SUMMARY
The late-acting endosomal sorting complex required for transport (ESCRT) machinery has been implicated in facilitating the resealing of the nuclear envelope (NE) after mitosis, enabling compartmentalization of the genome away from the cytoplasm. Here, we leverage the stereotypic first division of the C. elegans embryo to identify additional functions of the ESCRT machinery in maintaining the structure of the inner nuclear membrane. Specifically, impaired ESCRT function results in a defect in the pruning of inner nuclear membrane invaginations, which arise normally during NE reformation and expansion. Additionally, in combination with a hypomorphic mutation that interferes with assembly of the underlying nuclear lamina, inhibition of ESCRT function significantly perturbs NE architecture and increases chromosome segregation defects, resulting in penetrant embryonic lethality. Our findings highlight links between ESCRT-mediated inner nuclear membrane remodeling, maintenance of nuclear envelope morphology, and the preservation of the genome during early development.
Graphical Abstract
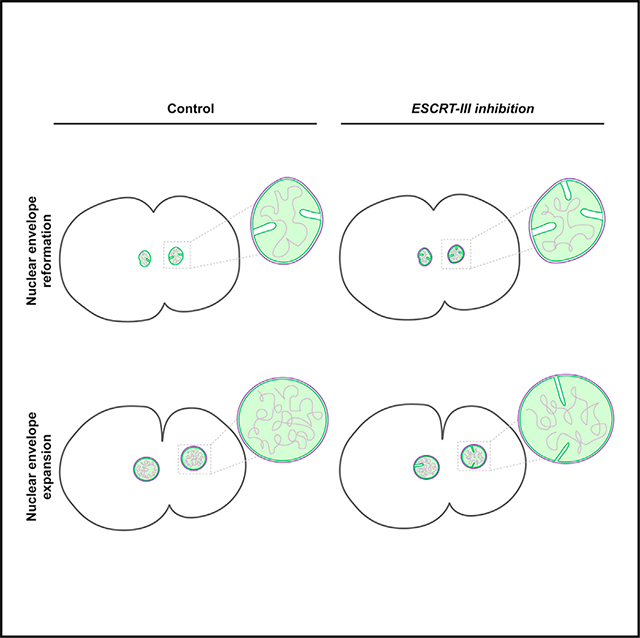
In brief
In this study, Shankar et al. demonstrate that defects in ESCRT machinery functions impair pruning of inner nuclear membrane invaginations that form normally after mitotic exit as the nuclear envelope undergoes expansion. These findings highlight a critical role for the ESCRT machinery in the maintenance of inner nuclear membrane morphology.
INTRODUCTION
The nuclear envelope (NE) is composed of two distinct lipid bilayers, an outer nuclear membrane (ONM) and an inner nuclear membrane (INM), that form an effective barrier between the genome of eukaryotic cells and cytoplasmic factors that might otherwise cause DNA damage and lead to genome instability (Martins et al., 2020; Ungricht and Kutay, 2017). The ONM is continuous with the endoplasmic reticulum (ER), harboring many of the same proteins and lipids, although their morphologies differ substantially. The ONM is also continuous with the INM; they share a similar surface topology and are joined at small pores that mediate nucleocytoplasmic exchange. Nonetheless, numerous studies have highlighted that these connected bilayers exhibit unique proteomes and lipidomes, contributing to their distinct cellular functions (Ungricht and Kutay, 2015; Schirmer et al., 2013; Romanauska and Köhler, 2018). In particular, the INM plays a key role in regulating genome organization by facilitating the separation of peripheral heterochromatic DNA away from actively transcribing euchromatic DNA (Mekhail and Moazed, 2010; Cabianca et al., 2019). The mechanisms underlying this phenomenon are not entirely clear, although specific chromatin-INM protein interactions likely play an important role (Barrales et al., 2016; van Steensel and Belmont, 2017; Iglesias et al., 2020). Consistent with this idea, impaired function of the nuclear lamina, which underlies the INM and contacts DNA directly, results in altered chromatin organization and gene transcription, as well as chromosome missegregation during mitosis (Smith et al., 2018; Kuga et al., 2014; Liu et al., 2000). Similarly, the loss of LEM (LAP2, emerin, MAN1) domain family members that also decorate the INM and associate with DNA-binding proteins, including barrier-to-autointegration factor (BAF), can lead to disruptions in gene silencing and perturbations to chromatin architecture, which may ultimately contribute to chromosome segregation defects observed during cell division (Buchwalter et al., 2019).
Beyond a role in linking chromatin to the INM, the LEM domain protein LEMD2 has also been implicated in recruiting components of the endosomal sorting complex required for transport (ESCRT) machinery to gaps that remain in the NE after initial steps of its reformation during telophase (Gu et al., 2017; Halfmann et al., 2019; Webster et al., 2016). At this phase of the cell cycle, LEMD2 binds to the ESCRT-III subunit CHMP7 (Thaller et al., 2019; Capella et al., 2020), which has been implicated in the nucleation of heteropolymeric filaments composed of other ESCRT-III proteins, including Did2/CHMP1, Vps2/CHMP2, Vps24/CHMP3, Vps32/CHMP4, and Ist1, at NE holes to promote the membrane remodeling necessary for NE sealing (Vietri et al., 2015, 2020a; Olmos et al., 2015). The precise mechanism by which ESCRT-III promotes membrane closure in this context remains unknown, although rapid assembly and dynamic restructuring of Vps32 spiral filaments have been implicated in nearly all other ESCRT-mediated scission events that take place on endosomes, lysosomes, autophagosomes, the ER, and the plasma membrane (Vietri et al., 2020a; Shen et al., 2014). In the majority of these cases, the ESCRT-III complex, together with the Vps4 ATPase, promotes membrane bending away from the cytoplasm, functioning at the curved inner surface of bud necks to facilitate the close apposition of two bilayers, which ultimately enables spontaneous fission (Vietri et al., 2020a). This topology contrasts more canonical budding events that promote the formation of cytoplasmic transport intermediates, including those mediated by COPI, COPII, and clathrin. However, several recent studies have argued that the ESCRT machinery also participates in a subset of outward membrane bending and scission events, raising the possibility that ESCRT-III activity may exhibit more plasticity than previously imagined (Allison et al., 2013; McCullough et al., 2015; Mast et al., 2018; Bertin et al., 2020).
Ruptures in the NE that occur during interphase as a result of mechanical stress or other insults similarly result in ESCRT recruitment (Halfmann et al., 2019; Denais et al., 2016; Raab et al., 2016). Under these conditions, current models suggest that LEMD2 present on the INM becomes exposed to the cytoplasm, enabling the capture of CHMP7 present on ER membranes (Thaller et al., 2019; Capella et al., 2020; Gu et al., 2017; Vietri et al., 2015). Additionally, phosphatidic acids present in the INM may further facilitate this redistribution of CHMP7 (Thaller et al., 2021; Olmos et al., 2016). Subsequent recruitment of additional ESCRT subunits enables rapid NE repair. Somewhat surprisingly, inhibition of ESCRT function following NE rupture fails to significantly impact cell viability, suggesting that multiple parallel systems are in place to ensure NE repair and limit DNA damage (Halfmann et al., 2019; Denais et al., 2016; Raab et al., 2016). The LEM domain proteins, including LEMD2, emerin, MAN1, and Ankle2, may function in this context, acting downstream of BAF, which coats chromatin and establishes a foundation upon which NE reformation occurs (Halfmann et al., 2019; Young et al., 2020). However, a mechanism by which the LEM proteins could drive NE sealing independently of ESCRT function remains undefined. Lastly, the underlying nuclear lamina also contributes to maintenance of the NE in the face of damage. In the absence of DNA repair machinery, mechanically strained cells rely on both a functional lamin mesh-work and the ESCRT machinery for viability, even in the presence of LEM domain proteins (Denais et al., 2016; Raab et al., 2016). These studies collectively suggest that several partially redundant systems exist, which likely work in a coordinated manner to ensure rapid NE sealing and maintenance of genome integrity.
To date, most studies examining ESCRT function at the NE have used mammalian tissue culture cells or yeast, with relatively little information available in developing animal models. Here, we leverage the C. elegans early embryo, which features a highly stereotyped first mitotic division, enabling quantitative measurements of NE dynamics. We demonstrate that animals lacking CHMP7 exhibit minimal defects in NE resealing after mitosis but instead accumulate membrane invaginations emanating from the INM into the nucleoplasm. This perturbation is further exacerbated by a hypomorphic mutation in the single C. elegans lamin gene that underlies dilated cardiomyopathy (DCM) (Wiesel et al., 2008), with double mutants exhibiting penetrant embryonic lethality that results from catastrophic chromosome missegregation events during mitosis. Additionally, we identify Vps60/CHMP5 as another component of the ESCRT machinery that associates with LEM domain proteins and functions redundantly with CHMP7 and Vps32/CHMP4 to maintain nuclei within the germline. Taken together, our data are consistent with a model in which the ESCRT machinery acts within the nucleus to perform quality control over the INM.
RESULTS
Deletion of CHMP7 impairs NE function and exacerbates chromosome misalignment and missegregation caused by a hypomorphic lamin allele during mitosis
To determine the roles for the ESCRT-III subunit CHMP7 during early animal development, we identified the single C. elegans homolog encoded by open reading frame T24B8.2 and used CRISPR-Cas9 technology to engineer an insertion/deletion (indel) mutation immediately downstream of the initiation codon to generate the chmp7 (hz12) allele, which eliminates more than 95% of the native locus based on sequence analysis (Figure S1A). Animals were subsequently backcrossed 4 times to reduce the potential presence of off-target edits. In parallel, we generated affinity-purified rabbit polyclonal antibodies directed against full-length CHMP7 and confirmed by immunoblot analysis that the deletion mutant no longer produced the protein (Figure 1A). To determine whether the loss of CHMP7 affects development, we examined the relative rate at which animals progress through each stage of the C. elegans life cycle, calculated the brood size of gravid adults, and measured embryo viability. In each case, we found no significant differences between control and chmp7 mutant animals (Figure S1B). Together, these studies demonstrate that CHMP7 is not essential in C. elegans unlike most other ESCRT-III subunits, including CHMP1, CHMP2, CHMP3, CHMP4, and CHMP6 (Frankel et al., 2017).
Figure 1. Loss of CHMP7 does not impair MVE biogenesis or ESCRT-mediated endosomal protein sorting.
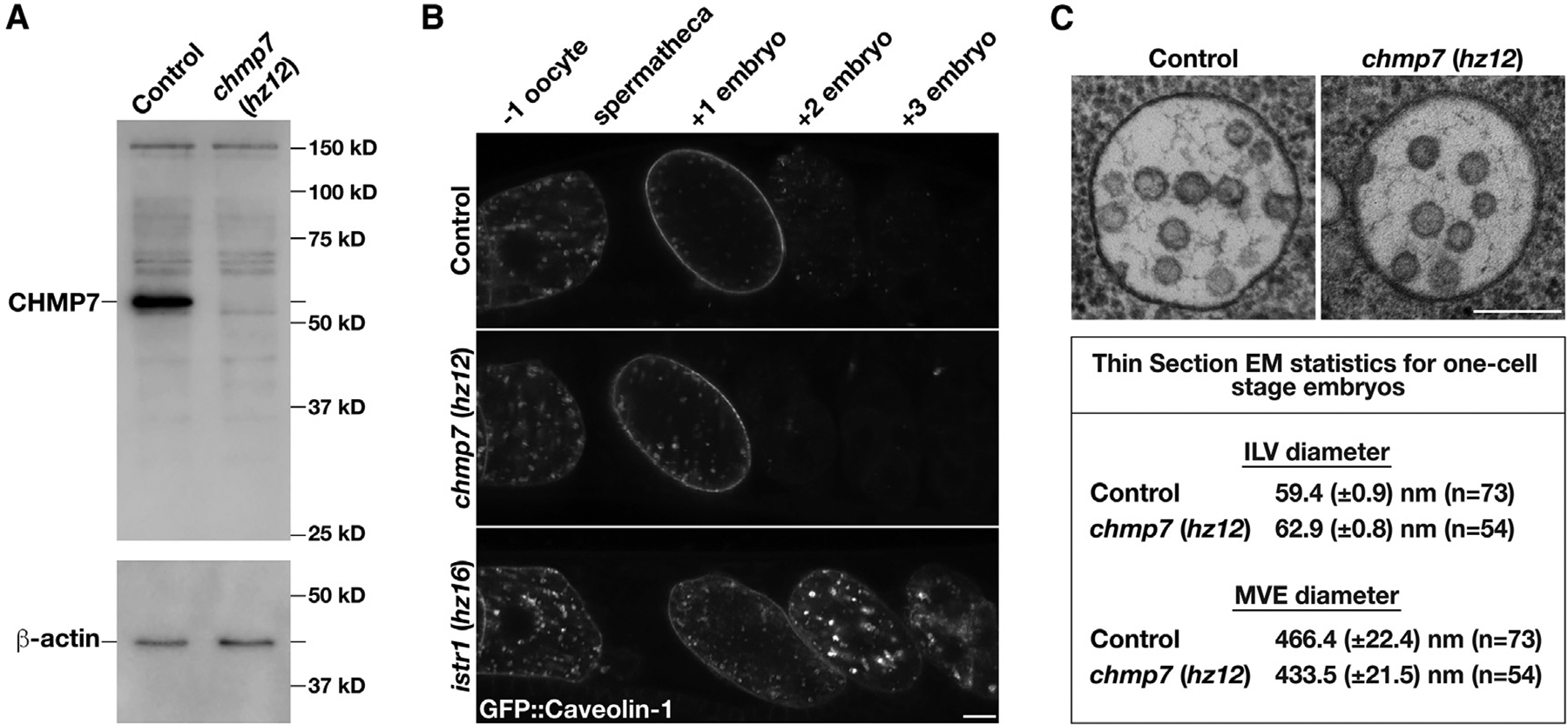
(A) Representative immunoblot of extracts generated from control and chmp7 (hz12) mutant animals (n = 3 each) using antibodies directed against CHMP7 (top) and beta-actin (bottom; load control).
(B) Anesthetized control and CRISPR-Cas9-edited animals harboring deletion mutations in CHMP7 or ISTR1 and expressing a GFP fusion to C. elegans caveolin-1 were imaged using swept-field confocal microscopy. Caveolin-1 is normally degraded prior to embryos reaching the two-cell stage (Frankel et al., 2017). The relative positions of oocytes and embryos are highlighted. Images are representative of at least 10 animals of each genotype. Bar, 10 μm.
(C) High-pressure frozen control and chmp7 (hz12) mutant animals were processed for thin section electron microscopy to visualize MVEs within the one-cell stage embryo. Micrographs are representative of at least 50 MVEs. Bar, 200 nm. A summary of MVE and ILV sizes is also shown.
See also Figure S1.
Since our previous work highlighted important roles for the ESCRT-III machinery during de novo multivesicular endosome (MVE) biogenesis that occurs in newly fertilized one-cell stage embryos and mediates the degradation of numerous integral membrane proteins, we next examined whether CHMP7 functions in the ESCRT-mediated transport of a well-characterized, ubiquitin-modified cargo, caveolin-1 (Frankel et al., 2017). In contrast to the impacts of depleting other components of the ESCRT machinery, an examination of caveolin-1 trafficking in chmp7 knockout animals revealed no visible delays in cargo degradation (Figure 1B). Nevertheless, similar to prior observations made in mammalian cells (Horii et al., 2006), we found that endogenous CHMP7 localizes to endosomes that are positive for the ESCRT subunit Vps4 when its ATPase function is impaired, although this may be the result of a titration effect caused by the aberrant accumulation of other ESCRT proteins (Figure S1C). To determine whether CHMP7 contributes directly to MVE biogenesis, we conducted a series of electron-microscopy-based studies in control and chmp7 knockout embryos. In contrast to the effects of depleting other ESCRT-III subunits (Frankel et al., 2017), deletion of CHMP7 failed to impact intralumenal vesicle formation within MVEs (Figure 1C). These data strongly suggest that CHMP7 does not normally function on endosomes with other components of the ESCRT machinery, which is consistent with prior studies (Vietri et al., 2015; Bauer et al., 2015).
ESCRT-III has also been implicated in the regulation of NE sealing, both during its reformation following anaphase and its recovery after mechanical damage (Gu et al., 2017; Halfmann et al., 2019; Denais et al., 2016; Raab et al., 2016). In each case, CHMP7 has been suggested to play a key role in recruiting Vps32 to initiate ESCRT-III filament assembly and membrane closure. To determine whether C. elegans CHMP7 may similarly play a role in regulating NE integrity, we leveraged a sensitized genetic background in which the nuclear lamina underlying the NE is compromised. Specifically, we used a strain that expresses a mutant form of the single C. elegans lamin gene (lmn1N209K), which corresponds to the N195K mutation in lamin A that underlies DCM and reduces the stability of the nuclear lamina (Wiesel et al., 2008; Penfield et al., 2018). Animals solely expressing this form of lamin exhibited elevated embryo lethality (~47% ± 13%) as compared with control animals. When additionally combined with a deletion in CHMP7, embryo lethality increased significantly (~90% ± 7%) (Figure 2A). This synthetic genetic interaction suggests that CHMP7 functions synergistically with the nuclear lamina to maintain NE structure and function. To our surprise, however, the penetrant embryo lethality was not accompanied by an increase in NE permeability, as quantitatively assayed by fluorescence intensity measurements of a GFP fusion to nuclear localized LacI following the first mitotic division (Figure 2B; Figures S1D and S1E). These findings argue against a solitary role for ESCRT function at the NE in membrane sealing.
Figure 2. CHMP7 becomes essential under conditions where the nuclear lamina is weakened.
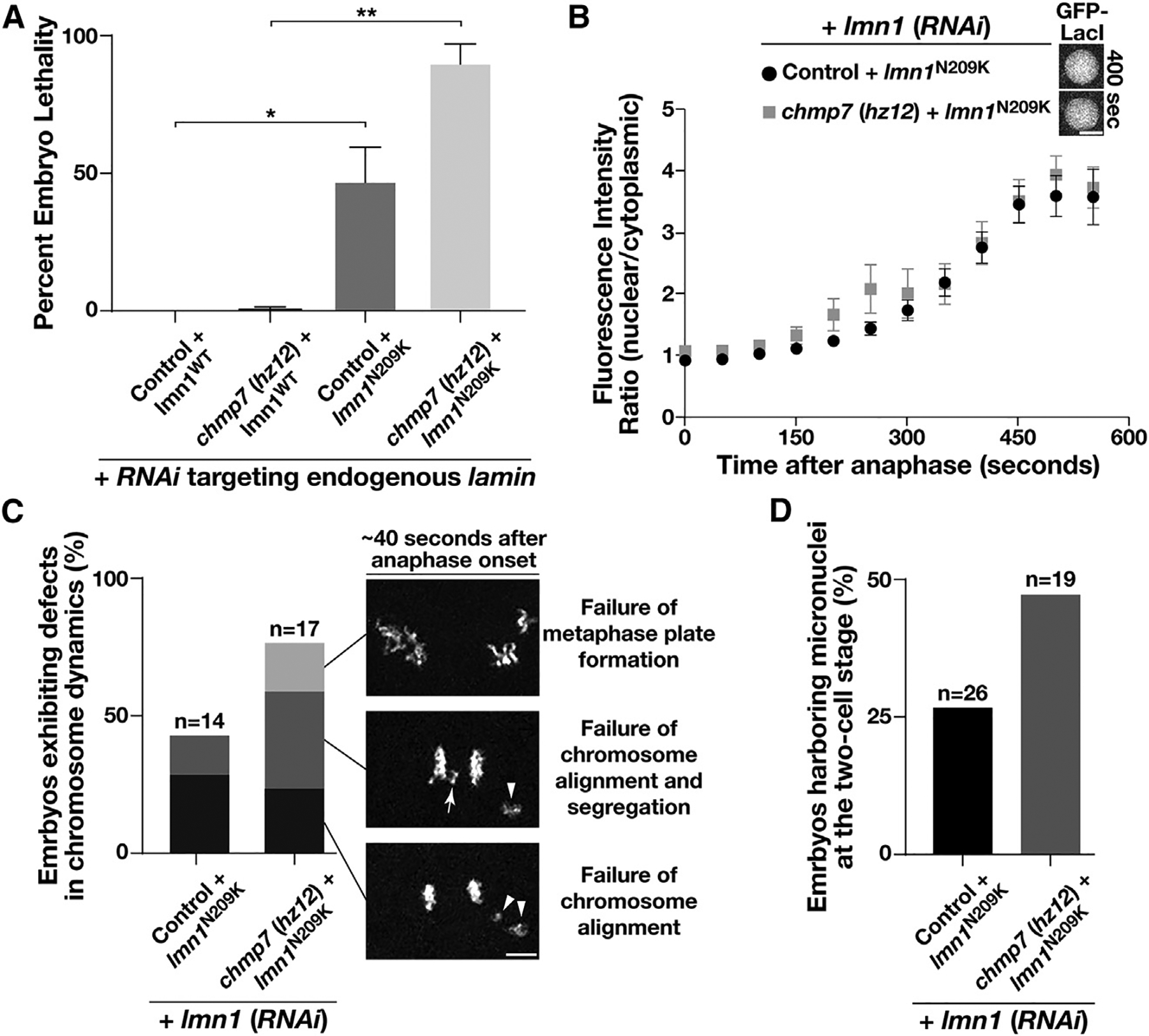
(A) Control and chmp7 (hz12) mutant animals expressing a RNAi-resistant form of lamin (either wild-type or mutant N209K) were depleted of endogenous lamin. After 48 h, hermaphrodites were moved to individual plates and allowed to lay eggs. The percentage of embryos that failed to hatch in each case was calculated, and all experiments were repeated at least two times. Error bars represent mean ± SEM. **p < 0.01 and *p < 0.05, compared with controls using an ANOVA with Tukey’s multiple comparison test.
(B) The relative nuclear versus cytoplasmic fluorescence ratio of a GFP fusion to LacI was measured at different timepoints following anaphase onset in control and mutant embryos as shown. Error bars represent mean ± SEM. No statistically significant differences were observed at any time point. Bar, 5 μm.
(C) Chromosome dynamics were analyzed in control and chmp7 (hz12) mutant embryos expressing an mCherry fusion to histone H2B and a RNAi-resistant mutant form of lamin (N209K) following depletion of endogenous lamin. Images shown are representative of the major defects identified, the frequencies of which are highlighted in the bar graph. Bar, 5 μm.
(D) The percentages of two-cell stage embryos harboring micronuclei following the first mitotic division were calculated for control and chmp7 (hz12) mutant animals expressing the mutant, RNAi-resistant form of lamin (N209K) following depletion of endogenous lamin.
See also Figure S1.
Beyond providing rigidity to the NE, the nuclear lamina has also been shown to regulate chromatin dynamics and promote organization of the genome. In C. elegans, RNA interference (RNAi)-mediated depletion of lamin leads to chromosome misalignment and missegregation during mitosis, resulting in embryo lethality (100%) (Liu et al., 2000). Animals expressing lmn1N209K also exhibited partial defects in mitotic chromosome dynamics, which correlates precisely with the level of embryo lethality we determined (Figure 2C) (Penfield et al., 2018). Strikingly, the additional absence of CHMP7 exacerbated mitotic chromosome alignment and segregation defects in animals expressing lmn1N209K (Figure 2C), leading to the accumulation of micronuclei in two-cell stage embryos more frequently (Figure 2D). Additionally, lmn1N209K mutant nuclei lacking CHMP7 appeared more elongated as compared with controls, particularly during migration ahead of the first mitotic division, consistent with a worsened defect in nuclear structure (Figure S1F). Together, these data suggest that CHMP7 possesses additional functions at the NE beyond regulating NE sealing.
CHMP7 acts at the INM in C. elegans embryos
Our data suggesting that there are coordinated roles for CHMP7 and the nuclear lamina to maintain genome integrity support a model in which both factors act within the nucleus. However, in mammalian tissue culture cells, overexpressed GFP-tagged CHMP7 localizes to the ER during most of the cell cycle and only transiently associates with the reforming NE after anaphase onset (Gu et al., 2017; Olmos et al., 2016; Vietri et al., 2020a). Similarly, when overexpressed in yeast, GFP-Chm7 localizes largely within the cytoplasm and is actively exported from the nucleus under steady-state conditions (Webster et al., 2016). To determine the localization of native C. elegans CHMP7 during early development, we immunostained embryos using our affinity-purified antibodies and demonstrated that it accumulates specifically at the NE throughout interphase (Figure 3A) despite the presence of a conserved nuclear export signal within its carboxyl-terminus (Figure S1G). To further refine our understanding of its distribution in living embryos, we used CRISPR-Cas9-mediated genome editing to append a GFP tag onto the amino-terminus of endogenous CHMP7. Live cell imaging revealed an identical distribution as our immunostaining studies showed but also allowed us to demonstrate that CHMP7 transiently leaves the NE during NE breakdown (~180 s prior to anaphase onset) and associates in part with the ER until NE reassembly initiates (~45–50 s after anaphase onset) and the NE reseals (Figures 3B and 3C). These data indicate that CHMP7 paralogs exhibit identical distributions in C. elegans embryos and human tissue culture cells during mitosis, transitioning from the broad ER network to the NE. However, during late telophase and the subsequent interphase, CHMP7 remains associated with the NE in C. elegans, while it returns to the ER in human cells.
Figure 3. CHMP7 localizes to the inner nuclear membrane in a manner that depends upon multiple LEM domain proteins.
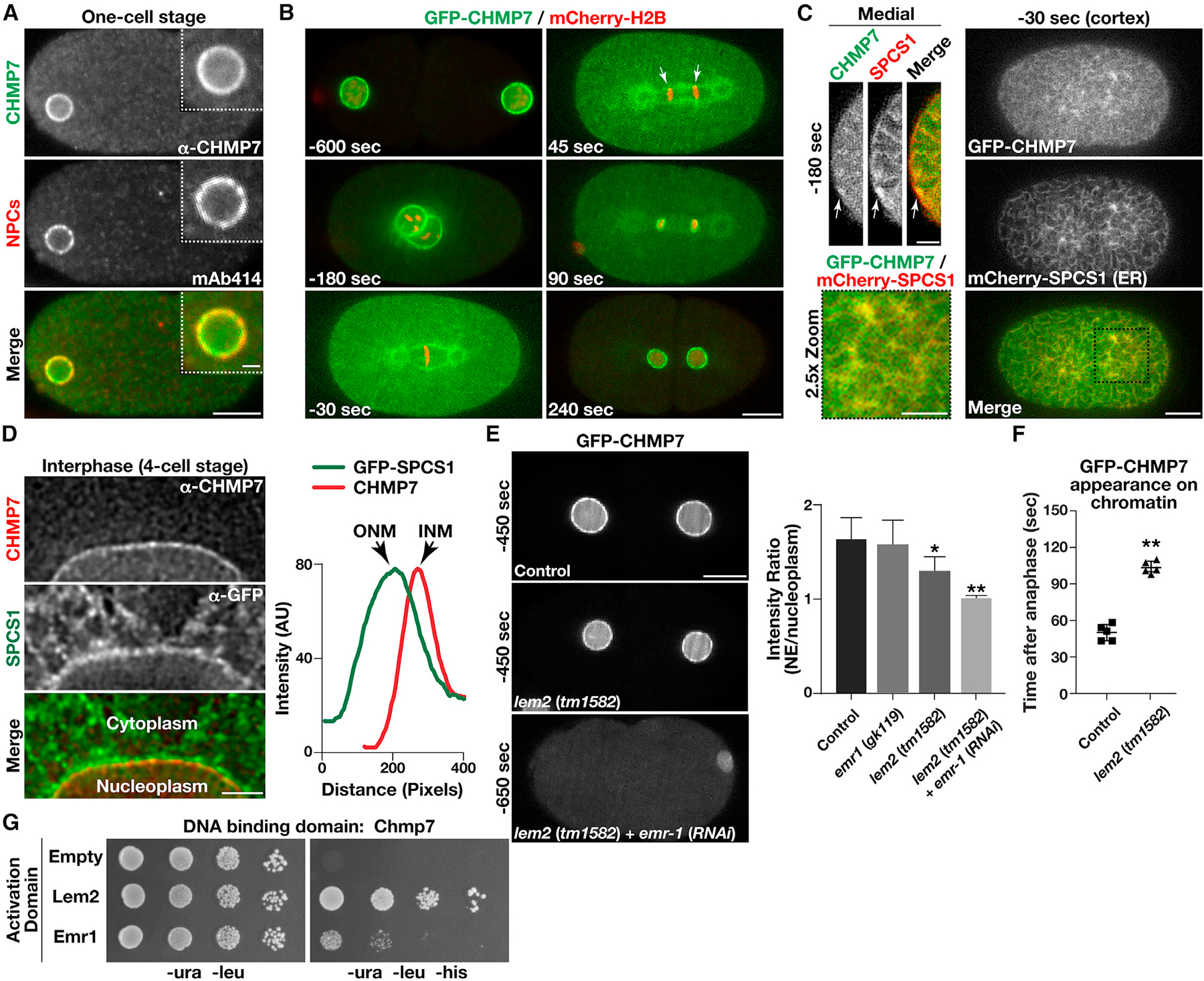
(A) Wild-type embryos were fixed and stained using antibodies directed against CHMP7 (green) and nuclear pore complexes (NPCs; recognized by monoclonal antibody mAb414; red) and imaged using confocal microscopy. Images are representative of more than 10 embryos analyzed. Bar, 10 μm; inset bar, 2 μm.
(B) Genome-edited embryos expressing native levels of a GFP fusion to CHMP7 (green) and a mCherry fusion to histone H2B (red) were imaged using time-lapse confocal microscopy (n = 10 animals). Time relative to anaphase onset is shown (bottom left of each image). Arrows highlight the initial concentration of CHMP7 on chromatin discs following anaphase onset based on line scan analysis. Bar, 10 μm.
(C) Genome-edited embryos expressing native levels of a GFP fusion to CHMP7 (green) and a mCherry fusion to the ER protein SPCS1 (red) were imaged using confocal microscopy (n = 10 animals). Representative cortical (right and bottom left) and medial (top left) sections are shown (time relative to anaphase onset). Bar (cortex), 10 μm; bar (medial), 5 μm; bar (2.5× zoom), 5 μm.
(D) Embryos expressing a GFP fusion to SPCS1 (ER marker) were fixed and stained using antibodies directed against CHMP7 (red) and GFP (green) and imaged using STED microscopy. Images are representative of more than 10 embryos analyzed. Line scan analysis was used to define the distribution of CHMP7 on the inner nuclear membrane (INM) relative to SPSC1, which is present on the outer nuclear membrane (ONM). Bar, 2 μm.
(E) Genome-edited embryos expressing native levels of a GFP fusion to CHMP7 (green) in the presence or absence of LEM domain proteins were imaged using confocal microscopy (left). Times indicated are relative to anaphase onset. The ratio of fluorescence intensities at the nuclear membrane as compared with the nucleoplasm is shown (right; prior to pronuclear meeting and before nuclear envelope (NE) structure is severely perturbed). Error bars represent mean ± SEM. **p < 0.01 and *p < 0.05, as compared with control, using an ANOVA with Dunnett’s test. Bar, 10 μm.
(F) The timing of CHMP7 accumulation on chromatin after anaphase onset is shown in control embryos and embryos lacking LEM2. Error bars represent mean ± SEM. **p < 0.01, as compared with control, using a t test.
(G) Yeast co-expressing plasmids encoding CHMP7 (bait fusion) and two distinct prey constructs were plated (10-fold dilutions, left to right) on either selective (−ura, −leu, −his) or histidine-supplemented medium for 72 h (n = 3).
See also Figures S1 and S2.
To determine whether CHMP7 associates with the ONM or the INM during interphase, we conducted a series of super-resolution imaging studies, leveraging stimulated emission depletion (STED) microscopy. Using animals expressing a GFP fusion to signal peptidase (SPCS1), which is concentrated specifically on the ONM and ER (Poteryaev et al., 2005), we co-immunostained embryos at various stages of development with antibodies directed against endogenous CHMP7. In all cases, line scan analysis clearly demonstrated a juxtaposed localization of SPCS1 relative to CHMP7 around the nuclear periphery, with CHMP7 distributed on the inner face of the NE (Figure 3D). Consistent with these data, STED imaging also revealed a precise co-localization of CHMP7 with a functional GFP fusion to lamin, which decorates the INM (Figure S1H).
Previous work has highlighted a role for the INM protein LEMD2 in recruiting CHMP7 to the NE during late anaphase (Gu et al., 2017). To determine the role of the C. elegans LEMD2 paralog (LEM2) in regulating CHMP7 localization, we examined the distribution of GFP-CHMP7 in the tm1582 mutant, which introduces a large deletion into LEM2, eliminating the majority of exons 1 and 2 and causing a frameshift (Barkan et al., 2012). The resulting allele is predicted to produce only the first 21 amino acids of LEM2, truncating more than 95% of the protein. Using quantitative fluorescence imaging in one-cell stage embryos, we found that the absence of LEM2 resulted in a decline in CHMP7 accumulation at the NE as compared with control embryos prior to NE breakdown (Figure 3E). Moreover, we found that the recruitment of CHMP7 to the reforming NE during anaphase was delayed significantly, but not eliminated, in the absence of LEM2. Unlike control embryos in which CHMP7 and LEM2 both arrive at NE membranes ~45 s after anaphase onset, embryos lacking LEM2 accumulated CHMP7 at the NE ~105 s after anaphase onset (Figure 3F). In contrast, deletion of CHMP7 did not have an impact on the timing of LEM2 recruitment to the NE after anaphase (Figure S2A). Remarkably, the timing of CHMP7 accumulation in lem2 knockout embryos correlated precisely with the normal timing of C. elegans emerin (EMR1) accumulation at the NE during its reformation (Figure S2B). To determine whether EMR1 may function with LEM2 to recruit CHMP7 onto the INM, we first measured the fluorescence intensity of NE-localized GFP-CHMP7 in embryos harboring a null allele of EMR1 (gk119). These studies revealed that EMR1 plays a relatively modest role in recruiting CHMP7 as compared with LEM2 (Figure 3E). However, following RNAi-mediated depletion of EMR1 in mutant embryos lacking LEM2, which was sufficient to result in penetrant embryo lethality, we found a dramatic reduction in CHMP7 localization to the NE (Figures 3E and S2C). These data strongly suggest partially redundant functions for EMR1 and LEM2 in recruiting CHMP7 to the INM, although their affinities appear distinct. This difference may be a consequence of some LEM2 paralogs potentially harboring multiple CHMP7-binding regions both in their carboxyl-terminal winged-helix (WH) domains (Gu et al., 2017) and in their amino-termini (Webster et al., 2014, 2016), while EMR1 lacks a WH domain. Despite full-length EMR1 and LEM2 not being amenable to biochemical purification, we successfully demonstrated associations between CHMP7 and both LEM domain proteins using yeast 2-hybrid studies, further supporting a model in which CHMP7 exploits multiple factors to associate with the INM (Figure 3G). Notably, CHMP7 also harbors a conserved hydrophobic motif near its amino-terminus, which was shown previously to mediate direct membrane association (Olmos et al., 2016), potentially accounting for residual binding of CHMP7 with the NE following inhibition of LEM2 and EMR1 (Figure S2D).
The ESCRT machinery regulates INM morphology
Our finding that CHMP7 localizes to the INM during interphase raises the intriguing possibility that it may function there to facilitate ESCRT-mediated membrane remodeling after NE resealing is complete. To explore this possibility, we first used animals expressing a functional, GFP-tagged form of lamin to examine INM morphology during NE reformation and expansion that accompanies the first C. elegans mitotic division (Link et al., 2018). In control embryos, we found that lamin begins to concentrate on segregated chromatin discs as cleavage furrow ingression initiates (starting ~50 s after anaphase onset), nearly equivalent to the timing of CHMP7, LEM2, and Nup54/NPP1 accumulation at the reforming NE (Figures 4A and S2E). Over the subsequent ~300 s, the NE expanded dramatically, increasing in surface area by approximately 4-fold. During early phases of this expansion process, we reproducibly identified numerous invaginations emanating from the INM (greater than 400 nm in length) that penetrated the nucleoplasm in control embryos (Figures 4A and 4B). However, the presence of these invaginations declined significantly as NE expansion proceeded (Figure 4B). By contrast, in the absence of CHMP7, the presence of lamin-positive NE invaginations persisted throughout NE expansion, continuing even after the NE reached it maximal size in two-cell stage embryos (Figures 4B and 4C). Using electron tomography, we analyzed NE morphology, revealing numerous small (on average, 336 ± 84 nm in length) and narrow (25 ± 3 nm in width) invaginations, which decorated the INM in control embryos, while the ONM exhibited a relatively smooth topology (Figure 4D). In animals lacking CHMP7, INM tubulation was exacerbated, highlighted by INM tubules that were up to nearly 1 μm in length and 34 nm (±5 nm) wide (Figure 4E). These data indicate that the ESCRT machinery plays a role in regulating INM morphology, akin to a quality control mechanism.
Figure 4. Inhibition of ESCRT-III function leads to increased persistence of nuclear membrane invaginations.
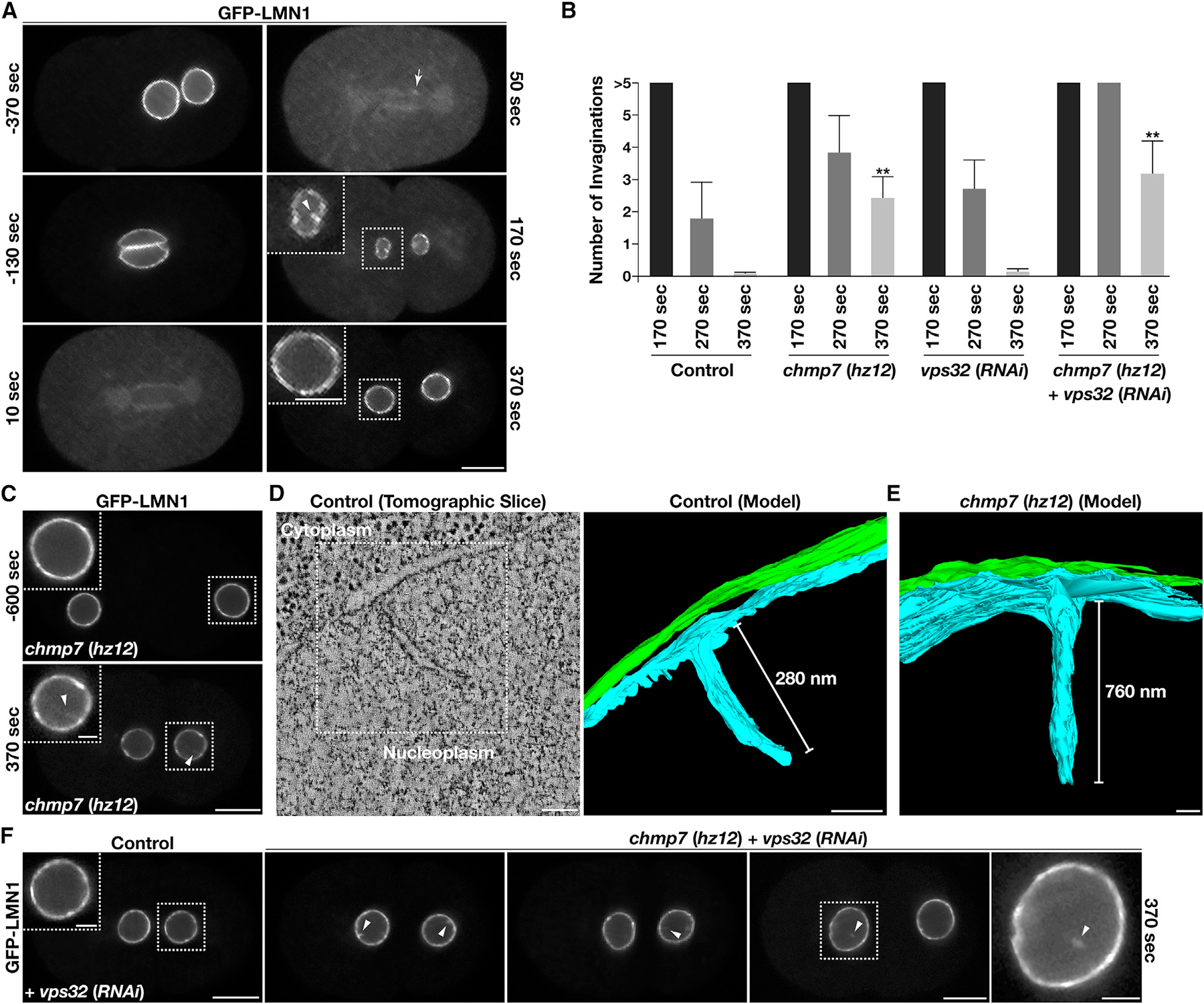
(A) Embryos expressing a functional GFP fusion to lamin were imaged using time-lapse confocal microscopy (n = 10 animals). Time relative to anaphase onset is shown. An arrow highlights the initial concentration of lamin on chromatin discs following anaphase onset based on line scan analysis, and an arrowhead indicates a nuclear membrane invagination that forms normally during NE reformation and expansion. Bar, 10 μm; inset bar, 5 μm.
(B) The number of nuclear membrane invaginations was quantified during NE expansion in control two-cell stage embryos and embryos lacking components of the ESCRT machinery. Time shown is relative to anaphase onset. Error bars represent mean ± SEM. **p < 0.01, as compared with control at an identical time point after anaphase onset, using an ANOVA with Dunnett’s test.
(C–F) Embryos expressing a functional GFP fusion to lamin in the presence or absence of specific ESCRT-III components were imaged using time-lapse confocal microscopy (n = 10 animals each at a minimum). Time relative to anaphase onset is shown. Arrowheads indicate nuclear membrane invaginations or severed lamin-positive nuclear membrane tubules that persist during NE reformation and expansion. Bars, 10 μm; boxed inset bars, 2 μm.
(D and E) Representative nuclear invaginations identified in control embryos and embryos lacking CHMP7 were imaged using electron tomography (more than 5 embryos examined each). An individual slice and reconstructed models (bars, 100 nm) are shown.
See Figure S2.
We also tested the impact of depleting the core ESCRT-III subunit Vps32/CHMP4 on NE morphology. To our surprise, Vps32/CHMP4 knock down that was sufficient to cause 100% embryo lethality failed to impact INM quality control as compared with control embryos (Figure 4B). However, depletion of Vps32/CHMP4 in animals lacking CHMP7 resulted in more frequent INM invaginations, as observed by fluorescence light microscopy, many of which appeared fragmented (12 out of 27 invaginations examined using multiplane imaging), resulting in the accumulation of untethered, lamin-positive tubules within the nucleoplasm (Figure 4F). Depletion of the Vps4 ATPase was also sufficient to cause an increased number of INM invaginations, which were further exacerbated in size and frequency by the additional loss of CHMP7 (Figures S2F and S2G). Together, these data suggest that multiple ESCRT components function in parallel to restrict the prolonged presence of INM invaginations.
Since weakening of the nuclear lamina is incompatible with the viability of embryos lacking CHMP7 and results in penetrant chromosome segregation defects, we additionally determined how NE morphology was affected under these conditions. For this work, we examined the distributions of two integral membrane proteins, SPCS1 to mark the ONM and SUN1, a component of the linker of nucleoskeleton and cytoskeleton (LINC) complex, to mark the INM (Malone et al., 2003). Based on diffraction-limited confocal imaging, one-cell stage embryos expressing lmn1N209K failed to exhibit nucleoplasmic membrane tubules that harbored either marker (Figures 5A and 5B). When CHMP7 was additionally eliminated however, we reproducibly identified SUN1-positive NE invaginations (Figure 5B). In contrast, SPCS1 was not found on membrane tubules penetrating the nucleoplasm under these conditions, suggesting that the invaginations form specifically from the INM (Figure 5A). These studies contrast our prior work highlighting the impact of excess phosphatidylinositol (PI) synthesis, which leads to the invasion of ER membranes into the nucleoplasm (Penfield et al., 2020). To further confirm this distinction, we depleted enzymes that mediate PI production in animals lacking either CHMP7 or LEM2 and demonstrated that stabilization of nuclear membrane invaginations resulting from impaired ESCRT function was unaffected (Figure S2H).
Figure 5. ESCRT-III specifically remodels the INM and promotes the formation of intranuclear vesicles.
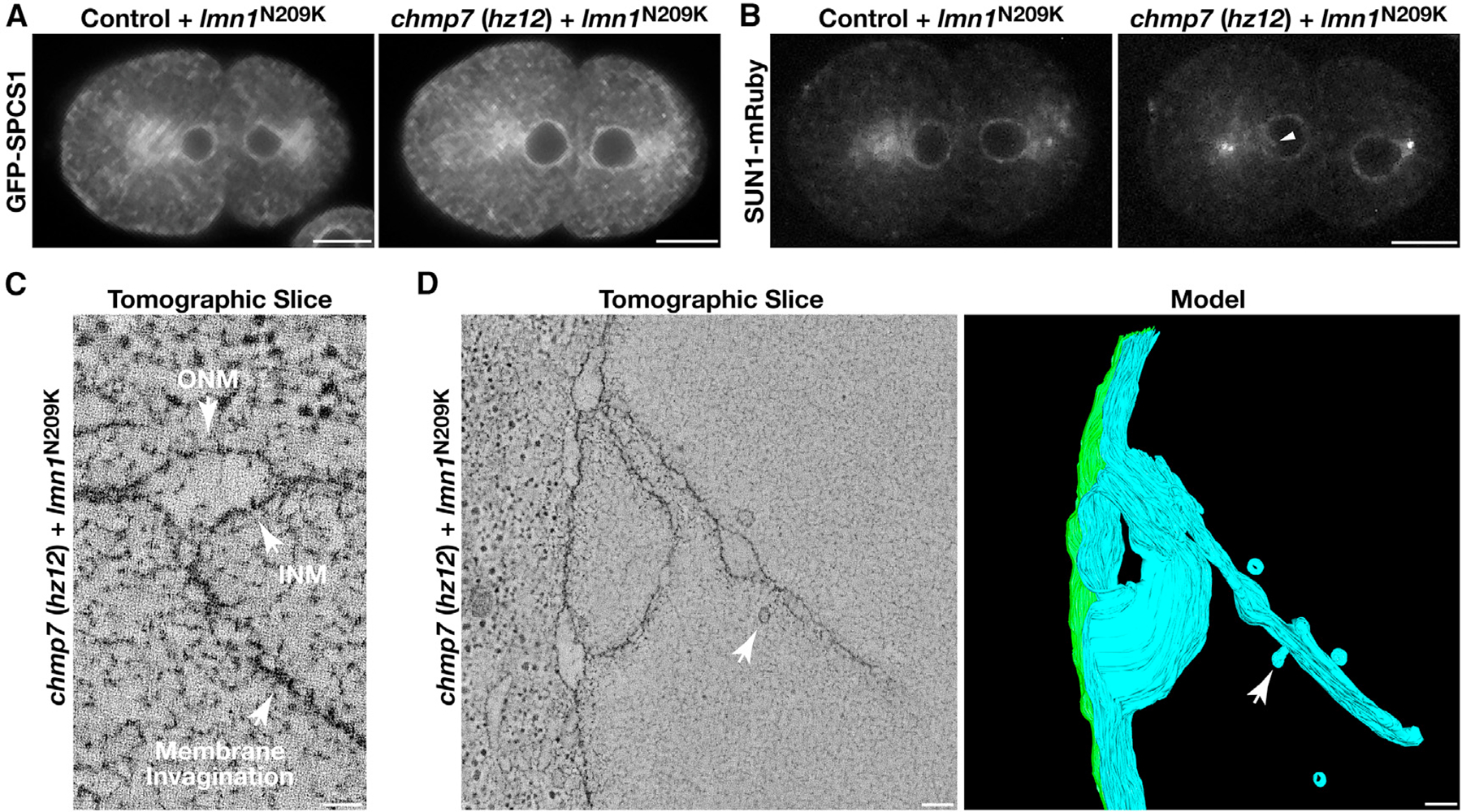
(A and B) Control and chmp7 (hz12) null embryos expressing a GFP fusion to SPCS1 (A) or a mRuby fusion to SUN1 (B) and the mutant, RNAi-resistant form of lamin (N209K) were imaged using confocal microscopy 370 s after anaphase onset following depletion of endogenous lamin (n = 10 embryos each). An arrowhead indicates an INM invagination (B). Bars, 10 μm.
(C and D) Representative INM invaginations identified in embryos lacking CHMP7 and expressing a mutant, RNAi-resistant form of lamin (N209K) were imaged using electron tomography following depletion of endogenous lamin (more than 5 embryos examined). Individual slices (bars, 50 nm in C and 100 nm in D) and a reconstructed model (bar, 100 nm) are shown. Arrows in (D) highlight the formation of intranuclear vesicles from membrane invaginations.
To better understand the impact to NE morphology in mutant animals that lack CHMP7 and express lmn1N209K as their sole source of lamin, we conducted a series of electron-tomography-based studies, which highlighted the presence of complex networks of membrane tubules emanating specifically from the INM (Figures 5C and 5D). Moreover, we reproducibly identified nascent budding structures and intranuclear vesicles approximately 55.4 nm (±2.6) in diameter at the termini of multiple tubules, suggesting that impaired ESCRT function results in kinetically delayed vesiculation of INM invaginations (Figure 5D; Video S1). These data strongly suggest that the ESCRT machinery remodels the INM to regulate its morphology.
Multiple ESCRT-III subunits act redundantly at the INM to facilitate nuclear maintenance
To determine which components of the late-acting ESCRT machinery function at the INM, we leveraged an assay developed previously in which the NE is acutely perturbed by partial RNAi-mediated depletion of lamin (Penfield et al., 2018). Under these conditions, LEM2 hyperaccumulates at one or more of the subdomains of the NE, promoting local ESCRT-III recruitment. Using our toolbox of reagents directed against the endogenous ESCRT-III subunits, we systematically demonstrated that CHMP1/Did2, CHMP2/Vps2, CHMP3/Vps24, CHMP4/Vps32, CHMP5/Vps60, CHMP7, Ist1, and Vps4 are recruited to LEM2-positive structures at the NE (Figures 6A and 6B). In contrast, we failed to find evidence for CHMP6/Vps20 accumulation at these sites, consistent with the idea that CHMP7 substitutes for it in promoting ESCRT-III complex assembly at the NE. In all cases, hyperaccumulation of ESCRT-III subunits at the NE was at least partially dependent upon the presence of CHMP7, emphasizing its importance to downstream ESCRT-III assembly (Figures 6B, 6C, and S3A). Notably, embryos lacking CHMP7 failed to accumulate LEM2 at NE subdomains to a similar extent as found in control embryos (Figure 6D). These data suggest that the ESCRT machinery and LEM domain proteins are at least partially interdependent for their localized accumulation at the NE under reduced levels of lamin expression. In addition, similar to the impact of inhibiting CHMP7, partial lamin depletion also results in an increased frequency of NE invaginations (Figure S3B), and we found that components of the ESCRT machinery associated with these nucleoplasmic membrane tubules, consistent with their function in membrane remodeling within the nucleus (Figures 6A and 6E).
Figure 6. ESCRT-III accumulates on INM invaginations.
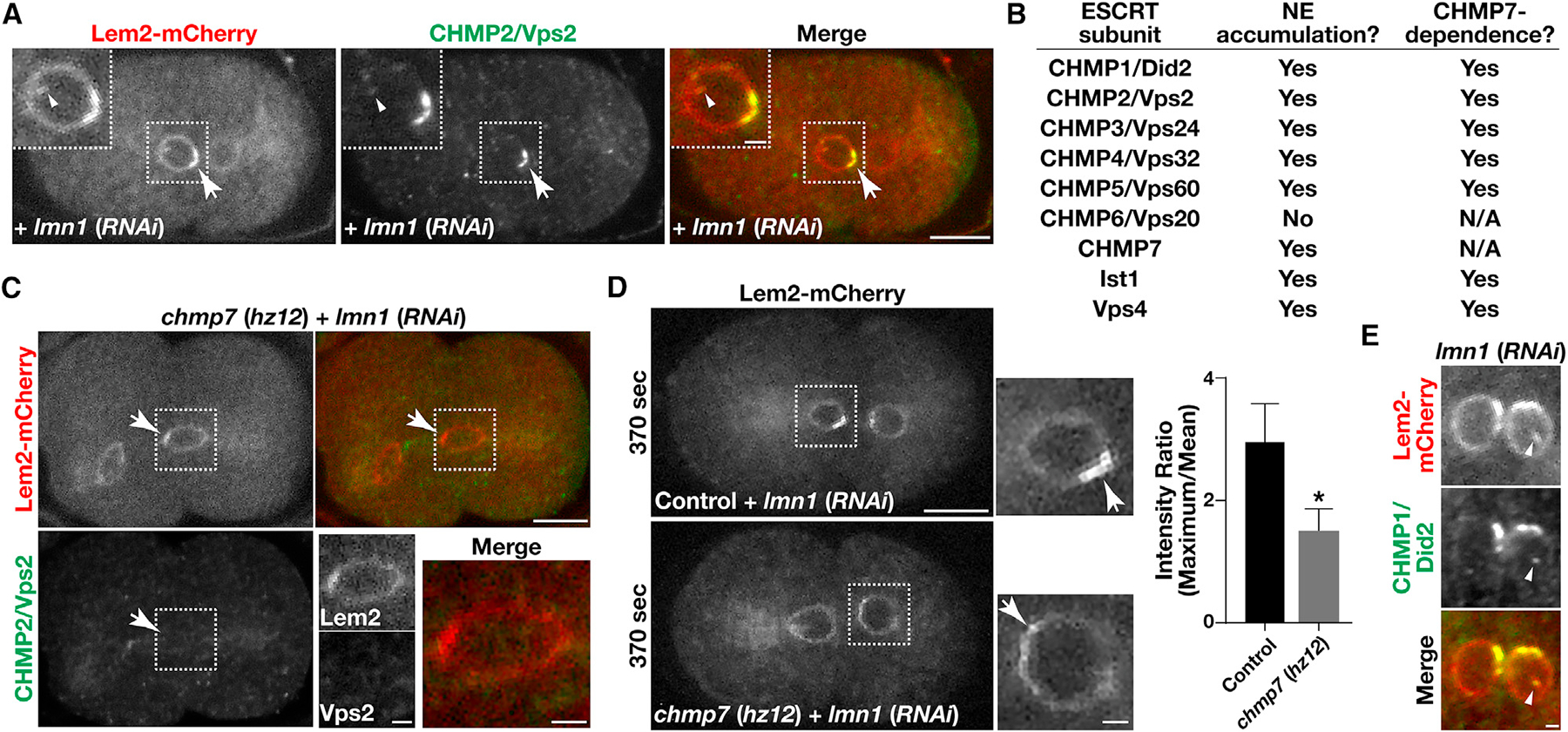
(A, C, and E) Embryos expressing a mCherry fusion to LEM2 (red) were fixed and stained using antibodies directed against CHMP2/Vps2 (green; A and C) or CHMP1/Did2 (green; E) and imaged using confocal microscopy following partial depletion of lamin in the presence (A and E) or absence (C) of CHMP7. Images are representative of more than 10 embryos analyzed. Bars, 10 μm (A and C) or 2 μm (E); inset bars, 2 μm.
(B) A summary highlighting the ESCRT-III components that accumulate on the NE together with LEM2 and their dependence on CHMP7.
(D) Embryos expressing a mCherry fusion to LEM2 were imaged live using confocal microscopy 370 s after anaphase onset, following partial depletion of lamin in the presence and absence of CHMP7. Images are representative of more than 10 embryos analyzed. Bar, 10 μm; inset bar, 2 μm. Hyperaccumulation of LEM2 on NE subdomains was quantified under both conditions (right). Error bars represent mean ± SEM. *p < 0.05, as compared with control, using a t test.
See also Figure S3.
Based on the large number of ESCRT-III subunits involved in INM quality control and their potential for redundancy in membrane remodeling processes, we questioned whether the relatively subtle phenotypes associated with the deletion of CHMP7 may result from the existence of redundant mechanisms underlying ESCRT-III complex assembly. To test this idea, we first used yeast 2-hybrid studies to screen for additional components of the ESCRT machinery that could associate with the LEM domain proteins LEM2 and EMR1. In addition to CHMP7 and Vps32/CHMP4, which have both been previously shown to associate with LEM2 homologs in other systems (Gu et al., 2017; Webster et al., 2016), we found that Vps60/CHMP5 also bound to both LEM domain proteins, while none of the other ESCRT-III subunits exhibited this capability (Figures 7A and S3C). Additionally, yeast 2-hybrid studies showed that Vps60/CHMP5 binds to CHMP7, similar to Vps32/CHMP4 (Figures 7A and S3C), and appears to be highly promiscuous in binding other ESCRT-III subunits (Figure S3D), consistent with previous immunoprecipitation studies performed from yeast extracts (Heinzle et al., 2019).
Figure 7. Inhibition of ESCRT-III function leads to an increase in germline apoptosis.
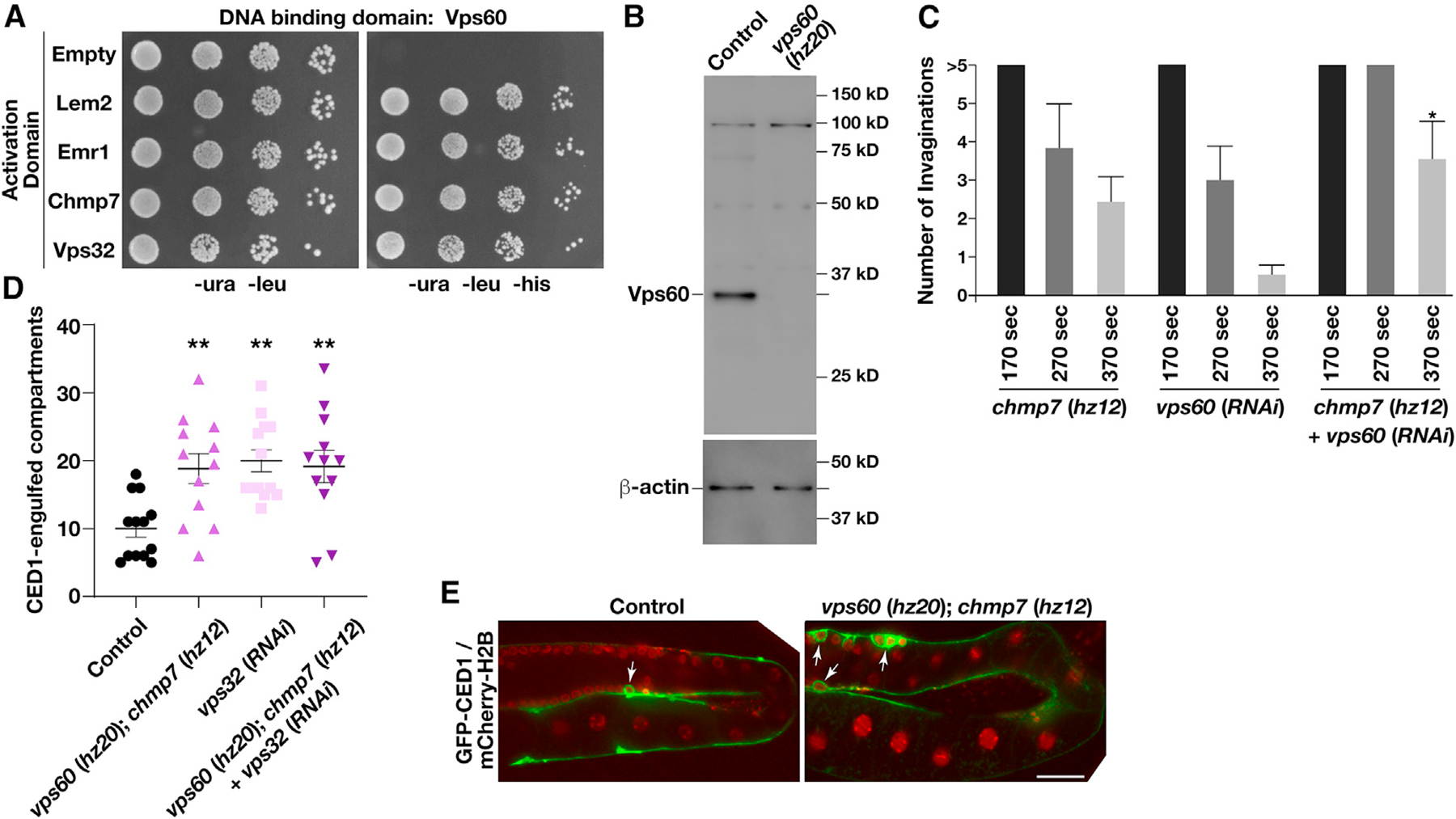
(A) Yeast co-expressing plasmids encoding VPS60 (bait fusion) and different prey constructs as indicated were plated (10-fold dilutions, left to right) on either selective (−ura, −leu, −his) or histidine-supplemented medium for 72 h (n = 2). In all cases, constructs were shown not to promote auto-activation.
(B) Representative immunoblot of extracts generated from control and vps60 (hz20) mutant animals (n = 2 each) using antibodies directed against VPS60 (top) and beta-actin (bottom; load control).
(C) The number of nuclear membrane invaginations was quantified during NE expansion in embryos lacking CHMP7 and/or VPS60. Time shown is relative to anaphase onset. Error bars represent mean ± SEM. *p < 0.05, as compared with single mutants at an identical time point after anaphase onset, using an ANOVA with Dunnett’s test.
(D and E) The germlines of animals co-expressing a GFP fusion to CED1 and a mCherry fusion to histone H2B were imaged in the presence and absence of different ESCRT-III subunits. Quantification of the number of CED1-positive compartments (D) and representative images (E) are shown. Error bars represent mean ± SEM. **p < 0.01, as compared with control, using an ANOVA with Tukey’s test. Bar, 50 μm.
See also Figure S3.
To determine the role of Vps60/CHMP5 at the NE, we first used CRISPR-Cas9 to incorporate an indel just downstream of its initiation codon, causing an early frameshift predicted to result in truncation of the protein immediately upstream of its first helical domain (Figure S3E). Immunoblot analysis confirmed the absence of Vps60/CHMP5 at the protein level in knockout animals (Figure 7B). Similar to one-cell stage embryos lacking CHMP7, we failed to identify a function for Vps60/CHMP5 in cargo sorting at MVEs based on the trafficking and degradation rate of caveolin-1, and elimination of Vps60/CHMP5 did not dramatically impact embryo viability or brood size relative to control animals (Figures S3F and S3G). To determine whether Vps60/CHMP5 acts redundantly with other components of the ESCRT machinery, we depleted it in animals lacking CHMP7 and created a double mutant lacking both Vps60/CHMP5 and CHMP7. These animals were fertile and continued to produce viable progeny, although we consistently observed an increased frequency of NE invaginations (Figures 7C and S3B), similar to that observed in animals lacking CHMP7 and depleted of Vps32. Additionally, we identified a significant increase in germline apoptosis, as indicated by an elevated number of compartments encircled by the engulfment protein CED1/SREC (Figures 7D and 7E). Inhibition of Vps32/CHMP4 resulted in a similar increase in germline apoptosis, which was not further exacerbated by the additional removal of Vps60/CHMP5 and CHMP7 (Figure 7D). These data suggest that all three ESCRT-III subunits function in a common pathway to regulate germline apoptosis. In contrast, depletion of Vps20/CHMP6, which functions on endosomes but not the NE, failed to increase the frequency of germline apoptosis as compared with controls (Figure S3H). Taken together, these data further indicate that the ESCRT machinery plays a critical function to ensure the proper maintenance of nuclear architecture.
DISCUSSION
The human ESCRT-III components are sometimes referred to as chromatin-modifying proteins (CHMPs), based largely on the identification of CHMP1/Did2 as a putative binding partner for Polycomb-group protein Polycomblike (Pcl), a factor important for gene silencing during development (Stauffer et al., 2001). Subsequently, another screen implicated both CHMP1/Did2 and CHMP5 in chromatin remodeling, but these studies provided limited mechanistic insights into how the ESCRT machinery contributes to chromatin organization (Tsang et al., 2006). Most recently, ESCRT-III and Vps4 were implicated in resolving attachments between heterochromatin and a LEM2 complex in fission yeast by promoting the disengagement of CHMP7 from LEM2 (Pieper et al., 2020). As release of membrane-chromatin contacts has been demonstrated to be essential for normal chromosome segregation during mitosis, these findings provide one possible explanation for genomic instability that arises following inhibition of ESCRT function (Champion et al., 2019). Other work has suggested that a complex of CHMP1B and Ist1 can associate directly with nucleic acids, although it remains unclear whether this charge-based interaction holds physiological relevance in cells (Talledge et al., 2018).
Our findings support a distinct model to describe how the ESCRT machinery regulates chromatin organization, which does not rely on a direct association between these membrane remodeling factors and DNA or DNA-associated proteins. Instead, through its ability to promote the vesiculation and clearance of nucleoplasmic membrane invaginations, ESCRT-III complexes together with Vps4 to ensure the normal topology of the INM, which is critical for heterochromatic gene regulation (Buchwalter et al., 2019). Our work is consistent with a model in which INM tubules penetrate persistently into the nuclear interior in the absence of ESCRT function. As the membrane tubules contain factors that bind and organize heterochromatic DNA, this abnormal topology may disrupt the normal separation of heterochromatic and euchromatic DNA. This impact is further exacerbated under conditions where the nuclear lamina is weakened. Based on prior work, such a disruption to chromatin organization may impact its ability to compact properly during mitosis, leading to defects in chromosome segregation and cell death (Batty and Gerlich, 2019). Notably, inhibition of ESCRT-III or Vps4 has also been shown to affect centrosomes, causing the accumulation of multipolar or monopolar spindles, which would further contribute to chromosome missegregation during mitosis (Morita et al., 2010). Together, our studies combined with previous work in the field highlight the multiple integral roles that the ESCRT machinery plays to maintain genome integrity. Alternatively, as loss of ESCRT function causes significant perturbations to nuclear membrane morphology, especially when combined with impaired nuclear lamina assembly, the effect on chromatin organization may be indirect. Instead, the irregular INM topology following inhibition of the ESCRT machinery may impact the ability of the NE to withstand dynein-mediated pulling forces required to position nuclei during zygotic development (Penfield et al., 2018), which could promote increased chromosome missegregation observed during the first embryonic division.
Despite these important nuclear functions, the major ESCRT-III nucleator at the NE, CHMP7, is not essential for development or viability in C. elegans, unlike its endosomal counterpart Vps20/CHMP6 (Frankel et al., 2017). This finding strongly suggests a redundancy in processes regulated by the ESCRT machinery at the NE, including resealing and INM remodeling. In particular, several LEM-domain-containing proteins have been implicated in resolving disruptions to the NE, and their collective inhibition has been shown to strongly perturb NE architecture (Halfmann et al., 2019). In contrast, loss of individual ESCRT-III components, including the key scaffolding protein Vps32/CHMP4 that plays a central role in all other ESCRT-mediated membrane remodeling events (Vietri et al., 2020a), results in relatively minor impacts to the NE in embryos. Only upon co-depletion of multiple ESCRT-III subunits did we identify severe consequences to NE morphology, which were limited to the INM. These data highlight an important contrast between INM remodeling during embryogenesis, in which different ESCRT-III subunits likely act in a partially redundant manner, and intralumenal vesicle (ILV) formation at endosomes that requires non-redundant activities of multiple individual ESCRT-III components (Frankel et al., 2017). Topologically, membrane scission at these two sites occurs in opposite manners. At the INM, the ESCRT machinery likely functions at the outer surface of membrane tubules, akin to the localization of dynamin during endocytosis from the plasma membrane, while ILV formation requires ESCRT activity on the inner surface of a bud neck (Vietri et al., 2020a; Mettlen et al., 2018). The differing complexities associated with these unique membrane rearrangements likely necessitate distinct requirements for ESCRT-III assembly and activity, with ILV formation imposing stricter demands for polymer nucleation and dynamic restructuring to ultimately achieve the close membrane apposition needed for spontaneous vesicle fission. In contrast, since a majority of ESCRT-III subunits, including CHMP1, CHMP2, CHMP3, CHMP4, CHMP7, and Ist1, exhibit an ability to oligomerize on bent membranes and alter bilayer organization (McCullough et al., 2015; Pfitzner et al., 2020; Nguyen et al., 2020; von Filseck et al., 2020; von Appen et al., 2020; Saksena et al., 2009; Mierzwa et al., 2017), our findings suggest that they can act redundantly to facilitate INM tubule vesiculation. Given their overlapping roles, it is perhaps unsurprising that the activities of ESCRT-III complexes on the INM have been overlooked until now. Notably, our data further support a role for Vps60/CHMP5 at the INM, likely via an ability to polymerize on membranes and facilitate scission, although this remains to be demonstrated.
In contrast to embryonic cells, which are constantly undergoing cycles of NE assembly and disassembly, germline nuclei that share a common syncytium and only divide in the mitotic distal tip appear highly vulnerable to the loss of nuclear ESCRT-III activity. One explanation for this finding relates to the susceptibility of pachytene-stage germ cells to undergo DNA-damage-induced apoptosis, unlike somatic cells (Gartner et al., 2000). The absence of even a single nuclear ESCRT-III subunit such as Vps32/CHMP4 may increase the likelihood of genotoxic stress within the germline either due to an impaired ability to reseal the NE in response to damage that occurs normally within the syncytium or a defect in INM morphology that results in perturbations to heterochromatin organization (Link et al., 2018). Irrespective of the specific pathway leading to increased apoptosis, however, the large number of germ nuclei produced in C. elegans limits the overall impact of ESCRT-III inhibition on the ability to continue embryo production.
At a mechanistic level, CHMP7 has been suggested to undergo LEM2-stimulated polymer formation to promote NE sealing after its assembly following chromosome segregation (von Appen et al., 2020). In contrast, C. elegans embryos lacking CHMP7 failed to exhibit a delay in the timing of NE compartmentalization, indicating a more limited role for the ESCRT machinery during this essential process in some animal cells. Instead, under these conditions, NE invaginations into the nucleoplasm persisted, suggesting a kinetic delay in the pruning of INM invaginations by ESCRT-III during interphase, a finding that is consistent with prior work conducted using mammalian cells (Arii et al., 2018). Surprisingly, a recent study found that constitutive co-assembly of LEM2-CHMP7 complexes at the INM results in exaggerated membrane distortions that compromise nuclear integrity (Vietri et al., 2020b). However, this effect may be a consequence of CHMP7 overexpression within the nucleus, which may promote ESCRT-III mediated membrane remodeling in a non-physiological manner (Vietri et al., 2020b). Nevertheless, this finding raises the interesting question of how nuclear ESCRT-III activity is properly tuned to enable maintenance of INM topology. One possibility is that the CHMP7-LEM2 interaction is tightly regulated to prevent hyperactivity during interphase. Consistent with this idea, CHMP7 has been shown to be phosphorylated in a cell-cycle-dependent manner, which limits its ability to co-polymerize with LEM2 (Gatta et al., 2021). Additionally, CHMP7 association with phosphatidic acid and/or the ESCRT-III regulator CC2D1B may also affect its function (Thaller et al., 2021; Ventimiglia et al., 2018). Alternatively, the presence of highly bent membranes within the nucleus may be sufficient to activate the ESCRT machinery (Fyfe et al., 2011). Based on our finding that co-inhibition of CHMP7 and CHMP4 results in the accumulation of deep INM invaginations, it is unlikely that ESCRT-III activity is responsible for their formation. Instead, these tubules may form during the initial stages of NE reformation when heterochromatin and euchromatin remain intertwined and INM proteins begin to associate with heterochromatic factors scattered throughout the decondensing DNA mass (Buchwalter et al., 2019). In this model, the ESCRT machinery acts to vesiculate remaining tubules as the chromatin becomes organized within the expanding nucleus. Based on cryoelectron microscopy (cryo-EM) studies, small ER-derived intranuclear vesicles are routinely observed in mammalian cells (Hoffman et al., 2020), which may be a product of ESCRT-III activity within the nucleus. Future studies aimed at defining the origin of these vesicles will be necessary to resolve this question unambiguously.
Limitations of the study
Although our data strongly support a model in which the ESCRT machinery functions at the INM to prune membrane invaginations that form during NE reformation, the precise mechanisms by which ESCRT-III complexes promote membrane scission at this site remain unclear. Future studies leveraging high-resolution cryo-EM will likely be necessary to define the precise distribution of ESCRT-III filaments during this process. Additionally, reconstituting the interactions between CHMP7, Vps60/CHMP5, and the LEM-domain-containing proteins will help to determine how these factors co-assemble to promote efficient ESCRT-III polymer formation and membrane remodeling. Unfortunately, we were unable to biochemically purify soluble Vps60/CHMP5, LEM2, and EMR1 to resolve this question. This limits our ability to understand whether Vps60/CHMP5 polymers form at the INM during its formation and expansion after chromosome segregation. Additionally, a clear understanding of the exacerbation of mitotic chromosome alignment and segregation caused by CHMP7 deletion in the lmn1N209K mutant background remains to be defined. This is complicated by defects in NE morphology observed in the double mutant animals analyzed, leaving open questions around the mechanism by which ESCRT-III directly facilitates genome maintenance.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Anjon Audhya (audhya@wisc.edu).
Materials availability
All reagents generated in this study are available from the Lead Contact with a Material Transfer Agreement.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains were grown on Nematode Growth Media (NGM) agar plates seeded with E. coli OP50 and maintained at 20°C. Genome editing to create chmp7 (hz12), istr1 (hz16) and vps60 (hz20) deletion alleles, as well as an amino-terminal GFP fusion with endogenous CHMP7, was carried out in the Bristol strain N2 (or a derivative) as described previously (Paix et al., 2015). Alleles were combined using standard genetic methods.
METHOD DETAILS
Larval development assay
Larval development was assayed by monitoring the growth rate of individual 2-cell stage embryos through adulthood. Embryo lethality was calculated during a 24-hour period after animals began egg laying, and brood size was measured based on the sum of hatched and unhatched embryos laid during that time interval.
RNA interference (RNAi)
Templates for double stranded RNAs used in RNAi experiments were generated by PCR from N2 genomic DNA or cDNA. L4 larvae were incubated in dsRNA for 24 hours in humidified chambers and subsequently recovered for approximately 48 hours before imaging or used for brood size and lethality assays. For VPS32, EMR1, and CDGS1 depletions, worms were recovered onto plates seeded with bacteria expressing the corresponding dsRNA. For all other depletions, worms were recovered onto plates seeded with OP50 or HT115.
Antibody production
Production of rabbit polyclonal antiserum directed against CHMP7 and Vps60/CHMP5 was outsourced (Pacific Immunology, Ramona, CA). Rabbits were immunized with GST fusions to each protein, which were purified from Escherichia coli. Affinity purification was carried out as described previously (Mayers et al., 2013). Antibodies directed against other C. elegans ESCRT proteins have been described previously (Shen et al., 2014; Frankel et al., 2017; Fyfe et al., 2011; Schuh et al., 2015), and antibodies against β-actin (Sigma-Aldrich, St. Louis, MO) and GFP (Rockland Immunochemicals Inc, Pottstown, PA) were obtained from commercial sources. Antibodies were used at a final concentration of 1 μg/mL for immunofluorescence and immunoblotting studies.
Fluorescence imaging studies
Confocal imaging was conducted either using a Prairie Technologies swept-field confocal scanhead mounted onto a Nikon Eclipse Ti-E, equipped with Planapo oil immersion objectives and a CoolSNAP HQ2 CCD camera or a Yokogawa W1 confocal scanhead mounted onto a Nikon Ti2 microscope with a Hamamatsu Orca Flash 4 camera. STED microscopy was performed on a Leica TCS SP8 using a 775 nm depletion laser, a 100x, 1.4NA Planapo oil immersion objective, and HyD detectors. For immunofluorescence studies, embryos were fixed using cold methanol and stained as described previously (Audhya et al., 2007). Alternatively, for live imaging, dissected embryos were mounted onto 2% agarose pads and imaged under minimal compression in M9 minimal medium (Wang and Audhya, 2014). For germline examination, animals were anesthetized for 15–30 minutes in freshly prepared 0.1% Tricaine and immobilized in polystyrene beads on 4% agarose pads. For assessment of nuclear envelope permeability after mitotic exit, data analysis was performed using ImageJ. Gross chromosomal segregation defects were scored manually.
Electron microscopy studies
C. elegans were loaded into 100 μm deep aluminum sample holders (Technotrade, Manchester, NH) coated with 1-hexadecene, and a suspension of OP50 bacteria was used as a cryoprotectant. High pressure freezing was carried out using a Balzers HPM 010 or Leica EM ICE. After freezing, samples were substituted into a solution of 1% OsO4 and 1% H20 in acetone over 3–4 hours (McDonald, 2014), followed by centrifugation into increasing concentrations of epoxy EMbed 812 resin (Electron Microscopy Sciences, Hat-field, PA) to allow resin infiltration and polymerization (60°C for 24 hours). Animals were mounted on BEEM capsules and sectioned along the longitudinal axis. Sections were collected on copper slot grids coated with Pioloform or Formvar film for transmission electron microscopy (~80 nm) or electron tomography (~300 nm). Samples were post-stained with 8% uranyl acetate in 50% ethanol, followed by Reynold’s lead citrate. For tomography, sections were additionally floated on drops containing 10 nm colloidal gold particles, to aid in tomographic reconstruction (Frankel et al., 2017). Samples (~80 nm sections) were imaged using a Philips CM120 TEM operated at 80 kV and equipped with an AMT Biosprint 12 series digital camera. Dual-axis tilt series were acquired on a 300 kV Tecnai TF-30 equipped with a Gatan 2k × 2k Ultrascan camera using SerialEM software. Tilt series images were captured at 23000X (0.467 nm/pixel) from −60° to +60° at 1° increments (Frankel et al., 2017). Reconstruction and modeling of tilt series was carried out using IMOD and 3dmod software.
Yeast two-hybrid studies
All plasmids used were sequence verified. Transformed yeast strains (James et al., 1996) were grown overnight at 30°C in selective media, diluted to an optical density of 0.25 in minimal media, and subsequently 10-fold serial dilutions were prepared. Each dilution was spotted onto selective plates and incubated at 30°C for ~72 hours before imaging.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was conducted using Graphpad Prism 8.0 software. Linescan analyses and other fluorescence intensity measurements were conducted using Nikon Elements or ImageJ software, following background subtraction. A paired t-test was used to compare two conditions, while multiple conditions were compared using a two-way ANOVA followed by Tukey’s multiple comparison test or Dunnett’s test. See figure legends for details.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-actin | Sigma-Aldrich | Cat# A1978-200UL Lot# 087M4850V; RRID:AB_476692 |
| Anti-GFP | Rockland Immunochemicals Inc | Cat# 600-101-215 Lot# 33301; RRID:AB_218182 |
| Anti-chmp7 | This study | N/A |
| Anti-vps32 | Shen et al. (2014) | N/A |
| Anti-vps60 | This study | N/A |
| Anti-vps2 | Frankel et al. (2017) | N/A |
| Anti-did2 | Frankel et al. (2017) | N/A |
| Anti-vps24 | Frankel et al. (2017) | N/A |
| Anti-vps20 | Schuh et al. (2015) | N/A |
| Anti-ist1 | Frankel et al. (2017) | N/A |
| Anti-vps4 | Frankel et al. (2017) | N/A |
| ECL Anti-mouse IgG, Horseradish Peroxidase linked whole antibody from donkey | GE Healthcare | Cat# NA934V Lot# 17197685; RRID:AB_772206 |
| ECL Anti-mouse IgG, Horseradish Peroxidase linked whole antibody from sheep | GE Healthcare | Cat# NA931V Lot# 16908225; RRID:AB_772210 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Thermo Fisher Scientific | Cat# A-11058; RRID:AB_2534105 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31573; RRID:AB_2536183 |
| Bacterial and virus strains | ||
| Escherichia coli OP50 | Caenorhabditis Genetics Center (CGC) | N/A |
| Escherichia coli HT115 | Caenorhabditis Genetics Center (CGC) | N/A |
| Escherichia coli HT115 RNAi library | Caenorhabditis Genetics Center (CGC) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Cas9 | Integrated DNA Technologies, Inc. | Cat# 1081059 I |
| Tricaine | Sigma-Aldrich | Cat# E10521 |
| EMbed 812 | Electron Microscopy Sciences | Cat# 14120 |
| Pierce Anti-HA Agarose | Thermo Fisher Scientific | Cat# 26182 |
| Critical commercial assays | ||
| SuperSignal West Femto Maximum Sensitivity Substrate | Fisher Scientific | Cat# PI34095 |
| MEGAclear Kit | Invitrogen | Cat# AM1908 |
| MEGAscript T7 | Invitrogen | Cat# AM1334 |
| MEGAscript T3 | Invitrogen | Cat# AM1338 |
| Experimental models: Organisms/strains | ||
| N2 (Caenorhabditis elegans wild type) | Caenorhabditis Genetics Center (CGC) | N/A |
| chmp-7(hz12) II | This study | MSN772 |
| istr-1(hz16) V; unc-119(ed3) III; hzIs100[pie-1p-GFP::CAV-1; unc-119 + ] | This study | MSN836 |
| vps-60(hz20) V; unc-119(ed3) III; hzIs100[pie-1p-GFP::CAV-1; unc-119 + ] | This study | MSN912 |
| gfp::chmp-7 II | This study | MSN793 |
| unc-119(ed3) III; hzIs100[pie-1p-GFP::CAV-1; unc-119 + ] | Sato et al. (2006) | OD176 |
| unc-119(ed3) III; ltIs156 [pie-1/VPS-4(EQ)::GFP; unc-119 (+)] | Frankel et al. (2017) | MSN431 |
| unc-119(ed3) III; ltSi491 [pRB10; lmn-1 3kb::LMN-1 (WT) RR (AA:105–176); cb-unc-119(+)] II | Penfield et al. (2018) | OD1004 |
| unc-119(ed3) III; ltSi493 [pRB19; lmn-1 3kb::LMN-1 (N209K) RR (AA:105–176); cb-unc-119(+)] II | Penfield et al. (2018) | OD1323 |
| unc-119(ed3) III; ltIs76 [pie-1/mCherry::SP-12; unc-119 (+)] | Joseph-Strauss et al. (2012) | MSN99 |
| unc-119(ed3) III; ItIs51 [pie-1/SP12::GFP; unc-119(+)] | Poteryaev et al. (2005) | MSN67 |
| lem-2(tm1582) II | Poteryaev et al. (2005) | MSN244 |
| emr-1(gk119) I | Poteryaev et al. (2005) | MSN203 |
| bqSi242 [lem-2p::lem-2::mCherry + unc-119(+)] IV | Morales-Martínez et al., 2015 | MSN1160 (Derived from BN243) |
| bqSi235 [emr-1p::emr-1::GFP + unc-119 (+)] II | Morales-Martínez et al., 2015 | MSN1164 (Derived from BN243) |
| unc-119(ed3) III; ltIs37 [pie-1/mCHERRY::HIS-58; unc-119 (+)] | McNally et al. (2006) | OD56 |
| lmn-1(tm1502) I; Jfsi68[lmn-1(4kb 5’ UTR)::GFP cb-unc-119 (+)] II | (Franz et al., 2005) | UV120 |
| unc-119(ed3) III; ltIs37 [pie-1/mCHERRY:: HIS-58; unc-119 (+)]; qaIs3546 [pie-1p/GFP::npp-8; unc-119(+)] | Ródenas et al. (2009) | BN13 |
| unc-119(ed3) III; ltIs37 [(pAA64) pie-1p::mCherry::his-58 + unc-119(+)] IV; jjIs1092 [(pNUT1) npp-1::GFP + unc-119(+)] | Golden et al. (2009) | OCF3 |
| unc-119(ed3) III; ieSi21 [sun-1p::sun-1::mRuby::sun-1 3’UTR + Cbr-unc-119(+)] IV | Rog and Dernburg, 2015 | CA1219 |
| unc-119(ed3) III; ltIs75 [pie-1/ GFP::LacI; unc-119 (+)];ltIs37 [pie-1/mCHERRY::HIS-58; unc-119 (+)] | Yuen et al. (2011) | MSN147 |
| bcIs39 [lim-7p::ced-1::GFP + lin-15(+)];ltIs37 [pie-1/mCHERRY::HIS-58; unc-119 (+)] | Zhou et al. (2001) | MSN388 |
| Saccharomyces cerevisiae MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | James et al. (1996) | PJ69-4A |
| Oligonucleotides | ||
| chmp-7 sgRNA1: 5’ CTTCCTATCTCCCTTCCGAA 3’ | This study | N/A |
| chmp-7 sgRNA2: 5’ CAAAGAAGTAACGCCAGAAG 3’ | This study | N/A |
| istr-1 sgRNA1: 5’ TGAAAACTAATCTTCGATTG 3’ | This study | N/A |
| vps-60 sgRNA1: 5’ GCATTGTTGAGATCTGGTGG 3’ | This study | N/A |
| vps-60 sgRNA2: 5’ GAAGCGGAATTGGCGATGCT 3’ | This study | N/A |
| Recombinant DNA | ||
| pDBD-Chmp7 | This study | N/A |
| pAD-Empty | James et al. (1996) | pGAD-C1 |
| pAD-Lem2 | This study | N/A |
| pAD-Emr1 | This study | N/A |
| pDBD-Vps60 | This study | N/A |
| pAD-Chmp7 | This study | N/A |
| pAD-Vps32 | This study | N/A |
| pAD-Vps20 | This study | N/A |
| pDBD-Empty | James et al. (1996) | pGBDU-C1 |
| Software and algorithms | ||
| FIJI v1.53c | https://imagej.net/software/fiji/ | N/A |
| IMOD and 3dmod v4.9.12 | https://bio3d.colorado.edu/imod/ | N/A |
| Graphpad Prism v8.4.3 | https://www.graphpad.com/ | N/A |
| NIS Elements AR and Analysis v4.30.02 | Nikon Instruments Inc | N/A |
| SerialEM v3.6.22 | https://bio3d.colorado.edu/SerialEM/ | N/A |
| Other | ||
| Polybead Polystyrene 0.10micron Microspheres |
Polyscience, Inc | Cat# 00876–15 Lot# A789968 |
Highlights.
Chmp7 becomes essential when the nuclear lamina is partially compromised
Chmp7 localization is mediated by two LEM domain proteins
Impaired ESCRT function increases inner nuclear membrane invaginations
Multiple ESCRT-III subunits function to maintain inner nuclear membrane morphology
ACKNOWLEDGMENTS
This work was supported in part by NIH grant GM134865 (to A.A.) and NSF grant DMS1661900 (to A.A.). E.B.F. previously received support from NIH T32 GM007215. We thank David Dyer for initial assistance with the purification of C. elegans CHMP7 antibodies, Aryel Clarke and Daniel Kieffer for conducting preliminary biochemical studies, and members of the Audhya lab for critically reading this manuscript.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110263.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Allison R, Lumb JH, Fassier C, Connell JW, Ten Martin D, Seaman MN, Hazan J, and Reid E (2013). An ESCRT-spastin interaction promotes fission of recycling tubules from the endosome. J. Cell Biol 202, 527–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arii J, Watanabe M, Maeda F, Tokai-Nishizumi N, Chihara T, Miura M, Maruzuru Y, Koyanagi N, Kato A, and Kawaguchi Y (2018). ESCRT-III mediates budding across the inner nuclear membrane and regulates its integrity. Nat. Commun 9, 3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, McLeod IX, Yates JR, and Oegema K (2007). MVB-12, a fourth subunit of metazoan ESCRT-I, functions in receptor downregulation. PLoS One 2, e956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan R, Zahand AJ, Sharabi K, Lamm AT, Feinstein N, Haithcock E, Wilson KL, Liu J, and Gruenbaum Y (2012). Ce-emerin and LEM-2: essential roles in Caenorhabditis elegans development, muscle function, and mitosis. Mol. Biol. Cell 23, 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrales RR, Forn M, Georgescu PR, Sarkadi Z, and Braun S (2016). Control of heterochromatin localization and silencing by the nuclear membrane protein Lem2. Genes Dev. 30, 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty P, and Gerlich DW (2019). Mitotic chromosome mechanics: how cells segregate their genome. Trends Cell Biol. 29, 717–726. [DOI] [PubMed] [Google Scholar]
- Bauer I, Brune T, Preiss R, and Kolling R (2015). Evidence for a nonendosomal function of the Saccharomyces cerevisiae ESCRT-III-like protein Chm7. Genetics 201, 1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, de Franceschi N, de la Mora E, Maiti S, Alqabandi M, Miguet N, di Cicco A, Roos WH, Mangenot S, Weissenhorn W, and Bassereau P (2020). Human ESCRT-III polymers assemble on positively curved membranes and induce helical membrane tube formation. Nat. Commun 11, 2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter A, Kaneshiro JM, and Hetzer MW (2019). Coaching from the sidelines: the nuclear periphery in genome regulation. Nat. Rev. Genet 20, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabianca DS, Muñoz-Jiménez C, Kalck V, Gaidatzis D, Padeken J, Seeber A, Askjaer P, and Gasser SM (2019). Active chromatin marks drive spatial sequestration of heterochromatin in C. elegans nuclei. Nature 569, 734–739. [DOI] [PubMed] [Google Scholar]
- Capella M, Caballero LM, Pfander B, Braun S, and Jentsch S (2020). ESCRT recruitment by the S. cerevisiae inner nuclear membrane protein Heh1 is regulated by Hub1-mediated alternative splicing. J. Cell Sci 133, jcs250688. [DOI] [PubMed] [Google Scholar]
- Champion L, Pawar S, Luithle N, Ungricht R, and Kutay U (2019). Dissociation of membrane–chromatin contacts is required for proper chromosome segregation in mitosis. Mol. Biol. Cell 30, 427–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, and Lammerding J (2016). Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel EB, Shankar R, Moresco JJ, Yates JR 3rd, Volkmann N, and Audhya A (2017). Ist1 regulates ESCRT-III assembly and function during multivesicular endosome biogenesis in Caenorhabditis elegans embryos. Nat. Commun 8, 1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C, Askjaer P, Antonin W, Iglesias CL, Haselmann U, Schelder M, de Marco A, Wilm M, Antony C, and Mattaj IW (2005). Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J 24, 3519–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyfe I, Schuh AL, Edwardson JM, and Audhya A (2011). Association of the endosomal sorting complex ESCRT-II with the Vps20 subunit of ESCRT-III generates a curvature-sensitive complex capable of nucleating ESCRT-III filaments. J. Biol. Chem 286, 34262–34270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Milstein S, Ahmed S, Hodgkin J, and Hengartner MO (2000). A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5, 435–443. [DOI] [PubMed] [Google Scholar]
- Gatta AT, Olmos Y, Stoten CL, Chen Q, Rosenthal PB, and Carlton JG (2021). CDK1 controls CHMP7-dependent nuclear envelope reformation. Elife 10, e59999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden A, Liu J, and Cohen-Fix O (2009). Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J. Cell Sci 122, 1970–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, LaJoie D, Chen OS, von Appen A, Ladinsky MS, Redd MJ, Nikolova L, Bjorkman PJ, Sundquist WI, Ullman KS, and Frost A (2017). LEM2 recruits CHMP7 for ESCRT-mediated nuclear envelope closure in fission yeast and human cells. Proc. Natl. Acad. Sci. U S A 114, E2166–E2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann CT, Sears RM, Katiyar A, Busselman BW, Aman LK, Zhang Q, O’Bryan CS, Angelini TE, Lele TP, and Roux KJ (2019). Repair of nuclear ruptures requires barrier-to-autointegration factor. J. Cell Biol 218, 2136–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzle C, Mucke L, Brune T, and Kolling R (2019). Comprehensive analysis of yeast ESCRT-III composition in single ESCRT-III deletion mutants. Biochem. J 476, 2021–2046. [DOI] [PubMed] [Google Scholar]
- Hoffman DP, Shtengel G, Xu CS, Campbell KR, Freeman M, Wang L, Milkie DE, Pasolli HA, Iyer N, Bogovic JA, et al. (2020). Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science 367, eaaz5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii M, Shibata H, Kobayashi R, Katoh K, Yorikawa C, Yasuda J, and Maki M (2006). CHMP7, a Novel ESCRT-III-related protein, associates with CHMP4b and functions in the endosomal sorting pathway. Biochem. J 400, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N, Paulo JA, Tatarakis A, Wang X, Edwards AL, Bhanu NV, Garcia BA, Haas W, Gygi SP, and Moazed D (2020). Native chromatin proteomics reveals a role for specific nucleoporins in heterochromatin organization and maintenance. Mol. Cell 77, 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, and Craig EA (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph-Strauss D, Gorjánácz M, Santarella-Mellwig R, Voronina E, Audhya A, and Cohen-Fix O (2012). Sm protein down-regulation leads to defects in nuclear pore complex disassembly and distribution in C. elegans embryos. Dev. Biol 365, 445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuga T, Nie H, Kazami T, Satoh M, Matsushita K, Nomura F, Maeshima K, Nakayama Y, and Tomonaga T (2014). Lamin B2 prevents chromosome instability by ensuring proper mitotic chromosome segregation. Oncogenesis 3, e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link J, Paouneskou D, Velkova M, Daryabeigi A, Laos T, Labella S, Barroso C, Piñol SP, Montoya A, Kramer H, et al. (2018). Transient and partial nuclear lamina disruption promotes chromosome movement in early meiotic prophase. Dev. Cell 45, 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rolef Ben-Shahar T, Riemer D, Treinin M, Spann P, Weber K, Fire A, and Gruenbaum Y (2000). Essential roles for caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol. Biol. Cell 11, 3937–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Bot N, Tsai MC, Campbell JM, Ahringer J, and White JG (2003). The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115, 825–836. [DOI] [PubMed] [Google Scholar]
- Martins F, Sousa J, Pereira CD, da Cruz E Silva OAB, and Rebelo S (2020). Nuclear envelope dysfunction and its contribution to the aging process. Aging Cell 19, e13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast FD, Herricks T, Strehler KM, Miller LR, Saleem RA, Rachubinski RA, and Aitchison JD (2018). ESCRT-III is required for scissioning new peroxisomes from the endoplasmic reticulum. J. Cell Biol 217, 2087–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Wang L, Pramanik J, Johnson A, Sarkeshik A, Wang Y, Saengsawang W, Yates JR 3rd, and Audhya A (2013). Regulation of ubiquitin-dependent cargo sorting by multiple endocytic adaptors at the plasma membrane. Proc. Natl. Acad. Sci. U S A 110, 11857–11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Clippinger AK, Talledge N, Skowyra ML, Saunders MG, Naismith TV, Colf LA, Afonine P, Arthur C, Sundquist WI, et al. (2015). Structure and membrane remodeling activity of ESCRT-III helical polymers. Science 350, 1548–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KL (2014). Out with the old and in with the new: rapid specimen preparation procedures for electron microscopy of sectioned biological material. Protoplasma 251, 429–448. [DOI] [PubMed] [Google Scholar]
- McNally K, Audhya A, Oegema K, and McNally FJ (2006). Katanin controls mitotic and meiotic spindle length. J. Cell Biol 175, 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K, and Moazed D (2010). The nuclear envelope in genome organization, expression and stability. Nat. Rev. Mol. Cell Biol 11, 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettlen M, Chen PH, Srinivasan S, Danuser G, and Schmid SL (2018). Regulation of clathrin-mediated endocytosis. Annu. Rev. Biochem 87, 871–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzwa BE, Chiaruttini N, Redondo-Morata L, von Filseck JM, König J, Larios J, Poser I, Müller-Reichert T, Scheuring S, Roux A, and Gerlich DW (2017). Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodeling during cytokinesis. Nat. Cell Biol 19, 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Martínez A, Dobrzynska A, and Askjaer P (2015). Inner nuclear membrane protein LEM-2 is required for correct nuclear separation and morphology in C. elegans. J. Cell Sci 128, 1090–1096. [DOI] [PubMed] [Google Scholar]
- Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, and Sundquist WI (2010). Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc. Natl. Acad. Sci. U S A 107, 12889–12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HC, Talledge N, McCullough J, Sharma A, Moss FR 3rd, Iwasa JH, Vershinin MD, Sundquist WI, and Frost A (2020). Membrane constriction and thinning by sequential ESCRT-III polymerization. Nat. Struct. Mol. Biol 27, 392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y, Hodgson L, Mantell J, Verkade P, and Carlton JG (2015). ESCRT-III controls nuclear envelope reformation. Nature 522, 236–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y, Perdrix-Rosell A, and Carlton JG (2016). Membrane binding by CHMP7 coordinates ESCRT-III-dependent nuclear envelope reformation. Curr. Biol 26, 2635–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield L, Wysolmerski B, Mauro M, Farhadifar R, Martinez MA, Biggs R, Wu HY, Broberg C, Needleman D, and Bahmanyar S (2018). Dynein pulling forces counteract lamin-mediated nuclear stability during nuclear envelope repair. Mol. Biol. Cell 29, 852–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Rasoloson D, and Seydoux G (2015). High Efficiency, Homology-Directed Genome Editing in Caenorhabditis elegans Using CRISPR-Cas9 Ribonucleoprotein Complexes. Genetics 20, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield L, Shankar R, Szentgyorgyi E, Laffitte A, Mauro MS, Audhya A, Muller-Reichert T, and Bahmanyar S (2020). Regulated lipid synthesis and LEM2/CHMP7 jointly control nuclear envelope closure. J. Cell Biol 219, e201908179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfitzner AK, Mercier V, Jiang X, von Filseck JM, Baum B, Šarić A, and Roux A (2020). An ESCRT-III polymerization sequence drives membrane deformation and fission. Cell 182, 1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper GH, Sprenger S, Teis D, and Oliferenko S (2020). ESCRT-III/Vps4 controls heterochromatin-nuclear envelope attachments. Dev. Cell 53, 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev D, Squirrell JM, Campbell JM, White JG, and Spang A (2005). Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol. Biol. Cell 16, 2139–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Lennon-Duménil AM, Manel N, and Piel M (2016). ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352, 359–362. [DOI] [PubMed] [Google Scholar]
- Ródenas E, Klerkx EP, Ayuso C, Audhya A, and Askjaer P (2009). Early embryonic requirement for nucleoporin Nup35/NPP-19 in nuclear assembly. Dev. Biol 327, 399–409. [DOI] [PubMed] [Google Scholar]
- Rog O, and Dernburg AF (2015). Direct visualization reveals kinetics of meiotic chromosome synapsis. Cell Rep. 10, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanauska A, and Köhler A (2018). The inner nuclear membrane is a meta-bolically active territory that generates nuclear lipid droplets. Cell 174, 700–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, and Emr SD (2009). Functional reconstitution of ESCRT-III assembly and disassembly. Cell 136, 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Sato M, Audhya A, Oegema K, Schweinsberg P, and Grant BD (2006). Dynamic regulation of caveolin-1 trafficking in the germ line and embryo of Caenorhabditis elegans. Mol. Biol. Cell 17, 3085–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer EC, Florens L, Guan T, Yates JR 3rd, and Gerace L (2013). Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301, 1380–1382. [DOI] [PubMed] [Google Scholar]
- Schuh AL, Hanna M, Quinney K, Wang L, Sarkeshik A, Yates JR 3rd, and Audhya A (2015). The VPS-20 subunit of the endosomal sorting complex ESCRT-III exhibits an open conformation in the absence of upstream activation. Biochem. J 466, 625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QT, Schuh AL, Zheng Y, Quinney K, Wang L, Hanna M, Mitchell JC, Otegui MS, Ahlquist P, Cui Q, and Audhya A (2014). Structural analysis and modeling reveals new mechanisms governing ESCRT-III spiral filament assembly. J. Cell Biol 206, 763–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Capo-chichi CD, and Xu XX (2018). Defective nuclear lamina in aneuploidy and carcinogenesis. Front. Oncol 8, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer DR, Howard TL, Nyun T, and Hollenberg SM (2001). CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J. Cell Sci 114, 2383–2393. [DOI] [PubMed] [Google Scholar]
- Talledge N, McCullough J, Wenzel D, Nguyen HC, Lalonde MS, Bajorek M, Skalicky J, Frost A, and Sundqust WI (2018). The ESCRT-III proteins IST1 and CHMP1B assemble around nucleic acids. bioRxiv, 386532. 10.1101/386532. [DOI] [Google Scholar]
- Thaller DJ, Allegretti M, Borah S, Ronchi P, Beck M, and Lusk CP (2019). An ESCRT-LEM protein surveillance system is poised to directly monitor the nuclear envelope and nuclear transport system. Elife 8, e45284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaller DJ, Tong D, Marklew CJ, Ader NR, Mannino PJ, Borah S, King MC, Ciani B, and Lusk CP (2021). Direct binding of ESCRT protein Chm7 to phosphatidic acid-rich membranes at nuclear envelope herniations. J. Cell Biol 220, e202004222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang HTH, Connell JW, Brown SE, Thompson A, Reid E, and Sanderson CM (2006). A systematic analysis of human CHMP protein interactions: additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics 88, 333–346. [DOI] [PubMed] [Google Scholar]
- Ungricht R, and Kutay U (2015). Establishment of NE asymmetry - targeting of membrane proteins to the inner nuclear membrane. Curr. Opin. Cell Biol 34, 135–141. [DOI] [PubMed] [Google Scholar]
- Ungricht R, and Kutay U (2017). Mechanisms and functions of nuclear envelope remodeling. Nat. Rev. Mol. Biol 18, 229–245. [DOI] [PubMed] [Google Scholar]
- van Steensel B, and Belmont AS (2017). Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventimiglia LN, Cuesta-Geijo MA, Martinelli N, Caballe A, Macheboeuf P, Miguet N, Parnham IM, Olmos Y, Carlton JG, Weissenhorn W, and Martin-Serrano J (2018). CC2D1B coordinates ESCRT-III activity during the mitotic reformation of the nuclear envelope. Dev. Cell 47, 547–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, Christ L, Thoresen SB, Brech A, Raiborg C, and Stenmark H (2015). Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 522, 231–235. [DOI] [PubMed] [Google Scholar]
- Vietri M, Radulovic M, and Stenmark H (2020a). The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol 21, 25–42. [DOI] [PubMed] [Google Scholar]
- Vietri M, Schultz SW, Bellanger A, Jones CM, Petersen LI, Raiborg C, Skarpen E, Pedurupillay CRJ, Kjos I, Kip E, et al. (2020b). Unrestrained ESCRT-III drives micronuclear catastrophe and chromosome fragmentation. Nat. Cell Biol 22, 856–867. [DOI] [PubMed] [Google Scholar]
- von Appen A, LaJoie D, Johnson IE, Trnka MJ, Pick SM, Burlingame AL, Ullman KS, and Frost A (2020). LEM2 phase separation promotes ESCRT-mediated nuclear envelope reformation. Nature 582, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Filseck JM, Barberi L, Talledge N, Johnson IE, Frost A, Lenz M, and Roux A (2020). Anisotropic ESCRT-III architecture governs helical membrane tube formation. Nat. Commun 11, 1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, and Audhya A (2014). In vivo imaging of C. elegans endocytosis. Methods 68, 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BM, Colombi P, Jager J, and Lusk CP (2014). Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell 159, 388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BM, Thaller DJ, Jäger J, Ochmann SE, Borah S, and Lusk CP (2016). Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. EMBO J. 35, 2447–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesel N, Mattout A, Melcer S, Melamed-Book N, Herrmann H, Medalia O, Aebi U, and Gruenbaum Y (2008). Laminopathic mutations interfere with the assembly, localization, and dynamics of nuclear lamins. Proc. Natl. Acad. Sci. U S A 105, 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AM, Gunn AL, and Hatch EM (2020). BAF facilitates interphase nuclear membrane repair through recruitment of nuclear transmembrane proteins. Mol. Biol. Cell 31, 1551–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen KWY, Nabeshima K, Oegema K, and Desai A (2011). Rapid de novo centromere formation occurs independently of heterochromatin protein 1 in C. elegans embryos. Curr. Biol 21, 1800–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hartwieg E, and Horvitz HR (2001). CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104, 43–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


