Abstract
Exercise provides a robust physiological stimulus that evokes cross-talk among multiple tissues that when repeated regularly (i.e., training) improves physiological capacity, benefits numerous organ systems and decreases the risk for premature mortality. However, a gap remains in identifying the detailed molecular signals induced by exercise that benefit health and prevent disease. The Molecular Transducers of Physical Activity Consortium (MoTrPAC) was established to address this gap and generate a molecular map of exercise. Preclinical and clinical studies will examine the systemic effects of endurance and resistance exercise across a range of ages and fitness levels by molecular probing of multiple tissues before and after acute and chronic exercise. From this multi-omic and bioinformatic analysis, a molecular map of exercise will be established. Altogether, MoTrPAC will provide a public database that is expected to enhance our understanding of the health benefits of exercise and to provide insight into how activity mitigates disease.
eTOC blurb Walsh
The Molecular Transducers of Physical Activity Consortium (MoTrPAC) aims to create a comprehensive, integrative multi-omic map of the effects of exercise across tissues in rodents and healthy people.
Introduction
Exercise perturbs multiple systems from the whole body to the molecular level in an integrated manner (Hawley et al., 2014). However, in-depth fundamental knowledge into the molecular and cellular mechanisms that are responsible for physical activity’s benefits on multiple organ systems and the diseases and disorders that derive from inactivity is incomplete (Booth et al., 2017; Neufer et al., 2015). A better understanding of these biological processes and pathways would allow for the development of targeted exercise interventions and prescriptions and provide a foundation for developing exercise-mimetic pharmacologic interventions.
The Molecular Transducers of Physical Activity Consortium (MoTrPAC) was established to elucidate how exercise improves health and ameliorates diseases by building a map of the molecular responses to acute and chronic exercise. In 2014, a portfolio analysis of NIH grants revealed that most research regarding physical activity involved disease prevention or treatment. In fact, almost all the grants which employed an exercise intervention only addressed health outcomes and adherence issues. The MoTrPAC initiative provides a much-needed comprehensive program to understand the interplay between these biological systems with the goal of improving the design of physical activity interventions. In addition, there is a potential to identify molecular targets that can be manipulated to mimic the effects of exercise in persons unable to do so for a variety of reasons, such as physical disability, coma, or paralysis.
To address the gaps in knowledge about how exercise enhances health and ameliorates disease, multiple agencies at the NIH—including the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), the National Institute on Aging (NIA) and other institutes and centers who participated in the trans-NIH Exercise Interest Group—proposed the Common Fund program supporting MoTrPAC. To create a substantive complex map of molecular transducers in diverse populations across the lifespan, MoTrPAC established a multi-site collaboration across the United States encompassing various scientific disciplines: preclinical animal study sites and human clinical exercise sites to perform the exercise testing, exercise interventions and biospecimen collection; a consortium coordinating center to manage sample collection, distribution of samples and consortium logistics; chemical analysis sites to perform ‘omics analysis from the samples collected; and a bioinformatics center to collaboratively facilitate data quality control, bioinformatics analysis and dissemination to make the data and related resources available to the broad scientific community (Figure 1). The animal studies will enable analysis of the effects of exercise on many different tissues that are not readily obtainable in humans, thereby enabling a broad view of the systemic effects of exercise. The collection of human specimens (blood, muscle and adipose) will permit the analysis of these critical systems, which are central to the energetics of exercise and appear to interact in a coordinated manner to improve overall metabolic health (Pedersen and Febbraio, 2012; Romijn et al., 1993; Stanford and Goodyear, 2018). In addition to providing information concerning the effects of exercise at different physiological and molecular levels, the large scope of this study (humans: N=~2,600; rats: N=820) will create a complex, integrative dataset that will be available to the scientific community. This dataset and some associated biospecimens can be leveraged by other groups by proposing ancillary studies to MoTrPAC.
Figure 1. General overview of MoTrPAC.
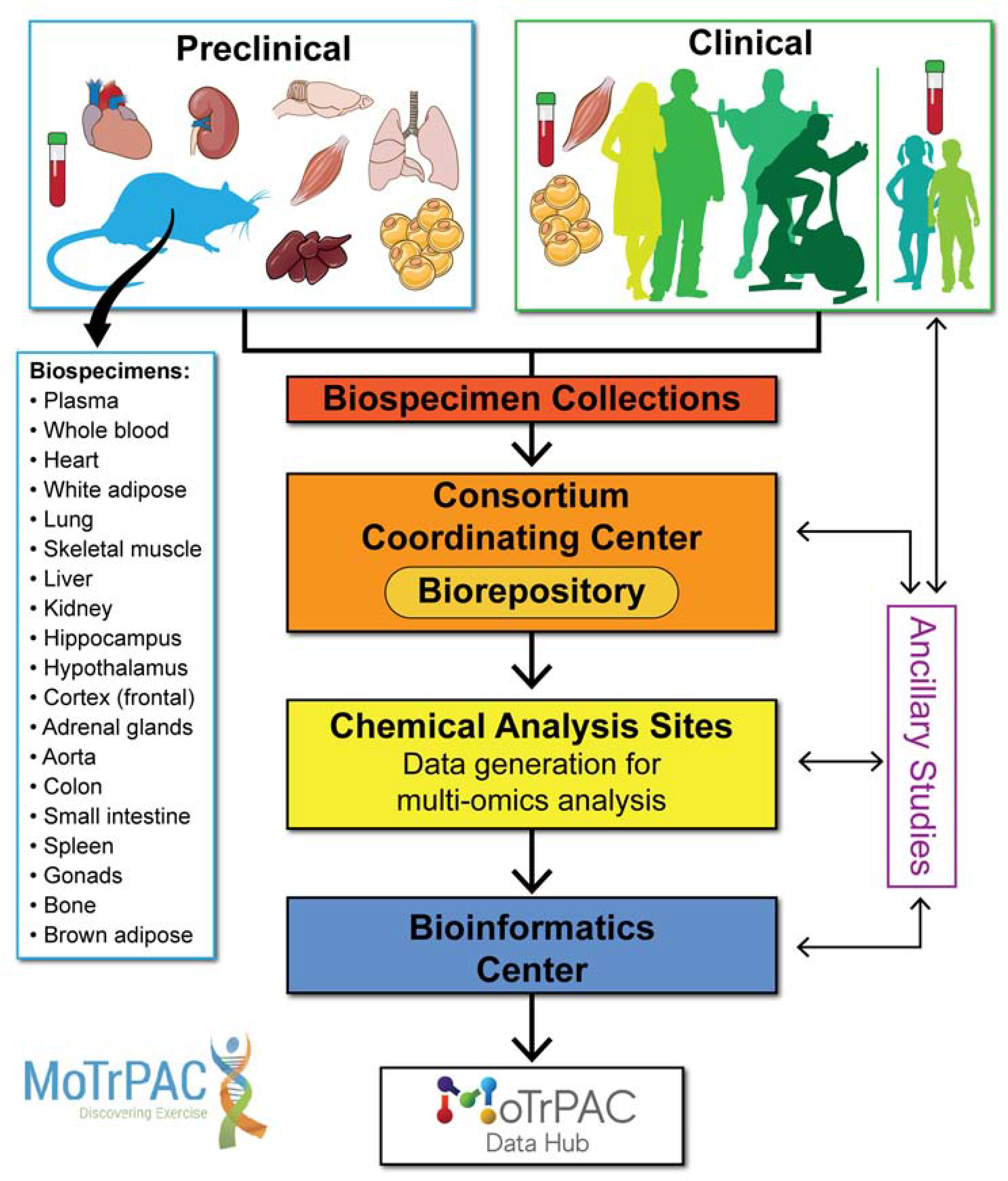
Preclinical Animal Study Sites (PASS) (rats) and Human Clinical Exercise Sites will collect biospecimen samples after acute and chronic exercise. The biospecimen samples will be sent to a central biorepository where they will be logged, processed and distributed to various Chemical Analysis Sites (CAS) for ‘omics analysis. A portion of the biospecimen samples will be kept at the biorepository for future ancillary study opportunities by the scientific community. Data generated from the CAS sites will be transferred to the Bioinformatics Center (BIC) for a multi-omics, multi-species, multi-tissue and multi-time point integration of the data with the goal of generating a molecular map of exercise. The data will be available to the scientific community via the MoTrPAC Data Hub: https://motrpac-data.org.
Preclinical Animal Study Sites
A primary goal of the Preclinical Animal Study Sites (PASS) investigations is to enable the analysis of the systemic effects of exercise across many different organs and blood, most of which cannot be collected in humans. The first phase of the PASS studies is being conducted across three separate sites, with each collecting numerous biospecimens after acute (i.e. single bout) treadmill exercise or chronic treadmill training of male and female F344 rats at 6 and 18 months of age (Figure 2). For both acute and chronic exercise studies, the same biospecimens are being collected from non-exercised, control animals; for the acute studies this includes groups to account for potential effects of time of day and time of feeding. This rat strain was selected for MoTrPAC because there is a large body of previous exercise research utilizing this strain, and rats provide larger amounts of tissue in contrast to mice. Larger tissue samples allow for multiple assays to be performed on the same sample, which supports informatics analysis for the development of the molecular map of exercise. Also, larger tissues will provide additional material for ancillary studies. To control variation across the three PASS sites, the rats are being provided from the same NIA colony, and the housing and feeding conditions are standardized across the sites. Furthermore, because rats are most active during the “dark” light phase, all rats are first being acclimated to a reverse light cycle for a minimum of 10 days, and with all exercise bouts conducted during the dark cycle.
Figure 2. A general schematic of the preclinical and clinical studies.
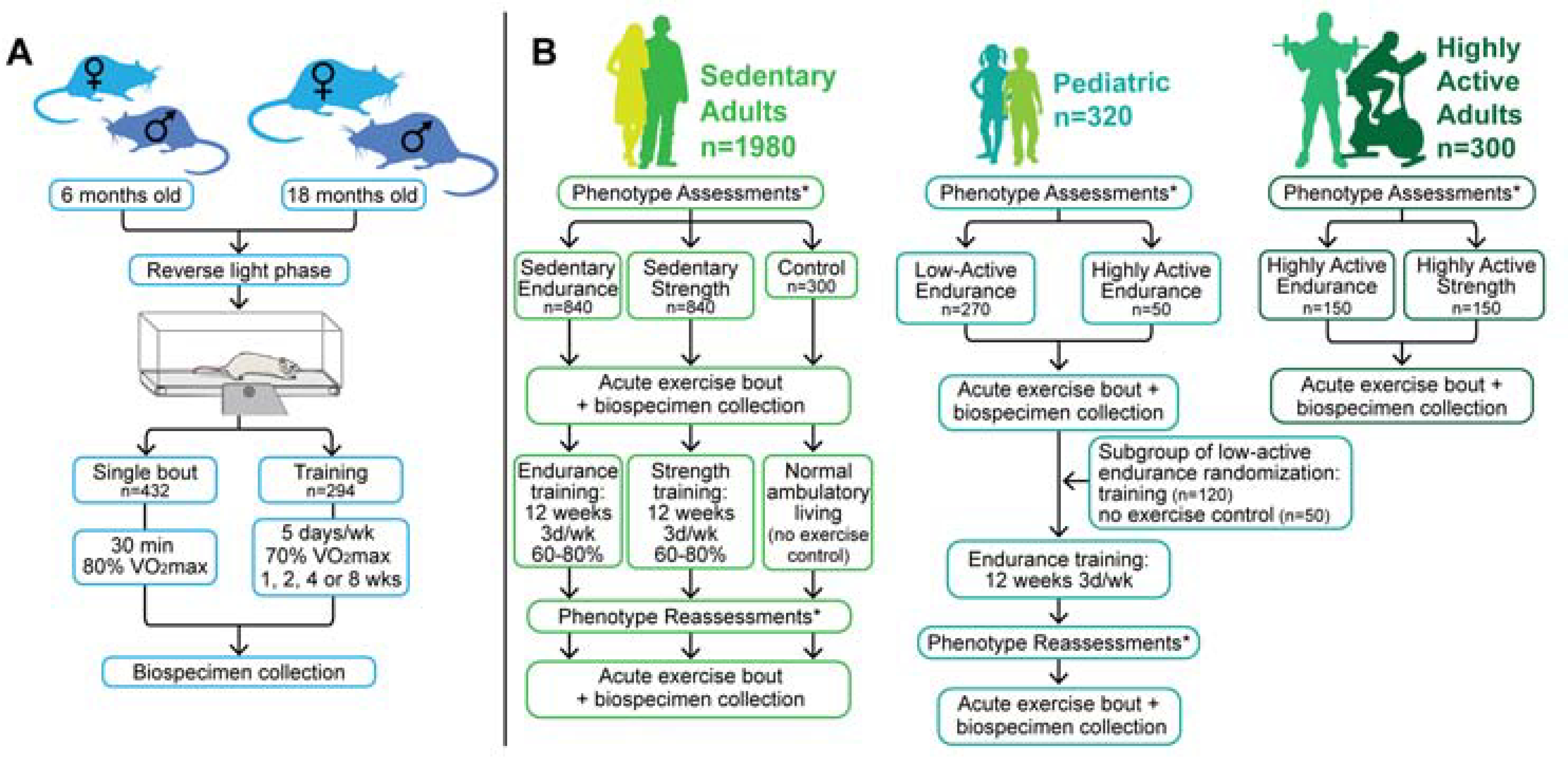
A) Biospecimen collection for the rat studies (N=820, including non-exercised controls) is ongoing and will be completed in Fall 2020. For both the acute (i.e. single bout; n=526) and chronic phases (i.e. training; n=294), six- and eighteen-month old male and female rats are being studied, and both exercise phases include a cohort of non-exercised controls. B) ~2600 healthy individuals will be evaluated physiologically, and biospecimens will be collected to accommodate molecular probing of various tissues before and after acute and chronic endurance or resistance exercise. For the adult cohort, the control participants will rest quietly (i.e., no exercise) during the acute bout with biospecimen collections.
*Assessments include blood profile, body composition, heart health, VO2max and strength.
The PASS study design for the acute response to exercise entails a single 30-minute bout of treadmill running (Intensity: ~80–90% VO2max; Incline: 5°; Speed: 6 months: Male – 16.8 m/min, Female – 18.0 m/min; 18 months: Male – 12.0 m/min, Female – 13.8 m/min), with tissue collections occurring immediately post-exercise and at six additional times up to 48 hours after the exercise bout. This sampling series is weighted toward early time points (0, 0.5, 1 and 4 hours post-exercise) to capture the temporal dynamics of the molecular responses, but it also includes later time points (7, 24 and 48 hours post-exercise) to capture long-duration primary responses as well as secondary molecular events (detailed protocols are available at https://motrpac.org/protocols.cfm). To study the biological events that occur during the early, intermediate and later stages of endurance training, the PASS study design for the chronic response to exercise, which has been completed, entailed up to 8 weeks of treadmill training (5 days per week at ~70% VO2max), with tissues collected 48 hours after 1, 2, 4 and 8 weeks of training; incline, duration and speed of exercise progressively increased on a daily-to-weekly basis during the initial 6 weeks of training. Sex differences in both the acute exercise response and the training response are being investigated along with other study aims.
The most powerful aspect of the PASS design is the breadth of tissues collected. In addition to being studied in the context of MoTrPAC, these will serve as a data resource for generating hypotheses for future studies. For both the acute exercise and chronic exercise training studies (including the non-exercise controls), as many as 27 biospecimens per rat are being collected for potential analysis. In addition to biospecimen collection, other phenotypic outcomes are being collected, including blood lactate concentration, maximal oxygen consumption (VO2max) and body composition. At Chemical Analysis Sites, initial biospecimens of focus will include those that overlap with the human studies (i.e., plasma, skeletal muscle, white adipose), as well as liver, heart, kidney, lungs, brain and brown adipose. It is expected that nucleic acid, proteomic, and targeted metabolomic assays will be performed only on tissues where the amount is non-limiting, whereas transcriptomics, non-targeted metabolomics and non-targeted lipidomics will be performed on all tissues. Together, these assays are expected to provide molecular and physiological insights about the effect of exercise on many different organs. Ultimately, MoTrPAC should begin to explain how molecular transducers function across an entire mammal (Pedersen and Febbraio, 2012).
After preliminary characterization of the changes that occur in the initial set of analyses, a second phase of the PASS will include mechanistic studies of exercise-induced molecules that transduce stress resistance and circulating factors that might be implicated in the health benefits of exercise. Additional studies will focus on the adaptation to chronic resistance exercise and the impact of age and sex on these responses, as well as other studies that have yet to be determined.
Human Clinical Exercise Sites
The human component of MoTrPAC is an in-depth study of the effects of two different forms of exercise (endurance and resistance training) across multiple individuals of different ages (including children) and sexes, as well as sedentary and highly active individuals. This large cohort will be used to study the response to exercise at the whole body and cellular levels and attempt to identify the molecular underpinnings that might be responsible for the adaptive process and variation among individuals. Several traditional methods from the field of exercise physiology will be combined with novel biospecimen sampling and high-throughput molecular analytical approaches that will likely yield important insights into the effects of exercise on health. The human study has many unique aspects that are highlighted below and will be conducted as a randomized controlled trial (RCT) with an intent-to-treat design.
Participants.
The goal is to recruit 270 children and adolescents (10–17 years of age) who are low-active in endurance-type exercise and 1980 healthy sedentary adults (age 18 years or greater) who will be medically screened and randomly assigned to endurance training (170 youth, 840 adults), resistance training (840 adults), or non-exercise control (50 youth, 300 adults) (Figure 2). An additional group of highly active endurance (50 youth, 150 adults) and resistance (150 adults) trained individuals will serve as comparators and will not participate in the MoTrPAC exercise training programs. The recruitment and enrollment approach will be sex balanced and provide participants across a wide range of ages (10–17, 18–39, 40–59 and ≥60 year age groups) and of different races.
Exercise Training Program - Adults.
The sedentary adult participants randomized to endurance or resistance exercise training will perform 12 weeks of supervised exercise, three days per week, with progression in both volume and intensity. Each endurance training session will be ~1 hour in duration and be evenly split between cycling and treadmill (walking/running) exercise with intensity set to ~60–80% of heart rate reserve and monitored in real-time during each session. Each resistance training session will target the whole body and consist of eight total exercises (five upper body: chest press, military press, seated row, triceps extension, biceps curl; three lower body: leg press, leg curl, knee extension) at a prescribed plan of 3 sets of 8–12 repetitions at an intensity of ~60–80% of maximum for each exercise. These exercise protocols are well-known to improve clinically relevant parameters (i.e., VO2max and muscular strength and hypertrophy) via alterations in metabolic, biochemical and molecular signatures (Coggan et al., 1990; Gollnick et al., 1973; Raue et al., 2012; Rönn et al., 2014; Timmons et al., 2010).
Acute Exercise Bout and Biospecimens Collections - Adults.
A unique feature of the MoTrPAC adult protocol will be the integration of strategic biospecimen collections (blood, muscle and adipose) before, during and after standardized bouts of acute exercise. Participants will perform a 40–45-minute bout of exercise (exercise-mode specific; rest for the non-exercise controls) with biospecimens collected before and after 12 weeks of training. The highly active group will perform the exercise-mode specific bout only once. Compared to resting homeostasis, these types of exercise challenges are expected to dramatically increase metabolic rate ~5–10-fold (Coggan et al., 1990; Farinatti and Castinheiras Neto, 2011; Mulla et al., 2000; Romijn et al., 1993), bioenergetic flux >10-fold (Kjaer et al., 1991; Romijn et al., 1993; Steensberg et al., 2000) and large dynamic range in gene expression from small to >100-fold changes (Louis et al., 2007; Radom-Aizik et al., 2013, 2014), and likely enhance cross-talk among many organs (Pedersen and Febbraio, 2012).
Standardized conditions that control for physical activity and time of day, as well as take dietary intake into account, will be implemented prior to the acute exercise bout. On the day of an acute exercise bout with biospecimen collections, volunteers will arrive at a Human Clinical Center in the morning after an overnight fast and rest comfortably for 0.5 hour prior to obtaining baseline blood (antecubital vein), skeletal muscle (vastus lateralis) and adipose (periumbilical region) samples. Participants will then perform the standardized acute exercise bout (or rest for the non-exercise control group) with additional biospecimen samples (blood, muscle, adipose) obtained ~0.5 hour (early), ~4 hours (middle) and ~24 hours (late) after exercise. These time points were chosen to capture the dynamic changes in the response to exercise as metabolic, post-translational modification and epigenetic modifications can occur quite rapidly (Barrès et al., 2012; Bolster et al., 2003; Hoffman et al., 2015; Romijn et al., 1993), while mRNA induction generally peaks a few hours after exercise (Louis et al., 2007; Yang et al., 2005) and increases in protein synthesis rates are detectable in the hours and days following exercise (Phillips et al., 1997) (Figure 3). Additional blood samples will be collected during the endurance exercise bout (20- and 40-minute timepoints) and shortly after (10 minutes) both endurance and resistance exercise bouts. All participants will have pre-exercise biospecimen collections, but to reduce participant burden in the post-exercise phase, sedentary participants will undergo skeletal muscle and adipose biopsies at one of three time points (early, middle, late). The highly active participants will have muscle biopsies and blood at all timepoints while adipose biopsies will be collected at the pre and middle timepoints.
Figure 3. Overview of biospecimen collections and integrated analysis plan.

Preclinical studies: For the acute exercise arm, biospecimens (N=25 per animal) are being collected 0 (i.e., immediately post), 0.5, 1, 4, 7, 24 or 48 hours following exercise. For the chronic exercise training arm, which has been completed, biospecimens (N=28) were collected 48 hours after rats completed 1, 2, 4 or 8 weeks of treadmill training. For both study arms, the same biospecimens are being collected from an unexercised group of rats. Clinical Studies: Human biospecimens (blood, muscle, adipose; blood only for pediatrics) will be obtained before and at 0.5, 4 and 24 hours following endurance or resistance exercise. The sedentary participants will perform the acute exercise bout with biospecimen collections twice (before and after 12 weeks of training) while the highly active participants will perform this one time (see Human Clinical Sites description in the text for more detail). The ‘omics data generated from multiple different genomics, proteomics and metabolomics assays will be processed in assay-specific pipelines. Omic specific analyses followed by state-of-the-art integrative methods will be applied for a multi-omic analysis of the multiple timepoints and tissues collected in MoTrPAC with the goal of creating a map of the molecular transducers of exercise. All data (pipelines, raw and processed data, results, and integrative analyses) will be made publicly available through the MoTrPAC Data Hub: https://motrpac-data.org.
Acute Exercise Bout and Biospecimens Collections - Pediatrics.
Children and adolescents undergo critical periods of growth and development which are distinct from adult physiology. Pediatric studies must also comply with additional ethical considerations (Radom-Aizik and Cooper, 2016). Consequently, while the pediatric arm of the study will mimic the adult protocol as closely as possible, there are a few notable exceptions including: (1) children who are low-active in endurance-type exercise will be recruited (vs. sedentary adults) to account for the fact that children are naturally more active than adults and also participate in mandatory physical education classes; (2) no tissue biopsies (only blood will be collected); (3) an acute bout of endurance exercise with blood collection will be performed in both the training intervention and no-exercise control groups; (4) blood samples will be collected in all participants before, 20 and 40 minutes during exercise and 10 minutes, 0.5 hour and 3.5 hours into recovery; and (5) for a subgroup of 170 low-active endurance exercise children and adolescents who will be randomized to receive 12-week endurance training (N=120) or continue their standard practice (N=50) (Figure 2), the endurance exercise intervention will be modified to provide an exercise intervention that is appropriate to the pediatric participants’ age group. For middle and high school students the modes of endurance exercise will include cycling and treadmill, and also include an option for elliptical and rowing machines. Elementary school students will be trained in a form of circuit training (e.g., endurance activity stations: cycle ergometer, steppers, individual jump rope, pacer and sliders) to keep the younger participants engaged.
Phenotype Measures - Pediatrics and Adults.
To complement the molecular map, selected phenotypic measures will be obtained. Participants will be assessed before and after the 12-week intervention period, while the highly active adult comparator group members will be assessed once. These measurements include maximal oxygen consumption on a cycle ergometer (VO2max), grip strength, maximal isometric knee extension strength, body composition (DXA, dual-energy X-ray absorptiometry), clinical blood profiles, heart rate profiles during the acute endurance and resistance exercise bouts, substrate utilization (carbohydrate and fat) during the acute endurance exercise bout at ~65% VO2max (adult endurance participants only) and upper and lower body strength (one-repetition maximum; adult resistance participants only). Most of the adult participants also will provide information on self-reported health outcomes using PROMIS measures (www.nihpromis.org) that will provide opportunities to investigate the effect of exercise on mood, anxiety and depression. In a subgroup of adult participants, skeletal muscle and adipose histology (cell type, size, capillarization) and single cell analysis across various ‘omics platforms will be conducted. Investigators are exploring the potential for collecting and analyzing microbiome samples from a subgroup of adult participants.
Implementation.
To optimize this complex protocol, the adult component of the study will be implemented in two phases. The first phase will involve ~150 adult participants and will require ~4–6 months, enabling assessment of participant and clinical burden and feasibility, as well as allowing for refinement of the MoTrPAC protocol. In phase two, the remainder of the project with a target of over 2000 participants will be implemented.
Consortium Coordinating Center
The Consortium Coordinating Center (CCC) is comprised of four parts: an Administrative Coordinating Center (ACC), a Data Management and Quality Control (DMAQC) Core, an Exercise Intervention Core (EIC) and a central Biorepository. The role of the ACC is to enable the organization and governance of MoTrPAC by facilitating key processes such as meeting logistics, IRB submission and preparation of Manuals of Operations.
The Biorepository, working with the preclinical and clinical sites, the DMAQC and the Chemical Analysis Sites, oversees sample collection, shipping, archiving and distribution of human and animal samples. This includes ensuring that homogenous cryo-pulverization of tissue samples occurs prior to distribution of aliquots to the various Chemical Analysis Sites. Uniform sample processing is important to ensure that diverse data types can be directly compared. Each tissue sample will also be stored for future use by MoTrPAC and non-MoTrPAC investigators. Samples include serum, EDTA plasma, PAXgene-protected whole blood and peripheral blood mononuclear cells, and vastus lateralis skeletal muscle and subcutaneous abdominal adipose tissue from humans and >20 different tissues from the preclinical animals. Each sample will be analyzed by the Chemical Analysis Sites, and additional material will be archived for future use. The Biorepository inventory system interacts with the DMAQC to enable sample tracking, quality control and other process support systems.
Chemical Analysis Sites
To understand the exercise response in detail, an in-depth analysis of molecular and ‘omic assays will be performed using state-of-the-art laboratory techniques. Technologies include genomics, transcriptomics, DNA methylomics, targeted and untargeted proteomics and targeted and untargeted metabolomics.
Genomic, transcriptomic and regulatory analyses
Evidence has shown, through more than 150 small cohort studies (typically with under 50 participants analyzed) (Bouchard et al., 2011; Pacheco et al., 2018), that exercise is accompanied with massive changes at both the transcriptional and epigenomic levels in muscle, adipose and most other tissue systems (Lindholm et al., 2014; Ling and Rönn, 2014; Rönn and Ling, 2013) with the poorly understood influence of the underlying human genetic/environmental variation that exists between and within populations (Leońska-Duniec et al., 2016). Therefore, recent scientific studies have been conducted generating data reflecting some of the underlying genetic and epigenetic basis for responses to exercise, physical activity and training linking specific targets to benefits (Carter et al., 2017). While several studies [for review see: (Loos et al., 2015; Warburton et al., 2006)] have provided a rich source of information to develop a foundation for larger and more comprehensive genomic, epigenomic and transcriptomic analyses, sufficiently powered studies with the complementary detailed study design to make accurate predictions as machine-learned models remain underdeveloped. Moreover, the information needed to understand the role genetic variation plays in the response of individuals to acute and chronic exercise remains limited. MoTrPAC, while predicted to be statistically underpowered for a genome-wide association study (GWAS), should be able to be statistically organized and prioritized (Cantor et al., 2010) so that there is potential benefit from the orthogonal measurements assessed through other ‘omes, leading to improved mechanistic insight. Such information can begin as a knowledge base for enabling better treatment considerations for a variety of diseases (whether acute or chronic) through recognizing potential genetic and epigenetic differences in responses to exercise and training. This could be accomplished through identifying novel gene/genetic network involvement, their corresponding changes in RNA transcripts and how such genes are regulated at the epigenetic level from adult and adolescent and between athletic and sedentary individuals and associated sex differences in response to acute and chronic exercise.
The goal of the genomic, epigenomic and transcriptomic (GET) assays are to map and measure changes in the 1) RNA transcriptome and transcript isoforms including small and micro RNA using RNA-Seq; 2) DNA methylation and chromatin accessibility from rat and human tissues using reduced representation bisulfite (RRBS) for rat or methyl CpG hybrid capture for human specimens and ATAC-Seq, respectively; and 3) genomic sequence and structure of all human participants. The assays are expected to provide insights into changes in biological processes as well as gene regulatory networks that occur in response to acute and chronic exercise. The GET assay component of MoTrPAC will involve comprehensive analyses of extensively curated rat and human MoTrPAC samples with an exercise intervention, contribute these data to public databases, help identify candidate molecular transducers, elucidate new mechanisms that might explain the human response to exercise and cooperate with the Bioinformatics Center (BIC) to develop predictive models of the individual response to physical activity.
Proteomics
Proteins are important drivers of cellular structure, function and signal mediation (Cox and Mann, 2011); thus, uncovering the pathways through which physical activity influences health requires analysis of the proteome and the critical signaling-associated post-translational modifications of the proteome in various tissues. To date, a number of proteomic studies have shown important changes influenced by exercise (Burniston, 2008; Hoffman et al., 2015; Magherini et al., 2012; Sollanek et al., 2017). The majority of this work has focused on skeletal muscle, which is the tissue that actively performs the motions involved in exercise, and blood and plasma, which circulates signals systemically through the body and may be responsible for facilitating cross-talk between organ systems. Furthermore, this research is often performed in the context of diabetes due to the role of muscle and the interplay of exercise with insulin-resistance (Kleinert et al., 2018). While these studies have largely been constrained to experimental models of exercise in animals or very small cohorts of human subjects, the results are tantalizing and have identified several proteins and signaling molecules that potentially play a key role in the response to exercise. The large-scale and well-controlled preclinical and clinical protocols adopted for MoTrPAC will allow for expansion of this knowledge by providing a deeper interrogation of the proteomic response to acute and chronic exercise in numerous tissues from individuals across a range of fitness levels.
Importantly, proteomic analyses should be inclusive of not only protein expression, but also the state of protein post-translational modifications, such as phosphorylation or acetylation, as these chemical moieties can act as rapid integrators by dictating protein localization and enzymatic activity (Brandes et al., 2009; Choudhary et al., 2014; Emmerich et al., 2011; Hunter, 1995). Primarily, untargeted mass spectrometry methods and targeted aptamer-based detection techniques will be employed to probe changes in protein abundance and modifications induced by exercise. As distinct tissues present technological challenges to discovery-based proteomic analysis (e.g., dynamic range in skeletal muscle), state-of-the-art instrumentation and protocols, including tandem mass tag labelling and fractionation (Mertins et al., 2018), will be employed. Indeed, pilot discovery-based proteomics efforts with muscle and other tissues have yielded robust datasets with levels of protein coverage exceeding previous studies, presenting a wealth of opportunities to elucidate proteomic response to exercise and integrate these findings with data obtained from GET and metabolomic studies of the same tissues.
Metabolomics
Complementing genomics, transcriptomics, epigenomics and proteomic studies, MoTrPAC will also carry out a highly comprehensive mapping of exercise-associated alterations in the metabolome of both rats and humans. The metabolome is the total collection of biologically active small molecules in a given organism (Nicholson and Wilson, 2003). This includes endogenous molecules that are biosynthesized by metabolic networks in primary metabolism, molecules derived from diet or environmental exposures (the exposome (Wild, 2005)) and molecules derived from the biosynthetic interactions with the microbiome. Metabolomics can either be “targeted” to a set of known compounds (e.g., certain acylcarnitines), or “non-targeted”, which attempts to detect and relatively quantify as many metabolites as possible (Dettmer et al., 2007). In the context of acute and chronic exercise, metabolomics can provide sensitive and dynamic phenotypic patterns that closely reflect cellular and molecular changes and will likely improve our understanding of the effects of exercise beyond the individual pathway level (Heaney et al., 2017).
A number of studies have documented profound metabolomic alterations associated with exercise (Fukai et al., 2016; Heaney et al., 2017; Lewis et al., 2010; Xiao et al., 2016), but these typically involve smaller cohorts (n<100), are limited to only one (or a handful of) metabolomics assays, or focus primarily on alterations in energy production pathways. The vast chemical diversity of the metabolome, which includes lipids, sugars, amino acids and myriads of other molecule types, and its wide dynamic range (sub-nM to mM) implies that no single chemical assay can adequately profile all metabolites in one experiment (Smilde et al., 2005). To this end, MoTrPAC will employ a combination of non-targeted and targeted approaches for mapping the broader effects of exercise on both the metabolome and lipidome. These will range from triple-quadrupole-based liquid chromatography-mass spectrometry (LC-MS) using stable isotope labelled internal standards for absolute quantification, to high resolution MS and tandem MS using reversed phase and hydrophilic interaction LC for mapping relative changes in both known and unknown molecular transducers. Targeted and non-targeted LC-MS assays that focus on the non-polar fraction of the metabolome (the lipidome) will also be leveraged to map exercise effects on lipid metabolism and oxidation (Nieman et al., 2013, 2014). It is expected that these studies will provide insights into energy metabolism and signaling molecules involved in the response to exercise.
Exosomes
Exercise is a potent stimulus that has broad ranging systemic effects that are indubitable (Egan and Zierath, 2013). One prevailing hypothesis is that circulating extracellular vesicles termed exosomes play an important role in carrying training-induced protein, mRNA and miRNA cargo between organs as a means of integrating responses to exercise (Safdar and Tarnopolsky, 2018) (Figure 4). Many techniques have been developed for isolating exosomes from plasma to analyze their molecular cargo (Barrachina et al., 2019). Importantly, these techniques have been used to demonstrate that acute exercise increases the abundance of a wide variety of exosome-associated proteins related to metabolic and immune regulation (Whitham et al., 2018). Exosome isolation and analysis of MoTrPAC specimens will further investigate these effects by describing how exosome content is modulated in response to endurance and resistance exercise. The identification of protein and RNA signatures associated with exercise will shed light on exosome-mediated inter-organ crosstalk and provide a framework for studies to characterize the systemic response to physical activity.
Figure 4. Role of exosomes in integrating the exercise response across organ systems.
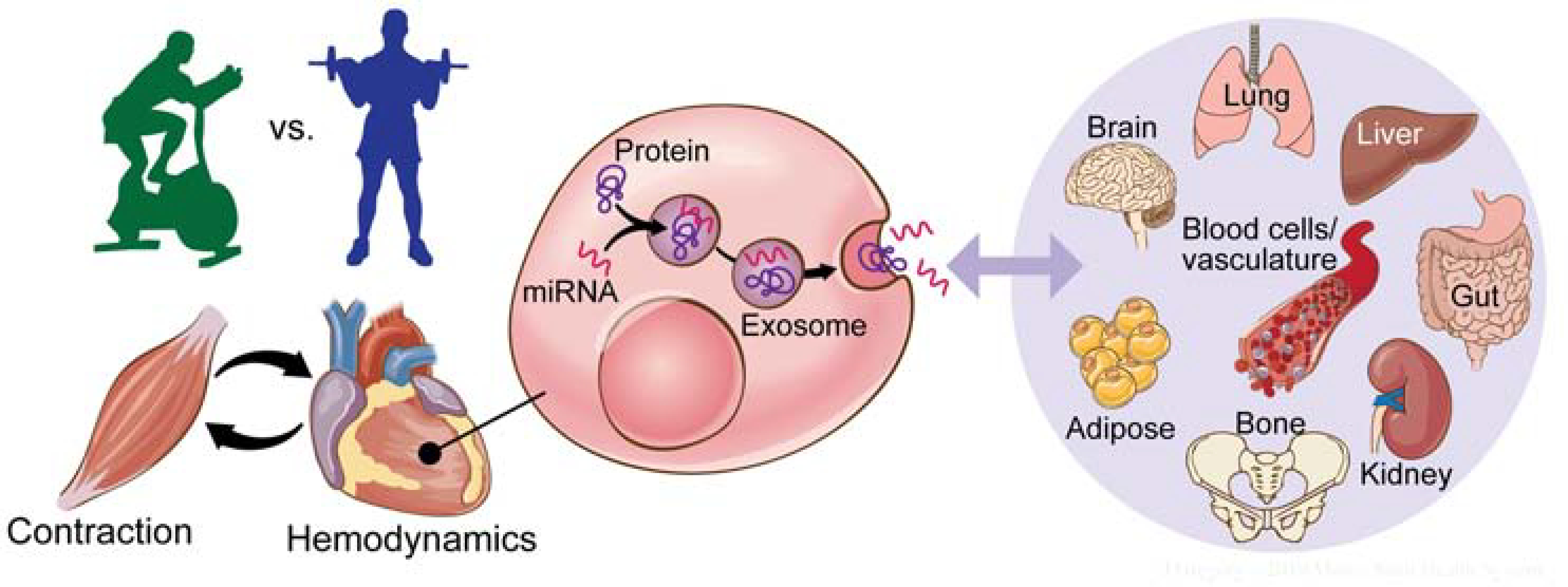
Exosomes are small extracellular vesicles (EVs) that are packaged with functional molecular cargo and released from most cell types. Exosomes released in response to exercise can be transported in the blood to various other tissues (adipose, brain, liver, etc.) where their contents can be released and have biological impact. Protein and miRNA cargo of circulating EVs from MoTrPAC samples will be analyzed to provide additional insight into this emerging biological phenomenon.
Bioinformatics Center
The immediate goals of MoTrPAC will be vested in the ‘omic platforms used and the data being generated, the quality of this data, both meta and experimental, and how it will be utilized to map the molecular transducers involving the responses to acute and chronic exercise. Data from each assay will be collected at the Bioinformatics Center and analyzed using consistent bioinformatic and analytic pipelines, whenever possible. This will improve reproducibility, interpretability and ease in data harmonization across sites. Assay-specific quantitative data will undergo quality control assessment and be normalized to reduce undesirable sample-to-sample variation, minimize batch effects and deal with analyte heteroskedasticity typically observed in molecular abundance datasets. Relative levels of analytes will be determined and the changes in molecules and pathways in response to exercise deduced.
Investigators across the consortium will conduct a series of integrative analyses with the end goal of creating a map of molecular transducers of physical activity and understanding the dynamic biological changes that occur in response to acute and chronic exercise (Figure 3). Unbiased statistical analysis will be performed to understand the variability in the data, thereby revealing significant differential responses possibly interacting with other clinical variables (e.g., sex, age, and VO2max). Systems biology methods and network algorithms will be used to correlate the different ‘omics and provide interpretable modules, focusing on regulatory processes. This will include basic interpretation of differential abundance patterns via network and pathway enrichment analyses. Additional analysis will be performed to understand the correlation structure between ‘omics, detecting latent subject classes with different temporal responses to exercise, and network propagation techniques to highlight tissue-specific response (Amar and Shamir, 2014; Amar et al., 2015; Cowen et al., 2017; Gallant et al., 2013; Hofree et al., 2013; Jo et al., 2016; Schulz et al., 2012). In addition to identifying differential molecular responses and their associations with physiological changes (e.g., molecules and pathways that are associated with VO2max), MoTrPAC will attempt to build predictive models of the effects of exercise on change in these parameters. Plans are in place to study the effects of age, sex, race and exercise type on these changes. To enhance interpretability, the results from all analyses above will be summarized using multiple visualization techniques.
Data and Resources Dissemination
As an NIH Common Fund Program, MoTrPAC aims to provide a foundation for further research to be used by the broad biomedical research community. The goal is to share all MoTrPAC data that does not compromise Personal Health Information (PHI) for General Research Use. All of the produced raw and processed data, analysis pipelines and results will be made rapidly available to the scientific community through the MoTrPAC Data Hub (https://motrpac-data.org) and, whenever suitable, in the appropriate public repositories (e.g., Metabolomics Workbench for metabolomics data and dbGaP for sequencing data). We plan to follow the FAIR (findable, accessible, interoperable and reusable) data principles (Wilkinson et al., 2016) to ensure that the data are widely available to and usable by the scientific community. In addition, we will be collaborating with the Common Fund Data Ecosystem (https://commonfund.nih.gov/dataecosystem) team on tools, techniques and external user training to maximize the utility of the MoTrPAC data to the scientific community. The MoTrPAC Data Hub itself is a cloud-native application which uses a service-oriented architecture. It is being hosted on the Google Cloud Platform (GCP). In addition to providing direct access to data, there will be tools for customizable web-based data visualizations provided on the site.
Scientific Opportunities/Ancillary Studies
Although many activities are occurring within MoTrPAC, there will be opportunities to expand the breadth and depth of MoTrPAC. These opportunities include: 1) additional interrogation of clinical metadata derived from animal and human studies and through the ‘omics applications and the interpretation of the data beyond what has been proposed and approved through the main MoTrPAC study, 2) addition or expansion of data collection to the current parent protocol and 3) accession of the biorepository for remaining biospecimen samples obtained from the animal and human studies to perform complementary analyses on these tissues. The MoTrPAC Ancillary Study policy and proposal template are available at https://motrpac.org/ancillarystudyguidelines.cfm.
Study Challenges
There are many challenges associated with a large multicenter project generating a wide variety of data types. Standardization procedures, operation manuals and quality control steps are in place to reduce the variation in exercise performance and evaluation and sample collections of the animal and human samples. Participant and clinic burden are a concern given the large scope and complexity of MoTrPAC; the consortium has made strategic choices regarding the protocol design and biospecimens sampling to help mitigate these challenges. Similarly, standardization and quality control steps are in place for the data generated using multiple ‘omics platforms even for similar data types (e.g., metabolomics and proteomics). An initial implementation phase (described above) was also put in place for the human studies to further evaluate the adult protocol during the early stages of recruitment, data collection and analysis to identify any unforeseen issues.
Integrating heterogeneous data types across MoTrPAC will require sophisticated tracking, data normalization and analytic approaches. MoTrPAC will generate large amounts of data, and many measurements may not meet statistical significance at the individual molecule level but may do so at the pathway level. State-of-the art analytic tools are in place to help manage the depth and breadth of data analysis, and these tools will continue to evolve as MoTrPAC progresses. Finally, the data are complex; ensuring that they are accessible and understandable to the broad scientific community in a timely fashion is essential for this project to be successful. Protection of participant clinical data (PHI, genomics data) will be given the highest priority to protect each individual’s identity. Incorporation of useful visualization tools as well as active engagement with the broader scientific community will be equally important to fully capitalize on the MoTrPAC project.
Summary
When complete, MoTrPAC will deliver a map of the biological molecules and pathways underlying the systemic effects of acute and chronic exercise. The data, which will ultimately be made freely available to the scientific community, will provide unprecedented opportunities to begin to understand the pathways by which physical activity influences health. In the future, it is expected that the knowledge gained will allow researchers and health professionals to develop personalized exercise recommendations and provide insights into molecular targets that could be manipulated to mimic some of the effects of exercise in persons unable to do so.
Supplementary Material
Acknowledgments
The authors would like to gratefully acknowledge Jill K. Gregory, CMI, FAMI (Certified Medical Illustrator) of the Icahn School of Medicine at Mount Sinai for working with the writing group to generate the figures enclosed within this article. Additionally, the authors would like to gratefully acknowledge the expert administrative functions by Heather Kiesel of the University of Florida for help organizing efforts by the MoTrPAC Writing Group to better enable the completion of this article. The MoTrPAC Study is supported by NIH grants U24OD026629 (Bioinformatics Center), U24DK112349, U24DK112342, U24DK112340, U24DK112341, U24DK112326, U24DK112331, U24DK112348 (Chemical Analysis Sites), U01AR071133, U01AR071130, U01AR071124–01, U01AR071128, U01AR071150, U01AR071160, U01AR071158 (Clinical Centers), U24AR071113 (Consortium Coordinating Center), U01AG055133, U01AG055137 and U01AG055135 (PASS/Animal Sites). The views expressed are those of the authors and do not necessarily reflect those of the NIH or the Department of Health and Human Services of the United States.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amar D, and Shamir R (2014). Constructing module maps for integrated analysis of heterogeneous biological networks. Nucleic Acids Res. 42, 4208–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amar D, Yekutieli D, Maron-Katz A, Hendler T, and Shamir R (2015). A hierarchical Bayesian model for flexible module discovery in three-way time-series data. Bioinformatics 31, i17–i26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrachina MN, Calderón-Cruz B, Fernandez-Rocca L, and García Á (2019). Application of Extracellular Vesicles Proteomics to Cardiovascular Disease: Guidelines, Data Analysis, and Future Perspectives. Proteomics 19, e1800247. [DOI] [PubMed] [Google Scholar]
- Barrès R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, Caidahl K, Krook A, O’Gorman DJ, and Zierath JR (2012). Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 15, 405–411. [DOI] [PubMed] [Google Scholar]
- Bolster DR, Kubica N, Crozier SJ, Williamson DL, Farrell PA, Kimball SR, and Jefferson LS (2003). Immediate response of mammalian target of rapamycin (mTOR)-mediated signalling following acute resistance exercise in rat skeletal muscle. J. Physiol 553, 213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth FW, Roberts CK, Thyfault JP, Ruegsegger GN, and Toedebusch RG (2017). Role of Inactivity in Chronic Diseases: Evolutionary Insight and Pathophysiological Mechanisms. Physiol. Rev 97, 1351–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Rankinen T, and Timmons JA (2011). Genomics and genetics in the biology of adaptation to exercise. Compr. Physiol 1, 1603–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes N, Schmitt S, and Jakob U (2009). Thiol-based redox switches in eukaryotic proteins. Antioxid. Redox Signal 11, 997–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burniston JG (2008). Changes in the rat skeletal muscle proteome induced by moderate-intensity endurance exercise. Biochim. Biophys. Acta 1784, 1077–1086. [DOI] [PubMed] [Google Scholar]
- Cantor RM, Lange K, and Sinsheimer JS (2010). Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am. J. Hum. Genet 86, 6–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AC, Chang HY, Church G, Dombkowski A, Ecker JR, Gil E, Giresi PG, Greely H, Greenleaf WJ, Hacohen N, et al. (2017). Challenges and recommendations for epigenomics in precision health. Nat. Biotechnol 35, 1128–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Weinert BT, Nishida Y, Verdin E, and Mann M (2014). The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol 15, 536–550. [DOI] [PubMed] [Google Scholar]
- Coggan AR, Kohrt WM, Spina RJ, Bier DM, and Holloszy JO (1990). Endurance training decreases plasma glucose turnover and oxidation during moderate-intensity exercise in men. J. Appl. Physiol 68, 990–996. [DOI] [PubMed] [Google Scholar]
- Cowen L, Ideker T, Raphael BJ, and Sharan R (2017). Network propagation: a universal amplifier of genetic associations. Nat. Rev. Genet 18, 551–562. [DOI] [PubMed] [Google Scholar]
- Cox J, and Mann M (2011). Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem 80, 273–299. [DOI] [PubMed] [Google Scholar]
- Dettmer K, Aronov PA, and Hammock BD (2007). Mass spectrometry-based metabolomics. Mass Spectrom. Rev 26, 51–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan B, and Zierath JR (2013). Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 17, 162–184. [DOI] [PubMed] [Google Scholar]
- Emmerich CH, Schmukle AC, and Walczak H (2011). The emerging role of linear ubiquitination in cell signaling. Sci. Signal 4, re5. [DOI] [PubMed] [Google Scholar]
- Farinatti PTV, and Castinheiras Neto AG (2011). The effect of between-set rest intervals on the oxygen uptake during and after resistance exercise sessions performed with large- and small-muscle mass. J. Strength Cond. Res 25, 3181–3190. [DOI] [PubMed] [Google Scholar]
- Fukai K, Harada S, Iida M, Kurihara A, Takeuchi A, Kuwabara K, Sugiyama D, Okamura T, Akiyama M, Nishiwaki Y, et al. (2016). Metabolic Profiling of Total Physical Activity and Sedentary Behavior in Community-Dwelling Men. PLoS One 11, e0164877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant A, Leiserson MDM, Kachalov M, Cowen LJ, and Hescott BJ (2013). Genecentric: a package to uncover graph-theoretic structure in high-throughput epistasis data. BMC Bioinformatics 14, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollnick PD, Armstrong RB, Saltin B, Saubert CW 4th, Sembrowich WL, and Shepherd RE (1973). Effect of training on enzyme activity and fiber composition of human skeletal muscle. J. Appl. Physiol 34, 107–111. [DOI] [PubMed] [Google Scholar]
- Hawley JA, Hargreaves M, Joyner MJ, and Zierath JR (2014). Integrative biology of exercise. Cell 159, 738–749. [DOI] [PubMed] [Google Scholar]
- Heaney LM, Deighton K, and Suzuki T (2017). Non-targeted metabolomics in sport and exercise science. J. Sports Sci 1–9. [DOI] [PubMed] [Google Scholar]
- Hoffman NJ, Parker BL, Chaudhuri R, Fisher-Wellman KH, Kleinert M, Humphrey SJ, Yang P, Holliday M, Trefely S, Fazakerley DJ, et al. (2015). Global Phosphoproteomic Analysis of Human Skeletal Muscle Reveals a Network of Exercise-Regulated Kinases and AMPK Substrates. Cell Metab. 22, 922–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofree M, Shen JP, Carter H, Gross A, and Ideker T (2013). Network-based stratification of tumor mutations. Nat. Methods 10, 1108–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T (1995). Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80, 225–236. [DOI] [PubMed] [Google Scholar]
- Jo K, Jung I, Moon JH, and Kim S (2016). Influence maximization in time bounded network identifies transcription factors regulating perturbed pathways. Bioinformatics 32, i128–i136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaer M, Kiens B, Hargreaves M, and Richter EA (1991). Influence of active muscle mass on glucose homeostasis during exercise in humans. J. Appl. Physiol 71, 552–557. [DOI] [PubMed] [Google Scholar]
- Kleinert M, Parker BL, Jensen TE, Raun SH, Pham P, Han X, James DE, Richter EA, and Sylow L (2018). Quantitative proteomic characterization of cellular pathways associated with altered insulin sensitivity in skeletal muscle following high-fat diet feeding and exercise training. Sci. Rep 8, 10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leońska-Duniec A, Ahmetov II, and Zmijewski P (2016). Genetic variants influencing effectiveness of exercise training programmes in obesity - an overview of human studies. Biol. Sport 33, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis GD, Farrell L, Wood MJ, Martinovic M, Arany Z, Rowe GC, Souza A, Cheng S, McCabe EL, Yang E, et al. (2010). Metabolic signatures of exercise in human plasma. Sci. Transl. Med 2, 33ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm ME, Marabita F, Gomez-Cabrero D, Rundqvist H, Ekström TJ, Tegnér J, and Sundberg CJ (2014). An integrative analysis reveals coordinated reprogramming of the epigenome and the transcriptome in human skeletal muscle after training. Epigenetics 9, 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, and Rönn T (2014). Epigenetic adaptation to regular exercise in humans. Drug Discov. Today 19, 1015–1018. [DOI] [PubMed] [Google Scholar]
- Loos RJF, Hagberg JM, Pérusse L, Roth SM, Sarzynski MA, Wolfarth B, Rankinen T, and Bouchard C (2015). Advances in exercise, fitness, and performance genomics in 2014. Med. Sci. Sports Exerc 47, 1105–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis E, Raue U, Yang Y, Jemiolo B, and Trappe S (2007). Time course of proteolytic, cytokine, and myostatin gene expression after acute exercise in human skeletal muscle. J. Appl. Physiol 103, 1744–1751. [DOI] [PubMed] [Google Scholar]
- Magherini F, Abruzzo PM, Puglia M, Bini L, Gamberi T, Esposito F, Veicsteinas A, Marini M, Fiorillo C, Gulisano M, et al. (2012). Proteomic analysis and protein carbonylation profile in trained and untrained rat muscles. J. Proteomics 75, 978–992. [DOI] [PubMed] [Google Scholar]
- Mertins P, Tang LC, Krug K, Clark DJ, Gritsenko MA, Chen L, Clauser KR, Clauss TR, Shah P, Gillette MA, et al. (2018). Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat. Protoc 13, 1632–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulla NA, Simonsen L, and Bülow J (2000). Post-exercise adipose tissue and skeletal muscle lipid metabolism in humans: the effects of exercise intensity. J. Physiol 524 Pt 3, 919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufer PD, Bamman MM, Muoio DM, Bouchard C, Cooper DM, Goodpaster BH, Booth FW, Kohrt WM, Gerszten RE, Mattson MP, et al. (2015). Understanding the Cellular and Molecular Mechanisms of Physical Activity-Induced Health Benefits. Cell Metab. 22, 4–11. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, and Wilson ID (2003). Opinion: understanding “global” systems biology: metabonomics and the continuum of metabolism. Nat. Rev. Drug Discov. 2, 668–676. [DOI] [PubMed] [Google Scholar]
- Nieman DC, Gillitt ND, Knab AM, Shanely RA, Pappan KL, Jin F, and Lila MA (2013). Influence of a polyphenol-enriched protein powder on exercise-induced inflammation and oxidative stress in athletes: a randomized trial using a metabolomics approach. PLoS One 8, e72215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman DC, Shanely RA, Luo B, Meaney MP, Dew DA, and Pappan KL (2014). Metabolomics approach to assessing plasma 13- and 9-hydroxy-octadecadienoic acid and linoleic acid metabolite responses to 75-km cycling. Am. J. Physiol. Regul. Integr. Comp. Physiol 307, R68–R74. [DOI] [PubMed] [Google Scholar]
- Pacheco C, Felipe S.M. da S., Soares M.M.D. de C., Alves JO, Soares PM, Leal-Cardoso JH, Loureiro ACC, Ferraz ASM, de Carvalho DP, and Ceccatto VM (2018). A compendium of physical exercise-related human genes: an ‘omic scale analysis. Biol. Sport 35, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BK, and Febbraio MA (2012). Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat. Rev. Endocrinol 8, 457–465. [DOI] [PubMed] [Google Scholar]
- Phillips SM, Tipton KD, Aarsland A, Wolf SE, and Wolfe RR (1997). Mixed muscle protein synthesis and breakdown after resistance exercise in humans. Am. J. Physiol 273, E99–E107. [DOI] [PubMed] [Google Scholar]
- Radom-Aizik S, and Cooper DM (2016). Bridging the Gaps: the Promise of Omics Studies in Pediatric Exercise Research. Pediatr. Exerc. Sci 28, 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radom-Aizik S, Zaldivar F, Haddad F, and Cooper DM (2013). Impact of brief exercise on peripheral blood NK cell gene and microRNA expression in young adults. J. Appl. Physiol 114, 628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radom-Aizik S, Zaldivar FP Jr, Haddad F, and Cooper DM (2014). Impact of brief exercise on circulating monocyte gene and microRNA expression: implications for atherosclerotic vascular disease. Brain Behav. Immun 39, 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raue U, Trappe TA, Estrem ST, Qian H-R, Helvering LM, Smith RC, and Trappe S (2012). Transcriptome signature of resistance exercise adaptations: mixed muscle and fiber type specific profiles in young and old adults. J. Appl. Physiol 112, 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romijn JA, Coyle EF, Sidossis LS, Gastaldelli A, Horowitz JF, Endert E, and Wolfe RR (1993). Regulation of endogenous fat and carbohydrate metabolism in relation to exercise intensity and duration. Am. J. Physiol 265, E380–E391. [DOI] [PubMed] [Google Scholar]
- Rönn T, and Ling C (2013). Effect of exercise on DNA methylation and metabolism in human adipose tissue and skeletal muscle. Epigenomics 5, 603–605. [DOI] [PubMed] [Google Scholar]
- Rönn T, Volkov P, Tornberg A, Elgzyri T, Hansson O, Eriksson K-F, Groop L, and Ling C (2014). Extensive changes in the transcriptional profile of human adipose tissue including genes involved in oxidative phosphorylation after a 6-month exercise intervention. Acta Physiol. 211, 188–200. [DOI] [PubMed] [Google Scholar]
- Safdar A, and Tarnopolsky MA (2018). Exosomes as Mediators of the Systemic Adaptations to Endurance Exercise. Cold Spring Harb. Perspect. Med 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz MH, Devanny WE, Gitter A, Zhong S, Ernst J, and Bar-Joseph Z (2012). DREM 2.0: Improved reconstruction of dynamic regulatory networks from time-series expression data. BMC Syst. Biol 6, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilde AK, van der Werf MJ, Bijlsma S, van der Werff-van der Vat BJC, and Jellema RH (2005). Fusion of mass spectrometry-based metabolomics data. Anal. Chem. 77, 6729–6736. [DOI] [PubMed] [Google Scholar]
- Sollanek KJ, Burniston JG, Kavazis AN, Morton AB, Wiggs MP, Ahn B, Smuder AJ, and Powers SK (2017). Global Proteome Changes in the Rat Diaphragm Induced by Endurance Exercise Training. PLoS One 12, e0171007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford KI, and Goodyear LJ (2018). Muscle-Adipose Tissue Cross Talk. Cold Spring Harb. Perspect. Med 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensberg A, van Hall G, Osada T, Sacchetti M, Saltin B, and Klarlund Pedersen B (2000). Production of interleukin-6 in contracting human skeletal muscles can account for the exercise-induced increase in plasma interleukin-6. J. Physiol 529 Pt 1, 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons JA, Knudsen S, Rankinen T, Koch LG, Sarzynski M, Jensen T, Keller P, Scheele C, Vollaard NBJ, Nielsen S, et al. (2010). Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J. Appl. Physiol 108, 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton DER, Nicol CW, and Bredin SSD (2006). Health benefits of physical activity: the evidence. CMAJ 174, 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham M, Parker BL, Friedrichsen M, Hingst JR, Hjorth M, Hughes WE, Egan CL, Cron L, Watt KI, Kuchel RP, et al. (2018). Extracellular Vesicles Provide a Means for Tissue Crosstalk during Exercise. Cell Metab. 27, 237–251.e4. [DOI] [PubMed] [Google Scholar]
- Wild CP (2005). Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomarkers Prev 14, 1847–1850. [DOI] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE, et al. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3, 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Moore SC, Keadle SK, Xiang Y-B, Zheng W, Peters TM, Leitzmann MF, Ji B-T, Sampson JN, Shu X-O, et al. (2016). Objectively measured physical activity and plasma metabolomics in the Shanghai Physical Activity Study. Int. J. Epidemiol 45, 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Creer A, Jemiolo B, and Trappe S (2005). Time course of myogenic and metabolic gene expression in response to acute exercise in human skeletal muscle. J. Appl. Physiol 98, 1745–1752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


