Abstract
Background:
While hidradenitis suppurativa (HS) shares some transcriptomic and cellular infiltrate features with psoriasis, their skin proteome remains unknown.
Objective:
To define and compare inflammatory protein biomarkers of HS and psoriasis skin.
Methods:
We assessed 92 inflammatory biomarkers in HS (n=13), psoriasis (n=11) and control skin (n=11) using Olink high-throughput proteomics. We also correlated HS skin and blood biomarkers using proteomics and RNA-sequencing.
Results:
We identified 57 differentially expressed proteins (DEPs) in lesional psoriasis and 64 DEPs in lesional HS skin, compared to healthy controls. Both HS and psoriasis lesional skin demonstrated a significant upregulation of Th1 and Th17 proteins. Healthy-appearing perilesional HS skin had 63 DEPs compared to healthy controls. Nonlesional HS and psoriasis skin had 24 and 7 DEPs, respectively, compared to healthy controls. TNF and 8 other proteins were significantly correlated with clinical severity in perilesional skin (2cm from a nodule).
Limitations:
Inclusion of only moderate-to-severe patients and the cohort size.
Conclusion:
HS has a higher inflammatory profile and is more diffusely distributed versus psoriasis. Proteins correlated with disease severity are potential disease mediators. Perilesional skin is comparably inflamed to lesional skin, suggesting the need to treat beyond skin nodules.
Keywords: Hidradenitis Suppurativa, psoriasis, biomarkers, Olink proteomics, lesional, perilesional, nonlesional, inflammation
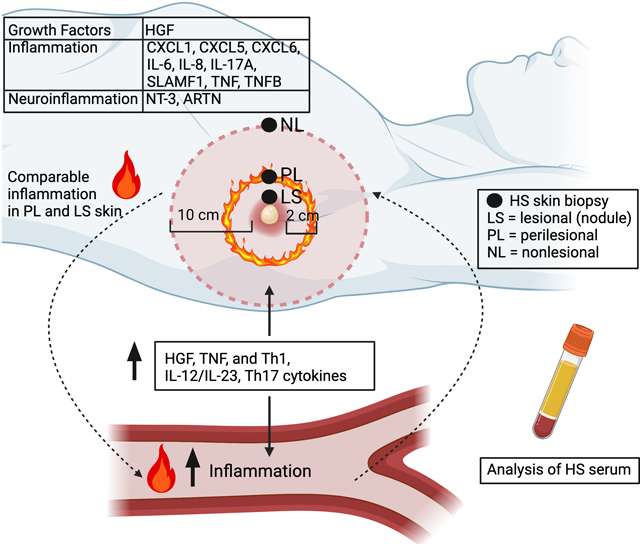
Graphical Abstract
Capsule summary
(First bullet) How does this article integrate into what was already known?
Hidradenitis suppurativa and psoriasis share some overlap in transcriptomic profiles, but a comparison of the skin proteomes has not been performed.
(Second bullet) How does it change practice? That is, what does the article mean to the practice of dermatology and what should you do as a result of having read this article? What should change in the way you practice?
This study identifies unique proteins in hidradenitis suppurativa that could be future therapeutic targets. The inflammation extends to perilesional and nonlesional skin suggesting that this is a large-field disease.
INTRODUCTION
Hidradenitis suppurativa (HS) and psoriasis are inflammatory skin diseases with a significant burden on the quality of life [1, 2]. HS and psoriasis share mutual pathophysiology marked by dysregulation of keratinocytes, up-regulation of type I interferon (IFN) signature, tumor necrosis factor (TNF) and IL-1 signaling, as well as hyperactivation of the Th17 and IL-12/IL-23 axis [3–10]. The relatively-recent discovery of these pathways in HS has stimulated numerous ongoing trials studying psoriasis-approved therapeutics in HS, including TNF inhibitors (adalimumab, infliximab), and anti IL-17 (brodalumab, secukinumab, ixekizumab, bimekizumab), IL-12/IL-23 (guselkumab, rizakizumab, ustekinumab) and IL-1 therapies (anakinra, bermekimab) [11–24]. While adalimumab is the only FDA-approved biologic for HS, near half of the patients either fail to respond or lose response efficacy overtime [25].
Despite the growing research efforts in HS, the pathophysiology of HS remains incompletely understood [26]. While both HS and psoriasis share similar epidermal alterations (e.g. epidermal hyperplasia), HS also manifests with unique dermal inflammation including inflammatory-active epithelialized dermal tunnels [27]. Majority of studies in HS have thus far focused on transcriptomic profiling but have not examined the proteomic profile of skin, and therefore have not demonstrated whether the mRNA is actively translated into functional protein on a proteomic level [3, 4, 28]. Furthermore, recent studies of cytokines and biomarkers in HS blood have demonstrated an ongoing systemic inflammation further giving credence to HS as a systemic disease [29]. However, a study comparing the protein expression of inflammatory markers in HS compared to both healthy control and psoriasis skin is lacking. We thus sought to characterize the inflammatory proteome in skin and correlate these biomarkers with clinical severity in HS.
METHODS
Patient Enrollment:
This study was approved by the Institutional Review Board and written informed consent was obtained. Untreated patients with moderate-to-severe HS (n=13, Hurley Stage 2 and 3), psoriasis (n=11, PASI ≥12) and age/BMI- matched healthy controls (n=11) were enrolled in the study (Supplementary Table 1).
Skin and serum sample collection:
HS lesional punch biopsies were obtained at an edge of an active inflammatory lesion, while perilesional and nonlesional skin biopsies were taken from visually healthy-appearing skin 2cm and 10cm away from the lesion, respectively [30]. Lesional psoriasis punch biopsies were obtained from an active plaque, and nonlesional biopsies were obtained from healthy-appearing skin. Skin tissue and serum were processed as previously described [31–34].
Statistical Analysis
Analysis was performed with R language (www.R-project.org) using publicly available Bioconductor packages (www.bioconductor.org). Olink data was subjected to quality control (QC) using the QC pipeline [35]. mRNA and protein (Olink) expression were modeled under the R limma framework using the mixed-effect linear model [32, 36]. Correlation between clinical parameters and protein levels were assessed using Pearson correlation. Gene-set variation analysis (GSVA) was performed, and z-score was quantified for previously published pathways [33, 34]. The correlation between gene expression (mRNA) and protein levels was evaluated by Pearson correlation.
RESULTS
The proteomic profile of HS and psoriasis skin has an increased inflammatory tone
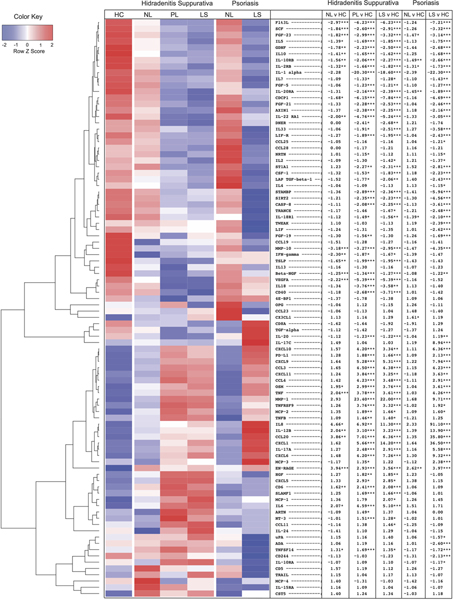
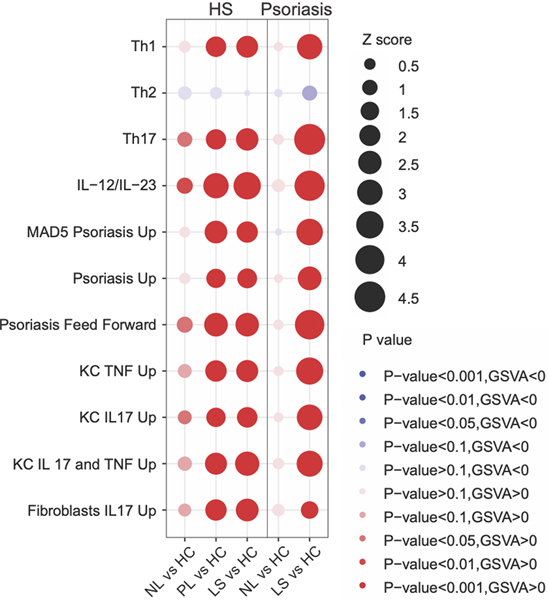
Principal component analysis demonstrated that lesional skin in HS and psoriasis clustered separately from nonlesional skin and healthy controls (Supplementary Figure 1). An overall narrower inflammatory tone was observed in psoriasis compared to HS (Supplementary Figure 2). Using a criterion of |FCH|≥1.2 and p≤0.05, we identified 57 differentially expressed proteins (DEPs) in lesional psoriasis skin and 64 DEPs in lesional HS skin, compared to healthy controls (Figure 1). Lesional skin in both dermatoses showed a significant upregulation of Th1(IL-8, CCL3, CCL4, CXCL9, CXCL10, CXCL11), IL-12/IL-23 (CCL3, CXCL9, TNF, IL-17A, IL-12B), Th17 (CXCL1, CCL20, IL-17A), neutrophil-related (CXCL1, CXCL6, IL-8) and cardiovascular-related (EN-RAGE, OSM, TNF, MMP-1, IL-8) proteins compared to healthy controls [34]. Anti-inflammatory cytokine IL-10, which prevents excessive Th1 response and limits the production of multiple inflammatory cytokines including TNF, IL-6 and IL-1α/β, was significantly decreased in both diseases [37]. Consistent with this, GSVA demonstrated a comparable up-regulation of Th1 and IL-12/IL-23 pathways but higher Th17 and known psoriasis-signatures in psoriasis (Figure 2).
Figure 1:
Heatmap of proteins in healthy controls (HC), hidradenitis suppurativa (HS) and psoriasis skin. The table shows fold change (FCH) of nonlesional (NL), perilesional (PL) and lesional (LS) skin relative to HC with stars denoting significance. * ≤0.05 ** ≤0.01 ***≤0.001.
Figure 2:
Bubble plot comparing Gene Set Variation Analysis (GSVA) scores between hidradenitis suppurativa (HS) and psoriasis skin relative to healthy controls. Bubble diameter is proportional to the Z score while the color of the bubble reflects the significance (p value). Abbreviations: nonlesional (NL), perilesional (PL), lesional (LS), healthy controls (HC).
Among proteins uniquely up-regulated in HS were mesenchymal-cell produced Hepatocyte Growth Factor (HGF), which has been shown to play a role in hair follicle growth, re-epithelialization during skin wound healing and induction of epithelial cell migration, and CXCL5, a neutrophil chemoattractant activated by both IL-1 and TNF [38–41]. Both Artemin (ARTN), a member of the glial cell line-derived neutrophilic factor related family which has been reported to be elevated in deep epidermis and dermis and involved in neuropathic and inflammatory pain, and Neutrophin-3 (NT-3), involved in the neuro-immune interface of allergic skin disease and diabetic polyneuropathy skin, were elevated in HS [42, 43] [44, 45]. Proteins involved in continued T cell development and activation (CD6), eosinophil recruitment (CCL11) and monocyte recruitment (MCP-1) were also upregulated in HS [46–48].
To better understand how HS inflammation compares to psoriasis, we then directly compared HS to psoriasis lesional skin and identified an upregulation of 18 DEPs. In addition to CXCL5 and HGF, we observed a significant upregulation of IL-6 and IL-6 family cytokine Leukemia Inhibitory Factor (LIF). HS had an upregulation of TNF superfamily TNFSF14/LIGHT expressed on immature dendritic cells (DCs), and TWEAK, which has been implicated in chronic inflammation [49–51]. IL-24, expressed by keratinocytes during wound healing, was also elevated in HS. [52, 53]. Psoriasis, on the other hand, showed upregulation of 14 proteins compared to HS, including IL-17 cytokines (IL-17A/C and CCL20), CXCL1, IL-8, IL-12B and IL-20. HS perilesional and lesional skin shared a highly similar proteomic profile. In a direct comparison of HS lesional to perilesional skin, lesional skin had only 1 significantly up-(CXCL1) and 1 down-regulated (TNFSF14) protein.
Nonlesional HS skin is more inflamed than nonlesional psoriasis skin
We next compared the proteomic profile of nonlesional skin in HS and psoriasis [54, 55]. Compared to healthy control skin, HS and psoriasis had 24 and 7 DEPs, respectively. Unlike psoriasis nonlesional skin, HS had a significant upregulation of TNF, CCL20, IL-6, IL-8 and IL-12B cytokines. GSVA analysis demonstrated a higher dysregulation of Th17 and IL-12/IL-23 pathways in HS versus psoriasis nonlesional skin (Figure 2).
mRNA expression and protein levels in HS tissue are significantly correlated
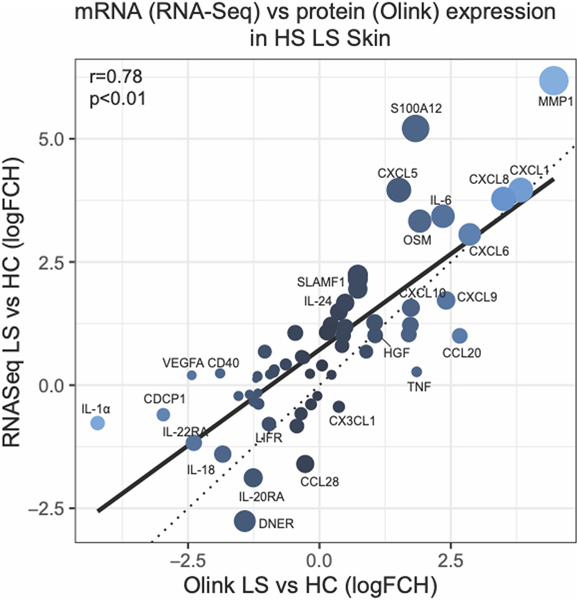
Majority of previous studies have focused on the transcriptomic analysis of HS skin however whether these transcripts are actively translated into protein products has not been determined. We therefore assessed if there was a correlation between mRNA levels and the protein levels in the skin. We first limited the RNA-sequencing (RNA-seq) analysis to only those transcripts that overlapped with the Olink inflammation panel. There was a significant correlation between mRNA and protein levels within lesional skin and perilesional as compared to healthy control skin (r=0.78, p<0.01, and r=0.76, p<0.01, respectively) (Figure 3). The correlation between nonlesional skin was also significant but less pronounced (r=0.57, p<0.01). We note that in both perilesional and lesional HS skin several neutrophil-associated proteins (MMP1, CXCL1, CXCL5, CXCL6, IL-8, S100A12) had a higher mRNA expression compared to protein levels, both relative to healthy controls.
Figure 3:
Protein levels (NPX units, Olink) versus gene expression (mRNA, RNA-sequencing) in hidradenitis suppurativa (HS) lesional as compared to healthy control (HC) skin. The size of the circle depicts the difference in log FCH between HS and HC skin by Olink and RNA-sequencing. R is Pearson correlation.
Protein expression in the skin is correlated with clinical parameters
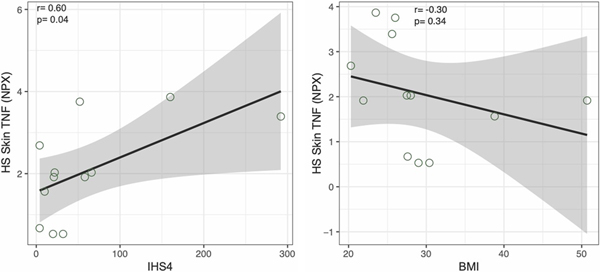
We then correlated clinical parameters (absolute granulocyte, lymphocyte, neutrophil counts and IHS4) with protein biomarkers in the skin (Pearson correlation (r) ≥ 0.5 and p value of ≤ 0.1) (Supplementary Table 2). TNF protein levels in nonlesional (r=0.75, p=0.01), perilesional (r=0.60, p=0.04) and lesional skin (r=0.64, p=0.02) significantly correlated with IHS4 score (Figure 4). TNF levels were not correlated with BMI or other clinical parameters (absolute granulocyte, lymphocyte and neutrophil counts), suggesting that the correlation is specific to clinical severity of HS. In perilesional HS skin, levels of 9 proteins (CXCL10, CXCL11, FGF23, IFNγ, IL-10, SLAMF1, ST1A1, TNF and TRANCE) correlated with IHS4 score. None of these proteins in the skin correlated with BMI or absolute lymphocyte count. In nonlesional skin, expression of 20 proteins correlated with IHS4. Among these, members of Th1 family (CXCL9, CXCL10 and CXCL11), TNF and IL-17A had a positive correlation with IHS4. In lesional HS skin, only protein levels of TNF had a significant correlation with IHS4 score, likely due to the highly neutrophilic environment and protease associated degradation, which may affect the stability of proteins.
Figure 4:
Correlation of clinical parameters (IHS4 and BMI) and protein expression of TNF in perilesional hidradenitis suppurativa (HS) skin. r is Pearson correlation.
Protein expression in HS blood and skin is significantly correlated
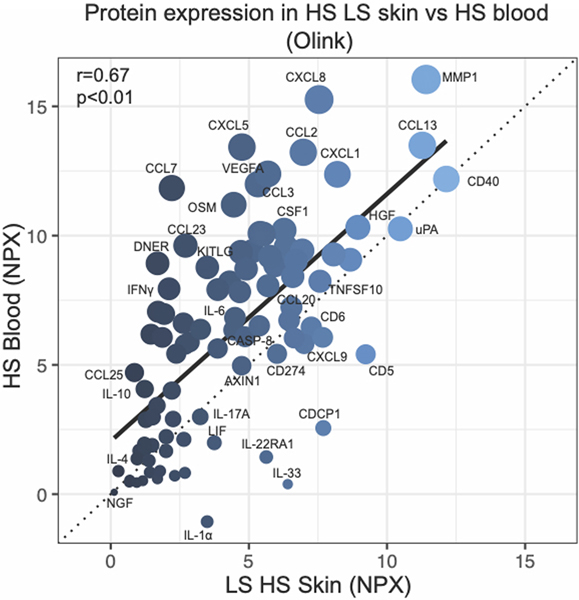
We also correlated protein expression between serum and skin in HS patients (Figure 5). There was a significant correlation between protein levels in blood and lesional (r=0.67, p<0.01) and perilesional (r=0.61, p<0.01) skin. In total, 26 proteins were up-regulated, and 26 proteins were down-regulated in HS lesional skin and blood, both relative to healthy controls. Common proteins that were up-regulated in HS blood/skin include Th1 cytokines ((CXCL9 (FCHskin=5.32, FCHblood=1.48), CXCL11 (FCHskin=3.25, FCHblood=1.49)), IL-12/IL-23 cytokines ((CXLC1 (FCHskin=14.19, FCHblood=1.07), TNF (FCHskin=3.61, FCHblood=1.26)), the Th17 cytokine CCL20 (FCHskin=6.37, FCHblood=1.29), and HGF (FCHskin=1.85, FCHblood=1.16). Not all proteins were overexpressed in both tissue compartments. We detected increased expression of 15 proteins including MCP-1 and MMP-1 uniquely in skin. Conversely, expressions of 25 proteins, including VEGF-A, immunoregulatory IL-10RB, and growth factors TGF-alpha and FGF isoforms, were upregulated only in blood.
Figure 5:
Olink proteomics in hidradenitis suppurativa (HS) blood versus lesional skin. r is Pearson correlation.
DISCUSSION
This is the first study comparing the proteomic profile of nonlesional, perilesional, and lesional HS skin versus psoriasis and healthy controls. One previous study identified 17 proteins upregulated in HS, 14 of which overlapped with the 64 DEPs we identified in our study [56]. However, that study did not detect TNF, or neutrophil-associated (CXCL1, IL-8) proteins that were detected in our analysis. In our study, we extracted proteins directly from frozen tissue and were able to quantify all 92 analytes within the Olink inflammation panel whereas only 75/92 proteins were detected after ex vivo tissue culture by other groups [31, 32, 56]. Other studies of select proteins reported an increased expression of IL-1β, IL-17A, TNF, IL-8, CXCL16 and RANTES in HS skin, consistent with our findings [6, 57–59].
We demonstrate skewing towards Th1 (IL-8, CXCL9, CXCL10, CXCL11) and Th17 (IL-17A, CCL20, CXCL1) in HS, as well as an increase of TNF which was previously identified by transcriptomic analysis [3, 28, 60, 61]. We also detect neutrophil-related proteins (CXCL5, CXCL6, CXCL1, IL-8). While HS and psoriasis share epidermal features, HS has discernable dermal inflammation and the presence of immunologically active dermal tunnels [27]. We identify novel proteins previously not reported in HS which may reflect tunnel biology. Artemin (ARTN), whose elevated expression in skin has been associated with hyperalgesia and Neutrophin-3 (NT-3), whose levels are increased in skin of patients with diabetic neuropathy, are both elevated in HS [45, 62, 63]. This suggests neuroinflammation might contribute to pain common in HS patients [42, 43] [44, 45, 64, 65]. Tunnels are inflammatory-active, epithelialized structures unique to HS, and their development is not well understood [27]. Hepatocyte Growth Factor (HGF) and epithelial-derived IL-24, both elevated in HS skin, might influence formation of epithelialized dermal tunnels [52, 53, 66, 67].
We show that expression of TNF in the skin is significantly correlated with IHS4. The role of TNF as a disease driver is firmly established through antagonism in clinical studies [9, 19, 20, 28, 60]. Previous work has shown that elevations of TNF in HS skin may be correlated with Hurley staging [6, 28, 60]. We found that TNF protein levels in both skin and blood were significantly correlated with IHS4 but were not correlated with other clinical parameters (BMI, absolute granulocyte, lymphocyte, or neutrophil counts), establishing a direct association with disease activity. While TNF could originate either from innate inflammatory pathways or activated T cells, our data presents evidence that activated polar T cell subsets including Th1 (CXCL9, CXCL10, CXCL11, IFNγ) and Th17 (IL-17A, CXCL1) may correlate with disease activity in perilesional and nonlesional skin.
To our knowledge, this is the largest study integrating the inflammatory cytokines in HS serum and skin using an unbiased panel of 92 biomarkers. While one group compared levels of 30 protein between HS skin and blood, they did not find strong skin-blood correlations [59]. We found a correlation between skin mRNA and protein expression, as well as significant correlation between protein levels in HS skin and blood. This suggests elevated mRNAs are being actively translated into protein products in the skin, and that resulting proteins may be released into skin and potentially diffuse into the blood, as has also been proposed for elevated cytokines measured in the blood of psoriasis patients.
Our findings that inflammation extends to perilesional and nonlesional skin have importance for conceptualizing HS pathogenesis and for adopting scoring systems that incorporate measures beyond inflammatory nodules (currently considered the main “lesion” in affected HS regions). In previous work, we found that perilesional skin may contain deep dermal tunnels with active inflammation despite appearing visually healthy on the surface [68]. This is coupled with intense production of inflammatory transcripts in deep dermis of HS skin, especially in skin with tunnels [27]. Therapeutic modulation of dermal inflammation has been demonstrated by Doppler ultrasound and by reduced drainage from surface ostia that are termini of dermal tunnels at the epidermis [14, 27, 68]. The differences in inflammatory gene expression in nonlesional HS skin compared to healthy controls also supports the view of HS pathology as not discreetly localized within the visibly affected regions of the skin. Accordingly, HS might be better characterized as a ‘field’ disease rather than focused only on localized nodules. This view is reflected in alternative scoring systems such as the IHS4, which considers both inflammatory nodules and draining tunnels/fistulae. Measurement approaches that do not integrate perilesional involvement may provide a limited measurement of disease.
Limitations of our study include the use of a biomarker panel with only 92 inflammatory proteins as other proteins beyond those examined may also play a critical role in HS pathogenesis. We also note that HS lesions are heavily infiltrated by activated neutrophils, thus there is high production of many proteases that can degrade extracellular proteins [27]. Therefore, detection of mRNA transcripts without detecting elevated protein levels could reflect more rapid degradation of that protein in a highly inflammatory environment. As such, some overproduced proteins in HS lesions might be rapidly degraded and would not be measured in our assay. Another limitation is the inclusion of only moderate-to-severe (Hurley stage 2–3) patients, and we believe that patients with new onset or Hurley Stage 1 disease should also be studied in the future.
Taken together, our results highlight the need for further biomarker studies in HS that integrate the skin and blood proteome, and associate measured abnormalities with clinical disease activity scores and changes with disease improvement induced by a range of established and evolving therapeutic approaches. We identify a number of new proteins that are dysregulated in HS, and which could contribute to overall disease activity, as well as the complexity of the therapeutic approaches. This may explain the difficulty of treating HS with ‘narrow’ cytokine antagonists, as is highly successful with psoriasis that shows predominantly Th17 pathway activation.
ACKNOWLEDGEMENTS
We thank members of the Krueger laboratory for thoughtful discussions. Graphical abstract was created with BioRender.com.
IRB approval status: The study was approved by the Institutional Review Board of the Rockefeller University.
Funding sources:
K.N. was supported by a MSTP grant from the National Institute of General Medical Sciences of the NIH under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program. J.W.F. and J.G.K. were supported in part by grant # UL1 TR001866 from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program. J.W.F. was supported by the Shapiro-Silverberg Fund for the Advancement of Translational Research and the Hidradenitis Suppurativa Foundation Danby Grant.
Conflicts of Interest: J.G.K. has received research support (grants paid to the institution) from AbbVie, Amgen, BMS, Boehringer, EMD Serono, Innovaderm, Kineta, LEO Pharma, Novan, Novartis, Paraxel, Pfizer, Regeneron, and Vitae and personal fees from AbbVie, Acros, Allergan, Aurigne, BiogenIdec, Boehringer, Escalier, Janssen, Lilly, Novartis, Pfizer, Roche, and Valeant. E.G.Y. is an employee of Mount Sinai and has received research funds (grants paid to the institution) from AbbVie, Celgene, Eli Lilly, Janssen, Medimmune/Astra Zeneca, Novartis, Pfizer, Regeneron, Vitae, Glenmark, Galderma, Asana, Innovaderm, Dermira, and UCB and is also a consultant for Sanofi Aventis, Regeneron, Stiefel/GlaxoSmithKline, MedImmune, Celgene, Anacor, AnaptysBio, Dermira, Galderma, Glenmark, Novartis, Pfizer, Vitae, LEO Pharma, AbbVie, Eli Lilly, Kyowa, Mitsubishi Tanabe, Asana Biosciences, and Promius. The other authors declare they have no relevant conflicts of interest.
ABBREVIATIONS
- BMI
Body Mass Index
- DC
Dendritic Cell
- FCH
Fold Change
- FDA
Food Drug Administration
- GSVA
Gene Set Variation Analysis
- HiSCR
Hidradenitis Suppurativa Clinical Response
- HS
Hidradenitis Suppurativa
- IFNγ
Interferon gamma
- IHS4
International Hidradenitis Suppurativa Severity Score System
- IL
Interleukin
- MAD3
Meta-analysis derived psoriasis dataset
- mRNA
Messenger RNA
- PASI
Psoriasis Area and Severity Index
- TH
T helper cell
- TNF
Tumor necrosis factor
- WBC
White Blood Cell Count
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTARY DATA
All supplementary data is available on Mendeley (http://dx.doi.org/10.17632/s99c7tmmmg.1)
REFERENCES
- 1.Basra MK, et al. , The Dermatology Life Quality Index 1994–2007: a comprehensive review of validation data and clinical results. Br J Dermatol, 2008. 159(5): p. 997–1035. [DOI] [PubMed] [Google Scholar]
- 2.Kridin K, et al. , Psoriasis and Hidradenitis Suppurativa: A Large-scale Population-based Study. J Am Acad Dermatol, 2018. [DOI] [PubMed] [Google Scholar]
- 3.Schlapbach C, et al. , Expression of the IL-23/Th17 pathway in lesions of hidradenitis suppurativa. J Am Acad Dermatol, 2011. 65(4): p. 790–798. [DOI] [PubMed] [Google Scholar]
- 4.Kelly G, et al. , Dysregulated cytokine expression in lesional and nonlesional skin in hidradenitis suppurativa. Br J Dermatol, 2015. 173(6): p. 1431–9. [DOI] [PubMed] [Google Scholar]
- 5.Navrazhina K, Frew JW, and Krueger JG, Interleukin 17C is elevated in lesional tissue of hidradenitis suppurativa. Br J Dermatol, 2020. 182(4): p. 1045–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Zee HH, et al. , Elevated levels of tumour necrosis factor (TNF)-α, interleukin (IL)-1β and IL-10 in hidradenitis suppurativa skin: a rationale for targeting TNF-α and IL-1β. Br J Dermatol, 2011. 164(6): p. 1292–8. [DOI] [PubMed] [Google Scholar]
- 7.Hotz C, et al. , Intrinsic Defect in Keratinocyte Function Leads to Inflammation in Hidradenitis Suppurativa. J Invest Dermatol, 2016. 136(9): p. 1768–1780. [DOI] [PubMed] [Google Scholar]
- 8.Wolk K, et al. , Deficiency of IL-22 contributes to a chronic inflammatory disease: pathogenetic mechanisms in acne inversa. J Immunol, 2011. 186(2): p. 1228–39. [DOI] [PubMed] [Google Scholar]
- 9.van der Zee HH, et al. , Adalimumab (antitumour necrosis factor-α) treatment of hidradenitis suppurativa ameliorates skin inflammation: an in situ and ex vivo study. Br J Dermatol, 2012. 166(2): p. 298–305. [DOI] [PubMed] [Google Scholar]
- 10.Banerjee A, McNish S, and Shanmugam VK, Interferon-gamma (IFN-γ) is Elevated in Wound Exudate from Hidradenitis Suppurativa. Immunol Invest, 2017. 46(2): p. 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovacs M and Podda M, Guselkumab in the treatment of severe hidradenitis suppurativa. J Eur Acad Dermatol Venereol, 2019. 33(3): p. e140–e141. [DOI] [PubMed] [Google Scholar]
- 12.Kearney N, et al. , Successful use of guselkumab in the treatment of severe hidradenitis suppurativa. Clin Exp Dermatol, 2020. 45(5): p. 618–619. [DOI] [PubMed] [Google Scholar]
- 13.Montero-Vilchez T, et al. , The use of guselkumab 100 mg every 4 weeks on patients with hidradenitis suppurativa and a literature review. Dermatol Ther, 2020. 33(3): p. e13456. [DOI] [PubMed] [Google Scholar]
- 14.Frew JW, et al. , The effect of subcutaneous brodalumab on clinical disease activity in hidradenitis suppurativa: An open-label cohort study. J Am Acad Dermatol, 2020. 83(5): p. 1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reguiai Z, et al. , Effectiveness of secukinumab in hidradenitis suppurativa: an open study (20 cases). J Eur Acad Dermatol Venereol, 2020. 34(11): p. e750–e751. [DOI] [PubMed] [Google Scholar]
- 16.Casseres RG, et al. , Secukinumab in the treatment of moderate to severe hidradenitis suppurativa: Results of an open-label trial. J Am Acad Dermatol, 2020. 82(6): p. 1524–1526. [DOI] [PubMed] [Google Scholar]
- 17.Prussick L, et al. , Open-label, investigator-initiated, single-site exploratory trial evaluating secukinumab, an anti-interleukin-17A monoclonal antibody, for patients with moderate-to-severe hidradenitis suppurativa. Br J Dermatol, 2019. 181(3): p. 609–611. [DOI] [PubMed] [Google Scholar]
- 18.Ghias MH, et al. , High-dose, high-frequency infliximab: A novel treatment paradigm for hidradenitis suppurativa. J Am Acad Dermatol, 2020. 82(5): p. 1094–1101. [DOI] [PubMed] [Google Scholar]
- 19.Prens LM, et al. , Drug survival of Adalimumab and Infliximab in Hidradenitis Suppurativa Patients: A Daily Practice Cohort Study. Br J Dermatol, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimball AB, et al. , Two Phase 3 Trials of Adalimumab for Hidradenitis Suppurativa. N Engl J Med, 2016. 375(5): p. 422–34. [DOI] [PubMed] [Google Scholar]
- 21.Frew JW, et al. , A Systematic Review of Promising Therapeutic Targets in Hidradenitis Suppurativa: A Critical Evaluation of Mechanistic and Clinical Relevance. J Invest Dermatol, 2021. 141(2): p. 316–324.e2. [DOI] [PubMed] [Google Scholar]
- 22.Tzanetakou V, et al. , Safety and Efficacy of Anakinra in Severe Hidradenitis Suppurativa: A Randomized Clinical Trial. JAMA Dermatol, 2016. 152(1): p. 52–59. [DOI] [PubMed] [Google Scholar]
- 23.Leslie KS, et al. , An open-label study of anakinra for the treatment of moderate to severe hidradenitis suppurativa. J Am Acad Dermatol, 2014. 70(2): p. 243–51. [DOI] [PubMed] [Google Scholar]
- 24.Ribero S, et al. , Effectiveness of Secukinumab in the treatment of moderate-severe hidradenitis suppurativa: results from an Italian multicentric retrospective study in a real-life setting. J Eur Acad Dermatol Venereol, 2021. [DOI] [PubMed] [Google Scholar]
- 25.Zouboulis CC, et al. , Long-term adalimumab efficacy in patients with moderate-to-severe hidradenitis suppurativa/acne inversa: 3-year results of a phase 3 open-label extension study. J Am Acad Dermatol, 2019. 80(1): p. 60–69.e2. [DOI] [PubMed] [Google Scholar]
- 26.Naik HB and Lowes MA, A Call to Accelerate Hidradenitis Suppurativa Research and Improve Care-Moving Beyond Burden. JAMA Dermatol, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navrazhina K, et al. , Epithelialized Tunnels are a Source of Inflammation in Hidradenitis Suppurativa. J Allergy Clin Immunol, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe MM, et al. , Immunopathogenesis of hidradenitis suppurativa and response to anti-TNF-α therapy. JCI Insight, 2020. 5(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navrazhina K, et al. , In-Depth Analysis of the Hidradenitis Suppurativa Serum Proteome Identifies Distinct Inflammatory Subtypes. J Invest Dermatol, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frew JW, et al. , Defining lesional, perilesional and unaffected skin in hidradenitis suppurativa: proposed recommendations for clinical trials and translational research studies. Br J Dermatol, 2019. 181(6): p. 1339–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He H, et al. , Tape-Strip Proteomic Profiling of Atopic Dermatitis on Dupilumab Identifies Minimally Invasive Biomarkers. Front Immunol, 2020. 11: p. 1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pavel AB, et al. , The proteomic skin profile of moderate-to-severe atopic dermatitis patients shows an inflammatory signature. J Am Acad Dermatol, 2020. 82(3): p. 690–699. [DOI] [PubMed] [Google Scholar]
- 33.Visvanathan S, et al. , Psoriatic skin molecular and histopathologic profiles after treatment with risankizumab versus ustekinumab. J Allergy Clin Immunol, 2019. 143(6): p. 2158–2169. [DOI] [PubMed] [Google Scholar]
- 34.Glickman JW, et al. , Cross-sectional study of blood biomarkers of patients with moderate to severe alopecia areata reveals systemic immune and cardiovascular biomarker dysregulation. J Am Acad Dermatol, 2021. 84(2): p. 370–380. [DOI] [PubMed] [Google Scholar]
- 35.Lind L, et al. , Use of a proximity extension assay proteomics chip to discover new biomarkers for human atherosclerosis. Atherosclerosis, 2015. 242(1): p. 205–10. [DOI] [PubMed] [Google Scholar]
- 36.Smyth GK, Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol, 2004. 3: p. Article3. [DOI] [PubMed] [Google Scholar]
- 37.Couper KN, Blount DG, and Riley EM, IL-10: the master regulator of immunity to infection. J Immunol, 2008. 180(9): p. 5771–7. [DOI] [PubMed] [Google Scholar]
- 38.Lee YR, et al. , Hepatocyte growth factor (HGF) activator expressed in hair follicles is involved in in vitro HGF-dependent hair follicle elongation. J Dermatol Sci, 2001. 25(2): p. 156–63. [DOI] [PubMed] [Google Scholar]
- 39.Li JF, et al. , HGF accelerates wound healing by promoting the dedifferentiation of epidermal cells through β1-integrin/ILK pathway. Biomed Res Int, 2013. 2013: p. 470418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spix JK, et al. , Hepatocyte growth factor induces epithelial cell motility through transactivation of the epidermal growth factor receptor. Exp Cell Res, 2007. 313(15): p. 3319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metzemaekers M, Gouwy M, and Proost P, Neutrophil chemoattractant receptors in health and disease: double-edged swords. Cell Mol Immunol, 2020. 17(5): p. 433–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikeda-Miyagawa Y, et al. , Peripherally increased artemin is a key regulator of TRPA1/V1 expression in primary afferent neurons. Mol Pain, 2015. 11: p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elitt CM, et al. , Artemin overexpression in skin enhances expression of TRPV1 and TRPA1 in cutaneous sensory neurons and leads to behavioral sensitivity to heat and cold. J Neurosci, 2006. 26(33): p. 8578–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quarcoo D, et al. , High abundances of neurotrophin 3 in atopic dermatitis mast cell. J Occup Med Toxicol, 2009. 4: p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kennedy AJ, et al. , Neurotrophin-3 is increased in skin in human diabetic neuropathy. J Neurol Neurosurg Psychiatry, 1998. 65(3): p. 393–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teixeira AL, et al. , Revisiting the Role of Eotaxin-1/CCL11 in Psychiatric Disorders. Front Psychiatry, 2018. 9: p. 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deshmane SL, et al. , Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res, 2009. 29(6): p. 313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orta-Mascaró M, et al. , CD6 modulates thymocyte selection and peripheral T cell homeostasis. J Exp Med, 2016. 213(8): p. 1387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donohue PJ, et al. , TWEAK is an endothelial cell growth and chemotactic factor that also potentiates FGF-2 and VEGF-A mitogenic activity. Arterioscler Thromb Vasc Biol, 2003. 23(4): p. 594–600. [DOI] [PubMed] [Google Scholar]
- 50.Tamada K, et al. , LIGHT, a TNF-like molecule, costimulates T cell proliferation and is required for dendritic cell-mediated allogeneic T cell response. J Immunol, 2000. 164(8): p. 4105–10. [DOI] [PubMed] [Google Scholar]
- 51.Burkly LC, TWEAK/Fn14 axis: the current paradigm of tissue injury-inducible function in the midst of complexities. Semin Immunol, 2014. 26(3): p. 229–36. [DOI] [PubMed] [Google Scholar]
- 52.Poindexter NJ, et al. , IL-24 is expressed during wound repair and inhibits TGFalpha-induced migration and proliferation of keratinocytes. Exp Dermatol, 2010. 19(8): p. 714–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin SH, et al. , Keratinocyte-derived IL-24 plays a role in the positive feedback regulation of epidermal inflammation in response to environmental and endogenous toxic stressors. Toxicol Appl Pharmacol, 2014. 280(2): p. 199–206. [DOI] [PubMed] [Google Scholar]
- 54.Chiricozzi A, et al. , Increased expression of interleukin-17 pathway genes in nonlesional skin of moderate-to-severe psoriasis vulgaris. Br J Dermatol, 2016. 174(1): p. 136–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keermann M, et al. , Transcriptional landscape of psoriasis identifies the involvement of IL36 and IL36RN. BMC Genomics, 2015. 16(1): p. 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vossen A, et al. , Apremilast for moderate hidradenitis suppurativa: no significant change in lesional skin inflammatory biomarkers. J Eur Acad Dermatol Venereol, 2019. 33(4): p. 761–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marzano AV, et al. , Autoinflammation in pyoderma gangrenosum and its syndromic form (pyoderma gangrenosum, acne and suppurative hidradenitis). Br J Dermatol, 2017. 176(6): p. 1588–1598. [DOI] [PubMed] [Google Scholar]
- 58.Witte-Händel E, et al. , The IL-1 Pathway Is Hyperactive in Hidradenitis Suppurativa and Contributes to Skin Infiltration and Destruction. J Invest Dermatol, 2019. 139(6): p. 1294–1305. [DOI] [PubMed] [Google Scholar]
- 59.Vossen A, et al. , Novel cytokine and chemokine markers of hidradenitis suppurativa reflect chronic inflammation and itch. Allergy, 2019. 74(3): p. 631–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moran B, et al. , Hidradenitis Suppurativa Is Characterized by Dysregulation of the Th17:Treg Cell Axis, Which Is Corrected by Anti-TNF Therapy. J Invest Dermatol, 2017. 137(11): p. 2389–2395. [DOI] [PubMed] [Google Scholar]
- 61.Hoffman LK, et al. , Integrating the skin and blood transcriptomes and serum proteome in hidradenitis suppurativa reveals complement dysregulation and a plasma cell signature. PLoS One, 2018. 13(9): p. e0203672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murota H, et al. , Artemin causes hypersensitivity to warm sensation, mimicking warmth-provoked pruritus in atopic dermatitis. J Allergy Clin Immunol, 2012. 130(3): p. 671–682.e4. [DOI] [PubMed] [Google Scholar]
- 63.Albers KM, et al. , Artemin growth factor increases nicotinic cholinergic receptor subunit expression and activity in nociceptive sensory neurons. Mol Pain, 2014. 10: p. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krajewski PK, et al. , Pain in Hidradenitis Suppurativa: A Cross-sectional Study of 1,795 Patients. Acta Derm Venereol, 2021. 101(1): p. adv00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matusiak Ł, et al. , Clinical Characteristics of Pruritus and Pain in Patients with Hidradenitis Suppurativa. Acta Derm Venereol, 2018. 98(2): p. 191–194. [DOI] [PubMed] [Google Scholar]
- 66.Nakamura T, et al. , Molecular cloning and expression of human hepatocyte growth factor. Nature, 1989. 342(6248): p. 440–3. [DOI] [PubMed] [Google Scholar]
- 67.Phan LM, et al. , Hepatocyte Growth Factor/cMET Pathway Activation Enhances Cancer Hallmarks in Adrenocortical Carcinoma. Cancer Res, 2015. 75(19): p. 4131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grand D, et al. , Doppler ultrasound-based noninvasive biomarkers in hidradenitis suppurativa: evaluation of analytical and clinical validity. Br J Dermatol, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All supplementary data is available on Mendeley (http://dx.doi.org/10.17632/s99c7tmmmg.1)