Abstract
Polyserositis caused by Haemophilus parasuis is an important disease that affects mostly weaned pigs. Recent studies have shown that virulence can differ among strains recovered from distinct body sites and also that it may be related to the presence of certain outer membrane proteins (OMPs). The objective of this study was to compare the OMP and DNA profiles of H. parasuis strains isolated from systemic and respiratory sites from diseased and healthy pigs. Strains evaluated in this study were processed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and repetitive-PCR techniques. Two experiments were conducted in order to better define the relationship among genotype, phenotype, and site of isolation. Experiment 1 included 53 H. parasuis isolates recovered from healthy and diseased pigs from unrelated herds. Experiment 2 included 31 isolates of H. parasuis obtained from diseased pigs involved in an outbreak in a large, multifarm system. Results showed that strains recovered from systemic sites had more homogeneous OMP and DNA profiles than those isolated from respiratory sites. Evaluation of isolates involved in the multifarm outbreak showed that only two H. parasuis strains were causing disease. These strains had homogeneous OMP and DNA profiles. However, it was noted that these two parameters were unrelated, since strains classified in the same genotype group expressed different OMP profiles. The homogeneity of OMP and DNA profiles of strains isolated from systemic sites strongly suggests the existence of clonal relationships between virulent strains and also suggests that expression of certain OMP profiles may be related to virulence.
Infection by Haemophilus parasuis has been increasingly diagnosed in the last few years. Work has attempted to establish and compare the prevalence of different serovars of H. parasuis in the United States and Canada (13). However, a successful correlation among serovars, site of isolation, and disease has not been established (7). Also, the distribution of H. parasuis serovars has been studied in strains isolated mostly from the respiratory tract and mucosal surfaces of healthy piglets, which are probably different from those isolated from systemic sites of sick pigs (19). Recent work done with Streptococcus suis showed that isolates recovered from tonsils of healthy pigs are genetically different from isolates recovered from the brains or joints of infected pigs (21).
Records from the Veterinary Diagnostic Laboratory at the University of Minnesota showed that H. parasuis was isolated from 85% of the case samples submitted but that only 2% of the samples included isolates from pigs with generalized cases of polyserositis. Additionally, very few isolates were recovered from the brains of pigs with clinical meningitis (22).
Studies of the virulence factors for H. parasuis as well as its pathogenicity have been inconsistent. Some studies have demonstrated virulence to be associated with capsule expression (4), outer membrane protein (OMP) profiles (10, 12), or whole-cell protein profiles (7). In contrast, other studies have demonstrated that some of these apparent virulence factors can also be found in isolates recovered from healthy piglets (9, 17). Although the factors that enable the organism to be virulent and therefore to cause clinical disease have not been established, there may be an association between serovars and presentation of severe disease (14). This suggests that heat-stable capsular or lipooligosaccharide antigens may be partially associated with virulence. However, both virulent and avirulent strains of the same serovar have also been reported previously (15), suggesting that other factors may be involved in virulence. The present study investigated both DNA and OMP profiles of strains recovered from different body sites of healthy and clinically sick pigs. In addition, the relationship of protein profiles to site of isolation was examined in an effort to indirectly associate certain protein profiles with virulence. This information will be relevant to the swine industry, since appropriate control of H. parasuis disease will likely be achieved only when the organism's pathogenicity and virulence factors are well understood.
The objectives of this study were (i) characterization and comparison of the OMP and DNA profiles of H. parasuis strains recovered from systemic and respiratory sites of pigs originating from different herds and (ii) characterization of a multifarm H. parasuis outbreak, based on genotypic and phenotypic features of the strains involved.
MATERIALS AND METHODS
Bacterial isolates.
Experiment 1 was conducted using H. parasuis isolates recovered from pigs either submitted to the Veterinary Diagnostic Laboratory at the University of Minnesota or obtained by our laboratory from field cases. A total of 53 H. parasuis isolates obtained from different farms and regions in the United States were evaluated. Twenty-eight isolates had been recovered from pigs with systemic polyserositis, seven isolates were recovered from pneumonic lesions, and 18 isolates were recovered from the respiratory tract of healthy pigs. In experiment 2, 31 H. parasuis isolates recovered from diseased pigs involved in a multifarm outbreak were evaluated. Twenty of the 31 isolates were recovered from respiratory sites (lung), and 11 were recovered from systemic sites (joint, brain, heart, or spinal fluid). Most of the lung isolates were from pneumonic lungs, with the exception of two isolates from lungs without lesions. All H. parasuis isolates were thawed and cultured in chocolate agar plates for 24 h at 37°C. Single colonies were then recultured in PPLO medium supplemented with 5% horse serum and 40 μg of NAD/ml. Biochemical tests were performed to confirm the identity of the strains (13).
OMP profiles.
In order to avoid possible variation between isolates due to growth conditions, all H. parasuis isolates for each experiment were grown under the same culture conditions. The method used for OMP isolation has been described by Carlone et al. (3). Briefly, an overnight culture of H. parasuis was centrifuged for 20 min at 2,500 × g. The pellet was resuspended in 10 mM HEPES buffer (pH 7.4), and the suspension was subjected to sonication. Cellular debris was removed by centrifugation at 26,450 × g for 3 min. The supernatant was then removed and centrifuged at 26,450 × g for 40 min at 4°C. The pellet containing the cell membrane material was resuspended in HEPES buffer and sodium lauryl sarcosinate (2% in HEPES buffer) for 30 min. After that, each preparation was centrifuged at 26,450 × g for 40 min at 4°C. The resulting pellet was washed once with 10 mM HEPES buffer, without resuspending the pellet. The membrane pellet was then resuspended in 10 mM HEPES buffer, and the membrane suspension was diluted in distilled water to a concentration of 1,400 μg of protein per ml. These solutions were then stored at −70°C until all samples were processed. Twenty-five microliters of each membrane suspension was added to 50 μl of sample buffer containing 50% bidistilled water, 12.6% 0.5 M Tris-HCl (pH 6.8), 10.5% glycerol, 21% sodium dodecyl sulfate (SDS) (10% solution), and 5% bromophenol blue (0.1% solution). The final solution was boiled for 5 min. Samples were loaded onto SDS-polyacrylamide gels (4% stacking gel and 10% separating gel), prepared, and run according to the method of Laemmli (5) as modified by Carlone et al. (2).
DNA profiles.
H. parasuis was cultured in chocolate agar for 24 h and harvested in 1.5 ml of phosphate-buffered saline. After that, the culture was pelleted in a microcentrifuge (30,600 × g for 5 min) and resuspended in 250 μl of phosphate-buffered saline. The solution was then boiled for 5 min, and 250 μl of phenol-isoamyl chloroform was added. The mixture was centrifuged at 30,600 × g for 2 min, and 250 μl of isoamyl chloroform was added to the supernatant. The mixture was centrifuged at 30,600 × g for a further 2 min, and the supernatant was incubated on ice for 10 min with 12 μl of sodium acetate (3 M) and 400 μl of 100% alcohol. The mixture was pelleted in a microcentrifuge at 30,600 × g for 15 min and washed once with 70% alcohol. The DNA pellet was dried, resuspended in 40 μl of Tris-EDTA buffer, and stored at −70°C until all samples were processed (18). For the repetitive element-based PCR (Rep-PCR), the primers used were ERIC-1R (5′-ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC-2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′) (23). Three microliters of the DNA preparation was used as PCR template in the reaction. The amplifications were performed in 25 μl of reaction mixture containing 0.4 μM (each) primer, 0.3 mM nucleotides, 1× PCR buffer, 3 mM MgCl2, and 1 U of Taq polymerase. The reaction was run in a thermocycler for 30 cycles with the following steps: denaturation at 94°C for 30 s, primer annealing at 40°C for 2 min, and extension at 72°C for 2 min. Ten-microliter aliquots of the amplified samples were loaded onto a 2% agarose gel with 0.5 μg of ethidium bromide/ml and run at 70 V for 75 min. Gels were visualized and photographed using the Eagle Eye system (Stratagene, La Jolla, Calif.).
RESULTS
Experiment 1.
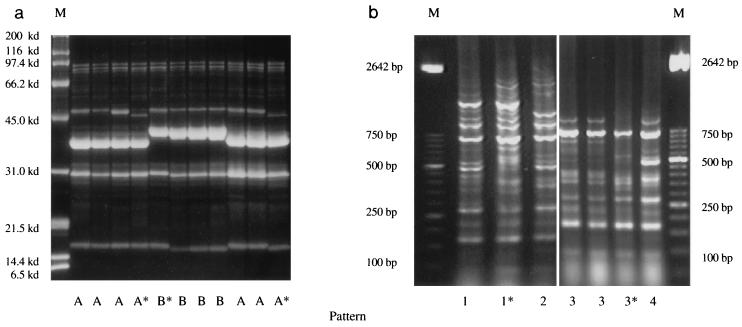
SDS-polyacrylamide gel electrophoresis (PAGE) clearly showed the presence of different OMP profiles between the isolates recovered from healthy pigs and those of sick pigs. The protein profiles of the strains isolated from pigs with polyserositis were clearly similar although not identical (Fig. 1a). Only two OMP profiles were identified among these 28 polyserositis isolates, which had a slight variation in the 36-kDa band level. These strains were characterized by eight different protein bands of different molecular sizes and intensities, with the following molecular masses: two bands between 97.4 and 85 kDa, two very light bands with a molecular mass between 62 and 50 kDa, one light band of approximately 46 kDa, one dense band between 36.6 and 38.5 kDa, one band of approximately 31 kDa, and one light band of approximately 15 kDa.
FIG. 1.
OMP (a) and DNA (b) profiles of the strains isolated from pigs with polyserositis based on SDS-PAGE and Rep-PCR results. Lanes M, molecular size markers. Asterisks indicate isolates showing slight differences within a group.
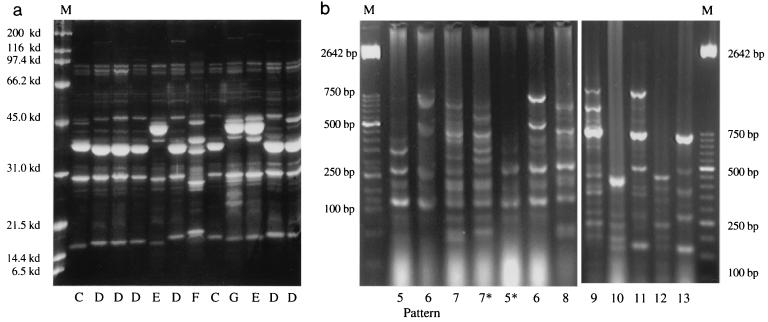
In contrast, protein profiles of isolates from the respiratory tract of healthy pigs showed very heterogeneous patterns, which were distinct from those observed for the systemic strains (Fig. 2a). Five OMP profiles were identified among the 18 isolates. These patterns had eight or more bands, with molecular sizes distinct from those observed for isolates from the pigs with polyserositis. These bands had molecular masses ranging from 116 to 14 kDa.
FIG. 2.
OMP (a) and DNA (b) profiles of the strains isolated from the respiratory tract of pigs without pneumonia based on SDS-PAGE and Rep-PCR results. Lanes M, molecular size markers. Asterisks indicate isolates showing slight differences within a group.
Protein profiles of strains isolated from the respiratory tract of pigs with pneumonia were less heterogeneous than those for strains isolated from the respiratory tracts of healthy pigs (Fig. 3a). A total of four patterns were observed among these seven isolates; two of these patterns were similar to those found for the isolates from systemic sites, while the other patterns were observed in one strain each. These strains had eight or more protein bands with a molecular size range of 116 to 14 kDa. Thirty-four percent of these strains were similar to the ones isolated from systemic sites, 27% had minor differences, and 39% were totally different.
FIG. 3.
OMP (a) and DNA (b) profiles of the strains isolated from pigs with pneumonia based on SDS-PAGE and Rep-PCR results. Lanes M, molecular size markers. Asterisks indicate isolates showing slight differences within a group.
Similarly to the SDS-PAGE results, the Rep-PCR DNA patterns clearly showed differences between the isolates from pigs with polyserositis and those isolated from the respiratory tract of pigs. The strains isolated from pigs with polyserositis were in general homogeneous, but we could recognize four different genetic patterns among the 28 isolates (Fig. 1b). These patterns consisted of eight or more DNA bands with molecular sizes ranging from 2,642 to 50 bp.
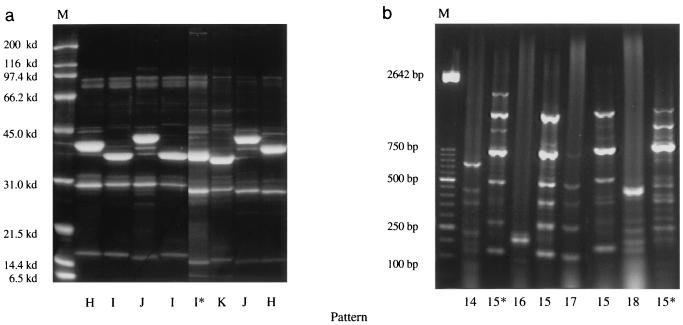
Genetic patterns from isolates from the respiratory tract of pigs without pneumonia were very heterogeneous (Fig. 2b). We identified nine patterns, consisting of 5 to 13 different bands, for the 18 isolates.
Genetic patterns from isolates from the respiratory tract of pigs with pneumonia also showed some heterogeneity (Fig. 3b). In these seven isolates, we identified five patterns, with 6 to 12 different DNA bands with different intensities.
Experiment 2.
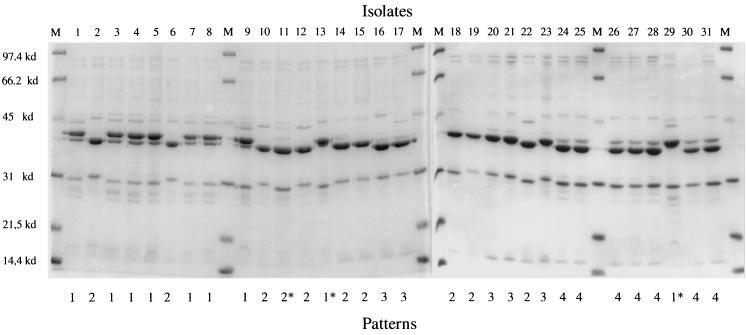
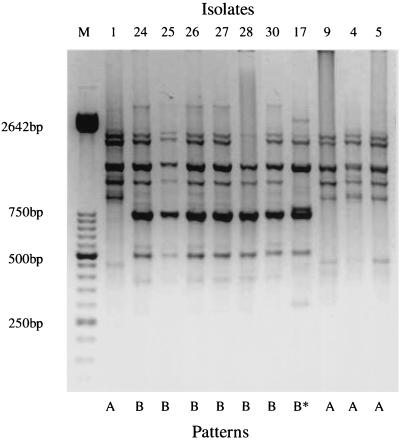
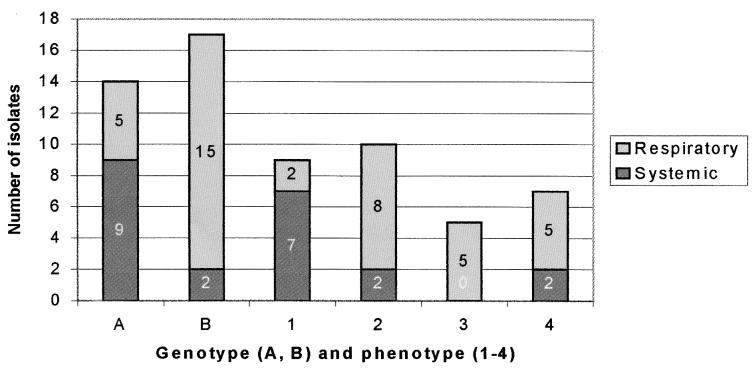
The evaluation of H. parasuis isolates involved in the multifarm outbreak showed considerable uniformity among OMP and DNA profiles. The 31 H. parasuis isolates were clustered into four phenotype (Fig. 4) and two genotype (A and B) (Fig. 5) groups based on OMP and Rep-PCR profiles, respectively. The association of OMP profiles and serotype is shown in Table 1. Fourteen isolates were classified as genotype A, and 17 isolates were classified as genotype B. Nine isolates were classified as OMP pattern 1, 10 were classified as pattern 2, five were classified as pattern 3, and seven were classified as pattern 4. Most of the systemic isolates were classified as genotype A (9 of 11) and OMP pattern 1 (7 of 11), and most of the lung isolates were classified as genotype B (15 of 20) (Fig. 6). Lung isolates had more heterogeneous OMP patterns. However, groups 2, 3, and 4 included most of the lung isolates (18 of 20). Differences among isolates clustered in different groups were very slight, especially isolates classified as OMP groups 2 and 3. The major OMP in all isolates had a molecular size between 31 and 45 kDa (Fig. 4).
FIG. 4.
OMP patterns (1, 2, 3, and 4) of 31 H. parasuis isolates recovered from diseased pigs, based on SDS-PAGE results. Lanes M, molecular size markers. Asterisks indicate isolates showing slight differences within a group.
FIG. 5.
Genomic patterns (A and B) of H. parasuis isolates recovered from diseased pigs, based on Rep-PCR results. Lane M, molecular size markers. The asterisk indicates an isolate showing slight differences from isolates from group B.
TABLE 1.
Genotype and phenotype classification of 31 H. parasuis isolates recovered from different sites of diseased pigs involved in a multifarm outbreak
| Isolate | Identification | Site of isolation | Genotype | Phenotype (OMP) |
|---|---|---|---|---|
| 1 | 32735 | Joint | A | 1 |
| 2 | 35594-1 | Joint | A | 2 |
| 3 | 35594-2 | Brain | A | 1 |
| 4 | 35594 sfa | Spinal fluid | A | 1 |
| 5 | 35594-BrE | Brain | A | 1 |
| 6 | 35594 H+H | Heart | Aa | 2 |
| 7 | 35594 JTB sat | Joint | A | 1 |
| 8 | 35595 JTB sun | Joint | A | 1 |
| 9 | 35566 | Lung | A | 1 |
| 10 | 34082-10 | Lung | A | 2 |
| 11 | 34082-11 | Lung | A | 2c |
| 12 | 33768 L2 | Lung | Aab | 2 |
| 13 | 33768 L4 | Lung | Aab | 1c |
| 14 | 9861-3 | Lung | B | 2 |
| 15 | 36836 JT2 | Lung | B | 2 |
| 16 | 35567 | Lung | B | 3 |
| 17 | 32957 | Lung | B | 3 |
| 18 | 33550 L7 | Lung | Ba | 2 |
| 19 | 34082-7 | Lung | B | 2 |
| 20 | 34082-8 | Lung | B | 3 |
| 21 | 33768 L1 | Lung | B | 3 |
| 22 | 33768 L3 | Lung | B | 2 |
| 23 | 33768 L7 | Lung | B | 3 |
| 24 | 36836 LH | Lung | B | 4 |
| 25 | 36836 LJ | Lung | B | 4 |
| 26 | 36836 LK | Lung | B | 4 |
| 27 | 36836 LL | Lung | B | 4 |
| 28 | 36836 L6 | Lung | B | 4 |
| 29 | 35594 H+F | Heart | A | 1 |
| 30 | 36836 JT1 | Joint | B | 4 |
| 31 | 36836 JT2 | Joint | B | 4 |
Clonally related (similar but not identical genotype).
Lung without lesion.
Very similar OMP patterns.
FIG. 6.
Distribution of systemic and respiratory isolates recovered from diseased pigs involved in a multifarm outbreak according to the genotype (A and B) and phenotype (1, 2, 3, and 4).
DISCUSSION
The SDS-PAGE results for the polyserositis isolates showed that they were clearly similar. In experiment 1, we identified two different patterns whose principal variation was the location of a dense protein band with a molecular size range of 36.3 to 38.5 kDa. This band had a molecular size similar to that of a protein that had been previously described as being associated with virulence in H. parasuis (16, 20).
Strains isolated from the respiratory tract of pigs without pneumonia showed considerable heterogeneity. Only 34% of the strains were similar to the systemic strains. Another 27% showed minor differences, especially around the 40-kDa band. Finally, 39% of the strains had a pattern that was totally different from those of systemic strains. Most importantly, these strains lacked the protein with a molecular size of 36 to 39 kDa, which had been previously associated with virulence.
Strains isolated from pneumonic lungs also showed some heterogeneity, but less so than the respiratory strains from pigs without pneumonia. All of these strains had patterns that could be seen in the other two types of isolates. However, 29% of the isolates had patterns different from those seen in the septicemic cases.
The Rep-PCR patterns were more variable than the OMP patterns. Systemic isolates, either septicemic or pneumonic, were remarkably similar to each other, even though they were isolated from different farms and regions. These findings suggest a clonal relationship between virulent (systemic) strains. The remarkable similarity of the OMP profiles of systemic isolates also suggests that these proteins may be associated with virulence or that they are virulence markers.
In experiment 2, 31 H. parasuis isolates recovered from clinical cases in a multifarm outbreak were compared based on DNA and OMP profiles. Our results showed that even in large, multifarm companies few H. parasuis strains are involved in clinical outbreaks. In this case, only two genetic patterns were identified. Evaluation of genomic fingerprints showed that the systemic and respiratory isolates tended to be clustered in distinct genotypic and phenotypic groups, indicating again that virulent strains may be clonally related. Similar results were described by Musser et al. (8), who found by multilocus enzyme electrophoresis that pathogenic isolates form a distinct cluster of closely related clones. Additionally, they reported that isolates of the same electrophoretic type generally showed similar OMP patterns. Nevertheless, some electrophoretic types were represented by isolates of different OMP patterns. Musser et al. (8) also showed evidence that chromosomal recombination among cell lines is very infrequent. Based on the OMP and DNA profiles of the second experiment, the genotype A group and OMP group 1 were the groups that included the highest number of systemic isolates. These results support those obtained in experiment 1, where systemic isolates were found to be closely related based on OMP and DNA profiles. Most of the isolates classified as genotype B based on Rep-PCR profiles were lung isolates (15 of 17). However, two systemic isolates were also included in this group. Similar results could be observed for isolates clustered in the genotype A group, where 5 of 14 isolates were recovered from the lung. These results suggest a relationship between lung and systemic isolates. However, the factors involved in systemic spread of respiratory H. parasuis strains still remain to be identified. Most of the lung isolates (18 of 20) were recovered from pneumonic lungs. As experiment 1 demonstrated, pneumonic isolates are more homogeneous and more similar to systemic isolates than are isolates obtained from lungs without lesions. These results are in agreement with those described by Blackall et al. (1), who were able to demonstrate the clonal relationship of different field isolates despite the diversity that exists within serovars. Additionally, they found some electrophoretic types that contained isolates of different serovars.
Experiment 2 also showed that both techniques could successfully be used to characterize virulent strains. There was no clear relationship between Rep-PCR and SDS-PAGE profiles. It was noted that some strains that were clustered in the same genotype group showed different OMP profiles. The repetitive sequences that are used as targets in the Rep-PCR represent noncoding regions, and their role in gene expression has not been clearly defined (6). This can explain why strains with similar genetic fingerprints can express different OMP profiles. Different environmental conditions also could be influencing the expression of different sets of proteins by genetically similar strains. However, we were not able to evaluate these variables in this study. Nevertheless, Rapp et al. (11) did not find an appreciable effect of colony type, growth medium, time of harvest, and in vitro or in vivo passage on the protein profiles. The comparison of the Rep-PCR and OMP profiles showed that these techniques are complementary in strain differentiation. The procedure used for Haemophilus OMP preparation has been used in previous researches (3, 11), and it was shown that this method allows selective solubilization of the inner membrane with sodium N-lauryl sarcosinate.
Results obtained in this study suggest the importance of establishing OMP patterns from strains isolated from different organs. H. parasuis is part of the common flora of the upper respiratory tract of pigs, and its isolation from this site may be clinically irrelevant, since most pigs are colonized. However, some of these isolates appeared to be potentially virulent.
The OMP homogeneity observed among the systemic isolates suggests that only some strains of H parasuis are able to successfully cause disease in pigs. Similar findings were described by Musser et al. (8), who found a close genetic relationship of isolates 1, 5, and 7 in the midwestern United States. This close homogeneity of the OMP patterns also suggests a common clonal origin for these strains. Since DNA patterns were more variable than were protein profiles in the systemic isolates, this also suggests that some OMPs may be involved in virulence either as virulence factors or as markers. It is these systemic strains that need to be detected in order to achieve a correct diagnosis and then perform relevant antibiotic sensitivity and serotyping tests.
REFERENCES
- 1.Blackall P J, Trott D J, Rapp-Gabrielson V, Hampson D J. Analysis of Haemophilus parasuis by multilocus enzyme electrophoresis. Vet Microbiol. 1997;56:125–134. doi: 10.1016/S0378-1135(96)01342-9. [DOI] [PubMed] [Google Scholar]
- 2.Carlone G, Sottnek F, Plikaytis B. Comparison of outer membrane protein and biochemical profiles of Haemophilus aegyptius and Haemophilus influenzae biotype III. J Clin Microbiol. 1985;22:708–713. doi: 10.1128/jcm.22.5.708-713.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlone G, Thomas M, Rumschlag H, Sottnek F. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J Clin Microbiol. 1986;24:330–332. doi: 10.1128/jcm.24.3.330-332.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kielstein P, Rosner H, Muller W. Typing of heat stable soluble Haemophilus parasuis antigen by means of Agargel precipitation and dot blot procedure. J Vet Med. 1991;38:315–320. doi: 10.1111/j.1439-0450.1991.tb00877.x. [DOI] [PubMed] [Google Scholar]
- 5.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 6.Lupski J R, Weinstock G S. Interspersed repetitive DNA sequences in prokaryotic genomes. J Bacteriol. 1992;174:4525–4529. doi: 10.1128/jb.174.14.4525-4529.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morozumi T, Nicolet J. Morphological variations of Haemophilus parasuis strains. J Clin Microbiol. 1986;23:138–142. doi: 10.1128/jcm.23.1.138-142.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Musser J M, Rapp V J, Selander R K. Clonal diversity in Haemophilus pleuropneumoniae. Infect Immun. 1987;55:1207–1215. doi: 10.1128/iai.55.5.1207-1215.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen R. Pathogenicity and immunity studies of Haemophilus parasuis serotypes. Acta Vet Scand. 1993;34:193–198. doi: 10.1186/BF03548209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rapp V, Ross R, Nicolet J. Proceedings of the 9th International Pig Veterinary Society Congress. 1986. Characterization of the outer membrane proteins of Haemophilus parasuis; p. 262. [Google Scholar]
- 11.Rapp V, Munson R S, Ross R F. Outer membrane protein profiles of Haemophilus pleuropneumoniae. Infect Immun. 1986;52:414–420. doi: 10.1128/iai.52.2.414-420.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rapp-Gabrielson V, Gabrielson D, Musser J. Proceedings of the 12th International Pig Veterinary Society Congress. 1992. Phenotypic and genotypic diversity of Haemophilus parasuis; p. 334. [Google Scholar]
- 13.Rapp-Gabrielson V J, Gabrielson D. Prevalence of Haemophilus parasuis serovars among isolates from swine. Am J Vet Res. 1992;53:659–664. [PubMed] [Google Scholar]
- 14.Rapp-Gabrielson V J, Kocur G J, Muir S K, Clark T. Proceedings of the American Association of Swine Practices. 1994. Haemophilus parasuis: immunogenicity and cross protection between different serovars; pp. 26–28. Chicago, Ill. [Google Scholar]
- 15.Rapp-Gabrielson V J, Kocur G J, Clark J T, Muir S K. Haemophilus parasuis: immunity in swine after vaccination. Vet Med. 1997;92:83–90. [Google Scholar]
- 16.Rapp-Gabrielson V J. Haemophilus parasuis. In: Straw B, et al., editors. Diseases of swine. 8th ed. Ames: Iowa State University Press; 1999. pp. 475–481. [Google Scholar]
- 17.Rosner H, Kielstein P, Muller W, Rohrmann B. Relationship between serotype, virulence and SDS-PAGE protein patterns of Haemophilus parasuis. Dtsch Tieraerztl Wochenschr. 1991;98:327–330. [PubMed] [Google Scholar]
- 18.Sambroook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Smart N, Hurmik D, MacInnes J. An investigation of enzootic Glasser's disease in a specific pathogen free grower finisher facility using restriction endonuclease analysis. Can Vet J. 1993;34:487–490. [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor D J. Bacterial diseases. In: Taylor D J, editor. Pig diseases. 7th ed. Glasgow, United Kingdom: Glasgow University; 1999. pp. 218–221. [Google Scholar]
- 21.Torremorell M, Pijoan C. Prolonged persistence of an epidemic Streptococcus suis strain in a closed swine population. Vet Rec. 1998;143:394–395. doi: 10.1136/vr.143.14.394. [DOI] [PubMed] [Google Scholar]
- 22.University of Minnesota. Annual report of the Minnesota Veterinary Diagnostic Laboratory. St. Paul: University of Minnesota; 1999. [Google Scholar]
- 23.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]