Abstract
The investigation of conventional complete blood-count (CBC) data for classifying the SARS-CoV-2 infection status became a topic of interest, particularly as a complementary laboratory tool in developing and third-world countries that financially struggled to test their population. Although hematological parameters in COVID-19-affected individuals from Asian and USA populations are available, there are no descriptions of comparative analyses of CBC findings between COVID-19 positive and negative cases from Latin American countries. In this sense, machine learning techniques have been employed to examine CBC data and aid in screening patients suspected of SARS-CoV-2 infection. In this work, we used machine learning to compare CBC data between two highly genetically distinguished Latin American countries: Brazil and Ecuador. We notice a clear distribution pattern of positive and negative cases between the two countries. Interestingly, almost all red blood cell count parameters were divergent. For males, neutrophils and lymphocytes are distinct between Brazil and Ecuador, while eosinophils are distinguished for females. Finally, neutrophils, lymphocytes, and monocytes displayed a particular distribution for both genders. Therefore, our findings demonstrate that the same set of CBC features relevant to one population is unlikely to apply to another. This is the first study to compare CBC data from two genetically distinct Latin American countries.
Keywords: COVID-19, SARS-CoV-2, Machine learning, Hematological data, Brazil, Ecuador
1. Introduction
Since the end of 2019, the Coronavirus disease (COVID-19) has impacted nearly all branches of society, especially the global economy and health services (Nicola et al., 2020; Pak et al., 2020). This reality was particularly noticeable in developing and third-world countries, which struggled with financial resources and administration problems, leading to inefficient population testing and poorly devised strategies to manage the crisis (Hotez et al., 2020).
As expected and heavily warned, SARS-CoV-2 mutated, and now a myriad of variants, including rapidly expanding virus lineages since December 2020, designated variants of concern (VOCs) were identified in distinct populations (Tao et al., 2021). In this sense, some VOCs became more threatening than others due to their higher infectability, such as Alpha (initially identified in the UK), Beta (South Africa), Gamma (Brazil), and the Delta (India) variants (Faria et al., 2021; Nonaka et al., 2021; Singh et al., 2021; Tao et al., 2021). In addition to the high genetic variability of the virus, the extensively diverse genetic background of the human host also contributes to the complexity of COVID-19 clinical outcomes. Thus, comparing clinical and laboratory data between affected and non-affected individuals from extensive admixture-Latin American populations is essential to better understand the effect of the interaction between host genetic ancestry and exposure to specific SARS-CoV-2 VOCs on the susceptibility to infection and disease severity (Fricke-Galindo and Falfán-Valencia, 2021; Harvey et al., 2021). In particular, Brazilian and Ecuadorian populations have heterogeneous genetic constitutions with a predominant three-hybrid composition (European, African, and Native American). Nonetheless, the contribution of each ancestry component is markedly different between these countries (de Moura et al., 2015; Santangelo et al., 2017; de Souza et al., 2019; Zambrano et al., 2019).
Unfortunately, the current scenario is that numerous countries are still struggling with high SARS-CoV-2-associated infection and mortality rates, mainly because of the new VOCs that can infect fully vaccinated individuals (Gupta, 2021). Thus, more efforts should be made to properly recognize the differences between the populations to understand the distinct hematological and/or immunological reactions to SARS-CoV-2 infection. The hematological manifestations of COVID-19, mainly due to the exacerbated release of inflammatory mediators ("cytokine storm"), include blood cell count alterations, particularly lymphopenia, neutrophilia, and thrombocytopenia exhibiting potential for early diagnosis of the disease and prognostic significance (Agbuduwe and Basu, 2020; Li et al., 2020; Terpos et al., 2020; Zhu et al., 2020). Interestingly, despite several studies describing the profile of hematological parameters in COVID-19-affected cohorts predominantly from Asian and USA populations (Elshazli et al., 2020; Koç et al., 2021; Stegeman et al., 2020), no works compared these laboratory data from COVID-19 patients between countries in Latin America. Given that population-specific genetic ancestry influences on hematopoiesis and in turn on Complete Blood Count (CBC)-associated traits (Chen et al., 2020; Vuckovic et al., 2020), as well as the high genetic variability observed in Latin America (Adhikari et al., 2016), it is imperative to learn the differences in hematological profiles to SARS-CoV-2 infection between individuals from distinct Latin countries. Being this a challenging task, employing advanced computational techniques that could automatically detect patterns buried within the large amount of available data obtained from clinical data is gradually gaining more ground in the Medical field.
In this sense, Machine Learning (ML) techniques have been employed to investigate CBC data and aid in the screening of patients suspected of SARS-CoV-2 infection (Alimadadi et al., 2020; Avila et al., 2020; Brinati et al., 2020; Gong et al., 2020; Imran et al., 2020; Wu et al., 2020; Yan et al., 2020; Yao et al., 2020). Briefly ML algorithms perform computational tasks by relying on learning patterns from data samples to automate inferences. In classification tasks, the aim is to induce a model that can distinguish pathological from non-pathological (healthy) samples from the population. Some variables used in the induction of the classification model are more relevant than others and may reveal important characteristics about the set of samples analyzed. These variables can change from one cohort to another, revealing relevant characteristics and comparing populations.
In this work, we used ML to compare CBC data between two highly genetically distinct Latin American countries: Brazil and Ecuador. We found striking differences in CBC profiles from the two countries, especially in red-blood-cell counts. Moreover, white-blood-cell counts displayed distinguished patterns, specific for males and females, positive and negative cases. Therefore, our data showed that features relevant to one population are unlikely to apply to another, which agrees with the genetic ancestry background of both countries.
2. Material and methods
2.1. Datasets
Datasets were obtained from an open repository, the COVID-19 Data Sharing/BR initiative (FAPESP, 2020; Mello et al., 2020), of COVID-19-related cases in Brazil, which is comprised by nearly 177,000 clinical cases. Datasets originate from the Albert Einstein Hospital.1 The data from these datasets were collected from February 26th, 2020, to June 30th, 2020, and the control data (individuals without COVID-19) was collected from November 1st, 2019, to June 30th, 2020. Patient data is anonymized, demographic data (including sex, year of birth, and residence zip code) is also described. Other clinical results are also available, such as pulmonary function tests, urinalysis parameters, and pulmonary imaging results. COVID-19 detection by RT-PCR tests is described for all patients, and serology diagnosis (IgG and IgM antibody detection) is provided for some samples. Information is not complete for all patients, with a distinct mixture of results offered individually.
Twenty distinct CBC test parameters were obtained from the dataset, including hematocrit (%), hemoglobin (g/dl), platelets (×103 μl), mean platelet volume (MPV) (fl), red blood cells (RBCs) (×106 μl), white blood cells or leukocytes (×103 μl), lymphocytes (×103 μl), basophils (×103 μl), eosinophils (×103 μl), monocytes (×103 μl), neutrophils (×103 μl), mean corpuscular volume (MCV) (fl), mean corpuscular hemoglobin (MCH) (pg), mean corpuscular hemoglobin concentration (MCHC) (g/dl), red blood cell distribution width (RDW) (%), % Basophils, % Eosinophils, % Lymphocytes % Monocytes, and % Neutrophils. Since elevated neutrophil/lymphocyte ratio (NLR) has been described as a hematologic biomarker in COVID-19 positive patients (Alkhatip et al., 2021), it was calculated from the result of absolute counts of these respective types of leukocytes from each patient of the datasets. Overall baseline characteristics can be found on the complete dataset, available at the FAPESP COVID-19 Data Sharing/BR. 2 Patients with incomplete (missing data) or lacking the above parameters were not included. A unique CBC test was used for patients with multiple test results, with the selection based on the blood test date. In this sense, same-day, or the day closest to the test, results to the PCR-test collection date were utilized as a reference.
For Ecuador, the CBC data was obtained between March 3rd, 2021, to August 9th, 2021. Exams were carried out by Segurilab3 and Previne Salub4 laboratories. The data set is comprised by nearly 400 clinical cases and each patient were agree to participate in the study by sharing your PCR test results along with a CBC. The features in the CBC data from Ecuador are the same as the Brazilian one. We used three datasets in our experiments (Table 1 ): Br-v1 contains data from 3108 patients from Brazil; Ecu-v1 contains data from 251 patients from Ecuador, and Br-v2 contains a reduced version of the Br-v1 dataset considering the same number of samples as in Ecu-v1. By constructing the Br-v2 dataset, we ensured that the age and sex of the patients were compatible with the data from Ecu-v1 (Table 2 ), thus avoiding any bias in the subsequent analyses. All datasets are available at https://github.com/sbcblab/CBCBrazilEcuador.
Table 1.
Data summary of the datasets.
| Dataset |
Samples |
PCR Positive |
PCR Negative |
Shannon entropy |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Selected | Male | Female | Total | Male | Female | Total | Male | Female | Total | |
| Br-v1 | 4473 | 744 | 621 | 1365 | 1440 | 1668 | 3108 | 0.93 | 0.84 | 0.89 |
| Ecu-v1 | 375 | 75 | 49 | 124 | 160 | 91 | 251 | 0.90 | 0.93 | 0.92 |
| Br-v2 | 375 | 75 | 49 | 124 | 160 | 91 | 251 | 0.90 | 0.93 | 0.92 |
Table 2.
Dataset preprocessing, displaying the mean age of males and females individuals in positive and negative samples.
| Dataset |
PCR Positive |
PCR Negative |
||||
|---|---|---|---|---|---|---|
| Male | Female | Total | Male | Female | Total | |
| Br-v1 | 53.27±15.91 | 52.07±17.28 | 52.72±16.55 | 46.60±19.16 | 45.44±17.65 | 45.98±18.37 |
| Ecu-v1 | 38.40±14.20 | 35.00±14.30 | 37.10±14.30 | 37.10±11.30 | 36.20±13.50 | 36.80±12.10 |
| Br-v2 | 38.40±14.25 | 35.02±14.29 | 37.06±14.31 | 37.09±11.26 | 36.16±13.48 | 36.76±12.09 |
The number of negative samples for all datasets exceeds the positive samples. This is expected from disease data since the number of infections will be small compared to the entire population. Class imbalance is common in many real-world applications and affects the quality and reliability of ML approaches (Johnson and Khoshgoftaar, 2019; Leevy et al., 2018; López et al., 2013). We use Shannon's entropy to measure the imbalance of each dataset (Table 5). On a dataset of n instances, k classes of size c i we compute the entropy using Eq. (1), and then the balance is measured of each dataset using Eq. (2) getting 0 for an unbalanced dataset and 1 for a balanced dataset.
Finally, the datasets do not provide viral sequencing data to investigate which SARS-CoV-2 variant was identified in positive cases (i.e., specific VOC/viral lineage of each patient, or dominant transmission lineage in a subset of samples, were not available).
| (1) |
| (2) |
2.2. Feature selection and feature extraction
Feature selection (FS) is part of the tasks involving dimensionality reduction, one of the fundamental data pre-processing steps for building ML models (Ang et al., 2015). ML models assume that all features are relevant for the task at hand. However, the computational cost of inducing the model increases with the number of features. Irrelevant attributes can also impair the predictive ability of the model, as there is evidence that the same learning algorithm achieves better performance by inducing on a subset of the same data where some of the original features are discarded (Cilia et al., 2019; Dash and Liu, 1997; Utans et al., 1995). In most problems, the relevant attributes are unknown, and FS can discover knowledge from the collected data. In different tasks, the collection, maintenance, and provision of input data present an economic cost to be minimized and more stringent performance requirements. In CBC data, FS is performed as a selection of the most representative variables in the dataset, taking into account the values of each feature in the different samples. With this, it can assist in biological pattern discovery, class prediction, and recognition of data that are not directly associated or known to the problem but may play a key role (Ang et al., 2015; Mirza et al., 2019).
FS methods are grouped into four types (Ang et al., 2016; Lazar et al., 2012): (i) Filter, (ii) Wrapper, (iii) Embedded and (iv) Hybrid. While the evaluation of the filter-based methods is independent of any classifier, the wrapper and embedded methods use the accuracy of the classifier itself. However, they also require strategies to search the feature space to perform the selection (Lazar et al., 2012). In Wrapper type methods, the selection is done using some optimization algorithm (particularly Metaheuristics) and then wrapping a classifier around the selected features, using their accuracy as an evaluation criterion. The set of the most discriminative features is found by minimizing the classification error, which often generates better results than filters. In this paper, we use a wrapper FS algorithm based on Recursive Feature Elimination (RFE) and Support Vector Machines (SVM-RFE) (Guyon et al., 2002), in which SVM is the classifier employed to assess the quality of inputs subsets.
The main group of algorithms for dimensionality reduction is Feature Extraction (FE), a set of methods that transforms the original feature space into different spaces with a new set of axis by combining their features and finding the ones that preserve the original information (Varshavsky et al., 2006). In our experiments, we used two features extractors to compare the population of Brazil and Ecuador: Principal Component Analysis (PCA) (Jolliffe and Cadima, 2016) and t-Distributed Stochastic Neighbor Embedding (t-SNE) (van der Maaten and Hinton, 2008). FS techniques are used to filter irrelevant or redundant features from the datasets. FE methods are used for dimensionality reduction, in which an initial set of raw data is reduced into more manageable groups. The main difference between these approaches is that FS keeps a subset of the original features, while FE creates new ones.
2.3. Statistical analysis
The Mann-Whitney U (MWU) statistical test (3) was used to obtain the statistical significance between COVID-19 positive and negative groups for each component of the CBC tests (Table 3 ). The MWU test is a non-parametric statistical technique employed to analyze differences between the medians of two distinct datasets (Milenovic, 2011). A p-value < 0.05 was considered statistically significant.
| (3) |
where n 1 and n 2 are the number of the score in the 1st and 2nd condition, respectively. R 1 is the sum of the ranks in the 1st condiction.
Table 3.
Statistical significance (Mann-Whitney U test) of the CBC parameters' differences between COVID-19 positive and negative individuals considering gender and country.
| Feature | ECU-F | ECU-M | ECU-MF | BR-v2-F | BR-v2-M | BR-v2-MF |
|---|---|---|---|---|---|---|
| Age | (U = 2126.0 p < 0.654) | (U = 6235.5 p < 0.630) | (U = 15,587.5 p < 0.980) | (U = 2128.0 p < 0.660) | (U = 6245.5 p < 0.616) | (U = 15,596.0 p < 0.973) |
| Leukocytes | (U = 1496.5 p < 0.001) | (U = 3840.5 p < 0.000) | (U = 9861.0 p < 0.000) | (U = 885.5 p < 0.000) | (U = 4572.5 p < 0.003) | (U = 9595.0 p < 0.000) |
| %Neutrophils | (U = 2366.0 p < 0.554) | (U = 7215.5 p < 0.012) | (U = 18,012.0 p < 0.013) | (U = 2226.0 p < 0.990) | (U = 6973.0 p < 0.045) | (U = 17,257.5 p < 0.086) |
| %Lymphocytes | (U = 2238.0 p < 0.972) | (U = 4874.5 p < 0.020) | (U = 13,868.5 p < 0.087) | (U = 2068.0 p < 0.483) | (U = 4796.5 p < 0.013) | (U = 13,224.0 p < 0.018) |
| %Monocytes | (U = 2303.0 p < 0.751) | (U = 6123.0 p < 0.801) | (U = 15,707.5 p < 0.884) | (U = 2978.5 p < 0.001) | (U = 5768.0 p < 0.634) | (U = 17,052.0 p < 0.132) |
| %Eosinophils | (U = 1197.5 p < 0.000) | (U = 3331.5 p < 0.000) | (U = 8477.5 p < 0.000) | (U = 1301.0 p < 0.000) | (U = 4998.5 p < 0.039) | (U = 11,594.0 p < 0.000) |
| %Basophils | (U = 1470.0 p < 0.001) | (U = 3910.0 p < 0.000) | (U = 10,174.5 p < 0.000) | (U = 1541.0 p < 0.002) | (U = 4435.0 p < 0.001) | (U = 11,269.5 p < 0.000) |
| Neutrophils | (U = 1724.0 p < 0.027) | (U = 5606.0 p < 0.419) | (U = 13,425.0 p < 0.003) | (U = 1140.0 p < 0.000) | (U = 5280.0 p < 0.139) | (U = 11,550.0 p < 0.000) |
| Lymphocytes | (U = 1277.0 p < 0.000) | (U = 2142.5 p < 0.000) | (U = 6835.0 p < 0.000) | (U = 721.5 p < 0.000) | (U = 3406.0 p < 0.000) | (U = 7469.5 p < 0.000) |
| NLR | (U = 2360.5 p < 0.571) | (U = 7502.5 p < 0.002) | (U = 18,377.0 p < 0.004) | (U = 2337.0 p < 0.641) | (U = 7007.0 p < 0.038) | (U = 17,538.0 p < 0.045) |
| Monocytes | (U = 1627.5 p < 0.008) | (U = 4708.5 p < 0.008) | (U = 11,818.0 p < 0.000) | (U = 1467.0 p < 0.001) | (U = 4510.0 p < 0.002) | (U = 10,929.5 p < 0.000) |
| Eosinophils | (U = 819.5 p < 0.000) | (U = 2505.0 p < 0.000) | (U = 6223.5 p < 0.000) | (U = 993.5 p < 0.000) | (U = 4276.0 p < 0.000) | (U = 9694.5 p < 0.000) |
| Basophils | (U = 1384.5 p < 0.000) | (U = 3733.0 p < 0.000) | (U = 9654.0 p < 0.000) | (U = 744.0 p < 0.000) | (U = 3756.0 p < 0.000) | (U = 8269.5 p < 0.000) |
| RBCs | (U = 2756.0 p < 0.021) | (U = 7481.0 p < 0.002) | (U = 18,081.0 p < 0.011) | (U = 2370.0 p < 0.542) | (U = 5081.0 p < 0.059) | (U = 13,944.5 p < 0.102) |
| MCV | (U = 2527.0 p < 0.195) | (U = 6843.5 p < 0.083) | (U = 17,815.5 p < 0.022) | (U = 2056.0 p < 0.451) | (U = 6362.5 p < 0.458) | (U = 15,729.5 p < 0.866) |
| MCH | (U = 2074.0 p < 0.500) | (U = 4330.0 p < 0.001) | (U = 12,445.5 p < 0.002) | (U = 1918.5 p < 0.176) | (U = 5899.5 p < 0.838) | (U = 14,695.0 p < 0.381) |
| MCHC | (U = 1882.5 p < 0.131) | (U = 3944.0 p < 0.000) | (U = 11,411.5 p < 0.000) | (U = 2100.0 p < 0.574) | (U = 5378.0 p < 0.201) | (U = 14,256.5 p < 0.187) |
| RDW | (U = 2042.0 p < 0.415) | (U = 4767.5 p < 0.011) | (U = 13,157.0 p < 0.015) | (U = 2592.5 p < 0.114) | (U = 6420.5 p < 0.389) | (U = 17,302.5 p < 0.078) |
| Hemoglobin | (U = 2648.5 p < 0.068) | (U = 6539.5 p < 0.268) | (U = 16,643.0 p < 0.274) | (U = 2239.0 p < 0.969) | (U = 4707.0 p < 0.008) | (U = 12,953.5 p < 0.008) |
| Hematocrit | (U = 2769.5 p < 0.018) | (U = 7649.5 p < 0.001) | (U = 18,608.5 p < 0.002) | (U = 2164.5 p < 0.781) | (U = 5113.0 p < 0.068) | (U = 13,327.5 p < 0.024) |
| Platelets | (U = 1365.5 p < 0.000) | (U = 2945.0 p < 0.000) | (U = 8551.0 p < 0.000) | (U = 1298.5 p < 0.000) | (U = 4352.0 p < 0.001) | (U = 10,301.5 p < 0.000) |
| MPV | (U = 2591.0 p < 0.115) | (U = 6391.0 p < 0.422) | (U = 17,147.0 p < 0.109) | (U = 2102.0 p < 0.580) | (U = 7118.5 p < 0.021) | (U = 17,038.0 p < 0.135) |
3. Results
3.1. CBC differences between Brazilian and Ecuadorian populations
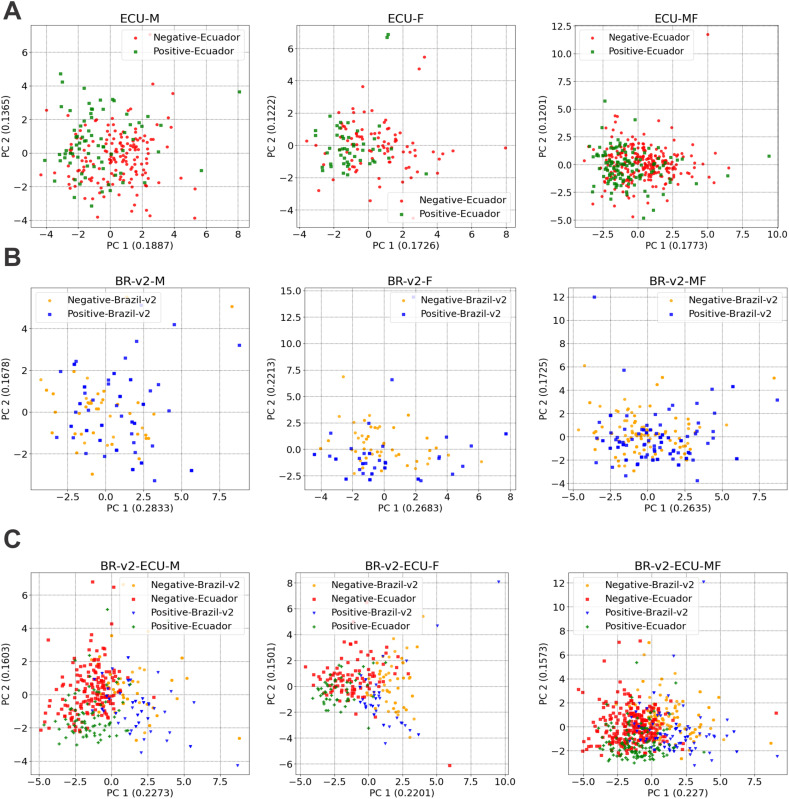
Intra- and inter-populational variations were spotted in our PCA and t-SNE analyzes, with a clear division observed in the PCA between positive and negative cases of both genders between Brazilian and Ecuadorian populations.
Positive and negative cases in male and female individuals, when looked at individually, were distinctively distributed in the Ecuadorian population. In contrast, the Brazilian cases were more mixed (Fig. 1A-B). However, when comparing male and female positive and negative cases together from both populations, an explicit division was observed (Fig. 1C). The same tendency is noticed in the t-SNE (Fig. 2C). The results indicate that samples from male and female individuals might show a mixed distribution when observed individually in the context of their own countries. However, when analyzed together, Brazilian and Ecuadorian positive and negative cases have a distinct distribution, suggesting particularities in CBC patterns.
Fig. 1.
PCA analysis, displaying the distribution of negative and positive cases, divided by male (M) and female (F) individuals, and by population. A) Distribution for M, F and MF individuals, respectively, in Ecuador. B) Distribution for M, F and MF individuals, respectively, in Brazil, after data treatment; and C) Distribution for M, F and MF, respectively, individuals in Brazil and Ecuador.
Fig. 2.
T-SNE analysis, displaying the distribution of negative and positive cases, divided by male (M) and female (F) individuals, and by population. A) Distribution for M, F and MF individuals, respectively, in Ecuador. B) Distribution for M, F and MF individuals, respectively, in Brazil, after data treatment; and C) Distribution for M, F and MF, respectively, individuals in Brazil and Ecuador.
Fig. 3 and Fig. 4 display the CBC data of positive and negative cases for female and male individuals, respectively, between the two Southern American populations. The figures are also divided into white blood cell-derived parameters (WBCP) and red blood cell-derived parameters (RBCP), along with two platelet-derived variables (MPV and platelet count).
Fig. 3.
Distributions of white and red blood cells as well as platelets related variables for positive (red) and negative (green) classes of the datasets: Brazil-V2 and Ecuador - Gender Feminine. The central white dot is the median. A) Violin charts for white blood cell-derived parameters. B) Violin charts for red blood cell-derived parameters and platelet-derived variables. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Distributions of white and red blood cells as well as platelets related variables for positive (red) and negative (green) classes of the datasets: Brazil-V2 and Ecuador - Gender Masculine. The central white dot is the median. A) Violin charts for white blood cell-derived parameters. B) Violin charts for red blood cell-derived parameters and platelet-derived variables. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Regarding inter-populational differences for female negative cases, a remarkable distinction was observed between basophils, eosinophils, and all RBCP, except for MCH and RDW. The same pattern was evident for female positive cases; besides that, only MCH exhibited similar distribution Fig. 3. In contrast, inter-populational distinctions between positive male patients, basophils, neutrophils, lymphocytes, and all RBCP, except MCH, can be noted. The same pattern was sustained in negative cases Fig. 4. Hence, CBC patterns are characteristic of each country, especially the red blood cell ones. Likewise, basophils appear as significant features in male and female individuals regardless of SARS-CoV-2 infection status.
When mixing male and female positive and negative cases from Brazilian (Brazil-v2) and Ecuadorian datasets Fig. 5 , we also discerned a similar pattern, where RBCP remain as key separation feature, including an intriguing double profile of RDW found only in Brazilian negative cases. However, neutrophils, lymphocytes, and monocytes display particular distributions in negative and positive patients, especially in the Brazilian population. In addition, no recognizable intra- or inter-population patterns of platelet-derived variables was observed between positive and negative cases of both genders.
Fig. 5.
Distributions of white and red blood cells as well as platelets related variables for positive (red) and negative (green) classes of the datasets: Brazil-V2 and Ecuador - Gender Masculine and Feminine. The central white dot is the median. A) Violin charts for white blood cell-derived parameters. B) Violin charts for red blood cell-derived parameters and platelet-derived variables. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Distinct CBC parameters classify SARS-CoV-2 infection status in Brazil and Ecuador
Lastly, a FS approach (RFE-SVM) was used to identify the most relevant parameters (features) that could better classify the CBC profiles between positive and negative cases for COVID-19 of the different genders and population groups analyzed (Fig. 6 ). Briefly, a smaller number of CBC features were found to explain the CBC differences that better classified positive and negative cases for COVID-19 in the Ecuadorian population (n features = 2–3), except for the analysis involving only female individuals from the same population (n features = 12), in comparison with the Brazilian dataset (BR-V2, n = 11–16 features depending on gender) (Fig. 6A and B). Concerning the analysis of the Ecuadorian population involving male individuals and both genders together, the selected features were the lymphocyte and neutrophil counts and two specific RBCP. In general, focusing on the Brazilian dataset, monocyte and lymphocyte counts exhibited a more central representative role. The results from ML/FS approaches were supported by the statistical analysis (Supp. Material Table 2), which also showed that despite the statistical significance of several CBC parameters in both males and females from both populations, they still present heterogeneous patterns (Supp. Material Table 2). For instance, some CBC features were significant only for a specific gender in one of the populations, such as monocyte counts, which were exclusively relevant in females from the BR-v2 (Brazil) dataset. It must be noted that the MWU statistical test (Table 3) provides a uni-dimensional selection of the most relevant CBC features for discriminating COVID-19 positive and negative cases from both populations. At the same time, the FS approach is a multidimensional analysis considering the global set of features included in the study.
Fig. 6.
Feature Selection (RFE-SVM) for each dataset. A) FS results for Brazil (BR-V2); B) FS results for Ecuador (ECU); and C) FS results for Ecuador + Brazil (ECU + BR-v2). The results consider gender (M,F) and the population (M + F).
Moreover, when considering both populations together in FS analyses (Fig. 6C), lymphocyte and neutrophil counts remained the most relevant features when comparing male and female individuals separately. These findings suggest that the number and types of CBC parameters conferring decisive differences to distinguish SARS-CoV-2 infection status tend to be population-specific. Hence, the same set of features (CBC variables) relevant to one population is unlikely to apply to another.
It is also interesting to highlight that three features were deemed enough to classify individuals with COVID-19 in the Ecuadorian population (Fig. 6B), excluding analysis restricted to females. In contrast, in the Brazilian one, at least eleven features were needed (Fig. 6A). This explains why, when combining the datasets, the classifier required numerous features to classify the inter-populational CBC data correctly (Fig. 6C).
4. Discussion
Recently, given the scenario of limited resources for large-scale testing of individuals for COVID-19 using molecular biology techniques (mainly RT-PCR), a wide range of studies have explored the importance of identifying blood cell alterations from conventional CBC data as a complementary laboratory tool for the classification of the SARS-CoV-2 infection status in different populations (Alimadadi et al., 2020; Avila et al., 2020; Brinati et al., 2020; Gong et al., 2020; Imran et al., 2020; Wu et al., 2020; Yan et al., 2020; Yao et al., 2020). Nonetheless, there are no descriptions in the literature of comparative analyses of CBC findings between COVID-19 positive and negative cases from certain Latin American countries, such as Ecuador, as well as a scarcity of comparisons of CBC profiles from these countries concerning other populations with different genetic background on the same continent.
In the present study, ML/FS approaches were applied to identify intra- and inter-populational CBC alterations in datasets from Brazil and Ecuador, two Latin American countries with marked differences in their genetic ancestry composition. In this sense, we notice a clear distribution pattern in the PCA and t-SNE analysis of positive and negative cases between the two countries. Such cases were distinctly grouped, with only a mild overlap. Almost all RBCP were strikingly divergent in the two populations; however, for WBCP, the results were mixed, with few patterns emerging. Nevertheless, basophils surfaced an important differential feature for both males and females in both populations. For males, neutrophil and lymphocyte counts were distinct between Brazil and Ecuador, while eosinophils profiles were distinguished for females. Finally, when combining male and female samples, neutrophils, lymphocytes, and monocytes displayed a particular distribution. Previous findings support our results. For instance, WBCP alterations, specifically of neutrophils and lymphocytes counts, have been reported in numerous studies from different populations (Agbuduwe and Basu, 2020; Terpos et al., 2020; Zhu et al., 2020). Remarkably, a recent work using ML and Artificial Intelligence approaches to predict SARS-CoV-2 positive patients from the full CBC dataset originated from the Albert Einstein Hospital, the same dataset from the Brazilian population queried here, also identified WBCP alterations in basophils, lymphocytes, eosinophils, and monocytes, as well as in the count of RBCs (Banerjee et al., 2020). These similar results for the WBCP group validate the computational strategy applied in the present study and support our comparison of CBC patterns between the Brazilian population and the Ecuadorian one. Nonetheless, when comparing affected and unaffected Brazilian individuals in our analyses, almost all RBCP had divergent patterns, not exclusively the RBCs count reported in the previous study. Lastly, unlike the study conducted by Banerjee and colleagues (2020) (Banerjee et al., 2020), we did not identify a clear, recognizable pattern of decrease in platelets in Brazilian individuals with COVID-19.
Although Brazilian and Ecuadorian populations have a similar three-hybrid genetic composition (European, African, and Native American), the fraction of each ancestry component significantly differs between the two countries. Briefly, the European and the"Mestizo" ancestry (an admixture of Spanish and Indigenous American ancestry) are the most predominant contributions in Brazil and Ecuador, respectively (de Moura et al., 2015; Santangelo et al., 2017; de Souza et al., 2019; Zambrano et al., 2019). These distinct genetic ancestries' profiles may explain, at least in part, some differences found in the present study comparing CBC patterns between affected and unaffected individuals. In this context, our results from the FS approach (RFE-SVM), employed to identify the most relevant CBC variables that could better distinguish the SARS-CoV-2 infection status, indicated that only three CBC features successfully classify Ecuadorian individuals with COVID-19. In comparison, more than ten features were required for the same classification purpose in the Brazilian population. Of note, all these CBC features' differences derived from RFE-SVM analyses were statistically significant using the MWU test (Table 3), reinforcing the findings obtained through FS approach. We hypothesize that the more significant number of CBC features needed to distinguish positive from negative cases for COVID-19 in Brazil may be explained by the fact that this country has one of the most heterogeneous genetic constitutions in the world with a highly extensive admixture (Pena et al., 2020; Ruiz-Linares et al., 2014; de Souza et al., 2019). Two additional factors should also be pointed out. Firstly is the exposure to different predominant VOCs or viral lineages of SARS-CoV-2 in the pandemic epidemiological week period in which the CBC data was collected in each country. For example, the dominant transmission lineage in Ecuador throughout 2020 was B.1.1.74 (previously designated as an Alpha VOC, one of the lineages descended from B.1.1, which became one of the most dominant lineages during the early phase of the pandemic in Europe and North America) (Gutierrez et al., 2021). This lineage differs from those more commonly identified as circulating in most regions of Brazil in the same year (B.1.1.28 and B.1.1.33, as well as P.1 and P.2 lineages, previously designated as Alpha and Gamma VOCs, respectively) (Faria et al., 2021; Franceschi et al., 2021; Nonaka et al., 2021; Resende et al., 2020). Secondly is the influence of altitude differences (Ecuadorians are located at a higher altitude than Brazilians) on each population's CBC constitutive profiles (baseline or healthy condition). It has been widely described that the rate of erythropoiesis is altitude-dependent (Ge et al., 2002; Robach et al., 2004), as well as the occurrence of human genetic adaptations to higher altitudes for maintaining an adequate efficiency of oxygen delivery (Julian and Moore, 2019; Murray et al., 2018), which ultimately can explain different RBCP patterns between the two populations studied.
The results of the current study must be interpreted in the context of the following limitations: (i) small sample number in the Ecuador dataset (Ecu-v1, n = 375); (ii) lack of data regarding the results of other clinical and laboratory tests in the Ecuadorian sampling, in addition to the CBC-associated hematological parameters analyzed here, such as biochemical and immunological tests, pulmonary function and imaging tests; and (iii) temporal difference in the period of the pandemic in which the datasets from the two countries were collected, being those of the Brazilian population obtained at an earlier time of the pandemic (November 1st, 2019 to June 30th, 2020) and those of Ecuador (March 3rd, 2021 to August 9th, 2021). On the other hand, a positive aspect of our study is the importance of the CBC profiles' comparison between COVID-19 positive and negative individuals from Latin American countries, providing additional evidence that the same hematological parameters considered in a specific population context cannot be used in the complementary differential diagnosis of SARS-CoV-2 infection in another population with a distinct genetic ancestry profile. Importantly, this is the first study to explore CBC patterns among infected and uninfected individuals from Ecuador.
5. Conclusions
In conclusion, the current study highlights the importance of applying ML/FS approaches to identify changes in peripheral blood parameters derived from CBC in positive and negative individuals for SARS-CoV-2 infection and prioritize more relevant parameters of prediction infection status in different populations. In summary, there was a distinct RBCP pattern between the two populations in almost all parameters for both male and female individuals. However, WBCP had no clear patterns. This result strengthens the need to evaluate and find these population differences since it directly impacts the understanding of treatment options, outcomes, and preventive measures.
Additionally, the interpretation of our results was in agreement with the genetic ancestry background behind the CBC data of Brazil and Ecuador. Our findings demonstrate that the same CBC parameters filtered by ML/FS approaches that allow distinguishing positive from negative cases for COVID-19 in a given population will not necessarily be able to discriminate infection status in another population. These approaches should be employed in a population-specific context, considering the differential aspects of genetic ancestry, evolutionary adaptations (e.g., high altitudes and oxygenation levels), and/or exposure to different environmental factors (e.g., infection by predominant SARS-CoV-2 genetic lineages in specific periods of the pandemic), which make these CBC patterns as complementary laboratory biomarkers for COVID-19 diagnosis that are unique to each population.
Acknowledgments
This work was supported by grants from the Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul - FAPERGS [19/2551-0001906-8], Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq [311611 / 2018-4], and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - STICAMSUD [88881.522073 / 2020-01]. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001. Authors thanks the Universität St.Gallen (HSG) through the Seed Money Grants 2020 Program under the project “A hemogram-data-based machine learning approach to optimize the use of COVID-19 diagnostic tests”.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2022.105228.
Appendix A. Supplementary data
Supplementary material
References
- Adhikari Kaustubh, Mendoza-Revilla Javier, Chacón-Duque Juan Camilo, Fuentes-Guajardo Macarena, Ruiz-Linares Andrés. Admixture in latin america. Curr. Opin. Genet. Dev. 2016;41:106–114. doi: 10.1016/j.gde.2016.09.003. [DOI] [PubMed] [Google Scholar]
- Agbuduwe Charles, Basu Supratik. Haematological manifestations of covid-19: from cytopenia to coagulopathy. Eur. J. Haematol. 2020;105(5):540–546. doi: 10.1111/ejh.13491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alimadadi Ahmad, Aryal Sachin, Manandhar Ishan, Munroe Patricia B., Joe Bina, Cheng Xi. 2020. Artificial Intelligence and Machine Learning to Fight Covid-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhatip Ahmed Abdelaal Ahmed Mahmoud M., Kamel Mohamed Gomaa, Hamza Mohamed Khaled, Farag Ehab Mohamed, Yassin Hany Mahmoud, Elayashy Mohamed, Naguib Amr Ahmed, Wagih Mohamed, Abd-Elhay Fatma Abd-Elshahed, Algameel Haytham Zien, et al. Expert Review of Molecular Diagnostics. 2021. The diagnostic and prognostic role of neutrophil-to-lymphocyte ratio in covid-19: a systematic review and meta-analysis; pp. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang Jun Chin, Mirzal Andri, Haron Habibollah, Hamed Haza Nuzly Abdull. Supervised, unsupervised, and semi-supervised feature selection: a review on gene selection. IEEE/ACM Trans. Comput. Biol. Bioinform. 2015;13:971–989. doi: 10.1109/TCBB.2015.2478454. [DOI] [PubMed] [Google Scholar]
- Ang J.C., Mirzal A., Haron H., et al. Supervised, unsupervised, and semi-supervised feature selection: a review on gene selection. IEEE/ACM Trans. Comput. Biol. Bioinform. 2016;13(5):971–989. doi: 10.1109/TCBB.2015.2478454. [DOI] [PubMed] [Google Scholar]
- Avila Eduardo, Kahmann Alessandro, Alho Clarice, Dorn Marcio. Hemogram data as a tool for decision-making in covid-19 management: applications to resource scarcity scenarios. PeerJ. 2020;8 doi: 10.7717/peerj.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee Abhirup, Ray Surajit, Vorselaars Bart, Kitson Joanne, Mamalakis Michail, Weeks Simonne, Baker Mark, Mackenzie Louise S. Use of machine learning and artificial intelligence to predict sars-cov-2 infection from full blood counts in a population. Int. Immunopharmacol. 2020;86:106705. doi: 10.1016/j.intimp.2020.106705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinati Davide, Campagner Andrea, Ferrari Davide, Locatelli Massimo, Banfi Giuseppe, Cabitza Federico. Detection of covid-19 infection from routine blood exams with machine learning: a feasibility study. J. Med. Syst. 2020;44(8):1–12. doi: 10.1007/s10916-020-01597-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Ming-Huei, Raffield Laura M., Mousas Abdou, Sakaue Saori, Huffman Jennifer E., Moscati Arden, Trivedi Bhavi, Jiang Tao, Akbari Parsa, Vuckovic Dragana, et al. Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell. 2020;182(5):1198–1213. doi: 10.1016/j.cell.2020.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilia N., De Stefano C., Fontanella F., Raimondo S., di Freca A. An experimental comparison of feature-selection and classification methods for microarray datasets. Information (Switzerland) 2019 [Google Scholar]
- Dash M., Liu H. Feature selection for classification, in intelligent data analysis. Elsevier. Intell. Data Anal. 1997 [Google Scholar]
- Elshazli Rami M., Toraih Eman A., Elgaml Abdelaziz, El-Mowafy Mohammed, El-Mesery Mohamed, Amin Mohamed N., Hussein Mohammad H., Killackey Mary T., Fawzy Manal S., Kandil Emad. Diagnostic and prognostic value of hematological and immunological markers in covid-19 infection: a meta-analysis of 6320 patients. PLoS One. 2020;15(8) doi: 10.1371/journal.pone.0238160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAPESP FAPESP COVID-19 Data Sharing/BR. 2020. https://repositoriodatasharingfapesp.uspdigital.usp.br
- Faria Nuno R., Mellan Thomas A., Whittaker Charles, Claro Ingra M., Candido Darlan S., Mishra Swapnil, Crispim Myuki A.E., Sales Flavia C.S., Hawryluk Iwona, McCrone John T., et al. Genomics and epidemiology of the p. 1 sars-cov-2 lineage in Manaus, Brazil. Science. 2021;372(6544):815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi Vincius Bonetti, Ferrareze Patrícia Aline Gröhs, Zimerman Ricardo Ariel, Cybis Gabriela Bettella, Thompson Claudia Elizabeth. Mutation hotspots and spatiotemporal distribution of sars-cov-2 lineages in brazil, february 2020–2021. Virus Res. 2021;304:198532. doi: 10.1016/j.virusres.2021.198532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke-Galindo Ingrid, Falfán-Valencia Ramcés. Genetics insight for covid-19 susceptibility and severity: a review. Front. Immunol. 2021;12:1057. doi: 10.3389/fimmu.2021.622176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Ri-Li, Witkowski Sarah, Yu Zhang, Alfrey Clarence, Sivieri Mark, Karlsen Trine, Resaland Geir K., Harber Matthew, Stray-Gundersen James, Levine B.D. Determinants of erythropoietin release in response to short-term hypobaric hypoxia. J. Appl. Physiol. 2002;92(6):2361–2367. doi: 10.1152/japplphysiol.00684.2001. [DOI] [PubMed] [Google Scholar]
- Gong Jiao, Jingyi Ou, Qiu Xueping, Jie Yusheng, Chen Yaqiong, Yuan Lianxiong, Cao Jing, Tan Mingkai, Wenxiong Xu, Zheng Fang, et al. A tool for early prediction of severe coronavirus disease 2019 (covid-19): a multicenter study using the risk nomogram in Wuhan and Guangdong, China. Clin. Infect. Dis. 2020;71(15):833–840. doi: 10.1093/cid/ciaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta Ravindra K. Will sars-cov-2 variants of concern affect the promise of vaccines? Nat. Rev. Immunol. 2021;21(6):340–341. doi: 10.1038/s41577-021-00556-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez Bernardo, Márquez Sully, Prado-Vivar Belén, Becerra-Wong Mónica, Guadalupe Juan José, da Silva Candido Darlan, Fernandez-Cadena Juan Carlos, Morey-Leon Gabriel, Armas-Gonzalez Rubén, Andrade-Molina Derly Madeleiny, et al. Genomic epidemiology of sars-cov-2 transmission lineages in ecuador. medRxiv. 2021 doi: 10.1093/ve/veab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon Isabelle, Weston Jason, Barnhill Stephen, Vapnik Vladimir. Gene selection for Cancer classification using support vector machines. Mach. Learn. 2002;46(1):389–422. [Google Scholar]
- Harvey William T., Carabelli Alessandro M., Jackson Ben, Gupta Ravindra K., Thomson Emma C., Harrison Ewan M., Ludden Catherine, Reeve Richard, Rambaut Andrew, Peacock Sharon J., et al. Sars-cov-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotez Peter J., Huete-Perez Jorge A., Bottazzi Maria Elena. 2020. Covid-19 in the Americas and the Erosion of Human Rights for the Poor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imran Ali, Posokhova Iryna, Qureshi Haneya N., Masood Usama, Riaz Muhammad Sajid, Ali Kamran, John Charles N., Hussain M.D. Iftikhar, Nabeel Muhammad. Ai4covid-19: Ai enabled preliminary diagnosis for covid-19 from cough samples via an app. Inform. Med. Unlock. 2020;20:100378. doi: 10.1016/j.imu.2020.100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson Justin M., Khoshgoftaar Taghi M. Survey on deep learning with class imbalance. J. Big Data. 2019;6(1):27. [Google Scholar]
- Jolliffe I.T., Cadima J. Principal component analysis: a review and recent developments. Phil. Trans. R. Soc. A. 2016;374(2065):20150202. doi: 10.1098/rsta.2015.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian Colleen G., Moore Lorna G. Human genetic adaptation to high altitude: evidence from the andes. Genes. 2019;10(2):150. doi: 10.3390/genes10020150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koç Esin Merve Erol, Fındık Rahime Bedir, Akkaya Hatice, Karadag Işılay, Tokaloglu Eda Özden, Tekin Özlem Moraloglu. Comparison of hematological parameters and perinatal outcomes between covid-19 pregnancies and healthy pregnancy cohort. J. Perinat. Med. 2021;49(2):141–147. doi: 10.1515/jpm-2020-0403. [DOI] [PubMed] [Google Scholar]
- Lazar Cosmin, Taminau Jonatan, Meganck Stijn, Steenhoff David, Coletta Alain, Molter Colin, de Schaetzen Virginie, Duque Robin, Bersini Hugues, Nowe Ann. A survey on filter techniques for feature selection in gene expression microarray analysis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2012;9(4):1106–1119. doi: 10.1109/TCBB.2012.33. [DOI] [PubMed] [Google Scholar]
- Leevy Joffrey L., Khoshgoftaar Taghi M., Bauder Richard A., Seliya Naeem. A survey on addressing high-class imbalance in big data. J. Big Data. 2018;5(1):42. [Google Scholar]
- Li Qiubai, Cao Yulin, Chen Lei, Di Wu, Jianming Yu, Wang Hongxiang, He Wenjuan, Chen Li, Dong Fang, Chen Weiqun, et al. Hematological features of persons with covid-19. Leukemia. 2020;34(8):2163–2172. doi: 10.1038/s41375-020-0910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López Victoria, Fernández Alberto, García Salvador, Palade Vasile, Herrera Francisco. An insight into classification with imbalanced data: empirical results and current trends on using data intrinsic characteristics. Inf. Sci. 2013;250:113–141. [Google Scholar]
- van der Maaten Laurens, Hinton Geoffrey. Visualizing data using t-sne. J. Mach. Learn. Res. 2008;9(Nov):2579–2605. [Google Scholar]
- Mello Luiz E., Suman Andrea, Medeiros Claudia Bauzer, Prado Claudio Almeida, Rizzatti Edgar Gil, Nunes Fatima L.S., Barnabé Gabriela F., Ferreira Joào Eduardo, Sá José, Reis Luiz F.L., Rizzo Luiz Vicente, Sarno Luzia, de Lamonica Raphael, de Barros Maciel Rui Monteiro, Cesar-Jr Roberto Marcondes, Carvalho Rodrigo. 2020. Opening Brazilian COVID-19 Patient Data to Support World Research on Pandemics. [Google Scholar]
- Milenovic Zivorad M. Application of mann-Whitney u test in research of professional training of primary school teachers. Metod. Obzori. 2011;6(1):73–79. [Google Scholar]
- Mirza Bilal, Wang Wei, Wang Jie, Choi Howard, Chung Neo Christopher, Ping Peipei. Machine learning and integrative analysis of biomedical big data. Genes. 2019;10:87. doi: 10.3390/genes10020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moura Ronald Rodrigues, Coelho Antonio Victor Campos, de Queiroz Balbino Valdir, Crovella Sergio, Brandão Lucas André Cavalcanti. Meta-analysis of brazilian genetic admixture and comparison with other latin america countries. Am. J. Hum. Biol. 2015;27(5):674–680. doi: 10.1002/ajhb.22714. [DOI] [PubMed] [Google Scholar]
- Murray Andrew J., Montgomery Hugh E., Feelisch Martin, Grocott Michael P.W., Martin Daniel S. Metabolic adjustment to high-altitude hypoxia: from genetic signals to physiological implications. Biochem. Soc. Trans. 2018;46(3):599–607. doi: 10.1042/BST20170502. [DOI] [PubMed] [Google Scholar]
- Nicola Maria, Alsafi Zaid, Sohrabi Catrin, Kerwan Ahmed, Al-Jabir Ahmed, Iosifidis Christos, Agha Maliha, Agha Riaz. The socio-economic implications of the coronavirus pandemic (covid-19): a review. Int. J. Surg. 2020;78:185–193. doi: 10.1016/j.ijsu.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka Carolina Kymie Vasques, Gräf Tiago, de Lorenzo Barcia Camila Araújo, Costa Vanessa Ferreira, de Oliveira Janderson Lopes, da Hora Passos Rogério, Bastos Iasmin Nogueira, de Santana Maria Clara Brito, Santos Ian Marinho, de Sousa Karoline Almeida Felix, et al. Sars-cov-2 variant of concern p. 1 (gamma) infection in young and middle-aged patients admitted to the intensive care units of a single hospital in salvador, northeast brazil, february 2021. Int. J. Infect. Dis. 2021;111:47–54. doi: 10.1016/j.ijid.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak Anton, Adegboye Oyelola A., Adekunle Adeshina I., Rahman Kazi M., McBryde Emma S., Eisen Damon P. Economic consequences of the covid-19 outbreak: the need for epidemic preparedness. Front. Public Health. 2020;8:241. doi: 10.3389/fpubh.2020.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena Sergio D.J., Santos Fabrício R., Tarazona-Santos Eduardo. Genetic admixture in brazil. Am. J. Med. Genet. Part C. 2020;184:928–938. doi: 10.1002/ajmg.c.31853. Wiley Online Library. [DOI] [PubMed] [Google Scholar]
- Resende Paola Cristina, Delatorre Edson, Gräf Tiago, Mir Daiana, Motta Fernando Couto, Appolinario Luciana Reis, da Paixão Anna Carolina Dias, da Fonseca Mendonça Ana Carolina, Ogrzewalska Maria, Caetano Braulia, et al. Evolutionary dynamics and dissemination pattern of the sars-cov-2 lineage b. 1.1. 33 during the early pandemic phase in brazil. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.615280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robach Paul, Fulla Yvonne, Westerterp Klaas R., Richalet Jean-Paul. Comparative response of epo and soluble transferrin receptor at high altitude. Med. Sci. Sports Exerc. 2004;36(9):1493–1498. doi: 10.1249/01.mss.0000139889.56481.e0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Linares Andrés, Adhikari Kaustubh, Acuña-Alonzo Victor, Quinto-Sanchez Mirsha, Jaramillo Claudia, Arias William, Fuentes Macarena, Pizarro María, Everardo Paola, De Avila Francisco, et al. Admixture in latin america: geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet. 2014;10(9) doi: 10.1371/journal.pgen.1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo Roberta, González-Andrade Fabricio, Børsting Claus, Torroni Antonio, Pereira Vania, Morling Niels. Analysis of ancestry informative markers in three main ethnic groups from Ecuador supports a trihybrid origin of ecuadorians. Foren. Sci. Int. 2017;31:29–33. doi: 10.1016/j.fsigen.2017.08.012. [DOI] [PubMed] [Google Scholar]
- Singh Jasdeep, Rahman Syed Asad, Ehtesham Nasreen Z., Hira Subhash, Hasnain Seyed E. Sars-cov-2 variants of concern are emerging in india. Nat. Med. 2021:1–3. doi: 10.1038/s41591-021-01397-4. [DOI] [PubMed] [Google Scholar]
- de Souza Aracele Maria, Resende Sarah Stela, de Sousa Taís Nóbrega, de Brito Cristiana Ferreira Alves. A systematic scoping review of the genetic ancestry of the brazilian population. Genet. Mol. Biol. 2019;42:495–508. doi: 10.1590/1678-4685-GMB-2018-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegeman Inge, Ochodo Eleanor A., Guleid Fatuma, Holtman Gea A., Yang Bada, Davenport Clare, Deeks Jonathan J., Dinnes Jacqueline, Dittrich Sabine, Emperador Devy, et al. Routine laboratory testing to determine if a patient has covid-19. Cochrane Database Syst. Rev. 2020;11 doi: 10.1002/14651858.CD013787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Kaiming, Tzou Philip L., Nouhin Janin, Gupta Ravindra K., de Oliveira Tulio, Kosakovsky Sergei L., Pond Daniela Fera, Shafer Robert W. The biological and clinical significance of emerging sars-cov-2 variants. Nat. Rev. Genet. 2021:1–17. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpos Evangelos, Ntanasis-Stathopoulos Ioannis, Elalamy Ismail, Kastritis Efstathios, Sergentanis Theodoros N., Politou Marianna, Psaltopoulou Theodora, Gerotziafas Grigoris, Dimopoulos Meletios A. Hematological findings and complications of covid-19. Am. J. Hematol. 2020;95(7):834–847. doi: 10.1002/ajh.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utans J., Moddy J., Rehfuss S. IEEE/IAFE Conference on Computational Intelligence for Financial Engineering, Proceedings (CIFEr) 1995. Input variable selection for neural networks: application to predicting the u.s. business cycle. [Google Scholar]
- Varshavsky R., Gottlieb A., Linial M., et al. Novel unsupervised feature filtering of biological data. Bioinformatics. 2006;22(14):e507–e513. doi: 10.1093/bioinformatics/btl214. [DOI] [PubMed] [Google Scholar]
- Vuckovic Dragana, Bao Erik L., Akbari Parsa, Lareau Caleb A., Mousas Abdou, Jiang Tao, Chen Ming-Huei, Raffield Laura M., Tardaguila Manuel, Huffman Jennifer E., et al. The polygenic and monogenic basis of blood traits and diseases. Cell. 2020;182(5):1214–1231. doi: 10.1016/j.cell.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Guangyao, Yang Pei, Xie Yuanliang, Woodruff Henry C., Rao Xiangang, Guiot Julien, Frix Anne-Noelle, Louis Renaud, Moutschen Michel, Li Jiawei, et al. Development of a clinical decision support system for severity risk prediction and triage of covid-19 patients at hospital admission: an international multicentre study. Eur. Respir. J. 2020;56(2) doi: 10.1183/13993003.01104-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Li, Zhang Hai-Tao, Goncalves Jorge, Xiao Yang, Wang Maolin, Guo Yuqi, Sun Chuan, Tang Xiuchuan, Jing Liang, Zhang Mingyang, et al. An interpretable mortality prediction model for covid-19 patients. Nat. Mach. Intell. 2020;2(5):283–288. [Google Scholar]
- Yao Haochen, Zhang Nan, Zhang Ruochi, Duan Meiyu, Xie Tianqi, Pan Jiahui, Peng Ejun, Huang Juanjuan, Zhang Yingli, Xiaoming Xu, et al. Severity detection for the coronavirus disease 2019 (covid-19) patients using a machine learning model based on the blood and urine tests. Front. Cell Dev. Biol. 2020;8:683. doi: 10.3389/fcell.2020.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano Ana Karina, Gaviria Aníbal, Cobos-Navarrete Santiago, Gruezo Carmen, Rodríguez-Pollit Cristina, Armendáriz-Castillo Isaac, García-Cárdenas Jennyfer M., Guerrero Santiago, López-Cortés Andrés, Leone Paola E., et al. The three-hybrid genetic composition of an ecuadorian population using aims-indels compared with autosomes, mitochondrial dna and y chromosome data. Sci. Rep. 2019;9(1):1–8. doi: 10.1038/s41598-019-45723-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Yihua, Cao XingJian, Yonghui Lu, Dongsheng Xu, Renfei Lu, Li Xinling. Lymphocyte cell population as a potential hematological index for early diagnosis of covid-19. Cell. Mol. Biol. 2020;66(7):202–206. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material