Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread worldwide. The pathophysiological mechanisms linking gut dysbiosis and severe SARS-CoV-2 infection are poorly understood, although gut microbiota disorders are related to severe SARS-CoV-2 infections. The roles of the gut microbiota in severe SARS-CoV-2 infection were compared with those in respiratory viral infection, which is an easily understood and enlightening analogy. Secondary bacterial infections caused by immune disorders and antibiotic abuse can lead to dysregulation of the gut microbiota in patients with respiratory viral infections. The gut microbiota can influence the progression of respiratory viral infections through metabolites and the immune response, which is known as the gut–lung axis. Angiotensin-converting enzyme 2 is expressed in both the lungs and the small intestine, which may be a bridge between the lung and the gut. Similarly, SARS-CoV-2 infection has been shown to disturb the gut microbiota, which may be the cause of cytokine storms. Bacteria in the gut, lung, and other tissues and respiratory viruses can be considered microecosystems and may exert overall effects on the host. By referencing respiratory viral infections, this review focused on the mechanisms involved in the interaction between SARS-CoV-2 infections and the gut microbiota and provides new strategies for the treatment or prevention of severe SARS-CoV-2 infections by improving gut microbial homeostasis.
Keywords: SARS-CoV-2, Cytokine storms, Gut microbiota, Gut–lung axis
Graphical Abstract
1. Introduction
An unexplained case of pneumonia was discovered in Wuhan, Hubei Province, China, on 29 December 2019. A new coronavirus, which was subsequently named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the World Health Organisation, was soon isolated from a patient [1]. The epidemic of SARS-CoV-2 infection has increased rapidly, with cases spreading across China and other countries since the first case was confirmed.
SARS-CoV-2 infection often cooccurs with gastrointestinal symptoms [2] that were also commonly associated with other known human coronaviruses (e.g., SARS-CoV and Middle East respiratory syndrome-CoV) [3], [4]. In addition, secondary infection with bacteria after viral infection is very common. Furthermore, secondary infection with respiratory bacteria often impairs the balance of the pulmonary microbiota [5]. However, the largest bacterial pool in the human body is in the intestinal tract, which leads to the vital question of whether the gut microbiota plays an important role in secondary infection. The gut microbiota, which is a general term for the microorganisms that colonise the human intestine, is the second genome in humans and plays a crucial role in maintaining the physiological balance of metabolism and immunity. The roles of gut microbiota dysbiosis are increasingly understood in metabolic diseases (e.g., diabetes and obesity) [6]. Furthermore, increasing evidence suggests that viral infection may disturb gut microbiota homeostasis, resulting in an increase in harmful bacteria and a decrease in beneficial bacteria [7]. Bacteria and viruses can sometimes cooperate to exacerbate illness [8]. Respiratory viral infections can trigger a series of immunologic reactions, which may be caused by the gut microbiota, resulting in cytokine storms in severe cases [9]. SARS-CoV-2 has many similarities with other respiratory viruses in the context of secondary bacterial infection, immunity, and other aspects. Many studies have been conducted examining the relationship between other respiratory viruses and the gut microbiota, although few studies have been conducted on the relationship between SARS-CoV-2 and the gut microbiota. Many mechanisms of mortality caused by SARS-CoV-2 infection remain unclear because studies have shown that there is a relationship between respiratory viruses and the gut microbiota, and it is necessary to explore the potential mechanisms of the gut microbiota in severe SARS-CoV-2 infection. Thus, this study aimed to investigate whether severe SARS-CoV-2 infection is not only caused the virus and subsequent bacterial secondary infections in the respiratory and intestinal tracts but is also closely related to gut microbiota dysbiosis.
2. Cytokine storms in severe SARS-CoV-2 infection
Cytokine storms manifest as extremely increases in inflammatory cytokines and are important causes of severe disease or mortality in patients with coronavirus disease-19 (COVID-19) [10]. Patients with COVID-19 experience a large amount of elevated cytokines. As reported, D-dimer levels were increased in patients with COVID-19, and plasma concentrations of interleukin (IL)-2, IL-6, IL-7, IL-10, granulocyte colony-stimulating factor, INF-inducible protein-10, macrophage inflammatory protein (MIP) 1, MIP1A, and tumour necrosis factor-alpha (TNF-α) were higher in ICU patients than in non-ICU patients with SARS-CoV-2 infection [2], [11]. Among patients, IL-6 levels gradually increased over time and were associated with the highest mortality. In addition, the proportion of CD14 + CD16 + inflammatory monocytes in the peripheral blood of patients with severe disease was significantly higher than that of patients with mild disease [12].
Cytokine storms often occur in patients with severe COVID-19, followed by multiple organ failure and possibly death [11]. A recent study showed that SARS-CoV-2 infection and lung cell destruction first cause a local immune response accompanied by the accumulation of macrophages and monocytes and then cause immune responses in adaptive T and B cells, among which CD8 + T cells and CD4 + T cells play important roles. Viral infection causes a highly inflammatory pyroptosis response and triggers a subsequent inflammatory response, including the secretion of proinflammatory cytokines into the circulatory system. Infection with microorganisms causes rapid and massive production of various cytokines in body fluids (e.g., TNF-α, IL-1, IL-6, IL-12, interferon (IFN)-α, IFN-β, IFN-γ, MCP-1, and IL-8), ultimately leading to a cytokine storm, which is an important cause of acute respiratory distress syndrome and multiple organ failure [12]. An excessive inflammatory response worsens lung pathology and attracts additional inflammatory cells to the site, and these cells increased amounts of cytokines and ultimately lead to a cytokine storm [13]. In addition, acute respiratory distress syndrome (ARDS), which is a clinical symptom resulting from the excessive release of local cytokines, occurred in 67·3% of patients with COVID-19. Furthermore, serum cytokine levels were significantly increased in patients with ARDS, and the degree of this increase correlated positively with mortality [11]. Thus, the occurrence of ARDS is closely related to cytokine storms.
This evidence suggests that patients with severe COVID-19 experience a series of inflammatory responses, which are often followed by cytokine storms. Thus, cytokine storms are one of the characteristics of severe SARS-CoV-2 infection. However, the pathogenesis of cytokine storms following severe SARS-CoV-2 infection is poorly understood. The current study considers the inflammatory response in respiratory viral infections as a reference to better understand this mechanism.
3. SARS-CoV-2 and secondary bacterial infection: Learning from respiratory viral infections
Respiratory viruses often increase the host’s susceptibility to secondary bacterial pneumonia [14]. Bacterial infections further increase the inflammatory response if the bacteria persist [14], [15]. The type 1 interferon response occurs in SARS-CoV-2 infections, as well as in other respiratory viral infections [15]. IFN-I is produced after viral infection and negatively regulates T cells in the lung, inhibits IL-17 production and leukocyte recruitment, and increases the possibility of Streptococcus pneumoniae infection [16]. In influenza-infected animals, Ifnar−/− animals were able to produce the more robust neutrophil chemoattractants KC and Mip2 in response to secondary S. pneumoniae challenge than Ifnar+/+ animals [17]. Thus, IFN-I attenuates early KC and Mip2 chemokine production in response to secondary bacterial infections in the lungs of influenza-infected hosts. IFN-I significantly increases the susceptibility of influenza-infected hosts to secondary bacterial infections in the lung [18]. Similarly, secondary bacterial infections are also observed in SARS-CoV-2 infection [19], suggesting that bacterial infections secondary to respiratory viruses can be used for reference.
A series of studies in recent years have confirmed that the human lower respiratory tract is colonised with microbes [20]. In addition, the impact on the composition of the pulmonary microbiota depends in part on the immune response [21]. Low diversity in the pulmonary microbiota was associated with reduced immune responses [22]. A recent study on SARS-CoV-2 reported that the expression of inflammatory factors was significantly increased by the SARS-CoV-2 S protein and that the compound RS5645 (a new and potent inhibitor) inhibited the expression of a number of inflammatory factors. Mice were individually treated with the S protein plus LPS (M group) and with RS-5645 (R group). The relative abundances of Porphyromonas, Rothia, Streptococcus and Neisseria in the alveoli of Group R mice increased significantly, while the relative abundances of Psychrobacter, Shimia, and Sporosarcina decreased. The alveolar microbiota of Group R mice had increased translation and decreased amino acid metabolic pathway activity. RS-5645 attenuated lung inflammatory cell infiltration and S-protein- and LPS-induced inflammation by modulating the lung microbiota [23]. SARS-CoV-2 infection causes changes in the lung microbiota, but it is not known whether this change leads to secondary bacterial infections or worsened cytokine storms by affecting the lung microbiota. We used other respiratory viruses as examples to explore the underlying relationship between the lung microbiota, secondary infections and cytokine storms. Significant changes in the composition and species diversity of the pulmonary microbiota could be observed in a mouse model of the influenza virus. Among them, the dominant bacterial class changed from Alphaproteobacteria to Gammaproteobacteria and Actinobacteria, and there was a significant increase in the relative abundances of anaerobes and facultative anaerobes (e.g., Streptococcus and Staphylococcus) [24]. Secondary bacterial infections caused by respiratory viruses can cause lung microbiota imbalances. Influenza viruses increase host susceptibility to secondary lung bacterial colonisation [24]. The lung microbiota provides the necessary signals for the secretion of inflammatory cytokines when mice are infected with a virus, resulting in inflammasome activation and IL-1β and IL-18 release. Inflammasome activation is associated with the migration of lung dendritic cells (DCs) to draining lymph nodes, where these cells stimulate innate and adaptive immune responses to promote the priming of T cells. Notably, both signal expression and migratory activity require commensal bacteria. This mechanism exerts beneficial immunomodulatory effects on the respiratory mucosa [25]. Interestingly, innate immunity acts as the first defence against viral infection, and immune cells are recruited to the infected airway by the production of inflammatory cytokines early during viral infection. Inflammatory cells induce phagocytosis by secreting inflammatory cytokines [26]. Macrophages bind RNA from the influenza A virus by secreting the pattern recognition receptor NLRP3 to form inflammasomes during infection with the influenza virus. Inflammasomes prompt macrophages to secrete IL-1β and IL-18, leading to an inflammatory response [27], [28]. This immune response controls viral replication and spread during the early infection stage, but this response can trigger a cytokine storm when the inflammatory response is too strong, leading to lung tissue damage [26], [27], [28]. Therefore, secondary bacterial pneumonia is highly likely to occur after respiratory viral infection. Respiratory viral infections can exert a direct effect on the respiratory microbiota, increasing the risk of exogenous pathogen colonisation in the lungs and resulting in secondary bacterial infection. Cytokine storms may also occur in the context of severe respiratory viral infections. We are trying to identify clues to the mechanism of the interaction between the lung microbiota and viruses to help us better understand SARS-CoV-2 infections and the lung microbiota.
As mentioned previously, severe SARS-CoV-2 infections often cause cytokine storms, which are very complex conditions leading to death in patients [10]. Therefore, we can examine the relationship between other respiratory viral infections and cytokine storms for clues. A cytokine storm often originates from bacterial invasion. Previous studies have shown that Streptococcus pyogenes can invade the skin and mucous membranes, while superantigenic streptococcal pyrogenic exotoxins can contribute to tissue invasion and trigger cytokine storms [29]. In addition, staphylococcal and streptococcal superantigens rapidly elicit IL-17A responses in humans, which cause hyperinflammatory responses and toxic shock syndrome, as represented by cytokine storms [30]. IL-18 may be involved in these excessive inflammatory responses because IL-18 also plays a role in haemophagocytic lymphohistiocytosis, which is characterised by a cytokine storm that may be secondary to bacterial infection. IL-18-mediated inflammation has been shown in animal studies of bacterial infection to reduce IL-18-induced IFN-γ release. Mice treated with anti-IL-18 antibodies showed lower IFN-γ levels and were more susceptible to infection caused by intracellular bacteria than control mice. Thus, IL-18 may be involved in bacterial clearance, especially in infections caused by intracellular bacteria [31].
Respiratory viral infection induces the production of various cytokines, occasionally leading to cytokine storms. Cytokines can be produced by viruses or bacteria. Toll-like receptors (TLRs), which receive signals from inflammatory cytokines, may be responsible for cytokine production in bacterial- or viral-infected epithelial and immune cells [32]. TLR-4 is a ligand for the RSV F protein, which activates the MyD88-dependent signalling pathway and leads to the production of Th1 cytokines [33]. However, TLR-4 is more likely to act as a receptor for LPS than for other factors, activating the innate immune system [14], [34]. Thus, respiratory viral infections accompanied by bacterial infections may activate receptors that cause the secretion of inflammatory cytokines [14]. This effect is also observed in SARS-CoV-2 infections. A number of indicators associated with bacterial infection were shown to be elevated in patients with severe SARS-CoV-2 who presented with cytokine storms [35].
Analogous to other respiratory viral infections, SARS-CoV-2 infections can also predispose the lungs to secondary bacterial infections. In a survey of 1495 hospitalised patients with COVID-19, 102 (6.8%) patients suffered from secondary bacterial infections, and nearly half of patients (50/102) expired due to bacterial infections dominated by gram-negative bacteria. A greater incidence of SBIs results in more severe illness [19]. In severe COVID-19 cases, the affected lung tissue is very conducive to the growth of opportunistic pathogens, including Pseudomonas aeruginosa and Staphylococcus aureus [36]. Critically ill patients with COVID-19 often experience cytokine storms, sharp decreases in lymphocytes and natural killer cells and increases in D-dimer, C-reactive protein, ferritin, and procalcitonin levels [35], which are usually considered indicators of bacterial infection. In patients with severe and critical COVID-19, a highly impaired interferon type I response occurs, and the degree of impairment is associated with increased blood viral load and increased inflammatory responses. The increases in TNF-α and IL-6, which affect the transcription factor nuclear factor-κB, lead to an inflammatory response [15]. IL-18 may be involved in secondary COVID-19 bacterial infection. Either viruses or bacteria are possible agents contributing to severe COVID-19 progression [36]. Thus, the aforementioned evidence indicate that SARS-CoV-2 infection easily leads to secondary bacterial infection, and bacteria may lead to cytokine storms through a series of immune effects, in addition to the direct stimulation of cytokine storms by SARS-CoV-2.
4. Gut microbiota dysbiosis in COVID-19 and cytokine storms: Learning from respiratory viral infections
Antibiotic abuse occurs during the treatment of SARS-CoV-2 infections [19] and has been observed in the treatment of other respiratory viral infections. Antibiotic abuse can lead to microbiota depletion and a decrease in immune antibody production. Many patients can improve with the combination of hydroxychloroquine and streptomycin, but 30–40% of common bacteria are resistant to streptomycin [37]. Therefore, antibiotic overuse may result in bacterial resistance, which leads to poor antibiotic efficacy. Another unfortunate outcome of antibiotic abuse is subsequent gut microbiota dysbiosis. Although the abuse of antibiotics in the treatment of SARS-CoV-2 infections has not been reported to cause gut microbiota dysbiosis, such an incorrect treatment for other respiratory viruses can damage the structure of the gut microbiota. This evidence serves as a warning for the appropriate use of antibiotics and maintaining intestinal homeostasis in the clinical management of SARS-CoV-2 infections. A significant increase in resistance genes was observed in the gut microbiota after amoxicillin administration in mice. The abundance of certain opportunistic pathogens (e.g., Klebsiella and Escherichia–Shigella) significantly increased, while potential beneficial gut bacteria (e.g., Bifidobacterium and Lactobacillus) significantly decreased when antibiotics were used [38]. In addition, the use of antibiotics can affect bile acid metabolism and cause inflammation [37]. Antibiotic use leads to disturbances in the gut commensal microbiota, which inhibits commensal bacteria-mediated immune regulation and exacerbates respiratory symptoms [25]. Gut microbiota dysbiosis caused by antibiotic use weakens the immune response to the influenza virus. Mice with dysregulated gut microbiotas showed prolonged viral clearance in vivo, resulting from reduced expression of IFN-I and IFN-II, reducing the ability of these animals to limit viral replication during antibiotic use [39].
Intestinal secondary infection with exogenous bacteria may lead to gut microbiota dysbiosis in patients infected with respiratory viruses. Mice with an influenza virus that had been treated with streptomycin were administered Salmonella by gavage, and the results showed that IFN-I induced in the lung during Salmonella-induced colitis inhibited the antibacterial and inflammatory responses of the intestine, disrupted the balance of the gut microbiota, and promoted Salmonella colonisation in the intestine and systemic transmission. IFN-I produced in the lung was transmitted to the intestine, promoting Salmonella growth in other parts of the body, such as the intestine, causing secondary infections [40]. Although the disruption of intestinal homeostasis after exogenous bacterial invasion has not been reported in SARS-CoV-2 infections, this possibility still needs to be considered in the future.
The gut microbiota may also play an important role in SARS-CoV-2 infections, but the exact mechanisms have not been elucidated. We can gain some insight from the role of the gut microbiota in some inflammatory lung diseases. Respiratory viruses can change the gut microbiota through the IFN-I pathway. The abundance of Proteobacteria, especially Escherichia coli, was increased in influenza A4-infected mice, indicating that IFN-I produced in the lung can lead to changes in the gut microbiota [40]. In addition, lung effector T cells and CCR9 + CD4 + T cells are recruited to the intestine, where they cause changes in the gut microbiota by secreting IFN-I. The gut microbiota is then characterised by E. coli growth, and this change in the microbiota promotes IL-15 expression, resulting in Th17 cell polarization and IL-17 production, ultimately leading to immune damage in the intestine [41]. Moreover, gut microbiota dysbiosis following respiratory viral infections may lead to susceptibility to viruses due to the loss of intestinal immune homeostasis, resulting in potential secondary infection [42]. After acute influenza A virus infection, the commensal gut microbiota will undergo transient but significant depletion, and pathogens occupy the vacant niche, increasing susceptibility to intestinal bacterial infection [43]. Therefore, respiratory viral infections may change the composition of the gut microbiota through immunomodulation and predispose patients to secondary bacterial superinfection, which is the main cause of mortality in a substantial number of patients [42]. As with other respiratory viruses, patients with SARS-CoV-2 infections also present with gut microbiota dysbiosis. Disturbances in the gut microbiota can also lead to secondary intestinal inflammation [44], [45]. Moreover, the direct invasion of the intestine by SARS-CoV-2 leads to increased intestinal permeability, allowing LPS produced by harmful bacteria to invade the body and stimulate the production of certain cytokines, such as interleukins and interferons, during respiratory viral infections [9]. Additionally, some inflammatory markers associated with bacterial infections are elevated in patients with SARS-CoV-2 infection [46]. Previous studies showed that respiratory viral infections may lead to disturbances in inflammatory cytokines by influencing the gut microbiota [22]. Beneficial gut bacteria are capable of producing anti-inflammatory metabolites such as SCFAs, which can increase IL-10 production and reduce inflammation. Harmful gut bacteria produce proapoptotic metabolites such as biogenic amines, which reduce the levels of IL-4, IL-5, and IL-13 in lung homogenates [47]. The activity of alveolar macrophages, blood neutrophils, and inflammatory cytokines was also reduced when the abundances of gut and lung bacteria were reduced [42]. In addition, in mice with influenza virus and intestinal anaerobic dysregulation, the expression levels of the inflammatory cytokines IL-4 and IL-10 decreased, and the expression levels of IFN-γ and IL-17 increased in the lung [48]. Similarly, SARS-CoV-2 stimulates the production of harmful metabolites by disturbing the gut microbiota, which stimulates the production of cytokines that play roles in cytokine storms [9]. Cytokine storms are usually defined as extremely elevated cytokine levels associated with infection and immunity. The dramatic increase in cytokines leads to complement activation and immune thrombosis and ultimately causes disseminated intravascular coagulation, multiple organ failures, and other serious clinical manifestations.
SARS-CoV-2 infection may also affect the gut microbiota through secondary bacterial infection and antibiotic abuse. Patients with COVID-19 with secondary bacterial infections are often treated with antibiotics. More than 90% of patients with COVID-19, whether they suffer from secondary bacterial infections or not, are currently taking antibiotics [19]. However, antibiotic overuse leading to gut microbiota dysregulation is observed in other respiratory viral infections but not in COVID-19. This finding suggests that antibiotic abuse after SARS-CoV-2 infection can easily lead to dysbiosis of the immune and gut microbiota, resulting in secondary infection. The aforementioned evidence indicated that the role of gut microbiota in SARS-CoV-2 infection might affect immune response similar to that in other respiratory viral infections [49].
In cases of SARS-CoV-2 infection, patients have not only fever, cough, and dyspnoea but also gastrointestinal symptoms (e.g., diarrhoea and vomiting). Notably, SARS-CoV-2 nucleic acid can be detected in the stool of some patients [44]. In addition, SARS-CoV-2 can be detected in both the small and large intestines. Consequently, SARS-CoV-2 infection can lead to gut microbiota dysbiosis [44], [45]. A study recruited 56 patients with COVID-19 and 47 healthy subjects of similar age and sex and found that purine metabolites in the faeces of patients with COVID-19 were significantly reduced, and these reduction in purine metabolites was significantly associated with alterations in the gut microbiota, including Ruminococcaceae, Actinomyces, Sphingomonas, and Aspergillus. This alteration may be the cause of malnutrition and intestinal inflammation in some patients with COVID-19 [50]. In a comparison of the gut microbiota composition of patients with severe COVID-19, patients with common COVID-19, and healthy individuals using 16 S rRNA gene sequencing methods revealed that the abundance of the gut microbiota was lower in patients with severe COVID-19. The number of Proteobacteria was increased in patients with common COVID-19 compared with healthy individuals. The abundance of Fusobacteria and Spirochetes was reduced in patients with severe COVID-19 compared to healthy individuals [51]. Therefore, the composition of the gut microbiota is different in patients with different COVID-19 severities, and changes in the composition of the gut microbiota may indicate severe COVID-19. Gut microbiota dysbiosis may cause increased cytokines, resulting in severe COVID-19 [45].
A disturbance in the gut microbiota may be a potential cause of cytokine storms. Critically ill patients with COVID-19 experience increased levels of cytokines, which may eventually lead to cytokine storms. This interaction is not only the result of SARS-CoV-2 infection but also an important factor in patients with severe COVID-19. SARS-CoV-2 infection may trigger the host’s inflammatory immune response. Bacterial translocation across the intestinal epithelium may be increased, resulting in intestinal epithelial damage and ultimately leading to intestine–blood barrier disruption and systemic endotoxaemia, which may lead to cytokine storms in severe SARS-CoV-2 infection. A close link between the gut microbiota and systemic inflammation may exist [45]. Based on data from patients with COVID-19, Guo et al. developed a blood proteome risk score (PRS) to predict SARS-CoV-2 progression to clinically severe stages. A higher blood PRS was significantly associated with higher hsCRP and TNF-α serum concentrations in older healthy individuals without COVID-19, suggesting that the PRS was positively correlated with proinflammatory cytokines. In addition, a set of gut bacteria was able to accurately predict blood proteome biomarkers in healthy individuals. Bacteroides, Streptococcus, and Lactobacillus were negatively correlated with most of the target inflammatory cytokines (including IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12, IL-13, TNF-α, and INF-γ), while Ruminaceae, Lactariaceae, and Portunales were positively correlated with most of the target inflammatory cytokines. Moreover, amino acid metabolism-related pathways may play crucial roles in crosstalk between the gut microbiota and inflammation. This evidence suggests that disordered composition of the gut microbiota may be the underlying factor in severe SARS-CoV-2 infection and contributes to disease severity in patients with COVID-19 (especially the elderly population) [10]. The invasion of the intestine by SARS-CoV-2 may lead to an increase in mesenteric permeability, and LPS can enter the circulatory system and stimulate the production of IL-6, IL-1, IL-8, TNF-α, and INF-γ. These cytokines, which are greatly increased in critical patients with COVID-19, can induce an inflammatory response and play roles in cytokine storms. A study also suggested that Proteobacteria in the intestine produce LPS, which can affect the inflammatory response in SARS-CoV-2 infection [9]. The gut microbial composition and plasma concentrations of inflammatory cytokines and blood markers in the blood and stool specimens of 100 patients with COVID-19 were measured. In these patients, the gut microbiota had very low numbers of Faecalibacterium prausnitzii, Eubacterium rectale, and Bifidobacteria, and these gut commensal bacteria all have potential immunomodulatory abilities. Furthermore, the decreased numbers of these gut commensal bacteria was consistent with disease severity, and the increases in the concentrations of inflammatory cytokines and blood markers, including C-reactive protein, lactate dehydrogenase, aspartate aminotransferase, and γ-glutamyltransferase, were closely related to cytokine storms [46].
This evidence shows that the gut microbiota may be involved in the inflammatory response to SARS-CoV-2 infection and may lead to cytokine storms [9]. SARS-CoV-2, like many other respiratory viruses, may lead to intestinal dysregulation in patients through secondary intestinal infection with exogenous bacteria, although no study has been reported. However, whether the increase in harmful bacteria is caused by intestinal disturbances in exogenous bacteria or by gut microbiota dysbiosis itself remains to be further studied. Notably, gut microbiota dysbiosis may also play an important role in cytokine storms, in addition to secondary infections of the lung. Based on the aforementioned evidence, the gut microbiota may mediate the susceptibility of normal subjects to severe SARS-CoV-2 infection and have some regulatory effect on SARS-CoV-2 infection. Importantly, no mechanism explains the dysregulation of the gut microbiota in patients with COVID-19.
SARS-CoV-2 infection, like respiratory viral infections, also causes gut microbiota dysregulation in general. In addition, changes in the composition of the gut microbiota may indicate the severity of SARS-CoV-2 infection. Cytokine storms are also manifestations of severe SARS-CoV-2 infection, and disturbances in the gut microbiota may also cause cytokine storms. However, this mechanism of action has not been thoroughly studied, in COVID-19 or other respiratory viruses. In addition, whether such changes in the gut microbiota are a direct result of COVID-19 infection or due to secondary bacterial infection has not been further investigated.
5. Gut–lung axis dysbiosis in severe COVID-19: Learning from respiratory viral infections
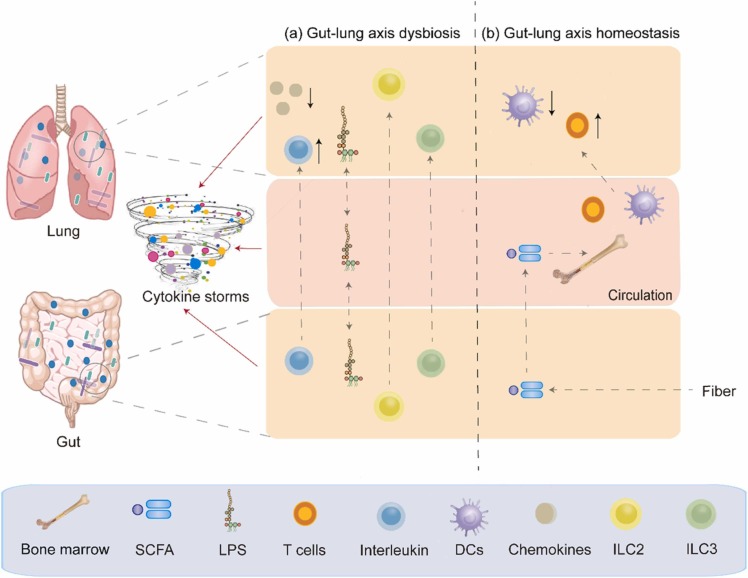
The interaction between the gut and the lung may be inextricably linked in SARS-CoV-2 infections. The role of gut–lung axis dysbiosis in SARS-CoV-2 infections is also a point of great interest. Therefore, we have summarised the relationship between other respiratory viruses and the gut–lung axis as a reference. Researchers have proposed the idea of the gut–lung axis to explain how the gut microbiota affects the function of the lung [50]. Microbial interactions in the gut and lung and their products locally affect immune function; for example, the gut microbiota affects susceptibility to asthma and pneumonia [52]. Dysbiosis of both the gut microbiota [51] and the lung microbiota [23] in patients with SARS-CoV-2 infections also confirms the inseparability of the gut and the lung. The gut–lung axis can transport LPS via the circulation. LPS injection into the rectum of antibiotic-treated mice restored the ability of the mice to produce an effective immune response to influenza virus infection in the lung. In addition, the host can sense SCFAs and deaminotyrosine produced by the gut microbiota. An immune response occurs locally in the intestine when SCFAs are released into the intestine. When unmetabolized SCFAs are transferred to bone marrow, they affect bone marrow immune cell development and the immune response. Moreover, butyrate from dietary fiber can also control the immunopathology caused by neutrophil infiltration by promoting haematopoiesis of antipneumonic macrophages in the bone marrow, thereby achieving a protective effect against influenza virus infection. In addition, deaminotyrosine can protect mice against influenza virus infection by enhancing IFN-I production [21]. Importantly, the same is true of SARS-CoV-2 infection, which can damage intestinal integrity, allowing bacterial toxins and metabolites to transfer to the lung and even the whole body, leading to immune disorders [53]. In addition to the role of metabolites produced by the gut microbiota, the gut–lung axis can also enhance resistance to infection by the migration of IL-C2-producing IL-C3 immune cells directly from the intestine to the respiratory tract through the effects of intestinal DCs on commensal bacteria [21]. This finding also validates the idea that the gut–lung axis affects the immune response in the lung and gut microbiota and that gut dysbacteriosis affects lung homeostasis.
SARS-CoV-2 infection disturbs lung and gut microbial homeostasis. Studies have compared the respiratory and intestinal microbial communities of 35 patients with COVID-19 and 19 healthy adults, and the results showed that the bacterial diversity in patients with SARS-CoV-2 was lower than that in healthy individuals. In addition, the upper respiratory tract and the gut microbiota of patients with mild COVID-19 can synchronize from dysregulation to recovery. This study suggests a microbial interaction in the gut–lung axis [54]. SARS-CoV-2 infections that increase inflammation levels may lead to intestinal leakage, allowing bacterial toxins and metabolites to be transferred to systemic circulation, exacerbating the sepsis and multiple organ failure in patients. In addition, intestinal leakage can lead to microbial translocation to the lungs, causing secondary infection and even sepsis. Moreover, SARS-CoV-2 may be transferred through the gut–lung axis via systemic circulation, resulting in immune disorders leading to cytokine storms in patients with COVID-19 [53]. ( Fig. 1).
Fig. 1.
Impairment and regulation of Gut-lung axis in severe SARS-CoV-2 infection. (a) Dysbiosis of the gut–lung axis, which can lead to cytokine storms, can be caused by disordered immune cells and increased bacteria-derived LPS. (b) Immune homeostasis can be maintained by regulating the gut microbiota, which may lead to gut-lung axis balance.
6. Angiotensin-converting enzyme 2: A bridge between the gut and lung
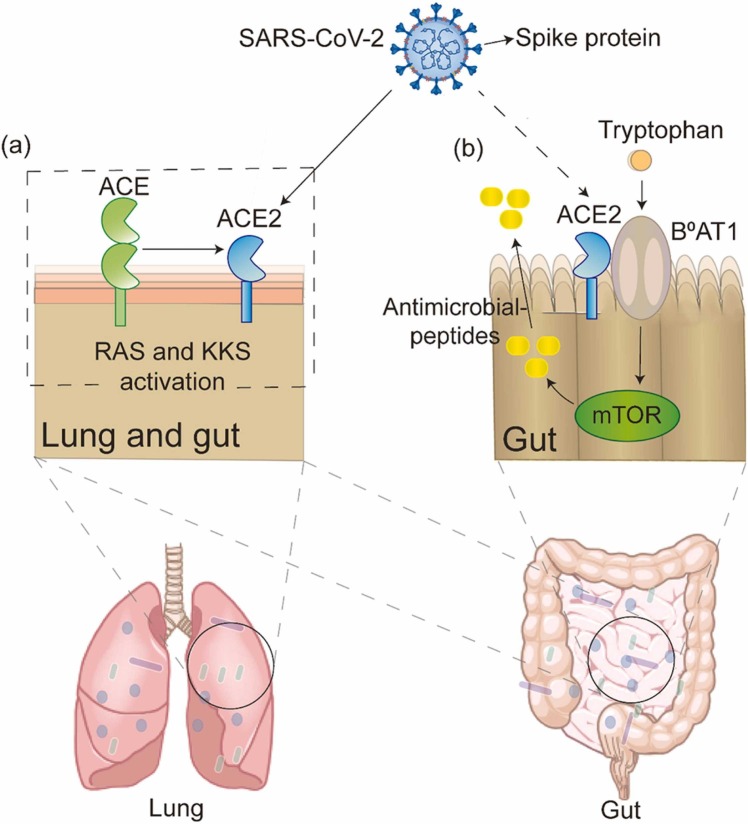
Angiotensin-converting enzyme 2 (ACE2) is a common target of both SARS-CoV-2 and other coronaviruses. ACE2 has been shown to play a crucial role in preventing acute lung injury induced by certain respiratory viruses and is expected to be a potential therapeutic target [55], [56], [57]. Coronaviruses (e.g., SARS-CoV and the known human coronavirus-NL63 (HCoV-NL63)) efficiently bind to ACE2 [58], [59]. Some studies have shown that SARS-CoV-2 can bind to human ACE2 and thus invade the human body [60], [61]. SARS-CoV-2 infections lead to decreases in all ACE2 effects [62], [63]. ACE2 can affect bradykinin levels through the renin–angiotensin system (RAS) and kinin-releasing enzyme kinin system (KKS), and higher bradykinin levels may lead to increased systemic inflammation, influencing the lung and gastrointestinal systems and ultimately leading to multiple organ failure [64], [65].
ACE2 is abundantly expressed in the human lung and small intestinal epithelial cells [66]. The ACE2 gene is also highly expressed in the ileum, suggesting that the intestine is likely to be invaded by SARS-CoV-2 [67]. Thus, it is thought that SARS-CoV-2 not only invades lung tissue through ACE2 but may also bind to intestinal epithelial receptors.
Since high expression of ACE2 was found in the intestine of patients with SARS-CoV-2 infection with gut microbiota dysbiosis, it is not difficult to hypothesize that there may be some link between ACE2 and the gut microbiota. However, there is currently insufficient evidence to verify this point of view. In previous studies, ACE2 was shown to affect the expression of antimicrobial substances in the intestine, thereby affecting the gut microbiota. Collectrin is a renal amino acid transporter regulator that was shown to share high homology with ACE2 in the membrane-proximal domain. In addition, collectrin can bind B0AT1 family amino acid transporters [68]. Moreover, collectrin can stabilise the expression of amino acid transporters [69]. Collectrin and ACE2 have similar transcription factor binding sites and transport functions. Protein expression of the neutral amino acid transporter B0AT1 was absent. However, serum tryptophan levels and the expression of multiple antimicrobial peptides decreased significantly in ACE2-KO mice. A decrease in antimicrobial peptides was also observed in mice that received a tryptophan-free diet [70]. ACE2 can bind to B0AT1 in small intestinal epithelial cells, affecting mTOR pathway activity and antimicrobial peptide expression by altering the level of the essential amino acid tryptophan, ultimately leading to intestinal microbiota dysregulation [69]. In addition, ACE2 has also been shown to interact with SIT1 transporters of amino acids and alkaloids, which have antibacterial effects on the intestine [69]. Tryptophan is absorbed through the B0AT1/ACE2 transport pathway in small intestinal epithelial cells, activating mTOR and affecting antimicrobial peptide expression and the composition of the gut microbiota. In addition, ACE2 may lead to local intestinal inflammation. A lack of ACE2 can lead to severe damage to local tryptophan homeostasis and affect intestinal inflammation susceptibility [70].
Patients with COVID-19 who have gastrointestinal symptoms currently have a poor prognosis. Thus, it is thought that ACE2 expression in the gastrointestinal tract is regulated by the gut microbiota. ACE2 expression in mice was examined by quantitative polymerase chain reaction revealed, and the results showed that the intestinal and respiratory ACE2 expression levels in germ-free mice were significantly higher than those in conventional mice. The expression of intestinal ACE2 correlated with higher proteolytic and peptidase activities in the gut microbiota [71]. Therefore, the gut microbiota may be an important factor in determining COVID-19 severity, while ACE2 is expected to be a target for severe COVID-19 prevention.
In summary, SARS-CoV-2 may be involved in the RAS and KKS, leading to the deregulated action of ACE2 and causing local or systemic inflammatory responses, which may lead to cytokine storms in severe cases. Moreover, ACE2 can cause local intestinal inflammation, inevitably bringing about an imbalance in the gut microbiota. This evidence suggests that ACE2 links the gut microbiota and SARS-CoV-2. ( Fig. 2).
Fig. 2.
ACE2: a bridge between gut and lung. (a) In the lung and gut, SARS-CoV-2 binds to the ACE2 and activates renin–angiotensin system (RAS) and kinin-releasing enzyme kinin system (KKS). (b) After tryptophan intake, ACE2 binds to B0AT1 to produce antimicrobial peptides via the mTOR pathway in the gut, which may be mediated by SARS-CoV-2 infection.
7. Microbiological therapy
Given that the gut microbiota may play an important role in SARS-CoV-2 infections, maintaining gut homeostasis is a key point. The use of some microbiological therapies should then be effective in maintaining intestinal homeostasis in patients with SARS-CoV-2, as has been reported in other respiratory viruses. Many microbiological therapies [e.g., probiotics and prebiotics, dietary fiber, traditional Chinese medicine, and faecal microbial transplantation (FMT)] can treat respiratory viral infections by regulating the gut microbiota. Probiotics and prebiotic interventions can reduce the incidence of viral respiratory tract infections and improve the clinical symptoms of viral infections [72]. An experiment showed that probiotics and prebiotics could improve the structure of the gut microbiota in patients with H7N9, thereby reducing the incidence of secondary intestinal infections [73]. Furthermore, Co et al. demonstrated that treatment with dietary fiber increased the abundance of Lactobacillus and Bifidobacterium in the gut microbiota of healthy people [74]. In addition, the metabolism of dietary fiber by the gut microbiota leading to the production of SCFAs can reduce bacterial lung inflammation and the occurrence of allergic inflammation in the lungs [75], [76]. However, several microbiological therapies mentioned previously are not yet being effectively used to treat SARS-CoV-2 infection. Interestingly, traditional Chinese medicine can also improve viral infection symptoms by regulating the gut microbiota. In an animal study, H2N9 AIV infection disrupted the intestinal mucosa in mice, resulting in the spread of Staphylococcus and E. coli in the mouse intestine to the liver and lungs. No bacterial transfer was found in the parenteral tissues of mice in the ageratum liquid (AL)-treated group, indicating that AL could effectively prevent and reverse the imbalance in the gut microbiota in mice infected with H9N2 AIV [77]. In addition, Houttuynia cordata polysaccharide (HCP) improved the changes in gut microbiota composition caused by infection with the A (H1N1) virus, significantly reducing the relative abundances of the pathogenic bacteria Vibrio and Bacillus and improving intestinal homeostasis, thereby reducing lung function damage caused by infection. In addition, the effect of HCP is mainly reflected in the decreases in intestinal TLRs and IL-1β and the increases in IL-10 levels [78]. Chinese herbal medicine is by far the most common type of microbial therapy used in China and has also played a role in the treatment of SARS-CoV-2 infection. Two excellent methods are the use of Lianhuaqingwen capsules [79] and ShufengJiedu capsules [80]. Additional herbal treatments can refer to other respiratory viruses. FMT has also been useful in the treatment of relapsed and refractory Clostridium difficile infections and is much more effective than antibiotic therapy. In addition, FMT can increase the abundance of lung bacteria and reduce the abundance of the gut microbiota [73]. This outcome inhibits the immune response, reduces lung injury, decreases LPS, and affects the TGFERK1/Smads/β pathway [81]. FMT is effective in treating antibiotic-associated diarrhoea. Similarly, FMT has been useful in treating the gastrointestinal symptoms of SARS-CoV-2 infection [82].
Some microbial therapies have been currently used in COVID-19 treatment. At present, traditional Chinese medicine is the most clinically used microbial therapy in China and has been proven to be effective against COVID-19, and a relatively extensive clinical study has been conducted. After the administration of Lianhuaqingwen capsules, components were found to be absorbed by the gastrointestinal tract and transformed. In addition, components including rhein, forsythoside A, forsythoside I, neochlorogenic acid, and their isomers can greatly reduce ACE2 function [79]. Another study showed that ShufengJiedu capsules (SFJDCs) may inhibit SARS-CoV-2, and the active components of SFJDCs can participate in immune regulation and anti-inflammation through related targets [80]. Substantial evidence showed that the interaction of the gut microbiota with Chinese herbal medicines could directly convert molecules (e.g., polysaccharides, oligosaccharides, saponins, and phenolic compounds) into beneficial metabolites, which can improve oral availability [83]. FMT was also used as adjuvant therapy during COVID-19 disease. In a recent study, two patients with COVID-19 with recurrent C. difficile infection were treated with FMT without adverse effects, which suggested that FMT might contribute to COVID-19 treatment [84]. In addition, 11 patients with COVID-19 were treated with FMT for 4 days. Five of the 11 patients had experienced gastrointestinal symptoms that significantly improved with FMT. Naive B cells decreased and memory B cells and nonswitched B cells increased after FMT in all patients. FMT improved actinobacterial abundance and reduced proteobacterial abundance at the phylum level. FMT can significantly increase Bifidobacterium and Faecalibacterium levels [82].
In summary, microbiological therapy can improve the symptoms associated with respiratory viral infections to some extent, and some agents can also improve the systemic and local immune response by regulating the gut microbiota to improve lung symptoms. In addition, these microbial therapies reflect not only improvements in the gut microbiota and regulation of the gut–lung axis in SARS-CoV-2 treatment but also the potential pathogenesis of immune dysregulation associated with SARS-CoV-2 through the gut–lung axis. This finding also showed that more microbial therapies could be used to achieve the effect of adjuvant therapy in the treatment of SARS-CoV-2 infection.
8. Conclusion
Bacterial infection is often secondary to respiratory viral infection, including infection by not only lung bacteria but also intestinal bacteria. SARS-CoV-2 infection is often accompanied by gastrointestinal symptoms, which suggests that viral infection may be associated with intestinal microbial imbalance. Moreover, gut microbiota imbalance may cause vulnerable healthy individuals to be infected with SARS-CoV-2 and develop severe symptoms. When abundant inflammatory factors are induced by dysregulation of the gut microbiota, the immune response may be impaired, thereby affecting the immune regulation of the gut microbiota and causing a cytokine storm. However, the mechanism of the immune effect of the gut microbiota on cytokine storms needs to be further studied. In addition to immune mechanisms, ACE2, a receptor of SARS-CoV-2, plays an important role in preventing acute lung injury caused by respiratory viruses and is expected to be a potential therapeutic target in recent years. ACE2 was also shown to be related to the gut microbiota, resulting in changes in the levels of antimicrobial peptides, intestinal amino acids, and other substances. However, relevant mechanistic studies are also limited. In summary, a close relationship exists between respiratory viral infection and gut microbiota dysregulation. Moreover, symptoms can be improved by microbial treatments, which may provide a new cost-effective treatment for SARS-CoV-2 infection.
Many questions still deserve further exploration. More attention should be focused not only on exogenous bacterial infections in the intestine other than SARS-CoV-2 infection but also on severe infections caused by immune dysregulation leading to gut microbiota dysbiosis. The mechanisms involved in the dysregulation of the gut microbiota that may lead to cytokine storms remain to be investigated. The gut–lung axis may also refer to the connection between the gut and lung microbiota. Whether the gut microbiota and lung microbiota have a synergistic or mutual effect on disease progression needs to be explored. Respiratory viruses, bacteria, and the gut microbiota constitute the external environment of the microbiota, which can be considered a microecosystem that is closely related to each other. Therefore, the balance of the entire system should be considered when improving the overall microbial system environment.
Funding
This work was supported by the National Natural Science Foundation of China (81870544 and 81870594); the Natural Science Foundation of Jiangsu Province (BK20181132, BK20210060); Scientific Research Project of Jiangsu Commission of Health (M2021055); Jiangsu Scientific Research Project of Elderly Health (LK2021035); Jiangsu Scientific Research Project of Women’s and Children’s Health (F201741); Scientific Research Project of Wuxi Commission of Health (ZZ003 and Q201762); Wuxi Scientific and Technological Development Project (N20192024, N20191001 and N2020X007). Translational Medicine Research Program of Wuxi Translational Medicine Center (2020ZHYB08).
Declaration of interests
There are no conflicts of interest.
Biographies

Hong Cao, Doctor of Medicine, Professor, Chief physician, Vice President of Affiliated Hospital of Jiangnan University and senior visiting scholar at Ohio State University Medical Center. She focuses on the alleviative effect of nutritional intervention on metabolic diseases via improving the structure of the gut microbiota. Since 2020, she also has devoted herself to the study on roles of the gut microbiota in SARS-CoV-2 infection.

Feng Zhang is the director of Nutrition Department (Functional Food Clinical Evaluation Center). He studies roles of gut microbiota in metabolic diseases, including obesity, type 2 diabetes, prediabetes, gestational diabetes mellitus, and polycystic ovarian syndrome. He focused on the effects of dietary fiber on the enrichment of SCFA-producing bacteria, which could enhance GLP-1 levels and thereafter alleviate type 2 diabetes. Recently, he has been interested in roles of the gut microbiota in the alleviation of metabolic diseases by functional foods.
References
- 1.J.M. Read, J.R. Bridgen, D.A. Cummings, A. Ho, C.P. Jewell, Novel Coronavirus 2019-nCoV: Early Estimation Of Epidemiological Parameters and Epidemic Predictions, (2020). [DOI] [PMC free article] [PubMed]
- 2.Wu J., Li J., Zhu G., Zhang Y., Bi Z., Yu Y., Huang B., Fu S., Tan Y., Sun J., Li X. Clinical features of maintenance hemodialysis patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. Clin. J. Am. Soc. Nephrol. 2020;15(8):1139–1145. doi: 10.2215/CJN.04160320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leung W., To K., Chan P., Chan H., Wu A., Lee N., Yuen K., Sung J. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125(4):1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Assiri A., Al-Tawfiq J., Al-Rabeeah A., Al-Rabiah F., Al-Hajjar S., Al-Barrak A., Flemban H., Al-Nassir W., Balkhy H., Al-Hakeem R., Makhdoom H., Zumla A., Memish Z. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet. Infect. Dis. 2013;13(9):752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray J., Oehrle K., Worthen G., Alenghat T., Whitsett J., Deshmukh H. Intestinal commensal bacteria mediate lung mucosal immunity and promote resistance of newborn mice to infection. Sci .Transl. Med. 2017;9(376) doi: 10.1126/scitranslmed.aaf9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao L. The gut microbiota and obesity: from correlation to causality. Nat. Rev. Microbiol. 2013;11(9):639–647. doi: 10.1038/nrmicro3089. [DOI] [PubMed] [Google Scholar]
- 7.Qin N., Zheng B., Yao J., Guo L., Zuo J., Wu L., Zhou J., Liu L., Guo J., Ni S., Li A., Zhu Y., Liang W., Xiao Y., Ehrlich S.D., Li L. Influence of H7N9 virus infection and associated treatment on human gut microbiota. Sci. Rep. 2015;5:14771. doi: 10.1038/srep14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iizasa H., Ishihara S., Richardo T., Kanehiro Y., Yoshiyama H. Dysbiotic infection in the stomach. World J. Gastroenterol. 2015;21(40):11450–11457. doi: 10.3748/wjg.v21.i40.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onishi J., Häggblom M., Shapses S. Can dietary fatty acids affect the COVID-19 infection outcome in vulnerable populations? mBio. 2020;11(4) doi: 10.1128/mBio.01723-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.W. Gou, Y. Fu, L. Yue, G.D. Chen, J.S. Zheng, Gut Microbiota May Underlie the Predisposition of Healthy Individuals to COVID-19, (2020).
- 11.Ye Q., Wang B., Mao J. The pathogenesis and treatment of the `Cytokine Storm’ in COVID-19. J. Infect. 2020;80(6):607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.An P.J., Zhu Y.Z., Yang L.P. Biochemical indicators of coronavirus disease 2019 exacerbation and the clinical implications. Pharmacol. Res. 2020;159 doi: 10.1016/j.phrs.2020.104946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellinghausen C., Rohde G.G.U., Savelkoul P.H.M., Wouters E.F.M., Stassen F.R.M. Viral-bacterial interactions in the respiratory tract. J. Gen. Virol. 2016;97(12):3089–3102. doi: 10.1099/jgv.0.000627. [DOI] [PubMed] [Google Scholar]
- 15.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pene F., Marin N., Roche N., Szwebel T.A., Merkling S.H., Treluyer J.M., Veyer D., Mouthon L., Blanc C., Tharaux P.L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kerneis S., Terrier B. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W., Moltedo B., Moran T.M. Type I interferon induction during influenza virus infection increases susceptibility to secondary Streptococcus pneumoniae infection by negative regulation of gammadelta T cells. J. Virol. 2012;86(22):12304–12312. doi: 10.1128/JVI.01269-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garvy B.A., Harmsen A.G. The importance of neutrophils in resistance to pneumococcal pneumonia in adult and neonatal mice. Inflammation. 1996;20(5):499–512. doi: 10.1007/BF01487042. [DOI] [PubMed] [Google Scholar]
- 18.Shahangian A., Chow E.K., Tian X., Kang J.R., Ghaffari A., Liu S.Y., Belperio J.A., Cheng G., Deng J.C. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J. Clin. Investig. 2009;119(7):1910–1920. doi: 10.1172/JCI35412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J., Wang J., Yang Y., Cai P., Cao J., Cai X., Zhang Y. Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with COVID-19 in Wuhan, China: a retrospective analysis. Antimicrob. Resist. Infect. Control. 2020;9(1):153. doi: 10.1186/s13756-020-00819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gollwitzer E.S., Marsland B.J. Microbiota abnormalities in inflammatory airway diseases - potential for therapy. Pharmacol. Ther. 2014;141(1):32–39. doi: 10.1016/j.pharmthera.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Wypych T.P., Wickramasinghe L.C., Marsland B.J. The influence of the microbiome on respiratory health. Nat. Immunol. 2019;20(10):1279–1290. doi: 10.1038/s41590-019-0451-9. [DOI] [PubMed] [Google Scholar]
- 22.Marsland B.J., Gollwitzer E.S. Host-microorganism interactions in lung diseases. Nat. Rev. Immunol. 2014;14(12):827–835. doi: 10.1038/nri3769. [DOI] [PubMed] [Google Scholar]
- 23.Liu T., Wu J., Han C., Gong Z., Regina G., Chen J., Dou F., Silvestri R., Chen C., Yu Z. RS-5645 attenuates inflammatory cytokine storm induced by SARS-CoV-2 spike protein and LPS by modulating pulmonary microbiota. Int. J. BIol. Sci. 2021;17(13):3305–3319. doi: 10.7150/ijbs.63329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu L., Deng H., Ren Z., Zhao Y., Yu S., Guo Y., Dai J., Chen X., Li K., Li R., Wang G. Dynamic changes in the microbiome and mucosal immune microenvironment of the lower respiratory tract by influenza virus infection. Front. Microbiol. 2019;10:2491. doi: 10.3389/fmicb.2019.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichinohe T., Pang I.K., Kumamoto Y., Peaper D.R., Ho J.H., Murray T.S., Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA. 2011;108(13):5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu Y., Hsu A.C., Pang Z., Pan H., Zuo X., Wang G., Zheng J., Wang F. Role of the innate cytokine storm induced by the influenza A virus. Viral Immunol. 2019;32(6):244–251. doi: 10.1089/vim.2019.0032. [DOI] [PubMed] [Google Scholar]
- 27.Allen I., Scull M., Moore C., Holl E., McElvania-TeKippe E., Taxman D., Guthrie E., Pickles R., Ting J. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30(4):556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas P.G., Dash P., Aldridge J.R., Jr., Ellebedy A.H., Reynolds C., Funk A.J., Martin W.J., Lamkanfi M., Webby R.J., Boyd K.L., Doherty P.C., Kanneganti T.D. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30(4):566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bisno A.L., Brito M.O., Collins C.M. Molecular basis of group A streptococcal virulence. Lancet Infect. Dis. 2003;3(4):191–200. doi: 10.1016/s1473-3099(03)00576-0. [DOI] [PubMed] [Google Scholar]
- 30.Szabo P.A., Goswami A., Mazzuca D.M., Kim K., O’Gorman D.B., Hess D.A., Welch I.D., Young H.A., Singh B., McCormick J.K., Haeryfar S.M. Rapid and rigorous IL-17A production by a distinct subpopulation of effector memory T lymphocytes constitutes a novel mechanism of toxic shock syndrome immunopathology. J. Immunol. 2017;198(7):2805–2818. doi: 10.4049/jimmunol.1601366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vecchie A., Bonaventura A., Toldo S., Dagna L., Dinarello C.A., Abbate A. IL-18 and infections: is there a role for targeted therapies? J. Cell. Physiol. 2021;236(3):1638–1657. doi: 10.1002/jcp.30008. [DOI] [PubMed] [Google Scholar]
- 32.Rizvi M.S., Gallo De Moraes A. New decade, old debate: blocking the cytokine pathways in infection-induced cytokine cascade. Crit. Care Explor. 2021;3(3) doi: 10.1097/CCE.0000000000000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimura H., Yoshizumi M., Ishii H., Oishi K., Ryo A. Cytokine production and signaling pathways in respiratory virus infection. Front. Microbiol. 2013;4:276. doi: 10.3389/fmicb.2013.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Candelli M., Franza L., Pignataro G., Ojetti V., Covino M., Piccioni A., Gasbarrini A., Franceschi F. Interaction between lipopolysaccharide and gut microbiota in inflammatory bowel diseases. Int. J. Mol. Sci. 2021;22(12) doi: 10.3390/ijms22126242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J., Jiang M., Chen X., Montaner L. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J. Leukoc. Biol. 2020;108(1):17–41. doi: 10.1002/JLB.3COVR0520-272R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manohar P., Loh B., Nachimuthu R., Hua X., Welburn S., Leptihn S. Secondary bacterial infections in patients with viral pneumonia. Front. Med. 2020;7:420. doi: 10.3389/fmed.2020.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirzaei R., Goodarzi P., Asadi M., Soltani A., Aljanabi H.A.A., Jeda A.S., Dashtbin S., Jalalifar S., Mohammadzadeh R., Teimoori A., Tari K., Salari M., Ghiasvand S., Kazemi S., Yousefimashouf R., Keyvani H., Karampoor S. Bacterial co-infections with SARS-CoV-2. IUBMB Life. 2020;72(10):2097–2111. doi: 10.1002/iub.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin H., Wang Q., Yuan M., Liu L., Chen Z., Zhao Y., Das R., Duan Y., Xu X., Xue Y., Luo Y., Mao D. The prolonged disruption of a single-course amoxicillin on mice gut microbiota and resistome, and recovery by inulin, Bifidobacterium longum and fecal microbiota transplantation. Environ. Pollut. 2020;265(Pt A) doi: 10.1016/j.envpol.2020.114651. [DOI] [PubMed] [Google Scholar]
- 39.Abt M.C., Osborne L.C., Monticelli L.A., Doering T.A., Alenghat T., Sonnenberg G.F., Paley M.A., Antenus M., Williams K.L., Erikson J., Wherry E.J., Artis D. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37(1):158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deriu E., Boxx G.M., He X., Pan C., Benavidez S.D., Cen L., Rozengurt N., Shi W., Cheng G. Influenza virus affects intestinal microbiota and secondary salmonella infection in the gut through type I interferons. PLoS Pathog. 2016;12(5) doi: 10.1371/journal.ppat.1005572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J., Li F., Wei H., Lian Z.X., Sun R., Tian Z. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J. Exp. Med. 2014;211(12):2397–2410. doi: 10.1084/jem.20140625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanada S., Pirzadeh M., Carver K.Y., Deng J.C. Respiratory viral infection-induced microbiome alterations and secondary bacterial pneumonia. Front. Immunol. 2018;9:2640. doi: 10.3389/fimmu.2018.02640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yildiz S., Mazel-Sanchez B., Kandasamy M., Manicassamy B., Schmolke M. Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Microbiome. 2018;6(1):9. doi: 10.1186/s40168-017-0386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cha M.H., Regueiro M., Sandhu D.S. Gastrointestinal and hepatic manifestations of COVID-19: a comprehensive review. World J. Gastroenterol. 2020;26(19):2323–2332. doi: 10.3748/wjg.v26.i19.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferreira C., Viana S.D., Reis F. Gut microbiota dysbiosis-immune hyperresponse-inflammation triad in coronavirus disease 2019 (COVID-19): impact of pharmacological and nutraceutical approaches. Microorganisms. 2020;8(10) doi: 10.3390/microorganisms8101514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeoh Y.K., Zuo T., Lui G.C., Zhang F., Liu Q., Li A.Y., Chung A.C., Cheung C.P., Tso E.Y., Fung K.S., Chan V., Ling L., Joynt G., Hui D.S., Chow K.M., Ng S.S.S., Li T.C., Ng R.W., Yip T.C., Wong G.L., Chan F.K., Wong C.K., Chan P.K., Ng S.C. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70(4):698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barcik W., Boutin R.C.T., Sokolowska M., Finlay B.B. The role of lung and gut microbiota in the pathology of asthma. Immunity. 2020;52(2):241–255. doi: 10.1016/j.immuni.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu S., Yan Y., Zhang M., Shi S., Jiang Z. [Intestinal disorder of anaerobic bacteria aggravates pulmonary immune pathological injury of mice infected with influenza virus] Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2016;32(4):433–436. [PubMed] [Google Scholar]
- 49.Mangalmurti N., Hunter C.A. Cytokine storms: understanding COVID-19. Immunity. 2020;53(1):19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lv L., Jiang H., Chen Y., Gu S., Xia J., Zhang H., Lu Y., Yan R., Li L. The faecal metabolome in COVID-19 patients is altered and associated with clinical features and gut microbes. Anal. Chim. Acta. 2021;1152 doi: 10.1016/j.aca.2021.338267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mazzarelli A., Giancola M.L., Farina A., Marchioni L., Rueca M., Gruber C.E.M., Bartolini B., Ascoli Bartoli T., Maffongelli G., Capobianchi M.R., Ippolito G., Di Caro A., Nicastri E., Pazienza V., I.C.-s. group 16S rRNA gene sequencing of rectal swab in patients affected by COVID-19. PLoS One. 2021;16(2) doi: 10.1371/journal.pone.0247041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baindara P., Chakraborty R., Holliday Z.M., Mandal S.M., Schrum A.G. Oral probiotics in coronavirus disease 2019: connecting the gut-lung axis to viral pathogenesis, inflammation, secondary infection and clinical trials. New Microbes New Infect. 2021;40 doi: 10.1016/j.nmni.2021.100837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vignesh R., Swathirajan C.R., Tun Z.H., Rameshkumar M.R., Solomon S.S., Balakrishnan P. Could perturbation of gut microbiota possibly exacerbate the severity of COVID-19 via cytokine storm? Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.607734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu R., Lu R., Zhang T., Wu Q., Cai W., Han X., Wan Z., Jin X., Zhang Z., Zhang C. Temporal association between human upper respiratory and gut bacterial microbiomes during the course of COVID-19 in adults. Commun. Biol. 2021;4(1):240. doi: 10.1038/s42003-021-01796-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou Z., Yan Y., Shu Y., Gao R., Sun Y., Li X., Ju X., Liang Z., Liu Q., Zhao Y., Guo F., Bai T., Han Z., Zhu J., Zhou H., Huang F., Li C., Lu H., Li N., Li D., Jin N., Penninger J.M., Jiang C. Angiotensin-converting enzyme 2 protects from lethal avian influenza A H5N1 infections. Nat. Commun. 2014;5:3594. doi: 10.1038/ncomms4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gu H., Xie Z., Li T., Zhang S., Lai C., Zhu P., Wang K., Han L., Duan Y., Zhao Z., Yang X., Xing L., Zhang P., Wang Z., Li R., Yu J.J., Wang X., Yang P. Angiotensin-converting enzyme 2 inhibits lung injury induced by respiratory syncytial virus. Sci. Rep. 2016;6:19840. doi: 10.1038/srep19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang P., Gu H., Zhao Z., Wang W., Cao B., Lai C., Yang X., Zhang L., Duan Y., Zhang S., Chen W., Zhen W., Cai M., Penninger J.M., Jiang C., Wang X. Angiotensin-converting enzyme 2 (ACE2) mediates influenza H7N9 virus-induced acute lung injury. Sci. Rep. 2014;4:7027. doi: 10.1038/srep07027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14(8) doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hofmann H., Simmons G., Rennekamp A., Chaipan C., Gramberg T., Heck E., Geier M., Wegele A., Marzi A., Bates P., Pöhlmann S. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J. Virol. 2006;80(17):8639–8652. doi: 10.1128/JVI.00560-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grimes J.M., Grimes K.V. p38 MAPK inhibition: a promising therapeutic approach for COVID-19. J. Mol. Cell. Cardiol. 2020;144:63–65. doi: 10.1016/j.yjmcc.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dijkman R., Jebbink M.F., Deijs M., Milewska A., Pyrc K., Buelow E., van der Bijl A., van der Hoek L. Replication-dependent downregulation of cellular angiotensin-converting enzyme 2 protein expression by human coronavirus NL63. J. Gen. Virol. 2012;93(Pt 9):1924–1929. doi: 10.1099/vir.0.043919-0. [DOI] [PubMed] [Google Scholar]
- 63.Annweiler C., Cao Z., Wu Y., Faucon E., Mouhat S., Kovacic H., Sabatier J. Counter-regulatory ‘Renin-Angiotensin’ system-based candidate drugs to treat COVID-19 diseases in SARS-CoV-2-infected patients. Infect. Disord. Drug Targets. 2020;20(4):407–408. doi: 10.2174/1871526520666200518073329. [DOI] [PubMed] [Google Scholar]
- 64.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Polidoro R.B., Hagan R.S., de Santis Santiago R., Schmidt N.W. Overview: systemic inflammatory response derived from lung injury caused by SARS-CoV-2 infection explains severe outcomes in COVID-19. Front. Immunol. 2020;11:1626. doi: 10.3389/fimmu.2020.01626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14(2):185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuba K., Imai Y., Ohto-Nakanishi T., Penninger J.M. Trilogy of ACE2: a peptidase in the renin-angiotensin system, a SARS receptor, and a partner for amino acid transporters. Pharmacol. Ther. 2010;128(1):119–128. doi: 10.1016/j.pharmthera.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perlot T., Penninger J.M. ACE2 - from the renin-angiotensin system to gut microbiota and malnutrition. Microbes Infect. 2013;15(13):866–873. doi: 10.1016/j.micinf.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hashimoto T., Perlot T., Rehman A., Trichereau J., Ishiguro H., Paolino M., Sigl V., Hanada T., Hanada R., Lipinski S., Wild B., Camargo S.M., Singer D., Richter A., Kuba K., Fukamizu A., Schreiber S., Clevers H., Verrey F., Rosenstiel P., Penninger J.M. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487(7408):477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koester S., Li N., Lachance D., Morella N., Dey N. Variability in digestive and respiratory tract Ace2 expression is associated with the microbiome. PLoS One. 2021;16(3) doi: 10.1371/journal.pone.0248730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luoto R., Ruuskanen O., Waris M., Kalliomaki M., Salminen S., Isolauri E. Prebiotic and probiotic supplementation prevents rhinovirus infections in preterm infants: a randomized, placebo-controlled trial. J. Allergy Clin. Immunol. 2014;133(2):405–413. doi: 10.1016/j.jaci.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu H., Zhang C., Qian G., Hu X., Zhang H., Chen C., Liang W., Gao H., Yang Y., Li L. An analysis of microbiota-targeted therapies in patients with avian influenza virus subtype H7N9 infection. BMC Infect. Dis. 2014;14:359. doi: 10.1186/1471-2334-14-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.So D., Whelan K., Rossi M., Morrison M., Holtmann G., Kelly J.T., Shanahan E.R., Staudacher H.M., Campbell K.L. Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis. Am. J. Clin. Nutr. 2018;107(6):965–983. doi: 10.1093/ajcn/nqy041. [DOI] [PubMed] [Google Scholar]
- 75.Vaughan A., Frazer Z.A., Hansbro P.M., Yang I.A. COPD and the gut-lung axis: the therapeutic potential of fibre. J. Thorac. Dis. 2019;11(Suppl 17):S2173–S2180. doi: 10.21037/jtd.2019.10.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trompette A., Gollwitzer E.S., Yadava K., Sichelstiel A.K., Sprenger N., Ngom-Bru C., Blanchard C., Junt T., Nicod L.P., Harris N.L., Marsland B.J. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014;20(2):159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 77.Lu H., Zhang L., Xiao J., Wu C., Zhang H., Chen Y., Hu Z., Lin W., Xie Q., Li H. Effect of feeding Chinese herb medicine ageratum-liquid on intestinal bacterial translocations induced by H9N2 AIV in mice. Virol. J. 2019;16(1):24. doi: 10.1186/s12985-019-1131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen M.Y., Li H., Lu X.X., Ling L.J., Weng H.B., Sun W., Chen D.F., Zhang Y.Y. Houttuynia cordata polysaccharide alleviated intestinal injury and modulated intestinal microbiota in H1N1 virus infected mice. Chin. J. Nat. Med. 2019;17(3):187–197. doi: 10.1016/S1875-5364(19)30021-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen X., Wu Y., Chen C., Gu Y., Zhu C., Wang S., Chen J., Zhang L., Lv L., Zhang G., Yuan Y., Chai Y., Zhu M., Wu C. Identifying potential anti-COVID-19 pharmacological components of traditional Chinese medicine Lianhuaqingwen capsule based on human exposure and ACE2 biochromatography screening. Acta Pharm. Sin. B. 2021;11(1):222–236. doi: 10.1016/j.apsb.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tao Z., Zhang L., Friedemann T., Yang G., Li J., Wen Y., Wang J., Shen A. Systematic analyses on the potential immune and anti-inflammatory mechanisms of Shufeng Jiedu Capsule against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)-caused pneumonia. J. Funct. Foods. 2020;75 doi: 10.1016/j.jff.2020.104243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li B., Yin G.F., Wang Y.L., Tan Y.M., Huang C.L., Fan X.M. Impact of fecal microbiota transplantation on TGF-beta1/Smads/ERK signaling pathway of endotoxic acute lung injury in rats. 3 Biotech. 2020;10(2):52. doi: 10.1007/s13205-020-2062-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu F., Ye S., Zhu X., He X., Wang S., Li Y., Lin J., Wang J., Lin Y., Ren X., Li Y., Deng Z. Gastrointestinal disturbance and effect of fecal microbiota transplantation in discharged COVID-19 patients. J. Med. Case Rep. 2021;15(1):60. doi: 10.1186/s13256-020-02583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang F., He F., Li L., Guo L., Zhang B., Yu S., Zhao W. Bioavailability based on the gut microbiota: a new perspective. Microbiol. Mol. Biol. Rev. 2020;84(2) doi: 10.1128/MMBR.00072-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ianiro G., Bibbo S., Masucci L., Quaranta G., Porcari S., Settanni C.R., Lopetuso L.R., Fantoni M., Sanguinetti M., Gasbarrini A., Cammarota G. Maintaining standard volumes, efficacy and safety, of fecal microbiota transplantation for C. difficile infection during the COVID-19 pandemic: A prospective cohort study. Dig. Liver Dis. 2020;52(12):1390–1395. doi: 10.1016/j.dld.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]