Abstract
The emergence of Variants of Concern (VOC) presenting an unusual number of new mutations is one of the most remarkable features of SARS-CoV-2. The Delta variant, since its appearance, replaced the VOC Gamma, which was responsible for the major COVID-19 wave in Brazil. In this study, we performed a Delta whole-genome sequencing of 183 samples as part of a major genomic surveillance study performed since the beginning of the pandemic. Here, we showed an emergence, widespread dispersion and consolidation of the Delta variant in Rio Grande do Sul State, completely replacing the Gamma variant in a four to five months period. Performing the phylogenetic and phylodynamic analysis, the majority of the sequences generated herein were classified as AY.99.2, AY.99.2-like and AY.101. AY.99.2 Delta-related lineage has been widely reported in Brazil and in the Americas as well. Altogether, our findings provided a mutational profile of the sequences and presented high substitutions per site in the root-to-tip phylogenetic tree, corroborating studies that show the high mutational rate of SARS-CoV-2 over time.
Keywords: SARS-CoV-2, B.1.617.2, AY.99.2, AY.101, Delta variant, Southern Brazil
1. Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is an enveloped RNA virus that has been proved to change over time. Some mutations may affect the virus's properties interfering with how easily it spreads, the associated disease severity, and the efficiency of vaccines, diagnostic tools, or other public health and social efforts (World Health Organization, 2021). Therefore, quick spread, genetic viral evolution, and incomplete vaccination schedules in many countries have been driving the emergence of SARS-CoV-2 variants that may interfere in the vaccine-derived immune response protective effectiveness (Kuzmina et al., 2021).
SARS-CoV-2 variant B.1.617.2, also known as the Delta variant, has been quickly spread and detected in many countries (Bolze et al., 2021; Li et al., 2021; Public Health England, 2021). The first samples were documented from India in October of 2020, and it has been classified as a variant of concern (VOC) by WHO since May 11, 2021 World Health Organization (2021). As main aspects that support the increase in the transmissibility of the Delta variant and consequently a replacement of other variants in circulation worldwide, it is possible to highlight a fitness advantage led by higher viral loads and shorter incubation time (Grant et al., 2021; Li et al., 2021).
In Brazil, the first community sustained transmission chains were reported in Rio de Janeiro in June of 2021 (Lamarca et al., 2021) and it has been widely detected in other Brazilian states over time (Ministério da Saúde (MS), 2021). In Rio Grande do Sul (RS), the southernmost state of Brazil, the first case was officially reported by local health authorities from a clinical specimen collected on July 12, 2021 (Secretaria da Saúde do Rio Grande do Sul, 2021a). An analysis of data provided by GISAID from October to November of 2021 in Brazil showed that the most common lineages circulating in Brazil are Delta-related variants (AY.99.2, AY.43, AY.101, AY.34.1, AY.43.1, AY.43.2, AY.46.3, AY.100, AY.99.1, AY.36) (Latif et al., 2021a). Analyzing S-gene mutations in > 75% of sequences for lineages found in Brazil in the same period, the most common mutations mentioned are T19R, T95I, E156G, DEL157/158, L452R, T478K, D614G, Q677H, P681R, D950N, V1104L and L1265F (Latif et al., 2021a). Some of these mutations have been related to viral fitness advantages such as enhanced viral entry, infectivity, pathogenesis, and immune escape (Khan et al., 2021; Korber et al., 2020; Li et al., 2020; Saito et al., 2021).
Tracking SARS-CoV-2 variants through sequencing becomes a key part of a better understanding of viral evolution, especially after the beginning of the vaccination programs. Therefore, the main objective of this work was to describe the appearance, dissemination, and predominance of the Delta variant over time in the state of Rio Grande do Sul in southern Brazil. Besides, through the genomic analysis, we aimed to report which Delta related lineages are circulating as well as the mutational distribution and signatures across the SARS-CoV-2 genomes.
2. Material and methods
2.1. Sampling
A total of 183 complete Delta variant SARS-CoV-2 genomes were sequenced. These samples were collected from June to October of 2021 covering 20 cities of RS State and two patients from other Brazilian states that were hospitalized in RS. The cities and the corresponding number of samples are described in Supplementary Figure 1. Male patients corresponded to 93 samples and female to 91 with age varying from 4 months to 93 years old. A retrospective study was also performed in order to observe the lineages fluctuation in 2021. To understand the Delta variant emergence context, we analyzed 429 complete genomes sequenced (including 183 samples mentioned above) in our laboratory since the beginning of 2021.
2.2. Samples screening and genome sequencing
Naso-oropharyngeal swab samples were received from suspected patients of COVID-19 at Laboratório de Microbiologia Molecular of Universidade Feevale, Novo Hambugo, Brazil. After the diagnosis confirmation, a pre-selection was carried out, choosing samples that presented Ct below 30. The commercial MagMAX™ CORE Nucleic Acid Purification Kit (Applied biosystems™, Thermo Fisher Scientific, Waltham, MA, USA) kit was used to perform viral RNA extraction using the automated equipment KingFisher™ Duo Prime (Thermo Fisher Scientific™). As previously described by Silva et al. (2021), viral genome sequencing and phylogenetic analysis were carried out. Briefly, reverse transcription reaction was carried out in RNA extracted by SuperScript IV reverse transcriptase kit (Thermo Fisher Scientific, Waltham, MA, USA). According to the manufacturer instructions (QIAGEN, Hilden, Germany), viral genome library preparation was carried out using the QIAseq SARS-CoV-2 Primer Panel paired for library enrichment and QIAseq FX DNA Library UDI Kit. Also, Illumina MiSeq platform (Foster City, CA, USA), using MiSeq Reagent Kit v3 (600-cycle) was used.
2.3. Phylogenetic analysis
The Geneious Prime™ suite was used for genome assembly and editing. Briefly, FASTQ reads were imported, trimmed to remove low quality sequences and primers used for library generation (BBDuk 37.25), and mapped against the reference sequence hCoV-19/Wuhan/WIV04/2019 (EPI_ISL_402124) available in the EpiCoV database from GISAID (Shu and McCauley, 2017). PANGO and Nextstrain lineage assignments were applied to characterize the consensus sequences. Phylogenetic tree was constructed including all SARS-CoV-2 complete genomes available from GISAID (between November 2019 and December 2021) through the Nextclade tool on Nextstrain server (Aksamentov et al., 2021). Mutational sequence profiles and signatures were also analyzed in order to better understand the lineage sequences.
Nextstrain workflow (https://github.com/nextstrain/ncov) (Nextstrain, 2021a) was used for the temporal analysis of amino acid changes in the Delta variant genomes sequenced in this study. The 183 Delta variant genomes were aligned against the Wuhan-Hu-1 reference through Nextclade tool, and maximum-likelihood phylogenetic tree was constructed in IQ-TREE33. The molecular clock branch lengths and amino acid changes were obtained via TreeTime.
3. Results
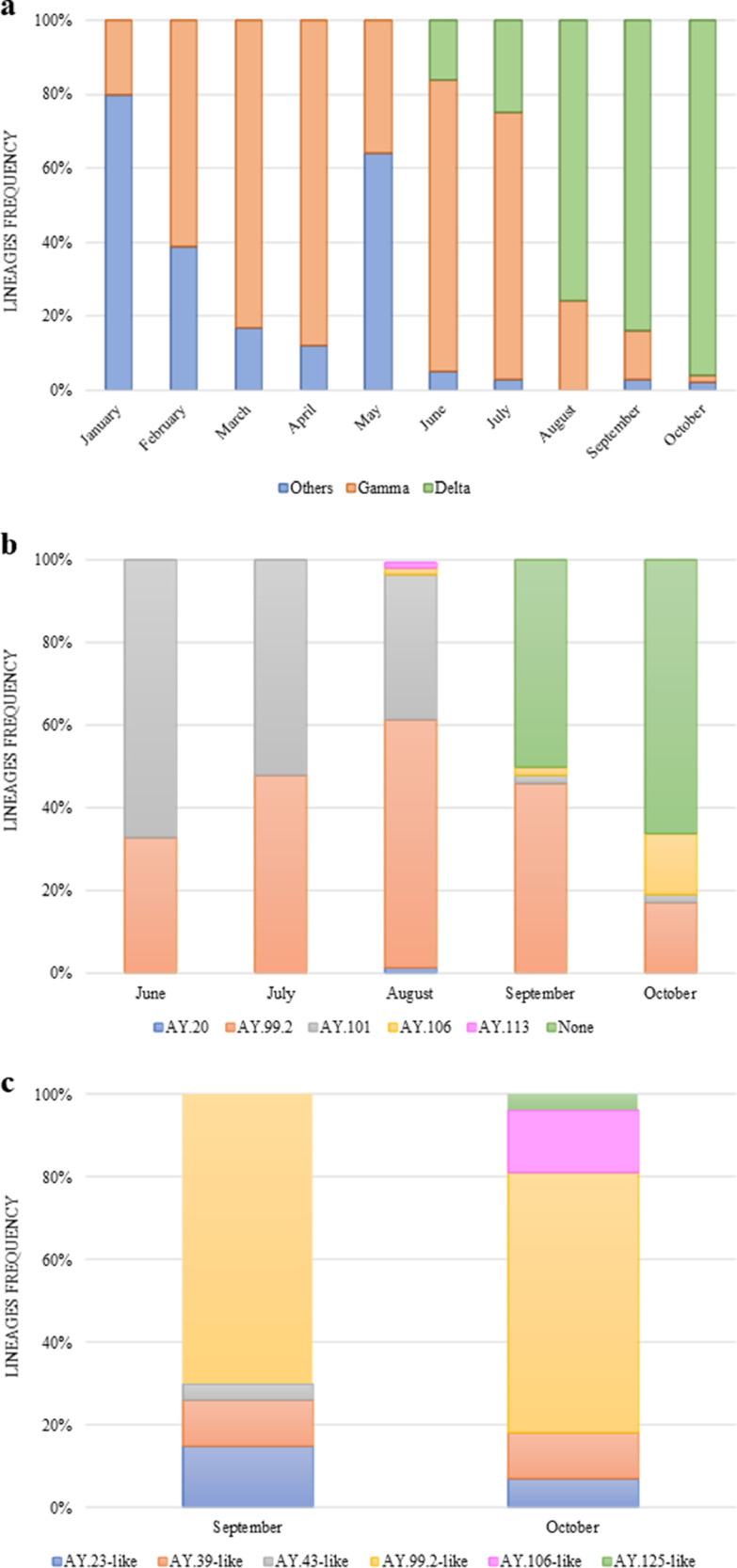
Analyzing our data from 429 COVID-19 positive samples since the beginning of 2021, Delta variant, which appeared in June of 2021, overcome the frequency of Gamma and other lineages showing an exponential increase in June (16%), July (25%), August (76%), September (84%), and October (96%) (Fig. 1 a). In the first months of the Delta lineage entrance, Gamma lineage was still predominant until July (72%). However, in the coming months that followed, it was possible to observe the significant growth of Delta lineages circulation (Fig. 1a). Among these samples, a total of 183 high-quality complete genomes from Delta sequences were retrieved with a breadth coverage of 99% considering the totality of coding regions sequences and the depth coverage was between 200X and 2000X, with an average of 800X.
Fig. 1.
SARS-CoV-2 lineage's frequencies in Rio Grande do Sul State (RS), Brazil. Images show the lineages frequencies from genomic surveillance (1a), the Delta “AY.XX” 21 J frequencies (1b) and AY.XX-like sub-lineage frequencies (1c). These analyzes were performed with samples of our laboratory since the beginning of 2021.
Through PANGO lineage assignment tool, these sequences were classified into five different Delta sub-lineages (AY.99.2, AY.101, AY.106, AY.20, AY.113) and the most prevalent sub-lineages were AY.99.2 (63%) and AY.101 (29%) (Fig. 1b). It's important to note that 54 sequences were classified as “none” and the analysis showed that they were more related to specific sub-lineages, so they were putatively classified as AY.XX-like (Fig. 1c). The majority of the “none” sequences were classified as AY.99.2-like (67%). The other sequences classified as "none" (AY.39-like, AY.23-like, AY.106-like, AY.43-like, and AY.125-like) were not very considerable, corresponding together to 33%. Thus, AY.99.2 and AY.99.2-like together corresponded to 64% of the obtainable frequency in all samples being the most significant found.
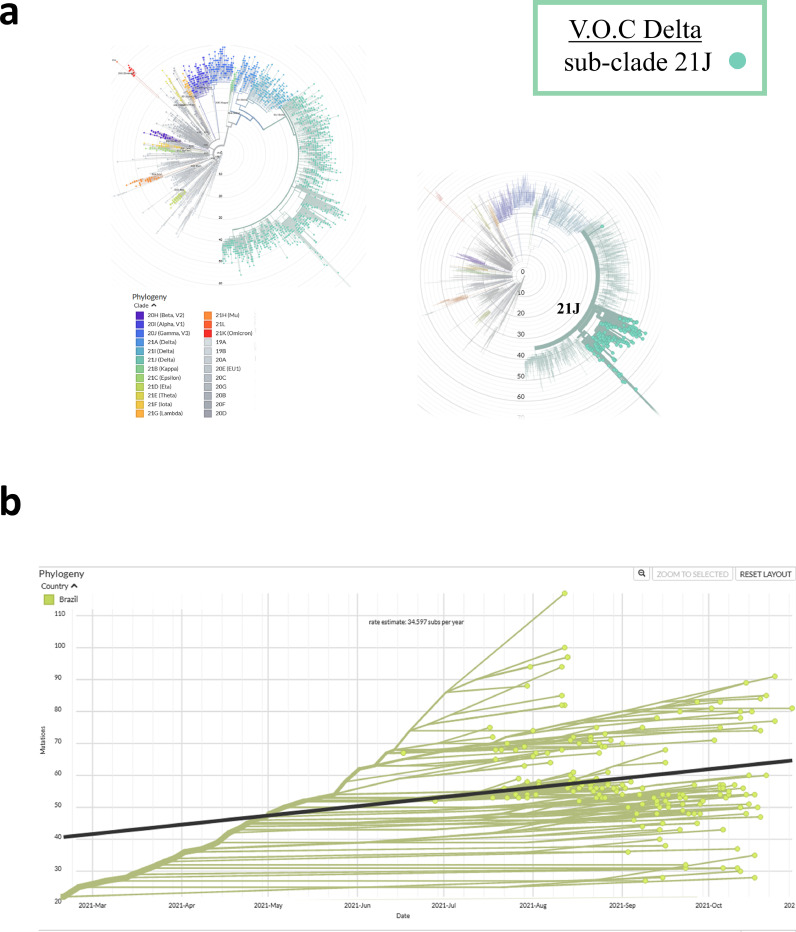
The Nexstrain phylogenetic tree showed that the sequences clustered in a single Delta cluster, belonging to 21 J clade (Fig. 2 a). Using root-to-tip regression, an estimated high rate of 34 substitutions per year was suggested through molecular clock analysis (Fig. 2b). Compared to the reference strain, it was observed in the 21 J clade sequences a wide range (from 15 to 108) in the quantification of genome nucleotide mutations. Also, a subcluster above the regression line suggested a mutation rate above the average.
Fig. 2.
Phylogenetic and phylodynamic Delta SARS-CoV-2 variant analysis in Rio Grande do Sul State. Panel A shows the phylogenetic tree of all circulating lineages, developed through the Nextstrain server (Nextclade tool), using GISAID data. The sequences generated herein are clustered into a single Delta lineage, marked in green (21 J). Panel B shows the phylogenetic tree of 183 SARS-CoV-2 Delta variants. The phylogeny is embedded as a root-to-tip plot, in which the x axis represents the date of sample collection, and the y axis represents the number of genomewide mutations that have occurred since the phylogeny root. The regression line represents the average mutation rate of the SARS-CoV-2 sequences in the tree (34.5 substitutions/year).
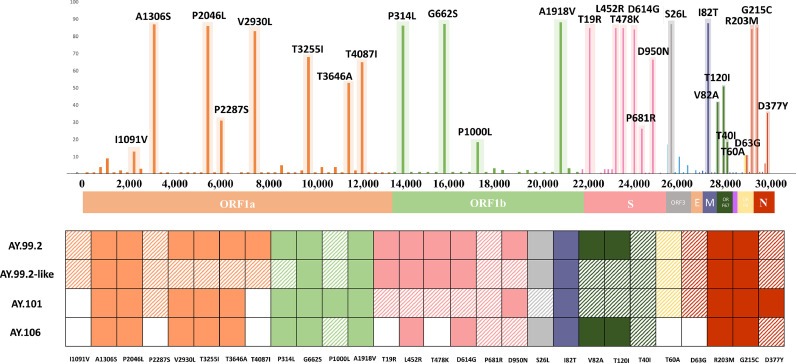
In order to better understating Delta sub-lineages, a complete mutational profile along the genome of SARS-CoV-2, including all coding regions, was described. The mutations, analyzed in each of the most prevalent sub-lineages (AY.99.2, AY.99.2-like, AY.101 and AY.106), presenting more than 10%, were highlighted and compared according to each sub-lineage. The most common mutations, as the complete mutational profile and information's could be observed in Fig. 3 .
Fig. 3.
Mutational profile and frequency of mutations in complete genome sequences of SARS-CoV-2. Mutations with more than 10% of frequency were described. The presence of mutations along the genome of SARS-CoV-2, in each of the most prevalent sub-lineages, was described as follows: Fully painted squares correspond to 100% frequency of a given mutation. Hatched squares correspond to at least one sequence with the given mutation although it has never been found with 100% frequency.
4. Discussion
COVID-19 epidemic in Brazil, during 2021, was driven by the spread and predominance of the VOC Gamma (lineage P.1) which caused the highest wave of SARS-CoV-2, considering the number of cases, deaths, and hospitalization per day (Demoliner et al., 2021). The first Delta variant local case described in Brazil was in July 2021, by Lamarca et al. (2021). Posteriorly, through retrospective analysis, we described three earlier cases in late June, showing that the Delta variant was circulating in the State before that. Since the Delta lineage started to appear in RS State, the cases increase monthly as observed in our results. Despite this, in contrast to the huge COVID-19 epidemic wave associated with the substitution of B.1.1.28 by the VOC Gamma (Demoliner et al., 2021; Faria et al., 2021), Delta viral lineage replacement occurred without the SARS-CoV-2 cases increase, probably due to the high immunity levels (natural and/or vaccinated) in the RS State population (Secretaria da Saúde do Rio Grande do Sul, 2021b). This scenario is similar to what has happened in the European Union (EU) since its emergence in March 2021. A significant transmission advantage was observed when compared to the previously circulating SARS-CoV-2 strains therefore Delta variant quickly became predominant. However, immunity, especially through vaccination, remains protective against severe outcomes (European Centre for Disease Prevention and Control, 2021).
The totality of the sequences generated herein clustered into the Delta 21 J. It is a major sub-clade of the Delta variant, which has grown worldwide, particularly in Europe, Americas, Africa, and Oceania. The 21J sub-clade has all the mutations of21A, in addition to an extra mutation in N at position N:G215C, which was described in all our sequences. It also has additional amino-acid mutations in ORF1a, ORF1b, and ORF7b: ORF1a:A1306S, ORF1a:V2930L, ORF1a:T3255I, ORF1a:T3646A, ORF1b:A1918V, and ORF7b:T40I (CoVariants, 2021a). Except for the ORF1a:V2930L which was interestingly not detected in AY.XX-like sequences and the ORF7b:T40I that was not described in the entirety of the sequences, all of the other mutations were present. In summary, most Delta samples from RS shared the same combination of mutations.
AY.99.2 and AY.99.2-like were the most frequent Delta sub-lineages found in our study. As of 15 December 2021, more than 16 thousand sequences of AY.99.2 lineage have been detected in at least 28 countries representing < 0.5% cumulative prevalence Worldwide (Latif et al., 2021b). This new sub-lineage has been mostly detected in the United States of America, Chile, Portugal, France, and especially in Brazil, which represents 98% of the described AY.99.2 (PANGO Lineages, 2021). In Brazil, it is possible to observe an expressive difference in the frequency of detection between the states as well. The cumulative prevalence of AY.99.2 is 78% in Distrito Federal followed by Paraíba (63%) and Rio de Janeiro (39%) (Latif et al., 2021b). Although the southernmost state in Brazil, RS, shows 10% of cumulative prevalence (Latif et al., 2021b), our data detected 63% of this sub-lineage in samples analyzed in this study. It's possible that the sequences assigned as “none”, putatively named as AY.XX-like, according to the most related group, are likely to be classified later, as databases are routinely updated. They can represent a separated group with a high rate and speed of diversification within the cited Delta sub-lineages.
Preliminary molecular clock estimates suggested that the overall rate of evolution of SARS-CoV-2 in 2020 was 8 × 10−4 substitutions/site/year, which equates to 24 substitutions per year (Duchene et al., 2020). The current global estimate (Nextstrain, 2021b) including multiple variants of concern/interest suggests a similar rate of approximately 25.2 substitutions per year. Our Delta analysis presented a higher clock rate of 34.5 substitutions per year, showing a higher substitution rate than the majority of other sequences. It is important to point out that this higher rate was probably overestimated as there is a smaller number of sequences analyzed when compared to the global analysis. Also, the analyzed sequences are genetically divergent from reference lineages, which influenced the mutation rate to a higher number.
The Delta variant spread was remarkable in Brazil as in other countries, becoming the predominant variant in a matter of months since its first descriptions in the middle of 2021 (Nextstrain, 2021b). In December of 2021, a new VOC had emerged with several new genome mutations and higher rates of infection, and in a shorter period became present all over the world (Brandal et al., 2021; Centers for Disease Control and Prevention, 2021; Wang and Cheng, 2021). This variant, called Omicron by WHO and clade 21 K by Nextstrain, is already the predominant variant in some countries despite its recent first description (late November in South Africa). Updated Nexstrain data (Nextstrain, 2021c) shows that, in contrast to Delta, this variant does not descend from recent variants and it's phylogenetically closer to viruses circulating in 2020. By looking into Nextstrain updated molecular clock analysis (Nextstrain, 2021c), all the so far sequenced Omicron viruses have numbers of mutations higher than the average. By looking at our temporal analysis, many of Delta sequences have genome mutation numbers higher than the average for SARS-CoV-2, but other ones have less mutations than average (Fig. 2b). A matter of concern is that, of the at least 30 amino acid substitutions that characterize the variant, 15 of them are in the receptor binding domain (RBD) (Centers for Disease Control and Prevention, 2021). As comparison, the Delta variant has 9 substitutions/deletions in the spike protein (CoVariants, 2021b).
The Omicron variant has already been described in Brazil a few weeks after the first report in South Africa, from an airplane passenger that arrived in São Paulo state from south Africa in late November. Our analysis contains sequences collected in RS state in the year of 2021 up to the last week of October, and no Omicron sequence was detected since the first case in RS state was detected on December 3rd. This fact suggests that there was no previous introduction of Omicron in the RS state before the first official detection in Brazil.
In conclusion, we have described the appearance and consolidation of the SARS-CoV-2 Delta variant in RS State, Brazil, replacing the variant Gamma. Although the accelerated dispersion pattern was very similar to what happened with the introduction of the Gamma variant in RS, one very important point needs to be highlighted. The Delta variant did not cause an increase in the number of hospitalizations, and deaths as Gamma did in the region. We emphasize that the difference between these two moments of the pandemic is mainly related to the increase in the number of people currently vaccinated/immunized. Therefore, the vaccination coverage in RS has so far limited the number of serious cases relative to the unvaccinated population. Phylogenetic analysis showed that the sequences generated therein belonged to the 21 J Delta major clade, and most of the sequences were classified as AY.99.2, AY.99.2-like, and AY.101 sub-lineages. In order to avoid new health crises, continuous surveillance to monitor the spread of SARS-CoV-2 is extremely necessary, thus being able to assess the effectiveness of immunization against the emergence of new variants.
Ethical aspects
Project approved by the Research Ethics Committee (CEP) at Feevale University. Process number: CAAE: 33202820.7.1001.5348.
CRediT authorship contribution statement
Juliana Schons Gularte: Conceptualization, Formal analysis, Investigation, Writing – original draft. Mariana Soares da Silva: Conceptualization, Formal analysis, Investigation, Writing – original draft. Ana Cristina Sbaraini Mosena: Formal analysis, Investigation. Meriane Demoliner: Formal analysis, Investigation. Alana Witt Hansen: Formal analysis, Investigation. Micheli Filippi: Formal analysis, Investigation. Vyctoria Malayhka de Abreu Góes Pereira: Formal analysis, Investigation. Fágner Henrique Heldt: Formal analysis, Investigation. Matheus Nunes Weber: Formal analysis, Investigation, Resources. Paula Rodrigues de Almeida: Formal analysis, Investigation. Andressa Taiz Hoffmann: Formal analysis, Investigation. Andreia Rosane de Moura Valim: Formal analysis, Investigation. Lia Gonçalves Possuelo: Formal analysis, Investigation. Juliane Deise Fleck: Funding acquisition, Resources, Supervision. Fernando Rosado Spilki: Conceptualization, Writing – review & editing, Funding acquisition, Resources, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the Brazilian Coordination for the Improvement of Higher-Level Personnel (CAPES), Foundation for Research Support of the State of Rio Grande do Sul (FAPERGS), and Brazilian National Council for Scientific Development (CNPq) for scholarships. This work is an initiative of Rede Corona-ômica BR MCTI/FINEP affiliated to RedeVírus/MCTI (FINEP = 01.20.0029.000462/20, CNPq = 404096/2020–4), and FAPERGS (grant 21/2551–0000081–3).
References
- Aksamentov I., Roemer C., Hodcroft E., Neher R. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021;6:3773. doi: 10.21105/joss.03773. [DOI] [Google Scholar]
- Bolze A., Cirulli E.T., Luo S., White S., Wyman D., Rossi A.D., Machado H., Cassens T., Jacobs S., Barrett K.M.S., Tsan K., Nguyen J., Ramirez J.M., Sandoval E., Wang X., Wong D., Becker D., Laurent M., Lu J., Isaksson M., Washington N.L., Lee W. SARS-CoV-2 variant Delta rapidly displaced variant Alpha in the United States and led to higher viral loads. medRxiv (Preprint) 2021 doi: 10.1101/2021.06.20.21259195. https://www.medrxiv.org/content/10.1101/2021.06.20.21259195v3 (acessed 12.10.21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandal L.T., MacDonald E., Veneti L., Ravlo T., Lange H., Naseer U., Feruglio S., Bragstad K., Hungnes O., Ødeskaug L.E., Hagen F., Hanch-Hansen K.E., Lind A., Watle S.V., Taxt A.M., Johansen M., Vold L., Aavitsland P., Nygård K., Madslien E.H. Outbreak caused by the SARS-CoV-2 Omicron variant in Norway, November to December 2021. Eurosurveillance. 2021;26:1–5. doi: 10.2807/1560-7917.ES.2021.26.50.2101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention, 2021. SARS-CoV-2 B.1.1.529 (Omicron) Variant. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-omicron-variant.html (accessed 12.18.21).
- CoVariants Variant: 21J (Delta) CoVariants.org. 2021 https://covariants.org/variants/21J.Delta (accessed 12.8.21) [Google Scholar]
- CoVariants Variant : 21A ( Delta ) CoVariants.org. 2021 https://covariants.org/variants/21A.Delta (accessed 12.10.21) [Google Scholar]
- Demoliner M., da Silva M.S., Gularte J.S., Hansen A.W., de Almeida P.R., Weber M.N., Heldt F.H., Silveira F., Filippi M., de Abreu Góes Pereira V.M., da Silva F.P., Mallmann L., Fink P., de Moura Valim A.R., Possuelo L.G., Fleck J.D., Spilki F.R. Predominance of SARS-CoV-2 P1 (Gamma) lineage inducing the recent COVID-19 wave in Southern Brazil and the finding of an additional S: D614A mutation. Infect. Genet. Evol. 2021;96 doi: 10.1016/j.meegid.2021.105134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchene S., Featherstone L., Haritopoulou-Sinanidou M., Rambaut A., Lemey P., Baele G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020;6:1–8. doi: 10.1093/ve/veaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control . ECDC; Stockholm: 2021. Assessing SARS-CoV-2 circulation, variants of concern, non-pharmaceutical interventions and vaccine rollout in the EU/EEA, 16th update. [Google Scholar]
- Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., Hulswit R.J.G., Franco L.A.M., Ramundo M.S., de Jesus J.G., Andrade P.S., Coletti T.M., Ferreira G.M., Silva C.A.M., Manuli E.R., Pereira R.H.M., Peixoto P.S., Kraemer M.U.G., Gaburo N., Camilo C.C., Hoeltgebaum H., Souza W.M., Rocha E.C., de Souza L.M., de Pinho M.C., Araujo L.J.T., Malta F.S.V., de Lima A.B., Silva J.P., Zauli D.A.G., Ferreira A.C.S., Schnekenberg R.P., Laydon D.J., Walker P.G.T., Schlüter H.M., dos Santos A.L.P., Vidal M.S., Del Caro V.S., Filho R.M.F., dos Santos H.M., Aguiar R.S., Proença-Modena J.L., Nelson B., Hay J.A., Monod M., Miscouridou X., Coupland H., Sonabend R., Vollmer M., Gandy A., Prete C.A., Nascimento V.H., Suchard M.A., Bowden T.A., Pond S.L.K., Wu C.H., Ratmann O., Ferguson N.M., Dye C., Loman N.J., Lemey P., Rambaut A., Fraiji N.A., Carvalho M.D.P.S.S., Pybus O.G., Flaxman S., Bhatt S., Sabino E.C. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. (80-.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant R., Charmet T., Schaeffer L., Galmiche S., Madec Y., Von Platen C., Chény O., Omar F., David C., Rogoff A., Paireau J., Cauchemez S., Carrat F., Septfons A., Levy-Bruhl D., Mailles A., Fontanet A. Impact of SARS-CoV-2 Delta variant on incubation, transmission settings and vaccine effectiveness: results from a nationwide case-control study in France. Lancet Reg. Health Eur. 2021;1–13 doi: 10.1016/j.lanepe.2021.100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.I., Baig M.H., Mondal T., Alorabi M., Sharma T., Dong J., Cho J.Y. Impact of the double mutants on spike protein of SARS-CoV-2 B1.617 lineage on the human ACE2 receptor binding: a structural insight. Viruses. 2021;13:1–19. doi: 10.3390/v13112295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Angyal A., Brown R.L., Carrilero L., Green L.R., Groves D.C., Johnson K.J., Keeley A.J., Lindsey B.B., Parsons P.J., Raza M., Rowland-Jones S., Smith N., Tucker R.M., Wang D., Wyles M.D., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmina A., Wattad S., Khalaila Y., Ottolenghi A., Rosental B., Engel S., Rosenberg E., Taube R. SARS CoV-2 Delta variant exhibits enhanced infectivity and a minor decrease in neutralization sensitivity to convalescent or post-vaccination sera. iScience. 2021 doi: 10.1016/j.isci.2021.103467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarca A.P., Almeida L.G.P.De, Francisco S., Machado D.T., Brustolini O., Gerber A.L., De A.P., Policarpo C., Oliveira G.S.De, Boullosa L.T., Souza I.V.De, Carvalho E.M.De, Ribeiro M.S., Souza D., Gomes C., Silva D., Luiz C., Ribeiro P., Cavalcanti A.C., Maria C., Mello B.De, Tanuri A., Vasconcelos A.T.R. Genomic surveillance tracks the first communitary outbreak of Delta (B.1.617.2) variant in Brazil. Virological.org. 2021 https://virological.org/t/genomic-surveillance-tracks-the-first-communitary-outbreak-of-delta-b-1-617-2-variant-in-brazil/733 (accessed 11.15.21) [Google Scholar]
- Latif, A.A., Mullen, J.L., Alkuzweny, M., Tsueng, G., Cano, M., Haag, E., Zhou, J., Zeller, M., Hufbauer, E., Matteson, N., Wu, C., Andersen, K.G., Su, A.I., Gangavarapu, K., Hughes, L.D., Biology., and the C. for V.S., 2021a. Brazil Variant Report. https://outbreak.info/location-reports?loc=BRA&alias=Iota&alias=Mu&pango=P.2&dark=true&selected=P.2&selected=Iota&selected=Mu&selected=Delta&selected=Alpha&sele (accessed 11.30.21).
- Latif, A.A., Mullen, J.L., Alkuzweny, M., Tsueng, G., Cano, M., Haag, E., Zhou, J., Zeller, M., Hufbauer, E., Matteson, N., Wu, C., Andersen, K.G., Su, A.I., Gangavarapu, K., Hughes, L.D., Biology, C. for V.S., 2021b. AY.99.2 Lineage Report. https://outbreak.info/situation-reports?pango=AY.99.2&loc=USA&loc=BRA&selected=BRA&overlay=false (accessed 12.16.21).
- Li, B., Deng, A., Li, K., Hu, Y., Li, Z., Xiong, Q., Liu, Z., Guo, Q., Zou, L., Zhang, H., Zhang, M., Ouyang, F., Su, J., Su, W., Xu, J., Lin, H., Sun, J., Peng, J., Jiang, H., Zhou, P., Hu, T., Luo, M., Zhang, Y., Zheng, H., Xiao, J., Liu, T., Che, R., Zeng, H., Zheng, Z., Huang, Y., Yu, J., Yi, L., Wu, J., Chen, J., Zhong, H., Deng, X., Kang, M., Pybus, O.G., Hall, M., Lythgoe, K.A., Li, Y., Yuan, J., He, J., Lu, J., 2021. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. medRxiv (Preprint). https://www.medrxiv.org/content/10.1101/2021.07.07.21260122v2 (acessed 12.10.21). doi: 10.1101/2021.07.07.21260122.
- Li Q., Wu J., Nie J., Zhang Li, Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Zhang L., Li X., Huang W., Wang Y. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministério da Saúde (MS), 2021. Boletim epidemiológico especial doença pelo novo coronavírus – COVID-19. https://covid.saude.gov.br (acessed 12.01.21).
- Nextstrain, 2021a. Nextstrain build for novel coronavirus SARS-CoV-2. https://github.com/nextstrain/ncov (accessed 11.10.21).
- Nextstrain, 2021b. Genomic epidemiology of novel coronavirus - Global subsampling. https://nextstrain.org/ncov/global (accessed 12.8.21).
- Nextstrain, 2021c. Genomic epidemiology of novel coronavirus - Africa. https://nextstrain.org/ncov/gisaid/africa (accessed 12.8.21).
- PANGO Lineages, 2021. Lineage List. https://cov-lineages.org/lineage_list.html (accessed 12.10.21.
- Public Health England . Public Health England; 2021. SARS-CoV-2 Variants of Concern and Variants Under Investigation in England - Technical Briefing 17. [Google Scholar]
- Saito A., Irie T., Suzuki R., Maemura T., Nasser H., Uriu K., Kosugi Y., Shirakawa K., Sadamasu K., Kimura I., Ito J., Wu J., Iwatsuki-Horimoto K., Ito M., Yamayoshi S., Loeber S., Tsuda M., Wang L., Ozono S., Butlertanaka E.P., Tanaka Y.L., Shimizu R., Shimizu K., Yoshimatsu K., Kawabata R., Sakaguchi T., Tokunaga K., Yoshida I., Asakura H., Nagashima M., Kazuma Y., Nomura R., Horisawa Y., Yoshimura K., Takaori-Kondo A., Imai M., Genotype to Phenotype Japan (G2P-Japan) Consortium. Tanaka S., Nakagawa S., Ikeda T., Fukuhara T., Kawaoka Y., Sato K. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. 2021 doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secretaria da Saúde do Rio Grande do Sul . 2021. Confirmados dois primeiros casos da variante Delta no RS.https://saude.rs.gov.br/confirmados-dois-primeiros-casos-da-variante-delta-no-rs (accessed 11.12.21) [Google Scholar]
- Secretaria da Saúde do Rio Grande do Sul . 2021. Monitoramento da Imunização Covid-19. https://vacina.saude.rs.gov.br/ (accessed 12.10.21) [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Eurosurveillance. 2017;22:2–4. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva da M.S., Demoliner M., Hansen A.W., Gularte J.S., Silveira F., Heldt F.H., Filippi M., Pereira da V.M.A.G., Silva da F.P., Mallmann L., Fink P., Silva da L.L., Weber M.N., Almeida da P.R., Fleck J.D., Spilki F.R. Early detection of SARS-CoV-2 P1 variant in Southern Brazil and reinfection of the same patient by P.2. Rev. Inst. Med. Trop. 2021;63:1–8. doi: 10.1590/s1678-9946202163058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Cheng G. Sequence analysis of the emerging SARS-CoV-2 variant Omicron in South Africa. J. Med. Virol. 2021;26:1–5. doi: 10.1002/jmv.27516. [DOI] [PubMed] [Google Scholar]
- World Health Organization, 2021. Tracking SARS-CoV-2 variants. URL https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed 8.18.21).