Lee C.-S., Kim S., Park E.C., Park D., Lee J.-H., Byeon C.W., Lee J.J., Maeng J.-S., Kim S.-J., Kim S.I., Kim B.-T., Lee M.J., Kim H.G. Comparative analysis of primer–probe sets for RT-qPCR of COVID-19 causative virus (SARS-CoV-2) ACS Infect. Dis. 2020;6(9):2513–2523. doi: 10.1021/acsinfecdis.0c00464. [DOI] [PubMed] [Google Scholar]
- Kilic T., Weissleder R., Lee H. Molecular and immunological diagnostic tests of COVID-19: current status and challenges. iScience. 2020;23(8):101406. doi: 10.1016/j.isci.2020.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-Y., Lee J.-H., Kim M.J., Park S.C., Choi M., Lee W., Ku K.B., Kim B.T., Changkyun Park E., Kim H.G., Kim S.I. Development of a SARS-CoV-2-specific biosensor for antigen detection using scFv-Fc fusion proteins. Biosens. Bioelectron. 2021;175 doi: 10.1016/j.bios.2020.112868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakidis N.C., López-Cortés A., González E.V., Grimaldos A.B., Prado E.O. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. NPJ Vaccines. 2021;6(1):28. doi: 10.1038/s41541-021-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lee J.-H., Choi M., Jung Y., Lee S.K., Lee C.-S., Kim J., Kim J., Kim N.H., Kim B.-T., Kim H.G. A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2) Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Nie J., Wu J., Zhang L., Ding R., Wang H., Zhang Y., Li T., Liu S., Zhang M., Zhao C., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Liu J., Liang H., Shi Y., Shen Y., Xie L., Zhang L., Qu X., Xu W., Huang W., Wang Y. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184(9):2362–2371. doi: 10.1016/j.cell.2021.02.042. e2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Fang Y.-Y., Deng Y., Liu W., Wang M.-F., Ma J.-P., Xiao W., Wang Y.-N., Zhong M.-H., Li C.-H., Li G.-C., Liu H.-G. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chinese Med J. 2020;133(9) doi: 10.1097/CM9.0000000000000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., Wang D.-Q., Hu Y., Ren J.-H., Tang N., Xu Y.-Y., Yu L.-H., Mo Z., Gong F., Zhang X.-L., Tian W.-G., Hu L., Zhang X.-X., Xiang J.-L., Du H.-X., Liu H.-W., Lang C.-H., Luo X.-H., Wu S.-B., Cui X.-P., Zhou Z., Zhu M.-M., Wang J., Xue C.-J., Li X.-F., Wang L., Li Z.-J., Wang K., Niu C.-C., Yang Q.-J., Tang X.-J., Zhang Y., Liu X.-M., Li J.-J., Zhang D.-C., Zhang F., Liu P., Yuan J., Li Q., Hu J.-L., Chen J., Huang A.-L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Luan B., Wang H., Huynh T. Enhanced binding of the N501Y-mutated SARS-CoV-2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2021;595(10):1454–1461. doi: 10.1002/1873-3468.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozono S., Zhang Y., Ode H., Sano K., Tan T.S., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y., Suzuki T., Tokunaga K. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021;12(1):848. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan M., Ferguson I.D., Miao W., Khavari P.A. SARS-CoV-2 B.1.1.7 and B.1.351 spike variants bind human ACE2 with increased affinity. Lancet Infect. Dis. 2021;21(8):1070. doi: 10.1016/S1473-3099(21)00262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I.C., Tiu C., Hu Z., Chen V.C.-W., Young B.E., Sia W.R., Tan Y.-J., Foo R., Yi Y., Lye D.C., Anderson D.E., Wang L.-F. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. 2020;38(9):1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- Taylor P.C., Adams A.C., Hufford M.M., de la Torre I., Winthrop K., Gottlieb R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21(6):382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904. doi: 10.1016/j.cell.2020.03.045. e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.