Highlights
-
•
HNF1A-AS1 promotes 5-FU resistance in gastric cancer cells.
-
•
HNF1A-AS1 regulates EIF5A2 expression through sponging miR-30b-5p.
-
•
HNF1A-AS1 promotes EMT process and 5-FU resistance through miR-30b-5p/EIF5A2 pathway.
-
•
HNF1A-AS1 promotes 5-FU resistance in gastric cancer in vivo.
Keywords: Gastric cancer, Chemoresistance, HNF1A-AS1, miR-30b-5p, EIF5A2, Epithelial mesenchymal transition
Abbreviations: GC, Gastric cancer; lncRNA, Long non-coding RNA; HNF1A-AS1, LncRNA HNF1A antisense RNA 1; 5-FU, 5-fluorouracil; ceRNA, Competitive endogenous RNA; EMT, Epithelial mesenchymal transition; EIF5A2, Eukaryotic translation initiation factor 5A2; AJCC, American Joint Committee on Cancer; CAP, College of American Pathologists
Abstract
Background
Gastric cancer (GC) is one of the leading causes of cancer-related deaths worldwide and chemoresistance is a major cause for its poor prognosis. Long non-coding RNAs (lncRNAs) are associated with cancer chemoresistance. The current study sought to explore the mechanism of lncRNA HNF1A antisense RNA 1 (HNF1A-AS1) in mediating 5-fluorouracil (5-FU) resistance of GC.
Methods
qRT-PCR was performed to detect the expression level of HNF1A-AS1 in GC tissues and cells. Abnormal expression of HNF1A-AS1 in GC cells was induced by lentivirus infection. Protein levels of EIF5A2, E-Cadherin, Vimentin and N-Cadherin were detected using western blot. Competitive endogenous RNA (ceRNA) mechanisms were explored through luciferase assays and RNA immunoprecipitation (RIP) assays. Functional experiments of chemoresistance were performed by CCK-8 assays, colony formation assays and flow cytometry with the treatment of 5-FU. Mouse tumor xenograft assays were performed to verify the findings in vivo.
Results
The findings showed HNF1A-AS1 was significantly upregulated in GC tissues especially in chemoresistance group. Findings from in vitro and in vivo experiments showed HNF1A-AS1 increased cell viability and proliferation, repressed apoptosis and promoted xenograft tumors growth in the presence of 5-FU. Mechanistic studies revealed HNF1A-AS1 promoted chemoresistance by facilitating epithelial mesenchymal transition (EMT) process through upregulating EIF5A2 expression and HNF1A-AS1 acted as a sponge of miR-30b-5p.
Conclusions
The findings from the current study showed HNF1A-AS1 promoted 5-FU resistance by acting as a ceRNA of miR-30b-5p and promoting EIF5A2-induced EMT process in GC. This indicates that HNF1A-AS1 is a potential therapeutic target for alleviating GC chemoresistance.
Introduction
Gastric cancer (GC) is a common malignant tumor in the world. It is the fifth most common cancer type and the third leading cause of cancer-related deaths globally [1]. Combination of perioperative chemotherapy and surgery is currently the first-line treatment for advanced GC. Although several novel drugs have been developed for patients with advanced GC, 5-fluorouracil (5-FU) is the most widely used basic drug in perioperative chemotherapy and palliative chemotherapy as it improves prognosis of GC patients. However, the global 5-year survival rate of GC is approximately 20–40% [2]. Chemoresistance is one of the main causes of the low survival rate of GC patients. Development of drug resistance significantly limits efficacy of chemotherapy and ultimately leads to chemotherapy failure, tumor progression or recurrence [3]. Therefore, exploring the mechanism of chemoresistance and increasing chemosensitivity is crucial to improve treatment efficacy in GC.
Long non-coding RNAs (lncRNAs) are a family of non-coding RNAs with lengths of more than 200 nucleotides and lack protein-coding capacity [4]. Accumulating studies have reported that lncRNAs play important roles in carcinogenesis, cancer progression and drug resistance [5], [6], [7]. In addition, some lncRNAs are implicated in chemoresistance such as HOTAIR which promotes 5-FU resistance in colorectal cancer through the miR-218/NF-κB pathway [8]. Overexpression of UCA1 promotes development of chemoresistance to cisplatin through the miR-495/NRF2 signaling pathway in patients with non‐small‐cell lung cancer [9]. Moreover, MALAT1 plays a key role in docetaxel resistance in prostate cancer by modulating miR-145–5p/ AKAP12 axis [10]. Currently, studies on the role of lncRNAs in GC 5-FU resistance are limited.
LncRNA HNF1A antisense RNA 1 (HNF1A-AS1) is a 2.455 kb lncRNA located on chromosome 12q24.31. It is transcribed from the opposite strand of HNF1A gene [11]. HNF1A-AS1 is highly expressed in various cancers and is implicated in their progression, including esophageal adenocarcinoma [12], hepatocellular carcinoma [13], colon cancer [14], and non-small-cell lung cancer [15]. In addition, previous studies have reported that HNF1A-AS1 promotes proliferation, invasion and metastasis of GC [16,17]. However, the potential involvement of HNF1A-AS1 in chemoresistance of GC has not been fully elucidated.
Eukaryotic translation initiation factor 5A2 (EIF5A2) is a member of the EIF5A gene family [18]. EIF5A2 is implicated in the progression of various cancers, including colorectal cancer [19], ovarian cancer [20], hepatocellular cancer [21], and bladder cancer [22]. In addition, findings from our previous study showed that EIF5A2 upregulation played an important oncogenic role in GC [23]. Another study demonstrated that EIF5A2 promoted chemoresistance to doxorubicin through regulation of epithelial mesenchymal transition (EMT) in colon cancer cells [24], implying that EIF5A2 may participate in the chemoresistance of cancer. Therefore, whether EIF5A2 is involved in chemoresistance of GC is worth studying.
In the present study, in vitro and in vivo experiments were performed to explore the role of HNF1A-AS1 in 5-FU resistance in GC. Findings from clinical samples showed that among GC patients receiving 5-FU based chemotherapy, HNF1A-AS1 was highly upregulated in chemotherapy resistant patients. Findings from functional experiments showed that HNF1A-AS1 enhanced 5-FU resistance in GC cells. In addition, findings from mechanistic studies indicated that HNF1A-AS1 promoted EMT through EIF5A2 and HNF1A-AS1 acted as a competing endogenous RNA (ceRNA) of miR-30b-5p, which was a suppressive microRNA (miRNA) of EIF5A2. Moreover, knockdown of HNF1A-AS1 reduced tumor volume and weight in vivo. Taken together, our present work reveals a novel regulatory pathway of HNF1A-AS1/miR-30b-5p/EIF5A2/EMT in 5-FU resistance of GC, indicating that HNF1A-AS1 is a potential therapeutic target in GC.
Materials and methods
Clinical samples
A total of 30 gastric cancer tissues and 5 normal gastric mucosal tissues were harvested by endoscopic biopsy from thirty-five individuals at Peking Union Medical College Hospital between 2017 and 2019. The 30 gastric cancer tissues were obtained from 30 gastric cancer patients who were pathologically diagnosed as gastric adenocarcinoma and the 5 normal gastric mucosal tissues were obtained from 5 healthy people for medical examination. The clinical TNM (cTNM) stages of gastric cancer patients were determined as IIB or III by contrast-enhanced computed tomography (CT) based on the 8th edition American Joint Committee on Cancer (AJCC) Staging Manual [25]. All patients initially received 5-FU based neoadjuvant chemotherapy (Oxaliplatin plus S-1, SOX), then underwent radical gastrectomy for cancer. The 30 gastric cancer tissues used in the current study were harvested before neoadjuvant chemotherapy by endoscopic biopsy to ensure reliability of the findings. Pathological tumor response after neoadjuvant chemotherapy was evaluated by analyzing postoperative pathological specimens based on the guidelines of College of American Pathologists (CAP) [26]. In the current study, CAP 0, CAP 1 and CAP 2 were defined as pathological response whereas CAP 3 was defined as no pathological response. In addition, some clinicopathological data of these patients were collected. The study was reviewed and approved by the Institutional Review Board of Peking Union Medical College Hospital and all participants provided written informed consent. The ethical approval code of this study is JS-2587.
Cell culture
Four human gastric cancer cell lines HGC-27, MKN-45, AGS, NCI-N87 and human embryonic kidney (HEK) 293T cells were purchased from China Infrastructure of Cell Line Resource (Beijing, China). Immortalized gastric mucosal epithelial cell line GES-1 was purchased from Beijing ComWin Biotech Co., Ltd (Beijing, China). HEK 293T cells were cultured in Dulbecco's modified eagle medium (DMEM; Gibco, Carlsbad, CA, USA). HGC-27, MKN-45, AGS, NCI-N87 and GES-1 were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA). All media were supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) and 1% penicillin/streptomycin (Gibco, Carlsbad, CA, USA). Cells were cultured at 37 °C in a humidified incubator with 5% CO2.
RNA extraction and quantitative real-time PCR analyses (qRT-PCR)
Total RNA was extracted from tissues or cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). For lncRNA and mRNA detection, cDNA was synthesized from 100 ng extracted RNA using the PrimeScript RT reagent Kit (Takara, Dalian, China) and amplified by qRT-PCR using TB Green Premix Ex Taq II (Takara, Dalian, China). GAPDH was used as internal control. In addition, 1ug total RNA was reverse-transcribed with the Mir-X miRNA First-Strand Synthesis Kit (Clontech, Mountain View, CA, USA) and amplified using the TB Green Advantage qPCR Premix (Clontech, Mountain View, CA, USA) to explore expression of miR-30b-5p. U6 was used as internal control. Real-time PCR was performed using QuantStudio3 System (Applied Biosystems, Foster City, CA, USA). The 2−ΔΔCt method was used to determine the relative expression levels of genes. Primer sequences used in the current study are presented in Supplementary Table S1.
Cell transfection
Stable cell lines with silenced or overexpressed HNF1A-AS1 were established through lentiviral transfection. Lentiviral vectors were packaged with full-length HNF1A-AS1 gene or shRNA targeting HNF1A-AS1 by Sangon Biotech (Shanghai, China). Three sequences of shRNA targeting HNF1A-AS1 are listed in Supplementary Table S2. MKN-45 cells were transfected with shRNA targeting HNF1A-AS1 lentivirus (sh-HNF1A-AS1) or its negative control (sh-NC) and HGC-27 cells were transfected with HNF1A-AS1 overexpression lentivirus (LV-HNF1A-AS1) or its negative control (LV-NC) based on the relative expression level of HNF1A-AS1 in four gastric cancer cell lines. Multiplicity of infection (MOI) value was set to 10 based on the results from preliminary experiments. All transfections were supplemented with 5 μg/mL polybrene. Screening was conducted with 2 μg/mL puromycin (Sangon Biotech, Shanghai, China) for 2 weeks to obtain stable cell lines. Abnormal expression of miR-30b-5p was achieved through transfection with miR-30b-5p mimic or inhibitor (RiboBio, Guangzhou, China) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Negative controls miR-NC and anti-miR-NC were also synthesized by RiboBio. Their sequences are presented in Supplementary Table S2.
Cell viability assays
Cell viability was evaluated using a Cell Counting Kit-8 kit (CCK-8; Dojindo, Kumamoto, Japan). Cells were seeded in 96-well plates at a density of 5000 cells per well. After incubation for 24 h, cells were treated with 5-FU at different concentrations for another 24 h. Then 10ul CCK-8 reagent was added to each well. After incubation for 2 h at 37 °C, the optical density (OD) value was measured at 450 nm using a microplate reader, and the relative cell viability was calculated.
Colony formation assays
Cells were seeded into 6-well plates at 500 cells per well and incubated for 24 h. Cells were then treated with 5 μg/mL 5-FU for another 24 h. After being rinsed with fresh RPMI-1640 medium, cells were then incubated for 2 weeks. At last, the colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet at room temperature. Cell colonies were quantified and recorded.
Apoptosis analyses
Cells transfected with lentivirus used in the current study expressed green fluorescent protein (GFP). Thus, apoptosis assay was performed using an Annexin V, 633 Apoptosis Detection Kit (Dojindo, Kumamoto, Japan). Cells were seeded into 6‐well plates at 1 × 106 cells per well and incubated overnight. Cells were treated with 12.5 μg/mL 5-FU for 24 h. Then cells were harvested, washed with phosphate buffer saline (PBS) and resuspended in binding buffer. Further, cells were stained with 5 μl Annexin V and 5 μl propidium iodide (PI) for 15 min under dark conditions. At last, cell samples were analyzed using BD Accuri C6 Plus flow cytometer (BD Biosciences, San Jose, CA, USA) within 1 h.
Luciferase reporter assays
The psiCHECK-2-HNF1A-AS-1-wt vector/psiCHECK2-HNF1A-AS-1-mut vector and psiCHECK-2-EIF5A2-wt vector/psiCHECK-2-EIF5A2-mut vector were obtained from Promega (Madison, WI, USA). 293T cells were transiently transfected with psiCHECK-2-HNF1A-AS-1-wt vector or psiCHECK2-HNF1A-AS-1-mut vector together with miR-30b-5p mimic or miR-NC in 96-well plates using Lipofectamine 2000. Similarly, psiCHECK-2-EIF5A2 3′UTR-wt vector or psiCHECK-2-EIF5A2 3′UTR-mut vector was co-transfected with miR-30b-5p mimic or miR-NC into 293T cells. Luciferase and Renilla signals were measured after 48 h using a Dual Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) according to the manufacturer's protocol.
RNA immunoprecipitation (RIP)
RIP assays were performed using Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) according to the manufacturer's instructions. Argonaute 2 (AGO2) antibody and immunoglobulin G (IgG) used for RIP assays were purchased from Abcam (Cambridge, UK). Expression levels of HNF1A-AS1 and miR-30b-5p were detected by real-time PCR in MKN-45 cells.
Western blot and antibodies
Total protein was lysed and extracted using RIPA Lysis and Extraction Buffer (Thermo Scientific, Rockford, IL, USA) with Halt Protease and Phosphatase inhibitor Cocktail (Thermo Scientific, Rockford, IL, USA) for 15 min on ice. The lysates were centrifuged at 14,000 g for 15 min. Protein concentration was measured by Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, IL, USA). Approximately 30 µg of extracted protein was separated by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto 0.45 μm polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Membranes were blocked with 5% non-fat milk in tri-buffered saline plus Tween (TBS-T) at room temperature for 2 h. Further, membranes were incubated with primary antibodies against EIF5A2 (1:1000, Abcam), E-Cadherin (1:1000, Cell Signaling Technology, MA, USA), Vimentin (1:1000, Cell Signaling Technology), N-Cadherin (1:1000, Cell Signaling Technology), β-actin (1:1000, Cell Signaling Technology) overnight at 4 °C. β-actin was used as internal control. Membranes were then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:10,000, Cell Signaling Technology) at room temperature for 1 h. After incubation with antibodies, membranes were washed and immunoblots were visualized using SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Scientific, Rockford, IL, USA).
Mouse tumor xenograft assays
Male athymic nude BALB/c mice were purchased from Charles River (Beijing, China). Mice were randomly divided into four groups at 5 weeks of age (n = 5/group) based on the different types of injected cells. (1) MKN-45 cells transfected with negative control lentivirus (sh-NC), (2) MKN-45 cells transfected with shRNA targeting HNF1A-AS1 lentivirus (sh-HNF1A-AS1), (3) HGC-27 cells transfected with negative control lentivirus (LV-NC) and (4) HGC-27 cells transfected with HNF1A-AS1 overexpression lentivirus (LV-HNF1A-AS1). 1 × 107 cells were subcutaneously injected into the armpit of the mice. 10 days after administration of GC cells, all the mice were intraperitoneally administered with 5-FU (30 mg/kg) every 3 days for eight cycles. Tumor lengths and widths were measured during this period, and tumor volumes were calculated as follows: tumor volume = (length × width2)/2. Tumors were then harvested and weighed after the eighth cycle. Animal procedures were in line with the National Institutes of Health guide for the care and use of Laboratory animals and approved by the Ethics Committee of Animal Experiments of Peking Union Medical College Hospital.
Statistical analysis
Data were expressed as mean ± standard deviation (SD) of at least three separate experiments. Differences between groups were analyzed using Student's t-test or one-way analysis of variance (ANOVA). Chi-square (χ2) test was used to analyze the association between HNF1A-AS1 expression and clinicopathological factors. Spearman correlation analysis was performed to explore the relationship between expression level of HNF1A-AS1 and miR-30b-5p. Statistical analyses were performed using GraphPad Prism 8 (GraphPad Prism Software, Inc., San Diego, CA, USA). p < 0.05 was considered statistically significant.
Results
High HNF1A-AS1 expression level is associated with poor pathological response to 5-FU based neoadjuvant chemotherapy in GC patients
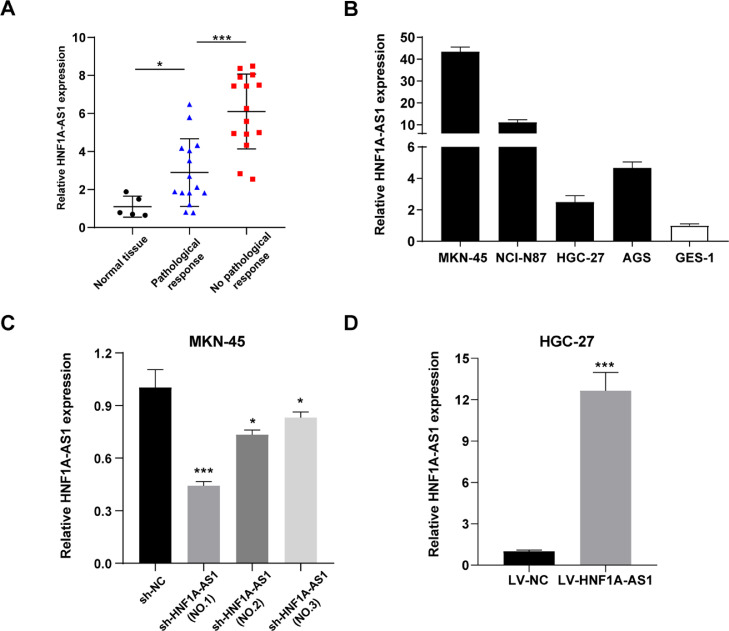
The current study sought to explore the clinical value of HNF1A-AS1 in 30 GC patients receiving 5-FU based neoadjuvant chemotherapy. The findings showed that expression of HNF1A-AS1 was significantly higher in GC tissues compared with that in normal tissues by qRT-PCR (Fig. 1A). Patients were then grouped into high or low expression groups based on the median score of HNF1A-AS1 expression level. And clinicopathological characteristics of these patients are summarized in Supplementary Table S3. Further analysis showed that patients who did not respond to chemotherapy had significantly higher HNF1A-AS1 expression level compared with patients who responded to chemotherapy as shown by postoperative pathology analysis (CAP grading, Fig. 1A). These findings indicated that HNF1A-AS1 was upregulated in GC tissues especially in the chemoresistance group.
Fig. 1.
HNF1A-AS1 expression was upregulated in GC tissues and cell lines. (A) Expression levels of HNF1A-AS1 in normal gastric mucosal tissues (n = 5) and in GC tissues of patients with pathological response (n = 15) and no pathological response (n = 15) detected by qRT-PCR. (B) Relative expression level of HNF1A-AS1 in GC cell lines and the normal gastric mucosal epithelial cell line GES-1 detected by qRT-PCR. (C) Validation of knockdown efficacy of HNF1A-AS1 in MKN-45 cell line by qRT-PCR. (D) Validation of overexpression efficacy of HNF1A-AS1 in HGC-27 cell line by qRT-PCR. Data were presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
HNF1A-AS1 is overexpressed in GC cells
qRT-PCR was performed to detect HNF1A-AS1 expression levels in GC cell lines and normal gastric mucosal epithelial cell line GES-1. The findings showed that the four GC cell lines had higher HNF1A-AS1 expression levels compared with GES-1 cell line. Notably, expression of HNF1A-AS1 was highest in MKN-45 cells and lowest in HGC-27 cells (Fig. 1B). Therefore, HNF1A-AS1 was downregulated in MKN-45 cells (sh-HNF1A-AS1) and upregulated in HGC-27 cells (LV-HNF1A-AS1). The findings showed that knockdown effect was more effective using sh-HNF1A-AS1 (NO.1) compared with sh-HNF1A-AS1 (NO.2) and sh-HNF1A-AS1 (NO.3) in MKN-45 cells (Fig. 1C). Therefore, sh-HNF1A-AS1 (NO.1) was used for further experiments. In addition, a HNF1A-AS1 overexpression model was constructed using HGC-27 cells (Fig. 1D).
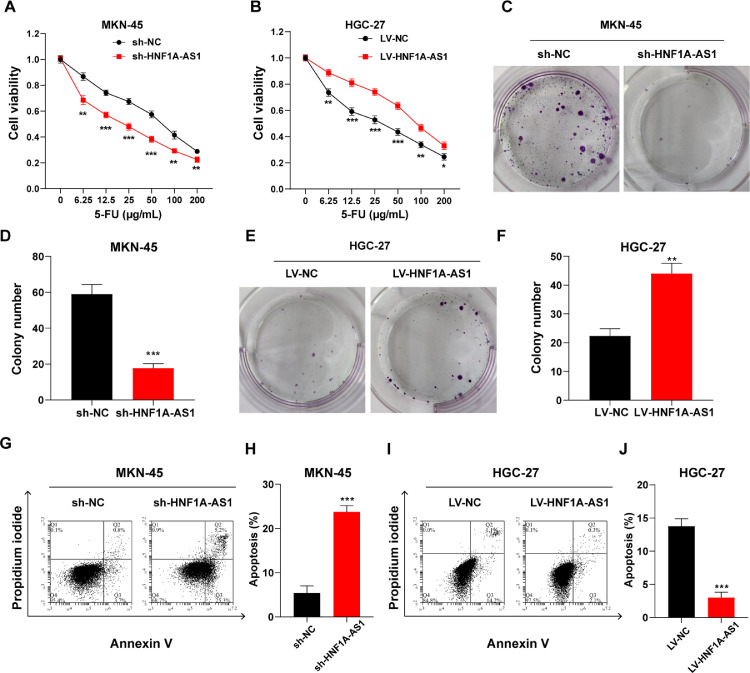
Knockdown/overexpression of HNF1A-AS1 was inversely/positively correlated with the 5-FU resistance of GC cells
Further, sh-HNF1A-AS1 MKN-45 cells/LV-HNF1A-AS1 HGC-27 cells and their negative controls sh-NC MKN-45 cells/LV-NC HGC-27 cells were treated with 5-FU to explore the effect of HNF1A-AS1 on chemoresistance of GC cells. CCK-8 assays showed that knockdown of HNF1A-AS1 decreased cell viability of MKN-45 cells (Fig. 2A). Plate colony formation assays indicated that knockdown of HNF1A-AS1 inhibited colony formation of MKN-45 cells (Fig. 2C and D). Moreover, flow cytometry detection showed suppression of HNF1A-AS1 significantly increased apoptosis of MKN-45 cells (Fig. 2G and H). On the contrary, overexpressing HNF1A-AS1 increased cell viability, promoted colony formation, and inhibited apoptosis in HGC-27 cells (Fig. 2B, E, F, I and J). These findings indicate that knockdown of HNF1A-AS1 decreased chemoresistance of GC cells to 5-FU. However, overexpression of HNF1A-AS1 increased chemoresistance of GC cells.
Fig. 2.
Knockdown/Overexpression of HNF1A-AS1 decreased/increased 5-FU resistance of GC cells. (A) Cell viability of MKN-45 cells transfected with sh-NC or sh-HNF1A-AS1 and treated with 5-FU for 24 h at the indicated concentrations detected by CCK-8 assay. (B) Cell viability of HGC-27 cells transfected with LV-NC or LV-HNF1A-AS1 and treated with 5-FU for 24 h at the indicated concentrations detected by CCK-8 assay. (C and D) Colony formation assay of MKN-45 cells transfected with sh-NC or sh-HNF1A-AS1 and pretreated with 5-FU. (E and F) Colony formation assay of HGC-27 cells transfected with LV-NC or LV-HNF1A-AS1 and pretreated with 5-FU. (G and H) Apoptosis rate of MKN-45 cells transfected with sh-NC or sh-HNF1A-AS1 and treated with 5-FU for 24 h detected by flow cytometry. (I and J) Apoptosis rate of HGC-27 cells transfected with LV-NC or LV-HNF1A-AS1 and treated with 5-FU for 24 h detected by flow cytometry. Data were presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
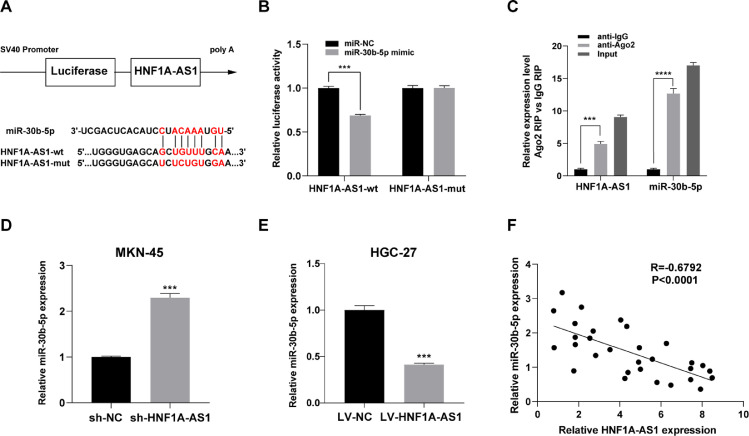
HNF1A-AS1 inhibits miR-30b-5p expression by acting as a sponge
Potential binding sites between HNF1A-AS1 and miR-30b-5p were predicted based through bioinformatic analysis (Fig. 3A). Findings from dual luciferase reporter assay showed that co-transfection with HNF1A-AS1-wt and miR-30b-5p mimic significantly inhibited luciferase activity, whereas co-transfection with HNF1A-AS1-mut and miR-30b-5p mimic had no effect on luciferase activity in 293T cells (Fig. 3B). RIP assay was performed in MKN-45 cells to further explore the potential binding of HNF1A-AS1 and miR-30b-5p. The findings showed that both HNF1A-AS1 and miR-30b-5p were significantly enriched in AGO2 complex, indicating that HNF1A-AS1 was part of the RNA-induced silencing complex (RISC), probably through binding to miR-30b-5p (Fig. 3C). Moreover, analysis of sh-HNF1A-AS1 MKN-45 cells showed that HNF1A-AS1 knockdown increased expression level of miR-30b-5p (Fig. 3D). On the contrary, LV-HNF1A-AS1-mediated overexpression of HNF1A-AS1 downregulated miR-30b-5p expression in HGC-27 cells (Fig. 3E). In addition, Spearman correlation analysis of expression levels in clinical GC samples indicated that HNF1A-AS1 expression was negatively correlated with miR-30b-5p (Fig. 3F). These findings indicated that HNF1A-AS1 acted as a sponge of miR-30b-5p.
Fig. 3.
HNF1A-AS1 suppressed miR-30b-5p expression by acting as a sponge. (A) Schematic diagram showing the predicted binding sites between HNF1A-AS1 and miR-30b-5p. (B) Dual luciferase reporter assay on HEK 293T cells with HNF1A-AS1-wt or HNF1A-AS1-mut was performed to confirm the interaction between HNF1A-AS1 and miR-30b-5p. (C) Cell lysate from MKN-45 cells incubated with anti-Ago2 antibody or IgG antibody for RIP. Expression levels of HNF1A-AS1 and miR-30b-5p were detected by qRT-PCR. (D) Effect of HNF1A-AS1 knockdown on miR-30b-5p expression level in MKN-45 cells analyzed by qRT- PCR. (E) Effect of HNF1A-AS1 overexpression on miR-30b-5p expression level in HGC-27 cells analyzed by qRT- PCR. (F) Correlation between HNF1A-AS1 and miR-30b-5p expression levels in GC tissues. Data were presented as mean ± SD. **P < 0.01, ***P < 0.001, ****P < 0.0001.
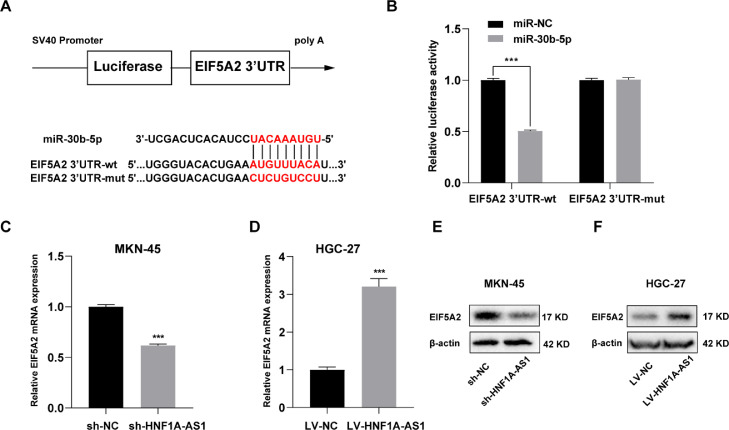
HNF1A-AS1 promoted EIF5A2 expression through miR-30b-5p
A dual luciferase reporter assay was performed to explore the interaction between miR-30b-5p and EIF5A2. Binding site of miR-30b-5p to EIF5A2 3′ UTR were predicted using TargetScan database (Fig. 4A). EIF5A2 3′ UTR-wt/mut were cloned into dual-luciferase vectors and transfected to 293T cells along with miR-30b-5p mimic or miR-NC. The findings showed that co-transfection with EIF5A2 3′ UTR-wt and miR-30b-5p mimic significantly reduced luciferase activity, whereas co-transfection with EIF5A2 3′ UTR-mut mimic and miR-30b-5p had no effect on luciferase activity (Fig. 4B). Contrary to the expression of miR-30b-5p, mRNA and protein levels of EIF5A2 were decreased in sh-HNF1A-AS1 MKN-45 cells after downregulation of HNF1A-AS1 (Fig. 4C and E). And overexpression of HNF1A-AS1 significantly upregulated mRNA and protein levels of EIF5A2 in LV-HNF1A-AS1 HGC-27 cells (Fig. 4D and F).
Fig. 4.
HNF1A-AS1 promoted EIF5A2 expression through miR-30b-5p. (A) Schematic diagram showing the predicted binding sites between EIF5A2 3′UTR and miR-30b-5p. (B) Dual luciferase reporter assay on HEK 293T cells with EIF5A2 3′UTR-wt or EIF5A2 3′UTR-mut was performed to confirm the interaction between EIF5A2 3′UTR and miR-30b-5p. (C) Effect of HNF1A-AS1 knockdown on EIF5A2 mRNA expression level in MKN-45 cells analyzed by qRT- PCR. (D) Effect of HNF1A-AS1 overexpression on EIF5A2 mRNA expression level in HGC-27 cells analyzed by qRT- PCR. (E) Effect of HNF1A-AS1 knockdown on EIF5A2 protein expression level in MKN-45 cells detected by western blot. (F) Effect of HNF1A-AS1 overexpression on EIF5A2 protein expression level in HGC-27 cells detected by western blot. Data were presented as mean ± SD. ***P < 0.001.
Knockdown/overexpression of HNF1A-AS1 alleviated/aggravated 5-FU resistance through the miR-30b-5p/EIF5A2 axis in GC cells
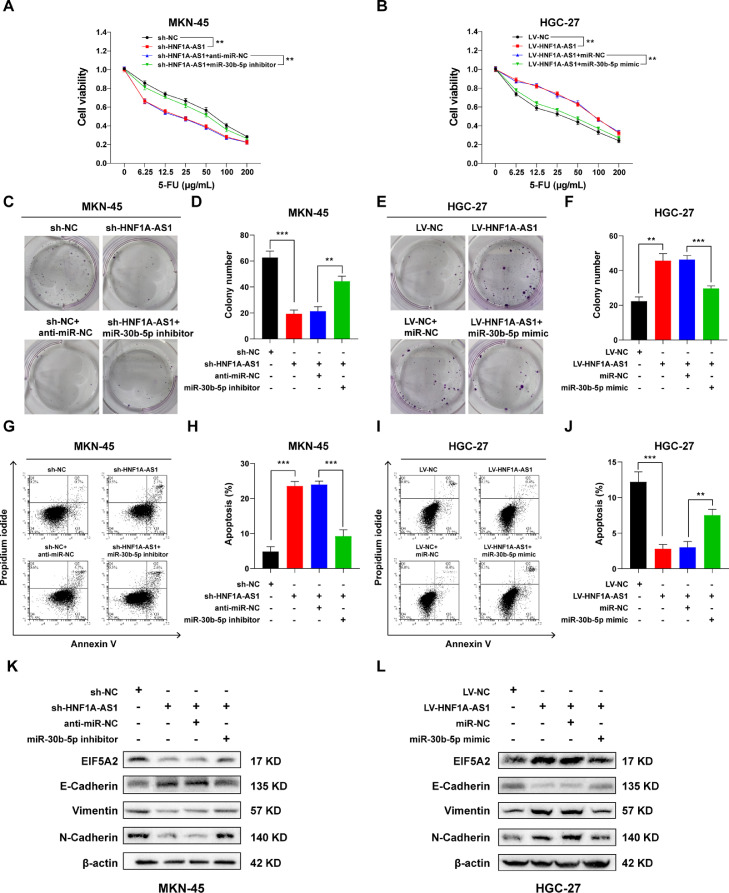
Rescue experiments were conducted to explore the role of miR-30b-5p/EIF5A2 axis in HNF1A-AS1-mediated 5-FU resistance. The sh-HNF1A-AS1 MKN-45 cells were transfected with miR-30b-5p inhibitor or anti-miR-NC whereas LV-NC HGC-27 cells were transfected with miR-30b-5p mimic or miR-NC and then treated with 5-FU for further experiments. The findings showed that miR-30b-5p inhibitor reversed the suppressive effect of HNF1A-AS1 knockdown on cell viability (Fig. 5A) and colony formation (Fig. 5C and D) and decreased cell apoptosis (Fig. 5G and H) in MKN-45 cells, indicating that miR-139–5p inhibitor reversed the suppressive effect of sh-HNF1A-AS1 on 5-FU resistance. On the contrary, miR-30b-5p mimic reversed the stimulative effect of HNF1A-AS1 overexpression on cell viability (Fig. 5B) and colony formation (Fig. 5E and F) and increased cell apoptosis (Fig. 5I and J) in HGC-27 cells, indicating that miR-139–5p mimic reversed the stimulative effect of LV-HNF1A-AS1 on 5-FU resistance. In addition, HNF1A-AS1 knockdown decreased protein expression level of EIF5A2, Vimentin, N-Cadherin and increased protein expression level of E-Cadherin in MKN-45 cells. However, miR-30b-5p inhibitor reversed these effects (Fig. 5K). Moreover, upregulation of HNF1A-AS1 increased protein expression level of EIF5A2, Vimentin, N-Cadherin and decreased protein expression level of E-Cadherin in HGC-27 cells. Notably, miR-30b-5p mimic reversed these effects (Fig. 5L). These findings suggested that HNF1A-AS1 promoted 5-FU resistance of GC cells through EMT by regulating miR-30b-5p/EIF5A2 axis.
Fig. 5.
HNF1A-AS1 induced 5-FU resistance of GC cells through miR-30b-5p/EIF5A2 axis. (A) CCK-8 assay showed miR-30b-5p inhibitor reversed the suppressive effect of HNF1A-AS1 knockdown on cell viability of MKN-45 cells treated with 5-FU. (B) CCK-8 assay showed miR-30b-5p mimic reversed the promotion effect of HNF1A-AS1 overexpression on cell viability of HGC-27 cells treated with 5-FU. (C and D) The miR-30b-5p inhibitor reversed the suppressive effect of HNF1A-AS1 knockdown on colony formation of MKN-45 cells pretreated with 5-FU. (E and F) The miR-30b-5p mimic reversed the promotion effect of HNF1A-AS1 overexpression on colony formation of HGC-27 cells pretreated with 5-FU. (G and H) Flow cytometry showed miR-30b-5p inhibitor reversed the promotion effect of HNF1A-AS1 knockdown on apoptosis of MKN-45 cells treated with 5-FU. (I and J) Flow cytometry showed miR-30b-5p mimic reversed the suppressive effect of HNF1A-AS1 overexpression on apoptosis of HGC-27 cells treated with 5-FU. K Western blot showed HNF1A-AS1 knockdown downregulated EIF5A2, Vimentin, N-Cadherin protein expression and upregulated E-Cadherin protein expression, whereas miR-30b-5p inhibitor reversed these effects in MKN-45 cells. L Western blot showed HNF1A-AS1 overexpression upregulated EIF5A2, Vimentin, N-Cadherin protein expression and downregulated E-Cadherin protein expression, whereas miR-30b-5p mimic reversed these effects in HGC-27 cells. Data were presented as mean ± SD. **P < 0.01, ***P < 0.001.
Knockdown/overexpression of HNF1A-AS1 was inversely/positively correlated with 5-FU resistance of GC in vivo
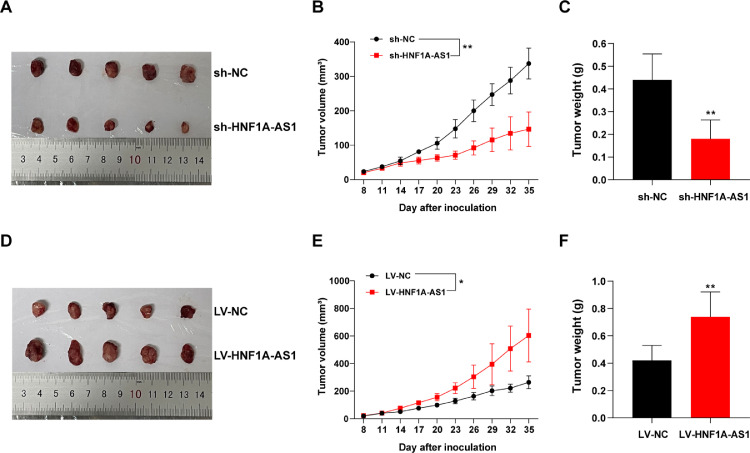
The biological role of HNF1A-AS1 was further explored in vivo. The sh-HNF1A-AS1 MKN-45 cells/LV-HNF1A-AS1 HGC-27 cells or their negative controls sh-NC MKN-45 cells/LV-NC HGC-27 cells were subcutaneously injected into nude mice. Mice were then treated with 5-FU as described in the “Methods” section. The findings showed that HNF1A-AS1 knockdown significantly suppressed chemoresistance to 5-FU compared with the effect in control mice. Volume and weight of tumors derived from MKN-45 cells with HNF1A-AS1 knockdown were significantly decreased compared with those obtained from control xenografts in response to 5-FU (Fig. 6A–C). On the contrary, HNF1A-AS1 overexpression significantly promoted chemoresistance compared with the control group, as indicated by significant increase in tumor volume and weight (Fig. 6D–F).
Fig. 6.
Knockdown/Overexpression of HNF1A-AS1 decreased/increased 5-FU resistance of GC in vivo. (A–C) Tumor volume and weight in nude mice injected with sh-NC MKN-45 cells or sh-HNF1A-AS1 MKN-45 cells. (D-F) Tumor volume and weight in nude mice injected with LV-NC HGC-27 cells or LV-HNF1A-AS1 HGC-27 cells. n = 5/group. Data were presented as mean ± SD. *P < 0.05, **P < 0.01.
Discussion
GC is one of the leading causes of cancer-related deaths around the world and 5-FU based chemotherapy is widely used to treat different types of cancer including GC. However, efficacy of 5-FU is limited by chemoresistance. Therefore, there is a need to explore the mechanism of 5-FU resistance in GC to improve prognosis of GC patients.
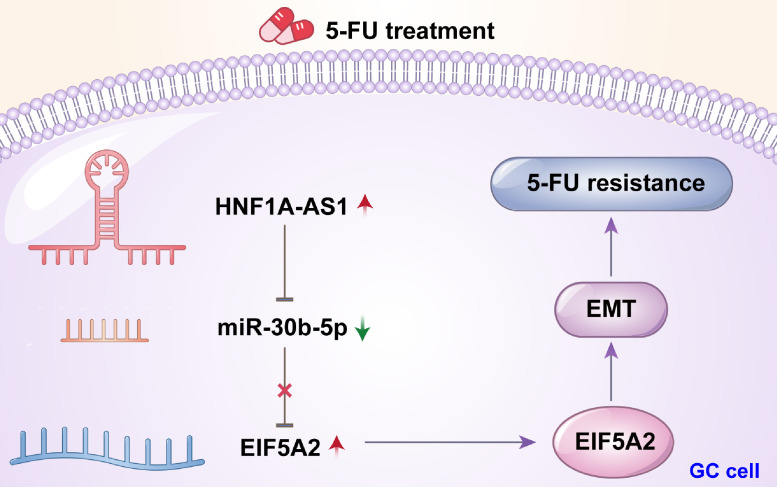
LncRNAs have been demonstrated to be involved in several aspects of cancer development and chemoresistance [5], [6], [7], [8], [9], [10]. Findings from our present study showed that HNF1A-AS1 was significantly upregulated in GC tissues especially in the chemoresistance group. In vitro and in vivo experiments showed that HNF1A-AS1 increased cell viability and proliferation, repressed apoptosis and promoted growth of xenograft tumors in the presence of 5-FU, indicating that HNF1A-AS1 enhanced 5-FU resistance in GC. Mechanistic researches revealed that HNF1A-AS1 promoted chemoresistance by facilitating EMT process through upregulation of EIF5A2 expression and HNF1A-AS1 acted as a sponge of miR-30b-5p. These findings suggested that HNF1A-AS1 induced 5-FU resistance by modulating miR-30b-5p/EIF5A2/EMT signaling pathway in GC (Fig. 7).
Fig. 7.
A schematic model depicting the molecular mechanism of HNF1A-AS1 in 5-FUresistance of GC.
Emerging evidences have revealed that aberrant expression of lncRNAs are major contributors to carcinogenesis, cancer development, and cancer progression [27]. Some lncRNAs are implicated in important biological processes in GC. For example, abnormal histone modification-activated HOXC-AS3 promotes tumorigenesis of GC by binding to YBX1 [28]. In addition, NEAT1 acts as a sponge of miR-506 to increase proliferation, invasion and migration of GC by inducing STAT3 expression [29]. Moreover, BLACAT1 induces oxaliplatin resistance of GC by promoting ABCB1 protein expression through miR-361 [30]. The findings of our current study showed that HNF1A-AS1 was upregulated in GC tissues and cells, and it exhibited a higher expression level in 5-FU based chemotherapy-resistant tissues, implying that HNF1A-AS1 played an important role in 5-FU resistance of GC. Recent studies have demonstrated that lncRNAs can act as ceRNAs of miRNAs [31]. LncRNAs can block the repression effect of miRNAs on their targets by competitively binding their common miRNA responsive elements (MREs) [32].
In the present study, bioinformatics analysis showed that HNF1A-AS1 and EIF5A2 shared the similar binding sites of miR-30b-5p, which acted as a tumor suppressor in various cancers including GC [33]. Luciferase reporter assays showed that both HNF1A-AS1 and EIF5A2 were physically associated with miR-30b-5p. RIP assays further demonstrated that HNF1A-AS1 acted as a sponge of miR-30b-5p. In addition, findings from rescue experiments showed that downregulation of HNF1A-AS1 decreased expression of EIF5A2 whereas miR-30b-5p inhibitor reversed inhibition of EIF5A2 expression and promoted 5-FU resistance of GC cells. On the contrary, miR-30b-5p mimic significantly alleviated HNF1A-AS1-induced 5-FU resistance by downregulating expression of EIF5A2 in GC cells. These findings demonstrated that HNF1A-AS1 promoted 5-FU resistance, at least partially, through sponging miR-30b-5p and thus regulating EIF5A2. These components formed a ceRNA network in GC.
EMT is a biological process in which epithelial cells lose their polarity and acquire mesenchymal morphology [34]. Previous studies have reported that EMT plays a critical role in chemoresistance in various cancers including GC [35], [36], [37]. Therefore, our current study sought to explore the relationship between EIF5A2 and EMT in 5-FU resistance of GC. The findings showed that overexpression of EIF5A2 induced by upregulation of HNF1A-AS1 significantly decreased protein level of E-Cadherin and increased protein level of Vimentin and N-Cadherin in HGC-27 cells, implying the occurrence of EMT. On the contrary, HNF1A-AS1 knockdown-induced downregulation of EIF5A2 reversed EMT process in MKN-45 cells. These findings indicated that HNF1A-AS1 promoted 5-FU resistance of GC by modulating EMT through upregulation of EIF5A2.
Conclusions
In summary, the findings of our current study shows that HNF1A-AS1 is upregulated in GC tissues especially in non-response group receiving 5-FU based chemotherapy. Moreover, EIF5A2-induced EMT process promotes chemoresistance to 5-FU in GC cells, which is modulated by HNF1A-AS1/miR-30b-5p axis. In addition, suppression of HNF1A-AS1 inhibits tumor growth and alleviates chemoresistance in vivo. Therefore, HNF1A-AS1 is a promising therapeutic target against 5-FU resistance in GC. Further studies should be performed to explore precise strategies targeting HNF1A-AS1/miR-30b/EIF5A2 pathway.
CRediT authorship contribution statement
Lin Jiang: Conceptualization, Visualization, Methodology, Data curation, Formal analysis, Writing – original draft, Writing – review & editing, Validation. Yingjing Zhang: Methodology, Writing – review & editing, Validation. Pengfei Su: Methodology, Writing – review & editing, Validation. Zhiqiang Ma: Data curation, Writing – review & editing, Validation. Xin Ye: Data curation, Writing – review & editing, Validation. Weiming Kang: Data curation, Writing – review & editing, Validation. Yuqin Liu: Supervision, Writing – review & editing, Validation. Jianchun Yu: Supervision, Writing – review & editing, Validation.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
Acknowledgment
Not applicable.
Funding
This work was supported by Beijing Municipal Science and Technology Commission (D171100006517002, D171100006517004) and CAMS Innovation Fund for Medical Sciences (2020-I2M-C&T-B-017). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101351.
Appendix. Supplementary materials
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Allemani C., Matsuda T., Di Carlo V., Harewood R., Matz M., Nikšić M., Bonaventure A., Valkov M., Johnson C.J., Estève J., CONCORD Working Group Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391:1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biagioni A., Skalamera I., Peri S., Schiavone N., Cianchi F., Giommoni E., Magnelli L., Papucci L. Update on gastric cancer treatments and gene therapies. Cancer Metastasis Rev. 2019;38:537–548. doi: 10.1007/s10555-019-09803-7. [DOI] [PubMed] [Google Scholar]
- 4.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartonicek N., Maag J.L., Dinger M.E. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol. Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M., Han D., Yuan Z., Hu H., Zhao Z., Yang R., Jin Y., Zou C., Chen Y., Wang G., et al. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9:1149. doi: 10.1038/s41419-018-1187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi X., Sun M., Liu H., Yao Y., Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Li P., Zhang X., Wang L., Du L., Yang Y., Liu T., Li C., Wang C. lncRNA HOTAIR contributes to 5FU resistance through suppressing miR-218 and activating NF-κB/TS signaling in colorectal cancer. Mol. Ther. Nucleic Acids. 2017;8:356–369. doi: 10.1016/j.omtn.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C., Fan K., Qu Y., Zhai W., Huang A., Sun X., Xing S. Deregulation of UCA1 expression may be involved in the development of chemoresistance to cisplatin in the treatment of non-small-cell lung cancer via regulating the signaling pathway of microRNA-495/NRF2. J. Cell Physiol. 2020;235:3721–3730. doi: 10.1002/jcp.29266. [DOI] [PubMed] [Google Scholar]
- 10.Xue D., Lu H., Xu H.Y., Zhou C.X., He X.Z. Long noncoding RNA MALAT1 enhances the docetaxel resistance of prostate cancer cells via miR-145-5p-mediated regulation of AKAP12. J. Cell Mol. Med. 2018;22:3223–3237. doi: 10.1111/jcmm.13604. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Chambers J.C., Zhang W., Sehmi J., Li X., Wass M.N., Van der Harst P., Holm H., Sanna S., Kavousi M., Baumeister S.E., et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat. Genet. 2011;43:1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X., Song J.H., Cheng Y., Wu W., Bhagat T., Yu Y., Abraham J.M., Ibrahim S., Ravich W., Roland B.C., et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–890. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C., Mou L., Chai H.X., Wang F., Yin Y.Z., Zhang X.Y. Long non-coding RNA HNF1A-AS1 promotes hepatocellular carcinoma cell proliferation by repressing NKD1 and P21 expression. Biomed. Pharmacother. 2017;89:926–932. doi: 10.1016/j.biopha.2017.01.031. [DOI] [PubMed] [Google Scholar]
- 14.Fang C., Qiu S., Sun F., Li W., Wang Z., Yue B., Wu X., Yan D. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. 2017;410:50–62. doi: 10.1016/j.canlet.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Liu L., Chen Y., Li Q., Duan P. lncRNA HNF1A-AS1 modulates non-small cell lung cancer progression by targeting miR-149-5p/Cdk6. J. Cell. Biochem. 2019;120:18736–18750. doi: 10.1002/jcb.29186. [DOI] [PubMed] [Google Scholar]
- 16.Liu H.T., Liu S., Liu L., Ma R.R., Gao P. EGR1-mediated transcription of lncRNA-HNF1A-AS1 promotes cell-cycle progression in gastric cancer. Cancer Res. 2018;78:5877–5890. doi: 10.1158/0008-5472.CAN-18-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H.T., Ma R.R., Lv B.B., Zhang H., Shi D.B., Guo X.Y., Zhang G.H., Gao P. LncRNA-HNF1A-AS1 functions as a competing endogenous RNA to activate PI3K/AKT signalling pathway by sponging miR-30b-3p in gastric cancer. Br. J. Cancer. 2020;122:1825–1836. doi: 10.1038/s41416-020-0836-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan X.Y., Fung J.M., Ma N.F., Lau S.H., Tai L.S., Xie D., Zhang Y., Hu L., Wu Q.L., Fang Y., Sham J.S. Oncogenic role of eIF-5A2 in the development of ovarian cancer. Cancer Res. 2004;64:4197–4200. doi: 10.1158/0008-5472.CAN-03-3747. [DOI] [PubMed] [Google Scholar]
- 19.Zhu W., Cai M.Y., Tong Z.T., Dong S.S., Mai S.J., Liao Y.J., Bian X.W., Lin M.C., Kung H.F., Zeng Y.X., et al. Overexpression of EIF5A2 promotes colorectal carcinoma cell aggressiveness by upregulating MTA1 through C-myc to induce epithelial-mesenchymaltransition. Gut. 2012;61:562–575. doi: 10.1136/gutjnl-2011-300207. [DOI] [PubMed] [Google Scholar]
- 20.Yang G.F., Xie D., Liu J.H., Luo J.H., Li L.J., Hua W.F., Wu H.M., Kung H.F., Zeng Y.X., Guan X.Y. Expression and amplification of eIF-5A2 in human epithelial ovarian tumors and overexpression of EIF-5A2 is a new independent predictor of outcome in patients with ovarian carcinoma. Gynecol. Oncol. 2009;112:314–318. doi: 10.1016/j.ygyno.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 21.Tang D.J., Dong S.S., Ma N.F., Xie D., Chen L., Fu L., Lau S.H., Li Y., Li Y., Guan X.Y. Overexpression of eukaryotic initiation factor 5A2 enhances cell motility and promotes tumor metastasis in hepatocellular carcinoma. Hepatology. 2010;51:1255–1263. doi: 10.1002/hep.23451. [DOI] [PubMed] [Google Scholar]
- 22.Luo J.H., Hua W.F., Rao H.L., Liao Y.J., Kung H.F., Zeng Y.X., Guan X.Y., Chen W., Xie D. Overexpression of EIF-5A2 predicts tumor recurrence and progression in pTa/pT1 urothelial carcinoma of the bladder. Cancer Sci. 2009;100:896–902. doi: 10.1111/j.1349-7006.2009.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng Q.B., Kang W.M., Yu J.C., Liu Y.Q., Ma Z.Q., Zhou L., Cui Q.C., Zhou W.X. Overexpression of eukaryotic translation initiation factor 5A2 (EIF5A2) correlates with cell aggressiveness and poor survival in gastric cancer. PLoS One. 2015;10 doi: 10.1371/journal.pone.0119229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bao Y., Lu Y., Wang X., Feng W., Sun X., Guo H., Tang C., Zhang X., Shi Q., Yu H. Eukaryotic translation initiation factor 5A2 (eIF5A2) regulates chemoresistance in colorectal cancer through epithelial mesenchymal transition. Cancer Cell Int. 2015;15:109. doi: 10.1186/s12935-015-0250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amin M.B., Edge S.B., Greene F.L., Byrd D.R. 8th Ed. Springer; New York, NY: 2017. AJCC Cancer Staging Manual. [Google Scholar]
- 26.Tang L.H., Berlin J., Branton P., Burgart L.J., Carter D.K., Compton C.C., Fitzgibbons P., Frankel W.L., Jessup J., Kakar S., et al. Protocol for the examination of specimens from patients with carcinoma of the stomach. Coll. Am. Pathol. 2014:1–19. v3.3.0.0. [Google Scholar]
- 27.Bhan A., Soleimani M., Mandal S.S. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang E., He X., Zhang C., Su J., Lu X., Si X., Chen J., Yin D., Han L., De W. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19:154. doi: 10.1186/s13059-018-1523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan H.Y., Wang C., Liu G., Zhou X. Long noncoding RNA NEAT1-modulated miR-506 regulates gastric cancer development through targeting STAT3. J. Cell Biochem. 2019;120:4827–4836. doi: 10.1002/jcb.26691. [DOI] [PubMed] [Google Scholar]
- 30.Wu X., Zheng Y., Han B., Dong X. Long noncoding RNA BLACAT1 modulates ABCB1 to promote oxaliplatin resistance of gastric cancer via sponging miR-361. Biomed. Pharmacother. 2018;99:832–838. doi: 10.1016/j.biopha.2018.01.130. [DOI] [PubMed] [Google Scholar]
- 31.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin C., Yang L. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 2018;28:287–301. doi: 10.1016/j.tcb.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian S.B., Yu J.C., Liu Y.Q., Kang W.M., Ma Z.Q., Ye X., Yan C. MiR-30b suppresses tumor migration and invasion by targeting EIF5A2 in gastric cancer. World J. Gastroenterol. 2015;21:9337–9347. doi: 10.3748/wjg.v21.i31.9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dongre A., Weinberg R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 35.van Staalduinen J., Baker D., Ten Dijke P., van Dam H. Epithelial-mesenchymal-transition-inducing transcription factors: new targets for tackling chemoresistance in cancer? Oncogene. 2018;37:6195–6211. doi: 10.1038/s41388-018-0378-x. [DOI] [PubMed] [Google Scholar]
- 36.Dudas J., Ladanyi A., Ingruber J., Steinbichler T.B., Riechelmann H. Epithelial to mesenchymal transition: a mechanism that fuels cancer radio/chemoresistance. Cells. 2020;9:428. doi: 10.3390/cells9020428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marin J.J.G., Perez-Silva L., Macias R.I.R., Asensio M., Peleteiro-Vigil A., Sanchez-Martin A., Cives-Losada C., Sanchon-Sanchez P., Sanchez De Blas B., Herraez E., et al. Molecular bases of mechanisms accounting for drug resistance in gastric adenocarcinoma. Cancers. 2020;12:2116. doi: 10.3390/cancers12082116. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.