Abstract
Background
Carbapenemase-producing, carbapenem-resistant Pseudomonas aeruginosa (CP-CRPA) is a global challenge. However, detection efforts can be laborious because numerous mechanisms produce carbapenem resistance. A minimum inhibitory concentration–based algorithm (imipenem- or meropenem-resistant plus ceftazidime-nonsusceptible plus cefepime-nonsusceptible) was proposed to identify the isolates most likely to harbor a carbapenemase; however, prospective validation in geographies displaying genotypic diversity and varied carbapenemase prevalence is warranted.
Methods
CRPA isolates were collected during the Enhancing Rational Antimicrobials for P. aeruginosa (ERACE-PA) global surveillance program from 17 sites in 12 countries. Isolates underwent susceptibility testing following local standards to ceftazidime, cefepime, and ceftolozane/tazobactam. Isolates underwent initial phenotypic carbapenemase screening followed by molecular testing if positive. The primary algorithm criteria were applied, and results were compared with phenotypic carbapenemase results to assess the performance of the algorithm. A secondary criterion, the algorithm criterion or imipenem- or meropenem-resistant plus ceftolozane/tazobactam-nonsusceptible, was assessed.
Results
A total of 807 CRPA were assessed, and 464 isolates met the algorithm criteria described above. Overall, testing was reduced by 43% compared with testing all CRPA. Carbapenemase-positive isolates missed by the algorithm were largely driven by Guiana extended spectrum (GES). Addition of the criterion of imipenem- or meropenem-resistant plus ceftolozane/tazobactam-nonsusceptible decreased the number of CP-CRPA missed by the algorithm (21 vs 40 isolates, respectively), reducing number of isolates tested by 39%.
Conclusions
Application of the initial algorithm (imipenem- or meropenem-resistant plus ceftazidime-nonsusceptible plus cefepime-nonsusceptible) performed well in a global cohort, with 33% phenotypically carbapenemase-positive isolates. The addition of imipenem- or meropenem-resistant plus ceftolozane/tazobactam-nonsusceptible reduced the number of phenotypically carbapenemase-positive isolates missed and may be useful in areas with a prominence of GES.
Keywords: algorithm, carbapenemase, genotypic, molecular diagnostics, Pseudomonas aeruginosa
Carbapenem-resistant Pseudomonas aeruginosa (CRPA) is a well-described nosocomial pathogen that is associated with limited treatment options and substantial morbidity and mortality. Numerous mechanisms of resistance can cause carbapenem resistance in P. aeruginosa [1]. In the United States, the most common mechanisms of carbapenem resistance are alterations in porins coupled with overexpression of cephalosporinases and/or efflux pumps [2]. However, globally carbapenemase-producing CRPA (CP-CRPA) are common, and the underappreciation of their contribution in the United States may be due to limited testing [3]. Detection of CP-CRPA is important because these carbapenemases are typically encoded by genes on plasmids that can be transferred among bacteria and often confer resistance to new β-lactam/β-lactamase inhibitors [4].
A number of phenotypic and genotypic tests are available for carbapenemase detection in P. aeruginosa [5–7]. However, carbapenem resistance alone may be indiscriminate for CP-CRPA vs CRPA due to noncarbapenemase mechanisms [8]. Additionally, carbapenemase testing may have a low yield relative to the resources needed to test all carbapenem-resistant P. aeruginosa isolates [4]. Strategies to identify the isolates that are most likely to harbor carbapenemases may streamline detection workflows as well as improve the cost-effectiveness of additional screening procedures for P. aeruginosa.
We previously described the derivation of a pragmatic, minimum inhibitory concentration (MIC)–based algorithm to guide definitive carbapenemase testing in carbapenem-resistant P. aeruginosa [8]. This algorithm was further optimized in a separate cohort from German medical centers and modified to the following: imipenem- or meropenem-resistant (-R) plus ceftazidime-nonsusceptible (-NS) plus cefepime-NS [9]. This algorithm represents a starting point to guide carbapenemase testing while reducing overall workload that would encompass testing all CRPA. The evaluation of these criteria in a global cohort that represents regions with varying prevalence and diversity of carbapenemase genes would further define the utility of this algorithmic approach. Herein, we describe the application of the algorithm in the previously described Enhancing Rational Antimicrobials for P. aeruginosa (ERACE-PA) Global Surveillance Program, which included 33% of isolates testing phenotypically positive for carbapenemase production, of which 86% were genotypically confirmed for carbapenemases [10].
METHODS
Bacterial Isolates
Isolates were compiled as part of the ERACE-PA Global Surveillance Program, which enrolled 17 sites from 12 countries spanning Europe, the Middle East, the United States, South America, and Africa [10]. Isolates were sent to a central laboratory (Center for Anti-Infective Research and Development, Hartford, CT, USA) for storage and frozen at –80°C in skim milk until assessment.
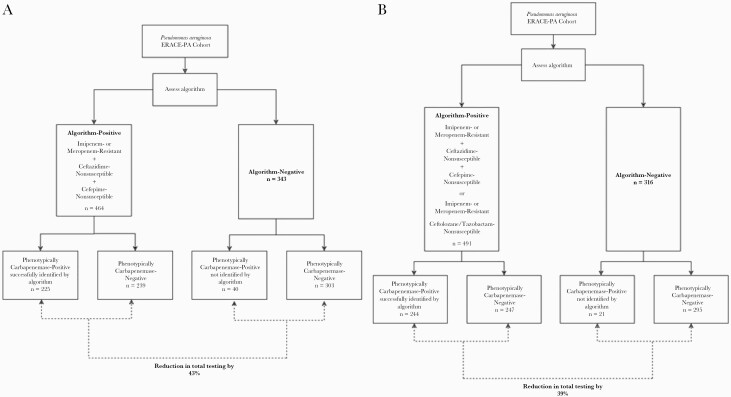
Isolates were eligible for inclusion if they were identified as P. aeruginosa by local standards of practice and determined to be carbapenem-resistant at the submitting site. Nonduplicate isolates were included from any age patient or any anatomical source over 2019–2021 [10]. Figure 1, A and B outlines the application of the primary and primary or secondary algorithm criteria, respectively.
Figure 1.
Phenotypic carbapenemase designation of algorithm-positive and -negative isolates using (A) imipenem- or meropenem-R plus ceftazidime-NS plus cefepime-NS and (B) imipenem- or meropenem-R plus ceftazidime-NS plus cefepime-NS OR imipenem- or meropenem-R plus ceftolozane/tazobactam-NS. Abbreviations: ERACE-PA, Enhancing Rational Antimicrobials for P. aeruginosa; NS, nonsusceptible; R, resistant.
Algorithm Criteria
The primary phenotypic algorithm of imipenem or meropenem resistance plus ceftazidime-NS plus cefepime-NS to guide carbapenemase testing in carbapenem-resistant P. aeruginosa was assessed [8, 9]. To limit the number of carbapenemase producers missed by the algorithm, a secondary criterion was evaluated to assess the performance of testing isolates that meet the algorithm criterion or that meet a second criterion: imipenem- or meropenem-resistant plus ceftolozane/tazobactam-nonsusceptible. Ceftazidime/avibactam was not assessed as many serine carbapenemase–producing P. aeruginosa may test susceptible, and thus this would have poor sensitivity [8].
In Vitro Susceptibility Testing
In vitro susceptibility testing was conducted by the submitting sites per local standards of practice including automated susceptibility systems and disk diffusion. If the susceptibility test result was not available due to the laboratory’s standard of care, the susceptibility testing was conducted using broth microdilution, per Clinical and Laboratory Standards Institute (CLSI) standards at the central laboratory. MICs were assessed for ceftazidime, cefepime, and ceftolozane/tazobactam and interpreted per CLSI and European Committee on Antimicrobial Susceptibility Testing (EUCAST) standards as the MIC breakpoints that constitute nonsusceptibility match between both interpretations [5, 11]. For EUCAST, the “I” interpretation was considered susceptible as it implies susceptible increased exposure [11].
Phenotypic and Genotypic Carbapenemase Categorization
Carbapenemase testing as defined previously was used in the present analysis [10]. Briefly, isolates were tested for carbapenemase production by local genotypic testing, if available. All isolates underwent testing with the modified carbapenem inactivation method (mCIM) as previously described [5]. Phenotypically carbapenemase-positive isolates were then tested genotypically using the Carba-R, Carba-R NxG (Cepheid, Sunnyvale, CA, USA) and whole-genome sequencing (if negative on the first 2 platforms) [10]. In total, 265 of the 807 isolates (33%) tested phenotypically positive for carbapenemase activity [10]. Of these, 228 had genotypically confirmed carbapenemase genes including blaVIM, blaIMP, blaNDM, blaKPC, blaGES, and multiple carbapenemases (blaVIM and blaIMP, blaVIM and blaKPC, blaVIM and blaOXA-48) [10]. The remaining 37 isolates did not have detection of any previously defined carbapenemase genes on polymerase chain reaction (PCR). Whole-genome sequencing was conducted on 21 of these 37 isolates to assess for other carbapenemases, of which none were detected. The other 16 isolates had similar phenotypes and similar geographic origins as isolates that were assessed via whole-genome sequencing.
Analysis
Isolates were categorized as algorithm-positive (meeting the criteria) or algorithm-negative (not meeting algorithm criteria) as they correspond to the primary and secondary criteria. To analyze the performance of each algorithm criterion, phenotypic carbapenemase testing was utilized as the reference standard to which the algorithm results were compared. This was selected as a conservative assessment as genotypic testing may be limited by the included genotypic targets but phenotypic testing is not target specific and thus represents a global assessment of carbapenemase activity. The mCIM has a test performance for P. aeruginosa of sensitivity 93%–98% and specificity 85%–95% [5–7]. The number (%) of isolates that would be tested for carbapenemase genes (meeting algorithm criteria) was compared with all included carbapenem-resistant P. aeruginosa isolates. The number of isolates missed by the algorithm (algorithm-negative) but positive by phenotypic carbapenemase testing was categorized. Sensitivity and specificity were calculated for each algorithm criterion. Positive and negative predictive values were calculated using the 33% phenotypically carbapenemase-positive isolates from the cohort to describe prevalence.
RESULTS
Eight hundred seven CRPA were assessed, and 464 isolates met the initial algorithm criterion (imipenem- or meropenem-R plus ceftazidime-NS plus cefepime-NS) and thus would have been selected for carbapenemase testing, resulting in a 43% reduction in testing compared with testing all carbapenem-resistant isolates. When tested against phenotypically carbapenemase-positive isolates, a total of 40 (15% of phenotypically carbapenemase-positive) isolates would have not been tested due to not meeting algorithm criteria. Figure 1A depicts the number of phenotypically carbapenemase-positive isolates that were algorithm-positive and -negative. The summary of test performance and genotypic characteristics of isolates that were algorithm-negative but phenotypically carbapenemase-positive is reported in Table 1. Notably, 20 of the 40 (50%) phenotypically carbapenemase-positive isolates missed by the algorithm lacked a known carbapenemase gene by PCR or whole-genome sequencing. The most commonly missed carbapenemase class was Guiana extended spectrum (GES)–harboring isolates (n = 13 of the 20 isolates), including isolates from 4 different countries (Turkey, Kuwait, Saudi Arabia, and Italy). Table 2 describes the genotypic carbapenemase targets that were true positives and false negatives by each algorithm assessed. The 40 algorithm false negatives were also assessed by broth microdilution, which confirmed algorithm false negativity in 83% of these isolates.
Table 1.
Performance Characteristics of the 2 Evaluated Algorithm Criteria Against a Cohort of 807 Globally Collected Carbapenem-Resistant P. aeruginosa That Included 265 Phenotypically Carbapenemase-Positive Isolates
| Criteria | No. of Isolates Tested | % Reduction in Carbapenemase Tests | No. of Phenotypically CP-CRPA Successfully Identified (% of All CP-CRPA) | No. of Phenotypically CP-CRPA Missed (% of All CP-CRPA) | Genotype of Phenotypically Carbapenemase-Positive Isolates Missed |
|---|---|---|---|---|---|
| Primary algorithm criterion: imipenem- or meropenem-R plus ceftazidime-NS plus cefepime-NS |
464 | 43 | 225 (85) | 40 (15) | Isolates with classically described carbapenemase gene, n = 20 -GES, n = 13 -VIM, n = 6 -KPC, n = 1 Isolates without classically described carbapenemase genes after whole-genome sequencing, n = 20 -OXA-10-like + OXA-50-like + PDC, n = 2 -n = 11, not sequenced but have similar phenotypes and same site -OXA-2-like + OXA-50-like + PDC, n = 3 -OXA-50-like + PDC, n = 2 -n = 2, not sequenced but have similar phenotypes and same site |
| Primary algorithm criterion OR secondary algorithm criterion: imipenem- or meropenem-R plus ceftazidime-NS plus cefepime-NS (algorithm) OR imipenem- or meropenem-R plus ceftolozane/tazobactam-NS |
491 | 39 | 244 (92) | 21 (8) | Isolates with classically described carbapenemase gene, n = 4 -GES, n = 1 -VIM, n = 2 -KPC, n = 1 Isolates without classically described carbapenemase genes after whole-genome sequencing, n = 17 -OXA-10-like + OXA-50-like + PDC, n = 1 -n = 11, not sequenced but have similar phenotypes and same site -OXA-2-like + OXA-50-like + PDC, n = 1 -OXA-50-like + PDC, n = 2 -n = 2, not sequenced but have similar phenotypes and same site |
Abbreviations: CP-CRPA, carbapenem-resistant Pseudomonas aeruginosa; GES, Guiana extended spectrum; KPC, Klebsiella pneumoniae carbapenemase; NS, nonsusceptible; OXA, oxacillinase; PDC, Pseudomonas-derived cephalosporinase; R, resistant; VIM, Verona integron-encoded metallo-β-lactamase.
Table 2.
Genotypic Carbapenemase Targets Identified in Phenotypically Carbapenemase Positive Isolates
| Primary Algorithm Criterion: Imipenem- or Meropenem-R Plus Ceftazidime-NS Plus Cefepime-NS |
Primary Algorithm Criterion OR Secondary Algorithm Criterion: Imipenem- or Meropenem-R Plus Ceftazidime-NS Plus Cefepime-NS (Algorithm) OR Imipenem- or Meropenem-R Plus Ceftolozane/Tazobactam-NS |
|||
|---|---|---|---|---|
| Genotypic Target | True Positive | False Negative | True Positive | False Negative |
| VIM | 130 | 6 | 134 | 2 |
| IMP | 15 | 0 | 15 | 0 |
| NDM | 13 | 0 | 13 | 0 |
| KPC | 7 | 1 | 7 | 1 |
| GES | 31 | 13 | 43 | 1 |
| VIM and IMP | 3 | 0 | 3 | 0 |
| VIM and OXA | 1 | 0 | 1 | 0 |
| VIM and KPC | 8 | 0 | 8 | 0 |
| No targets identified | 17 | 20 | 20 | 17 |
Abbreviations: GES, Guiana extended spectrum; IMP, imipenemase; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-beta-lactamase; NS, nonsusceptible; OXA, oxacillinase; R, resistant; VIM, Verona integron-encoded metallo-β-lactamase.
The addition of a secondary carbapenemase testing criterion to the algorithm—imipenem- or meropenem-R plus ceftolozane/tazobactam-NS—resulted in 491 isolates tested and thus a 39% decrease in testing compared with testing all CRPA. Figure 1B depicts the number of phenotypically carbapenemase-positive isolates that were algorithm-positive and -negative with the addition of the ceftolozane/tazobactam-based criteria. The number of phenotypically carbapenemase-positive isolates that were missed by the addition of this criterion decreased to 21 isolates (8% of all carbapenemase-positive). Similarly, 81% of phenotypically carbapenemase-positive isolates missed lacked classic carbapenemases when tested by PCR or whole-genome sequencing (Table 1).
The test performance of each algorithm is presented in Table 3. Indeed, despite the addition of the secondary algorithm criterion reducing the number of false-negative isolates, the increase in sensitivity and negative predicted value was marginal due to the high performance of the primary algorithm.
Table 3.
Test Performance of Each Algorithm Criteria
| Primary Algorithm Criterion: Imipenem- or Meropenem-R Plus Ceftazidime-NS Plus Cefepime-NS |
Primary Algorithm Criterion OR Secondary Algorithm Criterion: Imipenem- or Meropenem-R Plus Ceftazidime-NS Plus Cefepime-NS (Algorithm) OR Imipenem- or Meropenem-R Plus Ceftolozane/Tazobactam-NS |
|
|---|---|---|
| Sensitivity (95% CI), % | 85 (80–89) | 92 (88–95) |
| Specificity (95% CI), % | 56 (52–60) | 54 (50–59) |
| Positive predictive value, % | 49 | 50 |
| Negative predictive value, % | 88 | 93 |
Abbreviations: NS, nonsusceptible; R, resistant.
DISCUSSION
Application of our previously derived, clinically applicable algorithm to guide definitive carbapenemase testing among P. aeruginosa performed well to reduce the number of unnecessary carbapenemase tests compared with testing all carbapenem-resistant isolates in a diverse population of CRPA. Importantly, a high percentage of isolates that tested phenotypically positive for carbapenemase were captured by the primary algorithm criterion. The addition of the secondary testing criterion of imipenem- or meropenem-resistant plus ceftolozane/tazobactam-nonsusceptible further increased the number of phenotypically carbapenemase-positive isolates that would be selected for testing by the algorithm criteria, although the improvement in sensitivity was only marginal. Indeed, both approaches presented above have value to both CLSI and EUCAST interpretations as both were considered in the development. Using this algorithmic approach may help streamline carbapenemase detection efforts.
Some isolates were missed by the algorithm that tested phenotypically positive for carbapenemase production. Introduction of the second step (ie, ceftolozane/tazobactam-nonsusceptible) reduced the number of isolates missed, adding sensitivity to the previously defined criterion. GES-positive isolates accounted for 13 of the isolates missed by the primary algorithm due to cefepime susceptibility. The addition of the secondary criteria reduced the number of GES isolates missed to 1 isolate. This is not unlike our previous findings where the addition of ceftolozane/tazobactam failed to capture 2 GES-harboring isolates and may be due to the variable hydrolytic potentials of this diverse class of enzymes [9, 12]. Importantly, the addition of the ceftolozane/tazobactam criterion represents an alternative, as not all institutions have these MIC data readily available. The base algorithm still performed well to streamline carbapenemase testing while not excluding a high proportion of phenotypically carbapenemase-positive isolates. Interestingly, over half of phenotypically carbapenemase-positive isolates missed by the algorithm criteria lacked classically categorized carbapenemase genes. This was similar to our previous findings, where phenotypic susceptibility to ceftolozane/tazobactam was noted in an OXA-2- and OXA-50-like harboring isolate that tested mCIM-positive [8]. These mismatched results of carbapenemase detection that were phenotypically susceptible to various β-lactam agents represent a distinct clinical challenge, as the best therapeutic strategy is unclear, although when using phenotypic detection it is plausible that the test is detecting an enzyme(s) that breaks down the test compound (ie, meropenem in mCIM) but lacks viable hydrolytic activity against other agents (ie, antipseudomonal cephalosporins) [13]. Clinical and in vivo data using human-simulated exposures are needed to help categorize the clinical relevance of these findings to guide treatment decisions [14]. While ceftazidime/avibactam plays an important clinical role in the management of CRPA inclusive of GES- and Klebsiella pneumoniae carbapenemase (KPC)–producing strains, inclusion of this compound’s phenotypic profile in the algorithm criterion was not undertaken as the high proportion testing susceptible would result in a false-negative screen by the criterion [8, 10]. In the context of the algorithmic carbapenemase screening, the limited number of phenotypically carbapenemase-positive isolates missed by the algorithm further supports its utility in the clinical laboratory.
Our work builds upon our previous studies as well as those recently reported using a similar approach [8, 9, 15]. Vallabhaneni and colleagues recently reported data that showed that adding the criteria of cefepime- or ceftazidime-NS reduced the number needed to test for detection of CP-CRPA, which was further decreased if ceftolozane/tazobactam-NS was employed in CRPA isolates from the United States [15]. The differences in our study designs are notable as we used phenotypic detection of carbapenemases as the standard identification, to which we compared algorithm performance rather than genotypic carbapenemases. Both approaches have their own benefits and limitations, although genotypic detection is limited to the targeted enzymology. In our approach, all CRPA were systematically assessed using phenotypic carbapenemase detection, and positive isolates underwent genotypic testing with Carba-R and Carba-R NxG, which enhances the carbapenemase detection profile in our study to include GES and certain IMP subtypes, among others [16]. Similarly, isolates that were phenotypically carbapenemase-positive but genotypically negative by PCR underwent whole-genome sequencing to further evaluate the underlying mechanisms [10]. Although mCIM has been noted to have excellent sensitivity (93%–98% for P. aeruginosa), it is liable to false-positive results, and thus the present study represents a conservative assessment of our proposed algorithm’s performance [6, 7]. The number of isolates testing mCIM-positive, genotypically carbapenemase-negative further highlights the need for confirmatory genotypic testing of phenotypically carbapenemase-positive isolates in the clinic. Additionally, our data come from a global collection of CRPA that assessed regions with both high and low prevalence of CP-CRPA including an overall prevalence of phenotypically defined carbapenemase activity in 33% of the CRPA assessed. This compares to the ~3%–10% of CRPA testing positive for carbapenemases from our previous and other studies [9, 15]. These data provide important insight into the algorithm’s performance when the baseline prevalence of CP-CRPA is high and thus adds translatability of this approach to areas with higher prevalence of CP-CRPA than previously assessed. Another strength of the present study included that MICs were determined by the standard methods of study sites, further adding to the applicability of this study to the clinic. Indeed, the algorithm false negatives were assessed by reference broth microdilution, revealing that 83% were confirmed to be algorithm-negative (ie, susceptible to either cefepime or ceftazidime). This is not unexpected; particularly when isolates’ MICs are at or around the susceptibility breakpoint, there is a high potential for categorical disagreement as the acceptable variability of MIC testing can vary by 100% on either side of the MIC result [17]. In the analysis of the algorithm, we utilized the MIC result from the submitting sites as it represents (1) a worst-case assessment of the proposed algorithm and (2) is clinically applicable as it reflects the methods utilized regularly in clinical practice.
In conclusion, the application of our phenotypic algorithm to guide definitive carbapenemase testing among carbapenem-resistant P. aeruginosa decreased the number of tests conducted compared with testing all carbapenem-resistant isolates. Even in the high-prevalence global cohort with diverse genotypic profiles, a high proportion of phenotypically carbapenemase-positive isolates were captured by the primary algorithm criteria. The addition of imipenem- or meropenem-R plus ceftolozane/tazobactam-NS further improved the algorithm’s performance and may be most useful in regions with high prevalence of GES enzymes. These data support the utilization of this approach in regions with varying prevalence and diversity of CP-CRPA. Clinical implementation will aid in streamlining of carbapenemase detection workflow.
Ackowledgments
We would like to acknowledge all members of the ERACE-PA Global Study Group: Julia Wille, Thais Teles Freitas Rezende, Zuhal Cekin, Gulsah Malkocoglu, Desirèe Gijón, Layla Abdullah Tarakmeh, Chun Yat Chu, Christoffel Johannes Opperman, Hafsah Deepa Tootla, Clinton Moodley, Jennifer Coetzee, Sophia Vourli, George Dimopolus, Dalya M. Attallah, Giusy Tiseo, Alessandro Leonildi, Cesira Giordano, Simona Barnini, Francesco Menichetti, Vincenzo Di Pilato, Giulia Codda, Antonio Vena, Daniele Roberto Giacobbe, Lars Westblade, Armando Cardona, Lauren Curtis, Ferric Fang, and Gina Thomson. We would like to thank the staff from the Center for Anti-Infective Research and Development for their assistance in the conduct of this study.
Financial support. This study was internally funded by the Center for Anti-Infective Research and Development.
Potential conflicts of interest. A.B. is a speaker bureau member of Merck and Pfizer and has received research support from FIND. H.S. has received grants or research support from the German Research Foundation (DFG) and the German Centre for Infection Research (DZIF). H.S. is a consultant or speaker bureau member for Basilea, Entasis, Eumedica, Gilead, MSD, and Shionogi. D.P.N. is a consultant, speaker bureau member, or has received research support from AbbVie, Cepheid, Merck, Paratek, Pfizer, Wockhardt, Shionogi, and Tetraphase. All other authors have no conflicts to declare. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Patient consent. The present study was institutional review board approved under exempt status as not human subjects research, and thus informed consent was not required.
Prior presentation. This was presented in part at World Microbe Forum 2021.
References
- 1. Lister PD, Wolter DJ, Hanson ND.. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 2009; 22:582–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Woodworth KR, Walters MS, Weiner LM, et al. . Vital signs: containment of novel multidrug-resistant organisms and resistance mechanisms - United States, 2006-2017. MMWR Morb Mortal Wkly Rep 2018; 67:396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hammoudi Halat D, Ayoub Moubareck C.. The current burden of carbapenemases: review of significant properties and dissemination among gram-negative bacteria. Antibiotics (Basel) 2020; 9:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nordmann P, Poirel L.. Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin Infect Dis 2019; 69:S521–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. Clinical and Laboratory Standards Institute; 2020. [Google Scholar]
- 6. Tamma PD, Simner PJ.. Phenotypic detection of carbapenemase-producing organisms from clinical isolates. J Clin Microbiol 2018; 56:e01140–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gill CM, Lasko MJ, Asempa TE, et al. . Evaluation of the EDTA-modified carbapenem inactivation method for detecting metallo-β-lactamase-producing Pseudomonas aeruginosa. J Clin Microbiol 2020; 58:e02015–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gill CM, Asempa TE, Nicolau DP.. Development and application of a pragmatic algorithm to guide definitive carbapenemase testing to identify carbapenemase-producing Pseudomonas aeruginosa. Antibiotics (Basel) 2020; 9:738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gill CM, Kresken M, Seifert H, et al. . Evaluation of a phenotypic algorithm to direct carbapenemase testing in Pseudomonas aeruginosa: validation in a multicenter German cohort. Microb Drug Resist. 2021;27:1243–8. [DOI] [PubMed] [Google Scholar]
- 10. Gill CM, Aktaþ E, Alfouzan W, et al. ; on behalf of the ERACE-PA Global Study Group. The ERACE-PA global surveillance program: ceftolozane/tazobactam and ceftazidime/avibactam in vitro activity against a global collection of carbapenem-resistant Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis. 2021;40:2533–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 11.0. 2021. Available at: http://www.eucast.org. Accessed November 17, 2021.
- 12. Naas T, Dortet L, Iorga BI.. Structural and functional aspects of class a carbapenemases. Curr Drug Targets 2016; 17:1006–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yee R, Dien Bard J, Simner PJ.. The genotype to phenotype dilemma: how should laboratories approach discordant susceptibility results? J Clin Microbiol 2021; 13:JCM.00138-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gill CM, Brink A, Chu CY, et al. . Phenotypic/genotypic profile of OXA-10-like-harboring, carbapenem-resistant Pseudomonas aeruginosa: using validated pharmacokinetic/pharmacodynamic in vivo models to further evaluate enzyme functionality and clinical implications. Antimicrob Agents Chemother 2021; 65:e0127421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vallabhaneni S, Huang JY, Grass JE, et al. Antimicrobial susceptibility profiles to predict the presence of carbapenemase genes among carbapenem-resistant Pseudomonas aeruginosa. J Clin Microbiol. 2021;59:e02874–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gill CM, Asempa TE, Tickler IA, et al. . Evaluation of the Xpert Carba-R NxG assay for detection of carbapenemase genes in a global challenge set of Pseudomonas aeruginosa isolates. J Clin Microbiol 2020; 58:e01098–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lasko MJ, Huse HK, Nicolau DP, Kuti JL.. Contemporary analysis of ETEST for antibiotic susceptibility and minimum inhibitory concentration agreement against Pseudomonas aeruginosa from patients with cystic fibrosis. Ann Clin Microbiol Antimicrob 2021; 20:9. [DOI] [PMC free article] [PubMed] [Google Scholar]