Abstract
Actin cytoskeletal reorganization plays an important role in regulating smooth muscle contraction, which is essential for the modulation of various physiological functions including airway tone. The adapter protein Abi1 (Abelson interactor 1) participates in the control of smooth muscle contraction. The mechanisms by which Abi1 coordinates smooth muscle function are not fully understood. Here, we found that contractile stimulation elicited Abi1 acetylation in human airway smooth muscle (HASM) cells. Mutagenesis analysis identified lysine‐416 (K416) as a major acetylation site. Replacement of K416 with Q (glutamine) enhanced the interaction of Abi1 with neuronal Wiskott‐Aldrich syndrome protein (N‐WASP), an important actin‐regulatory protein. Moreover, the expression of K416Q Abi1 promoted actin polymerization and smooth muscle contraction without affecting myosin light chain phosphorylation at Ser‐19 and vimentin phosphorylation at Ser‐56. Furthermore, p300 is a lysine acetyltransferase that catalyzes acetylation of histone and non‐histone proteins in various cell types. Here, we discovered that a portion of p300 was localized in the cytoplasm of HASM cells. Knockdown of p300 reduced the agonist‐induced Abi1 acetylation in HASM cells and in mouse airway smooth muscle tissues. Smooth muscle conditional knockout of p300 inhibited actin polymerization and the contraction of airway smooth muscle tissues without affecting myosin light chain phosphorylation and vimentin phosphorylation. Together, our results suggest that contractile stimulation induces Abi1 acetylation via p300 in smooth muscle. Acetylation at K416 promotes the coupling of Abi1 with N‐WASP, which facilitates actin polymerization and smooth muscle contraction. This is a novel acetylation‐dependent regulation of the actin cytoskeleton in smooth muscle.
Keywords: actin cytoskeleton, adapter protein, contraction, smooth muscle
Abbreviations
- Abi1
Abelson interactor 1
- ACh
acetylcholine
- GAPDH
glyceraldehyde 3‐phosphate dehydrogenase
- HASM
human airway smooth muscle
- KD
knockdown
- MLC
myosin light chain
- N‐WASP
neuronal Wiskott‐Aldrich Syndrome Protein
- siRNA
small interfering RNA
- WT
wild type
1. INTRODUCTION
Smooth muscle contractility plays a critical role in regulating a variety of physiological functions of human body including airway tone and blood pressure. Contractile activation leads to the phosphorylation of 20‐kDa myosin light chain (MLC20), which triggers cross‐bridge cycling and myofilament sliding. In addition, contractile stimulation elicits actin polymerization and reorganization, which facilitates smooth muscle contractility by promoting force transmission between the contractile units and the extracellular matrix, 1 , 2 , 3 , 4 and enhancing the number of contractile units. 3 , 5 , 6 Furthermore, contractile stimulation increases the phosphorylation of the intermediate filament protein vimentin, which facilitates the contractility of smooth muscle and endothelial cells. 7 , 8 , 9 , 10 Despite a wealth of information available for myosin and vimentin regulation, the cellular and molecular mechanisms that control actin polymerization in smooth muscle are not fully elucidated.
The adapter protein Abi1 (Abelson interactor 1) participates in the regulation of actin polymerization, 11 intercellular adhesion, 12 cardiovascular development, 13 and cell migration. 14 , 15 , 16 In smooth muscle, knockdown of Abi1 reduces force development upon contractile stimulation. 17 Abi1 complexes with neuronal Wiskott‐–Aldrich syndrome protein (N‐WASP), which subsequently promotes actin filament branching and growth in smooth muscle, 17 which facilitates smooth muscle contraction. 1 , 2 , 3 , 4
Molecular mapping suggests that the interaction of Abi1 with N‐WASP is mediated by the SH3 domain of Abi1 and the proline‐rich domain of N‐WASP. 3 , 11 , 18 The binding of Abi1 to N‐WASP is sufficient to activate N‐WASP and initiate actin polymerization in in vitro biochemical studies. 11 The cellular mechanism that modulates the complexing of Abi1 with N‐WASP is largely unknown.
Nε‐lysine acetylation of nuclear proteins including histone regulates chromatin remodeling, DNA replication, gene transcription, and cell proliferation. 19 Recent studies suggest that cellular proteins also undergo lysine acetylation/deacetylation in mammalian cells in response to intracellular and extracellular cues. For instance, the actin‐binding protein cortactin undergoes deacetylation in cells, which affects its binding to F‐actin and cell migration 20 and contraction. 21
p300 is a histone acetyltransferase that plays a role in regulating gene transcription and cell proliferation. 22 , 23 , 24 p300 promotes histone acetylation, which induces chromatin unpacking and modulates gene expression. 22 In addition, p300 induces acetylation of the non‐histone protein Nrf in macrophages, which has been implicated in the pathogenesis of chronic obstructive pulmonary disease. 24 The functional role of p300 in smooth muscle is largely unknown.
Here, we find that Abi1 undergoes acetylation in smooth muscle in response to contractile stimulation, which regulates actin cytoskeletal reorganization and smooth muscle contraction. These processes are regulated by p300 lysine acetyltransferase during contractile activation.
2. MATERIALS AND METHODS
2.1. Cell culture
Human airway smooth muscle (HASM) cells were prepared from human bronchi and adjacent tracheas obtained from the International Institute for Advanced Medicine. 17 , 25 , 26 , 27 , 28 Human tissues were non‐transplantable, and informed consents were obtained from all subjects for research. This study was approved by the Albany Medical College Committee on Research Involving Human Subjects. Briefly, muscle tissues were incubated for 20 minutes with dissociation solution [130 mM NaCl, 5 mM KCl, 1.0 mM CaCl2, 1.0 mM MgCl2, 10 mM Hepes, 0.25 mM EDTA, 10 mM D‐glucose, 10 mM taurine, pH 7, 4.5 mg collagenase (type I), 10 mg papain (type IV), 1 mg/ml BSA, and 1 mM dithiothreitol]. All enzymes were purchased from Sigma‐Aldrich. The tissues were then washed with Hepes‐buffered saline solution (composition in mM: 10 Hepes, 130 NaCl, 5 KCl, 10 glucose, 1 CaCl2, 1 MgCl2, 0.25 EDTA, and 10 taurine, pH 7). The cell suspension was mixed with Ham's F12 medium supplemented with 10% (v/v) fetal bovine serum (FBS) and antibiotics (100 units/mL penicillin and 100 µg/mL streptomycin). Cells were cultured at 37℃ in the presence of 5% CO2 in the same medium. The medium was changed every 3‐4 days until cells reached confluence, and confluent cells were passaged with trypsin/EDTA solution. 25 , 29 Smooth muscle cells within passage 5 were used for the studies. 30 Smooth muscle cells were serum starved for 20 hours before experiments. Cells from three donors were used for the experiments. In some cases, duplicated experiments were performed for cells from a donor. 30 , 31
2.2. Immunoblot analysis and co‐immunoprecipitation
Western blotting of cell lysis and co‐immunoprecipitation were performed using the experimental procedures as previously described. 8 , 26 , 30 , 32 , 33 Antibodies used were anti‐Abi1 (1:1500, Sigma #A5106‐200UL, L/N 076M4842V), anti‐GAPDH (1:1500, Ambion #AM4300, L/N 1311029), and anti‐α‐actin (Sigma, A2647, #22190320, 1:3000), anti‐phospho‐myosin light chain (Ser‐19, Santa Cruz Biotechnology, 1:500), Ac‐lysine Antibody (Santa Cruz Biotechnology, S7F8, sc‐81623), anti‐N‐WASP (Santa Cruz Biotechnology, #sc‐10121, L/N G2211 1:500), anti‐myosin light chain (1:1000, a gift of Dr Gunst), anti‐phospho‐N‐WASP (Y256) (EMD Millipore, #AB1966, L/N 2795491,2838736, 1:1000), and anti‐vimentin (BD Biosciences, #550515, L/N 3214517, 1:10 000). Phospho‐vimentin (Ser56) antibody (1:500) was produced as previously described. 29 , 34 The antibodies were validated by examining the molecular weight of target proteins. In addition, anti‐Abi1 was validated using Abi1 KD cells. 17 Finally, vendors have provided datasheet to show that antibodies were validated by positive controls. The levels of proteins were quantified by scanning densitometry of immunoblots (Fuji Multi Gauge Software or GE IQTL software). The luminescent signals from all immunoblots were within the linear range.
2.3. Immunofluorescence microscopy
Cells were plated in dishes containing coverslips followed by fixation and permeabilization. 25 , 29 , 34 , 35 These cells were immunofluorescently stained using primary antibody followed by appropriate secondary antibody conjugated to Alexa‐488 or Alexa‐555 (Invitrogen, Thermo Fisher Scientific). The cellular localization of fluorescently labeled proteins was viewed using a Leica DMI600 microscope or a Leica TCS SPE confocal microscope (Buffalo Grove, IL, United States). Image analysis for protein localization was performed using the previously described method with minor modifications. 36 Using Leica DMI 6000 software, the pixel intensity was quantified for minimal five line scans across the periphery of cells (nuclei excluded). Ratios of pixel intensity at the cell edge to pixel intensity at the cell interior were determined for each line scan as follows: ratios of the average maximal pixel intensity at the cell periphery to minimal pixel intensity in the cell interior. The ratios of pixel intensity at the cell border to that in the cell interior for all the line scans performed on a given cell were averaged to obtain a single value for the ratio of each cell.
2.4. Site‐directed mutagenesis
Wild‐type Abi1 construct is a gift of Dr Pendergast of Duke University. Different Abi1 acetylation mimic mutants were generated using a Quick change II XL site‐directed mutagenesis kit (Agilent Technologies) as previously described. 37 , 38 For K388Q mutant, the sequence of forward primer was 5′‐CCA AGA ATT ATA TTG AGC AAG TTG TTG CAA TAT ATG‐3′. The sequence of reverse primer was 5′‐CAT ATA TTG CAA CAA CTT GCT CAA TAT AAT TCT TGG‐3′. For K397Q mutant, the sequence of forward primer was 5′‐CAA TAT ATG ATT ATA CAC AAG ACA AGG ATG ATG AGC TG‐3′. The sequence of reverse primer was 5′‐CAG CTC ATC ATC CTT GTC TTG TGT ATA ATC ATA TAT TG‐3′. For K399Q mutant, the sequence of forward primer was 5′‐GAT TAT ACA AAA GAC CAG GAT GAT GAG CTG TC‐3′. The sequence of reverse primer was 5′‐GAC AGC TCA TCA TCC TGG TCT TTT GTA TAA TC‐3′. For K415Q mutant, the sequence of forward primer was 5′‐GCA ATC ATT TAT GTT ATA CAG AAG AAT GAT GAT GGC TGG‐3′. The sequence of reverse primer was 5′‐CCA GCC ATC ATC ATT CTT CTG TAT AAC ATA AAT GAT TGC‐3′. For K416Q mutant, the sequence of forward primer was 5′‐CAT TTA TGT TAT AAA GCA GAA TGA TGA TGG CTG G‐3′. The sequence of reverse primer was 5′‐CCA GCC ATC ATC ATT CTG CTT TAT AAC ATA AAT G‐3′. Plasmids were purified using the QIAPrep Spin Miniprep kit (Qiagen, Germany). DNA sequencing was performed by Genewiz. For Abi1 overexpression experiment, Abi1 KD cells were transfected with WT Abi1 and various mutants using the Fugene HD transfection reagent kit (Promega) according to the manual of the manufacture.
2.5. Precisioncut lung slices
Human lungs were obtained from the International Institute for Advanced Medicine. 17 , 25 , 26 , 27 , 28 Again, human tissues were non‐transplantable, and informed consents were obtained from all subjects for research. This study was approved by the Albany Medical College Committee on Research Involving Human Subjects. Bronchi (Generation 2‐3) were slowly filled with warm 2% low melting point agarose (type VII) prepared in Hank's Balanced Salt Solution (HBSS). Once the lungs were filled with agarose, bronchi were clamped to prevent leakage. Lungs were rinsed with ice‐cold HBSS supplemented with HEPES and placed at 4℃ overnight to allow the agarose to cool completely. Cubes (1‐1.5 cm) of lung tissue were prepared and cut transversely at 300 µm in HBSS supplemented with HEPES using a Vibratome (Leica VT1200S). The precisioncut lung slices (PCLS) were placed in warm Dulbecco's Modified Eagle Medium (DMEM) supplemented with antibiotics and incubated at 37°C with 5% CO2. Prior to experiments, PCLS were placed in the modified physiological saline solution (PSS; 110 mM NaCl, 3.4 mM KCl, 0.8 mM MgSO4, 4.8 mM CaCl2, 25 mM HEPES, and 1 g/L dextrose, pH 7.4) for 30 minutes in an incubator and subsequently placed in a 35‐mm glass bottom culture dish with the modified PSS. PCLS were treated with 100 µM ACh, and images of airway lumen were captured using a time‐lapse microscopy (Leica DMI 6000, 10× object, phase‐contrast). Change in lumen area was examined relative to that induced by stimulation of PCLS with ACh.
2.6. Transfection
For p300 knockdown, control siRNA (SC‐37007/Lot# K2320) and p300 siRNA (SC‐29431/Lot # L1918) were purchased from Santa Cruz Biotechnology. HASM cells were transfected with siRNA according to the manual of the manufacture (Santa Cruz Biotechnology). For tissue transfection, PCLS were treated with plasmids encoding WT or mutant Abi1 premixed with the FuGene HD transfection reagent for 48 hours according to the instruction of the manufacture (Promega).
2.7. Animals
All animal protocols were reviewed and approved by the Institutional Animal Care and Usage Committee (IACUC) of Albany Medical College. All experiments were strictly performed in accordance with approved protocols and regulations of IACUC. Animals were bred in the specific‐pathogen‐free housing of the Animal Research Facility, Albany Medical College. Male and female mice were randomly assigned to the experimental or control groups.
To generate smooth muscle conditional p300 knockout mice (p300smko mice), p300‐lox (B6.129P2Ep30/J) mice (from Jackson Laboratory; genetic background, C57BL/6) were crossed with B6. Cg‐Tg (Myh11‐cre,‐EGFP) 2Mik/J mice (from Jackson Laboratory, genetic background, C57BL/6J). p300‐lox and p300smko mice aging 12‐16 weeks were used for the experiments.
2.8. Assessment of tracheal ring contraction
Mice were euthanized by intraperitoneal injection of euthanasia solution (VEDCO, 0.1 mL/25 g). All experimental protocols were approved by the Institutional Animal Care and Usage Committee. A segment of tracheas (4‐5 mm in length) was immediately removed and placed in PSS containing 110 mM NaCl, 3.4 mM KCl, 2.4 mM CaCl2, 0.8 mM MgSO4, 25.8 mM NaHCO3, 1.2 mM KH2PO4, and 5.6 mM glucose. The solution was aerated with 95%O2‐5%CO2 to maintain a pH of 7.4. Two stainless steel wires were passed through the lumen of tracheal rings. One of the wires was connected to the bottom of organ baths and the other was attached to a Grass force transducer that had been connected to a computer with A/D converter (Grass). Tracheal segments were then placed in PSS at 37°C. At the beginning of each experiment, 0.5 g passive tension was applied to tracheal rings. After 60‐min equilibrium, they were stimulated with 80 mM KCl repeatedly until contractile responses and passive tension reached a steady state. Contractile force in response to acetylcholine was then measured.
2.9. Statistical analysis
All statistical analysis was performed using Prism software (GraphPad Software, San Diego, CA). Differences between pairs of groups were analyzed by Student's t test. A comparison among multiple groups was performed by one‐way or two‐way ANOVA followed by a post hoc test (Tukey's multiple comparisons). Values of n refer to the number of experiments used to obtain each value. P < .05 was considered to be statistically significant.
3. RESULTS
3.1. Contractile stimulation enhances Abi1 acetylation and the association of Abi1 with N‐WASP in smooth muscle cells
Because the state of cortactin acetylation has been implicated in cell contraction and migration, 20 , 21 we evaluated whether contractile stimulation alters the acetylation of Abi1 in smooth muscle. We used anti‐acetyl lysine antibody to blot extracts of smooth muscle cells treated with acetylcholine (ACh) for different time points. We found that Abi1 acetylation was increased in response to ACh stimulation, which peaked at 10 minutes after stimulation (Figure 1A). In addition, we found that ACh‐induced Abi1 acetylation was dose‐dependent (Figure 1B). Because there is no Abi1 acetylation‐specific antibody available, we immunoprecipated Abi1 from cell extracts, and immunoblotted with antibodies against Abi1 and acetyl lysine. We verified that ACh stimulation enhanced Abi1 acetylation in smooth muscle cells (Figure 1C).
FIGURE 1.
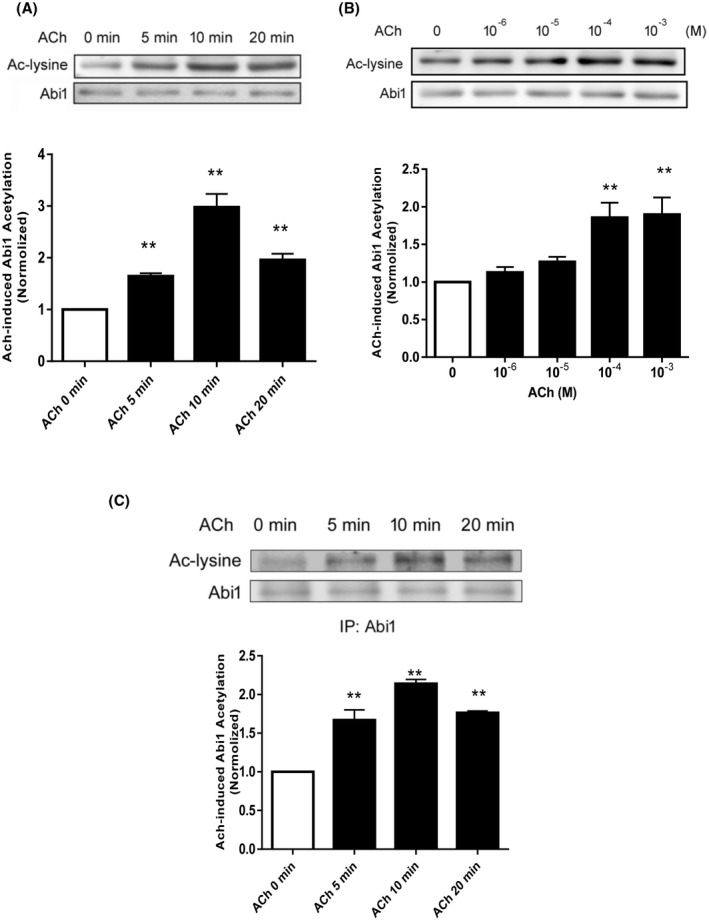
Contractile stimulation induces Abi1 acetylation in smooth muscle cells. A, Human airway smooth muscle (HASM) cells were stimulated with 10‐4 M acetylcholine (ACh) for different time points. Abi1 acetylation was evaluated by immunoblot analysis using acetyl lysine (Ac‐lysine) antibody and Abi1 antibody. Data are mean values of experiments from six batches of cell culture. Error bars indicate SE. B, HASM cells were treated with different doses of ACh for 10 minutes followed by immunoblotting with antibodies against acetyl lysine and Abi1. Data are mean values of experiments from four batches of cell culture. Error bars indicate SE. C, Extracts of cells treated with 10‐4 M ACh were immunoprecipitated with Abi1 antibody and blotted with antibodies against acetyl lysine and Abi1. Data are mean values of experiments from three batches of cell culture. Error bars indicate SE. **P < .01 compared to ACh, 0 minutes or 0 dose
We also assessed whether ACh stimulation affects the interaction of Abi1 with N‐WASP, one of the major targets for Abi1. 11 , 17 Coimmunoprecipitation analysis showed that the amount of N‐WASP in Abi1 precipitates was greater in ACh‐stimulated cells as compared to control cells (Figure 2A). Furthermore, ACh stimulation induced the redistribution of Abi1 and pN‐WASP (phospho‐N‐WASP (Y256), activated form) 39 from the myoplasm to the cell periphery (Figure 2B). Total N‐WASP also translocated to the cell edge (Figure 2C). These results suggest that contractile activation promotes Abi1 acetylation and the Abi1/N‐WASP coupling and translocation.
FIGURE 2.
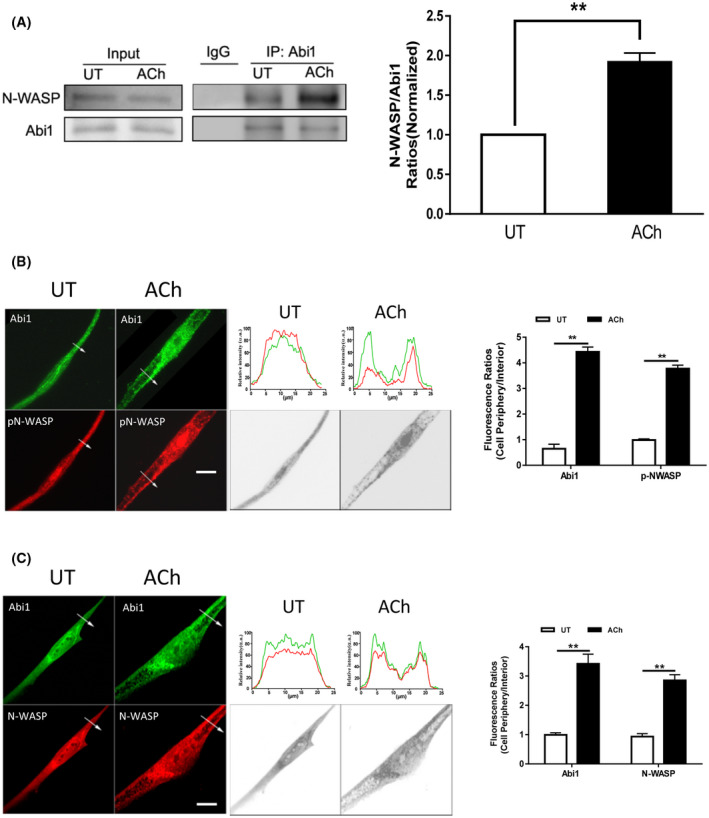
Contractile stimulation promotes Abi1/N‐WASP coupling and translocation. A, Input or Abi1 precipitates of untreated (UT) or ACh‐treated HASM cells were immunoblotted with antibodies against Abi1 or N‐WASP. ACh treatment increases the ratios of N‐WASP/Abi1 immunoprecipitates in cells. Data are mean values of experiments from four batches of cell culture. Error bars indicate SE. B and C, Representative micrographs illustrating the effects of ACh (10−4 M, 5 minutes) on the spatial localization of Abi1 and pN‐WASP or N‐WASP in HASM cells. The arrows indicate a single line scan to quantify the fluorescent signals in cells. The line scan graphs show relative fluorescent intensity. Black/white images show the shape and edge of these cells. Image analysis for protein localization was described in the section of Materials and Methods. Scale bar, 10 µm. Protein distribution in cells is expressed a ratio of the intensity at the cell periphery to cell interior. Data are mean values of experiments from three batches of cell culture. Error bars indicate SE. **P < .01
3.2. Acetylation of Abi1 at K416 regulates its interaction with N‐WASP
Previous studies demonstrate that the SH3 domain of Abi1 is a key motif that interacts with the proline domain of N‐WASP. 11 Because the acetylation of cortactin (an actin‐regulatory protein) regulates its binding to F‐actin, 20 we speculated that acetylation of the Abi1 SH3 domain may affect its interaction with N‐WASP. We analyzed the amino acid sequence of Abi1 SH3 domain (NP_001171592.1) and found that there are five potential acetyl‐lysine sites: K388, K397, K399, K415, and K416 (Figure 3A). To assess which lysine is the most important for its interaction with N‐WASP, we used glutamine (Q) to replace lysine at these five sites and generated the acetylation mimic mutants K388Q, K397Q, K399Q, K415Q, and K416Q. Coimmunoprecipitation analysis showed that the expression of K416Q, but not other mutants, enhanced its interaction with N‐WASP (Figure 3B). In addition, we assessed the effects of various Abi1 mutants on N‐WASP phosphorylation at Y256. The expression of K416Q enhanced N‐WASP phosphorylation at this residue in smooth muscle cells (Figure 3C). These results suggest that Abi1 acetylation at K416 modulates its interaction with N‐WASP and N‐WASP activation.
FIGURE 3.
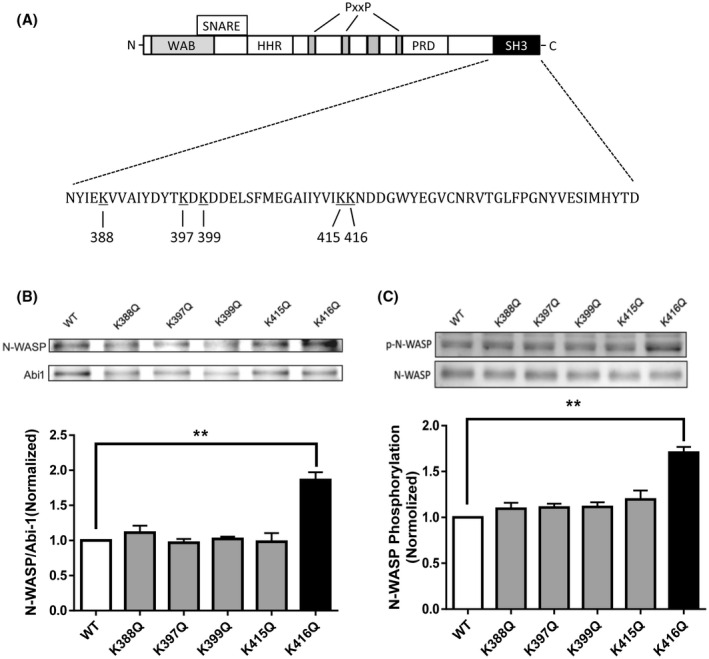
Acetylation of K416 increases the interaction of Abi1 with N‐WASP and N‐WASP phosphorylation in smooth muscle cells. A, Schematic diagram of Abi1 structural domains. Abi1 consists of several domains including WAB (WAVE‐binding domain), SNARE (SNARE‐binding domain), HHR (homeodomain homologous region), PRO (proline‐rich domain), and SH3 (Src homology 3). There are five lysines in the SH3 domain. PxxP, proline. B, Abi1 KD cells were transfected with wild‐type (WT) or various Abi1 mutants, and cell extracts were immunoprecipitated with Abi1 antibody and blotted with antibodies against Abi1 and N‐WASP. Data are mean values of three batches of cell culture. Error bars indicate SE. C, Extracts of cells expressing wild‐type (WT) or various mutants were blotted with antibodies against pN‐WASP (Y256) and total N‐WASP. Data are mean values of three batches of cell culture. Error bars indicate SE. *P < .01
3.3. Abi1 acetylation at K416 regulates its translocation and actin polymerization in smooth muscle cells
It has been shown that actin‐regulatory proteins translocate to the membrane, which facilitates cortical actin polymerization. 3 , 40 Thus, we used immunofluorescence microscopy to assess the cellular localization of K416Q. As shown in Figure 4A, WT Abi1 was largely distributed in the cytoplasm, whereas K416Q was localized at the cell periphery. In addition, pN‐WASP was largely positioned at the cytoplasm of cells expressing WT Abi1. In contrast, pN‐WASP was predominantly localized at the edge of cells expressing K416Q. Furthermore, we also found that the expression of K416Q was sufficient to increase F/G‐actin ratios in smooth muscle cells (Figure 4B). These results suggest that Abi1 acetylation at K416 modulates the translocation of Abi1, activated N‐WASP, and actin polymerization.
FIGURE 4.
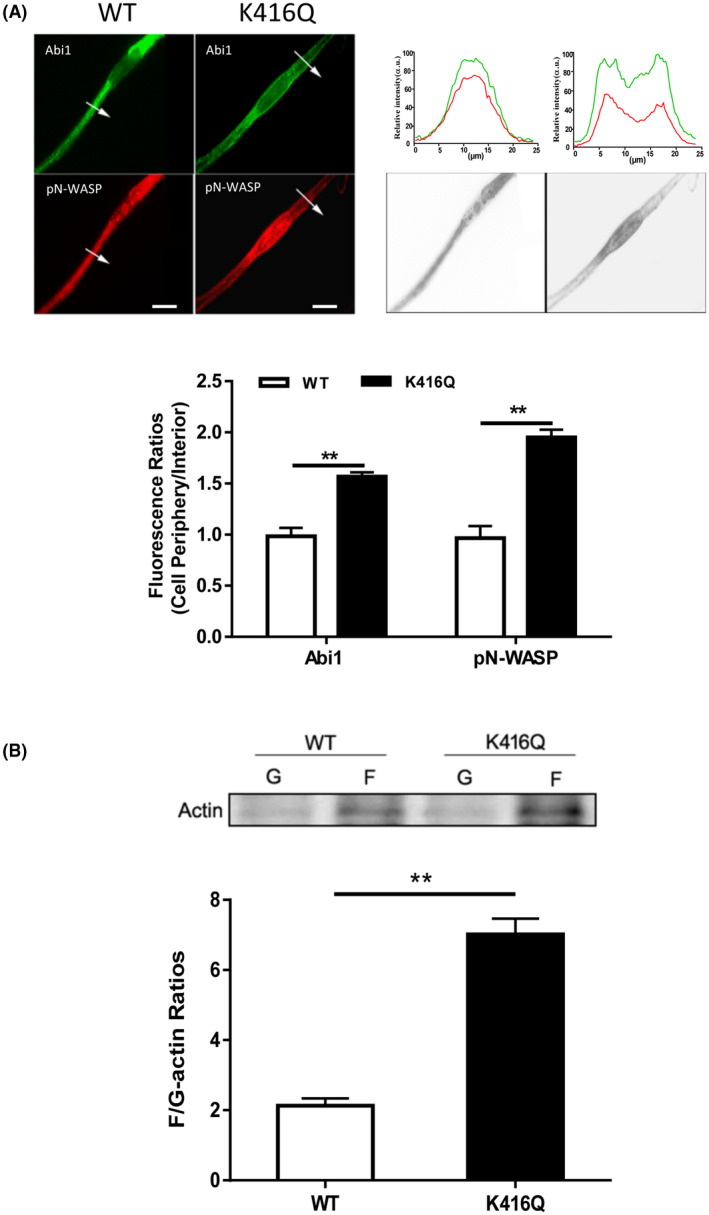
Acetylation of K416 enhances the translocation of Abi1 and pN‐WASP and actin polymerization. A, Cells expressing WT or K416Q Abi1 were immunostained for Abi1 and pN‐WASP. Data are mean values of three batches of cell culture. The arrows indicate a single line scan to quantify the fluorescent signals in cells. The line scan graphs show relative fluorescent intensity. Image analysis for protein localization was described in the section of Materials and Methods. Black/white images show the shape and edge of these cells. Scale bar, 10 µm. Error bars indicate SE. B, F/G‐actin ratios of cells expressing WT or K416Q Abi1 were evaluated using the immunoblot analysis. Data are mean values of four batches of cell culture. Error bars indicate SE. **P < .01
3.4. Abi1 acetylation at K416 modulates smooth muscle contraction without affecting myosin light chain phosphorylation and vimentin phosphorylation
To determine the functional consequence of Abi1 acetylation, we evaluated the effects of K416Q on smooth muscle contraction. Immunoblot analysis verified similar expression of WT and K416Q Abi1 in smooth muscle (Figure 5A). The expression of K416Q enhanced the contraction of human airways (Figure 5B). Since phosphorylation of myosin light chain and vimentin participates in the regulation of smooth muscle contraction, 8 , 9 we also assessed the effects of K416Q expression on these cellular processes. MLC phosphorylation at Ser‐19 and vimentin phosphorylation at Ser‐56 were not different between cells expressing WT and K416Q Abi1 (Figure 5C,D). These findings suggest that Abi1 acetylation at this residue does not modulate the activation of myosin and vimentin.
FIGURE 5.
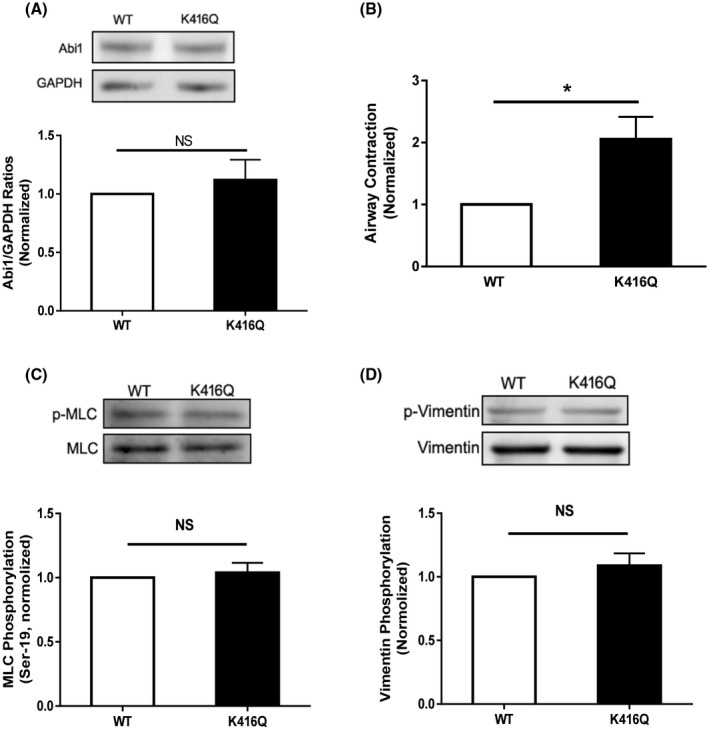
Acetylation of K416 regulates airway constriction without affecting myosin light chain phosphorylation at Ser‐19 and vimentin phosphorylation at Ser‐56. A, Human lung slices were treated with constructs for WT or K416Q Abi1 for 2 days. Immunoblot analysis showed similar protein expression in these tissues. Data are mean values of four lung slices. Error bars indicate SE. B, Airway constriction of lung slices expressing K416Q is greater than WT Abi1 in response to ACh treatment. Data are mean values of four lung slices. Error bars indicate SE. C and D, Myosin light chain (MLC) phosphorylation and vimentin phosphorylation are similar in cells expressing WT or K416Q Abi1. Data are mean values of four batches of cell culture. **P < .01
3.5. p300 regulates Abi1 acetylation and contraction in smooth muscle
Because p300 has a role in regulating acetylation of histone and non‐histone proteins, we sought to understand whether p300 regulates Abi1 acetylation in smooth muscle. First, we found that p300 was localized both in the nucleus and in the myoplasm as evidenced by immunofluorescence microscopy (Figure 6A). Second, p300 knockdown reduced the agonist‐induced Abi1 acetylation in HASM cells (Figure 6B). To further assess the role of p300 in smooth muscle, we generated p300 smooth muscle conditional knockout (p300smko) mice and assessed the acetylation of Abi1 ex vivo. ACh‐induced Abi1 acetylation was significantly reduced in tracheal rings from p300smko mice as compared to p300flox mice (p300‐flox mice, control) (Figure 6C). More importantly, contractile response to ACh stimulation was diminished in tracheal rings from p300smko mice (Figure 6D). These results suggest that p300 modulates Abi1 acetylation and smooth muscle contraction.
FIGURE 6.
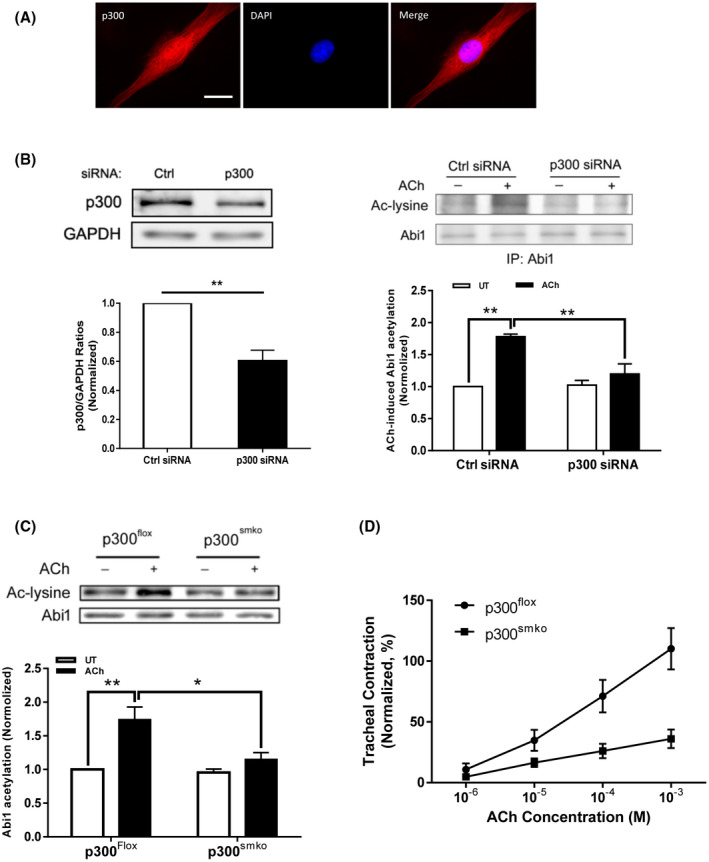
p300 mediates Abi1 acetylation and smooth muscle contraction. A, p300 localizes both in the myoplasm and the nucleus. HASM cells were immunofluorescently stained for p300 and the nucleus was visualized by DAPI. Scare bar. 10 µm. B, p300 knockdown (KD) inhibits Abi1 acetylation in HASM cells. Left panel, p300 knockdown in HASM cells treated with p300 siRNA. Data are mean values of four batches of cell culture. Error bars indicate SE. Right panel, the ACh‐induced Abi1 acetylation is reduced in p300 KD cells. Data are mean values of four batches of cell culture. Error bars indicate SE. C, Smooth muscle conditional p300 knockout reduces ACh‐induced Abi1 acetylation in airway tissues. Tracheal rings were stimulated with 10−4 M ACh for 5 minutes. Abi1 acetylation was evaluated using immunoblot analysis. Data are mean values of four tracheal tissues. D, Smooth muscle conditional p300 knockout reduces airway smooth muscle contraction. Data are mean values of five tracheal tissues. **P < .01; *P < .05
3.6. p300 modulates actin polymerization without affecting myosin light chain phosphorylation and vimentin phosphorylation
We also assessed the effects of p300 KO on actin polymerization, myosin light chain phosphorylation, and vimentin phosphorylation. The ACh‐induced increases in F/G‐actin ratios were reduced in tissues from p300smko mice as compared to p300flox mice (Figure 7A). However, p300 KO did not affect myosin light chain phosphorylation at Ser‐19 and vimentin phosphorylation at Ser‐56 in the tissues (Figure 7B,C).
FIGURE 7.
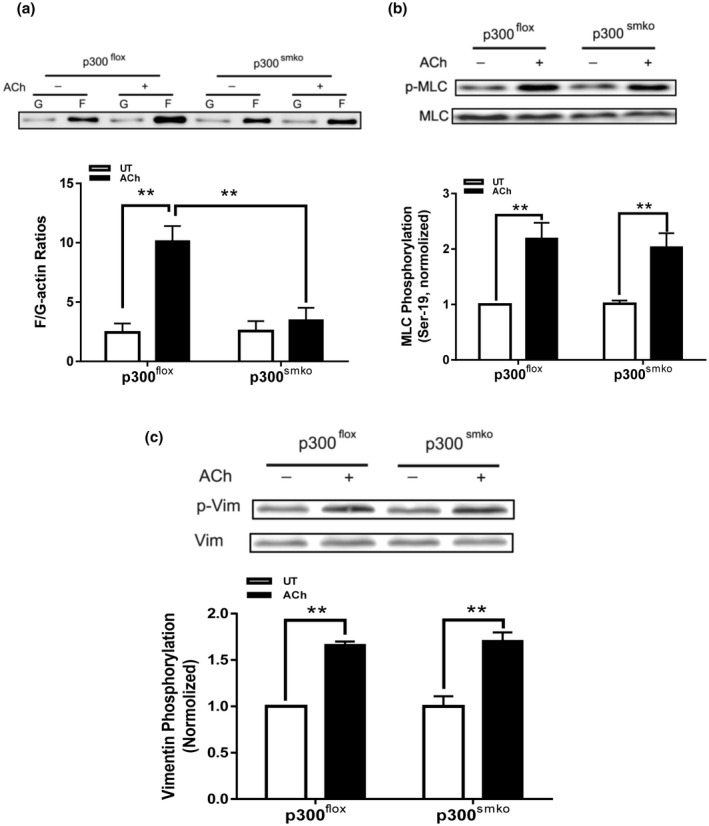
p300 mediates actin polymerization in airway smooth muscle without affecting phosphorylation of myosin light chain and vimentin. A, Smooth muscle conditional p300 knockout inhibits ACh‐elicited F/G‐actin ratios in airway smooth muscle. Data are mean values of four tracheal tissues. B, Myosin light chain (MLC) phosphorylation at Ser‐19 is not affected by smooth muscle conditional p300 knockout. Data are mean values of four tracheal tissues. C, Vimentin phosphorylation at Ser‐56 is not modulated by smooth muscle p300. Data are mean values of four tracheal tissues
4. DISCUSSION
The adapter protein Abi1 plays a role in regulating smooth muscle contraction and cell migration. However, the mechanisms by which Abi1 modulates the cellular processes are incompletely elucidated. In this study, we discovered that Abi1 underwent acetylation at lysines in smooth muscle cells. In addition, contractile stimulation increased the Abi1/N‐WASP coupling, which is consistent with our previous finding. 17 Moreover, Abi1 and N‐WASP translocated to the cell periphery upon contractile activation. Cortical actin polymerization is important for force transmission between the contractile units and the extracellular matrix. 3 , 40 To the best of our knowledge, this is the first evidence to demonstrate that Abi1 undergoes acetylation in smooth muscle during contractile activation. Abi1 acetylation may regulate its interaction with N‐WASP, which facilitates juxtamembrane actin polymerization and smooth muscle contraction. 3 , 4 , 17
In this report, we used cultured HASM cells as primary experimental model. The cell model retains cytoskeletal and contractile signaling, and is excellent to investigate the spatial distribution of proteins. 3 , 8 , 17 , 26 , 27 , 41 , 42 , 43 The cell model also avoids potential interactions of smooth muscle cells with other cell types (eg, epithelial cells and fibroblasts) that occur in bronchial tissues and limited availability of fresh bronchial tissues from donors. Although actin polymerization, myosin light chain phosphorylation, and vimentin phosphorylation are indications of contractile state of smooth muscle, we used human lung slices and mouse tracheal tissues to verify the roles of Abi1 and p300 in contractile responses. The combination of cell model and tissue model provides a unique opportunity to enable us to investigate the role of Abi1 and p300 at gene, cell, and tissue levels.
Using mutagenesis, we further identify that K416 is a major acetylation residue on Abi1. K416Q Abi1 increased its binding to N‐WASP. K416 is localized in the SH3 domain of Abi1, which is responsible for the binding to the proline‐rich domain of N‐WASP. Lysine is a basic amino acid, which is polar and positively charged. The transfer of an acetyl group from acetyl‐coenzyme A to the primary amine in the ε‐position of the lysine side chain leads to neutralization of the position's positive electrostatic charge. 22 Therefore, acetylation at K416 may neutralize the position's positive charge, which leads to conformation changes of Abi1 and increases its interaction with N‐WASP.
In this report, the acetylation mimic mutant K416Q Abi1 was positioned on the cell edge, whereas wild‐type Abi1 was localized in the myoplasm. These findings suggest that acetylation at K416 regulates the spatial distribution of Abi1 in cells. Lysine acetylation has been reported to regulate the spatial distribution of PD‐L1 in a breast cancer cell line. 44 Moreover, N‐WASP activation, actin polymerization, and contraction were enhanced in smooth muscle expressing K416Q Abi1. However, myosin light chain phosphorylation and vimentin phosphorylation were not affected by K416Q Abi1. Thus, it is likely that the acetylation of Abi1 enhances binding to N‐WASP, which activates N‐WASP, actin polymerization, and smooth muscle contraction.
The tyrosine phosphorylation of N‐WASP is catalyzed by several tyrosine kinases including c‐Abl (Abelson tyrosine kinase). 45 c‐Abl is activated in smooth muscle during contractile stimulation. 17 , 46 N‐WASP phosphorylation promotes actin comet tail elongation in Shigella. 45 In smooth muscle, N‐WASP phosphorylation and activation promotes the Arp2/3‐mediated actin network branching and polymerization. 2 , 3 Therefore, c‐Abl may mediate N‐WASP tyrosine phosphorylation and promote actin polymerization in smooth muscle in response to contractile stimulation.
p300 predominantly resides in the nucleus, and catalyzes the acetylation of histone and non‐histone proteins in various cell types, which regulates gene transcription and cell proliferation. 22 , 23 , 24 In this report, we discovered that p300 was positioned in the nucleus of human smooth muscle cells, which supports the concept that p300 regulates the acetylation of histone and transcription factors in nuclei. Furthermore, we noticed that p300 was also found in the cytoplasm of human smooth muscle cells. Our novel finding was supported by a previous study, in which p300 was found in the cytoplasm of breast cancer cells. 47 This raised the possibility that p300 may mediate Abi1 acetylation in smooth muscle. Our studies on human smooth muscle cells and p300 smooth muscle conditional knockout mice confirm this possibility. Thus, p300 regulates Abi1 acetylation and actin polymerization in smooth muscle during contractile stimulation. It is currently unknown how contractile stimulation activates p300 in smooth muscle. p300 is phosphorylated by p38 MAPK in skeletal muscle upon cancer‐induced activation of Toll‐like receptor 4. 48 It is possible that contractile stimulation induces phosphorylation and activation of p300 in smooth muscle. Future studies are required to test the possibility.
Our previous studies show that contractile stimulation induces cortactin deacetylation in smooth muscle, which is mediated by histone deacetylase 8. 21 Deacetylated cortactin enhances its binding to F‐actin and promotes actin polymerization. 20 In this report, we uncover the role of p300/Abi1 in smooth muscle. It is likely that these two pathways occur in parallel to regulate actin polymerization upon contractile stimulation.
In summary, we unveil a novel mechanism for the regulation of smooth muscle contraction. Contractile stimulation induces Abi1 acetylation via p300 in smooth muscle. Acetylation at K416 promotes the coupling of Abi1 with N‐WASP, which facilitates actin polymerization and smooth muscle contraction (Figure 8).
FIGURE 8.
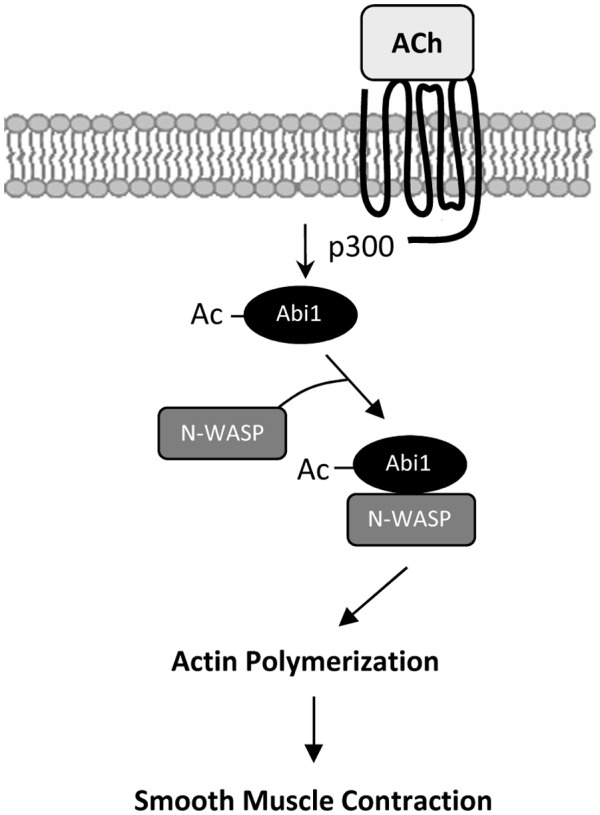
Proposed mechanism. Contractile stimulation elicits Abi1 acetylation in smooth muscle via p300, which enhances the coupling and translocation of Abi1 and N‐WASP promoting actin polymerization and contraction
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
AUTHOR CONTRIBUTIONS
Y. Wang performed cellular, biochemical, and physiological experiments; G. Liao performed molecular experiments; R. Wang performed cell culture experiments; D.D. Tang conceived the research direction, coordinated the study, and wrote the manuscript. All authors approved the final version of the manuscript.
ACKNOWLEDGMENTS
The authors thank Eylon Arbel and Saiyang Hu for technical assistance. This work was supported by NHLBI Grants HL‐110951, HL‐130304, and HL‐145392 from the National Institutes of Health (to Dale D. Tang).
Wang Y, Liao G, Wang R, Tang DD. Acetylation of Abelson interactor 1 at K416 regulates actin cytoskeleton and smooth muscle contraction. FASEB J. 2021;35:e21811. 10.1096/fj.202100415R
REFERENCES
- 1. Kim HR, Graceffa P, Ferron F, et al. Actin polymerization in differentiated vascular smooth muscle cells requires vasodilator‐stimulated phosphoprotein. AJP Cell Physiol. 2010;298:C559‐C571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gunst SJ, Zhang W. Actin cytoskeletal dynamics in smooth muscle: a new paradigm for the regulation of smooth muscle contraction. AJP Cell Physiol. 2008;295:C576‐C587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tang DD. The dynamic actin cytoskeleton in smooth muscle. Adv Pharmacol. 2018;81:1‐38. [DOI] [PubMed] [Google Scholar]
- 4. Tang DD. Critical role of actin‐associated proteins in smooth muscle contraction, cell proliferation, airway hyperresponsiveness and airway remodeling. Respir Res. 2015;16:134‐147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cipolla MJ, Gokina NI, Osol G. Pressure‐induced actin polymerization in vascular smooth muscle as a mechanism underlying myogenic behavior. FASEB J. 2002;16:72‐76. [DOI] [PubMed] [Google Scholar]
- 6. Herrera AM, Martinez EC, Seow CY. Electron microscopic study of actin polymerization in airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2004;286:L1161‐L1168. [DOI] [PubMed] [Google Scholar]
- 7. Li J, Wang R, Tang DD. Vimentin dephosphorylation at ser‐56 is regulated by type 1 protein phosphatase in smooth muscle. Respir Res. 2016;17:91‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li J, Wang R, Gannon OJ, et al. Polo‐like kinase 1 regulates vimentin phosphorylation at Ser‐56 and contraction in smooth muscle. J Biol Chem. 2016;291:23693‐23703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tang DD. Invited review: intermediate filaments in smooth muscle. Am J Physiol Cell Physiol. 2008;294:C869‐C878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu T, Ghamloush MM, Aldawood A, et al. Modulating endothelial barrier function by targeting vimentin phosphorylation. J Cell Physiol. 2014;229:1484‐1493. [DOI] [PubMed] [Google Scholar]
- 11. Innocenti M, Gerboth S, Rottner K, et al. Abi1 regulates the activity of N‐WASP and WAVE in distinct actin‐based processes. Nat Cell Biol. 2005;7:969‐976. [DOI] [PubMed] [Google Scholar]
- 12. Ryu JR, Echarri A, Li R, Pendergast AM. Regulation of cell‐cell adhesion by Abi/Diaphanous complexes. Mol Cell Biol. 2009;29:1735‐1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ring C, Ginsberg MH, Haling J, Pendergast AM. Abl‐interactor‐1 (Abi1) has a role in cardiovascular and placental development and is a binding partner of the alpha4 integrin. Proc Natl Acad Sci U S A. 2011;108:149‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu W, Sun X, Clough N, Cobos E, Tao Y, Dai Z. Abi1 gene silencing by short hairpin RNA impairs Bcr‐Abl‐induced cell adhesion and migration in vitro and leukemogenesis in vivo. Carcinogenesis. 2008;29:1717‐1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stradal T, Courtney KD, Rottner K, Hahne P, Small JV, Pendergast AM. The Abl interactor proteins localize to sites of actin polymerization at the tips of lamellipodia and filopodia. Curr Biol. 2001;11:891‐895. [DOI] [PubMed] [Google Scholar]
- 16. Wang R, Liao G, Wang Y, Tang DD. Distinctive roles of Abi1 in regulating actin‐associated proteins during human smooth muscle cell migration. Sci Rep. 2020;10:10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang T, Cleary RA, Wang R, Tang DD. Role of the adapter protein Abi1 in actin‐associated signaling and smooth muscle contraction. J Biol Chem. 2013;288:20713‐20722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451‐477. [DOI] [PubMed] [Google Scholar]
- 19. Ali I, Conrad RJ, Verdin E, Ott M. Lysine acetylation goes global: from epigenetics to metabolism and therapeutics. Chem Rev. 2018;118:1216‐1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang X, Yuan Z, Zhang Y, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell. 2007;27:197‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li J, Chen S, Cleary RA, et al. Histone deacetylase 8 regulates cortactin deacetylation and contraction in smooth muscle tissues. Am J Physiol Cell Physiol. 2014;307:C288‐C295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dancy BM, Cole PA. Protein lysine acetylation by p300/CBP. Chem Rev. 2015;115:2419‐2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jin L, Garcia J, Chan E, et al. Therapeutic targeting of the CBP/p300 bromodomain blocks the growth of castration‐resistant prostate cancer. Can Res. 2017;77:5564‐5575. [DOI] [PubMed] [Google Scholar]
- 24. Nakamura N. Cigarette smoke attenuates p300‐mediated Nrf2 acetylation in macrophages: Is stabilizing Nrf2 enough to halt COPD progression? Respirology. 2021;26:19‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang R, Mercaitis OP, Jia L, Panettieri RA, Tang DD. Raf‐1, actin dynamics and Abl in human airway smooth muscle cells. Am J Respir Cell Mol Biol. 2013;48:172‐178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang T, Wang R, Cleary RA, Gannon OJ, Tang DD. Recruitment of beta‐Catenin to N‐Cadherin is necessary for smooth muscle contraction. J Biol Chem. 2015;290:8913‐8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang R, Cleary RA, Wang T, Li J, Tang DD. The association of cortactin with profilin‐1 is critical for smooth muscle contraction. J Biol Chem. 2014;289:14157‐14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang T, Cleary RA, Wang R, Tang DD. Glia maturation factor‐gamma phosphorylation at Tyr‐104 regulates actin dynamics and contraction in human airway smooth muscle. Am J Respir Cell Mol Biol. 2014;51:652‐659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li QF, Spinelli AM, Wang R, Anfinogenova Y, Singer HA, Tang DD. Critical role of vimentin phosphorylation at Ser‐56 by p21‐activated kinase in vimentin cytoskeleton signaling. J Biol Chem. 2006;281:34716‐34724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liao G, Wang R, Rezey AC, Gerlach BD, Tang DD. MicroRNA miR‐509 regulates ERK1/2, the vimentin network, and focal adhesions by targeting Plk1. Sci Rep. 2018;8:12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liao G, Panettieri RA, Tang DD. MicroRNA‐203 negatively regulates c‐Abl, ERK1/2 phosphorylation, and proliferation in smooth muscle cells. Physiol Rep. 2015;3:e12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Y, Rezey AC, Wang R, Tang DD. Role and regulation of Abelson tyrosine kinase in Crk‐associated substrate/profilin‐1 interaction and airway smooth muscle contraction. Respir Res. 2018;19:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rezey AC, Gerlach BD, Wang R, Liao G, Tang DD. Plk1 mediates Paxillin phosphorylation (Ser‐272), centrosome maturation, and airway smooth muscle layer thickening in allergic asthma. Sci Rep. 2019;9:7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tang DD, Bai Y, Gunst SJ. Silencing of p21‐activated kinase attenuates vimentin phosphorylation on Ser‐56 and reorientation of the vimentin network during stimulation of smooth muscle cells by 5‐hydroxytryptamine. Biochem J. 2005;388:773‐783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jia L, Wang R, Tang DD. Abl regulates smooth muscle cell proliferation by modulating actin dynamics and ERK1/2 activation. Am J Physiol Cell Physiol. 2012;302:C1026‐C1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang R, Li QF, Anfinogenova Y, Tang DD. Dissociation of Crk‐associated substrate from the vimentin network is regulated by p21‐activated kinase on ACh activation of airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2007;292:L240‐L248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiang S, Tang DD. Plk1 regulates MEK1/2 and proliferation in airway smooth muscle cells. Respir Res. 2015;16:93‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Long J, Liao G, Wang Y, Tang DD. Specific protein 1, c‐Abl and ERK1/2 form a regulatory loop. J Cell Sci. 2019;132:jcs222380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gerlach BD, Tubbesing K, Liao G, et al. Phosphorylation of GMFgamma by c‐Abl coordinates lamellipodial and focal adhesion dynamics to regulate airway smooth muscle cell migration. Am J Respir Cell Mol Biol. 2019;61:219‐231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu Y, Gunst SJ. Vasodilator‐stimulated phosphoprotein (VASP) regulates actin polymerization and contraction in airway smooth muscle by a vinculin‐dependent mechanism. J Biol Chem. 2015;290:11403‐11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. An SS, Mitzner W, Tang WY, et al. An inflammation‐independent contraction mechanophenotype of airway smooth muscle in asthma. J Allergy Clin Immunol. 2016;138:294‐297.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Y, Wang R, Tang DD. Ste20‐like kinase‐mediated control of actin polymerization is a new mechanism for thin filament‐associated regulation of airway smooth muscle contraction. Am J Respir Cell Mol Biol. 2020;62:645‐656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Michael JV, Gavrila A, Nayak AP, et al. Cooperativity of E‐prostanoid receptor subtypes in regulating signaling and growth inhibition in human airway smooth muscle. FASEB J. 2019;33:4780‐4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gao Y, Nihira NT, Bu X, et al. Acetylation‐dependent regulation of PD‐L1 nuclear translocation dictates the efficacy of anti‐PD‐1 immunotherapy. Nat Cell Biol. 2020;22:1064‐1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burton EA, Oliver TN, Pendergast AM. Abl kinases regulate actin comet tail elongation via an N‐WASP‐dependent pathway. Mol Cell Biol. 2005;25:8834‐8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Anfinogenova Y, Wang R, Li QF, Spinelli AM, Tang DD. Abl silencing inhibits CAS‐Mediated process and constriction in resistance arteries. Circ Res. 2007;101:420‐428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fermento ME, Gandini NA, Lang CA, et al. Intracellular distribution of p300 and its differential recruitment to aggresomes in breast cancer. Exp Mol Pathol. 2010;88:256‐264. [DOI] [PubMed] [Google Scholar]
- 48. Sin TK, Zhang G, Zhang Z, et al. Cancer‐induced muscle wasting requires p38beta MAPK activation of p300. Can Res. 2021;81:885‐897. [DOI] [PMC free article] [PubMed] [Google Scholar]


