Abstract
Antigen-specific T cells can serve as a response biomarker in non-clinical or clinical immunotherapy studies in autoimmune disease. There are protocols with optimized multimer staining methods to detect peptide (p)MHCII+ CD4+ T cells, and some qualified and validated protocols for pMHCI+ CD8+ T cells. However, no protocol is fully or partially qualified to enumerate and characterize antigen-specific pMHCII+ CD4+ T cells from patient samples. Implementing such an assay requires a desired level of specificity and precision, in terms of assay repeatability and reproducibility. In transgenic type II collagen (CII)-immunized HLA-DR1/DR4 humanized mouse models of collagen-induced arthritis (CIA), CII259-273-specific T cells dominantly expand. Therefore antigen-specific T cells recognizing this epitope presented by rheumatoid arthritis (RA)-associated risk HLA-DR allomorphs are of interest to understand disease progression and responses to immunotherapy in RA patients. Using HLA-DRB1∗04:01 or ∗01:01-collagen type II (CII)259–273 tetramers, we evaluated parameters influencing precision and reproducibility of an optimized flow cytometry–based method for antigen-specific CD4+ T cells and eight specific subpopulations with and without tetramer positivity. We evaluated specificity, precision, and reproducibility for research environments and non-regulated laboratories. The assay has excellent overall precision with %CV<25% for intra-assay repeatability, inter-analyst precision, and inter-assay reproducibility. The precision of the assay correlated negatively with the cell viability after thawing, indicating that post-thaw viability is a critical parameter for reproducibility. This assay is suitable for longitudinal analysis of treatment response and disease activity outcome in RA patients, and adaptable for translational or immunotherapy clinical trial settings.
Keywords: CD4+ T cells, tetramer, immunomonitoring, precision, reproducibility, antigen, rheumatoid arthritis, flow cytometry
Introduction
Despite the differences in the underlying pathological mechanisms, autoimmune diseases are characterized by the ability of autoreactive T or B cells to damage self-tissues [1]. Antigen-specific CD4+ T helper cells recognize peptide-major histocompatibility complex (pMHC) II molecules of antigen-presenting cells (APCs) [2]. They activate and sustain autoimmunity by cross-talk with APCs to activate antigen-specific CD8+ cytotoxic T cells (CTL) and B cells producing autoantibodies [3]. Longitudinal enumeration and phenotypic analysis of antigen-specific CD4+ T cells can elucidate their contribution to disease pathology and evaluate their response to therapeutic intervention, for example, novel antigen-specific immune tolerizing therapies (ASIT) that target antigen-specific CD4+ T cells for the treatment of autoimmune diseases. ASIT show promise in preclinical models [4–7], but there is a need for better biomarker assays reflecting antigen-specific immune function in clinical trials [8–10].
Immune response biomarkers that demonstrate the immunoregulatory effects of ASIT during early-phase clinical trials are a major challenge in the development of novel tolerizing immunotherapies. Such assays could also be used to understand potential immunoregulatory effects of existing disease modifying therapies [11, 12]. Clear and reproducible biomarker outcomes from preclinical animal studies or small dose-ranging mechanistic clinical trials can guide the outcome assessment and the drug development process. Given their central role from the beginning of the autoimmune pathogenesis, antigen-specific CD4+ T cells are an ideal candidate for monitoring the outcome of tolerizing immunotherapies. Soluble pMHCII tetramers are an important flow cytometry–based tool to enumerate and phenotype antigen-specific T cells from peripheral blood (PB) or tissues [13–17]. However, analysis of pMHCII+ CD4+ T cells with flow cytometry is challenging for many reasons, including sensitivity and specificity [18–22]. pMHCII+ CD4+ T cells circulate in low frequency, and autoreactive T-cell receptors (TCR) typically have lower avidity in PB T cells, when compared with T-cell clones or CD8+ viral-specific T cells [18].
While previous studies have optimized methods for pMHCII tetramer staining techniques [13, 14, 16] and qualified or validated pMHCI multimer methods [17, 23, 24], no studies have qualified pMHCII multimer methods to date with systematic precision and reproducibility outcomes for clinical use. Qualification of methods for specificity, accuracy, precision, and reproducibility is important in the drug development process to improve reliability and confidence where data are used to support assessment of the mechanism of action of an emerging therapy. Most qualification studies use samples from mice, healthy donor, or T cell lines to measure optimal detection. While these may achieve qualification criteria, it may then be very difficult to achieve the same in clinical PB samples. Challenges include low cell counts, high flow cytometric background, and compromised quality of thawed cryopreserved peripheral blood mononuclear cell (PBMC) samples [17, 23, 24].
Most clinical trials cryopreserve patient PBMC samples longitudinally to enable analysis at a later stage. Collagen type II (CII) is exclusively expressed in articular cartilage. Immunization of mice, rats, and monkeys with bovine, chick, or human CII leads to the development of collagen-induced arthritis (CIA). CII-autoreactive CD4+ T cells and B cells have been demonstrated in mice and humans. The dominant T-cell epitope (CII259-273) overlaps in H-2q and DR4 or DR1 transgenic mice immunized with bovine or human CII [25, 26], and in CII-immunized HLA-DR1 humanized mouse models of CIA, CII259-273-specific T cells dominantly expand [27]. Therefore antigen-specific T cells recognizing this epitope presented by the rheumatoid arthritis (RA)-associated risk HLA-DR allomorphs, HLA-DRB1∗04:01 and ∗01:01 [28–30], are of interest to understand disease progression and responses to immunotherapy in RA patients. Furthermore, previous studies suggest that self-antigen-specific T cells in RA blood are present at a frequency of 1–100/106 CD4+ T cells and enriched in memory cells relative to healthy control blood [31, 32]. Building on our previously published protocol for HLA-DRB1∗04:01 or ∗01:01-collagen type II (CII)259–273 tetramers (20), with improvements to the flow panel and gating strategy, we evaluated parameters influencing precision and reproducibility of this method as a fit-for-purpose assay to enumerate antigen-specific and total CD4+ T cells as well as eight subpopulations, using a panel of 11 cell surface markers for analysis of thawed cryopreserved PBMC samples from RA patients. For this purpose, we also identified a HLA-DRB1∗01:01-CII259–273-specific TCR and transduced this into a T cell line for use as a standard positive control. We assessed qualification parameters of specificity, repeatability and reproducibility for research environments and non-regulated laboratories as per recent guidelines for flow cytometry assays [33]. This assay provides the basis for development of similar flow cytometric assays with additional pMHCII tetramers and phenotypic markers.
Materials and methods
Samples
Previously frozen (10% dimethyl sulfoxide and 90% fetal bovine serum) PBMC collected from HLA-DRB1∗04:01 or HLA-DRB1∗01:01 RA patients visiting the outpatient clinic of the Princess Alexandra Hospital in Brisbane, Australia were used for this study. The study was approved by the Metro South and UQ Human Research Ethics Committee, and informed consent was obtained from all the patients. Buffy coat samples were obtained from the Red Cross Blood Transfusion Service in Brisbane within 48 h of the blood draw from healthy human volunteers. PBMC were separated from buffy coats and frozen in liquid nitrogen.
pMHCII tetramers
Streptavidin-conjugated phycoerythrin (PE Streptavidin, BD Pharmingen)-labeled HLA-DRB1∗04:01 or HLA-DRB1∗01:01 CII259–273 (GIAGFKGEQGPKGEP) tetramers were generated by co-transfection of HEK 293 cells with the extracellular domains of HLA-DRA and either HLA-DRB1∗04:01-CII259–273 or HLA- DRB1∗01:01 CII259–273 cloned into the pHLsec vector [34]. The constructs encoded the CII259–273 peptide coupled to the N-terminus of the HLA-DR β-chain via flexible Glycine-Serine linker. The HLA-DRA and DRB constructs encoded C-terminal enterokinase cleavable fos/jun zippers to promote dimerization. The β-chain also contained a BirA site for biotinylation and tetramer generation and a Histidine tag for IMAC purification. After biotinylation of pHLA-DR samples, the percentage biotinylation was determined by native gel electrophoresis and complexation with avidin. Tetramers were generated within one week of staining by the addition of streptavidin-PE in a 1:8 ratio with biotinylated monomer as previously described [14, 31]
Monoclonal antibodies
Depending on HLA type, PBMC were stained for dump (CD19/14/16/11c)/CD3/CD4/PD-1/CD45RO/CCR7/CD25/CD127/DRB1∗04:01 or DRB1∗01:01-CII259-273/Live-Dead. Monoclonal antibodies (mAb) used in the assays were selected after titration for optimal concentration and incubation time, and multiple runs were repeated for consistent results in different RA samples. The final selected antibodies for the base panel were—CD19 FITC (0.5 µl; Biolegend; HIB19), CD14 FITC (0.5 µl; Biolegend; HC14), CD16 FITC (0.5 µl; Biolegend; 3.9), CD11c FITC (0.5 µl; Biolegend; 3G8), CD3 BUV737 (1 µl; BD Biosciences; UCHT1), CD4 BUV395 (1 µl; BD Biosciences; SK3). For Panel-1—CD25 BV650 (5 µl; Biolegend; BC96), CD127 BV421 (2.5 µl; Biolegend; A019D5), CCR7 (CD197) BV510 (5 µl; BD Biosciences; 2-L1-A), PD-1 (CD279) BB700 (5 µl; BD Biosciences; EH12.1), CD45RO APC-H7 (2.5 µl; BD Biosciences; UCHL1).
Tetramer staining procedure
Frozen vials of PBMC were rapidly thawed and washed with RPMI 1640 Complete Medium (Gibco) supplemented with 5% Human AB serum (Sigma-Aldrich), 1× Penicillin/Streptomycin/Glutamate (Invitrogen) and 1 mM sodium pyruvate (Gibco) in presence of DNase I (Sigma-Aldrich) at 12.5 and 6.25 µg/ml using a two-step thawing procedure as previously described [14]. Cells were counted using the Vi-Cell XR Cell Viability Analyzer (Beckman Coulter). The viable cell concentration and the percent viability were recorded. The cells were then suspended in FACS buffer (0.1% BSA, 2 mM EDTA in 0.01M PBS pH7.4) at 15 × 106 cells/ml. Non-specific binding sites were blocked using 2 µl FcR Blocking Reagent (Miltenyi Biotech) up to 10 × 106 cells. Without washing, 3 × 106 (200 µl) cells were aliquoted for each stain in labeled 5 ml FACS tubes as required for fluorescence minus one (FMO) and tetramer staining samples. PE-labeled DRB1∗01:01 or ∗04:01 tetramers were added at a final concentration of 6.2 µg/ml, except for the FMO where no DRB1∗01:01 or ∗04:01 tetramers were added. Cells were mixed and incubated at 4°C for 1 h in the dark with intermittent mixing at 30 min. Cells were washed by adding 3 ml 1× PBS (0.01M pH7.4) and centrifugation at 4°C, 450 g for 5 min. The supernatant was discarded, and cells were resuspended in 100 µl residual volume. A BD Horizon™ Fixable Viability Stain 700 (FVS700) (BD Biosciences) was reconstituted according to the manufacturer’s instruction, diluted 1:1000 in saline, and 10 µl added to ~100 µl of cells to a final dilution of 1:10,000. Cells were gently mixed, incubated in the fridge for 20 min in the dark, then washed with FACS buffer, centrifuged as before, and resuspended in 100 µl residual volume. For surface marker staining, a master mix of mAb (listed above) was prepared depending upon the number of tubes, added as 24 µl/tube, giving a final volume of antibody mix of 124 µl/tube. The cells were incubated in the antibody mix at 4°C for 20 min in the dark. After incubation, cells were washed with FACS buffer and resuspended in 200 µl FACS buffer. A minimum of 1 × 106 events were acquired in the lymphocyte gate. All tubes including controls were stored on ice until ready for acquisition.
Single stain controls and FMOs
Due to limitations of cell numbers from patient samples, FMOs for gating controls and single stain (SS) controls for flow cytometer setup were prepared from buffy coat samples. For SS controls, briefly, a frozen buffy coat PBMC aliquot was thawed, washed, and resuspended at 5 × 106/ml in FACS buffer. One hundred microliters of cell aliquots were divided into 11 pre-labeled tubes (0.5 × 106/100 µl/tube), one for each fluorescently labeled antibody from the panel listed in 2.3 including CD4-PE, Live/Dead FVS700, and a no-stain control. Respective antibodies were added as indicated except in no-stain and the Live/Dead FVS700 tube, performed in PBS as described before. Cells were mixed and refrigerated in the dark for 20 min. After incubation, cells were washed and resuspended in 200 µl of FACS buffer. A minimum of 20,000 total events were acquired for each control.
The FMOs for CD25, CD127, CCR7, PD-1, CD45RO, and Live/Dead FVS700 for the gating control were included in the staining set as described below. The buffy coat PBMC were divided among six pre-labeled FACS tubes (0.5 × 106/100 µl/tube) to be used as FMO-A to FMO-F for CD25-BV650, CD127-BV421, CCR7 -BV510, PD1-BB700, CD45RO-APC-H7 mAb, and Live/Dead FVS700 dye. For all FMOs, base panel mAb were added and then all four mAb except the one for which the FMO was used e.g. in FMO-A of CD25-BV650, all mAb except CD25-BV650 were added as indicated. All five mAb were added in FMO-F, but not the Live/Dead FVS-700 dye. Cells were incubated at 4°C for 20 min in the dark then washed with PBS and resuspended in 100 µl. Live/Dead FVS700 dye was then added to all FMOs, except the FMO-F, incubated and washed as before. Cells were resuspended in 200 µl FACS buffer. A minimum of 100,000 events were acquired in the lymphocyte gate.
Acquisition of samples by flow cytometry and flow data analysis
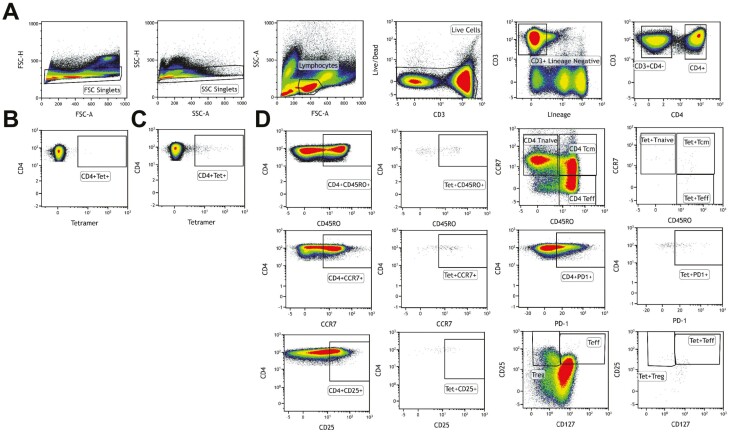
All stained samples and controls were acquired on a BD LSR FortessaX20 (Becton Dickinson) and analyzed using Kaluza Software Version 2. The acquisition limits and voltages were set using unstained cells and kept constant for all samples. Compensation was performed for all the fluorochromes, single cells gated and dead cells excluded for analysis. The cells were gated as shown in Fig. 1.
Fig. 1.
Gating strategy for tetramer analysis. (A) Live cells were gated using forward scatter (FSC) and side scatter (SSC) followed by singlets and lymphocytes. Live CD3+ cells were gated using Lineage/LIVE/DEAD marker negative and CD3+ cells. CD4+ T cells were gated from CD3+ T cells. Tet+ T cells were gated from CD4+ T cells as double positives. (B) Tetramer-negative FMO sample used to determine gating for Tet+ cells. (C) Representative sample with Tet+ staining (patient R3). (D) Based on surface markers used, subpopulation within CD4+ and Tet+ CD4+ cells were gated. The figure is representative of the evaluation assays.
Evaluation procedure
A formal evaluation plan was prepared with a detailed standard operating procedure and work flow (Fig. 2). Frozen samples from two RA patients R1 (HLA-DRB1∗04:01) and R2 (HLA-DRB1∗01:01) were selected. These samples had multiple cryopreserved PBMC aliquots each that were prepared and stored on the same day. On day 1, one analyst thawed the required vials of both the samples (R1 and R2) and set up Assay 1. The second analyst followed the same procedure to set up Assay 2 (Fig. 2). The procedure was repeated on day 2 for Assay 3 and Assay 4 from R1 and R2. Based on the results obtained from the initial four assays, additional assays were conducted by two analysts using PBMC from the buffy coat sample (Assay 5 and 6 on day 3) for the surface markers without tetramer. This was followed by two more assays by two analysts (Assay 7 and 8 on day 4) with RA sample R3 (HLA-DRB1∗04:01). A specificity test was included on each day, along with the precision assays, by one of the analysts. At the end of the evaluation process, we assayed two additional samples with viability >85% to confirm reproducibility of replicate samples.
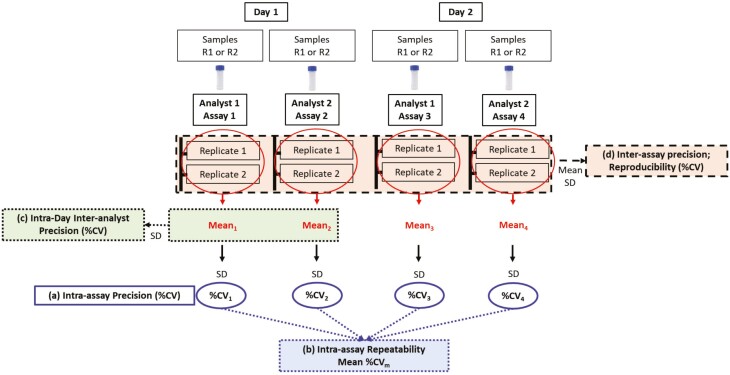
Fig. 2.
Assay qualification plan. Four assays were performed using the frozen PBMC of two RA patients R1 (HLA-DRB1∗04:01) and R2 (HLA-DRB1∗01:01), one assay from one sample each by two analysts on two different days. Each analyst independently thawed frozen vials of each RA sample on day 1 and day 2 and performed assays and flow cytometry analysis separately. Mean, SD, and %CV were calculated for different precision measurements. (A) Intra-assay precision of each assay was measured as %CV1-4 from individual assay replicates. (B) Intra-assay repeatability was measured as mean %CV (%CVm) of all four assays of each sample. (C) Intra-day Inter-analyst precision was measured as %CV from the mean value of assays performed by two analysts using the same sample on the same day. (D) Inter-assay precision was calculated from all replicates of all four assays performed from the same sample on both the days.
System suitability and assay acceptance criteria
Only those assays that satisfied the system suitability criteria along with acceptance criteria were considered acceptable. To pass the system suitability criteria, the flow cytometer instrument calibration test must have demonstrated that the flow cytometer is functioning appropriately. Initially, we included cells with viability ≥75% as a system suitability criterion. However, we subsequently accepted samples with lower viability. This allowed us to calculate the threshold of required viability and its impact on the precision outcome. The assay acceptance criteria included the presence of tetramer-positive cells in the positive control sample, which was achieved in all assays. The evaluation exercise was intended to understand the precision of the assay to assess rare CII-specific tetramer positive (tet+) T-cell populations. Therefore, we did not set any acceptance or rejection criteria based on the coefficient of variations (%CV) calculated from replicates. All assays performed during evaluation passed system suitability and assay acceptance criteria except viability.
Staining for specificity test
The specificity of HLA-DRB1∗04:01 or ∗01:01 tetramers was tested using PBMC (3 × 106) spiked with in-house generated HLA-DRB1∗01:01-CII259-273-specific SKW3 cell line (1 × 105) (method of clone generation is described below) when stained with HLA-DRB1∗01:01-CII259-273 tetramer, which served as a positive control (PC). The negative control (NC) sample was prepared similarly by spiking HLA-DRB1∗01:01-CII259-273-specific SKW3 cells and staining with HLA-DRB1∗04:01 CII259-273 tetramer. On day 3, the specificity was assessed by staining HLA-DRB1∗04:01+ PBMC with HLA-DRB1∗01:01 CII259-273 tetramer (NC). All specificity samples with or without SKW3 cells were subjected to the same procedure of tetramer staining. NC/PC were set up as a part of the precision assay by one of the two analysts on each day of the assay. The percentage of non-specific binding in NC samples was calculated by considering PC as 100% specific binding, using formula (% tet+ cells in NC x 100) / % tet+ cells in PC.
Generation of HLA-DRB1∗01:01-CII259-273-specific TCR transductants
To identify HLA-DRB1∗01:01-restricted CII-specific TCRs, HLA-DR1 transgenic mice were primed s.c. with bovine type II collagen emulsified in Complete Freund’s adjuvant. It should be noted that mouse TCRs autoreactive with pHLA-II tend to bind with higher affinity than human TCRs autoreactive with pHLA-II. Splenocytes and draining inguinal lymph node cells were harvested 12 days post-prime, pooled and cultured for 7 days with CII259-273 peptide (30 µg/ml). Cultured cells were stained with a PE-conjugated HLA-DRB1∗01:01-CII259-273 tetramer (6.2 µg/ml) and sorted as single tet+ or tet- CD4+ T cells. After multiplex-nested PCR [35] and Sanger sequencing we identified preferential usage of TRAV19 and TRBV31 gene families by CII259-273-specific CD4+ T cells, which arose from vigorous clonal expansion (Supplementary Fig. 1A).
To generate HLA-DRB1∗01:01-CII259-273 TCR transductants, we stably transduced the TCRαβ-deficient SKW-3 cell line with a HLA-DRB1∗01:01-CII259-273-restricted TCR expressing TRBV31 identified at a frequency of 24% after in vitro re-stimulation, using lentiviral transduction. Briefly, full-length TCRα and TCRβ cDNA (Supplementary Table 1) was cloned into a pELNs third-generation lentivirus expression vector [36]. HEK293T packaging cells were co-transfected with the pELN-αβTCR lentivirus construct and packaging plasmids pMDLg/pRRE, pRSV-Rev and pMD2-G using FuGENE6 (Promega) (Supplementary Fig. 1B). HEK293T cell culture supernatant containing virus particles carrying the TCR transgene was then used to stably transduce SKW-3 cells in the presence of polybrene. TCR transductants were bulk sorted using a MoFlo Astrios (Beckman Coulter) cell sorter as live hCD3+/mTCRβ+ cells (Supplementary Fig. 1C) and expanded in culture. Within 2 weeks of culture, the enriched transduced cells were stained with HLA-DRB1∗01:01-CII259-273 tetramer, in the absence or presence of 50 nM Dasatinib (Selleck Chemicals) at 37°C (added 30 min prior) to prevent rapid TCR internalization triggered by cognate antigen signaling of the cell line [14, 37] (Supplementary Fig. 1C).
To confirm the specificity of the SKW-3 TCR for HLA-DRB1∗01:01-CII259-273, TCR transductants were 1. stained with HLA-DRB1∗04:01-CII259-273 tetramer, and 2. cultured for 18 h with gamma irradiated healthy human HLA-DRB1∗01:01 PBMCs as APCs and varying concentrations of CII259-273 peptide, irrelevant human Aggrecan89-103 (ATEGRVRVNSAYQDK) peptide or PMA (10 ng/ml)/Ionomycin (1 µg/ml) as a positive control. The response to stimulation was measured by change in MFI of CD69 on the SKW-3 TCR transduced cells, a marker of early T cell activation following TCR engagement. To test the sensitivity of the tetramer in PBMC, 105 CII-TCR-transduced SKW-3 cells were spiked into 3 × 106 healthy control buffy coat or RA patient PBMC, then stained with relevant HLA-DRB1∗01:01-CII259-273 or irrelevant HLA-DRB1∗04:01-CII259-273 tetramer (Supplementary Fig. 1D). Cells were acquired on a Gallios flow cytometer (Beckman Coulter) and data analyzed using Kaluza software V2 (Supplementary Fig. 1E).
Calculations and statistical analysis
From each test sample, surface marker positivity was reported as the percentage gated of CD4+ T cells or CD4+tet+ population. Mean, standard deviation (SD), and precision in terms of the coefficient of variation (% CV), where CV is (standard deviation/mean) × 100, were calculated. The intra-assay precision (%CV) was calculated from the assay replicates. Intra-assay repeatability (mean %CV) was calculated as the mean of %CVs of individual assays. The intra-day inter-analyst precision was calculated from the assays performed by two analysts on the same day from the same sample. The inter-assay precision was calculated by pooling all replicates of all the assays performed from the same sample during the qualification process. The schematic of the qualification plan and precision calculations is presented in Fig. 2.
All data obtained during evaluation, including failed assays and outliers, identified by the ROUT test (Q = 5%), are included in the precision estimates. Identification of outliers and correlation analysis were performed using GraphPad Prism 8 software. The correlation analysis used the Pearson Correlation test and calculated the Pearson coefficient ‘r’ value and significance by a two-tailed P test with a significance threshold of P ≤ 0.05.
The method described here can be categorized as quasi-quantitative i.e. produces continuous numeric results expressed in terms of the test sample but not derived from a calibration curve or reference standard [38]. The evaluation data are analyzed and discussed according to the recommendations for the precision analysis described for quantitative assays in the white papers [33, 39].
Results
Modification of flow panel to improve fluorochrome compatibility and CD4 gating
Our previously developed flow cytometric assay methodology for pMHCII tetramer staining, for the quantification and phenotyping of antigen-specific CD4+ T cells [14], was not yet qualified for use in a clinical trial setting. As a first step, we adapted our panel to improve the separation of CD4 and CD3 cells, while retaining other methodologies. Our initial panel design used CD3 BUV737 and CD4 Alexa Fluor700 fluorochrome combinations which, although excited by different lasers, still yield significant fluorescence spillover. Although trivial when assessing healthy individuals, in RA patients with activated PB CD4+ T cells, surface expression of CD3 and CD4 often changes, impairing the reliable definition of CD3 and CD4 expression (Supplementary Fig. 2). We carefully redesigned the panel, following good flow cytometry panel design principles, employing antibody/fluorochrome combinations with low spillover and with high detection levels to ensure robust marker separation. We evaluated this flow cytometry–based method for precision and reproducibility to detect total and HLA-DRB1∗04:01- and HLA-DRB1∗01:01-CII259-273-specific CD4+ T cells and T-cell subsets, including 16 subpopulations of CD4+ and CD4+ tet+ T cells: CD127- CD25+, CD127+ CD25+, PD1+, CD45RO+, CCR7+, T naive (CCR7+ CD45RO-), T effector memory (Tem) (CCR7- CD45RO+), and T central memory (Tcm) (CCR7+ CD45RO+) subpopulations. Ten assays were performed using samples from five RA patients and one buffy coat, including one assay comprising two RA samples (R1 and R2) on day 1 and day 2 performed together by one analyst (Table 1). From collated results, we calculated specificity, intra-assay precision, repeatability, intra-day inter-analyst precision, and inter-assay reproducibility. All individual assay data, replicates and calculations are presented in Supplementary Tables 2–5. Comparative tetramer staining for patient R1 and R2 are depicted in Supplementary Figs 3 and 4, respectively.
Table 1.
Evaluation summary with cell recovery and viability. Ten assays were performed using three samples from RA patients (days 1, 2, and 4) and one buffy coat (day 3)
| Assays performed | Sample code | HLA Type | No of Vials (Cells per vial at the time of freezing) | Frozen on | Cell count after thawing | Cell recovery after thawing (%) | Cell viability (%) |
|---|---|---|---|---|---|---|---|
| Day 1 | |||||||
| Assay 1 Analyst 1 | R1 | DRB1∗04:01DRB1∗03:01 | 2 vials (17 × 106) | 21/07/2016 | 3.56 × 106 | 10.47 | 72.00 |
| R2 | DRB1∗01:01 DRB1∗16:01 |
2 vials (18 × 106) | 15/06/2016 | 11.15 × 106 | 30.97 | 66.90 | |
| Assay 2 Analyst 2 | R1 | DRB1∗04:01DRB1∗03:01 | 1 vial (17 × 106) | 21/07/2016 | 1.38 × 106 | 8.12 | 70.30 |
| R2 | DRB1∗01:01 DRB1∗16:01 |
1 vial (18 × 106) | 15/06/2016 | 3.93 × 106 | 21.83 | 84.10 | |
| Day 2 | |||||||
| Assay 3 Analyst 1 | R1 | DRB1∗04:01DRB1∗03:01 | 3 vials (17 × 106) | 21/07/2016 | 2.5 × 106 | 4.90 | 73.40 |
| R2 | DRB1∗01:01 DRB1∗16:01 |
2 vials (18 × 106) | 15/06/2016 | 6.48 × 106 | 18.00 | 87.10 | |
| Assay 4 Analyst 2 | R1 | DRB1∗04:01DRB1∗03:01 | 3 vials (17 × 106) | 21/07/2016 | 3.88 × 106 | 7.61 | 88.00 |
| R2 | DRB1∗01:01 DRB1∗16:01 |
1 vials (17 × 106) | 15/06/2016 | 3.56 × 106 | 20.94 | 91.80 | |
| Day 3 | |||||||
| Assay 5 Analyst 1 | Buffy Coat | ND | 36 × 106 | 16/10/2016 | 17.63 × 106 | 48.97 | 92.70 |
| Assay 6 Analyst 2 | Buffy Coat | ND | 36 × 106 | 16/10/2016 | 17.26 × 106 | 43.53 | 92.30 |
| Day 4 | |||||||
| Assay 7 Analyst 1 | R3 | DRB1∗04:01 DRB1∗04:04 |
2 vials (18 × 106) | 12/03/2020 | 21.93 × 106 | 60.92 | 93.40 |
| Assay 8 Analyst 2 | R3 | DRB1∗04:01 DRB1∗04:04 |
2 vials (18 × 106) | 12/03/2020 | 26.13 × 106 | 72.58 | 92.00 |
| Replication | |||||||
| Assay 9 Analyst 1 | R4 | DRB1∗04:01 DRB1∗04:04 |
2 vials (20 × 106) | 22/11/2019 | 25.7 × 106 | 68.75 | 98.40 |
| Assay 10 Analyst 1 | R5 | DRB1∗04:01 DRB1∗04:01 |
2 vials (22 × 106) | 15/06/2018 | 37.3 × 106 | 84.77 | 96.60 |
Cell count and viability determined with the Vi-Cell XR Cell Viability Analyzer.
ND, not determined.
Specificity
The specificity of each tetramer was tested by staining the HLA-DRB1∗01:01-CII(259-273)-specific SKW3 cell line with HLA-DRB1∗04:01- relative to HLA-DRB1∗01:01-CII259-273 tetramers. Non-specific binding in NC on days 1 and 2 was 5.56% and 1.08%, respectively, demonstrating the high level of specificity of the antigen-specific T cell line (Table 2). On day 4, we used cells from HLA-DRB1∗04:01+ patient R3 for specificity testing by similar staining with HLA-DRB1∗01:01-CII259-273 tetramers. We observed 34.48% non-specific binding in NC (Table 2). The dot plots with gating for specificity measures are presented in Supplementary Fig. 5.
Table 2.
Specificity of HLA-DRB1∗04:01- or HLA-DRB1∗01:01-CII259–273 tetramers for CII-specific cells
| Day | Controls/Specificity samples | # CD4+ T cells |
# Tet+ cells |
% Tet-positive cells | MFI CD4+ Tet+ | % Binding considering PC as 100% binding | % Specificity |
|---|---|---|---|---|---|---|---|
| 1 | SKW3 PC | 9900 | 415 | 4.1919 | 3.770 | 100 | 100 |
| SKW3 NC | 10295 | 24 | 0.2331 | 2.920 | 5.56 | 94.44 | |
| 2 | SKW3 PC | 12166 | 283 | 2.3262 | 3.182 | 100 | 100 |
| SKW3 NC | 11914 | 3 | 0.0252 | 2.825 | 1.08 | 98.91 | |
| 4 | R3 PC | 282856.8 | 167.4 | 0.0580 | 3.616 | 100 | 100 |
| R3 NC | 211768 | 44 | 0.0200 | 3.790 | 34.48 | 65.52 |
Negative (NC) and positive (PC) controls were set up as a part of the precision assay by one analyst on each day of assay (days 1-4), except day 3 (buffy coat). The PC sample consists of (n = 1) HLA-DRB1∗01:01+ PBMC spiked with 105 HLA-DRB1∗01:01-CII259-273-specific SKW3 cells (1 × 105) when stained with HLA-DRB1∗01:01-CII259-273 tetramer. The NC sample consists of (n=1) HLA-DRB1∗01:01+ PBMC spiked with 105 HLA-DRB1∗01:01-CII259-273-specific SKW3 cells (1x105) when stained with HLA-DRB1∗04:01-CII259-273 tetramer. On day 4, HLA-DRB1∗04:01+ RA sample R3 was stained with mismatched HLA-DRB1∗01:01-collagen II259-273 tetramer without spiking SKW3 cells, as NC. The PC value of R3 was calculated as a mean of replicates (n = 5) from the assay of HLA-DRB1∗04:01-CII259-273 tetramer staining without spiking, conducted as part of precision. Representative flow cytometry plots in Supplementary Fig. 3.
Intra-assay precision repeatability
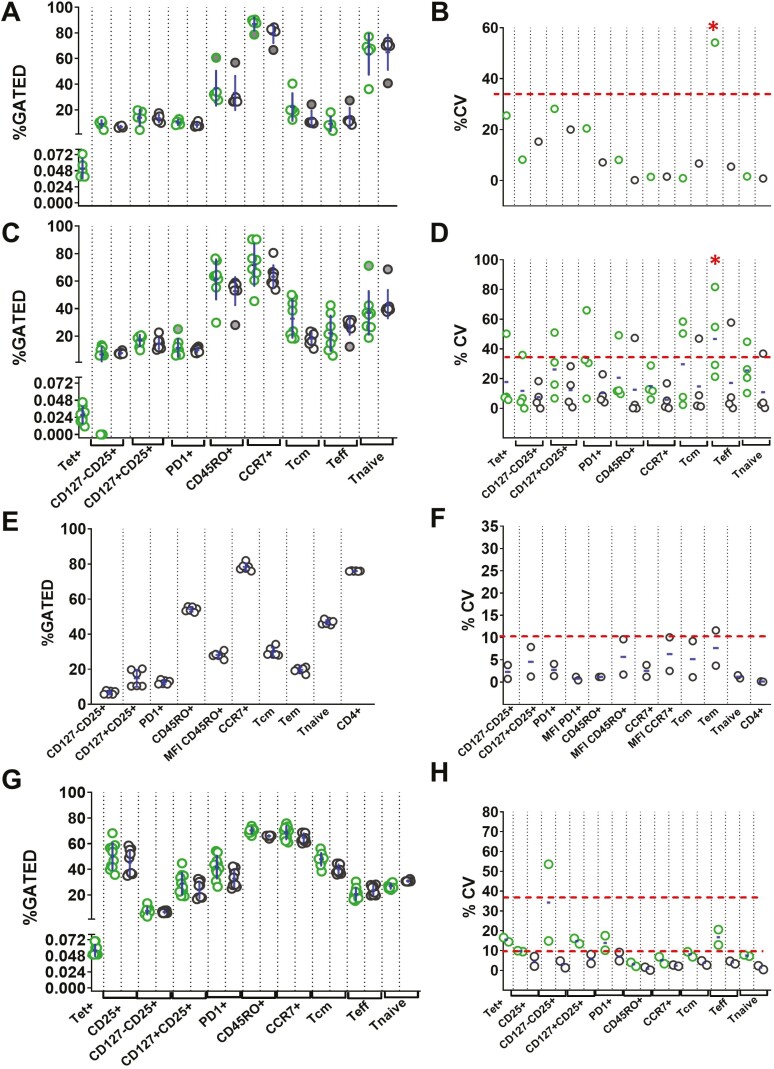
For initial evaluation, we phenotyped the CD4+ and CD4+tet+ subpopulations as indicated in Fig. 1. HLA-DRB1∗04:01 (R1) and HLA-DRB1∗01:01 (R2) samples were assessed by two analysts on two separate days (Table 1). There was no significant difference in percent gated expression of any subpopulations over the four assays (Fig. 3A and C).
Fig. 3.
RA samples R1 (HLA-DRB1∗04:01), R2 (HLA-DRB1∗01:01), buffy coat sample and RA sample R3 (HLA-DRB1∗04:01). Assays were conducted as shown in Figure 2. (A) Percent of gated subpopulations of CD4+ T cells with (green symbols) and without tetramer (black symbols) from RA sample R1 (day 1, two analysts). (B) Intra-assay precision (%CV) and intra-assay repeatability (mean %CV) from R1 (two assays, two analysts). (C) Percent of gated CD4+ T cell subpopulations with and without tetramer in RA sample R2 (day 2, two analysts). (D) Intra-assay precision and Intra-assay repeatability from R2 (two assays, two analysts). (E) Percent of gated subpopulations of CD4+ T cells from the buffy coat sample (day 3, two analysts). (F) Intra-assay precision and intra-assay repeatability from the buffy coat sample (two assays, two analysts). (G) Percent of gated subpopulations of CD4+ T cells with and without tetramer in RA sample R3 (day 4, two analysts). (H) Intra-assay precision and intra-assay repeatability from R3 (two assays, two analysts). Datapoints in (A), (C), (E), and (G) represent replicate results. The filled data points were identified as outliers from combined data of two assays. Outliers are included in all calculations. Each data point in (B), (D), (F), and (H) represents %CV from one assay and the mean bar indicates Intra-assay repeatability (mean %CV) from two assays. The dotted line at 35% represents the recommended precision. Red ∗ indicates markers failing to meet intra-assay repeatability of %CV>35.
One analyst ran replicates for sample R1 (Supplementary Table 2), allowing estimation of intra-assay precision for that day. This was found to be well within an acceptable range (%CV≤35), apart from the tet+ CD4+ Tem (%CV of 54.18%) (Fig. 3B). For sample R2, (Supplementary Table 3), one or two populations showed intra-assay precision outside the acceptable limit, out of four assays. However, all markers were well within acceptance criteria for repeatability, ranging from 5.79% (CCR7+) to 33.93% (tet+ PD1+), again with the exception of tet+ CD4+ Tem (%CV 46.70%) (Fig. 3D). As per recommendations, for intra-assay repeatability, if the mean value of %CV from all the assays meet the criteria, it is not necessary for each individual assay to have %CV ≤35% [33]. Accordingly, the intra-assay precision result fulfills the criteria of intra-assay repeatability for the haplotype HLA-DRB1∗01:01.
However, we recorded poor cell recovery and viability (<75%) in four of eight thaws of R1 and R2 (Table 1). Thus, we hypothesized that low numbers, especially of antigen-specific T cells, contributed the main source of variability. To test this, we repeated the precision experiments with a healthy control buffy coat sample (Assay 5 and 6; day 3) without tetramer staining, maintaining three sample replicates per assay (Supplementary Table 4), and with an additional RA sample (R3; HLA-DRB1∗04:01; Assay 7 and 8, day 4) with a higher cell recovery and >90% viability. For this RA sample we included tetramer staining and five replicates per assay (Supplementary Table 5). From the buffy coat sample, all gated populations were identified with a precision within %CV of 10%, except for CD4+ Tem cells (%CV of 11.62%). The intra-assay repeatability was also within the mean 10%CV for all the markers (Fig. 3E and F). Similar results were obtained for R3 (Fig. 3G and H and Supplementary Table 5). The intra-assay repeatability was well within an acceptable range from 0.86% to 16.72%, with the tet+ CD127-CD25+ subpopulation higher but still acceptable, at a mean %CV of 34.22% (Fig. 3G and H). These results indicate that the desired intra-assay precision and repeatability was achieved for all gated populations with cell recovery and viability of >90%.
Intra-day inter-analyst precision, inter-assay precision, and reproducibility
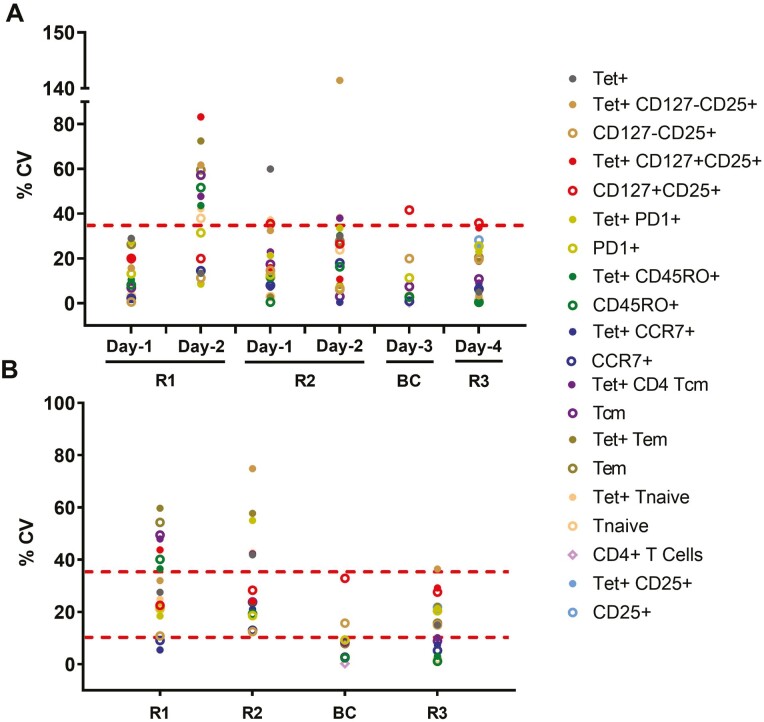
The intra-day inter-analyst precision was calculated as %CV from the mean of individual assays performed by two analysts on the same day and same sample, as depicted in Fig. 2.
The inter-analyst precision on day 1 for haplotype HLA-DRB1∗04:01 (R1) was within the acceptable limit for all subpopulations. On day 2, 10 subpopulations showed %CV greater than 35% (Fig. 4A). The overall low reproducibility in this sample could be attributed to low cell numbers, poor quality, and few replicates included per experiment.
Fig. 4.
Intra-day inter-analyst precision (intra-day reproducibility) and inter-assay reproducibility from day 1 to day 4. One assay from each sample (R1 and R2 on day 1 and 2, buffy coat on day 3 and R3 on day 4) was conducted by two analysts. (A) The %CV of two assays from the same sample on the same day by different analysts was calculated as intra-day inter-analyst precision or intra-day reproducibility. Each data point represents %CV for an indicated subpopulation. The dotted line at 35% represents recommended precision for rare cell populations. (B) Inter-assay precision from all replicates from all assays. The dotted line at 10% represents the desired precision and at 35% represents recommended precision for rare cell populations.
For haplotype HLA-DRB1∗01:01 (R2), % CV values for inter-analyst precision on day 1 and day 2 were mostly within the acceptable limit, below 35%. Results improved when samples with >90% cell recovery and viability (buffy coat and R3) were assayed on day 3 and 4, respectively. From the buffy coat sample, all subpopulations except CD127+CD25+ (41.62%) were within the recommended %CV limit. For sample R3, all subpopulations showed acceptable %CV (Fig. 4A). Notably, even though measuring rare tet+ cells, only three subpopulations had %CV > 25%.
The inter-assay precision was calculated as %CV from the replicates of all assays performed from the same sample on different days by different analysts, representing the overall reproducibility of the assay (Fig. 1). Similar to the previous calculations, the inter-assay precision analysis for R1 and R2 failed to meet the %CV cutoff criteria in 7 and 6 subpopulations, respectively, but were within the 35% range for the buffy coat sample and R3, except tet+ CD127-CD25+ cells at 36.43% (Fig. 4B).
Correlation of intra-assay precision with cell recovery and viability
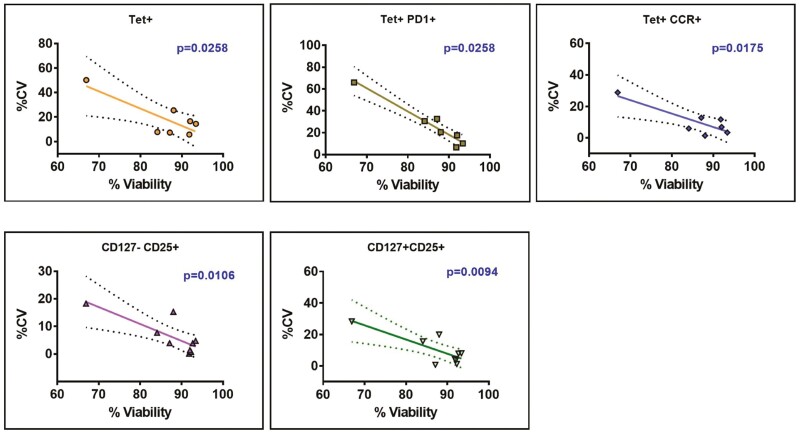
Given these observations, we correlated intra-assay precision (%CV) with viability of sample and recovery of cells from all assay samples. Cell viability was negatively correlated (P ≤ 0.05) for five subpopulations: tet+, tet+PD1+, tet+CCR7+, CD127-CD25+, and CD127+CD25+ (Fig. 5). The linear regression equations generated from these significantly correlated subpopulations were used to extrapolate the minimum viability required to achieve %CV of 10%, 25% and 35% respectively (Supplementary Table 6). According to their individual linear equations, the highest % viability required to meet %CV value of 25% is 86.78% (for Tet+PD1+), indicating that samples with % viability of ≥85% should meet acceptance criteria of %CV≤35% for the five significantly correlated subpopulations.
Fig. 5.
Linear correlation analysis of intra-assay precision to cell viability. The degree of linear correlation between intra-assay %CV and %sample viability. From all the subpopulations tested across all assays, five subpopulations - tet+ (n = 7), tet+PD1+ (n = 7), tet+CCR7+ (n = 7), CD127-CD25+ (n = 9), and CD127+CD25+ (n = 9) cells – were significantly negatively correlated (P ≤ 0.05), with Pearson r values of −0.8142, −0.9684, −0.8421, −0.794, and −0.8012, respectively. The dotted lines in the graph indicate 95% confidence intervals.
Reproducibility experiments after completion of the evaluation exercise
After completing the formal evaluation, we tested two more samples, R4 and R5, from RA patients with post-thaw viability of 98.4% and 96.5%, respectively to assess reproducibility. After exclusion of outliers ascertained using Dixon’s test (for a single outlier in a small data set), only one %CV value was >35% (Supplementary Table 7). These data confirm that reproducibility is high for replicate samples with viability >85%. Under these conditions, intra-assay repeatability, inter-analyst precision, and inter-assay reproducibility for the tetramer assays were well within the %CV ≤35% acceptable range, for all the tested rare subpopulations except tet+ CD127-CD25+ [33].
Discussion
Clinical validation of antigen-specific T cells is greatly needed as an immunological biomarker tool to monitor antigen-specific T-cell frequency or phenotype before or at clinical diagnosis in natural history studies or association with clinical response to immunotherapy [11, 40]. Flow cytometry–based soluble pMHC tetramer techniques are the most reliable methods for monitoring antigen-specific T or B cells [41]. Implementing flow cytometry methods according to the regulatory requirements of drug development and as established validated clinical or diagnostic markers is challenging due to the lack of reference standards, inherent biological lymphocyte variability, limited stability of samples, cytometer set-up requirements, the complexity of data output and the interpretation of results with lack of relevant guidelines [42–45]. Recently, guidelines for validating flow methods were published as white papers [33, 39]. Apart from recent successful validation and application at multicenter level of a flow cytometry method for abundant peripheral blood cell populations [46], few fit-for-purpose qualification studies have been validated for relevant clinical settings. Qualifying a flow-based method for detection of rare antigen-specific T cells (~1–100 in 106 CD4+ T cells) with low affinity TCRs, is more challenging than phenotyping abundant cells. Some self-epitopes have very low affinity for HLA-II. Moreover, self-reactive TCRs bind to self-epitopes with lower affinity, and with shorter dwell times than TCRs specific for foreign, pathogen-derived peptides, which makes their detection challenging [47, 48]. This instability has been the focus of recent protein engineering strategies [49]. Several methods are qualified or validated for antigen-specific CD8+ T cells, which are detected at higher frequencies [22, 50, 51], and are generally less difficult to quantify than antigen-specific CD4+ T cells [47, 48]. Circulating antigen-specific CD4+ T cells are harder to identify, due to the relatively low affinity and avidity of their TCRs to cognate pMHC, as well as cross-reactivity to environmental antigens [18, 19, 21, 22, 51, 52]. For example, virus-specific CD4 memory T cells are present in circulation of unexposed adults and expand after influenza immunization. These T cells recognize multiple microbial epitopes, due to cross-reactivity of the TCRs for other peptide-MHC combinations [21]. Similarly, blood from both HLA-DRB1∗04:01+ healthy donors and RA patients contains circulating CD4+ T cells that recognize multiple self-antigens [31, 53, 54]. Thus, it has been challenging to develop tetramer reagents and protocols for flow cytometry and other relevant technologies [18, 55]. Furthermore, autoreactive CD4+ T cells may be compartmentalized in tissues, resulting in low frequencies in PB [18]. Attempts to enrich them may lead to cell death and autofluorescence [14, 18].
Tetramer staining methods can be classified as quasi-quantitative or qualitative, as there are no calibration standards available. The most important parameters that can be improved in flow-based methods are specificity, precision and reproducibility, based on repetitive analysis within one or multiple laboratories. In the current studies, we evaluated our optimized assay for CII259-273-specific CD4+ T cells from RA patients using pMHCII tetramers to develop biomarkers assessing the longitudinal impact of therapy, particularly antigen-specific immunotherapies. We expected to see high %CVs, because antigen-specific T cells are rare (1–100 per million cells) and susceptible to cell death in thawed PBMC from patients with active disease [22, 56, 57]. To increase the yield of phenotypic information, we endeavored to identify multiple subpopulations of these rare antigen-specific T cells from frozen PBMC. Therefore, rather than set %CV assay acceptance criteria, we wished to understand limitations and identify critical parameters influencing overall assay reproducibility. The negative correlation between cell viability and %CV provides important insight into cell viability as one of the most critical parameters affecting overall precision of the assay. Furthermore, as antigen-specific T cells are generally very infrequent, a reduction in cells per test reduces the chance of detection and increases variability between assays. Thus, optimizing the number of cells per test and acquiring a pre-set minimum number of cells is important when measuring rare tetramer subpopulations. Attention to PBMC preparation and post-thaw viability will enhance the quality of outputs from clinical trials of antigen-specific immunotherapy.
The specificity assays showed high tetramer specificity for clonal T cell lines expressing high levels of TCR, but a degree of cross-reactivity to TCR from PBMC with mis-matched HLA haplotype. This is not unexpected for pMHCII tetramers, due to TCR-tetramer complex instability (low ‘functional avidity’), and cross-reactivity to other low avidity alloreactive T cells [21, 58, 59]. From the perspective of the TCR, HLA-DRB1∗01:01-CII259-273 and HLA-DRB1∗04:01-CII259-273 have somewhat similar architecture [60], which might increase cross-reactivity.
CD25 had consistently high %CV in all subpopulations. A previous multicenter validation study phenotyping PB cells showed a consistent reduction in %CD4+CD25hiCD127lo Treg after cryopreservation, indicating particular sensitivity to loss of these surface markers [46]. This is an important limitation of the use of CD25 in panels to quantify total or antigen-specific Treg, which should be factored into decisions about which outcomes will assess the immune regulatory response to ASIT.
The quasi-quantitative tetramer assay described here was optimized for the detection of CII259-273-specific CD4+ T cells in patients with HLA-DRB1∗01:01 or HLA-DRB1∗04:01 and was suitable as a fit-for-purpose assay in research environments. We show that a precision outcome with recommended repeatability and reproducibility levels is achievable for rare antigen-specific T-cell subpopulations, provided that the cellular viability is ≥90% after thaw, and that a minimum number of cells is assessed. These measures increase confidence in subgating strategies. The precision level of 10% CV in healthy control PBMC indicates that the assay is probably feasible for application in clinical trials, if validated on a larger scale at multiple centers. In RA patients, the precision level near 25% indicates applicability of the assay for fit-for-purpose biomarker studies [33]. Previous studies demonstrated that titration of tetramer concentration, antibody concentration and standardized gating strategy using FMOs are also critical parameters for assay reproducibility [20]. Full qualification and validation that include linearity, specificity, sensitivity, LLD, and addressing limitations, are now feasible in future studies using relevant acceptance criteria in a patient population.
Supplementary Material
Acknowledgments
We would like to acknowledge the contribution of David Sester, Dalia Khalil and Yitian Ding at the Translational Research Institute (TRI) Flow Cytometry Facility for invaluable help and technical assistance. We thank Kylie Loh for technical assistance in generating pMHC-II tetramers.
Glossary
Abbreviations
- APCs
antigen presenting cells
- ASIT
antigen-specific immunotherapy
- CII
type II collagen
- CIA
collagen-induced arthritis
- CTL
cytotoxic T cells
- FMO
fluorescence minus one
- mAb
monoclonal antibody
- MHCII
class II major histocompatibility complex
- NC
negative control
- p
peptide
- PBMC
peripheral blood mononuclear cells
- PC
positive control
- Tcm
central memory T cells TCR, T cell receptor
- tet
tetramer
- Tem
effector memory T cells
Funding
This manuscript is part of a project that has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No 777357. This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA. RT is supported by Arthritis Queensland and a Fellowship from NHMRC.
Conflict of interest
R.T. reports past funding from Janssen Biotech Inc to Uniquest, related to the submitted work, and patent 9,017,697 B2: 2006 issued. K.C. is an employee and stock holder of Janssen Research & Development.
Author contributions
S.P. prepared qualification plan and analyzed qualification data; H.N., N.R. and P.W. performed experiments and analyzed data; H.H.R. and J.R. provided the tetramers: R.T. reviewed overall research and analyzed data; S.P. prepared the manuscript; H.N., N.R., P.W., H.R., K.C., and R.T. reviewed the manuscript.
Data availability
Data and protocols are available from corresponding author upon request.
References
- 1. Sinha AA, Lopez MT, McDevitt HO.. Autoimmune diseases: the failure of self tolerance. Science 1990, 248, 1380–8. [DOI] [PubMed] [Google Scholar]
- 2. Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J.. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol 2015, 33, 169–200. [DOI] [PubMed] [Google Scholar]
- 3. Ara A, Ahmed KA, Xiang J.. Multiple effects of CD40-CD40L axis in immunity against infection and cancer. Immunotargets Ther 2018, 7, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Law SC, Benham H, Reid HH, Rossjohn J, Thomas R.. Identification of self-antigen-specific T cells reflecting loss of tolerance in autoimmune disease underpins preventative immunotherapeutic strategies in rheumatoid arthritis. Rheum Dis Clin North Am 2014, 40, 735–52. [DOI] [PubMed] [Google Scholar]
- 5. Galea R, Nel HJ, Talekar M, et al. PD-L1- and calcitriol-dependent liposomal antigen-specific regulation of systemic inflammatory autoimmune disease. JCI Insight 2019, 4, e126025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Capini C, Jaturanpinyo M, Chang HI, et al.. Antigen-specific suppression of inflammatory arthritis using liposomes. J Immunol 2009, 182, 3556–65. [DOI] [PubMed] [Google Scholar]
- 7. Bergot AS, Buckle I, Cikaluru S, et al.. Regulatory T cells induced by single-peptide liposome immunotherapy suppress islet-specific T cell responses to multiple antigens and protect from autoimmune diabetes. J Immunol 2020, 204, 1787–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Riou C, Jhilmeet N, Rangaka MX, Wilkinson RJ, Wilkinson KA.. Tuberculosis antigen-specific T-cell responses during the first 6 months of antiretroviral treatment. J Infect Dis 2020, 221, 162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pol JG, Bridle BW, Lichty BD.. Detection of tumor antigen-specific T-cell responses after oncolytic vaccination. Methods Mol Biol 2020, 2058, 191–211. [DOI] [PubMed] [Google Scholar]
- 10. Palata O, Podzimkova Hradilova N, Mysiková D, et al.. Detection of tumor antigens and tumor-antigen specific T cells in NSCLC patients: correlation of the quality of T cell responses with NSCLC subtype. Immunol Lett 2020, 219, 46–53. [DOI] [PubMed] [Google Scholar]
- 11. Ahmed S, Cerosaletti K, James E, et al. Standardizing T-cell biomarkers in type 1 diabetes: challenges and recent advances. Diabetes 2019, 68, 1366–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mathieu C, Lahesmaa R, Bonifacio E, Achenbach P, Tree T.. Immunological biomarkers for the development and progression of type 1 diabetes. Diabetologia 2018, 61, 2252–8. [DOI] [PubMed] [Google Scholar]
- 13. Abdelaal HM, Cartwright EK, Skinner PJ.. Detection of antigen-specific Tcells using in situ MHC tetramer staining. Int J Mol Sci 2019, 20, 5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jansen DTSL, Ramnoruth N, Loh KL, et al.. Flow cytometric clinical immunomonitoring using peptide-MHC class II tetramers: optimization of methods and protocol development. Front Immunol 2018, 9, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wölfl M, Schalk S, Hellmich M, Huster KM, Busch DH, Berthold F.. Quantitation of MHC tetramer-positive cells from whole blood: evaluation of a single-platform, six-parameter flow cytometric method. Cytometry A 2004, 57, 120–30. [DOI] [PubMed] [Google Scholar]
- 16. Pastore G, Carraro M, Pettini E, Nolfi E, Medaglini D, Ciabattini A.. Optimized protocol for the detection of multifunctional epitope-specific CD4+ T cells combining MHC-II tetramer and intracellular cytokine staining technologies. Front Immunol 2019, 10, 2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maecker HT, Hassler J, Payne JK, et al.. Precision and linearity targets for validation of an IFNgamma ELISPOT, cytokine flow cytometry, and tetramer assay using CMV peptides. BMC Immunol 2008, 9, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vollers SS, Stern LJ.. Class II major histocompatibility complex tetramer staining: progress, problems, and prospects. Immunology 2008, 123, 305–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Andreatta M, Karosiene E, Rasmussen M, Stryhn A, Buus S, Nielsen M.. Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification. Immunogenetics 2015, 67, 641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dolton G, Zervoudi E, Rius C, et al.. Optimized peptide-MHC multimer protocols for detection and isolation of autoimmune T-cells. Front Immunol 2018, 9, 1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM.. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity 2013, 38, 373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wooldridge L, Lissina A, Cole DK, van den Berg HA, Price DA, Sewell AK.. Tricks with tetramers: how to get the most from multimeric peptide-MHC. Immunology 2009, 126, 147–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chandran PA, Laske K, Cazaly A, et al.. Validation of immunomonitoring methods for application in clinical studies: the HLA-peptide multimer staining assay. Cytometry B Clin Cytom 2018, 94, 342–53. [DOI] [PubMed] [Google Scholar]
- 24. Shen C, Xu T, Wu Y, et al.. Frequency and reactivity of antigen-specific T cells were concurrently measured through the combination of artificial antigen-presenting cell, MACS and ELISPOT. Sci Rep 2017, 7, 16400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holmdahl R, Andersson EC, Andersen CB, Svejgaard A, Fugger L.. Transgenic mouse models of rheumatoid arthritis. Immunol Rev 1999, 169, 161–73. [DOI] [PubMed] [Google Scholar]
- 26. Rosloniec EF, Whittington KB, He X, Stuart JM, Kang AH.. Collagen-induced arthritis mediated by HLA-DR1 (∗0101) and HLA-DR4 (∗0401). Am J Med Sci 2004, 327, 169–79. [DOI] [PubMed] [Google Scholar]
- 27. Qian Z, Latham KA, Whittington KB, Miller DC, Brand DD, Rosloniec EF.. An autoantigen-specific, highly restricted T cell repertoire infiltrates the arthritic joints of mice in an HLA-DR1 humanized mouse model of autoimmune arthritis. J Immunol 2010, 185, 110–8. [DOI] [PubMed] [Google Scholar]
- 28. Raychaudhuri S, Sandor C, Stahl EA, et al.. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet 2012, 44, 291–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scally SW, Law SC, Ting YT, et al.. Molecular basis for increased susceptibility of Indigenous North Americans to seropositive rheumatoid arthritis. Ann Rheum Dis 2017, 76, 1915–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ting YT, Petersen J, Ramarathinam SH, et al.. The interplay between citrullination and HLA-DRB1 polymorphism in shaping peptide binding hierarchies in rheumatoid arthritis. J Biol Chem 2018, 293, 3236–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Scally SW, Petersen J, Law SC, et al.. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J Exp Med 2013, 210, 2569–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. James EA, Rieck M, Pieper J, et al.. Citrulline-specific Th1 cells are increased in rheumatoid arthritis and their frequency is influenced by disease duration and therapy. Arthritis Rheumatol 2014, 66, 1712–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Selliah N, Eck S, Green C, et al.. Flow cytometry method validation protocols. Curr Protoc Cytom 2019, 87, e53. [DOI] [PubMed] [Google Scholar]
- 34. Aricescu AR, Lu W, Jones EY.. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr 2006, 62, 1243–50. [DOI] [PubMed] [Google Scholar]
- 35. Wang GC, Dash P, McCullers JA, Doherty PC, Thomas PG.. T cell receptor αβ diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection. Sci Transl Med 2012, 4, 128ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dull T, Zufferey R, Kelly M, et al.. A third-generation lentivirus vector with a conditional packaging system. J Virol 1998, 72, 8463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lissina A, Ladell K, Skowera A, et al.. Protein kinase inhibitors substantially improve the physical detection of T-cells with peptide-MHC tetramers. J Immunol Methods 2009, 340, 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee JW, Weiner RS, Sailstad JM, et al.. Method validation and measurement of biomarkers in nonclinical and clinical samples in drug development: a conference report. Pharm Res 2005, 22, 499–511. [DOI] [PubMed] [Google Scholar]
- 39. Piccoli S, Mehta D, Vitaliti A, et al.. 2019 White Paper on Recent Issues in Bioanalysis: FDA immunogenicity guidance, gene therapy, critical reagents, biomarkers and flow cytometry validation (Part 3 - Recommendations on 2019 FDA immunogenicity guidance, gene therapy bioanalytical challenges, strategies for critical reagent management, biomarker assay validation, flow cytometry validation & CLSI H62). Bioanalysis 2019, 11, 2207–44. [DOI] [PubMed] [Google Scholar]
- 40. Thomas JW. Antigen-specific responses in autoimmunity and tolerance. Immunol Res 2001, 23, 235–44. [DOI] [PubMed] [Google Scholar]
- 41. Altman JD, Moss PA, Goulder PJ, et al.. Phenotypic analysis of antigen-specific T lymphocytes. Science 1996, 274, 94–6. [PubMed] [Google Scholar]
- 42. der Strate BV, Longdin R, Geerlings M, et al.. Best practices in performing flow cytometry in a regulated environment: feedback from experience within the European Bioanalysis Forum. Bioanalysis 2017, 9, 1253–64. [DOI] [PubMed] [Google Scholar]
- 43. Du L, Grover A, Ramanan S, Litwin V.. The evolution of guidelines for the validation of flow cytometric methods. Int J Lab Hematol 2015, 37Suppl 1, 3–10. [DOI] [PubMed] [Google Scholar]
- 44. Green CL, Brown L, Stewart JJ, Xu Y, Litwin V, Mc Closkey TW.. Recommendations for the validation of flow cytometric testing during drug development: I instrumentation. J Immunol Methods 2011, 363, 104–19. [DOI] [PubMed] [Google Scholar]
- 45. O’Hara DM, Xu Y, Liang Z, Reddy MP, Wu DY, Litwin V.. Recommendations for the validation of flow cytometric testing during drug development: II assays. J Immunol Methods 2011, 363, 120–34. [DOI] [PubMed] [Google Scholar]
- 46. Ivison S, Malek M, Garcia RV, et al.. A standardized immune phenotyping and automated data analysis platform for multicenter biomarker studies. JCI Insight 2018, 3, e121867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Aleksic M, Liddy N, Molloy PE, et al.. Different affinity windows for virus and cancer-specific T-cell receptors: implications for therapeutic strategies. Eur J Immunol 2012, 42, 3174–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cole DK, Pumphrey NJ, Boulter JM, et al.. Human TCR-binding affinity is governed by MHC class restriction. J Immunol 2007, 178, 5727–34. [DOI] [PubMed] [Google Scholar]
- 49. Serra P, Garabatos N, Singha S, et al.. Increased yields and biological potency of knob-into-hole-based soluble MHC class II molecules. Nat Commun 2019, 10, 4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bentzen AK, Hadrup SR.. Evolution of MHC-based technologies used for detection of antigen-responsive T cells. Cancer Immunol Immunother 2017, 66, 657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dolton G, Tungatt K, Lloyd A, et al.. More tricks with tetramers: a practical guide to staining T cells with peptide-MHC multimers. Immunology 2015, 146, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Petersen J, Ciacchi L, Tran MT, et al.. T cell receptor cross-reactivity between gliadin and bacterial peptides in celiac disease. Nat Struct Mol Biol 2020, 27, 49–61. [DOI] [PubMed] [Google Scholar]
- 53. Rims C, Uchtenhagen H, Kaplan MJ, et al.. Citrullinated aggrecan epitopes as targets of autoreactive CD4+ T cells in patients with rheumatoid arthritis. Arthritis Rheumatol 2019, 71, 518–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pieper J, Dubnovitsky A, Gerstner C, et al.. Memory T cells specific to citrullinated α-enolase are enriched in the rheumatic joint. J Autoimmun 2018, 92, 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dey S, Vaidyanathan R, Reza KK, et al.. A microfluidic-SERSplatform for isolation and immuno-phenotyping of antigen specific T-cells. Sensor Actuat B-Chem 2019, 284, 281–8. [Google Scholar]
- 56. Nepom GT. MHC class II tetramers. J Immunol 2012, 188, 2477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sims S, Willberg C, Klenerman P.. MHC-peptide tetramers for the analysis of antigen-specific T cells. Expert Rev Vaccines 2010, 9, 765–74. [DOI] [PubMed] [Google Scholar]
- 58. Moris A, Teichgräber V, Gauthier L, Bühring HJ, Rammensee HG.. Cutting edge: characterization of allorestricted and peptide-selective alloreactive T cells using HLA-tetramer selection. J Immunol 2001, 166, 4818–21. [DOI] [PubMed] [Google Scholar]
- 59. Mallet-Designe VI, Stratmann T, Homann D, Carbone F, Oldstone MB, Teyton L.. Detection of low-avidity CD4+ T cells using recombinant artificial APC: following the antiovalbumin immune response. J Immunol 2003, 170, 123–31. [DOI] [PubMed] [Google Scholar]
- 60. Rosloniec EF, Whittington KB, Zaller DM, Kang AH.. HLA-DR1 (DRB1∗0101) and DR4 (DRB1∗0401) use the same anchor residues for binding an immunodominant peptide derived from human type II collagen. J Immunol 2002, 168, 253–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and protocols are available from corresponding author upon request.