Graphical abstract

Keywords: SARS-CoV-2, COVID-19, Nucleocapsid protein, Monoclonal antibody, Antigen detection, Chemiluminescence enzyme immunoassay
Abstract
Background
Recently, the Coronavirus Disease 2019 (COVID-19) caused by SARS-CoV-2 infection has spread rapidly around the world, becoming a new global pandemic disease. Nucleic acid detection is the primary method for clinical diagnosis of SARS-CoV-2 infection, with the addition of antibody and antigen detection. Nucleocapsid protein (NP) is a kind of conservative structural protein with abundant expression during SARS-CoV-2 infection, which makes it an ideal target for immunoassay.
Methods
The coding sequence for SARS-CoV-2-NP was obtained by chemical synthesis, and then inserted into pET28a(+). The soluble recombinant NP (rNP) with an estimated molecular weight of 49.4 kDa was expressed in E. coli cells after IPTG induction. Six-week-old BALB/c mice were immunized with rNP, and then their spleen cells were fused with SP2/0 cells, to develop hybridoma cell lines that stably secreted monoclonal antibodies (mAbs) against NP. The mAbs were preliminarily evaluated by enzyme-linked immunosorbent assay (ELISA), and then used to develop a magnetic particle-based chemiluminescence enzyme immunoassay (CLEIA) for measurement of SARS-CoV-2-NP.
Results
mAb 15B1 and mAb 18G10 were selected as capture and detection antibody respectively to develop CLEIA, due to the highest sensitivity for rNP detection. The proposed CLEIA presented a good linearity for rNP detection at a working range from 0.1 to 160 μg/L, with a precision coefficient of variance below 10 %.
Conclusion
The newly developed mAbs and CLEIA can serve as potential diagnostic tools for clinical measurement of SARS-CoV-2-NP.
1. Introduction
Although most common cold infections caused by coronaviruses don’t have significant clinical sequelae, several recently identified human coronaviruses have posed serious threats to public health, including severe acute respiratory syndrome-CoV (SARS-CoV), Middle East respiratory syndrome-CoV (MERS-CoV) and SARS-CoV-2. SARS-CoV-2 was the cause of the current COVID-19 pandemic, with a major clinical feature of pneumonitis (Li et al., 2020; Zhu et al., 2020). According to the collected data from World Health Organization, more than 326 million cases of COVID-19 have been confirmed and over 5.5 million people died from the disease until January 2022.
SARS-CoV-2 was mainly transmitted by droplet, mucosa contact and aerosol transmission routes (Huang et al., 2020; Chen et al., 2020), but studies found the virus could also be detected in non-respiratory specimens from COVID-19 patients, such as feces and urine (Wang et al., 2020). Its genome is closely related to a bat coronavirus (Zhou et al., 2020). Although it shares a close evolutionary relationship with SARS-CoV (Lu et al., 2020), the receptor-binding domain of SARS-CoV-2 presented a stronger binding affinity with human ACE2 receptor (Wrapp et al., 2020), which might account for its characteristics of being highly contagious.
Similar to other beta-coronavirus, the virion of SARS-CoV-2 possesses a nucleocapsid composed of genomic RNA and phosphorylated nucleocapsid protein. The genome size of virus is about 29.9 kb and encodes four major structural proteins, including the spike (S), membrane (M), envelope (E) and nucleocapsid (N) proteins (Jin et al., 2020). The clinical diagnosis for SARS-CoV-2 infection is primarily based on detection of viral nucleic acid by the method of real-time RT-PCR, and serological antibody tests (Wölfel et al., 2020).
Several studies found that about 2 weeks after the virus infection, the conversion of serum specific antibody against N and S protein was observed in most people (Guo et al., 2020; Xiang et al., 2020). While in the first week of onset of the disease with high viral loads, the antigen detection in clinical samples presents higher sensitivity than serum antibody detection, which would be beneficial for the rapid diagnosis of SARS-CoV-2 infection (Paules et al., 2020; Li and Li, 2021). N protein (NP) only has a molecular weight of about 40 kDa and is abundantly expressed during viral infection, which makes it an ideal target for antigen detection. In a preliminary clinical study, with Ct value 40 as the cutoff of nucleic acid test, the sensitivity, specificity and percentage agreement of a fluorescence immunochromatographic (FIC) assay for NP detection was about 75 %, 100 % and 80 % respectively (Diao et al., 2021). The current immunoassays for NP had a high positive predictive value for SARS-CoV-2 infection. However, compared with the nucleic acid tests, the rapid antigen detection tests still lacks sensitivity, which would lead to an increased risk of false-negative results (Vandenberg et al., 2021; Scohy et al., 2020). The present study aimed to develop a magnetic particle-based chemiluminescence enzyme immunoassay (CLEIA) with high sensitivity and specificity for the measurement of SARS-CoV-2-NP.
2. Materials and methods
2.1. Materials
The expression plasmid (pET-28a), Escherichia coli cells (BL21) and myeloma cell line (SP2/0) were preserved in our lab. RPMI 1640 medium, penicillin- streptomycin was purchased from Gibco. Fetal bovine serum (FBS) was purchased from ExCell. Hypoxanthine and Thymidine medium (HT), hypoxanthine- aminopterin-thymidine medium (HAT), polyethylene glycol (PEG), complete and incomplete Freund’s adjuvant were purchased from Sigma-Aldrich. Ni-NTA Sefinose Resin and Protein A/G Prepacked chromatographic column, ammonium sulfate, BCA protein assay kit and TMB Chromogenic Reagent kit were purchased from Sangon Biotech. Horseradish peroxidases (HRP), alkaline phosphatase (AP), Dynabeads™ M-280, EDC and Sulfo-NHS were purchased from Thermo fisher. All chemicals used were of molecular biology grade or higher.
2.2. Animals and ethics approval
Six to eight-week-old BALB/c mice were purchased from Hunan SJA laboratory animal company. The manipulation of animals was approved by Ethics Committee Board of Hunan Normal University, and performed according to the guidelines in accordance with National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978).
2.3. Expression and purification of recombinant NP
Based on the gene information published in GenBank (Gene ID: 43740575), the full-length coding sequence for SARS-CoV-2 nucleocapsid protein was chemically synthesized with endonuclease digestion sites of EcoR I and Xho I at two terminals. The recombinant plasmid pET28a(+)-SARS-CoV-2-NP was transformed into Escherichia coli strain BL21, and recombinant his-tag fusion NP was expressed after induction with 1 mmol/L isopropyl-β-d-thiogalactoside (IPTG). The cell pellet was harvested and then lysed by sonication. The saturated ammonium sulfate solution (pH 7.4) was slowly added into the supernatant of bacterial lysate, to get a concentration of 30 %. Stood at 4℃ for more than 2 h, and then centrifuged at 12,000 g for 10 min. The protein precipitates was collected and the leftover supernatant was further treated with saturated ammonium sulfate solution at the concentration of 40 % and 50 % in succession. The collected protein samples were respectively dissolved in sterile Binding/Wash buffer (0.05 mol/L NaH2PO4, 0.3 mol/L NaCl, pH 8.0), and identified by 12 % SDS-PAGE.
The protein sample which contained most of recombinant NP was further purified by affinity chromatography. The protein sample was slowly added onto the prepared Ni-NTA resin column, and the outflow was collected. Then the resin was washed with two resin-bed volumes of Binding/Wash buffer, and the outflow was collected. His-tagged proteins were gradient eluted from the resin with two resin-bed volumes of Binding/Wash buffer containing imidazole with the concentration varied from 10 to 200 mmol/L. The outflow and gradient eluent solutions were identified by 12 % SDS-PAGE. The elute solutions which contained most of recombinant NP were collected and dialyzed with 10 times volume of phosphate buffer (pH 7.45) at 4 °C for 12 h. The purified recombinant NP was stored at -80℃ and its concentration was determined by BCA kit.
2.4. Preparation and purification of mAbs
The recombinant NP (rNP) was used as antigen to immunize six-week-old BALB/c mice, with the dose of 100 μg per mice and an interval of 2 weeks. The titer of serum antibody to rNP in immunized mice was determined by indirect enzyme-linked immunosorbent assay (ELISA), and the mice were undertaken 4 rounds of subcutaneous immunization and a final immunization via spleen. 3 days after the final immunization, the spleen cells of immunized mice were collected and fused with myeloma cell strain SP2/0 using PEG. The cells were cultured in RPMI-1640 medium supplemented with HAT and 20 % FBS for a week, and then cultured in RPMI-1640 medium supplemented with HT and 10 % FBS. The hybridoma cell colonies which secrete antibodies against NP were identified by indirect ELISA. After four rounds of subcloning by limiting dilution, several hybridoma cell lines that stably secreted anti-NP mAbs were obtained and stored in liquid nitrogen.
Eight-week-old BALB/c mice mouse was primed by intraperitoneally injection with 0.5 mL of mineral oil a week before, and then inoculated with 1 × 106 hybridoma cells to generate mAbs. 8–10 days later, ascites fluid was collected by abdominal paracentesis, and then immediately centrifuged at 3000 g for 10 min to collect the supernatant containing antibodies. The antibody solution was purified through the treatment of 4% caprylic acid precipitation, 50 % saturated ammonium sulfate precipitation and Protein A/G affinity chromatography in succession. The antibody solution was further dialyzed with phosphate buffer (pH 7.45) at 4 °C for 12 h. The purified antibody solution was stored at −20 °C and its concentration was determined by BCA kit.
2.5. Western blot
The sample of rNP was separated by sodium dodecyl sulfate poly-acrylamide gel electrophoresis and electrophoretically transferred to polyvinylidene fluoride membrane. After blocking, the membranes were incubated with the newly developed mAb against NP at a dilution of 1:1000. After washing, the membranes were further incubated with HRP-conjugated second antibody at a dilution of 1:5000 (goat anti-mouse IgG, abs20001, Absin Bioscience Co. Ltd., China). The immune complexes were detected with ECL reagent (130231, Monad Biotech. Co. Ltd., China), and the chemiluminescence signal was imaged by Tanon 5500 system (Tanon Co. Ltd., Shanghai, China).
2.6. Indirect enzyme-linked immunosorbent assay
Each well of 96-well microplate was coated with 100 μL of 1 μg/mL purified rNP at 4 °C overnight, and then blocked with 200 μL of TBST buffer (0.2 mol/L Tris−HCl, 0.9 % NaCl, 0.05 % Tween-20, pH 7.4) containing 0.25 % casein at 37 °C for 2 h. The prepared microplate was incubated with 100 μL of sample per well at 37 °C for 60 min, and then washed 3 times with TBST by an automatic plate washer. The plate was further incubated with 100 μL of diluted HRP-conjugated anti-mouse IgG antibody solution per well at 37℃ for 30 min, and washed 3 times with TBST. The enzymatic chromogenic reaction was performed with TMB Reagent kit according to the manufacturer’s instructions. The absorbance was recorded at 450 nm/630 nm by a microplate reader (Biotek Elx800).
2.7. Double antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA)
Each well of 96-well microplate was coated with 100 μL of 10 μg/mL capture mAb at 4 °C overnight, and then blocked with TBST buffer containing 0.25 % casein at 37 °C for 2 h. The prepared plates were store at 4 °C until needed. 5 mg of detection mAb was conjugated to activated horseradish peroxidase (HRP), and the antibody precipitate was dissolved in 1 mL of 50 % glycerol and stored at −20 °C.
The capture antibody-coated microplate was incubated with 100 μL of sample per well at 37 °C for 60 min, and then washed 3 times with TBST buffer. The plate was further incubated with 100 μL of diluted HRP-conjugated detection antibody at 37℃ for 30 min, and then washed 3 times with TBST buffer. The enzymatic chromogenic reaction was performed with substrate TMB, and the absorbance was recorded at 450 nm/630 nm as mentioned in the part 2.6.
2.8. Chemiluminescence enzyme immunoassay (CLEIA)
The capture mAb was coupled with magnetic particles, usually 0.1 mg antibody per 10 mg Dynabeads. The prepared beads-conjugated mAb was suspended with a desired buffer and stored at 2−8 °C until needed. 0.1 mg of detection mAb was conjugated with alkaline phosphatase (AP), and the antibody precipitate was dissolved in 0.25 mL of 50 % glycerol and stored at −20 °C. The purified rNP was used as calibration substance and its concentration was determined by BCA protein assay kit. The measurement of calibrators and samples was performed on RangeCL-1200i chemiluminescence immunoanalyzer (Yuan Jing Biotechnology Co. Ltd., Changsha, Hunan, China).
20 μL of sample, 30 μL of magnetic particle-conjugated antibody (0.1 or 0.2 mg/mL) and 50 μL of AP-labeled antibody (diluted as 1:1000 or higher) were added into reaction tube. The tube was incubated at 37 °C for 5 min, and then washed 3 times. 200 μL of chemiluminescence substrate solution (LOT 20180802, Yuan Jing Biotech.) was added into the tube, and the total relative luminous unit (RLU) was recorded in 10 s. Measurements of NP calibrators were all done in triplicates. Logistic four-parameter fitting was carried out by Zecen software from RangeCL immunoanalyzer, based on the concentration of rNP calibrators and the corresponding mean value of obtained RLU reads. The values of NP samples were calculated based on the calibration curve from same batch.
2.9. Data analysis
Calculation of mean values and standard deviation (SD), analysis of linear regression and correlation were all carried out by GraphPad PRISM 7 software. Individual values were plotted as scatter with lines showing the mean value and SD. Calibration curves of CLEIA were plotted by Zecen software from RangeCL immunoanalyzer. p < 0.05 was considered statistically significant.
3. Results
3.1. The expression of recombinant SARS-CoV-2-NP
The recombinant NP had an estimated molecular weight of 49.4 kDa, and was abundantly expressed in Escherichia coli strain BL21(DE3) with the induction of isopropyl-β-d-thiogalactoside (IPTG). As shown in Fig. 1 , the target protein was mostly existed in the supernatant of bacterial lysate, suggesting the rNP was expressed in a soluble pattern. After the treatment of 30 % saturated ammonium sulfate solution and Ni-chelating affinity chromatography, purified rNP was obtained for further use.
Fig. 1.
The expression and purification of recombinant NP. (a) pET28a(+)-SARS-CoV-2-NP was transformed into E. coli cells, and the recombinant NP was expressed by the induction of IPTG. Lane M indicated the molecule weight marker. Lane 1 and Lane 2 respectively represented the bacterial lysate without or with the induction of IPTG. Lane 3 and Lane 4 respectively represented the supernatant and sediment of IPTG-induced bacterial lysate after centrifugation. The supernatant of bacterial lysate was gradient treated with 30 %, 40 % and 50 % saturated ammonium sulfate in succession, while Lane 5, Lane 6 and Lane 7 respectively represented the protein precipitate samples by the treatment. (b) The protein precipitate sample by the treatment of 30 % saturated ammonium sulfate was further purified by affinity chromatography. Lane M indicated the molecule weight marker. Lane 1 represented the protein sample prepared for chromatography. Lane 2 represented the outflow of protein sample through the chromatography column, while Lane 3 represented the subsequent outflow of washing buffer. Lane 4 to Lane 14 respectively represented the eluent of washing buffer containing 10, 20, 30, 40, 50, 60, 70, 80, 90, 100 and 200 mM imidazole.
3.2. Development of monoclonal antibodies against rNP
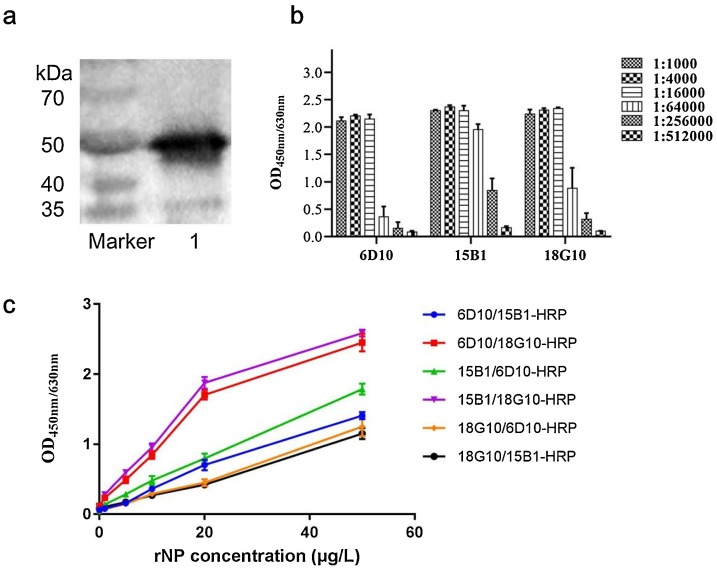
Several cell lines that stably secrete monoclonal antibody (mAb) against SARS-CoV-2-NP were obtained by hybridoma technique. The immunoreactivity of developed mAbs to NP was confirmed by western blot, and as shown in Fig. 2 a, the developed mAb could specifically recognize rNP and form an immune complex band at 50 kDa.
Fig. 2.
Evaluation of developed anti-NP monoclonal antibodies. (a) The immunoreactivity of developed mAb to SARS-CoV-2-NP was identified by western blot. Lane 1 represented the immune complex band of rNP and mAb 18G10. (b) The titer of prepared mAbs was determined by indirect ELISA. (c) mAb 6D10, 15B1 and 18G10 were cross-paired to compare the detection effectiveness of antibody pairs by DAS-ELISA. The recombinant NP was used as antigen sample and its concentration varied from 0 to 50 μg/L.
The prepared mAb solution was adjusted to 1 mg/mL, and the antibody titer was determined by indirect ELISA. As illustrated in Fig. 2b, three developed mAbs (6D10, 15B1 and 18G10) which were all characterized as subtype of IgG1, presented a good sensitivity for rNP detection at the titer of 1:64000 or higher in indirect ELISA. As illustrated in Fig. 2c, the three developed mAbs were further cross-paired to perform the measurement of rNP by DAS-ELISA. The mAb pair of 15B1/18G10-HRP presented best sensitivity for rNP detection, and its detection threshold in DAS-ELISA could reach as low as 1.0 μg/L.
3.3. Analysis performance of CLEIA based on mAb pair of 15B1/18G10-AP
Based on the detection effectiveness of the mAb pairs in DAS-ELISA, we selected mAb 15B1 as magnetic particles-conjugated antibody and mAb 18G10 as AP-labeled antibody to establish the proposed chemiluminescence enzyme immunoassay (CLEIA) for rNP measurement.
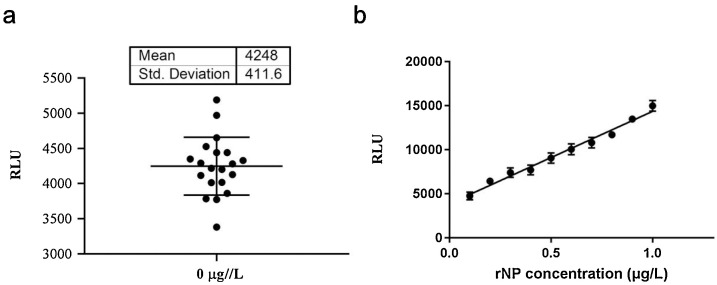
Limit of detection (LoD) was used to evaluate the analytical sensitivity of proposed assay. It usually requires the analyte signal to be three to ten times larger than the background signal, and it defines the point at which the analysis becomes possible. As illustrated in Fig. 3 a, the measurement of blank control (rNP: 0 μg/L) was performed in twenty replicates, and then the mean and standard deviation (SD) of obtained RLU reads were calculated. The LoD was defined as the concentration converted from mean RLU plus three SD (n = 20) of blank control. The LoD of proposed CLEIA was calculated according to the calibration curve in the same batch, which was 0.1 μg/L. Then rNP was diluted to ten gradient concentrations ranged from 0.1 to 1 μg/L, and measured by proposed CLEIA. As illustrated in Fig. 3b, the RLU reads presented a good correlation with the rNP concentrations (r = 0.992, p < 0.0001), suggesting the proposed CLEIA had a good sensitivity for the measurement of NP at low concentrations.
Fig. 3.
Sensitivity analysis of proposed CLEIA. Magnetic particles-conjugated mAb 15B1 (0.1 mg/mL) and AP-labeled mAb 18G10 (diluted as 1:1000) were used to establish the proposed CLEIA for SARS-CoV-2-NP measurement. (a) The RLU reads obtained in 20 tests of blank control (rNP concentration assigned as 0 μg/L). (b) Ten gradient diluted rNP samples with the concentration varied from 0.1 to 1 μg/L were measured by proposed CLEIA. Linear regression was performed with the RLU reads and assigned rNP concentration.
Dose-response is an important indicator for quantitative measurement. As illustrated in the Fig. 4 , the proposed CLEIA showed a good linearity in the measurement of rNP ranged from 0 to 160 μg/L (R 2>0.98, p<0.0001). 3 rNP samples with the concentration of 0.5, 1 and 5 μg/L were respectively measured in ten replicates per day for three days. Coefficient of variations (CV) is defined as standard variation divided by the mean, and used to evaluate the precision of assay. As shown in Fig. 5 , the precision CV for rNP measurement was below 10 %, indicating a satisfactory reproducibility of the proposed assay.
Fig. 4.
Linearity analysis of proposed CLEIA. Several rNP samples ranged from 0 to 160 μg/L were measured by proposed CLEIA based on mAb pair of 15B1/18G10-AP. Linear regression analysis was carried out based on the RLU reads and rNP concentrations. (a) rNP concentrations assigned as 0, 1, 2, 4, 8 and 16 μg/L. (b) rNP concentrations assigned as 0, 5, 10, 20, 80 and 160 μg/L.
Fig. 5.
Precision analysis of proposed CLEIA. (a∼c) Three rNP samples at the concentration of 0.5, 1 and 5 μg/L were measured by proposed CLEIA in ten times per day for 3 days. (d) The coefficient of variations was calculated based on daily RLU reads of each sample.
4. Discussions
In the case of COVID-19, testing capacity at the primary care level is a key step for the prevention of disease transmission. Though the nucleic acid detection is taken as a gold standard method for early screening and diagnosis of virus infection, there were many challenges for the developing regions to implement the testing, including the lack of trained personnel, state-of-the-art equipment and the reagents. Development of highly sensitive and specific immunoassays with low costs would be beneficial for early clinical diagnosis of SARS-CoV-2 infection, especially in low-resource settings (Younes et al., 2020).
A few Point-of-Care (POC) tests for rapid antigen detection were developed during the first global wave of COVID-19, but further optimization and clinical validation of these tests are still needed (Mak et al., 2020; Dinnes et al., 2020). Compared with conventional immunochromatographic assays, the newly developed CLEIA for SARS-CoV-2-NP had shown a better sensitivity and a wider linear detection range (between 0.1 and 160 μg/L). Recently, a nanoenzyme linked immunochromatographic sensor was reported, with a LoD of 0.026 μg/L NP (Liang et al., 2021). The LoD of proposed CLEIA in present study was about 0.1 μg/L, suggesting the sensitivity needs to be further improved. The mean RLU reads of blank control in our previous developed CLEIA for PCT measurement was not higher than 2000 (Liao et al., 2021). But in present study, the mean RLU reads of blank control was higher than 4000, suggesting the optimization of dilution buffer and antibody preparation is needed to further reduce the background signal. The analysis performance of developed mAbs and CLEIA also needs to be fully evaluated in clinical specimens and compared with other existing immunoassays and molecular diagnostic tools.
Nowadays, with the introduction of vaccines, SARS-CoV-2 is likely to be recognized as one of respiratory pathogens routinely detected in clinic. Chemiluminescence immunoassay (CLIA) is a promising choice for antigen detection in clinical laboratory, which possessed the characteristics of fast, sensitive, automated and high-throughput quantitative detection. A recently developed CLIA for SARS-CoV-2 antigen test presented a sensitivity of 94 % and specificity of 100 % in samples with high viral load (corresponding Ct values ≤ 25 by RT-PCR) (Paul et al., 2021). The performance of CLIA would not permit it to replace nuclear acid testing for SARS-CoV-2, but may enable rapid and efficient detection of people with high viral load who are responsible for the infectious clusters (Salvagno et al., 2021; Basso et al., 2021; Krüttgen et al., 2021).
Author statement
Ye Xu, Chuan Xia, Xuan Zeng and Qing Jiang contributed to the investigation. Yilan Qiu and Meifang Quan contributed to the funding acquisition and resources. Minjing Liao contributed to the writing. Rushi Liu contributed to the conceptualization and funding acquisition.
Declaration of Competing Interest
None
Acknowledgements
We thank the support of grants from National Natural Science Foundation of China (No. 31072141); Key R&D Program of Hunan Province (No. 2019SK2042); Science and Technology Program of Changsha City (Nos. KQ1801039, KQ1907130).
Data availability
Data will be made available on request.
References
- Basso D., Aita A., Padoan A., et al. Salivary SARS-CoV-2 antigen rapid detection: a prospective cohort study. Clin. Chim. Acta. 2021;517:54–59. doi: 10.1016/j.cca.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wen K., Zhang J., et al. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin. Microbiol. Infect. 2021;27(2) doi: 10.1016/j.cmi.2020.09.057. 289.e1-289.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnes J., Deeks J.J., Adriano A., et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2020;8(8) doi: 10.1002/14651858.CD013705. Cd013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Ren L., Yang S., et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Yang H., Ji W., et al. Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses. 2020;12(4):372. doi: 10.3390/v12040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüttgen A., Cornelissen C.G., Dreher M., et al. Comparison of the SARS-CoV-2 Rapid antigen test to the real star Sars-CoV-2 RT PCR kit. J. Virol. Methods. 2021;288 doi: 10.1016/j.jviromet.2020.114024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Li J. Immunologic testing for SARS-CoV-2 infection from the antigen perspective. J. Clin. Microbiol. 2021;59(5) doi: 10.1128/JCM.02160-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Liu B., Li J., et al. A nanoenzyme linked immunochromatographic sensor for rapid and quantitative detection of SARS-CoV-2 nucleocapsid protein in human blood. Sens. Actuators B Chem. 2021;349 doi: 10.1016/j.snb.2021.130718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Zheng J., Xu Y., et al. Development of magnetic particle-based chemiluminescence immunoassay for measurement of human procalcitonin in serum. J. Immunol. Methods. 2021;488 doi: 10.1016/j.jim.2020.112913. [DOI] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak G.C., Cheng P.K., Lau S.S., et al. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Gupta A., Rooge S., et al. Performance evaluation of automated chemiluminescence immunoassay based antigen detection - moving towards more reliable ways to predict SARS-CoV-2 infection. J. Virol. Methods. 2021;298 doi: 10.1016/j.jviromet.2021.114299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections-more than just the common cold. Jama. 2020;323(8):707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- Salvagno G.L., Gianfilippi G., Fiorio G., et al. Clinical assessment of the DiaSorin LIAISON SARS-CoV-2 Ag chemiluminescence immunoassay. Ejifcc. 2021;32(2):216–223. [PMC free article] [PubMed] [Google Scholar]
- Scohy A., Anantharajah A., Bodéus M., et al. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg O., Martiny D., Rochas O., et al. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2021;19(3):171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., et al. Detection of SARS-CoV-2 in different types of clinical specimens. Jama. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang F., Wang X., He X., et al. Antibody detection and dynamic characteristics in patients with COVID-19. Clin. Infect. Dis. 2020;71(8):1930–1934. doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes N., Al-Sadeq D.W., Al-Jighefee H., et al. Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2. Viruses. 2020;12(6):582. doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.