Abstract
In recent years, mass spectrometry (MS) is increasingly attracting interests for clinical applications, which also calls for technical innovations to make a transfer of MS from conventional analytical laboratories to clinics. The system design and analysis procedure should be friendly for novice users and appliable for on-site clinical diagnosis. In addition, the analysis result should be auto-interpreted and reported in formats much simpler than mass spectra. This motivates new ideas for developments in all the aspects of MS. In this review, we report recent advances of direct sampling ionization and miniature MS system, which have been developed targeting clinical and even point-of-care analysis. We also discuss the trend of the development and provide perspective on the technical challenges raised by diseases such as coronavirus SARS-CoV-2.
Keywords: Ambient ionization, Direct sampling, Point-of-care testing, Clinical diagnostics, Miniature mass spectrometry
1. Introduction
Mass spectrometry (MS) is a powerful analytical technology with high sensitivity and high specificity. For over a century, MS system has evolved as a major tool in analytical chemistry for manipulating ions and analyzing chemical and biological species. Analyte ions are processed in the electromagnetic fields of a mass spectrometer to measure their mass-to-charge ratio (m/z), while fragmentation information of the ions can facilitate structure identification and confirmation via tandem mass spectrometry (MS/MS). Owing to the high specificity and high sensitivity, the scope of MS is extended to the field of clinical applications in past decades [[1], [2], [3], [4]]. Since then, MS is increasingly applied to deal with clinical samples containing complicated matrices, such as tissues [5] and biofluids [6], and provides high-quality quantitative analysis even at low concentrations.
In analytical laboratories, MS is often regarded as a universal tool, applicable for any possible applications. To extract more sample information from an unknown sample, the sample has to be subjected to various separation and extraction treatments, such as gas chromatography (GC) [7] and liquid chromatography (LC) [8], prior to the MS analysis. The MS systems employed are expected to have higher resolutions, wider mass ranges, in addition to higher sensitivities. While the LC-MS method has been regarded as a gold standard in analytical laboratories. However, the rigorous sample preparation or purification processes required in the chromatography-based methods are labor- and time-consuming and the dedicate equipment that can provide high-performance analysis required are expensive. These limitations mean that the gold-standard LC-MS method is unsuitable for the real-time on-site clinical testing.
Technical barrier and high cost of MS are two major difficulties for its on-site clinical translation. During the past two decades, two remarkable techniques, ambient ionization [[9], [10], [11]] and miniature MS [12,13], emerged and their combination became a major solution to point-of-care (POC) MS analysis. The concept of ambient ionization was introduced as a solution for direct mass analysis with minimal sample preparation. This appeared and evolved independently from miniaturization of mass spectrometer; however, it had a significant implication to the research direction for miniature MS systems. If the focus of the traditional MS is to make MS a versatile tool, applicable for any fields of study, the goal of the miniature MS system is, or should be to allow the MS to be applied anywhere and by anybody who needs an answer related to chemical analysis right away. Here, we review recent advances on clinical MS and discuss its application potential for on-site testing.
2. MS analysis using ambient ionization: transferring MS from laboratories to clinics
In the early 2000s, ambient ionizations [[9], [10], [11]], including desorption electrospray ionization (DESI) in 2004 [14] and direct analysis in real time (DART) in 2005 [15], were introduced to enable direct sampling ionization of biological samples in their native environment without pretreatment. For the first time, complex mixtures, such as whole blood, urine and tissue samples, can be analyzed on-site directly without the need of sample preparation and chromatographic separation. Ambient ionization MS is, therefore, widely employed for analysis of therapeutic drugs, metabolites, and other bioactive species in biofluids. This technology has advanced rapidly in the last decade and presented a huge implication on transferring MS technology to non-expert users, such as nurses and physicians for clinical and POC disease diagnosis. For instance, towards rapid quantitative analysis, a series of direct sampling methods have been developed [[16], [17], [18]], including paper spray (PS) [19,20], extraction spray [21], and slug flow microextraction [22]. For instance, paper spray has shown a good performance in analysis of therapeutic drug monitoring, especially for those in whole bloods [23], dry blood spots [24], and urines [25]. The raw samples are first deposited onto a triangle paper substrate; then, by using a small amount of solvent and a high voltage, analytes in samples are desorbed and ionized through spray. A paper-based immunoassay platform that can generate stable and cleavable ionic probes as mass reporters has also been developed [18]. Patient--friendly MS analysis of specific biomarkers has been achieved with a high sensitivity. Note that ambient technologies used for chemical analysis have been extensively reviewed in literatures [[26], [27], [28], [29]]. This review will focus on the translation of the direct sampling method for on-site clinical use.
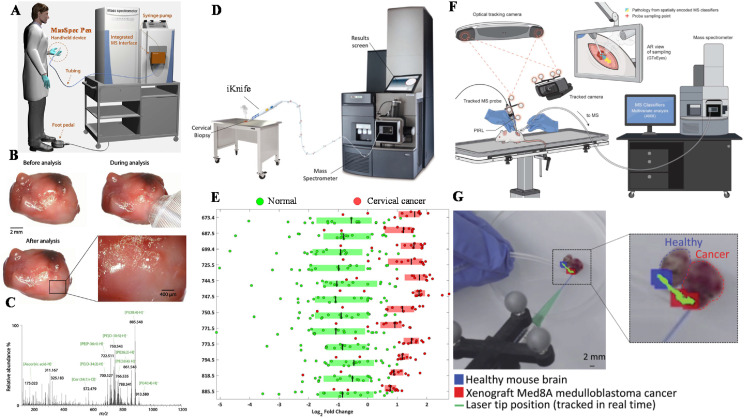
Ambient MS produces minimal damages to the sample, which also shows exceptional potential for tissue analysis. For instance, DESI is one of the ambient ionization methods mostly employed for this purpose. DESI employs a spray of charged micro-droplets to form a thin solvent film to extract the analytes from the sample, which are subsequently desorbed and ionized in the gas phase. DESI is powerful for molecular profiling in tissues and has been used for ex vivo cancer diagnosis and surgical margin evaluation of tissue sections and tissue smears [30]. Recently, a direct sampling probe, named MassSpec Pen [31], was developed to further enable on-site tissue profiling. Real-time nondestructive sampling and molecular diagnosis of tissues have been achieved (Fig. 1 a) [32]. The probe used water droplets to extract the sample analytes, thereby enabling gentle and controllable MS analysis of the tissue samples. From the optical images of a lung adenocarcinoma tissue sample, no observable damage to the tissue could be found before, during, and after the MasSpec Pen analysis (Fig. 1b). Abundant information including diagnostic metabolites, lipids, and proteins were obtained (Fig. 1c). Every three mass spectra were averaged for subsequent data processing processes. Through statistical analysis of human normal and cancerous tissue samples from 253 patients (95 lung samples, 57 ovary samples, 56 thyroid samples, and 45 breast samples), prediction of cancer with high sensitivity and specificity has been achieved. A rapid evaporative ionization MS (REIMS) method [33,34], also known as the intelligent knife (iKnife) [35], was also developed for the chemical analysis of electrosurgery-generated aerosols (Fig. 1d). The surgical aerosol is generated directly from tissue surfaces using standard electrosurgery devices. Through the analysis of dysregulation of lipid metabolism, successful discrimination of cancer from normal tissue at different tumor sites including the brain, breast, colon, ovary, and cervix have been achieved (Fig. 1e). A hand-held mass spectrometry desorption probe using picosecond infrared laser (PIRL) was developed to couple with an optical surgical tracking system (Fig. 1f) [36]. This direct sampling MS system enabled in situ tissue pathology and the histopathological analysis of neuronal tissues in a mouse model showed comparable results between PIRL and the surgical scalpel method (Fig. 1g). Rapid classification of medulloblastoma subgroups based on small molecule signatures have been achieved subjected to 10- to 15-s principal component analysis of PIRL-MS data [37,38].
Fig. 1.
Surgical MS systems for intraoperative diagnosis. (a) Schematic of an MasSpec Pen MS system. (b) Optical images of a lung adenocarcinoma tissue sample before, during, and after using the MasSpec Pen. The inset shows no observable damage to the tissue sample due to the MasSpec Pen. (c) Negative ion mode mass spectrum of the tissue sample using the MasSpec Pen. Reprinted from Ref. [32] with permission from American Association for the Advancement of Science. (d) Schematic of an iKnife MS system. (e) Classification of the normal and cancerous cervical tissue via univariate analysis of mass spectra collected by the iKnife MS system. Reprinted from Ref. [35] with permission from National Academy of Sciences. (f) Schematic of a PIRL MS system. (g) PIRL direct sampling of normal and cancer tissues for in situ pathological MS analysis. Reprinted from Ref. [36] with permission from The Royal Society of Chemistry.
3. Mass spectrometry imaging (MSI) of tissues using direct sampling
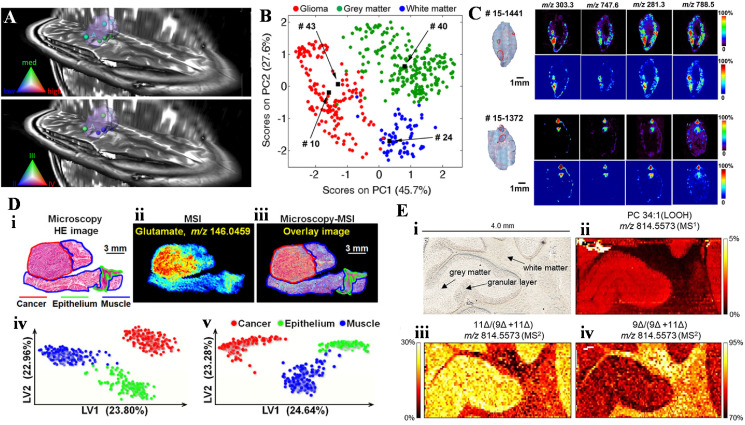
In addition to the molecular information of MS analysis, mass spectrometry imaging (MSI) further offers the information of spatial distribution of the molecules in tissue [39]. This additional information could facilitate the division of the diseased and normal tissue and shows value for clinical use. For instance, in brain tumor surgery, MSI can guide accurate delineation of tumor boundaries to perform maximal tumor resection while preserving brain function. Methods most employed for MSI for clinical samples include DESI [40,41], laser desorption ionization (LDI) [42], and more [43]. As a direct sampling ionization method, DESI-MSI has been used to visualize the tumor grades, tumor cell concentrations, and the definition of margins as the tumor infiltrates into surrounding normal brain (Fig. 2 a) [40]. As shown in Fig. 2a, Positions A and B, classified as grade III, are from the margins of the tumor, while position C and D, classified as grade II, are from the tumor mass center. Using principal component analysis, normal, tumor, and the margins of the tumor can be further differentiated (Fig. 2b) [41]. Through analysis of lipids and metabolites, DESI-MSI has also been successfully identified a common skin cancer, basal cell carcinoma (BCC), and distinguish them from adjacent normal skin (Fig. 2c) [44]. Using gas flow assisted DESI-MSI (Fig. 2d), tumor-associated metabolites from 256 esophageal cancer patients were identified [45]. From the MS images of both positive (case iv) and negative ion modes (case v), cancer (red), epithelial (green) and muscular regions in esophageal squamous cell carcinoma (ESCC) tissues were discriminated via partial least squares discriminant analysis. Isomeric structures of lipids, such as C C bond positional isomers, could reveal information of diseases. C C bond positions could be identified through fragmentations, such as ozone-induced dissociation [46], reaction-based dissociation [47,48], and ultraviolet photodissociation [49], and derivations, such as Paternò-Bǜchi [50], ozonolysis [51], epoxidation [52], and thiol-ene reactions [53]. Together with tandem MS and MSI, MS2I, it revealed the spatial localization of isomeric lipids in biological systems otherwise invisible with traditional MSI. As shown in Fig. 2e, MS image of lipid hydroperoxides (LOOH), indicated that PC 34:1 (LOOH) was enhanced in the gray matter (case ii); however, MS2I indicated that more 9Δ isomer than the 11Δ isomer existed in the white matter (cases iii and iv) [54].
Fig. 2.
(a) Glioma tumor cell concentration (upper) and grade classification (lower) visualized by using segmented preoperative 3D MRI volume reconstruction. Tumor volume is marked in light purple. Reprinted from Ref. [40] with permission from National Academy of Sciences. (b) Chemical predictions of disease state by using principal component analysis. Reprinted from Ref. [41] with permission from National Academy of Sciences. (c) Optical and MS images of skin sections containing miniscule basal cell carcinoma (BCC) aggregates. BCC regions are marked in red. Reprinted from Ref. [44] with permission from National Academy of Sciences. (d) Optical and MS images in heterogeneous ESCC tissue: (i) optical image, (ii) MSI of glutamate, (iii) optical and MSI overlayed, classifications of the cancer (red), epithelial (green) and muscular tissues (blue) in (iv) positive and (v) negative ion modes. Reprinted from Ref. [45] with permission from National Academy of Sciences. (e) Optical and MS images of rat cerebellum: (i) optical image, (ii) MSI of PC 34:1 (LOOH), and MS2I of two C C bond positional isomers, iii) 11Δ/(9Δ+11Δ) and iv) 9Δ/(9Δ+11Δ). Reprinted from Ref. [54] with permission from Wiley-VCH.
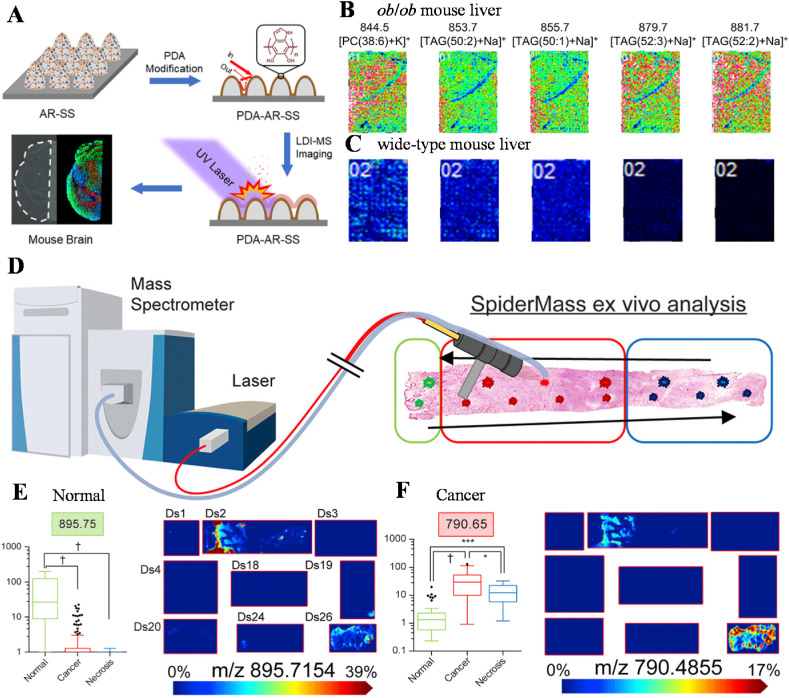
Matrix-assisted laser desorption ionization (MALDI) is another soft ionization method, which has been widely used for imaging clinical samples [55,56], such as metabolites in single cells [57] and drug release in tissues [58]. The matrix compounds absorb the energy from the laser to ionize the sample analytes. However, the matrix may complicate the sample preparation process and result in spectral interferences in the low mass range; hence, great efforts have been done to enable matrix-free LDI through approaches such as nanostructures [59,60] and nanoparticles [61]. For instance, an antireflection (AR) material was used to fabricate micro- and nanostructures on a copper substrate, which achieved a UV absorption up to 97% and a high light-to-heat conversion efficiency for LDI-MS analysis [62]. Using polydopamine (PDA)-coated AR surface on a polished 304 stainless steel (SS), PDA-AR-SS, matrix-free LDI-MSI of around 30 lipid species of mouse brain tissues was performed with a spatial resolution of 50 m (Fig. 3 a) [63]. The method was applied to determine mouse fatty liver disease by monitoring the abundances and distributions of triacylglycerols and glycerophospholipids. Dramatic differences in lipid profiles have been found between the obese (Fig. 3b) and the normal mice (Fig. 3c). Alternatively, excitation of endogenous water molecules as matrix has been used for fast ex vivo LDI-MSI diagnosis of sarcomas [64]. The MSI system, called SpiderMass, included an optical fiber (red line, Fig. 3d) for and a handpiece for sampling and an ion transfer line foe collection of the ions for MS analysis (blue line, Fig. 3d). Normal region and cancer were discriminated via MS imaging of specific biomarkers, e.g., m/z 895.75 (Fig. 3e) and 790.65 (Fig. 3f).
Fig. 3.
(a) Workflow of a PDA-AR LDI-MS analysis and imaging. LDI-MSI of liver samples from (b) ob/ob mouse and (c) wild-type mouse using the PDA-AR substrate. Reprinted from Ref. [63] with permission from American Chemical Society. (d) Schematic of a water-assisted LDI-MS analysis and imaging system, SpiderMass. MSI of normal and cancer tissues of sarcoma using specific biomarkers, m/z 895.75 (normal, e) and 790.65 (cancer, f). Reprinted from Ref. [64] with permission from Elsevier.
4. MS miniaturization: a necessary step to on-site applications
Miniature MS systems are designed to perform on-site bioanalysis distinct from those done in laboratory environment. During the clinical translation, the end user of a miniature instrument could be a nurse or a physician rather than an analytical chemist. Miniature MS systems are, therefore, expected to be adequately small to fit on the scenario in clinics and simple enough for the end users without analytical trainings. The small systems could automatically take raw samples, such as biofluids and tissues, and generate the analytical reports understandable to the end users, instead of MS spectra or chromatograms. Other than the clinical use, miniature MS has also been widely used in on-site applications, such as environmental monitoring [65], food safety [66,67], homeland security [68,69], and space exploration [70,71].
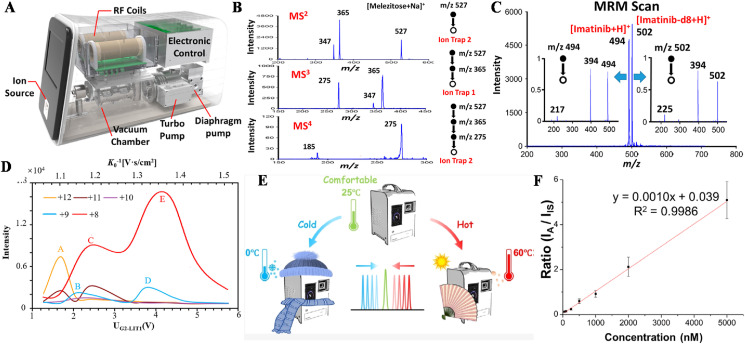
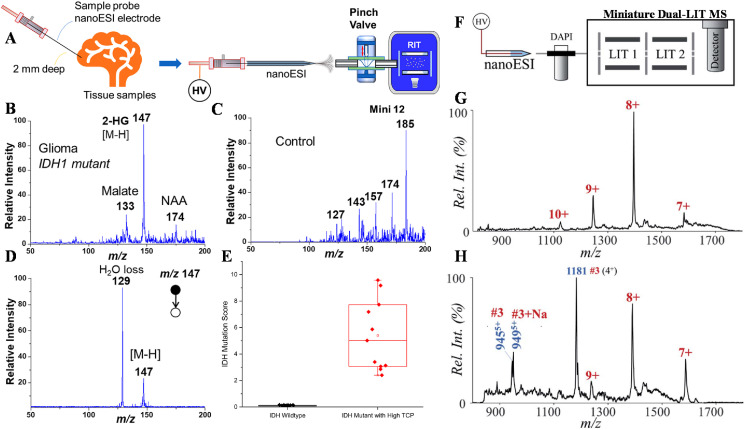
The development of miniature MS systems has experienced three stages. The first one is the miniaturization of mass analyzers. For instance, geometrical optimization of the original quadrupole ion trap leads to the generation of cylindrical ion trap (CIT) [72,73], linear ion trap (LIT) [74], rectilinear ion trap (RIT) [75], and toroidal ion trap (TIT) [76]. The second stage aims on make a small instrument, where all the components, such as the ionization source, mass analyzer, and pump are integrated into a small compartment, such as the Mini 10 [77], Mini 11 [78]. The box-size Mini 10 system weighted 10 kg and dimensions of 12.6 × 8.6 × 7.5 in. (length × width × height) and the Mini 11 weighted 5 kg and smaller dimensions of 8.6 × 4.7 × 7.1 in. However, accompanied with the size miniaturization, the analytical performances of a mass spectrometer were compromised. To meet the demands of clinical and POC applications, a POC Mini 12 system [79], as a representative third-stage prototype, was developed, where the entire MS system and analytical procedure were both miniature. The Mini 12 desktop had sizes of 19.6 × 22.1 × 16.5 in., a little larger than the Mini 10 and Mini 11, weighed 25 kg and consumed a power of less than 100 W. By coupling with an ambient ionization source, paper spray cartridge, it could perform MS and MS/MS analysis of organic compounds in complex samples. Recently, on the basis of the Mini 12 system, its analytical performance was further strengthened with a configuration of dual linear ion traps (dual-LIT) system, also known as Mini (Fig. 4 a) [80]. The dual-LIT system had similar sizes and weight as the Mini 12 system; however, the latest dual-LIT system allowed ion processing between the two LITs, thus enabling comprehensive scan modes for tandem MS analysis. For instance, MS/MS analysis could be performed in either of the dual LITs, thus enabling multiple MS/MS analysis, e.g., the MS2 to MS4 in the dual-LIT system (Fig. 4b). Triple quadrupole function, multiple-reaction monitoring (Fig. 4c), has also been achieved. As shown in Fig. 4c, the protonated imatinib, m/z 494 and m/z 502, were sequentially transferred from LIT 1 to LIT 2, during which beam-type MS/MS analysis was performed to obtain the fragment ion, m/z 394. Additionally, using a dynamic gas flow introduced by the discontinuous atmospheric pressure interface (DAPI), the dual-LIT system could perform ion mobility analysis by using the existing mass analyzers without complicating the overall instrument configuration [81]. Conformers of cytochrome c ions at different charge states were separated according to their collisional cross-sections (Fig. 4d). Generally, MS systems are operated in a well-controlled environment and standard analysis protocols as in laboratory. However, for on-site analysis using miniature MS systems, environmental conditions such as temperature, humidity, vibration, and air pressure may cause fatal effects on the performance of mass spectrometers. A mini 14 system capable of automatically adapting to complex conditions for on-site and POC analysis was developed [82]. Using machine learning, the Mini 14 system was able to establish a model for autocorrecting the mass offsets due to temperature variations from 0°C to 60°C (Fig. 4e). In addition, a gas leaking method was also developed to allow flexible real-time adjustment of gas pressure for MS and tandem MS analyses [83]. The intelligent MS system with comprehensive scan modes facilitated high-quality qualitative and quantitative analysis of clinic samples, such as drugs in biofluids and biomarker analysis. For analysis of peptides as potential biomarkers, a calibration curve was established for Met peptide by using 100 ppb of trypsin digested mouse liver protein (MLP) as background peptides (Fig. 4f) [84]. The Met peptide concentration varied from 50 nM to 5 M and a good linearity was observed throughout all concentrations with R-squared values > 0.99.
Fig. 4.
Instrumentation and analytical performances of state-of-the-art miniature MS systems. (a) Instrumental setup of a dual-LIT miniature mass spectrometer. (b) Tandem MS analysis, MS2 to MS4, of melezitose in the dual LITs. (c) Multiple reaction monitoring (MRM) analysis of drugs. Reprinted from Ref. [80] with permission from American Chemical Society. (d) Ion mobility spectrum of cytochrome c for charge states between +8 and +12. Reprinted from Ref. [81] with permission from American Chemical Society. (e) Schematic of the Mini 14 system with intelligent adaptability for on-site and POC analysis. Reprinted from Ref. [82] with permission from American Chemical Society. (f) Quantitation analysis of the Met peptide. Reprinted from Ref. [84] with permission from American Chemical Society.
The latest miniature Mini MS systems, such as the Mini 12 [79] and the dual-LIT MS [80], present a huge opportunity for POC applications [85,86]. Unlike unknown profiling using lab-scale instruments, the POC miniature MS systems aim on targeted analysis, such as analyzing biomarkers directly from biological samples, and the whole analysis procedures are technically accessible to nurse and physician in office or the patients themselves. For instance, a stainless-steel sampling probe was developed for direct sampling of organ tissue. The probe was also be used as an ionization source of the Mini 12 MS system for fast analysis of metabolite biomarkers, 2-hydroxylglutarate (2-HG), for clinical diagnosis of glioma (Fig. 5 a) [87]. The 2-HG had a significant increase in glioma tissue with isocitrate dehydrogenase (IDH) mutation; hence, fast analysis of 2-HG in glioma tissue allows real-time surgery decision making. For the glioma (IDH1 mutant, grade IV) sample, a significant peak of 2-HG was detected at m/z 147 in the MS spectrum (Fig. 5b), while the peak at m/z 147 was negligible in the control (Fig. 5c). Tandem MS analysis of the m/z 147 further confirmed the presence of 2-HG in the glioma tissue sample (Fig. 5d). The concentration of 2-HG was used to calculate IDH mutation scores, which could facilitate the accurate determination of IDH mutation status. From Fig. 5e, it was observed that IDH mutant with high tumor cell percentages had a much higher score than the IDH wild-type samples [88,89]. As another example, POC analysis of protein biomarkers, such as ESAT-6, was performed by coupling a high-temperature micro-reactor with a miniature dual-LIT MS system (Fig. 5f) [90]. The ESAT-6 is a biomarkers of Mycobacterium tuberculosis (Mtb) infections for the diagnosis of tuberculosis (TB) disease. From the mass spectrum, the small protein, ESAT-6, had a charge state distribution centered at +8 (Fig. 5g). After high-temperature hydrolysis using the micro-reactor for 2 min, some major peptide fragments of ESAT-6 were observed, e.g., m/z 944, m/z 949 and m/z 1181 (Fig. 5h).
Fig. 5.
Application demonstrations of miniature MS systems. (a) Direct analysis of glioma tissues using a sampling probe and Mini 12 MS system. Mass spectra of 2-HG in the (b) glioma tissue and (c) normal brain tissue. (d) Tandem MS spectra of the 2-HG for the glioma tissue. Reprinted from Ref. [87] with permission from American Chemical Society. Box-and-whisker plot of IDH mutation scores in IDH mutant (11 samples, red) and IDH wild-type glioma tissues (16 samples, black). Here, boxes show median, lower, and upper quartiles, and whiskers are at minimum and maximum values. Reprinted from Ref. [88] with permission from Springer Nature. (f) Analysis of ESAT-6 by high-temperature hydrolysis and a miniature dual-LIT MS system. Mass spectra of ESAT-6 (g) before and (h) after the high-temperature hydrolysis. Characteristic peptides of ESAT-6 are marked in dark blue. Reprinted from Ref. [90] with permission from The Royal Society of Chemistry.
5. MS solution to the challenge of pandemic diseases
Since its outbreak in 2019, an epidemic disease, COVID-19 caused by the SARS-CoV-2 coronavirus, has become a severe public health concern worldwide. SARS-CoV-2 has high transmission rate and mortality rate and has a long incubation period prior to visible symptoms; hence, timely detection of SARS-CoV-2 is thus critical for the control of the spread of the disease. In response to the unknown challenge, MS provides its solution as a complementary approach to the mostly employed polymerase chain reaction (PCR) method.
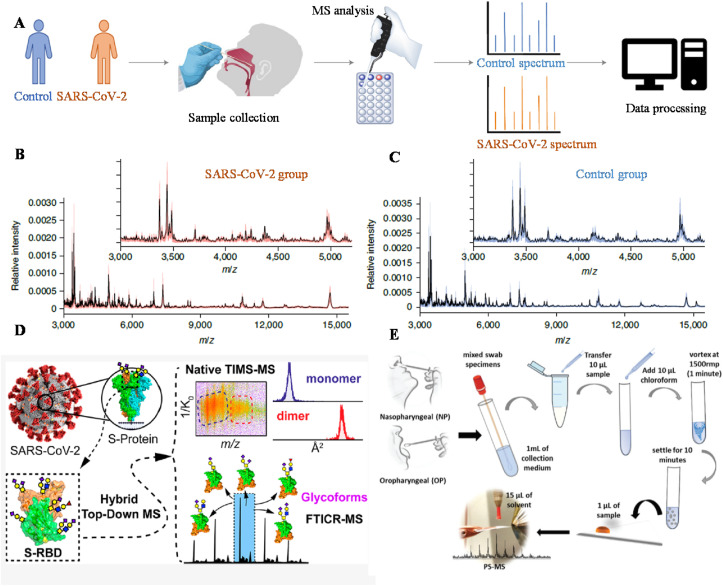
MS analysis of potential biomarkers, including characteristic proteins [[91], [92], [93], [94]], lipids, and metabolites [[95], [96], [97]], have been found to be effective for the diagnosis of SARS-CoV-2. For instance, a MALDI-MS platform was developed for detecting proteomic features that could be used for pathogen identification of large-scale SARS-CoV-2 cases (Fig. 6 a) [93]. Nasal swab samples collected from 362 patients with 211 positive and 151 negative, confirmed by the PCR, were processed by the MALAI-MS analysis. The obtained spectra were then trained through machine learning to identify positive (Fig. 6b) and negative groups (Fig. 6c). Machine learning revealed an accuracy of 93.9% with 7% false positives and 5% false negatives. An LC-MS platform was also used to identify proteomic and metabolic signatures in sera and obtained an accuracy of 93.5% with a training set of 31 patients [96]. By using a MasSpec Pen probe to extract lipids and metabolites in nasopharyngeal swabs, MS-based diagnostic results could achieve a cross-validation accuracy up to 89.6% [95]. The spike (S) protein of SARS-CoV-2 has been found to be critical for viral binding and infectivity. Using top-down and ion mobility analysis, the structures of intact O-glycan proteoforms on the S protein regional-binding domain (RBD) has been elucidated, which reveals potential relation between the heterogeneity of O-glycans and SARS-CoV-2 S protein RBD variants (Fig. 6d) [94]. Direct sampling ionization can minimize the efforts in sample preparation to allow rapid diagnosis of SARS-CoV-2. Using paper spray, diagnostic MS results based on the analysis of lipids and metabolites can be achieved within 1 min (Fig. 6e) [97]. Using a robotic handler, SARS-CoV-2 samples were treated automatically to enable MS analysis of more than 500 samples per day [98]. It is also exciting to see that a Cov-MS consortium, consisting of 15 academic laboratories and several industrial partners, has been established, promoting technical translation from analytical laboratories to clinics [99].
Fig. 6.
MS platforms for SARS-CoV-2 diagnosis. (a) Scheme for SARS-CoV-2 detection by using MALDI-MS. Characteristic mass spectra of (b) SARS-CoV-2 positive and (c) SARS-CoV-2 negative. Reprinted from Ref. [93] with permission from Springer Nature. (d) Structural analysis of O-glycans on the S protein RBD. Reprinted from Ref. [94] with permission from American Chemical Society. (e) Scheme for SARS-CoV-2 detection by using paper spray MS. Reprinted from Ref. [97] with permission from The Royal Society of Chemistry.
6. Conclusion
MS is powerful tool in analytical chemistry and is experiencing a translation from analytical chemistry to clinical applications. Ambient ionizations allow biological samples to be sampled directly in their native environment without pretreatment. Meanwhile, the soft nature of ambient ionization guarantees minimal destructions to the biological samples and invasions to the patients. This is a prerequisite that MS technique could be transferred to non-expert users and used in clinical settings, such as nurses and physicians in clinical office and operation room. Another difficulty for the technical translation is the size and mobility of the MS systems, which prevent the application of MS in on-site scenarios, such as ambulances and outdoors for POC diagnosis. Miniature MS systems with an adequate performance can well fill the gap. Instead of pursuing ultimate high performance as large-scale instruments, the system design for miniature MS systems is intended to achieve a balance among the size reduction, ease of use and required performance of specific applications. With highly specific molecular information in hand, miniature MS systems incorporated with direct sampling methods are expected to provide POC service and personalized treatment for patients in future.
Declaration of competing interest
Zheng Ouyang is the founder of PURSPEC Technologies Inc. that develops miniature mass spectrometer.
Acknowledgments
This work was supported by National Natural Science Foundation of China (Project No. 21627807, 21874081 and 21934003) and National Key R&D Program of China (2018YFF0109500).
References
- 1.Lee S., Chintalapudi K., Badu-Tawiah A.K. Clinical chemistry for developing countries: mass spectrometry. Annu. Rev. Anal. Chem. 2021;14:437. doi: 10.1146/annurev-anchem-091520-085936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferreira C.R., Yannell K.E., Jarmusch A.K., Pirro V., Ouyang Z., Cooks R.G. Ambient ionization mass spectrometry for point-of-care diagnostics and other clinical measurements. Clin. Chem. 2016;62:99. doi: 10.1373/clinchem.2014.237164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jannetto P.J., Fitzgerald R.L. Effective use of mass spectrometry in the clinical laboratory. Clin. Chem. 2016;62:92. doi: 10.1373/clinchem.2015.248146. [DOI] [PubMed] [Google Scholar]
- 4.Swiner D.J., Jackson S., Burris B.J., Badu-Tawiah A.K. Applications of mass spectrometry for clinical diagnostics: the influence of turnaround time. Anal. Chem. 2020;92:183. doi: 10.1021/acs.analchem.9b04901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma X., Zhao X., Li J., Zhang W., Cheng J.-X., Ouyang Z., Xia Y. Photochemical tagging for quantitation of unsaturated fatty acids by mass spectrometry. Anal. Chem. 2016;88:8931. doi: 10.1021/acs.analchem.6b02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren Y., Chiang S., Zhang W., Wang X., Lin Z., Ouyang Z. Paper-capillary spray for direct mass spectrometry analysis of biofluid samples. Anal. Bioanal. Chem. 2016;408:1385. doi: 10.1007/s00216-015-9129-9. [DOI] [PubMed] [Google Scholar]
- 7.Shi L., Bucknall M.P., Young T.L., Zhang M., Hu L., Bing J., Lee D.S., Kim J., Wu T., Takamure N., McKenzie D.R., Huang S., Green M.A., Ho-Baillie A.W.Y. Gas chromatography-mass spectrometry analyses of encapsulated stable perovskite solar cells. Science. 2020;368 doi: 10.1126/science.aba2412. [DOI] [PubMed] [Google Scholar]
- 8.Wolfender J.-L., Nuzillard J.-M., van der Hooft J.J.J., Renault J.-H., Bertrand S. Accelerating metabolite identification in natural product research: toward an ideal combination of liquid chromatography–high-resolution tandem mass spectrometry and NMR profiling, in silico databases, and chemometrics. Anal. Chem. 2019;91:704. doi: 10.1021/acs.analchem.8b05112. [DOI] [PubMed] [Google Scholar]
- 9.Cooks R.G., Ouyang Z., Takats Z., Wiseman J.M. Ambient mass spectrometry. Science. 2006;311:1566. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- 10.Monge M.E., Harris G.A., Dwivedi P., Fernández F.M. Mass spectrometry: recent advances in direct open air surface sampling/ionization. Chem. Rev. 2013;113:2269. doi: 10.1021/cr300309q. [DOI] [PubMed] [Google Scholar]
- 11.Takyi-Williams J., Liu C.-F., Tang K. Ambient ionization MS for bioanalysis: recent developments and challenges. Bioanalysis. 2015;7:1901. doi: 10.4155/bio.15.116. [DOI] [PubMed] [Google Scholar]
- 12.Ouyang Z., Cooks R.G. Miniature mass spectrometers. Annu. Rev. Anal. Chem. 2009;2:187. doi: 10.1146/annurev-anchem-060908-155229. [DOI] [PubMed] [Google Scholar]
- 13.Snyder D.T., Pulliam C.J., Ouyang Z., Cooks R.G. Miniature and fieldable mass spectrometers: recent advances. Anal. Chem. 2016;88:2. doi: 10.1021/acs.analchem.5b03070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takáts Z., Wiseman J.M., Gologan B., Cooks R.G. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 15.Cody R.B., Laramée J.A., Durst H.D. Versatile new ion source for the analysis of materials in open air under ambient conditions. Anal. Chem. 2005;77:2297. doi: 10.1021/ac050162j. [DOI] [PubMed] [Google Scholar]
- 16.Yang Q., Wang H., Maas J.D., Chappell W.J., Manicke N.E., Cooks R.G., Ouyang Z. Paper spray ionization devices for direct, biomedical analysis using mass spectrometry. Int. J. Mass Spectrom. 2012;312:201. doi: 10.1016/j.ijms.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C., Manicke N.E. Development of a paper spray mass spectrometry cartridge with integrated solid phase extraction for bioanalysis. Anal. Chem. 2015;87:6212. doi: 10.1021/acs.analchem.5b00884. [DOI] [PubMed] [Google Scholar]
- 18.Chen S., Wan Q., Badu-Tawiah A.K. Mass spectrometry for paper-based immunoassays: toward on-demand diagnosis. J. Am. Chem. Soc. 2016;138:6356. doi: 10.1021/jacs.6b02232. [DOI] [PubMed] [Google Scholar]
- 19.Wang H., Liu J., Cooks R.G., Ouyang Z. Paper spray for direct analysis of complex mixtures using mass spectrometry. Angew. Chem. Int. Ed. 2010;49:877. doi: 10.1002/anie.200906314. [DOI] [PubMed] [Google Scholar]
- 20.Skaggs C., Kirkpatrick L., Wichert W.R.A., Skaggs N., Manicke N.E. A statistical approach to optimizing paper spray mass spectrometry parameters. Rapid Commun. Mass Spectrom. 2020;34 doi: 10.1002/rcm.8601. [DOI] [PubMed] [Google Scholar]
- 21.Espy R.D., Teunissen S.F., Manicke N.E., Ren Y., Ouyang Z., van Asten A., Cooks R.G. Paper spray and extraction spray mass spectrometry for the direct and simultaneous quantification of eight drugs of abuse in whole blood. Anal. Chem. 2014;86:7712. doi: 10.1021/ac5016408. [DOI] [PubMed] [Google Scholar]
- 22.Ren Y., McLuckey M.N., Liu J., Ouyang Z. Direct mass spectrometry analysis of biofluid samples using slug-flow microextraction nano-electrospray ionization. Angew. Chem. Int. Ed. 2014;53:14124. doi: 10.1002/anie.201408338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Espy R.D., Manicke N.E., Ouyang Z., Cooks R.G. Rapid analysis of whole blood by paper spray mass spectrometry for point-of-care therapeutic drug monitoring. Analyst. 2012;137:2344. doi: 10.1039/c2an35082c. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z., Xu W., Manicke N.E., Cooks R.G., Ouyang Z. Silica coated paper substrate for paper-spray analysis of therapeutic drugs in dried blood spots. Anal. Chem. 2012;84:931. doi: 10.1021/ac202058w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossini E.L., Kulyk D.S., Ansu-Gyeabourh E., Sahraeian T., Pezza H.R., Badu-Tawiah A.K. Direct analysis of doping agents in raw urine using hydrophobic paper spray mass spectrometry. J. Am. Soc. Mass Spectrom. 2020;31:1212. doi: 10.1021/jasms.0c00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X., Liu J., Cooks R.G., Ouyang Z. Development of miniature mass spectrometry systems for bioanalysis outside the conventional laboratories. Bioanalysis. 2014;6:1497. doi: 10.4155/bio.14.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma X., Ouyang Z. Ambient ionization and miniature mass spectrometry system for chemical and biological analysis. TrAC Trends Anal. Chem. (Reference Ed.) 2016;85:10. doi: 10.1016/j.trac.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W., Wang X., Xia Y., Ouyang Z. Ambient ionization and miniature mass spectrometry systems for disease diagnosis and therapeutic monitoring. Theranostics. 2017;7:2968. doi: 10.7150/thno.19410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pu F., Chiang S., Zhang W., Ouyang Z. Direct sampling mass spectrometry for clinical analysis. Analyst. 2019;144:1034. doi: 10.1039/c8an01722k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ifa D.R., Eberlin L.S. Ambient ionization mass spectrometry for cancer diagnosis and surgical margin evaluation. Clin. Chem. 2016;62:111. doi: 10.1373/clinchem.2014.237172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sans M., Zhang J., Lin J.Q., Feider C.L., Giese N., Breen M.T., Sebastian K., Liu J., Sood A.K., Eberlin L.S. Performance of the MasSpec pen for rapid diagnosis of ovarian cancer. Clin. Chem. 2019;65:674. doi: 10.1373/clinchem.2018.299289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J., Rector J., Lin J.Q., Young J.H., Sans M., Katta N., Giese N., Yu W., Nagi C., Suliburk J., Liu J., Bensussan A., DeHoog R.J., Garza K.Y., Ludolph B., Sorace A.G., Syed A., Zahedivash A., Milner T.E., Eberlin L.S. Nondestructive tissue analysis for ex vivo and in vivo cancer diagnosis using a handheld mass spectrometry system. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aan3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strittmatter N., Rebec M., Jones E.A., Golf O., Abdolrasouli A., Balog J., Behrends V., Veselkov K.A., Takats Z. Characterization and identification of clinically relevant microorganisms using rapid evaporative ionization mass spectrometry. Anal. Chem. 2014;86:6555. doi: 10.1021/ac501075f. [DOI] [PubMed] [Google Scholar]
- 34.Schäfer K.-C., Dénes J., Albrecht K., Szaniszló T., Balog J., Skoumal R., Katona M., Tóth M., Balogh L., Takáts Z. In vivo, in situ tissue analysis using rapid evaporative ionization mass spectrometry. Angew. Chem. Int. Ed. 2009;48:8240. doi: 10.1002/anie.200902546. [DOI] [PubMed] [Google Scholar]
- 35.Tzafetas M., Mitra A., Paraskevaidi M., Bodai Z., Kalliala I., Bowden S., Lathouras K., Rosini F., Szasz M., Savage A., Manoli E., Balog J., McKenzie J., Lyons D., Bennett P., MacIntyre D., Ghaem-Maghami S., Takats Z., Kyrgiou M. The intelligent knife (iKnife) and its intraoperative diagnostic advantage for the treatment of cervical disease. Proc. Natl. Acad. Sci. U.S.A. 2020;117:7338. doi: 10.1073/pnas.1916960117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woolman M., Qiu J., Kuzan-Fischer C.M., Ferry I., Dara D., Katz L., Daud F., Wu M., Ventura M., Bernards N., Chan H., Fricke I., Zaidi M., Wouters B.G., Rutka J.T., Das S., Irish J., Weersink R., Ginsberg H.J., Jaffray D.A., Zarrine-Afsar A. In situ tissue pathology from spatially encoded mass spectrometry classifiers visualized in real time through augmented reality. Chem. Sci. 2020;11:8723. doi: 10.1039/d0sc02241a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woolman M., Ferry I., Kuzan-Fischer Claudia M., Wu M., Zou J., Kiyota T., Isik S., Dara D., Aman A., Das S., Taylor M.D., Rutka J.T., Ginsberg H.J., Zarrine-Afsar A. Rapid determination of medulloblastoma subgroup affiliation with mass spectrometry using a handheld picosecond infrared laser desorption probe. Chem. Sci. 2017;8:6508. doi: 10.1039/c7sc01974b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woolman M., Kuzan-Fischer C.M., Ferry I., Kiyota T., Luu B., Wu M., Munoz D.G., Das S., Aman A., Taylor M.D., Rutka J.T., Ginsberg H.J., Zarrine-Afsar A. Picosecond infrared laser desorption mass spectrometry identifies medulloblastoma subgroups on intrasurgical timescales. Cancer Res. 2019;79:2426. doi: 10.1158/0008-5472.CAN-18-3411. [DOI] [PubMed] [Google Scholar]
- 39.Buchberger A.R., DeLaney K., Johnson J., Li L. Mass spectrometry imaging: a review of emerging advancements and future insights. Anal. Chem. 2018;90:240. doi: 10.1021/acs.analchem.7b04733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eberlin L.S., Norton I., Orringer D., Dunn I.F., Liu X., Ide J.L., Jarmusch A.K., Ligon K.L., Jolesz F.A., Golby A.J., Santagata S., Agar N.Y.R., Cooks R.G. Ambient mass spectrometry for the intraoperative molecular diagnosis of human brain tumors. Proc. Natl. Acad. Sci. U.S.A. 2013;110:1611. doi: 10.1073/pnas.1215687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pirro V., Alfaro C.M., Jarmusch A.K., Hattab E.M., Cohen-Gadol A.A., Cooks R.G. Intraoperative assessment of tumor margins during glioma resection by desorption electrospray ionization-mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2017;114:6700. doi: 10.1073/pnas.1706459114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang Y., Sun J., Xiong C., Liu H., Li Y., Wang X., Nie Z. Mass spectrometry imaging reveals in situ behaviors of multiple components in aerosol particles. Angew. Chem. Int. Ed. 2021;60:23225. doi: 10.1002/anie.202103874. [DOI] [PubMed] [Google Scholar]
- 43.Angerer T.B., Dowlatshahi Pour M., Malmberg P., Fletcher J.S. Improved molecular imaging in rodent brain with time-of-flight-secondary ion mass spectrometry using gas cluster ion beams and reactive vapor exposure. Anal. Chem. 2015;87:4305. doi: 10.1021/ac504774y. [DOI] [PubMed] [Google Scholar]
- 44.Margulis K., Chiou A.S., Aasi S.Z., Tibshirani R.J., Tang J.Y., Zare R.N. Distinguishing malignant from benign microscopic skin lesions using desorption electrospray ionization mass spectrometry imaging. Proc. Natl. Acad. Sci. U.S.A. 2018;115:6347. doi: 10.1073/pnas.1803733115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun C., Li T., Song X., Huang L., Zang Q., Xu J., Bi N., Jiao G., Hao Y., Chen Y., Zhang R., Luo Z., Li X., Wang L., Wang Z., Song Y., He J., Abliz Z. Spatially resolved metabolomics to discover tumor-associated metabolic alterations. Proc. Natl. Acad. Sci. U.S.A. 2019;116:52. doi: 10.1073/pnas.1808950116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marshall D.L., Criscuolo A., Young R.S.E., Poad B.L.J., Zeller M., Reid G.E., Mitchell T.W., Blanksby S.J. Mapping unsaturation in human plasma lipids by data-independent ozone-induced dissociation. J. Am. Soc. Mass Spectrom. 2019;30:1621. doi: 10.1007/s13361-019-02261-z. [DOI] [PubMed] [Google Scholar]
- 47.Randolph C.E., Foreman D.J., Betancourt S.K., Blanksby S.J., McLuckey S.A. Gas-phase ion/ion reactions involving tris-phenanthroline alkaline earth metal complexes as charge inversion reagents for the identification of fatty acids. Anal. Chem. 2018;90:12861. doi: 10.1021/acs.analchem.8b03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Born M.-E.N., Prentice B.M. Structural elucidation of phosphatidylcholines from tissue using electron induced dissociation. Int. J. Mass Spectrom. 2020;452:116338. doi: 10.1016/j.ijms.2022.116998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klein D.R., Feider C.L., Garza K.Y., Lin J.Q., Eberlin L.S., Brodbelt J.S. Desorption electrospray ionization coupled with ultraviolet photodissociation for characterization of phospholipid isomers in tissue sections. Anal. Chem. 2018;90:10100. doi: 10.1021/acs.analchem.8b03026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma X., Xia Y. Pinpointing double bonds in lipids by paternò-büchi reactions and mass spectrometry. Angew. Chem. Int. Ed. 2014;53:2592. doi: 10.1002/anie.201310699. [DOI] [PubMed] [Google Scholar]
- 51.Zhang J.I., Tao W.A., Cooks R.G. Facile determination of double bond position in unsaturated fatty acids and esters by low temperature plasma ionization mass spectrometry. Anal. Chem. 2011;83:4738. doi: 10.1021/ac1030946. [DOI] [PubMed] [Google Scholar]
- 52.Tang S., Cheng H., Yan X. On-demand electrochemical epoxidation in nano-electrospray ionization mass spectrometry to locate carbon–carbon double bonds. Angew. Chem. Int. Ed. 2020;59:209. doi: 10.1002/anie.201911070. [DOI] [PubMed] [Google Scholar]
- 53.Adhikari S., Zhang W., Xie X., Chen Q., Xia Y. Shotgun analysis of diacylglycerols enabled by thiol–ene click chemistry. Anal. Chem. 2018;90:5239. doi: 10.1021/acs.analchem.8b00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Unsihuay D., Su P., Hu H., Qiu J., Kuang S., Li Y., Sun X., Dey S.K., Laskin J. Imaging and analysis of isomeric unsaturated lipids through online photochemical derivatization of carbon–carbon double bonds. Angew. Chem. Int. Ed. 2021;60:7559. doi: 10.1002/anie.202016734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kompauer M., Heiles S., Spengler B. Autofocusing MALDI mass spectrometry imaging of tissue sections and 3D chemical topography of nonflat surfaces. Nat. Methods. 2017;14:1156. doi: 10.1038/nmeth.4433. [DOI] [PubMed] [Google Scholar]
- 56.Kompauer M., Heiles S., Spengler B. Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-μm lateral resolution. Nat. Methods. 2017;14:90. doi: 10.1038/nmeth.4071. [DOI] [PubMed] [Google Scholar]
- 57.Amantonico A., Urban P.L., Fagerer S.R., Balabin R.M., Zenobi R. Single-cell MALDI-MS as an analytical tool for studying intrapopulation metabolic heterogeneity of unicellular organisms. Anal. Chem. 2010;82:7394. doi: 10.1021/ac1015326. [DOI] [PubMed] [Google Scholar]
- 58.Xue J., Liu H., Chen S., Xiong C., Zhan L., Sun J., Nie Z. Mass spectrometry imaging of the in situ drug release from nanocarriers. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aat9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei J., Buriak J.M., Siuzdak G. Desorption–ionization mass spectrometry on porous silicon. Nature. 1999;399:243. doi: 10.1038/20400. [DOI] [PubMed] [Google Scholar]
- 60.Stopka S.A., Rong C., Korte A.R., Yadavilli S., Nazarian J., Razunguzwa T.T., Morris N.J., Vertes A. Molecular imaging of biological samples on nanophotonic laser desorption ionization platforms. Angew. Chem. Int. Ed. 2016;55:4482. doi: 10.1002/anie.201511691. [DOI] [PubMed] [Google Scholar]
- 61.Kurczy M.E., Zhu Z.-J., Ivanisevic J., Schuyler A.M., Lalwani K., Santidrian A.F., David J.M., Giddabasappa A., Roberts A.J., Olivos H.J., O'Brien P.J., Franco L., Fields M.W., Paris L.P., Friedlander M., Johnson C.H., Epstein A.A., Gendelman H.E., Wood M.R., Felding B.H., Patti G.J., Spilker M.E., Siuzdak G. Comprehensive bioimaging with fluorinated nanoparticles using breathable liquids. Nat. Commun. 2015;6:5998. doi: 10.1038/ncomms6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang J., Zhang H., Jia J., Zhang X., Ma X., Zhong M., Ouyang Z. Antireflection surfaces for biological analysis using laser desorption ionization mass spectrometry. Research. 2018;2018:5439729. doi: 10.1155/2018/5439729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang J., Zhang W., Zhang H., Zhong M., Cao W., Li Z., Huang X., Nie Z., Liu J., Li P., Ma X., Ouyang Z. Polydopamine-modified substrates for high-sensitivity laser desorption ionization mass spectrometry imaging. ACS Appl. Mater. Interfaces. 2019;11:46140. doi: 10.1021/acsami.9b16260. [DOI] [PubMed] [Google Scholar]
- 64.Saudemont P., Quanico J., Robin Y.-M., Baud A., Balog J., Fatou B., Tierny D., Pascal Q., Minier K., Pottier M., Focsa C., Ziskind M., Takats Z., Salzet M., Fournier I. Real-time molecular diagnosis of tumors using water-assisted laser desorption/ionization mass spectrometry technology. Cancer Cell. 2018;34:840. doi: 10.1016/j.ccell.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 65.Mach P.M., Winfield J.L., Aguilar R.A., Wright K.C., Verbeck G.F. A portable mass spectrometer study targeting anthropogenic contaminants in Sub-Antarctic Puerto Williams, Chile. Int. J. Mass Spectrom. 2017;422:148. [Google Scholar]
- 66.Gerbig S., Neese S., Penner A., Spengler B., Schulz S. Real-time food authentication using a miniature mass spectrometer. Anal. Chem. 2017;89:10717. doi: 10.1021/acs.analchem.7b01689. [DOI] [PubMed] [Google Scholar]
- 67.Zhai Y., Zhang X., Xu H., Zheng Y., Yuan T., Xu W. Mini mass spectrometer integrated with a miniature ion funnel. Anal. Chem. 2017;89:4177. doi: 10.1021/acs.analchem.7b00195. [DOI] [PubMed] [Google Scholar]
- 68.Virgen C.A., Fox J.D., Santariello P., Winfield J.L., Wright K.C., Verbeck G.F. Portable membrane inlet mass spectrometric detection and analysis of chemical warfare agent simulants at the U.S. Army Dugway Proving Ground S/K challenge event. Int. J. Mass Spectrom. 2021;468:116635. [Google Scholar]
- 69.Kelly J.T., Qualley A., Hughes G.T., Arroyo J.W., Malloy T.A., Piatkowski T., Russell M., Lewis D., Rubenstein H.M. Anchoring the quantification of VX and Russian VX using portable gas chromatography-mass spectrometry and focusing agents. Int. J. Mass Spectrom. 2021;468:116659. [Google Scholar]
- 70.Mahaffy P., Webster C., Cabane M., Conrad P., Coll P., Atreya S., Arvey R., Barciniak M., Benna M., Bleacher L., Brinckerhoff W., Eigenbrode J., Carignan D., Cascia M., Chalmers R., Dworkin J., Errigo T., Everson P., Franz H., Farley R., Feng S., Frazier G., Freissinet C., Glavin D., Harpold D., Hawk D., Holmes V., Johnson C., Jones A., Jordan P., Kellogg J., Lewis J., Lyness E., Malespin C., Martin D., Maurer J., McAdam A., McLennan D., Nolan T., Noriega M., Pavlov A., Prats B., Raaen E., Sheinman O., Sheppard D., Smith J., Stern J., Tan F., Trainer M., Ming D., Morris R., Jones J., Gundersen C., Steele A., Wray J., Botta O., Leshin L., Owen T., Battel S., Jakosky B., Manning H., Squyres S., Navarro-Gonzalez R., McKay C., Raulin F., Sternberg R., Buch A., Sorensen P., Kline-Schoder R., Coscia D., Szopa C., Teinturier S., Baffes C., Feldman J., Flesch G., Forouhar S., Garcia R., Keymeulen D., Woodward S., Block B., Arnett K., Miller R., Edmonson C., Gorevan S., Mumm E. The sample analysis at mars investigation and instrument suite. Space Sci. Rev. 2012;170:401. [Google Scholar]
- 71.Grubisic A., Trainer M.G., Li X., Brinckerhoff W.B., van Amerom F.H., Danell R.M., Costa J.T., Castillo M., Kaplan D., Zacny K. Laser desorption mass spectrometry at saturn's moon titan. Int. J. Mass Spectrom. 2021;470:116707. [Google Scholar]
- 72.Wells J.M., Badman E.R., Cooks R.G. A quadrupole ion trap with cylindrical geometry operated in the mass-selective instability mode. Anal. Chem. 1998;70:438. doi: 10.1021/ac971198h. [DOI] [PubMed] [Google Scholar]
- 73.Blakeman K.H., Wolfe D.W., Cavanaugh C.A., Ramsey J.M. High pressure mass spectrometry: the generation of mass spectra at operating pressures exceeding 1 Torr in a microscale cylindrical ion trap. Anal. Chem. 2016;88:5378. doi: 10.1021/acs.analchem.6b00706. [DOI] [PubMed] [Google Scholar]
- 74.Clare A.T., Gao L., Brkic B., Chalker P.R., Taylor S. Linear ion trap fabricated using rapid manufacturing technology. J. Am. Soc. Mass Spectrom. 2010;21:317. doi: 10.1016/j.jasms.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 75.Ouyang Z., Wu G., Song Y., Li H., Plass W.R., Cooks R.G. Rectilinear ion trap: concepts, calculations, and analytical performance of a new mass analyzer. Anal. Chem. 2004;76:4595. doi: 10.1021/ac049420n. [DOI] [PubMed] [Google Scholar]
- 76.Hettikankanange P.M., Austin D.E. Varying the aspect ratio of toroidal ion traps: implications for design, performance, and miniaturization. Int. J. Mass Spectrom. 2021;470:116703. [Google Scholar]
- 77.Gao L., Song Q., Patterson G.E., Cooks R.G., Ouyang Z. Handheld rectilinear ion trap mass spectrometer. Anal. Chem. 2006;78:5994. doi: 10.1021/ac061144k. [DOI] [PubMed] [Google Scholar]
- 78.Gao L., Sugiarto A., Harper J.D., Cooks R.G., Ouyang Z. Design and characterization of a multisource hand-held tandem mass spectrometer. Anal. Chem. 2008;80:7198. doi: 10.1021/ac801275x. [DOI] [PubMed] [Google Scholar]
- 79.Li L., Chen T.-C., Ren Y., Hendricks P.I., Cooks R.G., Ouyang Z. Mini 12, miniature mass spectrometer for clinical and other applications - introduction and characterization. Anal. Chem. 2014;86:2909. doi: 10.1021/ac403766c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu X., Wang X., Bu J., Zhou X., Ouyang Z. Tandem analysis by a dual-trap miniature mass spectrometer. Anal. Chem. 2019;91:1391. doi: 10.1021/acs.analchem.8b03958. [DOI] [PubMed] [Google Scholar]
- 81.Fan J., Lian P., Li M., Liu X., Zhou X., Ouyang Z. Ion mobility separation using a dual-LIT miniature mass spectrometer. Anal. Chem. 2020;92:2573. doi: 10.1021/acs.analchem.9b04271. [DOI] [PubMed] [Google Scholar]
- 82.Jiao B., Ye H., Liu X., Bu J., Wu J., Zhang W., Zhang Y., Ouyang Z. Handheld mass spectrometer with intelligent adaptability for on-site and point-of-care analysis. Anal. Chem. 2021;93:15607. doi: 10.1021/acs.analchem.1c02508. [DOI] [PubMed] [Google Scholar]
- 83.Li N., Zhou X., Ouyang Z. Tandem-in-time mass spectrometry analysis facilitated by real-time pressure adjustments. Int. J. Mass Spectrom. 2021;462:116523. [Google Scholar]
- 84.Chiang S., Zhang W., Farnsworth C., Zhu Y., Lee K., Ouyang Z. Targeted quantification of peptides using miniature mass spectrometry. J. Proteome Res. 2020;19:2043. doi: 10.1021/acs.jproteome.9b00875. [DOI] [PubMed] [Google Scholar]
- 85.Kang M., Zhang W., Dong L., Ren X., Zhu Y., Wang Z., Liang L., Xue J., Zhang Y., Zhang W., Ouyang Z. On-site testing of multiple drugs of abuse in urine by a miniature dual-LIT mass spectrometer. Anal. Chim. Acta. 2020;1101:74. doi: 10.1016/j.aca.2019.12.028. [DOI] [PubMed] [Google Scholar]
- 86.Kang M., Lian R., Zhang X., Li Y., Zhang Y., Zhang Y., Zhang W., Ouyang Z. Rapid and on-site detection of multiple fentanyl compounds by dual-ion trap miniature mass spectrometry system. Talanta. 2020;217:121057. doi: 10.1016/j.talanta.2020.121057. [DOI] [PubMed] [Google Scholar]
- 87.Zou R., Cao W., Chong L., Hua W., Xu H., Mao Y., Page J., Shi R., Xia Y., Hu T.Y., Zhang W., Ouyang Z. Point-of-Care tissue analysis using miniature mass spectrometer. Anal. Chem. 2019;91:1157. doi: 10.1021/acs.analchem.8b04935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pu F., Alfaro C.M., Pirro V., Xie Z., Ouyang Z., Cooks R.G. Rapid determination of isocitrate dehydrogenase mutation status of human gliomas by extraction nanoelectrospray using a miniature mass spectrometer. Anal. Bioanal. Chem. 2019;411:1503. doi: 10.1007/s00216-019-01632-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brown H.M., Pu F., Dey M., Miller J., Shah M.V., Shapiro S.A., Ouyang Z., Cohen-Gadol A.A., Cooks R.G. Intraoperative detection of isocitrate dehydrogenase mutations in human gliomas using a miniature mass spectrometer. Anal. Bioanal. Chem. 2019;411:7929. doi: 10.1007/s00216-019-02198-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Y., Zhang W., Ouyang Z. Fast protein analysis enabled by high-temperature hydrolysis. Chem. Sci. 2020;11:10506. doi: 10.1039/d0sc03237a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ihling C., Tänzler D., Hagemann S., Kehlen A., Hüttelmaier S., Arlt C., Sinz A. Mass spectrometric identification of SARS-CoV-2 proteins from gargle solution samples of COVID-19 patients. J. Proteome Res. 2020;19:4389. doi: 10.1021/acs.jproteome.0c00280. [DOI] [PubMed] [Google Scholar]
- 92.Yan L., Yi J., Huang C., Zhang J., Fu S., Li Z., Lyu Q., Xu Y., Wang K., Yang H., Ma Q., Cui X., Qiao L., Sun W., Liao P. Rapid detection of COVID-19 using MALDI-TOF-based serum peptidome profiling. Anal. Chem. 2021;93:4782. doi: 10.1021/acs.analchem.0c04590. [DOI] [PubMed] [Google Scholar]
- 93.Nachtigall F.M., Pereira A., Trofymchuk O.S., Santos L.S. Detection of SARS-CoV-2 in nasal swabs using MALDI-MS. Nat. Biotechnol. 2020;38:1168. doi: 10.1038/s41587-020-0644-7. [DOI] [PubMed] [Google Scholar]
- 94.Roberts D.S., Mann M., Melby J.A., Larson E.J., Zhu Y., Brasier A.R., Jin S., Ge Y. Structural O-glycoform heterogeneity of the SARS-CoV-2 spike protein receptor-binding domain revealed by top-down mass spectrometry. J. Am. Chem. Soc. 2021;143:12014. doi: 10.1021/jacs.1c02713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Garza K.Y., Silva A.A.R., Rosa J.R., Keating M.F., Povilaitis S.C., Spradlin M., Sanches P.H.G., Varão Moura A., Marrero Gutierrez J., Lin J.Q., Zhang J., DeHoog R.J., Bensussan A., Badal S., Cardoso de Oliveira D., Dias Garcia P.H., Dias de Oliveira Negrini L., Antonio M.A., Canevari T.C., Eberlin M.N., Tibshirani R., Eberlin L.S., Porcari A.M. Rapid screening of COVID-19 directly from clinical nasopharyngeal swabs using the MasSpec pen. Anal. Chem. 2021;93:12582. doi: 10.1021/acs.analchem.1c01937. [DOI] [PubMed] [Google Scholar]
- 96.Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L., Ge W., Liu W., Liang S., Chen H., Zhang Y., Li J., Xu J., He Z., Chen B., Wang J., Yan H., Zheng Y., Wang D., Zhu J., Kong Z., Kang Z., Liang X., Ding X., Ruan G., Xiang N., Cai X., Gao H., Li L., Li S., Xiao Q., Lu T., Zhu Y., Liu H., Chen H., Guo T. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.De Silva I.W., Nayek S., Singh V., Reddy J., Granger J.K., Verbeck G.F. Paper spray mass spectrometry utilizing Teslin® substrate for rapid detection of lipid metabolite changes during COVID-19 infection. Analyst. 2020;145:5725. doi: 10.1039/d0an01074j. [DOI] [PubMed] [Google Scholar]
- 98.Cardozo K.H.M., Lebkuchen A., Okai G.G., Schuch R.A., Viana L.G., Olive A.N., Lazari C.d.S., Fraga A.M., Granato C.F.H., Pintão M.C.T., Carvalho V.M. Establishing a mass spectrometry-based system for rapid detection of SARS-CoV-2 in large clinical sample cohorts. Nat. Commun. 2020;11:6201. doi: 10.1038/s41467-020-19925-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Van Puyvelde B., Van Uytfanghe K., Tytgat O., Van Oudenhove L., Gabriels R., Bouwmeester R., Daled S., Van Den Bossche T., Ramasamy P., Verhelst S., De Clerck L., Corveleyn L., Willems S., Debunne N., Wynendaele E., De Spiegeleer B., Judak P., Roels K., De Wilde L., Van Eenoo P., Reyns T., Cherlet M., Dumont E., Debyser G., t'Kindt R., Sandra K., Gupta S., Drouin N., Harms A., Hankemeier T., Jones D.J.L., Gupta P., Lane D., Lane C.S., El Ouadi S., Vincendet J.-B., Morrice N., Oehrle S., Tanna N., Silvester S., Hannam S., Sigloch F.C., Bhangu-Uhlmann A., Claereboudt J., Anderson N.L., Razavi M., Degroeve S., Cuypers L., Stove C., Lagrou K., Martens G.A., Deforce D., Martens L., Vissers J.P.C., Dhaenens M., Cov M.S. A community-based template Assay for mass-spectrometry-based protein detection in SARS-CoV-2 patients. JACS Au. 2021;1:750. doi: 10.1021/jacsau.1c00048. [DOI] [PMC free article] [PubMed] [Google Scholar]