Abstract
The genotypes of 52 strains of Aspergillus fumigatus isolated from 12 patients with invasive aspergillosis were investigated using three typing methods (random amplified polymorphic DNA, sequence-specific DNA polymorphism, and microsatellite polymorphism) combined with multilocus enzyme electrophoresis. Isolates were from patients hospitalized in three different geographic areas (Lyon, France; Grenoble, France; and Milan, Italy). In each case, the genetic polymorphism of several colonies (two to five) within the first respiratory clinical sample was studied. For the 52 isolates tested, random amplified polymorphic DNA identified 8 different genotypes, sequence-specific DNA polymorphism identified 9 different types, and microsatellite polymorphism identified 14 types. A combination of these results with multilocus enzyme electrophoresis study identified 25 different types within the sample studied. We identified 3 patients (of the 12 studied) who carried a single genotype; 6 patients were infected by two genotypes, 1 patient had four genotypes, while the last patient had five. A combination of typing methods provided better discrimination than the use of a single method. Typing methods revealed a population structure within each geographical site, suggesting that the epidemiology of A. fumigatus should be considered separately for each of these geographic areas. This study demonstrates the usefulness of combining several typing methods in reaching an understanding of the epidemiology of A. fumigatus and clarifies whether it is sufficient to type one isolate from each specimen to determine the strain involved in invasive aspergillosis.
Aspergillus fumigatus is an opportunistic fungal pathogen responsible for most cases of invasive aspergillosis (IA), the most common systemic filamentous fungal infection worldwide (15). IA affects immunocompromised patients, particularly those who are suffering from severe hematological malignancies (18), and even with treatment the mortality is as high as 90% (14, 39). The incidence of IA has increased during the last two decades (37) because of the growing number of patients undergoing bone marrow or solid organ transplantation and because of the increasingly aggressive treatment of malignancies (13, 41). A. fumigatus is an airborne fungus, and infection occurs by inhaling conidia which may colonize airways prior to invasion. Epidemiologic studies of IA are essential to better understand this infection, to determine risk factors, and to improve preventative measures (2, 8, 21, 25, 38). Since A. fumigatus is responsible for about 80% of IA cases, subspecific typing methods are required to discriminate between isolates and to investigate their epidemiology (7). In recent years, several molecular methods have been developed to explore the genetic diversity of A. fumigatus, including random amplified polymorphic DNA (RAPD), sequence-specific DNA primer (SSDP), multilocus enzyme electrophoresis (MLEE), fingerprinting with moderately repeated DNA sequences, and microsatellite polymorphism (MSP) analysis. RAPD analysis can be performed quickly, a large number of isolates can be analyzed, and it can have a high discriminatory power. While the intra- and interlaboratory reproducibility of RAPD has not yet been proved, it has been validated in several studies (1, 20, 22, 35, 40). SSDP is a PCR typing method developed from RAPD and using specific primers. This method uses PCR at a higher stringency, thus avoiding the drawback of the lack of reproducibility of the RAPD method (9, 27). MLEE has provided good results when used for typing A. fumigatus (6, 26, 36) and combines both genotypic and population genetic information (5, 31, 36). MSP analysis has been used in several studies and has proved useful in the furthering our understanding of IA (3, 4); this approach also combines both genotypic and population genetic information. Fingerprinting by hybridization with a moderately repeated sequence provides a very high level of discrimination but is time-consuming (11, 32). Each of these methods thus has advantages and disadvantages. There is no “gold standard” typing method for investigating the genetic polymorphism of A. fumigatus. A combination of several different typing methods provides better discrimination than using a single method (22, 34, 35).
The European Research Group on Biotype and Genotype of Aspergillus (EBGA) was formed to study the genetic and phenotypic diversity of A. fumigatus in patients with IA in five European centers. One of the aims of the present study was to better understand the epidemiology of IA by using different methods to type isolates of the causal agents. The first step in any study of the molecular epidemiology of a pathogen is to determine its genetic diversity within a single clinical sample. This approach indicates whether the infection is due to one or more strains and so dictates the number of isolates per specimen that are necessary to type. In this study, the MLEE, SSDP, RAPD, and MSP methods were applied to determine the genetic variability of 52 isolates of A. fumigatus from 12 patients with IA. In each case two to five colonies of A. fumigatus from the first respiratory sample were typed by the four methods to investigate genetic polymorphism among multiple isolates within the first specimen. This has already been done using MLEE; the present study, using MLEE together with, SSDP, MSP, and RAPD analyses, demonstrates the importance of combining different typing methods to obtain a better understanding of the epidemiology of A. fumigatus.
MATERIALS AND METHODS
Isolates.
The EBGA collected A. fumigatus clinical strains isolated at three European hospital centers. This collection was maintained by the Scientific Institute of Public Health (IHEM Collection), Brussels, Belgium. A. fumigatus isolates from the first respiratory samples, containing at least two colonies, were collected and typed. The study was performed on 52 A. fumigatus isolates from 12 patients suffering from hematological malignancies and/or receiving long-term corticotherapy with or without neutropenia. Clinical, radiologic, and mycologic investigations and diagnoses of IA were carried out in three hospital centers (Lyon, France; Grenoble, France; and Milan, Italy). Isolates from these hospital centers were identified by classical techniques based on conventional morphologic identification in each center. For each patient, probable IA was diagnosed according to recently published criteria (S. Asciogliu, B. de Pauw, J. E. Bennett, J. Bille, F. Crokaert, D. W. Denning, P. Donnelly, J. E. Edwards, Z. Erjavec, D. Fiere, O. Lortholary, J. Maertens, J. Meis, T. Patterson, J. H. Rex, J. Ritter, D. Selleslag, P. M. Shah, D. A. Stevens, and T. J. Walsh, Abstr. 39th Int. Conf. Antimicrob. Agents Chemother., abstr. 159.J, 1999). The origin of each isolate of A. fumigatus is summarized in Table 1.
TABLE 1.
Characteristics of the strains of A. fumigatus studied
| Patient code | Origin | Underlying disease and/or outcome | Presence of neutropenia | Sample
|
IHEM no. | |
|---|---|---|---|---|---|---|
| Typea | Date obtained | |||||
| LY/07 | Lyon | Lymphoma | Yes | BAL | 11/16/94 | 9378–9382 |
| LY/20 | Lyon | Lymphoma | Yes | BAL | 02/17/95 | 9508–9512 |
| LY/28 | Lyon | Lymphoma, leukemia, solid tumor | Yes | Sputum | 01/10/95 | 9447–9451 |
| LY/25 | Lyon | Bone marrow transplant | Yes | BAL | 04/04/95 | 9595–9598 |
| MI/03 | Milan | Lymphoma | Yes | BA | 04/21/94 | 9025–9029 |
| MI/12 | Milan | Long-term corticotherapy | Yes | RA | 10/24/97 | 14202–14206 |
| MI/02 | Milan | Lymphoma | No | Sputum | 03/25/94 | 14317–14318 |
| MI/05 | Milan | Long-term corticotherapy | No | BA | 05/09/96 | 10054–10056 |
| GR/02 | Grenoble | Lymphoma; bone marrow transplant | Yes | BAL | 01/13/95 | 9418–9420 |
| GR/01 | Grenoble | Leukemia; long-term corticotherapy | Yes | BAL | 10/04/94 | 9347–9351 |
| GR/06 | Grenoble | Lymphoma; kidney transplant | Yes | BA | 05/29/95 | 9720–9724 |
| GR/04 | Grenoble | Leukemia | Yes | BA | 04/18/95 | 9600–9604 |
BAL, bronchoalveolar lavage; BA, bronchial aspiration; RA, rhinopharyngeal aspirate.
Date specimen was taken (month/day/year).
RAPD analysis.
Preparation of fungal genomic DNA and RAPD analysis with the primer NS3 (5′-GCAAGTCTGGTGCCAGCAGCC-3′) were performed as previously described (35, 38). The criterion used to establish different types was the presence or absence of an extra consistent band in the RAPD pattern. This work was done in Brussels, Belgium, at the Scientific Institute of Public Health (Mycology Section).
MSP analysis.
These assays were performed at the Department of Microbiology, Leeds, United Kingdom, using the method of Bart-Delabesse et al. (4), with the modifications described below. DNA was extracted as previously described (27). PCR was performed with primer pairs to amplify the B and D locus microsatellite loci (4). These two loci were selected to maximize the discrimination as described previously (4). Reactions contained 1.5 mM MgCl2, 10 mM Tris-HCl, 50 mM KCl, 1× Q solution (Qiagen), primers at 100 mM, nucleoside triphosphates at 100 μM each, 0.5 U of Taq polymerase (Qiagen), and ca. 50 ng DNA in a final volume of 50 μl. Amplification was performed in a Techne Progene Thermal Cycler for 5 min at 94°C, with 30 cycles of 30 s at 94°C, 30 s at 59°C, and 30 s at 72°C and a final extension step of 72°C for 30 min. PCR products were run on 3.5% Metaphor agarose gels (FMC) at 250 V for 3 h using 0.5× TBE buffer (Tris-borate [89 mM]–EDTA [2 mM]) cooled to 14°C and recirculated during the run. The gel was stained for 30 min in 0.5 μg of ethidium bromide per ml, and the product sizes were estimated to the nearest 5 bp using a 10-bp molecular weight marker (Gibco-BRL).
SSDP analysis.
Isolates were typed by sequence-specific DNA primers with the five sets of primers decribed by Mondon et al. (27). The primers used were Afp1 (5′-TTGGGGAGATTACCGAACTGG-3′) and Afp2 (5′-CCTTGACAACCGTCCCATTTC-3′) for marker SSDP 1, AfMp1 (5′-CAGTTCCCGTCTTGACCTC-3′) and AfMp2 (5′-CAGTTCCCGTTTTCCTAGTA-3′) for marker SSDP 2, Afd1 (5′-GTATTGCCCTATAACTTCTT-3′) and Afd2 (5′-GTATTGCCCTATTCCCAAAG-3′) for marker SSDP 3, Afs4 (5′-GTATTGCCCTAGCTTACTAA-3′) and Afr4 (5′-GTATTGCCCTATTACTAAAG-3′) for marker SSDP 4, and Afs5 (5′-GTATTGCCCTAAGGATTCTA-3′) and Afr5 (5′-GTATTGCCCTAGCTTGCTAA-3′) for marker SSDP 5.
The results (Table 2) are presented for SSDP1, SSDP2, SSDP3, SSDP4, and SSDP5 according to the presence (indicated by “1”) or absence (indicated by “0”) of the specific band and conform to previous studies (27, 35). This work was done in Grenoble, France, at the Interactions Cellulaires Parasite-Hôte, Université Joseph Fourier.
TABLE 2.
Allelic profiles of isolates of A. fumigatus obtained by the combination of RAPD, MSP, SSDP, and MLEE techniques
| Center, patient code, and IHEM no. | RAPDa (NS3) | MSP
|
SSDPb
|
MLEE at locusc
|
Final type | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus B | Locus D | Type | 1 | 2 | 3 | 4 | Type | Pep1 | Pep4 | Aat | Fum | Gpi | Pgm | Pgd | Type | |||
| Grenoble | ||||||||||||||||||
| GR/02 | ||||||||||||||||||
| 9418 | 1 | 110 | 95 | 1 | 0 | 0 | 0 | 0 | 24 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 2/2 | 7 | 1 |
| 9419 | 1 | 110 | 95 | 1 | 0 | 0 | 0 | 0 | 24 | 1/1 | 1/1 | 1/1 | 2/2 | 2/2 | 1/1 | 2/2 | 8 | 2 |
| 9420 | 1 | 110 | 95 | 1 | 0 | 0 | 0 | 0 | 24 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 2/2 | 7 | 1 |
| GR/04 | ||||||||||||||||||
| 9600 | 6 | 130 | 85 | 2 | 0 | 0 | 0 | 1 | 32 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1 | 3 |
| 9601 | 7 | 110 | 100 | 3 | 0 | 0 | 0 | 0 | 24 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1 | 4 |
| 9602 | 4 | 110 | 95 | 1 | 0 | 0 | 0 | 1 | 32 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1 | 5 |
| 9603 | 4 | 110 | 110 | 4 | 0 | 1 | 0 | 1 | 31 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1 | 6 |
| 9604 | 6 | 110 | 110 | 4 | 0 | 0 | 0 | 0 | 24 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1 | 7 |
| GR/06 | ||||||||||||||||||
| 9720 | 3 | 105 | 85 | 5 | 0 | 0 | 0 | 0 | 24 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1 | 8 |
| 9721 | 3 | 105 | 100 | 6 | 0 | 0 | 0 | 0 | 24 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1 | 9 |
| 9722 | 3 | 105 | 95 | 7 | 0 | 0 | 0 | 0 | 24 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1 | 10 |
| 9723 | 3 | 105 | 110 | 8 | 0 | 0 | 0 | 0 | 24 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1 | 11 |
| 9724 | 3 | 105 | 110 | 8 | 0 | 0 | 0 | 0 | 24 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1 | 11 |
| GR/01 | ||||||||||||||||||
| 9347 | 3 | 110 | 100 | 3 | 0 | 0 | 0 | 0 | 24 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1 | 12 |
| 9348 | 3 | 110 | 100 | 3 | 0 | 0 | 0 | 0 | 24 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1 | 12 |
| 9349 | 3 | 110 | 100 | 3 | 0 | 0 | 0 | 1 | 32 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1 | 13 |
| 9350 | 3 | 110 | 100 | 3 | 0 | 0 | 0 | 0 | 24 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1 | 12 |
| 9351 | 3 | 110 | 100 | 3 | 0 | 0 | 0 | 1 | 32 | 1/1 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1 | 13 |
| Lyon | ||||||||||||||||||
| LY/07 | ||||||||||||||||||
| 9378 | 2 | 110 | 115 | 9 | 1 | 0 | 0 | 1 | 30 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 14 |
| 9379 | 2 | 110 | 115 | 9 | 1 | 0 | 0 | 1 | 30 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 14 |
| 9380 | 2 | 110 | 115 | 9 | 1 | 0 | 0 | 1 | 30 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 14 |
| 9381 | 2 | 110 | 115 | 9 | 1 | 0 | 0 | 1 | 30 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 14 |
| 9382 | 2 | 110 | 115 | 9 | 1 | 0 | 0 | 1 | 30 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 14 |
| LY/20 | ||||||||||||||||||
| 9508 | 3 | 110 | 85 | 10 | 1 | 0 | 1 | 1 | 26 | 2/2 | 2/2 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 4 | 15 |
| 9509 | 3 | 110 | 85 | 10 | 0 | 0 | 1 | 1 | 28 | 2/2 | 2/2 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 4 | 16 |
| 9510 | 3 | 110 | 85 | 10 | 0 | 0 | 1 | 1 | 28 | 2/2 | 2/2 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 4 | 16 |
| 9511 | 3 | 110 | 85 | 10 | 0 | 0 | 1 | 1 | 28 | 2/2 | 2/2 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 4 | 16 |
| 9512 | 3 | 110 | 85 | 10 | 0 | 0 | 1 | 1 | 28 | 2/2 | 2/2 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 4 | 16 |
| LY/25 | ||||||||||||||||||
| 9595 | 2 | 110 | 140 | 11 | 0 | 0 | 1 | 1 | 28 | 2/2 | 2/2 | 2/2 | 1/1 | 1/1 | 1/2 | 1/1 | 2 | 17 |
| 9596 | 2 | 110 | 140 | 11 | 0 | 0 | 1 | 1 | 28 | 2/2 | 2/2 | 2/2 | 1/1 | 1/1 | 1/2 | 1/1 | 2 | 17 |
| 9597 | 2 | 110 | 140 | 11 | 0 | 0 | 1 | 1 | 28 | 2/2 | 2/2 | 2/2 | 1/1 | 1/1 | 1/2 | 1/1 | 2 | 17 |
| 9598 | 2 | 110 | 140 | 11 | 0 | 0 | 1 | 1 | 28 | 2/2 | 2/2 | 2/2 | 1/1 | 1/1 | 1/2 | 1/1 | 2 | 17 |
| LY/28 | ||||||||||||||||||
| 9447 | 4 | 110 | 115 | 9 | 1 | 0 | 0 | 1 | 30 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 18 |
| 9448 | 4 | 110 | 115 | 9 | 1 | 0 | 0 | 1 | 30 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 18 |
| 9449 | 4 | 110 | 115 | 9 | 1 | 0 | 0 | 1 | 30 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 18 |
| 9450 | 4 | 110 | 115 | 9 | 1 | 0 | 0 | 1 | 30 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 18 |
| 9451 | 4 | 105 | 110 | 8 | 1 | 0 | 0 | 1 | 30 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 19 |
| Milan | ||||||||||||||||||
| MI/02 | ||||||||||||||||||
| 14317 | 3 | 115 | 90 | 12 | 0 | 0 | 1 | 1 | 28 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 20 |
| 14318 | 3 | 110 | 100 | 3 | 1 | 0 | 0 | 1 | 30 | 2/2 | 2/2 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 4 | 21 |
| MI/05 | ||||||||||||||||||
| 10054 | 5 | 120 | 80 | 13 | 1 | 1 | 1 | 1 | 25 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 22 |
| 10055 | 5 | 120 | 80 | 13 | 1 | 1 | 1 | 1 | 25 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 22 |
| 10056 | 5 | 120 | 80 | 13 | 1 | 1 | 1 | 1 | 25 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 22 |
| MI/03 | ||||||||||||||||||
| 9025 | 8 | 130 | 105 | 14 | 1 | 1 | 0 | 1 | 29 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 23 |
| 9026 | 8 | 130 | 105 | 14 | 1 | 1 | 0 | 1 | 29 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 23 |
| 9027 | 8 | 130 | 105 | 14 | 1 | 1 | 0 | 1 | 29 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1/1 | 5 | 24 |
| 9028 | 8 | 130 | 105 | 14 | 1 | 1 | 0 | 1 | 29 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 23 |
| 9029 | 8 | 130 | 105 | 14 | 1 | 1 | 0 | 1 | 29 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 3 | 23 |
| MI/12 | ||||||||||||||||||
| 14202 | 3 | 110 | 95 | 1 | 1 | 0 | 1 | 0 | 18 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1/2 | 1/1 | 6 | 25 |
| 14203 | 3 | 110 | 95 | 1 | 1 | 0 | 1 | 0 | 18 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1/2 | 1/1 | 6 | 25 |
| 14204 | 3 | 110 | 95 | 1 | 1 | 0 | 1 | 0 | 18 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1/2 | 1/1 | 6 | 25 |
| 14205 | 3 | 110 | 95 | 1 | 1 | 0 | 1 | 0 | 18 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1/2 | 1/1 | 6 | 25 |
| 14206 | 3 | 110 | 95 | 1 | 1 | 0 | 1 | 0 | 18 | 1/1 | 1/1 | 2/2 | 1/1 | 1/1 | 1/2 | 1/1 | 6 | 25 |
Different types were established by the presence or absence of an extra consistent band on the RAPD pattern.
The presence or absence of SSDP1 to SSDP4 is indicated by “1” or “0,” respectively.
1/2, alleles 1 and 2 presented by heterozygotes. Alleles are encoded in decreasing order of their anodal modality.
For each method (RAPD, MSP, SSDP, and MLEE), the stability of the types was evaluated by testing 10 monosporous strains (IHEM 8065, IHEM 8070, IHEM 8075, IHEM 8080, IHEM 8085, IHEM 8086, IHEM 8090, IHEM 9000, IHEM 9005, and IHEM 9079), and each experiment was performed at least three times to evaluate the reproducibility (27, 36; F. Symoens, B. Van Vaerenbergh, P. Mondon, E. Rodriguez, J. M. Bastide, and the EBGA Network, Abstr. Soc. Francaise Mycol. Med., abstr. 29, 1995).
Hierarchical cluster analysis.
Hierarchical clustering displays clusters that are displayed in the form of a tree. Initially, each type is considered a separate cluster. This analysis begins by joining the two “closest” objects as a cluster and continues (in a stepwise manner), joining an object with another object, an object with a cluster, or a cluster with another cluster, until all objects are combined into one cluster. In this study, a complete linkage was used. It maximizes the differences between different types. Since genotypic types are qualitative data, a χ2 metric value was used in this study. Distances were computed as the χ2 measure of independence of rows and columns on 2-by-n frequency tables, formed by pairs of types (17). This study was performed with Systat 8.0 statistics software package (SPSS, Inc.).
RESULTS
Genetic diversity. (i) Enzyme typing method.
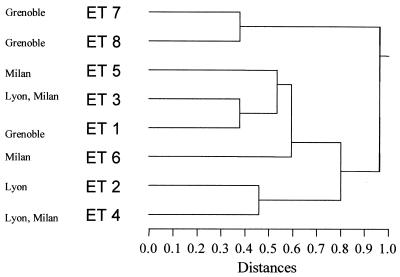
In the case of MLEE, the study has already been performed (6). The results are presented in Fig. 1.
FIG. 1.
Hierarchical classification of electrophoretic types of A. fumigatus analyzed by MLEE.
(ii) DNA typing methods.
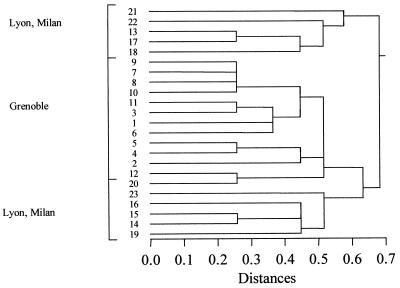
For SSDP analysis, four loci were polymorphic in the 52 isolates studied, each with two alleles. The study identified eight alleles, giving rise to nine different types. The remaining locus (SSDP 5) was highly conserved. For RAPD analysis, only one RAPD marker was studied for the 52 isolates; it yielded eight different amplification products, discriminating between eight types. For MSP analysis, two loci (B and D) were studied, one with five alleles and the other with nine alleles. The study identified 14 types in the isolates tested. For the combination of DNA typing methods, there were eight markers. The study identified 23 different types in the 52 isolates tested (Table 2). A hierarchical cluster analysis was performed on the types defined by the molecular methods. This demonstrated the extensive heterogeneity of the strains studied. All of the strains from Grenoble and genotype 20 (Milan) were grouped together on this tree (Fig. 2).
FIG. 2.
Hierarchical classification of genotypes of A. fumigatus obtained by three molecular typing techniques (RAPD, MSP, and SSDP.)
(iii) Combination of the four typing methods.
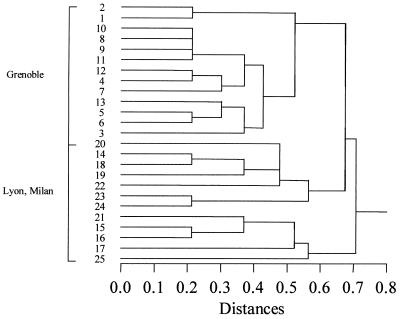
The combination of the three typing methods with MLEE results previously published (6) revealed 22 markers, 14 of which were polymorphic. The present study identified 25 overall genotypes in the 52 isolates tested here (Table 2). A hierarchical cluster analysis showed a particular genetic structure to the genotypes (Fig. 3). All of the strains from Grenoble (types 1 to 13) were clustered, whereas the strains from Lyon (types 14 to 19) were mixed up on the tree with those from Milan (types 20 to 25). This revealed an extensive variety in the genotypes of the strains from these two centers. The various types of diversity are discussed below.
FIG. 3.
Hierarchical classification of the genotypes of A. fumigatus obtained by a combination of different typing techniques (RAPD, MSP, SSDP, and MLEE).
(a) Strain diversity within the same sample.
Colonies from the same specimen displayed the same genotypic pattern in 4 patients (LY/07, LY/25, MI/05, and MI/12) of the 12 studied. Six patients (GR/01, GR/02, LY/20, LY/28, MI/02, and MI/12) exhibited two different strains within the same sample, and two patients from Grenoble (GR/06 and GR/04) exhibited four and five different genotypes, respectively. Thus, in Grenoble, types 12 and 13 were distinguishable for GR/01; types 1 and 2 were distinguishable for GR/02; types 4, 5, 6, and 7 were distinguishable for GR/04; and types 8, 9, 10, and 11 were distinguishable for GR/06. In Lyon, LY/20 exhibited types 15 and 16, while LY/28 exhibited types 18 and 19. In Milan, the four typing methods separated types 20 and 21 within the first specimen isolated from MI/02 and types 23 and 24 within isolates from MI/03.
(b) Diversity within the same geographic area.
There is extensive genetic variability within geographic sites. In Grenoble, of 18 isolates tested from four patients, 13 different genotypes were found; pairs of genotypes differed by between one and six markers. In Lyon, six genotypes were identified among 19 isolates tested from four patients; the genotypes differed by one to nine markers. In Milan, six genotypes were identified from 15 isolates tested from four patients; the genotypes differed by between one and nine markers.
(c) Diversity between geographic sites.
Each genotype was found in a given area. Genotypes 1 to 13 were found in Grenoble, genotypes 14 to 19 were found only in Lyon, and genotypes 20 to 25 were found only in Milan. These results suggest the existence of extensive intra- and intersite heterogeneity and, thus, the relative independence of each area. Moreover, there is a lack of correlation between genotypes and geographic distance. Hierarchical cluster analysis showed that strains from Grenoble can be grouped together, whereas strains from Lyon and Milan seem to be indistinguishable (Fig. 3).
DISCUSSION
MLEE is one of many typing methods developed for investigating the diversity of A. fumigatus (6, 34, 36). It has good discriminatory power and allows assessment of the structure and differentiation of A. fumigatus populations, including their genetic diversity (6). Other methods based on the investigation of DNA variations have been developed to type A. fumigatus in recent years. The present study analyzed 52 isolates of A. fumigatus from three geographic centers using three DNA typing methods (RAPD, SSDP, and MSP) combined with MLEE. RAPD has been widely used in Aspergillus epidemiologic studies (1, 20, 22, 32, 35, 41). Despite the difficulty of achieving reproducibility with this method (30), the primer NS3 used in the present study has been validated for Aspergillus spp. (38). The discriminating RAPD products have been sequenced, and specific primers have been developed for use in high-stringency PCR conditions for the SSDP method. This method has always been used and validated for investigating A. fumigatus genetic diversity (9, 27, 35). Microsatellite markers have proved their usefulness in several studies and provide a powerful tool for developing a better understanding of the local epidemiology of A. fumigatus. (3).
Several studies have reported the extensive variation of A. fumigatus. Mondon et al. found a total of 22 DNA types in 51 isolates of A. fumigatus studied by the SSDP technique (27). Bart-Delabesse et al. found that 102 isolates could be placed into 80 different types by using four microsatellite markers (4). Debeaupuis et al. found 424 different profiles among 879 isolates studied by fingerprinting with a moderately repeated sequence, the λ3.9 probe (11); and another study demonstrated that 252 isolates studied could be placed into 157 different genotypes by the same method (10). Thirty-five isolates exhibited 22 DNA types when five RAPD markers were used in the study of Lin et al. (22). Rodriguez et al. used MLEE and found 48 allelic combinations among 71 isolates when testing 14 different enzymes (36).
Despite the great homogeneity of the samples studied, our use of four different typing methods revealed 25 genotypes within 52 isolates tested. Even though some studies have suggested the existence of recombination in A. fumigatus, this variability remains unexplained. Indeed, Bart-Delabesse et al. and Debeaupuis et al. have suggested the existence of a sexual stage for A. fumigatus as one hypothesis to explain this variation (4, 11). If this is true, the teleomorph of this fungus has gone undetected. Recombinations might have occurred through a parasexual stage of the fungus (18). This would explain the apparently heterozygous patterns obtained by MLEE for strains from patients LY/25 and MI/12, which may be due to a duplication of the Pgm gene (6) or to heterocaryon variation.
We investigated the genetic polymorphism of A. fumigatus isolates within the same specimens from patients with IA. This type of study on a very structured sample has never been done before. Several previous studies have demonstrated that only one or two strains are responsible for infecting patients with IA (3, 11). However, only one colony was typed for a given specimen in these studies. Girardin et al., using fingerprinting with the λ3.9 probe, found that only one genotype was recovered among 53% of patients with IA (16). The present study also demonstrated, in most of cases, one or two genotypes within the same respiratory specimen for a given patient. Even though we could not investigate the overall variability within the same patient (using a follow-up of several colonies within the same sample), this finding is in agreement with previous works (3, 11, 16). This suggests that an infection caused by A. fumigatus is very different from one caused by C. albicans (19, 33). Indeed, up to seven different strains can be found in the oral cavity of patients in the early stage of candidosis, whereas only a single strain of A. fumigatus was found in most of the cases within samples of patients with IA.
Surprisingly, we found two patients from Grenoble who were infected with multiple strains: one by four strains and the other by five strains. Isolates of subject GR/06 differed by only one microsatellite locus, suggesting a variation within the same strain; but isolates of GR/04 were distinguished by three typing methods (RAPD, SSDP, and MSP), suggesting the existence of five different strains. It has been demonstrated that 5 to 10 different strains of A. fumigatus can be isolated in cases of bronchial colonization in patients with cystic fibrosis (28). However, these results have never been observed in the case of IA. This result is unlikely to be due to the contamination of the sample because bronchial aspiration (BA) is a very safe sampling method and the strains were verified throughout the study. This great heterogeneity within the same sample may be due to the number of isolates (up to two) studied for each patient in the present study.
Chazalet et al. showed that 85% of the strains of the airborne A. fumigatus population in a given geographic area are of a single genotype (10). A study on patients with aspergilloma showed that identical genotypes between two different geographic sites are rare (16). In the present study each genotype was found in a given area. These results are in agreement with previous conclusions (6, 35). Thus, the global diversity suggests that strains are adapted to a local environment and that the epidemiology of A. fumigatus should be studied independently within each geographic area. This situation may be the result of local selective pressure (24), including climate, environmental adaptation, and therapeutic selection, as previously suggested (12, 23).
A combination of several typing methods was able to discriminate among strains to a greater extent than the same typing methods used separately. This has always been found (22, 34, 35) to be true. The results of the present study suggest that using multiple methods yields increased discriminatory power. Indeed, the enzyme typing method and molecular typing methods used alone were not able to discriminate between strains from different geographic areas, whereas a combination of the four methods gave a cluster that included every strain from Grenoble. Strains from Lyon and Milan were indistinguishable on the cluster hierarchical analysis. This suggests that strains from Grenoble were different from those from Lyon and Milan. This is in agreement with previous studies (6, 35). Moreover, only 3 patients (of the 12 studied) were infected by a single genotype, whereas other methods used separately demonstrated a greater proportion of patients with IA to be infected by a single strain (3, 6, 11). Indeed, the present study demonstrated that discriminatory power increases when multiple methods are applied. This suggests that typing only one isolate is not enough in cases of IA. Nevertheless, typing multiple colonies with several methods in a first respiratory clinical sample would be very difficult to carry out in a clinical routine.
The present study has demonstrated the value of a structured sample and different typing methods for investigating the epidemiology of IA. Nevertheless, it remains very difficult to obtain multiple colonies within the first specimen, and it is not possible routinely to assess strains of A. fumigatus by several typing methods. In conclusion, we demonstrate here the complementarity of using several typing methods to determinate the genetic variability of A. fumigatus colonies within the same sample in cases of IA. A combination of different typing methods, including enzyme and molecular methods, is useful for improving our understanding of the epidemiology of A. fumigatus.
REFERENCES
- 1.Anderson M J, Gull K, Denning D W. Molecular typing by random amplification of polymorphic DNA and M13 Southern hybridization of related paired isolates of Aspergillus fumigatus. J Clin Microbiol. 1996;34:87–93. doi: 10.1128/jcm.34.1.87-93.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnow P M, Sadigh M, Costas C, Weil D, Chudy R. Endemic and epidemic aspergillosis associated with in-hospital replication of Aspergillus organisms. J Infect Dis. 1991;164:998–1002. doi: 10.1093/infdis/164.5.998. [DOI] [PubMed] [Google Scholar]
- 3.Bart-Delabesse E, Cordonnier C, Bretagne S. Usefulness of genotyping with microsatellite markers to investigate hospital-acquired invasive aspergillosis. J Hosp Infect. 1999;42:321–327. doi: 10.1053/jhin.1998.0590. [DOI] [PubMed] [Google Scholar]
- 4.Bart-Delabesse E, Humbert J F, Delabesse E, Bretagne S. Microsatellite markers for typing Aspergillus fumigatus isolates. J Clin Microbiol. 1998;36:2413–2418. doi: 10.1128/jcm.36.9.2413-2418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertout S, Renaud F, Swinne D, Mallié M, Bastide J M. Genetic multilocus studies of different strains of Cryptococcus neoformans: taxonomy and genetic structure. J Clin Microbiol. 1999;37:715–720. doi: 10.1128/jcm.37.3.715-720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertout S, Renaud F, de Meeüs T, Piens M A, Lebeau B, Viviani M A, Mallié M, Bastide J M the EBGA Network. Multilocus enzyme electrophoresis of Aspergillus fumigatus strains isolated from the first clinical sample from patients with invasive aspergillosis. J Med Microbiol. 2000;49:375–381. doi: 10.1099/0022-1317-49-4-375. [DOI] [PubMed] [Google Scholar]
- 7.Birch M, Anderson M J, Denning D W. Molecular typing of Aspergillus species. J Hosp Infect. 1995;30:339–351. doi: 10.1016/0195-6701(95)90037-3. [DOI] [PubMed] [Google Scholar]
- 8.Birch M, Nolard N, Shankland G S, Denning D W. DNA typing of epidemiologically-related isolates of Aspergillus fumigatus. Epidemiol Infect. 1995;114:461–168. doi: 10.1017/s0950268800052018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenier-Pinchart M P, Lebeau B, Devouassoux G, Mondon P, Pison C, Ambroise-Thomas P, Grillot R. Aspergillus and lung transplant recipients: a mycologic and molecular epidemiologic study. J Heart Lung Transplant. 1998;17:972–979. [PubMed] [Google Scholar]
- 10.Chazalet V, Debeaupuis J P, Sarfati J, Lortholary J, Ribaud P, Shah P, Cornet M, Vu Thien H, Gluckman E, Brücker G, Latgé J P. Molecular typing of environmental and patient isolates of Aspergillus fumigatus from various hospital settings. J Clin Microbiol. 1998;36:1494–1500. doi: 10.1128/jcm.36.6.1494-1500.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debeaupuis J P, Sarfati J, Chazalet V, Latgé J P. Genetic diversity among clinical and environmental isolates of Aspergillus fumigatus. Infect Immun. 1997;65:3080–3085. doi: 10.1128/iai.65.8.3080-3085.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denning D W, Clemons K V, Hansonand L H, Stevens D A. Restriction endonuclease analysis of total cellular DNA of Aspergillus fumigatus isolates of geographically and epidemiologically diverse origin. J Infect Dis. 1990;162:1151–1158. doi: 10.1093/infdis/162.5.1151. [DOI] [PubMed] [Google Scholar]
- 13.Denning D W. Invasive aspergillosis. Clin Infect Dis. 1998;26:781–803. doi: 10.1086/513943. [DOI] [PubMed] [Google Scholar]
- 14.Derouin F. Diagnostic, traitement et prévention de l′aspergillose invasive. Pathog Biol. 1994;42:629–631. [PubMed] [Google Scholar]
- 15.Fridkin S A, Jarvis W R. Epidemiology of fungal nosocomial infections. Clin Microbiol Rev. 1996;9:499–511. doi: 10.1128/cmr.9.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girardin H, Sarfati J, Kobayashi H, Bouchara J P, Latgé J P. Use of DNA moderately repetitive sequence to type Aspergillus fumigatus isolates from aspergilloma patients. J Infect Dis. 1994;169:683–685. doi: 10.1093/infdis/169.3.683. [DOI] [PubMed] [Google Scholar]
- 17.Hartigan J A. Clustering algorithms. New York, N.Y: John Wiley & Sons, Inc.; 1975. [Google Scholar]
- 18.Latgé J P. Aspergillus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.le Guennec R, Reynes J, Mallié M, Pujol C, Janbon F, Bastide J M. Fluconazole- and itraconazole-resistant Candida albicans strains from AIDS patients: multilocus enzyme electrophoresis analysis and antifungal susceptibilities. J Clin Microbiol. 1995;33:2732–2737. doi: 10.1128/jcm.33.10.2732-2737.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leenders A, van Belkum A, Janssen S, de Marie S, Kluytmans J, Wielenga J, Lowenberg B, Verbrugh H. Molecular epidemiology of apparent outbreak of invasive aspergillosis in a hematology ward. J Clin Microbiol. 1996;34:345–351. doi: 10.1128/jcm.34.2.345-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leenders A C, van Belkum A, Behrendt M, Luijendijk A, Verbrugh H A. Density and molecular epidemiology of Aspergillus in air and relationship to outbreaks of Aspergillus infection. J Clin Microbiol. 1999;37:1752–1757. doi: 10.1128/jcm.37.6.1752-1757.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin D, Lehmann P F, Hamory B H, Padhye A A, Durry E, Pinner R W, Lasker B A. Comparison of three typing methods for clinical and environmental isolates of Aspergillus fumigatus. J Clin Microbiol. 1995;33:1596–1601. doi: 10.1128/jcm.33.6.1596-1601.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loudon K W, Coke A P, Burnie J P, Lucas G S, Liu Yin J A. Invasive aspergillosis: clusters and sources? J Med Vet Mycol. 1994;32:217–224. doi: 10.1080/02681219480000281. [DOI] [PubMed] [Google Scholar]
- 24.Manly B J F. The statistics of natural selection on animal populations. London, England: Chapman and Hall; 1985. [Google Scholar]
- 25.Manuel R J, Kibbler C C. The epidemiology and prevention of invasive aspergillosis. J Hosp Infect. 1998;39:95–109. doi: 10.1016/s0195-6701(98)90323-1. [DOI] [PubMed] [Google Scholar]
- 26.Matsuda H, Kohno S, Maesaki B, Yamada H, Koga H, Tamura M, Kuraishi H, Sugiyama J. Application of ubiquinone system and electrophoretic comparison of enzymes to identification of clinical isolates of A. fumigatus and several other species of Aspergillus. J Clin Microbiol. 1992;30:1999–2005. doi: 10.1128/jcm.30.8.1999-2005.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mondon P, Brenier M P, Symoens F, Rodriguez E, Coursange E, Chaib F, Lebeau B, Piens M A, Tortorano A M, Mallié M, Chapuis F, Carlotti A, Villard J, Viviani M A, Nolard N, Bastide J M, Ambroise-Thomas P, Grillot R. Molecular typing of Aspergillus fumigatus strains by sequence-specific DNA primer (SSDP) analysis. FEMS Immunol Med Microbiol. 1997;17:95–102. doi: 10.1111/j.1574-695X.1997.tb01001.x. [DOI] [PubMed] [Google Scholar]
- 28.Neuveglise C, Sarfati J, Latgé J P, Paris S. Afut1, a retrotransposon-like element from Aspergillus fumigatus. Nucleic Acids Res. 1996;24:1428–1434. doi: 10.1093/nar/24.8.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasteur N, Pasteur G, Bonhomme F, Catalan J, Britton-Davidian J. Manuel technique de génétique par électrophorèse des protéines. Paris, France: Lavoisier; 1987. [Google Scholar]
- 30.Perez T, Albornoz J, Dominguez A. An evaluation of RAPD fragment reproducibility and nature. Mol Ecol. 1998;7:1347–1357. doi: 10.1046/j.1365-294x.1998.00484.x. [DOI] [PubMed] [Google Scholar]
- 31.Pujol C, Reynes J, Renaud F, Raymond M, Tibayrenc M, Ayala F J, Janbon F, Mallié M, Bastide J M. The yeast Candida albicans has a clonal mode of reproduction in the population of infected HIV-positive patients. Proc Natl Acad Sci USA. 1993;90:9456–9459. doi: 10.1073/pnas.90.20.9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rath P M, Marggraf G, Dermoumi H, Ansorg R. Use of phenotypic and genotypic fingerprinting methods in the strain identification of Aspergillus fumigatus. Mycoses. 1995;38:429–434. doi: 10.1111/j.1439-0507.1995.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 33.Reynes J, Pujol C, Moreau C, Mallié M, Renaud F, Janbon F, Bastide J M. Simultaneous carriage of Candida albicans strains from HIV-infected patients with oral candidiasis: multilocus enzyme electrophoresis analysis. FEMS Microbiol Lett. 1996;137:269–273. doi: 10.1111/j.1574-6968.1996.tb08117.x. [DOI] [PubMed] [Google Scholar]
- 34.Rinyu E, Varga J, Ferenczy M. Phenotypic and genotypic analysis of variability in Aspergillus fumigatus. J Clin Microbiol. 1995;33:2567–2575. doi: 10.1128/jcm.33.10.2567-2575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez E, Symoens F, Mondon P, Mallié M, Piens M A, Lebeau B, Tortorano A M, Chaib F, Carlotti A, Villard J, Viviani M A, Chapuis F, Nolard N, Grillot R, Bastide J M. Combination of three typing methods for the molecular epidemiology of Aspergillus fumigatus infections. J Med Microbiol. 1999;48:181–194. doi: 10.1099/00222615-48-2-181. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez E, de Meeüs T, Mallié M, Renaud F, Symoens F, Mondon P, Piens M A, Lebeau B, Viviani M A, Grillot R, Nolard N, Chapuis F, Tortorano A M, Bastide J M. Multicentric epidemiological study of Aspergillus fumigatus isolates by multilocus enzyme electrophoresis. J Clin Microbiol. 1996;34:2559–2568. doi: 10.1128/jcm.34.10.2559-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers T R. Epidemiology and control of nosocomial fungal infections. Curr Opin Infect Dis. 1995;8:287–290. [Google Scholar]
- 38.Symoens F, Bouchara J P, Heinemann S, Nolard N. Molecular typing of Aspergillus terreus isolates by RAPD. J Hosp Infect. 2000;44:273–278. doi: 10.1053/jhin.1999.0707. [DOI] [PubMed] [Google Scholar]
- 39.Verweij P E, Denning D W. Diagnostic and therapeutic strategies for invasive aspergillosis. Respir Crit Care Med. 1997;18:203–215. [Google Scholar]
- 40.Verweij P E, Meis J F, Sarfati J, Hoogkamp-Korstanje J A, Latgé J P, Melchers W J. Genotypic characterization of sequential Aspergillus fumigatus isolates from patients with cystic fibrosis. J Clin Microbiol. 1996;34:2595–2597. doi: 10.1128/jcm.34.10.2595-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wald A, Leisenring W, van Burik J A, Bowden R A. Epidemiology of Aspergillus infections in a large cohort of patients undergoing bone marrow transplantation. J Infect Dis. 1997;175:1459–1466. doi: 10.1086/516480. [DOI] [PubMed] [Google Scholar]