Abstract
Objectives
The World Health Organization has promoted the use of serological testing as a rapid and accurate technique for the detection of immunity against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In Lebanon, a better understanding of the immune response against SARS-CoV-2 is needed to develop effective measures for prevention and to plan an appropriate national vaccination program. This study aimed to measure the immunity status in Lebanon.
Methods
In this cross-sectional study, the population comprised male and female Lebanese and non-Lebanese residents of Lebanon between the ages 15 and 75. The exclusion criteria included: same household, symptomatic individuals, and extremes of age (< 15 and > 75). Representative testing for SARS-CoV-2 antibodies (anti-SARS-CoV-2 electrochemiluminescence immunoassay/ECLIA) was used to assess the prevalence of SARS-CoV-2 infection in Lebanon.
Results
In total, 13 755 participants were recruited over a 6-month period. Of these, 3168 (23.03%) individuals tested positive for anti-SARS-CoV-2, with levels of positivity varying among districts. A higher level of seropositivity was detected in the female participants.
Conclusion
Seroprevalence against SARS-CoV-2 varied within Lebanon, but was comparable to the levels reported in the MENA region at the time of the study. The seroprevalence documented in this study represents a level of immunity that is not protective at the national level.
Funding
This study was funded by the Lebanese American University School of Medicine.
Keywords: infectious disease, seroprevalence, COVID-19, medicine, epidemiology, cross-sectional
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused a major spread of coronavirus disease 2019 (COVID-19) globally, which was declared a pandemic by the World Health Organization (WHO) on March 21, 2020. Several studies started to assess its viral pathogenesis, genetic shifts, and variations, in addition to the acquired immunity status against SARS-CoV-2. Such immunity is usually acquired at an individual level via natural infection or through vaccination (Randolph and Barreiro, 2020).
Following the identification of the virus, many rapid tests became available, including antigen detection and polymerase chain reaction (PCR) testing, as well as highly specific and well-validated serological assays (Beavis et al., 2020; GeurtsvanKessel et al., 2020; Meyer et al., 2020). Highly specific assays have been used to screen populations with low seroprevalence (Gudbjartsson et al., 2020). Large-scale serology prevalence studies have provided important estimates of the fraction of a population that has developed antibodies against SARS-CoV-2 (Kritsotakis, 2020). Indeed, WHO has promoted the use of serological testing as a rapid and accurate technique for detecting immunity against SARS-CoV-2 (WHO, n.d.).
COVID-19 antibody testing is based on detecting antibodies against the spike protein (S), the nucleocapsid protein (N), and other proteins that elicit high antigenicity (Liu et al., 2020). Several serology methods have been developed to detect immunoglobulin M (IgM) and IgG antibodies targeted against SARS-COV-2 antigens in the serum of patients (Xiang et al., 2020). In the summer of 2020, some studies raised concerns on the waning of antibody immunity, especially in patients who were mildly symptomatic (Ibarrondo et al., 2020; Long et al., 2020). However, it was later suggested that studies conducted after the first COVID-19 wave probably assessed transient waning of antibody titers, whereas studies performed after the second wave reflected a more factually correct state of longer-lasting antibody titers (Alter and Seder, 2020). Overall, antibody assays remain a more cost-effective and reliable representation of collective immunity in populations (Alter and Seder, 2020).
Since April 2020, seroprevalence studies have been conducted in numerous countries, including China, France, Iran, Spain, Brazil, Peru, and the USA (Álvarez-Antonio et al., 2021; Gallian et al., 2020; Hallal et al., 2020; Havers et al., 2020; Pollán et al., 2020; Shakiba et al., 2020; Xu et al., 2020). Such studies have provided insights into the levels of immunity in these countries, and have highlighted the statistically significant variables that have contributed to them. One meta-analysis studied SARS-CoV-2 seroprevalence globally, and concluded that it varies across different regions in the world depending on several factors, including human development indices, income levels, and geographic latitudes and/or climate (Rostami et al., 2021).
In Lebanon, 547 497 infected cases had been reported by July 11, 2021, with 7873 individuals losing their lives due to COVID-19. Despite an incremental increase in cured cases, the rapid spread and transmission of SARS-CoV-2 remain a major problem. Consequently, more information on the immune response against this virus is needed for better infection control and treatment management.
To date, no national serology prevalence study for the residents of Lebanon has been published. Our study measured the titers of SARS-CoV-2 antibodies in a cross-sectional sample of the population in Lebanon to assess levels of SARS-CoV-2 immunity. Greater understanding of collective immunity against SARS-CoV-2 can help steer public-health policies for better disease control, and will provide insight into the seroprevalence rates in other countries of the Middle East and North Africa (MENA) region.
Methods
Study design, population, and sampling
In this population-based cross-sectional study, serological testing for anti-SARS-CoV-2 antibodies was used to assess the prevalence of SARS-CoV-2 infection across all eight Lebanese governorates, 26 Lebanese districts, and 138 Lebanese municipalities. A minimum sample size of 12 372 participants was determined to be required for significance in this study; ultimately, 13 755 participants were tested. The sample included male and female Lebanese and non-Lebanese residents, as well as Syrian and Palestinian refugees living in Lebanon, between 15 and 75 years of age. Individuals of the same household, symptomatic individuals (for consistency and the safety of participants), individuals of extreme ages (< 15 and > 75 years old) due to sample acquiring difficulty, and vaccinated individuals were excluded.
Collaboration was established with municipality officials within the Lebanese governorates and districts represented in this study. The municipalities selected and invited eligible participants, based on official records. Participants were contacted via telephone to document any exclusion criteria and to schedule appointments. Major municipalities with high population densities were selected to be representative, while maintaining spatial diversity.
Case ascertainment rates have varied drastically over time in Lebanon. However, accurate estimates from this seroprevalence study were achieved by recruiting a representative sample of the population of interest. Furthermore, congregate settings (e.g. nursing homes, prisons, same household, or same building) were avoided to decrease bias.
Procedures
Participants were screened and recruited based on the inclusion criteria. These individuals signed consent forms and were guided to fill an online questionnaire about their adherence to social distancing and mask wearing, COVID-19-related symptoms since the beginning of the pandemic, exposures, and travel history. Five mL of venous blood was collected into a standard sampling tube. Next, the Elecsys Anti-SARS-CoV-2 assay (Roche Diagnostics) was used to analyze each serum sample, with the manufacturer's data stating a specificity of 99.81% (95% CI: 99.67%-99.90%) and sensitivities of 60.2% (95% CI: 52.3%-67.8%)on days 0–6, 85.3% (95% CI: 78.6%-90.6%)on days 7–13, and 99.5% (95% CI: 97.0%-100%) from day 14 of PCR confirmation. The manufacturer validated the clinical sensitivity of this assay by analysing 496 samples from 102 positive-PCR symptomatic patients, validated its analytical specificity by analyzing a cohort of 792 samples, and validated its clinical specificity by analysing 10 453 samples according to routine diagnoses (Hallal et al., 2020; Kronbichler et al., 2020).
The Elecsys Anti‐SARS‐CoV‐2 assay uses a recombinant protein representing the nucleocapsid (N) antigen for the determination of antibodies against SARS‐CoV‐2. During the first incubation, 20 μL of biotinylated SARS‐CoV‐2‐specific recombinant antigen and SARS‐CoV‐2‐specific recombinant antigen labeled with a ruthenium complex form a sandwich complex. During the second incubation, after the addition of streptavidin-coated microparticles, the complex becomes bound to the solid phase via the interaction of biotin and streptavidin. Then the reaction mixture is aspirated into the measuring cell, where the microparticles are magnetically captured onto the surface of the electrode. Unbound substances are removed using ProCell/ProCell M. A voltage is applied to the electrode, inducing chemiluminescent emission, which is measured by a photomultiplier (Elecsys® Anti-SARS-CoV-2, 2022).
Finally, the results are determined automatically using software that compares the electrochemiluminescence signal obtained from the reaction product with the signal of the cut-off value previously obtained by calibration. If the COI is more than 1.0, the sample is considered reactive and positive for anti-SARS-CoV-2 antibodies.
The first blood samples were collected on September 21, 2020, in the Jezzine district, with the last collected on April 8, 2021, in the Nabatieh district. Sample collection visits ranged between two and five trips weekly. The number of municipalities visited depended on the logistical feasibility and spread status of SARS-CoV-2 in the targeted areas. Our study took course across seven partial lockdowns and four complete lockdowns. During periods of lockdown, our trips were limited or adjusted as much as possible to comply with public health measures and to prevent superspreader events. The study design was not affected. Our blood sample collection took place over 103 days, with an average of 265 collected samples per day.
Patients’ identities were kept confidential. Only the investigators and research team had access to these data. The data were not shared with an entities, persons, or organisations outside of those with access to the Lebanese American University Medical Centre-Rizk Hospital (LAUMC-RH) confidential records. Analyses and publications did not include any medical numbers or social security numbers. All study personnel had been trained in maintaining the confidentiality of health information. All data with patient identifiers will be destroyed 5 years after publication of the final study report.
Covariates
Demographic variables included sex, age, municipality, district, governorate, and date of sample collection. Participants’ ages were grouped into 5-year intervals (15–19, 20–24, 25–29, 30–34, 35–39, 40–44, 45–49, 50–54, 55–59, 60–64, 65–69, and 70–74 years). COVID-19-related exposure was defined as exposure to individuals who had recent travels, infectious symptoms, or a confirmed positive COVID-19 PCR test. Exposure was defined as any violation of social distancing or absence of face mask. If exposure was confirmed, patients were asked about the time of exposure and the total number of individuals they were exposed to. Individuals were also asked about their own recent travel history, time of travel, and country of travel origin.
Sample size calculation
The minimum sample size was computed while assuming an infinite population size, and using the following formula:
where p is the seroprevalence rate and d is the precision.
A sample size of 5379 was initially required for our study. This was calculated while considering a type I error rate of 5% and a precision (margin of error) of 1%, and assuming a seroprevalence rate of 20%. The 26 districts and the combined refugee camps were then considered as the 27 clusters in the study, and used to compute the design effect while assuming an intraclass correlation of 0.05, as follows:
Applying the design effect to correct the estimated sample size, resulted in a total of 12 372 (5379 × 2.3) individuals.
The required sample size for each age-group and sex in each Lebanese governorate and district, as well as in the refugee camps, was proportional to their population relative to the total population of all registered residents in Lebanon, as per the ‘Presidency of the Council of Ministers Central Administration of Statistics CAS’ (http://www.cas.gov.lb/images/Publications/LFHLCS2018-2019/LFHLCS_2018_2019_Demography.xls) and the detailed analytical report ‘Population and Housing Census in Palestinian Camps and Gatherings in Lebanon’ (http://www.lpdc.gov.lb/DocumentFiles/8-10-2019-637068152405545447.pdf).
Statistical analysis
Baseline characteristics, sex, and age-group were described for each district. Crude SARS-CoV-2 seroprevalence was computed for the general population as well as by sex, district, and age-group. The overall crude frequencies of positive tests were calculated and stratified by age and sex. The 95% CIs for seroprevalence were constructed using the bootstrap method in ci_proportion from the ‘confintr’ package in R, with 10 000 repetitions. The seroprevalence rate adjusted by the test sensitivity and specificity was also computed using the following formula from Sempos et al. (Sempos and Tian, 2021): adjusted prevalence = (crude prevalence + specificity − 1)/(sensitivity + specificity − 1)
The specificity and sensitivity for the ECLIA test were assumed to be 99.81% and 99.5%, respectively. Given that sensitivity changes relative to the number of days since infection, the weighted average was computed based on the number of samples reporting as being exposed – within 6 days, between 7 and 13 days, and after 14 days.
The chi-squared test of independence was used to compare seroprevalence rates between the different sex and age groups and districts. The Pearson correlation coefficient was computed to assess the relationship between our daily SARS-CoV-2 seroprevalence rates, and the daily percentage of COVID-19 PCR-positive cases reported by the Lebanese Ministry of Public Health. A p-value < 0.05 was deemed statistically significant.
The quantitative antibody titers for SARS-CoV-2 among the different sex and age groups were also compared. The Shapiro test was first used to to check for normality, followed by the Wilcoxon rank sum and Kruskal-Wallis tests to assess the differences between the sex and age groups, respectively. The quantitative measures are reported as mean ± standard deviation. The odds ratios (OR) of having a positive seroprevalence, along with their 95% confidence intervals, were computed using the logistic regression function glm, and were adjusted to the sex, age group, being exposed, and symptomatic covariates. The ORs were computed relative to individuals who were 15–19 years old, female, non-exposed, and non-symptomatic.
Bias
The response rate was very high in our study, which limited response bias. However, there may have been potential for mild sampling bias because the municipalities had varying access to their citizens in times of quarantine. This bias was mostly negligible due to the efforts to maintain a representative sample. In addition, recruited individuals were subjective in reporting their symptoms. This might have added to the limitations of this study, since there was no documentation of symptoms by clinicians at the time of disease manifestation.
Role of the funding source
The sponsor of this study was the Lebanese American University (LAU). LAU administration had no intervening role in the study design, data analysis or interpretation, or writing of the report. In addition, the corresponding author had full access to all the data used in this study, and had final responsibility for the decision to submit this study for publication.
Results
For this population-based cross-sectional study, 13 843 individuals were initially recruited, but 88 were excluded for not meeting the eligibility criteria. Blood samples were collected from the 13 755 remaining individuals for testing with the Elecsys Anti-SARS-CoV-2 assay (Figure 1). Of the included individuals, 7200 were males (52.34%) and 6555 were females (47.66%), giving a male-to-female ratio of 1.098. Only 22.64% (3144/13 755) of participants had experienced symptoms since the beginning of the pandemic, 86.56% (11906/13 755) applied quarantine/social distancing, 33.38% (4592/13 755) reported a previous exposure to a patient who wad COVID-19 PCR-positive, 18.91% (2600/13 755) were exposed to symptomatic individuals, 1.87% (257/13 755) had travelled within a month of enrolment in the study, and 15.93% (2191/13 755) reported contact with a recent traveller since enrolling in the study.
Figure 1.
Participant selection.
Using the ECLIA technique, 3168 individuals tested positive for anti-SARS-CoV-2 antibodies, which had a crude seroprevalence of 23.03% (22.35–23.74). Table 1 shows that 21.53% (20.57–22.47) (1550/7200) of males and 24.68% (23.66–25.75) (1618/6555) of females tested positive, and seroprevalence rates varied for members of different age groups.
Table 1.
Seroprevalence of Participants Stratified by Sex and Age-Group.
| Sample size | Seropositive participants | Seroprevalence, % (95% CI) | Adjusted Seroprevalence, % | |
|---|---|---|---|---|
| Total | 13755 | 3168 | 23.03% (22.35-23.74) | 24.60% |
| Sex | ||||
| Male | 7200 | 1550 | 21.53% (20.57-22.47) | 22.98% |
| Female | 6555 | 1618 | 24.68% (23.66-25.75) | 26.38% |
| Age, Years | ||||
| 15-19 | 1,250 | 349 | 27.92% (25.44-30.40) | 29.87% |
| 20-24 | 1,474 | 385 | 26.12% (23.88-28.36) | 27.93% |
| 25-29 | 1,258 | 312 | 24.80% (22.41-27.19) | 26.51% |
| 30-34 | 1,188 | 282 | 23.74% (21.30-26.18) | 25.36% |
| 35-39 | 1,230 | 321 | 26.10% (23.66-28.54) | 27.91% |
| 40-44 | 1,281 | 292 | 22.79% (20.53-25.14) | 24.35% |
| 45-49 | 1,318 | 311 | 23.60% (21.32-25.87) | 25.21% |
| 50-54 | 1,447 | 308 | 21.29% (19.07-23.43) | 22.72% |
| 55-59 | 1,320 | 253 | 19.17% (17.05-21.21) | 20.44% |
| 60-64 | 942 | 159 | 16.88% (14.54-19.32) | 17.97% |
| 65-69 | 611 | 106 | 17.35% (14.40-20.29) | 18.48% |
| 70-74 | 436 | 90 | 20.64% (16.97-24.31) | 22.03% |
Seroprevalences were stratified by Lebanese district (Table 2); for example, the highest level of 60.26% (47.43–69.23) seropositivity was detected in Hasbaya, 40.46% (35.04–45.29) was recorded in Minieh-Danieh, and the lowest seropositivities of 1.76% (0.50–3.01) and 1.04% (0.00–2.59) in Jubail and Marjaayoun, respectively.
Table 2.
Seroprevalence of Participants Stratified by District.
| Sample Size | Seropositive pariticipants, n | Seroprevalence % (95% CI) | Adjusted Seroprevalence, % | |
|---|---|---|---|---|
| Total | 13755 | 3168 | 23.03% (22.35-23.74) | 24.60% |
| District | ||||
| Akkar | 806 | 116 | 14.39% (12.03-16.87) | 15.29% |
| Aley | 844 | 298 | 35.31% (31.99-38.39) | 37.83% |
| Baabda | 1,416 | 375 | 26.48% (24.22-28.74) | 28.32% |
| Baalbeck | 596 | 176 | 29.53% (25.83-33.22) | 31.61% |
| Batroun | 141 | 25 | 17.73% (11.34-23.40) | 18.89% |
| Bcharre | 93 | 17 | 18.28% (10.75-25.80) | 19.48% |
| Beirut | 1,085 | 285 | 26.27% (23.59-28.84) | 28.09% |
| Bint Jbayl | 229 | 8 | 3.49% (1.31-6.11) | 3.55% |
| Chouf | 745 | 164 | 22.01% (19.06-24.83) | 23.51% |
| El Metn | 1,538 | 558 | 36.28% (33.87-38.68) | 38.88% |
| Hasbaya | 78 | 47 | 60.26% (47.43-69.23) | 64.72% |
| Hermel | 96 | 4 | 4.17% (1.04-8.33) | 4.27% |
| Jezzine | 97 | 3 | 3.09% (0.00-7.21) | 3.12% |
| Jubail | 398 | 7 | 1.76% (0.50-3.01) | 1.68% |
| Kasrouane | 697 | 89 | 12.77% (10.33-15.20) | 13.54% |
| Kou ra | 257 | 38 | 14.79% (10.50-19.06) | 15.72% |
| Marjaayoun | 193 | 2 | 1.04% (0.00-2.59) | 0.90% |
| Minieh-Danieh | 351 | 142 | 40.46% (35.04-45.29) | 43.38% |
| Nabatiyeh | 520 | 51 | 9.81% (7.30-12.30) | 10.35% |
| Rachiaya | 98 | 31 | 31.63% (22.44-39.79) | 33.87% |
| Refugees | 447 | 164 | 36.69% (32.21-41.16) | 39.32% |
| Saida | 791 | 43 | 5.44% (3.91-6.95) | 5.64% |
| Sour | 634 | 70 | 11.04% (8.67-13.56) | 11.68% |
| Tripoli | 688 | 247 | 35.90% (32.26-39.38) | 38.47% |
| West Bekaa | 220 | 40 | 18.18% (13.18-23.18) | 19.38% |
| Zahleh | 461 | 117 | 25.38% (21.25-29.28) | 27.13% |
| Zgharta | 236 | 51 | 21.61% (16.10-26.69) | 23.07% |
The seroprevalence rate was found to be significantly associated with the three covariates: sex, age group, and district (chi-squared, p < 0.0001 for the three variables). For example, 24.68% (1618/6555) of females tested positive for SARS-CoV-2 antibodies, while the prevalence was only 21.43% (1550/7200) in males (Table 1).
A statistically significant difference was recorded between males and females with respect to positive seroprevalence (chi-squared, p = 1.24e−05), with 24.68% (1618/6555) of females and 21.43% (1550/7200) of males testing positive for SARS-CoV-2 antibodies (Table 1).
Out of all positive-testing participants, 51% (1618/3168) were females and 49% (1550/3168) were males (Table 3). In addition, 87% reported applying quarantine and social distancing, 57% reported having prior symptoms since the beginning of the pandemic, 54% reported history of exposure to patients with COVID-19, 15% reported a history of exposure to symptomatic individuals, and 11% reported contact with a recent traveller. Of participants who tested negative for anti-SARS-CoV-2 antibodies, 47% were females and 53% were males, 86% reported applying quarantine and social distancing, 12% reported having prior symptoms since the beginning of the pandemic, 27% reported a history of exposure to patients with COVID-19, 31% reported a history of exposure to symptomatic individuals, and 17% reported contact with a recent traveller.
Table 3.
Exposure History to PCR positive or Symptomatic Individuals and Symptomatology of Male and Female Seropositive Participants.
| Serology Positive 23% (3168) | Serology Negative 77% (10587) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Males 49% (1550) | Females 51% (1618) | Males 53% (5650) | Females 47% (4937) | ||||||
| Symptomatic 56% (863) | Asymptomatic 44% (687) | Symptomatic 58% (945) | Asymptomatic 42% (673) | ||||||
| Exposed 72% (620) | Not Exposed 28% (243) | Exposed 41% (280) | Not Exposed 59% (407) | Exposed 74% (698) | Not Exposed 26% (247) | Exposed 36% (244) | Not Exposed 64% (249) | ||
Table 3 shows the symptomatology and exposure history of male and female seropositive participants to PCR-positive or symptomatic individuals. Of the seropositive male participants, 56% (863/1550) reported being symptomatic at least once since the pandemic, whereas 44% (687/1550) were asymptomatic. Of the symptomatic participants, 72% (620/863) reported an exposure history, whereas 28% (243/863) reported no exposure history. Of the asymptomatic participants, 41% (280/687) had an exposure history, whereas 59% (407/687) denied having an exposure history. With regard to female seropositive participants, 58% (945/1618) reported being symptomatic at least once since the pandemic, whereas 42% (673/1618) reported being asymptomatic. Of the symptomatic participants, 74% (698/945) reported an exposure history, whereas 26% (247/945) reported no exposure history. Of the asymptomatic participants, 36% (244/673) reported an exposure history, whereas 64% (429/673) denied having an exposure history.
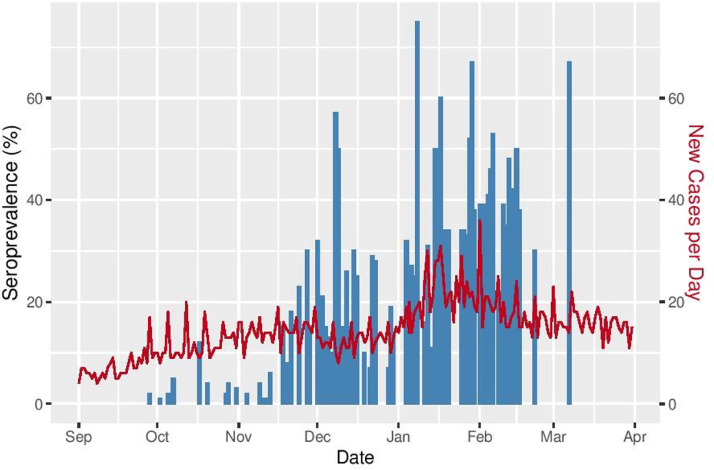
A statistically significant positive correlation (correlation coefficient = 0.402; p < 0.0001) was also found between our daily SARS-CoV-2 seroprevalence rates and daily percentages of COVID-19 PCR-positive cases as reported by the Lebanese Ministry of Public Health (Figure 2).
Figure 2.
Daily SARS-CoV-2 seroprevalence rates and daily percentages of COVID-19 PCR-positive cases, as reported by the Lebanese Ministry of Public Health.
The positive seroprevalence adjusted odds ratios (OR) are presented in Table 4. There was a statistically significant increase in positive seroprevalence among individuals who were exposed to an infected individual. Among those exhibiting symptoms, seroprevalence was 1.76 and 7.78 times higher in the exposed and symptomatic individuals, respectively, compared with non-exposed and non-symptomatic 15–19 years old females (exposed: OR = 1.76, 95% CI = 1.6–1.93; symptomatic: OR = 7.78, 95% CI = 7.07–8.57; both p < 0.0001). On the other hand, positive seroprevalence was 0.87 times lower in male individuals (OR = 0.87, 95% CI = 0.8–0.96; p = 0.003), and was also statistically significantly lower in all age groups except for those aged 35–39, 45–49, and 70–74 years (Table 4).
Table 4.
Positive seroprevalence adjusted odds ratios and their 95% CI.
| Odds-Ratio | 95% CI | P-values | |
|---|---|---|---|
| Age 20-24 | 0.78 | 0.64 - 0.94 | 0.01* |
| Age 25-29 | 0.71 | 0.58-0.87 | 0.001* |
| Age 30-34 | 0.74 | 0.6 - 0.91 | 0.003** |
| Age 35-39 | 0.84 | 0.69 - 1.03 | 0.097 |
| Age 40-44 | 0.75 | 0.61 -0.92 | 0.005** |
| Age 45-49 | 0.84 | 0.68 - 1.02 | 0.083 |
| Age 50-54 | 0.74 | 0.6 - 0.9 | 0.002** |
| Age 55-59 | 0.72 | 0.59-0.89 | 0.002* |
| Age 60-64 | 0.68 | 0.54 -0.86 | 0.001* |
| Age 65-69 | 0.76 | 0.58 -0.99 | 0.045** |
| Age 70-74 | 0.98 | 0.73 - 1.31 | 0.914 |
| Male | 0.87 | 0.8 -0.96 | 0.003** |
| Exposed | 1.76 | 1.6 - 1.93 | <0.0001*** |
| Symptomatic | 7.78 | 7.07 - 8.57 | <0.0001*** |
Discussion
In this population-based cross-sectional study, the seroprevalence of anti-SARS-CoV-2 antibodies was estimated for all Lebanese governorates and districts, including refugee camps. This was performed through 13 755 tests conducted in 138 randomly selected municipalities. No seroprevalence study of anti-SARS-CoV-2 antibodies to estimate the immunity status in Lebanon had previously been published. A predictive model estimating a 27% immunity status by May 1, 2021 was reported; however, such a predication is prone to shifting due to multiple variables, including social distancing measures and vaccination rates (Institute for Health Metrics and Evaluation, 2022). In our study, 23.037% of the tested population was found to be immune. This rate was higher than other countries’ reported rates for the same time period in-spite of the early lockdown in Lebanon Institute for Health Metrics and Evaluation, 2022). Notably, mask-wearing and social distancing were not properly practiced because of social norms.
Further analysis of the results revealed that females were more exposed, infected, and immune to SARS-CoV-2 than males (24.68% vs 21.53%). However, different age groups had different immunity statuses. Younger age groups (15–19 years and 35–39 years) had the highest positive rates, whereas the older age groups (e.g. 55–64 years) had the lowest. This might have been a result of the belief that the disease is selectively worse for older individuals, making the young less committed to mask-wearing and social-distancing measures, whereas older individuals took stricter precautions. In addition, the majority of those who tested positive denied exposure to symptomatic individuals. Hence, most exposure probably involved asymptomatic patients prior to their disease manifestation. Moreover, only 11% of individuals who tested positive reported recent exposure to a recent traveller. Thus, most of the SARS-CoV-2 spread was local. In addition, most of those who tested positive reported practicing quarantine or social distancing measures, which most likely were carried out inefficiently, failing to yield the desired disease protection.
With regard to geographic distribution, our results showed that the highest prevalence of SARS-CoV-2 antibodies was found in the Hasbaya (60%) and Minieh-Danieh (40%) districts, which may be attributed to potential socioeconomic confounding factors in the mentioned locations, where members interact more closely and disregard preventive measures in the face of social norms that encourage physical contact. In addition, the positive correlation between the daily SARS-CoV-2 cases reported by the Lebanese Ministry of Public Health and SARS-CoV-2-immune individuals in this study confirms the correlation between the rate of infectivity and the rate of development of immunity against the virus (Figure 2). It also serves to externally validate our study.
Seroprevalence studies serve to estimate COVID-19 infections, since most asymptomatic and presymptomatic patients do not usually get tested (Kronbichler et al., 2020). One study in the USA revealed a total number of cases that was 6–24 times the previously confirmed number after accounting for the untested asymptomatic and presymptomatic patients (Havers et al., 2020). Seroprevalence studies are also important in predicting second and third waves in all countries, as suggested by a recent meta-analysis (Rostami et al., 2021). When compared with global results, the seroprevalence rate in Lebanon (23.03% at 95% CI) was one of the highest in the world. It was significantly above the global average (3.38% at 95% CI), and very similar to previously reported results in Central and Southern Asia (22.16% at 95% CI in Iran, for instance) (Rostami et al., 2021).
To our knowledge, no other SARS-CoV-2 seroprevalence study in Lebanon has offered an insight into anti-SARS-CoV-2 immunity status (Bizri et al., 2021). This study was conducted at a time when the Institute for Health Metrics and Evaluation (IHME) noted a three-fold increase in the death rate in the MENA region, between September and December 2020 (Institute for Health Metrics and Evaluation, 2020). Therefore, the results of our study are essential for understanding the initial spatial spread of SARS-CoV-2 and the immunity status for different subpopulations in Lebanon. This study will help guide necessary interventions where needed, such as allocating vigorous testing and vaccination in districts where these are most needed. This, along with adjusting quarantine measures, would increase the efficiency of lockdown measures without heavily stressing the economy.
There were some limitations to this study. First, some participants may have belonged to the same household. This task was allocated to contacted municipalities, some of which could have allowed a few violations. However, our research team double-checked on this issue and made sure that no members of the same family were included. Second, our selection criteria excluded individuals < 15 years of age and > 74 years of age, as well as symptomatic individuals regardless of their age. This may have neglected a sector of society that contributed to the overall seroprevalence status, but this was compensated by adding more numbers in the other age groups similar, as has been done in other epidemiological studies carried out at a national level. Third, recruitment frequency was partially limited during lockdown periods, although all efforts were made to compensate for the period of lockdown. Fourth, the study period also served as a limitation of this study; however, the study was completed within 6 months, decreasing bias and serving as an acceptable estimation of cross-sectional seroprevalence. Another limitation was the hook effect of the test used, which could not be adjusted for. Finally, limited medical history appropriate for the study was collected. However, the results can be generalized to the studied population, because the sample size was representative of the population of interest, the response rate was very high among recruited individuals, and the technique used had high sensitivity and specificity.
Conclusion
In conclusion, this study examined both nationwide and regional estimates of SARS-CoV-2 spread in Lebanon. It demonstrated different seroprevalence rates in different districts, and according to age group and sex. It highlighted how transmission most likely occurred through asymptomatic locals. In addition, it showed that the seroprevalence rate was not sufficient to achieve a high immunity status, thus reemphasizing the importance of adhering to social distancing and wearing of face masks to avoid transmission and infection.
Acknowledgments
Author contributions
Ahmad Mahdi was responsible for data curation, investigation, methodology, project administration, resources supervision, writing the original draft, and reviewing and editing the manuscript. George Khazen was responsible for data curation, formal analysis, methodology, software, validation, visualization, writing the original draft, and reviewing and editing the manuscript. Nivine Azziz was responsible for data curation, formal analysis, methodology, investigation, software, validation, visualization, and reviewing and editing the manuscript. Jonathan Mina was responsible for data curation, investigation, methodology, and reviewing and editing the manuscript. Aram Papazian was responsible for data curation, formal analysis, methodology, investigation, software, validation, and visualization. Leonardo Daou was responsible for data curation, formal analysis, methodology, investigation, software, validation, and visualization. Jana Ahmar was responsible for data curation and investigation. Nour Assaf was responsible for data curation and investigation. Anjy Abdulkhalek was responsible for data curation and investigation. Hussein Farhat was responsible for methodology and resources. Jacques Mokhbat was responsible for reviewing and editing the manuscript. Anna Farra was responsible for reviewing and editing the manuscript. Rola Husni was responsible for conceptualization, funding acquisition, methodology, project administration, resources, supervision, and reviewing and editing the manuscript. All authors had full access to all the data in the study, and had final responsibility for the decision to submit for publication.
Data sharing
All the individual participant data collected during the trial, after deidentification, along with the study protocol, will be available starting 3 months and ending 5 years after publication via a link to be revealed at a later date.
Conflicts of interest
The authors declare no competing interests.
Acknowledgements
This study was funded by the Lebanese American University School of Medicine (LAU SOM). The authors would like to thank the Lebanese American University Medical Center-Rizk Hospital laboratory technicians and LAU drivers for helping them conduct this study, and all municipality employees for collaborating with them in conducting the participant recruitment events.
Ethical approval
This study was reviewed and approved by the Lebanese American University (LAU) Institutional Review Board (IRB), reference number LAUMCRH.RH5.24/Apr/2020.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijregi.2022.01.011.
Appendix. Supplementary materials
References
- Alter G., Seder R. The power of antibody-based surveillance. New England Journal of Medicine. 2020;383(18):1782–1784. doi: 10.1056/nejme2028079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Antonio C., Meza-Sánchez G., Calampa C., Casanova W., Carey C., Alava F., Rodríguez-Ferrucci H., Quispe A.M. Seroprevalence of anti-SARS-CoV-2 antibodies in Iquitos, Peru in July and August, 2020: a population-based study. The Lancet Global Health. 2021 doi: 10.1016/S2214-109X(21)00173-X. 0(0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis K.G., Matushek S.M., Abeleda A.P.F., Bethel C., Hunt C., Gillen S., Moran A., Tesic V. Evaluation of the EUROIMMUN Anti-SARS-CoV-2 ELISA Assay for detection of IgA and IgG antibodies. Journal of Clinical Virology. 2020;129 doi: 10.1016/j.jcv.2020.104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizri A.R., Khachfe H.H., Fares M.Y., Musharrafieh U. COVID-19 pandemic: an insult over injury for Lebanon. Journal of Community Health. 2021;46(3):487–493. doi: 10.1007/s10900-020-00884-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elecsys® Anti-SARS-CoV-2. Diagnostics. Retrieved January 3, 2022, from https://diagnostics.roche.com/us/en/products/params/elecsys-anti-sars-cov-2.html
- Gallian P., Pastorino B., Morel P., Chiaroni J., Ninove L., de Lamballerie X. Lower prevalence of antibodies neutralizing SARS-CoV-2 in group O French blood donors. Antiviral Research. 2020;181 doi: 10.1016/j.antiviral.2020.104880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., Okba N.M.A., Igloi Z., Bogers S., Embregts C.W.E., Laksono B.M., Leijten L., Rokx C., Rijnders B., Rahamat-Langendoen J., van den Akker J.P.C., van Kampen J.J.A., van der Eijk A.A., van Binnendijk R.S., Haagmans B., Koopmans M. An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nature Communications. 2020;11(1) doi: 10.1038/s41467-020-17317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E., Arnthorsson A.O., Helgason D., Bjarnadottir K., Ingvarsson R.F., Thorsteinsdottir B., Kristjansdottir S., Birgisdottir K., Kristinsdottir A.M., Sigurdsson M.I., Arnadottir G.A., Ivarsdottir E.V., Andresdottir M., Jonsson F.…Stefansson K. Humoral immune response to SARS-CoV-2 in Iceland. New England Journal of Medicine. 2020;383(18):1724–1734. doi: 10.1056/nejmoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallal, P. C., Hartwig, F. P., Horta, B. L., Victora, G. D., Silveira, M. F., Struchiner, C. J., Vidaleti, L. P., Neumann, N. A., Pellanda, L. C., Dellagostin, O. A., Burattini, M. N., Menezes, A. M. B., Barros, F. C., Barros, A. J. D., & Victora, C. G. (2020). Remarkable variability in SARS-CoV-2 antibodies across Brazilian regions: nationwide serological household survey in 27 states. In MedRxiv (p. 2020.05.30.20117531). medRxiv. doi: 10.1101/2020.05.30.20117531. [DOI]
- Havers F.P., Reed C., Lim T., Montgomery J.M., Klena J.D., Hall A.J., Fry A.M., Cannon D.L., Chiang C.F., Gibbons A., Krapiunaya I., Morales-Betoulle M., Roguski K., Rasheed M.A.U., Freeman B., Lester S., Mills L., Carroll D.S., Owen S.M.…Thornburg N.J. Seroprevalence of antibodies to SARS-CoV-2 in 10 sites in the United States, March 23–May 12, 2020. JAMA Internal Medicine. 2020;180(12):1776–1786. doi: 10.1001/jamainternmed.2020.4130. [DOI] [PubMed] [Google Scholar]
- Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A., Ferbas K.G., Tobin N.H., Aldrovandi G.M., Yang O.O. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild COVID-19. New England Journal of Medicine. 2020;383(11):1085–1087. doi: 10.1056/nejmc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute for Health Metrics and Evaluation . 2020. COVID-19 results briefing: Eastern Mediterranean Region.http://www.healthdata.org/sites/default/files/files/Projects/COVID/briefing_EMRO_20201223.pdf [Google Scholar]
- Institute for Health Metrics and Evaluation, (2022). Institute for Health Metrics and Evaluation. Retrieved February 9, 2022, from https://www.healthdata.org/institute-health-metrics-and-evaluation.
- Kritsotakis E.I. On the importance of population-based serological surveys of SARS-CoV-2 without overlooking their inherent uncertainties. Public Health in Practice. 2020;1 doi: 10.1016/j.puhip.2020.100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronbichler A., Kresse D., Yoon S., Lee K.H., Effenberger M., Shin J., II Asymptomatic patients as a source of COVID-19 infections: a systematic review and meta-analysis. International Journal of Infectious Diseases. 2020;98:180–186. doi: 10.1016/j.ijid.2020.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Xiong Z., Zheng S. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. Journal of Clinical Microbiology. 2020;58(6) doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., Hu J.L., Xu W., Zhang Y., Lv F.J., Su K., Zhang F., Gong J., Wu B., Liu X.M., Li J.J., Qiu J.F., Chen J., Huang A.L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nature Medicine. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Meyer B., Torriani G., Yerly S., Mazza L., Calame A., Arm-Vernez I., Zimmer G., Agoritsas T., Stirnemann J., Spechbach H., Guessous I., Stringhini S., Pugin J., Roux-Lombard P., Fontao L., Siegrist C.A., Eckerle I., Vuilleumier N., Kaiser L. Validation of a commercially available SARS-CoV-2 serological immunoassay. Clinical Microbiology and Infection. 2020;26(10):1386–1394. doi: 10.1016/j.cmi.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollán M., Pérez-Gómez B., Pastor-Barriuso R., Oteo J., Hernán M.A., Pérez-Olmeda M., Sanmartín J.L., Fernández-García A., Cruz I., Fernández De Larrea N., Molina M., Rodríguez-Cabrera F., Martín M., Merino-Amador P., Paniagua J.L., Muñoz-Montalvo J.F., Blanco F., Yotti R. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. The Lancet. 2020;396:535–544. doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph H.E., Barreiro L.B. Herd immunity: understanding COVID-19. Immunity. 2020;52(5):737–741. doi: 10.1016/j.immuni.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostami A., Sepidarkish M., Leeflang M.M.G., Riahi S.M., Nourollahpour Shiadeh M., Esfandyari S., Mokdad A.H., Hotez P.J., Gasser R.B. SARS-CoV-2 seroprevalence worldwide: a systematic review and meta-analysis. Clinical Microbiology and Infection. 2021;27(Issue 3):331–340. doi: 10.1016/j.cmi.2020.10.020. Elsevier B.V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempos C.T., Tian L. Adjusting coronavirus prevalence estimates for laboratory test kit error. American Journal of Epidemiology. 2021;190(1):109. doi: 10.1093/AJE/KWAA174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakiba, M., Nazari, S. S. H., Mehrabian, F., Rezvani, S. M., Ghasempour, Z., & Heidarzadeh, A. (2020). Seroprevalence of COVID-19 virus infection in Guilan province, Iran. In MedRxiv (p. 2020.04.26.20079244). medRxiv. 10.1101/2020.04.26.20079244. [DOI]
- WHO (n.d.)2022 Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. Retrieved April 23, 2021, from https://www.who.int/publications/i/item/10665-331501.
- Xiang, J., Yan, M., Li, H., Liu, T., Lin, C., Huang, S., & Shen, C. (2020). Evaluation of enzyme-linked immunoassay and colloidal gold-immunochromatographic assay kit for detection of novel coronavirus (SARS-Cov-2) causing an outbreak of pneumonia (COVID-19). In MedRxiv (p. 2020.02.27.20028787). medRxiv. 10.1101/2020.02.27.20028787. [DOI]
- Xu X., Sun J., Nie S., Li H., Kong Y., Liang M., Hou J., Huang X., Li D., Ma T., Peng J., Gao S., Shao Y., Zhu H., Lau J.Y.N., Wang G., Xie C., Jiang L., Huang A.…Hou F.F. Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nature Medicine. 2020;26(8):1193–1195. doi: 10.1038/s41591-020-0949-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.