Abstract
Background
Heat-clearing and detoxifying herbs (HDHs) play an important role in the prevention and treatment of coronavirus infection. However, their mechanism of action needs further study. This study aimed to explore the anti-coronavirus basis and mechanism of HDHs.
Material/Methods
Database mining was performed on 7 HDHs. Core ingredients and targets were screened according to ADME rules combined with Neighborhood, Co-occurrence, Co-expression, and other algorithms. GO enrichment and KEGG pathway analyses were performed using the R language. Finally, high-throughput molecular docking was used for verification.
Results
HDHs mainly acts on NOS3, EGFR, IL-6, MAPK8, PTGS2, MAPK14, NFKB1, and CASP3 through quercetin, luteolin, wogonin, indirubin alkaloids, β-sitosterol, and isolariciresinol. These targets are mainly involved in the regulation of biological processes such as inflammation, activation of MAPK activity, and positive regulation of NF-κB transcription factor activity. Pathway analysis further revealed that the pathways regulated by these targets mainly include: signaling pathways related to viral and bacterial infections such as tuberculosis, influenza A, Ras signaling pathways; inflammation-related pathways such as the TLR, TNF, MAPK, and HIF-1 signaling pathways; and immune-related pathways such as NOD receptor signaling pathways. These pathways play a synergistic role in inhibiting lung inflammation and regulating immunity and antiviral activity.
Conclusions
HDHs play a role in the treatment of coronavirus infection by regulating the body’s immunity, fighting inflammation, and antiviral activities, suggesting a molecular basis and new strategies for the treatment of COVID-19 and a foundation for the screening of new antiviral drugs.
Keywords: 3a Protein, Severe Acute Respiratory Syndrome Coronavirus; COVID-19; Molecular Docking Simulation; Molecular Mechanisms of Pharmacological Action
Background
In the 21st century, 3 unprecedented coronaviruses have spread worldwide: severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002, Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012, and the novel coronavirus (SARS-CoV-2) in 2019. All 3 epidemics caused a large number of human deaths, resulting in panic and economic regression. Traditional Chinese medicines (TCM) have exerted significant clinical treatment effects in the fight against the epidemic, and sequelae and drug resistance have rarely occurred. In 2020, the National Administration of TCM issued a clinical screening of the “3 medicines and 3 prescriptions” with definite curative effects against COVID-19. According to the characteristics of “heat and toxic” in the pathogenesis of coronavirus pneumonia [1], heat-clearing and detoxifying herbs (HDHs) of TCM have considerable clinical therapeutic effects in fighting COVID-19. However, their underlying molecular mechanism remains unknown.
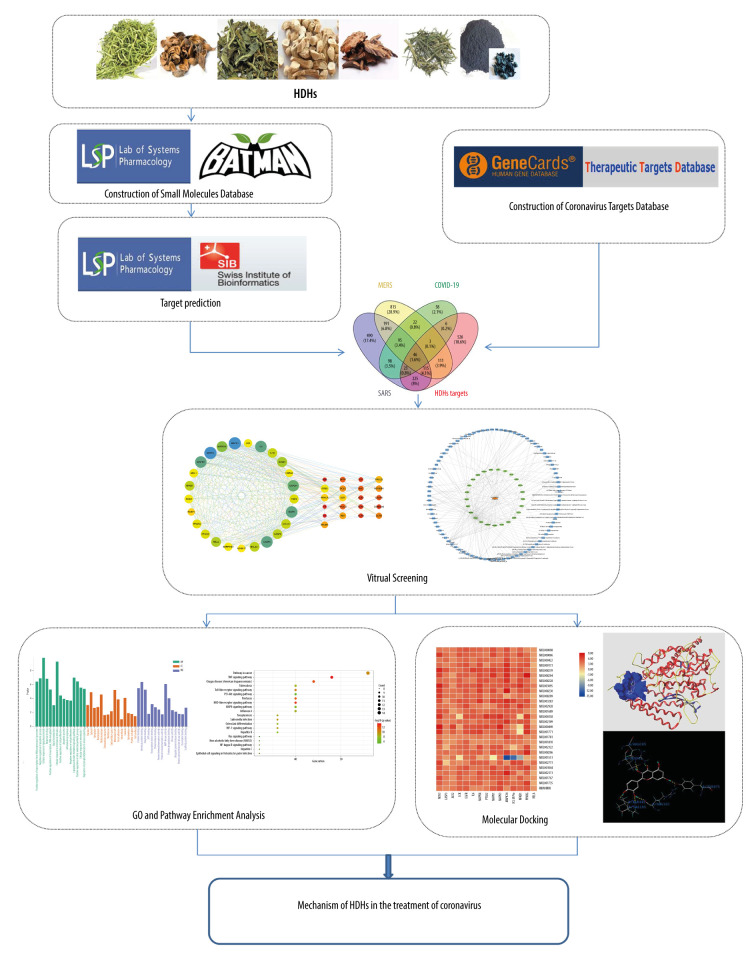
In this study, based on the theory of TCM, Scheme for Diagnosis and Treatment of 2019 Novel Coronavirus Pneumonia, and literature review, 7 HDHs, including Lonicerae Japonicae Flos, Forsythiae Fructus, Isatidis Folium, Isatidis Radix, Indigo Naturalis, Dryopteridis Crassirhizomatis Rhizoma, and Andrographis Herba, were selected to study the molecular mechanism of TCMs based on 2 aspects of heat-clearing and detoxifying. Network pharmacology, ADME (Absorption, Distribution, Metabolism and Excretion) rule virtual screening, Tanimoto coefficient, neighborhood, co-occurrence, co-expression, and other algorithms were used to establish databases of 7 HDHs active ingredient small molecules and their corresponding targets. GO and KEGG enrichment methods were used to study the molecular pathways and mechanisms of action of the targets. Finally, molecular docking was used to verify the degree of binding between the main active ingredients and the related targets. This study aimed to elucidate the mechanisms of HDHs from a molecular perspective to provide an alternative therapeutic strategy for the prevention and treatment of epidemics caused by coronaviruses. The Graphic abstract of this study is shown in Figure 1.
Figure 1.
The integrated workflow. (Microsoft Word 2010).
Material and Methods
Construction of HDHs Small Molecule Database
The chemical ingredients of Lonicerae Japonicae Flos (Jinyinhua), Forsythiae Fructus (Lianqiao), Isatidis Folium (Daqingye), Isatidis Radix (Banlangen), Indigo Naturalis (Qingdai), Dryopteridis Crassirhizomatis Rhizoma (Guanzhong) and Andrographis Herba (Chuanxinlian) were searched in the TCMSP database (http://tcmspw.com/tcmsp.php) and BATMAN database (http://bionet.ncpsb.org/batman-tcm/), and screened according to the oral bioavailability (OB) and drug-likeness (DL) of ADME rule. In ADME, OB is the amount of the drug that actually enters the circulation when it enters the body. A high OB value is usually a key indicator to determine the drug-like property of bioactive molecules as therapeutic agents. DL is a qualitative concept for drug design, the DL index (see formula below), which is calculated using the Tanimoto coefficient:
In the formula, x represents the ingredients from 3 herbal medicines, and y represents the average drug-likeness index. Setting OB ≥30% and DL ≥0.18 as parameters [2,3], combined with results from our literature review, we established the above 7 HDHs small molecule database. Taking the intersection of 7 HDHs ingredients, the ingredients with higher repetition frequency were selected as the important HDHs ingredients for subsequent analysis.
Construction and Screening of Coronavirus Targets Database
The targets informations of the above 7 HDHs ingredients were inquired by TCMSP and Swiss TargetPrediction database (http://www.swisstargetprediction.ch/). The target information of the above 7 HDHs ingredients were inquired by TCMSP and Swiss TargetPrediction database. Databases such as Genecards (https://www.genecards.org/) and TTD (http://db.idrblab.net/ttd/) were used to search “Coronavirus”, “SARS”, and “MERS” as keywords to collect the targets of the 3 diseases, and the intersection of HDHs targets and the common targets of the 3 diseases was obtained by using the DAVID platform. Thus, a database of 7 HDHs targets for the treatment of coronavirus was constructed.
Construct PPI Network of Targets
We imported the targets from the 7 HDHs target databases for the treatment of coronaviruses constructed above into the STRING data analysis platform (https://www.string-db.org/), and used Neighborhood, Co-occurrence, Co-expression, and other algorithms for PPI network analysis. The data analysis mode was set as “Multiple proteins”, and the Organism was selected as “Homo sapiens”, with the medium confidence of ≥0.40. The TSV files were imported into Cytoscape 3.6.1, and the MCODE plug-in was used to conduct module analysis on the targets, and the targets with degree value greater than 2 times its median were selected to obtain the important target set of HDHs for the treatment of coronavirus, and for further screening and optimization of the target database.
Construction of the Ingredients-Targets-Diseases Network
Using Cytoscape 3.7.1 to construct a “ingredients-targets-diseases” network, we performed reverse screening of the set of important targets screened out by the PPI network, and screened out the ingredients corresponding to the important targets. Through visual analysis, we selected targets and ingredients with a degree value greater than twice the median to explore the potential material basis of HDHs [4].
Enrichment Analysis of GO and KEGG pathways
To further understand the gene functions of core targets and the main pathways of HDHs in the treatment of COVID-19, SARS, and MERS, we selected the species as humans, set the threshold P<0.05 for the enrichment analysis of GO and KEGG pathways, and finally used the R language visually to analyze the analysis results.
Molecular Docking Verification
Based on the above analysis results, the active ingredients and key targets were screened for molecular docking. First, the PDB-ID of the corresponding targets were downloaded from the PDB database and the 3D structure PDB Format files of targets containing original ligands were downloaded. Next, we downloaded the SDF format files of the ingredients from the PubChem database, and used Chemoffice to convert them into the mol2 format files. The Surflex-Dock in SYBYL software is a fast and automatic docking method. It uses unique empirical scoring functions and a search engine based on molecular similarity to dock the ligand molecule with the binding site of the protein, and considers ligand flexibility through fragment growth method. Firstly, SYBYL-X2.0 software was used for protein pretreatment, including extraction of ligand small molecules, removal of water molecules, and hydrogenation. Then, SYBYL-X2.0 was used to conduct molecular docking between key ingredients and proteins, and the interaction between small molecules and targets was scored by combining the CSCORE and Total-score scoring functions of Surflex-Dock molecular docking module results, and the molecular docking results of ribavirin were used as the control group [5,6]. The docking results were visualized using TBTools software and Discovery Studio 2016 Client.
Results
Confirmation of Core Ingredients and Core Targets
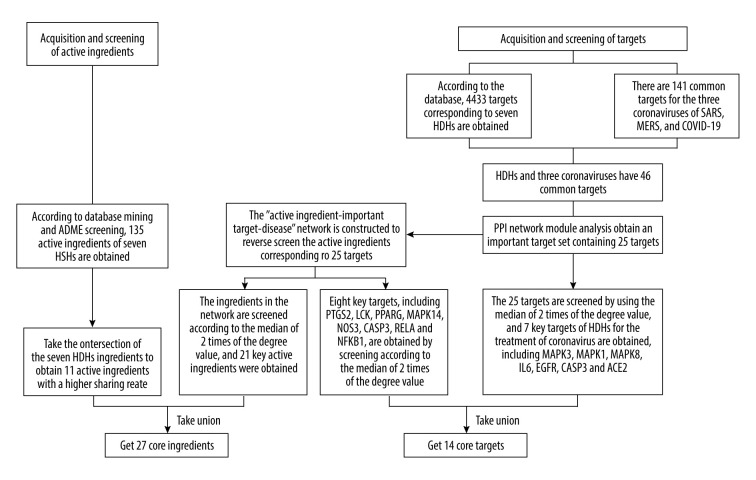
The Schematic diagram of confirmation of core ingredients and core targets is shown in Figure 2. Using the TCMSP and BATMAN databases by screening according to the conditions of DL ≥0.18 and OB ≥30%, 135 active ingredients were screened from the 7 HDHs small molecule databases constructed. They were the same ingredients as in HDHs. Through the intersection of 7 HDHs, the results showed that kaempferol was found in Lonicerae Japonicae Flos, Forsythiae Fructus, and Dryopteridis Crassirhizomatis Rhizoma; quercetin and luteolin were found in Lonicerae Japonicae Flos and Forsythiae Fructus; β-sitosterol was found in Lonicerae Japonicae Flos, Isatidis Folium, Forsythiae Fructus, Isatidis Radix, and Indigo Naturalis; indirubin was found in Isatidis Folium and Indigo Naturalis; wogonin was found in Forsythiae Fructus and Andrographis Herba; stigmasterol was found in Isatidis Radix and Lonicerae Japonicae Flos; poriferast-5-en-3beta-ol was found in Isatidis Radix and Isatidis Folium; and indigo, candidine, and isovitexin were found in Isatidis Folium, Isatidis Radix, and Indigo Naturalis.
Figure 2.
Schematic diagram of the screening of core ingredients and core targets. (Microsoft Word 2010).
Based on the TCMSP and SwissTargetPrediction databases, 4433 targets corresponding to 7 HDHs were obtained. The intersection of the 3 coronaviruses, SARS-CoV, MERS-CoV, and SARS-CoV-2, showed that there were 141 common targets, indicating that the 3 coronaviruses might have similar pathogenic characteristics. The corresponding targets of HDHs and the 3 coronaviruses showed 46 common targets, indicating that HDHs and the 3 coronaviruses had potential corresponding relationships.
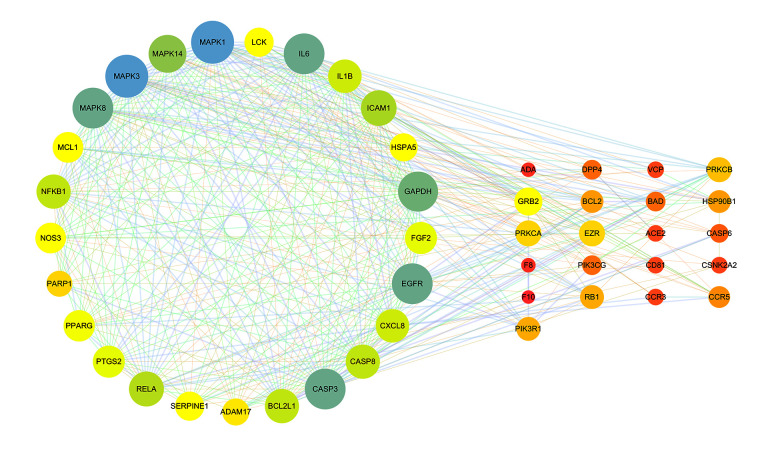
The STRING database was used to construct a PPI network for 46 common targets. MCODE software was used to analyze 46 targets and obtain an important target set containing 25 targets. The 25 targets were screened with a median of 2 times the degree value, and 7 key targets were obtained, including MAPK3, MAPK1, MAPK8, IL-6, EGFR, CASP3, and ACE2.
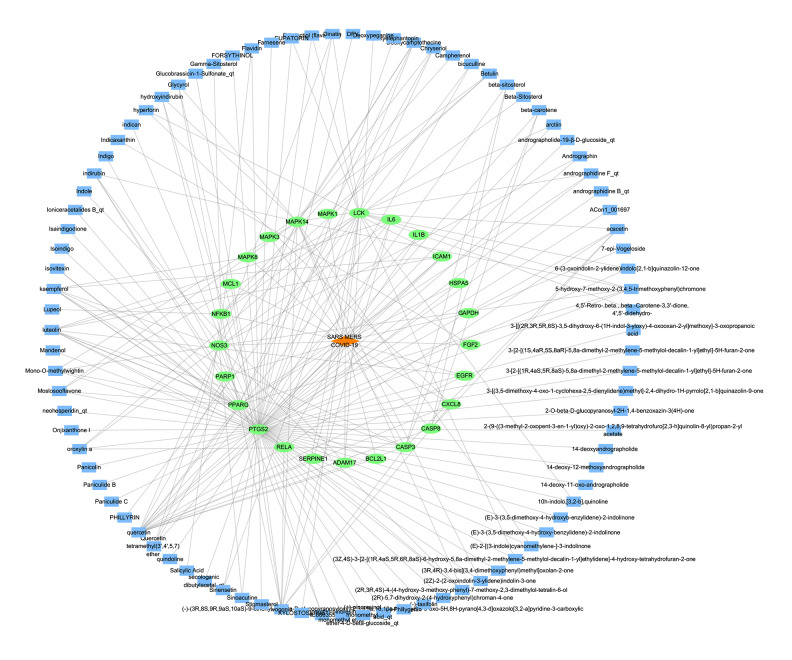
To supplement and improve the target database and further optimize the active ingredients and their corresponding targets, as shiwn in Figure 3. The 25 targets were screened for their ingredients in reverse, and the “ingredients-targets-diseases” network was constructed (Figure 4). Screening was performed according to a median of 2 times the degree value, and 21 active ingredients were obtained, including quercetin, luteolin, wogonin, kaempferol, acacetin, betulin, dinatin, glycerol, chryseriol, 5-hydroxy-7-methoxy-2-(3,4,5-trimethoxyphenyl)chromone, β-sitosterol, (2R,3R,4S)-4-(4-hydroxy-3-methoxy-phenyl)-7-methoxy-2,3-dimethylol-tetralin-6-ol, oroxylin a, mono-O-methylwightin, moslosooflavone, deoxycamptothecine, quercetin tetramethyl(3′,4′,5,7) ether, indirubin, β-carotene, and rographidine F, and hydroxyindirubin, acting on 8 key targets, including PTGS2, LCK, PPARG, MAPK14, NOS3, CASP3, RELA, and NFKB1.
Figure 3.
Protein interaction networks of related genes in the treatment of coronavirus infection with HDHs. (Cytoscape 3.7.1).
Figure 4.
Active ingredient-target-disease network diagram. (Cytoscape 3.7.1). There are 115 nodes and 211 edges in the network, among which blue rectangles represent ingredients, green ellipses represent targets, and orange diamonds represent disease.
After using Neighborhood, Co-occurrence, Co-expression, and other algorithms, module analysis, and “ingredients-targets-diseases” network screening, a total of 27 major active ingredients were obtained (Table 1). The 14 targets included MAPK3, MAPK1, MAPK8, IL-6, EGFR, CASP3, ACE2, PTGS2, LCK, PPARG, MAPK14, NOS3, RELA, and NFKB1, which are the core targets for HDHs acting on diseases caused by coronavirus.
Table 1.
Active ingredient list of HDHs in the treatment of coronavirus infection.
| MOL ID | Ingredient | Classification | Chem 2D | Core target |
|---|---|---|---|---|
| MOL000098 | Quercetin | Flavonoid |
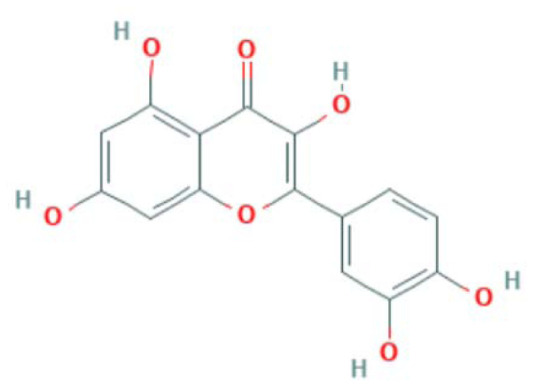
|
IL6, EGFR, CASP3, RELA, PTGS2, PPARG, NOS3 |
| MOL000006 | Luteolin | Flavonoid |
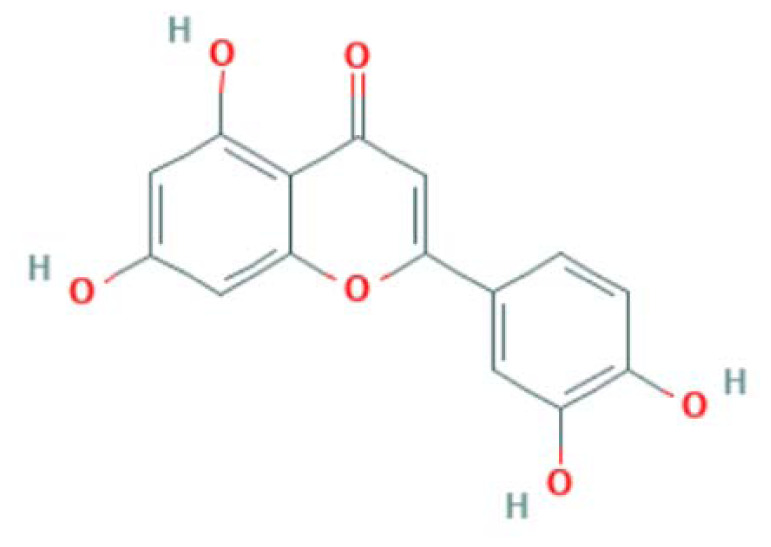
|
IL6, EGFR, CASP3, RELA, PTGS2, PPARG, |
| MOL000173 | Wogonin | Flavonoid |
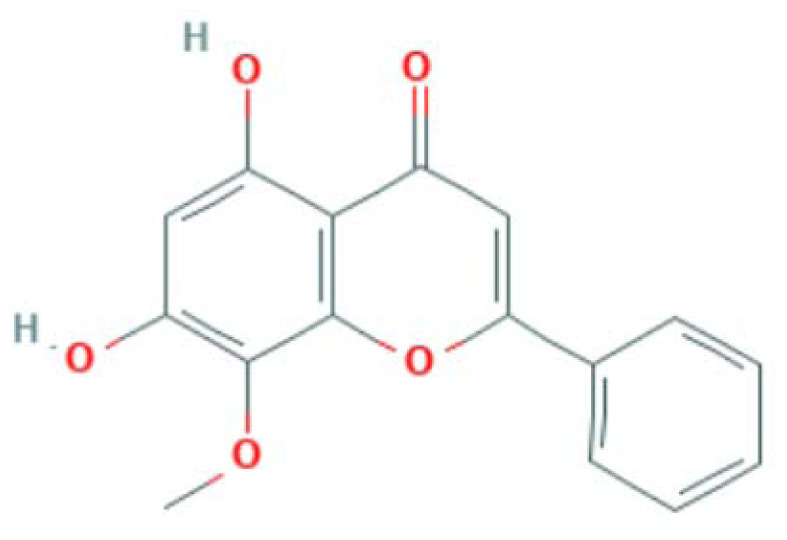
|
IL6, CASP3, MAPK14, RELA, PTGS2, PPARG, LCK |
| MOL000422 | Kaempferol | Flavonoid |
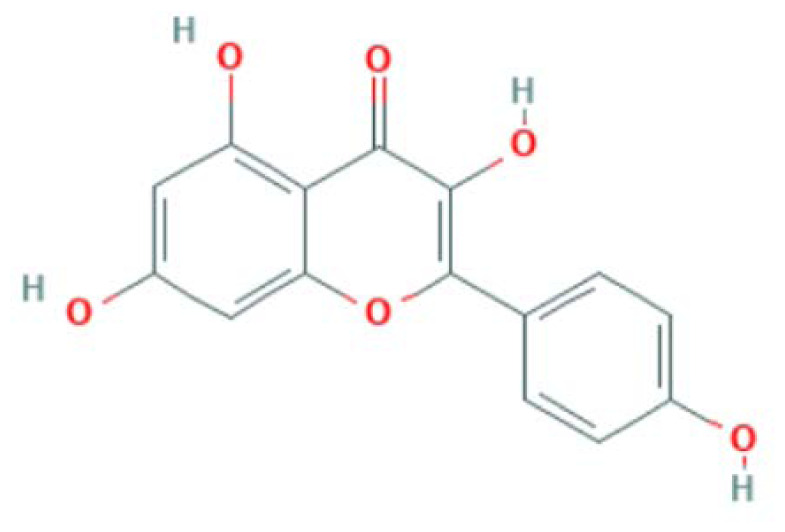
|
CASP3, MAPK8, RELA, PTGS2, PPARG, NOS3 |
| MOL008228 | Andrographin | Flavonoid |
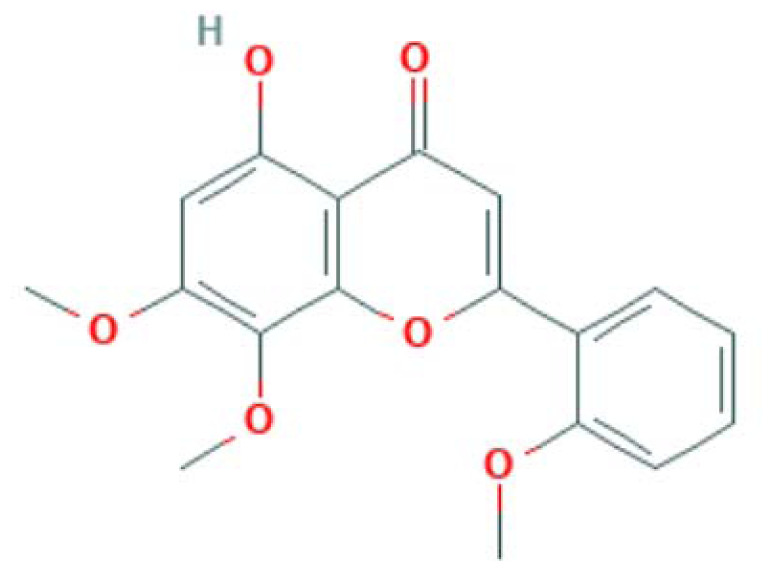
|
PTGS2, NOS3 |
| MOL003095 | 5-hydroxy-7-methoxy-2-(3,4,5-trimethoxyphenyl)chromone | Flavonoid |
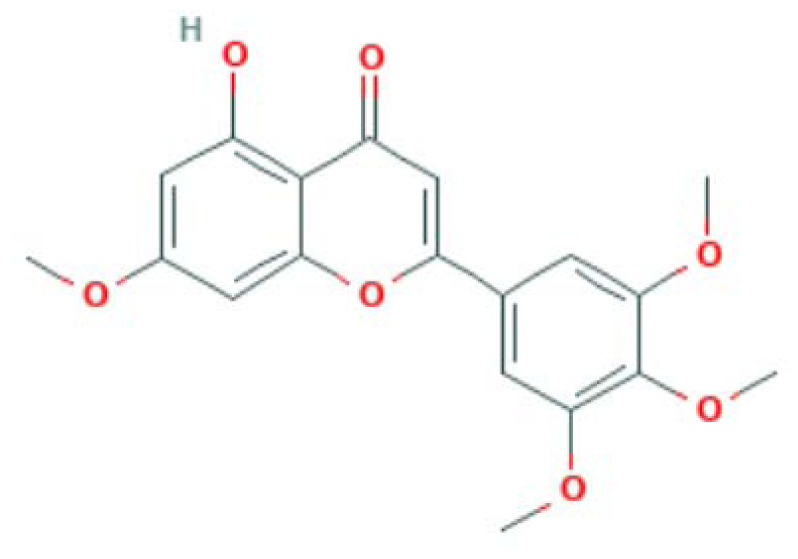
|
MAPK14, PTGS2, PPARG, NOS3, LCK |
| MOL001689 | Acacetin | Flavonoid |
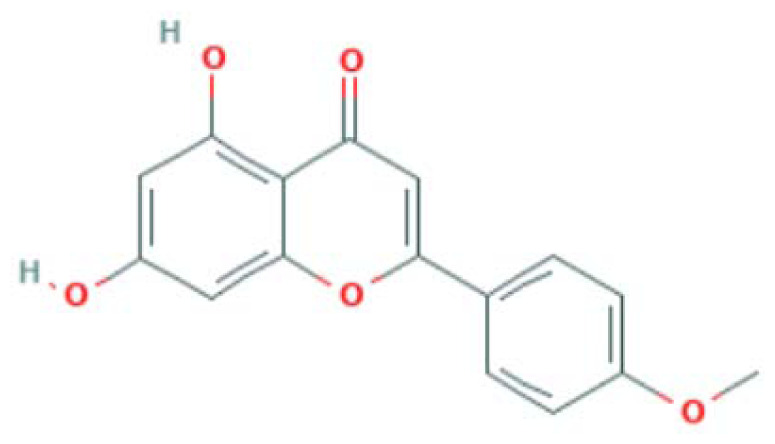
|
CASP3, RELA, PTGS2, LCK |
| MOL008239 | Quercetin tetramethyl(3′,4′,5,7) ether | Flavonoid |
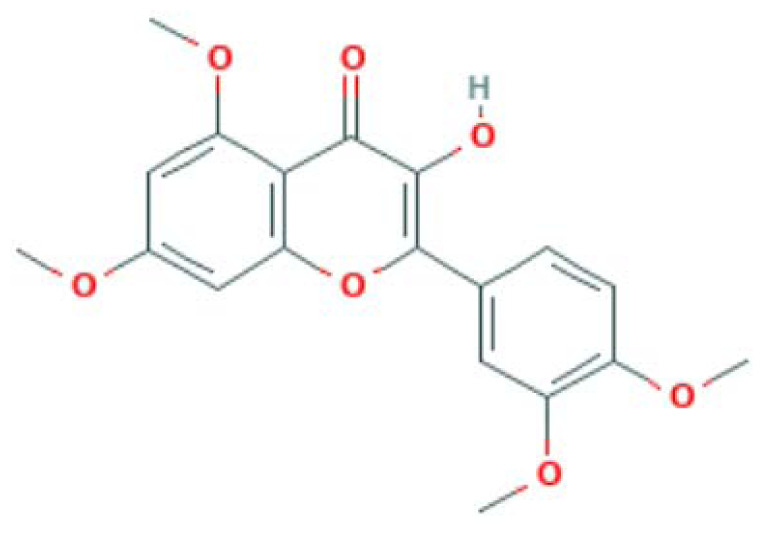
|
MAPK14, PTGS2, PPARG, NOS3 |
| MOL008204 | Mono-O-methylwightin | Flavonoid |
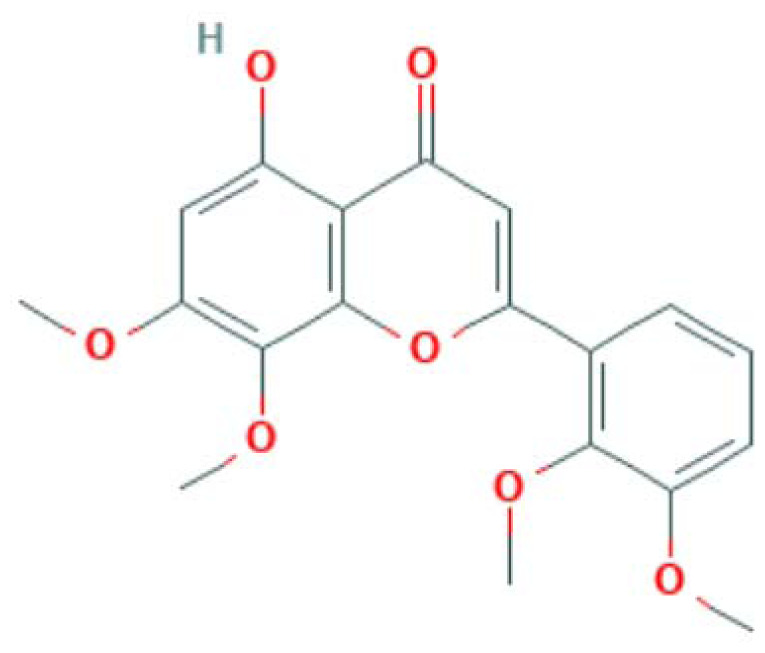
|
MAPK14, PTGS2, PPARG, NOS3 |
| MOL008209 | Deoxycamptothecine | Alkaloid |
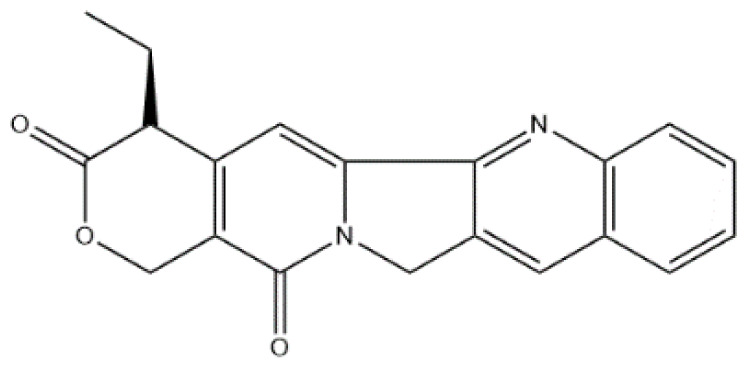
|
MAPK14, PTGS2, PPARG, NOS3 |
| MOL003283 | (2R,3R,4S)-4-(4-hydroxy-3-methoxy-phenyl)-7-methoxy-2,3-dimethylol-tetralin-6-ol (Isolariciresinol) | Lignans |
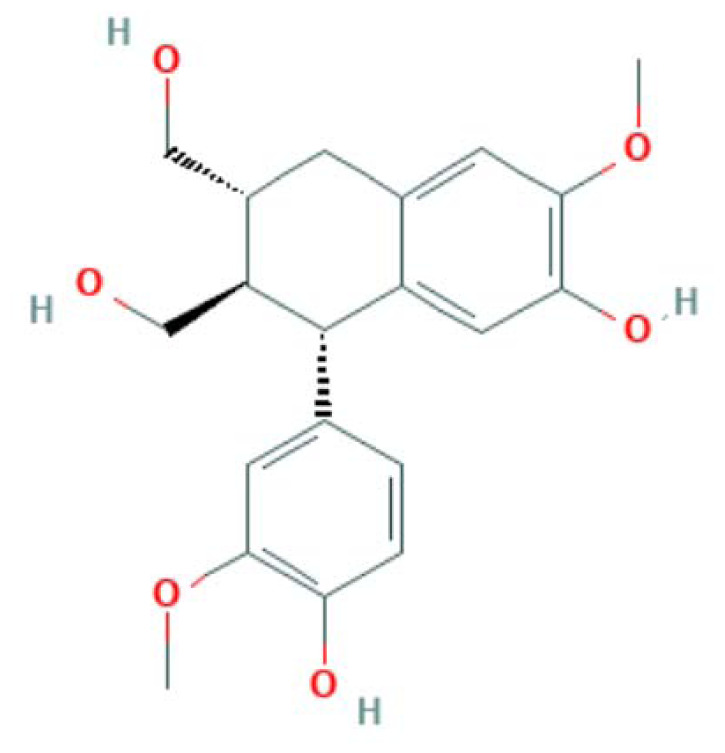
|
MAPK14, PTGS2, PPARG, NOS3 |
| MOL002928 | Oroxylin a | Flavonoid |
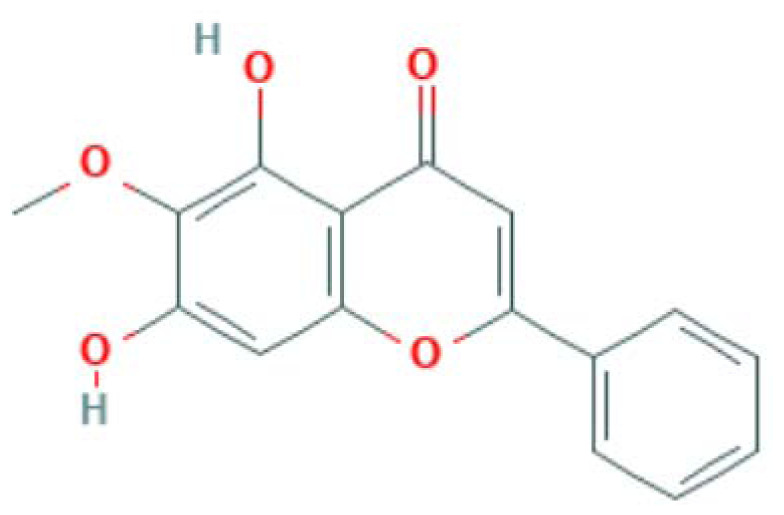
|
IL6, CASP3, PTGS2, LCK |
| MOL002309 | Indirubin | Alkaloid |
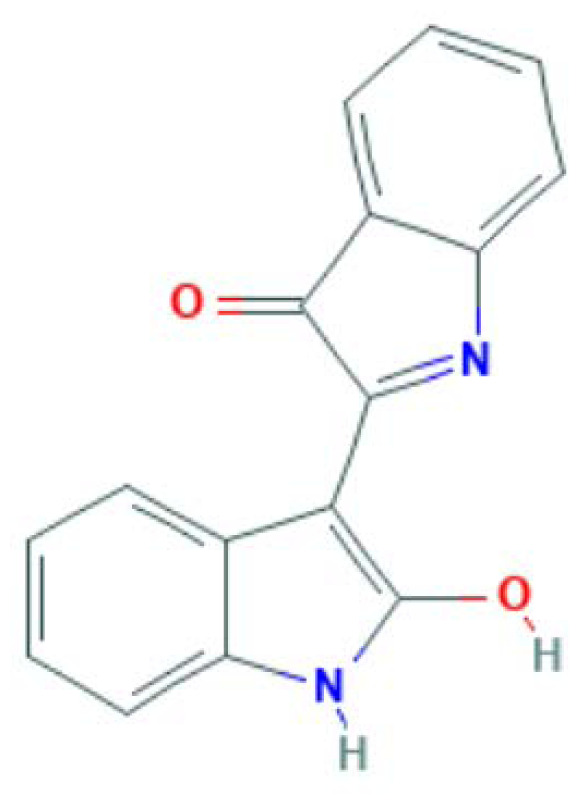
|
MAPK14, PTGS2, PPARG, NOS3 |
| MOL008206 | Moslosooflavone | Flavonoid |
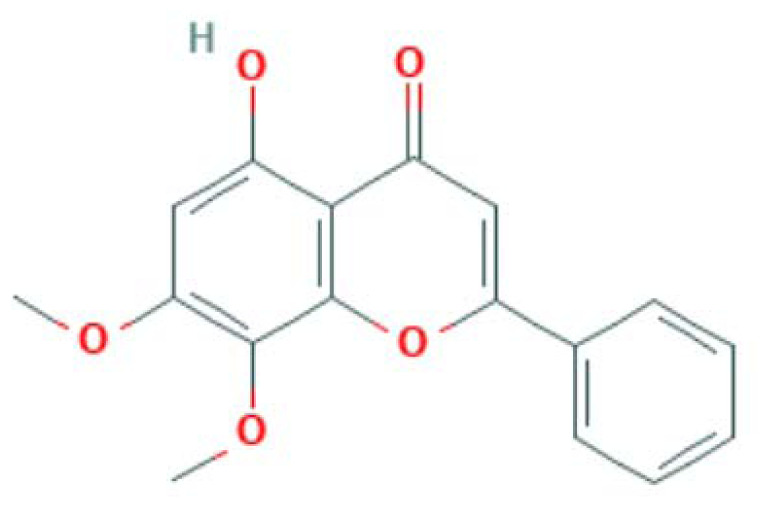
|
MAPK14, PTGS2, PPARG, LCK |
| MOL008230 | Andrographidine F | Flavonoid |
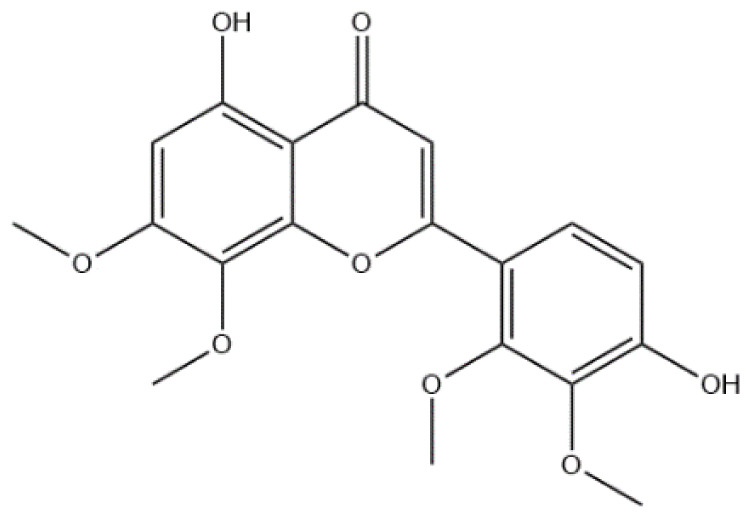
|
MAPK14, PTGS2, PPARG |
| MOL000358 | β-sitosterol | Steroid |
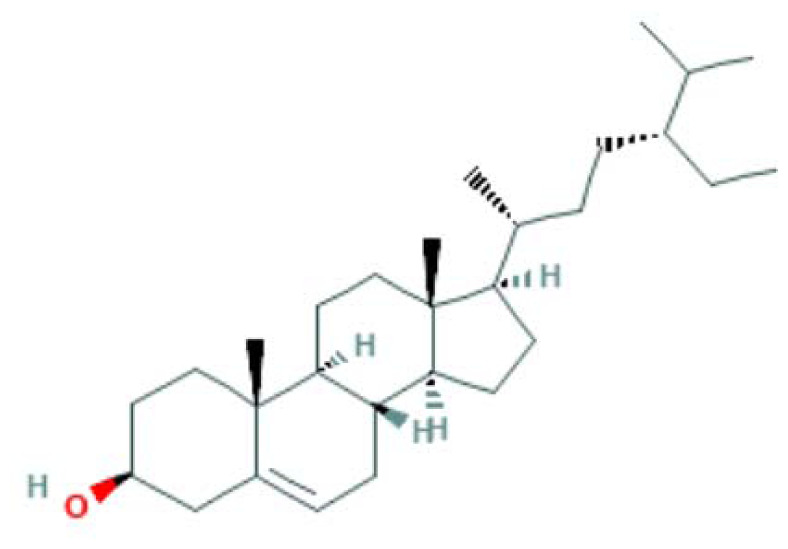
|
CASP3, NFKB1, PTGS2 |
| MOL001551 | Betulin | Terpenoid |
|
MAPK3, MAPK1, NFKB1 |
GO and Pathway Enrichment Analysis for Targets
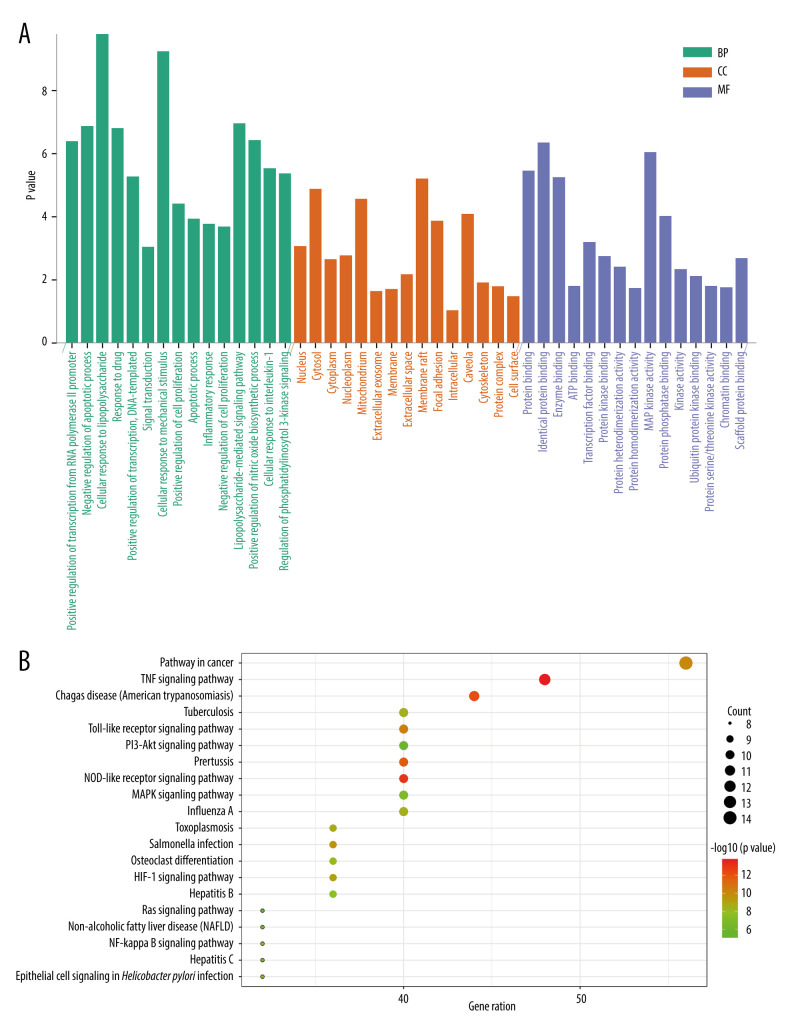
To further analyze the main pathways of HDHs for the treatment of coronavirus infections, functional enrichment and pathway enrichment of the targets were carried out. Figure 5A shows that 246 GO entries were obtained (P<0.05), of which there were 184 biological processes (BP), mainly involving cellular response to lipopolysaccharide, inflammatory response, cellular response to interleukin-1, regulation of phosphatidylinositol 3-kinase signaling, activation of MAPK activity, positive regulation of NF-κB transcription factor activity, and Fc-epsilon receptor signaling pathway. There were 24 items involved in cell composition (CC). The top 5 cell components were the nucleus, cytosol, cytoplasm, nucleoplasm, and mitochondrion. In addition, 38 items were involved in molecular functions (MFs) such as protein, enzyme, ATP, transcription factor, and protein kinase bindings.
Figure 5.
GO and KEGG pathway enrichment analysis. (clusterProfiler software package on the R 4.0.3 and bioinformatics platform). (A) GO enrichment analysis diagram. (B) Functional annotation diagram.
The results of KEGG pathway enrichment analysis are shown in Figure 5B. Among the top 20 pathways, pathways related to viral and bacterial infections included Chagas disease (American trypanosomiasis), Pertussis, Influenza A, Tuberculosis, Salmonella infection, Toxoplasmosis, Hepatitis B, epithelial cell signaling in Helicobacter pylori infection, Hepatitis C, and Ras signaling pathway. Inflammation-related pathways included Toll-like receptor (TLR), TNF, MAPK, HIF-1, NF-κB, and PI3K-AKT signaling pathways. Immune-related pathways included the NOD-like receptor signaling pathway.
Validation of Surflex-Dock Ingredient-Target Interaction
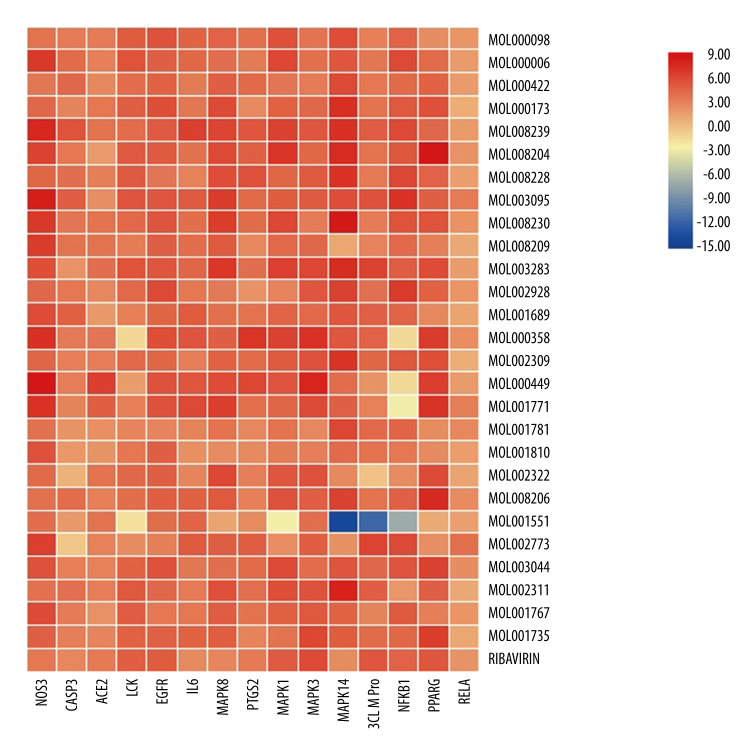
To verify the ability of the active ingredients to bind to core targets, the core active chemical ingredients in Table 1 were used as small molecule ligands in HDHs to select the 14 key targets in combination with the latest known coronavirus targets for receptors [7]. The molecular docking results of ribavirin were used as a control [8,9], and SYBYL-X 2.0 was used for molecular docking verification. The docking results of CSCORE ≥4 are generally more reliable, while the total-score function considers factors such as polarity, hydrophobicity, enthalpy, and solvation. The greater the value, the more stable the docking complex, indicating better matching and binding between small molecule ingredients and proteins. Therefore, in this experiment, ingredients with CS-SCORE ≥4 and total score greater than ribavirin total score were selected. The visualized heat map results are presented in Figure 6.
Figure 6.
Heat map of molecular docking results. (TBTools).
Assuming that C represents the number of ingredients (C=27), P represents the number of targets (P=15), the total score of the docking result of ribavirin and 15 targets is represented by Ti, Ni and represents the number of targets with a total score o Ti in each row, and Ng represents the number of ingredients with a total score o Ti in each column. The core active ingredients with Ng ≥50%C and the core targets with Ni ≥50% P are stable in docking. The docking results of the above compounds were 51.85% Ng ≥50%C, better than the docking scores of ribavirin and core targets. Among them, 14 small molecule ligands, including quercetin, luteolin, wogonin, indirubin, chryseriol, isolariciresinol, 5-hydroxy-7-methoxy-2-(3,4,5-trimethoxyphenyl) chromone, andrographidine F, poriferast-5-en-3beta-ol, β-sitosterol, stigmasterol, and mother-nuclear similar moslosooflavone, mono-o-methylwightin, and quercetin tetramethyl(3′,4′,5,7) ether, could be stably docked to NOS3, EGFR, and IL-6, MAPK8, PTGS2, MAPK14, NFKB1, and CASP3 active pockets of the protein structures (Figure 7A–7C).
Figure 7.
Schematic diagram of molecular docking. (SYBYL-X 2.0). (A) With the small molecule ligand of wogonin as the center, the amino acid residues of ACE2 within the range of 5A form an added structure to the surface of the interface pocket; (B) With the small molecule ligand of isolariciresinol as the center, the amino acid residues of ACE2 within the range of 5A form an added structure to the surface of the interface pocket; (C) With the small molecule ligand of quercetin as the center, the amino acid residues of NOS3 within the range of 5A form an added structure to the surface of the interface pocket.
The molecular docking results (Table 2) showed that the total score of ribavirin and NOS3 was 3.37, and the total scores of core ingredients and NOS3 were both greater than 3.37. The docking score of ribavirin and CASP3 was 2.71, and the docking scores of core ingredients and CASP3 were w 2.71, accounting for 77.78%. The docking score of ribavirin with EGFR was 4.93, 44.44% of the core ingredients had a docking score with EGFR ≥4.93, and 51.85% of the core ingredients had docking scores between 3.00 and 4.93. The docking score of ribavirin and IL-6 was 2.54, and the docking scores of core ingredients and IL-6 were ≥2.54, accounting for 96.30%. The docking score of ribavirin and MAPK14 was 2.43, and the docking scores of core ingredients and MAPK14 were ≥2.43, accounting for 88.89%. The docking score of ribavirin with NFKB1 was 4.52, 55.56% of the core ingredients had docking scores of NFKB1 ≥4.52, and 22.22% of the core ingredients had docking scores between 3.00 and 4.52. The docking score of ribavirin and PTGS2 was 3.35, and the docking scores of core ingredients and PTGS2 were ≥3.35, accounting for 62.96%. In addition, all coronaviruses invaded human cells by attaching to the surface of the receptor ACE2 [5], and the total score of ribavirin docking with ACE2 was 3.35. Among the key active ingredients, the total scores of wogonin, quercetin tetramethyl(3′,4′,5,7) ether, andrographidine F, isolariciresinol, and β-sitosterol with ACE2 were all greater than 3.35, indicating that these small molecular ingredients had good binding activity with ACE2.
Table 2.
The molecular docking results of the main active ingredients and the core targets.
| Compound | Total-Score | |||||||
|---|---|---|---|---|---|---|---|---|
| NOS3 | CASP3 | ACE2 | EGFR | IL6 | PTGS2 | MAPK14 | NFKB1 | |
| MOL000098 | 3.70 | 3.34 | 3.30 | 5.46 | 4.48 | 3.82 | 5.63 | 4.53 |
| MOL000006 | 6.64 | 4.08 | 3.05 | 4.68 | 4.21 | 3.28 | 5.22 | 5.87 |
| MOL000422 | 3.51 | 4.24 | 2.67 | 4.46 | 3.26 | 4.12 | 5.88 | 4.13 |
| MOL000173 | 4.24 | 2.82 | 3.43 | 5.59 | 3.42 | 2.54 | 7.23 | 5.11 |
| MOL008239 | 7.57 | 5.34 | 3.72 | 4.96 | 6.37 | 5.18 | 7.11 | 5.85 |
| MOL008204 | 6.15 | 3.36 | 1.66 | 5.03 | 3.76 | 4.64 | 7.42 | 5.17 |
| MOL008228 | 4.29 | 3.85 | 3.09 | 3.59 | 2.99 | 5.36 | 7.00 | 6.03 |
| MOL003095 | 8.02 | 4.77 | 2.27 | 5.20 | 4.94 | 4.13 | 5.65 | 7.02 |
| MOL008230 | 6.66 | 3.56 | 3.70 | 5.26 | 3.86 | 4.04 | 8.46 | 5.32 |
| MOL008209 | 6.57 | 3.62 | 3.60 | 4.71 | 3.99 | 2.58 | 1.10 | 4.17 |
| MOL003283 | 5.60 | 2.05 | 3.94 | 5.22 | 4.35 | 3.94 | 7.32 | 4.84 |
| MOL002928 | 4.24 | 3.44 | 2.63 | 5.90 | 3.38 | 2.04 | 6.25 | 6.60 |
| MOL001689 | 5.69 | 4.56 | 1.72 | 4.38 | 4.89 | 3.65 | 5.21 | 4.42 |
| MOL000358 | 7.08 | 3.24 | 3.58 | 5.60 | 5.29 | 6.86 | 5.23 | −1.45 |
| MOL002309 | 4.36 | 3.08 | 3.13 | 4.30 | 3.15 | 4.08 | 6.89 | 5.16 |
| MOL000449 | 8.76 | 3.12 | 6.37 | 5.44 | 5.21 | 6.01 | 4.01 | −1.39 |
| MOL001771 | 7.03 | 2.85 | 4.74 | 5.36 | 5.81 | 3.80 | 4.69 | −2.77 |
| MOL001781 | 3.77 | 2.00 | 2.24 | 2.75 | 2.83 | 2.64 | 6.08 | 4.41 |
| MOL001810 | 5.30 | 1.82 | 2.06 | 4.69 | 2.18 | 2.53 | 4.10 | 3.39 |
| MOL002322 | 4.13 | 0.56 | 3.71 | 4.69 | 2.77 | 3.12 | 2.58 | 2.48 |
| MOL008206 | 3.74 | 3.98 | 3.04 | 4.73 | 4.57 | 3.01 | 6.21 | 4.61 |
| MOL001551 | 4.01 | 1.72 | 3.62 | 3.95 | 4.39 | 2.41 | −13.95 | −6.90 |
| MOL002773 | 6.36 | −0.50 | 2.94 | 3.10 | 5.03 | 4.78 | 2.20 | 5.76 |
| MOL003044 | 5.46 | 3.04 | 2.98 | 5.46 | 3.49 | 4.04 | 5.27 | 5.29 |
| MOL002311 | 3.77 | 3.84 | 3.14 | 4.25 | 3.34 | 3.94 | 7.90 | 1.85 |
| MOL001767 | 5.73 | 3.30 | 2.12 | 3.40 | 3.46 | 3.62 | 4.46 | 5.01 |
| MOL001735 | 4.69 | 3.15 | 2.78 | 4.58 | 4.48 | 2.92 | 4.81 | 4.21 |
| RIBAVIRIN | 3.37 | 2.71 | 3.35 | 4.93 | 2.54 | 3.35 | 2.43 | 4.52 |
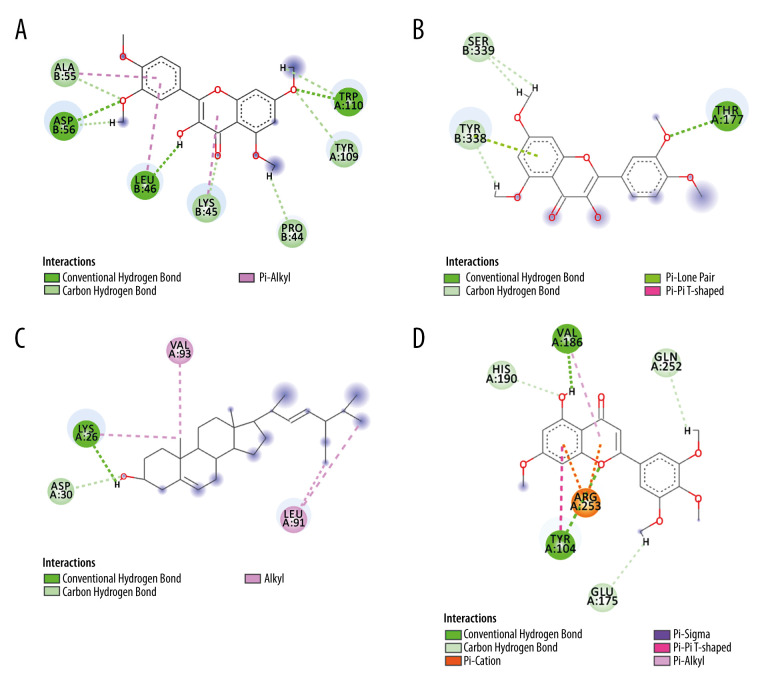
For the trusted target proteins in Tabel2 such as NOS3, NFKB1, ACE2, CASP3, EGFR, MAPK14, PTGS2, IL6, and the ingredients with the highest total-score in the docking results, the structure was visualized using Discovery Studio 2016 Client. Figure 8 shows the 2D structure of some proteins and ingredients. After analysis, it can be seen that quercetin tetramethyl(3′,4′,5,7) ether interacts with tyrosine (TYR) residues in IL6 and CASP3. It interacts with A/TYR109 in IL6 and B/TYR338 in CASP3 through intermolecular van der Waals forces. In addition, quercetin tetramethyl(3′,4′,5,7) ether also forms intermolecular hydrogen bond with tryptophan (TRY) residue A/TRP110, aspartic acid (ASP) residue B/ASP56, leucine (LEU) residue B/LEU46 and π-alkyl interaction with alanine (ALA) residue B/ALA55, B/LEU46, and lysine residue B/LYS45. Quercetin tetramethyl(3′,4′,5,7) ether also interacts with a variety of other amino acid residues in CASP3. There is van der Waals force between it and the serine (SER) residue B/SER339 in CASP3. There is a hydrogen bond between amino acid (THR) residue A/THR177. Stigmasterol forms strong intermolecular hydrogen bonds with amino acid residues in ACE2 and NOS3 through a hydroxyl group. The H+ on the hydroxyl group in stigmasterol forms a hydrogen bond with A/LYS26 in ACE2, and has van der Waals force with A/ASP30. The H+ on the hydroxyl group in stigmasterol forms a hydrogen bond with the glutamic acid (GLU) residue A/GLU361 in NOS3, and the oxygen anion forms a hydrogen bond with the arginine (ARG) residue A/ARG365. β-sitosterol interacts with leucine residues in EGFR and PTGS2, and multiple alkyl groups in β-sitosterol interact with A/LEU844, A/LEU718, A/LEU792 in EGFR, and A/LEU145 and B/LEU238 in PTGS2 all form π-alkyl interaction. Andrographidine F interacts with multiple amino acids in MAPK14. For example, the electron cloud formed by the benzene ring in the andrographidine F structure and the benzene ring in the phenylalanine residue A/PHE169 are parallel to each other to form π-π conjugation. The hydroxyl group forms an intermolecular hydrogen bond with the glycine residue A/GLY170, and has A π-σ bond with A/THR106. Van der Waals forces and π-alkyl interactions are also available for alkyl interactions with multiple amino acid residues. 5-hydroxy-7-methoxy-2-(3,4,5-trimethoxyphenyl)chromone has intermolecular hydrogen bonding interaction with A/VAL186 and A/TYR104 in NFKB1, and it also has π-alkyl interaction with A/VAL186. A/TYR104 also forms a π-π T-shaped interaction with the benzene ring in the flavonoid structure. 5-hydroxy-7-methoxy-2-(3,4,5-trimethoxyphenyl)chromone has van der Waals force with glutamine residue A/GLN252 and histidine residue A/HIS190. It is worth noting that there is a π-cation bond between the benzene ring in its ingredient structure and the cation in A/ARG253.
Figure 8.
Typical binding mode of the trusted target proteins with the active ingredients of HDHs by molecular docking. (Discovery Studio 2016 Client) (A) Quercetin tetramethyl(3′,4′,5,7) ether and IL6; (B) Quercetin tetramethyl(3′,4′,5,7) ether and CASP3; (C) Stigmasterol and ACE2; (D) 5-hydroxy-7-methoxy-2-(3,4,5-trimethoxyphenyl)chromone and NFKB1. (The rest of the figures have been uploaded to the attachment).
Discussion
The holistic theory of TCM and treatment based on syndrome differentiation have unique advantages in the prevention and treatment of epidemic diseases. The Huangdi Neijing holds: the healthy-qi is stored in the inside, and pathogenic-qi cannot be interfered with. Healthy-qi refers to the ability of the human body to defend against viral and bacterial infections, maintaining the internal balance of the body and immune supervision [10,11]. The purpose of TCM treatment is to balance the physiological homeostasis of the body to fight coronavirus [12]. The first pathological changes caused by coronavirus are mainly in the lung, leading to immune system damage and injury to other organs. In this study, 27 active ingredients of 7 HDHs and their corresponding 14 targets were obtained. The heat-clearing effect mainly relieves the inflammatory storm through the anti-inflammatory pathway, while the detoxifying effect prevents the reproduction and transmission of coronavirus in vivo.
The first way is to alleviate the inflammatory storm caused by coronavirus infection
Studies have shown that plasma inflammatory factors such as IL-2, IL-7, IL-10, G-CSF, IP10, MCP-I, MIP-Ia, and TNF-α are significantly increased in patients with severe COVID-19 [13]. The reason for this is the inflammatory storm caused by viral infection, which is caused by the overreaction of the immune system caused by some pathogens, resulting in the overexpression of inflammatory cytokines, leading to large-scale inflammatory cascade reaction, and subsequent damage to multiple organs of the body [14]. Wogonin, chryseriol, andrographidine F, moslosooflavone, quercetin tetramethyl(3′,4′,5,7) ether, mono-o-methylwightin, and other similar flavonoids, as well as indirubin and isolariciresinol, affect the TNF, TLR, and MAPK receptor signaling pathways. MAPK14 and PTGS2 act on the signaling pathway as well. MAPK14 plays an important role in the cellular response cascade caused by pro-inflammatory cytokines or physiological stress that directly activates the activation of transcription factors [15]. PTGS2 is a cyclooxygenase 2 (COX-2) that converts arachidonic acid into prostaglandin H2 (PGH2) [16] and mediates inflammatory reactions [17]. The TNF and MAPK signaling pathways cause damage to the lungs during activation [18]. The MAPK pathway is an important potential target for inflammation and viral infection [19]. After pathogen antigen stimulation, TLRs recruit downstream adaptor molecules. TLRs such as TLR-2 and TLR-4 activate intracellular signal transduction cascades, upregulate gene transcription, increase the release of inflammatory mediators, and produce pro-inflammatory molecules, intensifying the body inflammatory response [20]. In addition, wogonin can reduce the production of TNF-α, CASP3, IL-6, and other pro-inflammatory factors, weaken the nuclear localization of NF-κB in macrophages, inhibit the mRNA transcription of the corresponding inflammatory molecules, reduce lung inflammatory cell infiltration, and improve the acute lung injury induced by influenza A virus [21].
Quercetin reduces the release of pro-inflammatory factors by acting on inflammation-related factors such as IL-6, CASP3, PTGS2, NOS3, and EGFRA. At the same time, quercetin can inhibit the activation of inflammation-related pathways such as TNF signal transduction, TLR, MAPK receptor, PI3K-AKT, and HIF-1 signaling pathways. Among them, IL-6 can activate the NF-κB signaling pathway, promote the release of inflammatory factors, and promote the inflammatory response, leading to the production of cytokines. The higher the IL-6 index, the worse the lung function [22]. Downregulation of EGFR can inhibit activation of the PI3K-AKT signaling pathway, promote cell apoptosis, inhibit secondary inflammation, reduce pneumonia symptoms, and delay acute lung injury [23]. Quercetin can also act on CASP3 and inhibit the MAPK and TNF signaling pathways. PTGS2 can convert arachidonic acid into PGH2, which is closely related to airway inflammation, regulates the TNF signaling pathway, inhibits inflammation, reduces inflammatory factors, and protects lung tissue. NO produced by the NOS3 reaction can participate in the inflammatory response, affect pro-inflammatory mediators, and regulate the PI3K-AKT and HIF-1 signaling pathways. The PI3K-AKT signaling pathway plays an important negative regulatory role in the pulmonary inflammatory response [24]. Activation of the HIF-1α signaling pathway can ameliorate acute lung injury induced by lipopolysaccharide by reducing inflammation and inhibiting oxidative stress [25,26].
Both luteolin and indirubin can regulate the TNF, TLR, MAPK receptor, PI3K-AKT, and HIF-1 signaling pathways, thereby inhibiting inflammatory cytokines and expression of inflammatory mediators [27]. The TLR4/NF-κB signaling pathway can stimulate the release of a large number of inflammatory factors, causing macrophages and neutrophils to accumulate in the lungs, leading to lung tissue damage and accelerating progression of disease. Studies have shown that indirubin can reduce the expression of inflammatory factors TNF-α, IL-1β, and IL-6, increase the production of anti-inflammatory factor IL-10, and reduce secondary toxic pneumonia [28].
β-Sitosterol can exert anti-inflammatory effects by inhibiting the release of inflammatory factors NF-κB, PTGS2, and CASP3. It can also inhibit the activation of NF-κB, TNF, and MAPK signaling pathways, thereby protecting against acute lung injury caused by lipopolysaccharide [29]. NFKB1 is involved in the inflammatory response, regulation of immunity, and transcriptional regulation of related genes [30]. By inhibiting the expression of NFkB1 in pulmonary microvascular endothelial cells, it can inhibit inflammation and protect against acute lung injury [31]. The lignan ingredient isolariciresinol can also regulate the HIF-1 and PI3K-AKT signaling pathways by acting on NOS3, reducing inflammation, and protecting against lung injury.
The second way directly acts on all links of the virus infection pathway to prevent the reproduction and spread of coronavirus in the body
Quercetin can inhibit infection by influenza A virus, Salmonella, Mycobacterium tuberculosis, Bordetella pertussis, Helicobacter pylori, and hepatitis C virus (HCV) by acting on EGFR and RELA. Reducing viral intracellular replication inhibits virus-induced apoptosis, thereby achieving antiviral effects. Quercetin can also act on RELA and EGFR to regulate the Ras signaling pathway. EGFR is related to the receptor for virus entry into host cells [32], Ras is the renin-angiotensin system, and ACE2 is a protective factor that inhibits lung injury and multiple-organ failure caused by the classic Ras axis. The treatment of coronavirus infection occurs in vivo through biaxial regulatory pathways: ACE/AngII/AT1R axis and ACE2/Ang(1–7)/Mas axis [33]. According to the molecular docking results, the total score of quercetin combined with ACE2 was 3.30, similar to the total score of 3.35 of ribavirin and ACE2. Both SARS-CoV-2 and SARS-CoV combine their expressed S-protein with ACE2 in the human body, causing the virus to invade the body and cause disease [7,34,35]. Luteolin can also act on EGFR through the Ras signaling pathway and can regulate the expression of apoptosis-related genes in exogenous pathways and mitochondrial pathways, thereby inhibiting cell apoptosis induced by influenza virus infection, with a broad-spectrum antiviral effect [36]. Quercetin tetramethyl(3′,4′,5,7) ether, mono-o-methylwightin, moslosooflavone, andrographidine F, chryseriol, and other similar flavonoids can inhibit influenza A virus, Bordetella pertussis, Mycobacterium tuberculosis, Salmonella, Helicobacter pylori, and other viruses by acting on MAPK14. In addition, chryseriol exhibits antibacterial properties [37]. Quercetin, indirubin, and isolariciresinol also act on NOS3 to participate in the ACE protein inhibitor pathway of the spike protein target of SARS-CoV and SARS-CoV-2 [38].
Third, the prevention and treatment of coronavirus infection by regulating the body immunity
Quercetin and luteolin both act on IL-6 through the NOD-like receptor signaling pathway to regulate the immune response. IL-6 can induce B cells to differentiate into immunoglobulin secreting cells, produce antibodies, induce T-cell proliferation, and participate in the immune response [39]. In addition, quercetin acts on NOS3 to participate in the physiological processes of the immune system and regulation of biological signals. Wogonin acts on IL-6 and MAPK14 through the NOD-like receptor signaling pathway. Abnormal expression of MAPK14 can cause physiological dysfunction in the body and autoimmune dysfunction.
Conclusions
In summary, the 7 HDHs, Lonicerae Japonicae Flos, Forsythiae Fructus, Isatidis Folium, Isatidis Radix, Indigo Naturalis, Dryopteridis Crassirhizomatis Rhizoma, and Andrographis Herba, are mainly composed of flavonoids, quercetin, luteolin, wogonin, mother-nuclear similar moslosooflavone, mono-o-methylwightin, quercetin tetramethyl(3′,4′,5,7) ether, andrographidine F, chryseriol, alkaloid ingredient indirubin, steroid ingredient β-sitosterol, and lignan ingredient isolariciresinol to act on NOS3, EGFR, IL-6, MAPK8, PTGS2, MAPK14, NFKB1, CASP3, and ACE2. Multiple pathways and multiple targets are involved in inflammatory responses, activation of MAPK activity, positive regulation of NF-κB transcription factor activity, protein binding, ATP binding, and other biological processes, thereby regulating inflammation-related pathways, including TLR, TNF, MAPK, HIF-1, NF-κB, and PI3K-AKT signaling pathways. Pathways related to viral and bacterial infections include Chagas disease (American trypanosomiasis), Pertussis, Influenza A, Tuberculosis, Salmonella infection, Toxoplasmosis, Hepatitis B, epithelial cell signaling in Helicobacter pylori infection, Hepatitis C, and Ras signaling pathway. Immune-related pathways include NOD-like receptor signaling pathways. They correspond to the “lung”, which is the first onset of coronavirus infection, thus exerting synergistic effects such as anti-inflammatory, immune regulation, and antiviral effects. Viruses mutate rapidly, and the design of new drugs is time-consuming and labor-intensive. In the prevention and treatment of diseases caused by viruses, TCM are remarkably effective, have rare sequelae, and are not easily affected by viral mutations. The characteristics of TCM, including multi-pathway, multi-target, and integrity, can effectively deal with the high variability of viruses. In this study, a variety of algorithms combined with virtual screening and computer simulation of molecular docking were used to screen and verify the ingredients of HDHs that can be used to prevent and treat coronavirus infections, and explain its mechanism of action. The systematic elucidation of the molecular mechanism of Chinese medicine to prevent and treat viral infections can help in the development of clinical drugs for the prevention and treatment of viral infections and provide alternative treatment strategies for the treatment of epidemics caused by viruses.
Acknowledgements
We acknowledge and thank the Collaborative Innovation Center of Shaanxi University of Chinese Medicine, Tianjin Institute of Pharmaceutical Research, and the Center of Bioinformatics, Northwest A&F University supports Research work.
Abbreviations
- HDHs
heat-clearing and detoxifying herbs
- TCM
Traditional Chinese medicines
- ADME
absorption, distribution, metabolism, and excretion
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OB
Oral Bioavailability
- DL
drug-likeness
- IL-6
Interleukin-6
- NOS3
nitric oxide synthase, endothelial
- EGFR
epidermal growth factor receptor
- MAPK1
mitogen-activated protein kinase 1
- MAPK3
mitogen-activated protein kinase 3
- MAPK8
mitogen-activated protein kinase 8
- MAPK14
mitogen-activated protein kinase 14
- PTGS2
prostaglandin G/H synthase 2
- NFKB1
nuclear factor NF-κ-B p105 subunit
- CASP3
Caspase-3
- ACE2
angiotensin-converting enzyme 2
- LCK
tyrosine-protein kinase LCK
- PPARG
peroxisome proliferator-activated receptor gamma
- RELA
transcription factor p65
- NF-κB
nuclear factor kappa-B
Footnotes
Conflict of interest: None declared
Declaration of Figures’ Authenticity
All figures submitted have been created by the authors who confirm that the images are original with no duplication and have not been previously published in whole or in part.
Financial support: This study was supported by the National Natural Science Foundation of China (No. 81973754), School-level Scientific Research Project of Shaanxi University of Chinese Medicine (2020GP09), and the 2018 Public Health Service Subsidy Fund Special “The project of national survey on Chinese material medica resources” (CS [2018] No. 43)
References
- 1.Zhou SS, Li WN, Ai ZZ, et al. Investigating mechanism of Qingfei Dayuan Granules for treatment of COVID-19 based on network pharmacology and molecular docking. Zhong Cao Yao. 2020;51(7):1804–13. [Google Scholar]
- 2.Pei L, Shen X, Yan Y, et al. Virtual screening of the multi-pathway and multi-gene regulatory molecular mechanism of dachengqi decoction in the treatment of stroke based on network pharmacology. Comb Chem High Throughput Screen. 2020;23(8):775–87. doi: 10.2174/1386207323666200311113747. [DOI] [PubMed] [Google Scholar]
- 3.Pei L, Shen X, Qu K, et al. Exploration of the two-way adjustment mechanism of rhei radix et rhizoma for cardiovascular diseases. Comb Chem High Throughput Screen. 2020;23(10):1100–12. doi: 10.2174/1386207323666200521120308. [DOI] [PubMed] [Google Scholar]
- 4.Tahir Ul Qamar M, Alqahtani SM, et al. Structural basis of SARS-CoV-2 3CL(pro) and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal. 2020;10(4):313–19. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unal MA, Bitirim CV, Summak GY, et al. Ribavirin shows antiviral activity against SARS-CoV-2 and downregulates the activity of TMPRSS2 and the expression of ACE2 in vitro. Can J Physiol Pharmacol. 2021;99(5):449–60. doi: 10.1139/cjpp-2020-0734. [DOI] [PubMed] [Google Scholar]
- 6.Tong S, Su Y, Yu Y, et al. Ribavirin therapy for severe COVID-19: A retrospective cohort study. Int J Antimicrob Agents. 2020;56(3):106114. doi: 10.1016/j.ijantimicag.2020.106114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–80. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu J. The latest progress in research on clinical therapeutic agents of COVID-19. Zhong Cao Yao. 2021;30(02):154–61. [Google Scholar]
- 9.Hung IF, Lung KC, Tso EY, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: An open-label, randomised, phase 2 trial. Lancet. 2020;395(10238):1695–704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan YP, Wang YP, Zhang HM, Wang YY. Analysis on the treatment of New Coronavirus Pneumonia (COVID-19) from the cold epidemic treatment. Journal of Traditional Chinese Medicine. 2020;61(5):369–74. [Google Scholar]
- 11.Lee DYW, Li QY, Liu J, Efferth T. Traditional Chinese herbal medicine at the forefront battle against COVID-19: Clinical experience and scientific basis. Phytomedicine. 2021;80:153337. doi: 10.1016/j.phymed.2020.153337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan QZ, Huang YJ, Lin SH, Chu QM. Study on active compounds of Yupingfeng San for prevention of coronavirus disease 2019(COVID-19) based on network pharmacology and molecular dockin. Zhong Cao Yao. 2020;51(07):1731–40. [Google Scholar]
- 13.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H, Wang F, Zhang P, et al. Management of cytokine release syndrome related to CAR-T cell therapy. Front Med. 2019;13(5):610–17. doi: 10.1007/s11684-019-0714-8. [DOI] [PubMed] [Google Scholar]
- 15.Xu P, Derynck R. Direct activation of TACE-mediated ectodomain shedding by p38 MAP kinase regulates EGF receptor-dependent cell proliferation. Mol Cell. 2010;37(4):551–66. doi: 10.1016/j.molcel.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orlando BJ, Malkowski MG. Crystal structure of rofecoxib bound to human cyclooxygenase-2. Acta Crystallogr F Struct Biol Commun. 2016;72(Pt 10):772–76. doi: 10.1107/S2053230X16014230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucido MJ, Orlando BJ, Vecchio AJ, Malkowski MG. Crystal structure of aspirin-acetylated human cyclooxygenase-2: Insight into the formation of products with reversed stereochemistry. Biochemistry. 2016;55(8):1226–38. doi: 10.1021/acs.biochem.5b01378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen LY, Yi L, Zhou MJ, et al. Effect of Euphorbia helioscopia alcohol extract on MAPK/NF-κB inflammation pathway on mice with acute lung injury induced by LPS. Zhongguo Shi Yan Fang Ji Xue Za Zhi. 2020;26(20):46–51. [Google Scholar]
- 19.Meineke R, Rimmelzwaan GF, Elbahesh H. Influenza virus infections and cellular kinases. Viruses. 2019;11(2):171. doi: 10.3390/v11020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyavar SR, Singh R, Emani R, et al. Role of toll-like receptor 7/8 pathways in regulation of interferon response and inflammatory mediators during SARS-CoV2 infection and potential therapeutic options. Biomed Pharmacother. 2021;141:111794. doi: 10.1016/j.biopha.2021.111794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong Z, Shen Q, Jiang J, et al. Wogonin improves functional neuroprotection for acute cerebral ischemia in rats by promoting angiogenesis via TGF-β1. Ann Transl Med. 2019;7(22):639. doi: 10.21037/atm.2019.10.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Qin Y, Cui HS, et al. Clinical observation of Sangmei Zhike Granules in treating postinfectious cough with syndrome of yin deficiency by wind heat. Zhonghua Zhong Yi Yao Za Zhi. 2018;33(10):4752–55. [Google Scholar]
- 23.Wu S, Jin XH, Shi SS, et al. Antiviral effects of quercetin and andrographolide in vitro. Zhong Yao Cai. 2012;35(12):2003–6. [Google Scholar]
- 24.Cai B, Gan X, He J, et al. Morin attenuates cigarette smoke-induced lung inflammation through inhibition of PI3K/AKT/NF-κB signaling pathway. Int Immunopharmacol. 2018;63:198–203. doi: 10.1016/j.intimp.2018.07.035. [DOI] [PubMed] [Google Scholar]
- 25.Han F, Wu G, Han S, et al. Hypoxia-inducible factor prolyl-hydroxylase inhibitor roxadustat (FG-4592) alleviates sepsis-induced acute lung injury. Respir Physiol Neurobiol. 2020;281:103506. doi: 10.1016/j.resp.2020.103506. [DOI] [PubMed] [Google Scholar]
- 26.He C, Zhang W, Li S, et al. Edaravone improves septic cardiac function by inducing an HIF-1α/HO-1 pathway. Oxid Med Cell Longev. 2018;2018:5216383. doi: 10.1155/2018/5216383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aziz N, Kim MY, Cho JY. Anti-inflammatory effects of luteolin: A review of in vitro, in vivo, and in silico studies. J Ethnopharmacol. 2018;225:342–58. doi: 10.1016/j.jep.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 28.Wang Q, Yu J, Hu Y, et al. Indirubin alleviates bleomycin-induced pulmonary fibrosis in mice by suppressing fibroblast to myofibroblast differentiation. Biomed Pharmacother. 2020;131:110715. doi: 10.1016/j.biopha.2020.110715. [DOI] [PubMed] [Google Scholar]
- 29.Zhou BX, Li J, Liang XL, et al. β-sitosterol ameliorates influenza A virus-induced proinflammatory response and acute lung injury in mice by disrupting the cross-talk between RIG-I and IFN/STAT signaling. Acta Pharmacol Sin. 2020;41(9):1178–96. doi: 10.1038/s41401-020-0403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bryant VL, Tangye SG. The expanding spectrum of NFkB1 deficiency. J Clin Immunol. 2016;36(6):531–32. doi: 10.1007/s10875-016-0310-5. [DOI] [PubMed] [Google Scholar]
- 31.Tang J, Xu L, Zeng Y, Gong F. Effect of gut microbiota on LPS-induced acute lung injury by regulating the TLR4/NF-κB signaling pathway. Int Immunopharmacol. 2021;91:107272. doi: 10.1016/j.intimp.2020.107272. [DOI] [PubMed] [Google Scholar]
- 32.Miao ZW, Xu Y, Ning LQ, et al. Network pharmacological analysis and preliminary validation of mechanisms of Baitouweng Decoction in treatment of ulcerative colitis. Zhongguo Zhong Yao Za Zhi. 2020;45(08):1808–15. doi: 10.19540/j.cnki.cjcmm.20191112.406. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Wang X, Zhou X, et al. Intervention mechanism of traditional chinese medicine on Corona Virus Disease 2019 (COVID-19) based on RAS Axes. Chinese Archives of Traditional Chinese Medicine. 2020;38(8):12–16+259. [Google Scholar]
- 34.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5(4):562–69. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zong Y, Ding ML, Ma ST, Ju WZ. Investigation of potential Chinese materia medica and its monomers in treatment of coronavirus disease 2019(COVID-19) based on angiotensin converting enzyme II(ACE2) receptor. Zhong Cao Yao. 2020;51(5):1123–29. [Google Scholar]
- 36.Deng DY, Gu LG, Liu XT, et al. Luteolin’s intervention effect and its mechanism of apoptosis induced by H1N1 in vitro. Zhonghua Zhong Yi Yao Za Zhi. 2017;32(04):1524–27. [Google Scholar]
- 37.Jang YH, Park JR, Kim KM. Antimicrobial activity of chrysoeriol 7 and chochlioquinone 9, white-backed planthopper-resistant compounds, against rice pathogenic strains. Biology (Basel) 2020;9(11):382. doi: 10.3390/biology9110382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, Tian X, Wang X, et al. The discussion of material basis and mechanism of action of Qingre Huashi Kangdu Prescription on the treatment of COVID-19 based on network pharmacology and molecular docking. Zhong Yao Cai. 2021;(03):758–68. [Google Scholar]
- 39.Huang J, Zhang B, Lin ZJ. Intervention effect of Chinese Medicine on interleukin cytokines and thinking on its prevention and treatment for inflammatory storm of COVID-19. Zhong Yao Yao Li Yu Lin Chuang. 2020;36(2):23–28. [Google Scholar]