Abstract
Rapid and accurate identification of enterococci at the species level is an essential task in clinical microbiology since these organisms have emerged as one of the leading causes of nosocomial infections worldwide. Vibrational spectroscopic techniques (infrared [IR] and Raman) could provide potential alternatives to conventional typing methods, because they are fast, easy to perform, and economical. We present a comparative study using phenotypic, genotypic, and vibrational spectroscopic techniques for typing a collection of 18 Enterococcus strains comprising six different species. Classification of the bacteria by Fourier transform (FT)-IR spectroscopy in combination with hierarchical cluster analysis revealed discrepancies for certain strains when compared with results obtained from automated phenotypic systems, such as API and MicroScan. Further diagnostic evaluation using genotypic methods—i.e., PCR of the species-specific ligase and glycopeptide resistance genes, which is limited to the identification of only four Enterococcus species and 16S RNA sequencing, the “gold standard” for identification of enterococci—confirmed the results obtained by the FT-IR classification. These results were later reproduced by three different laboratories, using confocal Raman microspectroscopy, FT-IR attenuated total reflectance spectroscopy, and FT-IR microspectroscopy, demonstrating the discriminative capacity and the reproducibility of the technique. It is concluded that vibrational spectroscopic techniques have great potential as routine methods in clinical microbiology.
Enterococci are opportunistic human pathogens. The two most important species, E. faecalis and E. faecium, which are considered part of the normal intestinal flora, are among the leading causes of nosocomial infections and may cause severe infections, including endocarditis and septicemia (16). These infections are often difficult to treat due to the increased antibiotic resistance associated with this organism (4, 27, 29). The recent increase of vancomycin-resistant E. faecium (VRE) strains in clinical isolates is especially a cause of serious concern, because this glycopeptide-type antibiotic often remains the last treatment available in life-threatening infections (31). The situation is further complicated by the fact that enterococci have developed a number of mechanisms for the transfer of resistance genes (1). Therefore perhaps the greatest threat posed by VRE comes not from these organisms themselves but from the potential that they could transfer their resistance genes to other more pathogenic gram-positive bacteria, thus creating a highly dangerous pathogen difficult to treat with currently available antibiotics (15). Furthermore, recent studies have revealed that the incidence of more unusual species such as E. durans, E. hirae, E. gallinarum and E. casseliflavus has increased significantly in clinically isolated enterococci (32). Overall, this has resulted in an increased need for rapid and accurate identification of enterococci at the species and subspecies level as a means of effectively assisting infection control and epidemiological studies.
For most clinical microbiological laboratories, the primary method of identifying Enterococcus strains relies on phenotypic characterization. However, various studies have shown that an unequivocal species identification of enterococci by phenotypic means is a challenging procedure that can take several days to accomplish because of the phenotypic and biochemical similarities between many enterococci (5). In addition, the automated systems currently in use often fail to accurately identify rare species (3, 6, 11, 23, 26). Molecular genetic techniques, such as randomly amplified polymorphic DNA analysis, intergenic ribosomal PCR, or other PCR-based methods targeting various genes, have been successfully used to identify enterococci at the species level (6, 12, 13, 21, 22, 28). Although these techniques are specific and sensitive, it is difficult to adapt them for use in routine laboratories due to their high costs and the requirement for highly skilled personnel. Infection control and epidemiological studies primarily require rapid and simple means of identifying and typing clinical isolates. As a consequence, a variety of approaches have been developed. The application of vibrational spectroscopic techniques (Fourier transform-infrared [FT-IR] and near-IR Raman spectroscopies) is such an approach which may provide a potential alternative to conventional methods. These techniques are rapid because little biomass is needed, significantly reducing culturing time. Vibrational spectroscopies are also easy to use and may become very cost-effective, because they enable considerable reduction in sample handling and use of reagents and do not require highly skilled personnel. These methods allow the discrimination of intact microbial cells without their destruction and produce complex biochemical fingerprint-like spectra which are reproducible and distinct for different microorganisms. IR and Raman spectroscopy measure molecular vibrations on the basis of the absorption (IR) or scattering (Raman) of IR or near-IR radiation interacting with a sample. The observed microbial IR or Raman spectra are a complex composition of many different vibrational modes of all the cell components, i.e., DNA, RNA, proteins, and membrane and cell wall components. The applicability of FT-IR spectroscopy in the field of microbiology has already been persuasively demonstrated (2, 8–10, 17–19, 25). Various studies have shown that vibrational spectroscopy provides sufficient resolution power to distinguish microbial cells at different taxonomic levels, even at the strain level (10). Raman spectroscopy of microorganisms is a relatively new and promising approach, since recent studies revealed that it is possible to discriminate among various microorganisms at the species level based on Raman spectra of 6-h-old microcolonies (14). The two vibrational spectroscopic techniques provide complementary information. The combined use of IR and Raman spectroscopy could therefore offer a more complete approach for analyzing intact bacterial cells.
The purpose of this study was to evaluate the discriminatory power of vibrational spectroscopic techniques for accurately typing enterococci at the species level in direct comparison with phenotypic and genotypic methods.
MATERIALS AND METHODS
Strains and growth conditions.
A collection of 18 Enterococcus strains were used in this study. Strains were either food isolates, clinical isolates, or from the collection of the Pasteur Institute (CIP, Paris, France) as summarized in Table 1. The strains were stored in cryovials containing a cryopreservative (MICROBANK [Mast Diagnostica, Reinfeld, Germany]) at −70°C until use. Strains were streaked onto agar plates using a three-quadrant streak pattern. All strains were subcultured on casein peptone-soy meal peptone (CASO) agar plates (Merck, Darmstadt, Germany) for 24 h. The growth temperature was 37 ± 2°C.
TABLE 1.
Enterococcus strains used in this studya
| Strain no. | API identification | Specimen source | Wardc |
|---|---|---|---|
| 1 | E. faecium | Urine | MED |
| 2 | E. hiraeb | Urine | MED |
| 3 | E. hiraeb | Hemoc | ICU |
| 4 | E. faecalis | Urine | MED |
| 5 | E. faecium | Urine | SUR |
| 6 | E. duransb | Food | ICU |
| 8 | E. duransb | Food | ICU |
| 9 | E. faecalis | Carriage | PED |
| 10 | E. faecalis | Food | ICU |
| 11 | E. faecalis | CIP | |
| 12 | E. faecium | CIP | |
| 13 | E. faecalis | CIP | |
| 14 | E. gallinarum | Carriage | ELD |
| 15 | E. faecium | Carriage | ELD |
| 16 | E. casseliflavus | Carriage | ELD |
| 17 | E. hiraeb | Carriage | MED |
| 18 | E. faecalis | Carriage | PED |
| 19 | E. faecium | Food | ICU |
Strains were isolated from different sources, such as food, clinical isolates, or strain collections.
Strain was reidentified.
Abbreviations: MED, medical; ICU, intensive care unit; SUR, surgery; PED, pediatric; ELD, elderly.
Sample preparation.
For the IR absorbance measurements, bacterial cells from 24-h-old cultures were harvested and prepared as described earlier (9, 10, 17). Briefly, small amounts of late-exponential-phase cells (∼10 to 60 μg [dry weight]) were carefully removed with a platinum loop from regions of confluent colony growth in the third quadrant of the culture plate and suspended in 80 μl of distilled water. An aliquot (35 μl) of the suspension was transferred to a ZnSe optical plate in a multisampling cuvette and dried in a desiccator over a drying agent (P4O10 [Sicapent]; Merck) with the application of a moderate vaccum (2.5 to 7.5 kPa) to form a transparent film suitable for FT-IR measurements. Prior to spectral measurements, the sample holder was sealed with a KBr cover plate to control the humidity and to prevent the instrument from contamination.
For Raman studies, a loopful of biomass from 24-h cultures was transferred onto CaF2 substrate. From each strain, duplicate smears were made. The smears on CaF2 substrate were dried in a desiccator over drying beads for at least 15 min, prior to Raman measurements.
For the IR attenuated total reflectance (ATR) measurements, cells from 18-h-old cultures were carefully harvested from the solid agar plate and homogeneously spread over the whole ATR crystal surface. The substrate used for ATR measurements was a ZnSe crystal (50 by 10 by 1.5 mm; Specac, Orpington, United Kingdom) with a refractive index of 2.4 and an incidence angle of 45°, yielding a total of six internal reflections at the sample.
For the IR microspectroscopic studies, the bacterial cultures were incubated for 8 to 10 h and produced colonies of approximately 100 to 250 μm in diameter. The microcolonies were transferred manually from the agar plate to an IR-transparent ZnSe optical plate by gently pressing the plate onto the agar surface. Imprints were allowed to air dry prior to spectral measurement. Spectra were acquired from the dried microcolony imprints on this substrate.
Recording of spectra and data evaluation. (i) FT-IR spectroscopy.
Spectra were recorded in the region between 500 and 4,000 cm−1 on an IFS 28/B spectrometer (Bruker Optics, Karlsruhe, Germany) specially designed for the measurement of microorganisms and equipped with a deuterated triglycerine sulfate detector. For each FT-IR spectrum, 64 scans were coadded and averaged. Fourier transformation was done using a Blackmann-Harris 3-term apodization function and a zerofilling factor of 4, to give a nominal resolution of 6 cm−1. The spectrometer was continuously purged by dry air to reduce contributions from water vapor and CO2.
Evaluation of IR spectral data (calculation of derivatives, normalization, etc.) was performed using OPUS software (version 3.0; Bruker). First and second derivatives of the original IR spectra were calculated using a 9-point Savitzky-Golay filter to enhance the resolution of superimposed bands and to minimize problems from unavoidable baseline shifts. Multivariate statistical analysis was carried out using the cluster analysis module of OPUS (version 3.0). To compare spectra of the six different species, cluster analyses using the first derivatives of the original spectra as input were carried out for different wave number regions. Spectral distances, providing a measure of the similarity of the spectra, calculated from Pearson's correlation coefficient and Ward's algorithm, were used for hierarchical clustering analysis.
(ii) Raman spectroscopy.
Raman measurements were performed as described earlier (14), using a confocal Raman microspectrometer. Briefly, bacterial smears were placed under a microscope objective and excited with 100 mW of laser power (830 nm). At random locations in each smear, 10 spectra, each with a 30-s signal integration time, were collected. The 10 spectra thus obtained were averaged before being used in further analysis.
Evaluation of Raman spectra was accomplished as already described for the IR data. First-derivative spectra consisting of the spectral region 400 to 1,800 cm−1 were used. Cluster analysis was also performed, considering four spectral regions (400 to 980, 1,020 to 1,140, 1,190 to 1,500 and 1,550 to 1,800 cm−1) in order to exclude the intensive spectral features that are caused by the carotenoids of the pigmented E. casseliflavus strain 16 and E. hirae strain 6.
(iii) FT-IR attenuated total reflectance (ATR) spectroscopy.
Spectra were recorded using a Bomem Mb-100 (Vannier, Quebec, Canada) FT-IR spectrometer equipped with a KBr beam splitter and a DTGS detector. One-hundred interferograms were averaged per spectrum, at a resolution of 4 cm−1. For each strain, 10 spectra were recorded and averaged.
(iv) FT-IR microspectrometry.
FT-IR absorption spectra were collected using a UMA 500 IR microscope coupled to an FTS-40A spectrometer (Spectroscopy Division, Bio-Rad Cambridge, Mass.) equipped with a mercury cadmium telluride narrow-band detector. A microscope diaphragm size of 80 by 80 μm was used for spectral data acquisition. Measurements were performed in transmission mode using the following parameters: 4-cm−1 resolution, 5-kHz scan speed, 32 to 64 scans of coaddition, triangular apodization, and spectral range of 800 to 4,000 cm−1. No baseline correction or smoothing was applied to the data. First-derivative spectra were subjected to cluster analysis using Matlab's Statistics Toolbox (The Math Works, Inc., Natick, Mass.) employing Ward's algorithm and Euclidean distance measure.
Phenotypic and genotypic methods. (i) Phenotypic method: API and MICROSCAN.
Phenotypic identification of all strains was performed using the automated API (bioMérieux, Marcy I'Etoile, France) and the MicroScan (Dade International, MicroScan Inc., West Sacramento, Calif.) systems.
(ii) Genotypic methods. (a) PCR.
PCR analyses of species-specific ligase genes (ddl) and related glycopeptide enzymes were performed according to the method of Dukta-Malen et al. (7) for all Enterococcus strains investigated in this study.
(b) 16S RNA sequencing.
16S RNA sequencing was performed for the five equivocally typed strains, i.e., strains 2, 3, 6, 8, and 17, as well as for strains 10, 15, and 19. 16S RNA sequencing was performed using the MicroSeq 500 sequencing kit (Perkin-Elmer). The sequence data were analyzed with a Genetic ABI PRISM 310 sequencer (Perkin-Elmer).
RESULTS AND DISCUSSION
Phenotypic identification by the API test system.
Conventional identification was performed for all 18 strains used for vibrational spectroscopic analyses. Of the 18 isolates studied, five were identified as E. faecium (strains 1, 5, 12, 15, and 19) and six were identified as E. faecalis (strains 4, 9, 10, 11, 13, and 18). Three isolates were identified as E. hirae (strains 2, 3, and 17), and two were identified as E. durans (strains 6 and 8). Of the remaining two isolates one was identified as E. gallinarum (strain 14) and one was identified as E. casseliflavus (strain 16) (Table 1).
Comparison between phenotypic identification and FT-IR analysis.
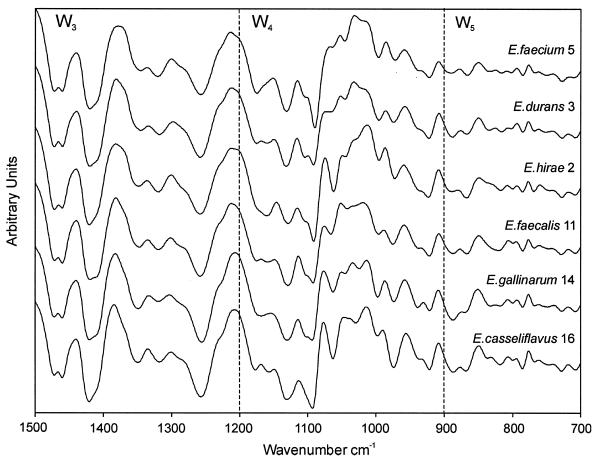
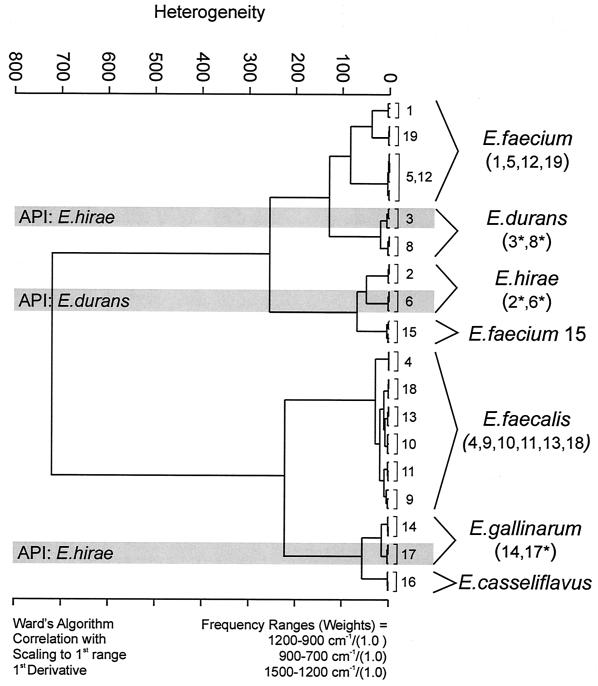
Seven repetitive measurements over a period of 6 months from independent sample preparations of all strains were performed, resulting in 126 spectra. The repetitive measurements, performed to judge the reproducibility of the IR technique, yielded strain-specific subclusters (data not shown for brevity), indicating that this method is highly reproducible and specific at the strain level. A representative data set consisting of three repetitive measurements including all strains was subjected to multivariate statistical analysis to explore the discriminative potential of the spectral information for this taxonomic task. Typical first-derivative IR spectra of the six different species plotted for the spectral ranges used to calculate spectral distances are displayed in Fig. 1. This figure reveals the spectral differences responsible for the separation of the six Enterococcus species, which, however, are not yet interpretable in terms of biochemical and/or chemical structures. Hierarchical clustering based on spectral information contained in three different spectral ranges—1,200 to 900 cm−1, the polysaccharide region; 900 to 700 cm−1, the fingerprint region; and 1,500 to 1,200 cm−1, the mixed region—was used as a classification method, resulting in the dendrogram displayed in Fig. 2. A clear discrimination could be observed and 6 distinct clusters were produced. However, the grouping of some strains was in contradiction to the routine phenotypic classification. Most noticeable is the separation between the two major enterococcal species into two clusters, with all but one E. faecium strain (strain 15) in one group and all E. faecalis strains (strains 4, 9, 10, 11, 13, and 18) in the other group. Upon closer inspection the inconsistencies with the phenotypic identification become apparent. The dendrogram suggests a close relatedness between strains 2 and 6 and also between strains 3 and 8, which is in contradiction to the phenotype-based identification results. Accordingly, the findings indicate either that the FT-IR classification is not useful for the differentiation of enterococci or that the four strains have not been typed accurately by the conventional methods. Further discrepancies between FT-IR and conventional methods were also observed for strain 17, which was typed as E. hirae by the API method but clustered together with E. gallinarum (strain 14) in the FT-IR analysis. From these findings we hypothesized that the identification of rare Enterococcus species by conventional identification systems like the API system might not be reliable, as described before (11, 27). In fact, studies evaluating the commonly used commercially available bacterial identification systems have repeatedly encountered problems associated with enterococcal species identification (23, 27). Error rates for enterococcal species identification of 2 to 21% for E. faecalis, 5 to 9% for E. faecium, and 14 to 79% for other species have been found for these systems (32). To give an example, Singer and coworkers reported a high percentage of misidentifications for the analysis of isolates from a VRE outbreak in a hospital after the introduction of an automated identification system software update (Vitek gram-positive identification card) (24). He concluded from the study that “automated microbial analysis is a potential source of error that is not easily recognized” (24).
FIG. 1.
Typical first-derivative IR spectra of the six different Enterococcus species depicted in the most-discriminatory spectral windows. The spectral windows are defined according to the classification of Helm et al. (10) as follows: W3, the window between 1,500 and 1,200 cm−1 (the mixed region), a spectral region containing information from proteins, fatty acids, and phosphate-carrying compounds; W4, the window between 1,200 and 900 cm−1 (the polysaccharide region), a spectral region dominated by the fingerprint-like absorption bands of the carbohydrates present within the cell wall; W5, the window between 900 and 700 cm−1 (the true fingerprint region), showing some remarkably specific spectral patterns, which are as yet unassigned to cellular components or to functional groups.
FIG. 2.
Classification scheme based on the FT-IR spectra of six different Enterococcus species. Cluster analysis of three repetitive measurements was performed using the first derivatives of the spectra, considering the spectral ranges from 1,200 to 900, 900 to 700, and 1,500 to 1,200 cm−1. All spectral ranges were equally weighted. Ward's algorithm was applied. The strains marked with an asterisk were not in accordance with the phenotypic identification by the API system. Shading highlights the identity of certain strains by the API system.
Species identification by Microscan, PCR, and 16S RNA sequencing.
To clarify the discrepancy between FT-IR spectroscopic and phenotypic classifications, further diagnostic evaluation, using another phenotype-based automated test (namely, MicroScan) and a species-specific PCR approach, were undertaken for all 18 strains. Only the identification results of the five equivocally typed strains (strains 2, 3, 6, 8, and 17) are summarized in Table 2. All the other strains yielded uniform identification results by the various methods used. As for the first automated test system (API), the MicroScan system also failed in identifying these obviously rare species unequivocally. All the analyzed strains were identified as E. durans, producing results in contradiction to both the API and the FT-IR classification. Subsequently, PCR analyses of the species-specific genes coding for the d-alanine:d-alanine (d-Ala:d-Ala) ligases and related glycopeptide resistance enzymes was performed for all 18 strains. This technique, although reliable, identifies only four species, i.e., E. faecium, E. faecalis, E. gallinarum, and E. casseliflavus, leaving the other Enterococcus species unidentified (7). Therefore, only 14 of the 18 strains investigated could be identified. The analysis produced an outcome in good agreement with the FT-IR classification results. Particularly interesting is the identification of one of the five equivocally typed strains (strain 17) as E. gallinarum by the PCR approach in accordance with the FT-IR analysis (strain 17 is grouped in one cluster with strain 14, an E. gallinarum species) (Fig. 2).
TABLE 2.
Identification of five equivocally typed strains based on phenotypic, genotypic, and vibrational spectroscopic data
| Strain no. | Identification by:
|
||||
|---|---|---|---|---|---|
| API | FT-IR and Raman clusteringa | MicroScan | PCR | 16S RNA sequencing | |
| 2 | E. hirae | 6 | E. durans | —b | E. hirae |
| 3 | E. hirae | 8 | E. durans | — | E. durans |
| 6 | E. durans | 2 | E. durans | — | E. hirae |
| 8 | E. durans | 3 | E. durans | — | E. durans |
| 17 | E. hirae | 14 (E. gallinarum) | E. durans | E. gallinarum | E. gallinarum |
Numbers indicate with which strain the indicated strain clustered.
—, not identifiable by PCR.
For the five Enterococcus species whose phenotypic and genotypic identification differed from the FT-IR identification, 16S RNA sequencing was performed to assess the accuracy of all methods involved. The 16S RNA sequencing is considered to be the “gold standard” for microbial identification. It is particularly encouraging that of the results of all methods used in this study only the FT-IR result was in accordance with the identification by the gold standard (Table 1). Specifically, 16S RNA sequencing identified strains 2 and 6 as E. hirae and strains 3 and 8 as E. durans, providing strong support for the FT-IR classification, which yielded two distinct clusters for these species. Furthermore, strain 17, originally classified as E. hirae, was determined to be E. gallinarum, again confirming the FT-IR analysis and the PCR result. Therefore, these findings demonstrate the superior discrimination ability of the FT-IR technique on the one hand and on the other hand demonstrate the weakness of the API and the MicroScan systems.
Classification by Raman spectroscopy.
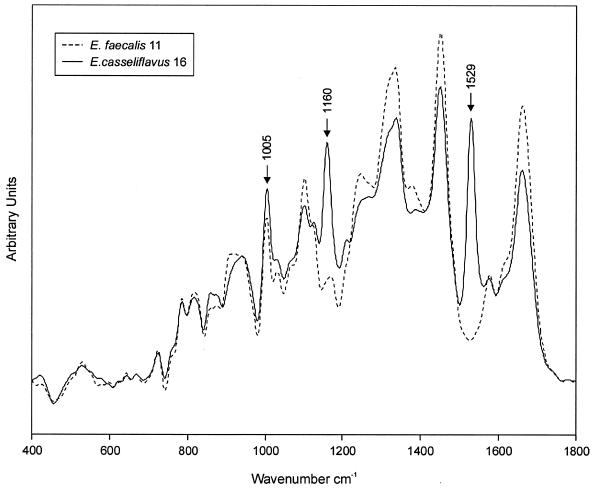
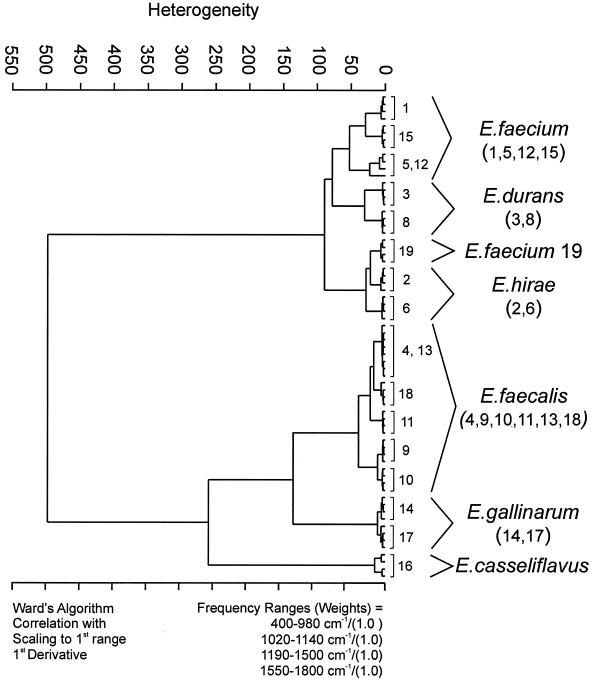
The classification results shown in Fig. 2 could be reproduced by three other laboratories using confocal near-IR–Raman microspectroscopy, FT-IR ATR spectroscopy, and FT-IR microspectroscopy. Raman spectroscopy produced correct species differentiation for all but one strain (E. hirae strain 6) when the analysis was performed considering the whole spectral range from 400 to 1,800 cm−1 (data not shown). Interestingly, this Raman dendrogram suggested, in contrast to the FT-IR dendrogram, that the E. casseliflavus strain shows only little similarity to the other strains. This finding can be attributed to additional peaks that occur at 1,005, 1,160, and 1,529 cm−1 in the Raman spectrum of the strain as shown in Fig. 3. Specific bacterial constituents such as pigments give rise to these additional Raman signals, which gain considerable intensity due to a preresonance effect. According to the diagnostic bands near 1,160 cm−1 [ν(C—C)] and 1,529 cm−1 [ν(C⩵C)] (ν, stretching vibrations), the yellow-to-orange pigment compound of E. casseliflavus strain 16 can be assigned to a carotenoid structure (30). Upon closer spectral inspection, the different clustering of E. hirae strain 6 by the Raman approach could be ascribed to the fact that this strain expresses low levels of carotenoid as well. On the basis of these findings we performed a cluster analysis of the Raman spectra, excluding the carotenoid-specific regions. This could be achieved by using the spectral information encoded in four spectral regions (400 to 980, 1020 to 1140, 1190 to 1500, and 1550 to 1800 cm−1) as input data. The dendrogram (Fig. 4) obtained on the basis of this wavelength selection is comparable to the FT-IR dendrogram. It was also very encouraging that replicate cultures of all strains turned out to be in strain-specific subclusters, illustrating the excellent reproducibility and strain specificity of the Raman technique, as was found with the IR measurements.
FIG. 3.
Raman spectra of E. casseliflavus strain 16 and E. faecalis strain 11 are depicted for spectral comparison. The Raman spectrum of the pigmented E. casseliflavus exhibits preresonance-enhanced bands of carotenoids. Three characteristic bands near 1005, 1155, and 1529 cm−1 are clearly visible.
FIG. 4.
Classification scheme based on the Raman spectra of six different Enterococcus species. Cluster analysis of four repetitive measurements was performed using the first derivatives of the spectra, considering the spectral ranges from 400 to 980, 1,020 to 1,140, 1,190 to 1500, and 1,500 to 1,800 cm−1, with the aim of excluding the spectral features that are caused by the carotenoid pigmentation of E. casseliflavus strain 16 and E. hirae strain 6. All spectral ranges were equally weighted. Ward's algorithm was applied.
However, both spectroscopic techniques did not clearly classify all five E. faecium species. A closer inspection of the E. faecium species group (20) shown in Fig. 2 demonstrates that the E. faecium strains cluster in two groups consisting of all but one strain (strain 15) for the IR data. This strain seems to be more closely related to the E. hirae strains (strains 2 and 6) than to the other E. faecium strains. Similarly for the clustering of the Raman data, again the E. faecium strains cluster into two groups; however, in this case, it is strain 19 that clusters apart. To further investigate the findings, isolates 15 and 19 were retyped by 16S RNA sequencing. The sequencing revealed high sequence identities for E. faecium, E. hirae, and E. durans, indicating a high relatedness within this group.
In conclusion we have shown the potential usefulness of vibrational spectroscopic techniques in the differentiation of enterococci from various sources, including isolates from food, patient material, and strain collections. The results of our study reflect the high discriminatory power of the IR and the Raman technique that allows accurate differentiation of closely related bacterial species such as enterococci. Comparison of FT-IR and Raman clustering showed that there was considerable consistency between both methods, since very similar classification schemes were obtained. This is most encouraging considering that IR and Raman methods “see” the total cell composition and structure on the basis of different molecular vibrational modes. In addition, both spectroscopic techniques proved to be capable of discriminating accurately at the strain level, which opens the door for using these physicochemical techniques as tools for epidemiological studies. In comparison to conventional automated identification systems which have been confirmed to be reliable only for the more common clinical isolates such as E. faecalis and E. faecium, the FT-IR and Raman methods proved to be also applicable for less frequently encountered Enterococcus species, for instance E. hirae and E. durans. Moreover, our study indicates that vibrational spectroscopic techniques might not only be superior to conventional phenotypic methods but also are more broadly applicable than genotypic identification by means of one or even a few very specific PCR analyses which are limited to the identification of only four Enterococcus species. Finally, the species differentiation based on the spectroscopic data is consistent only with the analysis results obtained by the 16S RNA sequencing. Though regarded as the gold standard, 16S RNA sequencing is not appropriate for routine analysis, due to its complexity and high costs. Because of this vibrational spectroscopy is not only advantageous as a tool for taxonomic studies but also proves to be very rapid and reliable as a potential routine classification method.
ACKNOWLEDGMENT
We gratefully acknowledge financial support from the European Union Biomed II program, project BMH4-97-2054.
REFERENCES
- 1.Bodnar U R, Noskin G A, Suriano T, Cooper I, Reisberg B E, Peterson L R. Use of in-house studies of molecular epidemology and full species identification for controlling spread of vancomycin-resistant Enterococcus faecalis isolates. J Clin Microbiol. 1996;34:2129–2132. doi: 10.1128/jcm.34.9.2129-2132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouhedja W, Sockalingum G D, Pina P, Allouch P, Bloy C, Labia R, Millot J M, Manfait M. ATR-FTIR spectroscopic investigation of E. coli transconjugants beta-lactams-resistance phenotype. FEBS Lett. 1997;412:39–42. doi: 10.1016/s0014-5793(97)00725-4. [DOI] [PubMed] [Google Scholar]
- 3.Bryce E A, Zemcov S J, Clarke A M. Species identification and antibiotic resistance patterns of the enterococci. Eur J Clin Microbiol Infect Dis. 1991;10:745–747. doi: 10.1007/BF01972500. [DOI] [PubMed] [Google Scholar]
- 4.Buschelman B J, Bale M J, Jones R N. Species identification and determination of high-level aminoglycoside resistance among enterococci. Comparison study of sterile body fluid isolates, 1985–1991. Diagn Microbiol Infect Dis. 1993;16:119–122. doi: 10.1016/0732-8893(93)90005-r. [DOI] [PubMed] [Google Scholar]
- 5.Cheng S, McCleskey F-K, Gress M J, Petroziello J M, Liu R, Namdari H, Beninga K, Salmen A, DelVecchio V G. A PCR assay for identification of Enterococcus faecium. J Clin Microbiol. 1997;35:1248–1250. doi: 10.1128/jcm.35.5.1248-1250.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devriese L A, Pot B, Van Damme L, Kersters K, Haesebrouck F. Identification of Enterococcus species isolated from foods of animal origin. Int J Food Microbiol. 1995;26:187–197. doi: 10.1016/0168-1605(94)00119-q. [DOI] [PubMed] [Google Scholar]
- 7.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodacre R, Timmins E M, Rooney P J, Rowland J J, Kell D B. Rapid identification of Streptococcus and Enterococcus species using diffuse reflectance-absorbance Fourier transform infrared spectroscopy and artifical neural networks. FEMS Microbiol Lett. 1996;140:233–239. doi: 10.1016/0378-1097(96)00186-3. [DOI] [PubMed] [Google Scholar]
- 9.Helm D, Labischinski H, Naumann D. Elaboration of a procedure for identification of bacteria using Fourier-Transform IR spectral libraries: a stepwise correlation approach. J Microbiol Methods. 1991;14:127–142. [Google Scholar]
- 10.Helm D, Labischinski H, Schallehn G, Naumann D. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. J Gen Microbiol. 1991;137:69–79. doi: 10.1099/00221287-137-1-69. [DOI] [PubMed] [Google Scholar]
- 11.Iwen P C, Kelly D M, Linder J, Hinrichs S H. Revised approach for identification and detection of ampicillin and vancomycin resistance in Enterococcus species by using MicroScan panels. J Clin Microbiol. 1996;34:1779–1783. doi: 10.1128/jcm.34.7.1779-1783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones R N, Marshall S A, Pfaller M A, Wilke W W, Hollis R J, Erwin M E, Edmond M B, Wenzel R P. Nosocomial enterococcal blood stream infections in the SCOPE Program: antimicrobial resistance, species occurrence, molecular testing results, and laboratory testing accuracy. SCOPE Hospital Study Group. Diagn Microbiol Infect Dis. 1997;29:95–102. doi: 10.1016/s0732-8893(97)00115-6. [DOI] [PubMed] [Google Scholar]
- 13.Ke D, Picard F, Martineau F, Menard C, Roy P, Ouellette M, Bergeron M. Development of a PCR assay for rapid detection of enterococci. J Clin Microbiol. 1999;37:3497–3503. doi: 10.1128/jcm.37.11.3497-3503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maquelin K, Choo-Smith L-P, van Vreeswijk T, Endtz H P, Smith B, Bennett R, Bruining H A, Puppels G J. Raman spectroscopic method for identification of clinically relevant microorganisms growing on solid culture medium. Anal Chem. 2000;72:12–19. doi: 10.1021/ac991011h. [DOI] [PubMed] [Google Scholar]
- 15.Moellering R C. Vancomycin-resistant enterococci. Clin Infect Dis. 1998;26:1196–1199. doi: 10.1086/520283. [DOI] [PubMed] [Google Scholar]
- 16.Monstein H-J, Quednau M, Samuelsson A, Ahrne S, Isaksson B, Jonasson J. Division of the genus Enterococcus into species groups using PCR-based molecular typing methods. Microbiology. 1998;144:1171–1179. doi: 10.1099/00221287-144-5-1171. [DOI] [PubMed] [Google Scholar]
- 17.Naumann D, Helm D, Labischinski H. Microbiological characterizations by FT-IR spectroscopy. Nature. 1991;351:81–82. doi: 10.1038/351081a0. [DOI] [PubMed] [Google Scholar]
- 18.Naumann D, Labischinski H. The characterization of microorganisms by Fourier-transform infrared spectroscopy. In: Nelson W H, editor. Modern techniques for rapid microbiological analysis. New York, N.Y: VCH-Publisher; 1991. pp. 43–96. [Google Scholar]
- 19.Naumann D, Schultz C P, Helm D. What can infrared spectroscopy tell us about the structure and composition of intact bacterial cell? In: Mantsch H H, Chapmann D, editors. Infrared spectroscopy of biomolecules. New York, N.Y: Wiley-Liss; 1996. pp. 279–310. [Google Scholar]
- 20.Ozawa Y, Courvalin P, Galimand M. Identification of enterococci at the species level by sequencing of the genes for D-alanine:D-alanine ligases. Syst Appl Microbiol. 2000;23:230–237. doi: 10.1016/s0723-2020(00)80009-0. [DOI] [PubMed] [Google Scholar]
- 21.Pryce T M, Wilson R D, Kulski J K. Identification of enterococci by ribotyping with horseradish-peroxidase-labelled 16S rDNA probes. J Microbiol Methods. 1999;36:147–155. doi: 10.1016/s0167-7012(99)00026-3. [DOI] [PubMed] [Google Scholar]
- 22.Roger M, Faucher M-C, Forest P, St.-Antoine P, Coutlee F. Evaluation of a vanA-Specific PCR assay for detection of vancomycin-resistant Enterococcus faecium during a hospital outbreak. J Clin Microbiol. 1999;37:3348–3349. doi: 10.1128/jcm.37.10.3348-3349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sader H S, Biedenbach D, Jones R N. Evaluation of Vitek and API 20S for species identification of enterococci. Diagn Microbiol Infect Dis. 1995;22:315–319. doi: 10.1016/0732-8893(95)00146-5. [DOI] [PubMed] [Google Scholar]
- 24.Singer D, Jochimsen E M, Gielerak P, Jarvis W R. Pseudo-outbreak of Enterococcus durans infections and colonization associated with introduction of an automated identification system software update. J Clin Microbiol. 1996;34:2685–2687. doi: 10.1128/jcm.34.11.2685-2687.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sockalingum G W, Bouhedja W, Pina P, Allouch P, Mandray C, Labia R, Millot J M, Manfait M. ATR-FTIR spectroscopic investigation of imipenem-susceptible and -resistant Pseudomonas aeruginosa isogenic strains. Biochem Biophys Res Commun. 1997;232:240–246. doi: 10.1006/bbrc.1997.6263. [DOI] [PubMed] [Google Scholar]
- 26.Tritz D M, Iwen P C, Woods G L. Evaluation of MicroScan for identification of Enterococcus species. J Clin Microbiol. 1990;28:1477–1478. doi: 10.1128/jcm.28.6.1477-1478.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsakris A, Woodford N, Pournaras S, Kaufmann M, Douboyas J. Apparent increased prevalence of high-level aminoglycoside-resistant Enterococcus durans resulting from false identification by a semiatomated software system. J Clin Microbiol. 1998;36:1419–1421. doi: 10.1128/jcm.36.5.1419-1421.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyrrell G J, Bethune R N, Willey B, Low D E. Species identification of enterococci via intergenic ribosomal PCR. J Clin Microbiol. 1997;35:1054–1060. doi: 10.1128/jcm.35.5.1054-1060.1997. . (Erratum, 35:2434.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vincent S, Knight R G, Green M, Sahm D F, Shlaes D M. Vancomycin susceptibility and identification of motile enterococci. J Clin Microbiol. 1991;29:2335–2337. doi: 10.1128/jcm.29.10.2335-2337.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner W D. Raman excitation profiles from pigments in vivo. J Raman Spectros. 1986;17:51. [Google Scholar]
- 31.Wilke W W, Marshall S A, Coffman S L, Pfaller M A, Edmund M B, Wenzel R P, Jones R N. Vancomycin-resistant Enterococcus raffinosus: molecular epidemiology, species identification error, and frequency of occurrence in a national resistance surveillance program. Diagn Microbiol Infect Dis. 1997;29:43–49. doi: 10.1016/s0732-8893(97)00059-x. [DOI] [PubMed] [Google Scholar]
- 32.Willey B M, Jones R N, McGeer A, Witte W, French G, Roberts R B, Jenkins S G, Nadler H, Low D E. Practical approach to the identification of clinically relevant Enterococcus species. Diagn Microbiol Infect Dis. 1999;34:165–171. doi: 10.1016/s0732-8893(99)00032-2. [DOI] [PubMed] [Google Scholar]