Abstract
Background:
Teeth have unique histology that make this biomatrix a time-capsule for retrospective exposure analysis of fetal and early life. However, most analytic methods require pulverizing the whole tooth, which eliminates exposure timing information. Further, the range of chemicals and endogenous exposures that can be measured in teeth has yet to be fully characterized.
Methods:
We performed untargeted metabolomics on micro-dissected layers from naturally shed deciduous teeth. Using four liquid-chromatography high-resolution mass spectrometry analytical modes, we profiled small molecules (<1000 Da) from prenatal and postnatal tooth fractions. In addition, we employed linear regression on the tooth fraction pairs from 31 children to identify metabolites that discriminate between prenatal and postnatal exposures.
Results:
Of over 10,000 features measured in teeth dentin, 390 unique compounds were annotated from 62 chemical classes. The class with the largest number of compounds was carboxylic acids and their derivatives (36%). Of the annotated exogenous metabolites (phthalates, parabens, perfluoroalkyl compounds, and cotinine) and endogenous metabolites (fatty acids, steroids, carnitines, amino acids, and others), 91 are linked to 256 health conditions through published literature. Differential analysis revealed 267 metabolites significantly different between the prenatal and the postnatal tooth fractions (adj. p-value < 0.05, Bonferroni correction), and 21 metabolites exclusive to the prenatal fraction.
Conclusions:
The prenatal and early postnatal exposome revealed from dental biomarkers represents a broad range of endogenous and exogenous metabolites for a comprehensive characterization in environmental health research. Most importantly, this technology provides a direct window into fetal exposures that is not possible by maternal biomarkers. Indeed, we identified several metabolites exclusively in the prenatal fraction, suggesting unique fetal exposures that are markedly different to postnatal exposures. Expansion of databases that include tooth matrix metabolites will strengthen biological interpretation and shed light on exposures during gestation and early life that may be causally linked with later health conditions.
Keywords: Tooth, Metabolomics, Prenatal, Exposome, Exposure, Environment, Fetal
1. Introduction
Early-life development is particularly vulnerable to environmental factors that may influence later health. In fact, a central tenet of the Developmental Origins of Health and Disease (DOHaD) is that many childhood and adult diseases, including cancer, arise early in life (Sun et al., 2016; Moore, 2016; Almeida et al., 2019). This phenomenon may result from increased susceptibility to exposures via greater absorption, nascent detoxification mechanisms, or heightened sensitivity to small homeostatic fluctuations that coincides with rapid organ and tissue development during prenatal development and early life (Landrigan and Goldman, 2011; Lu and Rosenbaum, 2014; van Anker and den; Reed et al., 2018). Indeed, epidemiological studies have identified associations between prenatal and early-life exposures to air pollution (Stephane et al. 2020; Hsu et al., 2015), endocrine disrupting chemicals (Braun, 2017; Tanner et al., 2020), trace metals (Valeri et al., 2017), and pesticides (von Ehrenstein et al., 2019; Hyland et al., 2018) and a range of adverse health outcomes from neurodevelopment to cancer. However, obtaining direct measurements of exposures to the developing fetus or child and related dysregulated biology during these critical time periods is challenging, and particularly so for studies of rare and low frequency diseases and disorders. Consequently, most epidemiological studies rely on questionnaire data and/or measurement of direct exposure in maternal prenatal matrices or from external exposure sources (e. g. regional air monitors, pesticide application maps, exposures measured at home or school), which may not accurately represent fetal or early life exposures.
Human teeth initiate development during second trimester gestation and naturally shed during childhood and adolescence. As teeth grow and form, many chemicals circulating in the blood are deposited in the organic matrix which later undergoes mineralization forming a crystalline structure that preserves the timing and intensity of the exposure (Andra et al., 2015; Arora and Austin, 2013). This deposition is progressive, similar to growth rings in a tree, creating a unique ‘biological hard drive’ that keeps a record of past exposures. Leveraging this record via chemical interrogation of tooth matrix provides a retrospective direct analysis of prenatal and early life exposures. Further, by measuring both exogenous chemicals and endogenous metabolites in teeth, data can be collected on exposure, metabolism, and biologic response, all at developmentally important time points for identifying causal risk factors and unraveling biological pathways.
Organic compounds directly measured in teeth include drugs, tobacco metabolites, plasticizers, and pesticides (Ottaviani et al., 2017; Palmer et al., 2015; Camann et al., 2013; Marchei et al., 2008; Pellegrini et al., 2006), demonstrating the broad range of chemical exposures preserved in teeth for exposome analysis. However, most teeth preparation protocols require pulverizing the whole tooth to analyze the powder, which results in a loss of temporal information for examining the role of exposure timing. We developed laser-ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) technologies to enable measurements of metals with weekly temporal resolution (Austin et al., 2013). This technique successfully identified key windows of susceptibility to metal exposures linked with adverse health outcomes such as autism spectrum disorder (ASD) (Arora et al., 2017) and amyotrophic lateral sclerosis (ALS) (Figueroa-Romero et al., 2020), and reinforces the importance of exposure timing in environmental health research. Further methods development can help to maximally leverage this unique exposure matrix for future discovery.
One approach to generate extensive exposure data is untargeted metabolomics, the unbiased measurement of thousands of small molecules, including both exogenous compounds and endogenous metabolites, that can be linked to exposures and health outcomes. To date, only a single proof-of-concept study has reported metabolite and environmental exposure measurements in teeth while maintaining temporal resolution between prenatal and postnatal tooth fractions (Andra et al., 2015). Here, we perform the first comprehensive untargeted profiling of deciduous teeth using liquid chromatography high-resolution mass spectrometry (LC-HRMS) to characterize the tooth exposome. Further, using 31 naturally shed deciduous teeth, we investigate differences in metabolome profiles between prenatal and postnatal fractions to demonstrate the utility of this method for molecular phenotyping to discriminate groups for population-based etiological research.
2. Methods
2.1. Tooth samples
Experimental deciduous teeth for pooled comprehensive metabolomics profiling across four analytical modes were anonymously donated to the Icahn School of Medicine at Mount Sinai (ISMMS).
Pediatric study teeth were collected from 31 pediatric subjects enrolled in the Early Autism Risk Longitudinal Investigation (EARLI Study). EARLI enrolls pregnant mothers, who have already had a child with ASD, and follows them prospectively throughout pregnancy and the child’s first 3 years of age. EARLI is a network of four recruitment sites across the country, leveraging a common protocol for phenotypic assessment and biosampling. EARLI sites on the East Coast (Baltimore, Philadelphia) recruited primarily through mailings to families in early intervention and special education systems. The EARLI site in Sacramento recruited families through records of the California Developmental Disabilities Service system, while the EARLI site in San Francisco (Kaiser Permanente) recruited through their membership. All cohorts advertised through autism service, community, and advocacy organizations in their regions.
Teeth were naturally shed between January 2017 and April 2018, collected at home, and were mailed to each EARLI collection site in polypropylene vials. Teeth were assigned a coded identifier and shipped at room temperature to ISMMS for untargeted chemical analysis.
2.2. Sampling pretreatment
Teeth were prepared including washing, mounting, and drilling as previously described to separate prenatal and postnatal fractions (Hare et al., 2011). At the time of collection, teeth were washed in distilled water, air-dried and placed individually in sterile plastic specimen containers (Sarstedt, USA). Specific regions of the tooth dentin are micro-dissected using a custom-built robotic platform that removes dentine fragments at pre-specified locations in prenatal and postnatal dentine. The prenatal and postnatal dentin fractions are then dried using a freeze-dryer and weighed before storage at – 20°C until extraction. Tooth dentin were then pooled and re-aliquoted before extraction. Details of the general analytical methods have been described elsewhere (Andra et al., 2015) and we summarize here.
For metabolomics profiling, pooled experimental tooth extracts and process blanks were analyzed on four LC-HRMS modes: ZIC-hydrophilic liquid chromatography (HILIC) in positive and negative electrospray ionization (ESI) mode (ZHP, ZHN) for analysis of polar metabolites, and reverse phase liquid chromatography in positive and negative ESI mode (RPP, RPN) for analysis of semi- and non-polar metabolites, using established methods (Yu et al., 2020; Hu et al., 2021). See Supplemental Materials for instrument details. Pooled experimental samples were analyzed in triplicate using MS full scan and MS/MS data-dependent analysis (DDA) mode for metabolite identification, in all four analytical modes.
Individual extracts of the pre- and postnatal tooth fractions of EARLI study subjects were analyzed in a randomized run order only on the RPP mode as a single injection using full MS1 mode. An external diluted reference plasma sample was prepared as a quality control (QC) and injected routinely throughout the batch. Process blanks were analyzed at the beginning and end of the analytical run. Remaining extracts from the EARLI samples were pooled to generate pre- and postnatal sets, and analyzed in MS/MS mode either as targeted MS/MS or as DDA.
2.3. Data analysis
2.3.1. Data preprocessing, visualization, and statistics analysis
Raw data from the LC-MS analyses were converted into mzXML format and analyzed using R programming platform (version 3.6.3) (R Core Team, 2020). The ‘XCMS’ package was used to extract peaks, align the peaks, group peaks into features, and fill the baseline for grouped peaks as a final peak table using optimized parameters by IPO package (Smith et al., 2006; Libiseller et al., 2015). Further preprocessing details can be found in the Supplemental Materials. For the comparison of overall feature numbers across analytical modes using experimental pooled teeth extracts, only peaks that appeared in all triplicate injections were retained for further analysis. For analysis of EARLI tooth samples, peaks that were missing in more than 50% of either prenatal or postnatal samples were removed before downstream analysis.
The extracted peaks list from MS1 was filtered by calculating the ratio of the average intensity in the sample: average intensity in the blank samples (fold change, FC). Retained features were those with a FC > 3 in the pooled samples compared to blanks for qualitative analysis, or FC > 3 in either the prenatal or postnatal groups compared to the blanks for differential analysis between the pre- and postnatal fractions.
To visualize the annotated compounds for qualitative analysis, tanimoto chemical similarity scores were calculated based on ECFP6 molecular fingerprints, which were generated by SMILES representation of each compound (Bajusz et al., 2015; Rogers and Hahn, 2010). Each compound was mapped as a node, with edges representing chemical similarity scores > 0.4.
Prenatal tooth fractions consistently weighed less than postnatal fractions. For comparisons between prenatal and postnatal metabolite profiles, tooth metabolite profiles were first normalized by dividing metabolite signal intensities by the original dried tooth mass. Further, missing peaks in the prenatal fraction may be a result of lower original tooth mass compared to the postnatal fraction and not as a result of peaks uniquely observed in the postnatal fraction. Therefore, we removed peaks that appeared in more than 15 postnatal samples and less than 16 prenatal samples. Furthermore, statistical analysis was performed only on peaks well-retained on the column (>25 s and <900 s).
Differential analysis between prenatal and postnatal fractions was performed based on a linear model for a paired study using the limma package (Ritchie et al., 2015). The differences between prenatal and postnatal samples from the same subjects were tested using an Empirical Bayesian method to provide a stable beta estimation. To control for false discovery rate (FDR), a Bonferroni correction was applied to the p- values. Statistically significant peaks were assessed visually, and only those representing high-quality peaks and integrations were reported (Schiffman et al., 2019; Chetnik et al., 2020).
Network analysis was performed to describe the overall relationship among unknown compounds that were significantly different between prenatal and postnatal fractions. To avoid redundant peaks from the same compounds such as isotopologue, adducts, or fragments, the GlobalStd algorithm (Yu et al., 2019) was applied to retain reduced independent peaks considering the Pearson’s correlation coefficient larger than 0.7 to filter the high frequency paired mass distances. Edges were built using the Pearson correlation coefficient among paired independent peaks larger than 0.9.
2.3.2. Metabolite annotation and identification
Detected metabolites were annotated and identified based upon in- house database matching considering retention time, accurate mass, and MS/MS matching (when available) with pure standards analyzed under the same conditions using the in-house Personal Chemical Database Library (PCDL) and Profinder software (Agilent Technologies, Santa Clara, USA) within a tolerance of 20 ppm and 0.3 min. Targeted MS/MS and DDA MS/MS data were exported from Masshunter software (Agilent Technologies, Santa Clara, USA), and matched to the Metlin database (Scripps Research, La Jolla, USA) (Xue et al., 2020) and the Global Natural Products Social Molecular Networking Database (GNPS) (Wang et al., 2016) for putative identifications. Annotations are Metabolomics Standard Initiative (MSI) level 1 or 2, as they were either matched to authentic standards by m/z and retention time (and MS/MS when available) or matched to a spectral database by m/z and MS/MS. Further details on the criteria used for spectral matching can be found in Table S1. Links to the GNPS results can be found in the Supplemental Materials. Metabolite classes were determined using ClassyFire (Djoumbou Feunang et al., 2016).
3. Results and discussion
3.1. Metabolite profiling of deciduous teeth
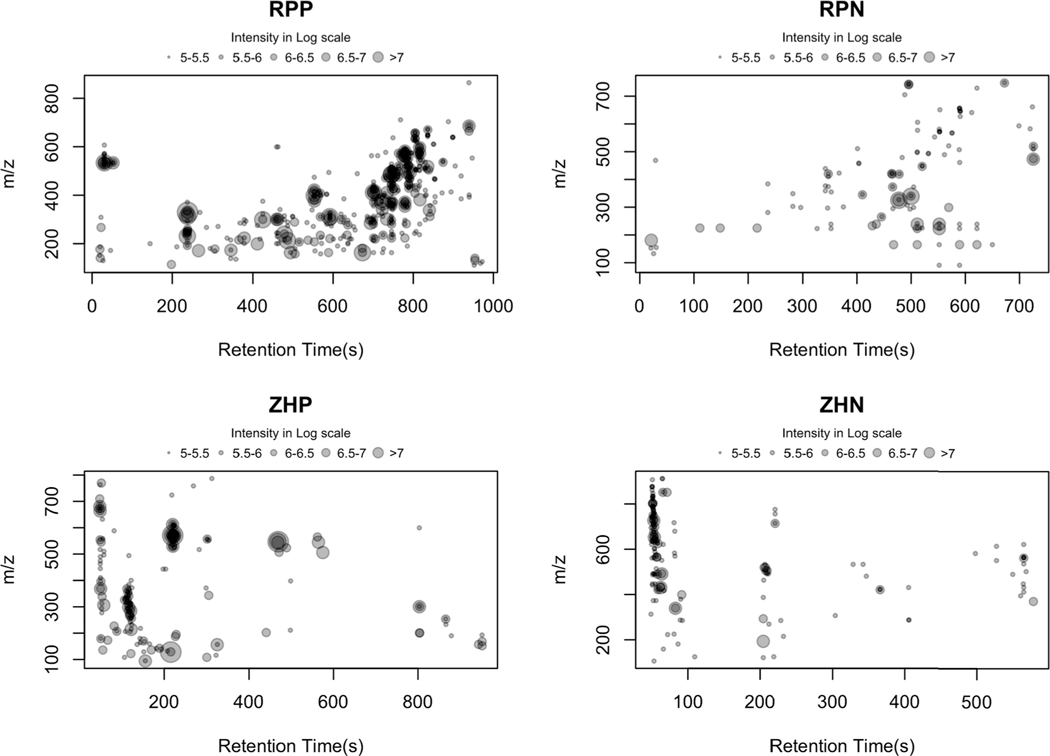
Untargeted chemical profiling of pooled prenatal experimental tooth dentine were performed across the four analytical modes. After filtering to remove signals < 3 fold greater in the tooth samples than procedural blanks, the total number of metabolite features for each analytical mode were 4579 and 2045 for reverse phase liquid chromatography in positive and negative mode, respectively (RPP and RPN), and 1503 and 2553 for ZIC-HILIC liquid chromatography in positive and negative mode, respectively (ZHP, ZHN). Bubble plots (Fig. 1) suggest that tooth metabolites measured with reverse-phase chromatography, in particular, positive mode, were well retained across the column compared to ZH chromatography. We further characterized the tooth exposome with respect to chemical parameters for each analytical mode, to prioritize analysis conditions for the limited sample available from teeth dentin. We then compared the molecular mass (m/z) and signal intensity (log scale) distribution of the tooth exposome (see Figs. S1 and S2). Molecules measured on the RPP mode were skewed toward smaller m/z distributions (median m/z 466.1250, 90% range of [189.0982, 774.4105], Fig. S1a) while molecules measured on the ZHN mode were skewed toward higher m/z distributions (median m/z 699.8351 [290.093, 1047.717], Fig. S1d). Distributions of measured molecules for RPN and ZHP modes were more gaussian, with median m/z of 555.4814 [221.2368, 970.4119] and median m/z 542.1089 [173.2469, 946.5747], respectively. In addition, median signal intensities (log scale) were similar for metabolites measured in positive mode (median 4.14 and 4.12) with a 90% range of [3.43, 5.42] and [3.48, 5.50], for RPP and ZHP, respectively (Fig. S2a,c). Mean signal intensities were slightly lower in negative mode for both chromatographies (median 3.90 and 4.00) with a range of [3.26, 5.08] and [3.43,5.08] for RPN and ZHN, respectively (Fig. S2b,d). These results suggest a broad range of small and larger molecules and a range of lipophilicity in the tooth exposome. Given the limited available sample volume, RPP mode was selected for analysis of the 31 study teeth.
Fig. 1. Bubble plots showing peaks and their relative abundances measured in each of four analytical modes: RPP, RPN, ZHP, and ZHN.
A broad range of small and larger molecules and a range of lipophilicity can be observed in the tooth exposome. The broad distribution of peaks across the RP modes, particularly for RPP, suggests better retention on the column.
Using a combination of mz, rt, and MS/MS matching to authentic standards or MS/MS matching to Metlin database or GNPS, 390 unique metabolites were identified in the tooth extracts from the 4 analytical modes (see Table S1). These metabolites comprised only 2.6% (177/ 4579) of the total features measured in RPP, 10.4% (157/1503) of the total features measured in ZHP, 3.7% (76/2045) of the total features in RPN, and 2.5% (64/2553) of the total features measured in ZHN, suggesting that current metabolite databases do not sufficiently cover the tooth exposome. Nevertheless, metabolites in teeth represented a broad range of structures and functions distributed across 62 chemical classes as determined by ClassyFire (Djoumbou Feunang et al., 2016). Carboxylic acids and their derivatives made up the largest proportion of metabolites (35.9%, 140/390); other common classes were benzene substituted derivatives (14.9%, 58/390), fatty acyls (11.3%, 44/390), organonitrogen compounds (3.3%, 13/390), organooxygen compounds (3.6%, 14/390) and steroids and their derivatives (3.1%, 12/390). The remaining 28.0% of identified metabolites are categorized into the other 56 classes.
3.2. Towards a novel approach to retrospective temporal biomarker phenotyping
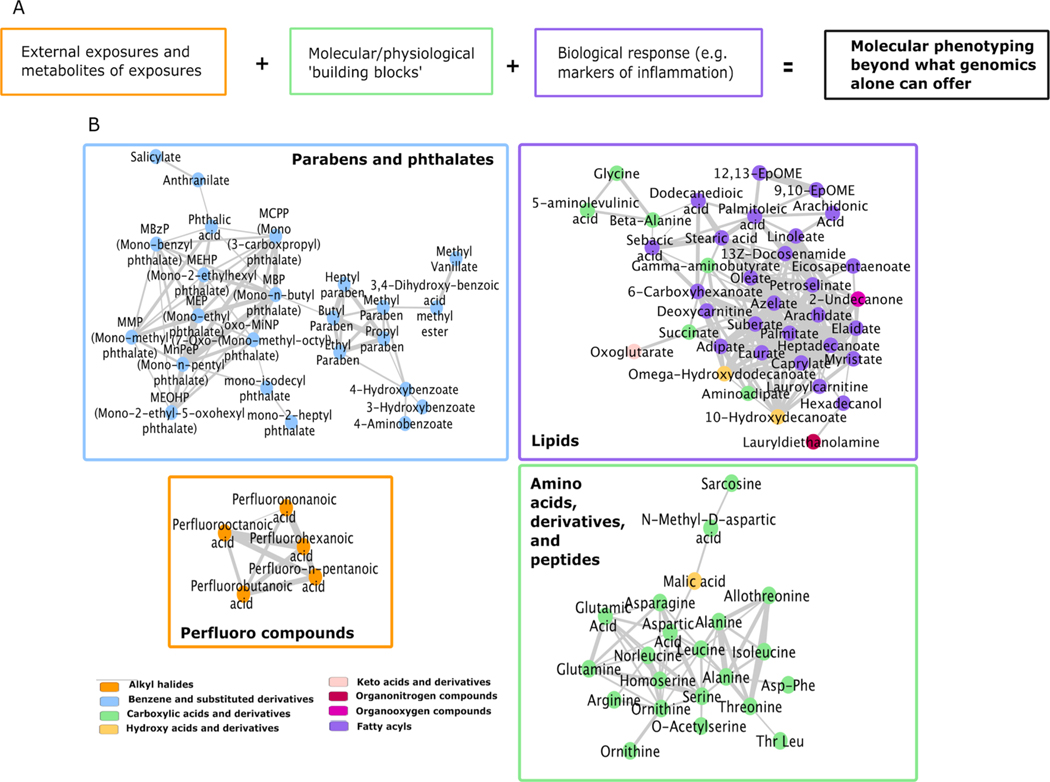
Metabolites annotated included both exogenous compounds (phthalates, parabens, perfluoroalkyl compounds, and cotinine) and endogenous metabolites (fatty acids, steroids, carnitines, amino acids, and other classes) detailed in Table S1. A subset of the annotated metabolites are visualized in Fig. 2, where several clusters of chemically similar compounds are formed. Therefore, our teeth exposome approach provides direct measurements across the continuum from external exposures and their metabolites, to molecular building blocks of physiological functions, and to biological response as a result of environmental factors interacting with our physiology (Fig. 2). Combined with the crucial component of being able to include developmental timing, we provide a new approach to molecular phenotyping beyond what genomics can offer.
Fig. 2. Retrospective temporal molecular phenotyping.
(A) The approach we are proposing provides a pathway from external exposures and their metabolites to the molecular architecture of physiology (the ‘building blocks of life’) to the biological response that is invoked when environmental factors interact with our physiology. (B) A subsample of the classes of compounds we have detected in prenatal and postnatal components of teeth to exemplify the categories in panel A. Clusters are generated by chemical similarity between metabolites.
To evaluate biological and health relevance of the metabolites measured in early-life tooth dentin, we then linked the 390 annotated compounds with disease-associated metabolite sets from the human metabolites database (HMDB) (Wishart et al., 2007). Of the identified metabolites, 91 compounds were found in HMDB and were linked with 256 health conditions through HMDB (Wishart et al., 2007). Diseases and disorders linked with at least 20 annotated tooth metabolites included colorectal cancer (70 metabolites), eosinophilic esophagitis (48 metabolites), ulcerative colitis (35 metabolites), Crohn’s disease (32 metabolites), schizophrenia (32 metabolites), and Alzheimer’s disease (21 metabolites). Schizophrenia (Modabbernia et al., 2016), Crohn’s disease, and ulcerative colitis (Nair et al., 2020) have been linked with deciduous tooth metal levels. Thus, our findings indicate that tooth metabolomics may be a particularly important resource to reveal early- life etiological factors, biological pathways, and biomarkers linked with adverse neurodevelopment and gastrointestinal health. In addition, nineteen metabolites measured in deciduous teeth—2-hydroxybutyrate, alanine, asparagine, citric acid, creatine, estriol, glutamine, glycine, arginine, isoleucine, leucine, phenylalanine, proline, serine, threonine, tyrosine, lauroylcarnitine, lysophosphatidylcholine (18:0), sphingosine, and succinate—have been linked with pregnancy complications, suggesting that teeth exposomics may facilitate understanding of complex relationships between maternal and fetal biological response. In essence, the disease-relevant signatures we are finding in prenatal and early postnatal fractions have the potential to serve as early warning indicators for diseases that currently are detected many years later.
3.3. Metabolites distinct to the prenatal fraction of the early-life tooth exposome
The fetal period is a time of rapid biological changes that may raise susceptibility to exposures that may initiate later adverse health outcomes. Capturing these fetal exposures is possible through leveraging the unique tooth histology. To measure the fetal exposome, we used 31 paired prenatal and postnatal tooth fractions from individual EARLI deciduous teeth and assessed whether any metabolites were distinct to the prenatal period. These metabolite features were those found in at least half of the prenatal fractions (>15 out of 31) but absent in the postnatal fraction, or were found in at least half of the prenatal fractions (>15 out of 31) but no more than half of the postnatal period fractions (<15 out of 31). We rationalized that such an approach would reveal direct measurements of fetal exposures that may be of particular interest for etiological studies of diseases hypothesized to have fetal origins.
After peak filtering, 4815 peaks were retained in all 62 tooth extracts analyzed on the RPP platform. We identified metabolite features that were present in a high proportion of prenatal fractions, and retained only those with peaks and peak integrations that were of high quality (Chetnik et al., 2020) (Table 1). There were 11 metabolite features present in at least 16 prenatal samples but not present in any postnatal samples, and 15 metabolite features present in at least 16 prenatal samples and present in only 2 or less postnatal samples (<10%). Of the 21 metabolites distinct to the prenatal fraction, only a single metabolite feature eluted early, at 0.57 min with an m/z of 294.9823, suggesting a highly hydrophilic compound. All other features distinct to the prenatal fraction were well retained on the column and eluted between 5.11 min and 7.55 min, with m/z ranging from 523.2758 to 683.4331. Confident annotations of these metabolites could not be made based on MS/MS spectral matching to the available databases. However, metabolite classes such as phthalates and fatty acyls elute from 5 to 8 min suggesting the analytes in question are possibly medium hydrophilicity compounds. While we were only able to annotate one of the metabolites (Tyr Leu Phe Asp), MS/MS fragments, when available, and m/z and retention time information is provided to enable future identifications (Table 1).
Table 1.
Summary of molecular features of the early-life tooth exposome that are found primarily in the prenatal fraction.
| Precursor (m/z) | Retention Time (min) | Annotation and Major MS/MS Fragmentsa | Number of Prenatal Samples | Number of Postnatal Samples |
|---|---|---|---|---|
| 294.9823 | 0.57 | NAb | 21 | 1 |
| 523.2758 | 5.11 | 523.2754 (392.1813, 293.1131, 231.1707, 136.0758, 72.0797) | 21 | 0 |
| 537.2915 | 5.45 | NA | 21 | 0 |
| 557.2597 | 5.70 | Tyr Leu Phe Asp, 557.2587 (86.0956, 136.0757, 249.1599, 277.1541, 281.1124) | 29 | 0 |
| 551.3071 | 5.97 | 551.3101 (420.2122, 293.1509) | 26 | 0 |
| 537.2914 | 5.98 | NA | 16 | 0 |
| 571.2758 | 6.06 | NA | 29 | 5 |
| 539.2497 | 6.12 | 539.2454 (249.1604, 136.0755) | 29 | 12 |
| 571.2755 | 6.46 | NA | 26 | 4 |
| 509.2757 | 6.52 | 509.2764 (410.2056, 313.1539, 197.1290, 64.9776) | 26 | 1 |
| 443.6955 | 6.57 | NA | 23 | 4 |
| 451.6842 | 6.57 | 451.6842 (333.5884, 248.2046) | 28 | 1 |
| 864.4130 | 6.57 | NA | 29 | 0 |
| 452.1867 | 6.58 | NA | 26 | 0 |
| 735.3702 | 6.68 | NA | 28 | 0 |
| 585.2911 | 6.76 | NA | 18 | 0 |
| 308.6307 | 7.05 | NA | 28 | 0 |
| 316.6199 | 7.05 | NA | 23 | 5 |
| 523.2915 | 7.12 | NA | 25 | 2 |
| 620.3446 | 7.38 | NA | 28 | 0 |
| 683.4331 | 7.55 | 683.4307 (328.2220, 197.1284) | 30 | 10 |
Precursor ion in MS/MS spectra with major fragment ions listed in parenthesis.
NA is listed for precursor ions that did not yield good-quality MSMS spectra and consistent fragment ions.
3.4. Metabolites differentially measured in prenatal versus postnatal tooth fractions
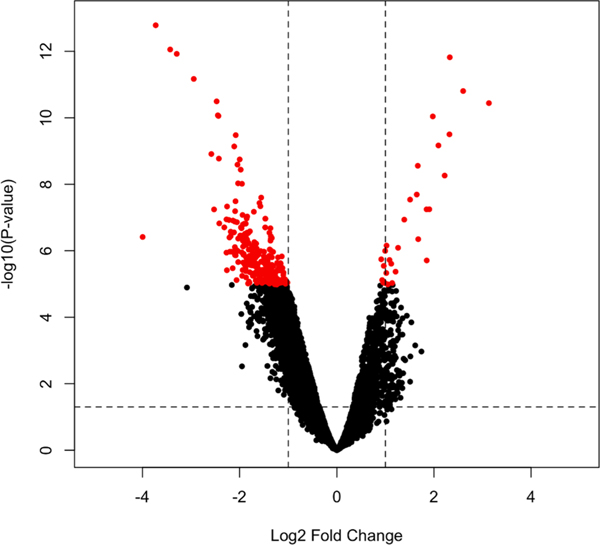
We then assessed whether there were significant differences in individual peak levels between the prenatal and postnatal tooth fractions. In total, 267 peaks show significant differences between pre- and postnatal dentin after Bonferroni correction (adjusted p-value < 0.05), with 237 metabolite features with higher abundances in the postnatal than the prenatal fraction, and 30 metabolite features with higher abundances in the prenatal fraction than the postnatal fraction (see Fig. 3, Table S2.) For metabolite features more abundant in the prenatal fraction, fold changes (prenatal abundance/postnatal abundance) ranged from 1.88 to 8.77, while for metabolites more abundant in the postnatal fraction fold changes (prenatal abundance/postnatal abundance) ranged from 0.06 to 0.49. These results are not surprising given that exposures (environmental, nutritional, psychosocial, etc.) are expected to differ between fetal life and infancy. Interestingly, however, the number of significantly increased metabolite features is smaller in the prenatal fraction (30 compared to 237), indicating possibly less diverse exposures, a role of maternal metabolism dampening exposures, a filtering effect of the placenta, or less developed metabolism during fetal life. In fact, qualitative comparison of untargeted profiles between prenatal and postnatal fractions suggests that features more abundant in the prenatal samples, but not reaching statistical significance, were eluted on the early or late ends of the chromatogram, which may reflect more easily excreted metabolites (very polar) or those very hard to metabolize (very non-polar) in the prenatal samples (Fig. S3). Further, the ability to detect a large number of statistically different features between the prenatal and postnatal fractions suggests that the tooth methodology described here is robust to performing relative quantification across groups for environmental health studies and etiological discovery. Interestingly, the 267 differentiating metabolites could not be annotated as the obtained MS/MS spectra did not match well to any databases. The use of acetic acid as a LC-buffer additive in this analysis could have hindered spectral matching to the databases. Acetate adducts may not match spectra obtained from the more conventionally used formic acid. More importantly, however, these results point to the vast chemical space in the tooth exposome that has yet to be fully characterized.
Fig. 3. Volcano plot of measured features.
Red features are those with p- values < 0.05 after Bonferroni correction. Significant peaks that were considered redundant peaks or those with unreliable peak integrations were removed. Fewer significant metabolites with positive fold changes (higher in prenatal tooth fraction than postnatal tooth fraction) were observed compared to those with negative fold changes.
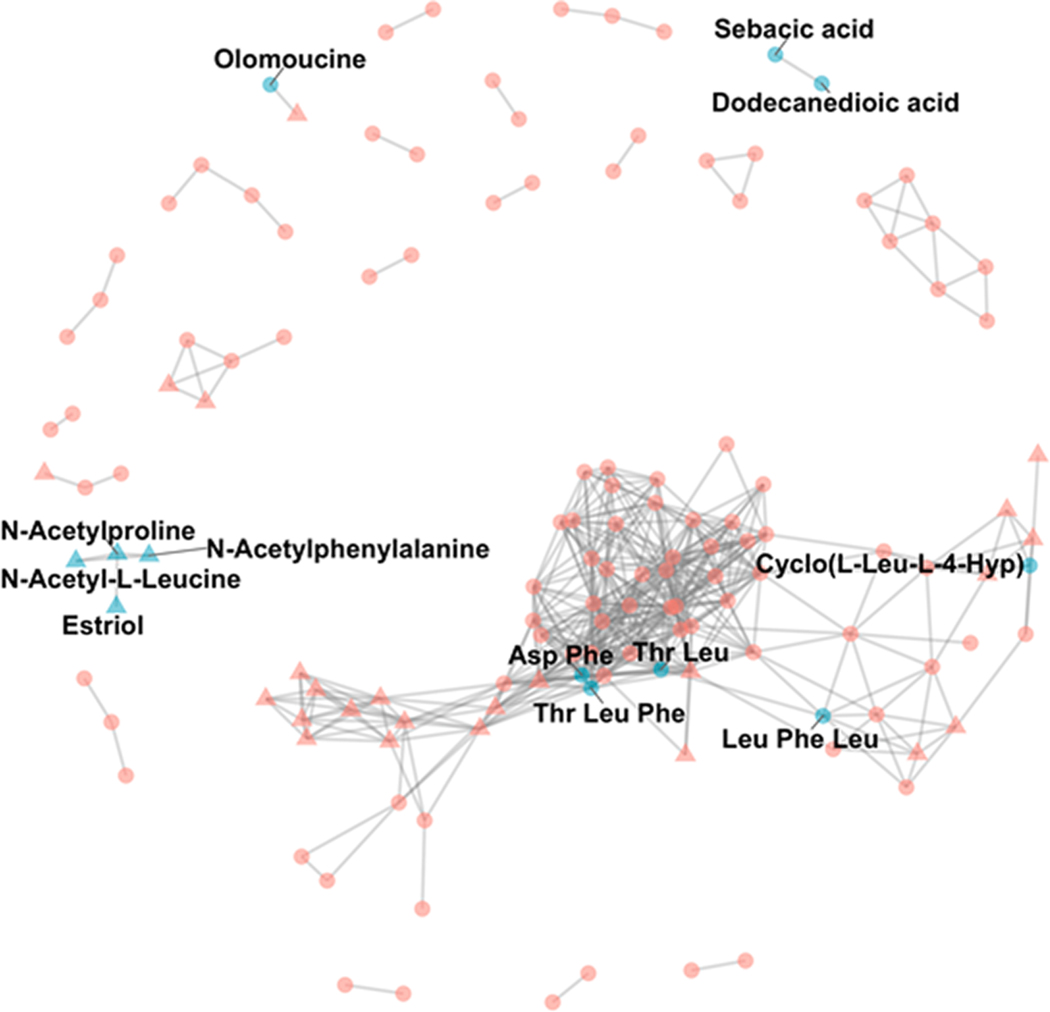
To obtain biological interpretation of the significant metabolites, we retained only the 195 independent peaks (Yu et al., 2019) after correlation filtering (see Methods). This reduces the total number of peaks for easier visualization without losing biological information. Since it has been shown in KEGG that many metabolites that are involved in similar biological pathways are highly correlated (Kanehisa et al., 2016), we generated a correlation map with edges representing those feature pairs with a Pearson’s correlation coefficient > 0.9 (see Fig. 4). We then mapped to the network the annotated metabolites from Table S1 (labels, Fig. 4), as well as those metabolites significantly different between pre- and postnatal fractions (triangles, Fig. 4) to infer biological information on the unknowns. Twenty metabolites with levels significantly different between the tooth fractions were located on the periphery of a large cluster of highly connected nodes that included dipeptides and tripeptides. In addition, four metabolites with levels significantly different between tooth fractions were correlated with acetylphenylalanine, acetylleucine, and acetylproline. Thus, we speculate that these 24 unknown differentially measured metabolites are functionally similar to small peptides or acetyl amino acids, respectively.
Fig. 4. Untargeted metabolite correlation network map.
Each node is an independent peak determined by the GlobalStd algorithm with correlation coefficient filtering. Edges are those peaks with Pearson correlation coefficient > 0.9. Independent peaks that could be matched to Table S1 are colored blue and labeled with annotations. Triangle nodes are those metabolites with significantly different abundances between paired pre- and postnatal samples (<0.05 adj. p-value, Bonferroni correction). Although not annotated, significantly different peaks may be functionally similar to small peptides or acetyl amino acids.
3.5. Considerations for future analysis
Although we demonstrated that a broad range of metabolites can be measured across the tooth exposome, there is a large chemical space of unidentified metabolites. This included all 267 significant peaks that discriminated the prenatal versus postnatal fraction and 20 of the 21 metabolites that were distinct in the prenatal tooth fraction. Even though we were able to generate MS/MS fragmentation data of several of these peaks (Table 1), they did not match any spectra in the Metlin or GNPS databases. This is surprising since both Metlin and GNPS are expansive databases with 4,000,000 and 221,000 reference MSMS spectra, respectively (Xue et al., 2020; Aron et al., 2020). It is possible that the complex processes involved in extracting small organic molecules from dentin tooth compared to simple protein precipitation that is commonly used in urine or plasma metabolomics produce unique adducts of metabolites that may limit spectral matching. Alternately, as this is the first untargeted analysis of tooth dentin, we may be observing small molecules (di- and tripeptides included) that are unique to this matrix. Future untargeted chemical studies on tooth dentin will enhance the breadth of annotations and identifications to facilitate deeper biological interpretation of early-life exposures and biology.
4. Conclusion
The tooth exposome represents an unparalleled landscape for etiological discovery of early life exposures and biological pathways. We performed the first untargeted assay to directly profile fetal and early postnatal exposures in tooth dentin. This approach identified several hundred small molecules including exogenous exposures and endogenous metabolites, many of which are biologically relevant and have been linked to adverse health outcomes. While this discovery study was limited to only 31 samples, we identified metabolites representing distinct fetal exposures and demonstrated the robustness of the methodology, which revealed over 250 metabolites significantly different between pre and postnatal tooth fractions. These compounds are unknown, but future work to identify these metabolites is warranted to define whether they play important roles in fetal and early-life programming, dysregulated physiology, and biological response linked with diseases and disorders.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health from National Institute of Environmental Health Sciences grants U2CES030859 (LP, MA), R21ES030882 (LP), and R01ES031117 (LP), the Eunice Kennedy Shriver National Institute of Child Health and Human Development grant R00HD087523 (CA), and the National Institute of Dental and Craniofacial Research grant DE029838 (LP, CA).
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2021.106849.
References
- Almeida DL, Pavanello A, Saavedra LP, Pereira TS, Castro-Prado MAAD, Mathias PCDF, 2019. Environmental Monitoring and the Developmental Origins of Health and Disease. J. Dev. Orig. Health Dis 10 (6), 608–615. 10.1017/S2040174419000151. [DOI] [PubMed] [Google Scholar]
- Andra SS, Austin C, Arora M, 2015. Tooth Matrix Analysis for Biomonitoring of Organic Chemical Exposure: Current Status, Challenges, and Opportunities. Environ. Res 142, 387–406. 10.1016/j.envres.2015.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andra SS, Austin C, Wright RO, Arora M, 2015. Reconstructing Pre-Natal and Early Childhood Exposure to Multi-Class Organic Chemicals Using Teeth: Towards a Retrospective Temporal Exposome. Environ. Int 83, 137–145. 10.1016/j.envint.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron AT, Gentry EC, McPhail KL, Nothias LF, Nothias-Esposito M, Bouslimani A, Petras D, Gauglitz JM, Sikora N, Vargas F, van Der Hooft JJ, 2020. Reproducible Molecular Networking of Untargeted Mass Spectrometry Data Using GNPS. Nat. Protoc 15 (6), 1954–1991. 10.1038/s41596-020-0317-5. [DOI] [PubMed] [Google Scholar]
- Arora M, Austin C, 2013. Teeth as a Biomarker of Past Chemical Exposure. Curr. Opin. Pediatr 25 (2), 261–267. 10.1097/MOP.0b013e32835e9084. [DOI] [PubMed] [Google Scholar]
- Arora M, Reichenberg A, Willfors C, Austin C, Gennings C, Berggren S, Lichtenstein P, Anckarsater H, Tammimies K, B¨ olte S, 2017. Fetal and Postnatal ¨ Metal Dysregulation in Autism. Nat. Commun 8 (1) 10.1038/ncomms15493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin C, Smith TM, Bradman A, Hinde K, Joannes-Boyau R, Bishop D, Hare DJ, Doble P, Eskenazi B, Arora M, 2013. Barium Distributions in Teeth Reveal Early-Life Dietary Transitions in Primates. Nature 498 (7453), 216–219. 10.1038/nature12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajusz D, Racz A, H´ éberger K, 2015. Why Is Tanimoto Index an Appropriate Choice for Fingerprint-Based Similarity Calculations? J. Cheminformatics 7 (1), 20. 10.1186/s13321-015-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JM, 2017. Early-Life Exposure to EDCs: Role in Childhood Obesity and Neurodevelopment. Nat. Rev. Endocrinol 13 (3), 161–173. 10.1038/nrendo.2016.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camann DE, Schultz ST, Yau AY, Heilbrun LP, Zuniga MM, Palmer RF, Miller CS, 2013. Acetaminophen, Pesticide, and Diethylhexyl Phthalate Metabolites, Anandamide, and Fatty Acids in Deciduous Molars: Potential Biomarkers of Perinatal Exposure. J. Expo. Sci. Environ. Epidemiol 23 (2), 190–196. 10.1038/jes.2012.71. [DOI] [PubMed] [Google Scholar]
- Chetnik K, Petrick L, Pandey G, 2020. MetaClean: A Machine Learning-Based Classifier for Reduced False Positive Peak Detection in Untargeted LC–MS Metabolomics Data. Metabolomics 16 (11). 10.1007/s11306-020-01738-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoumbou Feunang Y, Eisner R, Knox C, Chepelev L, Hastings J, Owen G, Fahy E, Steinbeck C, Subramanian S, Bolton E, Greiner R, Wishart DS, 2016. ClassyFire: Automated Chemical Classification with a Comprehensive, Computable Taxonomy. J. Cheminform 8, 61. 10.1186/s13321-016-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Romero C, Mikhail KA, Gennings C, Curtin P, Bello GA, Botero TM, Goutman SA, Feldman EL, Arora M, Austin C, 2020. Early Life Metal Dysregulation in Amyotrophic Lateral Sclerosis. Ann. Clin. Transl. Neurol 7 (6), 872–882. 10.1002/acn3.v7.610.1002/acn3.51006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare D, Austin C, Doble P, Arora M, 2011. Elemental Bio-Imaging of Trace Elements in Teeth Using Laser Ablation-Inductively Coupled Plasma-Mass Spectrometry. J. Dent 39 (5), 397–403. 10.1016/j.jdent.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Hsu H-H-L, Chiu Y-H-M, Coull BA, Kloog I, Schwartz J, Lee A, Wright RO, Wright RJ, 2015. Prenatal Particulate Air Pollution and Asthma Onset in Urban Children. Identifying Sensitive Windows and Sex Differences. Am. J. Respir. Crit. Care Med 192 (9), 1052–1059. 10.1164/rccm.201504-0658OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Lesseur C, Miao Y, Manservisi F, Panzacchi S, Mandrioli D, Belpoggi F, Chen J, Petrick L, 2021. Low-Dose Exposure of Glyphosate-Based Herbicides Disrupt the Urine Metabolome and Its Interaction with Gut Microbiota. Sci. Rep 11 (1), 3265. 10.1038/s41598-021-82552-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland C, Gunier RB, Metayer C, Bates MN, Wesseling C, Mora AM, 2018. Maternal Residential Pesticide Use and Risk of Childhood Leukemia in Costa Rica. Int. J. Cancer 143 (6), 1295–1304. 10.1002/ijc.31522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M, 2016. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 44 (D1), D457–D462. 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrigan PJ, Goldman LR, 2011. Children’s Vulnerability To Toxic Chemicals: A Challenge And Opportunity To Strengthen Health And Environmental Policy. Health Aff. (Millwood) 30 (5), 842–850. 10.1377/hlthaff.2011.0151. [DOI] [PubMed] [Google Scholar]
- Libiseller G, Dvorzak M, Kleb U, Gander E, Eisenberg T, Madeo F, Neumann S, Trausinger G, Sinner F, Pieber T, Magnes C, 2015. IPO: A Tool for Automated Optimization of XCMS Parameters. BMC Bioinformatics 16, 118. 10.1186/s12859-015-0562-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Rosenbaum S, 2014. Developmental Pharmacokinetics in Pediatric Populations. J. Pediatr. Pharmacol. Ther 19 (4), 262–276. 10.5863/1551-6776-19.4.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchei E, Joya X, Garcia-Algar O, Vall O, Pacifici R, Pichini S, 2008. Ultrasensitive Detection of Nicotine and Cotinine in Teeth by High-Performance Liquid Chromatography/Tandem Mass Spectrometry. Rapid Commun. Mass Spectrom 22 (16), 2609–2612. 10.1002/rcm.v22:1610.1002/rcm.3636. [DOI] [PubMed] [Google Scholar]
- Modabbernia A, Velthorst E, Gennings C, De Haan L, Austin C, Sutterland A, Mollon J, Frangou S, Wright R, Arora M, Reichenberg A, 2016. Early-Life Metal Exposure and Schizophrenia: A Proof-of-Concept Study Using Novel Tooth- Matrix Biomarkers. Eur. Psychiatry J. Assoc. Eur. Psychiatr 36, 1–6. 10.1016/j.eurpsy.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SE, 2016. Early Life Nutritional Programming of Health and Disease in The Gambia. J. Dev. Orig. Health Dis 7 (2), 123–131. 10.1017/S2040174415007199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair N, Austin C, Curtin P, Gouveia C, Arora M, Torres J, Dubinsky M, Colombel J-F, Peter I, 2020. Mount Sinai Road to Prevention Group. Association Between Early-Life Exposures and Inflammatory Bowel Diseases, Based on Analyses of Deciduous Teeth. Gastroenterology 159 (1), 383–385. 10.1053/j.gastro.2020.03.040. [DOI] [PubMed] [Google Scholar]
- Ottaviani G, Cameriere R, Cippitelli M, Froldi R, Tassoni G, Zampi M, Cingolani M, 2017. Determination of Drugs of Abuse in a Single Sample of Human Teeth by a Gas Chromatography-Mass Spectrometry Method. J. Anal. Toxicol 41 (1), 32–36. 10.1093/jat/bkw105. [DOI] [PubMed] [Google Scholar]
- Palmer RF, Heilbrun L, Camann D, Yau A, Schultz S, Elisco V, Tapia B, Garza N, Miller C, 2015. Organic Compounds Detected in Deciduous Teeth: A Replication Study from Children with Autism in Two Samples. J. Environ Public Health 2015, 1–9. 10.1155/2015/862414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Casa A, Marchei E, Pacifici R, Mayné R, Barbero V, Garcia-Algar O, Pichini S, 2006. Development and Validation of a Gas Chromatography-Mass Spectrometry Assay for Opiates and Cocaine in Human Teeth. J. Pharm. Biomed. Anal 40 (3), 662–668. 10.1016/j.jpba.2005.07.003. [DOI] [PubMed] [Google Scholar]
- R Core Team, 2020. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK, 2015. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. e47–e47 Nucleic Acids Res. 43 (7). 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers D, Hahn M, 2010. Extended-Connectivity Fingerprints. J. Chem. Inf. Model 50 (5), 742–754. 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- Schiffman C, Petrick L, Perttula K, Yano Y, Carlsson H, Whitehead T, Metayer C, Hayes J, Rappaport S, Dudoit S, 2019. Filtering Procedures for Untargeted LC-MS Metabolomics Data. BMC Bioinformatics 20 (1), 334. 10.1186/s12859-019-2871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G, 2006. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem 78 (3), 779–787. 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Stephane B, Sabrina B, Marianne B-B, Marianne H, Audrey S, Nathalie A, 2020. Association between Kawasaki Disease and Prenatal Exposure to Ambient and Industrial Air Pollution: A Population-Based Cohort Study. Environ. Health Perspect 128 (10) 10.1289/EHP6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Velazquez MA, Fleming TP, 2016. Chapter 3 - DOHaD and the Periconceptional Period, a Critical Window in Time. In: Rosenfeld CS (Ed.), The Epigenome and Developmental Origins of Health and Disease. Elsevier, pp. 33–47. 10.1016/B978-0-12-801383-0.00003-7. [DOI] [Google Scholar]
- Tanner EM, Hallerback MU, Wikström S, Lindh C, Kiviranta H, Gennings C, Bornehag C-G, 2020. Early Prenatal Exposure to Suspected Endocrine Disruptor Mixtures Is Associated with Lower IQ at Age Seven. Environ. Int 134, 105185. 10.1016/j.envint.2019.105185. [DOI] [PubMed] [Google Scholar]
- Valeri L, Mazumdar MM, Bobb JF, Claus Henn B, Rodrigues E, Sharif OIA, Kile ML, Quamruzzaman Q, Afroz S, Golam M, Amarasiriwardena C, Bellinger DC, Christiani DC, Coull BA, Wright RO, 2017. The Joint Effect of Prenatal Exposure to Metal Mixtures on Neurodevelopmental Outcomes at 20–40 Months of Age: Evidence from Rural Bangladesh. Environ. Health Perspect 125 (6), 067015. 10.1289/EHP614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Anker J, Reed MD, Allegaert K, Kearns GL, 2018. Developmental Changes in Pharmacokinetics and Pharmacodynamics. J. Clin. Pharmacol 58 (S10), S10–S25. 10.1002/jcph.1284. [DOI] [PubMed] [Google Scholar]
- von Ehrenstein OS, Ling C, Cui X, Cockburn M, Park AS, Yu F, Wu J, Ritz B, 2019. Prenatal and Infant Exposure to Ambient Pesticides and Autism Spectrum Disorder in Children: Population Based Case-Control Study. BMJ 364. 10.1136/bmj.l962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, Porto C, Bouslimani A, Melnik AV, Meehan MJ, Liu W-T, Crüsemann M, Boudreau PD, Esquenazi E, Sandoval-Calderon M, Kersten RD, Pace LA, Quinn RA, Duncan KR, Hsu C-´C, Floros DJ, Gavilan RG, Kleigrewe K, Northen T, Dutton RJ, Parrot D, Carlson EE, Aigle B, Michelsen CF, Jelsbak L, Sohlenkamp C, Pevzner P, Edlund A, McLean J, Piel Jorn, Murphy BT, Gerwick L, Liaw C-C, Yang Y-L, Humpf H-U, Maansson M, Keyzers RA, Sims AC, Johnson AR, Sidebottom AM, Sedio BE, Klitgaard A, Larson CB, Boya P CA, Torres- Mendoza D, Gonzalez DJ, Silva DB, Marques LM, Demarque DP, Pociute E, O’Neill EC, Briand E, Helfrich EJN, Granatosky EA, Glukhov E, Ryffel F, Houson H, Mohimani H, Kharbush JJ, Zeng Y, Vorholt JA, Kurita KL, Charusanti P, McPhail KL, Nielsen KF, Vuong L, Elfeki M, Traxler MF, Engene N, Koyama N, Vining OB, Baric R, Silva RR, Mascuch SJ, Tomasi S, Jenkins S, Macherla V, Hoffman T, Agarwal V, Williams PG, Dai J, Neupane R, Gurr J, Rodríguez Andrés.M.C., Lamsa A, Zhang C, Dorrestein K, Duggan BM, Almaliti J, Allard P-M, Phapale P, Nothias L-F, Alexandrov T, Litaudon M, Wolfender J-L, Kyle JE, Metz TO, Peryea T, Nguyen D-T, VanLeer D, Shinn P, Jadhav A, Müller R, Waters KM, Shi W, Liu X, Zhang L, Knight R, Jensen PR, Palsson BØ, Pogliano K, Linington RG, Gutiérrez M, Lopes NP, Gerwick WH, Moore BS, Dorrestein PC, Bandeira N, 2016. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol 34 (8), 828–837. 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly M-A, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, Macinnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L, 2007. HMDB: The Human Metabolome Database. Nucleic Acids Res. 35 (Database issue), D521–526. 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Guijas C, Benton HP, Warth B, Siuzdak G, 2020. METLIN MS 2 Molecular Standards Database: A Broad Chemical and Biological Resource. Nat. Methods 17 (10), 953–954. 10.1038/s41592-020-0942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Olkowicz M, Pawliszyn J, 2019. Structure/Reaction Directed Analysis for LC- MS Based Untargeted Analysis. Anal. Chim. Acta 1050, 16–24. 10.1016/j.aca.2018.10.062. [DOI] [PubMed] [Google Scholar]
- Yu M, Dolios G, Yong-Gonzalez V, Bjorkqvist O, Colicino E, Halfvarson J, ¨ Petrick L, 2020. Untargeted Metabolomics Profiling and Hemoglobin Normalization for Archived Newborn Dried Blood Spots from a Refrigerated Biorepository. J. Pharm. Biomed. Anal 191, 113574. 10.1016/j.jpba.2020.113574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.