Abstract
The emergence of the B.1.617.2 (Delta) variant of the severe acute syndrome coronavirus (SARS-CoV-2) that emerged in 2019 (COVID-19), resulted in a surge of cases in India and has expanded and been detected across the world, including in the United States. The B.1.617.2 (Delta) variant has been seen to be twice more transmissible coupled with potential increases in disease severity and immune escape. As a result, case numbers and hospitalisations are once again on the rise in the USA. On 16 July 2021, the Centers for Disease Control and Prevention (CDC) reported a 7-day average 69.3% increase in new cases and a 35% increase in hospitalisations. Although the gold standard for SARS-CoV-2 variants identification remains genomic sequencing, this approach is not accessible to many clinical laboratories. The main goal of this study was to validate and implement the detection of the B.1.617.2 (Delta) variant utilising an open reverse transcription polymerase chain reaction (RT-PCR) platform by explicitly detecting the S-gene target failure (SGTF) corresponding to the deletion of two amino acids (ΔE156/ΔF157) characteristic of B.1.617.2 (Delta) variant. This approach was conceived as a rapid screening of B.1.617.2 (Delta) variant in conjunction with CDC’s recommended N1 (nucleocapsid gene), N2, and RP (human RNase P) genes, as a pre-screening tool prior to viral genomic sequencing. We assessed 4,937 samples from 5 July to 5 September 2021. We identified the B.1.617.2 (Delta) variant in 435 of 495 positive samples (87.8%); the additional positive samples (7 samples, 1.4%) were found to belong to the B.1.1.7 (Alpha, UK) lineage and the remaining 53 samples (10.7%) were reported as ‘other’ lineages. Whole genome sequencing of 46 randomly selected samples validated the strains identified as positive and negative for the B.1.617.2 (Delta) variant and confirmed the S gene deletion in addition to B.1.617.2 characteristic mutations including L452R, T478K, P681R and D950N located in the spike protein. This modality has been used as routine testing at the Riverside University System Health (RUHS) Medical Center as a method for detection of B.1.617.2 (Delta) to pre-screen samples before genome sequencing. The assay can be easily implemented in clinical laboratories, most notably those with limited economic resources and access to genomic platforms.
Key words: SARS-CoV-2, RT-PCR test, Delta COVID-19 variant
Introduction
The increase of SARS-CoV-2 variants of concern (VOC) implicated in extensive outbreaks in South Africa (B.1.351, Beta), Brazil (P.1/Gamma), United Kingdom (B.1.1.7, Alpha), United States (B.1.427/429, Epsilon) and India (B.1.617.2, Delta) requires real-time surveillance and public health response.1, 2, 3, 4 In December 2020, the rapidly spreading SARS-CoV-2 virus B.1.617.2/Delta variant was first identified in India. The Delta variant is divided, according to the PANGO classification (https://www.pango.network/), into three sub-lineages including variants B.1.617.2, AY.1, AY.2 and AY.3, and has been classified as a variant of concern (VOC) by Public Health England (PHE), the World Health Organization (WHO) and the United States Centers for Disease Control and Prevention (CDC).5 Currently, the B.1.617.2 (Delta) lineage accounts for an increasing proportion of cases spreading in 98 countries around the world and has become the dominant variant responsible for more than 83% of COVID-19 reported cases in the US.5 , 6
It is known that the B.1.617.2 (Delta) variant has a distinct deletion (ΔE156/ΔF157) located at the Spike protein, that is uniquely present in this lineage and not in other variants including B.1.1.7 (Alpha), B.1.351 (Beta) and/or P.1 (Gamma). In addition, it shows multiple mutations including L452R, T478K, and D950N. L452R is known to increase affinity for ACE2 receptors found on the surface of a variety of human cells such as lung cells, while the T478K has been shown to increase receptor binding activity and to enable immune escape.7 Viral genome sequencing is currently the only method to reliably detect the rapidly emerging SARS-CoV-2 variants. Although genomic sequencing has the advantage of identifying new mutations, it is expensive and challenging to perform in real-time; in contrast, reverse transcription polymerase chain reaction (RT-PCR) testing for SARS-CoV-2 has become widespread. To support the detection of variants, the RT-PCR base assay can provide rapid results for known variants, as indicated by multiple reports directed to target specific variants of concern.8, 9, 10, 11, 12, 13
The main goal of this study was to validate and implement the detection of the B.1.617.2 (Delta) variant in an open RT-PCR platform by specifically detecting the S-gene target failure (SGTF) corresponding to the deletion of two amino acids, E156 and F157 (ΔE156/ΔF157). This approach was conceived as a rapid screening of the B.1.617.2 variant in conjunction with CDC’s recommended N1 (nucleocapsid gene), N2, and RP (human RNase P) genes, and prior to viral genomic sequencing. This modality has been used as routine testing at the Riverside University System Health (RUHS) Medical Center as a pre-screening method for detection of the B.1.617.2 (Delta) variant.
Materials and methods
Sample preparation and RT-qPCR assay
Total RNA was extracted from upper respiratory nasopharyngeal (NP) swab specimens collected from inpatients and outpatients at RUHS Medical Center during the study period from 5 July to 5 September 2021, in viral transport medium by using the MagMAX Viral/Pathogen Nucleic Acid Isolation Kit and KingFisher Flex Magnetic Particle Processor extraction system (ThermoFisher Scientific, USA), as per manufacturer’s instructions. For RNA extraction, 200 μL of the NP samples were used. Real-time RT-qPCR was performed using 5 μL of total RNA per well in a 96-well plate (Bio-Rad, USA). GoTaq probe RT-qPCR kit (Promega, USA) was used for the reaction mix; N1, N2 and RP primers/probes, as per CDC guidelines, were purchased from Biosearch Technologies (UK). S gene primers and 5'-FAM/3'-BHQ1-labelled probe were synthesisd by Genewiz (USA). In this assay, a FAM-labelled probe covering the region on the S gene where the ΔE156/ΔF157 deletion occurs in the B.1.617.2 (Delta) variant (position 22029 of NC_045512.2 reference genome) was chosen, together with the corresponding set of S gene forward and reverse primers (Fig. 1 ). They were used in an open platform real-time RT-PCR assay. The Delta SGTF primers/probe were added to those currently in use recommended by the US CDC, i.e., N1, N2 and RP genes (Bioresearch Technologies). The full description of the primers and probes used in this study are shown in Table 1 .
Fig. 1.
Primers and probes used to detect the SARS-CoV-2 SGTF. Regions targeted corresponding to variants B.1.1.7 (Alpha: upper set) and B.1.617.2 (Delta: lower set) are shown including specific forward/reverse primers and probes used on the open platform RT-PCR. Protein sequence shows the respective deletions (marked in blue).
Table 1.
Primers and probe sets used to test for the presence of SARS-CoV-2 virus RNA including those corresponding to B.1.617.2 (Delta) and B.1.1.7 (Alpha) detection from NP samples by real-time RT-PCR
| Name | Sequence | Source |
|---|---|---|
| 2019-nCoV_N1 Forward | GACCCCAAAATCAGCGAAAT | CDC |
| 2019-nCoV_N1 Reverse | TCTGGTTACTGCCAGTTGAATCTG | CDC |
| 2019-nCoV_N1 Probe | FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1 | CDC |
| 2019-nCoV_N2 Forward | TTACAAACATTGGCCGCAAA | CDC |
| 2019-nCoV_N2 Reverse | GCGCGACATTCCGAAGAA | CDC |
| 2019-nCoV_N2 Probe | FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ1 | CDC |
| 2019-nCoV_RP Forward | AGATTTGGACCTGCGAGCG | CDC |
| 2019-nCoV_RP Reverse | GAGCGGCTGTCTCCACAAGT | CDC |
| 2019-nCoV_RP Probe | FAM-TTCTGACCTGAAGGCTCTGCGCG-BHQ-1 | CDC |
| 2019-nCoV_S. Delta Forward | GTAATGATCCATTTTTGGGTG | This study |
| 2019-nCoV_S. Delta Reverse | GAAAAGGCTGAGAGACATATTC | This study |
| 2019-nCoV_S. Delta Probe | FAM-GGAAAGTGAGTTCAGAGTTTATTC-BHQ1 | This study |
| 2019-nCoV_S. Alpha Forward | TCAACTCAGGACTTGTTCTTAC | (17) and this study |
| 2019-nCoV_S. Alpha Reverse | TGGTAGGACAGGGTTATCAAAC | (17) and this study |
| 2019-nCoV_S. Alpha Probe | FAM-TGGTCCCAGAGACATGTATAGCAT-BHQ1 | (17) and this study |
Amplification was conducted in 96-well plates on a Bio-Rad CFX 96 Real-Time PCR Instrument. Thermocycling conditions consisted of 2 min at 25°C for uracil-DNA glycosylase incubation, 15 min at 50°C for reverse transcription, 2 min at 95°C for activation of the Taq enzyme, and 45 cycles of 3 s at 95°C and 30 s at 55°C. Assay controls were included in each run, as per CDC’s recommendations, as well as a positive template control (PTC), with an expected cycle threshold (CT) value range, and a negative template control (NTC) added during real-time RT-PCR reaction set-up.
Test algorithm
The threshold for the real-time RT-PCR was set in the middle of exponential amplification phase in log view. Under standard conditions, a positive SARS-CoV-2 test result was defined as an exponential fluorescent curve corresponding to N1, N2 and RP genes that crossed the threshold within 40 cycles (CT<40). B.1.617.2 (Delta) variant positive test was considered when N1, N2 and RP genes displayed CT values <35 with no detected S gene (containing ΔE156/ΔF157). This criterion was based on both previous experience with the B.1.1.7 (Alpha) STGF,6 and the values at which the probe directed to other regions of the S gene (positive control amplification) was not detected when the N1 and N2 CTs were above 35; this criterion was also corroborated by genome screening (Supplementary Table 1, Appendix A).
Next generation sequencing of SARS-COV2 variants
Whole genome sequencing and further validation of S-gene failure was performed in 46 randomly selected presumptive Delta samples. RNA from a volume of 200 μL NP swab sample was extracted with the QIAMP VIRAL RNA mini kit (Qiagen, USA) as per manufacturer’s instructions and quantified using a Qubit RNA High Sensitivity Kit (ThermoFisher Scientific). The extracted genomic RNAs were retro-transcribed accordingly to the Artic protocol.14 Briefly, 1 μL Random Primer Mix (ProtoScript II First Strand cDNA Synthesis Kit) and 1 μL 10 mM dNTP mix (NEB, USA) were added to 8 μL RNA and denatured on a BioRad CFX96 system at 65°C for 5 min and then incubated on ice. Then 12.5 μL 2X ProtoScript II Reaction Mix and 2.5 μL 10X ProtoScript II Enzyme Mix were added to the denatured sample and cDNA synthesis was performed using the following conditions: 25°C for 5 min, 42°C for 50 min and 80°C for 5 min. After cDNA synthesis, in a new PCR tube, 2.5 μL cDNA was combined with 12.5 μL Q5 High-Fidelity 2X Master Mix (NEB). To pool #1 mix, 5.87 μL nuclease free water (Thermo Fisher Scientific) and 4.13 μL of 10 μM ARTIC version 3 primer pool #1 was added. To pool #2 mix, 5.95 μL nuclease free water and 4.05 μL of 10 μM ARTIC version 3 primer pool #2 was added. PCR cycling was then performed as follows: 98°C for 30 s followed by 35 cycles of 98°C for 15 s and 65°C for 5 min. cDNA synthesis and PCR reactions were purified using RNAClean XP (Beckman Coulter, USA). Combined ARTIC amplicons were purified at 1.0× bead to amplicon ratio and eluted in 30 μL. Illumina sequencing libraries were prepared using Nextera DNA Flex Library Prep Kit and Nextera DNA CD Indexes (Illumina, USA) according to manufacturer’s instructions. Paired-end 150 bp sequencing was performed for each library on a MiniSeq with the 300-cycle mid output reagent kit (Illumina), multiplexed with targeted generation of ∼40,000 clusters per library. A negative control library with no input SARS-CoV-2 RNA extract was included using ARTIC amplification. Genomes were assembled, and SNPs were identified by comparison to the sequence of SARS-COV2 genome (GenBank accession number NC_045512.2) using the Lasergene (version 17) suite (DNASTAR, USA). Sequence assembly was done by SeqMan NGen and variant analyses with Seq Man Ultra (Lasergene 17). Sequence alignments were done by Clustal W and MUSCLE (Lasergene 17).
Results and discussion
The emergence of SARS-CoV2 VOC is of high relevance given the potential for increased transmission, disease severity and potential resistance to vaccine induced immunity.15 , 16 Since the inception of this approach, we have tested 4,937 samples from 5 July to 5 September 2021, with 495 testing positive, of which 435 samples were identified as B.1.167.2 (Delta) variant (87.8%), seven samples (1.4%) were B.1.1.7 (Alpha), while the remaining 53 (10.7%) were reported as ‘others’. A specimen was considered B.1.617.2 (Delta) variant positive when the real-time RT-PCR viral target(s) CT values of the N1-N2 targets were ≤35, and S target was not detected (Table 2 ). Negative B.1.617.2 variant samples displayed amplification of the S gene at similar CT levels as N1 and N2 (Table 2). Several controls were used to monitor the assay performance including positive template controls, no-template controls, and human specimen controls. In addition, we included as control a probe that was able to detect the B.1.1.7 (Alpha) variant previously validated in our laboratory targeting the deletion AH69/AV70, all routinely included in the runs. The design of the B.1.1.7, S-Alpha probe was based on the findings of Zhen and Berry who developed and validated a multiplex real-time RT-PCR assay for SARS-CoV-2 with primers designed to amplify a 108 bp target on the spike surface glycoprotein (S gene).17 This choice added additional confidence in the probe chosen in our approach. (Of note, the original purpose of the study by Zhen and Berry was different to ours as their design was intended for detection of SARS-COV2 and not for the SGTF used to identify SARS-CoV-2 Alpha VOC).
Table 2.
CT values corresponding to representative SARS-CoV-2 Delta variant detected in the present study (upper rows) and representative Delta variant-negative samples (lower rows)
| SARS-CoV-2 | B.1.617.2 variant | N1 gene | N2 gene | S-Alpha gene | S-Delta gene | RP gene |
|---|---|---|---|---|---|---|
| POS | YES | 22.61 | 22.73 | 22.79 | N/A | 29.18 |
| POS | YES | 29.03 | 29.38 | 29.19 | N/A | 27.02 |
| POS | YES | 23.94 | 24.61 | 22.66 | N/A | 27.64 |
| POS | YES | 20.83 | 20.37 | 22.19 | N/A | 25.35 |
| POS | YES | 16.05 | 14.41 | 16.48 | N/A | 26.24 |
| POS | YES | 25.4 | 25.2 | 26.23 | N/A | 26.72 |
| POS | YES | 24.73 | 24.06 | 26.01 | N/A | 27.19 |
| POS | YES | 15.81 | 16.56 | 18.02 | N/A | 22.21 |
| POS | YES | 20.79 | 22.32 | 23.01 | N/A | 23.06 |
| POS | YES | 15.42 | 14.93 | 17.75 | N/A | 27.13 |
| POS | YES | 23.25 | 23.2 | 23.07 | N/A | 27.46 |
| POS | YES | 16.33 | 15.93 | 16.38 | N/A | 25.06 |
| POS | NO | 21.64 | 22.3 | 22.22 | 26.75 | 28.25 |
| POS | NO | 27.36 | 28.82 | NA | 29.55 | 28.37 |
| POS | NO | 25.42 | 27.57 | NA | 28.94 | 27.41 |
| POS | NO | 26.55 | 27.36 | 20.13 | 25.65 | 22.50 |
B.1.1.7 (Alpha) S-gene target failure (SGTF; Alpha), S-gene SGTF (Delta); N1,N2, viral nucleocapsid gene; POS, positive for the presence of B.1.617.2 (Delta) SARS-CoV-2 virus; RP, human RNase P gene used as an endogenous internal control for specimen integrity, nucleic acid isolation, amplification and detection.
Inter-instrument comparison
Performance was compared between the four Bio-Rad CFX 96 instruments located at the Molecular Microbiology Lab at RUHS. Samples that tested positive in a representative number of 100 (n=100), tested positive using all four of the Bio-Rad systems. Similarly, all samples that tested negative in a representative number of 100, did also test negative in all the instruments. This shows excellent inter-instrument comparability approaching 100%.
Sensitivity and specificity
All known positive samples (n=495) were positive on this platform and all known negative samples (n=4,442) were negative. From the 495 positive samples, 435 were detected as B.1.617.2 (Delta) variant (87.8%) showing real-time RT-PCR viral target(s) N1 and N2 target CT≤35 and S target not detected. Specificity of the described set of primers (i.e., S-Delta/-Alpha probes and primers, Table 1) was examined using positive samples for the original SARS-CoV-2 and the B.1.1.7 (Alpha) variant (Supplementary Table 1, Appendix A). The aforementioned sets did not manifest a signal when tested with such negative controls and therefore were found to be highly specific. The sensitivity and specificity for this approach was 100%. The percent coefficient of variation (% CV) representing assay variability was calculated based on two samples tested in triplicate on three independent runs. For the wild-type and mutant S-Delta probe for the ΔE156/ΔF157 assay, inter-assay variability ranged from 0.4 to 0.9% and the intra-assay variability ranged from 0.09 to 1.18%.
Accuracies for the assay using a limited panel of 100 samples, including B.1.1.7 positive (#7), B.1.167.2 type positives (#88) and SARS-CoV-2 negative (#5) samples demonstrated values of 98.3% [95% confidence interval (CI) 95.8 to 99.9%] for the ΔE156/ΔF157 assay.
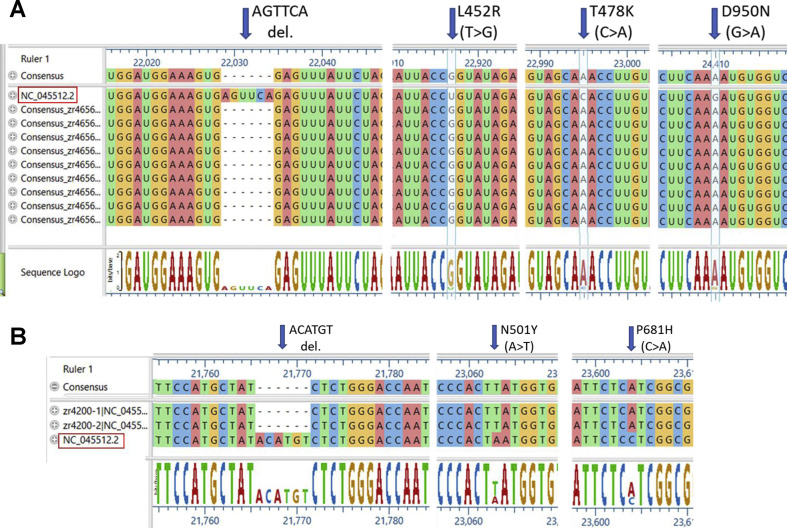
Randomly chosen 46 representative strains were whole genome sequenced in order to confirm the identity of the B.1.617.2 (Delta) variant (Fig. 2 A shows nine of those sequences). The analysis detected both the expected deletion and the additional characteristic N501Y and P681H mutations, as shown in Fig. 2A.
Fig. 2.
Sequence alignments of representative full genome sequencing of variants detected by real-time RT-PCR in samples from RUHS patient samples; genome reference NC_045512.2. Arrows indicate the deletion on the Spike protein at nucleotide positions 22029/71 (ΔE156/ΔF157), and representative mutations at positions 22917 (L452R), 22995 (T478K) and 24410 (D950N) of B.1.617.2 (Delta) variant (A), and the corresponding deletion and mutations corresponding to B.1.1.7 (Alpha) variant (B).
Precision
We identified the B.1.617.2 (Delta variant) in 435 samples out of 495 positives samples (87.8%); the additional positive samples were found to belong to other lineages including the B.1.1.7 (Alpha) UK lineage. Whole genome sequencing validated the strains identified as positive and negative for the B.1.617.2 (Delta) variant. Genome sequencing of 46 samples confirmed with 100% accuracy the S gene deletion in addition to B.1.617.2 characteristic L452R, T478K, P681R, and D950N mutations located in the spike protein (Fig. 2A), while six additional samples were categorised as B.1.1.7 variant (Fig. 2B). Considering the number of tested samples (n=4,937), we selected a representative number of randomly selected samples (n=50), both positive and negative, that were repeated daily for a total of 5 days. All expected positive and negative controls did result as expected. This represents an excellent precision approaching 100%. The characteristic AGTTCA deletion of the S gene (nucleotide genome position 22029–22034, ΔE156/ΔF157; reference sequence NC_045512.2) was readily present (Fig. 2). In addition, non-synonymous mutations C>G (T19R), T>G (L452R), C>A (T478K), C>G (P681R) and G>AǀG (D950N) were identified (Fig. 2, Table 3 ); additional mutations are depicted in Table 3. The characteristic variant mutations were seen in all the sequenced samples, supporting the accuracy and precision of the open platform method for a rapid identification of the variant described in this report.
Table 3.
Whole genome sequencing of samples identified as SARS-CoV-2 variant B.1.617.2 (Delta) by real-time RT-PCR: representative results of mutations found in the entire genome of RUHS-SARS-CoV-2 genome B.1.167.2 (Delta) variants with the corresponding nucleotide and protein amino acid changes
| Ref ID 045512.2 |
Ref pos 241 |
Gene name | Delta variant #1 called seq C>T | Delta variant #1 amino acid change |
|---|---|---|---|---|
| NC_045512.2 | 1191 | ORF1ab | C>T | p.P309L |
| NC_045512.2 | 1267 | ORF1ab | C>T | p.(=) |
| NC_045512.2 | 1877 | ORF1ab | T>G | p.S538A |
| NC_045512.2 | 3037 | ORF1ab | C>T | p.(=) |
| NC_045512.2 | 4666 | ORF1ab | A>C|A | p.R1467S, p.(=) |
| NC_045512.2 | 4668 | ORF1ab | C>G|C | p.S1468C, p.(=) |
| NC_045512.2 | 5184 | ORF1ab | C>T | p.P1640L |
| NC_045512.2 | 9891 | ORF1ab | C>T | p.A3209V |
| NC_045512.2 | 10870 | ORF1ab | G>T | p.(=) |
| NC_045512.2 | 11418 | ORF1ab | T>C | p.V3718A |
| NC_045512.2 | 12247 | ORF1ab | T>C | p.(=) |
| NC_045512.2 | 12480 | ORF1ab | T>C|T | p.V4072A, p.(=) |
| NC_045512.2 | 12946 | ORF1ab | T>C | p.(=) |
| NC_045512.2 | 14408 | ORF1ab | C>T | p.P4715L |
| NC_045512.2 | 15451 | ORF1ab | G>A | p.G5063S |
| NC_045512.2 | 16466 | ORF1ab | C>T | p.P5401L |
| NC_045512.2 | 18176 | ORF1ab | C>T | p.P5971L |
| NC_045512.2 | 19160 | ORF1ab | C>T | p.S6299F |
| NC_045512.2 | 20262 | ORF1ab | A>G | p.(=) |
| NC_045512.2 | 20718 | ORF1ab | G>T | p.M6818I |
| NC_045512.2 | 21618 | S | C>G | p.T19R |
| NC_045512.2 | 22029 | S | AGTTCA>del 6 | p.E156_F157delinsG |
| NC_045512.2 | 22917 | S | T>G | p.L452R |
| NC_045512.2 | 22995 | S | C>A | p.T478K |
| NC_045512.2 | 23403 | S | A>G | p.D614G |
| NC_045512.2 | 23604 | S | C>G | p.P681R |
| NC_045512.2 | 24410 | S | G>A|G | p.D950N, p.(=) |
| NC_045512.2 | 25469 | ORF3a | C>T | p.S26L |
| NC_045512.2 | 26767 | M | T>C | p.I82T |
| NC_045512.2 | 27638 | ORF7a | T>C | p.V82A |
| NC_045512.2 | 27739 | ORF7a | C>T | p.L116F |
| NC_045512.2 | 27752 | ORF7a | C>T | p.T120I |
| NC_045512.2 | 28248 | ORF8 | GATTTC>del 6 | p.D119_F120del |
| NC_045512.2 | 28271 | A>del 1 | ||
| NC_045512.2 | 28461 | N | A>G | p.D63G |
| NC_045512.2 | 28881 | N | G>T | p.R203M |
| NC_045512.2 | 29402 | N | G>T | p.D377Y |
| NC_045512.2 | 29742 | G>T |
The genetic variability of SARS-CoV-2 has imposed challenges for molecular diagnostics approaches, notably on the choice of primers and probes used in real-time RT-PCR-based methods.18 At this moment, identification of new variants including B.1.617.2 (Delta) is being performed by full genome sequencing. However, as is the case in our hospital, this implies increased time and costs associated with the detection of the variant, and more importantly, clinical patient care management as use of certain monoclonal antibodies have shown a reduced sensitivity to antibody neutralisation in the presence of the SARS-CoV-2 Delta variant.19 , 20 However, in the context of the SARS-CoV-2 B.1.617.2 (Delta), the use of a method based on an open platform real-time RT-PCR may serve as both an initial screening of presumptive variants to be further sequenced, considered as the gold standard, and for internal epidemiological information.
A similar approach based on the S deletion was developed and has been performed in our laboratory to identify the B.1.1.7 (Alpha) variant based on supporting information including the technical briefing published from Public Health England which stated that in the screening of 14,950 tested samples, the S gene failure detection was found in 99.5% of the samples (0.5% were negative). In the present study, the deletion of six bases existing in the SARS-CoV-2 B.1.617.2 (Delta) genome made it an obvious candidate for RT-PCR. Recently, a similar approach to the one described herein was developed to detect the Gamma (P.1, Brazilian) and Delta variants in municipal wastewater using an RT-qPCR assay.11
Given the fact that RT-PCR has become widely available for the SARS-CoV-2 diagnostics in clinical laboratories, the approach and findings described in this report, i.e., use of a set of primers/probe directed to detect the SGFT, makes this real-time RT-PCR-based assay an attractive modality that can be easily implemented in clinical laboratories. The rapid identification of B.1.617.2 (Delta) variant has served in our hospital to identify breakthrough infections that have occurred in vaccinated people and to assert the identification of this variant in unvaccinated patients.
Conclusion
In the present study we provide a simple approach that can be used in open platform RT-PCR for the rapid screening of both SARS-CoV-2 B.1.617.2 and B.1.1.7 variants. The simple addition of a specific set of primers/probe directed to the SGTF region allows the easy identification of the Alpha (B.1.1.7, UK) and Delta (B.1.617.2, India) variants.
Acknowledgements
The authors thank Ms Nerina Veliz, Mrs Beatriz Ramirez and Ms Syriphone Phommavanh for their assistance. The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. All data generated or analysed during this study are included in this published article.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pathol.2022.01.001.
Conflicts of interest and sources of funding
This study was funded by the Department of Pathology, Riverside University Health System Medical Center, Moreno Valley, CA, USA. The authors state that there are no conflicts of interest to disclose.
Appendix ASupplementary data
The following is the Supplementary data to this article:
References
- 1.Nonaka C.K.V., Franco M.M., Graf T., et al. Genomic evidence of SARS-CoV-2 reinfection involving E484K spike mutation, Brazil. Emerg Infect Dis. 2021;27:1522–1524. doi: 10.3201/eid2705.210191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iacobucci G. Covid-19: new UK variant may be linked to increased death rate, early data indicate. BMJ. 2021;372:n230. doi: 10.1136/bmj.n230. [DOI] [PubMed] [Google Scholar]
- 3.Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2 - what do they mean? JAMA. 2021;325:529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- 4.Galloway S.E., Paul P., MacCannell D.R., et al. Emergence of SARS-CoV-2 B.1.1.7 lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:95–99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention (CDC) 1 Dec 2021. COVID-19. SARS-CoV-2 variant classifications and definitions.https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html [Google Scholar]
- 6.Public Health England . 17 Sep 2021. Investigation of SARS-CoV-2 variants of concern: technical briefings.https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201 [Google Scholar]
- 7.Hagen A., American Society for Microbiology . Jul 2021. How Dangerous Is the Delta Variant (B.1.617.2)? 30.https://asm.org/Articles/2021/July/How-Dangerous-is-the-Delta-Variant-B-1-617-2 [Google Scholar]
- 8.Barreto H.G., de Padua Milagres F.A., de Araujo G.C., et al. Diagnosing the novel SARS-CoV-2 by quantitative RT-PCR: variations and opportunities. J Mol Med (Berl) 2020;98:1727–1736. doi: 10.1007/s00109-020-01992-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vega-Magana N., Sanchez-Sanchez R., Hernandez-Bello J., et al. RT-qPCR assays for rapid detection of the N501Y, 69-70del, K417N, and E484K SARS-CoV-2 mutations: a screening strategy to identify variants with clinical impact. Front Cell Infect Microbiol. 2021;11:672562. doi: 10.3389/fcimb.2021.672562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H., Miller J.A., Verghese M., et al. Multiplex SARS-CoV-2 genotyping reverse transcriptase PCR for population-level variant screening and epidemiologic surveillance. J Clin Microbiol. 2021;59 doi: 10.1128/JCM.00859-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yaniv K., Ozer E., Kushmaro A. SARS-CoV-2 variants of concern, Gamma (P.1) and Delta (B.1.617), sensitive detection and quantification in wastewater employing direct RT-qPCR. medRxiv. 2021; Jul 16 doi: 10.1101/2021.07.14.21260495. [DOI] [Google Scholar]
- 12.Gand M., Vanneste K., Thomas I., et al. Deepening of in silico evaluation of SARS-CoV-2 detection RT-qPCR assays in the context of new variants. Genes (Basel) 2021;12:565. doi: 10.3390/genes12040565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogels C.B.F., Breban M.I., Ott I.M., et al. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gohl D.M., Garbe J., Grady P., et al. A rapid, cost-effective tailed amplicon method for sequencing SARS-CoV-2. BMC Genomics. 2020;21:863. doi: 10.1186/s12864-020-07283-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubin R. COVID-19 Vaccines vs variants - determining how much immunity is enough. JAMA. 2021;325:1241–1243. doi: 10.1001/jama.2021.3370. [DOI] [PubMed] [Google Scholar]
- 16.Nelson G., Buzko O., Spilman P., et al. Molecular dynamic simulation reveals E484K mutation enhances spike RBD-ACE2 affinity and the combination of E484K, K417N and N501Y mutations (501Y.V2 variant) induces conformational change greater than N501Y mutant alone, potentially resulting in an escape mutant. bioRxiv. 2021 Jan 13: 2021.01.13.426558. [Google Scholar]
- 17.Zhen W., Berry G.J. Development of a new multiplex real-time RT-PCR assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection. J Mol Diagn. 2020;22:1367–1372. doi: 10.1016/j.jmoldx.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain A., Rophina M., Mahajan S., et al. Analysis of the potential impact of genomic variants in global SARS-CoV-2 genomes on molecular diagnostic assays. Int J Infect Dis. 2021;102:460–462. doi: 10.1016/j.ijid.2020.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Planas D., Veyer D., Baidaliuk A., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 20.Starr T.N., Greaney A.J., Dingens A.S., et al. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep Med. 2021;2:100255. doi: 10.1016/j.xcrm.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.