Abstract
Nearly all mammals display robust daily rhythms of physiology and behavior. These approximately 24-h cycles, known as circadian rhythms, are driven by a master clock in the suprachiasmatic nucleus (SCN) of the hypothalamus and affect biological processes ranging from metabolism to immune function. Perhaps the most overt output of the circadian clock is the sleep-wake cycle, the integrity of which is critical for health and homeostasis of the organism. In this review, we summarize our current understanding of the circadian regulation of sleep. We discuss the neural circuitry and molecular mechanisms underlying daily sleep timing, and the trajectory of circadian regulation of sleep across development. We conclude by proposing future research priorities for the field that will significantly advance our mechanistic understanding of the circadian regulation of sleep.
Keywords: Circadian rhythms, Sleep, Suprachiasmatic nucleus, Clock genes
1. Introduction
The success of a species is highly dependent on its ability to adapt to environmental pressures. As a consequence of the rotation of the Earth around its own axis and the sun, life evolved under multiple rhythmic environmental regimes, including both seasonal and daily changes in light exposure. As a result, nearly every species on Earth has developed a biological timekeeping system that can anticipate these changes and organize physiology and behavior in a way that is advantageous for the organism [1]. Biological rhythms of approximately 24 h, called circadian rhythms, are highly conserved among mammalian species and provide a temporal order for behavioral and physiological processes, the most overt of these being daily cycles of sleep and wake. Most mammals display bouts of sleep that are consolidated to a single phase of the environmental light-dark (LD) cycle, and this timing is highly influenced by the circadian clock. In this review, we provide a broad overview of the molecular, neural circuit and developmental mechanisms underlying the circadian regulation of sleep and pose several avenues for further investigation necessary to bridge critical gaps in our understanding.
1.1. The mammalian circadian clock
Circadian clocks in mammals, and indeed all species, have three key characteristics: they are endogenous to the organism, they can synchronize to environmental cycles (entrainable), and their intrinsic period remains relatively constant regardless of external temperature (temperature compensated) [1]. These characteristics allow organisms to both align their biological rhythms with environmental cycles, a process called entrainment, and to maintain rhythmicity in the absence of external time cues.
Virtually every mammalian cell contains its own molecular circadian clock constituted by a transcriptional-translational feedback loop (TTFL) in which translated proteins inhibit the activation of their own promoters, thus halting their continued production [2]. Eventually these proteins degrade and inhibition of their own transcription ceases, starting the loop over again. The proteins CLOCK and BMAL comprise the positive arm of the TTFL, and form heterodimers that promote the transcription of Period (PER1, PER2, PER3) and Cryptochrome (CRY1, CRY2) genes. PER and CRY proteins then heterodimerize and inhibit the activity of CLOCK/BMAL heterodimers, thus negatively regulating their own transcription. This primary clock mechanism is supported by a secondary feedback loop, in which retinoic acid receptor-related orphan receptors (RORs) and the transcriptional repressor REV-ERBα activate and inhibit the expression of BMAL1, respectively [2].
In mammals, the coordination of this symphony of circadian oscillators throughout the body is driven by the master circadian pacemaker housed in the suprachiasmatic nuclei (SCN), a small bundle of about 20,000 neurons in mice, and 50,000 neurons in humans, located in the ventral hypothalamus [3]. The molecular clockwork drives circadian rhythms in gene expression and excitability in SCN neurons [4,5], and these individual neuronal oscillators coordinate SCN-level rhythms through a combination of synaptic transmission and gap junction-mediated electrical coupling [3]. The SCN is synchronized to environmental LD cycles by way of light input from intrinsically photosensitive retinal ganglion cells (ipRCGs) in the retina [6]. These ipRGCs send direct projections to and increase activity of SCN neurons via release of glutamate and pituitary adenylate cyclase-activating peptide. This glutamatergic signaling underlies light-induced phase--shifting of SCN activity [7,8], and light exposure induces an acute increase in expression of the clock gene Per1, presumably leading to phase shifts in the molecular clockwork of SCN neurons [9]. Remarkably, the SCN is able to sustain circadian rhythmicity of neural activity in the absence of environmental cues or input from other regions of the brain [10].
Although the precise anatomy and cellular composition of the SCN differs slightly between mammalian species, the core features of the SCN network are thought to be well conserved across placental mammals [11]. Despite the fact that over 90% of SCN neurons express the inhibitory small neurotransmitter GABA, the SCN is highly heterogeneous, expressing a wide array of neuropeptides, cytokines and small neurotransmitters [3,12]. The SCN is roughly divided into two functional and anatomical subregions [13]: the ventro-lateral SCN (vlSCN) or “core”, which expresses the neuropeptide vasoactive intestinal polypeptide (VIP) and is highly light-responsive [14,15]; and the dorsal-medial SCN (dmSCN) or “shell”, containing arginine vasopressin (AVP) neurons that are strong intrinsic circadian oscillators. VIP neurons in the vlSCN receive dense glutamatergic inputs from ipRGCs, providing information about the presence or absence of environmental light. The vlSCN then relays light information to AVP neurons in the vlSCN, synchronizing their intrinsic oscillations to external environmental light and maintaining synchrony of the SCN neural network. Although this coupling mechanism between vl- and dmSCN is incompletely understood, evidence suggests that both GABA [16] and VIP [14,17] signaling are critical for maintaining network synchrony. Neurons in the dmSCN both send GABAergic and glutamatergic axonal projections to nuclei throughout the brain and release humoral factors such as Prokineticin-2 [18], signals which together provide a temporal order for daily cycles of sleep and wake. Although useful, recent work reveals the core-shell model to be an oversimplification [19,20]. Additional neuropeptides found in the SCN include gastrin-releasing peptide (GRP), Neuromedin-S (NMS) [21], substance P, somatostatin cholecystokinin and neurotensin, and it is typical for SCN neurons to co-express multiple neuropeptides [5].
2. Clocks, sleep and the mammalian brain
2.1. A mysterious state of being
The most overt output of the circadian clock is sleep, a reversible state of unconsciousness characterized by behavioral inactivity and reduced responsiveness to external sensory stimuli [22]. Although sleep is a highly conserved behavior in mammals and indeed virtually every animal species on earth, its precise functions and the evolutionary pressures that selected for it remain unclear [23]. Despite this fact, sleep has been shown to be critical in maintaining physiological processes including memory consolidation [24], immune function [25] and metabolism [26].
In mammals, sleep is typically classified as one of two primary stages that alternate throughout a sleep bout: rapid eye-movement (REM) sleep and non-rapid eye-movement (NREM) sleep. These two stages have distinct physiological signatures which can be measured using a combination of electroencephalography (EEG) and electromyography (EMG) in both humans and animals. NREM sleep is characterized by synchronous, high amplitude waves oscillating in the delta frequency range (0.5–4 Hz) as measured by EEG, and reduced muscle tone as measured by EMG. REM sleep is characterized by mixed frequency, low voltage amplitude waves, high power in the theta frequency range (6–10 Hz) and complete muscle atonia [22]. In humans, sleep is consolidated into a single primary bout happening at approximately the same time of day, in which NREM sleep is highly concentrated at the beginning of the sleep cycle, and REM sleep at the end [22]. In commonly studied mammalian model organisms such as mice and rats, sleep bouts are more fragmented and occur at multiple times throughout the day, although more sleep occurs during the day than at night [27]. The duration of the primary sleep bout and the timing of ultradian cycling between sleep stages varies between mammalian species, though in humans sleep duration is approximately 8 h and a full sleep stage cycle takes 90–110 min [22].
2.2. Sleep in the brain
The key brain regions involved in sleep and wake initiation and maintenance are well-studied. A brief overview of this neural circuitry is provided here and reviewed extensively by Scammell et al. [28].
There are two primary pathways involved in the initiation and maintenance of wakefulness, both of which originate from the brainstem and hypothalamus. The first is a dorsal pathway arising from cholinergic neurons in the pedunculopontine nucleus (PPT) and laterodorsal tegmental nuclei (LDT) of the pons [29,30]. These regions, which are highly active during wakefulness and REM but less so during NREM [31], project to and activate thalamic relay neurons that enable processing of sensory and motor information by the cortex. These neurons also send sparse projections to the cortex itself, and activation of PPT has been shown to promote fast EEG activity characteristic of wakefulness [32] and disrupt slow-wave EEG activity during NREM sleep [33]. Additionally, the cholinergic basal forebrain (BF) provides direct input to distinct subregions throughout the cortex and is implicated in initiating and maintaining arousal [34,35]. The second wake-promoting pathway arises from monoaminergic neurons in the rostral brainstem and caudal hypothalamus. Although each of these groups differs slightly in their targets and mechanisms of action, they share similar firing patterns such that they are highly active during wake, fire slowly during NREM and are quiescent during REM [28]. The first of these is the noradregenergic locus coeruleus (LC), which projects broadly to the thalamus, cortex and multiple hypothalamic areas and receives input from several wake-promoting areas [36], making it a critical node in the sleep-wake regulatory network. Wake-promoting serotoninergic neurons in the dorsal and median raphe send and receive projections from regions involved in sleep-wake maintenance [28,37]. Dopamine neurons in the ventral tegmental area (VTA) [38] and ventral periaqueductal gray (vPAG) [39] have also been shown to modulate arousal, and a recently-discovered population of dopaminergic neurons in the dorsal raphe promotes wakefulness [40]. Finally, wake-active histamine neurons in the tuberomammillary nucleus (TMN) of the posterior hypothalamus project widely throughout the brain and have been demonstrated to promote sustained arousal under certain behavioral conditions [41].
Among sleep-promoting brain regions, the ventrolateral preoptic area (VLPO) and median preoptic area (MnPO) stand out as key players. Both regions are highly active during NREM sleep, send GABAergic inhibitory projections to hypothalamic and brainstem nuclei that promote wakefulness as part of the ascending arousal system, and receive afferents from the wake-active monoaminergic brain regions described above [42,43]. Additionally, lesions of the VLPO and surrounding areas result in profound sleep loss [44] and activation of the VLPO has been demonstrated to promote NREM sleep [45]. Recent work has also identified a population of GABAergic neurons in the substantia nigra reticulata that promotes the transition from wake to sleep states, primarily by way of NREM sleep initiation [46]. Historically, the study of REM sleep-promoting brain regions has focused primarily on the pons, including subpopulations of the primarily cholinergic PPT and LDT [47, 48], the glutamatergic sublaterodorsal nucleus (SLD) [49] and the GABAergic ventral medulla [50]. More recently, researchers have identified REM-active neurons in subregions of the hypothalamus including melanin-concentrating hormone-expressing neurons in the lateral hypothalamus (LH) [51] and a subpopulation of galanin-expressing neurons in the largely GABAergic dorsomedial hypothalamus (DMH) involved in transitions between NREM and REM [52].
The reciprocal connections between nodes throughout the sleep-wake regulatory circuitry comprise a self-reinforcing loop where activity from wake-promoting regions inhibits sleep-promoting regions and vice versa. This observation led to the conceptual model of the sleep-wake regulatory network as a flip-flop switch, first proposed by Saper and colleagues [53]. In electrical engineering, a flip-flop switch can be used to produce sharp transitions between discrete states but requires a stabilizing force to prevent uncontrolled switching. Orexin-expressing neurons in the LH are critical for maintaining wakefulness and have been hypothesized to serve this role [54]. These neurons are wake-active [55] and send excitatory projections to cortex, brainstem and monoaminergic centers comprising the wake-promoting pathways described above [56], thus promoting arousal but not inhibiting the sleep-promoting action of the VLPO [54].
2.3. The two-process model of sleep regulation
Although the flip-flop switch model accounts for transitions between wakefulness and states of sleep with great precision, an additional framework is needed to describe patterns of sleep and wake on longer timescales. Our understanding of sleep timing is often framed in the context of the two-process model of sleep regulation, which posits that sleep is controlled by the interactions of separate circadian and homeostatic processes [57,58]. Circadian regulation of sleep (Process C) is driven by the circadian pacemaker, such that sleep drive oscillates with a periodicity that is typically entrained to the environmental LD cycle [57,59]. In humans, this is manifested as an increase in circadian arousal during the late afternoon and early evening, and as decrease in circadian wakefulness towards the end of the sleep bout.
Homeostatic regulation of sleep (Process S) on the other hand is driven by sleep debt, or how long an individual has been awake. Sleep pressure increases with time spent awake until the individual falls asleep and decreases the longer an individual has been asleep [57,60]. Indeed, it is well documented in both human [61,62] and animal models [63] that sleep drive increases during periods of sleep deprivation, Although sleep pressure may be hard to quantify, there are EEG signatures that have been validated as reliable markers of the Process S increase [58, 64]. First, during extended wakefulness, the EEG power within a range of frequencies that spans across theta and alpha waves (6.25–9 Hz) increases with the time of wakefulness [65]. Second, the EEG delta power (0.25 and 4.0 Hz) during NREM sleep at the beginning of the recovery sleep after extended wakefulness increases with the previous time spent [60]. This delta power at the initiation of sleep and its decay through recovery sleep have become signature metrics of the increase of process S during wakefulness and its decrease during recovery sleep, respectively.
Much of what we understand about the two-process model of sleep regulation was gleaned from landmark studies performed in the 1980’s investigating sleep in both rodents and human subjects. Seminal work by Tobler et al. [66], Mistlberger et al. [67] and Trachsel et al. [68] provided key evidence that these two processes are distinct by demonstrating that in SCN-lesioned nocturnal rats, the rhythmicity of sleep-wake cycles was abolished without altering the total amount of daily sleep time. Remarkably, these animals displayed increased NREM sleep during a rebound period following sleep deprivation, suggesting the integrity of Process S was left intact. Similar results were observed in studies of sleep and circadian behavior in diurnal squirrel monkeys with SCN lesions. Unlike nocturnal animals, SCN-ablated squirrel monkeys not only lost their circadian regulation of sleep but also slept more, providing evidence for the opponent sleep regulatory process model, in which process C induces wakefulness and process S sleep [69,70].
In humans, experiments in which subjects were placed in specialized laboratory conditions absent of environmental time cues established that sleep, body temperature and endocrine rhythms followed a circadian rhythm [71–74]. Additionally, while shifting the phase of the clock with light exposure affected times at which subjects were likely to fall asleep, these manipulations did not have an effect on the depth of sleep [75]. Although the contribution of the human SCN in circadian sleep regulation could not be directly established in these early studies, later work showed that human sleep patterns were consistent with a model in which Process C is driven by a single circadian pacemaker [76]. This work led to the development of a forced-desynchrony protocol in which participants were placed in an LD and rest-activity cycle that the circadian pacemaker cannot entrain to, resulting in the primary sleep bout occurring at different circadian times on each subsequent day of the experiment [61]. Similarly, nap protocols requiring that participants take 30-min naps followed by 60-min periods of wakefulness at different time points over 24–48 h established that wakefulness is also modulated in a circadian fashion [77]. These paradigms provided experimental frameworks in which to test predictions of the two-process model in humans, and demonstrated that the two processes make parallel contributions to sleep timing [71–73,75], including that the timing of REM sleep specifically is coupled to Process C [61,73,78].
While the two processes are clearly able to work independently, there is increasing evidence that they also influence each other and that Process S may have more influence over Process C than vice versa. For example, in both humans and rodent models, the magnitude of phase shifts induced by changes in the LD cycle are attenuated during periods of sleep deprivation [79,80]. Although some evidence indicates that clock genes may influence sleep depth and that SWS is modulated by circadian phase, more research is needed to determine if and how the circadian clock influences Process S [81].
2.4. Neuronal mechanisms of circadian sleep regulation
The SCN is the primary driver of Process C [59]. Following key experiments demonstrating the necessity of the SCN for circadian rhythmicity of sleep in both nocturnal rodents and diurnal primates, researchers developed a greater understanding of the mechanisms by which the SCN affects sleep timing. While there is heterogeneity in the activity rhythms of both single neurons and neuronal ensembles within the SCN [4], on average SCN neurons display peak firing rates during the subjective day in both diurnal and nocturnal mammals [82]. Simultaneous recording of EEG/EMG and SCN neural activity in rats revealed that the SCN is more active during REM sleep than NREM sleep [83], and SCN lesion studies suggest that the SCN facilitates transitions to REM sleep during the rest phase of the LD cycle [84]. Later work used long-term, continuous recording of EEG/EMG to evaluate sleep in a rodent model of the forced desynchrony protocols pioneered in humans [85]. Under this forced desynchrony protocol, animals were placed into a 22-h LD cycle that the circadian pacemaker is unable to entrain to, and as a result displayed two different rhythms of sleep, locomotor activity and core body temperature – one rhythm that was synchronized to the environmental LD cycle, and another that oscillated with a period longer than 24 h and did not entrain to the LD cycle. These rhythms were associated with rhythmic clock gene expression within the vl- and dmSCN, respectively. Under these conditions, researchers found that REM sleep was the sole sleep stage that oscillated with a > 24 h period [86]. Later experiments demonstrated that REM sleep propensity was associated with rhythmic clock gene expression in the dmSCN, specifically [87].
Given the clear role for the SCN in sleep timing, it is reasonable to suppose that the SCN should send strong projections to the sleep-wake regulating brain regions described above (Fig. 1). However, the SCN sends only sparse direct projections to sleep- and wake-active neuronal populations such as the VLPO [88] and orexinergic LH [89], but forms indirect contacts with these and other sleep-relevant regions via strong projections to the subparaventricular zone (SPZ) [90]. The SPZ densely innervates the heterogeneous DMH, and GABAergic DMH neurons project to and inhibit the VLPO, while glutamatergic DMH neurons excite the LH [91]. In this indirect fashion, the SCN is hypothesized to exert its influence over the timing of sleep-wake rhythms. Indeed, the relay stations provided by the SPZ are necessary for the circadian regulation of sleep, as lesions of the ventral SPZ eliminate circadian rhythmicity of sleep without altering total sleep time [90], and lesions of the DMH reduce the amplitude of circadian rhythms of sleep while slightly increasing total sleep time [91]. It has recently been shown that the SCN also promotes wakefulness via GABAergic projections to corticotropin-releasing factor neurons in the paraventricular nucleus of the hypothalamus (PVN), which then excite orexin neurons in the LH. Disruption of this circuitry has been shown to reduce wakefulness and amplitude of circadian sleep-wake rhythms [92]. Interestingly, stimulation of a subset of Brnb3-expressing ipRGCs that do not project to the SCN are sufficient to acutely promote sleep possibly via sparse projections to the VLPO but are not required for circadian photoentrainment of sleep [93], highlighting one avenue by which the environmental LD cycle can drive sleep timing while bypassing the master clock.
Fig. 1.
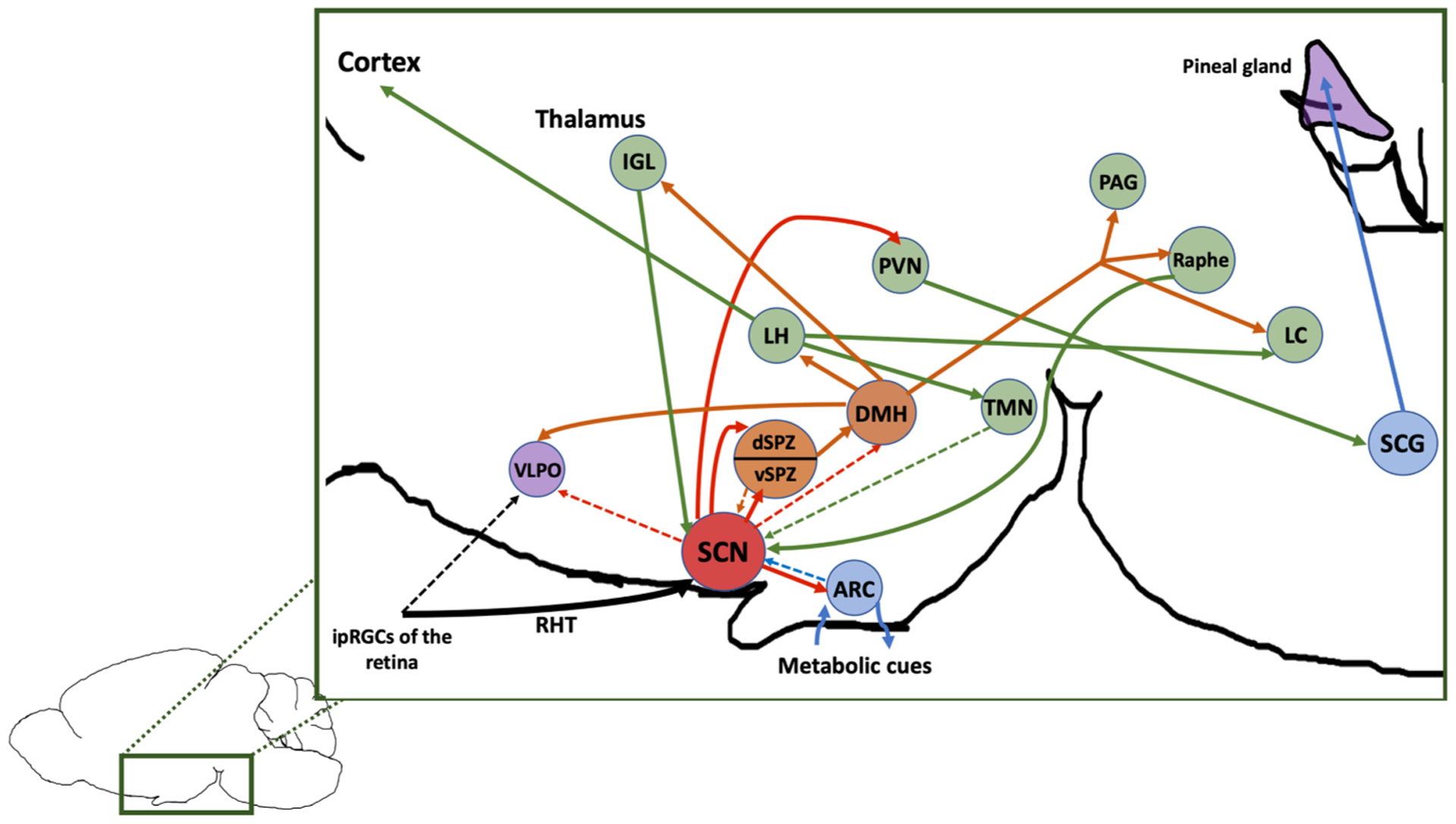
Simplified diagram of connections between the master circadian clock and sleep-wake circuitry. Diagram is color-coded as follows: Red = SCN, Green = generally wake-promoting, Purple = generally sleep promoting, orange = both, blue = neutral/other role, dotted lines = sparse projections. Abbreviations: ipRGCs, intrinsically photosensitive retinal ganglion cells; RHT, retinohypothalamic tract; SCN, suprachiasmatic nucleus; ARC, arcuate nucleus of the hypothalamus; VLPO, ventrolateral preoptic nucleus; dSPZ, dorsal subparaventricular zone; vSPZ, ventral subparaventricular zone; DMH, dorsomedial hypothalamus; TMN, tuberomammillary nucleus; LH, lateral hypothalamus; PVN, paraventricular nucleus; IGL, intergeniculate leaflet; PAG, periaqueductal gray; LC, locus coeruleus; SCG; spinal cervical ganglion.
In addition to primary connections from retinal ipRGCs to the ventral core of the SCN via the retinohypothalamic tract (RHT), there are two major afferent pathways leading to the SCN: the serotonergic median raphe and the intergeniculate leaflet (IGL). The median raphe plays a key role in maintaining wakefulness as described above, and its role in mediating circadian behavior is reviewed extensively elsewhere [94]. The neuropeptide Y (NPY)-expressing IGL projects to the SCN via the geniculohypothalamic tract and has been shown to regulate entrainment and phase resetting of the SCN [95]. However, the IGL also sends and receives projections between critical sleep-wake regulatory centers including the DMH, VLPO, SPZ and PAG, suggesting it may serve as an important point for relaying information between the SCN and the broader sleep-wake network [96]. The SCN also receives more modest direct projections from sleep-wake regulatory centers including the DMH, LC, SPZ, TMN and the arcuate nucleus of the hypothalamus (ARC), the latter of which has a prominent role in modulating circadian rhythms of sleep and metabolism [97].
In addition to synaptic transmission, humoral factors released by the SCN play a key role in maintaining circadian rhythmicity. In a landmark study by Silver et al., researchers demonstrated that SCN grafts housed in a semi-permeable capsule that prevented axonal outgrowth were able to restore behavioral circadian rhythmicity in SCN-lesioned animals, suggesting that diffusible factors play a key role in circadian sleep regulation [98].
2.5. The night hormone
The rhythmic release of the pineal hormone melatonin also plays a role in the timing of sleep in mammals [99]. The SCN regulates the timing of melatonin release via a multisynaptic projection connecting the PVN and the superior cervical ganglion of the spinal cord, and terminating in the pineal gland (Fig. 1). In both diurnal and nocturnal animals, melatonin release peaks during the night and is lowest during the day [100] and light inhibits its secretion, giving rise to the nickname the “night hormone.” Melatonin can phase shift and entrain behavioral circadian rhythms in both nocturnal rodent models [101] and humans [99], although contrary to popular belief, exogenous administration of melatonin is likely to only have a modest sleep-promoting effect [102]. Melatonin plays a direct role in rhythmically downregulating core body temperature (CBT) [103], and in humans low CBT is necessary for transitions from wakefulness to sleep [104]. Indeed, the acute effects of melatonin on sleepiness have been observed to be coincident with this dip in CBT [105]. Importantly, the effects of melatonin on sleep in humans are also time-dependent, such that administration of exogenous melatonin during the day when endogenous melatonin levels are low is sleep-promoting [106,107], whereas nighttime administration has been shown to depend more on dosage and vary between subjects [99]. Similar sleep-promoting effects have also been reported in three different species of diurnal non-human primates [108]. As a result, melatonin has been used to induce phase shifts of sleep in therapeutic contexts ranging from blindness [109], jet lag, shift work, insomnia and circadian rhythm disorders [110].
Despite modest sleep-promoting effects in diurnal animals, melatonin is unlikely to promote sleep in nocturnal animal models in fact may induce opposite effects. Melatonin has been shown to have wake-promoting effects when administered during the daytime in rats [111, 112], and there is evidence to suggest that endogenous melatonin is not necessary for proper timing and consolidation of sleep bouts in the rat [113]. Many strains of nocturnal mice commonly used as animal models in studies of circadian rhythms display little or no endogenous secretion of melatonin [114], and the effects of exogenous administration of melatonin on sleep in these mice remains controversial [115].
2.6. Sleep and the molecular circadian clockwork
In mammalian species from mice to humans, the core circadian clock genes comprising the molecular TTFL have been shown to be critical in the circadian regulation of sleep (Table 1). The first mammalian circadian clock gene to be identified, named Clock, was isolated and cloned in mice by Joseph Takahashi and colleagues [116] in the late 1990’s. This work opened the door to a flurry of discoveries about the roles of Clock and other mammalian circadian clock genes in assembling the molecular TTFL, and its essential nature in generating and sustaining circadian rhythms [117–119].
Table 1.
Summary of the effects of circadian clock gene mutations on sleep and circadian rhythms.
| Gene | Species | Manipulation or mutation | Region | Phenotype | Reference |
|---|---|---|---|---|---|
| Cry1/Cry2 | Mouse | Double knockout | Global |
|
[120] |
| Per1/Per2 | Mouse | Double knockout | Global |
|
[121, 122] |
| Per2 | Mouse | Human PER2 mutation associated with advanced sleep phase | Global | Shortened circadian period of activity | [139] |
| Per3 | Mouse | Human PER3 mutation associated with changes in sleep timing and sleep rebound | Global |
|
[126] |
| Clock | Mouse | Δ19 mutation | Global | Reduced NREM duration | [123] |
| Clock | Mouse | Knockout | Excitatory cortical neurons | Seizures during NREM sleep | [129] |
| Npas2 | Mouse | Knockout | Global |
|
[124] |
| Timeless | Mouse | Human TIM mutation associated with advanced sleep phase | Global |
|
[141] |
| Bmal1 | Mouse | Knockout | Global |
|
[125] |
| Bmal1 | Mouse | Knockout | Syt10-expressing neurons | Activity rhythms abolished | [130] |
| Bmal1 | Mouse | Knockout | Forebrain neurons including SCN | Activity rhythms abolished | [131] |
| Bmal1 | Mouse | Knockout | NMS SCN neurons | Activity rhythms abolished | [21] |
| Bmal1 | Mouse | Knockout | VIP SCN neurons | Reduced amplitude of circadian rhythms of sleep | [132] |
| Bmal1 | Mouse | Knockout | Forebrain neurons excluding SCN | Increased NREM during dark phase | [133] |
| Bmal1 | Mouse | Knockout | VIP SCN neurons | Disrupted daily siesta | [162] |
| Bmal1 | Mouse | Knockout | AVP SCN neurons | Loss of tissue-level Per2 expression rhythms in ex vivo SCN culture | [164] |
| Bmal1 | Mouse | Knockout, rescue | Expression rescued in skeletal muscle | Recovery of normal sleep duration but not timing | [134] |
| PER2 | Human | Missense mutation | Global | Advanced sleep phase syndrome (ASPS) | [138] |
| CRY2 | Human | Missense mutation | Global | ASPS | [140] |
| TIMELESS | Human | Nonsense mutation | Global | ASPS | [141] |
| PER3 | Human | Short length polymorphism | Global | Delayed sleep phase syndrome (DSPS) | [142] |
| PER3 | Human | Longer length polymorphism | Global | Extreme morningness | [142] |
| CRY1 | Human | Gain-of-function mutation | Global | DSPS | [143] |
Following these discoveries, researchers began to study the roles of clock genes in sleep regulation, although a clear picture of clock gene knockouts effects on circadian rhythms of sleep has yet to emerge. Mice containing a double knockout of the cryptochrome genes Cry1 and Cry2 not only lack circadian rhythmicity in constant conditions, they display a greater amount of NREM sleep and higher sleep consolidation, suggesting a role for clock genes in both homeostatic and circadian regulation of sleep [120]. Single and double knockouts of the Period genes Per1 and Per2 are similarly arrhythmic, but display typical amounts of NREM, REM and wake, as well as preserved homeostatic sleep regulation [121,122]. Mice with the Δ19 Clock mutation exhibit reduced amounts of NREM sleep but retain circadian rhythmicity of sleep [123]. Similarly, mice lacking the transcription factor Npas2, a paralog of Clock in brain regions outside of the SCN, display normal circadian rhythms of sleep but altered amounts of NREM sleep and sleep homeostasis [124]. Bmal1 knockout mice, which lack circadian rhythmicity under constant conditions [119], also display a multitude of sleep disturbances including reduced amplitude of circadian rhythms of sleep, sleep fragmentation, increased total sleep time and reduced homeostatic sleep response [125]. Interestingly, a primate-specific variable-number tandem-repeat polymorphism in PER3 is associated with an altered homeostatic sleep response but normal circadian rhythms of sleep in mice [126]. These seemingly contradictory results obviate the complex roles of individual TTFL components in regulating not only sleep timing but quality and duration.
It is important to note that these initial studies employed global deletion or mutations of core clock genes, which are not solely expressed in the SCN. Although the SCN coordinates cell- and tissue-level circadian rhythms throughout the entire organism, several other sleep-relevant brain regions display circadian oscillations in neuronal activity and gene expression themselves, including the ARC, DMH, and VTA [127]. Such oscillations, both in SCN and extra-SCN brain regions, drive rhythms of cellular processes including expression and modulation of receptors and ion channels that drive neuronal excitability [128], as illustrated by a recent study in which mice containing an excitatory cortical neuron-specific deletion of Clock displayed epileptic seizures during SWS [129]. Beyond the brain, clock gene expression in organs ranging from the lungs to the liver is necessary for a variety of physiological functions, the disruption of which may have marked effects on sleep.
Such observations make interpretation of results from global clock gene mutant mice more difficult and highlight the need for region and cell type-specific approaches to study the role of clock genes in the circadian regulation of sleep. For example, more recent studies have demonstrated that deletion of Bmal1 in Syt10-expressing neurons, which are abundant in the SCN [130], or the forebrain [131] is sufficient to abolish circadian rhythms of locomotor activity in mice. Similar behavior was observed following deletion of Bmal1 in NMS neurons [21], which are localized to the SCN. Finally, another study found that deletion of Bmal1 from VIP SCN neurons reduced the amplitude of circadian rhythms of sleep [132].
The importance of clock gene expression in extra-SCN brain regions was further highlighted by recent work demonstrating that deletion of Bmal1 in forebrain neurons outside of the SCN increased NREM sleep duration during the dark phase and altered daily timing of both NREM and REM sleep [133]. Clock gene expression in tissues outside of the brain likely also play a role in sleep regulation, as rescue of Bmal1 expression in skeletal muscle tissue in Bmal1 knockout mice was sufficient to curb associated deficits in sleep duration but not timing [134]. Further region-specific studies of clock gene expression and sleep, as well as investigation of the effects of different sleep states and perturbations on clock gene expression in regions beyond the SCN, will reveal much needed greater insights into how the molecular TTFL regulates sleep timing.
These characterizations of circadian clock genes in animal models also paved the way for greater understanding of their role in human sleep regulation [135]. Such insights first came from investigations of different chronotypes, which describe an individual’s preferred sleep timing. The distribution of chronotypes throughout the population is approximately normal, such that extreme morning “larks” will rise at the same time late “owls” fall asleep and most people display a more conventional sleep schedule, although these preferences change across the lifespan and under environmental influences like light exposure and timed feeding [136].
Familial advanced sleep phase syndrome (ASPS) is an inheritable disorder in which patients have an extreme early chronotype with a 4-h advance of sleep, melatonin and core body temperature rhythms [137]. In 2001, the first example of an association between a clock gene mutation and sleep timing in humans came to light when ASPS was attributed to a missense mutation in the human PER2 gene, saddling patients with an extremely short circadian period [138]. When researchers introduced a similar Per2 mutation in transgenic mice, the mice displayed the same shortened circadian period of behavior [139]. More recently, ASPS was associated with mutations in CRY2 [140] and TIMELESS, the latter of which has been studied extensively in Drosophila but is less well understood in mammals [141]. Delayed sleep phase syndrome (DSPS), in which people tend to be extreme late chronotypes, is associated with a short length polymorphism in the clock gene PER3, while the longer length allele was associated with morningness [142]. DSPS has also been associated with a gain-of-function mutation of CRY1, and patients with this mutation also displayed an elongated period of molecular circadian rhythms [143]. More recently, genome-wide association studies (GWAS) leveraging large genomic datasets and actigraphy-derived measures of sleep timing have suggested contributions for known clock genes, and several non-clock genes, to determining chronotype [144]. While GWAS studies are limited in their reproducibility and ability to establish causal relationships between loci and circadian behaviors, they provide potentially promising new lines of inquiry for researchers working to understand the genetics underlying the circadian regulation of sleep.
3. Sleep and the clock during development
Rhythms of sleep and wake, at both the circadian and ultradian scales, are hardly static throughout the lifespan. Indeed, both chronotype and the nature of ultradian sleep cycles undergo marked changes from infancy and early childhood to adolescence [145], and these changes are likely to play a significant role in several aspects of brain function and development [146]. While adult sleep duration is approximately 8 h per day, infants spend up to 16 h of their day sleeping with more rapid cycling between sleep stages. More typical ultradian rhythms of sleep do not emerge until children are school-age [145], such that ultradian cycle length increases while cycle number decreases as children progress through early childhood [147]. While circadian rhythms of core body temperature and melatonin secretion may emerge as early as 6 months [148], unlike adults toddlers may display REM sleep during daytime naps [149], suggesting that the development of circadian regulation of sleep during childhood may be more complex.
Surprisingly little is known about the mechanisms underlying these developmental transitions in sleep regulation [150]. A review from Blumberg & colleagues proposes a framework for studying the developmental trajectory of bidirectional interactions between brainstem and hypothalamic circuitry driving ultradian, circadian and homeostatic sleep regulation, using early postnatal rats as a model system [150]. While some qualitative differences in the manifestation of sleep exist at earlier developmental time points, such as the emergence of EEG delta activity at postnatal day 11 (P11) [151] and a reliance on the mother for entrainment of behavioral circadian rhythms until P8 [152], young rats display rapid ultradian cycling between sleep stages consistent with the flip-flop switch model, and evidence suggests these transitions are mediated by circuitry contained entirely within the brainstem during the early postnatal period [150]. Additionally, the distribution of sleep and wake bouts across the 24-h day display a predictable progression throughout development [150], with marked circadian regulation of sleep emerging by P17, well before sleep homeostasis [153].
These observations make young rats a highly tractable and fruitful model system in which to study the mechanisms of both circadian and ultradian sleep regulation. Researchers have found that rats display day-night cycling of sleep and wakefulness as early as P2, but unlike in adult rats, the duration of both sleep and wake bouts were shorter during the night [154]. Later work demonstrated that lesioning the SCN or DMH in rats at age P8 was associated with the fragmentation of wake bouts and disruption of wake bout distribution later in life, but the same was not true for sleep bouts [155]. Finally, differences in the developmental trajectory of connectivity between the SCN and ventral SPZ have been associated with differences in activity timing between diurnal and nocturnal rats [156].
One disadvantage of rats as model systems is the relative lack of readily available transgenic models and tools for the manipulation and imaging of neural activity. Mouse models meet this requirement, however due to technical difficulties in measuring sleep in perinatal mice, relatively few studies have characterized quantitative and qualitative changes in sleep across development. One notable exception demonstrated that orexin receptors were necessary for normal consolidation of sleep and wake bouts in mice between P12 and P21 [157]. Another study evaluated sleep architecture and homeostatic sleep regulation in adolescent mice and found a decrease in REM sleep and increase in NREM sleep relative to adults. Interestingly, while increases in delta sleep typical of a normal homeostatic sleep response were observed in adolescent mice, the magnitude of this increase showed high inter-individual variability until approximately P42 [158]. More recently, a comprehensive survey of sleep in mouse pups at different time points between P7 and P21 demonstrated that the emergence of both qualitative EEG features and consolidation of sleep stages largely mirrors the developmental trajectory of sleep features observed in rats and humans, although the study did not evaluate circadian regulation of sleep [159]. Increased understanding of sleep during development in the mouse, combined with the large transgenic toolbox available in mouse models, provides a new opportunity to characterize developmental mechanisms underlying the circadian regulation of sleep.
In addition to the behavioral and physiological studies described above, understanding the development of circadian sleep regulation calls for a comprehensive picture of the developing SCN. However, despite our knowledge of adult SCN anatomy and physiology, comparatively little attention is paid to unraveling the mechanisms of SCN development and its connectivity to sleep-relevant brain regions [160, 161]. The development of SCN cellular differentiation, patterning, rhythmicity, afferents and paracrine signaling are relatively well-studied, but little is known regarding the development of SCN efferents [160]. Because both direct and indirect projections from the SCN to brain regions in the sleep-wake regulatory network play key roles in sleep timing, characterizations of SCN efferents from a developmental perspective will be key to understanding the circadian regulation of sleep during development.
4. Advancing to the next phase – future directions for sleep and the circadian clock
4.1. Uncovering new roles for the master pacemaker in sleep regulation
In recent years, researchers have begun to elucidate mechanisms by which the SCN may exert a more direct influence over sleep timing. A report by Collins et al. [162] demonstrated that nighttime optogenetic stimulation of a subset night-active SCN neurons that co-express VIP and NMS is sufficient to acutely promote sleep in mice, while stimulating or inhibiting these neurons during the day has no effect on sleep. The peak of activity in these neurons was coincident with the daily “siesta” - a brief bout of sleep typically observed as a lull in locomotor activity occurring towards the end of the activity period in behavioral studies of circadian rhythms – and the timing of the siesta was dependent on an intact molecular clockwork in SCN VIP neurons. The authors thus hypothesized that SCN VIP neurons are responsible for timing the siesta such that this brief bout of sleep boosts wakefulness during the wake-maintenance zone, a brief period of wakefulness at the end of the activity bout [163]. Interestingly, another study found that while Bmal1 expression in SCN VIP neurons was necessary for normal amplitude of circadian sleep-wake rhythms, chemogenetic stimulation of these neurons during the night did not have an acute sleep- or wake-promoting effect [132]. These studies highlight the need for further inquiry into the role of these neurons in directly regulating sleep timing.
Elucidating a potential direct role for the SCN in circadian sleep regulation will be aided by better understanding of specific cell types within the SCN, and specific genetic tools with which to target them. A recent study employed single-cell RNA sequencing (scRNA-seq) of mouse SCN and identified 5 distinct SCN neuronal cell types, largely confirming our current understanding of the anatomy and function of the master clock [19] while simultaneously revealing greater heterogeneity than was previously appreciated [20]. Each cell type expressed genes encoding at least two primary neuropeptides and were demonstrated to play different roles in mediating circadian behavior. For example, Grp+/Vip+ neurons were highly photo-responsive but displayed weak intrinsic circadian oscillations in gene expression, while Avp+/Nms+ and Cck+/C1q13+ neurons displayed opposite behavior. Some cell types also oscillated at different phases with respect to other cell types, which the authors suggest could have a cell type-specific effect on the regulation of the SCN’s downstream neuronal targets. Based on these observations, the authors developed a Cre-recombinase reporter mouse in which Vip+/Nms+ SCN neurons specifically could be fluorescently labeled, thus allowing for a thorough characterization of their spatiotemporal distribution throughout the nucleus. Such emphasis on specific SCN cell types has already begun to reveal surprising insights about their differential contributions to regulating circadian rhythms. Shan et al. recently described the development of a Cre-inducible bioluminescent reporter mouse line in which rhythms of Per2 expression in AVP and VIP neurons of the SCN can be monitored simultaneously in different color channels [164]. Using this reporter mouse, the researchers found that in ex vivo SCN slice preparations, Bmal1 expression was necessary and sufficient for maintaining circadian rhythmicity of Per2 expression in AVP neurons, but not in VIP neurons.
Such lines of inquiry highlight the need for further investigation into the roles of specific SCN cell types in the circadian regulation of sleep. Evaluating sleep behavior in mice in which individual cell types are targeted for excitation, inhibition, imaging and/or genetic manipulation represents an especially promising direction for the field [165]. For example, recent work demonstrated that in vivo optogenetic manipulation of either the whole SCN or VIP neurons specifically is sufficient to entrain circadian rhythms of locomotor activity and CBT in mice [166]. Such cell type-specific manipulations of SCN neurons have also borne fruit in understanding how reciprocal connections between the SCN and downstream hypothalamic areas can acutely regulate behavior. In 2016, Gizowski et al. demonstrated that excitatory peptidergic signaling from SCN AVP neurons to the organum vasculosum lamina terminalis (OVLT) gates the timing of anticipatory water intake prior to sleep in mice. Optogenetic stimulation of SCN AVP neurons outside of the normal water intake period resulted in an acute increase in water drinking behavior [167]. Conversely, excitatory GABAergic signaling from osmo-sodium-sensing neurons in the OVLT to SCN AVP neurons conveyed information about systemic osmolality and could shift the phase of the circadian clock. Optogenetically stimulating this pathway phase shifted the locomotor activity bout and lowered CBT [168]. Similar characterizations of functional synaptic contacts between specific SCN cell types and sleep-relevant targets such as the SPZ, DMH and VLPO will deepen our understanding of how the circadian clock drives sleep timing. However, caution should be taken when interpreting the effects of manipulating SCN neuronal activity on sleep. The SCN is remarkable in its ability to regulate the 24-h timing of virtually all behavioral and physiological outputs, and its regulation of sleep could be indirect by affecting other outputs and not necessarily the generation of sleep or wakefulness. For instance, the activity modulation of SCN neurons essential to sustain CBT rhythms would not only change the circadian rhythm of CBT but also affect the temperature-dependent modulation of sleep.
4.2. Long-term monitoring of sleep architecture
Much of what we understand about the circadian regulation of sleep comes from animal studies measuring locomotor activity, as relatively few studies evaluate sleep rhythms for longer than a few days. In 1985, Richardson et al. employed continuous, tethered EEG/EMG recording in mice for durations of 60–280 days and studied sleep architecture under conditions including baseline LD cycles, constant darkness and following light pulses that shifted the phase of the circadian clock [169]. This work revealed great insight into the daily distribution of wakefulness, NREM and REM during these conditions, and largely reflected sleep behaviors observed in human forced desynchronization studies. Importantly, the rhythms of electrographically measured sleep differed from rhythms of locomotor activity as measured by wheel-running, highlighting the caveats of inferring sleep timing from locomotor activity alone, to say nothing of the ability to evaluate timing of sleep stages. Later work employed long-term sleep monitoring to demonstrate that the timing of REM sleep is coupled to an oscillator in the dmSCN in rats, as described above [87]. Other studies have used a combination of continuous and longitudinal monitoring of EEG/EMG to evaluate changes in sleep stage timing under conditions inducing chronic disruption to the circadian system such as LD cycles differing from 24 h [170] and serial jet lags [171] in mice. Most recently, continuous sleep monitoring in mice during a single jet lag paradigm revealed that the primary bout of sleep re-entrains to the new LD cycle faster than the daily siesta [172].
The paucity of studies employing long-term sleep monitoring likely stems from technical difficulties associated with chronic tethered recordings and a lack of standardization of methods for automatic sleep stage classification. This latter point is especially important, as manual sleep staging is labor-intensive and error prone. Automatic sleep stage classification has been an active area of research since the 1960’s, with proposed methods demonstrating a range of accuracy and level of adoption by the field [173]. More recent approaches leveraging machine learning and datasets from variety of animal models and experimental conditions have shown promise in both offline and real-time classification of sleep stages performed with user-friendly graphical interfaces [174–176]. Long-term, continuous sleep monitoring becomes far more practical when aided by these methods. Such experiments, when combined with systems neuroscience tools described above and newly described non-invasive methods for chronic imaging of peripheral clock gene expression in freely behaving mice [177] will be invaluable to furthering our understanding of how the circadian clock regulates sleep.
4.3. Bringing circadian regulation of sleep into the light
A key limitation in current approaches to understanding the circadian regulation of sleep is that nocturnal rodents are the most widely used animal models in the field. As a result, some insights we acquire with these models are unlikely to directly translate to humans and other diurnal animals. While sleep and circadian behavior in diurnal mammals such as the squirrel monkey and Nile grass rat [178] are well-studied, they are far less represented in the field than nocturnal mice, which have historically been more easily amenable to genetic manipulation. Although SCN physiology is similar in nocturnal and diurnal animals, it must regulate sleep in opposite directions during equivalent phases of the LD cycle. Similarly, light stimulates and inhibits sleep in nocturnal and diurnal species, respectively [179,180]. “ON/OFF” switches that distinguish these effects between diurnal and nocturnal models have been proposed. However, these switches remain to be identified and are likely a convenient oversimplification of how sleep is temporally organized.
With the advent of low-cost sequencing technologies like scRNA-seq and CRISPR-Cas-based gene editing methods, as well as behavioral paradigms that induce diurnality in nocturnal animals [181], the development of diurnal transgenic animal models becomes more tractable. Additionally, increasingly sophisticated wireless data loggers that can capture EEG, EMG, CBT and locomotor activity are allowing for sleep monitoring in unconventional model systems, including animals in the wild [182]. Leveraging these technologies to develop new model systems will fill crucial gaps in our understanding of circadian sleep regulation in diurnal mammals.
4.4. Conclusions
Many important questions remain unanswered in the field of circadian sleep regulation, from how the SCN drives sleep to occur at different times of day in diurnal versus nocturnal animals, to how the clock responds to internal and external environmental factors to modulate sleep timing in a flexible manner. While addressing these questions remains a tantalizing endeavor for basic researchers, the importance of developing a mechanistic understanding of Process C goes well beyond the bench, as we now know that disruptions of sleep and circadian rhythms are associated with metabolic disorders [26], cancer [183], mental health disorders [184], neurological disease (e.g., epilepsy), and neurodegenerative diseases (i.e, Parkinson’s and Alzheimer diseases) [185]. Additionally, there is increasing public understanding that artificial lighting [186], socioeconomic disparities [187], daylight savings time [188] and societal constraints [189] impact sleep timing. Recent advances described above promise to help resolve longstanding questions and reveal mechanistic insights into how and why the circadian clock gates the timing of sleep.
Funding
This work was supported by NIH awards R01 NS110012 and R01 NS108934 to HOD, a CURE Sleep & Epilepsy grant NINDS award R01 NS102796-01 to FK, and Washington Research Foundation Innovation Graduate Fellowship in Neuroengineering to RS.
References
- [1].Merrow M, Spoelstra K, Roenneberg T, The circadian cycle: daily rhythms from behaviour to genes, EMBO Rep. 6 (2005) 930–935, 10.1038/sj.embor.7400541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Partch CL, Green CB, Takahashi JS, Molecular architecture of the mammalian circadian clock, Trends Cell Biol. 24 (2014) 90–99, 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hastings MH, Maywood ES, Brancaccio M, Generation of circadian rhythms in the suprachiasmatic nucleus, Nat. Rev. Neurosci 19 (2018) 453–469, 10.1038/s41583-018-0026-z. [DOI] [PubMed] [Google Scholar]
- [4].Welsh DK, Logothetis DE, Meister M, Reppert SM, Individual Neurons Dissociated from Rat Suprachiasmatic Nucleus Express Independently Phased Circadian Firing Rhythms, 1995. [DOI] [PubMed] [Google Scholar]
- [5].Welsh DK, Takahashi JS, Kay SA, Suprachiasmatic nucleus: cell autonomy and network properties, Annu. Rev. Physiol 72 (2010) 551–577, 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hattar S, Liao HW, Takao M, Berson DM, Yau KW, Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity, Science 295 (80) (2002) 1065–1070, 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ding JM, Faiman LE, Hurst WJ, Kuriashkina LR, Gillette MU, Resetting the biological clock: mediation of nocturnal CREB phosphorylation via light, glutamate, and nitric oxide, J. Neurosci 17 (1997) 667–675, 10.1523/jneurosci.17-02-00667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mintz EM, Marvel CL, Gillespie CF, Price KM, Albers HE, Activation of NMDA receptors in the suprachiasmatic nucleus produces light-like phase shifts of the circadian clock in vivo, J. Neurosci 19 (1999) 5124–5130, 10.1523/jneurosci.19-12-05124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, Okamura H, Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript, Cell 91 (1997) 1043–1053, 10.1016/S0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- [10].Inouye ST, Kawamura H, Persistence of circadian rhythmicity in a mammalian hypothalamic “island” containing the suprachiasmatic nucleus, Proc. Natl. Acad. Sci. USA 76 (1979) 5962–5966. 〈 10.1073/pnas.76.11.5962〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cassone VM, Speh JC, Card JP, Moore RY, Comparative anatomy of the mammalian hypothalamic suprachiasmatic nucleus, J. Biol. Rhythms 3 (1988) 71–91, 10.1177/074873048800300106. [DOI] [PubMed] [Google Scholar]
- [12].Slat E, Freeman GM, Herzog ED, The clock in the brain: neurons, glia, and networks in daily rhythms, Handb. Exp. Pharmacol (2013) 105–123, 10.1007/978-3-642-25950-0_5. [DOI] [PubMed] [Google Scholar]
- [13].Morin LP, Shivers KY, Blanchard JH, Muscat L, Complex organization of mouse and rat suprachiasmatic nucleus, Neuroscience 137 (2006) 1285–1297, 10.1016/j.neuroscience.2005.10.030. [DOI] [PubMed] [Google Scholar]
- [14].Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, Kelly JS, Maywood ES, Hastings MH, The VPAC2 receptor is essential for circadian function in the mouse suprachiasmatic nuclei, Cell 109 (2002) 497–508, 10.1016/S0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- [15].Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED, Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons, Nat. Neurosci 8 (2005) 476–483, 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Albers HE, Walton JC, Gamble KL, McNeill JK, Hummer DL, The dynamics of GABA signaling: revelations from the circadian pacemaker in the suprachiasmatic nucleus, Front. Neuroendocrinol 44 (2017) 35–82, 10.1016/j.yfrne.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Maywood ES, Reddy AB, Wong GKY, O’Neill JS, O’Brien JA, McMahon DG, Harmar AJ, Okamura H, Hastings MH, Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling, Curr. Biol 16 (2006) 599–605, 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- [18].Cheng MY, Bullock CM, Li C, Lee AG, Bermak JC, Belluzzi J, Weaver DR, Leslie FM, Zhou QY, Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus, Nature 417 (2002) 405–410, 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- [19].Varadarajan S, Tajiri M, Jain R, Holt R, Ahmed Q, Lesauter J, Silver R, Connectome of the suprachiasmatic nucleus: New evidence of the core-shell relationship, ENeuro 5 (2018) 205–223, 10.1523/ENEURO.0205-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wen A, Ma D, Zhao M, Xie L, Wu Q, Gou L, Zhu C, Fan Y, Wang H, Yan J, Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus, Nat. Neurosci (n.d.). 〈 10.1038/s41593-020-0586-x〉. [DOI] [PubMed] [Google Scholar]
- [21].Lee IT, Chang AS, Manandhar M, Shan Y, Fan J, Izumo M, Ikeda Y, Motoike T, Dixon S, Seinfeld JE, Takahashi JS, Yanagisawa M, Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms, Neuron 85 (2015) 1086–1102, 10.1016/j.neuron.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Carskadon W, Dement MA, Normal human sleep: an overview, in: Principles and Practice of Sleep Medicine, fifth ed., 2004, pp. 16–26. 10.1016/j.mcna.2004.01.001. [DOI] [Google Scholar]
- [23].Joiner WJ, Current biology review unraveling the evolutionary determinants of sleep, Curr. Biol 26 (2016) R1073–R1087, 10.1016/j.cub.2016.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Watson BO, Buzsáki G, Sleep, memory & brain rhythms, Daedalus 144 (2015) 67–82, 10.1162/DAED_a_00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Besedovsky L, Lange T, Born J, Sleep and immune function, Pflug. Arch. Eur. J. Physiol 463 (2012) 121–137, 10.1007/s00424-011-1044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sharma S, Kavuru M, Sleep and metabolism: an overview, Int. J. Endocrinol 2010 (2010) 1–12, 10.1155/2010/270832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Campbell SS, Tobler I, Animal sleep: a review of sleep duration across phylogeny, Neurosci. Biobehav. Rev 8 (1984) 269–300, 10.1016/0149-7634(84)90054-X. [DOI] [PubMed] [Google Scholar]
- [28].Scammell TE, Arrigoni E, Lipton JO, Neural circuitry of wakefulness and sleep, Neuron 93 (2017) 747–765, 10.1016/j.neuron.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hallanger AE, Levey AI, Lee HJ, Rye DB, Wainer BH, The origins of cholinergic and other subcortical afferents to the thalamus in the rat, J. Comp. Neurol 262 (1987) 105–124, 10.1002/cne.902620109. [DOI] [PubMed] [Google Scholar]
- [30].HL W, M. M, Pedunculopontine and laterodorsal tegmental nuclei contain distinct populations of cholinergic, glutamatergic and GABAergic neurons in the rat, Eur. J. Neurosci 29 (2009), 10.1111/J.1460-9568.2008.06576.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].McCormick DA, Cholinergic and noradrenergic modulation of thalamocortical processing, Trends Neurosci. 12 (1989) 215–221, 10.1016/0166-2236(89)90125-2. [DOI] [PubMed] [Google Scholar]
- [32].Steriade M, Curro Dossi R, Pare D, Oakson G, Fast oscillations (20–40 Hz) in thalamocortical systems and their potentiation by mesopontine cholinergic nuclei in the cat, Proc. Natl. Acad. Sci. USA 88 (1991) 4396–4400, 10.1073/pnas.88.10.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kroeger D, Ferrari LL, Petit G, Mahoney CE, Fuller PM, Arrigoni E, Scammell TE, Cholinergic, glutamatergic, and GABAergic neurons of the pedunculopontine tegmental nucleus have distinct effects on sleep/wake behavior in mice, J. Neurosci 37 (2017) 1352–1366, 10.1523/JNEUROSCI.1405-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Buzsaki G, Bickford RG, Ponomareff G, Thal LJ, Mandel R, Gage FH, Nucleus basalis and thalamic control of neocortical activity in the freely moving rat, J. Neurosci 8 (1988) 4007–4026, 10.1523/jneurosci.08-11-04007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xu M, Chung S, Zhang S, Zhong P, Ma C, Chang W-C, Weissbourd B, Sakai N, Luo L, Nishino S, Dan Y, Basal forebrain circuit for sleep-wake control, Nat. Neurosci 18 (2015) 1641–1647, 10.1038/nn.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Luppi PH, Aston-Jones G, Akaoka H, Chouvet G, Jouvet M, Afferent projections to the rat locus coeruleus demonstrated by retrograde and anterograde tracing with cholera-toxin B subunit and Phaseolus vulgaris leucoagglutinin, Neuroscience 65 (1995) 119–160, 10.1016/0306-4522(94)00481-J. [DOI] [PubMed] [Google Scholar]
- [37].Weissbourd B, Ren J, DeLoach KE, Guenthner CJ, Miyamichi K, Luo L, Presynaptic partners of dorsal raphe serotonergic and GABAergic neurons, Neuron 83 (2014) 645–662, 10.1016/j.neuron.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Eban-Rothschild A, Rothschild G, Giardino WJ, Jones JR, De Lecea L, VTA dopaminergic neurons regulate ethologically relevant sleep-wake behaviors, Nat. Neurosci 19 (2016) 1356–1366, 10.1038/nn.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lu J, Jhou TC, Saper CB, Identification of wake-active dopaminergic neurons in the ventral periaqueductal gray matter, J. Neurosci 26 (2006) 193–202, 10.1523/JNEUROSCI.2244-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cho JR, Treweek JB, Robinson JE, Xiao C, Bremner LR, Greenbaum A, Gradinaru V, Dorsal raphe dopamine neurons modulate arousal and promote wakefulness by salient stimuli, Neuron 94 (2017) 1205–1219.e8, 10.1016/j.neuron.2017.05.020. [DOI] [PubMed] [Google Scholar]
- [41].Venner A, Mochizuki T, De Luca R, Anaclet C, Scammell TE, Saper CB, Arrigoni E, Fuller PM, Reassessing the role of histaminergic tuberomammillary neurons in arousal control, J. Neurosci 39 (2019) 8929–8939, 10.1523/JNEUROSCI.1032-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Szymusiak R, Alam N, Steininger TL, McGinty D, Sleep-waking discharge patterns of ventrolateral preoptic/anterior hypothalamic neurons in rats, Brain Res. 803 (1998) 178–188, 10.1016/S0006-8993(98)00631-3. [DOI] [PubMed] [Google Scholar]
- [43].Gaus SE, Strecker RE, Tate BA, Parker RA, Saper CB, Ventrolateral preoptic nucleus contains sleep-active, galaninergic neurons in multiple mammalian species, Neuroscience 115 (2002) 285–294, 10.1016/S0306-4522(02)00308-1. [DOI] [PubMed] [Google Scholar]
- [44].von Economo C, Sleep as a problem of localization, J. Nerv. Ment. Dis 71 (1930) 249–259. [Google Scholar]
- [45].Zhang Z, Ferretti V, Güntan I, Moro A, Steinberg EA, Ye Z, Zecharia AY, Yu X, Vyssotski AL, Brickley SG, Yustos R, Pillidge ZE, Harding EC, Wisden W, Franks NP, Neuronal ensembles sufficient for recovery sleep and the sedative actions of α 2 adrenergic agonists, Nat. Neurosci 18 (2015) 553–561, 10.1038/nn.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liu D, Li W, Ma C, Zheng W, Yao Y, Tso CF, Zhong P, Chen X, Song JH, Choi W, Paik SB, Han H, Dan Y, A common hub for sleep and motor control in the substantia nigra, Science 367 (2020) 440–445, 10.1126/science.aaz0956. [DOI] [PubMed] [Google Scholar]
- [47].Hobson JA, Mccarley RW, Wyzinski PW, Sleep cycle oscillation: reciprocal discharge by two brainstem neuronal groups, Science 189 (80) (1975) 55–58, 10.1126/science.1094539. [DOI] [PubMed] [Google Scholar]
- [48].Van Dort CJ, Zachs DP, Kenny JD, Zheng S, Goldblum RR, Gelwan NA, Ramos DM, Nolan MA, Wang K, Weng FJ, Lin Y, Wilson MA, Brown EN, Optogenetic activation of cholinergic neurons in the PPT or LDT induces REM sleep, Proc. Natl. Acad. Sci. USA 112 (2015) 584–589. 〈 10.1073/pnas.1423136112〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Krenzer M, Anaclet C, Vetrivelan R, Wang N, Vong L, Lowell BB, Fuller PM, Lu J, Brainstem and spinal cord circuitry regulating REM sleep and muscle atonia, PLoS One 6 (2011) 24998, 10.1371/journal.pone.0024998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Weber F, Chung S, Beier KT, Xu M, Luo L, Dan Y, Control of REM sleep by ventral medulla GABAergic neurons, Nature 526 (2015) 435–438, 10.1038/nature14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ferreira JGP, Bittencourt JC, Adamantidis A, Melanin-concentrating hormone and sleep, Curr. Opin. Neurobiol 44 (2017) 152–158, 10.1016/j.conb.2017.04.008. [DOI] [PubMed] [Google Scholar]
- [52].Chen KS, Xu M, Zhang Z, Chang WC, Gaj T, Schaffer DV, Dan Y, A hypothalamic switch for REM and Non-REM sleep, e4, Neuron 97 (2018) 1168–1176.e4, 10.1016/j.neuron.2018.02.005. [DOI] [PubMed] [Google Scholar]
- [53].Saper CB, Chou TC, Scammell TE, The sleep switch: hypothalamic control of sleep and wakefulness, Trends Neurosci. 24 (2001) 726–731, 10.1016/S0166-2236(00)02002-6. [DOI] [PubMed] [Google Scholar]
- [54].Saper CB, Scammell TE, Lu J, Hypothalamic regulation of sleep and circadian rhythms, Nature 437 (2005) 1257–1263, 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- [55].Lee MG, Hassani OK, Jones BE, Discharge of identified orexin/hypocretin neurons across the sleep-waking cycle, J. Neurosci 25 (2005) 6716–6720, 10.1523/JNEUROSCI.1887-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Peyron C, Tighe DK, Van Den Pol AN, De Lecea L, Heller HC, Sutcliffe JG, Kilduff TS, Neurons containing hypocretin (orexin) project to multiple neuronal systems, J. Neurosci 18 (1998) 9996–10015, 10.1523/jneurosci.18-23-09996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Borbély AA, Two Process A, Model of sleep regulation, Hum. Neurobiol 1 (1982). [PubMed] [Google Scholar]
- [58].Borbély AA, Daan S, Wirz-Justice A, Deboer T, The two-process model of sleep regulation: a reappraisal, J. Sleep Res 25 (2016) 131–143, 10.1111/jsr.12371. [DOI] [PubMed] [Google Scholar]
- [59].Mistlberger RE, Circadian regulation of sleep in mammals: role of the suprachiasmatic nucleus, Brain Res. Rev 49 (2005) 429–454, 10.1016/j.brainresrev.2005.01.005. [DOI] [PubMed] [Google Scholar]
- [60].Borbély AA, Neuhaus HU, Sleep-deprivation: effects on sleep and EEG in the rat, J. Comp. Physiol. A 133 (1979) 71–87, 10.1007/BF00663111. [DOI] [Google Scholar]
- [61].Dijk DJ, Czeisler CA, Contribution of the circadian pacemaker and the sleep homeostat to sleep propensity, sleep structure, electroencephalographic slow waves, and sleep spindle activity in humans, J. Neurosci 15 (1995) 3526–3538, 10.1523/jneurosci.15-05-03526.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Werth E, Dijk DJ, Achermann P, Borbély AA, Dynamics of the sleep EEG after an early evening nap: experimental data and simulations, Am. J. Physiol. Regul. Integr. Comp. Physiol 271 (1996) R501–R510, 10.1152/ajpregu.1996.271.3.r501. [DOI] [PubMed] [Google Scholar]
- [63].Huber R, Deboer T, Tobler I, Topography of EEG dynamics after sleep deprivation in mice, J. Neurophysiol 84 (2000) 1888–1893, 10.1152/jn.2000.84.4.1888. [DOI] [PubMed] [Google Scholar]
- [64].Deboer T, Behavioral and electrophysiological correlates of sleep and sleep homeostasis, Curr. Top. Behav. Neurosci 25 (2013), 10.1007/7854_2013_248. [DOI] [PubMed] [Google Scholar]
- [65].Cajochen C, Brunner DP, Krauchi K, Graw P, Wirz-Justice A, Power density in theta/alpha frequencies of the waking EEG progressively increases during sustained wakefulness, Sleep 18 (1995) 890–894, 10.1093/sleep/18.10.890. [DOI] [PubMed] [Google Scholar]
- [66].Tobler I, Borbély AA, Groos G, The effect of sleep deprivation on sleep in rats with suprachiasmatic lesions, Neurosci. Lett 42 (1983) 49–54, 10.1016/0304-3940(83)90420-2. [DOI] [PubMed] [Google Scholar]
- [67].Mistlberger RE, Bergmann BM, Waldenar W, Rechtschaffen A, Recovery sleep following sleep deprivation in intact and suprachiasmatic nuclei-lesioned rats, Sleep 6 (1983) 217–233, 10.1093/sleep/6.3.217. [DOI] [PubMed] [Google Scholar]
- [68].Trachsel L, Edgar DM, Seidel WF, Craig Heller H, Sleep homeostasis in suprachiasmatic nuclei-lesioned rats: effects of sleep deprivation and triazolam administration, Brain Res. 589 (1992) 253–261, 10.1016/0006-8993(92)91284-L. [DOI] [PubMed] [Google Scholar]
- [69].Edgar DM, Dement WC, Fuller CA, Effect of SCN Lesions on Sleep in Squirrel Monkeys: Evidence for Opponent Processes in Sleep-Wake Regulation, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Albers HE, Lydic R, Gander PH, Moore-Ede MC, Role of the suprachiasmatic nuclei in the circadian timing system of the squirrel monkey. I. The generation of rhythmicity, Brain Res. 300 (1984) 275–284, 10.1016/0006-8993(84)90837-0. [DOI] [PubMed] [Google Scholar]
- [71].Weitzman ED, Czeisler CA, Moore-Ede MC, Sleep-wake neuroendocrine and body temperature circadian rhythms under entrained and non-entrained (free running) conditions in man, Acta Endocrinol. Suppl 89 (1978) 25. [Google Scholar]
- [72].Czeisler CA, Weitzman ED, Moore-Ede MC, Zimmerman JC, Knauer RS, Human sleep: Its duration and organization depend on its circadian phase, Science 210 (1980) 1264–1267, 10.1126/science.7434029. [DOI] [PubMed] [Google Scholar]
- [73].Czeisler TA, Zimmerman JC, Ronda JM, Moore-Ede C, Weitzman ED, Timing of REM Sleep is Coupled to the Circadian Rhythm of Body Temperature in Man, 1980. 〈https://academic.oup.com/sleep/article/2/3/329/2750200〉. (Accessed 9 December 2020). [PubMed] [Google Scholar]
- [74].Aschoff J, Circadian rhythms in man, Science 148 (1965) 1427–1432, 10.1126/science.148.3676.1427. [DOI] [PubMed] [Google Scholar]
- [75].Dijk DJ, Visscher CA, Bloem GM, Beersma DGM, Daan S, Reduction of human sleep duration after bright light exposure in the morning, Neurosci. Lett 73 (1987) 181–186, 10.1016/0304-3940(87)90014-0. [DOI] [PubMed] [Google Scholar]
- [76].Daan S, Beersma DGM, Borbely AA, Timing of human sleep: recovery process gated by a circadian pacemaker, Am. J. Physiol. Regul. Integr. Comp. Physiol 15 (1984), 10.1152/ajpregu.1984.246.2.r161. [DOI] [PubMed] [Google Scholar]
- [77].Carskadon MA, Dement WC, Sleep studies on a 90-minute day, Electroencephalogr. Clin. Neurophysiol 39 (1975) 145–155, 10.1016/0013-4694(75)90004-8. [DOI] [PubMed] [Google Scholar]
- [78].Wyatt JK, Ritz-De Cecco A, Czeisler CA, Dijk DJ, Circadian temperature and melatonin rhythms, sleep, and neurobehavioral function in humans living on a 20-h day, Am. J. Physiol 277 (1999) 1152–1163, 10.1152/ajpregu.1999.277.4.r1152. [DOI] [PubMed] [Google Scholar]
- [79].Challet E, Turek FW, Laute MA, Van Reeth O, Sleep deprivation decreases phase-shift responses of circadian rhythms to light in the mouse: role of serotonergic and metabolic signals, Brain Res. 909 (2001) 81–91, 10.1016/S0006-8993(01)02625-7. [DOI] [PubMed] [Google Scholar]
- [80].Burgess HJ, Partial sleep deprivation reduces phase advances to light in humans, J. Biol. Rhythms 25 (2010) 460–468, 10.1177/0748730410385544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Deboer T, Sleep homeostasis and the circadian clock: do the circadian pacemaker and the sleep homeostat influence each other’s functioning? Neurobiol. Sleep Circadian Rhythm 5 (2018) 68–77, 10.1016/j.nbscr.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Schaap J, Pennartz CMA, Meijer JH, Electrophysiology of the circadian pacemaker in mammals, Chronobiol. Int 20 (2003) 171–188, 10.1081/CBI-120019311. [DOI] [PubMed] [Google Scholar]
- [83].Deboer T, Vansteensel MJ, Détári L, Meijer JH, Sleep states alter activity of suprachiasmatic nucleus neurons, Nat. Neurosci 6 (2003) 1086–1090, 10.1038/nn1122. [DOI] [PubMed] [Google Scholar]
- [84].Wurts SW, Edgar DM, Circadian and homeostatic control of rapid eye movement (REM) sleep: promotion of REM tendency by the suprachiasmatic nucleus, J. Neurosci 20 (2000) 4300–4310, 10.1523/jneurosci.20-11-04300.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].De La Iglesia HO, Cambras T, Schwartz WJ, Díez-Noguera A, Forced desynchronization of dual circadian oscillators within the rat suprachiasmatic nucleus, Curr. Biol 14 (2004) 796–800, 10.1016/j.cub.2004.04.034. [DOI] [PubMed] [Google Scholar]
- [86].Cambras T, Weller JR, Anglès-Pujoràs M, Lee ML, Christopher A, Díez-Noguera A, Krueger JM, De La Iglesia HO, Circadian desynchronization of core body temperature and sleep stages in the rat, Proc. Natl. Acad. Sci. USA 104 (2007) 7634–7639. 〈 10.1073/pnas.0702424104〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Lee ML, Swanson BE, de la Iglesia HO, Circadian timing of REM sleep is coupled to an oscillator within the dorsomedial suprachiasmatic nucleus, Curr. Biol 19 (2009) 848–852, 10.1016/j.cub.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Chou TC, Bjorkum AA, Gaus SE, Lu J, Scammell TE, Saper CB, Afferents to the ventrolateral preoptic nucleus, J. Neurosci 22 (2002) 977–990, 10.1523/jneurosci.22-03-00977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Sakurai T, Nagata R, Yamanaka A, Kawamura H, Tsujino N, Muraki Y, Kageyama H, Kunita S, Takahashi S, Goto K, Koyama Y, Shioda S, Yanagisawa M, Input of orexin/hypocretin neurons revealed by a genetically encoded tracer in mice, Neuron 46 (2005) 297–308, 10.1016/j.neuron.2005.03.010. [DOI] [PubMed] [Google Scholar]
- [90].Lu J, Zhang YH, Chou TC, Gaus SE, Elmquist JK, Shiromani R, Saper CB, Contrasting effects of ibotenate lesions of the paraventricular nucleus and subparaventricular zone on sleep-wake cycle and temperature regulation, J. Neurosci 21 (2001) 4864–4874, 10.1523/jneurosci.21-13-04864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Chou TC, Scammell TE, Gooley JJ, Gaus SE, Saper CB, Lu J, Critical role of dorsomedial hypothalamic nucleus in a wide range of behavioral circadian rhythms, J. Neurosci 23 (2003) 10691–10702, 10.1523/jneurosci.23-33-10691.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Ono D, Mukai Y, Hung CJ, Chowdhury S, Sugiyama T, Yamanaka A, The mammalian circadian pacemaker regulates wakefulness via CRF neurons in the paraventricular nucleus of the hypothalamus, Sci. Adv 6 (2020) eabd0384, 10.1126/SCIADV.ABD0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Rupp AC, Ren M, Altimus CM, Fernandez DC, Richardson M, Turek F, Hattar S, Schmidt TM, Distinct ipRGC subpopulations mediate light’s acute and circadian effects on body temperature and sleep, Elife 8 (2019), 10.7554/eLife.44358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Mistlberger RE, Antle MC, Glass JD, Miller JD, Behavioral and serotonergic regulation of circadian rhythms, Biol. Rhythm Res 31 (2000) 240–283, 10.1076/0929-1016(200007)31:3;1-K;FT240. [DOI] [Google Scholar]
- [95].Glass JD, Guinn J, Kaur G, Francl JM, On the intrinsic regulation of neuropeptide y release in the mammalian suprachiasmatic nucleus circadian clock, Eur. J. Neurosci 31 (2010) 1117–1126, 10.1111/j.1460-9568.2010.07139.x. [DOI] [PubMed] [Google Scholar]
- [96].Morin LP, Neuroanatomy of the extended circadian rhythm system, Exp. Neurol 243 (2013) 4–20, 10.1016/j.expneurol.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Padilla SL, Perez JG, Ben-Hamo M, Johnson CW, Sanchez REA, Bussi IL, Palmiter RD, de la Iglesia HO, Kisspeptin neurons in the arcuate nucleus of the hypothalamus orchestrate circadian rhythms and metabolism, Curr. Biol 29 (2019) 592–604, 10.1016/j.cub.2019.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Silver R, LeSauter J, Tresco PA, Lehman MN, A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms, Nature 382 (1996) 810–813, 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- [99].Scheer FAJL, Czeisler CA, Melatonin, sleep, and circadian rhythms, Sleep Med. Rev 9 (2005) 5–9, 10.1016/j.smrv.2004.11.004. [DOI] [PubMed] [Google Scholar]
- [100].Moore RY, Klein DC, Visual pathways and the central neural control of a circadian rhythm in pineal serotonin N-acetyltransferase activity, Brain Res. 71 (1974) 17–33, 10.1016/0006-8993(74)90188-7. [DOI] [PubMed] [Google Scholar]
- [101].Redman J, Armstrong S, Ng KT, Free-running activity rhythms in the rat: entrainment by melatonin, Science 219 (1983) 1089–1091, 10.1126/science.6823571. [DOI] [PubMed] [Google Scholar]
- [102].Brzezinski A, Vangel MG, Wurtman RJ, Norrie G, Zhdanova I, Ben-Shushan A, Ford I, Effects of exogenous melatonin on sleep: a meta-analysis, Sleep Med. Rev 9 (2005) 41–50, 10.1016/j.smrv.2004.06.004. [DOI] [PubMed] [Google Scholar]
- [103].Cagnacci A, Elliott JA, Yen SS, Melatonin: a major regulator of the circadian rhythm of core temperature in humans, J. Clin. Endocrinol. Metab 75 (1992) 447–452, 10.1210/jcem.75.2.1639946. [DOI] [PubMed] [Google Scholar]
- [104].Kräuchi K, Cajochen C, Werth E, Wirz-Justice A, Warm feet promote the rapid onset of sleep, Nature 401 (1999) 36–37, 10.1038/43366. [DOI] [PubMed] [Google Scholar]
- [105].Kräuchi K, Cajochen C, Wirz-Justice A, A relationship between heat loss and sleepiness: effects of postural change and melatonin administration, J. Appl. Physiol 83 (1997) 134–139, 10.1152/jappl.1997.83.1.134. [DOI] [PubMed] [Google Scholar]
- [106].Sack RL, Hughes RJ, Edgar DM, Lewy AJ, Sleep-promoting effects of melatonin: at what dose, in whom, under what conditions, and by what mechanisms? Sleep 20 (1997) 908–915, 10.1093/sleep/20.10.908. [DOI] [PubMed] [Google Scholar]
- [107].Van Den Heuvel CJ, Ferguson SA, Mila MacChi M, Dawson D, Melatonin as a hypnotic: Con, Sleep Med. Rev 9 (2005) 71–80, 10.1016/j.smrv.2004.07.001. [DOI] [PubMed] [Google Scholar]
- [108].Zhdanova IV, Geiger DA, Schwagerl AL, Leclair OU, Killiany R, Taylor JA, Rosene DL, Moss MB, Madras BK, Melatonin promotes sleep in three species of diurnal nonhuman primates, Physiol. Behav 75 (2002) 523–529, 10.1016/S0031-9384(02)00654-6. [DOI] [PubMed] [Google Scholar]
- [109].Skene DJ, Arendt J, Circadian rhythm sleep disorders in the blind and their treatment with melatonin, Sleep Med. 8 (2007) 651–655, 10.1016/j.sleep.2006.11.013. [DOI] [PubMed] [Google Scholar]
- [110].Arendt J, Melatonin: countering chaotic time cues, Front. Endocrinol 10 (2019) 391, 10.3389/fendo.2019.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Mendelson WB, Gillin JC, Dawson SD, Lewy AJ, Wyatt RJ, Effects of melatonin and propranolol on sleep of the rat, Brain Res. 201 (1980) 240–244, 10.1016/0006-8993(80)90793-3. [DOI] [PubMed] [Google Scholar]
- [112].Tobler I, Jaggi K, Borbély AA, Effects of melatonin and the melatonin receptor agonist S-20098 on the vigilance states, EEG spectra, and cortical temperature in the rat, J. Pineal Res 16 (1994) 26–32, 10.1111/j.1600-079X.1994.tb00078.x. [DOI] [PubMed] [Google Scholar]
- [113].Fisher SP, Sugden D, Endogenous melatonin is not obligatory for the regulation of the rat sleep-wake cycle, Sleep 33 (2010) 833–840, 10.1093/sleep/33.6.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Vivien-Roels B, Malan A, Rettori MC, Delagrange P, Jeanniot JP, Pévet P, Daily variations in pineal melatonin concentrations in inbred and outbred mice, J. Biol. Rhythms 13 (1998) 403–409, 10.1177/074873098129000228. [DOI] [PubMed] [Google Scholar]
- [115].Deboer T, Circadian regulation of sleep in mammals, Curr. Opin. Physiol 15 (2020) 89–95, 10.1016/j.cophys.2019.12.015. [DOI] [Google Scholar]
- [116].Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS, Functional identification of the mouse circadian Clock gene by transgenic BAC rescue, Cell 89 (1997) 655–667, 10.1016/S0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Sangoram AM, Saez L, Antoch MP, Gekakis N, Staknis D, Whiteley A, Fruechte EM, Vitaterna MH, Shimomura K, King DP, Young MW, Weitz CJ, Takahashi JS, Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription, Neuron 21 (1998) 1101–1113, 10.1016/S0896-6273(00)80627-3. [DOI] [PubMed] [Google Scholar]
- [118].Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacner LD, King DP, Takahashi JS, Weitz CJ, Role of the CLOCK protein in the mammalian circadian mechanism, Science 280 (1998) 1564–1569, 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- [119].Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA, Mop3 is an essential component of the master circadian pacemaker in mammals, Cell 103 (2000) 1009–1017, 10.1016/S0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Wisor JP, O’Hara BF, Terao A, Selby CP, Kilduff TS, Sancar A, Edgar DM, Franken P, A role for cryptochromes in sleep regulation, BMC Neurosci. 3 (2002) 20, 10.1186/1471-2202-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Kopp C, Albrecht U, Zheng B, Tobler I, Homeostatic sleep regulation is preserved in mPer1 and mPer2 mutant mice, Eur. J. Neurosci 16 (2002) 1099–1106, 10.1046/j.1460-9568.2002.02156.x. [DOI] [PubMed] [Google Scholar]
- [122].Shiromani PJ, Xu M, Winston EM, Shiromani SN, Gerashchenko D, Weaver DR, Sleep rhythmicity and homeostasis in mice with targeted disruption of mPeriod genes, Am. J. Physiol. Regul. Integr. Comp. Physiol 287 (2004) 47–57, 10.1152/ajpregu.00138.2004. [DOI] [PubMed] [Google Scholar]
- [123].Naylor E, Bergmann BM, Krauski K, Zee PC, Takahashi JS, Vitaterna MH, Turek FW, The circadian Clock mutation alters sleep homeostasis in the mouse, J. Neurosci 20 (2000) 8138–8143, 10.1523/jneurosci.20-21-08138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Franken P, Dudley CA, Estill SJ, Barakat M, Thomason R, O’Hara BF, McKnight SL, NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: genotype and sex interactions, Proc. Natl. Acad. Sci. USA 103 (2006) 7118–7123. 〈 10.1073/pnas.0602006103〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Laposky A, Easton A, Dugovic C, Walisser J, Bradfield C, Turek F, Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation, Sleep 28 (2005) 395–410, 10.1093/sleep/28.4.395. [DOI] [PubMed] [Google Scholar]
- [126].Hasan S, Van Der Veen DR, Winsky-Sommerer R, Hogben A, Laing EE, Koentgen F, Dijk DJ, Archer SN, A human sleep homeostasis phenotype in mice expressing a primate-specific PER3 variable-number tandem-repeat coding-region polymorphism, FASEB J. 28 (2014) 2441–2454, 10.1096/fj.13-240135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Begemann K, Neumann A, Oster H, Regulation and function of extra-SCN circadian oscillators in the brain, Acta Physiol. 229 (2020), 10.1111/apha.13446. [DOI] [PubMed] [Google Scholar]
- [128].Belle MDC, Diekman CO, Neuronal oscillations on an ultra-slow timescale: daily rhythms in electrical activity and gene expression in the mammalian master circadian clockwork (2018). 〈 10.1111/ejn.13856〉. [DOI] [PubMed] [Google Scholar]
- [129].Li P, Fu X, Smith NA, Ziobro J, Curiel J, Tenga MJ, Martin B, Freedman S, Cea-Del Rio CA, Oboti L, Tsuchida TN, Oluigbo C, Yaun A, Magge SN, O’Neill B, Kao A, Zelleke TG, Depositario-Cabacar DT, Ghimbovschi S, Knoblach S, Ho CY, Corbin JG, Goodkin HP, Vicini S, Huntsman MM, Gaillard WD, Valdez G, Liu JS, Loss of CLOCK results in dysfunction of brain circuits underlying focal epilepsy, Neuron 96 (2017) 387–401.e6, 10.1016/j.neuron.2017.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Husse J, Zhou X, Shostak A, Oster H, Eichele G, Synaptotagmin10-cre, a driver to disrupt clock genes in the SCN, J. Biol. Rhythms 26 (2011) 379–389, 10.1177/0748730411415363. [DOI] [PubMed] [Google Scholar]
- [131].Izumo M, Pejchal M, Schook AC, Lange RP, Walisser JA, Sato TR, Wang X, Bradfield CA, Takahashi JS, Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant, Elife 3 (2014), 10.7554/eLife.04617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Todd WD, Venner A, Anaclet C, Broadhurst RY, De Luca R, Bandaru SS, Issokson L, Hablitz LM, Cravetchi O, Arrigoni E, Campbell JN, Allen CN, Olson DP, Fuller PM, Suprachiasmatic VIP neurons are required for normal circadian rhythmicity and comprised of molecularly distinct subpopulations, Nat. Commun 11 (2020) 4410, 10.1038/s41467-020-17197-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Mieda M, Hasegawa E, Kessaris N, Sakurai T, Fine-tuning circadian rhythms: the importance of Bmal1 expression in the ventral forebrain, Front. Neurosci 11 (2017) 55, 10.3389/fnins.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Ehlen JC, Brager AJ, Baggs J, Pinckney L, Gray CL, DeBruyne JP, Esser KA, Takahashi JS, Paul KN, Bmal1 function in skeletal muscle regulates sleep, Elife 6 (2017), 10.7554/eLife.26557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Ashbrook LH, Krystal AD, Fu YH, Ptáček LJ, Genetics of the human circadian clock and sleep homeostat, Neuropsychopharmacology 45 (2020) 45–54, 10.1038/s41386-019-0476-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Roenneberg T, Wirz-Justice A, Merrow M, Life between clocks: daily temporal patterns of human chronotypes, J. Biol. Rhythms 18 (2003) 80–90, 10.1177/0748730402239679. [DOI] [PubMed] [Google Scholar]
- [137].Jones CR, Campbell SS, Zone SE, Cooper F, Desano A, Murphy PJ, Jones B, Czajkowski L, Ptáček LJ, Familial advanced sleep-phase syndrome: a short-period circadian rhythm variant in humans, Nat. Med 5 (1999) 1062–1065, 10.1038/12502. [DOI] [PubMed] [Google Scholar]
- [138].Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptáček LJ, Fu YH, An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome, Science 291 (2001) 1040–1043, 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- [139].Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptáček LJ, Modeling of a human circadian mutation yields insights into clock regulation by PER2, Cell 128 (2007) 59–70, 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Hirano A, Shi G, Jones CR, Lipzen A, Pennacchio LA, Xu Y, Hallows WC, McMahon T, Yamazaki M, Ptáček LJ, Fu YH, A cryptochrome 2 mutation yields advanced sleep phase in humans, Elife 5 (2016), 10.7554/eLife.16695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Kurien P, Hsu PK, Leon J, Wu D, McMahon T, Shi G, Xu Y, Lipzen A, Pennacchio LA, Jones CR, Fu YH, Ptáček LJ, TIMELESS mutation alters phase responsiveness and causes advanced sleep phase, Proc. Natl. Acad. Sci. USA 116 (2019) 12045–12053. 〈 10.1073/pnas.1819110116〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Dijk DJ, Archer SN, PERIOD3, circadian phenotypes, and sleep homeostasis, Sleep Med. Rev 14 (2010) 151–160, 10.1016/j.smrv.2009.07.002. [DOI] [PubMed] [Google Scholar]
- [143].Patke A, Murphy PJ, Onat OE, Krieger AC, Özçelik T, Campbell SS, Young MW, Mutation of the human circadian clock gene CRY1 in familial delayed sleep phase disorder, Cell 169 (2017) 203–215.e13, 10.1016/j.cell.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Jones SE, Lane JM, Wood AR, van Hees VT, Tyrrell J, Beaumont RN, Jeffries AR, Dashti HS, Hillsdon M, Ruth KS, Tuke MA, Yaghootkar H, Sharp SA, Jie Y, Thompson WD, Harrison JW, Dawes A, Byrne EM, Tiemeier H, Allebrandt KV, Bowden J, Ray DW, Freathy RM, Murray A, Mazzotti DR, Gehrman PR, Lawlor DA, Frayling TM, Rutter MK, Hinds DA, Saxena R, Weedon MN, Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms, Nat. Commun 10 (2019) 343, 10.1038/s41467-018-08259-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Jenni OG, Carskadon MA, Sleep behavior and sleep regulation from infancy through adolescence: normative aspects, Sleep Med. Clin 2 (2007) 321–329, 10.1016/j.jsmc.2007.05.001. [DOI] [Google Scholar]
- [146].Frank MG, The ontogenesis of mammalian sleep: form and function, Curr. Sleep Med. Rep 6 (2020) 267–279, 10.1007/s40675-020-00190-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Lopp S, Navidi W, Achermann P, LeBourgeois M, Behn CD, Developmental changes in ultradian sleep cycles across early childhood: preliminary insights, J. Biol. Rhythms 32 (2017) 64–74, 10.1177/0748730416685451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Guilleminault C, Leger D, Pelayo R, Gould S, Hayes B, Miles L, Development of circadian rhythmicity of temperature in full-term normal infants, Neurophysiol. Clin 26 (1996) 21–29, 10.1016/0987-7053(96)81531-0. [DOI] [PubMed] [Google Scholar]
- [149].Kurth S, Lassonde JM, Pierpoint LA, Rusterholz T, Jenni OG, McClain IJ, Achermann P, LeBourgeois MK, Development of nap neurophysiology: preliminary insights into sleep regulation in early childhood, J. Sleep Res 25 (2016) 646–654, 10.1111/jsr.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Blumberg MS, Gall AJ, Todd WD, The development of sleep-wake rhythms and the search for elemental circuits in the infant brain, Behav. Neurosci 128 (2014) 250–263, 10.1037/a0035891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Frank MG, Heller HC, Development of diurnal organization of EEG slow-wave activity and slow-wave sleep in the rat, Am. J. Physiol. Regul. Integr. Comp. Physiol 273 (1997) R472–R478, 10.1152/ajpregu.1997.273.2.r472. [DOI] [PubMed] [Google Scholar]
- [152].Duncan MJ, Banister MJ, Reppert SM, Developmental appearance of light-dark entrainment in the rat, Brain Res. 369 (1986) 326–330, 10.1016/0006-8993(86)90544-5. [DOI] [PubMed] [Google Scholar]
- [153].Frank MG, Ruby NF, Heller HC, Franken P, Development of circadian sleep regulation in the rat: a longitudinal study under constant conditions, Sleep 40 (2017), 10.1093/sleep/zsw077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Gall AJ, Todd WD, Ray B, Coleman CM, Blumberg MS, The development of day-night differences in sleep and wakefulness in Norway rats and the effect of bilateral enucleation, J. Biol. Rhythms 23 (2008) 232–241, 10.1177/0748730408316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Gall AJ, Todd WD, Blumberg MS, Development of SCN connectivity and the circadian control of arousal: a diminishing role for humoral factors? PLoS One 7 (2012) 45338, 10.1371/journal.pone.0045338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Todd WD, Gall AJ, Weiner JA, Blumberg MS, Distinct retinohypothalamic innervation patterns predict the developmental emergence of species-typical circadian phase preference in nocturnal Norway rats and diurnal nile grass rats, J. Comp. Neurol 520 (2012) 3277–3292, 10.1002/cne.23098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Blumberg MS, Coleman CM, Johnson ED, Shaw C, Developmental divergence of sleep-wake patterns in orexin knockout and wild-type mice, Eur. J. Neurosci 25 (2007) 512–518, 10.1111/j.1460-9568.2006.05292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Nelson AB, Faraguna U, Zoltan JT, Tononi G, Cirelli C, Sleep patterns and homeostatic mechanisms in adolescent mice, Brain Sci. 3 (2013) 318–343, 10.3390/brainsci3010318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Rensing N, Moy B, Friedman JL, Galindo R, Wong M, Longitudinal analysis of developmental changes in electroencephalography patterns and sleep-wake states of the neonatal mouse, PLoS One 13 (2018), 0207031, 10.1371/journal.pone.0207031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Bedont JL, Blackshaw S, Constructing the suprachiasmatic nucleus: a watchmaker’s perspective on the central clockworks, Front. Syst. Neurosci 9 (2015) 1–21, 10.3389/fnsys.2015.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Cheng AH, Cheng HYM, Genesis of the master circadian pacemaker in mice, Front. Neurosci 15 (2021), 10.3389/fnins.2021.659974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Collins B, Pierre-Ferrer S, Muheim C, Lukacsovich D, Cai Y, Spinnler A, Herrera CG, Wen SA, Winterer J, Belle MDC, Piggins HD, Hastings M, Loudon A, Yan J, Földy C, Adamantidis A, Brown SA, Circadian VIPergic neurons of the suprachiasmatic nuclei sculpt the sleep-wake cycle, Neuron 108 (2020) 486–499.e5, 10.1016/j.neuron.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].de Zeeuw J, Wisniewski S, Papakonstantinou A, Bes F, Wahnschaffe A, Zaleska M, Kunz D, Münch M, The alerting effect of the wake maintenance zone during 40 h of sleep deprivation, Sci. Rep 8 (2018) 11012, 10.1038/s41598-018-29380-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Shan Y, Abel JH, Li Y, Izumo M, Cox KH, Jeong B, Yoo SH, Olson DP, Doyle FJ, Takahashi JS, Dual-color single-cell imaging of the suprachiasmatic nucleus reveals a circadian role in network synchrony, Neuron 108 (2020) 164–179.e7, 10.1016/j.neuron.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Shiromani PJ, Peever JH, New neuroscience tools that are identifying the sleep-wake circuit, Sleep 40 (2017), 10.1093/sleep/zsx032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Jones JR, Tackenberg MC, Mcmahon DG, Manipulating circadian clock neuron firing rate resets molecular circadian rhythms and behavior, Nat. Neurosci 18 (2015) 373–377, 10.1038/nn.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Gizowski C, Zaelzer C, Bourque CW, Clock-driven vasopressin neurotransmission mediates anticipatory thirst prior to sleep, Nature 537 (2016) 685–688, 10.1038/nature19756. [DOI] [PubMed] [Google Scholar]
- [168].Gizowski C, Bourque CW, Sodium regulates clock time and output via an excitatory GABAergic pathway, Nature 583 (2020) 421–424, 10.1038/s41586-020-2471-x. [DOI] [PubMed] [Google Scholar]
- [169].Richardson GS, Moore-Ede MC, Czeisler CA, Dement WC, Circadian rhythms of sleep and wakefulness in mice: analysis using long-term automated recording of sleep, Am. J. Physiol. Regul. Integr. Comp. Physiol 17 (1985), 10.1152/ajpregu.1985.248.3.r320. [DOI] [PubMed] [Google Scholar]
- [170].Hasan S, Foster RG, Vyazovskiy VV, Peirson SN, Effects of circadian misalignment on sleep in mice, Sci. Rep 8 (2018) 15343, 10.1038/s41598-018-33480-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Brager AJ, Ehlen JC, Castanon-Cervantes O, Natarajan D, Delisser P, Davidson AJ, Paul KN, Sleep loss and the inflammatory response in mice under chronic environmental circadian disruption, PLoS One 8 (2013) 63752, 10.1371/journal.pone.0063752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Sanchez REA, Bussi IL, Ben-Hamo M, Caldart CS, Catterall WA, de la Iglesia HO, Circadian regulation of sleep in a pre-clinical model of dravet syndrome: dynamics of sleep stage and siesta re-entrainment, Sleep 42 (2019), 10.1093/sleep/zsz173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Fiorillo L, Puiatti A, Papandrea M, Ratti PL, Favaro P, Roth C, Bargiotas P, Bassetti C, Faraci FD, Automated sleep scoring: a review of the latest approaches, Sleep Med. Rev 48 (2019), 101204, 10.1016/j.smrv.2019.07.007. [DOI] [PubMed] [Google Scholar]
- [174].Allocca G, Ma S, Martelli D, Cerri M, Del Vecchio F, Bastianini S, Zoccoli G, Amici R, Morairty SR, Aulsebrook AE, Blackburn S, Lesku JA, Rattenborg NC, Vyssotski AL, Wams E, Porcheret K, Wulff K, Foster R, Chan JKM, Nicholas CL, Freestone DR, Johnston LA, Gundlach AL, Validation of ‘Somnivore’, a machine learning algorithm for automated scoring and analysis of polysomnography data, Front. Neurosci 13 (2019) 1–18, 10.3389/fnins.2019.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Yamabe M, Horie K, Shiokawa H, Funato H, Yanagisawa M, Kitagawa H, MC-SleepNet: large-scale sleep stage scoring in mice by deep neural networks, Sci. Rep 9 (2019) 1–12, 10.1038/s41598-019-51269-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Caldart C, Sanchez R, Ben-Hamo M, Beck A, Weil T, Perez J, Kalume F, Brunton BW, de la Iglesia H, Sleep Identification Enabled by Supervised Training Algorithms (SIESTA): an open-source platform for automatic sleep staging of rodent polysomnographic data, BioRxiv. (2020) 2020.07.06.186940. 〈 10.1101/2020.07.06.186940〉. [DOI] [Google Scholar]
- [177].Martin-Burgos B, Wang W, William I, Tir S, Mohammad I, Javed R, Smith S, Cui Y, Smith CB, Van Der Vinne V, Molyneux PC, Miller SC, Weaver DR, Leise TL, Harrington ME, Harrington M, Methods for detecting PER2:: LUCIFERASE bioluminescence rhythms in freely moving mice, BioRxiv (2020), 10.1101/2020.08.24.264531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Yan L, Smale L, Nunez AA, Circadian and photic modulation of daily rhythms in diurnal mammals, Eur. J. Neurosci 51 (2020) 551–566, 10.1111/ejn.14172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Chang AM, Scheer FAJL, Czeisler CA, Aeschbach D, Direct effects of light on alertness, vigilance, and the waking electroencephalogram in humans depend on prior light history, Sleep 36 (2013) 1239–1246, 10.5665/sleep.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Lupi D, Oster H, Thompson S, Foster RG, The acute light-induction of sleep is mediated by OPN4-based photoreception, Nat. Neurosci 11 (2008) 1068–1073, 10.1038/nn.2179. [DOI] [PubMed] [Google Scholar]
- [181].Riede SJ, Van Der Vinne V, Hut RA, The flexible clock: predictive and reactive homeostasis, energy balance and the circadian regulation of sleep-wake timing, J. Exp. Biol 220 (2017) 738–749, 10.1242/jeb.130757. [DOI] [PubMed] [Google Scholar]
- [182].Rattenborg NC, De La Iglesia HO, Kempenaers B, Lesku JA, Meerlo P, Scriba MF, Sleep research goes wild: new methods and approaches to investigate the ecology, evolution and functions of sleep, Philos. Trans. R. Soc. Lond. B Biol. Sci 372 (2017), 10.1098/rstb.2016.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Masri S, Sassone-Corsi P, The emerging link between cancer, metabolism, and circadian rhythms, Nat. Med 24 (2018) 1795–1803, 10.1038/s41591-018-0271-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Walker WH, Walton JC, DeVries AC, Nelson RJ, Circadian rhythm disruption and mental health, Transl. Psychiatry 10 (2020) 28, 10.1038/s41398-020-0694-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Hood S, Amir S, Neurodegeneration and the circadian clock, Front. Aging Neurosci 9 (2017) 170, 10.3389/fnagi.2017.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Wright KP, McHill AW, Birks BR, Griffin BR, Rusterholz T, Chinoy ED, Entrainment of the human circadian clock to the natural light-dark cycle, Curr. Biol 23 (2013) 1554–1558, 10.1016/j.cub.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Johnson DA, Billings ME, Hale L, Disorders C, Medical H, Disorders: Implications for Population Health, 5 (2019) 61–69. 10.1007/s40471-018-0139-y.Environmental. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Roenneberg T, Winnebeck EC, Klerman EB, Daylight saving time and artificial time zones - a battle between biological and social times, Front. Physiol 10 (2019), 10.3389/fphys.2019.00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Roenneberg T, Pilz LK, Zerbini G, Winnebeck EC, Chronotype and social jetlag: a (self-) critical review, Biology 8 (2019), 10.3390/biology8030054. [DOI] [PMC free article] [PubMed] [Google Scholar]


