Abstract
Objective
To determine the effects of using unstructured clinical text in machine learning (ML) for prediction, early detection, and identification of sepsis.
Materials and methods
PubMed, Scopus, ACM DL, dblp, and IEEE Xplore databases were searched. Articles utilizing clinical text for ML or natural language processing (NLP) to detect, identify, recognize, diagnose, or predict the onset, development, progress, or prognosis of systemic inflammatory response syndrome, sepsis, severe sepsis, or septic shock were included. Sepsis definition, dataset, types of data, ML models, NLP techniques, and evaluation metrics were extracted.
Results
The clinical text used in models include narrative notes written by nurses, physicians, and specialists in varying situations. This is often combined with common structured data such as demographics, vital signs, laboratory data, and medications. Area under the receiver operating characteristic curve (AUC) comparison of ML methods showed that utilizing both text and structured data predicts sepsis earlier and more accurately than structured data alone. No meta-analysis was performed because of incomparable measurements among the 9 included studies.
Discussion
Studies focused on sepsis identification or early detection before onset; no studies used patient histories beyond the current episode of care to predict sepsis. Sepsis definition affects reporting methods, outcomes, and results. Many methods rely on continuous vital sign measurements in intensive care, making them not easily transferable to general ward units.
Conclusions
Approaches were heterogeneous, but studies showed that utilizing both unstructured text and structured data in ML can improve identification and early detection of sepsis.
Keywords: sepsis, natural language processing, machine learning, electronic health records, systematic review
INTRODUCTION
Sepsis is a life-threatening illness caused by the body’s immune response to an infection that leads to multi-organ failure.1 Annually, there are 31.5 million sepsis cases, 19.4 million severe sepsis cases, and 5.3 million sepsis deaths estimated in high-income countries.2 Studies have shown that early identification of sepsis following rapid initiation of antibiotic treatment improves patient outcomes,3 and 6 h of treatment delay is shown to increase the mortality risk by 7.6%.4 Unfortunately, sepsis is commonly misdiagnosed and mistreated because deterioration with organ failure is also common in other diseases.5–8 The heterogeneity in infection source, immune responses, and pathophysiological changes make identification and therefore sepsis treatment difficult. Additionally, the diversity in age, gender, and comorbidities affect the symptoms and outcome of septic patients.7
Machine learning (ML) has been employed to improve sepsis outcomes through early detection. ML can utilize structured and unstructured data from electronic health records (EHRs).9–14 Structured clinical data come in a fixed format, such as age, vital signs, and laboratory data, which make data preprocessing easier. In contrast, clinical notes are in unstructured free-text form, such as progress notes, nursing notes, chief complaints, or discharge summaries. Clinical notes contain abbreviations, grammatical errors, and mis-spellings. Using clinical text is a complex, time-consuming process because it requires using natural language processing (NLP) to extract features that transform text into a machine-understandable representation.15–22 This usually requires assistance from clinical experts to convert text into machine-interpretable representations that capture clinical knowledge for specific clinical domains. The effort required to utilize unstructured clinical text can deter researchers; however, unstructured clinical text contains valuable information.16,22–25 Multiple studies and a review25 have shown that using unstructured clinical text has increased model performance to detect or predict colorectal surgical complications,26 postoperative acute respiratory failure,27 breast cancer,28 pancreatic cancer,29 fatty liver disease,30 pneumonia,31 inflammatory bowel disease,32,33 rheumatoid arthritis,34–36 multiple sclerosis,37 and acute respiratory infection.38,39
Prior reviews related to sepsis detection and prediction include: sepsis detection using Systemic Inflammatory Response Syndrome (SIRS) screening tools,40 sepsis detection using SIRS and organ dysfunction criteria with EHR vital signs and laboratory data,41 clinical perspectives on the use of ML for early detection of sepsis in daily practice,14 ML for diagnosis and early detection of sepsis patients,9–13 infectious disease clinical decision support,42 and healthcare-associated infections mentioning sepsis.43–45 However, to the best of our knowledge, no reviews focus on the effect of utilizing unstructured clinical text for sepsis prediction, early detection, or identification; this makes it challenging to assess and utilize text in future ML and NLP sepsis research.
OBJECTIVE
The review aims to gain an overview of studies utilizing clinical text in ML for sepsis prediction, early detection, or identification.
MATERIALS AND METHODS
This systematic review follows the Preferred Reporting Items for Systematic review and Meta-Analyses guidelines.46
Search strategy
Relevant articles were identified from 2 clinical databases (PubMed and Scopus) and 3 computer science databases (ACM DL, dblp, and IEEE Xplore) using defined search terms. The 3 sets of search terms included: (1) “sepsis,” “septic shock,” or “systemic inflammatory response syndrome”; (2) “natural language processing,” “machine learning,” “artificial intelligence,” “unstructured data,” “unstructured text,” “clinical note,” “clinical notes,” “clinical text,” “free-text,” “free text,” “record text,” “narrative,” or “narratives”; and (3) detect, identify, recognize, diagnosis, predict, prognosis, progress, develop, or onset. Searches on clinical databases were performed using all 3 sets of search terms and excluded animal-related terms. Whereas searches on computer science databases only used the first set of search terms. No additional search restrictions, such as date, language, and publication status, were included. Additional articles were identified from relevant review articles or backward reference and forward citation searches of eligible articles. Complete search strategies are in Supplementary Table S1.
The search was initially conducted using only computer science databases on December 10, 2019 and was updated to include clinical databases on December 14, 2020. The first search found that 4 of 454 articles met inclusion criteria,47–50 and the second search uncovered 2 more articles that met inclusion criteria (6 of 1335 articles).51,52 Those 2 searches did not contain the search terms: “systemic inflammatory response syndrome,” “artificial intelligence,” identify, recognize, diagnosis, prognosis, progress, develop, and onset. Hence, a search on May 15, 2021, including those terms, found 2 additional articles.53,54 To ensure inclusion of other relevant articles, a broader search was conducted on September 3, 2021 to include the following terms: “unstructured data,” “unstructured text,” “clinical note,” “clinical notes,” “clinical text,” “free-text,” “free text,” “record text,” “narrative,” or “narratives.” This resulted in 1 additional article.55
Study selection
Titles, abstracts, and keywords were screened using Zotero v5.0.96.3 (Corporation for Digital Scholarship, Vienna, VA) and Paperpile (Paperpile LLC, Cambridge, MA). Screening removed duplicates and articles that did not contain the following terms: (1) text, (2) notes, or (3) unstructured. Full-text articles were evaluated to determine if the study used unstructured clinical text for the identification, early detection, or prediction of sepsis onset in ML. Thus, selected articles had to rely on methods that automatically improve based on what they learn and not rely solely on human-curated rules. Additionally, articles solely focusing on predicting sepsis mortality were excluded as these articles are based on already established sepsis cases. Reviews, abstract-only articles, and presentations were removed. Additionally, a backward and forward search was performed on eligible full-text articles.
Data extraction
One author independently extracted data, which a second author verified. Any discrepancies were resolved either through discussion with the third author by assessing and comparing data to evidence from the studies or by directly communicating with authors from included articles. The following information was extracted: (1) general study information including authors and publication year, (2) data source, (3) sample size, (4) clinical setting, (5) sepsis infection definition, (6) task and objective, (7) characteristics of structured and unstructured data, (8) underlying ML and NLP techniques, and (9) evaluation metrics.
RESULTS
Selection process
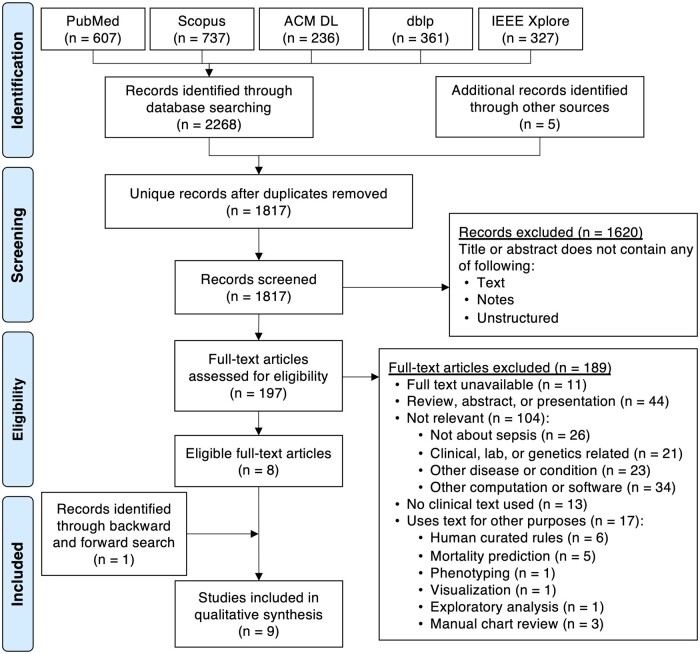
The initial search identified 2268 articles from 5 databases and 5 additional articles56–60 from 2 relevant review articles (Figure 1).43,44 From the 1817 unique articles, 1620 articles were excluded based on eligibility criteria described in the methods. After assessing the remaining 197 articles, most studies (189 of 197, ie, 96%) were excluded because they had not used or attempted to use unstructured clinical text in their ML models to identify, detect, or predict sepsis onset. For instance, there were sepsis-related studies that used text but for other purposes such as mortality prediction,61–65 phenotyping,66 visualization,67 exploratory data analysis,68 and manual chart review.69–71 Additionally, 6 articles about infection detection,60 central venous catheter adverse events,58 postoperative sepsis adverse events,72–74 and septic shock identification75 were excluded because they used manually human-curated rules instead of ML methods that automatically learn from data. The remaining 8 eligible articles were used to perform backward and forward searches,47–50,52–55 which led to the inclusion of 1 additional article.51 This resulted in 9 articles for synthesis.
Figure 1.
PRISMA (Preferred Reporting Items for Systemic reviews and Meta-Analyses) flowchart for study selection.
Study characteristics
Of the 9 identified articles, 2 studies aimed at identifying infection,47,48 6 studies focused on early detection of sepsis,51,53,55 severe sepsis,49 or septic shock,50,54 and 1 study considered both identification and early detection for a combination of sepsis, severe sepsis, and septic shock.52 Most studies focused on intensive care unit (ICU)48,50,52–55 or emergency department (ED)47,51 data; only 1 used inpatient care data.49 Four studies utilized data from hospitals,47,49,51,52 1 utilized MIMIC-II54 and 4 utilized MIMIC-III.48,50,53,55 MIMIC-II and MIMIC-III are publicly available ICU datasets created from Boston’s Beth Israel Deaconess Medical Center; MIMIC-II contains data from 2001–200776 and MIMIC-III contains data from 2001–2012.77 Eight studies used data from the United States47–51,53–55 and 1 study used data from Singapore.52 Sample sizes varied greatly in terms of the number of patients or notes used. To select patient cohorts or notes associated with sepsis, 3 studies used International Statistical Classification of Diseases and Related Health Problems (ICD) codes,47,49,52 5 applied sepsis definition criteria,49–51,53,55 1 utilized descriptions of antibiotics usage,48 and another54 applied criteria from Henry et al78 that include ICD codes, sepsis criteria, and notes mentioning sepsis or septic shock. Table 1 summarizes the study characteristics and additional details are in Supplementary Table S2 (for Culliton et al,49 the 8 structured variables for the Modified Baystate clinical definition of severe sepsis and 29 structured variables used in models were provided through personal communications with the corresponding author of Culliton et al,49 Steve Gallant, on June 4, 2021).
Table 1.
Study characteristics
| Study (year) | Clinical setting and data source | Sample sizea | Cohort criteria infection definition | Task and objective |
|---|---|---|---|---|
| Horng et al.47 (2017) |
|
230 936 patient visits
|
Angus Sepsis ICD-9-CM abstraction criteria79 | Identify patients with suspected infection to demonstrate benefits of using clinical text with structured data for detecting ED patients with suspected infection. |
| Apostolova and Velez48 (2017) |
|
634 369 nursing notes
|
Notes describing patient taking or being prescribed antibiotics for treating infection | Identify notes with suspected or presence of infection to develop a system for detecting infection signs and symptoms in free-text nursing notes. |
| Culliton et al.49 (2017) |
|
203 000 adult inpatient admission encounters
Test set: 2016 data:
|
Modified Baystate clinical definition of severe sepsis (8 structured variables) and severe sepsis ICD codes | Predict severe sepsis 4, 8, and 24 h before the earliest time structured variables meet the severe sepsis definition to compare accuracy of predicting patients that will meet the clinical definition of sepsis when using unstructured data only, structured data only, or both types. |
| Delahanty et al.51 (2019) |
|
2 759 529 patient encounters
Train: 1 839 503 E; 66.7%
Test: 920 026 E; 33.3%
|
Rhee’s modified Sepsis-3 definition80 | Predict sepsis risk in patients 1, 3, 6, 12, and 24 h after the first vital sign or laboratory result is recorded in the EHR to develop a new sepsis screening tool comparable to benchmark screening tools. |
| Liu et al.50 (2019) |
|
|
Sepsis-3 definition1 | Predict septic shock in sepsis patients before the earliest time septic shock criteria are met to demonstrate an approach using NLP features for septic shock prediction. |
| Amrollahi et al.53 (2020) |
|
40 175 adult patients
|
Sepsis-3 definition1 | Predict sepsis onset hours in advance using a deep learning approach to show a pre-trained neural language representation model can improve early sepsis detection. |
| Hammoud et al.54 (2020) |
|
17 763 patients
5-fold cross validation |
Sepsis definition based on what Henry et al78 used | Predict early septic shock in ICU patients using a model that can be optimized based on user preference or performance metrics. |
| Goh et al.52 (2021) |
|
Train and validation: 3722 P (80 162 N)
Test: 1595 P (34 440 N)
|
ICU admission with an ICD-10 code for sepsis, severe sepsis, or sepsis shock | Identify if a patient has sepsis at consultation time or predict sepsis 4, 6, 12, 24, and 48 h after consultation to develop an algorithm that uses structured and unstructured data to diagnose and predict sepsis. |
| Qin et al.55 (2021) |
|
Train: 33 434 P
Validation: 8358 P
Test: 7376 P
|
PhysioNet Challenge restrictive Sepsis-3 definition81 | Predict if a patient will develop sepsis to explore how numerical and textual features can be used to build a predictive model for early sepsis prediction. |
ED: emergency department; ICU: intensive care unit; ICD: International Classification of Diseases; ICD-9 CM: ICD Clinical Modification, 9th revision; ICD-10: ICD 10th revision; MIMIC-II: Multiparameter Intelligent Monitoring in Intensive Care II database; MIMIC-III: Medical Information Mart for Intensive Care dataset.
Sample size unit abbreviations: P: patients; N: notes; E: encounters.
Clinical text used in models
The 9 studies utilized narrative notes written by nurses,47–50,53–55 physicians,49–53,55 or specialists49–51,54,55 to document symptoms, signs, diagnoses, treatment plans, care provided, laboratory test results, or reports. EHRs contain various types of clinical notes. A note covers an implicit time period or activity and describes events, hypotheses, interventions, and observations within the health care provider’s responsibilities. The note’s form depends on its function: an order, a plan, a prescription, an investigation or analysis report, a narrative or log of events, information for the next shifts, or a requirement for legal, medical, or administrative purposes. An episode of care begins when a patient is admitted to the hospital and ends when the patient is discharged. Throughout a patient’s hospital stay, documentation can include chief complaints, history-and-physical notes, progress notes, reports, descriptions of various laboratory tests, procedures, or treatments, and a discharge summary. Chief complaints are the symptoms or complaints provided by a patient for why they are seeking care.82 History-and-physical notes can include history about the current illness, medical history, social history, family history, a physical examination, a chief complaint, probable diagnosis, and a treatment plan.83 Progress notes document care provided and a description of the patient’s condition to convey events to other clinicians.84 Free-text reports can include interpretations of echocardiograms, electrocardiograms (ECGs), or imaging results such as X-rays, computerized tomography scans, magnetic resonance imaging scans, and ultrasounds. At discharge, the health care personnel write a discharge summary note comprised of patient details, hospital admittance reason, diagnosis, conditions, history, progress, interventions, prescribed medications, and follow-up plans.85–87 The discharge summary letter is a formal document used to transfer patient care to another provider for further treatment and follow-up care.88–90
Studies have shown that nursing documentation differs from physician documentation.91,92 Nurses document more about a patient’s functional abilities than physicians,91 and the information from notes used and the frequency of viewing and documenting differs between health care personnel.92 Additionally, documentation varies between hospitals,93,94 hospitals have different resources and practices,95–97 and communicative behavior differs among professions in different wards.98 Hence, the type of notes used, who wrote the notes, and purpose of the note will play a role in how the documentation is interpreted.99
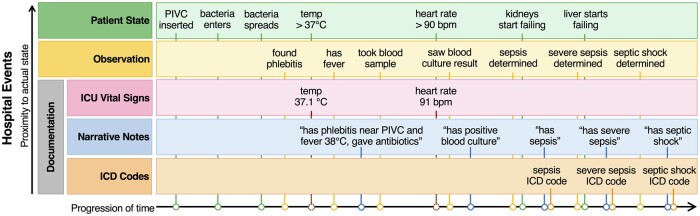
Table 2 provides information regarding documentation types, author of the note, time content of the data, time latency between documentation and availability in records, and the documentation frequency. In Figure 2, the relationship between hospital events and longitudinal data used to train models is shown. As sepsis develops in a patient over time, it shows there are typically delays between a patient’s actual state, clinical observations, and recorded documentation, such as ICU vital signs, narrative notes, and ICD codes.
Table 2.
Clinical documentation from electronic health records
| Documentation types | Author | Description | Temporal perspective | Record latencya | Frequency |
|---|---|---|---|---|---|
| Chief complaints |
|
Symptoms or complaints provided by a patient at start of care for why they are seeking care. | Current | Seconds to days | One per episode |
| History-and-physical notes |
|
Past medical history, family history, developmental history of present illness, problems about present illness, past medications or immunizations, allergies, or habits. | Retrospective | Immediately | One per episode |
| Progress notes |
|
|
|
4–8 h | One per shift |
| Reports | Specialist | Radiologist results and cardiology results. | Retrospective | Days | One to many per episode |
| Discharge summary notes | Health care personnel | Episode of care summary and follow-up plans. |
|
At discharge or days after | One per episode |
| Discharge summary letter | Physician | Formal required letter containing follow-up treatment plans. |
|
Days to months after episode | One per episode |
| Laboratory results | Laboratory technician | Laboratory test analysis results from provided samples (eg, blood, urine, skin, and device) based on the physician’s order. | Retrospective | Days | One to many per episode |
| ICD codes |
|
Diagnosis classification for billing. | Retrospective | Days to months | One per episode |
| Administrative |
|
Patient information such as name, age, gender, address, contact information, and occupation. |
|
Immediately | One per episode |
Record latency is defined as time between measurement/observation and the availability of the results in electronic health records.
Figure 2.
Overview of data from a patient timeline used to create models. The proximity of events toward a patient’s actual state and the actual documentation recorded in the electronic health records typically has delays. Green represents patient states as sepsis develops in a patient. Yellow are observations made by clinicians. Documentation includes ICU vital signsa in pink, narrative notes in blue, and ICD codes in orange. ICU vital signa documentation can be instantaneous, narrative notes can be written after observations are made, and ICD codes are typically registered after a patient is discharged. PIVC: peripheral intravenous catheter. aVital signs include temperature, pulse, blood pressure, respiratory rate, oxygen saturation, and level of consciousness and awareness.
The included studies utilized the following types of notes: 6 studies used unstructured nursing-related documentation,47,48,50,53–55 4 used physician notes,50,52,53,55 3 used radiology reports,50,54,55 3 used respiratory therapist progress notes,50,54,55 2 used ED chief complaints,47,51 2 used ECG interpretations,50,54 2 used pharmacy reports,50,54 2 used consultation notes,50,52 1 used discharge summaries,50 1 included mostly progress notes and history-and-physical notes,49 and 3 used additional unspecified notes.49,50,54 Not all notes used are listed. Liu et al50 used all MIMIC-III notes to build a vocabulary of unique words, and discharge summaries were likely not used in predictions because they are unlikely to occur before observations. Additionally, Hammoud et al54 used all MIMIC-II notes except discharge summaries.
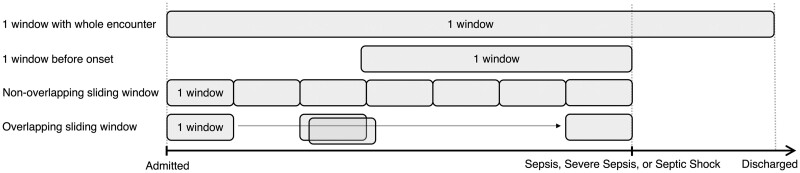
These 9 studies utilized clinical notes differently. For the unit of analysis, 6 studies used a single note,47,48,50,52–54 1 used a set of many notes from a patient encounter,49 1 used a set of many notes within a specific hour of consideration,55 and 1 used keywords from notes.51 To identify infection signs, Horng et al47 and Apostolova and Velez48 processed individual notes. While Goh et al52 used notes at each patient consultation instance to identify sepsis patients. For early detection, 5 studies defined onset time as the earliest time when definition criteria are met49,50,53–55 and 1 defined sepsis onset time as ICU ward admission time.52 Studies for early detection used varying windows with different durations. A window decides how and where to obtain longitudinal data, and duration is the length of time. As shown in Figure 3, studies can use windows differently, such as a window with the duration of the whole encounter, a window with a duration of hours before onset, non-overlapping sliding windows with a fixed duration until onset, or overlapping sliding windows with a fixed duration until onset. Culliton et al49 used a 4-, 8-, or 24-h duration window before severe sepsis, and concatenated all text within a window. Goh et al52 used a 4-, 6-, 12-, 24-, or 48-h duration window of before sepsis, severe sepsis, or septic shock onset. Liu et al50 used 10 data points within a 1-h duration window spanning 2 h before septic shock, and used the most recently entered note for a data point to predict septic shock. Hammoud et al54 binned data in 15-minute duration non-overlapping sliding windows to update septic shock predictions every 15 minutes, and used the last note within the window. Amrollahi et al53 binned data into 1-h duration non-overlapping sliding windows to provide hourly sepsis predictions, and used sentences within a note to capture the semantic meanings. Qin et al55 used 6-h duration overlapping sliding windows with 6 data points to predict sepsis; a data point was generated from each hour within the window and all clinical notes within the hour were concatenated in random-order. Delahanty et al51 used a 1-, 3-, 6-, 12-, or 24-h duration window after the first vial sign or laboratory result was documented in the EHR to identify patients at risk for sepsis, and utilized keywords.
Figure 3.
Different types of windows were used to obtain longitudinal data. Each gray box represents a single window, which can vary in duration (length of time) depending on the study. One window with the whole encounter means the study used a single window containing data with a duration of the whole encounter from admittance until discharge. One window before onset signifies data from a window with a duration of time before sepsis, severe sepsis, or septic shock onset. Sliding windows are consecutive windows until before sepsis, severe sepsis, or septic shock onset; this includes non-overlapping and overlapping sliding windows. Non-overlapping sliding windows indicate that data within one window of a fixed duration does not contain data in the next window. In contrast, overlapping sliding windows indicate windows of a fixed duration overlap, and data within one window will be partially in the next window.
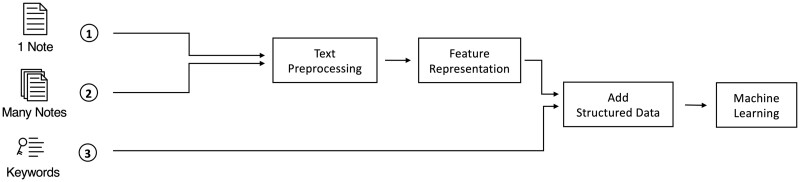
First 2 columns in Table 3 show the type of text and unit of analysis used. Additional details about variables and specific notes used are listed in Supplementary Table S3 (the types of notes and usage for Liu et al50 was confirmed through personal communications with Ran Liu on June 2, 2021, for Hammoud et al54 by Ibrahim Hammoud on May 29, 2021, and for Qin et al55 by Fred Qin on September 9, 2021. Additionally, the structured variables used in models for Culliton et al49 were provided through personal communications with Steve Gallant on June 4, 2021). In Figure 4, single notes or a set of many notes are preprocessed and represented to extract features, whereas keywords are used as is. Then structured data can be added, and the data are used to train ML models.
Table 3.
Text used in studies
| Study (year) | Free-text document type | Unit of analysis | Text processing |
|---|---|---|---|
| Horng et al.47 (2017) |
|
One note |
Representation:
Techniques:
|
|
Apostolova and Velez48 (2017) |
Nursing notes | One note |
Representation:
Techniques:
|
| Culliton et al.49 (2017) | Clinical notes (mostly progress notes and history-and-physical notes) | One patient encounter = many notes |
Representation:
Techniques:
|
| Delahanty et al.51 (2019) | ED chief complaints | Keywords | Other:
|
| Liu et al.50 (2019) | All MIMIC-III clinical notes, such as but not limited to:
|
One note |
Representation:
Techniques:
|
| Amrollahi et al.53 (2020) |
|
One note |
Representation:
Techniques:
|
| Hammoud et al.54 (2020) | All MIMIC-II notes except discharge summaries, such as but not limited to:
|
One note |
Representation:
Techniques:
|
| Goh et al.52 (2021) | Physician notes:
|
One note |
Representation:
Techniques:
|
| Qin et al.55(2021) |
|
Many notes |
Representation:
Techniques:
|
BoW: Bag-of-words; CBOW: Continuous bag-of-words; ClinicalBERT: Clinical Bidirectional Encoder Representations from Transformers; ED: emergency department; GloVe: Global Vectors for Word Representation; ICU: intensive care unit; LDA: Latent Dirichlet Allocation; POS tagging: Part-of-speech tagging; PV: paragraph vectors; tf-idf: term frequency-inverse document frequency.
Representation and technique details for Qin et al55 were provided through personal communications (with Fred Qin on September 7, 2021).
Figure 4.
The unit of analysis used to train machine learning models for the included studies was either (1) a single note, (2) a set of many notes, or (3) keywords. In general, text was preprocessed and represented as features interpretable by a computer, then structured data were added, and the data were used to fit machine learning models.
As shown in Figures 3 and 4 and listed in Tables 1 and 3 and Supplementary Tables S2 and S3, although all studies are related to sepsis, there are varying sample sizes, data types, inclusion criteria, and objectives. This heterogeneity makes it challenging to compare results for a meta-analysis.
Natural language processing and machine learning study outcomes
To utilize text in ML, it must be transformed into a representation understandable by computers. In order to do that, Bag-of-words (BoW),100 n-gram, term frequency-inverse document frequency (tf-idf), and paragraph vectors (PV)101 representations can be used. These representations can be improved using additional NLP techniques, such as stop word removal, lemmatization, and stemming. In addition, other useful features can be extracted from text using part-of-speech (POS) tagging, named entity recognition, or Latent Dirichlet Allocation (LDA) topic modeling.102 In recent years, neural networks (NNs) have shown high predictive performance. As a result, many state-of-the-art results have been achieved using NNs to learn a suitable representation of texts, often known as embeddings.103 Embedding techniques include Global Vectors for Word Representation (GloVe),104 Word2Vec as a continuous bag-of-words (CBOW) model or skip-gram model,105 Bidirectional Encoder Representations from Transformers (BERT),106 and ClinicalBERT.107 The advantage of using embeddings is that it retains the sequential information lost in a BoW representation and does feature extraction automatically.103
Utilized text processing operations are in Table 3. One study used keyword extraction instead of text processing operations.51 Six studies used tokenization of words for word-level representation,47–50,52,54 1 also tried PV for document-level representation,48 and another used the first 40 tokens in a sentence to get sentence-level representation and averaged sentence-level representations to provide document-level representation.53 The most common technique for improving representation was token removal, such as removing rare tokens,47,50,52–54 frequent tokens,48,50,53,54 punctuation or special characters,47,48,50,52,53 and stop words.52,53 The most frequently used representation was tf-idf,48,52–55 followed by BoW,47,48,50,54 LDA,47,52 GloVe,49,50 ClinicalBERT,53,55 bi-gram,47 CBOW,48 and PV.48 Three studies created a vocabulary of unique terms using BoW,50 CBOW,48 and tf-idf.53 Apostolova and Velez48 found that using structured data was inadequate for identifying infection in nursing notes, so they used antibiotic usage and word embeddings to create a labeled dataset of notes with infection, suspected infection, and no infection. Additionally, Horng et al47 and Liu et al50 listed predictive terms in their models, and Goh et al52 provided a list of categories used to classify the top 100 terms. Examples of predictive features are: (1) For sepsis, severe sepsis, or septic shock, Goh et al52 classified the top 100-topics into 7 categories: clinical condition or diagnosis, communication between staff, laboratory test order or results, non-clinical condition updates, social relationship information, symptoms, and treatments or medication. (2) Liu et al’s50 most predictive NLP terms for the pre-shock versus non-shock state include “tube,” “crrt,” “ards,” “vasopressin,” “portable,” “failure,” “shock,” “sepsis,” and “dl.” (3) Horng et al’s47 most predictive terms or topics for having an infection in the ED include “cellulitis,” “sore_throat,” “abscess,” “uti,” “dysuria,” “pneumonia,” “redness_swelling,” “erythema,” “swelling,” “redness, celluititis, left, leg, swelling, area, rle, arm, lle, increased, erythema,” “abcess, buttock, area, drainage, axilla, groin, painful, thigh, left, hx, abcesses, red, boil,” and “cellulitis, abx, pt, iv, infection, po, keflex, antibiotics, leg, treated, started, yesterday.” Whereas the least predictive terms or topics for not having an infection include “motor vehicle crash,” “laceration,” “epistaxis,” “pancreatitis”, “etoh”(ethanol for drunkenness), “etoh, found, vomiting, apparently, drunk, drinking, denies, friends, trauma_neg, triage,” and “watching, tv, sitting, sudden_onset, movie, television, smoked, couch, pt, pot, 5pm, theater.”
ML methods for detecting sepsis using clinical text included: ridge regression,49 lasso regression,54 logistic regression,47,48,52 Naïve Bayes (NB),47 support vector machines (SVMs),47,48 K-nearest neighbors (KNNs),48 random forest (RF),47,52 gradient boosted trees (GBTs),50–52,55 gated recurrent unit (GRU),50 and long short-term memory (LSTM).53 Although the methods are listed separately, 2 studies combined different ML methods48,52 (see Supplementary Table S4 for details). Ridge and lasso regression are linear regression methods that constrain the model parameters. A linear regression model is represented as , where is the predicted value, is the input variable and and are model parameters. Model parameters are estimated by minimizing , where is the label and is the number of training samples. In ridge and lasso regression, is minimized instead, where is a hyperparameter that trades-off between fitting the data and model complexity, and for ridge regression or for lasso regression. Logistic regression is a classification method that models , which is the probability of a class given the feature . The logistic regression model is defined as . NB is a Bayesian network that eases computation by assuming all input variables are independent given the outcome.108 SVM is an extension of a support vector classifier that separates training data points into 2 class regions using a linear decision boundary and classifies new data points based on which region they belong to. To accommodate for non-linearity in the data, SVM enlarges the feature space by applying kernels.109 KNNs assume similar data points are close together and use similarity measures to classify new data based on “proximity” to points in the training data.110 RF and GBT are ensemble models that use a collection of decision trees to improve the predictive performance of the models. RF classification takes the majority vote of a collection of trees to reduce the decision tree variance.111 GBT trains decision trees sequentially so that each tree trains based on information from previously trained trees.112,113 To avoid overfitting, each tree is scaled by a hyperparameter , often known as the shrinkage parameter or learning rate that controls the rate the model learns. Recurrent neural networks (RNNs) are a type of NN with recurrent connections and assume that the input data have an ordering, for example, words in a sentence.114–116 RNN can be seen as a feed-forward NN with a connection from output to input.115 GRU117 and LSTM118 are improved variations of RNN with gating mechanisms to combat the vanishing gradient problem. The improvements help the models to better model long-term temporal dependencies. To tune hyperparameters, grid-search and Bayesian optimization were used in the studies.47,48,50,53,54 The grid-search method iterates exhaustively through all hyperparameter values within a pre-defined set of values to find the optimal hyperparameter with respect to a validation set. In contrast, the Bayesian optimization method makes informed choices on which values to evaluate using the Bayes formula. The goal of using Bayesian optimization for hyperparameter tuning is to minimize the number of values to evaluate.
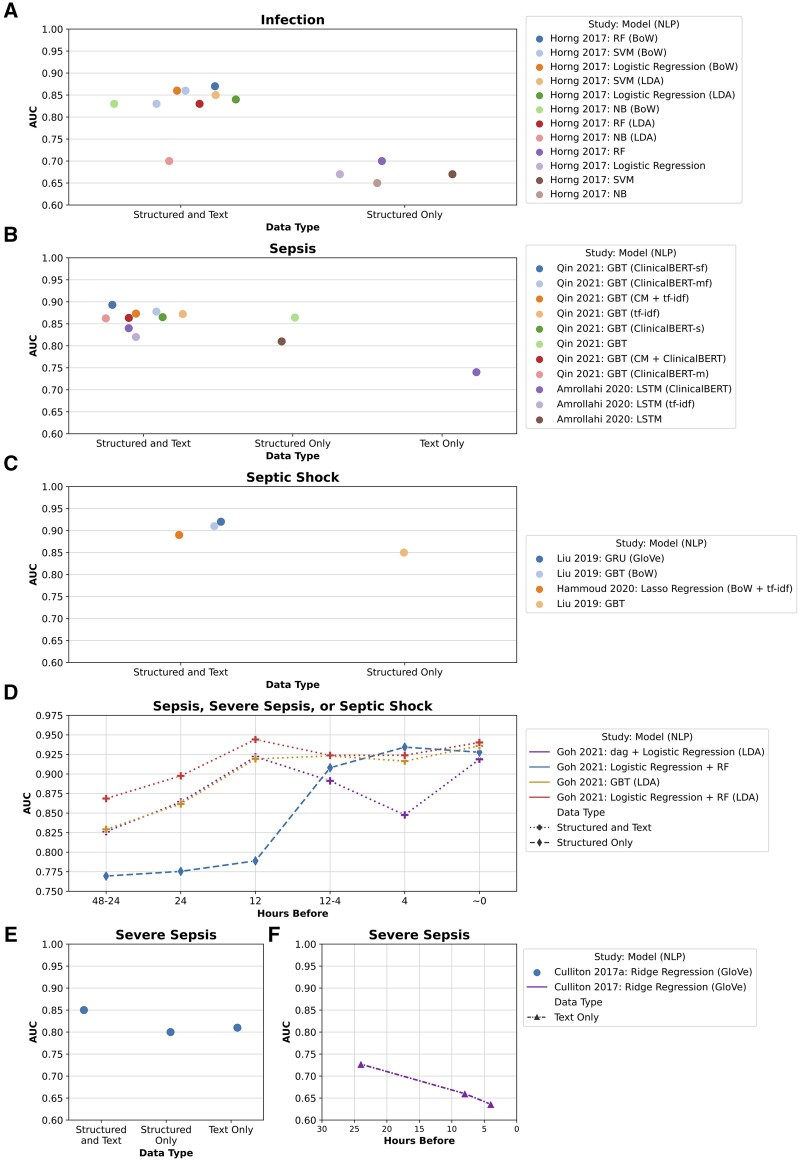
All studies reported evaluation results for different algorithms or data types and almost all reported area under the receiver operating characteristic curve (AUC) values except 1.48 Figure 5 shows differences in AUC values for infection (Figure 5A), sepsis (Figure 5B), septic shock (Figure 5C), and severe sepsis (Figure 5E) when using structured data only, text data only, or a combination of structured and text data. Studies that compared their methods for different hours prior to onset are also included (Figure 5D and F), the lines connecting the points are to visually separate the methods and do not indicate changing AUC values over time. This figure compares data type usage and model performance within an individual study; it should not be used to compare AUC values between subfigures and studies because the studies used different cohorts, sepsis definitions, and hours before onset. Additionally, sepsis, severe sepsis, and septic shock have different manifestations.119,120 Table 4 summarizes the best and worst AUC values for each study; a full table with additional evaluation metrics is available in Supplementary Table S4 (number of hours before onset for Amrollahi et al53 was confirmed through personal communications with Shamim Nemati on May 27, 2021 and Fatemeh Amrollahi on June 13, 2021). GBT was the most widely used ML method,50–52,55 followed by logistic regression,47,48,52 SVMs,47,48 RF,47,52 ridge regression,49 lasso regression,54 NB,47 KNNs,48 GRU,50 and LSTM.53 For hyperparameter tuning, 3 studies used the grid-search method47,48,54 and 2 used the Bayesian optimization method50,53 (hyperparameter tuning was provided by personal communication with Ran Liu on September 7, 2021 and Fatemeh Amrollahi on September 7, 2021). Delahanty et al,51 Hammoud et al,54 Goh et al,52 and Qin et al55 compared their algorithm to scoring systems used in clinical practice, such as SIRS,121 sequential organ failure assessment (SOFA),122 quick SOFA (qSOFA),123 modified early warning system (MEWS),124 or a targeted real-time early warning score (TREWScore).78 In addition, Apostolova and Velez48 evaluated their model on a ground truth set with 200 nursing notes that were manually reviewed by a qualified professional, and Goh et al52 compared their model with the Rhodes et al125 sepsis guidelines used by physicians. Furthermore, Horng et al47 performed additional tests on different patient cohorts for error analysis. Although results are difficult to compare directly because of study heterogeneity, most results suggest that utilizing both structured data and text generally results in better performance for sepsis identification and early detection.
Figure 5.
Overview of area under the curve (AUC) values for identification or early detection of infection, sepsis, septic shock, and severe sepsis using different data types (structured data and text, structured data only, and text only).∗ Each figure contains the study and year, machine learning model,a and natural language processing techniqueb. (A) AUC values for infection identification. Horng et al47 2017: SVM (BoW) has 2 AUC values; 0.86 when using chief complaints and nursing notes and 0.83 when using only chief complaints. (B) AUC values for early sepsis detection. Amrollahi et al53 AUC values are from detecting 4 h before sepsis onset, and Qin et al55 AUC values are the average from detecting 0 to 6 h before sepsis onset. (C) AUC values for early septic shock detection. Hammoud et al54 AUC values are from detecting 30.64 h before septic shock onset, and Liu et al50 AUC values are from detecting 6.0 to 7.3 h before septic shock onset. (D) AUC values for early sepsis, severe sepsis, or septic shock detection and sepsis identification in Goh et al.52 Different symbols separate data types. (E) AUC values for early septic shock detection for Culliton et al49 using results from the test set. (F) AUC values for early septic shock detection for Culliton et al49 using results from 3-fold validation. ∗Disclaimer: AUC values should not be directly compared between studies and different figures for infection, sepsis, severe sepsis, and septic shock. Additionally, the lines connecting points do not indicate AUC values changing over time (Figure 5D and 5F); lines only separate the different methods visually. aMachine learning models: dag: dagging (partition data into disjoint subgroups); GBT: gradient boosted trees; GRU: gated recurrent unit; LSTM: long short-term memory; NB: Naïve Bayes; RF: random forest; SVM: support vector machines. bNatural language processing techniques: BoW: Bag-of-words; ClinicalBERT: Clinical Bidirectional Encoder Representations from Transformers; ClinicalBERT-m: ClinicalBERT from merging all textual features to get embeddings; ClinicalBERT-sf; finetuned ClinicalBERT from concatenating individual embeddings of each textual feature; CM: Amazon Comprehend Medical service for named entity recognition; GloVe: Global Vectors for Word Representation; LDA: Latent Dirichlet Allocation; tf-idf: term frequency-inverse document frequency.
Table 4.
Study outcome overview of best and worst area under the curve values
| Study (year) | Hoursa | Data typesb |
Modelsd (NLP)e | AUCf | |
|---|---|---|---|---|---|
| DVLMC | Tc | ||||
| Horng et al.47 (2017) | Identify | DV- - - | CC + NN | RF (BoW) | 0.87 |
| DV- - - | – | NB | 0.65 | ||
| Apostolova and Velez48 (2017) | Identify | - - - - - | NN | SVM (BoW + tf-idf) | – |
| - - - - - | NN | Logistic regression + KNN + SVM (PV) | – | ||
| Culliton et al.49 (2017) | −4 | - - - - - | CN | Ridge regression (GloVe) | 0.64 |
| −8 | - - - - - | CN | Ridge regression (GloVe) | 0.66 | |
| −24 | - - - - - | CN | Ridge regression (GloVe) | 0.73 | |
| −24g | -V- -C | CN | Ridge regression (GloVe) | 0.85 | |
| -V- -C | – | Ridge regression (GloVe) | 0.80 | ||
| Delahanty et al.51 (2019) | +1 | -VL- - | – | GBT | 0.93 |
| +3 | -VL- - | – | GBT | 0.95 | |
| +6 | -VL- - | – | GBT | 0.96 | |
| +12 | -VL- - | – | GBT | 0.97 | |
| +24 | -VL- - | – | GBT | 0.97 | |
| Liu et al.50 (2019) | −7 | -VLM- | CN | GRU (GloVe) | 0.92 |
| −7.3 | -VLM- | CN | GBT (BoW) | 0.91 | |
| −6 | -VLM- | – | GBT | 0.85 | |
| Amrollahi et al.53 (2020) | −4h | -VL- - | PN + NN | LSTM (ClinicalBERT) | 0.84 |
| - - - - - | PN + NN | LSTM (ClinicalBERT) | 0.74 | ||
| Hammoud et al.54 (2020) | −30.6 | DVL- - | CN | Lasso regression (BoW + tf-idf) | 0.89 |
| Goh et al.52 (2021) | Identify | DVLM- | PN | Logistic regression + RF (LDA) | 0.94 |
| DVLM- | PN | dag + Logistic regression (LDA) | 0.92 | ||
| −4 | DVLM- | – | Logistic regression + RF | 0.93 | |
| DVLM- | PN | dag + Logistic regression (LDA) | 0.85 | ||
| −6 | DVLM- | PN | Logistic regression + RF (LDA) | 0.92 | |
| DVLM- | PN | dag + Logistic regression (LDA) | 0.89 | ||
| −12 | DVLM- | PN | Logistic regression + RF (LDA) | 0.94 | |
| DVLM- | – | Logistic regression + RF | 0.79 | ||
| −24 | DVLM- | PN | Logistic regression + RF (LDA) | 0.90 | |
| DVLM- | – | Logistic regression + RF | 0.78 | ||
| −48 | DVLM- | PN | Logistic regression + RF (LDA) | 0.87 | |
| DVLM- | – | Logistic regression + RF | 0.77 | ||
| Qin et al.55 (2021) | −6 to 0i | -VL- - | CN | GBT (ClinicalBERT-sf) | 0.89i |
| -VL- - | – | GBT (ClinicalBERT-m) | 0.86i | ||
Hours: Identify: not detecting hours before or after; –: hours before; +: hours after an event.
Data types: D: demographics; V: vitals; L: laboratory; M: medications; C: codes; T: text; -‘s position in DVLMC indicates which is not used.
Text data types: CC: chief complaints; CN: various types of clinical notes; NN: nursing notes; PN: physician notes; –: no notes.
Machine learning models: dag: dagging (partition data into disjoint subgroups); GBT: gradient boosted trees; GRU: gated recurrent unit; KNN: K-nearest neighbors; LSTM: long short-term memory; NB: Naïve Bayes; RF: random forest; SVM: support vector machines.
Natural language processing (NLP) techniques: BoW: Bag-of-words; ClinicalBERT: Clinical Bidirectional Encoder Representations from Transformers; ClinicalBERT-m: ClinicalBERT from merging all textual features to get embeddings; ClinicalBERT-sf: finetuned ClinicalBERT from concatenating individual embeddings of each textual feature; GloVe: Global Vectors for Word Representation; LDA: Latent Dirichlet Allocation; PV: paragraph vectors; tf-idf: term frequency-inverse document frequency.
Area under the curve (AUC). Apostolova and Velez48 did not provide metrics for AUC.
Culliton et al49 performed 2 experiments, these results are from using a test set instead of 3-fold validation.
Number of hours before onset for Amrollahi et al53 was confirmed through personal communications (with Shamim Nemati on May 27, 2021 and Fatemeh Amrollahi on June 13, 2021).
Qin et al55 AUC values are an average from 0 to 6 h before sepsis, not the specified hours.
DISCUSSION
Identification, early detection, prediction, and method transferability
Nine studies utilized clinical text for sepsis identification, early detection, or prediction. As all identified studies focus on the identification or early detection of sepsis within a fixed time frame, this indicates much work is still needed before sepsis prediction can use text from complete patient histories. Studies from this review focus mainly on the ICU and ED, and the addition of continuous measurements of vital signs for sepsis makes generalizability to the ward units limited. However, Culliton et al49 was successful in detecting sepsis early utilizing only the text from EHR clinical notes, which is a promising approach for all inpatients. Additionally, Horng et al47 showed that their ML model performed on subsets of specific patient cohorts like pneumonia or urinary tract infection. The different ML methods and NLP techniques from each study may be applicable for different retrospective cohort or case–control studies. Though the studies have varying sepsis definitions, cohorts, ML methods, and NLP techniques, overall, they show that using clinical text and structured data can improve sepsis identification and early detection. Unstructured clinical text predicts sepsis 48–12 h before onset, while structured data predicts sepsis closer to onset (<12 h before).
Sepsis definition impact
In ML, many studies rely heavily on sepsis definitions and ICD-codes to identify patient cohort datasets for sepsis studies.9,11,13 Among changing sepsis definitions over time are the 2001 Angus Sepsis ICD-9 abstraction criteria,79 2012 Surviving Sepsis Campaign Guidelines,126 2016 Sepsis-3 consensus definition,1 and 2017 Rhee’s modified Sepsis-3 definition.80 Although a consensus sepsis definition exists,1 not all definition elements will be present in a sepsis patient because sepsis is a very heterogeneous syndrome127 and the infection site is difficult to identify correctly.128 Many patients with sepsis are often misdiagnosed with other diseases such as respiratory failure129 and pneumonia.129,130 In practice, hospitals also have varying sepsis coding methods.131–135 As the sepsis definitions change, studies also tend to use the most current definition in their study. A recent study that used different sepsis definitions to generate patient cohorts found significant heterogeneous characteristics and clinical outcomes between cohorts.136 Similarly, previous work by Liu et al137 demonstrated that using different infection criteria resulted in a different number of patients and slightly different outcomes. Similar to how changes in the definition and varying coding methods can affect sepsis mortality outcomes,138 the sepsis definition and codes used in ML studies will likely change the outcome, results, and reporting methods. Thus, future studies should acknowledge that sepsis is a syndrome and clearly characterize each sign of sepsis to reflect the heterogeneity in the definition.
Suggestions for future studies
Predicting sepsis earlier than 12 h prior to sepsis onset can reduce treatment delays and improve patient outcomes.3,4 Because predictions 48–12 h before sepsis onset appear to rely more on clinical text than structured data, additional NLP techniques should be considered for future ML studies. Additionally, since the sepsis definition used will change the cohort, this indicates opportunities to expand the cohort. Like Apostolova and Velez,48 who determined their cohort by finding notes describing the use of antibiotics. It should be possible to determine cohorts by using notes describing infection signs (eg, fever, hypotension, or deterioration in mental status), indicators of diseases that sepsis is misdiagnosed with (eg, pulmonary embolism, adrenal insufficiency, diabetic ketoacidosis, pancreatitis, anaphylaxis, bowel obstruction, hypovolemia, colitis, or vasculitis), or medication effect and toxin ingestion, overdose, or withdrawal.139 NLP methods from infectious diseases known to trigger sepsis can be incorporated to extract infection signs and symptoms from the text for determining potential sepsis signs, patient groups, and risk factors. For instance, many sepsis patients are often admitted with pneumonia, and there are several studies about identifying pneumonia from radiology reports using NLP.23,140,141 Additionally, heterogeneous sepsis signs or symptoms might be identified by utilizing NLP features for detecting healthcare-associated infections risk patterns59 or infectious symptoms.142 Information from other NLP related reviews about using clinical notes can also be applied, such as: challenges to consider,16 clinical information extraction tools and methods,18 methods to overcome the need for annotated data,22 different embedding techniques,143,144 sources of labeled corpora,143 transferability of methods,145 and processing and analyzing symptoms.146 Moreover, heterogeneous or infectious diseases, with overlapping signs and symptoms of other diseases, can utilize similar sepsis ML and NLP methods to improve detection. The identified studies did not utilize complete patient history data. Thus, future research utilizing complete patient history data can study if sepsis risk can be predicted earlier than 48 h by incorporating sepsis risk factors, such as comorbidities,7 chronic diseases,147 patient trajectories,148 or prior infection incidents.149
Limitations
This review has several limitations. The narrow scope of including only studies about utilizing clinical text for sepsis detection or prediction could have missed studies that use other types of text for sepsis detection or prediction. For example, search terms did not include “early warning system,” “feature extraction,” and “topic modeling.” Additionally, search terms did not include possible sources of infection for sepsis, such as bloodstream infection, catheter-associated infection, pneumonia, and postoperative surgical complications. Further, the sensitivity to detect sepsis in text, structured data, or the combined data from these will depend on the timestamps these data recordings have in the EHR. These timestamps may vary depending on the data used to inform the study or the different systems implemented at different hospitals. The articles identified in this review had a homogenous choice of structured data (ie, demographics, vital signs, and laboratory measurements). Of those, laboratory test results have the largest time lag, around 1–2 h to obtain the blood test results.150 Thus, the good performance of text to detect sepsis in these articles are unlikely explained fully by the time lag between measurement and recording of the structured data. This review thus shows that it is possible to detect sepsis early using text, with or without the addition of structured data.
CONCLUSION
Many studies about sepsis detection exist, but very few studies utilize clinical text. Heterogeneous study characteristics made it difficult to compare results; however, the consensus from most studies was that combining structured data with clinical text improves identification and early detection of sepsis. There is a need to utilize the unstructured text in EHR data to create early detection models for sepsis. The lack of utilizing the complete patient history in early prediction models for sepsis is an opportunity for future ML and NLP studies.
FUNDING
Financial support for this study was provided by the Computational Sepsis Mining and Modelling project through the Norwegian University of Science and Technology Health Strategic Area.
AUTHOR CONTRIBUTIONS
MYY and ØN conceptualized the study and design with substantial clinical insight from LTG. MYY conducted the literature search and initial analysis, LTG verified results, and ØN resolved discrepancies. All authors participated in data analysis and interpretation. MYY drafted the manuscript, which LTG and ØN critically revised.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We thank those from the Gemini Center for Sepsis Research group for valuable discussions and recommendations related to clinical databases, missing search terms, and presenting results. Specifically, Ms Lise Husby Høvik (RN), Dr Erik Solligård, Dr Jan Kristian Damås, Dr Jan Egil Afset, Dr Kristin Vardheim Liyanarachi, Dr Randi Marie Mohus, and Dr Anuradha Ravi.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The data underlying this article are available in the article and in its online supplementary material.
Contributor Information
Melissa Y Yan, Department of Computer Science, Faculty of Information Technology and Electrical Engineering, Norwegian University of Science and Technology, Trondheim, Norway.
Lise Tuset Gustad, Department of Circulation and Medical Imaging, Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology, Trondheim, Norway; Department of Medicine, Levanger Hospital, Clinic of Medicine and Rehabilitation, Nord-Trøndelag Hospital Trust, Levanger, Norway.
Øystein Nytrø, Department of Computer Science, Faculty of Information Technology and Electrical Engineering, Norwegian University of Science and Technology, Trondheim, Norway.
REFERENCES
- 1. Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016; 315 (8): 801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fleischmann C, Scherag A, Adhikari NKJ, et al. ; International Forum of Acute Care Trialists. Assessment of global incidence and mortality of hospital-treated sepsis. Current estimates and limitations. Am J Respir Crit Care Med 2016; 193 (3): 259–72. [DOI] [PubMed] [Google Scholar]
- 3. Rivers E, Nguyen B, Havstad S, et al. Early goal-directed therapy in the treatment of severe sepsis and septic shock. N Engl J Med 2001; 345 (19): 1368–77. [DOI] [PubMed] [Google Scholar]
- 4. Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 2006; 34: 1589–96. [DOI] [PubMed] [Google Scholar]
- 5. Polat G, Ugan RA, Cadirci E, et al. Sepsis and septic shock: current treatment strategies and new approaches. Eurasian J Med 2017; 49 (1): 53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arnold C. News feature: the quest to solve sepsis. Proc Natl Acad Sci USA 2018; 115 (16): 3988–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iskander KN, Osuchowski MF, Stearns-Kurosawa DJ, et al. Sepsis: multiple abnormalities, heterogeneous responses, and evolving understanding. Physiol Rev 2013; 93 (3): 1247–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jawad I, Lukšić I, Rafnsson SB. Assessing available information on the burden of sepsis: global estimates of incidence, prevalence and mortality. J Glob Health 2012; 2 (1): 010404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Islam MM, Nasrin T, Walther BA, et al. Prediction of sepsis patients using machine learning approach: a meta-analysis. Comput Methods Programs Biomed 2019; 170: 1–9. [DOI] [PubMed] [Google Scholar]
- 10. Schinkel M, Paranjape K, Nannan Panday RS, et al. Clinical applications of artificial intelligence in sepsis: a narrative review. Comput Biol Med 2019; 115: 103488. [DOI] [PubMed] [Google Scholar]
- 11. Wulff A, Montag S, Marschollek M, et al. Clinical decision-support systems for detection of systemic inflammatory response syndrome, sepsis, and septic shock in critically Ill patients: a systematic review. Methods Inf Med 2019; 58 (S 02): e43–57. [DOI] [PubMed] [Google Scholar]
- 12. Teng AK, Wilcox AB. A review of predictive analytics solutions for sepsis patients. Appl Clin Inform 2020; 11 (3): 387–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fleuren LM, Klausch TLT, Zwager CL, et al. Machine learning for the prediction of sepsis: a systematic review and meta-analysis of diagnostic test accuracy. Intensive Care Med 2020; 46 (3): 383–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giacobbe DR, Signori A, Del Puente F, et al. Early detection of sepsis with machine learning techniques: a brief clinical perspective. Front Med (Lausanne) 2021; 8: 617486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Assale M, Dui LG, Cina A, et al. The revival of the notes field: leveraging the unstructured content in electronic health records. Front Med (Lausanne) 2019; 6: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tayefi M, Ngo P, Chomutare T, et al. Challenges and opportunities beyond structured data in analysis of electronic health records. Wiley Interdiscip Rev Comput Stat 2021; 13: e1549. doi:10.1002/wics.1549 [Google Scholar]
- 17. Sheikhalishahi S, Miotto R, Dudley JT, et al. Natural language processing of clinical notes on chronic diseases: systematic review. JMIR Med Inform 2019; 7 (2): e12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Y, Wang L, Rastegar-Mojarad M, et al. Clinical information extraction applications: a literature review. J Biomed Inform 2018; 77: 34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Datta S, Bernstam EV, Roberts K. A frame semantic overview of NLP-based information extraction for cancer-related EHR notes. J Biomed Inform 2019; 100: 103301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jackson RG, Patel R, Jayatilleke N, et al. Natural language processing to extract symptoms of severe mental illness from clinical text: the Clinical Record Interactive Search Comprehensive Data Extraction (CRIS-CODE) project. BMJ Open 2017; 7 (1): e012012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kreimeyer K, Foster M, Pandey A, et al. Natural language processing systems for capturing and standardizing unstructured clinical information: a systematic review. J Biomed Inform 2017; 73: 14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spasic I, Nenadic G. Clinical text data in machine learning: systematic review. JMIR Med Inform 2020; 8 (3): e17984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elkin PL, Froehling D, Wahner-Roedler D, et al. NLP-based identification of pneumonia cases from free-text radiological reports. AMIA Annu Symp Proc November 8–12, 2008; 2008: 172–6; Washington, DC. [PMC free article] [PubMed] [Google Scholar]
- 24. Jensen K, Soguero-Ruiz C, Oyvind Mikalsen K, et al. Analysis of free text in electronic health records for identification of cancer patient trajectories. Sci Rep 2017; 7: 46226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ford E, Carroll JA, Smith HE, et al. Extracting information from the text of electronic medical records to improve case detection: a systematic review. J Am Med Inform Assoc 2016; 23 (5): 1007–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Soguero-Ruiz C, Hindberg K, Mora-Jiménez I, et al. Predicting colorectal surgical complications using heterogeneous clinical data and kernel methods. J Biomed Inform 2016; 61: 87–96. [DOI] [PubMed] [Google Scholar]
- 27. Huddar V, Desiraju BK, Rajan V, et al. Predicting complications in critical care using heterogeneous clinical data. IEEE Access 2016; 4: 7988–8001. [Google Scholar]
- 28. Ribelles N, Jerez JM, Rodriguez-Brazzarola P, et al. Machine learning and natural language processing (NLP) approach to predict early progression to first-line treatment in real-world hormone receptor-positive (HR+)/HER2-negative advanced breast cancer patients. Eur J Cancer 2021; 144: 224–31. [DOI] [PubMed] [Google Scholar]
- 29. Friedlin J, Overhage M, Al-Haddad MA, et al. Comparing methods for identifying pancreatic cancer patients using electronic data sources. AMIA Annu Symp Proc November 13–17, 2010; 2010: 237–41; Washington, DC. [PMC free article] [PubMed] [Google Scholar]
- 30. Van Vleck TT, Chan L, Coca SG, et al. Augmented intelligence with natural language processing applied to electronic health records for identifying patients with non-alcoholic fatty liver disease at risk for disease progression. Int J Med Inform 2019; 129: 334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. DeLisle S, Kim B, Deepak J, et al. Using the electronic medical record to identify community-acquired pneumonia: toward a replicable automated strategy. PLoS One 2013; 8 (8): e70944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gundlapalli AV, South BR, Phansalkar S, et al. Application of natural language processing to VA electronic health records to identify phenotypic characteristics for clinical and research purposes. Summit Transl Bioinform March 10–12, 2008; 2008: 36–40; San Francisco, CA. [PMC free article] [PubMed] [Google Scholar]
- 33. Ananthakrishnan AN, Cai T, Savova G, et al. Improving case definition of Crohn’s disease and ulcerative colitis in electronic medical records using natural language processing: a novel informatics approach. Inflamm Bowel Dis 2013; 19 (7): 1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc 2012; 19 (e1): e162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liao KP, Cai T, Gainer V, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2010; 62 (8): 1120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carroll RJ, Eyler AE, Denny JC. Naïve electronic health record phenotype identification for Rheumatoid arthritis. AMIA Annu Symp Proc October 22–26, 2011; 2011: 189–96; Washington, DC. [PMC free article] [PubMed] [Google Scholar]
- 37. Xia Z, Secor E, Chibnik LB, et al. Modeling disease severity in multiple sclerosis using electronic health records. PLoS One 2013; 8 (11): e78927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. DeLisle S, South B, Anthony JA, et al. Combining free text and structured electronic medical record entries to detect acute respiratory infections. PLoS One 2010; 5 (10): e13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng H, Gaff H, Smith G, et al. Epidemic surveillance using an electronic medical record: an empiric approach to performance improvement. PLoS One 2014; 9 (7): e100845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bhattacharjee P, Edelson DP, Churpek MM. Identifying patients with sepsis on the hospital wards. Chest 2017; 151 (4): 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Despins LA. Automated detection of sepsis using electronic medical record data: a systematic review. J Healthc Qual 2017; 39 (6): 322–33. [DOI] [PubMed] [Google Scholar]
- 42. Peiffer-Smadja N, Rawson TM, Ahmad R, et al. Machine learning for clinical decision support in infectious diseases: a narrative review of current applications. Clin Microbiol Infect 2020; 26 (5): 584–95. [DOI] [PubMed] [Google Scholar]
- 43. Freeman R, Moore LSP, García Álvarez L, et al. Advances in electronic surveillance for healthcare-associated infections in the 21st century: a systematic review. J Hosp Infect 2013; 84 (2): 106–19. [DOI] [PubMed] [Google Scholar]
- 44. de Bruin JS, Seeling W, Schuh C. Data use and effectiveness in electronic surveillance of healthcare associated infections in the 21st century: a systematic review. J Am Med Inform Assoc 2014; 21 (5): 942–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luz CF, Vollmer M, Decruyenaere J, et al. Machine learning in infection management using routine electronic health records: tools, techniques, and reporting of future technologies. Clin Microbiol Infect 2020; 26 (10): 1291–9. [DOI] [PubMed] [Google Scholar]
- 46. Moher D, Liberati A, Tetzlaff J, et al. ; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009; 6 (7): e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Horng S, Sontag DA, Halpern Y, et al. Creating an automated trigger for sepsis clinical decision support at emergency department triage using machine learning. PLoS One 2017; 12 (4): e0174708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Apostolova E, Velez T. Toward automated early sepsis alerting: identifying infection patients from nursing notes. In: BioNLP 2017. Association for Computational Linguistics August 4, 2017: 257–62; Vancouver, Canada. doi:10.18653/v1/W17-2332
- 49. Culliton P, Levinson M, Ehresman A, et al. Predicting Severe Sepsis Using Text from the Electronic Health Record. In: Workshop on Machine Learning For Health at the Conference on Neural Information Processing Systems (NIPS ML4H 2017), December 8, 2017. arXiv: 1711.11536; Long Beach, CA.
- 50. Liu R, Greenstein JL, Sarma SV, et al. Natural language processing of clinical notes for improved early prediction of septic shock in the ICU. Annu Int Conf IEEE Eng Med Biol Soc July 23–27, 2019; 2019: 6103–8; Berlin, Germany. doi:10.1109/EMBC.2019.8857819 [DOI] [PubMed] [Google Scholar]
- 51. Delahanty RJ, Alvarez J, Flynn LM, et al. Development and evaluation of a machine learning model for the early identification of patients at risk for sepsis. Ann Emerg Med 2019; 73 (4): 334–44. [DOI] [PubMed] [Google Scholar]
- 52. Goh KH, Wang L, Yeow AYK, et al. Artificial intelligence in sepsis early prediction and diagnosis using unstructured data in healthcare. Nat Commun 2021; 12 (1): 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Amrollahi F, Shashikumar SP, Razmi F, et al. Contextual embeddings from clinical notes improves prediction of sepsis. AMIA Annu Symp Proc November 14–18, 2020; 2020: 197–202; Virtual event. [PMC free article] [PubMed] [Google Scholar]
- 54. Hammoud I, Ramakrishnan IV, Henry M, et al. Multimodal early septic shock prediction model using lasso regression with decaying response. In: 2020 IEEE International Conference on Healthcare Informatics (ICHI). IEEE November 30–December 3, 2020; Virtual conference in Oldenburg, Germany. doi:10.1109/ichi48887.2020.9374377
- 55. Qin F, Madan V, Ratan U, et al. Improving early sepsis prediction with multi modal learning, 2021. arXiv: 2107.11094
- 56. Bouam S, Girou E, Brun-Buisson C, et al. An intranet-based automated system for the surveillance of nosocomial infections: prospective validation compared with physicians’ self-reports. Infect Control Hosp Epidemiol 2003; 24 (1): 51–5. [DOI] [PubMed] [Google Scholar]
- 57. Koller W, Blacky A, Bauer C, et al. Electronic surveillance of healthcare-associated infections with MONI-ICU–a clinical breakthrough compared to conventional surveillance systems. Stud Health Technol Inform 2010; 160 (Pt 1): 432–6. [PubMed] [Google Scholar]
- 58. Penz JFE, Wilcox AB, Hurdle JF. Automated identification of adverse events related to central venous catheters. J Biomed Inform 2007; 40 (2): 174–82. [DOI] [PubMed] [Google Scholar]
- 59. Proux D, Marchal P, Segond F, et al. Natural language processing to detect risk patterns related to hospital acquired infections. In: Proceedings of the Workshop on Biomedical Information Extraction: Association for Computational Linguistics; September 18, 2009: 35–41; Borovets, Bulgaria. doi:10.5555/1859776.1859782
- 60. Bouzbid S, Gicquel Q, Gerbier S, et al. Automated detection of nosocomial infections: evaluation of different strategies in an intensive care unit 2000-2006. J Hosp Infect 2011; 79 (1): 38–43. [DOI] [PubMed] [Google Scholar]
- 61. Jo Y, Loghmanpour N, Rosé CP. Time series analysis of nursing notes for mortality prediction via a state transition topic model. In: Proceedings of the 24th ACM International on Conference on Information and Knowledge Management (CIKM). Association for Computing Machinery October 18–23, 2015: 1171–80; Melbourne, Australia. doi:10.1145/2806416.2806541
- 62. Wang T, Velez T, Apostolova E, et al. Semantically Enhanced Dynamic Bayesian Network for Detecting Sepsis Mortality Risk in ICU Patients with Infection, 2018. arXiv: 1806.10174
- 63. Baghdadi Y, Bourrée A, Robert A, et al. Automatic classification of free-text medical causes from death certificates for reactive mortality surveillance in France. Int J Med Inform 2019; 131: 103915. [DOI] [PubMed] [Google Scholar]
- 64. Guo W, Xu Z, Ye X, et al. A time-critical topic model for predicting the survival time of sepsis patients. Sci Program 2020; 2020: 1. doi:10.1155/2020/8884539 [Google Scholar]
- 65. Ribas Ripoll VJ, Vellido A, Romero E, et al. Sepsis mortality prediction with the Quotient Basis Kernel. Artif Intell Med 2014; 61: 45–52. [DOI] [PubMed] [Google Scholar]
- 66. Halpern Y, Horng S, Choi Y, et al. Electronic medical record phenotyping using the anchor and learn framework. J Am Med Inform Assoc 2016; 23 (4): 731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vilic A, Petersen JA, Hoppe K, et al. Visualizing patient journals by combining vital signs monitoring and natural language processing. Annu Int Conf IEEE Eng Med Biol Soc August 16–20, 2016; 2016: 2529–32; Orlando, FL. doi:10.1109/EMBC.2016.7591245 [DOI] [PubMed] [Google Scholar]
- 68. Zhu X, Plasek JM, Tang C, et al. Embedding, aligning and reconstructing clinical notes to explore sepsis. BMC Res Notes 2021; 14 (1): 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Marks M, Pollock E, Armstrong M, et al. Needles and the damage done: reasons for admission and financial costs associated with injecting drug use in a Central London Teaching Hospital. J Infect 2013; 66 (1): 95–102. [DOI] [PubMed] [Google Scholar]
- 70. Ippolito P, Larson EL, Furuya EY, et al. Utility of electronic medical records to assess the relationship between parenteral nutrition and central line-associated bloodstream infections in adult hospitalized patients. JPEN J Parenter Enteral Nutr 2015; 39 (8): 929–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu YZ, Chu R, Lee A, et al. A surveillance method to identify patients with sepsis from electronic health records in Hong Kong: a single centre retrospective study. BMC Infect Dis 2020; 20 (1): 652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Murff HJ, FitzHenry F, Matheny ME, et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA 2011; 306 (8): 848–55. [DOI] [PubMed] [Google Scholar]
- 73. FitzHenry F, Murff HJ, Matheny ME, et al. Exploring the frontier of electronic health record surveillance: the case of postoperative complications. Med Care 2013; 51 (6): 509–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Piscitelli A, Bevilacqua L, Labella B, et al. A keyword approach to identify adverse events within narrative documents from 4 Italian institutions. J Patient Saf. doi:10.1097/PTS.0000000000000783 [DOI] [PubMed] [Google Scholar]
- 75. Vermassen J, Colpaert K, De Bus L, et al. Automated screening of natural language in electronic health records for the diagnosis septic shock is feasible and outperforms an approach based on explicit administrative codes. J Crit Care 2020; 56: 203–7. [DOI] [PubMed] [Google Scholar]
- 76. Saeed M, Villarroel M, Reisner AT, et al. Multiparameter intelligent monitoring in intensive care II: a public-access intensive care unit database. Crit Care Med 2011; 39 (5): 952–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3: 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Henry KE, Hager DN, Pronovost PJ, et al. A targeted real-time early warning score (TREWScore) for septic shock. Sci Transl Med 2015; 7 (299): 299ra122. [DOI] [PubMed] [Google Scholar]
- 79. Angus DC, Linde-Zwirble WT, Lidicker J, et al. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med 2001; 29 (7): 1303–10. [DOI] [PubMed] [Google Scholar]
- 80. Rhee C, Dantes R, Epstein L, et al. ; CDC Prevention Epicenter Program. Incidence and trends of sepsis in US hospitals using clinical vs claims data, 2009-2014. JAMA 2017; 318 (13): 1241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Reyna MA, Josef C, Seyedi S, et al. Early prediction of sepsis from clinical data: the PhysioNet/Computing in Cardiology Challenge 2019. In: 2019 Computing in Cardiology (CinC) September 8–11, 2019: 1–4; Singapore. doi:10.23919/CinC49843.2019.9005736
- 82. Wagner MM, Hogan WR, Chapman WW, et al. Chief complaints and ICD codes. Handbook of Biosurveillance 2006: 333. [Google Scholar]
- 83. Lucian J., Davis JFM. History and physical examination. Murray and Nadel’s Textbook of Respiratory Medicine 2016: 263.doi:10.1016/B978-1-4557-3383-5.00016-6 [Google Scholar]
- 84. Aghili H, Mushlin RA, Williams RM, et al. Progress notes model. In: Proceedings of AMIA Annual Fall Symposium October 25–29, 1997: 12–6; Nashville, TN. [PMC free article] [PubMed]
- 85. Hellesø R. Information handling in the nursing discharge note. J Clin Nurs 2006; 15 (1): 11–21. [DOI] [PubMed] [Google Scholar]
- 86. Ioanna P, Stiliani K, Vasiliki B. Nursing documentation and recording systems of nursing care. Health Sci J 2007; (4): 7.https://www.hsj.gr/medicine/nursing-documentation-and-recording-systems-of-nursing-care.php?aid=3680 Accessed June 29, 2021. [Google Scholar]
- 87. Flink M, Ekstedt M. Planning for the discharge, not for patient self-management at home - an observational and interview study of hospital discharge. Int J Integr Care 2017; 17 (6): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Stopford E, Ninan S, Spencer N. How to write a discharge summary. BMJ 2015; 351: h2696. [Google Scholar]
- 89. Sørby ID, Nytrø Ø. Does the electronic patient record support the discharge process? A study on physicians’ use of clinical information systems during discharge of patients with coronary heart disease. Health Inf Manag 2006; 34 (4): 112–9. [DOI] [PubMed] [Google Scholar]
- 90. Kind AJH, Smith MA. Documentation of mandated discharge summary components in transitions from acute to subacute care. In: Henriksen K, Battles JB, Keyes MA, et al. , eds. Advances in Patient Safety: New Directions and Alternative Approaches (Vol. 2: Culture and Redesign. AHRQ Publication No. 08-0034-2). Rockville, MD: Agency for Healthcare Research and Quality (US; ); 2011. [PubMed] [Google Scholar]
- 91. Jensdóttir A-B, Jónsson P, Noro A, et al. Comparison of nurses’ and physicians’ documentation of functional abilities of older patients in acute care–patient records compared with standardized assessment. Scand J Caring Sci 2008; 22 (3): 341–7. [DOI] [PubMed] [Google Scholar]
- 92. Penoyer DA, Cortelyou-Ward KH, Noblin AM, et al. Use of electronic health record documentation by healthcare workers in an acute care hospital system. J Healthc Manag 2014; 59: 130–44. [PubMed] [Google Scholar]
- 93. Furniss D, Lyons I, Franklin BD, et al. Procedural and documentation variations in intravenous infusion administration: a mixed methods study of policy and practice across 16 hospital trusts in England. BMC Health Serv Res 2018; 18 (1): 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sohn S, Wang Y, Wi C-I, et al. Clinical documentation variations and NLP system portability: a case study in asthma birth cohorts across institutions. J Am Med Inform Assoc 2018; 25 (3): 353–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. McPherson K. International differences in medical care practices. Health Care Financ Rev 1989; 1989: 9–20. [PMC free article] [PubMed] [Google Scholar]
- 96. Georgopoulos BS. Organization structure and the performance of hospital emergency services. Ann Emerg Med 1985; 14 (7): 677–84. [DOI] [PubMed] [Google Scholar]
- 97. McKinlay J, Link C, Marceau L, et al. How do doctors in different countries manage the same patient? Results of a factorial experiment. Health Serv Res 2006; 41 (6): 2182–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sørby ID, Nytrø Ø. Analysis of communicative behaviour: profiling roles and activities. Int J Med Inform 2010; 79 (6): e144-51–e151. [DOI] [PubMed] [Google Scholar]
- 99. Røst TB, Tvedt CR, Husby H, et al. Identifying catheter-related events through sentence classification. Int J Data Min Bioinform 2020; 23 (3): 213–33. [Google Scholar]
- 100. Harris ZS. Distributional Structure. Word World 1954; 10 (2–3): 146–62. [Google Scholar]
- 101. Le Q, Mikolov T. Distributed representations of sentences and documents. In: Proceedings of the 31st International Conference on International Conference on Machine Learning - Volume 32. JMLR.org June 21–26, 2014: II-1188–96; Beijing, China. doi:10.5555/3044805.3045025
- 102. Blei DM, Ng AY, Jordan MI. Latent Dirichlet allocation. J Mach Learn Res 2003; 3: 993–1022. doi:10.5555/944919.944937 [Google Scholar]
- 103. Mikolov T, Sutskever I, Chen K, et al. Distributed Representations of Words and Phrases and their Compositionality. 2013. arXiv:1310.4546
- 104. Pennington J, Socher R, Manning C. GloVe: Global Vectors for Word Representation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP). Association for Computational Linguistics October 25–29, 2014: 1532–43; Doha, Qatar. doi:10.3115/v1/D14-1162
- 105. Mikolov T, Chen K, Corrado G, et al. Efficient Estimation of Word Representations in Vector Space. 2013. arXiv: 1301.3781
- 106. Devlin J, Chang M-W, Lee K, et al. BERT: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers). Association for Computational Linguistics June 2–7, 2019: 4171–86; Minneapolis, MN. doi:10.18653/v1/N19-1423
- 107. Alsentzer E, Murphy J, Boag W, et al. Publicly available clinical BERT Embeddings. In: Proceedings of the 2nd Clinical Natural Language Processing Workshop. Association for Computational Linguistics June 7, 2019: 72–8; Minneapolis, MN. doi:10.18653/v1/W19-1909
- 108. Koller D, Friedman N. Probabilistic Graphical Models: Principles and Techniques. Cambridge, MA and London, England: MIT Press, 2009. [Google Scholar]
- 109. Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995; 20 (3): 273–97. [Google Scholar]
- 110. Fix E, Hodges JL. Nonparametric discrimination: consistency properties. Randolph Field, Texas, Project 1951: 21–49.
- 111. Breiman L. Random forests. Mach Learn 2001; 45 (1): 5–32. [Google Scholar]
- 112. Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Stat 1999; 29: 1189–232. http://www.jstor.org/stable/2699986 Accessed May 3, 2021. [Google Scholar]
- 113. Friedman JH. Stochastic gradient boosting. Comput Stat Data Anal 2002; 38 (4): 367–78. [Google Scholar]
- 114. Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature 1986; 323 (6088): 533–6. [Google Scholar]
- 115. Rumelhart DE, Hinton GE, Williams RJ. Learning internal representations by error propagation In: Rumelhart DE, McClelland JL, eds. Parallel Distributed Processing: Explorations in the Microstructure of Cognition: Foundations. MIT Press; 1986: 318–62. https://ieeexplore.ieee.org/document/6302929 Accessed May 3, 2021. [Google Scholar]
- 116. Mikolov T, Karafiát M, Burget L, et al. Recurrent neural network based language model. In: INTERSPEECH 2010. ISCA September 26–30, 2010; Makuhari, Chiba, Japan.doi: 10.21437/Interspeech.2010-343
- 117. Cho K, van Merrienboer B, Gulcehre C, et al. Learning phrase representations using RNN Encoder-Decoder for Statistical Machine Translation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP). Association for Computational Linguistics October 25–29, 2014: 1724–34; Doha, Qatar. doi:10.3115/v1/D14-1179
- 118. Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput 1997; 9 (8): 1735–80. [DOI] [PubMed] [Google Scholar]
- 119. Cawcutt KA, Peters SG. Severe sepsis and septic shock: clinical overview and update on management. Mayo Clin Proc 2014; 89 (11): 1572–8. [DOI] [PubMed] [Google Scholar]
- 120. Hotchkiss RS, Moldawer LL, Opal SM, et al. Sepsis and septic shock. Nat Rev Dis Primers 2016; 2: 16045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Bone RC, Balk RA, Cerra FB, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest 1992; 101 (6): 1644–55. [DOI] [PubMed] [Google Scholar]
- 122. Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med 1996; 22 (7): 707–10. [DOI] [PubMed] [Google Scholar]
- 123. Seymour CW, Liu VX, Iwashyna TJ, et al. Assessment of clinical criteria for sepsis: for the third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016; 315 (8): 762–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Subbe CP, Kruger M, Rutherford P, et al. Validation of a modified early warning score in medical admissions. QJM 2001; 94 (10): 521–6. [DOI] [PubMed] [Google Scholar]
- 125. Rhodes A, Evans LE, Alhazzani W, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Med 2017; 43 (3): 304–77. [DOI] [PubMed] [Google Scholar]
- 126. Dellinger RP, Levy MM, Rhodes A, et al. ; Surviving Sepsis Campaign Guidelines Committee including the Pediatric Subgroup. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med 2013; 41 (2): 580–637. [DOI] [PubMed] [Google Scholar]
- 127. Vincent J-L. The clinical challenge of sepsis identification and monitoring. PLoS Med 2016; 13 (5): e1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Abe T, Tokuda Y, Shiraishi A, et al. ; JAAM SPICE Study Group. In-hospital mortality associated with the misdiagnosis or unidentified site of infection at admission. Crit Care 2019; 23 (1): 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Rothberg MB, Pekow PS, Priya A, et al. Variation in diagnostic coding of patients with pneumonia and its association with hospital risk-standardized mortality rates: a cross-sectional analysis. Ann Intern Med 2014; 160 (6): 380–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Bewick T, Simmonds M, Chikhani M, et al. Pneumonia in the context of severe sepsis: a significant diagnostic problem. Eur Respir J 2008; 32 (5): 1417–8. [DOI] [PubMed] [Google Scholar]
- 131. Rhee C, Murphy MV, Li L, et al. ; for the Centers for Disease Control and Prevention Epicenters Program. Comparison of trends in sepsis incidence and coding using administrative claims versus objective clinical data. Clin Infect Dis 2015; 60 (1): 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Tidswell R, Inada-Kim M, Singer M. Sepsis: the importance of an accurate final diagnosis. Lancet Respir Med 2021; 9 (1): 17–8. [DOI] [PubMed] [Google Scholar]
- 133. Lopansri BK, Miller RR III, Burke JP, et al. Physician agreement on the diagnosis of sepsis in the intensive care unit: estimation of concordance and analysis of underlying factors in a multicenter cohort. J Intensive Care 2019; 7: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Rhee C, Kadri SS, Danner RL, et al. Diagnosing sepsis is subjective and highly variable: a survey of intensivists using case vignettes. Crit Care 2016; 20: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Rhee C, Klompas M. Sepsis trends: increasing incidence and decreasing mortality, or changing denominator? J Thorac Dis 2020; 12 (Suppl 1): S89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Yu SC, Betthauser KD, Gupta A, et al. Comparison of sepsis definitions as automated criteria. Crit Care Med 2021; 49 (4): e433–43. [DOI] [PubMed] [Google Scholar]
- 137. Liu R, Greenstein JL, Granite SJ, et al. Data-driven discovery of a novel sepsis pre-shock state predicts impending septic shock in the ICU. Sci Rep 2019; 9 (1): 6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Walkey AJ, Shieh M-S, Liu VX, et al. Mortality measures to profile hospital performance for patients with septic shock. Crit Care Med 2018; 46 (8): 1247–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Vincent J-L. The challenge of early identification of the hospital patient at risk of septic complications. Ann Transl Med 2017; 5 (3): 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Mendonça EA, Haas J, Shagina L, et al. Extracting information on pneumonia in infants using natural language processing of radiology reports. J Biomed Inform 2005; 38 (4): 314–21. [DOI] [PubMed] [Google Scholar]
- 141. Dublin S, Baldwin E, Walker RL, et al. Natural language processing to identify pneumonia from radiology reports. Pharmacoepidemiol Drug Saf 2013; 22 (8): 834–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Matheny ME, Fitzhenry F, Speroff T, et al. Detection of infectious symptoms from VA emergency department and primary care clinical documentation. Int J Med Inform 2012; 81 (3): 143–56. [DOI] [PubMed] [Google Scholar]
- 143. Wu S, Roberts K, Datta S, et al. Deep learning in clinical natural language processing: a methodical review. J Am Med Inform Assoc 2020; 27 (3): 457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Khattak FK, Jeblee S, Pou-Prom C, et al. A survey of word embeddings for clinical text. J Biomed Informatics: X 2019; 100: 100057. [DOI] [PubMed] [Google Scholar]
- 145. Névéol A, Zweigenbaum P; Section Editors for the IMIA Yearbook Section on Clinical Natural Language Processing. Expanding the diversity of texts and applications: findings from the section on clinical natural language processing of the international medical informatics association yearbook. Yearb Med Inform 2018; 27 (1): 193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Koleck TA, Dreisbach C, Bourne PE, et al. Natural language processing of symptoms documented in free-text narratives of electronic health records: a systematic review. J Am Med Inform Assoc 2019; 26 (4): 364–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Wang HE, Shapiro NI, Griffin R, et al. Chronic medical conditions and risk of sepsis. PLoS One 2012; 7 (10): e48307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Prescott HC, Carmichael AG, Langa KM, et al. Paths into sepsis: trajectories of presepsis healthcare use. Ann Am Thorac Soc 2019; 16 (1): 116–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Delano MJ, Ward PA. The immune system’s role in sepsis progression, resolution, and long-term outcome. Immunol Rev 2016; 274 (1): 330–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Faisal M, Scally A, Richardson D, et al. Development and external validation of an automated computer-aided risk score for predicting sepsis in emergency medical admissions using the patient’s first electronically recorded vital signs and blood test results. Crit Care Med 2018; 46 (4): 612–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.