Abstract
A young immunocompetent patient was admitted for a febrile illness with malaise, arthralgias, painful leg swelling, and polyserositis. Shortly prior to becoming ill, the patient had traveled to the Northern African desert. The symptoms disappeared during treatment with antibiotics (doxycycline and ceftriaxone) but recurred twice after stopping therapy. A motile gram-negative fusiform rod was isolated from a blood culture taken on the first admission. Analysis of the 16S rRNA gene of the blood culture isolate revealed close similarity with Helicobacter sp. flexispira taxon 8 (99.9% identity), a species that was previously reported as “Flexispira rappini.” This is the first reported case of a recurrent Helicobacter sp. flexispira bacteremia in an adult, immunocompetent patient.
In recent years, several new members of the genus Helicobacter have been isolated from numerous human and animal sources. Members of the genus Helicobacter include Helicobacter pylori, the organism associated with atrophic gastritis and duodenal and gastric ulcers, as well as other, less-common organisms such as H. bilis (4), H. canis (21), H. acinonyx (21), and H. trogontum (14). Previously, a group of organisms closely related to the Helicobacter spp. was called provisionally “Flexispira rappini” (3, 16, 21, 25). These bacteria are gram negative, microaerophilic, and fusiform shaped, with periplasmic fibers and bipolar tufts of sheathed flagella (3; J. H. Bryner, 4th Int. Workshop Campylobacter Infections, p. 440–442, 1987; J. H. Bryner, 14th Int. Cong. Microbiol., 1986). In a recent study, the phylogenetic relationship of the strains that comprise the group “F. rappini” was determined and 10 different taxa were identified that were grouped with various Helicobacter species (3). The study proposed that the “F. rappini” strains should be referred to as Helicobacter sp. and the appropriate flexispira taxon number, for example, Helicobacter sp. flexispira taxon 1 (3).
Strains of Helicobacter sp. flexispira have been isolated from various animal sources, including aborted sheep fetuses (8), intestinal mucosae of laboratory mice (18), and stools of puppies (2, 17). The organism also has been rarely recovered from human clinical specimens. The initial report of Helicobacter sp. flexispira isolates from humans described two men with a mild chronic diarrhea, in whom stool cultures were positive for Helicobacter sp. flexispira (17). A case of Helicobacter sp. flexispira bacteremia has been described in a child with pneumonia (22). Two cases of recurrent Helicobacter sp. flexispira bacteremia have been reported: one in an adult patient undergoing hemodialysis (20) and another one in a patient with X-linked agammaglobulinemia (27). These three isolates belonged to the Helicobacter sp. flexispira taxon 8 (3). Other Helicobacter species, such as H. cinaedi and H. fennelliae, have been occasionally recovered from the blood of adult patients, mostly patients with human immunodeficiency virus (HIV) infection who presented with cellulitis (7, 13, 19, 24). One recent investigation isolated H. cinaedi from an immunocompetent patient with septic arthritis and bacteremia (10). In the present report, we describe the first immunocompetent adult with recurrent Helicobacter sp. flexispira taxon 8 bacteremia.
CASE REPORT
A 36-year-old female patient presented at the emergency department with a history of several days of fever and general malaise. She had abdominal pain, but no diarrhea. Except for a past history of malaria tropica 4 years earlier, she was in good health. One week before the current illness, she had returned after a 2-week period of travel in the Algerian desert, where she had observed a skin lesion on the lower limb compatible with the prick of an insect but was otherwise healthy. Laboratory investigations in the emergency department yielded a hemoglobin level of 140 g/liter, a leukocyte count of 11.4 × 109/liter (82.5% neutrophils with a left shift), and a platelet count of 190 × 109/liter. The C-reactive protein level was 15 mg/liter. Blood smears for malaria were negative, and no Plasmodium falciparum antigens were detected. A chest X-ray was normal. The patient was discharged.
The following day, the patient was admitted because of sweats, chills, fever, diffuse arthralgias, and painful legs. Physical examination showed a maximal temperature of 39.2°C and swelling of the legs. Abdominal examination showed no organomegaly. There was no lymphadenopathy. Laboratory investigations revealed a hemoglobin level of 135 g/liter, a leukocyte count of 18.7 × 109/liter (neutrophils at 88.0% with a left shift), and a platelet count of 183 × 109/liter. The C-reactive protein level was 183 mg/liter, and the creatinine kinase level was normal. A total of seven blood cultures were drawn on admission and during the next few days. All but the first blood culture remained sterile. Initial identification of the positive blood culture indicated a Campylobacter-like organism. Urine culture and microbiological examination of the stool (bacteria, parasites) were negative. Serologic tests were positive for past infections with Epstein-Barr virus and hepatitis A and were negative for HIV and H. pylori. The serology result for Campylobacter spp. was borderline positive.
The patient was treated empirically with ceftriaxone (2 g/day) and doxycycline (200 mg/day) intravenously. On day 9, a chest X-ray showed bilateral pleural effusion. The cultures of this exudate remained sterile. Echocardiography on day 10 showed pericardial effusion that was not hemodynamically relevant. An abdominal computed tomography scan was normal. The patient improved slowly with resolution of fever, pleuritis, and pericarditis. Ceftriaxone was stopped on day 16 because of a possible allergy (fever and rash), and doxycycline was stopped on day 23. The patient was discharged on day 24.
The patient was seen again in the outpatient clinic 14 days later. She was asymptomatic; her blood counts and C-reactive protein levels were normal, and a chest X-ray showed no abnormalities.
Nine days later, the patient was readmitted to the hospital because of recurrent fever and dyspnea. Laboratory tests showed a hemoglobin level of 131 g/liter, a leukocyte count of 14.9 × 109/liter (neutrophils at 85.3% with a left shift), a platelet count of 380 × 109/liter, and an increased C-reactive protein level of 136 mg/liter. Several blood cultures remained negative. A relapse of the pericardial and bilateral pleural effusions was documented. The pleural exudate cultures remained sterile for bacteria, mycobacteria, and fungi. Antinuclear antibodies and rheumatoid factors were negative, and protein electrophoresis was normal. Empirical intravenous antibiotic therapy was begun on the day of admission with levofloxacin (1,000 mg/day), to which doxycycline (200 mg/day) was added 2 days later. The patient improved rapidly with disappearance of pleural and pericardial effusion and normalization of laboratory tests. Levofloxacin was continued for a total of 9 days and doxycycline for a total of 6 weeks.
Two weeks after stopping doxycycline, the patient noted again sweats, fever, and dyspnea. Physical examination was normal. Blood tests showed an increased C-reactive protein level (142 mg/liter). Blood cultures remained negative. Echocardiography and a chest X-ray showed mild pericardial and pleural effusions. The patient was treated symptomatically without antibiotics. The patient recovered completely with no further recurrences of her symptoms during an 11-month follow-up period.
MATERIALS AND METHODS
Blood cultures.
Several blood samples of 10 ml each were obtained by venipuncture. Blood samples were inoculated into BacT/Alert FAN aerobe and anaerobic blood culture bottles. The vials were monitored with the BacT/Alert automatic blood culture system and routinely incubated for 5 days at 37°C. According our routine protocol for positive cultures, gram-stained smears and subcultures on chocolate and blood agar were done with the broth of vials with positive growth.
Growth conditions and microscopic examination.
Gram staining and dark-field microscopy of a wet-mount preparation were performed by standard methods. Subcultures were performed on Columbia sheep blood, chocolate, and anaerobe blood agars and incubated at 37°C in 5% CO2. The same media, as well as a Preston agar plate for the cultivation of Campylobacter species, were incubated in a microaerophilic atmosphere (CampyGen; Oxoid). In addition BSK II (for the cultivation of Borrelia and Spirochaeta species) and Korthof medium (for the cultivation of Leptospira species) were inoculated with several drops of blood culture broth. These media were incubated aerobically at 37°C.
Electron microscopy.
The bacteria were lifted off the agar and gently suspended in phosphate-buffered saline (PBS; 145 mM NaCl, 7.5 mM Na2HPO4, 3.2 mM NaH2PO4 [pH 7.1]). Two volumes of a 4% formaldehyde solution was added, and the sample was incubated at room temperature for 1 h. The sample was centrifuged at 5,000 × g for 10 min at 4°C and gently resuspended in PBS. The suspension was placed on a Formvar-coated grid for 2 min, and the cells adhering to the grid were stained with 1% uranyl acetate. The grids were examined with a Phillips EM300 electron microscope.
Biochemical characterization.
Catalase production was tested with 3% hydrogen peroxide. Oxidase activity was tested with the Pyo-Test (Medical Wire & Equipment Co.).
Chromosomal DNA isolation.
Chromosomal DNA was isolated with the QiaAmp DNA Minikit according to the manufacturer's instructions (Qiagen AG, Basel, Switzerland). Specifically, cells were recovered from the agar plates and resuspended in 180 μl of the buffer ATL. The sample was digested with proteinase K (Qiagen AG) overnight at 56°C and subsequently with RNase A (Boehringer Mannheim AG, Rotkreuz, Switzerland). The DNA was eluted from the columns twice with 200 μl of double-distilled water. The eluates were pooled and stored at −20°C until use.
PCR amplification and DNA sequence analysis of the 16S rRNA gene.
PCR amplification of the 16S rRNA gene was done using the eubacterium-specific primer 27f (5′-AGAGTTTGATCMTGGCTCAG) and the universal primer 1492r (5′-TACGGYTACCTTGTTACGACTT [9]). Each reaction contained 200 ng of DNA, 200 μM concentrations of each deoxynucleoside triphosphate, 0.5 μM concentrations of each primer, 2 μM MgSO4, and 1 U of Vent DNA polymerase (NEB, Inc., Beverly, Mass.) in a final volume of 100 μl. The amplification conditions were as follows: (i) 5 min at 95°C; (ii) 30 cycles of 1 min at 95°C, 1 min at 50°C, and 1.5 min at 75°C; and (iii) 5 min at 75°C (modified from Haygood and Davidson [6]). The PCR products were sequenced using the following primers: 27f, 519r, 530f, 907r, 926f, and 1492r (9). From each colony type, two DNA preparations and four PCR amplifications were done to sequence both strands. The PCR products were sequenced at the central sequencing facility of the Department of Clinical Research of the University of Berne. The sequences were compiled using SeqMan II (DNASTAR, Inc., Madison, Wis.). The consensus sequence was compared to the databases using BLAST 2.0 (1) and the Ribosomal Database Project II (12). The sequences of IMM 687835 and of related bacteria were aligned with MegAlign (DNASTAR). Phylogenetic trees were calculated and constructed using TreeCon (23). The distance estimation was done using three different algorithims: Jukes and Canters, Jin and Nei (with a = 0.5, 1, or 2), and Galtier and Gouy. The relationship of the strains was calculated using either neighbor-joining or cluster analysis using UPGMA. For each calculation, bootstrap analysis was done by resampling the data set 1,000 times. The secondary structure of the 16S rRNA was predicted using GeneQuest (DNASTAR).
Nucleotide sequence accession number.
The nucleotide sequence of the 16S rRNA gene of IMM 687835 was deposited in GenBank under accession no. AF302762.
RESULTS
Blood cultures.
The strain was recovered from the initial aerobic blood culture taken prior to antibiotic therapy. The bottle gave a positive signal after 2 days of incubation. Gram staining of this blood culture showed a curved, gram-negative organism. The blood culture was subcultured on Columbia sheep blood and Preston agar because, initially, a Campylobacter-like organism was suspected.
Growth requirements and microscopic examination.
The initial subcultures grew very slowly, only after 16 days colonies were visible. Best growth was observed on anaerobic blood agar. Good results were also obtained in the BSK II broth where the organism could be propagated most easily. In later subcultures, growth was already visible after 2 days of incubation at 37°C in a microaerophilic atmosphere. No growth was observed on Preston agar, in the Korthof medium, or on agar media incubated in 5% CO2.
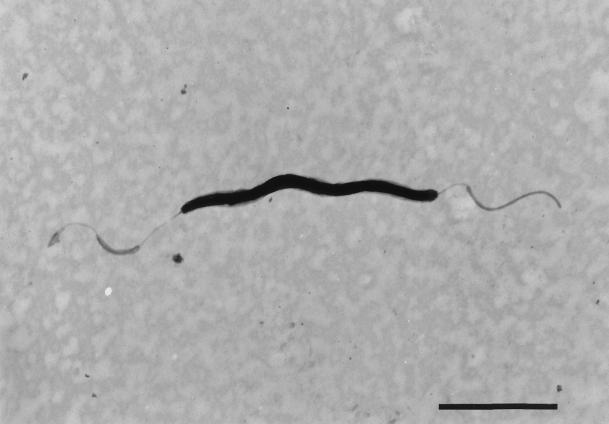
Two colony morphologies on anaerobic blood agar were observed: one was convex and undulate, and the other was flat and spreading. Colonies were alpha- or slightly beta-hemolytic. A particular smell was noted that was reminiscent of chlorine. Gram staining showed a slender, fusiform gram-negative rod. In the dark-field preparation, living bacteria showed a twisting motion. Electron microscopy revealed a thin, curved, fusiform bacterium up to 4 μm long, with bipolar sheathed flagella; interestingly, no periplasmic fibers were observed (Fig. 1).
FIG. 1.
Electron micrograph of a negatively stained sample. Single, sheathed bipolar flagella are apparent, and no periplasmic fibers were observed. Bar, 2 μm.
Phenotypic characterization.
The isolate was positive for oxidase and negative for the production of catalase. Further biochemical characterization could not be performed because the growth rate of the organism was too slow.
16S rRNA gene sequence analysis.
The DNA sequence of the 16S rRNA gene was identical for both colony morphologies, indicating that the different colonies types were variants of the same strain. Using BLAST 2.0 (1) and the Ribosomal Database Project II (12), the 16S rRNA gene sequence of the isolate was compared to the databases. This revealed that this strain was most similar to Helicobacter sp. flexispira taxon 8. The next-closest species, according to the 16S rRNA gene sequence, was a strain of H. cinaedi, followed by H. bilis. Alignment of the 16S rRNA genes revealed the presence of a 217-bp insert after nucleotide 156. Similar intervening sequences (IVS) at the same location in the 16S rRNA gene have been described in H. bilis (4), in H. canis (11), in H. cinaedi (26), in some “F. rappini” strains (4), and in a recently described new Helicobacter species (5). The IVS from IMM 687835 was most similar to the IVS from an H. cinaedi strain, CCUG 33887 (AF207738). The next most closely related IVS were from H. bilis. The IVS were shown to be highly variable in length and in base composition (4) and may alter the phylogenetic relationship between strains (15). Thus, we excluded the IVS from the 16S rRNA gene sequence for the subsequent phylogenetic analysis.
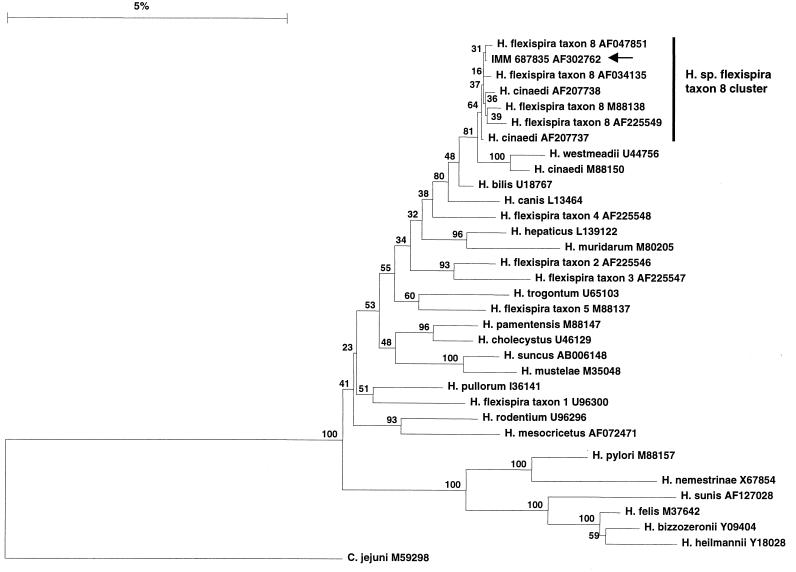
The phylogenetic relationship of IMM 687835 with other Helicobacter species was determined using TreeCon (23). The distance estimation was calculated using three different algorithms, and the phylogenetic relationship was determined either by neighbor-joining or clustering analysis. In all cases, IMM 687835 clustered with Helicobacter sp. flexispira taxon 8 strains (Fig. 2). Two nucleotide differences of 1,399 nucleotides were detected between IMM 687835 and the Helicobacter sp. flexispira taxon 8 strains ATCC 49317 (accession no. AF047851) and H1353 (accession no. AF118017). IMM 687835 was contained in a cluster that included four Helicobacter sp. flexispira taxon 8 strains and two H. cinaedi strains that had been isolated from dog feces (boot strap value of 64%). The type strain of H. cinaedi formed a cluster with H. westmeadii (bootstrap value of 100%) that was distinct from the Helicobacter sp. flexispira taxon 8 cluster. The further taxonomic characterization of the strains contained within the Helicobacter sp. flexispira taxon 8 cluster needs to be addressed in a study that includes numerous isolates of closely related species and employs several different methodologies (3, 26). The sequence similarity and the consistent placement in the phylogenetic trees indicate that the isolate described in our study is Helicobacter sp. flexispira taxon 8.
FIG. 2.
Phylogenetic trees of Helicobacter using the 16S rRNA gene sequence depict the close relationship of IMM 687835 (arrow) to Helicobacter sp. flexispira taxon 8. The distances between the strains were calculated using the Jukes and Canters algorithm, and the tree was constructed by neighbor-joining analysis. Bootstrap analysis was done with 1,000 resamplings; the percentage is shown at each node. The scale bar represents a 5% difference in the DNA sequence. The accession number for the sequences used in the study are shown in the figure.
DISCUSSION
In this report, we provide the first description of recurrent bacteremia with Helicobacter sp. flexispira taxon 8 in a young, healthy, immunocompetent woman without obvious risk factors. The bacteremia was associated with severe malaise, high fevers, arthralgias, painful leg swelling, and pericardial and pleural effusions. This is also the first patient with a Helicobacter sp. flexispira infection in whom polyserositis (pericardial and pleural effusion) has been observed. One previously described patient with agammaglobulinemia presented with persistent septicemia and leg swelling. Interestingly, the present patient also presented initially with bilateral painful leg swelling. The two other reported patients, a man undergoing hemodialysis and an otherwise healthy child, had evidence of lung infections as the most prominent organ involvement. In our patient, pulmonary involvement was limited to pleural effusions.
Response to antibiotic therapy was slow, and the patient had two episodes of relapsing symptoms after stopping antibiotic therapy, including recurrence of the pleural and pericardial effusions. The reason for the relapsing course is not known. During both relapses the blood cultures remained negative, and the second relapse was not treated with antibiotics and subsided spontaneously. The patient has since remained well. It is conceivable that relapsing symptoms were mediated by immunological phenomena. Alternatively, low-level recurrence of the bacteremia from an unrecognized persistant focus of infection may have escaped detection by blood cultures, since the organism is difficult to isolate (2, 20).
The susceptibility of our strain was not tested because of slow growth and lack of standardized procedures for this organism. Previous studies have reported that the organism is susceptible to doxycycline, ceftriaxone, and ciprofloxacine (20, 22). Our patient was thus treated with three antibiotics, all of which appear to be active in vitro against Helicobacter sp. flexispira taxon 8 strains. In contrast, in the agammaglobulinemic patient bacteremia persisted for months despite treatment with multiple antibiotics (including doxycycline, quinolones, and beta-lactams) and only cleared after prolonged therapy with imipenem and gentamicin. This may indicate that therapy for bacteremia is more difficult in immunocompromised patients.
The identification of Helicobacter sp. flexispira taxon 8 was difficult. As already described, the culture of Helicobacter sp. flexispira is fastidious. Because of the characteristic motility of these organisms, dark-field or phase-contrast microscopy of a wet-mount preparation can provide first clues as to its nature and help select the strategy to conclusively identify the pathogen. We and others used 16S rRNA gene sequencing. Phylogenetic analysis, independent of the algorithm used, grouped our isolate with Helicobacter sp. flexispira taxon 8. Interestingly, electron microscopy of our isolate did not reveal the typical flexispira morphology. No periplasmic fibers and no tufts of polar flagella were detected. Instead IMM 687835 had single bipolar flagella. This morphology is similar to that of an unpublished Helicobacter sp. flexispira taxon 8 strain, MIT 97-5078C, that was isolated from mice (F. E. Dewhirst, personal communication). This suggests that, although Helicobacter sp. flexispira taxon 8 has very similar 16S rRNA gene sequences, its morphology as determined by electron microscopy can vary.
Helicobacter sp. flexispira is considered a natural inhabitant of the intestinal mucosa of animals (18). In the present case, the source of the infection could not be identified. It is conceivable that the patient acquired the organism while traveling in the North-African desert. While an insect bite was reported by the patient, other sources of infection could not be excluded. In other patients, animal contacts have been implicated in the acquisition of Helicobacter sp. flexispira taxon 8 strains (17).
The present report indicates that immunocompetent adults can be infected with Helicobacter sp. flexispira taxon 8 and develop a prolonged and relapsing febrile illness with involvement of multiple organs.
ACKNOWLEDGMENTS
We thank S. Couzinet for help with the electron microscopy; T. Bodmer, F. E. Dewhirst, S. Droz, and K. Mühlemann for helpful discussion; and R. Troller for excellent technical assistance.
The Institute for Infectious Diseases of the University of Berne provided financial support for this work.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Archer J R, Romero S, Ritchie A E, Hamacher M E, Steiner B M, Bryner J H, Schell R F. Characterization of an unclassified microaerophilic bacterium associated with gastroenteritis. J Clin Microbiol. 1988;26:101–105. doi: 10.1128/jcm.26.1.101-105.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dewhirst F E, Fox J G, Mendes E N, Paster B J, Gates C E, Kirkbride C A, Eaton K A. ‘Flexispira rappini’ strains represent at least 10 Helicobacter taxa. Int J Syst Evol Microbiol. 2000;50:1781–1787. doi: 10.1099/00207713-50-5-1781. [DOI] [PubMed] [Google Scholar]
- 4.Fox J G, Yan L L, Dewhirst F E, Paster B J, Shames B, Murphy J C, Hayward A, Belcher J C, Mendes E N. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han S-R, Schindel C, Genitsariotis R, Märker-Hermann E, Bhakdi S, Maeurer M J. Identification of a unique Helicobacter species by 16S rRNA gene analysis in an abdominal abscess from a patient with X-linked hypogammaglobulinemia. J Clin Microbiol. 2000;38:2740–2742. doi: 10.1128/jcm.38.7.2740-2742.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haygood M G, Davidson S K. Small-subunit rRNA genes and in situ hybridization with oligonucleotides specific for the bacterial symbionts in the larvae of the bryozoan Bugula neritina and proposal of “Candidatus endobugula sertula.”. Appl Environ Microbiol. 1997;63:4612–4616. doi: 10.1128/aem.63.11.4612-4616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiehlbauch J A, Tauxe R V, Baker C N, Wachsmuth I K. Helicobacter cinaedi-associated bacteremia and cellulitis in immunocompromised patients. Ann Intern Med. 1994;121:90–93. doi: 10.7326/0003-4819-121-2-199407150-00002. [DOI] [PubMed] [Google Scholar]
- 8.Kirkbride C A, Gates C E, Collins J E, Ritchie A E. Ovine abortion associated with an anaerobic bacterium. J Am Vet Med Assoc. 1985;186:789–791. [PubMed] [Google Scholar]
- 9.Lane D S. 16S and 23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1990. [Google Scholar]
- 10.Lasry S, Simon J, Marais A, Pouchot J, Vinceneux P, Boussougant Y. Helicobacter cinaedi septic arthritis and bacteremia in an immunocompetent patient. Clin Infect Dis. 2000;31:201–202. doi: 10.1086/313930. [DOI] [PubMed] [Google Scholar]
- 11.Linton D, Clewley J P, Burnens A, Owen R J, Stanley J. An intervening sequence (IVS) in the 16S rRNA gene of the eubacterium Helicobacter canis. Nucleic Acid Res. 1994;22:1954–1958. doi: 10.1093/nar/22.11.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maidak B L, Cole J R, Lilburn T G, Parker C T, Jr, Saxman P R, Stredwick J M, Garrity G M, Li B, Olsen G J, Pramanik S, Schmidt T M, Tiedje J M. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 2000;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mammen M P, Jr, Aronson N E, Edenfield W J, Endy T P. Recurrent Helicobacter cinaedi bacteremia in a patient infected with human immunodeficiency virus: case report. Clin Infect Dis. 1995;21:1055. doi: 10.1093/clinids/21.4.1055. [DOI] [PubMed] [Google Scholar]
- 14.Mendes E N, Queiroz D M, Dewhirst F E, Paster B J, Moura S B, Fox J G. Helicobacter trogontum sp. nov., isolated from the rat intestine. Int J Syst Bacteriol. 1996;46:916–921. doi: 10.1099/00207713-46-4-916. [DOI] [PubMed] [Google Scholar]
- 15.Miller W L, Pabbaraju K, Sanderson K E. Fragmentation of 23S rRNA in strains of Proteus and Providencia results from intervening sequences in the rrn (rRNA) genes. J Bacteriol. 2000;182:1109–1117. doi: 10.1128/jb.182.4.1109-1117.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paster B J, Lee A, Fox J G, Dewhirst F E, Tordoff L A, Fraser G J, O'Rourke J L, Taylor N S, Ferrero R. Phylogeny of Helicobacter felis sp. nov., Helicobacter mustelae, and related bacteria. Int J Syst Bacteriol. 1991;41:31–38. doi: 10.1099/00207713-41-1-31. [DOI] [PubMed] [Google Scholar]
- 17.Romero S, Archer J R, Hamacher M E, Bologna S M, Schell R F. Case report of an unclassified microaerophilic bacterium associated with gastroenteritis. J Clin Microbiol. 1988;26:142–143. doi: 10.1128/jcm.26.1.142-143.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schauer D B, Ghori N, Falkow S. Isolation and characterization of “Flexispira rappini” from laboratory mice. J Clin Microbiol. 1993;31:2709–2714. doi: 10.1128/jcm.31.10.2709-2714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skirrow M B, Jones D M, Sutcliffe E, Benjamin J. Campylobacter bacteraemia in England and Wales, 1981–91. Epidemiol Infect. 1993;110:567–573. doi: 10.1017/s0950268800050986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorlin P, Vandamme P, Nortier J, Hoste B, Rossi C, Pavlof S, Struelens M J. Recurrent “Flexispira rappini” bacteremia in an adult patient undergoing hemodialysis: case report. J Clin Microbiol. 1999;37:1319–1323. doi: 10.1128/jcm.37.5.1319-1323.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanley J, Linton D, Burnens A P, Dewhirst F E, Owen R J, Porter A, On S L W, Costas M. Helicobacter canis sp. nov., a new species from dogs: an integrated study of phenotype and genotype. J Gen Microbiol. 1993;139:2495–2504. doi: 10.1099/00221287-139-10-2495. [DOI] [PubMed] [Google Scholar]
- 22.Tee W, Leder K, Karroum E, Dyall-Smith M. “Flexispira rappini” bacteremia in a child with pneumonia. J Clin Microbiol. 1998;36:1679–1682. doi: 10.1128/jcm.36.6.1679-1682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van de Peer Y, De Wachter R. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput Appl Biosci. 1997;13:227–230. doi: 10.1093/bioinformatics/13.3.227. [DOI] [PubMed] [Google Scholar]
- 24.Vandamme P, Falsen E, Pot B, Kersters K, De Ley J. Identification of Campylobacter cinaedi isolated from blood and feces of children and adult females. J Clin Microbiol. 1990;28:1016–1020. doi: 10.1128/jcm.28.5.1016-1020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vandamme P, Falsen E, Rossau R, Hoste B, Segers P, Tytgat R, De Ley J. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int J Syst Bacteriol. 1991;41:88–103. doi: 10.1099/00207713-41-1-88. [DOI] [PubMed] [Google Scholar]
- 26.Vandamme P, Harrington C S, Jalava K, On S L. Misidentifying helicobacters: the Helicobacter cinaedi example. J Clin Microbiol. 2000;38:2261–2266. doi: 10.1128/jcm.38.6.2261-2266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weir S, Cuccherini B, Whitney A M, Ray M L, MacGregor J P, Steigerwalt A, Daneshvar M I, Weyant R, Wray B, Steele J, Strober W, Gill V J. Recurrent bacteremia caused by a “Flexispira” -like organism in a patient with X-linked (Bruton's) agammaglobulinemia. J Clin Microbiol. 1999;37:2439–2445. doi: 10.1128/jcm.37.8.2439-2445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]