Abstract
Cold acclimation (CA) and deacclimation (DA), which are often accompanied by changes in freezing tolerance (FT), carbohydrates and hormones, are crucial for winter survival, especially under global warming. Plants with weak CA and premature DA caused by warm winters and/or unseasonal warm spells can be easily injured by adverse reactions to cold. Thus, understanding the molecular mechanisms of FT is imperative. In this study, we used high-throughput RNA-seq to profile the CA and DA of leaves of overwintering Rhododendron “Miyo-no-Sakae” over time; these leaves do not undergo dormancy but do undergo photoprotection during CA, and they do not grow during DA. Using Mfuzz and weighted gene coexpression network analysis, we identified specific transcriptional characteristics in each phase of CA and DA and proposed networks involving coexpressed genes and physiological traits. In particular, we discovered that the circadian rhythm is critical for obtaining the strongest FT, and high expression of circadian rhythm-related genes might be linked to sugar accumulation during winter. Furthermore, evergreen leaves exhibited robust photoprotection during winter, as revealed by high values of nonphotochemical quenching, high expression of transcripts annotated as “early light-induced proteins”, loss of granum stacks and destacking of thylakoids, all of which were alleviated during DA. The strong requirement of photoprotection could be the reason for decreased abscisic acid (ABA) and jasmonic acid (JA) contents during CA, and decreases in ABA and JA contents may contribute to decreases in lignin content. Our data suggest that the molecular mechanisms of FT in overwintering leaves are unique, which may be due to the high requirements for photoprotection during winter.
Introduction
Freezing tolerance (FT) is an acquired ability of plants native to boreal and temperate zones. However, the process by which woody perennials gradually acquire FT in response to a short photoperiod and low temperatures in the fall and winter is called “cold acclimation” (CA), while the process by which plants gradually lose their acquired FT in response to elevated temperatures is called deacclimation (DA) [1, 2]. In recent years, warm winters and unseasonal warming caused by climate change and global warming have occurred frequently, resulting in incomplete CA or premature DA of some plant species. For instance, peach, apple and pear trees exhibited early DA and vegetation development due to high late-winter temperatures in 2007 in Oak Ridge, Tennessee. As a result, adverse reactions to cold pose a serious threat to these plants [3]. Therefore, this highlights the imperative need to better understand the molecular mechanisms of CA and DA [4].
As early as in the 1980s, the short photoperiod in the fall was documented to induce FT in woody perennials, such as Haralson apple and rhododendrons [5, 6, 7, 8] However, many investigations on the molecular mechanisms of CA concentrated merely on the low temperature because the main cause of CA in herbaceous plants was low temperature and because the study of the underlying molecular mechanism in woody perennials has been slow. CA is typically induced at a temperature of 4°C with a constant photoperiod, light intensity, and/or light quality (applied via a growth chamber) [9, 10, 11]. As a result, researchers have identified classic cold response pathways involving C-repeat/DREB-binding factors (CBFs) [12] and largely confirmed the importance of CBFs for CA in various plant species and in various tissues, including the buds and leaves of woody perennials [13]. In addition to CBFs, abscisic acid (ABA) and jasmonic acid (JA) are also involved in CA in both herbaceous and woody species [14–17]. However, in the field, seasonal changes in temperature are accompanied by changes in light signals; e.g. as temperatures decrease from fall to winter, the photoperiod shortens, the light intensity decreases, and the ratio of red light to far-red light decreases [18], all of which are difficult to simulate artificially in growth chambers. Therefore, studying plant responses to a single stress—low temperature—has limitations for understanding the CA of plants under field conditions. As the recognition of such limitations grows, numerous investigations of CA have been carried out. It has been shown that a short photoperiod and light quality can affect CA by regulating circadian clock-associated genes such as CCA1 and LHY [19–21]. Moreover, circadian clock-related genes can influence the expression of cold-responsive genes (including CBFs and CORs) [22–24]. The circadian clock is thus the gateway of the cold response [23].
To date, most knowledge of plant CA is based on that from studies using Arabidopsis and other herbaceous model species [23]. Reviews on the molecular regulation of cold hardiness or CA of trees are still based on findings of Arabidopsis [4, 23]. Welling & Palva [25] reported that except for dormancy development during overwintering, buds of woody perennials shared similar signal-related and cold-regulated genes and mechanisms underlying CA with those of herbaceous annuals. For overwintering leaves that undergo no dormancy but do undergo photoprotection during CA, the mechanisms might not always follow the traditional regulatory pathways. For example, in our previous study on overwintering leaves of Rhododendron “Elsie Lee”, the ABA and JA contents, which usually exhibit a positive relationship with FT, decreased with increasing leaf FT (LFT) [18]. Furthermore, DA after CA is also crucial for plant winter survival but is largely neglected [26, 27]. In woody perennials, DA focuses mostly on buds, which are closely related to the release of endodormancy and growth [28–30], whereas no growth occurs for overwintering leaves during DA.
Here, we performed a transcriptomic analysis of an azalea cultivar of Rhododendron “Miyo-no-Sakae” across seasons to explore the molecular mechanisms of CA and DA in overwintering leaves. Using Mfuzz and weighted gene coexpression analysis (WGCNA), we found signature events in each phase of CA and DA. Our data reveal that some circadian rhythm-related genes are closely related to changes in LFT and that they are thought to regulate several photoprotective behaviors. Furthermore, photoprotection could be the reason for the decreased ABA and JA contents, thus decreasing lignin during CA. ABA and JA contents began to increase at the end of DA, but the lignin content did not.
Results
Plant growth and leaf freezing tolerance (LFT)
Seasonal changes in the air temperature and photoperiod during the study period are shown in Fig. 1 a and b. Stem growth gradually ceased in early November and did not resume until March (Fig. 1c). The amounts of stem growth from February 19 to March 20 and March 20 to April 18 were 7.3 mm and 20.9 mm, respectively (Fig. 1c). Flower buds gradually stopped growing in December and resumed growth in the following February (Fig. 1c). Flower bud growth significantly accelerated between February 19 and Mar.20 (Fig. 1c), and flowering occurred in late March and early April (Fig. S1). Leaf buds sprouted on Feb. 19, and some new leaves fully expanded on Mar. 20 (Fig. S2). These results indicate that the time of leaf bud emergence was essentially the same as the time of resumption of flower bud growth, which was approximately one month earlier than the resumption of stem growth (Fig. 1c; Fig. S2).
Figure 1.
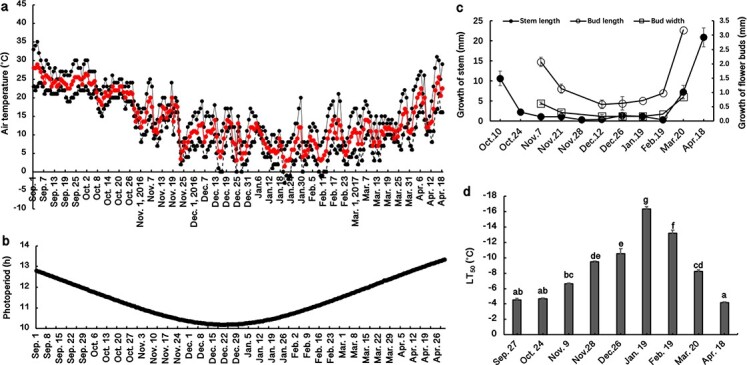
(a) Maximum (black solid line), minimum (black dashed line) and mean (red solid line) air temperatures at the experimental site from September 2016 to April 2017; (b) changes in photoperiod (h) from September 2016 to April 2017; (c) growth of stems and buds from October 2016 to April 2017; (d) seasonal changes in leaf freezing tolerance (indicated as the LT50 value, the temperature causing 50% injury); n = 4. The lowercase letters represent significant differences among sampling dates (p < 0.05), which were calculated using Fisher’s least significant difference test.
Generally, the LT50 at Sept. 27 was approximately −4.5°C, which did not significantly change from Sept. 27 to Oct. 24, despite the photoperiod being reduced by 0.8 h (Fig. 1b, d). The leaf freezing tolerance (LFT) began to increase (more negative LT50) from Oct. 24 to Jan. 19 as the temperature and photoperiod decreased (Fig. 1a, b, d) (i.e. CA) and then gradually decreased as the temperature and photoperiod increased from January to April (Fig. 1d) (i.e. DA). From Oct. 24 to Nov. 9, the LT50 decreased by ~2°C, during which the stems stopped growing (Fig. 1b, d). The strongest LFT occurred on Jan. 19, for which the LT50 was −16.4°C (Fig. 1d). From Jan. 19 to Feb. 19, the growth of new leaves seemed to occur simultaneously with the loss of LFT (Fig. 1d; Fig. S2). Our data suggest that the increase in LFT was accompanied by the cessation of stem growth during CA, and the loss of LFT was accompanied by the emergence of leaf buds and the regrowth of flower buds and stems during DA. After blossoming, the leaves completely lost their acquired LFT (Fig. 1d; Fig. S1).
Three phases of CA and two phases of DA
To fully explore the whole processes of CA and DA, we used all transcripts identified by RNA-seq to perform principal component analysis (PCA). As shown in Fig. 2a, the transcriptome data follow a circular trajectory according to the CA and DA over time. These results show that 1) the three biological replicates for each time point are similar and that 2) the relative proximity of the following pairs—Oct. 24 and Nov. 9, Nov. 28 and Dec. 26, and Feb. 19 and Mar. 20—are close, suggesting that the leaf physiological status in each pair was similar at the transcriptional level. Furthermore, the correlation matrix and hierarchical cluster analysis results using all transcripts were consistent with those of PCA (Fig. S3). According to these results, we selected several time points, i.e. Sept. 27, Nov. 9, Nov. 28, Jan. 19, Mar. 20, and Apr. 18, for further analysis and divided the whole experimental period into five phases—three phases for CA and two phases for DA—as follows: Phase I, the early phase of CA (Sept. 27–Nov. 9); Phase II, the middle phase of CA (Nov. 9–Nov. 28); Phase III, the late phase of CA (Nov. 28–Jan. 19); Phase IV, the early phase of DA (Jan. 19–Mar. 20); and Phase V, the late phase of DA (Mar. 20–Apr. 18).
Figure 2.
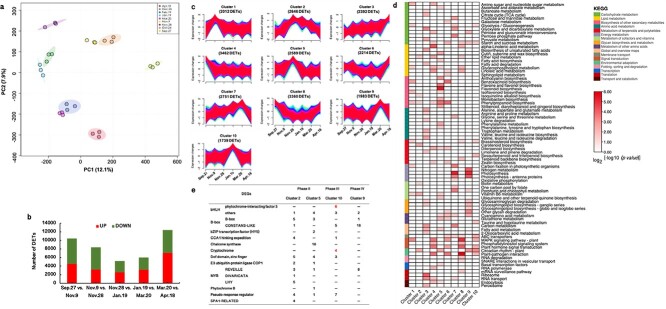
(a) Principal component analysis (performed by MetaboAnalyst, http://www.metaboanalyst.ca) results based on all transcripts identified by RNA-seq. PC1, principal component 1; PC2, principal component 2. Each treatment included three biological replicates; (b) Number of differentially expressed transcripts (DETs) (|fold change| ≥ 2; p < 0.01) identified by RNA-seq during cold acclimation and deacclimation over time; (c) In total, 25 148 DETs identified in this study were classified into ten clusters using the Mfuzz package in R; (d) KEGG pathway enrichment in each cluster (p < 0.05); (e) DETs annotated to the “circadian rhythm – plant” pathway of Clusters 2, 5, 9, and 10.
In total, we identified 25 148 transcripts that were differentially expressed in at least one phase through time-series pairwise comparisons. There were 10 435, 8404, 5201, 6012, and 12 386 DETs in Phases I, II, III, IV and V, respectively (Fig. 2b). To explore transcriptional changes in each phase during CA and DA, we performed a KEGG enrichment analysis. Six KEGG pathways, “phenylpropanoid biosynthesis”, “diterpenoid biosynthesis”, “starch and sucrose metabolism”, “MAPK signaling pathway - plant”, “circadian rhythm - plant”, and “plant-pathogen interaction”, were commonly enriched in each phase, suggesting that these biological processes were transcriptionally active in both CA and DA (Fig. S4).
Possible signature events in a certain phase, according to Mfuzz analysis
To define the temporal characteristics of the complete transcriptome dataset, using Mfuzz, we performed clustering analysis of 25 148 DETs across 18 samples, which divided all the DETs into 10 clusters (Fig. 2c). We searched for enriched KEGG pathways (p < 0.05) within each cluster and summarized these pathways via a heatmap (Fig. 2d). We found that some KEGG pathways were specific to one or two phases. For instance, the “α-linolenic acid metabolism” and “linoleic acid metabolism” pathways in Clusters 4, 6 and 8 were specific to Phases I and/or V; the “anthocyanin biosynthesis”, “flavone and flavonol biosynthesis”, and “flavonoid biosynthesis” pathways in Cluster 5 were specific to Phases II and III; “photosynthesis” and “photosynthesis - antenna proteins” in Clusters 3, 7 and 9 were specific to Phases IV and/or V; and the glycan biosynthesis and metabolism-related pathways in Clusters 3, 6 and 9 were specific to Phases IV and/or V (Fig. 2d).
The DETs in Cluster 10 (Data S1) could be candidates for affording the strongest LFT during winter, since the highest expression was exhibited on Jan. 19 with the strongest LFT (Fig. 1d; Fig. 2c). The top enriched KEGG pathway in this cluster was “circadian rhythm” (Fig. 2d) (p = 3.2e−9), which was also enriched in Clusters 2 (p = 4.7e−5), 5 (p = 4.1e−6), and 9 (p = 5.3e−8). Thus, the “circadian rhythm” pathway might represent different changes in Phases II, III and IV, all three of which are important for obtaining robust LFT in CA or an early DA response (Fig. 2e). We analyzed the DETs associated with “circadian rhythm” in each cluster and found that the DETs annotated as “cryptochrome 1 (CRY1)” and “bHLH/PIF3” were enriched only in Cluster 10 (Fig. 2e; Table S1). Thus, the upregulation of CRY1s and PIF3s could play important roles in obtaining the strongest LFT in overwintering leaves.
Coexpression networks of genes and relationships between physiological parameters and DETs
To explore coexpression networks of genes and possible relationships between the changes in physiological parameters and DETs, we performed a WGCNA, which revealed 20 modules (Fig. 3a; Fig. S5a). The expression trends in each module are shown in Fig. S5b. Three LT50-related modules, the “red” (1282 DETs) (r2 = −0.96), “blue” (3616 DETs) (r2 = −0.80) and “turquoise” (7049 DETs) (r2 = 0.65) modules, for which p was <0.01, were further analyzed (Fig. 3a). In the red module, DETs were generally up−/downregulated from Sept. 27 to Jan.19 and gradually down−/upregulated from Jan. 19 to Apr. 18 (Fig. 3b; Data S2). Genes coexpressed in this module were significantly enriched in “circadian rhythm”, “ABC transporters”, and “plant hormone signal transduction” (p < 0.05) (Fig. 3b). In the blue module, DETs up−/downregulated from Sept. 27 to Nov. 28, stayed at the upregulated levels until Jan. 19, and then progressively became down−/upregulated (Fig. 3b; Data S2). These DETs were significantly enriched in 8 KEGG pathways (p < 0.05), such as “circadian rhythm”, “vitamin B6 metabolism”, “ether lipid metabolism”, “basal transcription factor”, and “glutathione metabolism” (Fig. 3b). In the turquoise module, DETs were sharply downregulated/upregulated from Sept. 27 to Nov. 9, continuously downregulated/upregulated from Nov. 9 to Nov. 28, stayed at those levels, and then were upregulated/downregulated from Mar. 20 to Apr. 18 (Fig. 3b; Data S2). DETs were significantly enriched in 19 KEGG pathways (p < 0.05), such as “plant-pathogen interaction”, “MAPK signaling pathway-plant”, “α-linolenic acid metabolism”, “plant hormone signal transduction”, and “linoleic acid metabolism” (Fig. 3b).
Figure 3.
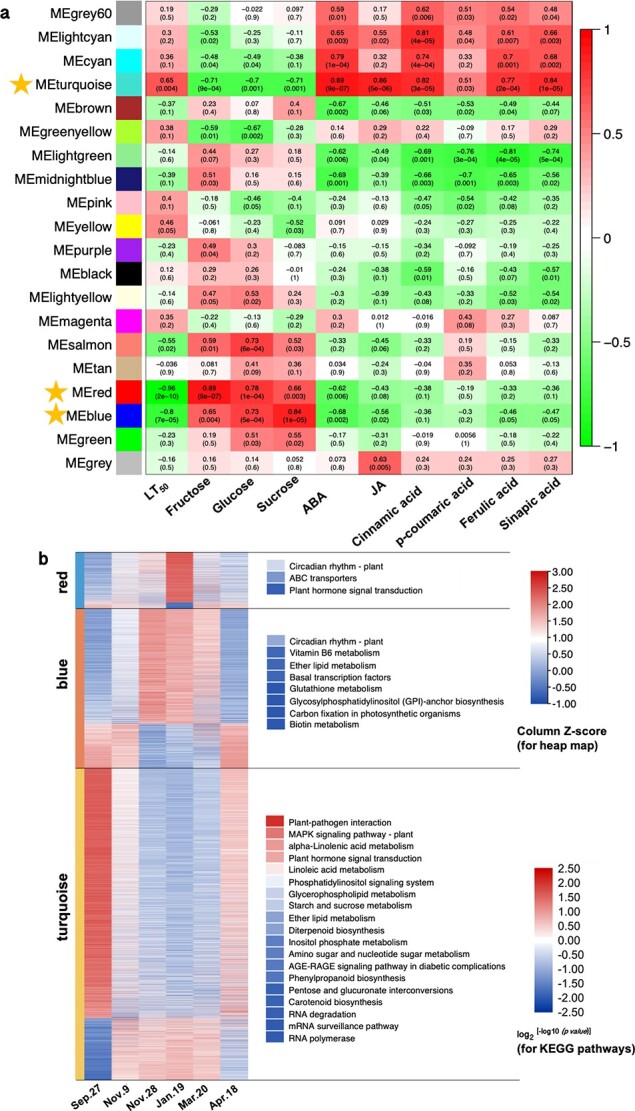
Analysis of gene coexpression and the relationships between genes and physiological parameters. (a) Module-trait correlations and gene coexpression network analysis performed by the weighted gene coexpression network analysis (WGCNA) package in R during cold acclimation and deacclimation in overwintering leaves of Rhododendron “Miyo-no-Sakae”. Module-trait correlations are shown, and the corresponding p values are shown in parentheses. The color scale on the right represents the module-trait correlations from −1 to 1. The red, blue and turquoise modules with stars closely related to the LT50 (p < 0.01). (b) Seasonal expression trends of DETs in the red, blue and turquoise modules and the KEGG pathway enrichment analysis (performed by the pHYPER function in R) for each module (p < 0.05).
“Circadian rhythm” was the most significantly enriched pathway, with 30 DETs (p = 1.3e−7) in the red module and 50 DETs in the blue module (p = 8.1e−5) (Table S2). In red module, the DETs were annotated as “transcription factor bHLH/phytochrome-interacting factor 3 (PIF3)”, “two-component response regulator APRR5/9 (APRR)”, “Cyclic dof factor 2/3 (C2C2-Dof)”, “zinc finger protein CONSTANS (CO)”, “E3 ubiquitin-protein ligase COP1 (COP1)”, and “cryptochrome 1 (CRY1)” (Table S2). In the blue module, DETs were annotated as “transcription factor MYBs”, “transcription factor TCPs”, “protein SPA1-RELATEDs”, “C2C2-Dofs”, “B-Box (BBXs)”, and “APRRs”. Module membership (MM) represents the degree of eigengene-based connectivity, which is a measurement of the membership of a gene with respect to a module and is used to screen hub genes. The highly connected hub genes with MM values higher than 0.9 included CRY1s, CO, C2C2-Dofs, bHLHs, and APRRs in the “red” variety and SPA1-RELATEDs, C2C2-Dofs, APRRs, BBXs, MYBs, and bHLHs in the “blue” variety (Table S2).
In the turquoise module, “starch and sucrose metabolism” is related to carbohydrate metabolism, “α-linolenic acid metabolism” and “linoleic acid metabolism” are upstream of JA biosynthesis, “carotenoid biosynthesis” is upstream of ABA biosynthesis, and “phenylpropanoid biosynthesis” is upstream of lignin biosynthesis. Thus, we measured carbohydrate (glucose, fructose and sucrose), ABA, JA and lignin contents across the six sampling dates (Fig. 4a, b, c, d, e, f, g). Relationships among modules and these physiological parameters were also determined via WGCNA (Fig. 3a). Fructose and glucose were highly correlated with the red module (r2 = 0.89, p = 9.0e−7 and r2 = 0.78, p = 1.0e−4, respectively) (Fig. 3a; Fig. 4a); sucrose was highly correlated with the blue module (r2 = 0.84, p = 1.0e−5) (Fig. 3a; Fig. 4a); ABA, JA, cinnamic acid, ferulic acid, and sinapic acid contents were highly correlated with the turquoise module (r2 = 0.89, p = 9.0e−7 and r2 = 0.86, p = 5.0e−6 and r2 = 0.82, p = 3.0e−5 and r2 = 0.77, p = 2.0e−4 and r2 = 0.84, p = 1.0e−5, respectively) (Fig. 3a; Fig. 4b, c, d, f, g); and no module was correlated with p-coumaric acid (Fig. 3a; Fig. 4e). Since the red, blue and turquoise modules were highly related to the LT50 (Fig. 3a), glucose, fructose, sucrose, ABA, JA and lignin contents were also correlated with LT50 values. Furthermore, “circadian rhythm” was the top pathway enriched in the red and blue modules (p = 1.3e−7 and p = 8.1e−5, respectively), which exhibited the highest relationships with sugars (Fig. 3a). Thus, there could be interactions between sugars and “circadian rhythm” during CA and DA. ABA, JA, and lignin contents showed the highest relationship with the turquoise module (Fig. 3a), suggesting that there could be interactions between ABA, JA, and lignin during CA and DA.
Figure 4.
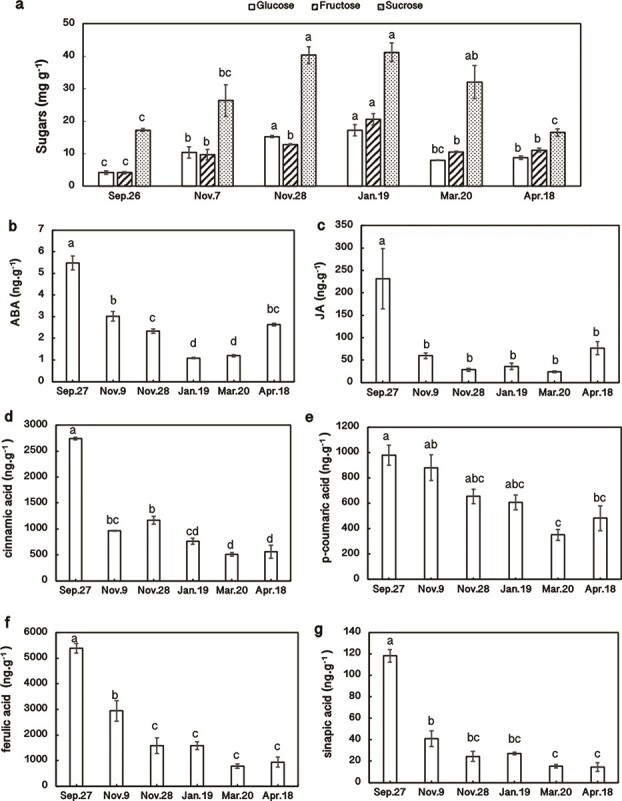
Seasonal changes in sugar (a), abscisic acid (ABA; b), jasmonic acid (JA; c), cinnamic acid (d), p-coumaric acid (e), ferulic acid (f), and sinapic acid (g) contents in overwintering leaf tissues during cold acclimation (from Sept. 27 to Jan. 19) and deacclimation (from Jan. 19 to Apr. 18); n = 3. The lowercase letters represent significant differences among sampling dates (p < 0.05), which were calculated using Fisher’s least significant difference test.
Photoprotection during CA and DA
Photosystem- or photoprotection-related DETs were enriched in the red and blue modules; these enriched DETs included “ribulose bisphosphate carboxylase small subunits (RBCSs)”, “early light-induced proteins (ELIPs)”, the “ATP-dependent zinc metalloprotease FTSH”, “chlorophyllase”, “low photosystem II (PSII) accumulation”, and the “PSII 22 kDa protein” (Table S3). Except for some RBCSs and one “low PSII accumulation” protein, others exhibited high expression levels in winter (Table S3). Photoprotection-related DETs were coexpressed with circadian rhythm-related genes in the red and blue modules, suggesting that there could be relationships between photoprotection and circadian rhythm during CA and DA.
To determine the PSII performance, we measured the fraction of absorbed light energy utilized by PSII photochemistry (ΦII) and the photochemical quenching (qP) (Fig. 5 a,b). ΦII and qP significantly declined during CA (p < 0.05), reached minimal values on Jan. 19, and then increased during DA (Fig. 5 a, b), suggesting that photosynthetic activities were gradually downregulated in CA and resumed in DA. Furthermore, we quantified the amount of absorbed light energy dissipated as heat by nonphotochemical quenching (NPQ) and the quantum yield of nonregulated or constitutive loss of energy (ΦNO) (Fig. 5c, d). NPQ significantly increased during CA (p < 0.05) and reached its maximal value on Jan. 19, which was almost twice that on Sept. 27 and Nov. 9. Afterward, it decreased during DA and returned to its original level on Apr. 18 (Fig. 5c). The accumulation of zeaxanthin is the main component of NPQ. Therefore, the trends of NPQ suggest that zeaxanthin gradually accumulated to dissipate excess light energy during CA. ΦNO, also called basal intrinsic nonradiative decay, exhibited its highest value on Nov. 9 but then gradually declined, reaching its minimal level on Apr. 18 (Fig. 5d). Furthermore, in terms of chloroplast ultrastructure, we found massive losses of granum stacks and destacking of thylakoids in the winter (Jan. 19) compared with the findings of chloroplasts after a 6-day recovery period with a photoperiod of 14 h under a light intensity of 80 μmol m−2 s−1 at 22°C (Fig. 5e, f). The destacking of thylakoids allows the formation of PSII and PSI complexes and allows the direct transfer of excess energy from PSII to PSI, which is a component of ΦNO [31].
Figure 5.
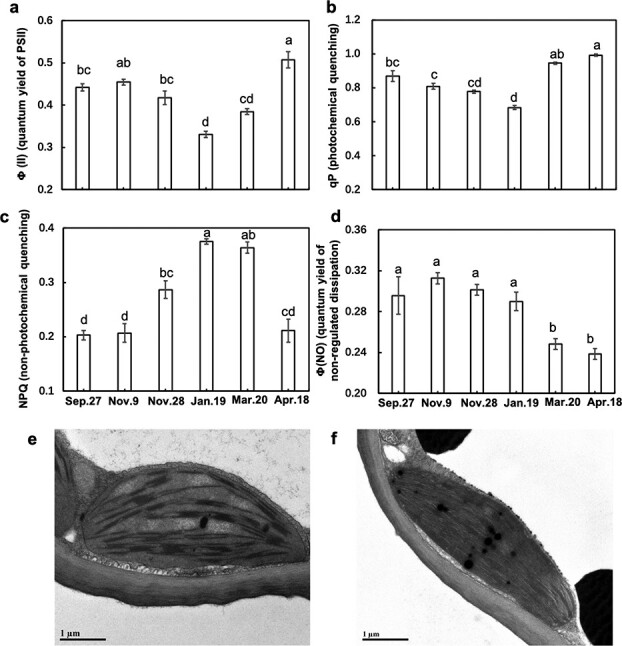
Seasonal dynamics of the photochemical performance of PSII by chlorophyll fluorescence (a, b, c, d) and transmission electron microphotographs of chlorophyll ultrastructure (e, f) in overwintering leaves of Rhododendron “Miyo-no-Sakae”. (a) Effective quantum yield of PSII (ΦII); (b) photochemical quenching (qP); (c) nonphotochemical quenching (NPQ); (d) quantum yield of nonregulated nonphotochemical quenching (ΦNO); n = 8. (e) Chlorophyll ultrastructure in January; (f) chlorophyll ultrastructure of January samples after recovery at 22°C for 6 days with a photoperiod of 14 h under a light intensity of 80 μmol m–2 s–1. The lowercase letters represent significant differences among sampling dates (p < 0.05), which were calculated using Fisher’s least significant difference test.
Changes in ABA-, JA-, and lignin-related genes during CA and DA
ABA content showed the highest level on Sept. 27 (Fig. 4b). The ABA content then gradually declined and reached its lowest level on Jan. 19, after which it progressively accumulated and reached approximately half of its original level on Apr. 18 (Fig. 4b). The changes in ABA content were negatively related to LFT (positive to LT50) (Fig. 1d; Fig. 3a; Fig. 4b), which was consistent with the results of our previous study [17]. From geranylgeranyl diphosphate to violaxanthin, most DETs showed high expression from Nov. 28 to Mar. 20 (Fig. 6a; Data S3). UDP-glucosyl transferase and ABA 8′-hydroxylase, two enzymes catabolizing ABA, exhibited low expression from Nov. 28 to Mar. 20 (Fig. 6a; Data S3). In ABA signaling, most PYLs showed high expression on Nov. 28 and Jan. 19, whereas most PP2Cs and SnRK2s showed high expression on Sept. 27 and Apr. 18 (Fig. 6a). The trends of most PP2Cs and SnRK2s were consistent with the changes in ABA content (Fig. 4b; Fig. 6a; Data S3).
Figure 6.
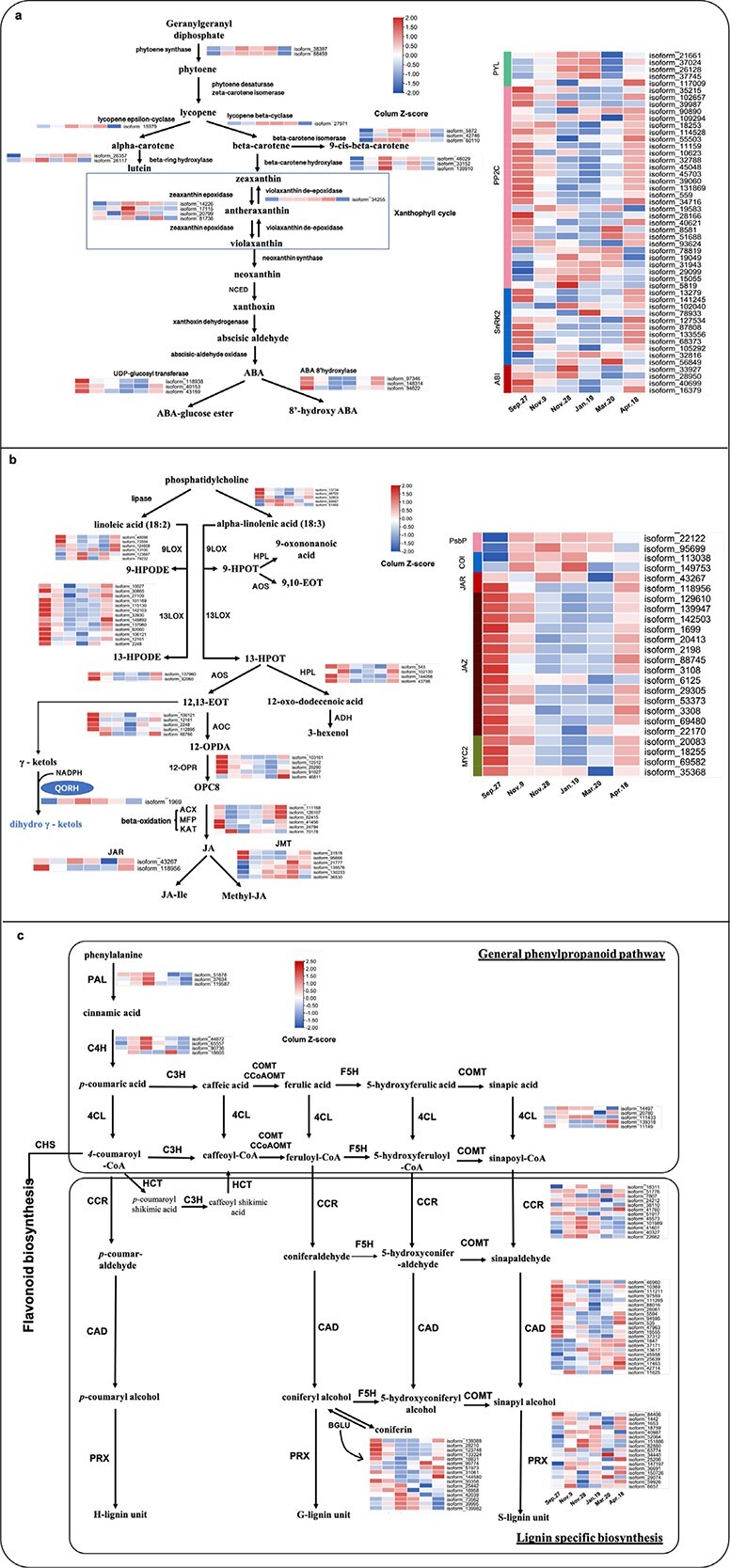
Pathways of abscisic acid (ABA), jasmonic acid (JA) and lignin. (a) Pathway of “carotenoid biosynthesis” in ABA biosynthesis and heat maps of differentially expressed transcripts (DETs) involved in this pathway and ABA signaling; NCED, 9-cis-epoxycarotenoid dioxygenase; (b) Pathways of “α-linolenic acid metabolism” and “linoleic acid metabolism” in JA biosynthesis and heat maps of DETs involved in these pathways and JA signaling; LOX, lipoxygenase; AOS, allene oxide synthase; AOC, allene oxide cyclase; HPL, fatty acid hydroperoxide lyase; 12-OPR, 12-oxophytodienoate reductase; ACX, peroxisomal acyl-coenzyme A oxidase; MFP, peroxisomal fatty acid beta-oxidation multifunctional protein; KAT, 3-ketoacyl-CoA thiolase; JAR, jasmonic acid-amino synthetase; JMT, jasmonate O-methyltransferase. (c) Pathway of “phenylpropanoid biosynthesis” in lignin biosynthesis and heat maps of DETs involved in this pathway. PAL, phenylalanine ammonia lyase; C4H, cinnamate 4- monooxygenase; 4CL, 4-coumarate-CoA ligase; CCR, cinnamoyl-CoA reductase; CAD, cinnamyl alcohol dehydrogenase; BGLU, beta-glycosidase; PRX, peroxidase.
The JA content was highest on Sept.27 (Fig. 4c). However, it sharply decreased from Sept. 27 to Nov. 9, stayed at a low level until Mar. 20, and then increased to approximately one-third of its original level on Apr. 18 (Fig. 4c). The changes in DETs related to JA biosynthesis were mostly consistent with the changes in JA content (Fig. 4c; Fig. 6b; Data S3). This is also consistent with the findings of our previous study [18]. The expression of DETs related to JA catabolism was mostly opposite that of the JA content (Fig. 6b). Furthermore, DETs annotated as “chloroplast envelope quinone oxidoreductase (QORH)” and “PsbP” were enriched in “α-linolenic acid metabolism” (Data S3), a JA biosynthesis pathway (Fig. 6b), and exhibited high expression from Nov. 9 to Mar. 20. QORH is located in the inner membrane of the chloroplast envelope and can detoxify the products of polyunsaturated fatty acid peroxides (γ-ketols [32]). PsbP is an extrinsic subunit of PSII that participates in the normal function of photosynthetic water oxidation. PSII lacking PsbP has been shown to be hypersensitive to light [33]. In JA signaling, except for COIs, which showed high expression from Nov. 9 to Jan. 19, the trends of most DETs annotated as JAZs and MYC2s were similar to those of the JA content (Fig. 6b). Seasonal changes in ABA and JA contents during CA were consistent with the findings of our previous study on overwintering leaves of Rhododendron “Elsie Lee” [18].
Although DETs annotated as “phenylalanine ammonia lyase (PAL)” and “cinnamate 4-monooxygenase (C4H)” were highly expressed on Nov. 28 (Fig. 6c), the cinnamic acid and p-coumaric acid contents decreased from Sept. 27 to Nov. 28 (Fig. 4 d, e). This suggests that changes in cinnamic acid and p-coumaric acid were not regulated at the transcriptional level during CA in the field. p-Coumaric acid, ferulic acid, and sinapic acid are precursors of three monolignols—p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol, which are incorporated into lignin polymers and represent p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) moieties, respectively (Fig. 6c). Ferulic acid and sinapic acid contents largely declined from Sept. 27 to Nov. 28. They then stayed at those levels until April (Fig. 4 f, g), suggesting that lignin contents decreased in CA and did not recover until the end of DA.
“4-Coumarate-CoA ligase (4CL)”, “cinnamoyl-CoA reductase (CCR), cinnamyl-alcohol dehydrogenase (CAD), and peroxidase (PRX)” were found to participate in lignin biosynthesis (Fig. 6c). Most CCRs showed high expression from Nov. 9 to Jan.19, and CADs exhibited high expression on Sept. 27 or during DA (Fig. 6c). Peroxidase catalysis is the last step for H-, G-, or S-lignin formation, and PRXs did not exhibit a noticeable trend during CA and DA (Fig. 6c). Furthermore, there were DETs annotated as “beta-glucosidase (BGLU)” enriched in the “phenylpropanoid biosynthesis” pathway (Data S3). Moreover, ten out of 16 BGLUs were downregulated during CA and upregulated during DA (Fig. 6c). BGLUs are capable of hydrolyzing coniferin to release coniferyl alcohol. Coniferin is the 4-O-glucoside of coniferyl alcohol and has been proposed to be either a transport or storage form of monolignols [34]. BGLU and PRX are the final steps of lignin biosynthesis, both of which occur in the cell wall [34, 35].
Discussion
Circadian rhythm could play a key role in the CA and DA of overwintering leaves of plants under field conditions
The accumulation of carbohydrate such as sucrose is often reported to be cryoprotectant in tissues in response to low temperatures during the winter [2, 36]. However, sugars can also be inducers of signals. Exogenous sucrose application could induce the expression of circadian clock-related genes such as AtCCA1, AtGIGANTEA (GI), and AtTOC1 [37]. Endogenous sugars produced by photosynthesis, including sucrose, glucose and fructose, could entrain circadian clock-related genes in Arabidopsis [38, 39]. These aforementioned studies support that sugars could regulate circadian-related function in plants. In the present study, seasonal changes in glucose, fructose, and sucrose levels were closely and significantly related to the red and blue modules according to the WGCNA (p < 0.01) (Fig. 3a), whose top KEGG pathway was “circadian rhythm” (p = 1.3e−7 and 8.1e−5 in the red and blue modules, respectively) (Fig. 3b). These findings suggest that sugars could be involved in regulating circadian rhythm-related genes during CA and DA and that their accumulation might induce the upregulation of circadian rhythm-related genes during winter.
Kidokoro et al. [40] reported that Arabidopsis plants recognize cold stress as two different signals—either rapid or gradual temperature decreases, resulting in the expression of AtCBF3, and the latter occurs through two circadian oscillators, CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY). In this study, the air temperature decreased from fall to winter, which corresponded to a gradual temperature decrease. “Circadian rhythm” was enriched in Phases II (p = 4.7e−5 and 4.1e−6 in Clusters 2 and 5, respectively) and III (p = 3.2e−9) during CA and Phase IV (p = 5.3e−8) during DA according to the Mfuzz analysis (Fig. 2d), suggesting that “circadian rhythm” is related to obtaining the hardiest LFT in winter and could be an early sensor of DA. Additionally, “circadian rhythm” was the top KEGG pathway in the red and blue modules of the WGCNA results (p < 0.05) (Fig. 3b), and these modules are closely related to LT50 values (r2 = −0.96 and p = 2e−10 for the red module and r2 = −0.8 and p = 7e−5 for the blue module) (Fig. 3a), suggesting again that the circadian rhythm is important for both CA and DA. Ramos et al [41] reported that the regular daily oscillation of CsTOC1 and CsLHY in chestnut was disrupted in tissues of stems and buds and showed constantly high expression during winter dormancy. CsPRR5/7/9 in the stems of chestnut is constantly and highly expressed at low temperature [42]. In Populus buds, continuously high expression of PttLHY1/2 was suggested to be essential for the clock’s adjustment to cold hardiness [43]. This information suggests that clock disruption under cold conditions could be part of an adaptive strategy that enables deciduous woody perennials to survive winter [44]. In this study, DETs related to “circadian rhythm” in Clusters 2, 5, and 10 and associated with the red and blue modules showed high expression in winter (on Nov. 28 or Jan. 19) (Fig. 2c; Fig. 3b), including DETs annotated as HY5s, LHYs, and APRRs (Fig. 2e; Data S2), which suggests that high expression also occurs in the tissues of overwintering leaves. Therefore, these DETs are important candidates for obtaining LFT and thus for winter survival.
In Arabidopsis, AtCCA1 and AtLHY bind to and induce the expression of AtCBFs [45, 46]. The induction of PttCBF1 was abolished in lhy mutant Populus buds [43]. In this study, sequences of transcripts in “Miyo-no-Sakae” leaves subjected to BLAST searches matching AtCBF3 with high bit scores (>100) were in the turquoise module and showed trends opposite those of LHYs and LFT (Table S2; Table S4; Fig. 3b; Fig. 1d). The sequences of five transcripts subjected to BLAST searches matching AtCBF3 with relatively low bit scores (<100) in the red or turquoise module, such as isoform_40839 and isoform_36666, were positively related to LHYs and LFT (Table S4). We then investigated trends of AtCBF downstream target genes, such as AtCOR15a, AtCOR47, and AtKIN1 [47], in Rhododendron “Miyo-no-Sakae”. There was no DET whose sequence in Rhododendron “Miyo-no-Sakae” matched those of AtCOR15a and AtKIN1 upon BLAST searches. The sequences of DETs that matched those of AtCOR47s via BLAST searches were mostly annotated as “dehydrins”, and they were downregulated during CA (Table S4). However, there were 6 DETs annotated as “dehydrins” but whose sequences were not homologous to that of AtCOR47 in the blue module, such as isoform_39935 and isoform_4588, which were upregulated during CA (Table S4). Jiang et al. [48] reported that PHYTOCHROME-INTERACTING FACTOR 3 (PIF3) negatively regulates FT in Arabidopsis, while in this study, PIF3s were positively correlated with LFT (p < 0.05) (Table S2; Fig. 3b). CRY1, a sensor of blue light, was reported to be negatively involved in AtCBF regulation in the early stage of CA [49] and could mediate plant responses to high irradiance by upregulating AtELIP1/2 in Arabidopsis [50]. In this study, CRY1s exhibited a positive relationship with LFT (p < 0.05) (Table S2, Fig. 3a) and was coexpressed with ELIPs in the red module (Table S2; Table S3).
Photoprotection and the necessity for photoprotection in overwintering leaves may be responsible for the decrease in ABA and JA during CA
In this study, as the air temperature decreased from fall to winter, the photosynthetic activity in terms of ΦII and qP significantly decreased (p < 0.05) (Fig. 5a, b). Therefore, overwintering leaves had a high demand for photoprotection under the combination of high light and low temperatures during CA. The photoprotection of overwintering leaves was multifaceted, which included the high expression of ELIPs, destacking of thylakoids, and a significant increase in NPQ (p < 0.05) (Table S3, Fig. 5c, e).
Harmer et al. [51] reported that the expression of genes implicated in light harvesting (LHCA and LHCB) and PSII reaction centers of photosynthesis cycled with the circadian rhythm, which suggests that the circadian clock regulates photosynthesis-related genes. In this study, we found that in the red and blue modules, many genes related to photoprotection were identified (Table S3). Among them, there were 37 DETs annotated as ELIPs. ELIPs are nuclear-encoded, thylakoid-bound proteins with a photoprotective function [52]. Moreover, they were proven to show diurnal circadian oscillations in pea and barley [53, 54], which suggested that they could be regulated by the circadian rhythm. Our data have proven that RhHY5 and RhBBX24, two circadian-related genes, can directly bind to and affect the expression of RhELIP3 in Rhododendron “Elsie Lee” (Fig. S6).
Thylakoid destacking was observed in January when the minimum air temperature was approximately 0°C (Fig. 5e; Fig. 1a). Bag et al. [31] reported that chloroplasts lost granum stacks during early spring. The average number of grana per chloroplast in Scots pine was shown to decline from autumn to winter and reached its minimal level in early spring. Evergreen conifers are mostly localized in boreal zones with a minimal temperature lower than −20°C during winter. Moreover, in early spring, the minimum air temperatures are also frequently lower than −10°C. Our data indicate that changes in chloroplast ultrastructure also occurred in overwintering leaves of plants inhabiting places with relatively warm winters.
NPQ is mainly dependent on the accumulation of zeaxanthin in the xanthophyll cycle [55, 56]. Zeaxanthin is proposed to be a quencher itself or plays an allosteric role; nonetheless, this compound can effectively inhibit the production of reactive oxygen species (ROS) [57, 58]. Oxylipins (including JA, its precursor 12-OPDA, and MeJA) are plant messengers derived from the oxidation of polyunsaturated fatty acids via the lipoxygenase (LOX) pathway (Fig. 6b) [59–60]. In this study, DETs annotated as LOXs were downregulated in the early stage of CA (Fig. 6b). Additionally, the significant upregulation of NPQ (p < 0.05) (Fig. 5c) (i.e. accumulation of zeaxanthin) and high expression of ELIPs (Table S3) during CA indicate the upregulation of photoprotection, which could effectively decrease the production of ROS, thus suppressing fatty acid peroxidation and oxylipin formation [61]. Wild-type Arabidopsis exhibited a much lower concentration of JA than did Atnpq1 mutants (which are deficient in zeaxanthin accumulation) after exposure to high light for 7 days [62]. Thus, zeaxanthin-dependent photoprotection prevented JA production under light stress [61]. The biosynthesis of zeaxanthin occurs upstream of ABA biosynthesis (Fig. 6a). The accumulation of zeaxanthin resulted in the deficiency of ABA biosynthesis, which has been observed in previous studies [18, 63, 64]. Previous reports also showed that exogenous application of MeJA significantly improved FT, while blocking JA biosynthesis led to hypersensitivity to freezing stress in Arabidopsis [65]. Endogenous JA increased under cold stress in Oryza sativa and Vitis amurensis calli, and JA biosynthesis-related genes were upregulated [17, 66]. Exogenous ABA increased bud FT, and the ABA-deficient mutant of Arabidopsis lost its capacity for CA [67, 68]. However, these results were obtained under cold stress only. Our data suggest that in overwintering evergreen leaves, the photoprotection caused by the combination of low temperatures and high light could inhibit the activity of the ABA- and JA-dependent pathways to increase the LFT in CA.
The decrease in ABA and JA could be partially responsible for the decrease in lignin contents during CA.
Lignin is the major component of plant secondary cell walls and is affected by various stresses. The PAL enzyme catalyzes the first step of the phenylpropanoid pathway to regulate the biosynthesis of lignin. Previous studies reported that Nicotiana tabacum and Brachypodium PAL-knockdown/downregulated plants presented significantly reduced lignin contents in their stems [69, 70], and Arabidopsis pal1 pal2 double mutants presented 30% less rosette leaf lignin content when compared with that of the wild type [71]. These previous results suggest that PALs positively regulate lignin biosynthesis under no abiotic or biotic stress. In this study, PALs were upregulated from Sept. 27 to Nov. 28 (Fig. 6c), while the cinnamic acid and precursors of lignin contents significantly decreased during winter (p < 0.05) (Fig. 4d, e, g, f). The decrease in lignin in the winter could be the following reasons:
(1) The phenylpropanoid pathway occurs upstream of both flavonoid and lignin biosynthesis (Fig. 6c). Cinnamic acid and p-coumaric acid could be mainly allocated to flavonoid accumulation, which could function as antioxidants during winter (Fig. S7) [72].
(2) Endogenous ABA biosynthesis and its associated signaling pathway are involved in lignin deposition in Arabidopsis through the core ABA-signaling component SnRK2 kinases [73]. Lignification was reduced in the aba2–1 and snrk2.2/3/6 triple mutants of Arabidopsis compared to the wild type [73]. In this study, SnRK2s were mostly downregulated during CA and upregulated during DA (Fig. 6a). WGCNA showed that seasonal changes in ABA and lignin contents were highly correlated with the same turquoise module, suggesting that the decreased ABA content and downregulation of SnRK2s during CA could lead to a decrease in lignin in overwintering leaves.
(3) Exogenous JA or its derivative methyl jasmonate (MeJA) treatment can stimulate the expression of genes related to the biosynthesis of cell wall components [74, 75] and MeJA induced cell wall in-growth in the phloem of leaves [76] which suggests that JA may be positively related to lignin. Thus, the decrease in JA during CA in this study may cause a decrease in lignin content (Fig. 4c, d, e, f, g).
(4) Ji et al. [71] revealed that the increase in FT is negatively correlated with lignin accumulation under CA in Arabidopsis, and this was regulated by the downregulation of the BLUE COPPER-BINDING PROTEIN (BCB) gene. We used the sequence of AtBCB (At5g20230) as a query to search BCBs in “Miyo-no-Sakae” via BLAST and found that most BCBs were in the turquoise or blue modules and were generally downregulated in CA but upregulated in DA (Table S5).
Conclusions
Based on the data analyzed during CA and DA in Rhododendron “Miyo-no-Sakae” in the field in the present work and those from our previous publication on the comparisons between field-based and artificial CA in overwintering evergreen leaves, we found that circadian rhythm might play a key role in obtaining robust LFT during winter. In this study, we proposed that the upregulation of some circadian rhythm-related genes during CA and their downregulation during DA could be related to the accumulation and catabolism of sugars, respectively, by our analysis of module-trait relationships via WGCNA (Fig. 7). The ABA and JA contents in overwintering leaves decreased during CA in both Rhododendron “Miyo-no-Sakae” and Rhododendron “Elsie Lee” and did not recover until the complete loss of the obtained LFT in Rhododendron “Miyo-no-Sakae”. The negative relationship between LFT and ABA or JA content was quite different from what has been reported for other tissues such as buds and stems or other plant species such as Arabidopsis and rice. The reason for this could be the induced photoprotection during CA under field conditions (Fig. 7). The decreased ABA and JA biosynthesis during winter could result in a decrease in lignin content (Fig. 7). We therefore propose that the mechanisms of acquiring LFT in overwintering leaves under both low temperature and high light stresses in CA under field conditions were unique, which occurred through the strong photoprotection and accumulation of carbohydrates in conjunction with decreases in ABA, JA and lignin contents.
Figure 7.
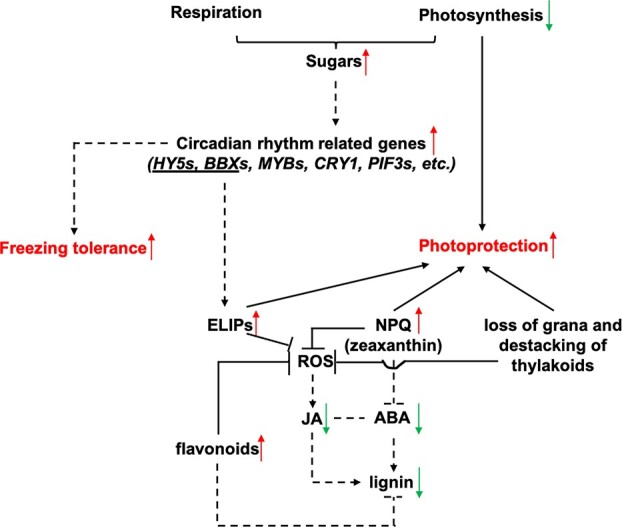
Schematic of cold acclimation (CA) in this study. The proposed model suggests that the demand for photoprotection in overwintering leaves during CA under field conditions could be the reason for decreased abscisic acid (ABA) and jasmonic acid (JA) contents and thus the decrease in lignin content. The upregulation of circadian rhythm-related genes could play key roles in increasing freezing tolerance. NPQ, photochemical quenching; ROS, reactive oxygen species. The dashed lines indicate that the inferences need further investigation.
Materials and methods
Plant materials and environmental conditions
Pots of 3-year-old cuttings of Rhododendron “Miyo-no-Sakae” (Hirado Hybrids) procured from Jinhua Yonggen Rhododendron Cultivation Co., China, were grown in a mixture of peat, pine needles, and yellow clay (3:1:1 by volume) and maintained outside under full sunlight for CA and DA in a nursery located in Hangzhou, Zhejiang Province (30° 15’N 120° 10’E, hardiness zone 10), for DA. The experimental period was from fall to the spring of the following year, with the specific sampling dates of Sept. 27, 2016, Oct. 24, Nov. 9, Nov. 28, Dec. 26, Jan. 19, 2017, Feb. 19, Mar. 20, and Apr. 18. Forty-eight pots were randomly divided into four biological replicates (12 pots per biological replicate). Fully expanded leaves from the current year of growth were collected for experiments. Leaf tissues (except for those used for leaf freezing tolerance measurements) collected on the sampling dates were flash frozen in liquid nitrogen and then stored at −80°C until analysis.
Plant growth measurements
Ten stems and flower buds were chosen at random and marked. Stem length from Sept. 26 to Apr. 18, as well as the length and width of flower buds from Oct. 24 to Mar. 20, were measured using a Vernier caliper approximately every 2 weeks. The difference between two consecutive measurements revealed the expansion of the stem or buds.
Leaf freezing tolerance (LFT) tests (determination of LT50)
The lab freeze–thaw protocol was used to investigate LFT [2]. Briefly, collected leaf tissues were placed in plastic bags and put into an ethanol freezing bath. The stepwise lowering of target temperatures was set according to its rough LT50 (the temperature causing 50% injury), e.g. 0, −5, −10, −15, −20, −25, and − 30°C for midwinter samples. The leaves were maintained for 4 h at each target temperature before thawing at 4°C overnight. Samples maintained at 4°C throughout the whole freeze–thaw protocol were considered unfrozen controls. After thawing at room temperature for 1 h, we immersed all samples in 15 mL of ultrapure water for 24 h after vacuum infiltration. First, ion leakage was measured after the samples were quickly vortexed, and a second measurement was conducted at room temperature after the tissues were destroyed by being subjected to 100°C. We used the Gompertz function to calculate LT50s [77].
Sample preparation for RNA-seq and ISO-seq
Total RNA was extracted from leaf tissues collected on each sampling date using an RNAprep Pure Plant Plus Kit (Polysaccharide & Polyphenolic-rich) (Tiangen, Beijing, China). For RNA-seq, each sampling date was considered one treatment, and each treatment included three biological replicates; for ISO-seq, one biological replicate in each treatment was mixed. The transcripts assembled by ISO-seq constituted the reference transcriptome used for RNA-seq. Unigenes with |log2(fold change) | ≥ 1 and p values < 0.01 were identified as differentially expressed transcripts (DETs). The ISO-seq and isoform annotations, RNA-seq and data analysis are described in the supporting methods.
Quantification of carbohydrates and the ABA, JA, and lignin contents
Glucose, fructose, and sucrose concentrations were measured by HPLC (Waters e2695) equipped with a refractive index detector (Waters 2414 RI Detector) using the method described in Liu et al. [18].
ABA, JA and lignin concentrations were measured by UPLC-ESI-MS/MS. The method for measuring ABA and JA was performed according to the methods of Liu et al [18]. Details of the method through which lignin was measured are provided in the supporting methods.
Modulated chlorophyll fluorescence measurements
Chlorophyll fluorescence parameters were measured in leaves after 20 min of dark adaptation with the MAXI version of an imaging pulse amplitude-modulated (PAM) fluorimeter (Heinz-Walz). The following equations were used: ΦII (the quantum yield of photochemical efficiency) = (Fm′-F)/Fm′, qP (photochemical quenching) = (Fm′-Fs)/(Fm′-Fo), NPQ (nonphotochemical quenching) = (Fm-Fm′)/Fm′, and ΦNO (the quantum yield of nonregulated dissipation) + ΦNPQ + ΦII = 1. The initial fluorescence (Fo) was determined after switching on the measuring beam, followed by a 0.8 s saturating pulse (4000 μmol m−2 s−1) to obtain the maximum fluorescence (Fm) [78]. Fm′ was the maximum fluorescence yield under 200 μmol m−2 s−1 actinic illumination, and Fs was the steady-state fluorescence during actinic illumination. We generated dark–light induction curves of leaves at 200 μmol m−2 s−1, and ΦII, qP, NPQ and ΦNO data were collected after 5 min of dark–light inductions.
Transmission electron microscopy (TEM)
Samples were first fixed in 2.5% glutaraldehyde. The samples were then thoroughly washed with 0.1 M phosphate buffer (pH 7.0) and postfixed in 1% osmium tetroxide. The fixed material was dehydrated in an ethanol series with increasing concentrations from 30% to 80% and acetone from 90% and 95%. The specimen was then infiltrated with a series of Spurr resin with increasing ratios from 1:1 to the final Spurr resin. After heating at 70°C for 9 h, the specimen was sectioned in a Leica EM UC7 ultratome. Ultrathin sections (70 nm) were stained with uranyl acetate and alkaline lead citrate and subsequently observed with a Hitachi H-7650 transmission electron microscope.
Statistical analysis
We used Fisher’s least significant difference test in R to calculate the statistical significance (p < 0.05), MetaboAnalyst (https://www.metaboanalyst.ca) to perform the principal component analysis, and the Mfuzz package in R to conduct the cluster analysis.
A gene coexpression network was established via the WGCNA package in R. Modules were identified using the “one-step network construction and module detection function” method with a soft thresholding power of 6 and a relatively large minimum module size of 30, and the threshold for merging modules was 0.25.
Analyses of KEGG pathway enrichment in this study were performed via the BGI Dr. Tom system, which uses the pHYPER function in R.
Supplementary Material
Acknowledgments
The authors are thankful to BGI Hudda for the transcriptome sequencing and subsequent analysis. The authors are grateful to the bioultrastructure analysis lab of the Analysis Center of Agrobiology and Environmental Sciences at Zhejiang University for performing the TEM. The authors also thank the Department of Horticulture, Zhejiang University, for loaning them the HPLC instrument to measure the carbohydrate concentrations. The authors thank Luming Biotechnology Company for measuring the ABA, JA, and lignin concentrations. We thank Dr. Danqing Li for suggestions concerning the WGCNA. This work was supported by funds from the National Natural Science Foundation of China (``Molecular regulation mechanism of R2R3 MYBs involved in cold acclimation to increase freezing tolerance of Rhododendron spp.'' - 31800597).
Author Contributions
BL, HZ and YX designed the research; BL, FZ, YC, ZL and SY performed the experiments; BL and XW analyzed and interpreted the data; BL wrote the paper; and YX and HZ revised the paper.
Data availability statement
The raw RNA-seq and ISO-seq data have been submitted to the Short Read Archive (SRA) data library under accession number PRJNA705504. The address is as follows: https://submit.ncbi.nlm.nih.gov/subs/.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Supplementary data
Supplementary data is available at Horticulture Research Journal online.
References
- 1. Oquist G, Huner NPA. 2003. Photosynthesis of overwintering evergreen plants. AnnuRevPlant Biol. 2003;54:329–55. [DOI] [PubMed] [Google Scholar]
- 2. Liu B, Zhou H, Cao Set al. Comparative physiology of natural deacclimation in ten azalea cultivars. HortScience. 2017;52:1451–7. [Google Scholar]
- 3. Gu L, et al. Eastern US spring freezing: increased cold damage in a warming world? Bioscience . 2008;58:253–62. [Google Scholar]
- 4. Wisniewski M, Nassuth A, Arora R. Cold hardiness in trees: a mini-review. Front Plant Sci. 2018;9:1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Howell GS, Weiser CJ. The environmental control of cold acclimation in apple. Plant Physiol. 1970;45:390–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Levitt J. Volume 1: chilling, freezing, and high temperature stresses. In: Responses of Plants to Environmental Stress. Elsevier: New York, 1980. [Google Scholar]
- 7. Sakai A, Larcher W. Frost Survival of Plants. Berlin: Springer Berlin Heidelberg; 1987. [Google Scholar]
- 8. Liu B, Xia YP, Krebs Set al. Seasonal responses to cold and light stresses by two elevational ecotypes of Rhododendron catawbiense: a comparative study of overwintering strategies. Environ Exp Bot. 2019;163:86–96. [Google Scholar]
- 9. Fowler S, Thomashow MF. 2002. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14:1675–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vogel JT, Zarka DG, Van Buskirk HAet al. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005;41:195–211. [DOI] [PubMed] [Google Scholar]
- 11. Ding Y, Jia Y, Shi Yet al. OST1-mediated BTF3L phosphorylation positively regulates CBFs during plant cold responses. EMBO J. 2018;37:e98228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi Y, Ding Y, Yang S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018;23:623–37. [DOI] [PubMed] [Google Scholar]
- 13. Benedict C, Skinner JS, Meng Ret al. The CBF1-dependent low temperature signaling pathway, regulon, and increase in freeze tolerance are conserved in Populus spp. Plant Cell Environ. 2006;29:1259–72. [DOI] [PubMed] [Google Scholar]
- 14. Mantyla E, Lang V, Palva ET. Role of abscisic acid in drought-induced freezing tolerance, cold acclimation, and accumulation of LT178 and RAB18 proteins in Arabidopsis thaliana. Plant Physiol. 1995;107:141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Welling A, Kaikuranta P, Rinne P. Photoperiiodic induction of dormancy and freezing tolerance in Betula pubescens. PhysiolPlant. 1997;100:119–25. [Google Scholar]
- 16. Pagter M, Jensen CR, Petersen KKet al. Changes in carbohydrates. Physiol Plant. 2008;134:473–85. [DOI] [PubMed] [Google Scholar]
- 17. Wang Z, Wong DCJ, Wang Jet al. GRAS-domain transcription factor PAT1 regulates jasmonic acid biosynthesis in grape cold stress response. Plant Physiol. 2021;186:1660–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu B, Wang X-Y, Cao Yet al. Factors affecting freezing tolerance: a comparative transcriptomics study between field and artificial cold acclimations in overwintering evergreens. Plant J. 2020;103:2279–300. [DOI] [PubMed] [Google Scholar]
- 19. Franklin KA, Whitelam GC. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat Genet. 2007;39:1410. [DOI] [PubMed] [Google Scholar]
- 20. Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–30. [DOI] [PubMed] [Google Scholar]
- 21. Lee CM, Thomashow MF. Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. PNAS. 2012;109:15054–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mikkelsen MD, Thomashow MF. A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 2009;60:328–39. [DOI] [PubMed] [Google Scholar]
- 23. Chang CY, Brautigam K, Huner NPAet al. Champions of winter survival: cold acclimation and molecular regulation of cold hardiness in evergreen conifers. New Phytol. 2021;229:675–91. [DOI] [PubMed] [Google Scholar]
- 24. Tominaga Y, Suzuki K, Uemura Met al. In planta monitoring of cold-responsive promoter activity reveals a distinctive photoperiodic response in cold acclimation. Plant Cell Physiol. 2021;62:43–52. [DOI] [PubMed] [Google Scholar]
- 25. Welling A, Palva ET. Molecular control of cold acclimation in trees. Physiol Plant. 2006;127:167–81. [Google Scholar]
- 26. Rapacz M, Jurczyk B, Sasal M. Deacclimation may be crucial for winter survival of cereals under warming climate. Plant Sci. 2017;256:5–15. [DOI] [PubMed] [Google Scholar]
- 27. Vyse K, Pagter M, Zuther Eet al. Deacclimation after cold acclimation -- a crucial, but widely neglected part of plant winter survival. J Exp Bot. 2019;70:4595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Basler D, Korner C. Photoperiod and temperature responses of bud swelling and bud burst in four temperature forest tree species. Tree Physiol. 2014;34:377–88. [DOI] [PubMed] [Google Scholar]
- 29. Pagter M, Andersen UB, Andersen L. Winter warming delays dormancy release, advances budburst, alters carbohydrate metabolism and reduces yield in a temperate shrub. AoB PLANTS. 2015;7: plv024, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Andersen UB, Pagter M, Andersen Let al. Impact of seasonal warming on overwintering and spring phenology of blackcurrant. Environ Exp Bot. 2017;140:96–109. [Google Scholar]
- 31. Bag P, Chukhutsina V, Zhang Zet al. Direct energy transfer from photosystem II to photosystem I confers winter sustainability in scots pine. Nat Commun. 2020;11:6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Curien G, Giustini C, Montillet J-Let al. The chloroplast membrane associated ceQORH putative quinone oxidoreductase reduces long-chain, stress-related oxidized lipids. Phytochemistry. 2016;122:45–55. [DOI] [PubMed] [Google Scholar]
- 33. Ifuku K, Yamamoto Y, Ono Tet al. PsbP protein, but bot PsbQ protein, is essential for the regulation and stabilization of photosystem II in higher plants. Plant Physiol. 2005;139:1175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fagerstedt KV, Kukkola EM, Koistinen VVTet al. Cell wall lignin is polymerized by class III secretable plant peroxidases in Norway spruce. J Integr Plant Biol. 2010;52:186–94. [DOI] [PubMed] [Google Scholar]
- 35. Dharmawardhana DP, Ellis BE, Carlson JE. A beta-glycosidase from Lodgepole pine xylem specific for the lignin precursor coniferin. Plant Physiol. 1995;107:331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stitt M, Hurry V. A plant for all seasons: alterations in photosynthetic carbon metabolism during cold acclimation in Arabidopsis. Curr Opin Plant Biol. 2002;5:199–206. [DOI] [PubMed] [Google Scholar]
- 37. Dalchau N, Baek SJ, Briggs HMet al. The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. PNAS. 2011;108:5104–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haydon M, Mielczarek O, Robertson FCet al. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature. 2013;502:689–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roman A, Li X, Deng Det al. Superoxide is promoted by sucrose and affects amplitude of circadian rhythms in the evening. PNAS. 2021;118:e2020646118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kidokoro S, Yoneda K, Takasaki Het al. Different cold-signaling pathway function in the responses to rapid and gradual decreases in temperature. Plant Cell. 2017;29:760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ramos A, Pérez-Solís E, Ibáñez Cet al. Winter disruption of the circadian clock in chestnut. PNAS. 2005;102:7037–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ibanez C, Ramos A, Acebo Pet al. Overall alteration of circadian clock gene expression in the chestnut cold response. PLoS One. 2008;3:e3567, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ibanez C, Kozarewa I, Johansson Met al. Circadian clock components regulate entry and affect exit of seasonal dormancy as well as winter hardiness in Populus trees. Plant Physiol. 2010;153:1823–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Johansson M, Ramons-Sanchez JM, Conde Det al. Role of the circadian clock in cold acclimation and winter dormancy in perennials plants. In: Anderson JV, ed. Advances in Plant Dormancy. Cham, Heidelberg, New York, Dordrecht, London: Springer, 2015. [Google Scholar]
- 45. Bieniawska Z, Espinoza C, Schlereth Aet al. Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 2008;147:263–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dong MA, Farre EM, Thomashow MF. CIRCADIAN CLOCK-ASSOCIATED 1 and LATE ELONGATED HYPOCOTYL regulate expression of the C-REPEAT BINDING FACTOR (CBF) pathway in Arabidopsis. PNAS. 2011;108:7241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee HG, Seo PJ. The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J. 2015;82:962–77. [DOI] [PubMed] [Google Scholar]
- 48. Jiang B, Shi Y, Zhang Xet al. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. PNAS. 2017;108:7241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Imai H, Kawamura Y, Nagatani Aet al. Effects of the blue light-cryptochrome system on the early process of cold acclimation of Arabidopsis thaliana. Environ Exp Bot. 2021;183:104340. [Google Scholar]
- 50. Kleine T, Kindgren P, Benedict Cet al. Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 2007;144:1391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Harmer SL, Hogenesch JB, Straume Met al. Orchestrated transcription of key pathway in Arabidopsis by the circadian clock. Science. 2000;290:2110–3. [DOI] [PubMed] [Google Scholar]
- 52. Peng Y, Lin W, Wei Het al. Phylogenetic analysis and seasonal cold acclimation-associated expression of early light-induced protein genes of Rhododendron catawbiense. Physiol Plant. 2008;132:44–52. [DOI] [PubMed] [Google Scholar]
- 53. Adamska I, Scheel B, Kloppstech K. Circadian oscillations of nuclear-encoded chloroplast proteins in pea (Pisum sativum). Plant Mol Biol. 1991;17:1055–65. [DOI] [PubMed] [Google Scholar]
- 54. Potter E, Kloppstech K. Effects of light stress on the expression of early light-inducible proteins in barley. Eur J Biochem. 1993;214:779–86. [DOI] [PubMed] [Google Scholar]
- 55. Jahns P, Holzwarth AR. The role of the xanthophyll cycle and of lutein in photoprotection of photosystem II. BBA Bioenergetics. 2012;1817:182–93. [DOI] [PubMed] [Google Scholar]
- 56. Van Amerongen H, Chmeliov J. Instantaneous switching between different modes of non-photochemical quenching in plants. Biochimica et biophysica acta Reviews on cancer. 2020;1861:148119. [DOI] [PubMed] [Google Scholar]
- 57. Holt NE, Zigmantas D, Valkunas Let al. Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science. 2005;307:433–6. [DOI] [PubMed] [Google Scholar]
- 58. Johnson MP, Goral TK, Duffy CDPet al. Photoprotective energy dissipation involves the reorganization of photosystem II light harvesting complexes in the grana membranes of spinach chloroplasts. Plant Cell. 2011;23:1468–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Berger S, Weichert H, Porzel Aet al. Enzymatic and non-enzymatic lipid peroxidation in leaf development. Biochim Biophys Acta. 2001;1533:266–76. [DOI] [PubMed] [Google Scholar]
- 60. Howe G. The role of hormones in defense against insects and disease. In: Davies PJ, ed. Plant Hormones Biosynthesis, Signal Transduction Action! Dordrecht, the. Kluwer Academic Publishers: Netherlands, 610,634–2004. [Google Scholar]
- 61. Demmig-Adams B, Cohu CM, Amiard Vet al. Energing trade-offs -- impact of photoprotectants (PsbS, xanthophylls, and vitamin E) on oxylipins as regulators of development and defense. New Phytol. 2013;197:720–9. [DOI] [PubMed] [Google Scholar]
- 62. Frenkel M, Kulheim C, Johanson Het al. Improper excess light energy dissipation in Arabidopsis results in a metabolic reprogramming. BMC Plant Biol. 2009;9:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Duckham SC, Linforth RST, Taylor IB. Abscisic-acid-deficient mutants at the aba gene locus of Arabidopsis thaliana are impaired in the epoxidation of zeaxanthin. Plant Cell Environ. 1991;14:601–6. [Google Scholar]
- 64. Marin E, Nussaume L, Quesada Aet al. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 1996;15:2331–42. [PMC free article] [PubMed] [Google Scholar]
- 65. Hu Y, Jiang Y, Han Xet al. Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J Exp Bot. 2017;68:1361–9. [DOI] [PubMed] [Google Scholar]
- 66. Du H, Liu H, Xiong L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front Plant Sci. 2013;4:397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rubio S, Perez FJ. ABA and its signaling pathway are involved in the cold acclimation and deacclimation of grapevine buds. Sci Hortic. 2019;256:108565–5. [Google Scholar]
- 68. Xiong L, Ishitani M, Lee Het al. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell. 2001;13:2063–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sewalt VJH, Ni W, Blount JWet al. Reduced lignin content and altered lignin composition in transgenic tobacco down-regulated in expression of L-phenylalanine ammonia-Lyase or Cinnamate 4-hydroxylase. Plant Physiol. 1997;115:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cass C, Peraldi A, Dowd PFet al. Effects of PHENYLALANINE AMMONIA LYASE (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. J Exp Bot. 2015;66:4317–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ji H, Wang Y, Cloix Cet al. The Arabidopsis RCC1 family protein TCF1 regulates freezing tolerance and cold acclimation through modulating lignin biosynthesis. PLoS Genet. 2015;11:e1005471, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hernandez I, Alegre L, Van Breusegem Fet al. How relevant are flavonoids as antioxidants in plants? Trends Plant Sci . 2009;14:125–32. [DOI] [PubMed] [Google Scholar]
- 73. Liu C, Yu H, Rao Xet al. Abscisic acid regulates secondary cell-wall formation and lignin deposition in Arabidopsis thaliana through phosphorylation of NST1. PNAS. 2021;118:e2010911118, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ellis C, Karagyllidis I, Wasternack Cet al. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell. 2002;14:1557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Uppalapati SR, Ayoubi P, Weng Het al. The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J. 2005;42:201–17. [DOI] [PubMed] [Google Scholar]
- 76. Amiard V, Demiming-Adams B, Mueh KEet al. Role of light and jasmonic acid signaling in regulating foliar phloem cell wall ingrowth development. New Physiol. 2007;173:772–31. [DOI] [PubMed] [Google Scholar]
- 77. Lim CC, Arora R. Comparing Gompertz and Richards functions to estimate freeze injury in rhododendron using electrolyte leakage. jashs. 1998;123:246–52. [Google Scholar]
- 78. Jiang X, Xu J, Lin Ret al. Light-induced HY5 functions as a systemic signal to coordinate the photoprotective response to light fluctuation. Plant Physiol. 2020;184:1181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA-seq and ISO-seq data have been submitted to the Short Read Archive (SRA) data library under accession number PRJNA705504. The address is as follows: https://submit.ncbi.nlm.nih.gov/subs/.