Abstract
A key element for the prevention and management of coronavirus disease 2019 is the development of effective therapeutics. Drug combination strategies offer several advantages over monotherapies. They have the potential to achieve greater efficacy, to increase the therapeutic index of drugs and to reduce the emergence of drug resistance. We assessed the in vitro synergistic interaction between remdesivir and ivermectin, both approved by the US Food and Drug Administration, and demonstrated enhanced antiviral activity against severe acute respiratory syndrome coronavirus-2. Whilst the in vitro synergistic activity reported here does not support the clinical application of this combination treatment strategy due to insufficient exposure of ivermectin in vivo, the data do warrant further investigation. Efforts to define the mechanisms underpinning the observed synergistic action could lead to the development of novel treatment strategies.
Keywords: SARS-CoV-2, COVID-19, Cytopathic activity, CPE, Combination therapy, Synergy
1. Introduction
At the time of writing, the World Health Organization (WHO) has reported more than 328 million cases of coronavirus disease 2019 (COVID-19) and more than 5.5 million deaths [1]. There remains a clear need for therapeutic strategies with activity against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Potential therapeutic strategies may include the repurposing of existing drugs as well as the discovery of novel therapies. Thousands of clinical trials are currently underway, with therapeutic approaches involving direct-acting antivirals for the prevention of viral replication, and host-directed therapies aimed at mitigating against the disease pathology [2,3].
Combination therapies can offer several advantages over monotherapies. They have the potential to achieve greater efficacy, to increase the therapeutic index of drugs and to reduce the emergence of drug resistance. Strategies to identify effective combination therapies are emerging, with several laboratories reporting in vitro combination screens [4] and in vivo animal combination studies [5]. In a recent clinical trial, baricitinib administered in combination with remdesivir was found to be superior, and to elicit fewer adverse effects, compared with either drug in isolation [6]. Importantly, even in the absence of synergistic activity, an additive interaction between two drugs with separate mechanisms of action may profoundly reduce the speed at which drug resistance is established.
Both remdesivir and ivermectin have received attention for the treatment of COVID-19. Remdesivir is a prodrug C-adenosine nucleoside analogue that inhibits the viral RNA-dependent, RNA polymerase. Early in the pandemic, remdesivir was shown to display in vitro antiviral efficacy against SARS-CoV-2 [7]. In a double-blind, randomized, placebo-controlled trial, intravenous administration of remdesivir showed superiority relative to placebo in shortening the time to recovery in adults who were hospitalized with COVID-19 [8]. However, other studies indicated that its impact was negligible [9], and on 20 November 2020, WHO issued a conditional recommendation against the use of remdesivir in hospitalized patients (irrespective of disease severity) due to and absence of evidence supporting an improvement in survival or other outcomes in patients.
Ivermectin is an antiparasitic which is active against a wide range of parasites, including gastrointestinal roundworms, lungworms, mites, lice, hornflies and ticks [10]. Ivermectin is reported to exhibit broad-spectrum antiviral activity against a wide range of RNA and DNA viruses [11]. Recently, ivermectin was also shown to display antiviral activity against SARS-CoV-2 [12], but approved doses are not expected to be high enough to achieve in vitro-defined target exposures systemically [13]. Several clinical trials are now evaluating the potential of ivermectin for both prophylaxis and treatment of COVID-19, but low exposures make the anti-inflammatory and/or immunomodulatory mechanisms of action more plausible than direct antiviral activity of the monotherapy [14], particularly as studies with SARS-CoV-2 in Syrian golden hamsters showed an impact upon disease pathology in the absence of any effect on viral titres [15].
The authors found a synergistic interaction between remdesivir and ivermectin resulting in improved in vitro antiviral activity against SARS-CoV-2 using two distinct methodologies – determination of the fractional inhibitory concentration index (FICI) with isobologram analyses, and checkerboard combinations with SynergyFinder analyses. The data are discussed in the context of current therapeutic efforts against COVID-19.
2. Materials and methods
2.1. SARS-CoV-2 strain
SARS-CoV-2/Human/Liverpool/REMRQ0001/2020 was isolated from a nasopharyngeal swab from a patient in Liverpool and passaged a further four times in Vero E6 cells. The mapped RNA sequence has been submitted to Genbank previously (Accession No. MW041156).
2.2. Vero E6 cell culture and plate preparation
Vero E6 cells were maintained in complete EMEM [EMEM supplemented with 10% heat-inactivated fetal bovine serum (Gibco; 10500-064) and 1% penicillin/streptomycin (Gibco; 15140-122)] in T175 flasks (Thermo Fisher Scientific, Waltham, MA, USA) at 37°C with 5% CO2. Cells were seeded in resting EMEM [EMEM supplemented with 10% heat-inactivated fetal bovine serum] at 1 × 105 cells/well in 96-well plates (Grenier Bio-one; 655090). Plates were incubated for 20 h at 37°C with 5% CO2 to allow the cells to reach 100% confluence. The resting minimal medium was removed, and the cells were used for downstream applications.
2.3. Concentration–response for remdesivir and ivermectin against SARS-CoV-2
Vero E6 cells were treated in triplicate with either drug in minimal medium at 25.00 µM, 8.33 µM, 2.78 µM, 0.93 µM, 0.31 µM, 0.10 µM and 0.03 µM (DMSO maintained at 0.25%) or control media, as appropriate. The plates were incubated at 37°C with 5% CO2 for 2 h. The minimal media containing the experimental compounds or the control media was then removed. Fifty microlitres of minimal media containing SARS-CoV-2 (MOI 0.05), 100 µL of 2× semi-solid media and 50 µL of minimal media containing experimental compounds and control media was added to each well, as appropriate. After 48 h, 4% v/v paraformaldehyde was added to each well and the plate was incubated for 1 h at room temperature. The medium was removed and cells were stained with crystal violet. Cells were washed three times with water, and cytopathic viral activity was determined by measuring the absorbance of each well at 590 nm using a Varioskan LUX microplate reader (Thermo Fisher Scientific).
Automated data quality control and data analyses were performed. For quality control, for the viral control, any well which had a log-transformed value that was 2 standard deviations above the mean of all log-transformed viral controls was excluded. Similarly, for the non-viral control, any well which had a log-transformed value that was 2 standard deviations below the mean of all log-transformed non-viral controls was excluded. If two or more wells were excluded on this basis for either control, the plate was voided and no further analysis was performed. Next, Z′ was calculated for each plate using the uninfected/untreated controls and infected/untreated controls according to Equation 1:
| (1) |
where and represent the standard deviation of the non-viral and viral controls respectively, while and represent the corresponding means of these controls. Drug activity was expressed as a percentage of inhibition of viral growth relative to the uninfected/untreated control (100% inhibition of viral cytopathic activity) and the infected/untreated control (0% inhibition of viral cytopathic activity) on that plate. Half maximal effective concentration (EC50) and 90% maximal effective concentration (EC90) were calculated for each compound that generated a robust, converged four-parameter fit according to Equation 2:
| (2) |
where E is the drug effect at any given concentration (C), Emax is the maximal level of viral inhibition (0–100%), EC50 is the concentration required to achieve half of this maximal inhibition, and h represents the hill slope which describes the steepness of the concentration–effect relationship.
Compounds that did not achieve ≥50% viral inhibition were deemed inactive without fitting. Concentrations that were deemed toxic, as evidenced by >20% (approximately two standard deviations of all data) drop in absorbance with concentration increase coupled with evidenced toxicity in drug controls, were excluded from fitting analysis.
2.4. FICI for remdesivir–ivermectin combinations against SARS-CoV-2
Following assessment of the inhibitory effect (EC50) of remdesivir and ivermectin monotherapy on the cytopathic viral activity of SARS-CoV-2, FICI was determined using the isobologram method developed by Berenbaum [16] using data from three independent biological replicates. Drug stocks were created in DMSO to provide a stock sufficient to produce a top concentration of 25 µM for each biological replicate. Drugs were combined to generate mixed ratios of 1:0, 0.8:0.2, 0.6:0.4, 0.4:0.6, 0.2:0.8 and 0:1.0. Fixed ratios were then diluted across a concentration range 1:2 (DMSO maintained at 1%) to generate concentration–response data for each ratio, as described previously. Ratio dilutions were performed in a single 2-mL deep-well plate, and added in parallel to three 96-well plates for each biological replicate. One additional plate which was not inoculated with virus was included to observe drug toxicity. Compound incubation and viral addition was performed as described above. Z′ was calculated and quality control was implemented as above. Interpretation of FICI (≤0.5 = synergistic; >4.0 = antagonistic; >0.5–4 = no interaction) was based on guidance provided by the Journal of Antimicrobial Chemotherapy [17].
2.5. Checkerboard combinations for remdesivir–ivermectin combinations against SARS-CoV-2
For robustness, a second method to assess pharmacodynamic drug combination interaction was utilized. Drug stocks were created by serial dilution. Compounds and controls were mixed 1:1 (DMSO maintained at 1%) to generate data for each combination alone and in combination. Remdesivir was studied at 10 µM, 5 µM, 2.5 µM, 1.25 µM and 0.63 µM, and ivermectin was studied at 5 µM, 2.5 µM, 1.25 µM, 0.63 µM and 0.31 µM. These concentrations were selected as they were determined not to cause cell toxicity to Vero E6 cells. Ratio dilutions were performed in a single 2-mL deep-well plate, and added in parallel to three 96-well plates for each biological replicate. Compound incubation and viral addition was performed as described above. Z′ was calculated and quality control was implemented as above. Data were analysed using SynergyFinder and a summary synergy score was generated (>10 synergistic, −10 to +10 additive, and <−10 antagonistic) [18].
3. Results
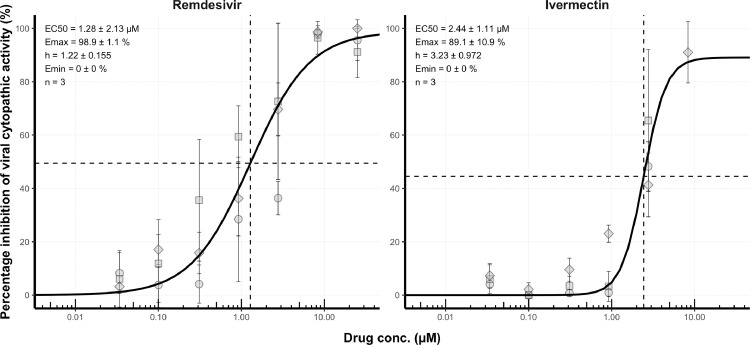
This study assessed the capacity of remdesivir and ivermectin combinations to inhibit the in vitro cytopathic activity of SARS-CoV-2. First, the activity of each compound in isolation was determined. For plates included in concentration–response analyses, the median signal to noise ratio was 29.3 and the median Z′ was 0.43 for concentration–response plates (Table 1). For each compound, a robust four-parameter fit was generated (Figure 1). EC50 was 2.4 ± 1.1 µM for ivermectin and 1.3 ± 2.1 µM for remdesivir (geometric mean ± geometric standard deviation).
Table 1.
Assay performance measures.
| Concentration–response | Isobologram | Checkerboard | |
|---|---|---|---|
| Total number of plates analysed | 6 | 9 | 9 |
| Signal to noise ratio (median [range]) | 29.3 (19.6–39.4) | 26.4 (13–37.3) | 23.6 (9.2–68.5) |
| Signal to background ratio (median [range]) | 2.6 (1.9–4.1) | 1.9 (1.6–2.2) | 2.7 (2.3–3.5) |
| Z′ (median [range]) | 0.43 (0.39–0.76) | 0.49 (0.18–0.7) | 0.62 (0.2–0.9) |
Fig. 1.
Concentration–effect relationship for the inhibition (%) of severe acute respiratory syndrome coronavirus-2 cytopathic activity for remdesivir and ivermectin. For each compound, activity was expressed relative to uninfected/untreated controls (100% inhibition of viral cytopathic activity) and infected/untreated controls (0% inhibition of viral activity). For each compound, activity was assessed at 25.00 µM, 8.33 µM, 2.78 µM, 0.93 µM, 0.31 µM, 0.10 µM and 0.03 µM in triplicate. Data points impacted by drug toxicity were removed automatically. Non-linear regression using an Emax model was performed on data taken from three independent biological replicates in order to generate concentration–effect predictions (solid black lines). For each compound, half maximal effective concentration (EC50) values, hillslope and replicate number (n) are shown. Dashed lines represent EC50 of each compound. Squares, diamonds and circles represent individual biological replicates, and error bars represent standard deviation calculated from technical triplicates.
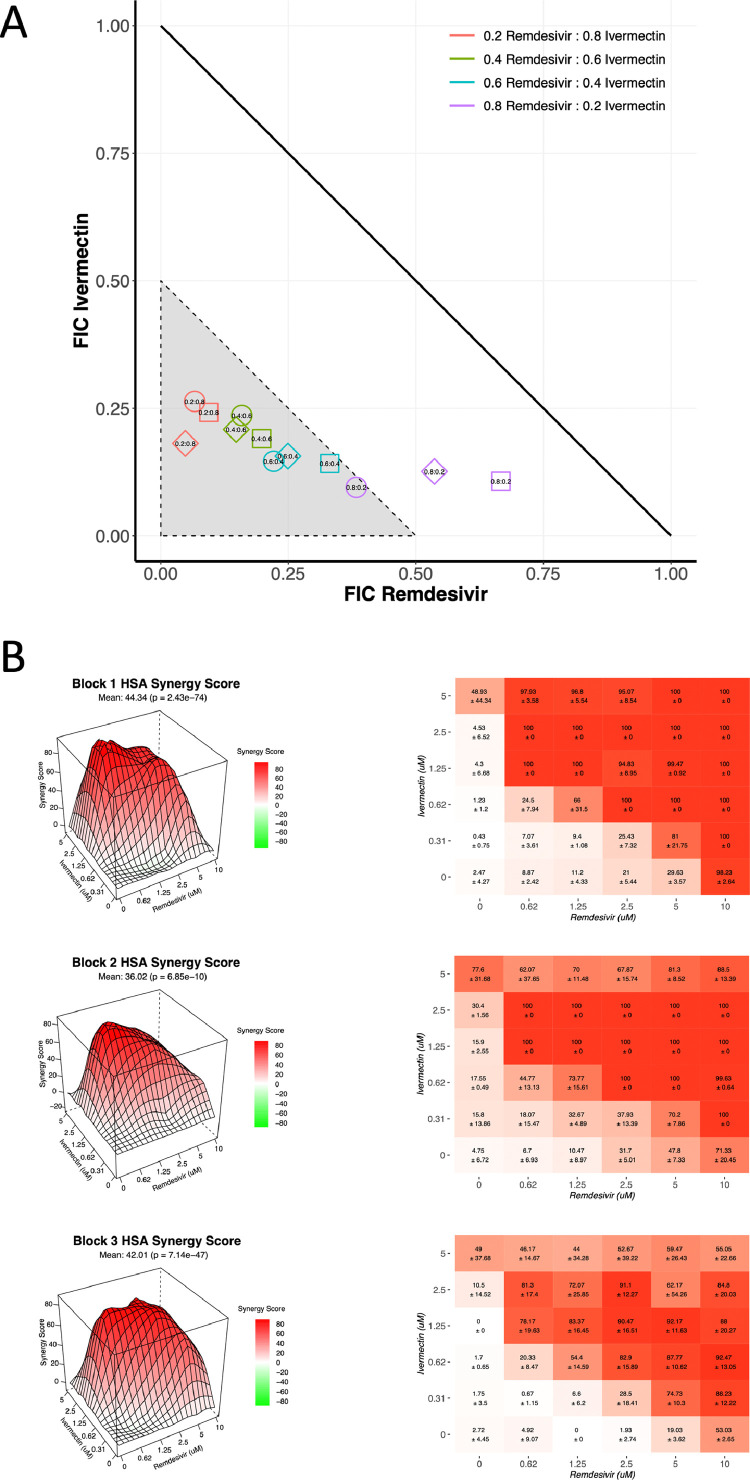
Next, the combination interaction between remdesivir and ivermectin was determined by isobologram. The median signal to noise ratio was 26.4 and the median Z′ was 0.49 for isobologram plates (Table 1). The 0.2:0.8 [remdesivir:ivermectin (5 µM:20 µM)] ratio, 0.4:0.6 ratio (10 µM:15 µM) and 0.6:0.4 (15 µM:10 µM) ratio demonstrated synergy (FICI<0.5) across all three biological replicates (Figure 2). For the 0.8:0.2 (20 µM:5 µM) ratio, just one biological replicate met the defined threshold of synergy (Figure 2). The other two biological replicates did, however, exceed the predicted effect assuming a purely additive relationship (Figure 2A).
Fig. 2.
Ivermectin and remdesivir display synergistic interaction. (A) Using half maximal effective concentration (EC50) values, ranges of ivermectin and remdesivir were analysed for synergy. Data are presented for fixed concentrations at 25 µM (corresponding to 1.0), 20 µM (0.8), 15 µM (0.6), 10 µM (0.4) and 5 µM (0.2). The area indicating synergy [fractional inhibitory concentration (FIC) ≤0.5] is shown in grey. Squares, diamonds and circles represent individual biological replicates, each derived from technical triplicates. (B) Three-dimensional (3D) visualization of compound integration based on the highest single agent (HSA) synergy score (left) alongside heatmap showing compound combination dose–response matrices (right). 3D visualizations and matrices are shown for individual biological replicates, each derived from technical triplicates.
The synergistic interaction was confirmed using interaction potency models using the SynergyFinder platform [18]. The median signal to noise ratio was 23.6 and the median Z′ was 0.62 for checkerboard plates (Table 1). All four integrated synergy models determined that interactions between remdesivir and ivermectin were synergistic with synergy scores that far exceeded the threshold for synergy (Table 2 and Figure 2B).
Table 2.
SynergyFinder synergy score summary table for remdesivir and ivermectin.
| Mean synergy score (median [range]) | |
|---|---|
| ZIP | 35.33 (28.01–40.84) |
| HSA | 40.25 (36.02–44.34) |
| Leowe | 26.34 (26.04–30.45) |
| Bliss | 37.77 (27.61–41.69) |
ZIP, zero interaction potency; HSA, highest single agent.
4. Discussion
This study found a synergistic interaction between remdesivir and ivermectin, both approved by the US Food and Drug Administration, resulting in enhanced in vitro antiviral activity against SARS-CoV-2. Although combination therapy offers a number of advantages compared with monotherapy, genuine descriptions of synergy are relatively infrequent [19]. Despite thousands of combination experiments having been performed, there have been very few reports of validated synergistic interactions against SARS-CoV-2 [4,20].
At this stage, the mechanism underpinning the synergistic interaction between remdesivir and ivermectin is unclear; however, both drugs have previously been shown to inhibit SARS-CoV-2 replication [7,12]. Given that remdesivir is known to inhibit the RNA-dependent, RNA polymerase [21], it will be of interest to investigate whether ivermectin confers synergy by inhibiting an undefined alternative but complimentary role in RNA synthesis. Ivermectin has been shown to inhibit replication of HIV-1 and dengue through inhibition of importin-β-mediated nuclear transport [22]. In silico predictions suggest that ivermectin may interact with host-cell proteins such as importins, which are required for nuclear transport, as well as viral proteins, including Nsp13 helicase and Mpro protease, which facilitate replication and translation of SARS-CoV-2 [23]. Further mechanistic studies will be required to determine the validity of in silico predictions.
Special care was taken to assess in vitro activity across concentrations that likely cover the physiological exposure of remdesivir and ivermectin in human plasma and lung tissue. In humans, a single 225-mg dose of remdesivir has been shown to produce a plasma Cmax of approximately 4000 ng/mL [24], exceeding its in vitro EC50 (1.3 ± 2.1 µM). In humans, a high dose of 600 µg/kg/day of ivermectin has been shown to produce a plasma Cmax of 120 ng/mL [25], which is much less than its in vitro EC50 (2.4 ± 1.1 µM). The Cmax of remdesivir in lung epithelial lining fluid (ELF) has not been established, and it is likely that these concentrations are important in terms of clinical activity. Poor exposure in lung ELF may well explain the limited impact of remdesivir in the clinic [8]. Interestingly, concentrations of ivermectin are predicted to be some three-fold higher in the lung than in plasma [26]; however, even at these levels, ivermectin fails to meet its in vitro EC50 and no data are presented here, or elsewhere, that would support the clinical application of ivermectin for the treatment of SARS-CoV-2 infection. Given that 88–93.6% of remdesivir [27] and 93.2% of ivermectin [28] is protein-bound, the availability of unbound drug at target sites is predicted to be considerably less than the reported values based on total drug concentrations.
Data presented here demonstrate that remdesivir administered in combination with ivermectin enhances in vitro antiviral activity. As described above, with respect to ivermectin, due to insufficient exposure of unbound drug at the target site, this combination strategy does not represent a clinically tractable therapeutic strategy. In addition, the differing routes of administration would likely impact the ability to achieve therapeutic concentrations of both drugs simultaneously. Further investigations are now required to determine whether the observed synergistic interaction can be replicated in animal disease models and with drugs that share similar modes of action, such as, for example, the orally bioavailable polymerase inhibitors, favipiravir or molnupiravir. The underpinning mechanisms for this synergy warrant further investigation so that this pharmacodynamic phenomenon can be exploited for the development of optimal drug combinations.
Acknowledgments
Funding
This study was supported by the Medical Research Council (MR/836 S00467X/1, GAB and SAW) and the UK Research and Innovation Strength in Places Fund (SIPF 20197, GAB, SAW and GH). AO acknowledges research funding from Unitaid (LONGEVITY) and EPSRC (EP/R024804/1). The authors also acknowledge funding by the National Institute for Health Research Health Protection Research Unit in Emerging and Zoonotic Infections, the Centre of Excellence in Infectious Diseases Research and the Alder Hey Charity. In addition, the authors wish to acknowledge support from Liverpool Health Partners and the Liverpool-Malawi-COVID-19 Consortium.
Competing interests
AO is a Director of Tandem Nano Ltd. AO has received research funding from ViiV, Merck and Janssen, and consultancy fees from Gilead. These associations had no influence on the content of the current manuscript. PON is currently engaged in a collaboration with Romark LLC, but this interaction had no influence on the content of the current manuscript. No other conflicts of interest are declared by the authors.
Ethical approval
Not required.
Editor: Dr Jim Gray
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijantimicag.2022.106542.
Contributor Information
Shaun H. Pennington, Email: shaun.pennington@lstmed.ac.uk.
Giancarlo A. Biagini, Email: giancarlo.biagini@lstmed.ac.uk.
Appendix. Supplementary materials
References
- 1.World Health Organization . WHO; Geneva: 2020. WHO coronovirus disease (COVID-19) dashboard. Available at https://covid19whoint/ [Google Scholar]
- 2.McKee DL, Sternberg A, Stange U, Laufer S, Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res. 2020;157 doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;323:1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 4.Ianevski A, Yao R, Biza S, Zusinaite E, Mannik A, Kivi G, et al. Identification and tracking of antiviral drug combinations. Viruses. 2020;12:1178. doi: 10.3390/v12101178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaptein SJF, Jacobs S, Langendries L, Seldeslachts L, Ter Horst S, Liesenborghs L, et al. Favipiravir at high doses has potent antiviral activity in SARS-CoV-2-infected hamsters, whereas hydroxychloroquine lacks activity. Proc Natl Acad Sci USA. 2020;117:26955–26965. doi: 10.1073/pnas.2014441117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalil AC, Patterson TF, Mehta AK, Tomashek KM, Wolfe CR, Ghazaryan V, et al. Baricitinib plus remdesivir for hospitalized adults with COVID-19. N Engl J Med. 2021;384:795–807. doi: 10.1056/NEJMoa2031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pizzorno A, Padey B, Dubois J, Julien T, Traversier A, Duliere V, et al. 2020. In vitro evaluation of antiviral activity of single and combined repurposable drugs against SARS-CoV-2. Antiviral Res. 2020;181 doi: 10.1016/j.antiviral.2020.104878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, et al. Remdesivir for the treatment of COVID-19 – final report. N Engl J Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan H, Peto R, Henao-Restrepo AM, Preziosi MP, Sathiyamoorthy V, WHO Solidarity Trial Consortium Repurposed antiviral drugs for COVID-19 – interim WHO Solidarity Trial results. N Engl J Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crump A, Omura S. Ivermectin, 'wonder drug' from Japan: the human use perspective. Proc Jpn Acad Ser B Phys Biol Sci. 2011;87:13–28. doi: 10.2183/pjab.87.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heidary F, Gharebaghi R. Ivermectin: a systematic review from antiviral effects to COVID-19 complementary regimen. J Antibiot. 2020;73:593–602. doi: 10.1038/s41429-020-0336-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arshad U, Pertinez H, Box H, Tatham L, Rajoli RKR, Curley P, et al. Prioritization of anti-SARS-CoV-2 drug repurposing opportunities based on plasma and target site concentrations derived from their established human pharmacokinetics. Clin Pharmacol Ther. 2020;108:775–790. doi: 10.1002/cpt.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinhoff M, Vocanson M, Voegel JJ, Hacini-Rachinel F, Schafer G. Topical ivermectin 10 mg/g and oral doxycycline 40 mg modified-release: current evidence on the complementary use of anti-inflammatory rosacea treatments. Adv Ther. 2016;33:1481–1501. doi: 10.1007/s12325-016-0380-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Melo GD, Lazarini F, Larrous F, Feige L, Kergoat L, Marchio A, et al. Anti-COVID-19 efficacy of ivermectin in the golden hamster. bioRxiv 2020.11.21.392639; doi:10.1101/2020.11.21.392639.
- 16.Berenbaum MC. A method for testing for synergy with any number of agents. J Infect Dis. 1978;137:122–130. doi: 10.1093/infdis/137.2.122. [DOI] [PubMed] [Google Scholar]
- 17.Odds FC. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother. 2003;52:1. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 18.Ianevski A, Giri AK, Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res. 2020;48:W488–W493. doi: 10.1093/nar/gkaa216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greco WR, Bravo G, Parsons JC. The search for synergy: a critical review from a response surface perspective. Pharmacol Rev. 1995;47:331–385. [PubMed] [Google Scholar]
- 20.Riva L, Yuan S, Yin X, Martin-Sancho L, Matsunaga N, Pache L, et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agostini ML, Andres EL, Sims AC, Graham RL, Sheahan TP, Lu X, et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9:e00221. doi: 10.1128/mBio.00221-18. -18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagstaff KM, Sivakumaran H, Heaton SM, Harrich D, Jans DA. Ivermectin is a specific inhibitor of importin alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem J. 2012;443:851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez-Paz L, Hurtado-Leon ML, Lossada C, Fernandez-Materan FV, Vera-Villalobos J, Lorono M, et al. Comparative study of the interaction of ivermectin with proteins of interest associated with SARS-CoV-2: a computational and biophysical approach. Biophys Chem. 2021;278 doi: 10.1016/j.bpc.2021.106677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humeniuk R, Mathias A, Cao H, Osinusi A, Shen G, Chng E, et al. Safety, tolerability, and pharmacokinetics of remdesivir, an antiviral for treatment of COVID-19, in healthy subjects. Clin Transl Sci. 2020;13:896–906. doi: 10.1111/cts.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smit MR, Ochomo EO, Waterhouse D, Kwambai TK, Abong'o BO, Bousema T, et al. Pharmacokinetics–pharmacodynamics of high-dose ivermectin with dihydroartemisinin-piperaquine on mosquitocidal activity and QT-prolongation (IVERMAL) Clin Pharmacol Ther. 2019;105:388–401. doi: 10.1002/cpt.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jermain B, Hanafin PO, Cao Y, Lifschitz A, Lanusse C, Rao GG. Development of a minimal physiologically-based pharmacokinetic model to simulate lung exposure in humans following oral administration of ivermectin for COVID-19 drug repurposing. J Pharm Sci. 2020;109:3574–3578. doi: 10.1016/j.xphs.2020.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Humeniuk R, Mathias A, Kirby BJ, Lutz JD, Cao H, Osinusi A, et al. Pharmacokinetic, pharmacodynamic, and drug-interaction profile of remdesivir, a SARS-CoV-2 replication inhibitor. Clin Pharmacokinet. 2021;60:569–583. doi: 10.1007/s40262-021-00984-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klotz U, Ogbuokiri JE, Okonkwo PO. Ivermectin binds avidly to plasma proteins. Eur J Clin Pharmacol. 1990;39:607–608. doi: 10.1007/BF00316107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.