Abstract
Ocean pollution is a worldwide environmental challenge that could be partially tackled through microbial applications. To shed light on the diversity and applications of the bacterial communities that inhabit the sediments trapped in artificial containers, we analyzed residues (polyethylene terephthalate [PET] bottles and aluminum cans) collected from the Mediterranean Sea by scanning electron microscopy and next generation sequencing. Moreover, we set a collection of culturable bacteria from the plastisphere that were screened for their ability to use PET as a carbon source. Our results reveal that Proteobacteria are the predominant phylum in all the samples and that Rhodobacteraceae, Woeseia, Actinomarinales, or Vibrio are also abundant in these residues. Moreover, we identified marine isolates with enhanced growth in the presence of PET: Aquimarina intermedia, Citricoccus spp., and Micrococcus spp. Our results suggest that the marine environment is a source of biotechnologically promising bacterial isolates that may use PET or PET additives as carbon sources.
Keywords: bioprospecting, bioremediation, marine sediments, marine waste, plastic‐degrading microorganisms, polyethylene terephthalate
Inner sediments from marine waste were analyzed using scanning electron microscopy, next generation sequencing, and culturomics. Proteobacteria, Bacteroidota, and Actinobacteria were predominant in all the samples. Enhanced growth on plastic by several microbial species was observed.

1. INTRODUCTION
Plastic production and, subsequently, plastic waste have increased exponentially through the last decades (Worm et al., 2017). The poor management of these residues, and their resistance to natural degradation (in some cases it comprises from hundreds to thousands of years) (Barnes et al., 2009), has resulted in a major, worldwide problem of plastic accumulation in all ecosystems on Earth. Even though the amount of recycled plastic has doubled from 2006 to 2018, the amount of post‐consumer waste plastic that is sent to landfills in Europe was still 25% in 2018 (PlasticsEurope, 2020).
Plastic residues in landfills are exposed to wind and water flows, which transport them into rivers and streams and, ultimately, into the oceans (Lebreton et al., 2017). Moreover, other direct sources such as beach littering, aquaculture, or fishing are also responsible for the accumulation of plastic in marine environments (GESAMP, 2016). Due to the generally low temperature and limited UV exposure in marine conditions, plastic degradation is considered to take longer in the sea (Gewert et al., 2015; Napper & Thompson, 2019). Plastic waste tends to fragment and spread in small particles (<5 mm) commonly known as microplastics (Arthur et al., 2009), which are easily ingested by marine wildlife, entering this way the trophic chain, and finally being ingested by humans (Setälä et al., 2014). Several studies have revealed the presence of plastic particles in fish, crustaceans, and mollusks (Neves et al., 2015; Van Cauwenberghe et al., 2015; Watts et al., 2014), and even in dietary salt (Iñiguez et al., 2017). This may have an impact on human health because of its physical accumulation as well as the toxicity of the additives used in plastic industries and the organic pollutants that plastic can adsorb in the marine environment (Bouwmeester et al., 2015; Rochman et al., 2013; Teuten et al., 2009). Moreover, not only the entrance of these microplastics on the trophic chain but also the enrichment of potentially pathogenic multidrug‐resistant bacterial strains in the plastisphere is a major health problem to face (Wang et al., 2021).
However, the amount of plastic estimated to enter into marine ecosystems does not correlate with the accumulation found by sampling techniques (Eriksen et al., 2014; Jambeck et al., 2015). Although there could be biases in sampling specific areas, this fact could also indicate that either physical or chemical plastic degradation is taking place in these ecosystems and/or microbial biodegradation is involved (Auta et al., 2017; Gewert et al., 2015; Sole et al., 2017; Zrimec et al., 2021). In recent years, plastic debris has proved a niche for specific plastic‐associated microbial communities to flourish, generally known as the “plastisphere” (Agostini et al., 2021; Zettler et al., 2013). Microbial growth on the plastisphere usually takes place in the shape of a biofilm on the plastic surface (Lobelle & Cunliffe, 2011). Although meta‐analyses are suggesting that a significant enrichment of potentially plastic biodegrading microorganisms in the plastisphere is detected (Wright, Langille, et al., 2021), there are still contradictory reports on the specificity of the composition of the microbial plastisphere. Specifically, some studies have shown that nonbiodegradable plastics, such as polyethylene terephthalate (PET), are colonized by a general biofilm rather than plastic‐specific species (Oberbeckmann et al., 2016; Pinnell & Turner, 2019). Therefore, microbial biofilms attached to plastic surfaces in the marine environment seem to be composed of complex communities where some microorganisms, although not being the primary producers, may have evolved or adapted to degrade plastic polymers or plasticizers (Pinnell & Turner, 2019).
In the last decades, there has been a rapid rise in the use of PET to produce disposable packaging, such as single‐use plastic bottles. This has led to a dramatic increase in PET waste generation, which is now one of the most common plastics polluting marine environments (PlasticsEurope, 2020; Ritchie & Roser, 2018). PET is a polymer made from raw petroleum‐derived monomers, terephthalic acid, and ethylene glycol. Its high content in aromatic compounds makes it chemically inert and subsequently very robust against biodegradation (Sinha et al., 2010).
In this context, bioprospecting microbial species able to in situ biodegrade plastic has arisen as a potentially useful tool for tackling the plastic contamination problem in the oceans (Danso et al., 2018). The first bacterium that demonstrated an effective PET‐degrading activity due to the expression of a lipase (PETase) was Ideonella sakaiensis, isolated from the sediments of a plastic‐recycling industry, which can hydrolyze this polymeric compound (Yoshida et al., 2016). However, these enzymes capable of PET hydrolysis have also been detected in other bacterial and fungal isolates, such as Thermobifida fusca, Streptomyces spp. or Fusarium solani, among others (Carr et al., 2020), and have been mainly described as cutinases, lipases, and esterases which are carboxylic ester hydrolases (Kawai et al., 2020).
Here, we show a complete characterization of the microbial communities associated with marine residues from the Mediterranean Western coast with a dual culture‐dependent and ‐independent approach. We have studied the biofilm morphology on plastic and aluminum debris through scanning electron microscopy (SEM), characterized the microbial communities of their inner sediments by 16S and 18S ribosomal RNA (rRNA) genes sequencing, and established a microbial collection of mainly culturable bacteria and some yeasts, whose ability to grow on media supplemented with PET as sole carbon source has been characterized.
2. MATERIALS AND METHODS
2.1. Sampling
Plastic residues and cans were collected from the Malva‐rosa beach (València, Spain; 39°27′48.3″N 0°19′07.6″W) in September 2017 (Figure 1). The sampling was carried out at 20 m from the coastline and 3 m in depth. Four PET plastic bottles (labeled as P1–4) and four metallic beverage cans (labeled as M10–13) were collected and transported to the laboratory into sterile plastic bags. All the residues were originally submerged or half‐buried in the marine sediments and they were thus partially filled with sand, mollusk shells, and marine plants (Posidonia oceanica) debris. Three samples of control seabed sediments (CS4–6) from the same area where plastic and aluminum residues were collected, which consisted of similar materials like sand, little stones, and shells, were also collected. Furthermore, some of the marine residues collected were still labeled with the expiration date of the product; therefore, an approximate age for these bottles or cans can be deduced: aluminum can M10 (expiration date 2003), aluminum can M12 and M13 (expiration date 2018), plastic bottle P1 (expiration date 2010).
Figure 1.

(a) Sampling location at the Mediterranean Western coast, Malva‐rosa beach, València (Spain). The specific sampling sites are pointed out with white arrows. (b) Examples of the samples collected, from left to right: PET plastic bottle P1; plastic bottle P2; aluminum can M10
Samples from the insides of each recipient (sediments) were collected under sterile conditions in the laboratory and stored at −20°C until required. To obtain samples from the plastic surface biofilms, recipients P1–4 were shortly rinsed with sterile water and then cut into small pieces which were shaken together with glass beads in phosphate‐buffered saline (PBS; pH 7.4; in g/L: 8.0 NaCl, 0.2 KCl, 1.42 Na2HPO4, 1.80 KH2PO4), at 500 rpm, for an hour. A total of 150 ml of the resulting suspension were collected and centrifuged at 4500 rpm for 15 min (sample P12) and stored at −20°C until required. Sample 12 was only analyzed in terms of culturable bacteria and it was not included in the high‐throughput 16S rRNA gene sequencing.
2.2. Isolation of microbial strains
Sediment samples from recipients P1–4, M10–13, biofilm sample P12, and control sediments CS4–6 were diluted in PBS at a final ratio of 1:4 (v:v). Serial dilutions were then prepared and four replicates of 50 µl aliquots were spread on commercial Marine Agar (MA) (Ref: 1059; Laboratorios Conda S.A.) and incubated at 18°C for 2 weeks. Two replicates were incubated under aerobic conditions and the other two replicates in anaerobic conditions by placing the dishes inside a hermetic container without oxygen (N2 atmosphere).
Individual colonies were picked according to morphological traits (color, shape, and size) and restreaked on fresh media until a pure culture was obtained. The strains were named after a code composed of a letter and a number associated with its origin (P1–4 and P12: plastic bottles; M10–13: aluminum cans; CS4–6: external sediments), followed by a unique number for each strain and a letter referring to the incubation conditions (X: aerobic conditions; A: anaerobic conditions). For example, P1.1X means the first colony isolated from bottle P1 that grew under aerobic conditions. The strains were stored in cryotubes with 20% glycerol at −80°C until used.
2.3. Molecular identification of isolates through 16S/18S rRNA gene sequencing
DNA extraction was carried out by using the protocol described by Latorre et al. (1986) and confirmed through electrophoresis in agarose gel (1.4% w/v). Strain identification was performed through 16S rRNA gene Sanger sequencing, by using the universal primers 8 F (5ʹ‐AGAGTTTGATCCTGGCTCAG‐3ʹ) and 1492 R (5ʹ‐CGGTTACCTTGTTACGACTT‐3ʹ). In the cases that the 16S rRNA gene amplification failed, 18S rRNA gene universal primers 86 F (5ʹ‐ACTGCGAATGGCTCATTAAATCAG‐3ʹ) and 1188 R (5ʹ‐AGTCAAATTAAGCCGCAG‐3ʹ) were used to verify whether the strains were eukaryotic. Amplicons were precipitated overnight in isopropanol 1:1 (v:v) and potassium acetate 3 M, pH 5, 1:10 (v:v) at −20°C. After centrifuging at 12,000 rpm for 10 min, DNA pellets were washed in 70% ethanol and resuspended in the required amount of sterile Milli‐Q water. BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) was used for amplicon tagging for Sanger sequencing, which was performed in the Sequencing Service (SCSIE) of the University of València (Spain). The Sequences were manually edited with Trev (Staden Package, 2002) to eliminate low‐quality base calls and compared by EzBioCloud 16S online tool (https://www.ezbiocloud.net/). The 16S rRNA genes of some interesting isolates holding an identity lower than 98.7% with the closest type strain were also sequenced with primers 341 R (5ʹ‐ CTGCTGCCTCCCGTAGG‐3ʹ) and 1055 F (5′‐ATGGCTGTCGTCAGCT‐3′) and complete 16S rRNA gene sequences were assembled with the MEGA10 tool and compared again by EzBioCloud 16S online tool.
2.4. Scanning electron microscopy
Plastic and aluminum samples were briefly washed with sterile distilled water and then pieces of ca. 0.25 cm2 were cut and fixed in Karnovsky's fixative (Karnovsky, 1965). The fixation solution was changed after five hours and samples were stored in this solution at 4°C until required. For SEM, the pieces were washed in phosphate buffer 0.1 M, pH 7.4 (PB, in g/L: 3.1 NaH2PO4·H2O, 10.9 Na2HPO4) to remove the fixative and progressively dehydrated in increasing ethanol concentrations. Samples were placed inside microporous specimen capsules (30 μm pore size) immersed in absolute ethanol, followed by critical point drying in an Autosamdri 814. The fragments were then arranged on SEM aluminum stubs using carbon tape and coated with Au/Pd sputtered in argon gas. The observation was carried out in a Scanning Electron Microscope Hitachi S‐4800 at the electron microscopy service of the University of València (SCSIE).
2.5. DNA purification and high‐throughput 16S rRNA gene sequencing
Internal sediments from the marine residues collected were subjected to DNA extraction. In particular, 1 g of sediments of each sample (Plastic bottles P1, P2, P3, and P4; Aluminum cans M10–M13; Control sediments CS4–CS6) were taken from 2 cm in depth from the inner sediments of each bottle/can. No replicates were performed. Metagenomic DNA extraction was carried out by using the Power Soil® DNA Isolation Kit (12888‐100; MoBio Laboratories Inc.) according to the manufacturer's instructions, but incubating at 65°C (10 min) after the addition of solution C1, and resuspending the extracted DNA in 25 µl of Milli‐Q water. The resulting DNA was quantified using the QUBIT dsDNA HS‐high sensitivity kit (Invitrogen). Then, primers 341 F (5′‐CCTAYGGGRBGCASCAG‐3′) and 806 R (5′‐GGACTACNNGGGTATCTAAT‐3′) were used to amplify the V3–V4 region of the 16S rRNA gene. All polymerase chain reactions (PCRs) were carried out with Phusion® High‐Fidelity PCR Master Mix (New England Biolabs). PCR products were mixed at equal density ratios. The pool was then purified with Qiagen Gel Extraction Kit (Qiagen). Sequencing libraries were generated with NEBNext® Ultra™ DNA Library Prep Kit for Illumina and quantified via Qubit and qPCR. Finally, the NovaSeq 6000 Sequencing System (2 × 250 bp) was employed for sequencing the samples. All the library preparation and sequencing steps were carried out by Novogene.
2.6. Bioinformatic analysis
Raw Illumina sequences were analyzed using Qiime2 (v. 2020.8) (Bolyen et al., 2019). Briefly, the quality of the reads was assessed with the Demux plugin, and the sequences were subsequently corrected, trimmed, and clustered into amplicon sequence variants (ASVs) via Dada2 (Callahan et al., 2016). The taxonomy of each sequence variant was assigned employing the classify‐Sklearn module from the feature‐classifier plugin (Bokulich et al., 2018). SILVA (v. 138) was used as a reference for the 16S rRNA gene assignment (Quast et al., 2013). The phyloseq R package (McMurdie & Holmes, 2013) was used for analyzing and visualizing the data. All the α‐diversity tests were carried out using ASVs and rarefying to the lowest library size (128,327 sequences). Principal coordinate analysis (PCoA) plots were created using Bray–Curtis as a dissimilarity measure. Finally, DESeq. 2 (Love et al., 2014) was used for differential abundance analyses).
2.7. Plastic degradation assay in solid medium
Plastic degradation was assessed through qualitative assays by comparing the growth of the bacterial strains on minimal marine medium (MMA), enriched marine medium (MME), and marine medium supplemented with plastic (MMP). MMA consisted of water from the Mediterranean Sea and 15 g/L agar, whereas MME consisted of seawater and, in g/L, 1.0 yeast extract, 5.0 bacteriological peptone, and 15 agar. MMP was prepared by using seawater, supplemented with 9.3 g/L of ground PET of approximately 0.5 mm in size, from a commercial PET water bottle (brand Cortes) and 15 g/L of agar, which was then sterilized at 121°C for 30 min. The PET bottle was ground in a coffee grinder for 5 min at maximum speed. As plastic particles tended to sediment on the bottom of the dishes, the media was stirred by using sterile spatulas before solidification.
Before the incubation with PET, bacterial isolates were grown on solid MMA for 4 days at room temperature. Cell suspensions with an Optical Density at 600 nm (OD600) of 1 were prepared in PBS and 4 µl of the suspensions were placed on Petri dishes containing MMA, MME, and MMP (in duplicate). The dishes were incubated for 16 days at 18°C. Isolates with a more vigorous growth (as determined by colony diameter and cell density) in MMP than in MMA were selected as potential plastic degrading bacteria and tested again in the same media conditions but using a 10‐fold dilution of the bacterial suspensions (OD600 of 0.1).
2.8. Plastic degradation assay in liquid medium
Assay tubes were prepared with 3 ml of seawater and 0.400 ± 0.001 g of particles of PET from a new water bottle (brand Cortes), of 3 mm in size (cut by hand to obtain homogeneous size), and sterilized by autoclaving at 121°C for 30 min. Bacterial strains were grown on solid MA for 4 days at room temperature. Cell suspensions were prepared in PBS and adjusted to a final OD600 of 0.05. The assay was carried out in duplicate by incubating the tubes at 18°C under shaking (200 rpm) for 3 months. Control tubes consisted of sterile seawater inoculated with the microbial cultures, as well as seawater and plastic particles but without inoculated bacteria.
At the end of the incubation period, PET fragments were rinsed with sterile water and vortexed for 2 min in distilled water. The process was repeated three times and the washed plastic particles were dried at 65°C for 48 h. Finally, the remaining plastic particles were weighted in a precision balance. To finally compare the colony‐forming units (CFU) in each condition, the recovered supernatants of each tube were diluted in serial dilutions and 50 µl of each dilution was inoculated in duplicate into MA plates.
3. RESULTS
3.1. Residue types and samples
Plastic PET bottles and aluminum cans were collected to study their associated microbiota as described in Section 2. The bacterial communities present in the inside‐sediments, coming from PET bottles and aluminum cans, were compared with control, non‐artificial residues‐associated sediments from the same area. Interestingly, some of the marine residues collected were still labeled with the expiration date of the product; therefore, an approximate age for these bottles or cans can be deduced: aluminum can M10 (expiration date 2003), aluminum can M12 and M13 (expiration date 2018), plastic bottle P1 (expiration date 2010).
3.2. Scanning electron microscopy
The SEM images of the surface of plastic and aluminum marine waste suggest a diverse microbial community attached to these surfaces (Figure 2). Different microbial morphologies could be differentiated in both cases, including rod‐ and coccus‐shaped cells as well as diatoms and filamentous microorganisms. In particular, spermatozoid‐shaped bacteria stood out in Figure 2c,e which may belong to prosthecate bacteria such as Hyphomonadaceae. Interestingly, several samples showed 2 µm fusiform bacilli firmly attached to the plastic surface, to which they were linked through polar fimbriae‐like structures (Figure 2a,b). In another plastic bottle, one of the most frequent morphologies was a square shape of around 0.6 µm in size which could not be attributed to a microorganism as it could instead correspond to mineral forms (Figure 2c,d). Finally, eukaryotic flagellated cells and diatoms were observed in the analyzed aluminum surfaces of cans (Figure 2e,f).
Figure 2.

Scanning electron microscopy images of microorganisms on the surface of different marine residues. Scale bar (a) 10 µm, (b) 2 µm, (c) 10 µm, (d) 3 µm, (e) 10 µm, and (F) 100 µm. (a, b) Microbial community on the plastic surface of sample P1. Fusiform bacilli‐like microorganisms attached to the surface by fimbriae‐like adhesion structures. (c, d) Biofilm on the plastic surface of sample P2. Square‐like nonidentified shapes of less than 1 µm are predominant in this sample. (e, f) The surface of aluminum cans with scattered microbial cells
3.3. Taxonomy of the waste‐associated bacterial communities
The bacterial community of marine waste was studied by high‐throughput 16S rRNA gene sequencing yielding the composition of the taxa in the inside sediments of four PET bottles, inner sediments of four aluminum cans, as well as three samples of control marine sediments. The shape of rarefaction curves revealed that sequencing was deep enough to cover all the microbial diversity for all samples (Figure A1). Furthermore, based on the comparison of the richness value (number of different AVSs; Figure 3a) and the diversity (Shannon index; Figure 3b and Simpson index; Figure 3c), the alpha diversity was not significantly different among samples (p > 0.1; Mann–Whitney U test).
Figure 3.
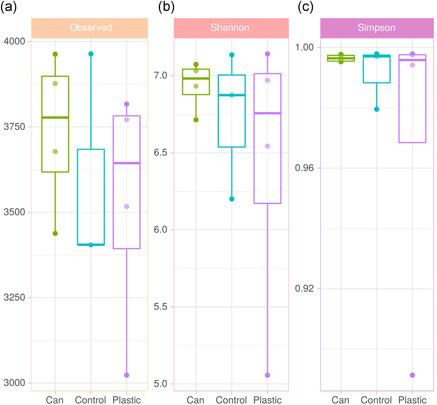
Representations of the values of alpha diversity indices in the (a) observed richness at the amplicon sequent variant (ASV) level (number of ASVs), (b) Shannon index of diversity, and (c) Simpson index of diversity. The 11 analyzed samples are represented: inside‐sediments of cans (green); polyethylene terephthalate inside‐sediments (purple); control‐sediments of the sea‐bed (blue)
A PCoA including samples P1, P3, P4, M10, M11, M12, and M13 revealed no significant difference between the composition of the bacterial communities of both (plastic and cans) inside waste sediments (p > 0.05; PERMANOVA) (Figure 4). Sample P2 was not included in Figure 4 due to its substantial difference in bacterial composition, which precluded its separation from the other samples in the PcoA and difficulted the interpretation of the figure (see Figure A2 for the complete analysis).
Figure 4.

Principal coordinates analysis (PCoA) based on Bray–Curtis dissimilarities at the genus level in bacterial populations of both inside‐sediments of marine residues, plastic (blue), and aluminum cans (red). Sample P2 not included
The representation of the relative abundance of the 20 most abundant phyla (Figure 5a) and the 20 most abundant classes (Figure 5b) showed that the microbial composition was similar among the different types of sediments. However, the comparison at the genus level of the 20 most abundant genera revealed some differences between samples (Figure 5c). At the phylum level, the bacteriomes of all the samples (mean relative abundance) were dominated by Proteobacteria (45.2%), followed by Bacteroidota (or Bacteroidetes) (11.9%), Actinobacteriota (or Actinobacteria) (11.2%), and Desulfobacterota (or Deltaproteobacteria) (7.3%). On top of that, other less frequent phyla that were present in all the samples were Campilobacterota (predominant in sample P2), Acidobacteriota, Firmicutes, Gemmatimonadota, Myxococcota, Crenarchaeota, and Calditrichota, among others. In terms of class, Gammaproteobacteria (27.2%), Alphaproteobacteria (18.0%), Acidimicrobiia (10.7%), and Bacteroidia (9.7%) comprised almost 50% of all the samples. Furthermore, at the genus level, high diversity was found in all the samples. On average, the top 10 genera described in these marine samples were: Unknown Rhodobacteraceae (9.0%), Woeseia (8.7%), uncultured Actinomarinales (8.0%), Vibrio (5.8%), Sulfurovum (4.7%), Gammaproteobacteria B2M28 (2.8%), unknown Gammaproteobacteria (2.3%), uncultured Saprospiraceae (1.9%), Desulfosarcinaceae Sva0081 (1.5%), and uncultured Syntrophobacterales (1.4%). Samples CS4 and P2 showed a similar taxa composition to the other samples, but clear differences in abundance, where Vibrio and Sulfurovum were the dominant genera in each sample, respectively. A test for differential abundance (Table A1) revealed that the phylum Caldatribacteriota was significantly more abundant in plastic sediments than in aluminum sediments. At the same time, it showed that when comparing debris sediments to control sediments, Cyanobacteria and Marinimicrobia were more abundant in can sediments as well as Campilobacteria, Cloacimonadota, and Acetothermia were significantly more abundant in inner plastic sediments.
Figure 5.

Barplots showing the taxonomic profiles at the phylum (a), class (b), and genus (c) level of the top 20 most abundant groups in terms of relative abundance of inside‐sediments from marine residues (plastic and aluminum cans) and control sediments by high‐throughput 16S ribosomal RNA gene sequencing
3.4. Strain collection and identification
Culturing the marine sediments associated with artificial residues yielded a large number of highly diverse microbial colonies, in terms of color and morphology. A total of 170 bacterial strains and one yeast were isolated. All the strains that grew at first under anaerobic conditions showed later the ability to grow in the presence of oxygen. In total, 142 out of 171 strains were identified through colony PCR and 16S and 18S rRNA gene sequencing (Table A2), whereas 29 remained unidentified due to the impossibility to carry out the amplification of these fragments through PCR. The identified bacteria were distributed into four phyla: Firmicutes, Proteobacteria, Bacteroidota, and Actinobacteriota (Figure 6). Bacillus spp. was by far the most abundant genus (33 species identified), followed by Vibrio spp. (9), Erythrobacter spp. (8), Planomicrobium spp (7), Sulfitobacter spp. (6) and Sphingorhabdus spp. (5) among other genera. Interestingly, the identification of a large fraction of the microorganisms in the collection revealed that some isolates could represent new species, as they held a percentage of identity with the closest type strain below the 98.7% threshold established to circumscribe a new bacterial species (Chun et al., 2018). In particular, isolates M10.2A, M10.9X, and P4.10X with the closest type strains belonging to the genera Gillisia, Sagittula, and Maritalea, respectively, are potentially new species. Further characterization is needed to determine it.
Figure 6.

Bar plots showing the distribution into four phyla of the isolated species within the collection. The different colors in each phylum represent one different genus and the numbers indicate the number of isolates identified, which are only written when the number of isolates per genus is greater than two (see Table A2 for detailed information about each strain identified)
3.5. PET degradation assays
To test the PET degrading activity of the microbial isolates obtained from marine waste, a preliminary qualitative screening was carried out consisting of a drop assay of bacterial culture in MMA and MMP to check differential growth when PET plastic was present (see Section 2.7). From this preliminary screening, differences in terms of growth after the drop assay performed as described in Section 2 are shown in Figure 7. In the first round of selection, 27 out of the 171 strains tested were selected as they showed increased growth in minimal medium supplemented with PET particles compared to the control medium without PET, after 28 days at 18°C. A second assay with the 27 selected strains was then carried out and led to the further selection of 16 strains with the more obvious differential growth on PET‐containing media. 16S rRNA complete gene sequences were obtained and compared using EzBioCloud thus allowing the identification at the species level (Table A3). A selection of eight of these isolates are shown in Figure 7 and they were identified as members of the species Bacillus algicola, Pseudomonas juntendi, Kocuria rosea, Aquimarina intermedia, Microbacterium aerolatum, Rhodotorula evergladensis, Citricoccus alkalitolerans, and Bacillus simplex.
Figure 7.

Differential growth of eight selected strains on minimal marine medium (MMA), minimal marine medium supplemented with polyethylene terephthalate (PET) (MMP), and enriched marine medium (MME). MMA was used as a control for the basal growth of the strain without any supplemented carbon source. MMP was used to compare the growth of the isolates in the presence of PET plastic. MME allowed the normal growth of the strain in a rich nutrient marine medium
The group of 16 strains selected in the previous assay was incubated for 3 months at 18°C in liquid MMP containing PET particles precisely weighted. The following controls were included in the assay: PET without inoculated bacteria; the medium without neither bacteria nor PET; and each bacterium incubated without plastic. The test resulted in no detectable weight loss of the plastic particles in any sample inoculated with any of the 16 strains. Surprisingly, a small weight loss was detected in the noninoculated controls, in which the liquid became cloudy, appearing a white precipitate (Figure A3). To discard microbial contamination of the controls, the commercial MA medium was inoculated with the cloudy supernatant, which was also observed under the microscope. Both experiments yielded negative results and contamination of the controls was thus discarded. There was one unit decrease in the pH of these control tubes (7.5 ± 0.1) compared with the tubes inoculated with a microorganism, all of which remained at a pH of 8.5 ± 0.3 and exhibited no turbidity in any inoculated tube.
Substantial differences in bacterial growth were found in the containing‐PET and non‐containing‐PET medium in four of the strains, by comparing cell number (CFU) of the supernatants inoculated in MA medium (Figure 8). The strains that showed an increased growth when PET was present were: Micrococcus luteus (CS5.4X, 20.8‐fold increased growth), Idiomarina piscisalsi (M11.3X, 4.7‐fold increase), Citricoccus alkalitolerans (P12.8X, 3.6‐fold increase), Aquimarina intermedia (M12.2X, 3.4‐fold increase), Microbacterium aerolatum (P12.4X, 2.4‐fold increase), Bacillus algicola (P1.1X, 2.1‐fold increase), and the yeast Rhodotorula evergladensis (P12.5X, 1.5‐fold increase) (Table A3).
Figure 8.

Comparison in colony‐forming units (CFU) count of selected isolates that showed increased growth in polyethylene terephthalate (PET)‐containing medium. Strains were incubated in a PET‐containing medium and control medium without PET at 18°C under shaking (200 rpm) for 3 months. Negative controls yielded no CFU count. Identification of strains based on 16S ribosomal RNA (rRNA) gene sequencing: CS5.4X: Micrococcus luteus; CS6.2X: Aurantimonas coralicida; M10.4A: Bacillus zhangzhouensis; M11.3X: Idiomarina piscisalsi; M12.2X: Aquimarina intermedia; M12.3A: Bacillus simplex; M13.5A: Pseudomonas juntendi; M13.7X: Kocuria rosea; P1.1X: Bacillus algicola; P12.4X: Microbacterium aerolatum; P12.5X: Rhodotorula evergladensis; P12.8X: Citricoccus alkalitolerans; P4.10X: Maritalea mobilis; P4.3X: Meridianimaribacter flavus; P4.7X: Microbacterium imperiale. Accession numbers of deposited 16S rRNA gene sequences and fold‐increase in CFU can be found in Table A3
4. DISCUSSION
Artificial residues hold great promise as a source of a huge variety of microorganisms for the bioremediation of plastic waste (Delacuvellerie et al., 2019; Yoshida et al., 2016). The interest in the study of the microbial communities associated with the plastisphere, as well as to other anthropic residues such as glass bottles or ceramic surfaces (McCormick et al., 2014; Oberbeckmann et al., 2016; Pinnell & Turner, 2019), has increased exponentially in the last years. The worldwide problem of plastic contamination in the oceans has led researchers to investigate the impact of these pollutants not only on the surfaces but also in deep‐sea areas (Woodall et al., 2018). Even though these studies shed light on the plastic degradation problem, there are still several questions that need further investigation in this field and more research focusing on other materials such as metal debris would be interesting.
Regarding the bacterial communities inhabiting the marine sediments studied in this study, at the β‐diversity level, the samples analyzed did not cluster together depending on the type of sediment (cans‐inner sediments, plastic‐inner sediments, and control‐external sediments) (Figures 4 and A2). This suggests that the bacterial profile of sediments trapped into artificial residues falls within the diversity of bacterial profiles of similar, natural environments. Interestingly, samples from each type (plastic or metal) displayed similar morphological features under SEM (Figure 2).
The morphology of the microorganisms in the biofilms we studied by SEM is in line with previous descriptions, in which a high diversity of microorganisms, both eukaryotes, and prokaryotes, were found (Bryant et al., 2016; Masó et al., 2016; Reisser et al., 2014). Interestingly, we found numerous fusiform bacteria attached to the plastic surface through fimbriae‐like structures (Figure 2b). Similar shapes have previously been described to inhabit plastic surfaces in marine environments. For example, Bryant et al. (2016), showed a similar microbial community and also reported a bacillary shape that is attached from one pole to the plastic surface. In another study on the plastisphere of microplastics from the Australian shores, the same bacillary shapes with fimbriae‐like structures adhering to the plastic surface were described (Reisser et al., 2014). Furthermore, the well‐known PET degrading bacteria Ideonella sakaiensis exhibits attaching appendages when growing on plastic (Figure 2f in Yoshida et al., 2016). Hence, the finding of microorganisms directly attached to the plastic surface points towards the possibility of these bacterial forms being anchored to the plastic substrate to allow its degradation by exoenzymes.
Another interesting morphological trait of the observed microorganisms is the presence of spermatozoid‐shaped bacteria (Figure 2c,e). This bacterial shape may correspond to prosthecate bacteria, particularly the genus Hyphomonadaceae, which is abundant in the microbial communities of plastic residues (fig. 5c,f from Bryant et al., 2016; fig. 2 from Zettler et al., 2013) and we have also detected this taxon, although in low abundance, in the marine debris analyzed through high‐throughput 16S rRNA gene sequencing.
In terms of microbiota, our results show that the bacterial profile is very similar between seafloor sediments and internal residue sediments. The microbial composition is characterized by a set of marine bacterial classes (Gammaproteobacteria, Alphaproteobacteria, Acidimicrobiia, and Bacteroidia) that belong to the phyla Proteobacteria, Actinobacteriota, and Bacteroidota which have widely been described in surface marine sediments (Hoshino et al., 2020). Indeed, Gammaproteobacteria and Alphaproteobacteria proved the dominant classes in all the samples analyzed, and they have been reported as the most abundant taxa in samples from pelagic to benthic locations (Petro et al., 2017; Zinger et al., 2011). Moreover, in our study, the phylum Desulfobacterota was detected in all the samples. This result correlates with the fact that sulfate concentrations are higher in the surface layers of seafloor sediments (Leloup et al., 2009; Pellerin et al., 2018), which allows the proliferation of species within this phylum, such as members of Desulfosarcinaceae, Syntrophobacterales, Desulfocapsaceae, and Desulfobulbaceae, all of which were found in the sediments analyzed in this study.
Interestingly, the abundance of the genus Vibrio is remarkable in all the samples. Pathogenic bacterial species belonging to Vibrio have been widely described in marine environments usually in low abundance and they have also been found in plastic debris (Delacuvellerie et al., 2019; Jacquin et al., 2019; Zettler et al., 2013). Vibrio is very resistant to hard conditions and can perform a rapid growth in marine environments in response to an increase of nutrients (Westrich et al., 2018). Another interesting fact is that PET bottle P2 was dominated by Sulfurovum while this genus remained in low abundance in the other samples. Species from the genus Sulfurovum are chemolithoautotrophic sulfur‐oxidizing bacteria that are primary producers in marine sediments communities (Mori et al., 2018) and even have been described to be the dominant taxon in seafloor sediments in some localizations (Sun et al., 2020).
The microbial composition we have found is similar to that reported in a variety of studies carried out on the biofilm that directly colonizes the plastic surface (Amaral‐Zettler et al., 2020; Delacuvellerie et al., 2019; Oberbeckmann et al., 2016). A recent review on colonization and plastic biodegradation in the marine environment (Jacquin et al., 2019), summarizes that the surface of plastic residues are generally quickly colonized by Gammaproteobacteria and Alphaproteobacteria, and then, with time, Bacteroidota also becomes an important group in the biofilm.
The microbial profiles observed in the collection of culturable strains we set are in accordance with the previous results reported by several authors. This collection of 171 microbial isolates includes strains of 53 different genera distributed among the phyla Firmicutes, Proteobacteria, Bacteroidota, and Actinobacteriota. Specifically, Proteobacteria, which is one of the most common phyla in most of the biomes, is also the most abundant phylum associated with plastic residues worldwide (Roager & Sonnenschein, 2019). Among the recurrent alphaproteobacterial families found in such environments are Erythrobacteraceae and Rhodobacteraceae, which in our collection are represented by the eight genera: two belonging to Erythrobacteraceae (Altererythrobacter, Erythrobacter) and six belonging to Rhodobacteraceae (Epibacterium, Maliponia, Ruegeria, Sagittula, Sulfitobacter, and Yoonia). Moreover, the eight representative genera of the phylum Bacteroidota belonged to the Flavobacteriaceae family, which is, again, a common plastic debris‐associated taxa (Amaral‐Zettler et al., 2020; Jacquin et al., 2019). The abundance of Firmicutes is linked to the high number of Bacillus spp, (33 species isolated in total) we found. This genus has been reported as a marine plastic colonizer and degrader (Delacuvellerie et al., 2019; Oberbeckmann et al., 2015; Ribitsch et al., 2011).
The diversity of microorganisms found on artificial debris, the presence of biofilms and plastic adhesion fimbriae‐like structures, and the taxonomic identity of some of the taxa suggest a possible role in plastic biodegradation of some of the bacteria of the collection we set and characterized. The quantitative PET degradation assay with the selected strains yielded no significant loss of non‐pretreated PET particles weight. However, this is not particularly surprising givien the fact that PET is very resistant to biodegradation due to its compact structure, hence heat or oxidative pretreatments are usually needed to enhance biodegradation (Gewert et al., 2015). Nevertheless, we observed an increased growth (measured as CFU count variation), of seven of the isolates when PET was present as the sole carbon source in the medium, suggesting the capability of some strains to degrade plastic or plastic additives, such as plasticizers, antioxidants, light and heat stabilizers, pigments or slip reagents that are usually added to plastics to enhance their structural properties. These compounds are commonly not covalently bonded to the plastic polymer; therefore, they can more easily leak out from the plastic structure to the liquid phase (Hahladakis et al., 2018). Remarkably, the strain of Micrococcus luteus we tested, showed a 20‐fold increase in CFUs when the minimal medium was supplemented with PET particles compared to a non‐supplemented‐PET medium. This is not the first time that Micrococcus luteus has been described to potentially degrade plastic (Montazer et al., 2018; Sivasankari & Vinotha, 2014), and its degrading ability seems to be associated with its ability to form biofilm in plastic surfaces (Blakeman et al., 2019; Feng et al., 2011). The isolates identified as Idiomarina piscisalsi, Citricoccus alkalitolerans, Aquimarina intermedia, and Microbacterium aerolatum which showed roughly a two‐ to four‐fold increase in growth in PET, have been sparsely studied in previous works regarding plastic‐degrading activity. Specifically, Idiomarina has been recently reported to possibly assist in the formation of biofilms on the surface of PET particles, although it showed no significant PET degradation (Gao & Sun, 2021). On the contrary, although there is no previous report on the ability of Bacillus algicola (which showed double CFU count when incubated with PET) to degrade plastic polymers, other species and strains within the genus have been described as degraders of polystyrene, polypropylene, polyethylene, and PET microplastic particles (Auta et al., 2017; Wright, Bosch, et al., 2021) as well as polyvinyl chloride (Giacomucci et al., 2019). Finally, the yeast Rhodotorula evergladensis, which showed a tiny increase in growth on PET in our study, has been previously reported to degrade plasticizers (Gartshore et al., 2003).
Taken together, our results suggest that the marine waste‐associated microbiota hold potential as a source of biotechnological interesting strains for plastic or plastic‐related compounds.
CONFLICT OF INTERESTS
None declared.
ETHICS STATEMENT
None required.
AUTHOR CONTRIBUTIONS
Àngela Vidal‐Verdú: Conceptualization (equal); Data curation (lead); Investigation (lead); Writing – original draft (lead); Writing – review and editing (equal); Adriel Latorre‐Pérez: Conceptualization (supporting); Formal analysis (lead); Writing – review and editing (equal); Esther Molina‐Menor: Investigation (supporting); Writing – original draft (supporting); Writing – review and editing (equal); Joaquin Baixeras: Investigation (Supporting); Supervision (Supporting); Writing – review and editing (equal); Juli Peretó: Conceptualization (equal); Writing – review and editing (equal); Manuel Porcar: Conceptualization (equal); Writing – review and editing (equal).
ACKNOWLEDGMENTS
We acknowledge Darwin Bioprospecting Excellence SL for the bioinformatic analysis and sequencing and the CSIC‐PTI Interdisciplinary Platform for Sustainable Plastics Towards a Circular Economy (SusPlast) for their support. Financial support by Ministerio de Ciencia e Innovación (grant SETH ref. RTI2018‐095584‐B‐C41‐42‐43‐44 co‐financed by FEDER), the European Union H2020 (BIOROBOOST project ID 210491758; Micro4Biogas project ref. 101000470), Agencia Estatal de la Innovación AEI (MIPLACE project ref. PCI2019‐111845‐2), and Agència Valenciana de la Innovación AVI (ENTOMOPLAST project ref. INNEST/2021/334) are acknowledged. Àngela Vidal‐Verdú and Esther Molina‐Menor are funded with a Formación del Profesorado Universitario grant from the Spanish Ministerio de Ciencia, Innovación y Universidades with references FPU18/02578 and FPU17/04184, respectively. Adriel Latorre‐Pérez is a recipient of a Doctorado Industrial fellowship from the Spanish Ministerio de Ciencia, Innovación y Universidades (reference DI‐17‐09613).
Figure A1.

Rarefaction curves
Figure A2.

Principal coordinate analysis including all samples
Figure A3.

Quantitative assays for polyethylene terephthalate (PET) degradation ability. (a) Negative control tubes at the end of the assay. On the left, the two replicates of the negative control consisting of marine water with PET fragments. A white precipitate of mineral nature appeared after the incubation time, probably due to the change in pH. The two tubes on the right contained only marine water. (b) A representative example of the assay with the isolate M11.3X. All the tubes were inoculated with the bacterium at the beginning of the assay in duplicate, on marine water supplemented with PET particles (left) and marine water without plastic as control (right)
Table A1.
Results of comparing plastic inner sediments, aluminum cans inner sediments, and control sediments from the seabed by DESeq. 2 test
| Phylum level | |||||||||||
| Base mean abundance | log2FoldChange | lfcSE | p value | FDR‐adjusted p value | Domain | Phylum | |||||
| Can sediment versus control sediment | |||||||||||
| Reference: Control sediment | |||||||||||
| Test: DESeq. 2 | |||||||||||
| 221.353 | 2.046 | 0.610 | 0.001 | 2.89E−02 | d__Bacteria | p__Cyanobacteria | |||||
| 128.105 | 1.612 | 0.463 | 0.000 | 2.89E−02 | d__Bacteria | p__Marinimicrobia_(SAR406_clade) | |||||
| Plastic sediment versus control sediment | |||||||||||
| Reference: Control sediment | |||||||||||
| Test: DESeq. 2 | |||||||||||
| 9090.508 | 5.066 | 1.36 | 1.97E−04 | 7.39E−03 | d__Bacteria | p__Campilobacterota | |||||
| 69.709 | 5.242 | 1.57 | 8.29E−04 | 2.07E−02 | d__Bacteria | p__Cloacimonadota | |||||
| 68.653 | 22.730 | 3.68 | 6.46E−10 | 4.84E−08 | d__Bacteria | p__Acetothermia | |||||
| Plastic sediment versus can sediment | |||||||||||
| Reference: Can sediment | |||||||||||
| Test: DESeq. 2 | |||||||||||
| 24.250 | 21.853 | 2.916 | 6.74E−14 | 5.12E−12 | d__Bacteria | p__Caldatribacteriota | |||||
| Genus level | |||||||||||
| Base mean abundance | Log2FoldChange | lfcSE | p value | FDR‐adjusted p value | Phylum | Family | Genus | ||||
| Can sediment versus control sediment | |||||||||||
| Reference: Control sediment | |||||||||||
| Test: DESeq. 2 | |||||||||||
| 172.967 | 23.916 | 3.678 | 7.93E−11 | 8.07E−09 | p__Firmicutes | f__Lachnospiraceae | g__Shuttleworthia | ||||
| 157.511 | 23.795 | 3.051 | 6.29E−15 | 3.20E−12 | p__Firmicutes | f__Hungateiclostridiaceae | g__Fastidiosipila | ||||
| 33.656 | −23.170 | 3.381 | 7.20E−12 | 9.16E−10 | p__Firmicutes | f__Clostridia_UCG‐014 | g__Clostridia_UCG‐014 | ||||
| 129.622 | 2.701 | 0.690 | 9.02E−05 | 5.10E−03 | p__Bacteroidota | f__Lentimicrobiaceae | g__Lentimicrobiaceae | ||||
| 103.293 | 3.529 | 0.936 | 1.64E−04 | 8.36E−03 | p__Bacteroidota | f__Prolixibacteraceae | g__uncultured | ||||
| 63.609 | 22.529 | 3.086 | 2.89E−13 | 4.90E−11 | p__Firmicutes | f__Hungateiclostridiaceae | g__Mageibacillus | ||||
| 10.991 | 20.088 | 3.682 | 4.87E‐08 | 3.54E−06 | p__Firmicutes | f__Veillonellaceae | g__Veillonella | ||||
| 55.211 | 22.335 | 3.036 | 1.89E−13 | 4.81E−11 | p__Firmicutes | f__Streptococcaceae | g__Lactococcus | ||||
| 21.466 | 7.658 | 2.282 | 7.91E−04 | 3.35E−02 | p__Proteobacteria | f__Rhodobacteraceae | g__Confluentimicrobium | ||||
| 23.666 | −2.979 | 0.855 | 4.94E−04 | 2.29E−02 | p__Proteobacteria | f__Kangiellaceae | g__Kangiella | ||||
| 21.844 | 21.042 | 3.680 | 1.08E−08 | 9.14E−07 | p__Firmicutes | f__Lachnospiraceae | g__[Ruminococcus]_torques_group | ||||
| 10.304 | 20.000 | 3.682 | 5.58E_08 | 3.55E−06 | p__Proteobacteria | f__Rhodobacteraceae | g__Pseudoruegeria | ||||
| Plastic sediment versus control sediment | |||||||||||
| Reference: Control sediment | |||||||||||
| Test: DESeq. 2 | |||||||||||
| 454.017 | 10.661 | 3.196 | 8.51E−04 | 2.66E−02 | p__Actinobacteriota | f__Propionibacteriaceae | g__Cutibacterium | ||||
| 18,607.676 | 6.694 | 2.024 | 9.41E−04 | 2.82E−02 | p__Campilobacterota | f__Sulfurovaceae | g__Sulfurovum | ||||
| 672.863 | 9.176 | 2.510 | 2.57E−04 | 8.41E−03 | p__Bacteroidota | f__Chlorobiaceae | g__Prosthecochloris | ||||
| 241.181 | 6.801 | 2.100 | 1.20E−03 | 3.46E−02 | p__Chloroflexi | f__661239 | g__661239 | ||||
| 1402.831 | 9.537 | 3.065 | 1.86E−03 | 4.79E−02 | p__Chloroflexi | f__GIF3 | g__GIF3 | ||||
| 41.074 | −23.332 | 3.632 | 1.32E−10 | 4.75E−08 | p__Firmicutes | f__Erysipelotrichaceae | g__Dubosiella | ||||
| 136.694 | 6.841 | 2.179 | 1.69E−03 | 4.51E−02 | p__Cloacimonadota | f__MSBL8 | g__MSBL8 | ||||
| 52.605 | 22.421 | 3.539 | 2.35E−10 | 5.65E−08 | p__Bacteroidota | f__SJA‐28 | g__SJA‐28 | ||||
| 44.815 | 22.197 | 3.679 | 1.61E−09 | 1.29E−07 | p__Actinobacteriota | f__Micrococcaceae | g__Rothia | ||||
| 15.691 | −22.020 | 3.633 | 1.35E−09 | 1.22E−07 | p__Bacteroidota | f__Rikenellaceae | g__Alistipes | ||||
| 12.107 | 20.379 | 3.682 | 3.12E−08 | 1.25E−06 | p__Halobacterota | f__Methanoregulaceae | g__Methanolinea | ||||
| 12.491 | 20.419 | 3.682 | 2.93E−08 | 1.24E−06 | p__Proteobacteria | f__Rhizobiaceae | g__Lentilitoribacter | ||||
| 143.561 | 23.346 | 3.599 | 8.74E−11 | 4.75E−08 | p__Acetothermia | f__Acetothermiia | g__Acetothermiia | ||||
| 23.229 | −22.563 | 3.632 | 5.24E−10 | 9.43E−08 | p__Bacteroidota | f__Muribaculaceae | g__Muribaculaceae | ||||
| 30.963 | 21.683 | 3.680 | 3.80E−09 | 2.11E−07 | p__Proteobacteria | f__Pasteurellaceae | g__Rodentibacter | ||||
| 30.963 | 21.683 | 3.680 | 3.80E−09 | 2.11E−07 | p__Actinobacteriota | f__Demequinaceae | g__Demequina | ||||
| 9.854 | 20.094 | 3.683 | 4.87E−08 | 1.67E−06 | p__Proteobacteria | f__Rhodobacteraceae | g__Tropicibacter | ||||
| 51.876 | 22.402 | 3.679 | 1.14E−09 | 1.22E−07 | p__Chloroflexi | f__FS117‐23B‐02 | g__FS117‐23B‐02 | ||||
| 60.568 | 22.616 | 3.679 | 7.87E−10 | 1.13E−07 | p__Chloroflexi | f__FW22 | g__FW22 | ||||
| 28.247 | 21.555 | 3.680 | 4.70E−09 | 2.42E−07 | p__Chloroflexi | f__uncultured | g__uncultured | ||||
| 11.692 | 20.331 | 3.682 | 3.36E−08 | 1.27E−06 | p__Proteobacteria | f__Rhodobacteraceae | g__Pseudophaeobacter | ||||
| 39.111 | 22.008 | 3.679 | 2.21E−09 | 1.57E−07 | p__Chloroflexi | f__AB‐539‐J10 | g__AB‐539‐J10 | ||||
| 49.975 | 22.348 | 3.679 | 1.24E−9 | 1.22E‐07 | p__Chloroflexi | f__Dehalococcoidia | g__Dehalococcoidia | ||||
| 18.741 | 20.981 | 3.681 | 1.20E−08 | 5.38E−07 | p__Aenigmarchaeota | f__Aenigmarchaeales | NA | ||||
| 12.303 | 7.012 | 2.206 | 1.48E−03 | 4.11E−02 | p__Acidobacteriota | f__Acanthopleuribacteraceae | g__Acanthopleuribacter | ||||
| 37.753 | 21.959 | 3.679 | 2.40E−09 | 1.57E−07 | p__Chloroflexi | f__GIF9 | g__GIF9 | ||||
| 10.548 | 20.188 | 3.683 | 4.21E−08 | 1.51E−06 | p__Proteobacteria | f__Caulobacteraceae | g__Brevundimonas | ||||
| 23.630 | 21.306 | 3.680 | 7.06E−09 | 3.39E−07 | p__Chloroflexi | f__Sh765B‐AG‐111 | g__Sh765B‐AG‐111 | ||||
| Plastic sediment versus can sediment | |||||||||||
| Reference: Can sediment | |||||||||||
| Test: DESeq. 2 | |||||||||||
| 420.185 | 8.385 | 2.380 | 4.26E−04 | 1.21E−02 | p__Actinobacteriota | f__Propionibacteriaceae | g__Cutibacterium | ||||
| 617.280 | 10.016 | 2.614 | 1.27E−04 | 3.81E−03 | p__Bacteroidota | f__Chlorobiaceae | g__Prosthecochloris | ||||
| 125.086 | −24.614 | 3.186 | 1.11E−14 | 5.99E−12 | p__Firmicutes | f__Hungateiclostridiaceae | g__Fastidiosipila | ||||
| 179.331 | −23.932 | 3.214 | 9.69E−14 | 1.74E−11 | p__Firmicutes | f__Veillonellaceae | g__Megasphaera | ||||
| 112.919 | 24.056 | 3.384 | 1.17E−12 | 1.26E−10 | p__Proteobacteria | f__Sedimenticolaceae | g__Candidatus_Thiodiazotropha | ||||
| 56.910 | −23.544 | 3.157 | 8.78E−14 | 1.74E−11 | p__Firmicutes | f__Erysipelotrichaceae | g__Dubosiella | ||||
| 48.636 | −23.324 | 3.199 | 3.10E−13 | 4.18E−11 | p__Firmicutes | f__Hungateiclostridiaceae | g__Mageibacillus | ||||
| 9.616 | −21.118 | 3.387 | 4.51E−10 | 1.74E−08 | p__Firmicutes | f__Veillonellaceae | g__Veillonella | ||||
| 56.335 | 23.106 | 3.384 | 8.64E−12 | 7.78E−10 | p__Caldatribacteriota | f__JS1 | g__JS1 | ||||
| 19.112 | −22.040 | 3.385 | 7.47E−11 | 3.36E−09 | p__Firmicutes | f__Lachnospiraceae | g__[Ruminococcus]_torques_group | ||||
| 41.129 | 22.668 | 3.385 | 2.12E−11 | 1.27E−09 | p__Actinobacteriota | f__Micrococcaceae | g__Rothia | ||||
| 10.782 | 20.807 | 3.388 | 8.16E−10 | 2.75E−08 | p__Halobacterota | f__Methanoregulaceae | g__Methanolinea | ||||
| 8.932 | −21.035 | 3.387 | 5.29E−10 | 1.90E−08 | p__Proteobacteria | f__Magnetospiraceae | g__Magnetovibrio | ||||
| 35.895 | 22.475 | 3.385 | 3.13E−11 | 1.69E−09 | p__Chloroflexi | f__AB‐539‐J10 | g__AB‐539‐J10 | ||||
| 45.865 | 22.821 | 3.384 | 1.55E−11 | 1.05E−09 | p__Chloroflexi | f__Dehalococcoidia | g__Dehalococcoidia | ||||
| 22.443 | 21.818 | 3.222 | 1.28E−11 | 9.89E−10 | p__Asgardarchaeota | f__Lokiarchaeia | g__Lokiarchaeia | ||||
| 34.648 | 22.428 | 3.385 | 3.44E−11 | 1.69E−09 | p__Chloroflexi | f__GIF9 | g__GIF9 | ||||
| 11.110 | −6.714 | 1.654 | 4.90E−05 | 156E−03 | p__Verrucomicrobiota | f__Rubritaleaceae | g__Rubritalea | ||||
| 21.686 | 21.623 | 3.386 | 1.69E−10 | 7.03E−09 | p__Chloroflexi | f__Sh765B‐AG‐111 | g__Sh765B‐AG‐111 | ||||
Note: Only significant results (FDR‐adjusted p < 0.05) are shown. Positive log2FoldChange values mean that the taxon is underrepresented in the reference, while negative values mean that the taxon is overrepresented in the reference.
Table A2.
List of the strains identified in the collection, with the closest type strain, accession number, ID percentage, and the GenBank accession number for the 16S or 18S rRNA gene sequences obtained in this study
| Sample | Closest type strain | Accession number | ID % | GenBank accession number |
|---|---|---|---|---|
| P1.1A | Marinobacter similis | CP007151 | 99.58 | MZ437807 |
| P1.2A | Aurantimonas coralicida | ATXK01000033 | 100 | MZ437808 |
| P1.3A | Sulfitobacter sabulilitoris | MK726099 | 98.71 | MZ437809 |
| P1.4A | Bacillus beringensis | FJ889576 | 99.1 | MZ437810 |
| P2.1A | Bacillus endophyticus | AF295302 | 99.67 | MZ437811 |
| P2.2A | Bacillus oryzaecorticis | KF548480 | 98.67 | MZ437812 |
| P2.3A | Bacillus altitudinis | ASJC01000029 | 99.58 | MZ437813 |
| P2.4A | Pontibacillus salipaludis | LN872943 | 99.73 | MZ437814 |
| P2.5A | Bacillus firmus | BCUY01000205 | 98.6 | MZ437815 |
| P3.1A | Erythrobacter arachoides | KU302715 | 98.36 | MZ437816 |
| P3.2A | Bacillus Altitudinis | ASJC01000029 | 99.86 | MZ437817 |
| P3.3A | Planomicrobium alkanoclasticum | AF029364 | 99.58 | MZ437818 |
| P3.4A | Bacillus drentensis | AJ542506 | 99.06 | MZ437819 |
| P3.5A | Erythrobacter longus | JMIW01000006 | 98.61 | MZ437820 |
| P4.1A | Bacillus wiedmannii | LOBC01000053 | 99.6 | MZ437821 |
| P4.2A | Jiella aquimaris | KJ620984 | 100 | MZ437822 |
| P4.3A | Aeromicrobium alkaliterrae | AY822044 | 97.57 | MZ437823 |
| P4.4A | Aurantimonas coralicida | ATXK01000033 | 99.54 | MZ437824 |
| P4.5A | Erythrobacter insulae | MK991775 | 98.91 | MZ437825 |
| M10.1A | Kocuria palustris | Y16263 | 100 | MZ437826 |
| M10.2A | Gillisia mitskevichiae | jgi.1107713 | 97.57 | MZ994595 |
| M10.3A | Sagittula stellata | AAYA01000003 | 97.81 | MZ437828 |
| M10.4A | Bacillus zhangzhouensis | JOTP01000061 | 99.60 | MZ437829 |
| M10.5A | Gramella sediminilitoris | KU696541 | 98.53 | MZ437830 |
| M11.1A | Planomicrobium alkanoclasticum | AF029364 | 99.85 | MZ437831 |
| M11.2A | Sulfitobacter undariae | KM275624 | 98.42 | MZ437832 |
| M11.3A | Hoeflea halophila | OCPC01000011 | 99.71 | MZ437833 |
| M11.4A | Pontixanthobacter luteolus | AY739662 | 99.61 | MZ437834 |
| M11.5A | Sulfitobacter mediterraneus | JASH01000023 | 98.48 | MZ437835 |
| M12.1A | Pseudomonas juntendi | MK680061 | 99.61 | MZ437836 |
| M12.3A | Bacillus simplex | BCVO01000086 | 100 | MZ437837 |
| M12.4A | Hoeflea halophila | OCPC01000011 | 99.67 | MZ437838 |
| M12.5A | Halobacillus locisalis | AY190534 | 99.83 | MZ437839 |
| M13.1A | Bacillus toyonensis | CP006863 | 99.87 | MZ437840 |
| M13.3A | Sulfitobacter undariae | KM275624 | 97.99 | MZ437841 |
| M13.4A | Bhargavaea beijingensis | EF371374 | 99.33 | MZ437842 |
| M13.5A | Pseudomonas juntendi | MK680061 | 99.87 | MZ437843 |
| CS5.1A | Sediminicola arcticus | KM576847 | 97.43 | MZ437844 |
| CS5.2A | Sediminicola arcticus | KM576847 | 98.29 | MZ437845 |
| CS5.3A | Planomicrobium alkanoclasticum | AF029364 | 99.36 | MZ437846 |
| CS5.4A | Pseudidiomarina aquimaris | PIPT01000016 | 98.98 | MZ437847 |
| CS5.5A | Erythrobacter citreus | AF118020 | 98.49 | MZ437848 |
| CS6.1A | Hoeflea halophila | OCPC01000011 | 98.98 | MZ437849 |
| CS6.2A | Aurantimonas coralicida | ATXK01000033 | 100.00 | MZ437850 |
| CS6.4A | Bacillus aciditolerans | MG589508 | 99.49 | MZ437851 |
| CS6.5A | Gramella salexigens | CP018153 | 98.67 | MZ437852 |
| P12.1A | Altererythrobacter luteolus | AY739662 | 99.54 | MZ437853 |
| P12.3A | Bacillus aciditolerans | MG589508 | 99.45 | MZ437854 |
| P12.4A | Bacillus zhangzhouensis | JOTP01000061 | 99.84 | MZ437855 |
| P12.5A | Bacillus megaterium | JJMH01000057 | 99.78 | MZ437856 |
| P12.6A | Bacillus filamentosus | KF265351 | 99.54 | MZ437857 |
| P1.1X | Bacillus algicola | NR_117184.1 | 100 | MZ604909 |
| P1.2X | Erythrobacter arachoides | KU302715 | 98.61 | MZ437858 |
| P1.4X | Terribacillus saccharophilus | AB243845 | 99.59 | MZ437859 |
| P1.5X | Sphingorhabdus flavimaris | AY554010 | 99.58 | MZ437860 |
| P1.7X | Halobacillus locisalis | AY190534 | 100 | MZ437861 |
| P1.8X | Sulfitobacter mediterraneus | JASH01000023 | 97.99 | MZ437862 |
| P2.1X | Bacillus oceanisediminis | GQ292772 | 99.14 | MZ437863 |
| P2.2X | Fictibacillus halophilus | KP265300 | 99.7 | MZ437864 |
| P2.3X | Bacillus firmus | BCUY01000205 | 98.61 | MZ437865 |
| P2.4X | Cytobacillus firmus | BCUY01000205 | 98.95 | MZ437866 |
| P2.5X | Alkalihalobacillus algicola | AY228462 | 99.13 | MZ437867 |
| P2.6X | Brevibacterium frigoritolerans | AM747813 | 100 | MZ437868 |
| P2.7X | Solibacillus isronensis | AMCK01000046 | 99.86 | MZ437869 |
| P3.2X | Bacillus algicola | AY228462 | 98.19 | MZ437870 |
| P3.4X | Pseudomonas juntendi | MK680061 | 99.82 | MZ437871 |
| P3.5X | Bacillus altitudinis | ASJC01000029 | 99.75 | MZ437872 |
| P3.7X | Actibacter haliotis | KC193210 | 98.83 | MZ437873 |
| P3.8X | Sphingorhabdus flavimaris | AY554010 | 99.31 | MZ437874 |
| P4.1X | Solibacillus isronensis | AMCK01000046 | 100 | MZ437875 |
| P4.3X | Meridianimaribacter flavus | jgi.1076156 | 99.71 | MZ437876 |
| P4.5X | Rhizobium marinum | JMQK01000051 | 97.87 | MZ437877 |
| P4.7X | Microbacterium imperiale | X77442 | 100.00 | MZ437878 |
| P4.8X | Rhizobium marinum | JMQK01000051 | 100.00 | MZ437879 |
| P4.9X | Nocardioides nitrophenolicus | AF005024 | 98.72 | MZ437880 |
| P4.10X | Maritalea myrionectae | AUHV01000006 | 98.25 | MZ994596 |
| M10.1X | Bacillus altitudinis | ASJC01000029 | 99.61 | MZ437882 |
| M10.2X | Gramella sediminilitoris | KU696541 | 98.68 | MZ437883 |
| M10.4X | Planomicrobium alkanoclasticum | AF029364 | 100 | MZ437884 |
| M10.5X | Piscibacillus halophilus | FM864227 | 98.78 | MZ437885 |
| M10.6X | Planomicrobium alkanoclasticum | AF029364 | 99.61 | MZ437886 |
| M10.7X | Sulfitobacter sabulilitois | MK726099 | 98.14 | MZ437887 |
| M10.8X | Gillisia hiemivivida | AY694006 | 97.45 | MZ437888 |
| M10.9X | Sagittula stellata | AAYA01000003 | 97.81 | MW785249 |
| M11.1X | Bacillus endophyticus | AF295302 | 100 | MZ437890 |
| M11.2X | Ruegeria atlantica | CYPU01000053 | 99.33 | MZ437891 |
| M11.3X | Idiomarina piscisalsi | AB619724 | 99.36 | MZ437892 |
| M11.4X | Bacillus pseudomycoides | ACMX01000133 | 100.00 | MZ437893 |
| M11.5X | Bacillus altitudinis | ASJC01000029 | 99.47 | MZ437894 |
| M11.6X | Erythrobacter arachoides | KU302715 | 99.16 | MZ437895 |
| M11.7X | Bacillus horikoshii | X76443 | 98.74 | MZ437896 |
| M11.8X | Sphingorhabdus flavimaris | AY554010 | 99.58 | MZ437897 |
| M11.9X | Altererythrobacter luteolus | AY739662 | 99.57 | MZ437898 |
| M11.10X | Planomicrobium alkanoclasticum | AF029364 | 99.6 | MZ437899 |
| M11.11X | Aquamicrobium lusatiense | AJ132378 | 90.26 | MZ437900 |
| M11.12X | Bacillus altitudinis | ASJC01000029 | 100.00 | MZ437901 |
| M12.1X | Erythrobacter arachoides | KU302715 | 98.64 | MZ437902 |
| M12.2X | Aquimarina intermedia | jgi.1107908 | 99.71 | MZ437903 |
| M12.3X | Lutimonas vermicola | EF108218 | 99.28 | MZ437904 |
| M12.4X | Parasphingorhabdus flavimaris | AY554010 | 99.71 | MZ437905 |
| M13.1X | Bacillus altitudinis | ASJC01000029 | 100.00 | MZ437906 |
| M13.2X | Halobacillus litoralis | X94558 | 99.86 | MZ437907 |
| M13.3X | Microbulbifer echini | KJ789957 | 99.76 | MZ437908 |
| M13.4X | Alkalihalobacillus algicola | AY228462 | 99.85 | MZ437909 |
| M13.7X | Kocuria salina | LT674162 | 99.31 | MZ437910 |
| CS4.1X | Yoonia litorea | jgi.1096519 | 99.74 | MZ437911 |
| CS4.3X | Vibrio comitans | DQ922915 | 99.45 | MZ437912 |
| CS4.4X | Mesobacillus subterraneus | RSFW01000004 | 99.52 | MZ437913 |
| CS4.5X | Agromyces indicus | HM036655 | 98.81 | MZ437914 |
| CS5.2X | Bacillus horikoshii | X76443 | 99.07 | MZ437915 |
| CS5.3X | Planomicrobium alkanoclasticum | AF029364 | 99.4 | MZ437916 |
| CS5.4X | Micrococcus luteus | CP001628 | 99.82 | MZ437917 |
| CS6.1X | Sphingorhabdus flavimaris | AY554010 | 99.32 | MZ437918 |
| CS6.2X | Aurantimonas coralicida | ATXK01000033 | 100.00 | MZ437919 |
| CS6.3X | Gramella salexigens | CP018153 | 98.87 | MZ437920 |
| CS6.4X | Microbulbifer echini | KJ789957 | 99.2 | MZ437921 |
| CS6.5X | Sphingorhabdus flavimaris | AY554010 | 99.31 | MZ437922 |
| CS6.6X | Gillisia mitskevichiae | jgi.1107713 | 99.74 | MZ437923 |
| CS6.7X | Erythrobacter citreus | AF118020 | 99.04 | MZ437924 |
| P12.4X | Microbacterium aerolatum | BJUW01000027 | 99.76 | MZ604910 |
| P12.5X | Rhodotorula mucilaginosa | KU167832.1 | 100.00 | MZ604692 |
| P12.6X | Yoonia rosea | jgi.1085777 | 99.06 | MZ437925 |
| P12.7X | Ruegeria arenilitoris | FXYG01000008 | 99.6 | MZ437926 |
| P12.8X | Citricoccus alkalitolerans | AY376164 | 99.21 | MZ437927 |
| P12.9X | Bacillus maritimus | KP317497 | 98.04 | MZ437928 |
| P12.10X | Tessaracoccus rhinocerotis | KT215777 | 97.89 | MZ437929 |
| P12.11X | Bacillus beringensis | FJ889576 | 98.98 | MZ437930 |
| P12.12X | Bacillus altitudinis | ASJC01000029 | 99.58 | MZ437931 |
| P12.13X | Bacillus firmus | BCUY01000205 | 97.85 | MZ437932 |
| P12.14X | Bacillus aryabhattai | EF114313 | 100.00 | MZ437933 |
| P12.15X | Ruegeria arenilitoris | FXYG01000008 | 100.00 | MZ437934 |
| P1.1D | Pseudoalteromonas piscicida | CP011925 | 100.00 | MZ437935 |
| P1.2D | Vibrio hyugaensis | LC004912 | 99.86 | MZ437936 |
| P1.5D | Vibrio azureus | LC004912 | 99.59 | MZ437937 |
| P6.1D | Vibrio alginolyticus | CP006718 | 99.34 | MZ437938 |
| P6.2D | Vibrio hyugaensis | LC004912 | 99.55 | MZ437939 |
| M10.3D | Epibacterium mobile | jgi.1108012 | 100 | MZ437940 |
| M10.5D | Vibrio azureus | BATL01000140 | 100.00 | MZ437941 |
| M10.6D | Vibrio shilonii | ABCH01000080 | 99.03 | MZ437942 |
| M10.7D | Vibrio hyugaensis | LC004912 | 99.86 | MZ437943 |
| M10.10D | Vibrio hyugaensis | LC004912 | 99.85 | MZ437944 |
| M10.11D | Tenacibaculum mesophilum | jgi.1107970 | 99.87 | MZ437945 |
Note: The identification code of the strains corresponds to the sediments from which it was isolated (CS, control sediments; M, can inside‐sediments; P, plastic inside‐sediments; and a number).
Table A3.
List of selected isolates that showed enhanced growth in PET‐containing medium, with the closest type strain, GenBank accession number for the 16S and 18 rRNA gene sequences, and results obtained in the quantitative assay
| Isolate | Closest Type Strain | Quantitative assay (CFU in minimal marine medium supplemented with PET/CFU in minimal marine medium) | GenBank accession number |
|---|---|---|---|
| P1.1X | Bacillus algicola | 2.1 | MZ604909 |
| P4.3X | Meridianimaribacter flavus | 0.8 | MZ437876 |
| P4.7X | Microbacterium imperiale | 0.5 | MZ437878 |
| M13.5A | Pseudomonas juntendi | 1.4 | MZ437843 |
| M13.7X | Kocuria rosea | 0.9 | MZ437910 |
| M11.3X | Idiomarina piscisalsi | 4.7 | MZ437892 |
| P4.10X | Maritalea mobilis | 0.8 | MZ437881 |
| P12.10X | Tessaracoccus rhinocerotis | * | MZ437929 |
| CS5.4X | Micrococcus luteus | 20.8 | MZ437917 |
| CS6.2X | Aurantimonas coralicida | 1.2 | MZ437919 |
| M12.2X | Aquimarina intermedia | 3.4 | MZ437903 |
| P12.4X | Microbacterium aerolatum | 2.4 | MZ604910 |
| P12.5X | Rhodotorula evergladensis | 1.5 | MZ604692 |
| P12.8X | Citricoccus alkalitolerans | 3.6 | MZ437927 |
| M10.4A | Bacillus zhangzhouensis | 0.9 | MZ437829 |
| M12.3A | Bacillus simplex | 0.5 | MZ437837 |
Abbreviations: CFU, colony‐forming units; PET, polyethylene terephthalate.
*Tessaracoccus rhinocerotis yielded an uncountable number of colonies; therefore, its differential growth was not measured.
Vidal‐Verdú, À. , Latorre‐Pérez, A. , Molina‐Menor, E. , Baixeras, J. , Peretó, J. , & Porcar, M. (2021). Living in a bottle: Bacteria from sediment‐associated Mediterranean waste and potential growth on polyethylene terephthalate. MicrobiologyOpen, 11, e1259. 10.1002/mbo3.1259
DATA AVAILABILITY STATEMENT
The datasets generated for this study can be found in online repositories. Raw reads are available in the NCBI repository (BioProject accession PRJNA704512: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA704512). 16S and 18S rRNA gene sequences are available in GenBank under accession numbers MZ437807‐MZ437945, MZ604909‐MZ604910, MZ604692, MZ994595‐MZ994596, and MW785249. The code and the results of the bioinformatics and statistical analyses (including taxonomy tables of absolute and relative abundances and the BIOM table) have been uploaded to GitHub (https://github.com/adlape95/Living-in-a-bottle).
REFERENCES
- Agostini, L. , Moreira, J. , Bendia, A. G. , Kmit, M. , Waters, L. G. , Santana, M. , Sumida, P. , Turra, A. , & Pellizari, V. H. (2021). Deep‐sea plastisphere: Long‐term colonization by plastic‐associated bacterial and archaeal communities in the Southwest Atlantic Ocean. Science of the Total Environment, 793, 148335. 10.1016/j.scitotenv.2021.148335 [DOI] [PubMed] [Google Scholar]
- Amaral‐Zettler, L. A. , Zettler, E. R. , & Mincer, T. J. (2020). Ecology of the plastisphere. Nature Reviews Microbiology, 18, 139–151. 10.1038/s41579-019-0308-0 [DOI] [PubMed] [Google Scholar]
- Arthur, C. , Baker, J. , & Bamford, H. (2009). Proceedings of the International Research Workshop on the Occurrence, Effects and Fate of Microplastic Marine Debris. Sept 9‐11, 2008. NOAA Technical Memorandum NOS‐OR&R‐30. National Oceanic and Atmospheric Administration. [Google Scholar]
- Auta, H. S. , Emenike, C. U. , & Fauziah, S. H. (2017). Screening of Bacillus strains isolated from mangrove ecosystems in Peninsular Malaysia for microplastic degradation. Environmental Pollution, 231, 1552–1559. 10.1016/j.envpol.2017.09.043 [DOI] [PubMed] [Google Scholar]
- Barnes, D. K. A. , Galgani, F. , Thompson, R. C. , & Barlaz, M. (2009). Accumulation and fragmentation of plastic debris in global environments. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 364(1526), 1985–1998. 10.1098/rstb.2008.0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeman, J. T. , Morales‐García, A. L. , Mukherjee, J. , Gori, K. , Hayward, A. S. , Lant, N. J. , & Geoghegan, M. (2019). Extracellular DNA provides structural integrity to a Micrococcus luteus biofilm. Langmuir, 35(19), 6468–6475. 10.1021/acs.langmuir.9b00297 [DOI] [PubMed] [Google Scholar]
- Bokulich, N. A. , Kaehler, B. D. , Rideout, J. R. , Dillon, M. , Bolyen, E. , Knight, R. , Huttley, G. A. , & Caporaso, J. G. (2018). Optimizing taxonomic classification of marker‐gene amplicon sequences with QIIME 2's q2‐feature‐classifier plugin. Microbiome, 6(90), 90. (2018) 10.1186/s40168-018-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen, E. , Rideout, J. R. , Dillon, M. R. , Bokulich, N. A. , Abnet, C. C. , Al‐Ghalith, G. A. , Alexander, H. , Alm, E. J. , Arumugam, M. , Asnicar, F. , Bai, Y. , Bisanz, J. E. , Bittinger, K. , Brejnrod, A. , Brislawn, C. J. , Brown, C. T. , Callahan, B. J. , Caraballo‐Rodríguez, A. M. , Chase, J. , … Caporaso, J. G. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology, 37(8), 852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester, H. , Hollman, P. C. H. , & Peters, R. J. B. (2015). Potential health impact of environmentally released micro‐ and nanoplastics in the human food production chain: Experiences from nanotoxicology. Environmental Science and Technology, 49, 8932–8947. 10.1021/acs.est.5b01090 [DOI] [PubMed] [Google Scholar]
- Bryant, J. A. , Clemente, T. M. , Viviani, D. A. , Fong, A. A. , Thomas, K. A. , Kemp, P. , Karl, D. M. , White, A. E. , & DeLong, E. F. (2016). Diversity and activity of communities inhabiting plastic debris in the North Pacific gyre. mSystems, 1(3):e00024‐16. 10.1128/msystems.00024-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan, B. , McMurdie, P. , Rosen, M. , Han, A. , Johnson, A. , & Holmes, S. (2016). DADA2: High‐resolution sample inference from Illumina amplicon data. Nature Methods, 13, 581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr, C. M. , Clarke, D. J. , & Dobson, A. D. W. (2020). Microbial polyethylene terephthalate hydrolases: Current and future perspectives. Frontiers in Microbiology, 11, 2825. 10.3389/fmicb.2020.571265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun, J. , Oren, A. , Ventosa, A. , Christensen, H. , Arahal, D. R. , da Costa, M. S. , Rooney, A. P. , Yi, H. , Xu, X. W. , de Meyer, S. , & Trujillo, M. E. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. International Journal of Systematic and Evolutionary Microbiology, 68(1), 461–466. 10.1099/ijsem.0.002516 [DOI] [PubMed] [Google Scholar]
- Danso, D. , Schmeisser, C. , Chow, J. , Zimmermann, W. , Wei, R. , Leggewie, C. , Li, X. , Hazen, T. , & Streit, W. R. (2018). New insights into the function and global distribution of polyethylene terephthalate (PET)‐degrading bacteria and enzymes in marine and terrestrial metagenomes. Applied and Environmental Microbiology, 84(8):e02773‐17. 10.1128/AEM.02773-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacuvellerie, A. , Cyriaque, V. , Gobert, S. , Benali, S. , & Wattiez, R. (2019). The plastisphere in marine ecosystem hosts potential specific microbial degraders including Alcanivorax borkumensis as a key player for the low‐density polyethylene degradation. Journal of Hazardous Materials, 380, 120899. 10.1016/j.jhazmat.2019.120899 [DOI] [PubMed] [Google Scholar]
- Eriksen, M. , Lebreton, L. C. , Carson, H. S. , Thiel, M. , Moore, C. J. , Borerro, J. C. , Galgani, F. , Ryan, P. G. , & Reisser, J. (2014). Plastic pollution in the world's oceans: More than 5 trillion plastic pieces weighing over 250,000 tons afloat at sea. PLOS One, 9(12), e111913. 10.1371/journal.pone.0111913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, L. , Li, X. , Song, P. , Du, G. , & Chen, J. (2011). Surface interactions and fouling properties of Micrococcus luteus with microfiltration membranes. Applied Biochemistry and Biotechnology, 165(5–6), 1235–1244. 10.1007/s12010-011-9341-9 [DOI] [PubMed] [Google Scholar]
- Gao, R. , & Sun, C. (2021). A marine bacterial community capable of degrading poly(ethylene terephthalate) and polyethylene. Journal of Hazardous Materials, 416, 125928. 10.1016/j.jhazmat.2021.125928 [DOI] [PubMed] [Google Scholar]
- Gartshore, J. , Cooper, D. G. , & Nicell, J. A. (2003). Biodegradation of plasticizers by Rhodotorula rubra . Environmental Toxicology and Chemistry, 22(6), 1244–1251. 10.1002/etc.5620220609 [DOI] [PubMed] [Google Scholar]
- GESAMP (2016). Sources, fate and effects of microplastics in the marine environment: part two of a global assessment, In Kershaw, P. J. , & Rochman, C. M. (Eds), Rep. Stud. GESAMP No. 93, (p. 220). IMO/FAO/UNESCO‐IOC/UNIDO/WMO/IAEA/UN/UNEP/UNDP Joint Group of Experts on the Scientific Aspects of Marine Environmental Protection. [Google Scholar]
- Gewert, B. , Plassmann, M. M. , & Macleod, M. (2015). Pathways for degradation of plastic polymers floating in the marine environment. Environmental Science: Processes & Impacts, 17, 1513–1521. 10.1039/c5em00207a [DOI] [PubMed] [Google Scholar]
- Giacomucci, L. , Raddadi, N. , Soccio, M. , Lotti, N. , & Fava, F. (2019). Polyvinyl chloride biodegradation by Pseudomonas citronellolis and Bacillus flexus. New Biotechnology, 52, 35–41. 10.1016/j.nbt.2019.04.005 [DOI] [PubMed] [Google Scholar]
- Hahladakis, J. N. , Velis, C. A. , Weber, R. , Iacovidou, E. , & Purnell, P. (2018). An overview of chemical additives present in plastics: Migration, release, fate and environmental impact during their use, disposal and recycling. Journal of Hazardous Materials, 344, 179–199. 10.1016/j.jhazmat.2017.10.014 [DOI] [PubMed] [Google Scholar]
- Hoshino, T. , Doi, H. , Uramoto, G. I. , Wörmer, L. , Adhikari, R. R. , Xiao, N. , Morono, Y. , D'Hondt, S. , Hinrichs, K. U. , & Inagaki, F. (2020). Global diversity of microbial communities in marine sediment. Proceedings of the National Academy of Sciences of the United States of America, 117(44), 27587–27597. 10.1073/pnas.1919139117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iñiguez, M. E. , Conesa, J. A. , & Fullana, A. (2017). Microplastics in Spanish table salt. Scientific Reports, 7(1), 8620. 10.1038/s41598-017-09128-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquin, J. , Cheng, J. , Odobel, C. , Pandin, C. , Conan, P. , Pujo‐Pay, M. , Barbe, V. , Meistertzheim, A. L. , & Ghiglione, J. F. (2019). Microbial ecotoxicology of marine plastic debris: A review on colonization and biodegradation by the “plastisphere. Frontiers in Microbiology, 10(Apr), 865. 10.3389/fmicb.2019.00865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambeck, J. R. , Geyer, R. , Wilcox, C. , Siegler, T. R. , Perryman, M. , Andrady, A. , Narayan, R. , & Law, K. L. (2015). Plastic waste inputs from land into the ocean. Science, 347(6223), 768–771. 10.1126/science.1260352 [DOI] [PubMed] [Google Scholar]
- Karnovsky, M. J. (1965). A formaldehyde‐glutaraldehyde fixative of high osmolality for use in electron microscopy. Journal of Cell Biology, 3(18), 200. [Google Scholar]
- Kawai, F. , Kawabata, T. , & Oda, M. (2020). Current state and perspectives related to the polyethylene terephthalate hydrolases available for biorecycling. ACS Sustainable Chemistry and Engineering, 8(24), 8894–8908. 10.1021/acssuschemeng.0c01638 [DOI] [Google Scholar]
- Latorre, A. , Moya, A. , & Ayala, F. J. (1986). Evolution of mitochondrial DNA in Drosophila subobscura . Proceedings of the National Academy of Sciences of the United States of America, 83(22), 8649–8653. 10.1073/pnas.83.22.8649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton, L. C. M. , van der Zwet, J. , Damsteeg, J. W. , Slat, B. , Andrady, A. , & Reisser, J. (2017). River plastic emissions to the world's oceans. Nature Communications, 8, 1–10. 10.1038/ncomms15611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leloup, J. , Fossing, H. , Kohls, K. , Holmkvist, L. , Borowski, C. , & Jørgensen, B. B. (2009). Sulfate‐reducing bacteria in marine sediment (Aarhus Bay, Denmark): Abundance and diversity related to geochemical zonation. Environmental Microbiology, 11(5), 1278–1291. 10.1111/j.1462-2920.2008.01855.x [DOI] [PubMed] [Google Scholar]
- Lobelle, D. , & Cunliffe, M. (2011). Early microbial biofilm formation on marine plastic debris. Marine Pollution Bulletin, 62(1), 197–200. 10.1016/j.marpolbul.2010.10.013 [DOI] [PubMed] [Google Scholar]
- Love, M. I. , Huber, W. , & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biology, 15(12), 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masó, M. , Fortuño, J. M. , de Juan, S. , & Demestre, M. (2016). Microfouling communities from pelagic and benthic marine plastic debris sampled across Mediterranean coastal waters. Scientia Marina, 80(S1), 117–127. 10.3989/scimar.04281.10a [DOI] [Google Scholar]
- McCormick, A. , Hoellein, T. J. , Mason, S. A. , Schluep, J. , & Kelly, J. J. (2014). Microplastic is an abundant and distinct microbial habitat in an urban river. Environmental Science and Technology, 48(20), 11863–11871. 10.1021/es503610r [DOI] [PubMed] [Google Scholar]
- McMurdie, P. J. , & Holmes, S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLOS One, 8(4), e61217. 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montazer, Z. , Habibi Najafi, M. B. , & Levin, D. B. (2018). Microbial degradation of low‐density polyethylene and synthesis of polyhydroxyalkanoate polymers. Canadian Journal of Microbiology, 65(3), 224–234. 10.1139/cjm-2018-0335 [DOI] [PubMed] [Google Scholar]
- Mori, K. , Yamaguchi, K. , & Hanada, S. (2018). Sulfurovum denitrificans sp. Nov., an obligately chemolithoautotrophic sulfur‐oxidizing epsilonproteobacterium isolated from a hydrothermal field. International Journal of Systematic and Evolutionary Microbiology, 68, 2183–2187. 10.1099/ijsem.0.002803 [DOI] [PubMed] [Google Scholar]
- Napper, I. E. , & Thompson, R. C. (2019). Marine plastic pollution: Other than microplastic, Waste: A handbook for managegement (2nd ed.). Academic Press. 10.1016/b978-0-12-815060-3.00022-0 [DOI] [Google Scholar]
- Neves, D. , Sobral, P. , Ferreira, J. L. , & Pereira, T. (2015). Ingestion of microplastics by commercial fish off the Portuguese coast. Marine Pollution Bulletin, 101(1), 119–126. 10.1016/j.marpolbul.2015.11.008 [DOI] [PubMed] [Google Scholar]
- Oberbeckmann, S. , Löder, M. G. J. , & Labrenz, M. (2015). Marine microplastic‐associated biofilms—A review. Environmental Chemistry 12(5), 551–562. 10.1071/EN15069 [DOI] [Google Scholar]
- Oberbeckmann, S. , Osborn, A. M. , & Duhaime, M. B. (2016). Microbes on a bottle: Substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PLOS One, 11(8), 159289. 10.1371/journal.pone.0159289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin, A. , Antler, G. , Røy, H. , Findlay, A. , Beulig, F. , Scholze, C. , Turchyn, A. V. , & Jørgensen, B. B. (2018). The sulfur cycle below the sulfate‐methane transition of marine sediments. Geochimica et Cosmochimica Acta, 239, 74–89. 10.1016/j.gca.2018.07.027 [DOI] [Google Scholar]
- Petro, C. , Starnawski, P. , Schramm, A. , & Kjeldsen, K. U. (2017). Microbial community assembly in marine sediments. Aquatic Microbial Ecology, 79(3), 177–195. 10.3354/ame01826 [DOI] [Google Scholar]
- Pinnell, L. J. , & Turner, J. W. (2019). Shotgun metagenomics reveals the benthic microbial community response to plastic and bioplastic in a coastal marine environment. Frontiers in Microbiology, 10, 1252. 10.3389/fmicb.2019.01252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- PlasticsEurope . (2020). Plastics—The facts 2020. Retrieved October 29, 2021, from https://plasticseurope.org/knowledge-hub/plastics-the-facts-2020
- Quast, C. , Pruesse, E. , Yilmaz, P. , Gerken, J. , Schweer, T. , Yarza, P. , Peplies, J. , & Glöckner, F. O. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web‐based tools. Nucleic Acids Research, 41, D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisser, J. , Shaw, J. , Hallegraeff, G. , Proietti, M. , Barnes, D. K. , Thums, M. , Wilcox, C. , Hardesty, B. D. , & Pattiaratchi, C. (2014). Millimeter‐sized marine plastics: A new pelagic habitat for microorganisms and invertebrates. PLOS One, 9(6), e100289. 10.1371/journal.pone.0100289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribitsch, D. , Heumann, S. , Trotscha, E. , Herrero Acero, E. , Greimel, K. , Leber, R. , Birner‐Gruenberger, R. , Deller, S. , Eiteljoerg, I. , Remler, P. , Weber, T. , Siegert, P. , Maurer, K. H. , Donelli, I. , Freddi, G. , Schwab, H. , & Guebitz, G. M. (2011). Hydrolysis of polyethyleneterephthalate by p‐nitrobenzylesterase from Bacillus subtilis . Biotechnology Progress, 27, 951–960. 10.1002/btpr.610 [DOI] [PubMed] [Google Scholar]
- Ritchie, H. , & Roser, M. (2018). Plastic pollution—Our world in data. Retrieved February 8, 2021, from https://ourworldindata.org/plastic-pollution
- Roager, L. , & Sonnenschein, E. C. (2019). Bacterial candidates for colonization and degradation of marine plastic debris. Environmental Science and Technology, 53, 11636–11643. 10.1021/acs.est.9b02212 [DOI] [PubMed] [Google Scholar]
- Rochman, C. M. , Hoh, E. , Kurobe, T. , & Teh, S. J. (2013). Ingested plastic transfers hazardous chemicals to fish and induces hepatic stress. Scientific Reports, 3(1), 1–7. 10.1038/srep03263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setälä, O. , Fleming‐Lehtinen, V. , & Lehtiniemi, M. (2014). Ingestion and transfer of microplastics in the planktonic food web. Environmental Pollution, 185, 77–83. 10.1016/j.envpol.2013.10.013 [DOI] [PubMed] [Google Scholar]
- Sinha, V. , Patel, M. R. , & Patel, J. V. (2010). PET waste management by chemical recycling: A review. Journal of Polymers and the Environment, 18, 8–25. 10.1007/s10924-008-0106-7 [DOI] [Google Scholar]
- Sivasankari, S. , & Vinotha, T. (2014). In vitro degradation of plastics (plastic cup) using Micrococcus luteus and Masoniella Sp. Scholars Academic Journal of Biosciences, 2(2), 85–89. [Google Scholar]
- Sole, R. , Fontich, E. , Vidiella, B. , Duran‐Nebreda, S. , Montanez, R. , Pinero, J. , & Valverde, S. (2017). The paradox of constant oceanic plastic debris: Evidence For evolved microbial biodegradation? BioRxiv, 457, e135582‐70. 10.1101/135582 [DOI] [Google Scholar]
- Sun, Q. L. , Zhang, J. , Wang, M. X. , Cao, L. , Du, Z. F. , Sun, Y. Y. , Liu, S. Q. , Li, C. L. , & Sun, L. (2020). High‐throughput sequencing reveals a potentially novel Sulfurovum species dominating the microbial communities of the seawater‐sediment interface of a deep‐sea cold seep in South China Sea. Microorganisms, 8(5), 687. 10.3390/microorganisms8050687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuten, E. L. , Saquing, J. M. , Knappe, D. R. U. , Barlaz, M. A. , Jonsson, S. , Björn, A. , Rowland, S. J. , Thompson, R. C. , Galloway, T. S. , Yamashita, R. , Ochi, D. , Watanuki, Y. , Moore, C. , Viet, P. H. , Tana, T. S. , Prudente, M. , Boonyatumanond, R. , Zakaria, M. P. , Akkhavong, K. , … Takada, H. (2009). Transport and release of chemicals from plastics to the environment and to wildlife. Philosophical Transactions of the Royal Society of London: Series B, Biological Sciences, 364, 2027–2045. 10.1098/rstb.2008.0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cauwenberghe, L. , Claessens, M. , Vandegehuchte, M. B. , & Janssen, C. R. (2015). Microplastics are taken up by mussels (Mytilus edulis) and lugworms (Arenicola marina) living in natural habitats. Environmental Pollution, 199, 10–17. 10.1016/j.envpol.2015.01.008 [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Gao, J. , Zhao, Y. , Dai, H. , Jia, J. , & Zhang, D. (2021). Plastisphere enrich antibiotic resistance genes and potential pathogenic bacteria in sewage with pharmaceuticals. Science of the Total Environment, 768, 144663. 10.1016/j.scitotenv.2020.144663 [DOI] [PubMed] [Google Scholar]
- Watts, A. J. R. , Lewis, C. , Goodhead, R. M. , Beckett, S. J. , Moger, J. , Tyler, C. R. , & Galloway, T. S. (2014). Uptake and retention of microplastics by the shore crab Circinus maenas. Environmental Science and Technology, 48(15), 8823–8830. 10.1021/es501090e [DOI] [PubMed] [Google Scholar]
- Westrich, J. R. , Griffin, D. W. , Westphal, D. L. , & Lipp, E. K. (2018). Vibrio population dynamics in mid‐Atlantic surface waters during Saharan dust events. Frontiers in Marine Science 5(Feb), 12. 10.3389/fmars.2018.00012 [DOI] [Google Scholar]
- Woodall, L. C. , Jungblut, A. D. , Hopkins, K. , Hall, A. , Robinson, L. F. , Gwinnett, C. , & Paterson, G. L. J. (2018). Deep‐sea anthropogenic macrodebris harbours rich and diverse communities of bacteria and archaea. PLOS One, 13(11), e0206220. 10.1371/journal.pone.0206220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worm, B. , Lotze, H. K. , Jubinville, I. , Wilcox, C. , & Jambeck, J. (2017). Plastic as a persistent marine pollutant. Annual Review of Environment and Resources 42, 1–26. 10.1146/annurev-environ [DOI] [Google Scholar]
- Wright, R. J. , Bosch, R. , Langille, M. G. I. , Gibson, M. I. , & Christie‐Oleza, J. A. (2021b). A multi‐OMIC characterisation of biodegradation and microbial community succession within the PET plastisphere. Microbiome, 9(1), 1–22. 10.1186/s40168-021-01054-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, R. J. , Langille, M. G. I. , & Walker, T. R. (2021a). Food or just a free ride? A meta‐analysis reveals the global diversity of the Plastisphere. ISME Journal, 15(3), 789–806. 10.1038/s41396-020-00814-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S. , Hiraga, K. , Takehana, T. , Taniguchi, I. , Yamaji, H. , Maeda, Y. , Toyohara, K. , Miyamoto, K. , Kimura, Y. , & Oda, K. (2016). A bacterium that degrades and assimilates poly(ethylene terephthalate). Science, 351(6278), 1196–1199. 10.1126/science.aad6359 [DOI] [PubMed] [Google Scholar]
- Zettler, E. R. , Mincer, T. J. , & Amaral‐Zettler, L. A. (2013). Life in the “plastisphere”: Microbial communities on plastic marine debris. Environmental Science and Technology, 47(13), 7137–7146. 10.1021/es401288x [DOI] [PubMed] [Google Scholar]
- Zinger, L. , Amaral‐Zettler, L. A. , Fuhrman, J. A. , Horner‐Devine, M. C. , Huse, S. M. , Welch, D. B. , Martiny, J. B. , Sogin, M. , Boetius, A. , & Ramette, A. (2011). Global patterns of bacterial beta‐diversity in seafloor and seawater ecosystems. PLOS One, 6(9), 24570. 10.1371/journal.pone.0024570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrimec, J. , Kokina, M. , Jonasson, S. , Zorrilla, F. , & Zelezniak, A. (2021). Plastic‐degrading potential across the global microbiome correlates with recent pollution trends. mBio, 26 12(5), e0215521. 10.1128/mBio.02155-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study can be found in online repositories. Raw reads are available in the NCBI repository (BioProject accession PRJNA704512: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA704512). 16S and 18S rRNA gene sequences are available in GenBank under accession numbers MZ437807‐MZ437945, MZ604909‐MZ604910, MZ604692, MZ994595‐MZ994596, and MW785249. The code and the results of the bioinformatics and statistical analyses (including taxonomy tables of absolute and relative abundances and the BIOM table) have been uploaded to GitHub (https://github.com/adlape95/Living-in-a-bottle).


