Abstract
Colon adenomas with proliferating mutant cells may progress to invasive carcinomas. Proliferation of cells in human colorectal tissue is circadian, greater in the interval 4 to 12 hours after midnight than 16 to 24 hours after midnight. We have tested the hypothesis that chemotherapy administered during the time of greater cell proliferation will be more effective than chemotherapy administered during the time of lesser proliferation. An agent-based computer model of cell proliferation in colon crypts was calibrated with measurements of cell numbers in human biopsy specimens. It was used to simulate cytotoxic chemotherapy of an early stage of colon cancer, adenomas with about 20% of mutant cells. Chemotherapy doses were scheduled at different 4-hour intervals during the 24-hour day, and repeated at weekly intervals. Chemotherapy administered at 4 to 8 hours after midnight cured mutant cells in 100% of 50 trials with an average time to cure of 7.82 days (s.e.m. = 0.99). In contrast, chemotherapy administered at 20 to 24 hours after midnight cured only 18% of 50 trials, with the average time to cure of 23.51 days (s.e.m. = 2.42). These simulation results suggest that clinical chemotherapy of early colon cancer may be more effective when given in the morning than later in the day.
Keywords: Chronotherapy, circadian rhythm, colorectal neoplasm, computer simulation, chemotherapy
Introduction
Colon cancer was diagnosed in more than 100 000 patients in the United States in 2019, and an estimated 50 000 people died of colon cancer. 1 For patients diagnosed with stage II or III colon cancer surgery with the intent to cure is recommended. 2 Adjuvant chemotherapy after surgery is given for the purpose of reducing the probability of recurrence. However, adjuvant therapy has had only a modest effect on the 5-year recurrence-free survival.3-7
Because of the modest effect of therapy on late stage colon cancer, we have considered therapy of early stages of colon cancer. Specifically, we propose to prevent the progression of mutant cells that first arise in the crypt from proliferating, filing the crypt, and developing adenomas, an early stage in the progression of colon cancer. This requires determining the highest chemotherapeutic drug doses, and related drug schedules, that are most effective in eliminating mutant cells while retaining the function of normal cells in the crypt.
In order to determine the highest dose it is not sufficient to just increase the dose of a chemotherapeutic drug since higher doses may have unacceptable adverse side effects. In order to increase the efficiency of chemotherapy without increasing the dose, several different strategies have been employed. An early suggestion was to use a combination of different drugs with the expectation of a synergistic effect at modest doses of each.8,9 This strategy has also been employed to overcome resistance that can develop to a single drug. 10
Another strategy to overcome the adverse effects high doses of anti-cancer drugs, is to modify the schedules in which drugs are administered. Such dose schedules include metronomic therapy in which low doses are given over a long time, 11 adaptive therapy in which the dose is modified depending on the tumor response, 12 and intermittent therapy in which high doses are alternated in time with low or no doses allowing recovery between doses. 13 Intermittent dose schedules have been proposed to be effective against drug-resistant early colon cancers, 14 especially when the dose schedules are optimized. 15
Modified dose schedule strategies have also been proposed to exploit the observation that in many tissues, 16 including gastrointestinal mucosa, 17 the rates of cell proliferation are different at different times of the day, they are circadian. The expectation is that more rapidly dividing cells at specific times of the day would be more sensitive to a cytotoxic drug and that less rapidly dividing cells would be less sensitive to a drug. Choosing doses during different times of the day, chronotherapy, could increase the efficiency of tumor treatment and reduce the toxic side effects to patients.18-20
However, chronotherapy has not been routinely adapted in clinical oncology protocols. 21 This may be due in part to the fact that chronomodulated doses in clinical trials of late stage metastatic colon cancer have shown statistically significant, but only modest clinical effects.22-25 Nevertheless, clinical trials of early, rather than late, colon cancer might reveal a larger effect of chronotherapy.
In this project we have investigated the efficiency of chronotherapy against early colon cancer. We have used a combination 2 strategies. First, dose schedules that are given at specific times of the day (circadian-timed) and that are repeated intermittently at weekly intervals. Second, we have targeted an early stage of colon cancer with the expectation that preventing its progression would prevent the development of invasive carcinoma. In this early stage of colon cancer there is an abnormal proliferation of mutant cells in the crypts of the colon. We have developed an agent-based computer model of cell dynamics in crypts that can simulate different circadian-timed chemotherapies. Computer simulations of doses at different times of day can indicate if there is a difference in the efficiency of eliminating mutants from the crypt while retaining normal function of the crypt. We have sought answers to the following questions:
Is there a difference in efficiency of eliminating mutants from crypts when chemotherapy is given at different times of the day?
If there is a difference, what time of day is chemotherapy most effective in curing mutants?
Methods
Figure 1 shows a flowchart of the major steps of the methods.
Figure 1.
Flowchart of the main steps in the modification of the agent-based model of cell proliferation in human colon crypts to account for circadian proliferation of mutant cells, and the response of mutant cells to cytotoxic therapy applied at different times of day. Simulation results indicate the probability to cure mutant cells, and the length of time to cure mutant cells, when cytotoxic therapy is applied at different times of day.
Computer model
Colon_Crypt_Model_041321.nlogo is an agent-based model of cell dynamics in human colon crypts. The model is available from the Rutgers University SOAR Repository at https://doi.org/doi:10.7282/t3-dj35-2d40. It was developed in the application NetLogo version 5.3.1, and revised to run in NetLogo version 6.2.0. It will not run in the Web version of NetLogo. NetLogo is a multi-agent programmable modeling environment. It is authored by Uri Wilenski and developed at The Center for Connected Learning (CCL) and Computer-Based Modeling. It is multi-platform (Mac, Windows, or Linux) open source application. NetLogo version 6.2.0, can be downloaded at http://ccl.northwestern.edu/netlogo/download.shtml. The model, and its biological basis, are described below.
Human colon crypts are test-tube shaped invaginations in the wall of the large intestine. Stem cells are located at the bottom of the crypt, proliferating cells are located above the stem cells in the bottom one-third of the crypt, and differentiated cells are located in the top two-thirds of the crypt. Stem cells may divide to produce more stem cells or stem cells and proliferating cells. The progeny of the proliferating cells move up the crypt, where they differentiate and have a reduced probability of dividing. Crypts maintain a quasi-stationary number of cells in a stochastic process as new cells are born from proliferating cells, and differentiated cells die and are removed at the top. 26
The current model, and an earlier version, 27 assumes that the probability that a cell will divide is determined, in part by its position in a microenvironment gradient along the crypt axis, with the probability of division higher at the bottom than at the top. The probability of a cell dying is determined by its position in gradient along the crypt axis, higher at the top than at the bottom. The stem cells at the bottom of the crypt are in a quiescent niche.
The model was calibrated with cell numbers measured in a human biopsy specimen. A histological slide containing sections from a biopsy of the sigmoid colon of a normal patient was stained with the anti-body MIB-1 to the proliferating antigen Ki-67, and counterstained with hematoxylin. Positively stained cells were considered proliferating cells. Unstained cells at the bottom of the crypt were considered quiescent stem cells. Unstained cells near the top of the crypt were considered differentiated cells. Model parameter values of the divide gradient and die gradient were determined that reproduced the measured mean and variance of each cell type in multiple crypts. 27 The “Info” tab accessible at the interface of the model contains additional descriptions of each feature of the model, and the bases for choosing the default model parameters.
The behavior of the model was verified as reproducing 3 features of the biological crypts that had previously been observed, that is, induction of adenomas by mutations occurring at the top or at the bottom of the crypts, 28 monoclonal conversion by neutral drift, 26 and robust recovery from perturbation by exposure to a cytotoxic agent. 29 Cytotoxic dose schedules were determined that could eliminate mutant cells while sparing normal crypt function.14,15
The current version of the model was developed to account for the observation that the probability of cell division in colon tissue is circadian. 30 In this model the probability of cell division of any cell in the crypt depends upon its position along the axis of the crypt, and is modified by the time of day. There are tabs to access 3 sections. Interface: a graphical user interface with dynamic representation of stem cells, proliferating cells, and differentiating cells as they divide and move, as well as plots of the number of each type of cell type. At the Interface, parameter values other than the default values may be entered, the switch “Circadian Proliferation” may be turned On, and the “Applies Chemotherapy” switch may be turned On. Information: extensive text describing the biological basis for the agent-based model, default parameter values, and description of each section of the model. Code: computer code with comments. Also, in the Tools menu, there is a selection for Behavior Space, which has code to enable multiple simulation runs with different parameter values.
Circadian simulation code
In order to simulate circadian proliferation of cell division the code was revised based upon published experimental data. 30 The incorporation of tritiated thymidine into DNA of mucosal biopsy specimens taken every 2 or 3 hours for a 24-hour span was measured, and reported as a circadian waveform. This experimental data was fitted by Sudeepti Vedula to a cosine function using the MATLAB routine “Cosinor Analysis, 2008” written by C. Cox, https://www.mathworks.com/matlabcentral/fileexchange/20329-cosinor-analysis/content/html/cosinor.html. Accessed Nov. 4, 2016. The MATLAB routine implemented “Cosinor-based Rhythmometry.” 31 The NetLogo code was revised so that the probability that a cell will divide is determined by its location in the crypt and, in addition, is multiplied by the following periodic function
Where 1 computer “step” is equal to 4 hours. This function is implemented at the NetLogo Interface by turning on the switch “Circadian Proliferation.” It reproduces both the phase and the 24-hour period of the experimental data. The coincidence of experimental and simulated phase is shown in Figure 2. The 24-hour periodicity was confirmed by spectral analysis using the statistical application JMP v.15.2.1 (Cary, NC. www.sas.com). Note that the argument of cosine in the NetLogo modeling application is in degrees, whereas the argument in the JMP statistical application is in radians. Simulations in NetLogo were done on a MacBook Pro 2017, using Mac OS v10.15.5.
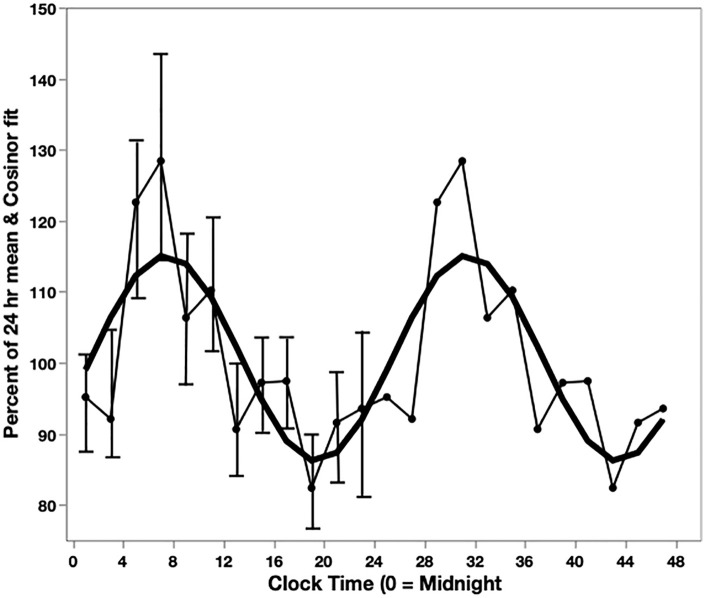
Figure 2.
Circadian rhythm of cell proliferation in humans, and fit to cosine function. The data for a 24-h interval is repeated for illustration. Light line: DNA synthesis determined by the incorporation of tritiated-thymidine in rectal mucosal samples from 16 human subjects. 30 Each point is the mean of samples in a 2-h interval. The s.e.m. are indicated for the 24-h on the left. Heavy line: Cosine function fit to the experimental data using the Cosinor method. 31 The cosine function was used to determine the rate of cell proliferation at different times of day in the model.
Chemotherapy parameters
Intermittent dose schedules of cytotoxic drugs are used with the goal of minimizing side effects while retaining anti-tumor effectiveness. 2 Use of intermediate dose schedules requires the choice of values for 3 parameters, the cytotoxic effect of the drug, duration of exposure to the drug, and intervals between repeated exposure to the drug. 14 In the code, these are referred to as Lethality, Duration, and Interval. In order to investigate the effect of doses at different times of day, we choose a Duration of 1 computer step, equal to 4-hour. A 4-hour duration allows investigation of the effect of drugs given at 6 different times during the day. In order to simulate a length of time between doses we choose an Interval commonly used in chemotherapy clinics, that is, 1 week, which is 42 computer steps. In order to minimize side effects, we choose a Lethality of 5. Lethality is a multiplication factor that increases the probability of a cell dying determined by its position along the crypt axis. It affects both normal cells and mutant cells. After choosing a Duration of 1 step and Interval of 42 steps, the value of Lethality of 5 was chosen because this combination was among the optimal dose schedules previously reported. 15 This value of lethality results in minimum side effect as indicated by the result that all crypts recovered from this cytotoxic dose in all 300 simulation runs (50 runs with different random seeds, at each of 6 different times of day).
Mutant cell parameters
In this project we are interested in the effect of circadian-timed dose schedules on an early colon cancer with the goal of intercepting the progression of adenomas (crypts with mutant cells) to invasive carcinomas. We previously investigated the effect of chemotherapy on mutant cells that divide at different rates than normal cells, and die at different rates. 14 Based upon that work, in this project we selected mutants that divide at 1.16X the rate of normal cells at the same place along the crypt axis, and die at 1.1X the rate of normal cells at the same place along the crypt axis. Mutants were placed in the region of proliferating cells because that is the region in which mutant cells are most efficient in proliferating and filling the crypt. 27 Cells in the proliferating region were induced to be mutant cells with 100% probability. The induction of mutants occurred at 6 time-steps before the beginning of chemotherapy resulting in the same proportion of mutant cells in the crypt (17.8 %, s.e.m. = 0.009) for each of the different times that chemotherapy started.
Results
Circadian cell proliferation
Colon crypts with few mutant cells are at an early stage in colon cancer. They may proliferate and progress to invasive colon cancer. In order to investigate the effect of circadian-timed chemotherapy on early colon cancer by computer simulation 2 items are needed. One is quantitative information on cell proliferation in human colorectal tissue at different times of day. The second is a computer model of cell proliferation in colon crypts that can simulate chemotherapy applied at different times of day, and that produces quantitative predictions on the efficiency of eliminating mutant cells.
As an indication of cell proliferation rates, the incorporation of tritiated thymidine into DNA of cells of human mucosa obtained at different times of day was measured. 30 A circadian rhythm was found with a maximum (acrophase) at 8-hour after midnight and a minimum (nadir) at 20 hours after midnight (Figure 2). These results were independently confirmed. 32 We used the Cosinar method 31 to fit the following function to the experimental data, Y = 1.00591 + 0.143376 × cos (60 × steps−175.574) (Figure 2), where 1 step equals 4-hour. This function was used to simulate different rates of cell division at different times of day.
We previously reported on the development of a model of cell proliferation in human colon crypts that was calibrated with measurements of stem cells, differentiated cells, and proliferating cells in normal human biopsy specimens taken at a single time of the day. 27 The model was realistic in that it could reproduce the average number and variance of each cell type in multiple crypts. Also, several emergent behaviors of the model reproduced experimentally observed properties of proliferating cells in human colon crypts. The model was used to demonstrate that intermittent chemotherapy could be effective in eliminating mutant cells in heterogenous and drug-resistant early colon cancer. 33
For this project we have revised the model so that the rates of cell proliferation have the same circadian period and phase as determined experimentally. 30 The rate of division of both mutant and normal cells is thus determined by their position in the crypt, multiplied by a factor determined by the time of day.
Probability of curing mutants with circadian-timed chemotherapy
Without chemotherapy mutant cells may continue to proliferate, fill the crypt, and progress to an invasive carcinoma. The goal of chemotherapy is to eliminate mutant cells before they fill the crypt, while allowing recovery of normal cells and normal crypt function. With cytotoxic chemotherapy, such as 5-fluoruracil, both dividing mutant cells and dividing normal cells may be killed. However, the rate of cell division of mutant cells is more rapid than normal cells, so they are more sensitive to cytotoxic chemotherapy. Quiescent stem cells are most resistant. Quiescent stem cells may survive chemotherapy and produce proliferating cells that replace any normal proliferating cells that are killed. An appropriate dose of chemotherapy may eliminate all mutant cells and maintain sufficient numbers of normal cells to allow crypts to recover and continue to function.14,27
In this project we have asked if the difference in cell proliferation at different times of day can affect the probability of eliminating all mutant cells from a crypt, while retaining normal crypt function. And if so, how long does it take to eliminate mutant cells.
Chemotherapy doses of 4-hours were started at 6 different times of day, and the probability of eliminating all mutants in each of 50 runs was determined. The probability of curing all mutant cells was not uniform for different times of day (Figure 3, Table 1). The probability of curing all mutants depended on the time of day that the chemotherapy was applied. For instance, chemotherapy started 4-hour after midnight eliminated all mutants in 100% of 50 runs. In contrast, chemotherapy started 20-hour after midnight cured all mutants in only 18% of 50 runs, while the remaining runs had mutant cells proliferate and fill the crypts.
Figure 3.

Probability that patients are cured with 4-h of chemotherapy starting at different times after midnight. For each time there were 50 simulation runs. The distribution is not uniform, chi-squared test, P < .0001.
Table 1.
Probability of curing all mutants and time to cure, for chemotherapy at different times of day.
| Circadian ON (step) a | Mutants induced (step) b | Chemotherapy ON (step) | Time of chemotherapy c (h after midnight) | Probability Cure d (n = 50 runs) | Time to Cure e (days: mean, s.e.m.) |
|---|---|---|---|---|---|
| 200 | 206 | 212 | 0-4 | 0.80 | 16.43, 1.45 |
| 200 | 207 | 213 | 4-8 | 1.00 | 7.82, 0.99 |
| 200 | 208 | 214 | 8-12 | 0.90 | 5.74, 0.76 |
| 200 | 209 | 215 | 12-16 | 0.90 | 16.34, 0.84 |
| 200 | 210 | 216 | 16-20 | 0.38 | 22.40, 8.08 |
| 200 | 211 | 217 | 20-24 | 0.18 | 23.51, 2.42 |
Each computer step = 4 h. Circadian ON after 200 steps allow crypts to reach quasi-stationary dynamics.
Probability of mutation = 1, at Rows = 55-60, yields 17.8% mutants, s.d. 0.009 mutants, allowing each run to start chemo with same number of mutants.
With no chemotherapy, all runs result in mutants filling the crypt and the crypt size overflows.
Cure means eliminate all mutants, and normal crypt recovers. The distribution of probabilities to cure is not uniform, chi-squared test, P < .0001.
The length of time to eliminate all mutants in a simulation run. Chemotherapy was repeated at weekly intervals until all mutants were cured, or a limit of 1000 steps, about 6 months. For chemotherapy starting at 8 to 12 h and at 20 to 24 h, the length of time to cure is significantly different, t-test assuming unequal variances, P < .0001.
Length of time to cure mutants for circadian-timed chemotherapy
Chemotherapy started at each time of day was repeated at weekly intervals at the same time of day, until all mutants were eliminated, or for a limit of 6 months. The average length of time to cure all mutants depended on the time of day that the chemotherapy was started. For instance, chemotherapy started in the morning at 4-hour or 8-hour after midnight cured mutants in a shorter average length of time than chemotherapy applied in the evening at 16- or 20-hour (Figure 4, Table 1).
Figure 4.

The length of time (days) that mutants are cured after 4-h of chemotherapy starting at different times after midnight. Mean and s.e.m., N = 50. The lengths of time to cure mutants for chemotherapy starting at 8- and 20-h after midnight are significantly different, t-test assuming unequal variances. P < .0001.
Chemotherapy applied at 20-hour after midnight took the longest average length of time to cure. But the distribution of lengths of time to cure was not unimodal. Rather, the lengths of time to cure occurred at weekly intervals. For chemotherapy applied at 20-hour after midnight mutants were cured from crypts at intervals of 14, 21, 28, and 35 days (Figure 5). In the remaining simulation runs, crypts were not cured of mutants in spite of weekly chemotherapy for 6 months.
Figure 5.

The length of time (days) to cure after 4-h of chemotherapy starting at 20-h after midnight (top) and at 8-h after midnight (bottom). For chemotherapy starting at 20-h, 38% (19/50) of the runs had all mutants cured, distributed at 14, 21, 28, and 35 days. For chemotherapy at 8-h after midnight, 90% (45/50) of the runs were cured distributed most within 1 day, with the remainder cured at 7 days, and 14 days.
In contrast, chemotherapy applied at 8-hour after midnight had the shortest average time to cure. And among those simulation runs that had mutants cured, 60.4% (20/45) were cured at the first application of chemotherapy. Subsequent weekly chemotherapy cured mutants at 7 and 14 days (Figure 5), with no remaining mutants.
Discussion
We have used computer simulation to determine if the efficiency of chemotherapy is different at different times of day. The results indicate that chemotherapy doses in the morning are more effective than later in the day. Chemotherapy at 4 to 8 hours after midnight eliminated all mutants from colon crypts with a higher probability, and in the shortest time. Chemotherapy at other times of day had a lower probability of eliminating mutants from all crypts, and in those crypts that did have mutants eliminated, it took a longer time and repeated doses.
The veracity of the simulation results reported here depends upon 2 items, a valid model and real data. We have previously described the development of an agent-based computer model of cell dynamics in the human colon crypt. 27 Its validity was indicated in the following ways. It reproduces the quasi-stationary stochastic dynamics of stem cells, proliferating cells, and differentiated cells in colon crypts that were measured in human biopsies. Also, the emergent behavior of the model was verified by its ability to reproduce experimentally observed monoclonal conversion by neutral drift, 26 formation of adenomas resulting from mutant cells initiated at the bottom or top of the crypt, 28 and by its robust ability to recover from perturbation by a cytotoxic chemotherapy. 29 The model has been useful in suggesting optimum therapeutic dose schedules, 15 and dose schedules that could be effective against heterogeneous and drug-resistant early colon cancers. 14 In addition to suggesting dose schedules for therapy, it has also been used to suggest dose schedules for primary chemo-prevention, 14 and secondary prevention of recurrence from minimal disease. 34
The second item that is necessary to assure the veracity of the simulation results is that the model was calibrated based upon real data. Numbers of stem cells, proliferating cells, and differentiated cells were measured in multiple crypts in microscopic sections of human biopsy specimens. 27 The average and variation in numbers of each cell type were used to calibrate gradients along the crypt axis. These gradients determined the probability that a cell would divide or die at any position as the cell moved up the crypt. The revised version of the model used in this project also had the probability of a cell dividing at any position in the crypt modified by a function that depends upon the time of day. This function was determined by fitting a cosine equation to experimental data from rectal mucosal biopsy specimens obtained every 2 to 3 hours during a 24-hour span, and measuring ex vivo the incorporation of tritiated thymidine into DNA. 30 The data indicated a circadian variation in cell division. Similar methods were used, and the results independently confirmed. 32
The simulation results reported here depend upon mutant cells having circadian rates of proliferation. It is likely that mutants in colon crypts have circadian rhythms of cell proliferation since circadian rhythms are found in the adjacent cells in normal colon tissue,30,32 in colon polyps which are another premalignant lesion, 32 and even in colorectal cancers. 35
Other computational and mathematical models that have been developed for chronotherapy have used different approaches than the agent-based computer modeling approach reported here. Six ordinary differential equations that described pharmacokinetics and pharmacodynamics, in addition to circadian cell proliferation, were simulated, and the relative toxicities and anti-tumor efficacies were determined. The results indicated the advantage of periodic time-schedules compared to continuous drug infusion of the same dose. 36 An automaton model was developed that described circadian transitions through the cell cycle, with cells most sensitive to a drug such as 5-fluoruracil during DNA synthesis. The conclusion was that the time of administration of the drug that is least toxic to the host is 4 AM, 37 consistent with our results. A differential equation model of proliferation of host and tumor cells was used to determine the parameters most influential in determining optimal chronomodulated treatment schedules. The conclusion was that the duration of the cell cycle phase targeted by the treatment and the cell proliferation rate is crucial in determining the best time to administer cell cycle-specific drugs. The efficacy of chronomodulated treatments with a 24-hour period administered at different times may depend upon the growth rate of the tumor, 38 and the host’s multi-scale systems and the drug pharmacokinetics.39,40
Chronomodulated dose schedules have been compared with constant doses in clinical trials.22,23 For instance, in a Phase III trial circadian schedules increased the median overall survival of metastatic colon cancer patients from 18.7 to 19.6 months. 24 And in a meta-analysis of 3 international Phase III trials, chronomodulated dose schedules increased the median overall survival from 17.5 to 20.8 months for men, and from 16.6 to 18.4 months for women. 25 Although a few months may seem modest, it should be noted that that these were patients with advanced stage cancer. Nevertheless, colorectal cancers, such as metastatic cancer, are modestly more responsive to circadian-modulated chemotherapy in the period of 1 to 4 AM. 22 This is similar to the time that we identified as most effective for early colon cancer. Our simulation results suggest that treatment of an early stage of colon cancer may be more effective than treatment of advanced colon cancer. It would be an example of interception and primary prevention of tumor progression. 41 An apt analogy is described by the aphorism “Locking the barn door is better than trying to round up horses that have already left the barn.”
There are some limitations to this study. This model, like all models, is limited in extent. It is focused on the cell proliferation and death of mutant cells in colon crypts. It does not include effects of drugs on other aspects of human physiology. 39 Therefore, the simulated result that 100% of mutant cells are cured when cytotoxic drugs are given in the early morning may not be fully achieved in the clinic.
Translation of the computer simulation results into clinical practice could be facilitated by technologies that are being developed to detect pre-invasive changes in the colon. For instance, mucin MUC5AC a specific marker of aberrant crypt foci, 42 autoantibodies to tumor-associated antigens on mutant cells, 43 and DNA in feces released from mutant cells when they undergo apoptosis at the top of the crypt.44,45 The simulation results reported here can provide an additional incentive to further develop these and other technologies for detection of early stages of colon cancer. 46 When these methods are established, our simulation results could help to guide the design of chronomodulated clinical trials.
In summary, results of computer simulation of chronotherapy of early colon cancer indicate (1) that doses scheduled in the early morning could be considerably more effective than doses scheduled later in the day, and (2) early morning doses might eliminate mutants from colon crypts with higher probability and shorter time than doses later in the day.
Acknowledgments
I thank Sudeepti Vedula, Department of Biomedical Engineering, Rutgers University for fitting a cosine function to experimental data, Dr. Chase Cockrell for discussions, Mr. Jeffrey Wood and the Rutgers University SOAR Repository staff for archive services
Footnotes
Funding: The author disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The Human Genetics Institute of New Jersey, the New Jersey Breast Cancer Fund, and the Rutgers Cancer Institute of New Jersey (grant no. PA 30CA072720).
Declaration of conflicting interests: The author declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: The author ran the simulations, interpreted the results, produced the figures, and wrote the manuscript.
ORCID iD: David E Axelrod  https://orcid.org/0000-0002-0912-3870
https://orcid.org/0000-0002-0912-3870
Supplementary Material: Model: The Netlogo model “Colon_Crypt_Model_041321.nlogo” contains a graphical user interface, explanatory information text, annotated code, and the Behavior Space Tool used to run the code. It is available from the Rutgers University SOAR Repository at https://doi.org/doi:10.7282/t3-dj35-2d40
Data: All simulation output data files and their descriptions are available from the Rutgers University SOAR Repository at https://doi.org/doi:10.7282/t3-c759-6p57
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34. [DOI] [PubMed] [Google Scholar]
- 2. Benson AB, III. NCCN guidelines version 1.2020, colon cancer. 2020. Accessed April 6, 2021 https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf
- 3. Moertel CG, Fleming TR, Macdonald JS, et al. Intergroup study of fluorouracil plus levamisole as adjuvant therapy for stage II/Dukes’ B2 colon cancer. J Clin Oncol. 1995;13:2936-2943. [DOI] [PubMed] [Google Scholar]
- 4. Schippinger W, Samonigg H, Schaberl-Moser R, et al. A prospective randomised phase III trial of adjuvant chemotherapy with 5-fluorouracil and leucovorin in patients with stage II colon cancer. Br J Cancer. 2007;97:1021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andre T, Colin P, Louvet C, et al. Semimonthly versus monthly regimen of fluorouracil and leucovorin administered for 24 or 36 weeks as adjuvant therapy in stage II and III colon cancer: results of a randomized trial. J Clin Oncol. 2003;21:2896-2903. [DOI] [PubMed] [Google Scholar]
- 6. Quasar Collaborative Group, Gray R, Barnwell J, McConkey C, Hills RK, Williams NS, et al. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370:2020-2029. [DOI] [PubMed] [Google Scholar]
- 7. Matsuda C, Ishiguro M, Teramukai S, et al. A randomised-controlled trial of 1-year adjuvant chemotherapy with oral tegafur-uracil versus surgery alone in stage II colon cancer: Sacura trial. Eur J Cancer. 2018;96:54-63. [DOI] [PubMed] [Google Scholar]
- 8. DeVita VT, Jr, Young RC, Canellos GP. Combination versus single agent chemotherapy: a review of the basis for selection of drug treatment of cancer. Cancer. 1975;35:98-110. [DOI] [PubMed] [Google Scholar]
- 9. Bayat Mokhtari R, Homayouni TS, Baluch N, et al. Combination therapy in combating cancer. Oncotarget. 2017;8:38022-38043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang L, Wang H, Song D, Xu M, Liebmen M. New strategies for targeting drug combinations to overcome mutation-driven drug resistance. Semin Cancer Biol. 2017;42:44-51. [DOI] [PubMed] [Google Scholar]
- 11. Pasquier E, Kavallaris M, André N. Metronomic chemo-therapy: new rationale for new directions. Nat Rev Clin Oncol. 2010;7:455-465. [DOI] [PubMed] [Google Scholar]
- 12. Gatenby RA, Silva AS, Gillies RJ, Frieden BR. Adaptive therapy. Cancer Res. 2009;69:4894-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leder K, Pitter K, LaPlant Q, et al. Mathematical modeling of PDGF-driven glioblastoma reveals optimized radiation dosing schedules. Cell. 2014;156:603-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Axelrod DE, Bravo R. Chemoprevention of colon cancer: advantage of intermittent pulse treatment schedules quantified by computer simulation of human colon crypts. Converg Sci Phys Oncol. 2017;3:035004. [Google Scholar]
- 15. Cockrell C, Axelrod DE. Optimization of dose schedules for chemotherapy of early colon cancer determined by high-performance computer simulations. Cancer Inform. 2019;18:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smaaland R. Circadian rhythm of cell division. Prog Cell Cycle Res. 1996;2:241-266. [DOI] [PubMed] [Google Scholar]
- 17. Bjarnason GA, Jordan R. Rhythms in human gastrointestinal mucosa and skin. Chronobiol Int. 2002;19:129-140. [DOI] [PubMed] [Google Scholar]
- 18. Hrushesky WJ, Bjarnason GA. Circadian cancer therapy. J Clin Oncol. 1993;11:1403-1417. [DOI] [PubMed] [Google Scholar]
- 19. Innominato PF, Roche VP, Palesh OG, Ulusakarya A, Spiegel D, Lévi FA. The circadian timing system in clinical oncology. Ann Med. 2014;46:191-207. [DOI] [PubMed] [Google Scholar]
- 20. Ruben MD, Smith DF, FitzGerald GA, Hogenesch JB. Dosing time matters. Science. 2019;365:547-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takimoto CH. Chronomodulated chemotherapy for colorectal cancer: failing the test of time? Eur J Cancer. 2006;42:574-581. [DOI] [PubMed] [Google Scholar]
- 22. Lévi F, Focan C, Karaboué A, et al. Implications of circadian clocks for the rhythmic delivery of cancer therapeutics. Adv Drug Deliv Rev. 2007;59:1015-1035. [DOI] [PubMed] [Google Scholar]
- 23. Lévi F, Okyar A, Dulong S, Innominato PF, Clairambault J. Circadian timing in cancer treatments. Annu Rev Pharmacol Toxicol. 2010;50:377-421. [DOI] [PubMed] [Google Scholar]
- 24. Giacchetti S, Bjarnason G, Garufi C, et al. Phase III trial comparing 4-day chronomodulated therapy versus 2-day conventional delivery of fluorouracil, leucovorin, and oxaliplatin as first-line chemotherapy of metastatic colorectal cancer: the European Organisation for Research and Treatment of Cancer Chronotherapy Group. J Clin Oncol. 2006;24:3562-3569. [DOI] [PubMed] [Google Scholar]
- 25. Giacchetti S, Dugué PA, Innominato PF, et al. Sex moderates circadian chemotherapy effects on survival of patients with metastatic colorectal cancer: a meta-analysis. Ann Oncol. 2012;23:3110-3116. [DOI] [PubMed] [Google Scholar]
- 26. Humphries A, Wright NA. Colonic crypt organization and tumorigenesis. Nat Rev Cancer. 2008;8:415-424. [DOI] [PubMed] [Google Scholar]
- 27. Bravo R, Axelrod DE. A calibrated agent-based computer model of stochastic cell dynamics in normal human colon crypts useful for in silico experiments. Theor Biol Med Model. 2013;10:66-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wright NA. Review article: is there a common principle in the development of gastrointestinal cancers? Stem cells in the origin of cancer. Aliment Pharmacol Ther. 2006;2(suppl 4):31-40. [Google Scholar]
- 29. Paulus U, Potten CS, Loeffler M. A model of the control of cellular regeneration in the intestinal crypt after perturbation based solely on local stem cell regulation. Cell Prolif. 1992;25:559-578. [DOI] [PubMed] [Google Scholar]
- 30. Buchi KN, Moore JG, Hrushesky WJM, Sothern RB, Rubin NH. Circadian rhythm of cellular proliferation in the human rectal mucosa. Gastroenterology. 1991;101:410-415. [DOI] [PubMed] [Google Scholar]
- 31. Cornelissen G. Cosinor-based rhythmometry. Theor Biol Med Model. 2014;11:16-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marra G, Anti M, Percesepe A, et al. Circadian variations of epithelial cell proliferation in human rectal crypts. Gastroenterology. 1994;106:982-987. [DOI] [PubMed] [Google Scholar]
- 33. Axelrod DE, Vedula S, Obaniyi J. Effective chemotherapy of heterogeneous and drug- resistant early colon cancer by intermittent dose schedules: a computer simulation study. Cancer Chemother Pharmacol. 2017;79:889-898. [DOI] [PubMed] [Google Scholar]
- 34. Cockrell C, Teague J, Axelrod DE. Prevention of colon cancer recurrence from minimal residual disease: Computer optimized dose schedules of intermittent apoptotic adjuvant therapy. JCO Clin Cancer Inform 2020;4:514-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mazzoccoli G, Panza A, Valvano MR, et al. Clock gene expression levels and relationship with clinical and pathological features in colorectal cancer patients. Chronobiol Int 2011;10:841-851. [DOI] [PubMed] [Google Scholar]
- 36. Clairambault J. Modeling oxaliplatin drug delivery to circadian rhythms in drug metabolism and host tolerance. Adv Drug Deliv Rev. 2007;59:1054-1068 [DOI] [PubMed] [Google Scholar]
- 37. Altinok A, Lévi F, Goldbeter A. A cell cycle automaton model for probing circadian patterns of anticancer drug delivery. Adv Drug Deliv Rev. 2007;59:1036-1053. [DOI] [PubMed] [Google Scholar]
- 38. Bernard S, Čajavec Bernard B, Lévi F, Herzel H. Tumor growth rate determines the timing of optimal chronomodulated treatment schedules. PLoS Comput Biol. 2010;6:e1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ballesta A, Innominato PF, Dallmann R, Rand DA, Lévi FA. Systems chronotherapeutics. Pharmacol Rev. 2017;69:161-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lemmer B. Chronopharmacokinetics: implications for drug treatment. J Pharm Pharmacol. 1999;51:887-890. [DOI] [PubMed] [Google Scholar]
- 41. Serrano MJ, Garrido-Navas MC, Diaz Mochon JJ, et al. Precision prevention and cancer interception: the new challenges of liquid biopsy. Cancer Discov. 2020;10:1635-1644. [DOI] [PubMed] [Google Scholar]
- 42. Rossez Y, Burtea C, Laurent S, et al. Early detection of colonic dysplasia by magnetic resonance molecular imaging with a contrast agent raised against the colon cancer marker MUC5AC. Contrast Media Mol Imaging. 2016;11:211-221. [DOI] [PubMed] [Google Scholar]
- 43. Werner S, Chen H, Butt J, et al. Evaluation of the diagnostic value of 64 simultaneously measured autoantibodies for early detection of gastric cancer. Sci Rep. 2016;6:25467-25477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. New Engl J Med. 2014;370:1287-1297. [DOI] [PubMed] [Google Scholar]
- 45. Cohen JD, Diergaarde B, Papadopoulos N, Kinzler KW, Schoen RE. Tumor DNA as a cancer biomarker through the lens of colorectal neoplasia. Cancer Epidemiol Biomarkers Prev. 2020;29:2441-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bresalier RS, Grady WM, Markowitz SD, Nielsen HJ, Batra SK, Lampe PD. Biomarkers for early detection of colorectal cancer: the early detection research network, a framework for clinical translation. Cancer Epidemiol Biomarkers Prev. 2020;29:2431-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]