Abstract
MYB transcription factors play important roles in plant responses to cold stress, but the associated underlying mechanisms remain unclear. In this study, a cold-induced MYB transcription factor, DgMYB2, was isolated from chrysanthemum (Chrysanthemum morifolium Ramat). DgMYB2 was localized to the nucleus and exhibited transactivational activity. Overexpression of DgMYB2 improved cold tolerance in chrysanthemum, while cold tolerance in the antisense suppression lines decreased compared to that of the wild type. Additionally, electrophoretic mobility shift assays, chromatin immunoprecipitation, luciferase complementary imaging analysis, and dual-luciferase reporter gene detection experiments confirmed that DgMYB2 directly targets DgGPX1 and increases the activity of glutathione peroxidase to reduce the accumulation of reactive oxygen species, thereby improving cold resistance in chrysanthemum.
Introduction
Cold stress is a common abiotic stress factor that negatively affects plant growth and is one of the leading factors restricting the distribution of plants. Cold stress can be classified as either chilling (0–15°C) stress or freezing (< 0°C) stress [1]. Plants have evolved defense mechanisms to tolerate cold stress. When plants are subjected to cold stress, transcription factors are activated through a series of signal transduction pathways, ultimately improving plant cold tolerance. Activated transcription factors specifically bind to their corresponding cis-acting elements and activate the expression of a series of downstream cold resistance-related genes, which then trigger a series of physiological and biochemical reactions, thereby improving plant cold resistance. Members of transcription factor families have been identified in plants, including members of NAC, WRKY, and AP2/ER families, which are some of the largest transcription factor families in plants. Specifically, members of the MYB transcription factor family are largely involved in plant cell morphology, plant physiological metabolism, and stress response processes [2–5].
Members of the MYB transcription factor family are named after their N-terminal conserved DNA-binding region, the MYB domain, which is composed of 1–4 incomplete repetitive (R) sequences consisting of 50–52 amino groups in a series [6]. Based on the different number of R repetitions in a sequence, MYB transcription factors can be divided into four subfamilies [7]. Members of the R1-MYB subfamily contain only one R domain and are referred to as R1-MYB/MYB-related transcription factors. The R2R3-MYB subfamily has the most members among the MYB transcription factor family, the members of which contain two R structures at the N-terminus. R1R2R3-MYB transcription factors contain three R domains and compose a relatively small MYB transcription factor subfamily, and research on the functions of R1R2R3-MYB transcription factors is scarce. The fourth subfamily, the 4R-MYB subfamily, has the fewest members, and their functions remain poorly understood [8, 9].
Of these subfamilies, the most widely studied is the R2R3-type MYB subfamily [10]. Research on the regulatory mechanisms of R2R3-MYBs in the plant cold tolerance process has mainly been conducted in apple and pear. Overexpression of the cold-inducible genes MdMYB88/124 significantly increased cold tolerance in apple, while a reduction in the transcript levels of these two genes weakened cold tolerance [11]. The expression of MdMYB23 was upregulated under cold stress, and its overexpression enhanced cold tolerance in transgenic apple calli and Arabidopsis plants [7]. Moreover, the overexpression of MdMYB308L significantly increased cold tolerance in apple plants [12]. In pear (Pyrus betulifolia Bunge), cold stress induced the transcription of PbrMYB5, and its overexpression improved the cold resistance of transgenic tobacco [13]. However, the mechanisms of R1-MYBs in response to cold stress in plants remain unclear, necessitating further research.
Reactive oxygen species (ROS) are the products of plant metabolism and have extremely high activity and toxicity [14]. Under normal growth conditions, ROS can be eliminated by the antioxidant defense mechanisms of plants. Under environmental stress, the balance between ROS production and elimination in plants is disrupted and leads to the rapid accumulation of ROS and oxidative damage [15]. Glutathione peroxidase (GPX) is an important enzyme in the ROS-scavenging system and plays a role in reducing the accumulation of ROS caused by abiotic stress and maintaining the integrity of the plasma membrane [16]. However, the regulatory mechanisms underlying the effects of MYB transcription factors on GPX remain unclear.
Chrysanthemums are often used as fresh cut flowers and have important ornamental and economic value [17]. As a common stress factor, cold stress hinders the productivity and economic value of chrysanthemum [18]. Studying the responses of chrysanthemum to cold stress will improve chrysanthemum production. In this study, a cold-induced MYB transcription factor, DgMYB2, was isolated from chrysanthemum. We found that DgMYB2 was localized in the nucleus and exhibited transcriptional activity. Moreover, overexpression of DgMYB2 enhanced cold tolerance in chrysanthemum, while reducing its expression reduced the survival rate of transgenic plants under cold stress. Based on the collective findings, we provide evidence that DgMYB2 plays a positive role in plants under cold stress by regulating the expression of DgGPX1 directly to increase GPX activity.
Results
Isolation and expression patterns of DgMYB2
Cold-treated (4°C for 24 h followed by −4°C for 4 h) and untreated (25°C) chrysanthemum seedlings were sampled for transcriptome analyses (NCBI accession No. GSE117262). The results revealed that 43 MYB genes were significantly (P < 0.05) induced at the transcriptional level in response to cold treatment (Table S1). Among these genes, DgMYB2 (GenBank accession No. MW853963) was notably induced (log2[fold change] = 2.7918) and was thus selected for further investigation. The full-length cDNA of DgMYB2 contained an open reading frame (ORF) of 885 bp and encoded a predicted protein of 294 amino acids with a molecular weight of 32.23 kDa. The amino acid sequence of DgMYB2 contained a SANT/MYB conserved domain, indicating that DgMYB2 is a typical R1-MYB transcription factor (Fig. S1a). Phylogenetic analysis revealed that the DgMYB2 sequence is highly homologous with other R1-MYB transcription factor sequences of various plant species and is closely related to AaMYB-like from Artemisia annua (Fig. S1b).
The expression patterns of DgMYB2 in different tissues showed that DgMYB2 was most highly expressed in the leaves, followed by the stems, and it was expressed the least in the roots (Fig. 1a). We also found that the expression of DgMYB2 increased under cold treatment, reaching the highest value after 12 h and then decreasing slightly (Fig. 1 b).
Figure 1.
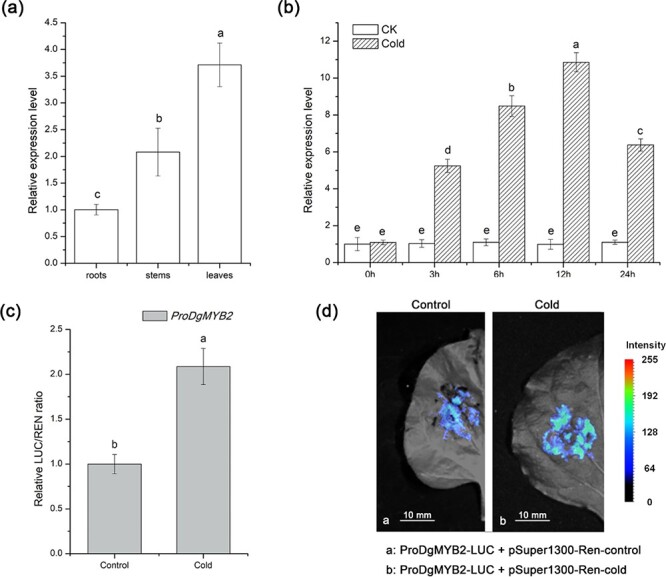
DgMYB2 response to cold stress. (a) Relative expression levels of DgMYB2 in the roots, stems, and leaves of wild-type (WT) chrysanthemum, as determined by qRT–PCR (P < 0.05; the data are presented as the means ± standard errors of 3 replicates; 20 plants per replicate). (b) Relative expression levels of DgMYB2 in WT chrysanthemum under cold conditions (4°C) (CK represents the control group under control conditions (25°C/22°C day/night). (c) Dual-LUC reporter gene assay (DLA) of the DgMYB2 native promoter after transient expression in tobacco; the results of the control (25°C/22°C day/night) and cold (4°C for 12 h) treatment groups are compared. (d) Luciferase complementation imaging (LCI) of the DgMYB2 native promoter after transient expression in tobacco.
A dual-LUC reporter gene assay (DLA) and luciferase complementation imaging (LCI) were performed to verify the function of the natural promoter of DgMYB2 (1500 bp). The results demonstrated that DgMYB2 responded to cold stress (Fig. 1c, d).
DgMYB2 was localized to the nucleus and had transactivational activity
To determine the subcellular localization of DgMYB2, pSuper1300-DgMYB2-GFP and the nuclear marker pSuper::NF-YA4-mCherry were transiently expressed in tobacco leaves, while pSuper1300-GFP and the nuclear marker pSuper::NF-YA4-mCherry were coexpressed in the epidermal cells of tobacco leaves as controls. The results indicated that DgMYB2 was localized within the nucleus (Fig. 2a).
Figure 2.
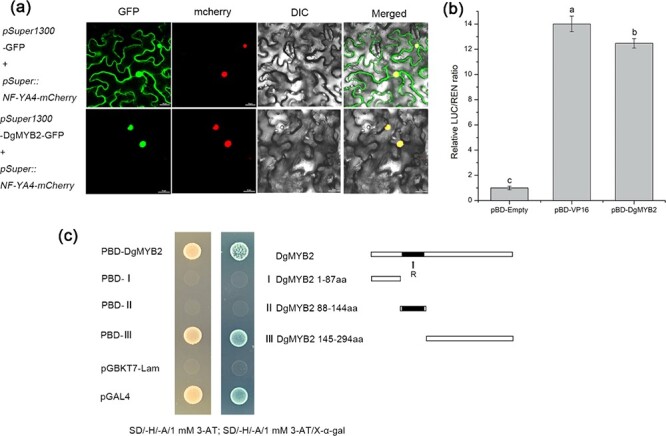
DgMYB2 is localized in the nucleus and has transactivational activity. (a) Subcellular localization of DgMYB2 in tobacco leaves; scale bars = 10 μm. (b) Transcriptional transactivation in tobacco. (c) Transcriptional transactivation in yeast; the right panel shows the DgMYB2 fragment used for testing.
A dual-luciferase reporter gene analysis system was used to evaluate the transcriptional activity of DgMYB2. The results revealed that the LUC/REN value of the experimental group whose members were coinjected with the reporter gene and pBD-DgMYB2 was significantly (P < 0.05) higher than that of the negative control (pBD-Empty) (Fig. 2b), indicating that DgMYB2 had transcriptional activation activity. To further analyze the location of the transcriptional activation region of DgMYB2, a recombinant plasmid containing the corresponding sequence fragment was transferred into yeast one-hybrid (Y1H) competent cells and then cultured on corresponding media. Yeasts transformed with the complete ORF sequence (1–294 aa) of DgMYB2 and yeasts transformed with the C-terminal sequence (145–294 aa) of DgMYB2 grew normally and appeared blue on SD/-H/−A plates including X-α-gal (Fig. 2c). Taken together, these results indicated that DgMYB2 had transcriptional activation activity and that the responsible region which was located within the C-terminal region (145–294 aa) of DgMYB2.
DgMYB2 played a positive regulatory role in the chrysanthemum response to cold stress
Transgenic chrysanthemum lines were obtained by Agrobacterium-mediated transformation of wild-type (WT) chrysanthemum. Two overexpression lines (DgMYB2 OE-6 and DgMYB2 OE-31) and two RNA interference (RNAi) lines (DgMYB2 Ri-11 and DgMYB2 Ri-45) were selected for further experimentation. Quantitative real-time PCR (qRT–PCR) was used to determine the expression level of DgMYB2 in WT and transgenic lines (Fig. 3a). Following cold treatment, the RNAi lines were the most severely withered, followed by the WT, while the overexpression lines maintained relatively good growth (Fig. 3b). Following cold treatment, plants were subjected to a 2-week recovery period at room temperature (25°C) to analyze the survival rate of the WT, overexpression, and RNAi lines. The survival rate of the overexpression lines was the highest, followed by the WT lines, while the RNAi lines had the lowest survival rate (Fig. 3c).
Figure 3.
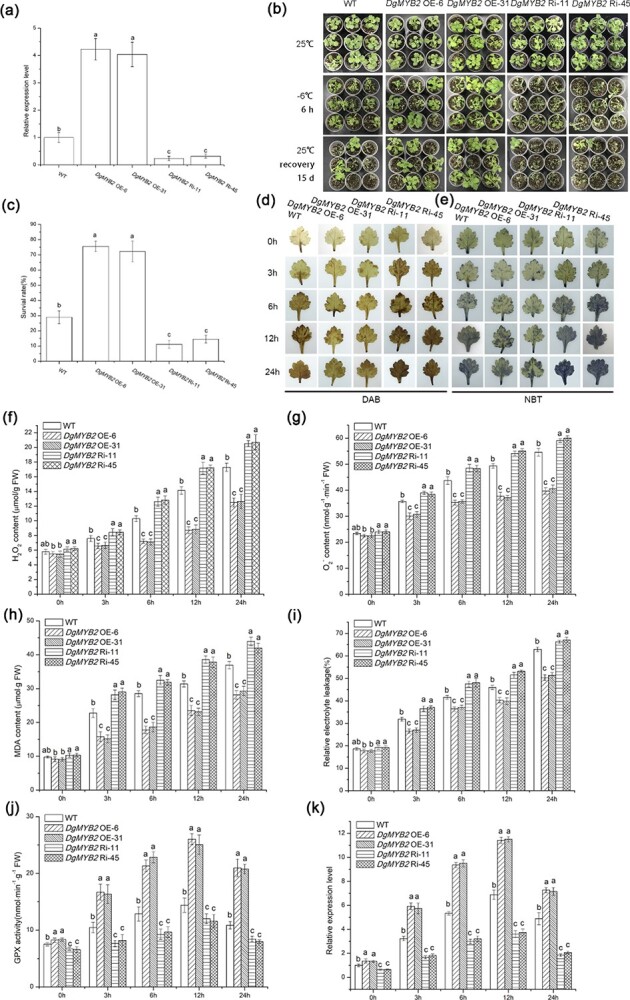
DgMYB2 plays a positive regulatory role in the response of chrysanthemum to cold stress. (a) Expression level of DgMYB2 in wild-type (WT) chrysanthemum and transgenic lines (DgMYB2 OE-6, DgMYB2 OE-31; DgMYB2 Ri-11, DgMYB2 Ri-45) under control conditions (25°C/22°C day/night), as determined by qRT–PCR (P < 0.05; 3 replicates; 20 plants per replicate). (b) Comparison phenotypes between transgenic lines (DgMYB2 OE-6, DgMYB2 OE-31; DgMYB2 Ri-11, DgMYB2 Ri-45) and WT chrysanthemum. (c) Survival rate of WT chrysanthemum and transgenic lines (DgMYB2 OE-6, DgMYB2 OE-31; DgMYB2 Ri-11, DgMYB2 Ri-45) after cold treatment (4°C for 24 h followed by −6°C for 6 h) followed by 15 d of recovery under control conditions (25°C/22°C day/night). (d, e) NBT and DAB histochemical staining. (f, g) Quantitative analysis of the hydrogen peroxide (H2O2) and superoxide anion (O2−) contents. (h) Malondialdehyde (MDA) content. (i) Relative electrolyte leakage. (j) GPX activity in WT chrysanthemum and transgenic lines (DgMYB2 OE-6, DgMYB2 OE-31; DgMYB2 Ri-11, DgMYB2 Ri-45). (k) Expression levels of DgGPX1 in WT chrysanthemum and transgenic lines (DgMYB2 OE-6, DgMYB2 OE-31; DgMYB2 Ri-11, DgMYB2 Ri-45).
The histochemical staining results showed that, compared with those of the WT, the spots on the leaves of the overexpression lines were fewer in number and lighter in color. In contrast, compared with those of the WT, the spots on the leaves of the RNAi lines were more numerous and darker (Fig. 3d, e). Quantitative analysis of the hydrogen peroxide (H2O2) and superoxide anion (O2−) contents was performed, and the results were consistent with the histochemical staining results. The results confirmed that the H2O2 and O2− contents were lower in the overexpression lines than in the WT under cold stress, while the RNAi lines had the highest H2O2 and O2− contents (Fig. 3f, g).
Under cold stress, the malondialdehyde (MDA) content and the relative electrolyte leakage in each line gradually tended to increase. Moreover, the MDA content and relative electrolyte leakage rate in the overexpression lines were the lowest, followed by the WT and RNAi lines (Fig. 3h, i). We measured the activities of antioxidant enzymes, including superoxide dismutase (SOD), peroxidase (POD), and glutathione peroxidase (GPX), which are components of the ROS-scavenging system. The results revealed that the activity of these enzymes increased to a certain degree when compared to values prior to cold treatment. No statistically significant differences in the SOD or POD activity between the transgenic, WT, and RNAi lines were detected (Fig. S2), but there were significant differences in GPX activity. The GPX activity of the overexpression lines was the highest, followed by that of the WT and RNAi lines (Fig. 3j). Furthermore, the qRT–PCR results were consistent with the GPX activity results. The overexpression lines had the highest expression level of DgGPX1, followed by the WT and RNAi lines (Fig. 3k).
Taken together, the results indicated that DgMYB2 positively affected the expression of DgGPX1 to regulate the activity of GPX, thereby reducing the accumulation of ROS to reduce plasma membrane damage, thus playing a positive regulatory role in the chrysanthemum response to cold stress.
DgMYB2 promoted DgGPX1 transcription by directly binding to the MYB-binding motif in the DgGPX1 promoter
The natural promoter sequence of DgGPX1 (1125 bp) contained a predicted MYB-binding site (CAACCA). The EMSA results confirmed that DgMYB2 was able to bind to the MYB-binding site (CAACCA) in the DgGPX1 promoter (Fig. 4a). The green fluorescent protein (GFP) expression levels in the transgenic lines were identified by western blots (Fig. S3). The transgenic lines were subjected to chromatin immunoprecipitation assay (ChIP)-qPCR experiments. ChIP-qPCR, LCI, and DLA experiments demonstrated that there was a protein-DNA interaction between DgMYB2 and the MYB-binding site (CAACCA) in the natural promoter sequence of DgGPX1 (Fig. 4b–d).
Figure 4.
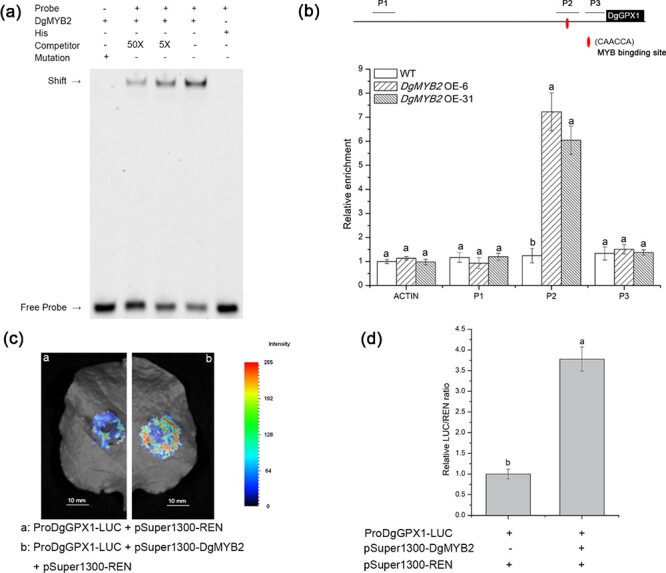
DgMYB2 binds to the promoter of DgGPX1. (a) EMSA, from left to right: DgMYB2 and 6-FAM labeled mutant probe; DgMYB2, 6-FAM labeled probe and 50x unlabeled probe; DgMYB2, 6-FAM labeled probe and 5x unlabeled probe; DgMYB2 and 6-FAM labeled probe; His protein and 6-FAM labeled probe. (b) ChIP-PCR assay; P1–P3: various segments of the promoter sequence, of which P2 contained the MYB-binding site (CAACCA). (c) LCI analysis results. (d) DLA analysis results.
Discussion
To date, many MYB transcription factors have been isolated from different species [19–21]. The genes encoding these proteins are widely involved in the abiotic and biotic stress responses of plants. In this study, we isolated a cold-induced MYB transcription factor, DgMYB2, which was upregulated at the transcriptional level under cold stress (Fig. 1b). Phylogenetic analysis revealed that DgMYB2 was highly homologous with other plant R1-MYB proteins and had the closest relationship with that of A. annua (Fig. S1a, b). The expression levels of many novel MYB transcription factors have been shown to be induced by low temperature. In rice, the expression of OsMYB3R-2 is induced by low temperature [22]. In sheepgrass, LcMYB4 is upregulated in response to cold induction [23]. In tomato, the R2R3-MYB transcription factor LeAN2 is induced by low temperature and oxidative stress [24]. However, the functions and mechanisms of MYB transcription factors in response to cold stress in plants remain poorly understood.
Previous research on the function of MYB transcription factors at low temperatures revealed that these proteins affect the cold resistance responses of plants by regulating the ROS-scavenging system. LcMYB4 overexpression in sheepgrass resulted in higher POD activity, suggesting that the antioxidant ability of transgenic plants improved [23]. A MYB-related transcription factor, AQUILO, was isolated from grapevine (Vitis vinifera L.), and overexpression of AQUILO in Arabidopsis increased the POD and SOD activities and upregulated the expression levels of their encoding genes [25]. To verify the function of DgMYB2 in chrysanthemum under cold stress, we obtained overexpression lines (DgMYB2 OE-6 and DgMYB2 OE-31) and RNAi lines (DgMYB2 Ri-11 and DgMYB2 Ri-45). The expression level of DgMYB2 in the overexpression lines was significantly (P < 0.05) higher than that in the WT and RNAi lines, while the expression level of DgMYB2 in the WT lines was significantly (P < 0.05) higher than that in the RNAi lines (Fig. 3a). Compared with the WT lines, the overexpression lines experienced less damage from cold stress, and their survival rate was significantly (P < 0.05) higher, while the RNAi lines had the lowest survival (Fig. 3c). The accumulation of ROS in the overexpression lines was significantly (P < 0.05) lower than that in the WT lines, while the RNAi lines accumulated more ROS than the WT lines (Fig. 3d–g). Under cold conditions, the relative electrolyte leakage and MDA content of the RNAi lines were the highest, followed by the WT and overexpression lines (Fig. 3h, i), indicating that, compared with the RNAi lines, the overexpression lines had a lower degree of membrane lipid peroxidation, experienced less peroxidation damage, and exhibited higher cold resistance. Additionally, the GPX activity of the overexpression lines was the highest, followed by the WT and RNAi lines (Fig. 3j). Moreover, the expression of DgGPX1 tended to be consistent with GPX activity (Fig. 3k). We found that DgMYB2 affected GPX activity by regulating the expression of DgGPX1, thereby influencing the ROS-scavenging process and degree of oxidative damage caused by ROS accumulation, thus positively regulating plant responses to cold stress.
MYB transcription factors bind to specific motifs in gene promoters to regulate the expression of cold stress-related genes to affect the cold stress resistance of plants [5]. In apple, the downstream target genes MdCCA1 and MdCSP3 of MdMYB88/124 were identified, and the overexpression of MdMYB88/124 contributed to increased cold tolerance [11]. MdMYB23 directly binds to the promoter regions of MdCBF1 and MdCBF2 and activates their expression to enhance plant cold resistance [7]. In pear, PbrMYB5 directly binds to the promoter region of PbrDHAR2 and activates its expression, which promotes the synthesis of ascorbic acid (AsA), in turn increasing plant cold resistance [13]. In this study, the EMSA, DLA, LCI, and ChIP-qPCR results demonstrated that DgMYB2 specifically bound to the MYB-binding site in the promoter region of DgGPX1. DgMYB2 directly targeted DgGPX1 to increase GPX activity, thus enhancing chrysanthemum cold resistance.
In conclusion, a cold-induced R1-MYB transcription factor, DgMYB2, was isolated from chrysanthemum, and its overexpression enhanced chrysanthemum cold resistance. DgMYB2 directly targeted DgGPX1 to increase GPX enzyme activity and thereby reduce ROS accumulation, thus improving chrysanthemum cold resistance (Fig. 5).
Figure 5.
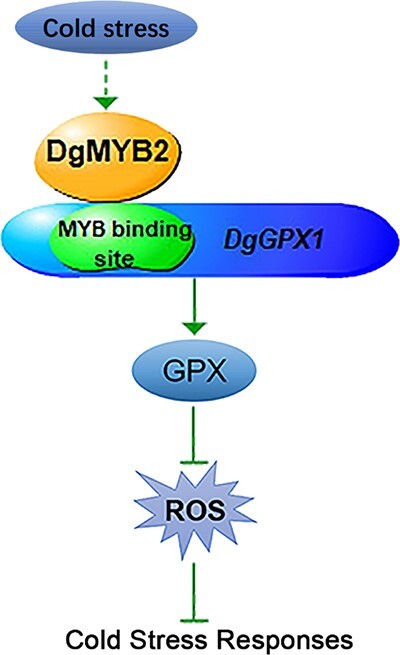
Proposed model through which DgMYB2 enhanced the cold tolerance of chrysanthemum by directly targeting DgGPX1. Cold stress induces the expression of DgMYB2. DgMYB2 directly targets DgGPX1 to positively regulate the expression of DgGPX1, which regulates GPX activity and reduces ROS accumulation, thereby playing a positive role in chrysanthemum responses to cold stress.
Materials and methods
Plant materials and cold treatment
WT chrysanthemum was cultured on Murashige and Skoog (MS) media for 30 d under normal conditions (25°C/22°C day/night temperature conditions with a 16/8-h photoperiod). Afterward, these plant materials were used to obtain transgenic plants using Agrobacterium-mediated methods. After 30 d of cultivation, the chrysanthemum plantlets were transplanted into pots with a 1:1 peat:perlite mixture and cultivated under the aforementioned conditions for 7 d. Then, these plant materials were used for subsequent physiological experiments.
Cold-treated (4°C for 24 h followed by −4°C for 4 h) and untreated (25°C) chrysanthemum seedlings were sampled for transcriptome analyses. The plant materials were subjected to 4°C for 24 h and sampled after 0, 3, 6, 12, and 24 h of cold treatment for target gene expression level determination; relative electrolyte leakage assays; MDA, H2O2, and O2− content determination; and GPX activity assays. The samples were flash-frozen in liquid nitrogen and used directly for subsequent experiments or otherwise stored at −80°C. To verify the survival rates of transgenic and WT lines and record phenotypic changes, plant materials were exposed to chilling (4°C for 24 h) and freezing (−6°C for 6 h) treatments, followed by a recovery period of 2 weeks at room temperature (25°C) to simulate the temperature conditions of the chrysanthemum production process.
RNA preparation and qRT–PCR
Chrysanthemum plants were sampled at the corresponding time points after cold stress treatment. An RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China) was used to obtain the total RNA from the chrysanthemum samples, and the obtained total RNA was reverse transcribed into cDNA using a One-Step gDNA Removal kit (TransGen Biotech, Beijing, China). The obtained cDNA was used as a template for qRT–PCR using a Two-Step qRT–PCR Kit (TransGen Biotech, Beijing, China); the elongation factor 1α gene (EF1α) was used as a stable reference gene. The results were detected using a CFX96 Real-Time PCR Detection System (Bio–Rad, Hercules, CA, USA). The 2-ΔΔCT method was used for analysis, and the primers used for qRT–PCR are listed in Table S1.
Construction of expression vectors and generation of transgenic chrysanthemum
The National Center for Biotechnology Information (NCBI) website was used for the conserved domain analysis of DgMYB2. Homology analysis of DgMYB2 was performed by DNAMAN (Lynnon Biosoft, San Ramon, CA, USA), and phylogenetic analysis was conducted using MEGA (Molecular Evolutionary Genetics Analysis) v7. The ORF sequence of DgMYB2 was amplified using the primer pair M2-F and M2-R (Table S2). Then, the DgMYB2 ORF sequence was inserted into a modified pSuper1300 plasmid together with GFP to obtain a 35S::DgMYB2-GFP recombinant plasmid. The 35S::DgMYB2-GFP plasmid was used in subsequent subcellular localization experiments and generation of DgMYB2-overexpressing lines. To obtain RNAi lines, a 300 bp fragment of the ORF region containing no structural domain was selected and inserted into the pCAMBIA2301-GW-RNAi vector to construct pCAMBIA2301-DgMYB2-RNAi vectors. Construction of the recombinant plasmids was performed by Shanghai Sangon Biotech Co., Ltd. (Shanghai, China).
To obtain transgenic chrysanthemum plants, the overexpression vector 35S::DgMYB2-GFP and RNAi vector pCAMBIA2301-DgMYB2-RNAi were transformed into WT chrysanthemum via Agrobacterium-mediated transformation. A DNAsecure Plant Kit (Tiangen, Beijing, China) was used to extract DNA from the obtained chrysanthemum plants to identify positive transgenic plants. Then, qRT–PCR-based detection of positive transgenic plants was performed. The operational steps were conducted following the manufacturer’s instructions.
Dual-LUC reporter gene assays
To verify the function of the natural promoter of DgMYB2, the sequence of the DgMYB2 promoter (1500 bp) was inserted into a pSuper1300 vector together with the LUC reporter gene to form the recombinant plasmid ProDgMYB2-LUC. Renilla luciferase (REN) was used as the internal reference gene and inserted into a pSuper1300 vector to form the recombinant plasmid pSuper1300-REN. Construction of the recombinant plasmids was performed by Shanghai Sangon Biotech Co., Ltd. (Shanghai, China). The recombinant plasmids ProDgMYB2-LUC and pSuper1300-REN were then transferred into Agrobacterium, which were subsequently coinjected into tobacco for transient expression. Samples were collected from the leaves after transient expression, and the LUC activity was detected. A Dual Luciferase Reporter Gene Assay Kit (Beyotime Biotechnology, Shanghai, China) was used for analysis.
The natural DgGPX1 promoter sequence (1250 bp) was cloned and inserted into a pSuper1300 vector together with the LUC reporter gene to form the recombinant plasmid ProDgGPX1-LUC. To verify the direct regulatory activity of DgMYB2 on DgGPX1, ProDgGPX1-LUC and 35S::DgMYB2 were coinjected into tobacco leaves as a test group; tobacco leaves injected with ProDgGPX1-LUC were used as a control group. As an internal control, pSuper1300-REN was injected into tobacco leaves for transient expression. Samples were collected 48 h after transient expression, and a Dual Luciferase Reporter Gene Assay Kit (Beyotime Biotechnology, Shanghai, China) was used for analysis.
Transient expression assays in tobacco and luciferase complementation imaging
To detect the response of DgMYB2 to cold stress, ProDgMYB2-LUC was transformed into Agrobacterium GV3101, which was subsequently coinjected into tobacco leaves. After 36 h, D-luciferin potassium salt was injected, and fluorescence detection was performed using a plant luciferase in vivo imaging system.
To detect the regulatory activity of DgMYB2 on the DgGPX1 promoter, ProDgGPX1-LUC and 35S::DgMYB2-GFP were first transformed into Agrobacterium GV3101, which was then coinjected into tobacco leaves as a test group; tobacco leaves injected with the ProDgGPX1-LUC vector served as the control group. After 36 h, D-luciferin potassium salt was injected, and fluorescence detection was performed using a plant luciferase in vivo imaging system.
Transient expression assays and subcellular localization of DgMYB2
Bacterial liquid containing 35S::DgMYB2-GFP and bacterial liquid containing the nuclear marker pSuper::NF-YA4-mCherry were mixed together at a 1:1 ratio, and then the combined bacterial liquid was injected into tobacco leaves. The injected leaves were used as the experimental group. Concurrently, tobacco leaves coinjected with a pSuper1300-GFP empty vector and the nuclear marker pSuper::NF-YA4-mCherry were used as the control group. The method used for transient expression in tobacco leaves was described in a previous study [26]. After the injected tobacco plants were cultured for 36 h (23°C ± 2°C, 75% relative humidity), their leaves were cut at the injection site in the form of a square fragment (2 mm × 2 mm). A laser scanning confocal microscope (LSCM; Nikon Corporation, Tokyo, Japan) was used to observe the experimental results.
Transactivation analysis and Y1H assays
A pGreenII0800-LUC vector was used to insert 5 GAL4 sequences and 1 TATA sequence in front of LUC to construct a reporter gene vector. The ORF region of DgMYB2 was inserted into a modified pSuper1300 vector containing the GAL4 BD to construct the recombinant plasmid pBD-DgMYB2. VP16 was inserted into a modified pSuper1300 vector containing GAL4 BD as a recombinant plasmid pBD-VP16, which was used as a positive control. The modified pSuper1300 empty vector pBD-Empty containing GAL4 BD was used as a negative control. The obtained recombinant plasmids were used to transform Agrobacterium GV3101 competent cells, and the transformed bacterial liquid containing the reporter gene and the transformed bacterial liquid containing pBD-DgMYB2, pBD-VP16, and pBD-Empty were mixed at a 1:1 ratio and injected into tobacco leaves. After culturing for 3 d, the LUC/REN value was determined using a dual-luciferase reporter gene detection kit (Beyotime Biotechnology, Shanghai, China) following the manufacturer’s instructions.
Three fragments of DgMYB2 were considered for measuring transcriptional activation activity: the N-terminal fragment (1–87 aa), the fragment containing the MYB domain R (88–144 aa), and the C-terminal fragment (145–294 aa). The full-length ORF sequence of DgMYB2 and the three sequence fragments were each ligated into the yeast expression vector pGBKT7. The resulting recombinant plasmids pGBKT7-DgMYB2 (1–294 aa), pGBKT7-DgMYB2N (1–87 aa), pGBKT7-DgMYB2R (88–144 aa), and pGBKT7-DgMYB2C (145–294 aa) were obtained. These plasmids were then transformed into the yeast strain Y1H and serially diluted. Then, the yeast cells were inoculated on two-deficiency (SD/−Ade/-His) culture media with or without X-α-gal for 3 d at 30°C. If yeast cells formed blue colonies, it was inferred that transactivation had occurred.
Electrophoretic mobility shift assays
The full-length ORF region of DgMYB2 was inserted into the pET28a expression vector to form a pET28a-DgMYB2 fusion construct. The recombinant plasmid was transformed into DE3 Escherichia coli competent cells, and 0.5 mM isopropyl bD-1 thiogalactopyranoside (IPTG) was added to induce the protein expression. Protein was extracted from the bacterial solution using a Bacterial Protein Extraction Kit (Sangon Biotech, Shanghai, China) following the manufacturer’s instructions. Afterward, a Ni-NTA Sefinose Resin Kit (Sangon Biotech, Shanghai, China) was used for protein purification.
To synthesize general probes, a 100 bp DNA fragment containing a MYB element and an A base at the 5′ end of the DgGPX1 promoter region was selected, and a 6-FAM tag was added at the 5′ end. The sequence of the competitive probe was the same as that of the general probe, but there was no 6-FAM tag at the 5′ end. When synthesizing the mutation probes, all sequences of the MYB site (CAACCA) were mutated to A bases, and the remaining sequences were the same as those of the general probe. Additionally, a His protein served as a negative control. The DNA probes used in the EMSA were all synthesized by Shanghai Sangon Biotech Co., Ltd. (Shanghai, China). The probes and samples were loaded onto a 6% polyacrylamide gel for the binding reaction and electrophoresis, and TBE buffer was used as the electrophoresis solution. The results were recorded using a charge-coupled device (CCD) camera.
Chromatin immunoprecipitation assays
The leaves of DgMYB2-overexpressing chrysanthemum plants were sampled and subjected to 1% formaldehyde cross-linking. Immunoprecipitation was performed using the SimpleChIP chromatin experimental procedure (magnetic beads) (Cell Signaling Technology, Danvers, Massachusetts, USA) following the manufacturer’s instructions. ChIP-qPCR analysis was performed after purifying the resulting chromatin preparation, and a fragment containing the MYB-binding site (CAACCA) in the DgGPX1 promoter region was considered the functional area. GFP tag monoclonal antibodies were produced by Proteintech Group, Inc. (Chicago, IL, USA). The primers used in the ChIP-qPCR analysis are listed in Table S1.
Histochemical staining and determination of H2O2 and O2−
To preliminarily detect H2O2 and O2− accumulation, histochemical staining assays were performed using nitro blue tetrazolium (NBT) and 3,3-diaminobenzidine (DAB). The process and operating steps were described in a previous publication [27].
Determination of the physiological data of transgenic chrysanthemum
A quantitative measurement kit (Keming Biological Co., Ltd., Suzhou, China) was used for the quantitative determination of the H2O2 and O2− contents. A GSH-PX assay kit (Jiancheng Bioengineering Institute, Nanjing, China) was used to determine the GPX activity. MDA accumulation was determined using an MDA assay kit (Jiancheng Bioengineering Institute, Nanjing, China), and relative electrolyte leakage was measured following previously described methods [28].
Acknowledgments
We acknowledge Dr. Zhizhong Gong (State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University) for providing the pSuper1300-GFP plasmids. This work was supported by grants from the National Natural Science Foundation of China (No. 31971707) and the Sichuan Agricultural University Student Innovation Training Program (No. 202110626017).
Author contributions
X.H.Y. designed and performed the experiments, analyzed the data, and wrote the manuscript. Y.C.L. and H.R.B. performed the experiments, analyzed the data, and wrote the manuscript. X.L., S.T. and X.Q.L. performed the experiments and analyzed the data. L.Z. analyzed the data. Q.L.L. designed the experiment, conceived the project, and supervised the study. All authors have read and approved the final manuscript. This work was supported by grants from the National Natural Science Foundation of China (No. 31971707) and the Sichuan Agricultural University Student Innovation Training Program (No. 202110626017).
Data availability
The raw sequence data have been submitted to the NCBI Sequence Read Archive database under accession number GSE117262. The sequence information of DgMYB2 has been submitted to GenBank under accession number MW853963.
The datasets generated and analyzed during this study are available from the corresponding author upon reasonable request.
Conflicts of interest statement
The authors declare they have no competing interests.
Supplementary data
Supplementary data is available at Horticulture Research Journal online.
Supplementary Material
References
- 1. Miura K, Furumoto T. Cold signaling and cold response in plants. Int J Mol Sci. 2013;14:5312–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McCarthy RL, Zhong R, Fowler Set al. The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 2010;51:1084–90. [DOI] [PubMed] [Google Scholar]
- 3. Stracke R, Ishihara H, Huep Get al. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling: TFs of a. thaliana R2R3-MYB subgroup 7. Plant J. 2007;50:660–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Su L-T, Li J-W, Liu D-Qet al. A novel MYB transcription factor, GmMYBJ1, from soybean confers drought and cold tolerance in Arabidopsis thaliana. Gene. 2014;538:46–55. [DOI] [PubMed] [Google Scholar]
- 5. Wang X, Niu Q-W, Teng Cet al. Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res. 2009;19:224–35. [DOI] [PubMed] [Google Scholar]
- 6. Kanei-Ishii C, Sarai A, Sawazaki Tet al. The tryptophan cluster: a hypothetical structure of the DNA-binding domain of the myb protooncogene product. J Biol Chem. 1990;265:19990–5. [PubMed] [Google Scholar]
- 7. An J-P, Li R, Qu F-Jet al. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 2018;96:562–77. [DOI] [PubMed] [Google Scholar]
- 8. Tombuloglu H. Genome-wide identification and expression analysis of R2R3, 3R- and 4R-MYB transcription factors during lignin biosynthesis in flax (Linum usitatissimum). Genomics. 2020;112:782–95. [DOI] [PubMed] [Google Scholar]
- 9. Haga N, Kato K, Murase Met al. R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development. 2007;134:1101–10. [DOI] [PubMed] [Google Scholar]
- 10. Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001;4:447–56. [DOI] [PubMed] [Google Scholar]
- 11. Xie Y, Chen P, Yan Yet al. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol. 2018;218:201–18. [DOI] [PubMed] [Google Scholar]
- 12. An J-P, Wang X-F, Zhang X-Wet al. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol J. 2020;18:337–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xing C, Liu Y, Zhao Let al. A novel MYB transcription factor regulates ascorbic acid synthesis and affects cold tolerance. Plant Cell Environ. 2019;42:832–45. [DOI] [PubMed] [Google Scholar]
- 14. Green R, Fluhr R. UV-B-induced PR-1 accumulation is mediated by active oxygen species. Plant Cell. 1995;7:203–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ahmad P, Jaleel CA, Salem MAet al. Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit Rev Biotechnol. 2010;30:161–75. [DOI] [PubMed] [Google Scholar]
- 16. Roxas VP, Lodhi SA, Garrett DKet al. Stress tolerance in transgenic tobacco seedlings that overexpress glutathione S-transferase/glutathione peroxidase. Plant Cell Physiol. 2000;41:1229–34. [DOI] [PubMed] [Google Scholar]
- 17. Wang X-G, Wang H-B, Chen F-Det al. Factors affecting quantity of pollen dispersal of spray cut chrysanthemum (Chrysanthemum morifolium). BMC Plant Biol. 2014;14:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang K, Bai Z-Y, Liang Q-Yet al. Transcriptome analysis of chrysanthemum (Dendranthema grandiflorum) in response to low temperature stress. BMC Genomics. 2018;19:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo H, Wang Y, Wang Let al. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol J. 2017;15:107–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mu R-L, Cao Y-R, Liu Y-Fet al. An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Res. 2009;19:1291–304. [DOI] [PubMed] [Google Scholar]
- 21. Campos JF, Cara B, Pérez-Martín Fet al. The tomato mutant ars1 (altered response to salt stress 1) identifies an R1-type MYB transcription factor involved in stomatal closure under salt acclimation. Plant Biotechnol J. 2016;14:1345–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dai X, Xu Y, Ma Qet al. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007;143:1739–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li X, Jia J, Zhao Pet al. LcMYB4, an unknown function transcription factor gene from sheepgrass, as a positive regulator of chilling and freezing tolerance in transgenic Arabidopsis. BMC Plant Biol. 2020;20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meng X, Yin B, Feng HLet al. Overexpression of R2R3-MYB gene leads to accumulation of anthocyanin and enhanced resistance to chilling and oxidative stress. Biol Plant. 2014;58:121–30. [Google Scholar]
- 25. Sun X, Matus JT, Wong DCJet al. The GARP/MYB-related grape transcription factor AQUILO improves cold tolerance and promotes the accumulation of raffinose family oligosaccharides. J Exp Bot. 2018;69:1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. An J-P, Yao J-F, Wang X-Net al. MdHY5 positively regulates cold tolerance via CBF-dependent and CBF-independent pathways in apple. J Plant Physiol. 2017;218:275–81. [DOI] [PubMed] [Google Scholar]
- 27. Wang K, Zhong M, Wu Y-Het al. Overexpression of a chrysanthemum transcription factor gene DgNAC1 improves the salinity tolerance in chrysanthemum. Plant Cell Rep. 2017;36:571–81. [DOI] [PubMed] [Google Scholar]
- 28. Kjellsen TD, Shiryaeva L, Schröder WPet al. Proteomics of extreme freezing tolerance in Siberian spruce (Picea obovata). J Proteome. 2010;73:965–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data have been submitted to the NCBI Sequence Read Archive database under accession number GSE117262. The sequence information of DgMYB2 has been submitted to GenBank under accession number MW853963.
The datasets generated and analyzed during this study are available from the corresponding author upon reasonable request.


