Abstract
Background
Propolis and honey have been studied as alternative treatments for patients with coronavirus disease 2019 (COVID-19). However, no study has yet summarized the full body of evidence for the use of propolis and honey in COVID-19 prevention and treatment.
Objective
This study systematically reviews the mechanisms of propolis and honey against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and current evidence for the use of propolis and honey in COVID-19 prevention and treatment.
Search strategy
A systematic search was conducted of electronic databases including PubMed, Scopus, ScienceDirect, and Cochrane Library from their inceptions to April 2021.
Inclusion criteria
Studies that evaluated the effect of propolis or bee products against SARS-CoV-2 using in silico methods, clinical studies, case reports and case series were included.
Data extraction and analysis
A standardized data extraction form was used, and data were extracted by two independent reviewers. Narrative synthesis was used to summarize study results concerning the use of propolis or honey in COVID-19 prevention and treatment and their potential mechanisms of action against SARS-CoV-2.
Results
A total of 15 studies were included. Nine studies were in silico studies, two studies were case reports, one study was a case series, and three studies were randomized controlled trials (RCTs). In silico studies, using molecular docking methods, showed that compounds in propolis could interact with several target proteins of SARS-CoV-2, including angiotensin-converting enzyme 2, the main protease enzyme, RNA-dependent RNA polymerase, and spike protein. Propolis may have a positive effect for clinical improvement in mild and moderate-to-severe COVID-19 patients, according to case reports and case series. The included RCTs indicated that propolis or honey could probably improve clinical symptoms and decrease viral clearance time when they were used as adjuvant therapy to standard of care.
Conclusion
In silico studies showed that compounds from propolis could interact with target proteins of SARS-CoV-2, interfering with viral entry and viral RNA replication, while clinical studies revealed that propolis and honey could probably improve clinical COVID-19 symptoms and decrease viral clearance time. However, clinical evidence is limited by the small number of studies and small sample sizes. Future clinical studies are warranted.
Keywords: Propolis, Honey, Coronavirus disease 2019, Systematic review
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic has affected people worldwide [1]. As of April 2021, approximately more than 200 million people were infected, resulting in over 4.9 million deaths [2], and the number of infections continues to rise. The numbers of infected patients have been increasing. Most COVID-19 patients develop mild to moderate respiratory symptoms, including dry cough, shortness of breath and sore throat. However, serious acute respiratory distress syndrome also develops in some patients, especially in the elderly or those with chronic diseases.
Health interventions have been implemented to reduce the rate of COVID-19 infection, including face masks, physical distancing, hand hygiene, and vaccines [3]. Medications have also been used to treat COVID-19-infected patients, such as antimalarial agents, antiviral therapy, immune-based therapy, and corticosteroids [4], [5]. Herbal medicine is a class of natural substances and are also used as adjuvant therapies for COVID-19. Some natural substances are reported to have inhibitory effects on coronavirus, such as psoralidin, silvestrol, quercetin, myricetin, flavonoids, and polyphenols [6], [7], [8].
Propolis, a resinous bee product, has been reported to have antimicrobial activities, based on its content of phenolic compounds, flavonoids, and esters of aromatic acids [9], [10], [11]. Specific to antiviral activities, propolis has been shown to inhibit varicella-zoster virus, herpes virus, and human immunodeficiency virus [12], [13], [14]. For COVID-19, pre-clinical studies report the interaction of propolis and some target proteins of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes COVID-19 [15], [16]. In addition, some clinical studies have shown a potential positive effect of propolis and honey products on SARS-CoV-2 viral clearance and patients’ symptoms [17], [18]. However, no study has yet summarized all of the available evidence concerning the use of propolis or honey for COVID-19 prevention and treatment.
This study systematically reviewed possible mechanisms of propolis and honey against SARS-CoV-2, and clinical evidence relevant to the effect of propolis and honey for COVID-19 prevention and treatment. This synthesis will be useful for supporting clinical decision making and the development of propolis and honey products for COVID-19 management.
2. Methods
2.1. Search and study selection
We systematically searched relevant studies from PubMed, Scopus, ScienceDirect, and Cochrane Library from their inceptions to April 2021. We also performed citation tracking from related articles to identify additional studies. Search terms were (“SARS-CoV-2” OR “severe acute respiratory syndrome coronavirus 2” OR “COVID-19” OR “coronavirus” OR “coronavirus disease” OR “novel coronavirus” OR “2019-nCoV” OR “COVID-2019 pneumonia”) AND (“propolis” OR “bee glue” OR “bee product” OR “honey”). Details of the search strategies are reported in Supplementary file Table S1.
Studies that met the following criteria were included: (1) studies on the effect of propolis and bee products against SARS-CoV-2, and (2) reports fitting the classifications of in silico studies, clinical studies, case report and case series. Studies reported in abstract form only and incomplete studies were excluded. Resources identified in the search were de-duplicated using EndNote™ version 20 (Clarivate Co. Ltd). All identified articles were independently reviewed by RW and RK. All disagreements were resolved through discussion with a third reviewer (WD).
2.2. Data extraction and quality assessment
A standardized data extraction form was developed. The extracted data included authors, year of publication, country, study design, intervention, comparator, inclusion and exclusion criteria, number of participants, methods, duration of assessment, and outcomes. For in silico studies, binding affinity was the primary outcome, while clinical symptom improvement was the primary outcome for clinical studies. Data extraction was performed by RW and RK and validated by WD and PD.
The quality of studies was assessed by RW and RK. Indicators of evidence quality and scoring system [19], [20], [21] were used to assess the quality of in silico studies. The JBI Critical Appraisal Checklist was used for case reports [22], the National Institutes of Health Quality Assessment Tool was used for case series [23], while version 2 of the Cochrane risk-of-bias tool was used to assess the quality of randomized controlled trials (RCTs) [24].
2.3. Data analysis
Various compounds from propolis and honey were reported to have activity against SARS-CoV-2. Study design, intervention, and outcomes of interest were different. Therefore, meta-analysis could not be performed. Narrative synthesis was used to summarize evidence on the use of propolis or honey for COVID-19 prevention and treatment. It was also used to summarize the potential mechanisms of these products against SARS-CoV-2.
3. Results
3.1. Study characteristics
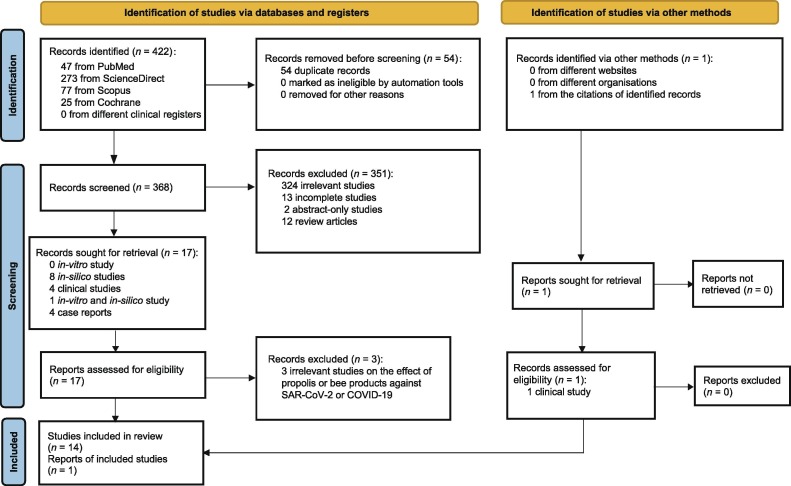
A total of 423 studies were retrieved from the database searches. Fifteen studies were included in this systematic review (Fig. 1 ). Nine studies were in silico studies [7], [8], [16], [25], [26], [27], [28], [29], [30], while three studies were case reports or case series [18], [31], [32], and the other three studies were RCTs [17], [33], [34]. Of the nine in silico studies, two studies determined the effect of propolis against SARS-CoV-2 angiotensin-converting enzyme 2 (ACE2) [7], [8], six studies used SARS-CoV-2 main protease enzyme as the target enzyme [16], [25], [26], [27], [28], [29], two studies used SARS-CoV-2 RNA-dependent RNA polymerase as the target enzyme [16], [29], three studies focused on spike protein subunit one [16], [27], [30], and one study focused on spike protein subunit two [25]. The molecular docking method was used to determine the binding affinity of propolis on the target protein.
Fig. 1.
Flow diagram. COVID-19: coronavirus disease 2019; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
Of the clinical studies, two were case reports [31], [32], while one was a case series [18]. The case reports followed the use of commercial propolis for COVID-19 treatment. One case was a patient with mild COVID-19 symptoms, while the other case was a patient with severe COVID-19 symptoms. The case series study reported a total of 40 subjects. Twenty subjects used propolis for COVID-19 prevention, while the rest used propolis for COVID-19 treatment.
Three RCTs [17], [33], [34] reported the effect of propolis or honey, as an adjuvant to standard care, on clinical symptoms of COVID-19. One study reported the effect of honey with Nigella sativa seeds on time of clinical symptom improvement and time to viral clearance [17]. One study reported the effect of propolis plus Hyoscyamus niger extract on clinical symptom scores [33]. Another study reported the effect of Brazilian green propolis extract on length of hospital stay, oxygen therapy dependency time, the number of acute kidney injuries, the use of renal replacement therapy, invasive ventilation, vasoactive agents, and intensive care unit (ICU) stay [34].
3.2. Quality of the included studies
Five in silico studies were assessed as having no discernable bias [16], [30], [26], [27], [28]. One study was assessed as having weak bias because the study did not report the reason for choosing the compounds [8]. Three studies were assessed as having moderate bias because of the discrepancy between their abstracts and study results. Another reason was because the study did not report the reason for choosing the compounds [7], [25], [29] (Supplementary file Table S2).
One case report was assessed as a good-quality study [32]. The study clearly described the patient’s demographic characteristics, diagnostic tests, assessment methods, and the treatment outcomes. Another case report was assessed as being of fair quality [31]. The study did not sufficiently provide patient’s history and clear takeaway lessons (Supplementary file Table S3). One case series [18] was assessed as a fair-quality study. The study did not clearly describe the study population, and participants were not comparable because they received different interventions (Supplementary file Table S4).
All included RCTs [17], [33], [34] were assessed as having high risk of bias. This was because of the lack of a clear randomization process, questionable outcome measures, or the selection of reported results (Supplementary file Table S5).
3.3. Results from in silico studies
3.3.1. Propolis and honey against SARS-CoV-2 ACE2
SARS-CoV-2 ACE2 was used as the target enzyme in two studies [7], [8]. One study [7] showed that 12 compounds from propolis had positive binding affinity to SARS-CoV-2 ACE2. It also indicated that rutin had the highest binding affinity with −8.04 kcal/mol, which was higher than its positive control (−7.24 kcal/mol). Another study found that 13 compounds from propolis had positive binding affinity to SARS-CoV-2 ACE2. Glyasperin A was reported to have the highest binding affinity to SARS-CoV-2 ACE2 (−10.8 kcal/mol) which was higher than its positive control (−9.2 kcal/mol) [8] (Table 1 and Table 2 ).
Table 1.
Baseline characteristics of in silico studies.
| Study | Country | Method | Target protein or enzyme | Source of study compound |
|---|---|---|---|---|
| Guler et al. [7] | Turkey | Molecular docking | Angiotensin-converting enzyme 2 | Raw propolis samples were obtained from experienced beekeepers in 2018 from Black Sea Region, Turkey |
| Khayrani et al. [8] | Indonesia | Molecular docking | Angiotensin-converting enzyme 2 | Sulawesi propolis compounds reported from North Luwu |
| Dewi et al. [26] | Indonesia | Molecular docking | Main protease | Propolis from stingless bees was used |
| Elwakil et al. [16] | Egypt | Molecular docking | Main protease, RNA-dependent RNA polymerase, and spike protein subunit 1 | Propolis collected by the authors from various Egyptian geographical areas: Alexandria, Tanta and Menoufia |
| Harisna et al. [25] | Indonesia | Molecular docking | Main protease, and spike protein subunit 2 | Propolis was supplied by PT Nano Herbaltama International, South Tangerang, Indonesia |
| Refaat et al. [27] | Egypt | Molecular docking | Main protease, and spike protein subunit 1 | Alcoholic extract of propolis purchased was from VACSERA-EGYPT (Cell Culture Department) |
| Sahlan et al. [28] | Indonesia | Molecular docking | Main protease | Sulawesi propolis compounds |
| Shaldam et al. [29] | Egypt | Molecular docking | Main protease, and RNA-dependent RNA polymerase | Not reported |
| Jain et al. [30] | Saudi Arabia | Molecular docking | Spike protein subunit 1 | PubChem database |
Table 2.
Molecular docking outcomes of in silico studies modeling the interaction of propolis constituents with SARS-CoV-2.
| Study | Comparator: binding affinity energy (kcal/mol) | Study compound (binding affinity energy [kcal/mol]) | Summary |
|---|---|---|---|
| Angiotensin-converting enzyme 2 | |||
| Guler et al. [7] | MLN-4760: −7.24 | Caffeic acid (−5.53), caffeic acid phenethyl ester (−7.58), chrysin (−7.17), galangin (−7.35), myricetin (−7.59), rutin (−8.04), hesperetin (−7.45), pinocembrin (−7.16), luteolin (−7.29), quercetin (−7.58), kaempferol (−7.23), and syringic acid (−4.49) | Rutin had the highest binding affinity compared to the other 11 compounds, which was higher than the reference compound MLN-4760 |
| Khayrani et al. [8] | MLN-4760: −9.2 | Sulabiroins A (−9.5), sulabiroins B (−8.8), 2,3-dihydro-3-hydroxypapuanic acid (−8.3), (−)-papuanic acid (−8.5), (−)-isocalolongic acid (−8.9), isopapuanic acid (−8.1), isocalopolyanic acid (−8.8), glyasperin A (−10.8), broussoflavonol F (−9.9), (2S)-5,7-dihydroxy-40-methoxy-8-prenylflavanone (−9.3), isorhamnetin (−9.2), (1S)-2-trans, 4-trans-abscisic acid (−7.3), and (1S)-2-cis, 4-trans-abscisic acid (−7.2) | Glyasperin A had the highest binding affinity compared to the other 12 compounds, which was higher than MLN-4760 |
| Main protease | |||
| Dewi et al. [26] | Native ligand (N3): −8.4 | Sulabiroins A (−8.1), sulabiroins B (−7.8), 2′,3′-dihydro-3′-hydroxypapuanic acid (−7.1), (−)-papuanic acid (−7.4), (−)-isocalolongic acid (−7.2), isopapuanic acid (−7.0), isocalopolyanic acid (−6.4), glyasperin A (−7.8), broussoflavonol F (−7.9), (2S)-5,7-dihydroxy-4′-methoxy-8-prenylflavanone (−7.9), isorhamnetinb (−7.3), (1′S)-2-trans,4-trans-abscisic acid (−6.5), (1′S)-2-cis,4-trans-abscisic acid (−6.0), curcumene (−5.5), thymol (−4.7), tetralin (−4.7), P-coumaric acid (−5.1), α-tocopherol succinate (−6.2), deoksi podophyllotoxin (−7.4), and xanthoxyletin (−6.7) | Sulabiroins A had the highest binding affinity compared to the other 19 compounds, which was lower than native ligand (N3) |
| Elwakil et al. [16] | Lopinavir: −8.18 | Acid: n-hexadecanoic acid (−6.28), benzoic acid (−3.70), trans-caffeic acid (−4.48), tetradecanoic acid (−5.84), and trans-13-octadecenoic acid (−6.06) | Octacosanol showed the highest binding affinity compared to the other 25 compounds, which was lower than lopinavir |
| Alkanes: heneicosane (−6.44), octacosane (−7.39), and heptacosane (−6.95) | |||
| Esters: hexadecanoic acid, methyl ester (−6.28), pinostrobin chalcone (−5.44), hexadecaneperoxoic acid, 1,1-dimethyl-3-[(1-oxohexadecyl)oxy]propyl ester (−7.35), oxalic acid, dodecyl 2-phenylethyl ester (−6.94), and methyl pentafluoropropionate (−3.40) | |||
| Triterpenoids: R1-barrigenol (−5.81), α-eudesmol (−4.61), and ß-eudesmol (−4.77) | |||
| Alcohol: octacosanol (−6.77) | |||
| Flavoniods: pinocembrin (−5.29) | |||
| Unspecified: bicyclo[2.2.2]octa-2,5-diene, 1,2,3,6-tetramethyl (−4.23), estra-1,3,5(10)-trien-17-one, 3-hydroxy-2-methoxy (−6.16), pregnan-20-one, 3,17-dihydroxy-,(3ß,5ß) (−5.53), octadecane, 3-ethyl-5-(2-ethylbutyl) (−6.55), 4ß-methylandrostane 2,3-diol-1,17-dione (−5.38), octatriacontyl pentafluoropropionate (−7.35), pregn-5-en-20-one, 11-(acetyloxy)-3,14-dihydroxy-12-(2-hydroxy-3-methyl-1-oxobutoxy)-,(3ß,11α,12ß,14ß) (−6.08), and cyclohexamine, N-n-butyl-1-[2-thionaphthenyl] (−5.33) | |||
| Harisna et al. [25] | Nelfinavir: −7.7 | Flavones: 3′-methoxydaidzein (−7.3), 3′-methoxydaidzin (−7.7), genistin (−7.6), xanthomicrol (−7.1), 3′,5,6,7-tetrahydroxy-4′-methoxyisofavone (−7.6), methylophiopogonone A (−7.6), 3′4′,7-trihydroxyfavanone (−7.5), moslosoofavone (−7.4), luteolin (−7.5), 2′,6′-dihydroxy-4′ methoxydihydrochalcone (−6.9), chrysoeriol (−7.3), jaceosidin (−7.2), (3R)-7,2′,3′-trihydroxy-4′-methoxyisofavanone (−7.1), and neobavaisofavone (−7.6) | 3′-Methoxydaidzin showed the highest binding affinity compared to the other 21 compounds but was similar to nelfinavir for the SARS-CoV-2 main protease |
| Flavonols: 3′-deoxysappanol (−7.0) | |||
| Phenolic acid: cinnamic acid (−5.4), caffeic acid (−5.9), 2,5-dimethyl-7-hydroxychromone (−6.2), isoferulic acid (−5.7), dimethylcaffeic acid (−5.7), benzyl caffeate (−7.1), and isoaloeresin D (−7.4) | |||
| Refaat et al. [27] | Native ligand (N3): −133.6; remdesivir: −136.4; favipiravir: −33.3; hydroxychloroquine: −65.9 | Rutin (−92.8), caffeic acid phenethyl ester (−67.8), quercetin (−57.5), kaempferol (−56.3), pinocembrin (−56.2), pinobanksin (−54.1), galangin (−53.2), chrysin (−52.9), p-cumaric acid (−45.5), and benzoic acid (−35.4) | Rutin had the highest binding affinity compared to the other 9 compounds, which was higher than favipiravir and hydroxychloroquine. However, it was lower than native ligand (N3) and remdesivir for the SARS-CoV-2 main protease |
| Sahlan et al. [28] | 13b: −8.2 | New podophyllotoxin derivative: sulabiroins A (−7.6) | Glyasperin A and broussoflavonol F had the highest binding affinity compared to the other 19 compounds, but they were lower than 13b for the SARS-CoV-2 main protease |
| Others: sulabiroins B (−7.0), 2′,3′-dihydro-3′-hydroxypapuanic acid (−6.7), (−)-papuanic acid (−6.6), (−)-isocalolongic acid (−6.7), isopapuanic acid (−6.8), isocalopolyanic acid (−6.8), glyasperin A (−7.8), broussoflavonol F (−7.8), (2s)-5,7-dihydroxy-4′-methoxy-8-prenylflavanone (−7.1), isorhamnetin (−7.5), (1′s)-2-trans,4 trans-abscisic acid (−6.1), (1′s)-2-cis,4 trans-abscisic acid (−5.9), a-tocopherol succinate (−5.1), xanthoxyletin (−6.2), P-coumaric acid (−4.9), curcumene (−4.7), thymol (−4.7), tetralin (−4.4), deoxypodophyllotoxin (−7.3), and 14b (−7.2) | |||
| Shaldam et al. [29] | Not available | 2,2-Dimethyl-8-prenylchromene (−6.8) | Kaempferol had the highest binding affinity compared to 13 other compounds |
| Phenylpropanes: artepillin C (−7.5), 3-prenyl cinnamic acid allyl ester (−6.2), isocupressic acid (−6.4), 13C-symphyoreticulic acid (−6.9), ellagic acid (−7.5), syringic acid (−5.6), caffeic acid phenethyl ester (−7.0), p-coumaric acid (−5.6), hesperetin (−7.4), naringenin (−6.5), kaempferol (−7.8), quercetin (−7.4), and chrysin (−7.2) | |||
| RNA-dependent RNA polymerase | |||
| Elwakil et al. [16] | Remdesivir: −6.77 | Acid: n-hexadecanoic acid (−5.70), benzoic acid (−3.91), trans-caffeic acid (−4.80), tetradecanoic acid (−5.34), and trans-13-octadecenoic acid (−5.97) | Octatriacontyl pentafluoropropionate had the highest binding affinity compared to the other 24 compounds, which was higher than the reference drug, remdesivir |
| Alkanes: heneicosane (−6.19), octacosane (−6.87), and heptacosane (−6.91) | |||
| Esters: hexadecanoic acid, methyl ester (−5.63), pinostrobin chalcone (−5.59), hexadecaneperoxoic acid, 1,1-dimethyl-3-[(1-oxohexadecyl)oxy]propyl ester (−8.04), oxalic acid, dodecyl 2-phenylethyl ester (−6.94), and methyl pentafluoropropionate (−3.54) | |||
| Triterpenoids: R1-barrigenol (−5.58), α-eudesmol (−4.47), and ß-eudesmol (−4.96) | |||
| Alcohol: octacosanol (−6.96) | |||
| Flavoniods: pinocembrin (−4.93) | |||
| Not specified: Bicyclo[2.2.2]octa-2,5-diene, 1,2,3,6-tetramethyl (−3.84), estra-1,3,5(10)-trien-17-one, 3-hydroxy-2-methoxy (−5.31), pregnan-20-one, 3,17-dihydroxy-, (3ß,5ß) (−5.06), octadecane, 3-ethyl-5-(2-ethylbutyl) (−6.11), 4ß-methylandrostane 2,3-diol-1,17-dione (−4.72), octatriacontyl pentafluoropropionate (−8.20), pregn-5-en-20-one,11-(acetyloxy)-3,14-dihydroxy-12-(2-hydroxy-3-methyl-1-oxobutoxy)-(3ß,11α,12ß,14ß) (−6.63), and cyclohexamine, N-n-butyl-1-[2-thionaphthenyl] (−4.90) | |||
| Shaldam et al. [29] | Not available | 2,2-Dimethyl-8-prenylchromene (−5.6) | Ellagic acid had the highest binding affinity compared to the other 13 compounds |
| Phenylpropanes: artepillin C (−5.9), 3-prenyl cinnamic acid allyl ester (−5.3), isocupressic acid (−5.8), 13C-symphyoreticulic acid (−5.7), ellagic acid (−6.4), syringic acid (−5.5), caffeic acid phenethyl ester (−5.4), p-coumaric acid (−5.3), hesperetin (−6.3), naringenin (−6.0), kaempferol (−6.2), quercetin (−6.1), and chrysin (−6.1) | |||
| Spike protein subunit 1 | |||
| Elwakil et al. [16] | Umifenovir: −5.56 | Acid: n-hexadecanoic acid (−5.64), benzoic acid (−4.16), trans-caffeic acid (−4.76), tetradecanoic acid (−5.23), and trans-13-octadecenoic acid (−5.29) | Octatriacontyl pentafluoropropionate showed the highest binding affinity, compared to the 25 other compounds, which was higher than umifenovir |
| Alkanes: heneicosane (−5.68), octacosane (−5.99), and heptacosane (−5.80) | |||
| Esters: hexadecanoic acid, methyl ester (−5.27), pinostrobin chalcone (−5.50), hexadecaneperoxoic acid,1,1-dimethyl-3-[(1-oxohexadecyl)oxy]propyl ester (−6.56), oxalic acid, dodecyl 2-phenylethyl ester (−5.95), and methyl pentafluoropropionate (−3.55) | |||
| Triterpenoids: R1-barrigenol (−4.44), α-eudesmol (−4.57), and ß-eudesmol (−4.96) | |||
| Alcohol: octacosanol (−6.26) | |||
| Flavoniod: pinocembrin (−4.78) | |||
| Not specified: bicyclo[2.2.2]octa-2,5-diene, 1,2,3,6-tetramethyl (−3.93), estra-1,3,5(10)-trien-17-one, 3-hydroxy-2-methoxy (−5.26), pregnan-20-one, 3,17-dihydroxy-, (3ß,5ß) (−4.36), octadecane, 3-ethyl-5-(2-ethylbutyl) (−5.83), 4ß-methylandrostane 2,3-diol-1,17-dione (−4.64), octatriacontyl pentafluoropropionate (−6.96), pregn-5-en-20-one, 11-(acetyloxy)-3,14-dihydroxy-12-(2-hydroxy-3-methyl-1-oxobutoxy)-,(3ß,11α,12ß,14ß) (−5.03), and cyclohexamine, N-n-butyl-1-[2-thionaphthenyl] (−5.5) | |||
| Jain et al. [30] | Dexamethasone: −7.9 | Flavonoids: chrysin (−8.1), and galangin (−8.2) | Chrysin and galangin showed higher binding affinity than dexamethasone |
| Refaat et al. [27] | Remdesivir: −165.9; faviripavir: −46.3; hydroxychloroquine: −79.8 | Rutin (−94.3), caffeic acid phenethyl ester (−77.8), quercetin (−67.8), kaempferol (−62.3), pinocembrin (−60.5), pinobanksin (−77.4), galangin (−59.5), chrysin (−66.2), p-cumaric acid (−56.5), and benzoic acid (−40.4) | Rutin had the highest binding affinity compared to the other 9 compounds which was higher than favipiravir and hydroxychloroquine but lower than remdesivir |
| Spike protein subunit 2 | |||
| Harisna et al. [25] | Pravastatin: −7.3 | Flavones: 3′-methoxydaidzein (−7.6), 3′-methoxydaidzin (−8.3), genistin (−8.3), xanthomicrol (−7.0), 3′,5,6,7-tetrahydroxy-4′-methoxyisofavone (−7.8), methylophiopogonone A (−8.2), 3′4′,7-trihydroxyfavanone (−7.6), moslosoofavone (−7.4), luteolin (−7.7), 2′,6′-dihydroxy-4′ methoxydihydrochalcone (−6.7), chrysoeriol (−7.7), jaceosidin (−7.3), (3R)-7,2′,3′-trihydroxy-4′-methoxyisofavanone (−7.5), and neobavaisofavone (−8.1) | 3′-Methoxydaidzin and genistin had the highest binding affinity compared to the other 20 compounds, which was higher than pravastatin |
| Flavonols: 3′-deoxysappanol (−7.0) | |||
| Phenolic acid: cinnamic acid (−5.3), caffeic acid (−5.5), 2,5-dimethyl-7-hydroxychromone (−6.1), isoferulic acid (−5.6), dimethylcaffeic acid (−5.7), benzyl caffeate (−6.5), and isoaloeresin D (−7.8) | |||
MLN-4760: (S,S)-2-{1-carboxy-2-[3-(3,5-dichlorobenzyl)-3H-imidazol4-yl]-ethylamino}-4-methylpentanoic acid; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
3.3.2. Propolis or honey against SARS-CoV-2 main protease enzyme
A total of six in silico studies were conducted using the SARS-CoV-2 main protease enzyme as the target enzyme [16], [25], [26], [27], [28], [29]. Several compounds from propolis had high binding affinity to the SARS-CoV-2 main protease enzyme, including octacosane (−7.39 kcal/mol) [16], 3′-methoxydaidzin (−7.7 kcal/mol) [25], sulabiroins A (−8.1 kcal/mol) [26], rutin (−92.8 kcal/mol) [27], glyasperin A (−7.8 kcal/mol) [28], broussoflavonol F (−7.8 kcal/mol) [28], and kaempferol (−7.8 kcal/mol) [29] (Table 2).
3.3.3. Interaction of propolis or honey with SARS-CoV-2 RNA-dependent RNA polymerase
SARS-CoV-2 RNA-dependent RNA polymerase was used as the target enzyme in two studies [16], [29]. One study reported that octatriacontyl pentafluoropropionate had the highest binding affinity to SARS-CoV-2 RNA-dependent RNA polymerase (−8.20 kcal/mol), which was higher than remdesivir (−6.77 kcal/mol) [16]. Another study indicated that ellagic acid (−6.4 kcal/mol) had the highest binding affinity to SARS-CoV-2 RNA-dependent RNA polymerase [29] (Table 2).
3.3.4. Propolis and honey against SARS-CoV-2 spike protein
Three studies used SARS-CoV-2 spike protein subunit 1 [16], [27], [30], while one study used SARS-CoV-2 spike protein subunit 2 as their target proteins [25]. Several compounds showed positive binding affinity to SARS-CoV-2 spike protein subunit 1. Octatriacontyl pentafluoropropionate had the highest binding affinity in one study [16], while rutin (−94.3 kcal/mol) had the highest binding affinity in another study [27]. Another study showed the positive binding affinities of chrysin (−8.1 kcal/mol) and galangin (−8.2 kcal/mol) [30] to SARS-CoV-2 spike protein subunit 1 (Table 2). A study that used SARS-CoV-2 spike protein subunit 2 as the target protein indicated that 3′-methoxydaidzin (−8.3 kcal/mol) and genistin (−8.3 kcal/mol) had positive binding affinities to the spike protein [25] (Table 2).
3.4. Results from clinical studies
3.4.1. Findings from case report or case series studies
Two case reports and one case series were identified in our literature search [18], [31], [32]. The case of a 52-year-old female patient with mild COVID-19 was reported in Brazil [31]. A 30% propolis solution was used as her main treatment, along with a healthy diet and adequate hydration. Forty-five drops, three times a day of the product were administered for 14 d. The patient was clinically improved after 12 d of treatment. In addition, the patient had a negative reverse transcription polymerase chain reaction test result for SARS-CoV-2 after 12 d of treatment (Table 3 and Table 4 ).
Table 3.
Baseline characteristics of clinical studies.
| Study | Country | Design | Sample size (intervention/comparator) | Characteristics | Intervention | Administration | Co-treatment | Comparator | Duration of assessment |
|---|---|---|---|---|---|---|---|---|---|
| Case report | |||||||||
| Fiorini et al. [31] | Brazil | Case report | 1/0 | 52 years old woman/mild | Brazilian green propolis | Brazilian green propolis at a dose of 45 drops, three times a day | At home, the patient maintained a healthy diet and adequate hydration | Not available | 14 days |
| Zorlu et al. [32] | Turkey | Case report | 1/0 | 38 years old male patient/severe | Anatolian propolis | Anatolian propolis given by dropping it into drinking water for 20 drops/d and increase to 80 drops/d | Hydroxychloroquine, favipiravir, steroid, nebulizer, oxygen support; IV moxifloxacin, and tocilizumab | Not available | 1 month |
| Case series | |||||||||
| El Sayed et al. [18] | Egypt | One arm retrospective study for treatment | 20/0 | COVID-19 positive patients | TaibUVID | Oral solution for asymptomatic case, and inhalation for moderate and severe case | Pharmacological protocol (undefined) for all, and one patient received oxygen therapy | Not available | > 10 days |
| One arm retrospective study for prevention | 20/0 | Non-COVID-19 subjects | TaibUVID | One TaibUVID once daily | None | Not available | 14 days | ||
| Randomized control trial | |||||||||
| Ashraf et al. [17] | Pakistan | Double-blinded two-arm design | 157/156 | Hospitalized patients | Honey (1 g/kg) plus encapsulated Nigella sativa seeds (80 mg/kg) | Orally in 2–3 divided doses daily for up to 13 d | Antipyretics, antibiotics, anticoagulants, steroids, supplemental oxygen, and mechanical ventilation | Standard care | 13 days |
| Kosari et al. [33] | Iran | Unspecified two-arm design | 25/25 | Adult outpatients | Syrup containing 1.6 mg of Hyoscyamus niger methanolic extract plus 450 mg of propolis per 10 mL | 10 mL, 3 times a day for 6 d | Routine medications (not well-defined) | Standard care | 6 days |
| Silveira et al. [34] | Brazil | Open-label three-arm design | 82/40/42 | Hospitalized adult or elderly patients | Standardized Brazilian green propolis extract | 400 and 800 mg/d plus standard care | Supplemental oxygen, noninvasive or invasive ventilation, corticosteroids, antibiotics and/or antiviral agents, vasopressor support, renal replacement therapy, intra-aortic balloon pump, extracorporeal membrane oxygenation | Standard care | 28 days |
Table 4.
Efficacy and safety of propolis or bee products to treat or prevent COVID-19 and related clinical outcomes.
| Study | Outcome | Duration of assessment | Comparator group | Intervention group | P value |
|---|---|---|---|---|---|
| Case report | |||||
| Fiorini et al. [31] | Clinical symptom improvement and RT-PCR | 12 days | Not applicable | After 12 days of treatment, the patient’s general clinical symptoms improved significantly, and the patient recovered with negative RT-PCR. | Not available |
| Zorlu et al. [32] | Clinical symptom improvement | 1 month | Not applicable | The patient’s clinical symptoms improved at day 7 and the patient was discharged at day 10. At the health check-up visit 1 month later, the patient had no complaint except the forced exertion dyspnea. Normal blood test was observed and abnormal thorax computed tomography completely regressed | Not available |
| Case series | |||||
| El Sayed et al. [18] | Treatment: clinical symptom improvement and RT-PCR | > 9 days | Not applicable | Clinical symptom improvement: 14 patients (70%) improved within 4 days; 5 patients (25%) improved in 5–10 days; 1 patient (5%) improved in > 10 days; all patients had negative RT-PCR after the treatment. | < 0.01 |
| Prevention: SARS-CoV2 infection | 14 days | Not applicable | 14 patients (70%) did not get infected; 6 patients (30%) got infected | NR | |
| Safety data | 3 patients (18.8%) reported non-serious side effects including sweating, hyperglycemia and diarrhea | ||||
| Randomized controlled trial | |||||
| Ashraf et al. [17] | Time taken for alleviation of symptoms (d, median [IQR]) | 13 days | Moderate: 7 (7–8) | Moderate: 4 (3–4) | < 0.0001 |
| Severe: 13 (9–15) | Severe: 6 (5–7) | < 0.0001 | |||
| Time taken for SARS-CoV-2 RT-PCR clearance (d, median [IQR]) | Moderate: 10 (9–12) | Moderate: 6 (6–7) | < 0.0001 | ||
| Severe: 12 (11–17) | Severe: 8.5 (8–9) | < 0.0001 | |||
| Clinical grading score at day 6 (median [IQR]) | Moderate: 1 (1–2) | Moderate: 0 (0–1) | < 0.0001 | ||
| Severe: 3 (3–4) | Severe: 1.5 (0–2) | < 0.0001 | |||
| Degree of fever at day 4 (median [IQR]) | Moderate: 2 (1–2) | Moderate: 0 (0–1) | < 0.0001 | ||
| Severe: 2 (1–3) | Severe: 2 (1–2) | 0.0001 | |||
| CRP level at day 6 (mg/L, mean ± SD) | Moderate: 9.44 ± 4.94 | Moderate: 6.15 ± 2.45 | < 0.0001 | ||
| Severe: 23.32 ± 8.73 | Severe: 15.83 ± 7.17 | < 0.0001 | |||
| Severity of symptoms at day 8 (median [IQR]) | Moderate: 0 (0–2) | Moderate: 0 (0–0) | < 0.0001 | ||
| Severe: 2 (1–3) | Severe: 0 (0–1) | < 0.0001 | |||
| Clinical grading score at day 10 (median [IQR]) | Moderate: 1 (1–2) | Moderate: 1 (1–2) | < 0.0001 | ||
| Severe: 4 (2–4) | Severe: 1 (1–1) | < 0.0001 | |||
| 30-day mortality (n) | Moderate: 1 | Moderate: 0 | 0.49 | ||
| Severe: 10 | Severe: 2 | 0.029 | |||
| Safety data | No adverse event was noted with intervention groups | ||||
| Kosari et al. [33] | Dry cough score (mean ± SD) | 2–6 day | Day 2: 1.0 | Day 2: 0.5 | < 0.05 |
| Day 4: 0.7 | Day 4: 0.2 | < 0.001 | |||
| Day 6: 0.4 | Day 6: 0.1 | < 0.01 | |||
| Shortness of breath score (mean ± SD) | Day 2: 0.7 | Day 2: 0.3 | < 0.05 | ||
| Day 4: 0.5 | Day 4: 0.0 | < 0.001 | |||
| Day 6: 0.2 | Day 6: 0.0 | < 0.01 | |||
| Sore throat score (mean ± SD) | Day 2: 0.5 | Day 2: 0.2 | < 0.05 | ||
| Day 4: 0.3 | Day 4: 0.1 | < 0.05 | |||
| Day 6: 0.3 | Day 6: 0.0 | < 0.01 | |||
| Chest pain score (mean ± SD) | Day 2: 0.5 | Day 2: 0.2 | NR | ||
| Day 4: 0.3 | Day 4: 0.1 | < 0.05 | |||
| Day 6: 0.2 | Day 6: 0.0 | < 0.05 | |||
| Fever score (mean ± SD) | Day 2: 0.2 ± 0.4 | Day 2: 0.0 | < 0.05 | ||
| Day 4: 0.0 | Day 4: 0.0 | NR | |||
| Day 6: 0.0 | Day 6: 0.0 | NR | |||
| Headache score (mean ± SD) | Day 2: 0.52 ± 0.7 | Day 2: 0.04 ± 0.2 | NR | ||
| Day 4: 0.5 ± 0.6 | Day 4: 0.0 | NR | |||
| Day 6: 0.2 ± 0.4 | Day 6: 0.0 | NR | |||
| Muscular pain score (mean ± SD) | Day 2: 0.7 ± 0.8 | Day 2: 0.4 ± 0.2 | NR | ||
| Day 4: 0.6 ± 0.7 | Day 4: 0.1 ± 0.3 | < 0.01 | |||
| Day 6: 0.4 ± 0.6 | Day 6: 0.0 | < 0.01 | |||
| Diarrhea score (mean ± SD) | Day 2: 0.2 ± 0.5 | Day 2: 0.0 | NR | ||
| Day 4: 0.1 ± 0.3 | Day 4: 0.0 | NR | |||
| Day 6: 0.1 ± 0.3 | Day 6: 0.0 | NR | |||
| Runny nose score (mean ± SD) | Day 2: 0.2 ± 0.4 | Day 2: 0.0 | < 0.05 | ||
| Day 4: 0.2 ± 0.4 | Day 4: 0.0 | < 0.05 | |||
| Day 6: 0.1 ± 0.3 | Day 6: 0.0 | NR | |||
| Sore throat and larynx score (mean ± SD) | Day 2: 0.5 ± 0.7 | Day 2: 0.2 ± 0.5 | < 0.05 | ||
| Day 4: 0.3 ± 0.6 | Day 4: 0.1 ± 0.2 | < 0.05 | |||
| Day 6: 0.2 ± 0.5 | Day 6: 0.0 | NR | |||
| Fatigue score (mean ± SD) | Day 2: 1.0 ± 0.9 | Day 2: 0.7 ± 0.6 | NR | ||
| Day 4: 0.9 ± 0.8 | Day 4: 0.4 ± 0.5 | < 0.05 | |||
| Day 6: 0.7 ± 0.8 | Day 6: 0.1 ± 0.3 | < 0.001 | |||
| Anorexia score (mean ± SD) | Day 2: 0.6 ± 0.7 | Day 2: 0.1 ± 0.2 | NR | ||
| Day 4: 0.4 ± 0.5 | Day 4: 0.0 | NR | |||
| Day 6: 0.2 ± 0.4 | Day 6: 0.0 | NR | |||
| Trembling score (mean ± SD) | Day 2: 0.2 ± 0.4 | Day 2: 0.0 | NR | ||
| Day 4: 0.1 ± 0.2 | Day 4: 0.0 | NR | |||
| Day 6: 0.1 ± 0.2 | Day 6: 0.0 | NR | |||
| Nausea score (mean ± SD) | Day 2: 0.1 ± 0.3 | Day 2: 0.0 | NR | ||
| Day 4: 0.1 ± 0.3 | Day 4: 0.0 | NR | |||
| Day 6: 0.1 ± 0.2 | Day 6: 0.0 | NR | |||
| Vomit score (mean ± SD) | Day 2: 0.0 | Day 2: 0.0 | NR | ||
| Day 4: 0.0 | Day 4: 0.0 | NR | |||
| Day 6: 0.0 | Day 6: 0.0 | NR | |||
| Dizziness score (mean ± SD) | Day 2: 0.2 ± 0.4 | Day 2: 0.1 ± 0.2 | NR | ||
| Day 4: 0.2 ± 0.4 | Day 4: 0.0 | < 0.05 | |||
| Day 6: 0.1 ± 0.2 | Day 6: 0.0 | NR | |||
| Abdominal pain score (mean ± SD) | Day 2: 0.4 ± 0.6 | Day 2: 0.0 | < 0.001 | ||
| Day 4: 0.3 ± 0.6 | Day 4: 0.0 | < 0.01 | |||
| Day 6: 0.2 ± 0.4 | Day 6: 0.0 | < 0.01 | |||
| Safety data | Not available | One patient reported hot flashes | |||
| Silveira et al. [34] | Length of hospital stay (d, median [IQR]) | 28 days | 12 (8–16) | 400 mg/d: 7 (5–12) | 0.049 |
| 800 mg/d: 6 (5–11) | 0.009 | ||||
| Oxygen therapy dependency time (d, median [IQR]) | 5 (3–11) | 400 mg/d: 3 (1–6) | 0.470 | ||
| 800 mg/d: 2 (1–5) | 0.710 | ||||
| Acute kidney injury (n) | 10 | 400 mg/d: 5 | 0.305 | ||
| 800 mg/d: 2 | 0.048 | ||||
| Renal replacement therapy (n) | 3 | 400 mg/d: 1 | 0.415 | ||
| 800 mg/d: 0 | 0.994 | ||||
| Invasive ventilation after randomization (n) | 8 | 400 mg/d: 2 | 0.065 | ||
| 800 mg/d: 3 | 0.107 | ||||
| Vasoactive agent (n) | 10 | 400 mg/d: 4 | 0.161 | ||
| 800 mg/d: 3 | 0.098 | ||||
| ICU after randomization (n) | 6 | 400 mg/d: 0 | 0.993 | ||
| 800 mg/d: 5 | 0.601 | ||||
| Safety data | No patient had propolis treatment discontinued due to side effects. The percentages of patients experiencing adverse events were not different significantly among the three groups. The most severe adverse event overall was shock/need for vasoactive drugs and acute respiratory failure | ||||
CRP: C-reactive protein; ICU: intensive care unit; IQR: inter-quartile range; NR: not reported; RT-PCR: real-time polymerase chain reaction; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; SD: standard deviation.
The case of a 38-year-old male patient with severe COVID-19 was reported in Turkey [32]. A 30% propolis solution was used as his adjuvant therapy along with hydroxychloroquine, favipiravir and tocilizumab. Twenty drops of the propolis solution were administered per day initially, with the dose increasing to 80 drops per day. The patient’s clinical symptoms were improved after seven days of treatment, and the patient was discharged on day 10 (Table 3 and Table 4).
A retrospective case series study was reported in Egypt [18]. In this report, 20 subjects used a natural honey product for COVID-19 prevention, and the other 20 subjects used a natural honey product for COVID-19 treatment. The natural honey product contained Nigella sativa seeds, cistus, clove oil, chamomile, natural honey, fennel, and senna. In the COVID-19 prevention group, six subjects (30.0%) tested positive for COVID-19, via nasopharyngeal swab and a PCR test, after using the product. In the COVID-19 treatment group, four patients (20.0%) with mild or no symptoms used an oral form of the product, while 16 patients (80.0%) with moderate to severe symptoms used an inhaled form of the product. According to the study, 14 patients (70.0%) clinically improved within four days of diagnosis, five patients (25.0%) improved within five to ten days, and one patient (5.0%) improved after more than ten days. In the end, all patients showed clinical improvement and had negative nasopharyngeal swab PCR tests (Table 3 and Table 4).
3.4.2. Results from RCTs
A total of three RCTs were identified in the literature search [17], [33], [34]. All studies used propolis or honey as an adjuvant therapy along with standard of care, such as antipyretics, corticosteroids, or supplemental oxygen.
A double-blinded RCT was conducted [17] to assess the effect of honey on COVID-19 symptoms. A total of 313 hospitalized patients received 1 g of honey plus 80 mg/(kg·d) of Nigella sativa seed with standard of care or standard of care with placebo. The study showed that patients with moderate symptoms receiving honey had a lower median time to alleviation of symptoms than patients with standard of care plus placebo (4 vs 7 d, P < 0.001). Similarly, the time to alleviation of symptoms was shorter in patients with severe cases who received honey than in those who received placebo (6 vs 13 d, P < 0.001). Patients receiving honey also had a lower median time to viral clearance than patients with placebo (P < 0.001 for both moderate and severe patients). In addition, patients who received honey products had improved clinical symptoms compared to those receiving placebo (Table 3 and Table 4).
An RCT was conducted to determine the efficacy of propolis in combination with H. niger extract, compared to the standard of care in COVID-19 patients visiting outpatient clinics [33]. Fifty COVID-19 patients aged 18–65 were assigned to receive 450 mg of propolis plus 1.6 mg of methanolic extract of H. niger three times/d for 6 d or standard of care. Clinical symptoms, including dry cough score, shortness of breath score, sore throat score, chest pain score, and other clinical symptoms, were assessed on days two, four, and six. The study found that patients who had received propolis with H. niger extract had lower scores for some clinical symptoms, including dry cough (P < 0.01), shortness of breath (P < 0.01), sore throat (P < 0.01), and chest pain (P < 0.05), at day six. No significant differences were observed for other clinical symptom scores (Table 3 and Table 4).
One open-label RCT was conducted in Brazil to compare the efficacy of Brazilian green propolis against the standard of care in hospitalized COVID-19 patients [34]. A total of 124 patients were assigned to receive 400 mg/d of Brazilian green propolis plus standard of care or 800 mg/d of Brazilian green propolis plus standard of care or just standard of care for 7 d. Length of stay (LOS), oxygen therapy dependency time, acute kidney injury, renal replacement therapy, invasive ventilation, vasoactive agents, and ICU admission were assessed. The study showed that patients who had received Brazilian green propolis had lower median LOS than patients receiving standard of care alone. The mean difference in LOS between patients with 400 mg/d of Brazilian green propolis and patients with standard of care only was −3.03 (−6.23, −0.07) days (P = 0.049), while the mean difference in LOS between patients with 800 mg/d of Brazilian green propolis and patients with standard of care only was −3.88 (−7.00, −1.09) days (P = 0.009). On the other hand, no statistically significant difference was observed for any comparisons of oxygen therapy dependency time. The mean difference in oxygen therapy dependency time between patients with 400 mg/d of Brazilian green propolis and patients with standard of care only was −2.13 (−7.84, 3.57) days (P = 0.470), while the mean difference in oxygen therapy dependency time between patients with 400 mg/d of Brazilian green propolis and patients with standard of care only was −0.99 days (P = 0.710; median 2 vs 5 d). No significant difference in other outcomes between Brazilian green propolis and standard of care was observed (Table 3 and Table 4).
3.5. Safety outcomes
Only four clinical studies reported safety outcomes [17], [18], [33], [34]. One study reported no adverse events related to propolis or honey [17], while three studies reported some adverse events related to propolis or honey. One study [18] reported non-serious adverse events from three patients. The adverse events were sweating, hyperglycemia, and diarrhea. The second study reported one patient experienced hot flashes [33], and the third reported no significant difference in adverse events between patients receiving propolis and standard of care [34].
4. Discussion
This study summarized possible active compounds and mechanisms of propolis and honey against SARS-CoV-2 infection and the current evidence concerning the effect of propolis and honey in COVID-19 prevention and treatment. Rutin, glyasperin A, 3′-methoxydaidzin, and octatriacontyl pentafluoropropionate were identified as the most likely compounds that could interact with SARS-CoV-2 through ACE2, main protease enzyme, RNA-dependent RNA polymerase or spike protein. The current limited clinical evidence shows that propolis and honey might improve clinical symptoms of COVID-19 and might decrease time to viral clearance. Clinical evidence concerning propolis and honey for COVID-19 prevention is sparse.
One possible mechanism of propolis against SARS-CoV-2 infection is to interfere with viral entry, an essential process for viral infection [35]. In silico studies [7], [8], [16], [25], [27], [30] showed higher binding affinity of some active compounds of propolis to spike proteins or human ACE2 than positive controls. SARS-CoV-2 invades host cells by engaging its spike protein S1 subunit with human ACE2. It also uses the spike protein S2 subunit to bind the virus to the host cell membrane [36], [37]. The interaction between propolis and human ACE2 or viral spike protein S1 and S2 could disrupt the viral entry process.
Another possible mechanism is to interfere with the viral replication process. SARS-CoV-2 uses the main protease to cleave the viral genome polyproteins, pp1a and pp1ab, into functional proteins which initiate viral replication by forming a replication complex with RNA-dependent RNA polymerase. Establishing the replication complex is crucial for the viral replication process [37], [38], [39]. Some studies revealed interactions between some chemical constituents of propolis and the SARS-CoV-2 main protease [16], [25], [26], [27], [28], [29]; some studies also showed that compounds present in propolis could inhibit RNA-dependent RNA polymerase activity [16], [29]. These interactions between propolis and SARS-CoV-2 proteins inhibit viral replication. Propolis contains compounds that interact with SARS-CoV-2 through several possible mechanisms, which might synergistically increase its antiviral activities.
Clinical studies have indicated the positive effect of propolis and honey for improvement of some clinical symptoms such as dry cough, sore throat, and fever in patients with both mild and moderate to severe symptoms. These benefits were due to propolis’s ability to hasten viral clearance, as observed in in silico studies. In addition, the clinical improvement might be due to other therapeutic activities of propolis. Some active compounds in propolis and honey have anti-inflammatory activities [40], [41]. Galangin, one of the active compounds in propolis, has been shown to inhibit tumor necrosis factor-α and interleukin-8, leading to a decrease in tissue inflammation and clinical symptoms.
The dose and dosage forms of propolis and honey used in adjuvant COVID-19 treatments varied among clinical studies. It varied from 400–800 mg/d of propolis or 1 g/d of honey. The dosage forms were oral or inhalation. In addition, propolis or honey was taken alone or in a combination with other herbs. Thus, it is still inconclusive which dose and dosage forms are the most effective. It is important for clinicians to critically assess whether propolis or honey is appropriate for their COVID-19 patients.
The clinical studies summarized here did not report which viral types were studied. Based on the time of the study conducted, the viral type could have been the wild type. The effect of propolis or honey could be different for other variants, because SARS-CoV-2 mutates its spike protein, which is one of the possible target proteins for propolis. The observed clinical effect might be different for other variants of concern, including α, β, γ, or δ variants.
Although we comprehensively searched through several databases, only three RCTs were identified. In addition, the RCTs were conducted on different populations, using different interventions and different outcome measures. Therefore, meta-analysis could not be performed. Moreover, all studies were assessed as having a high risk of bias, because they incompletely reported some important methodology and measured many clinical parameters, leading to the possibility of biases. These limitations suggest that readers should interpret the clinical findings with caution.
5. Conclusion
This review signifies a potential effect of propolis and honey on COVID-19 as an adjuvant treatment. In silico studies showed that compounds in propolis could interact with target proteins of SARS-CoV-2, potentially interfering with viral entry or viral RNA replication. Clinical studies revealed that propolis or honey could probably improve clinical COVID-19 symptoms and decrease viral clearance time. However, clinical evidence of the effect, appropriate dose, and suitable dosage forms is still limited by the small number of studies and small sample sizes. Future investigations should be undertaken to strengthen the understanding of potential benefits from honey and propolis on COVID-19 prevention and treatment.
Funding
This study has no funding support.
Authors’ contribution
All authors designed the study’s hypothesis and protocol. RK and RW were responsible for data collection. All authors participated in data analysis. All authors also participated in data interpretation, manuscript preparation and review, table and figure preparation, and the scientific discussion of the data. All authors approved the final version of this manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.joim.2022.01.008.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.World Health Organization. Timeline: WHO’s COVID-19 response (2021-10-25) [2021-10-26]. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/interactive-timeline.
- 2.World Health Organization. WHO coronavirus (COVID-19) dashboard. (2021-10-25) [2021-10-26]. https://covid19.who.int/.
- 3.Odusanya O., Odugbemi B., Odugbemi T., Ajisegiri W. COVID-19: a review of the effectiveness of non-pharmacological interventions. Niger Postgrad Med J. 2020;27(4):261–267. doi: 10.4103/npmj.npmj_208_20. [DOI] [PubMed] [Google Scholar]
- 4.National Institutes of Health. Coronavirus disease 2019 (COVID-19) treatment guidelines. (2021-10-19) [2021-10-26]. https://www.covid19treatmentguidelines.nih.gov/. [PubMed]
- 5.Saghir S.AM., AlGabri N.A., Alagawany M.M., Attia Y.A., Alyileili S.R., Elnesr S.S., et al. Chloroquine and hydroxychloroquine for the prevention and treatment of COVID-19: a fiction, hope or hype? An updated review. Ther Clin Risk Manag. 2021;17:371–387. doi: 10.2147/TCRM.S301817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Attia Y.A., Alagawany M.M., Farag M.R., Alkhatib F.M., Khafaga A.F., Abdel-Moneim A.-M., et al. Phytogenic products and phytochemicals as a candidate strategy to improve tolerance to coronavirus. Front Vet Sci. 2020;7:573159. doi: 10.3389/fvets.2020.573159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guler H.I., Tatar G., Yildiz O., Belduz A.O., Kolayli S. Investigation of potential inhibitor properties of ethanolic propolis extracts against ACE-II receptors for COVID-19 treatment by molecular docking study. Arch Microbiol. 2021;203(6):3557–3564. doi: 10.1007/s00203-021-02351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khayrani A.C., Irdiani R., Aditama R., Pratami D.K., Lischer K., Ansari M.J., et al. Evaluating the potency of Sulawesi propolis compounds as ACE-2 inhibitors through molecular docking for COVID-19 drug discovery preliminary study. J King Saud Univ Sci. 2021;33(2):101297. doi: 10.1016/j.jksus.2020.101297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachevski D., Damevska K., Simeonovski V., Dimova M. Back to the basics: propolis and COVID-19. Dermatol Ther. 2020;33(4) doi: 10.1111/dth.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pascoal A., Feás X., Dias T., Dias L.G., Estevinho L.M. In: Microbiology for surgical infections. Kon K., Rai M., editors. Academic Press; Amsterdam: 2014. The role of honey and propolis in the treatment of infected wounds; pp. 221–234. [Google Scholar]
- 11.Marcucci M.C. Propolis: chemical composition, biological properties and therapeutic activity. Apidologie. 1995;26(2):83–99. [Google Scholar]
- 12.Nolkemper S., Reichling J., Sensch K.H., Schnitzler P. Mechanism of herpes simplex virus type 2 suppression by propolis extracts. Phytomedicine. 2010;17(2):132–138. doi: 10.1016/j.phymed.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Ito J, Chang FR, Wang HK, Park YK, Ikegaki M, Kilgore N, et al. Anti-AIDS agents. 48.(1) Anti-HIV activity of moronic acid derivatives and the new melliferone-related triterpenoid isolated from Brazilian propolis. J Nat Prod 2001;64(10):1278–81. [DOI] [PubMed]
- 14.Vynograd N., Vynograd I., Sosnowski Z. A comparative multi-centre study of the efficacy of propolis, acyclovir and placebo in the treatment of genital herpes (HSV) Phytomedicine. 2000;7(1):1–6. doi: 10.1016/S0944-7113(00)80014-8. [DOI] [PubMed] [Google Scholar]
- 15.Berretta A.A., Silveira M.A.D., Cóndor Capcha J.M., De Jong D. Propolis and its potential against SARS-CoV-2 infection mechanisms and COVID-19 disease: running title: propolis against SARS-CoV-2 infection and COVID-19. Biomed Pharmacother. 2020;131:110622. doi: 10.1016/j.biopha.2020.110622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.H Elwakil B., Shaaban M.M., Bekhit A.A., El-Naggar M.Y., Olama Z.A. Potential anti-COVID-19 activity of Egyptian propolis using computational modeling. Future Virol. 2021;16(2):107–116. [Google Scholar]
- 17.Ashraf S, Ashraf S, Ashraf M, Imran MA, Kalsoom L, Siddiqui UN, et al. Honey and Nigella sativa against COVID-19 in Pakistan (HNS-COVID-PK): a multi-center placebo-controlled randomized clinical trial. medRxiv 2020.
- 18.El Sayed S.M., Aboonq M.S., El Rashedy A.G., Aljehani Y.T., Abou El-Magd R.M., Okashah A.M., et al. Promising preventive and therapeutic effects of TaibUVID nutritional supplements for COVID-19 pandemic: towards better public prophylaxis and treatment (a retrospective study) Am J Blood Res. 2020;10(5):266–282. [PMC free article] [PubMed] [Google Scholar]
- 19.Bowden RA. Building confidence in geological models. London: Special Publications; 2004.
- 20.Pollard S.J.T., Davies G.J., Coley F., Lemon M. Better environmental decision making—recent progress and future trends. Sci Total Environ. 2008;400(1-3):20–31. doi: 10.1016/j.scitotenv.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 21.Klimisch H.-J., Andreae M., Tillmann U. A systematic approach for evaluating the quality of experimental toxicological and ecotoxicological data. Regul Toxicol Pharmacol. 1997;25(1):1–5. doi: 10.1006/rtph.1996.1076. [DOI] [PubMed] [Google Scholar]
- 22.JBI. Checklist for case reports: critical appraisal tools for use in JBI systematic reviews. [2021-10-26]. https://jbi.global/critical-appraisal-tools.
- 23.National Heart, Lung, and Blood Institute. Quality assessment tools for case series studies. (2021-07) [2021-10-26]. https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools.
- 24.Sterne JAC, Savovic J, Page MJ, Elbers RG, Blencowe NS, Boutron I, et al. RoB 2: a revised tool for assessing risk of bias in randomised trials. BMJ 2019; 366: l4898. [DOI] [PubMed]
- 25.Harisna A.H., Nurdiansyah R., Syaifie P.H., Nugroho D.W., Saputro K.E., Firdayani, et al. In silico investigation of potential inhibitors to main protease and spike protein of SARS-CoV-2 in propolis. Biochem Biophys Rep. 2021;26:100969. doi: 10.1016/j.bbrep.2021.100969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dewi L.K., Sahlan M., Pratami D.K., Agus A., Agussalim S.A. Identifying propolis compounds potential to be COVID-19 therapies by targeting SARS-CoV-2 main protease. Int J Appl Pharm. 2021;13(Suppl 2):103–110. [Google Scholar]
- 27.Refaat H., Mady F.M., Sarhan H.A., Rateb H.S., Alaaeldin E. Optimization and evaluation of propolis liposomes as a promising therapeutic approach for COVID-19. Inter J Pharm. 2021;592:120028. doi: 10.1016/j.ijpharm.2020.120028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahlan M., Irdiani R., Flamandita D., Aditama R., Alfarraj S., Ansari M.J., et al. Molecular interaction analysis of Sulawesi propolis compounds with SARS-CoV-2 main protease as preliminary study for COVID-19 drug discovery. J King Saud Univ Sci. 2021;33(1):101234. doi: 10.1016/j.jksus.2020.101234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaldam M.A., Yahya G., Mohamed N.H., Abdel-Daim M.M., Al Naggar Y. In silico screening of potent bioactive compounds from honeybee products against COVID-19 target enzymes. Environ Sci Pollut Res Int. 2021;28(30):40507–40514. doi: 10.1007/s11356-021-14195-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain A.S., Sushma P., Dharmashekar C., Beelagi M.S., Prasad S.K., Shivamallu C., et al. In silico evaluation of flavonoids as effective antiviral agents on the spike glycoprotein of SARS-CoV-2. Saudi J Biol Sci. 2021;28(1):1040–1051. doi: 10.1016/j.sjbs.2020.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiorini A.C., Scorza C.A., de Almeida A.G., Fonseca M.C.M., Finsterer J., Fonseca F.L.A., et al. Antiviral activity of Brazilian green propolis extract against SARS-CoV-2 (severe acute respiratory syndrome-coronavirus 2) infection: case report and review. Clinics (Sao Paulo) 2021;76 doi: 10.6061/clinics/2021/e2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zorlu D. COVID-19 and anatolian propolis: a case report. Acta Medica Mediterranea. 2021;37(2):1229–1233. [Google Scholar]
- 33.Kosari M., Noureddini M., Khamechi S.P., Najafi A., Ghaderi A., Sehat M., et al. The effect of propolis plus Hyoscyamus niger L. methanolic extract on clinical symptoms in patients with acute respiratory syndrome suspected to COVID-19: a clinical trial. Phytother Res. 2021;35(7):4000–4006. doi: 10.1002/ptr.7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silveira M.A.D., De Jong D., Berretta A.A., Galvão E.B.D.S., Ribeiro J.C., Cerqueira-Silva T., et al. Efficacy of Brazilian green propolis (EPP-AF®) as an adjunct treatment for hospitalized COVID-19 patients: a randomized, controlled clinical trial. Biomed Pharmacother. 2021;138:111526. doi: 10.1016/j.biopha.2021.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni W., Yang X., Yang D., Bao J., Li R., Xiao Y., et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit Care. 2020;24(1):422. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg Med Chem Lett. 2020;30(17):127377. doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu W., Chen C.Z., Gorshkov K., Xu M., Lo D.C., Zheng W. RNA-dependent RNA polymerase as a target for COVID-19 drug discovery. SLAS Discov. 2020;25(10):1141–1151. doi: 10.1177/2472555220942123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pahlavani N., Malekahmadi M., Firouzi S., Rostami D., Sedaghat A., Moghaddam A.B., et al. Molecular and cellular mechanisms of the effects of propolis in inflammation, oxidative stress and glycemic control in chronic diseases. Nutri Metab. 2020;17(1):65. doi: 10.1186/s12986-020-00485-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramos A., Miranda J. Propolis: a review of its anti-inflammatory and healing actions. J Venom Anim Toxins Incl Trop Dis. 2007;13(4):697–710. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.