Abstract
Objective
Reducing suicidal behavior among patients in the healthcare system requires accurate and explainable predictive models of suicide risk across diverse healthcare settings.
Materials and Methods
We proposed a general targeted fusion learning framework that can be used to build a tailored risk prediction model for any specific healthcare setting, drawing on information fusion from a separate more comprehensive dataset with indirect sample linkage through patient similarities. As a proof of concept, we predicted suicide-related hospitalizations for pediatric patients in a limited statewide Hospital Inpatient Discharge Dataset (HIDD) fused with a more comprehensive medical All-Payer Claims Database (APCD) from Connecticut.
Results
We built a suicide risk prediction model for the source data (APCD) and calculated patient risk scores. Patient similarity scores between patients in the source and target (HIDD) datasets using their demographic characteristics and diagnosis codes were assessed. A fused risk score was generated for each patient in the target dataset using our proposed targeted fusion framework. With this model, the averaged sensitivities at 90% and 95% specificity improved by 67% and 171%, and the positive predictive values for the combined fusion model improved 64% and 135% compared to the conventional model.
Discussion and Conclusions
We proposed a general targeted fusion learning framework that can be used to build a tailored predictive model for any specific healthcare setting. Results from this study suggest we can improve the performance of predictive models in specific target settings without complete integration of the raw records from external data sources.
Keywords: suicide, suicide attempt prediction, predictive modeling, fusion learning, transfer learning, electronic healthcare record
INTRODUCTION
Rising rates of suicidal behavior among children and adolescents constitute one of the United States’ most critical public health challenges.1–5 Death by suicide has increased by over 30% over the past 20 years and has become the second leading cause of death among youth ages 10–24.6,7 In the past 5 years, a great deal of effort has been directed at improving the identification of individuals at risk of suicidal behavior using clinically derived risk algorithms. Although such efforts have generated a handful of viable predictive models,8,9 the vast majority have been focused on adults. To date, there were very limited published suicide risk prediction models for children and adolescents, with both achieving good predictive performance albeit in limited patient populations.10,11
However, the most daunting challenge facing those seeking to use pediatric suicide risk algorithms may not be the development of the algorithms themselves; rather, the primary barrier involves the limited data available to most healthcare providers with which to apply these predictive models to their patients. The rich datasets that have generated the most comprehensive and accurate suicide risk algorithms are derived from large and sophisticated integrated delivery systems, health plans, and research networks. Health system-wide medical records data of this nature are not, and may likely never be, available to the vast majority of healthcare providers in the United States.
Transfer learning,12 a learning mechanism that aims to leverage the shared knowledge emerging from similar tasks using data from different contexts and scenarios, provides a promising avenue for improving predictive analytics across different health research domains. Despite its promise, it has only recently been deployed for healthcare analytics yet with encouraging results.13 For example, the effectiveness of transfer learning techniques has been demonstrated with a diverse set of medical image analysis problems.14–16 Recent studies have shown that transfer learning can significantly improve the early identification of patients at risk of Alzheimer’s disease using on their longitudinal clinical records.17 In a direct application of this approach to hospital data, Wiens et al18 showed that health information from multiple hospitals could be used to improve hospital-specific predictions of the risk of hospital-associated infection with Clostridium difficile using a transfer learning framework. Two challenges in suicide risk prediction are particularly well-suited to transfer learning approaches: first, the fragmentation of relevant patient health information across the care spectrum (eg, primary care, behavioral health, and hospital-based care), and second, the rarity of suicidal behavior as an outcome, which requires large, integrated datasets under conventional analytic scenarios. As the attribute of data fusion, we used the terminologies, transfer learning and fusion learning synonymously.
In this analysis, we propose a general targeted fusion learning framework that can be used to build a tailored predictive model for any specific healthcare setting. As a proof of concept, we apply our model using data from a large statewide All-Payer Claims Database (APCD), integrated with statewide inpatient hospital claims data, to develop and test pediatric suicide risk algorithms using principles associated with transfer learning.12 Our approach, which uses comprehensive clinical data to develop both a robust risk prediction model and a similarity matrix to link patients in a more limited database to the features in the risk prediction model, is a dramatic departure from previous efforts to develop suicide risk algorithms using clinical data. The use of data fusion techniques allows us to expressly target these algorithms to clinical settings with access to sparse and limited data, that is, the settings in which the vast majority of patients in the United States receive their healthcare.
MATERIALS AND METHODS
Targeted fusion learning framework
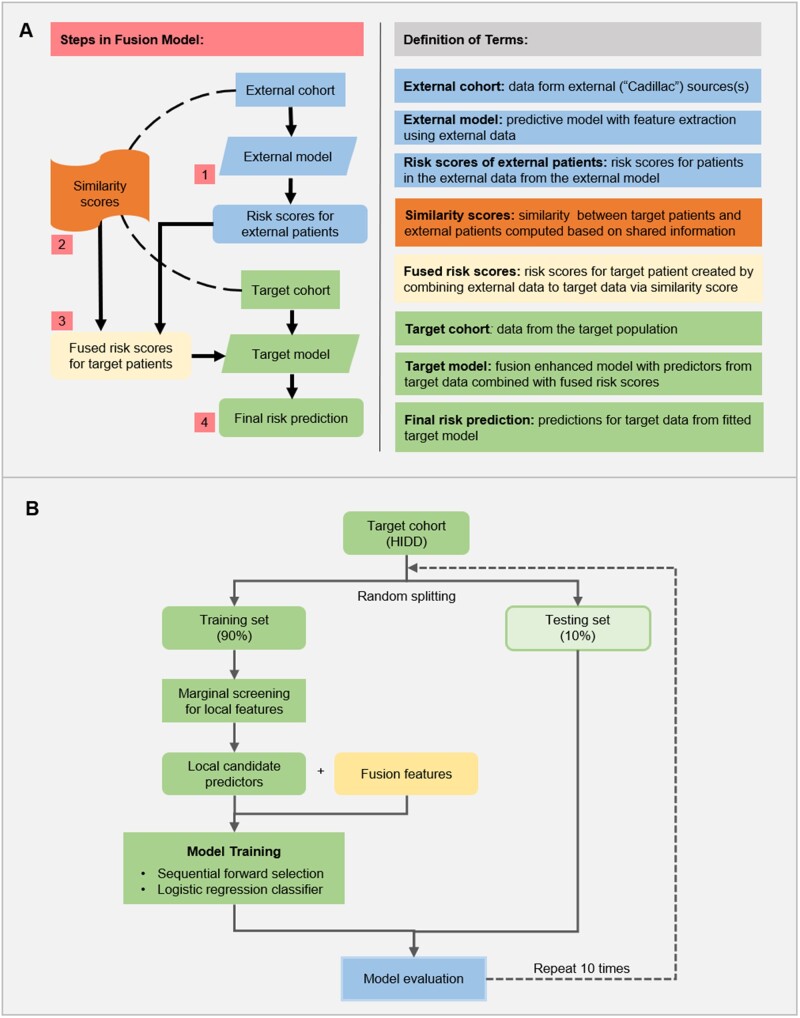
We describe our proposed targeted fusion learning framework that can be used to build a tailored predictive model for any specific healthcare setting. This approach is based on data from the patient population in the healthcare setting of interest (referred to as the target cohort), combined with data on patients from a larger and more comprehensive data source (referred to as the external cohort), such as a health information exchange or a research repository. Figure 1A illustrates our proposed approach. For generality, let and denote the response and predictor vector for the i-th target patient and the j-th external patient respectfully, where and for target sample size and external sample size
Figure 1.
Proposed targeted fusion learning framework. (A) The architecture of the general target fusion learning. (B) Model training and evaluation on the HIDD dataset.
Step 1
Generation of individual risk scores for external patients. Here, a model for predicting the risk levels of the patients from the external cohort is constructed. This model can incorporate different versions of risk scores that utilize multifold information and combine the strengths of different models. Validation of the initial risk model also typically includes review of the selected features by clinical experts.
Step 2
Construction of similarity scores between target and external patients, for . This step provides the foundation of information transfer and data fusion. The target and external cohorts do not necessarily have exactly the same candidate predictors. However, they generally have shared information domains which allow us to build a similarity measure to link the 2 sets of patients. For example, in our study of linking hospital inpatient data (target) and all-payers claims data (external), both datasets contained patient demographics and diagnosis codes. Different similarity/distance metrics, including Pearson correlation, Canberra distance, cosine distance, among others, could be considered in the computation of the pairwise similarity scores, based on the types of the available data.
Step 3
Computation of fused risk scores for target patients based on similarity scores and external risk scores. This step produces a set of projected risk scores for each patient in the target population, by combining his/her similarity measures with external patients obtained from Step 2 and the estimated risk scores of the external patients from Step 1:
| (1) |
where denotes a fusion function for integration of the target population and external population. As a simple example, the risk score can be computed as a similarity weighted average of the external scores, that is, an external patient with a higher risk score and is more similar to the target patient will contribute more, and vice versa.
Step 4
Predictive modeling for the target population assisted with the fused risk scores. This final step builds a predictive model for the target population, using both the target data and the fused risk scores from Step 3. Generally, the fused risk scores are used as predictors in the predictive model, which are subject to the same selection and shrinkage estimation process as the other predictors from the target data. The predicted value for i-th target patient can be formulated as:
| (2) |
Our targeted fusion learning approach provides a flexible way of utilizing available information from large external databases as an auxiliary information source for tailoring and improving the predictive model for a target population. An important benefit of this approach is that it does not require the difficult or even impossible task of fully integrating the target data with external data sources. Moreover, in our framework, different data sources may provide different sets of information, as long as there exists some overlap that allows for the calculation of a patient similarity measure; we do not require a unique identifier to link different datasets, and our approach can be implemented in a distributed fashion so that each provider does not have to gain full access of the external database.
Data and study cohorts
The 2 datasets used in this study were: the Connecticut APCD, which contained medical and pharmacy claims for Connecticut residents from January 1, 2012 to December 31, 2017; and the Connecticut Hospital Inpatient Discharge Dataset (HIDD), which contained inpatient hospitalizations from all acute care hospitals in the state from January 1, 2012 to September 30, 2017. The APCD contains both inpatient and outpatient encounters from approximately 35% of the commercially insured Connecticut population, while the HIDD contains a census of all inpatient hospitalizations across all insurers (including Medicaid and Medicare) during this period. Suicide attempts (SAs) were identified using ICD-9 diagnostic codes and code combinations, all the combinations are listed in Supplementary Table S1.19,20
Target cohort
The target cohort consisted of children, adolescents, and young adults aged 10–24 years who had at least one nonsuicide-related hospitalization from the HIDD data. This cohort had 38 806 patients with 485 suicide attempters. More specific preprocessing steps are included in Supplementary Table S2.
External cohort
The external cohort consisted of patients of the same age range 10–24 (in the year 2014) who had at least one nonsuicidal service claim within the recruiting window: January 1, 2014 to December 31, 2015 from the APCD data. Patients without continuous eligibility during the recruiting window, or with invalid enrollment were excluded.21 The illustration and the detailed description for the recruiting window are included in the Supplementary Material. This cohort had 155 486 patients with 2053 suicide attempters.
Event of interest
The event of interest was the first SA after the most recent nonsuicide-related hospitalization.
Candidate predictors
Historical information was aggregated from the first claim to the last nonsuicidal claim (or the last nonsuicidal within the recruiting window for APCD). Predictor variables included major demographic characteristics (age and gender) and ICD-9/10 diagnosis codes from each medical encounter. There were up to 10 primary and secondary diagnosis codes for each encounter or hospitalization. ICD-10 codes were converted back to ICD-9 codes using R package “touch” for consistency and were then grouped into larger categories using their first 3 digits.
Application of targeted fusion between HIDD and APCD
We have tailored the above general framework to build and validate setting-specific suicide prediction models, using the external data from APCD to improve the prediction of the target from HIDD (Figure 1B).
To build a suicide prediction model with external data, we focused on the prediction of the first SA using the APCD data. In particular, the risk scores for external patients were produced from an external model built by a “marginal screening + elastic-net regularized logistic regression” pipeline.5,21 Predictor screening was be done by fitting marginal models with each predictor and a set of control variables. Subsequentially, regularized statistical learning methods were used to conduct simultaneous risk factor identification and model estimation. The model hyperparameters were determined based on the Bayesian information criterion.22
- The similarities between the 2 sets of patients were computed based on demographics and diagnosis codes. We first require the exact match between the age group (10–14, 15–19, or 20–24) and gender (female or male), patients with different age group or gender are considered to have similarity score zero. The binary encoding of diagnosis codes (1 if the patients had this diagnosis code, 0 otherwise) is further used to compute the Pearson correlation coefficient. Without loss of generality, we assume that and are the common features between the target population and external population, recall Eq. (1) the similarity score for i-th target patient and j-th external patient is computed as:
(3) For each patient in the target cohort, we created a -order fused risk score—the summation of risk scores of the top most similar patients in the external cohort weighted by the calculated similarities. We choose from the grid of }. In other words, for each patient in the target cohort, we gathered information of his/her nearest neighbors in the external cohort to produce the -order fused risk score as a candidate predictor
Finally, we built a fusion enhanced model for the target HIDD cohort following the procedure illustrated in Figure 1B. (a) We first randomly divided the target cohort into 90% training and 10% testing. (b) On each training set, we perform marginal screening on the local features (demographics and diagnosis codes) first. The screening is based on Chi-square/Fisher’s exact test, the P values are further corrected to control the false discovery rate. The predictors with adjusted P values smaller than 0.1 are kept for the final modeling. (c) The marginal selected predictors combined with the k-order fused risk scores were used to build the predictive model. More specifically, we trained a logistic regression model with all candidate predictors available at the target and the augmented predictors derived by k-nearest neighbors-like risk scores from the external dataset, that is, , to predict the occurrence of SA via a forward selection procedure,23–25 which sequentially select predictor set to minimize the prediction error. In particular,
(i)
We initialized the predictor set as empty, that is, ;
-
(ii) In each step , we selected an optimal predictor , such that
(4) where is the prediction performance in terms of the area under the receiver operating characteristics curve (AUC) of the prediction model based on predictors from plus . To measure predictive performance, we introduced a 5-fold cross-validation strategy and calculated the mean of the AUC.
- (iii) Update
(5) (iv) go back to (ii).
Of note, as the number of selected predictors increased, the AUC first raised and then declined, because too many predictors will lead to overfitting of the model, that is, the model can well fit training set but fail to predict over test set. Therefore, we stopped the selection procedure when AUC begin to decline. We repeated the above random-splitting procedure 10 times to validate the effectiveness of our predictive model and identify the predictors with the strongest association with SAs.
Model evaluation
In order to estimate the proposed predictive model, we compared it with (1) what we refer to as the conventional model that was built based on local features of the target cohort only and (2) a model built using candidate predictors consisting of the fused risk scores only. To evaluate the predictive performance of the models, we examined out-of-sample performance metrics, including AUC, sensitivity, specificity, and positive predictive value (PPV). AUC is a broad metric of discrimination performance in the machine learning community that ranges from 0.5 (random guessing) to 1.0 (perfect prediction). Due to the high imbalance of the dataset, we calculated sensitivities when setting specificities to 90% and 95%, respectively. We also calculated PPV, which is the probability that predicted high-risk patients have actual SAs.
Model interpretation
To quantify the contributions to the final prediction from selected predictors, we counted the frequency that each predictor being selected among the 10 predictive models. The odds ratio of the top 30 most frequently selected risk factors as well as their averaged selection ranks are further computed. In order to explore how the fusion information has improved predictive modeling on the target cohort, we compared the logistic regression coefficients of the local predictors calculated with and without fusion features. Since we used the binary encoding of each predictor, the patient’s suicide risk score can be seen as the cumulation of coefficients of individual predictors associated with this patient. The suicide risk scores for patients in the testing sets were calculated, and the distribution of the scores among suicide attempters and nonattempters are compared under different modeling settings.
In addition to fitted coefficients, we performed statistical analysis to gain further insights into the reasons for the improved prediction performance. To be more specific, when comparing the predicted high-risk groups from the fusion enhanced model with actual suicide attempters, we examined whether there were any specific characteristics (ie, diagnosis codes as predictors) among the patients who were only correctly identified by the fusion model (“true” predictions). We computed the number of attempters with each diagnosis code who were identified correctly by both the conventional model and fusion enhanced model, or only by the fusion enhanced model. In other words, we developed a contingency table for each diagnosis code, where the rows correspond to the number of patients who had or did not have this code, while the columns indicate whether both the conventional and fusion enhanced models were correct or whether only the fusion enhanced model was correct. We further performed Fisher’s exact test to determine if the ratio of this code is significantly different between the 2 groups.26
RESULTS
Population demographics
Table 1 presents the distribution of demographic characteristics in the study population and methods used by attempters. The age reported for the HIDD was the age at the last non-SA record, while the age for the APCD was the age at the beginning of the recruiting window (January 1, 2014). Survival time was defined as the last non-SA record to the 1st SA, and the SA methods were derived from patients’ diagnosis codes at the 1st SA. The SA rates were similar in the HIDD (1.23%) and the APCD (1.32%); the HIDD had a higher proportion of females, and more patients in the 20–24 age group than the APCD. For suicide attempters, the age, gender, and SA method distributions were similar between the 2 cohorts.
Table 1.
Characteristics of the suicide attempt cases and controls for the study population
| Variable | HIDD (target cohort) |
APCD (external cohort) |
||
|---|---|---|---|---|
| Case | Control | Case | Control | |
| No. of patients | 485 | 38 806 | 2053 | 153 433 |
| Sex, N (%) | ||||
| Female | 308 (63.51) | 22 937 (59.11) | 1281 (62.4) | 76 533 (49.88) |
| Male | 177 (36.49) | 15 869 (40.89) | 772 (37.6) | 76 900 (50.12) |
| Age group, N (%) | ||||
| 10–14 years old | 72 (14.85) | 6266 (16.15) | 368 (17.92) | 46 374 (30.22) |
| 15–19 years old | 253 (52.16) | 12 798 (32.98) | 931 (45.35) | 53 184 (34.66) |
| 20–24 years old | 160 (32.99) | 19 742 (50.87) | 754 (36.73) | 53 875 (35.11) |
| Survival time, N (%) | ||||
| >1 year | 122 (25.15%) | – | 418 (20.36) | – |
| >3 years | 22 (4.54%) | – | <11 (<0.54) | – |
| Suicide attempt methods, N (%) | ||||
| Poisoning | 340 (70.1) | – | 1395 (67.95) | – |
| Cutting | 102 (21.03) | – | 546 (26.6) | – |
| Hanging | 9 (1.86) | – | 21 (1.02) | – |
| Firearm | <6 (<1.24) | – | <11 (<0.54) | – |
| Jumping | <6 (<1.24) | – | <11 (<0.54) | – |
| Others | 29 (5.98) | – | >61 (>2.97) | – |
APCD: All-Payer Claims Database.
Model performance
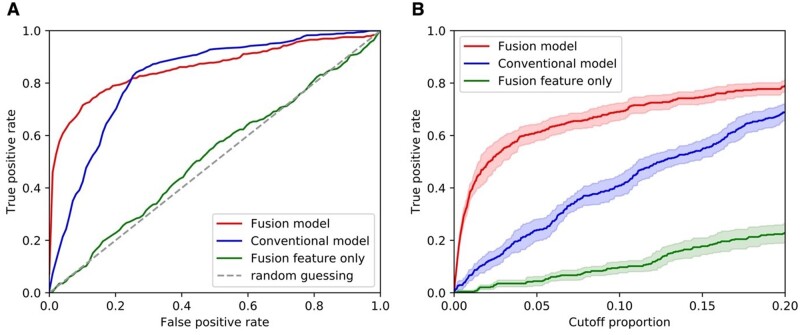
Results summarizing the quantitative performance of the conventional model and fusion enhanced model in predicting suicide risk in the target cohort, including receiver operating characteristic curves, AUC, and sensitivity and PPV at predefined specificity levels (90% specificity and 95% specificity) are shown in Table 2 and Figure 2. Overall, the proposed fusion enhanced model demonstrated much better predictive performance compared to the conventional model trained with local features only and the model trained with fused features only. Although the conventional model achieved good performance with an averaged AUC of 0.82 (95% CI [0.81, 0.84]), the proposed fusion enhanced model achieved an AUC of 0.86 (95% CI [0.84, 0.88]), and displayed sensitivities and PPVs at specific specificity levels that were significantly improved. In particular, the averaged sensitivities at 90% and 95% specificity improved by 67% and 171%, and the PPVs for the combined fusion model improved 64% and 135% compared to the conventional model (see Table 2 and Figure 2). In contrast, the model based on fused risk scores only showed poor prediction performance.
Table 2.
Performances of the predictive models
| Models | Candidate predictorsa | AUC (95% CI) | Sensitivity (SD) |
PPV (SD) |
||
|---|---|---|---|---|---|---|
| 90% specificity | 95% specificity | 90% specificity | 95% specificity | |||
| Fusion enhanced model | Local features + fused risk scores | 0.86 (0.84, 0.89) | 0.70 (0.03) | 0.65 (0.02) | 0.082 (0.008) | 0.134 (0.015) |
| Conventional model | Local features | 0.82 (0.81, 0.84) | 0.42 (0.07) | 0.24 (0.07) | 0.050 (0.009) | 0.057 (0.018) |
| Fusion only model | Fused risk scores | 0.52 (0.49, 0.55) | 0.10 (0.03) | 0.04 (0.03) | 0.012 (0.003) | 0.011 (0.006) |
AUC: area under the receiver operating characteristics curve; CI: confidence interval; PPV: positive predictive value; SD: standard deviation.
Local features included demographics and diagnosis codes collected in target cohort, that is, HIDD cohort.
Figure 2.
Prediction performances of the predictive models. A. The receiver operating characteristic curves of the predictive models. B. Curves showing true positive rate versus cutoff proportion of patients with top predicted risk scores.
Predictor importance
Table 3 depicts the top 30 predictors sorted by frequency and the average rank for being selected by the sequential forward selection procedure in our fusion enhanced model. Mental health-related diagnoses such as mood disorders, depressive disorders, other psychosocial circumstances, drug abuse, anxiety, drug dependence, and psychosocial circumstances, poisoning by analgesics antipyretics and antirheumatics, and age were the most important risk factors for suicidal behavior. Other diagnoses including acute appendicitis, general symptoms, and problems related to lifestyle were negatively associated with suicidal behavior. The 100- and 50-order fused risk scores were frequently selected as predictors, indicating that using information from the top 100 and 50 nearest neighbors in the external cohort improved the performance of the conventional model. As indicated in the right-hand column of Table 3, the signs of the predictors were uniformly positively or negatively associated with suicidal behavior across all the models in which they were selected.
Table 3.
Top 30 predictors selected by the proposed predictive model
| Predictors | Predictor category | Selection frequency | Averaged selection rank | Case exposed/case nonexposed | Control exposed/control nonexposed | Log odds ratio | Contribution +/− (count)a |
|---|---|---|---|---|---|---|---|
| Age 15–19 | Demographics | 1 | 10 | 253/232 | 12 798/26 008 | 0.8 | 10/0 |
| 100-order fused risk score | Fusion | 0.9 | 12.2 | – | – | – | 0/9 |
| 50-order fused risk score | Fusion | 0.7 | 3 | – | – | – | 0/7 |
| ICD-9 V27, Outcome of delivery | Medical condition | 1 | 4 | 11/474 | 8113/30 693 | −2.43 | 0/10 |
| ICD-9 540, Acute appendicitis | Medical condition | 1 | 8.4 | (1,6)/484 | 2003/36 803 | (−3.27, −1.48) | 0/10 |
| ICD-9473, Chronic sinusitis | Medical condition | 1 | 14.3 | 7/478 | 179/38 627 | 1.15 | 10/0 |
| ICD-9 780, General symptoms | Medical condition | 0.6 | 19.3 | 52/433 | 2902/35 904 | 0.4 | 0/6 |
| ICD-9 682, Other cellulitis and abscess | Medical condition | 0.4 | 12.7 | 5/480 | 1146/37 660 | −1.07 | 0/4 |
| ICD-9 648, Other current conditions in the mother classifiable elsewhere but complicating pregnancy childbirth or the puerperium | Medical condition | 0.4 | 19.7 | 10/475 | 4103/34 703 | −1.73 | 4/0 |
| ICD-9 659, Other indications for care or intervention related to labor and delivery not elsewhere classified | Medical condition | 0.3 | 17.3 | (1,6)/483 | 1986/36 820 | (−3.26, −1.47) | 1/2 |
| ICD-9 724, Other and unspecified disorders of back | Medical condition | 0.3 | 20 | 14/471 | 480/38 326 | 0.86 | 3/0 |
| ICD-9 296, Episodic mood disorders | Mental health | 1 | 1 | 334/151 | 8114/30 692 | 2.12 | 10/0 |
| ICD-9 311, Depressive disorder, not elsewhere classified | Mental health | 1 | 2 | 128/357 | 3560/35 246 | 1.27 | 10/0 |
| ICD-9 V62, Other psychosocial circumstances | Mental health | 1 | 5.2 | 214/271 | 3954/34 852 | 1.94 | 10/0 |
| ICD-9 300, Anxiety, dissociative and somatoform disorders | Mental health | 1 | 6.9 | 203/282 | 5713/33 093 | 1.43 | 10/0 |
| ICD-9298, Other nonorganic psychoses | Mental health | 0.8 | 13.2 | 34/451 | 1061/37 745 | 0.99 | 10/0 |
| ICD9-307, Special symptoms or syndromes not elsewhere classified | Mental health | 0.8 | 14.2 | 44/441 | 869/37 937 | 1.47 | 10/0 |
| ICD-9 308, Acute reaction to stress | Mental health | 0.8 | 17.3 | (1,6)/480 | 65/38 741 | (0.22, 2.01) | 10/0 |
| ICD-9 295, Schizophrenic disorders | Mental health | 0.6 | 14.1 | 19/466 | 672/38 134 | 0.84 | 6/0 |
| ICD-9 312, Disturbance of conduct not elsewhere classified | Mental health | 0.5 | 17.8 | 20/465 | 675/38 131 | 0.89 | 5/0 |
| ICD-9 309, Adjustment reaction | Mental health | 0.3 | 17.3 | 95/390 | 2035/36 771 | 1.48 | 3/0 |
| ICD-9 V17, Family history of certain chronic disabling diseases | Other | 0.4 | 19.2 | 47/438 | 1587/37 219 | 0.92 | 0/4 |
| ICD-9 V69, Problems related to lifestyle | SDoH | 0.5 | 16.8 | 9/476 | 282/38 524 | 0.95 | 0/5 |
| ICD-9 V60, Housing household and economic circumstances | SDoH | 0.5 | 20.6 | 14/471 | 294/38 512 | 1.36 | 5/0 |
| ICD-9 V61, Other family circumstances | SDoH | 0.3 | 20 | 67/418 | 1196/37 610 | 1.62 | 3/0 |
| ICD-9 304, Drug dependence | Substance use | 1 | 10 | 55/430 | 1457/37 349 | 1.19 | 10/0 |
| ICD-9 965, Poisoning by analgesics antipyretics and antirheumatics | Substance use | 1 | 10.7 | 15/470 | 205/38 601 | 1.79 | 10/0 |
| ICD-9 303, Alcohol dependence syndrome | Substance use | 0.5 | 18.6 | 18/467 | 411/38 395 | 1.28 | 5/0 |
| ICD-9 969, Poisoning by psychotropic agents | Substance use | 0.2 | 19 | 10/475 | 159/38 647 | 1.63 | 2/0 |
| ICD-9 305, Nondependent abuse of drug | Substance use | 1 | 6 | 158/327 | 5984/32 822 | 0.97 | 10/0 |
Note: Data are also available at: https://docs.google.com/spreadsheets/d/1TANDUqo0N6vLoA1BgsV1ZzbKVs0gsCiX/edit?usp=sharing&ouid=103028647972665991974&rtpof=true&sd=true.
SDoH: Social Determinants of Health.
The sign of coefficients of the predictors in each repeat in the predictive modeling.
To comply with cell suppression requirements, we present ranges for cell counts under 6 and their associated log odds calculations.
How does the fusion framework improve the predictive model?
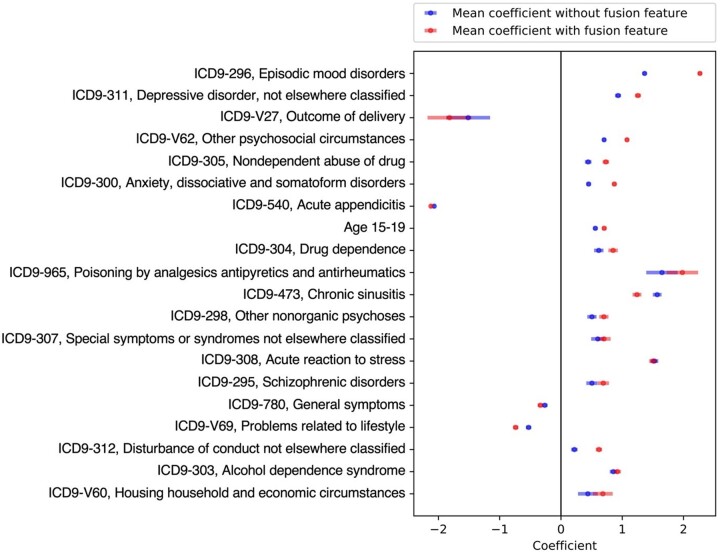
Further analyses were conducted to explore how the fused risk scores improved the prediction model for the target cohort. Figure 3 presents the logistic regression coefficients for the top 20 local predictors selected by the fusion enhanced model, with (red) and without (blue) the fused risk scores. Selected predictors were ordered by their frequency of being selected by the model. In almost every case, the magnitudes of the effects of the predictors (measured by the absolute values of their coefficients) were strengthened in the model including the fused risk scores. In some cases, the coefficients for the predictors were increased by a large extent: episodic mood disorders, the most prominent risk factor for suicidal behavior, almost doubled in magnitude when fusion predictors were added to the model (eg, for the predictor of ICD-9 296 [episodic mood disorders], without incorporating fused risk score vs in the fusion enhanced model). The only exception to this was ICD-9 473 [chronic sinusitis], where the average coefficients dropped from in the conventional model to in the fusion model.
Figure 3.
Coefficients of top 20 local predictors by logistic regression. Data were presented as mean value with standard deviation (SD).
In Figure 4, we illustrate how the fusion enhanced model improved the identification of high-risk patients. Figure 4 presents the distributions of individual cumulative risk score using all predictors selected by the fusion model, all local predictors selected by the fusion model, and all local predictors selected by the conventional model, respectively, in all 10 testing sets. Compared to the other 2 methods, the fusion model led to the improved separation of individual cumulative risk scores among the cases and controls. More specifically, it amplified the risk scores of the high-risk cases while reducing the risk scores of the controls. Such observations may explain why the fusion model resulted in large improvement in sensitivity (ie, the true positive rate) while maintaining a high level of specificity (see Figure 2).
Figure 4.
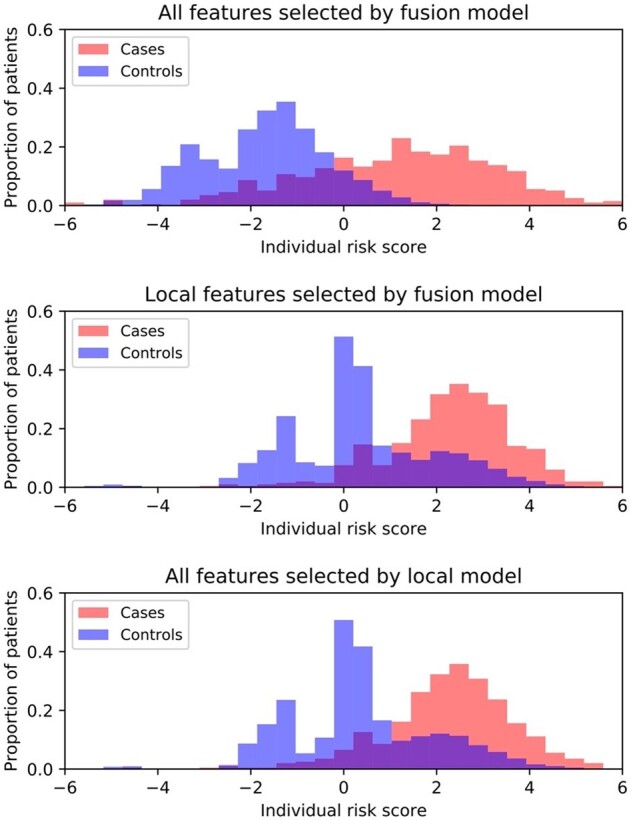
Distributions of individual suicide risk score by different models. X-axis indicates the individual risk score, which was calculated as the cumulation of coefficients of individual predictors associated with the patient. Y-axis denotes the proportion of patients (cases or controls).
The statistical analysis of the characteristics of fusion improved subjects who were incorrectly characterized as attempters/nonattempters in the conventional model but correctly characterized in the fusion enhanced model further confirmed these observations. Table 4 summarizes the prediction performance of the conventional model and fusion enhanced model across the 10 testing sets. The fusion model improved the identification of actual suicide attempters by 26.93–37.82%. In addition, we performed Fisher’s exact test between the “only fusion correct” and “both correct” groups, with the significant predictors and the corresponding percentage of patients that had that diagnosis presented in Table 5. The results are meant to be exploratory, and so we have listed all the potentially distinctive features based on unadjusted P values with a significance level of 5%. If a predictor was less frequently observed in the “only fusion correct” group, we refer to it as “Fusion Assisted.” For instance, the diagnosis of disorders involving the immune system (ICD-9 279) was only observed in the both correct group, which suggests that our fusion model managed to correctly identify patients who attempted suicide even when they did not have a high frequency of ICD-9 279. In other words, those codes were “assisted” by the power of the fusion model, which correctly identified the suicide attempters despite the low frequency of certain codes. In contrast, predictors that were more frequently observed in the only fusion correct group were labeled “Fusion Corrected.” They were underestimated in the conventional model and corrected by the fusion enhanced model.
Table 4.
Comparison of the conventional model and fusion enhanced model for different level high-risk groups
| High-risk group | Gender | Both correct, N (%) | Both incorrect, N (%) | Only conventional correct, N (%) | Only fusion enhanced correct, N (%) | Improvement a (%) |
|---|---|---|---|---|---|---|
| .05 | All | 123 (25.52) | 180 (37.34) | 2 (0.41) | 177 (36.72) | 36.31 |
| .05 | Female | 87 (27.88) | 107 (34.29) | 0 (0) | 118 (37.82) | 37.82 |
| .05 | Male | 36 (21.18) | 73 (42.94) | 2 (1.18) | 59 (34.71) | 33.53 |
| .1 | All | 198 (41.08) | 135 (28.01) | 9 (1.87) | 140 (29.05) | 27.18 |
| .1 | Female | 139 (44.55) | 79 (25.32) | 5 (1.6) | 89 (28.53) | 26.93 |
| .1 | Male | 59 (34.71) | 56 (32.94) | 4 (2.35) | 51 (30) | 27.65 |
Improvement is computed as: only fusion enhanced right (%)—only conventional right (%)
Table 5.
Summary of significant predictors for top 5% high-risk groups
| Predictors (ICD-9) | Only fusion enhanced correct (%) | Both correct (%) | P value | Case exposed/case nonexposed | Control exposed/control nonexposed | |
|---|---|---|---|---|---|---|
| Fusion assisted | 784, Symptoms involving head and neck | 0.56 | 5.69 | .01 | 14/471 | 919/37887 |
| 682, Other cellulitis and abscess | 0 | 4.07 | .01 | <6/480 | 1146/37660 | |
| 437, Other and ill-defined cerebrovascular disease | 0 | 4.07 | .01 | <6/483 | 38/38768 | |
| 054, Herpes simplex | 0 | 3.25 | .03 | <6/482 | 406/38400 | |
| 279, Disorders involving the immune mechanism | 0 | 3.25 | .03 | <6/483 | 233/38573 | |
| 656, Other known or suspected fetal and placental problems affecting management of mother | 0 | 3.25 | .03 | <6/482 | 1001/37805 | |
| E92, Other accidents | 0 | 3.25 | .03 | 8/477 | 603/38203 | |
| V63, Unavailability of other medical facilities for care | 0 | 3.25 | .03 | <6/481 | 25/38781 | |
| V27, Outcome of delivery | 0.56 | 4.07 | .04 | 11/474 | 8113/30693 | |
| Fusion corrected | 584, Acute kidney failure | 5.08 | 0 | .01 | 11/474 | 693/38113 |
| 427, Cardiac dysrhythmias | 5.08 | 0 | .01 | 16/469 | 963/37843 | |
| 285, Other and unspecified anemias | 3.95 | 0 | .04 | 11/474 | 2385/36421 | |
| 299, Pervasive developmental disorders | 6.78 | 1.63 | .05 | 23/462 | 960/37846 |
Notes: Both original and adjusted P values are reported, and the selected predictors with unadjusted P values less than .05 are listed.
DISCUSSION
To the best of our knowledge, our study is the first to combine the concepts of target learning and data fusion to address the rarity, uncertainty, and high-dimensionality of complex healthcare data in building actionable predictive models of suicide risk. Our conventional model, which was developed using 5 years of statewide inpatient hospitalization data, demonstrated good performance when compared with results generally observed in studies using clinical or claims data to predict suicide risk in the general population.27 However, our fusion enhanced model, which incorporated information from a complementary database containing a broader spectrum of healthcare encounters, substantially improved the predictive performance of the conventional model, surpassing the predictive performance reported in the literature for both adult and pediatric patients. The mechanism through which this improvement was achieved was tied to the strengthening of the selected predictors—which were mainly mental health and substance use diagnoses commonly associated with suicide—thereby clarifying their impact on suicidal behavior. This “tuning” of the model using fusion predictors derived from a more expansive, complementary database, resulted in a significant improvement in predictive performance and resulted in a dramatic reduction in the number of patients deemed to be at risk. Our results illustrate the potential of improved suicide risk prediction, as well as improved risk prediction generally, through the application of principles associated with transfer learning.
Suicide prediction represents a class of risk prediction scenarios in healthcare involving rare events. Although a variety of techniques have been developed to mitigate the class imbalance problem,28 robust improvements in both model performance as illustrated by improved PPV and AUC have been lacking. Our proposed fusion framework demonstrated, for the first time in suicide risk research, the ability to improve such predictive models by leveraging external large-scale patient datasets that incorporate patient similarities as additional covariates. While we provided quantitative and qualitative criteria to analyze the impact of those covariates, theory regarding exactly how they improve predictive performance is ongoing. Our research has shed light on the value of open data science, where large-scale, comprehensive clinical datasets can be used to improve the performance of prediction models trained on local patient populations. Importantly, our approach does not require the actual integration of identifiable patient data, thus freeing it from many of the limitations associated with the sharing and release of sensitive information.
As a proof-of-concept study, our statewide data do have certain limitations. First, the target data (HIDD) do not include a unique patient identification (ID) but rather was generated based on date of birth, gender, race and ethnicity, and zip code. The source data (APCD) were also deidentified and did not include patient race and ethnicity. Access to this additional information may improve the fusion model further by allowing us to construct similarity scores more accurately. Second, only demographic and medical history (ie, diagnosis codes) were included as predictors. The external data source used also included medication information, which if incorporated might help to build a more accurate external model. Third, the local cohort was restricted to inpatient settings with a limited number of suicidal events. Of the 39 291 patients eligible for analysis, only 485 (1.23%) had SAs. Specific strategies to address the positive-negative imbalance of SAs were not introduced. Consequently, certain risk factors, which have been associated with suicide risk but appear infrequently in this patient population, may not be identified. The need for methods addressing imbalanced clinical data analysis in the future is great.29,30 Finally, our analysis was confined to commercial claims data due to lack of access to Medicaid data, which may potentially limit the generalizability of our findings to these patient populations.
This analysis has also identified many important avenues for further research. One important avenue concerns the characteristics of the source and target data used for data fusion. In this study, there is some overlap in the kinds of data included in both datasets, as approximately 25% of the encounters in the target data would also be included in the source data. We view this as both a strength and a possible limitation. It is a strength as it presents a very common use case for a fusion approach, where limited data from a particular hospital are augmented and enhanced by fusion with a dataset containing ambulatory encounters. However, the overlap might actually raise the bar for demonstrating the utility of the fusion approach, as it may limit the benefits that could be reaped from data fusion given the source data are not completely distinct from the target data. This will be explored in future work. On the modeling side, it is worthwhile to explore other alternative statistical approaches in practice. Different similarity/distance metrics, including Pearson correlation, Canberra distance, cosine distance, among others, could be considered in the computation of the pairwise similarity scores, based on the types of the available data. In our study, alterative machine learning methods, including random forest, neural network, and support vector machines, did not lead to much improved performance comparing the reported regularized logistic regression. However, it is certainty worthwhile to explore different predictive modeling strategies under our proposed framework.
Finally, this study has substantial clinical relevance. By drawing on data commonly available in clinical data systems, the predictive model can be readily incorporated into existing electronic health record (EHR) platforms to support clinical decision-making without the need for additional data collection. The availability of such algorithms is of particular importance to facilities required to meet the Joint Commission’s National Patient Safety Goal (January 1, 2015): Reduce the risk for suicide.31 This required performance element for all JCOAH accredited hospitals and behavioral health care organizations has to date almost exclusively been met through the use of manual screening for patient suicide risk using tools such as the Columbia-Suicide Severity Risk Scale or the Ask Suicide-Screening Questions (ASQ) Toolkit.32,33 Although such screening tools have demonstrated good to excellent performance in identifying at-risk patients, the burden associated with screening patients is significant and demands clinical resources that could be allocated elsewhere if automated predictive algorithms could reduce or even eliminate the need for the collection of screening data.
FUNDING
The work was supported by NIH grant numbers R01MH124740 and R01MH112148.
AUTHOR CONTRIBUTIONS
FW, KC, and RA for conceptualization, investigation, writing, reviewing, and editing of the manuscript. WX and CS for investigation, data analysis, drafting, editing, and reviewing manuscript. YL for providing data analysis. SR for discussion, commenting, and editing the manuscript.
ETHICS APPROVAL
Our study was approved by the University of Connecticut Health Center Institutional Review Board and Weill Cornell Medical College Institutional Review Board, the CT Department of Public Health Human Investigations Committee, and the CT APCD Data Release Committee.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The data used in this analysis were obtained from the Connecticut Department of Public Health (DPH) and the Connecticut Office of Health Strategy (OHS). Neither agency endorses nor assumes any responsibility for any analyses, interpretations, or conclusions based on the data. The authors assume full responsibility for all such analyses, interpretations, and conclusions. The data use agreements governing access to and use of these datasets do not permit the authors to re-release the datasets or make the data publicly available. However, these data can be obtained by other researchers using the same application processes used by the authors.
Supplementary Material
Contributor Information
Wanwan Xu, Department of Statistics, University of Connecticut, Storrs, Connecticut, USA.
Chang Su, Department of Health Service Administration and Policy, Temple University, Philadelphia, Pennsylvania, USA.
Yan Li, Department of Statistics, University of Connecticut, Storrs, Connecticut, USA.
Steven Rogers, Department of Pediatrics, UCONN Health, Farmington, Connecticut, USA; Injury Prevention Center, Connecticut Children’s and Hartford Hospital, Hartford, Connecticut, USA.
Fei Wang, Department of Population Health Sciences, Weill Cornell Medicine, Cornell University, New York, New York, USA.
Kun Chen, Department of Statistics, University of Connecticut, Storrs, Connecticut, USA.
Robert Aseltine, Division of Behavioral Sciences and Community Health, UConn Health, Farmington, Connecticut, USA.
#Data analysis was done when Dr. Chang Su was in Department of Population Health Sciences, Weill Cornell Medicine of Cornell University.Wanwan Xu and Chang Su contributed equally to this work.
REFERENCES
- 1. Nock MK, Borges G, Bromet EJ, et al. Suicide and suicidal behavior. Epidemiol Rev 2008; 30: 133–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nock MK, Green JG, Hwang I, et al. Prevalence, correlates, and treatment of lifetime suicidal behavior among adolescents: Results from the national comorbidity survey replication adolescent supplement. JAMA Psychiatry 2013; 70 (3): 300–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kessler RC, Borges G, Walters EE. Prevalence of and risk factors for lifetime suicide attempts in the National Comorbidity Survey. Arch Gen Psychiatry 1999; 56 (7): 617–26. [DOI] [PubMed] [Google Scholar]
- 4. Voss C, Ollmann TM, Miché M, et al. Prevalence, onset, and course of suicidal behavior among adolescents and young adults in Germany. JAMA Netw Open 2019; 2 (10): e1914386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Doshi RP, Chen K, Wang F, et al. Identifying risk factors for mortality among patients previously hospitalized for a suicide attempt. Sci Rep 2020; 10 (1): 15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WISQARS (Web-based Injury Statistics Query and Reporting System)|Injury Center|CDC. https://www.cdc.gov/injury/wisqars/index.html Accessed March 29, 2021.
- 7. Stone DM, Simon TR, Fowler KA, et al. Vital signs: trends in state suicide rates—United States, 1999–2016 and circumstances contributing to suicide—27 states, 2015. MMWR Morb Mortal Wkly Rep 2018; 67 (22): 617–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barak-Corren Y, Castro VM, Javitt S, et al. Predicting suicidal behavior from longitudinal electronic health records. Am J Psychiatry 2017; 174 (2): 154–62. [DOI] [PubMed] [Google Scholar]
- 9. Harford TC, Yi H, Ye Chen CM, et al. Substance use disorders and self- and other-directed violence among adults: results from the National Survey on Drug Use and Health. J Affect Disord 2018; 225: 365–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Walsh CG, Ribeiro JD, Franklin JC. Predicting suicide attempts in adolescents with longitudinal clinical data and machine learning. J Child Psychol Psychiatry 2018; 59 (12): 1261–70. [DOI] [PubMed] [Google Scholar]
- 11. Su C, Aseltine R, Doshi R, et al. Machine learning for suicide risk prediction in children and adolescents with electronic health records. Transl Psychiatry 2020; 10 (1): 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng 2010; 22 (10): 1345–59. [Google Scholar]
- 13. Li Y, Vinzamuri B, Reddy CK. Constrained elastic net based knowledge transfer for healthcare information exchange. Data Min Knowl Disc 2015; 29 (4): 1094–112. [Google Scholar]
- 14. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 542 (7639): 115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016; 316 (22): 2402–10. [DOI] [PubMed] [Google Scholar]
- 16. Kermany DS, Goldbaum M, Cai W, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018; 172 (5): 1122–31.e9. [DOI] [PubMed] [Google Scholar]
- 17. Zhang XS, Tang F, Dodge H, et al. MetaPred: meta-learning for clinical risk prediction with limited patient electronic health records. In: Proceedings of the ACM SIGKDD International Conference of Knowledge Discovery Data Mining; 2019: 2487–95. http://arxiv.org/abs/1905.03218 Accessed March 29, 2021. [DOI] [PMC free article] [PubMed]
- 18. Wiens J, Guttag J, Horvitz E. A study in transfer learning: leveraging data from multiple hospitals to enhance hospital-specific predictions. J Am Med Inform Assoc 2014; 21 (4): 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patrick AR, Miller M, Barber CW, et al. Identification of hospitalizations for intentional self-harm when E-codes are incompletely recorded. Pharmacoepidemiol Drug Saf 2010; 19 (12): 1263–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen K, Aseltine RH. Using hospitalization and mortality data to identify areas at risk for adolescent suicide. J Adolesc Health 2017; 61 (2): 192–7. [DOI] [PubMed] [Google Scholar]
- 21. Wang W, Aseltine R, Chen K, et al. Integrative survival analysis with uncertain event times in application to a suicide risk study. Ann Appl Stat 2020; 14: 51–73. [Google Scholar]
- 22. Schwarz G. Estimating the dimension of a model. Ann Stat 1978; 1: 461–4. [Google Scholar]
- 23. Ferri FJ, Pudil P, Hatef M, et al. Comparative study of techniques for large-scale feature selection. Mach Intell Pattern Recogn 1994; 16: 403–13. [Google Scholar]
- 24. Raschka S. MLxtend: providing machine learning and data science utilities and extensions to Python’s scientific computing stack. J Open Source Softw 2018; 3 (24): 638. [Google Scholar]
- 25. Su C, Aseltine R, Doshi R, et al. Machine learning for suicide risk prediction in children and adolescents with electronic health records. Transl Psychiatry 2020; 10 (1): 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Agresti A. A Survey of Exact Inference for Contingency Tables. 1992. http://links.jstor.org/sici?sici=0883-4237%28199202%297%3A1%3C131%3AASOEIF%3E2.0.CO%3B2-A Accessed March 29, 2021. [Google Scholar]
- 27. Belsher BE, Smolenski DJ, Pruitt LD, et al. Prediction models for suicide attempts and deaths: a systematic review and simulation. JAMA Psychiatry 2019; 76 (6): 642–51. [DOI] [PubMed] [Google Scholar]
- 28. Xu Z, Feng Y, Li Y, et al. Predictive modeling of the risk of acute kidney injury in critical care: a systematic investigation of the class imbalance problem. AMIA Jt Summits Transl Sci 2019; 2019: 809–18. [PMC free article] [PubMed] [Google Scholar]
- 29. Khalilia M, Chakraborty S, Popescu M. Predicting disease risks from highly imbalanced data using random forest. BMC Med Inform Decis Mak 2011; 11: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mazurowski MA, Habas PA, Zurada JM, et al. Training neural network classifiers for medical decision making: the effects of imbalanced datasets on classification performance. Neural Netw 2008; 21 (2–3): 427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.R3 Report Issue 18: National Patient Safety Goal for Suicide Prevention | The Joint Commission. https://www.jointcommission.org/standards/r3-report/r3-report-issue-18-national-patient-safety-goal-for-suicide-prevention/ Accessed May 25, 2021.
- 32. Mundt JC, Greist JH, Jefferson JW, et al. Prediction of suicidal behavior in clinical research by lifetime suicidal ideation and behavior ascertained by the electronic Columbia-suicide severity rating scale. J Clin Psychiatry 2013; 74 (9): 887–93. [DOI] [PubMed] [Google Scholar]
- 33. Horowitz LM, Snyder DJ, Boudreaux ED, et al. Validation of the ask suicide-screening questions for adult medical inpatients: a brief tool for all ages. Psychosomatics 2020; 61 (6): 713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this analysis were obtained from the Connecticut Department of Public Health (DPH) and the Connecticut Office of Health Strategy (OHS). Neither agency endorses nor assumes any responsibility for any analyses, interpretations, or conclusions based on the data. The authors assume full responsibility for all such analyses, interpretations, and conclusions. The data use agreements governing access to and use of these datasets do not permit the authors to re-release the datasets or make the data publicly available. However, these data can be obtained by other researchers using the same application processes used by the authors.