Inhibition of a specific isoform of RON receptor tyrosine kinase (SF-RON) recruits stemlike CD4 T cells to the metastatic site and promotes strong antitumor immune responses that severely restrict growth of breast cancer metastases.
Abstract
Immunotherapy has potential to prevent and treat metastatic breast cancer, but strategies to enhance immune-mediated killing of metastatic tumors are urgently needed. We report that a ligand-independent isoform of Ron kinase (SF-Ron) is a key target to enhance immune infiltration and eradicate metastatic tumors. Host-specific deletion of SF-Ron caused recruitment of lymphocytes to micrometastases, augmented tumor-specific T-cell responses, and nearly eliminated breast cancer metastasis in mice. Lack of host SF-Ron caused stem-like TCF1+ CD4+ T cells with type I differentiation potential to accumulate in metastases and prevent metastatic outgrowth. There was a corresponding increase in tumor-specific CD8+ T cells, which were also required to eliminate lung metastases. Treatment of mice with a Ron kinase inhibitor increased tumor-specific CD8+ T cells and protected from metastatic outgrowth. These data provide a strong preclinical rationale to pursue small-molecule Ron kinase inhibitors for the prevention and treatment of metastatic breast cancer.
Significance:
The discovery that SF-Ron promotes antitumor immune responses has significant clinical implications. Therapeutic antibodies targeting full-length Ron may not be effective for immunotherapy; poor efficacy of such antibodies in trials may be due to their inability to block SF-Ron. Our data warrant trials with inhibitors targeting SF-Ron in combination with immunotherapy.
This article is highlighted in the In This Issue feature, p. 2945
INTRODUCTION
Breast cancer is the second leading cause of cancer-related death in women in the United States (1). Although early-stage primary breast cancer can be cured with surgery and systemic therapy, metastatic breast cancer remains incurable. Metastasis, the deadly spread of cancer to other organs, occurs in 20% to 30% of patients with breast cancer (2). In most of those cases, metastasis occurs prior to the primary tumor being detectable, and metastatic recurrences happen years after initial diagnosis and treatment (3). Thus, spontaneous outgrowth of initially undetectable micrometastatic disease is the largest contributor to breast cancer mortality. There is an urgent need to develop strategies that can prevent outgrowth of micrometastases and/or effectively treat metastatic disease. Because the immune system is able to protect against cancer in its early stages (4), immune stimulation is an attractive strategy to potentially eliminate micrometastatic and/or overtly metastatic cancer.
During tumor progression, the immune system becomes suppressed by multiple mechanisms, allowing tumor outgrowth. Continually evolving tumor–host interactions allow evasion from immune-mediated killing by cytotoxic T cells (CTL) or natural killer (NK) cells (4, 5). Immune evasion is further promoted by other immune cells, including monocytes, macrophages, dendritic cells, neutrophils, myeloid-derived suppressor cells, regulatory T cells (Treg), and B cells, all of which have been demonstrated to play an active role in promoting immune tolerance (6). Established tumors adopt features of chronic wounds that promote systemic inflammation, which in turn promotes distant metastasis (7, 8).
Infiltration of different immune cell populations into primary tumors and metastases can serve as a prognostic factor in various cancers, suggesting that the presence of a certain immune milieu can have beneficial antitumor effects (9). Reviving the immune system's ability to kill tumor cells is an exciting area of research, so that new targets may be identified to prevent or treat deadly metastatic disease. Cancer immunotherapy has brought some success to the treatment of several advanced cancers, including melanoma, renal cancer, and lung cancers, all of which have relatively high somatic mutation burden and are therefore somewhat immunogenic (10–13). Although breast cancer is recognized as poorly immunogenic and less responsive to immune checkpoint blockade, recent developments in high-throughput genomic and cellular analyses have revealed heterogeneous populations of immune cells in breast cancers and implied that breast cancers are not always “immune-cold” (14). Immunotherapies are not approved for most breast cancers; however, the FDA has approved atezolizumab, an anti–PD-L1 immune checkpoint inhibitor, in combination with nab-paclitaxel, for the treatment of advanced or metastatic triple-negative breast cancer (TNBC; ref. 15). Despite this clinical advancement, most overtly metastatic TNBC tumors do not respond to immunotherapy (16). The goal of our research is to identify pathways that can boost the immune response to tumors to eliminate micrometastatic disease and prevent metastatic recurrences.
Overexpression of the receptor tyrosine kinase Ron occurs in various epithelial cancers, including breast cancer, and is significantly associated with tumor progression and metastasis (17, 18). Ron is the cell surface receptor for macrophage-stimulating protein (MSP; ref. 19). In addition to its expression on tumor cells, Ron is found on tissue-resident macrophages (20); hence, Ron signaling has both tumor-intrinsic as well as tumor-extrinsic (host cell–mediated) effects on tumor progression and metastasis. Activation of Ron signaling specifically on tumor cells promotes tumor cell proliferation, survival, and invasion (21). Activation of Ron signaling on macrophages is reported to skew them toward an alternatively activated (M2-like) phenotype (22, 23). Genetically engineered mice that lack Ron tyrosine kinase activity (Ron TK−/−) fail to downregulate proinflammatory immune responses in response to infection, inflammation, and injury (24–26). We previously showed that Ron functions specifically in the host to facilitate mammary tumor metastasis to lungs in the MMTV-PyMT mouse breast cancer model and that blocking Ron tyrosine kinase activity, either genetically or pharmacologically, protected mice from metastasis through stimulation of antitumor immunity (27). Our preclinical studies also showed that Ron inhibition was effective alone, and even better in combination with anti-CTLA4 immune checkpoint blockade, to significantly shrink primary breast or colorectal tumors and to reduce lung metastatic tumor outgrowth (28). However, the mechanism by which Ron regulates the antitumor immune response is unknown and is an exciting area of investigation given the availability of Ron inhibitors (Roni) in clinical development.
More than a dozen small-molecule inhibitors and mAbs to target Ron are in clinical and preclinical development (29–32). Phase I trials have demonstrated that both categories of targeted therapeutics against Ron are well tolerated (33–35). So far, only one trial reported on efficacy and showed that the anti-Ron antibody narnatumab had only limited antitumor efficacy (33). This was perplexing given the strong preclinical genetic evidence that loss of Ron kinase activity protects from tumor growth and metastasis through both cell-intrinsic and immune-modulating effects.
Both breast cancer cells and macrophages, which are the only immune cells known to express Ron, express two isoforms of Ron with distinct functions: full-length Ron (FL-Ron) and short-form Ron (SF-Ron). SF-Ron is expressed from an alternative promoter located within intron 10 of the mouse Ron gene (Stk; refs. 36, 37). SF-Ron is also produced in human cells in a similar manner, where its production is thought to be regulated by methylation of the main Ron promoter (38). Importantly, due to its N-terminally truncated nature, SF-Ron protein lacks most of the extracellular domain, including the entire ligand binding domain. SF-Ron does contain the transmembrane and intracellular domains and hence retains the tyrosine kinase activity in a ligand-independent manner (39, 40). It has been demonstrated that FL-Ron and SF-Ron have distinct tumor-intrinsic roles (41) and have nonredundant inflammatory functions in a noncancer setting (42).
Although FL-Ron's function in macrophages has been studied extensively and its function in tumor immunity has been studied to some extent (22, 27, 28, 43), the role of SF-Ron in antitumor immune responses is entirely unknown. Distinguishing the relative function of Ron isoforms in antitumor immunity is a critical gap in our knowledge because although both isoforms can be targeted with small-molecule kinase inhibitors, only FL-Ron can be targeted with antibodies such as narnatumab due to lack of the extracellular domain in the SF-Ron protein. Likewise, only FL-Ron is responsive to the Ron ligand MSP. Here, we sought to define the function of SF-Ron in breast cancer metastasis and antitumor immunity. We unexpectedly found that SF-Ron, not FL-Ron, is the key isoform that suppresses immune responses against breast cancer metastasis. Examination of mice specifically lacking SF-Ron also revealed new insight into the role of naive, stem-like CD4+ T cells in control of micrometastatic outgrowth of breast cancer.
RESULTS
Mice Lacking SF-Ron Are Protected from Breast Cancer Lung Metastasis
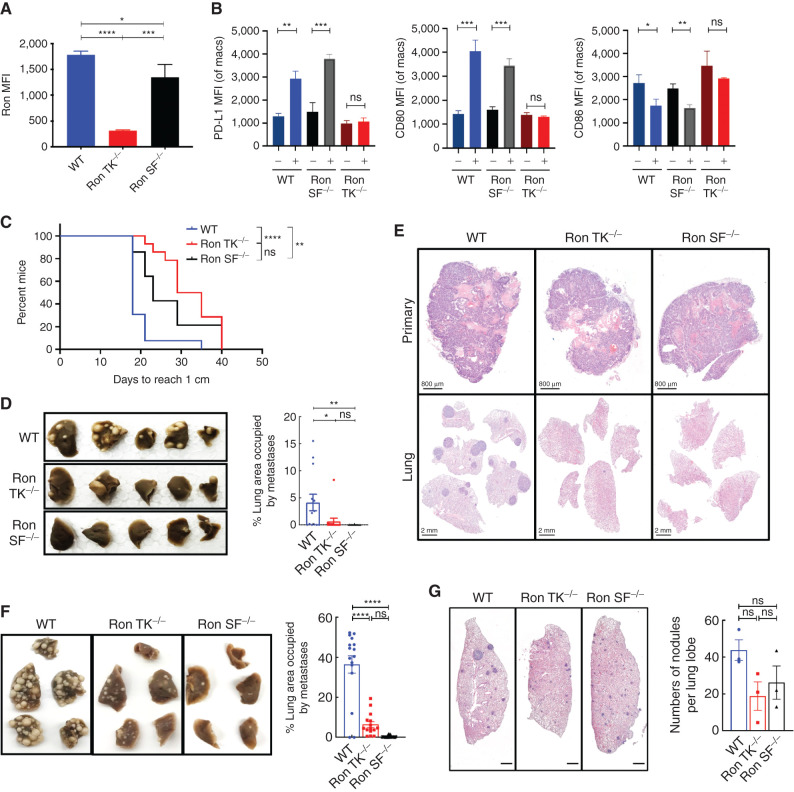
To determine whether the host SF-Ron plays a role in breast cancer progression, we used mice that were engineered to lack the SF-Ron isoform specifically (Ron SF−/−; ref. 42) on a pure FVB background, which is syngeneic with the MMTV-PyMT mammary tumor model (44). To verify the relative expression of different Ron isoforms in our model, we performed RT-PCR and flow cytometry on Ron-expressing peritoneal macrophages isolated from wild-type (WT), Ron tyrosine kinase deleted (Ron TK−/−; ref. 24), and Ron SF−/− mice. Ron TK−/− mice are engineered to lack exons 13 to 18 of Ron, which encode the entire tyrosine kinase domain and hence eliminate functional Ron tyrosine kinase activity from both isoforms (45). Using primers designed to specifically amplify cDNA encoding full-length Ron or primers specific for SF-Ron (36) for RT-PCR analysis, we verified that Ron SF−/− mice express FL-Ron mRNA but lack SF-Ron mRNA (Supplementary Fig. S1A). Analysis of FL-Ron protein by flow cytometry using an antibody against the extracellular domain of Ron (Supplementary Table S1) revealed that Ron SF−/− macrophages expressed FL-Ron protein at the cell surface at levels comparable to that of WT macrophages (Fig. 1A; Supplementary Fig. S1B and S1C). To ensure that FL-Ron was functional in Ron SF−/− macrophages, we examined three known downstream targets of Ron signaling: PD-L1, CD80, and CD86 (28). Stimulation of peritoneal macrophages from Ron SF−/− mice with MSP increased the expression of PD-L1 and CD80 and decreased the expression of CD86, similar to WT macrophages (Fig. 1B; Supplementary Fig. S1D). No significant change in any of these proteins was detected in Ron TK−/− macrophages in response to MSP. Together, these results verify that Ron SF−/− mice produce functional FL-Ron and specifically lack SF-Ron.
Figure 1.
Specific loss of host SF-Ron significantly protects mice from breast cancer metastasis: A, Quantification of mean fluorescence intensity (MFI) for FL-Ron protein on peritoneal macrophages by flow cytometry (n = 3 mice/group). B, Quantification of MFI by flow cytometry for PD-L1, CD80, and CD86 upon treatment of peritoneal macrophages from each group with or without MSP for 18 hours (n = 3 mice/group). C, Survival plot depicting significant delay in the primary tumors reaching 1 cm in diameter in Ron TK−/− and Ron SF−/− mice. D, Representative images of the fixed lungs from the spontaneous metastasis experiment (left) and quantification of the percent lung area occupied by metastasis by ImageJ (right; n = 13–14 mice/group). E, Representative images of orthotopic primary tumor (top) and spontaneous lung metastasis (bottom panel) tissue sections stained with hematoxylin and eosin (H&E). F, Representative images of the fixed lungs from all three genotypes of mice (left) and quantification of percent lung area occupied by metastasis at the endpoint by ImageJ (right; n = 16–23 mice/group). G, Histologic sections showing micrometastases across lungs of WT, Ron TK−/−, and Ron SF−/− mice. Sections were taken at 14 days after tumor cell injection. All scale bars correspond to 800 μm. Log-rank (Mantel–Cox) test was used for Kaplan–Meier analysis, and one-way ANOVA with Tukey correction was performed for statistical analysis of the MFI and lung tumor burden quantification; error bars represent SEM. ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
To test the role of host SF-Ron in spontaneous breast cancer metastasis, we orthotopically implanted MMTV-PyMT tumor cells into cleared mammary fat pads of WT or Ron SF−/− mice. Ron TK−/− mice were used as a positive control; we previously reported that lack of all host Ron tyrosine kinase activity reduces spontaneous and experimental metastasis (27). To control for any differences in primary tumor growth, we resected each primary tumor when they grew to 1 cm in diameter, then examined metastasis 4 weeks later. Loss of SF-Ron specifically (Ron SF−/−), or loss of function of both isoforms (Ron TK−/−), significantly delayed the primary breast tumor growth compared with WT mice (Fig. 1C). We also found that both Ron SF−/− and Ron TK−/− mice developed significantly lower metastatic tumor burden than WT mice, even though all primary tumors were allowed to grow to the same size prior to resection (Fig. 1D and E).
The most common scenario for human breast cancer metastasis is outgrowth of micrometastatic tumor cells at distant sites, usually long after the primary tumor has been resected. To determine whether SF-Ron plays a role in metastatic tumor outgrowth in the lungs, we performed experimental metastasis assays by injecting MMTV-PyMT tumor cells into the tail veins of WT, Ron TK−/−, and Ron SF−/− mice. As expected from our previous results, Ron TK−/− mice were protected from metastasis. Interestingly, we found that there were almost no lung metastases in Ron SF−/− mice (Fig. 1F). These results suggested that host SF-Ron, instead of FL-Ron, is required for lung metastasis of breast cancer.
To determine whether lack of SF-Ron prevented initial metastatic seeding versus metastatic outgrowth, we collected the lungs of WT, Ron TK−/−, and Ron SF−/− mice at an earlier time point, 2 weeks after tumor injection. Here, we found that Ron SF−/− mice had micrometastatic nodules (Fig. 1G), indicating that tumor cells are able to seed the lungs of Ron SF−/− mice and generate micrometastatic lesions, but that they disappear by the later time point. Therefore, SF-Ron is required for the critical step of micrometastatic outgrowth to overt metastasis in lungs, which is a disease stage amenable to metastatic tumor prevention in patients through the use of adjuvant therapy. We next investigated the mechanism by which metastatic outgrowth was prevented in these mice.
Lack of SF-Ron Stimulates Robust Systemic and Local Antitumor Immune Responses
FL-Ron is well known to regulate inflammation through its function on macrophages (22, 43, 46), and some data indicate that FL-Ron and SF-Ron have similar but nonredundant roles in inflammation in response to injury (42). However, the role of SF-Ron in regulating immune responses in cancer is unknown, so we focused efforts on determining how this isoform regulates metastatic outgrowth. To determine whether Ron SF−/− mice mount antitumor immune responses, we performed multicolor flow cytometry (Supplementary Fig. S2A–S2C) on splenocytes isolated from tumor-bearing Ron SF−/− mice and WT mice. These data revealed that Ron SF−/− mice produced significantly fewer CD4+ Tregs, F4/80+ macrophages, and CD11b+ Ly6G+ myeloid cells but produced more CD4+ and CD8+ T cells than WT mice (Fig. 2A–E). These data are consistent with the fact that Tregs, macrophages, and CD11b+ Ly6G+ myeloid cells can be immunosuppressive, facilitating tumor progression and metastasis (6, 47, 48), and that higher frequencies of Tregs also correlate with poor prognosis of patients with cancer (48, 49). One of the mechanisms by which T cells exhibit antitumor immunity is by secreting cytokines such as IFNγ and TNFα (50–53). We found that Ron SF−/− mice also have significantly more IFNγ-producing CD4+ and CD8+ T cells (Fig. 2F and G). We found no significant differences in B220+ B cells, CD11c+ dendritic cells, or NK1.1+ NK cells between WT and Ron SF−/− mice (Supplementary Fig. S3A–S3C).
Figure 2.
Lack of host SF-Ron promotes antitumor immune responses: A–G, Flow cytometry analysis of immune cell populations from the spleens of tumor-bearing WT and Ron SF−/− mice. Bar graphs show frequency of Ly6G+ CD11b+ myeloid cells (A), CD4+ CD25+ FOXP3+ splenic regulatory T cells (B), F4/80+ CD11b+ macrophages (C), CD4+ T cells (D), CD8+ T cells (E), and IFNγ-producing cells within the CD4+ and CD8+ compartment (F and G), with n = 16–23 mice/group. H–J, Quantification of metastatic tumor-infiltrating T cells. K, Representative IHC images showing lung metastatic nodules and T-cell infiltration into the metastatic nodules. All scale bars correspond to 100 μm. Unpaired t test was performed for statistical analysis; error bars represent SEM. ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Infiltration of immune cells into tumors is critical for tumor control and is prognostic for patient outcomes in multiple cancer types (9, 54). To determine the tumor-infiltrating lymphocyte (TIL) landscape in the metastases in our model, we performed IHC analysis on the lungs of tumor-bearing mice at the 2-week time point, when the micrometastases in Ron SF−/− mice were detected similar to those in WT mice. We found that TIL infiltration into metastases was much more robust in Ron SF−/− mice compared with WT mice (Fig. 2H–J). The metastases in Ron SF−/− mice were swarmed by CD3+ TILs (both CD4+ and CD8+ T cells; Fig. 2K).
To investigate the generality of the SF-Ron–regulated antitumor immune response, we used a second breast tumor model of a different subtype: the KPB1 (K14-Cre; p53f/f; Brca1f/f) basal-like mammary tumor model, also in the FVB/NJ background (55). We injected freshly isolated KPB1 tumor cells into tail veins of WT and Ron SF−/− mice, and the lung metastatic tumor burden was examined 24 days later. Again, lack of host SF-Ron significantly reduced the lung metastatic tumor burden when compared with WT mice (Supplementary Fig. S4A), although the lung metastases were not as efficiently cleared as in the MMTV-PyMT model. Immunofluorescence using lymphocyte markers (CD3e, CD4, and CD8a) and pan-cytokeratin as a tumor marker revealed a phenotype similar to what we found in the MMTV-PyMT metastasis model: significantly increased TIL infiltration in the tumor-bearing lungs of Ron SF−/− mice compared with WT mice (Supplementary Fig. S4B–S4E). Thus, loss of SF-Ron reproducibly promotes immune cell infiltration into micrometastases. We next set out to determine specific differences in the metastatic tumor immune microenvironment of Ron SF−/− mice compared with WT mice and whether the infiltrating immune cells were tumor specific.
Selective Loss of SF-Ron Results in Expansion of Stem Cell–Like T-cell Populations in Metastatic Lungs
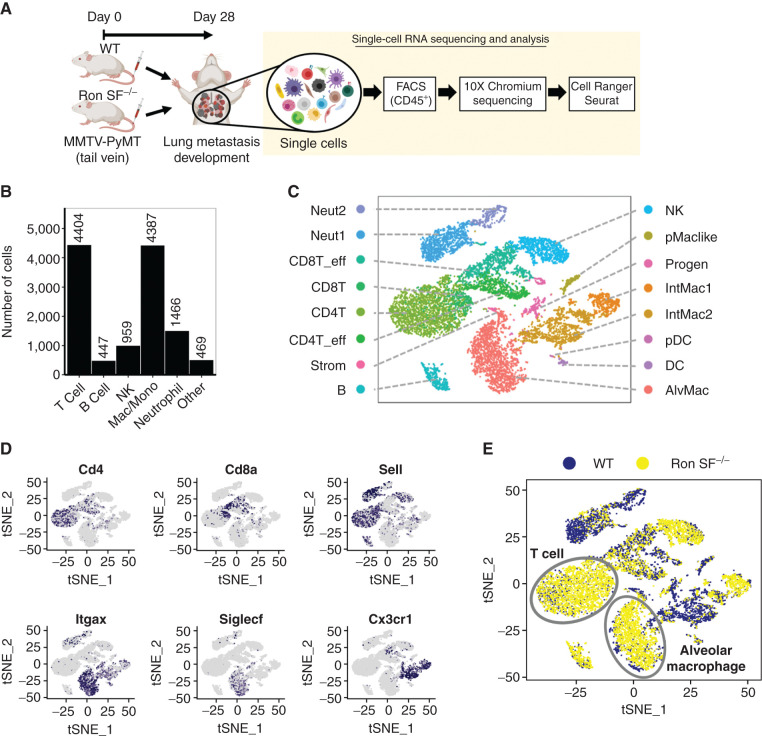
Tumors attract a variety of immune cells, giving rise to a complicated and heterogeneous tumor–immune microenvironment (56). To investigate the heterogeneity of the immune cell infiltrates resulting from the loss of SF-Ron, we performed single-cell RNA sequencing (scRNA-seq) on flow-sorted CD45+ immune cells from tumor-bearing lungs of WT and Ron SF−/− mice (Fig. 3A). Eight to 10 mice per group were pooled together as a single sample and around 16,000 sorted cells from each sample were loaded, to target 10,000 cells in the sequencing reaction. Expression data were recovered from 9,581 and 10,492 cells from WT and Ron SF−/− samples, respectively. The median numbers of genes detected in WT and Ron SF−/− samples were 1,511 and 1,515, respectively, which correspond to 22,399 and 18,684 mean reads per cell, respectively. Unbiased cell-type classification can be achieved with as few as 10,000 reads per cell (57, 58). Unbiased clustering after merging both data sets revealed the presence of 16 cell type clusters in both WT and Ron SF−/− mice, numbered 0 to 15 (Supplementary Fig. S5A). The two major cell types present in the metastatic tumor microenvironment in both groups of mice were T cells expressing Cd3e and macrophages expressing Cd68, followed by neutrophils expressing S100a8, NK cells expressing Ncr1, and B cells expressing Cd19 (Fig. 3B; Supplementary Fig. S5B). To further classify the cell clusters in detail and examine differences between genotypes, we used the web-based Cluster Identity Predictor (CIPR) tool, which compares the genome-wide expression signatures with those of known immune cell types from the publicly available Immunological Genome Project database (59, 60). This revealed that T cells and macrophages each can be separated into four distinct populations, with other cell types spreading out to a lesser extent (Fig. 3C). Using the expression of Cd4, Cd8a, and Sell, which encodes CD62L, the four clusters of T cells were further defined as conventional CD4+ and CD8+ T cells and their activated effector populations (Fig. 3D, top plots). Likewise, the macrophage populations were divided into alveolar and interstitial macrophages based on the expression of Itgax, Siglecf, and Cx3cr1 (61). In our data, we identified two interstitial macrophage clusters and one alveolar macrophage cluster (Fig. 3D, bottom plots). A full list of genes differentially expressed in each individual cluster compared with the rest of the data set is provided in Supplementary Table S2. Comparing immune cell clusters in metastatic lungs of WT and Ron SF−/− mice revealed significant differences in the immune microenvironment: Ron SF−/− lungs contained a large number of alveolar macrophages and T cells compared with WT lungs (Fig. 3E; Supplementary Fig. S5C), providing additional evidence that loss of SF-Ron modifies the immune microenvironment during metastatic outgrowth. These data, along with the swarming of early metastatic lesions by TILs (Fig. 2K), suggested that T cells might be more effective at eliminating metastases in mice lacking SF-Ron. Therefore, we next investigated the nature of the T cells in Ron SF−/− metastatic lungs.
Figure 3.
Single-cell transcriptomic analysis reveals robust immune cell infiltration in the metastatic tumor microenvironment (mTME) of Ron SF−/− mice. A, Workflow of scRNAseq of immune cells isolated from tumor-bearing lungs of WT and Ron SF−/− mice. CD45+ cells from at least eight mice per group were pooled together as a single sample, and an equal number of cells were processed for 10× scRNAseq. B, Number of immune cells of major cell populations present in the TME that were identified by typical markers as shown in Supplementary Fig. S5B. C, The t-distributed stochastic neighbor embedding (tSNE) plot of scRNA-seq data showing 16 distinct cell clusters identified by the CIPR tool. D, tSNE plots showing selected marker gene expression of infiltrating immune cells. E, tSNE plot depicting leukocytes in WT and Ron SF−/− mice, emphasizing the enrichment of T cells and alveolar macrophages in Ron SF−/− mice. AlvMac, alveolar macrophages; B, B cells; CD4T, CD4+ T cells; CD8T, CD8+ T cells; DC, dendritic cell; eff, effector; IntMac, interstitial macrophages; Mac, macrophage; Mono, monocyte; Neut, neutrophil; NK, natural killer cell; pDC, plasmacytoid DC; pMaclike, peritoneal macrophage–like cells; Progen, progenitors; Strom, stroma cells.
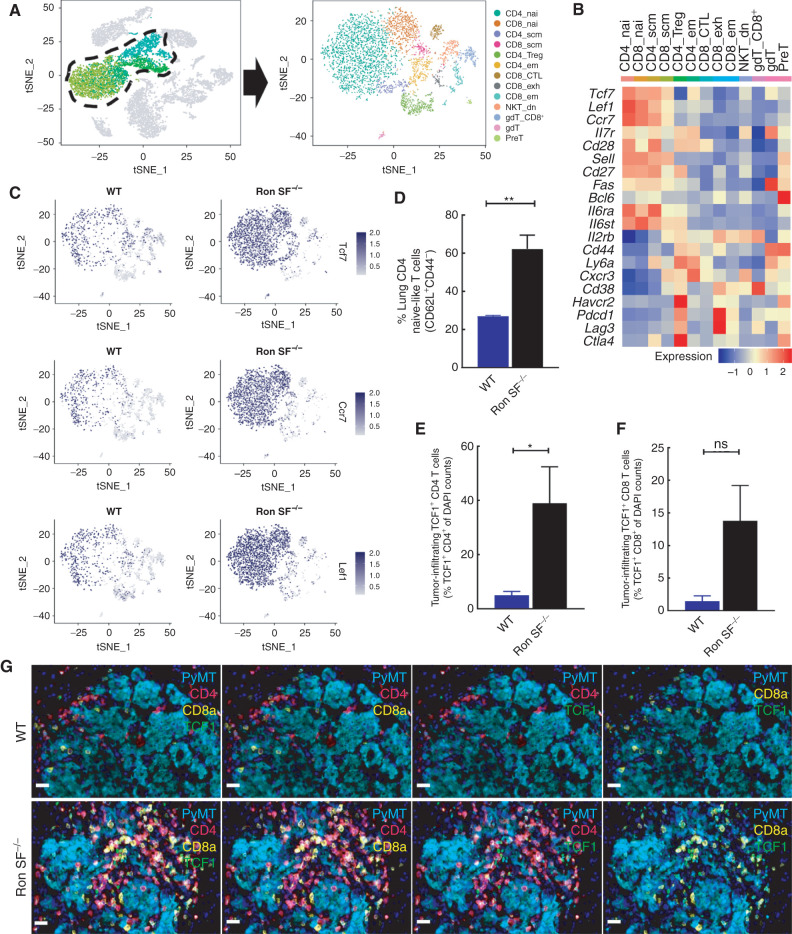
Analysis of the T cells in both mouse genotypes revealed the presence of 13 distinct T-cell clusters (Fig. 4A). Interestingly, the large proportion of CD4 T cells enriched in the Ron SF−/− lungs expressed Tcf7 (which encodes TCF1), Ccr7, and Lef1 (Fig. 4B and C), which together represent a less-differentiated or stem cell–like T-cell signature (62). There were higher frequencies of T cells with the less-differentiated phenotype (CD8+ or CD4+ naïve and stem cell–like) and lower frequencies of T cells with a terminally differentiated phenotype (Tregs and exhausted CD8 T cells) in Ron SF−/− lungs compared with WT lungs (Supplementary Fig. S5D). To validate the presence of these less-differentiated T cells in Ron SF−/− lungs, we isolated cells from the lungs of tumor-bearing WT and Ron SF−/− mice and performed flow cytometry analysis (Supplementary Fig. S6A). We found a significant enrichment of CD62L+CD44- CD4+ T cells in the lungs of Ron SF−/− mice (Fig. 4D), whereas CD62L-CD4+ T cells were more abundant in WT mice, either CD44+ or CD44−, indicating actively differentiated states (Supplementary Fig. S6B and S6C). CD62L+CD44+ resting central memory cells were rare and not significantly different between the two groups (Supplementary Fig. S6D). To validate presence of the stem-like CD4+ T cells in the tumor microenvironment, we conducted five-plex immunofluorescence staining and examined the expression of TCF1 in CD4+ or CD8+ T cells that had infiltrated into the metastatic tumor nodules at a 2-week time point when the tumor burden is comparable between WT and Ron SF−/− mice (Supplementary Fig. S7A–SE). Although the proportion of both TCF1-expressing CD4 or CD8 TILs was higher in Ron SF−/− lungs compared with the WT lungs, TCF1+ CD4+ T cells were the predominant population (Fig. 4E–G), consistent with the scRNA-seq data (Supplementary Fig. S5D). Maintaining a T-cell pool with a less-differentiated phenotype in nonlymphoid tissues, such as the microenvironment that harbors a tumor, provides the ability to replenish the effector T cells that may become exhausted or apoptotic (63, 64). A recently reported population of TCF1+ CD8+ stem-like T cells identified in human cancer was correlated with better treatment outcomes (65). Hence, we hypothesized that the presence of TCF1+ CD4+ T cells in Ron SF−/− lungs might be important in driving and/or maintaining the robust antitumor immune responses in Ron SF−/− mice during metastatic outgrowth.
Figure 4.
Loss of SF-Ron alters the composition of tumor-infiltrating T cells. A, tSNE plot showing 13 T-cell clusters extracted from the combined data set. B, Heatmap showing the average expression of selected genes in each cluster that are linked to memory, self-renewal, activation, and inhibitory T-cell functions. C, tSNE plots showing the Tcf7, Ccr7, and Lef1 expression in tumor-infiltrating T cells of WT and Ron SF−/− lungs. D, Flow cytometry quantification of lung CD4 T cells presenting naïve-like phenotype. E and F, Quantification of metastatic tumor-infiltrating TCF1 expressing CD4 (E) or CD8 T cells (F). G, Representative images of PyMT lung metastases from WT and Ron SF−/− mice subjected to a five-marker multiplexed immunofluorescence stain to demonstrate the TCF1-expressing CD4 or CD8 T-cell infiltration into tumor. All scale bars correspond to 20 μm. Statistical analysis was performed by two-tailed unpaired Student t test, and error bars represents the SEM. CTL, cytotoxic T lymphocyte; dn, double-negative; em, effector memory; exh, exhausted; nai, naive; scm, stem cell memory. ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Recruitment of CD4+ T Cells from Lymph Nodes Is Required for Clearance of Metastatic Lesions in Ron SF−/− Mice
To determine the importance of CD4+ T-cell enrichment in Ron SF−/− mice during metastatic outgrowth, we depleted CD4+ T cells in vivo using an anti-CD4 antibody and then injected tumor cells via tail vein. Compared with the isotype control, CD4+ T-cell depletion significantly enhanced lung metastasis in Ron SF−/− mice but had no significant effect in WT mice (Fig. 5A; Supplementary Fig. S8A). Given the high number of TCF1-expressing CD4+ TILs in Ron SF−/− lungs and their high expression of Ccr7 and Sell (Fig. 4B and C), which are known markers for naïve or resting memory T cells that home to lymph nodes (66), we hypothesized that cell trafficking between draining lymph nodes and lung was crucial for recruitment of those T cells into the tumor microenvironment and for suppression of metastatic growth. To determine whether circulating, rather than resident, T cells were important for controlling metastatic outgrowth, we treated WT and Ron SF−/− mice with the sphingosine 1 phosphate receptor inhibitor FTY-720, which prevents egress of T cells from the lymph node (67), beginning the day after tumor cell seeding. Inhibition of trafficking was verified by the significant decrease of T cells in the peripheral blood and spleens and retention of these cells in lymph nodes of tumor-bearing mice at the endpoint (Supplementary Fig. S8B and S8C). Sequestering T cells in the lymph node not only completely restored metastasis in Ron SF−/− mice but also resulted in increased metastasis burden in WT mice (Fig. 5B). IHC staining of T cells verified loss of tumor-infiltrating T cells (Supplementary Fig. S8D). These data demonstrate that recruitment of T cells from lymph nodes to the site of metastasis, as well as the presence of CD4+ T cells, is crucial to control metastasis in the Ron SF−/− mice.
Figure 5.
Recruitment of CD4 T cells from lymph nodes is required for control of metastasis in Ron SF−/− mice. A, Representative images of the fixed lungs from WT and Ron SF−/− mice treated with either isotype control or anti-CD4 antibody (left) and bar graphs representing the percent lung area occupied by metastasis at the endpoint after indicated treatment (right; n = 8 mice/group). B, Quantification of percentage lung area occupied by metastases in WT and Ron SF−/− mice treated with FTY720 or vehicle control at the endpoint of the experiment (n = 5 mice/group). C, Intracellular staining of master transcription factors denoting Th1 (T-BET), Th2 (GATA3), Th17 (RORγt), and Treg (FOXP3) on isolated CD4+ T cells from spleens of WT and Ron SF−/− mice at the endpoint. D, Boxplot analysis of each Th subtype in the CD4+ T-cell pool in spleens of WT and Ron SF−/− mice. The box presents the 25th and 75th percentiles of the variables, and the horizontal bar corresponds to the 50th percentile. Individual values from each sample were plotted as points. E, Flow plots demonstrating differentiation of different Th subtypes from CD4+ T cells isolated from lungs of tumor-bearing WT and Ron SF−/− mice. Purified CD4+ T cells were labeled with CTD and incubated with Th differentiation media for 96 hours. CTD versus major markers indicating Th1 subsets (Th1: T-BET, Th2: GATA3, Th17: IL17A, and iTreg: FOXP3) are shown. Data are representative of two individual experiments in which each sample represents a pool of cells from 5 to 10 mice per group. Statistical analysis was performed by one-way ANOVA with Tukey correction, and error bars represents the SEM. ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
CD4+ T Cells Are Skewed toward a Th1 Phenotype in Ron SF−/− Mice during Metastasis Elimination
Upon antigen encounter, naïve CD4+ T cells, which express TCF1, differentiate into at least four subsets to execute different immune functions: Th1, Th2, and Th17 T cells, as well as Tregs. This differentiation is determined by the signals received during antigen encounter (68–70). Expression of TCF1 on CD4+ T cells is also required for the generation of follicular Th cells (Tfh) in response to viral infection and the development of immunologic memory (71). How each of these CD4+ T-cell subsets contributes to antitumor immunity is still controversial. To determine the differentiation status of the CD4+ T cells present systemically in WT and Ron SF−/− mice, we isolated CD4+ T cells from the spleens of mice during metastatic outgrowth and performed flow cytometry to identify CD4+ T-cell subsets based on the expression of their master transcription factor proteins: T-BET for Th1, GATA3 for Th2, RORγT for Th17, and FOXP3 for Tregs (Fig. 5C; Supplementary Fig. S9A). We found that Ron SF−/− mice had significantly more Th1 and fewer Treg cells than WT mice; there was no significant difference in Th2 or Th17 cells (Fig. 5D). These data suggest that the increased antitumor immunity resulting from loss of SF-Ron may be associated with an increased systemic Th1 CD4+ T-cell response.
In influenza virus–infected animals and in autoimmune encephalomyelitis, TCF1+ CD4+ T cells are capable of differentiating into either Th1 cells in the presence of IL2 and IL12 or Th17 cells in a manner dependent on mTORC1 activity (72, 73). Because we observed robust enrichment of Tcf7/TCF1 in the naïve and stem cell–like CD4+ T cells in the lung of Ron SF−/− animals, which correlated with the elimination of metastasis, and because CD4+ T cells were required to suppress metastasis, we tested the differentiation potential of CD4+ T cells isolated from spleens and tumor-bearing lungs of WT and Ron SF−/− animals. Sorted CD4+ T cells from each organ were labeled with cell proliferation tracking dye before stimulation with plate-bound anti-CD3ε and anti-CD28 antibodies together with cytokines that are known to induce the four main Th subsets (74). We monitored both proliferation and differentiation of CD4+ T cells under each condition using the expression of T-BET, GATA3, IL17A, and FOXP3 as markers of Th1, Th2, Th17, and Treg differentiation, respectively. Cells that were positive for each marker were determined with no-antibody-control stains (Supplementary Fig. S9B and S9C). Relative to the control condition with only anti-CD3ε/CD28, CD4+ T cells isolated from the lungs of tumor-bearing WT mice proliferated dramatically and produced appropriately differentiated T cells under Th2-, Th17-, and Treg-inducing conditions but not under Th1-inducing conditions (Fig. 5E; Supplementary Fig. S9D and S9E). Expansion was most apparent in the Th17-inducing condition (Fig. 5E; Supplementary Fig. S9D and S9E). On the other hand, CD4+ T cells from tumor-bearing Ron SF−/− lungs were poised toward Th1 differentiation at baseline (in the anti-CD3ε/CD28 control setting), and with cytokine treatment, differentiation could be further enhanced to the Th1 state and induced to the Th17 or Treg state but not to the Th2 state (Fig. 5E; Supplementary Fig. S9D and S9E). The strong expansion of the Th17 subtype seen in cells isolated from WT animals was also not apparent in cells isolated from Ron SF−/− mice (Fig. 5E; Supplementary Fig. S9D and S9E). Resistance to Th17 differentiation was also observed using splenic CD4+ T cells from Ron SF−/− tumor-bearing mice (Supplementary Fig. S9F, top), suggesting that reduced Th17 differentiation of Ron SF−/− T cells might not depend on direct exposure to the tumor microenvironment. To address this question further, we also examined splenic CD4+ T-cell differentiation from WT or Ron SF−/− mice with no tumors. We found that splenic CD4+ T cells isolated from naive WT and Ron SF−/− mice were all able to differentiate appropriately; however, Th17 differentiation of CD4+ T cells from Ron SF−/− spleens was again reduced compared with WT (Supplementary Fig. S9F, bottom). These data suggest that loss of SF-Ron reduces Th17 differentiation of naïve CD4+ T cells, and this phenotype is enhanced in the presence of tumors, where Th1 differentiation is strongly favored in Ron SF−/− mice.
Taken together, these data show that Ron SF−/− animals effectively recruit T cells from lymph nodes into the lung metastatic microenvironment, that these T cells are predominantly a TCF1+ CD4+ population with increased potential for Th1 differentiation, and that CD4+ T cells are required to eliminate metastatic outgrowth.
Ron SF−/− Mice Promote CD8+ T Cell–Dependent, Tumor-Specific Immune Responses to Eliminate Lung Metastasis
A major role of Th1 CD4+ Th cells is to activate CD8+ cytotoxic T cells, which are the central effector cells for tumor surveillance due to their capacity for direct tumor cell killing (75). We therefore investigated the requirement for CD8+ T cells for clearance of metastasis, as well as their tumor specificity, in Ron SF−/− animals. To examine the tumor-specific CD8+ T-cell responses, we evaluated the response of splenocytes from tumor-bearing mice to a pool of three MHC-I (H-2q) PyMT-specific peptides (76) using IFNγ ELISpot assays. These peptides allow us to examine the PyMT tumor antigen–specific response without the need of exogenously expressed model antigen such as ovalbumin (77). Our results demonstrated that splenocytes isolated from tumor-bearing Ron SF−/− mice have significantly more tumor-specific, IFNγ-producing cells compared with those from WT mice (Fig. 6A and B). To further assess PyMT tumor-specific CD8+ T cells, we stained splenocytes with PyMT peptide-bound MHC-I tetramers (76) by flow cytometry. The results revealed that PyMT-specific T cells were rare among the entire CD8+ T-cell population in the spleen of WT mice, but the frequency was increased in the spleens of Ron SF−/− mice (Fig. 6C; Supplementary Fig. S10A and S10B). Interestingly, approximately 35% of the PyMT-specific (tetramer+) splenic CD8+ T cells from Ron SF−/− mice were CD62Llo and CD44hi, which define effector memory T cells known to be associated with antitumor response and responses to chronic infections (Fig. 6D; Supplementary Fig. S10C).
Figure 6.
Lack of host SF-Ron promotes tumor-specific immune responses, and depletion of CD8+ T cells rescues metastasis in Ron SF−/− mice. A, Representative images of the anti-IFNΥ ELISpot assay performed by stimulating splenocytes from tumor-bearing mice in vitro with tumor-specific MHC-I PyMT peptides. B, Quantification of anti-IFNΥ responses from the ELISpot assay. C, Flow cytometric quantification of frequency of tumor-specific CD8+ T cells after staining splenocytes with PyMT-Dq tetramer pool. D, Frequency of tumor-specific effector memory T cells within the CD8+ T-cell population quantified by flow cytometry. E, Representative images of the fixed lungs from WT and Ron SF−/− mice treated with either isotype control or anti-CD8 antibody. F, Quantification of the percent lung area occupied by metastasis at the endpoint after indicated treatment (n = 7–8 mice/group). G, H&E and PyMT immunohistochemistry stain of mammary tissue with or without tumor collected from tumor rechallenged site. Black arrow indicates a residual tumor cell cluster that is detected in one Ron SF−/− mouse. H, Numerical comparison of the presence of mammary tumor in WT and Ron SF−/− mice 2 weeks after tumor rechallenge. I, Quantification of PyMT peptide-specific IFNγ spots per million splenocytes isolated from WT or Ron SF−/− mice 2 weeks after tumor rechallenge. Statistical analysis of panels B–D and H was performed by two-tailed unpaired Student t test. One-way ANOVA with Tukey correction was performed for statistical analysis of panel F. Error bars represents the SEM. ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
We next asked whether CD8+ T cells were necessary for eliminating metastasis in Ron SF−/− mice. We depleted CD8+ T cells from WT and Ron SF−/− mice and then performed experimental metastasis assays. Flow cytometry confirmed complete depletion of CD8+ T cells in the spleen (Supplementary Fig. S10D). We analyzed the metastatic tumor burden at the endpoint and found that CD8+ T-cell depletion rescued metastasis in Ron SF−/− mice, while not significantly altering metastasis in WT mice (Fig. 6E and F).
T-cell memory is required to induce long-term protective immunity and rapid response upon reencounter with tumor antigens (78). To understand whether the lack of SF-Ron promotes T-cell memory development after the robust antitumor immune response seen in these mice, we performed a tumor rechallenge experiment. In this experiment, the lung metastases were first induced by tail vein injection of MMTV-PyMT tumor cells, followed by implantation 14 days later of the same batch of tumor cells into the inguinal mammary fat pads. Mice were harvested 4 weeks later, and the mammary tumor burden was assessed. Although all rechallenged WT mice had visible mammary tumor growth at the site of injection, rechallenged Ron SF−/− mice were almost completely protected. No palpable tumors were present or found by histology in Ron SF−/− mice, but IHC staining with PyMT antibody identified one small cluster of tumor cells remaining in one animal (Fig. 6G and H; Supplementary Fig. S10E). IFNγ ELISpot of splenocytes also showed a significant increase in the number of PyMT tumor–specific T cells in rechallenged Ron SF−/− mice compared with the rechallenged WT mice (Fig. 6I). Taken together, these data demonstrate that Ron SF−/− mice are better than WT mice at generating robust antitumor CD8+ immune responses, with effective immunologic memory that protects them from tumor outgrowth.
Ron SF−/− T Cells Are Sufficient to Protect from Metastatic Outgrowth When Transferred to WT Mice
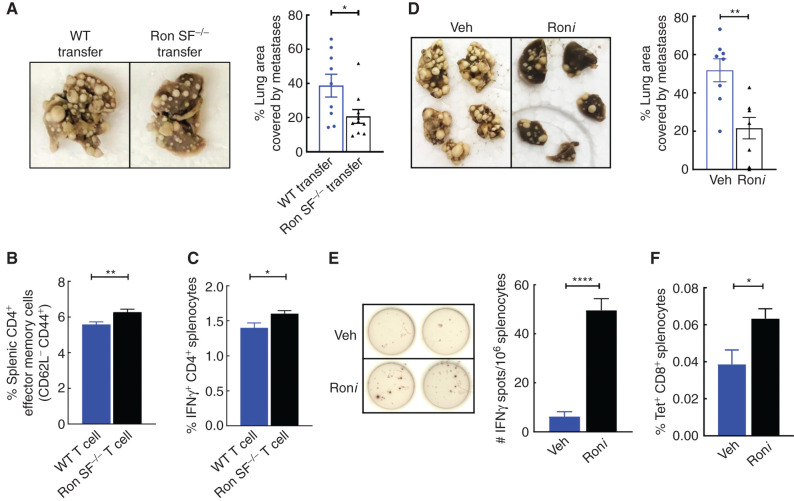
Although FL-Ron expression in the immune system is restricted to resident macrophages (Supplementary Fig. S11A and S11B), expression of SF-Ron has not been characterized. There are no antibodies that are specific for SF-Ron, so we used an SF-Ron–specific RT-PCR primer pair that takes advantage of the unique 5′ untranslated region of SF-Ron, derived from intron 10 (36), to examine SF-Ron expression in T cells. As expected, we found FL-Ron mRNA expressed in macrophages but not in T cells; however, SF-Ron mRNA was detected in both CD4+ and CD8+ T cells (Supplementary Fig. S11C). This raised the possibility that SF-Ron in T cells could be directly responsible for promoting increased antitumor activity. To test this idea, we examined whether T cells isolated from naïve Ron SF−/− mice could prevent metastasis when adoptively transferred to WT mice. We magnetically sorted T cells from either Ron SF−/− or WT mice and confirmed that the purity of magnetically sorted donor T cells from both genotypes was more than 97% (Supplementary Fig. S12A). These cells were adoptively transferred into WT hosts, and tumor cells were injected two days later. We isolated lungs three weeks after tumor injection and assessed metastasis burden and immunophenotype. As shown in Fig. 7A, WT mice that received T cells from naïve Ron SF−/− donor mice, and thus had both WT and Ron SF−/− T cells, had a significantly lower metastatic tumor burden compared with mice that received T cells from naïve WT donor mice. Moreover, we found more effector memory (CD62L−CD44+) and IFNγ-producing CD4+ T cells in spleens of tumor-bearing mice that received T cells from Ron SF−/− donors versus from WT donors (Fig. 7B and C; Supplementary Fig. S12B–S12D). No significant differences were observed in other CD4 subsets (Supplementary Fig. S12D). These data provide the first indication that SF-Ron functions in T cells to facilitate antitumor immune responses and can do so in the context of a WT host.
Figure 7.
SF-Ron−/− T cells are sufficient to protect from metastasis, and pharmaceutical inhibition of Ron suppresses metastatic progression by promoting tumor-specific immune responses. A, Representative images of the fixed lungs from WT mice that are adoptively transferred with T cells from WT mice or Ron SF−/− mice (left) and quantification of the percentage lung area occupied by metastasis at the endpoint of the experiment (right; n = 10 mice/group). B and C, Flow cytometric quantification of effector memory CD4 T cells (B) and the frequency of Th1 (characterized as IFNγ-producing CD4 T cells after 5-hour restimulation with phorbol myristate acetate and ionomycin, (C) in whole splenocytes isolated from adoptively transferred WT mice. D, Representative images of the fixed lungs from WT mice treated with either Ron inhibitor (Roni) or vehicle control (Veh; left), and the bar graph shows the quantification of the percentage lung area occupied by metastasis at the endpoint of the experiment (right; n = 8 mice/group). E, Representative images of the anti-IFNγ ELISpot assay performed by stimulating thawed splenocytes from tumor-bearing mice in vitro with tumor-specific MHC-I PyMT peptides. Data represent mean ± SEM (n = 5 per group). F, Flow cytometric quantification of frequency of tumor-specific CD8+ T cells after staining thawed splenocytes from Roni- or vehicle-treated mice with PyMT-Dq tetramer pool. Statistical analysis was performed by two-tailed unpaired Student t test, and error bars represent the SEM. ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Pharmacologic Inhibition of Ron Activity Boosts Tumor-Specific CD8+ T-cell Activity and Reduces Metastatic Outgrowth
The activation status and number of T cells found in the tumor microenvironment have been shown to dictate whether immunotherapy is effective (14). Because loss of SF-Ron significantly promoted robust T-cell infiltration and enhanced antitumor activity of T cells, targeting SF-Ron alone or in combination with immune checkpoint blockade to augment immunotherapy is an attractive approach. We previously found that a small-molecule kinase inhibitor of all Ron isoforms cooperates with anti-CTLA4, but not anti–PD-1, immunotherapy to abrogate breast cancer progression (28). The data presented here, which for the first time specifically implicate SF-Ron in the regulation of the antitumor T-cell response, provide a mechanistic explanation for how a Roni might boost tumor immunity. We therefore tested whether specific antitumor CD8+ T-cell responses and prevention of metastatic outgrowth could be achieved with a Ron kinase inhibitor. We established lung metastasis in WT mice via tail vein injection to mimic the clinical setting of already seeded breast cancer metastases and then, one week later, initiated treatment with either the Roni BMS-777607 (79) or vehicle control. After 4 weeks of treatment, mice were euthanized, and lung metastasis was quantified. Treatment of mice with BMS-777607 significantly reduced metastatic burden, to about half of that in control mice (Fig. 7D), and generated approximately 10-fold more IFNγ-producing T cells in response to stimulation with PyMT tumor–specific peptides (Fig. 7E). Tetramer staining with PyMT-specific MHC-I tetramers (76) also showed significantly more tumor-specific CD8+ T cells in mice treated with BMS-777607 (Fig. 7F). Together, these data show that Roni treatment can elicit tumor-specific CD8+ T-cell responses and results in reduced progression of already seeded metastatic lesions.
DISCUSSION
In this study, we provide the first evidence that deletion of host SF-Ron alone is sufficient to reverse the metastatic tumor microenvironment from a protumorigenic, immune-suppressive state to an immune-activated state in mammary tumor models that are classically recalcitrant to immunotherapy. Specifically, lack of SF-Ron redirected CD4+ T-cell differentiation and enhanced antitumor CD8+ T-cell activity. Enhanced immune activity in mice lacking SF-Ron culminated in robust infiltration of TILs into metastatic lesions, followed by clearance of lung metastases. Importantly, pharmaceutical inhibition of Ron kinase activity also manifested in enhanced tumor-specific immune responses and resulted in significantly less metastatic outgrowth. Our results provide a rationale for using Ron kinase inhibitors, at least two of which have completed phase I clinical trials, as a new form of immunotherapy to treat metastatic breast cancer.
We showed that loss of SF-Ron protects from metastatic outgrowth in two different mammary tumor types, representing the FVB genetic background. Our previous study showed that the combination of a Ron kinase inhibitor and anti-CTLA4 can shrink breast tumors, block metastatic outgrowth of breast cancer, and slow the growth of colorectal tumors in a host Ron-dependent manner (28), in two different models and two different mouse genetic backgrounds. Taken together, these data suggest that our findings can be extended to multiple cancer models and may have relevance for human cancer. Indeed, we showed in a phase I clinical trial in patients with cancer that a Ron kinase inhibitor blocked downstream effects of Ron in bone turnover (80). Although the latter study does not address the role of SF-Ron in immune responses to metastasis, it demonstrates that future studies to address this issue are feasible and can have an impact.
One particularly interesting finding was accumulation of less-differentiated naïve and stem cell–like CD4+ T-cell precursors that express Sell and Ccr7, but not traditional activation markers such as Pdcd1, in the lungs of Ron SF−/− mice with metastases. One possibility is that as metastases are eliminated in Ron SF−/− mice, the activated tumor-specific T cells may return to a resting phase; these T cells may be destined to become resident memory T cells, return to the lymph node, or patrol the periphery to prevent future relapse. It was recently reported that resident memory T cells (TRM) and stem cell–like memory T cells have superior antitumor immune responses compared with classic effector T cells, which are usually short-lived because of their terminal differentiated status (78). TRMs arise from a subset of antigen-experienced T cells and can be reactivated if the tumor relapses. Our data showing that Ron SF−/− mice are almost completely protected from tumor rechallenge supports the notion of a robust memory response when SF-Ron function is absent.
A similar T-cell population with self-renewal capacity and stem cell–like features, defined by the expression of Tcf7 (encoding TCF1), Lef1, and Ccr7 (65), was recently reported in tumors, albeit in the CD8+ subset. In kidney cancer, it was shown that this population of TCF1+ CD8+ TILs is present in regions dense with MHC-II+ antigen-presenting cells (APC), referred to as the APC niche, within the tumor. T cells in this APC niche continually differentiate to effector cells that express higher levels of immune checkpoint molecules and eventually become exhausted, which can be reversed by immune checkpoint inhibitor treatment (65). Likewise, patients with more APC niches in their tumors had better responses to immunotherapy (65, 81). Although the less-differentiated T cells identified in our study are CD4+ T cells that have not been previously described in tumors, we speculate that the TCF1+ CD4+ T cells present in the Ron SF−/− mice may have similar functions. In an influenza virus infection model, antigen-experienced TCF1+ CD4+ T cells were able to divide asymmetrically to give rise to a TCF1-negative Th1 effector cell population and maintain a TCF1+ CD4+ T-cell population for self-renewal (73). Moreover, in an experimental autoimmune encephalomyelitis (EAE) mouse model, a similar subset of TCF1+ cells was able to differentiate into Th17 cells that produced IL17 when the mTORC1 pathway was blocked (72). Our data show that CD4+ T cells are required to suppress metastasis in Ron SF−/− mice. Ron SF−/− mice have more Th1 cells and fewer Tregs compared with WT mice, which were skewed toward the Th2 subset. Importantly, although CD4+ T cells from metastatic lungs of both WT and Ron SF−/− mice were able to differentiate to Th17 cells in vitro, only CD4+ T cells from Ron SF−/− mice were capable of differentiation to Th1 cells that can enhance CD8+ cytotoxic T-cell activity.
Interestingly, a preclinical study in a melanoma model indicated that a subset of long-lived, less-differentiated Th17 cells that expressed TCF1, LEF1, and CCR7 are endowed with stem-like self-renewal potential and are able to repolarize to a Th1-like state and facilitate antitumor immune responses to eradicate tumors (82). The ability of less-differentiated Th17 cells to transdifferentiate to IFNγ-producing Th1-like cells was also shown in an EAE mouse model (72). Thus, the accumulation of naïve, stem cell–like CD4+ T cells in the lungs of Ron SF−/− mice with metastases, along with their ability to differentiate into Th17 cells but with a strong skew toward the Th1 phenotype, may contribute to elimination of metastasis. Our results raise the intriguing possibility that SF-Ron might act specifically to block the transdifferentiation of IL17-producing Th17 cells to antitumorigenic IFNγ-producing T-BET+ Th1-like effector cells that eradicate metastatic tumors. Indeed, CD4+ T cells from Ron SF−/− lungs were able to differentiate to a Th17 state but failed to accumulate as seen in the WT setting. These results warrant future studies to address the role of SF-Ron in mediating the differentiation and transdifferentiation potential of CD4+ T cells.
Our previous study investigating the combination of Ron kinase inhibition with immunotherapy (28) revealed that the Ron kinase inhibitor combined with anti–PD-1 immunotherapy provided no benefit, whereas the combination of Ron kinase inhibitor with anti-CTLA4 was beneficial in two different models and two different genetic backgrounds. However, it has been unclear whether or how SF-Ron contributes to this synergistic effect. The immune activation mechanisms of anti-CTLA4 and anti–PD-1 are distinct (83). CTLA4 blockade not only is important for early priming of T cells in the lymph node but also affects differentiation of CD4+ T cells. It has been shown that anti-CTLA4 monotherapy increased the frequency of CD4+ T cells and promoted them to differentiate to Th1-like ICOS1+ CD4+ T cells, whereas anti–PD-1 monotherapy only expanded the PD-1+ but not PD-1− CD8+ T cells. This indicates PD-1 blockade acts at later stages of T-cell activation, mainly in peripheral tissues, and has no effect on T-cell differentiation (84–86). The combination of both CTLA4 and PD-1 blockade on treating murine colon cancer increased Th1-like effectors and expanded both PD-1+ and PD-1 CD8+ T-cell effectors contributing to tumor elimination (87). Our new data reinforce the notion that inhibition of Ron, specifically SF-Ron, enriches naïve, stem-like CD4+ T cells that are then primed and kept from exhaustion when combined with anti-CTLA4. Adoptive transfer of Ron SF−/− T cells into WT hosts was sufficient to significantly reduce metastasis burden, further suggesting that T cells are the target of SF-Ron–associated immunosuppression. Interestingly, we found that SF-Ron, but not FL-Ron, is expressed in T cells, hinting that SF-Ron may dampen T-cell function by preventing differentiation of CD4+ T cells and therefore failing to recruit and revitalize CD8+ effectors. Inhibition of SF-Ron may maintain stem-like T-cell pools in the metastatic microenvironment with the potential to differentiate to tumoricidal effectors. However, pharmaceutical inhibition of Ron alone under the conditions we used did not provide complete protection from metastasis (Fig. 7; ref. 28), whereas mice genetically lacking SF-Ron nearly eliminated lung metastasis. Future work might entail how to achieve optimal inhibition of SF-Ron with the various Ron kinase inhibitors that are available.
Regulation of the expression of FL- and SF-Ron isoforms is not well studied. One study found that differences in the methylation of CpG islands in the main promoter region are responsible for the switch in the expression of FL- and SF-Ron isoforms (38). Importantly, many studies have identified nonredundant functions of FL- and SF-Ron in oncogenic processes and in regulating host immune responses to infection. In cancer cells, SF-Ron, but not FL-Ron, promotes epithelial–mesenchymal transition, invasive ability, anchorage-independent growth in vitro, tumor growth, and metastasis of human breast cancer cell lines (40, 41). Expression of SF-Ron activated the PI3K pathway and suppressed the MAPK pathway, whereas exogenous expression of FL-Ron activated the MAPK pathway in breast cancer cells (41, 88). Constitutively active SF-Ron, but not FL-Ron activated by MSP, was shown to induce resistance to MET inhibitor treatment in gastric cancer (89). Moreover, mice that specifically lack SF-Ron expression due to naturally occurring genetic polymorphisms are resistant to Friend virus–induced erythroleukemia (36) due to SF-Ron's ability to interact with viral protein gp55 in erythroid cells to induce an erythropoietin-independent signaling cascade (90). Moreover, mice genetically engineered to express only FL-Ron and lack SF-Ron, the same mouse strain used in our study, are more susceptible to concanavalin A–induced acute liver injury marked by enhanced IFNγ production (42). Thus, there is accumulating evidence in a variety of disease settings that FL-Ron and SF-Ron are functionally nonredundant and differentially orchestrate host immune responses. Although the role of host Ron in promoting breast cancer metastasis has been studied previously (27, 28), those experiments were carried out in mice that either express or lack both isoforms of Ron. Hence, the specific contribution of each of these isoforms for breast cancer progression was unclear and required further investigation. Our results reveal that SF-Ron is the major isoform regulating immune responses during breast cancer metastatic outgrowth: metastases are nearly eliminated from mice that lack SF-Ron, even though they still express functional FL-Ron.
The dual role of Ron and SF-Ron signaling in both tumors and host reinforces the potential of Ron kinase inhibitors for the treatment of metastatic breast cancer. Although mAbs can have fewer off-target effects, our data suggest that mAbs that target FL-Ron may not be effective for immunotherapy. Indeed, poor efficacy of anti-Ron mAbs such as narnatumab and others in clinical and preclinical studies (29, 33, 91) may be due to their inability to block SF-Ron. Our data warrant clinical investigations using small-molecule Ron kinase inhibitors that inhibit SF-Ron signaling, perhaps in combination with immunotherapy, for the prevention and treatment of metastatic breast cancer.
METHODS
Mice and Tumor Cells
All animal procedures were reviewed and approved by the University of Utah Institutional Animal Care and Use Committee. Ron TK−/− and Ron SF−/− mice were described previously (24, 42) and were backcrossed to the FVB genetic background. Six- to 8-week-old female WT, Ron TK−/−, and Ron SF−/− mice were used in all experiments unless otherwise specified. Spontaneous metastasis experiments were carried out in 3- to 5-week-old female WT, Ron TK−/−, and Ron SF−/− mice. Tumor cells were derived from spontaneous mammary tumors of transgenic MMTV-PyMT mice on the FVB background (44). Tumor cells were cultured short term (not as cell lines) in DMEM/F12 medium (Gibco, Invitrogen) supplemented with FBS (10%; Gibco, Invitrogen), insulin–transferrin–selenium–ethanolamine (1×; Gibco, Invitrogen), recombinant murine EGF (10 ng/mL; Invitrogen), hydrocortisone (1 μg/mL; Sigma), and penicillin–streptomycin–gentamycin (Gibco, Invitrogen; 1×) for a maximum of 2 days before injection into mice. In total, 500,000 MMTV-PyMT cells in 200 μL Hank's Balanced Salt Solution (HBSS) were injected via the lateral tail vein of mice for all experimental metastasis experiments. Lung metastases were allowed to develop for 3 to 4 weeks unless otherwise specified. Then, 100,000 MMTV-PyMT cells in 20 μL HBSS were injected orthotopically into the cleared mammary fat pads of mice for spontaneous metastasis experiments. For tumor rechallenge experiments, lung metastasis was first induced via tail vein injection of tumor cells as described above, followed by orthotopic implantation of the same tumor line as described above after 2 weeks. Mice were harvested 2 weeks after mammary tumor implantation for subsequent examination and analysis.
The basal-like KBP1 tumor line was kindly provided by Dr. Charles M. Perou of Lineberger Comprehensive Cancer Center at the University of North Carolina (55). In this model, freshly harvested cells from orthotopically grown mammary tumors were used for an experimental metastasis assay. Briefly, tumors were digested with collagenase for 45 minutes at 37°C and passed through the 40-μm cell strainer to obtain a single-cell suspension. Experimental metastasis assay was then carried out by intravenously injecting 150,000 KPB1 tumor cells resuspended in 200 μL 2% FBS containing HBSS via the lateral tail vein.
Macrophage Isolation, RT-PCR, and In Vitro MSP Stimulation
Peritoneal lavage fluid was collected by injecting 8 mL ice-cold DMEM+GlutaMAX (Gibco) medium supplemented with 10% heat-inactivated FBS into the peritoneal cavity of the WT, Ron TK−/−, and Ron SF−/− mice. The collected fluid was centrifuged for 5 minutes at 1,800 rpm, and the cell pellet was used for in vitro MSP stimulation, RT-PCR, and flow cytometry analysis. For FL- and SF-Ron mRNA expression analysis, total RNA was isolated using the RNeasy Plus Mini Kit (Qiagen), and 1 μg RNA was used to generate cDNA using the SuperScript IV VILO Master Mix Kit (Thermo Fisher Scientific) according to the manufacturer's protocol. PCR primers and conditions for SF-Ron expression analysis were described previously by Persons and colleagues (36). PCR primers for FL-Ron (which amplify the cDNA region between exons 9 and 13) are 5′–GATGGACAAAGTACAGTGGAGAG and 5′–GCAGCAGTGGGACACTTGTCC–3′, and SF-Ron–specific PCR primers are 5′ TCTGGCTGATCCTTCTGTCTG–3′ and 5′–GCAGCAGTGGGACACTTGTCC–3′. The PCR products were resolved using 2% agarose gel. For the determination of FL-Ron protein expression, flow cytometry was performed as described in the flow cytometry section. For FL-Ron function analysis, 500,000 peritoneal lavage cells per well were cultured for 18 hours in 12-well plates in 1 mL MSP-containing (100 ng/mL, recombinant human MSP; R&D Systems) or vehicle-containing DMEM supplemented with 10% FBS and penicillin/streptomycin (Gibco). At the endpoint, the wells were washed three times with 1× PBS (pH 7.4) and incubated for 30 minutes in 5 mmol/L EDTA in PBS (pH 7.4) at 37°C to dislodge and collect the cells. Flow cytometry staining was then performed as described in the flow cytometry section.
Tissue Processing
For quantification of metastatic lung tumor burden, mice were euthanized at their respective endpoints, and lungs were harvested and fixed for 48 hours in formalin-free zinc fixative (BD Pharmingen). Lung images were then captured and imported to ImageJ for quantification. The percentage of lung area occupied by the tumor was quantified by ImageJ software. For processing lungs for flow cytometry and scRNA-seq, lungs were perfused with 5 mL 1× PBS (pH 7.4). Lung lobes were then dissected, transferred to 5 mL Accumax cell/tissue dissociation solution (STEMCELL Technologies), and physically dissociated between frosted microscope slides. The suspension was then incubated at room temperature for 30 minutes with continuous agitation for enzymatic dissociation. After enzymatic dissociation, contaminating red blood cells were lysed using ammonium–chloride–potassium (ACK) lysis buffer and filtered through a 100-μm nylon mesh filter to obtain a single-cell suspension. For obtaining splenocytes for flow cytometry and in vitro differentiation assays, spleens harvested from mice were physically dissociated by pressing between frosted microscope slides. Contaminating red blood cells were then lysed using ACK buffer. The splenocytes were then filtered through a 100-μm nylon mesh filter to obtain a single-cell suspension. The single-cell suspension of lungs and spleens thus obtained was used for downstream processing accordingly (see below).
Flow Cytometry
Single-cell suspensions, as described above, were used to analyze several surface or intracellular markers, such as transcription factors (TF). For surface marker staining, cells were stained with fluorophore-conjugated antibodies on ice, in PBS supplemented with 2% FBS. For intracellular marker staining, cells were restimulated with phorbol myristate acetate (PMA; 50 ng/mL) and ionomycin (0.5 μg/mL) for 5 hours and stained with the BD Cytofix/Cytoperm Plus Kit according to the manufacturer's instructions. For TF staining, cells were stained with the eBioscience FoxP3/Transcription Factor staining buffer set according to the manufacturer's protocol. Prior to surface antibody staining in all surface/intracellular/TF staining protocols, cells were first stained with fixable viability dye in PBS to exclude dead cells and then incubated with anti-mouse CD16/32 Fc-blocking antibodies (Clone 93; BioLegend) to reduce nonspecific binding. For the list of the antibodies used, please refer to Supplementary Table S1. The stained samples were acquired using the BD LSRFortessa flow cytometer, and the analysis of different immune populations was carried out using FlowJo software. Briefly, for analysis, forward scatter versus side scatter was first used to exclude cellular debris and doublets. The single cells were then gated to exclude dead cells followed by gating on lineage-specific subpopulations. Frequencies of immune cell populations were compared using the Student t test or one-way ANOVA, where applicable.
Chromogenic IHC
The fixed lung tissues, as described above, were embedded in paraffin blocks. Four-micron–thick sections were obtained by manual rotary microtome (Leica Biosystems) and baked at 62°C for 1 hour. Deparaffinization and rehydration were carried out with CitriSolve solution and serial dilutions of ethanol followed by heat-inactivated epitope retrieval (HIER) with 10 mmol/L sodium citrate (pH 6.0). Tissue sections were incubated in 3% hydrogen peroxide diluted in methanol for 10 minutes to block endogenous peroxidase. Nonspecific blocking was performed by incubating the sections with PBS containing 5% BSA and 10% normal goat serum, together with mouse FcR blocking reagent (Miltenyi Biotec, GmbH). For antibodies raised from rat, the Mouse-On-Mouse blocking reagent (Vector Laboratories) was added into the blocking solution with the additional steps of incubating the sections with avidin/biotin blocking reagents (Vector Laboratories) to block endogenous avidin and biotin activity. After blocking, the sections were incubated with primary antibody overnight at 4°C, followed by corresponding secondary antibodies. EnVision+ goat anti-rabbit horseradish peroxidase (HRP) polymer reagent (Dako/Agilent) was used against primary antibodies raised in rabbit; biotinylated secondary antibodies were used for primary antibodies raised from rat, followed by the avidin–biotin HRP amplification system (Vector Laboratories). The chromogenic revelation was conducted with the 3,3′-diaminobenzidine system, and the nuclear counterstain was performed with Mayer's hematoxylin (Sigma-Aldrich). Stained tissue sections were then dehydrated with ethanol, infiltrated with CitriSolve solution, and mounted with Cytoseal Mounting medium (VWR). Image acquisition was performed with either an Olympus BX-50 microscope equipped with the Nikon camera or the Zeiss Axioscan Z1 slide scanner for whole-slide scan at 20× magnification. Quantification of TILs with IHC-stained samples was performed on five samples from each group, and at least 10 separate tumor fields of vision containing visible tumors were extracted from the whole section image. Computer-assisted quantification of CD3e, CD4, or CD8a signals from the extracted images was performed by setting a standard threshold for all images for the same markers by using ImageJ-based FIJI software.
Primary antibodies used for histology analysis in this study were all unconjugated and include CD3e (D4V8L; Cell Signaling Technology), CD4 (4SM95; eBioscience/Thermo Fisher), CD8a (4SM15; eBioscience/Thermo Fisher), B220 (CD45R; RA2–6B2; Santa Cruz Biotechnology), pan-cytokeratin (Agilent Technologies), TCF1 (C63-D9; Cell Signaling Technology), and PyMT (Novus Biologicals).
Multiplexed Immunofluorescence
After deparaffinization and rehydration of the slides, HIER was carried out with 10 mmol/L Tris-EDTA (pH 9.0) containing 0.05% Tween-20 followed by endogenous peroxidase blocking and nonspecific blocking as described above for IHC. Sections were then subjected to the consecutive staining procedure with primary antibodies and corresponding HRP-conjugated secondary antibodies followed by signal detection with the TSA-Opal detection system, including OPAL480, OPAL520, OPAL570, OPAL620, and OPAL690 (Akoya Bioscience). The primary antibody conditions used were the same as described for IHC. Antibody stripping with boiling 10 mmol/L sodium citrate (pH 6.0) was performed after signal detection of each primary antibody and before the application of the next primary antibody. Autofluorescence was removed by TrueBlack Lipofuscin Autofluorescence Quencher according to the manufacturer's protocol (Biotium). Following this, sections were stained with DAPI for nuclear counterstain and mounted in VECTASHIELD Vibrance Antifade mounting medium (Vector Laboratories). The staining panel in this study included a six-marker panel (B220, CD3e, CD8a, pan-CK, CD4, and DAPI) and a five-marker panel (CD8a, TCF1, CD4, CD8a, and DAPI), and the best sequence of staining with each antibody was established as addressed above. Upon completion of the staining process, the entire slides were scanned using the Vectra Polaris Multispectral Imaging system (Akoya Bioscience) and processed with either the Phenochart Whole Slide Viewer (Akoya Bioscience) or the QuPath open-source software for pseudo-coloring of each marker. The QuPath was also used for quantification of TILs in the tumor regions of the whole sections after defining the tumor regions by PyMT or pan-cytokeratin signal using the function of Object Classification by pixels, followed by Cell Detection with DAPI for cell number measurement and the Single Measurement Classifier function for defining each marker by thresholding. Once the parameters were set, machine-assisted automated quantification was performed for all slides.
scRNA-seq and Analysis
The graphic workflow of scRNA-seq is illustrated in Fig. 3A. Briefly, the lungs were harvested from mice and processed to obtain the single-cell suspension as described above. Lungs from at least eight mice were pooled together per group. Single-cell suspensions were stained with anti-CD45 (clone 30-F11; BD Biosciences) conjugated with phycoerythrin (PE) in 1:200 dilution for 30 minutes on ice and resuspended in PBS supplemented with 2% FBS. These samples were then incubated with DAPI (5 μg/mL) for 5 minutes before sorting with the BD FACSAria flow cytometer to obtain live CD45+ DAPI− immune cells. Sorted cells were washed three times with PBS and resuspended in 0.04% BSA containing PBS. Around 16,000 cells per sample were processed with droplet-based 3′-end scRNA-seq using the Chromium Single Cell 3′ Gene Expression Library Prep Kit V3 according to the manufacturer's protocol (10× Genomics), by which the sorted single cells were loaded into a Chromium Chip B along with partitioning oil, the reverse transcription reagents, and a collection of gel beads that contained 3,500,000 unique 10× barcodes. Only the droplets contain single cell and a gel bead with reverse transcription reagents will be subjected to libraries preparation. RNA libraries in paired-end format were then sequenced by the Illumina NovaSeq 6000 sequencer, and the sequence reads were preprocessed using the 10× Genomics CellRanger V3 pipeline and further analyzed with the Seurat R package V3. The output data from the CellRanger pipeline were processed to regress out mitochondrial genome representation and unique molecular identifier counts variance followed by integrating the two data sets, WT and Ron SF−/−, into a combined data set. The Seurat pipeline (92, 93) was then applied to the combined data set. Principal component analysis (PCA) and the t-distributed stochastic neighbor embedding dimensional reduction were performed using the first 30 PCA components to obtain a two-dimensional representation of the cell states. Cell clusters were identified using the Seurat's FindCluster function, which implements the shared nearest neighbor modularity optimization-based algorithm with a resolution of 0.4 to 0.7, leading to 16 to 24 clusters. A resolution of 0.4 was chosen for the analysis. The biological identities of cell clusters were annotated by using the web-based CIPR tool, which compares the cell cluster signatures with the publicly available Immunological Genome Project (ImmGen) database (59, 60). To examine T-cell subsets, clusters expressing Cd3e (Supplementary Fig. S4B) were extracted from the combined data set, and Seurat's pipeline with the resolution of 0.7 was reapplied on 30 PCA components for clustering. The mean expression of markers in each cluster was used for heat map representation.
CD4+, CD8+ T-cell Depletion and Drug Treatment
For CD4+ and CD8+ T-cell depletion, mice were injected with 100 μg anti-mouse CD4 (clone GK1.5; BioXCell) or anti-mouse CD8a (clones 53–6.7; Bio X Cell) or immunoglobulin G isotype control antibodies (BioXCell). Antibodies were injected intraperitoneally, once daily for 3 days, before tumor cell injection. Then, 500,000 MMTV-PyMT tumor cells were injected via the lateral tail vein of mice on the fourth day (day 0). Antibodies were then injected twice weekly until the endpoint. Mice were euthanized on day 28, and metastatic tumor burden was quantified as described earlier. For the treatment of the mice with Roni in the adjuvant setting, 500,000 MMTV-PyMT tumor cells were first injected via the tail vein on day 0 and were allowed to establish for 7 days. Starting on day 8, the mice were treated with either 50 mg/kg BMS-777607/ASLAN002 or 70% PEG-400 vehicle control, 5 days per week, until the endpoint. The mice were euthanized on day 32, and lungs and spleens were harvested for tumor burden quantification (as described above) and tumor-specific T-cell analysis (see below). For blocking lymphocyte egress, FTY-720 (Cayman Chemical) was dissolved in absolute ethanol to make a 100-mg/mL stock solution. The working solutions were then prepared in saline. Tumor cells were injected via tail vein on day 0, and beginning day 1, 25 μg FTY720 or saline vehicle control was injected intravenously for 3 days, followed by oral administration of 1 mg/kg daily until the endpoint.
In Vitro CD4+ T-cell Differentiation
Lungs and spleens were harvested 4 weeks after tumor cell injections via the tail vein and processed as described above. Tissues from at least five mice per group were pooled together. CD4+ T cells were sorted by magnetic cell separation using the EasySep Mouse CD4 Positive Selection Kit II according to the manufacturer's protocol (STEMCELL Technologies). Sorted CD4+ T cells were labeled with cell tracing violet dye (Invitrogen/Thermo Fisher) before adding to the subtype-specific differentiation medium. CD4+ T-cell in vitro differentiation protocol was described previously by Sekiya and Yoshimura (74). In brief, 200,000 CD4+ T cells per well were cultured for 96 hours in 24-well plates that were precoated with 2 μg/mL anti-CD3e and 0.5 μg/mL anti-CD28 in the presence of cytokines and antibodies described below, designed to facilitate differentiation of the sorted CD4+ T cells to four major subtypes. The differentiation media for specific subsets include Th1 (5 ng/mL mouse IL2, 10 ng/mL mouse IL12, and 1 μg/mL anti-IL4), Th2 (5 ng/mL mouse IL2, 10 ng/mL mouse IL4, and 1 μg/mL anti-IFNγ), Th17 (20 ng/mL mouse IL6, 1 ng/mL mouse TGFβ1, 1 μg/mL anti-IFNγ, 1 μg/mL anti-IL4, and 1 μg/mL anti-IL2), and Treg (2 ng/mL mouse TGFβ1, 1 μg/mL anti-IFNγ, and 1 μg/mL anti-IL4). At the endpoint, cells were restimulated with 50 ng/mL PMA and 0.5 μg/mL ionomycin for 5 hours. Cells were then harvested, stained, and analyzed as described above in the flow cytometry section.
Peptides
PyMT peptides (MPLTCLVNV, LPSLLSNPTY, YPRTPPELL) with previously established immunogenicity (76) were synthesized by Atlantic Peptides with at least 95% purity and used at a 10-μg/mL concentration for restimulation and for generating peptide-bound tetramers.
IFNγ ELISpot Assay
ELISpot assays were carried out on Multiscreen HTS IP 0.45-μm filter plates (Millipore Sigma). Briefly, the plates were coated with anti-IFNγ capture antibody (clone AN18; BioLegend) overnight at 4°C and blocked with RPMI1640 supplemented with 20% FBS for 2 hours at 37°C. Then, 500,000 splenocytes were then added, and the plates were incubated in the presence of 10 μg/mL pooled peptides or 25 ng/mL PMA and 0.5 μg/mL ionomycin in positive control wells or 0.1% DMSO in negative control wells. In addition, for freeze-thawed splenocytes (Fig. 7), 50 IU/mL mouse IL2 was added to the stimulation medium to enhance T-cell survival. After incubating the plate for 44 hours at 37°C, the wells were washed to remove the cells. The wells were then incubated with biotinylated anti-IFNγ detection antibody (clone R4–6A2; BioLegend) for 2 hours at 37°C followed by streptavidin-HRP for 1 hour at room temperature. IFNγ spots were developed using 3-amino-9-ethylcarbazole chromogen (Sigma), and images were captured by the ELISpot Reader (AID GmbH, Germany). Automatic spot counts were obtained using the AID ELISpot Reader software.
Tetramer Assay
The three PyMT peptides folded into MHC-I tetramers with the haplotype H-2Dq with human β2M and conjugated to R-PE were kindly provided by the NIH Tetramer Core Facility (Emory University). Whole splenocytes were obtained from tumor-bearing mice as described above, and flow cytometry staining was performed. Splenocytes were first stained with the fixable viability dye (Ghost Dye UV450; Tonbo Biosciences) for 30 minutes on ice to exclude dead cells. Cells were then stained with the tetramer pool containing all three tetramers (1:100 dilution each) for 30 minutes at room temperature. Mouse FcR blocking was performed by incubating with anti-CD16/CD32 for 10 minutes on ice followed by surface marker staining for 30 minutes on ice with a panel of antibodies including CD3e (APC, clone 145–2C11), CD4 (violetFluor450, clone GK1.5), CD8a (FITC, clone 53–6.7), CD62L (PE-Cy7, clone MEL-14), and CD44 (redFluor710, clone IM7). In addition, for compensation purposes in the tetramer-PE channel, anti-CD3e conjugated with PE was used. Data acquisition and analysis were performed as described above (see flow cytometry section).
Adoptive Transfer
For adoptive transfer, T cells isolated from spleens of 6- to 8-week-old naïve WT or Ron SF−/− donors were transferred into the age-matched WT recipients. Briefly, T cells were isolated from whole splenocytes of donor mice using the EasySep Mouse T cell Isolation kit (STEMCELL Technologies) following the manufacturer's recommendation. A total of 2 million T cells were then transferred to the recipient mice via the lateral tail vein, followed by the injection of 500,000 MMTV-PyMT tumor cells via the tail vein 48 hours later as described previously. Mice were then harvested 3 to 4 weeks after tumor injection; the lungs and spleens were collected for subsequent analysis of metastatic tumor burden and T-cell subset phenotyping.
Data Availability
scRNA-seq data from CD45+-sorted tumor-bearing lung samples are available on the NCBI Gene Expression Omnibus (GEO) database under the accession number GSE155011.
Authors' Disclosures
H. Gundlapalli is currently employed at Notable Labs, a precision oncology company. H. Ekiz reports grants from NIH during the conduct of the study. A.L. Welm reports grants from NCI, Susan G. Komen Foundation, Department of Defense, Aslan Pharmaceuticals, Zeno Pharmaceuticals, 3 Helix; grants and nonfinancial support from Aslan Pharmaceuticals; and nonfinancial support from NIH Tetramer Facility during the conduct of the study; other support from Thunder Biosciences, and other support from J. Michael Bishop Institute for Cancer Research outside the submitted work; in addition, A.L. Welm has a patent for U.S. Patent #9,907,791 issued and a patent for tangible property (not patented) licensed and with royalties paid from multiple. No disclosures were reported by the other authors.
Authors' Contributions
S.A. Lai: Conceptualization, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. H. Gundlapalli: Conceptualization, formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. H. Ekiz: Resources, software, formal analysis, investigation, methodology, writing–review and editing. A. Jiang: Investigation, methodology, writing–review and editing. E. Fernandez: Investigation, methodology, writing–review and editing. A.L. Welm: conceptualization, resources, supervision, funding acquisition, investigation, writing–original draft, project administration, Writing–review and editing.
Supplementary Material
Table S1 - List of antibodies and reagents
Table S2 - Genes in immune cell clusters
Supplementary Figures
Acknowledgments
This work was supported by the NIH/NCI (R01CA223245), the Department of Defense Breast Cancer Research Program (W81XWH-18–1-0616), the Susan G. Komen Foundation (SAC190078), 5 For the Fight, and the Huntsman Cancer Foundation. Shared resources such as High Throughput Genomics and Bioinformatics and Biostatistics Cores were supported by the HCI Cancer Center Support Grant (5P30CA042014). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCI or the NIH. We are grateful for use of the Flow Cytometry Facility (James Marvin) and the Cell Imaging Core Facility (Mike Bridge) at the University of Utah Health Sciences, both funded by the National Center for Research Resources of the NIH under Award Number 1S10RR026802–01. Research reported in this publication used the High-Throughput Genomics and Bioinformatic Analysis Shared Resource at Huntsman Cancer Institute at the University of Utah and was supported by the NCI of the NIH under award number P30CA042014. Microscopy equipment was obtained with a NCRR Shared Equipment Grant #1S10RR024761. We thank the NIH Tetramer Facility for the PE-labeled PyMT tetramers and the biotinylated PyMT monomers, Dr. Charles Perou for providing the KBP1 tumor line, Dr. Christa DeVette and Brock Johnson for technical advice and assistance, Dr. Yoko DeRose for her assistance with the Vectra Polaris Multispectral Scanning System, and Drs. Matthew Williams and Ryan O'Connell for helpful discussions and critical reading of the manuscript. We appreciate helpful editing suggestions by Life Science Editors.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
References
- 1. American Cancer Society. Cancer facts & figures 2020. Atlanta, GA: American Cancer Society; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen W, Hoffmann AD, Liu H, Liu X. Organotropism: new insights into molecular mechanisms of breast cancer metastasis. NPJ Precis Oncol 2018;2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weigelt B, Peterse JL, van 't Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer 2005;5:591–602. [DOI] [PubMed] [Google Scholar]
- 4. Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 2011;331:1565–70. [DOI] [PubMed] [Google Scholar]
- 5. Guerra N, Tan YX, Joncker NT, Choy A, Gallardo F, Xiong Net al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity 2008;28:571–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garner H, de Visser KE. Immune crosstalk in cancer progression and metastatic spread: a complex conversation. Nat Rev Immunol 2020;20:483–97. [DOI] [PubMed] [Google Scholar]
- 7. Dvorak HF. Tumors: wounds that do not heal-redux. Cancer Immunol Res 2015;3:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol 2014;16:717–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298–306. [DOI] [PubMed] [Google Scholar]
- 10. Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis Ret al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan Set al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJet al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Turajlic S, Litchfield K, Xu H, Rosenthal R, McGranahan N, Reading JLet al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol 2017;18:1009–21. [DOI] [PubMed] [Google Scholar]
- 14. Gatti-Mays ME, Balko JM, Gameiro SR, Bear HD, Prabhakaran S, Fukui Jet al. If we build it they will come: targeting the immune response to breast cancer. NPJ Breast Cancer 2019;5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adams S, Gatti-Mays ME, Kalinsky K, Korde LA, Sharon E, Amiri-Kordestani Let al. Current landscape of immunotherapy in breast cancer: a review. JAMA Oncol 2019;5:1205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmid P, Rugo HS, Adams S, Schneeweiss A, Barrios CH, Iwata Het al. Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2020;21:44–59. [DOI] [PubMed] [Google Scholar]
- 17. Kretschmann KL, Eyob H, Buys SS, Welm AL. The macrophage stimulating protein/Ron pathway as a potential therapeutic target to impede multiple mechanisms involved in breast cancer progression. Curr Drug Targets 2010;11:1157–68. [DOI] [PubMed] [Google Scholar]
- 18. Welm AL, Sneddon JB, Taylor C, Nuyten DS, van de Vijver MJ, Hasegawa BHet al. The macrophage-stimulating protein pathway promotes metastasis in a mouse model for breast cancer and predicts poor prognosis in humans. Proc Natl Acad Sci U S A 2007;104:7570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaudino G, Follenzi A, Naldini L, Collesi C, Santoro M, Gallo KAet al. RON is a heterodimeric tyrosine kinase receptor activated by the HGF homologue MSP. EMBO J 1994;13:3524–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iwama A, Wang MH, Yamaguchi N, Ohno N, Okano K, Sudo Tet al. Terminal differentiation of murine resident peritoneal macrophages is characterized by expression of the STK protein tyrosine kinase, a receptor for macrophage-stimulating protein. Blood 1995;86:3394–403. [PubMed] [Google Scholar]
- 21. Faham N, Welm AL. RON signaling is a key mediator of tumor progression in many human cancers. Cold Spring Harb Symp Quant Biol 2016;81:177–88. [DOI] [PubMed] [Google Scholar]
- 22. Sharda DR, Yu S, Ray M, Squadrito ML, De Palma M, Wynn TAet al. Regulation of macrophage arginase expression and tumor growth by the Ron receptor tyrosine kinase. J Immunol 2011;187:2181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chaudhuri A. Regulation of macrophage polarization by RON receptor tyrosine kinase signaling. Front Immunol 2014;5:546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Waltz SE, Eaton L, Toney-Earley K, Hess KA, Peace BE, Ihlendorf JRet al. Ron-mediated cytoplasmic signaling is dispensable for viability but is required to limit inflammatory responses. J Clin Invest 2001;108:567–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lutz MA, Gervais F, Bernstein A, Hattel AL, Correll PH. STK receptor tyrosine kinase regulates susceptibility to infection with Listeria monocytogenes. Infect Immun 2002;70:416–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lentsch AB, Pathrose P, Kader S, Kuboki S, Collins MH, Waltz SE. The Ron receptor tyrosine kinase regulates acute lung injury and suppresses nuclear factor kappaB activation. Shock 2007;27:274–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eyob H, Ekiz HA, Derose YS, Waltz SE, Williams MA, Welm AL. Inhibition of Ron kinase blocks conversion of micrometastases to overt metastases by boosting antitumor immunity. Cancer Discov 2013;3:751–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ekiz HA, Lai SA, Gundlapalli H, Haroun F, Williams MA, Welm AL. Inhibition of RON kinase potentiates anti-CTLA-4 immunotherapy to shrink breast tumors and prevent metastatic outgrowth. Oncoimmunology 2018;7:e1480286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yao HP, Suthe SR, Tong XM, Wang MH. Targeting RON receptor tyrosine kinase for treatment of advanced solid cancers: antibody-drug conjugates as lead drug candidates for clinical trials. Ther Adv Med Oncol 2020;12:1758835920920069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parikh PK, Ghate MD. Recent advances in the discovery of small molecule c-Met Kinase inhibitors. Eur J Med Chem 2018;143:1103–38. [DOI] [PubMed] [Google Scholar]
- 31. Koh XY, Koh XH, Hwang LA, Ferrer FJ, Rahmat SAB, Lama Det al. Therapeutic anti-cancer activity of antibodies targeting sulfhydryl bond constrained epitopes on unglycosylated RON receptor tyrosine kinase. Oncogene 2019;38:7342–56. [DOI] [PubMed] [Google Scholar]
- 32. Yao HP, Suthe SR, Hudson R, Wang MH. Antibody-drug conjugates targeting RON receptor tyrosine kinase as a novel strategy for treatment of triple-negative breast cancer. Drug Discov Today 2020;25:1160–73. [DOI] [PubMed] [Google Scholar]
- 33. LoRusso PM, Gounder M, Jalal SI, Andre V, Kambhampati SRP, Loizos Net al. Phase 1 study of narnatumab, an anti-RON receptor monoclonal antibody, in patients with advanced solid tumors. Invest New Drugs 2017;35:442–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roohullah A, Cooper A, Lomax AJ, Aung J, Barge A, Chow Let al. A phase I trial to determine safety and pharmacokinetics of ASLAN002, an oral MET superfamily kinase inhibitor, in patients with advanced or metastatic solid cancers. Invest New Drugs 2018;36:886–94. [DOI] [PubMed] [Google Scholar]
- 35. He AR, Cohen RB, Denlinger CS, Sama A, Birnbaum A, Hwang Jet al. First-in-human phase I study of merestinib, an oral multikinase inhibitor, in patients with advanced cancer. Oncologist 2019;24:e930–e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Persons DA, Paulson RF, Loyd MR, Herley MT, Bodner SM, Bernstein Aet al. Fv2 encodes a truncated form of the Stk receptor tyrosine kinase. Nat Genet 1999;23:159–65. [DOI] [PubMed] [Google Scholar]
- 37. Iwama A, Okano K, Sudo T, Matsuda Y, Suda T. Molecular cloning of a novel receptor tyrosine kinase gene, STK, derived from enriched hematopoietic stem cells. Blood 1994;83:3160–9. [PubMed] [Google Scholar]
- 38. Angeloni D, Danilkovitch-Miagkova A, Ivanova T, Braga E, Zabarovsky E, Lerman MI. Hypermethylation of Ron proximal promoter associates with lack of full-length Ron and transcription of oncogenic short-Ron from an internal promoter. Oncogene 2007;26:4499–512. [DOI] [PubMed] [Google Scholar]
- 39. Yao HP, Zhou YQ, Zhang R, Wang MH. MSP-RON signalling in cancer: pathogenesis and therapeutic potential. Nat Rev Cancer 2013;13:466–81. [DOI] [PubMed] [Google Scholar]
- 40. Bardella C, Costa B, Maggiora P, Patane S, Olivero M, Ranzani GNet al. Truncated RON tyrosine kinase drives tumor cell progression and abrogates cell-cell adhesion through E-cadherin transcriptional repression. Cancer Res 2004;64:5154–61. [DOI] [PubMed] [Google Scholar]
- 41. Liu X, Zhao L, Derose YS, Lin YC, Bieniasz M, Eyob Het al. Short-form Ron promotes spontaneous breast cancer metastasis through interaction with phosphoinositide 3-Kinase. Genes Cancer 2011;2:753–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wetzel CC, Leonis MA, Dent A, Olson MA, Longmeier AM, Ney PAet al. Short-form Ron receptor is required for normal IFN-gamma production in concanavalin A-induced acute liver injury. Am J Physiol Gastrointest Liver Physiol 2007;292:G253–61. [DOI] [PubMed] [Google Scholar]
- 43. Nikolaidis NM, Kulkarni RM, Gray JK, Collins MH, Waltz SE. Ron receptor deficient alveolar myeloid cells exacerbate LPS-induced acute lung injury in the murine lung. Innate Immun 2011;17:499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol 1992;12:954–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Waltz SE, Toms CL, McDowell SA, Clay LA, Muraoka RS, Air ELet al. Characterization of the mouse Ron/Stk receptor tyrosine kinase gene. Oncogene 1998;16:27–42. [DOI] [PubMed] [Google Scholar]
- 46. Chen YQ, Zhou YQ, Wang MH. Activation of the RON receptor tyrosine kinase protects murine macrophages from apoptotic death induced by bacterial lipopolysaccharide. J Leukoc Biol 2002;71:359–66. [PubMed] [Google Scholar]
- 47. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yan HH, Pickup M, Pang Y, Gorska AE, Li Z, Chytil Aet al. Gr-1+CD11b+ myeloid cells tip the balance of immune protection to tumor promotion in the premetastatic lung. Cancer Res 2010;70:6139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sasada T, Kimura M, Yoshida Y, Kanai M, Takabayashi A. CD4+CD25+ regulatory T cells in patients with gastrointestinal malignancies: possible involvement of regulatory T cells in disease progression. Cancer 2003;98:1089–99. [DOI] [PubMed] [Google Scholar]
- 50. Barth RJ Jr, Mule JJ, Spiess PJ, Rosenberg SA. Interferon gamma and tumor necrosis factor have a role in tumor regressions mediated by murine CD8+ tumor-infiltrating lymphocytes. J Exp Med 1991;173:647–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nelson N, Lopez-Pelaez M, Palazon A, Poon E, De La Roche M, Barry Set al. A cell-engineered system to assess tumor cell sensitivity to CD8(+) T cell-mediated cytotoxicity. Oncoimmunology 2019;8:1599635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Matsuzaki J, Tsuji T, Luescher IF, Shiku H, Mineno J, Okamoto Set al. Direct tumor recognition by a human CD4(+) T-cell subset potently mediates tumor growth inhibition and orchestrates anti-tumor immune responses. Sci Rep 2015;5:14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Quezada SA, Simpson TR, Peggs KS, Merghoub T, Vider J, Fan Xet al. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J Exp Med 2010;207:637–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Yang THOet al. The immune landscape of cancer. Immunity 2018;48:812–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hollern DP, Contreras CM, Dance-Barnes S, Silva GO, Pfefferle AD, Xiong Jet al. A mouse model featuring tissue-specific deletion of p53 and Brca1 gives rise to mammary tumors with genomic and transcriptomic similarities to human basal-like breast cancer. Breast Cancer Res Treat 2019;174:143–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad Met al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 2018;24:541–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky Iet al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 2014;343:776–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JHet al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol 2014;32:1053–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ekiz HA, Conley CJ, Stephens WZ, O'Connell RM. CIPR: a web-based R/shiny app and R package to annotate cell clusters in single cell RNA sequencing experiments. BMC Bioinformatics 2020;21:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ekiz HA, Huffaker TB, Grossmann AH, Stephens WZ, Williams MA, Round JLet al. MicroRNA-155 coordinates the immunological landscape within murine melanoma and correlates with immunity in human cancers. JCI Insight 2019;4:e126543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ural BB, Yeung ST, Damani-Yokota P, Devlin JC, de Vries M, Vera-Licona Pet al. Identification of a nerve-associated, lung-resident interstitial macrophage subset with distinct localization and immunoregulatory properties. Sci Immunol 2020;5:eaax8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gattinoni L, Speiser DE, Lichterfeld M, Bonini C. T memory stem cells in health and disease. Nat Med 2017;23:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MFet al. A human memory T cell subset with stem cell-like properties. Nat Med 2011;17:1290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gossel G, Hogan T, Cownden D, Seddon B, Yates AJ. Memory CD4 T cell subsets are kinetically heterogeneous and replenished from naive T cells at high levels. eLife 2017;6:e23013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jansen CS, Prokhnevska N, Master VA, Sanda MG, Carlisle JW, Bilen MAet al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 2019;576:465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999;401:708–12. [DOI] [PubMed] [Google Scholar]
- 67. Mandala S, Hajdu R, Bergstrom J, Quackenbush E, Xie J, Milligan Jet al. Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science 2002;296:346–9. [DOI] [PubMed] [Google Scholar]
- 68. Kim HJ, Cantor H. CD4 T-cell subsets and tumor immunity: the helpful and the not-so-helpful. Cancer Immunol Res 2014;2:91–8. [DOI] [PubMed] [Google Scholar]
- 69. Ruterbusch M, Pruner KB, Shehata L, Pepper M. In vivo CD4(+) T cell differentiation and function: revisiting the Th1/Th2 paradigm. Annu Rev Immunol 2020;38:705–25. [DOI] [PubMed] [Google Scholar]
- 70. Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood 2008;112:1557–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wu T, Shin HM, Moseman EA, Ji Y, Huang B, Harly Cet al. TCF1 is required for the T follicular helper cell response to viral infection. Cell Rep 2015;12:2099–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Karmaus PWF, Chen X, Lim SA, Herrada AA, Nguyen TM, Xu Bet al. Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity. Nature 2019;565:101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nish SA, Zens KD, Kratchmarov R, Lin WW, Adams WC, Chen YHet al. CD4+ T cell effector commitment coupled to self-renewal by asymmetric cell divisions. J Exp Med 2017;214:39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sekiya T, Yoshimura A. In vitro Th differentiation protocol. Methods Mol Biol 2016;1344:183–91. [DOI] [PubMed] [Google Scholar]
- 75. Apetoh L, Smyth MJ, Drake CG, Abastado JP, Apte RN, Ayyoub Met al. Consensus nomenclature for CD8(+) T cell phenotypes in cancer. Oncoimmunology 2015;4:e998538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. DeVette CI, Gundlapalli H, Lai SA, McMurtrey CP, Hoover AR, Gurung HRet al. A pipeline for identification and validation of tumor-specific antigens in a mouse model of metastatic breast cancer. Oncoimmunology 2020;9:1685300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. DeVette CI, Andreatta M, Bardet W, Cate SJ, Jurtz VI, Jackson KWet al. NetH2pan: a computational tool to guide MHC peptide prediction on murine tumors. Cancer Immunol Res 2018;6:636–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jameson SC, Masopust D. Understanding subset diversity in T cell memory. Immunity 2018;48:214–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schroeder GM, An Y, Cai ZW, Chen XT, Clark C, Cornelius LAet al. Discovery of N-(4-(2-amino-3-chloropyridin-4-yloxy)-3-fluorophenyl)-4-ethoxy-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide (BMS-777607), a selective and orally efficacious inhibitor of the Met kinase superfamily. J Med Chem 2009;52:1251–4. [DOI] [PubMed] [Google Scholar]
- 80. Andrade K, Fornetti J, Zhao L, Miller SC, Randall RL, Anderson Net al. RON kinase: a target for treatment of cancer-induced bone destruction and osteoporosis. Sci Transl Med 2017;9:eaai9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Siddiqui I, Schaeuble K, Chennupati V, Marraco SAF, Calderon-Copete S, Ferreira DPet al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 2019;50:195–211. [DOI] [PubMed] [Google Scholar]
- 82. Muranski P, Borman ZA, Kerkar SP, Klebanoff CA, Ji Y, Sanchez-Perez Let al. Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity 2011;35:972–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sharma P, Allison JP. Dissecting the mechanisms of immune checkpoint therapy. Nat Rev Immunol 2020;20:75–6. [DOI] [PubMed] [Google Scholar]
- 84. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wei SC, Levine JH, Cogdill AP, Zhao Y, Anang NAS, Andrews MCet al. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell 2017;170:1120–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wei SC, Sharma R, Anang NAS, Levine JH, Zhao Y, Mancuso JJet al. Negative co-stimulation constrains T cell differentiation by imposing boundaries on possible cell states. Immunity 2019;50:1084–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wei SC, Anang NAS, Sharma R, Andrews MC, Reuben A, Levine JHet al. Combination anti-CTLA-4 plus anti-PD-1 checkpoint blockade utilizes cellular mechanisms partially distinct from monotherapies. Proc Natl Acad Sci U S A 2019;116:22699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Moxley KM, Wang L, Welm AL, Bieniasz M. Short-form Ron is a novel determinant of ovarian cancer initiation and progression. Genes Cancer 2016;7:169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wu Z, Zhang Z, Ge X, Lin Y, Dai C, Chang Jet al. Identification of short-form RON as a novel intrinsic resistance mechanism for anti-MET therapy in MET-positive gastric cancer. Oncotarget 2015;6:40519–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nishigaki K, Thompson D, Hanson C, Yugawa T, Ruscetti S. The envelope glycoprotein of friend spleen focus-forming virus covalently interacts with and constitutively activates a truncated form of the receptor tyrosine kinase Stk. J Virol 2001;75:7893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yao HP, Zhou YQ, Ma Q, Guin S, Padhye SS, Zhang RWet al. The monoclonal antibody Zt/f2 targeting RON receptor tyrosine kinase as potential therapeutics against tumor growth-mediated by colon cancer cells. Mol Cancer 2011;10:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM IIIet al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 - List of antibodies and reagents
Table S2 - Genes in immune cell clusters
Supplementary Figures
Data Availability Statement
scRNA-seq data from CD45+-sorted tumor-bearing lung samples are available on the NCBI Gene Expression Omnibus (GEO) database under the accession number GSE155011.