Abstract
Specific adhesion (sequestration) of Plasmodium falciparum parasite-infected erythrocytes (IEs) in deep vascular beds can cause severe complications resulting in death. This review describes our work on the discovery, characterization, and optimization of novel inhibitors that specifically prevent adhesion of IEs to the host vasculature during severe malaria, especially its placental and cerebral forms. The main idea of using anti-adhesion drugs in severe malaria is to release sequestered parasites (or prevent additional sequestration) as quickly as possible. This may significantly improve the outcomes for patients with severe malaria by decreasing local and systemic inflammation associated with the disease and reestablishing the microvascular blood flow. To identify anti-malarial adhesion-inhibiting molecules, we have developed a high-throughput (HT) screening approach and found a number of promising leads that can be further developed into anti-adhesion drugs providing an efficient adjunct therapy against severe forms of malaria.
Keywords: Plasmodium falciparum, severe malaria, cytoadhesion, PfEMP1 proteins, high-throughput screening, anti-adhesion compounds
INTRODUCTION
Malaria caused by the parasite Plasmodium falciparum is one of the major devastating diseases worldwide, with half a billion cases and up to 0.5-1 million deaths, mostly among young children, yearly. According to the World Health Organization, most deaths result from severe malaria (SM) complications, the major forms of which include cerebral malaria (CM), placental malaria (PM), respiratory distress, and severe anemia [1]. Currently, there is a noticeable decline in the efficacy of commonly used anti-malarial drugs. It should be noted that although still efficient anti-malarial drugs rapidly kill the parasite, significant mortality (10-15%) ensues in SM, particularly within 24 h of hospital admission. This happens most likely due to the cytoadherence of infected erythrocytes (IEs) that continues long after the parasites inside the red blood cells have been killed [2]. Unfortunately, novel parasite-killing drugs will have the same problem if they do not affect the cytoadhesion of IEs. This strongly justifies the need for the development adjunctive therapies able to prevent and/or reverse the pathophysiological consequences of cytoadherence in SM. Our main objective was to discover, characterize, and optimize novel inhibitors of cytoadhesion of P. falciparum-infected erythrocytes to the host vasculature for the treatment of severe forms of malaria (including CM and PM). Below, we review our works in this area.
ADVANTAGES OF ANTI-ADHESION DRUGS IN SEVERE MALARIA
Most anti-malarial drugs target intracellular processes required for the parasite survival and propagation. Such drugs must penetrate into the parasite by crossing several membranes, including erythrocyte plasma membrane, parasitophorous vacuolar membrane, and parasite plasma membrane. These drugs also have potential side effects, since they may similarly penetrate into the host cells and interfere with the host cellular functions.
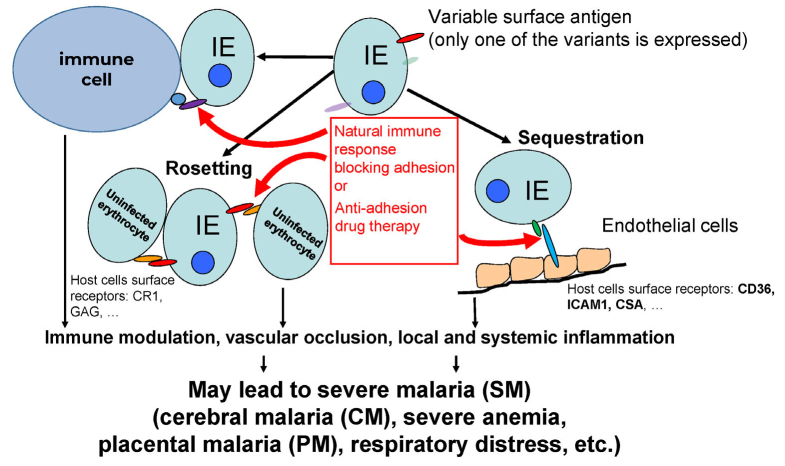
Cytoadherence plays an important role in the lifecycle and virulence of P. falciparum (reviewed in [3]), the deadliest of human malaria parasites, and is a cause of PM [4] and CM [5-7] (figure). It is reasonable to suggest that reducing cytoadherence of IEs to the host vasculature will have a strong beneficial effect on the host defense against the infection, a hypothesis that can only be tested experimentally. A simple fact that the appearance of anti-adhesion antibodies preventing IE cytoadhesion to the placental syncytiotrophoblast in multigravid women is associated with protection against PM pathology [8, 9] indicates that the anti-adhesion therapy might be effective against at least this severe disease (figure). The anti-adhesion activity is considered as an important factor in the development of successful vaccine against PM [10]. Furthermore, we found that >75% immune adults have high levels of antibodies inhibiting adhesion to the ICAM-1 receptor [11] involved in SM and CM (see below). This anti-adhesion activity may contribute (along with the ability to opsonize IEs) to the fast recovery of severely sick children injected with IgG from immune adults [12]. The time window for the pharmacological effect of drugs is extremely critical for the survival in SM. The anti-adhesion agents working from outside of IEs, will likely have a higher speed of action. In this respect, molecules with a limited ability to penetrate through the plasma membranes and, therefore, to interfere with the host functions, might be especially safe, although they may need to be delivered intravenously, which is well justified in the case of life-threatening SM episodes. This is extremely important in the treatment of malaria during pregnancy, as the safety of both mother and fetus is the main issue with currently used drugs [13]. Moreover, as IEs bind to functionally relevant parts of host receptors and may inhibit their normal physiological functions [14], anti-adhesion drugs would have an additional positive effect on the host. In our opinion, as well as in the opinion of other researchers, anti-adhesion drugs may be very efficient and safe for the anti-malarial therapy [15, 16] when used as an adjunctive therapy together with parasiticidal drugs (figure). If, by chance, an anti-adhesion drug also has a parasiticidal effect [16], it would be even more valuable.
Role of IE cytoadhesion mediated by the interactions of PfEMP1 proteins with host receptors in the pathology of malaria and its implications for anti-adhesion therapy
CYTOADHESION OF IEs IS MEDIATED BY A FAMILY OF PfEMP1 PROTEINS
P. falciparum is the only malaria parasite species that was clearly shown to efficiently sequester IEs in various organs using host cell surface receptors for adhesion to endothelial cells, uninfected red blood cells, and cells of immune system [17-21]. Cytoadhesion allows IEs to avoid clearance in the spleen (main innate anti-malarial mechanism). However, cytoadhesion also leads to the vascular occlusion that strongly correlates with disease severity [22] and inflammation of endothelial cells and various organs [23-28]. The main parasite ligand for the IE adhesion is a family (~60 members) of PfEMP1 proteins [29-31] that are expressed on the surface of IEs in specialized structures called knobs [32]. No parasite co-receptor molecules required for the IE adhesion have been discovered so far. PfEMP1 proteins are coded by var genes. Mature parasite expresses on the surface the product of only one of the var genes [33], but can switch to another variant, thus changing its antigenic and adhesion phenotype [30, 34] (figure). Each PfEMP1 protein consists of one intracellular, one trans-membrane, and several extracellular domains (~30-45 kDa, 2 to 7 domains per protein) heavily cross-linked by disulfide bonds [31, 35]. These domains have been classified using Greek letters for classes and numbers for sub-classes into Duffy binding-like (DBL) domains (α, β, γ, δ, ε, ζ, Χ) and cysteine-rich interdomain regions (CIDRs) (α, β, and γ) [31, 35, 36]. All DBL domains have a similar scaffold [37-40], and the C-terminal part of the CIDR scaffold is similar to the C-terminal part of the DBL domain [41]. These domains are responsible for the adhesion to various host receptors (reviewed in [42]), each domain representing a functional binding (with higher or lesser efficiency) unit. The specificity and combination of these domains in the expressed PfEMP1 may determine the final IE adhesion specificity, strength of binding, and, possibly, disease pathology [43]. We have cloned and expressed a library of PfEMP1 constructs (n > 180) that contained all domains (n = 272) or domain tandems (for example, many DBL domains are followed by the CIDR domains) from all PfEMP1 proteins (n = 59) from the NF54 parasite line, in their functional (receptor-binding) form, as has been demonstrated in a number of our publications [11, 44-49]. This proteome-wide collection of PfEMP1 constructs is an important tool for identification of various novel receptor-domain pairs and screening for anti-adhesion compounds against different binding IE specificities. Although the members of the PfEMP1 family belong to the variable class proteins (the sequences vary from parasite to parasite in the field), such variability may be not an insurmountable problem, as we discuss below.
SEVERE MALARIA SYNDROMES AND CYTOADHESION
As we mentioned above, cytoadhesion of IEs is either a cause or a prerequisite for SM, including CM [5-7, 27] and PM [4, 50]. The most studied syndrome is PM. Only one member of the PfEMP1 family, VAR2CSA, is involved in PM [51-53]. It has a characteristic sequence of three DBL-X domains followed by three DBLε domains. This architecture of VAR2CSA is conserved in all studied P. falciparum strains. Anti-adhesion antibodies, that appear after one or two successive pregnancies in malaria endemic regions, alleviate PM by inhibiting IE binding to the CSA receptor in the placenta [54]. Common drugs are not particularly effective in the prevention of PM and might be even harmful [13]. In light of this, a drug that does not penetrate cell membranes and, consequently, mother and fetus tissues, but interferes with the parasite adhesion (the key event in PM) would be particularly safe for the use in pregnant women.
As has been shown by us and other researchers, women that acquire resistance to pregnancy malaria, develop antibodies to VAR2CSA over successive pregnancies [44, 50]. Identification of a monoclonal antibody inhibiting the binding of several strains of PM parasites to the host ligand CSA indicates that the conserved features of these variable proteins are sufficient to provide a wide cross-strain effectiveness [55]. Such conserved structures of the VAR2CSA domains could be the targets for anti-adhesion compounds through the competitive or allosteric mode of action.
The main candidate for the host adhesion receptor in CM is ICAM-1 [6, 7, 27, 56]. However, it was suggested recently that PfEMP1 proteins containing a tandem of domains biding to ICAM-1 and endothelial protein C receptor (EPCR) are involved predominantly in CM pathogenesis [57, 58], potentially binding to one or both receptors [59, 60]. Only six DBLβ domains (out of more than 250 domains from ~60 PfEMP1 proteins) in P. falciparum IT4 strain have been identified as capable of binding to ICAM-1 [61], as well as only two DBLβ domain in the 3D7 strain [11, 62]. Similarly, only a limited set of domains in each genome variant bound EPCR [63, 64]. The fact that only a small number of PfEMP1 domains strongly bind to either of these receptors fits well with the epidemiology of CM, which is a rare but extremely deadly SM syndrome. Extensive epidemiological data [65, 66] support the hypothesis that only a limited number of parasite lines, possibly expressing particular variants of PfEMP1 (and containing particular domains), may be the source of severe disease, as it happens in PM.
VASCULAR INFLAMMATION IS A COMMON RESPONSE TO IE CYTOADHESION AND A KEY EVENT IN SEVERE MALARIA
Several studies have clearly demonstrated that IE cytoadhesion directly affects vascular inflammation and endothelial dysfunction [67, 68]. Vascular inflammation contributes to the disease severity [69], plays a key role in CM and PM, and is characterized by induction of adhesion molecules (e.g., ICAM-1) and pro-inflammatory cytokines, as well as by vascular permeability caused by the disruption of the endothelial barrier integrity [70-72]. Some of these inflammatory processes are initiated and stimulated by direct interactions between PfEMP1 domains and host receptors through a complex intracellular signaling networks [25, 26]. Consequently, anti-adhesion drugs that prevent or reverse these interactions may reduce inflammation, thus contributing to a better outcome of SM syndromes. Our recent preliminary results demonstrate that the anti-adhesion treatment may reduce vascular inflammation, although further studies on this topic are required to support this observation.
VARIABILITY OF PfEMP1 FAMILY MEMBERS IN THE FIELD AS A CHALLENGING FACTOR
The sequences of PfEMP1 family members significantly vary in the field. Nevertheless, this variability does not preclude specific groups of domains from binding to the corresponding conserved receptors. This was corroborated by the recent structural study that revealed conserved binding pockets in variant PfEMP1 domains that bind to the conserved receptors [14]. In this regard, all DBL and CIDR domains have a similar scaffold [37-41]. Theoretically, molecules that mimic the binding motifs of host receptors might be effective against multiple variants of relevant PfEMP1 proteins, as demonstrated for the small molecule that mimics the ICAM-1 loop and inhibits the binding of two parasite variants to the ICAM-1 receptor [73]. However, these types of anti-adhesion molecules might be problematic, since they may compete with the natural ligands of these receptors and produce harmful side effects. Therefore, molecules that bind directly to variable PfEMP1 domains and inhibit their binding to the specific receptors would be preferable. The above-mentioned structural study [14] strongly supports the idea of inhibiting the binding of variant domains to the corresponding receptor via interaction between a single molecule and the receptor-binding conserved pocket in the protein.
Importantly, the available data support the idea that the diversity of variants involved in SM cases is also significantly restricted. In malaria endemic areas, SM accounts for <2% of total malaria cases in young children and predominantly occurs only once or twice in a lifetime [65]. This indicates that the broad immunity against SM develops quickly in early childhood [66] despite variations in the IE surface proteins. The existence of a monoclonal antibody that inhibits the binding of several pregnancy malaria parasite strains to the host ligand chondroitin sulfate A (CSA) [55] further indicates that the conserved features of PfEMP1 proteins might be sufficient to provide the cross-strain effectiveness of anti-adhesion molecules. Hence, a single adhesion-inhibiting molecule might exhibit cross-strain/PfEMP1 activity for a particular host receptor.
STRATEGY FOR IDENTIFICATION OF ANTI-ADHESION COMPOUNDS
Direct screening of live laboratory or field isolates for identification of agents that would inhibit parasite adhesion to different host receptors is an extremely slow and tedious process not currently amenable to the use of high throughput (HT) protocols due to technical and biological limitations. Our two-step approach to identification of anti-adhesion molecules overcomes these limitations [47]. Moreover, it was designed to screen mixture-based libraries to allow fast deconvolution to individual active compounds in a few steps, because the first HT step is performed in vitro. Testing the sets of such compounds for the inhibition of live parasite binding is not feasible due to the toxic effects of these mixtures or of the solvent dimethyl sulfoxide (DMSO, tolerated up to 20% in our in vitro system), as well as due to the low throughput of live parasite assays.
First, mixture-based libraries of small molecules are screened and deconvoluted to single compounds in a HT testing for the inhibition of interactions between known host receptors involved in IE adhesion and various relevant PfEMP1 domains immobilized on BioPlex beads (Bio-Rad, USA). Second, molecules identified in the first step are validated for the adhesion-inhibiting activity using live heterologous IEs. The use of heterologous IEs, preferably from distant geographical regions, is important to demonstrate that identified anti-adhesion drugs inhibit the binding of IEs with diverse genetic background and are not specific to a single line only. Since the number of hits to test with IEs is low, the two-step procedure provides HT identification of anti-adhesion molecules.
DEVELOPMENT OF SCREENING PROCEDURE AND IDENTIFICATION OF NOVEL ANTI-ADHESION COMPOUNDS TO TREAT SM SYNDROMES CAUSED BY P. falciparum USING SMALL MOLECULE LIBRARIES
ChemBridge library. The development and proof-of-principle of our HT approach for the screening of anti-adhesion molecules have been performed [47] using a subset (n = 10,000) of the DIVERSet small molecule library from the ChemBridge Corporation. This library was selected based on its commercial availability, substantial size appropriate for identification of a sufficient number of active molecules, possibility to purchase separate subsets at affordable price for the initial development, as well as required physicochemical and pharmacological characteristics reflected in multiple publications. These characteristics include broad biologically relevant pharmacophore diversity space. For example, 50,000 drug-like small molecules of the DIVERSet library cover 60% of the total 3-point pharmacophore space of the entire EXPRESS-Pick™ Collection (internal) consisting of 500,000 compounds.
Before working with the small molecule libraries, we demonstrated the feasibility of our two-step approach using antibodies that inhibited the binding of three receptors, CD36 (the most common IE-binding receptor), CSA (receptor involved in PM), and ICAM-1 (receptor involved in CM) and erythrocytes infected with parasite lines binding to these receptors. When using PfEMP1 domains binding to these receptors and the corresponding binding parasite lines, we found a remarkable concordance between the in vitro molecular Bio-Plex assays and assays with live IEs, which strongly supported the feasibility of our two-step approach for screening anti-adhesion molecules. Since physical principles and readouts of the assay are the same for large (antibodies) and small anti-adhesion molecules, we expected to get similarly concordant results using the same two-step approach and small-molecule libraries.
As the first step, we used a sub-library of 10,000 compounds to prepare 125 pools, each containing 80 compounds (125 µM solution of each compound in DMSO) for the initial screening. In this way, the initial screening of all 10,000 compounds required less than two 96-well plates to assess their inhibitory effect on the binding of each of three endothelial receptors to the corresponding PfEMP1 domains.
We chose 75% inhibition of binding as an arbitrary threshold for our in vitro assay. Five different CD36-binding domains were selected for the screening of the CD36 binding inhibition. Interestingly, we did not find mixtures that inhibited CD36 binding over the selected threshold and, consequently, did not pursue further deconvolution. It is likely that identification of more efficient compounds for the inhibition of CD36 binding requires a larger library. For two other receptors (CSA and ICAM-1) and the corresponding three CSA-binding domains and one ICAM-1-binding domain (the only one identified by us in the NF54 line), we successfully identified inhibitory mixtures and deconvoluted them to three and two active individual compounds, respectively. Moreover, out of three compounds that inhibited the binding of the CSA-binding domain to CSA, two compounds shared the same sulfonyl-containing primary scaffold, which further strengthened the feasibility of our approach.
Using the in vitro assay, we demonstrated that the IC50 values for both receptors were in a low micromolar range: 1.73 µM for the full-length VAR2CSA PfEMP1 protein that contained six CSA-binding domains [47], and 18 µM for the ICAM-1 binding domain PF11_0521 DBL2β3. The latter binds ICAM-1 receptor with a very high avidity (~2 nM) [46, 59]. Therefore, identification of small molecular compounds that can compete in the domain–receptor interactions at low micromolar concentration is an important step toward identification of more potent compounds.
In accordance with our two-step approach, we demonstrated that compounds identified in our first in vitro step inhibited the binding of live IEs. In these experiments, which were performed at a single compound concentration of 100 µM as a proof-of-principle, we used the CS2 (CSA-binding) and 3G8 (ICAM-1-binding) parasite lines [74]. These lines have originated from a completely different genetic and geographic background (IT4 line from South-East Asia [75]) than the NF54 line (from Africa [76]) that was used as a source of our recombinant receptor-biding domains for the in vitro assays. Our experiments with live IEs clearly demonstrated that the identified active compounds inhibited the binding of CS2 and 3G8 lines to the corresponding binding receptors. This was a very important finding as it indicated that small molecule inhibitors may overcome a challenge presented by the domain diversity and can inhibit the binding of IEs of different genetic origin. Moreover, for the CS2 line, the identified compounds not only blocked, but also reversed the established interactions between IEs and CSA. This is also an extremely important feature, as reversing the established sequestration might have a significant impact on the SM pathology due to the removal of already sequestered parasites contributing significantly to the vascular inflammation.
Therefore, by using a subset (10,000 compounds) of the DIVERSet library, we not only successfully developed a two-step screening procedure, but identified three compounds that inhibited IE adhesion to the CSA receptor and two compounds that inhibited IE adhesion to the ICAM-1 receptor – two principal receptors involved in the sequestration of IEs in PM and CM [47]. This indicates that screening of currently commercially available full DIVERSet collection of small molecules from ChemBridge (n = 150,000) may reveal up to 30-50 individual active compounds/hits to select for the best leads based on their activity, cellular and animal toxicity, and pharmacokinetics/pharmacodynamics (PK/PD) in order to further develop them into anti-adhesion drugs.
Natural product library (conopeptides). Other libraries may provide an additional opportunity for screening for anti-adhesion compounds. In this respect, collections of natural products might be a good source for this type of screening. As a proof-of-principal, we used [77] a library of conotoxins/conopeptides, which are compounds expressed in the venom of marine mollusks belonging to the Conus genus. There are hundreds of thousands of these novel compounds, mostly disulfide-rich and well-structured peptides, expressed by more than 850 species of Conus mollusks [78-80]. Their presence in the venoms of mollusks does not necessarily mean that they all are toxic or harmful for humans. In addition, they can be modified to abolish the toxicity, while preserving their biological activity [81]. This class of molecules is actively pursued for the discovery of novel drugs to treat various diseases [82].
The high specificity and affinity of conotoxins for many ubiquitous somatic receptors suggest that they might be powerful compounds to prevent or reverse interactions between domains of malaria adhesins, PfEMP1 proteins, and host receptors involved in SM. In our work [77], we extracted venom samples from 460 specimens of Conus nux and fractionated them by size-exclusion (SE) high performance liquid chromatography (HPLC), followed by reverse phase (RP) HPLC. The obtained fractions were tested for the inhibition of binding between three vascular receptors (CD36, CSA, and ICAM-1) and the corresponding receptor-binding PfEMP1 domains immobilized on BioPlex beads using the above-described in vitro approach [47]. Remarkably, six individual C. nux venom fractions, which contained either single peptides or a very limited set of peptides, affected the binding of several domains with different receptor specificity to the corresponding receptors, including proteins CD36 and ICAM-1 and polysaccharide CSA. These results suggest that either the peptides in these fractions bind to common structural elements in different PfEMP1 domains with a similar general scaffold (as we discussed above) or a few different peptides in the fraction interact with different domains (or receptors) and block the domain–receptor interactions. The ability of conotoxin fractions to inhibit the binding of IE adhesins with multiple binding specificity to a broad set of receptors can be extremely useful in the development of efficient and safe anti-adhesion drugs against SM [15, 16], because IEs isolated from SM patients often bind multiple receptors [57, 83]. Therefore, natural products, such as conopeptides, might be a promising source of anti-adhesion compounds that can be used for identification of leads for their development into anti-adhesion anti-malarial drugs.
TPIMS libraries. The above results demonstrate that screening of large libraries may provide a sufficient number of hits with low IC50 values for their development into leads and, further, into anti-adhesion drugs; and that peptides and peptidomimetics might be extremely useful classes of molecules to identify these leads. Such large mixture-based libraries, ideally suitable for our two-step approach, have been developed at the Torrey Pines Institute for Molecular Studies (TPIMS) in a very convenient format allowing analysis of millions of compounds using just hundreds to thousands of samples [84-86]. The principle of this method is in the preparation of positional scanning libraries, each representing a different scaffold with several replacement groups, using the tea-bag approach [87]. The sub-libraries of each scaffold have one fixed (defined) replacement group and combinations of other replacement groups. The number of these sub-libraries is equal to the number of variants for the replacement groups. Analysis of screening results allows identification of the most active replacement groups. Then, a new sub-library(s) containing all combinations of these most active groups is prepared and tested for its activity. In 2-3 iterations (depending on the complexity of the initial library), a library of individual compounds is prepared based on the activity of mixtures with the defined replacement groups, and the most active individual compounds are identified. As we mentioned above, the screening of mixture-based libraries requires considerably less effort, time, and resources compared to the traditional HT screening of individual compounds.
TPIMS libraries only partially overlap with the structural space of currently approved drugs and, therefore, further add to the molecular complexity of libraries commonly used in HT screening [88, 89] and expand the traditional relevant space of medicinal chemistry [89]. In addition, large mixture-based libraries designed around a limited number of molecular scaffolds provide an opportunity for rapid elucidation of the structure–activity relationships (SARs) during screening. TPIMS collection is characterized by a larger degree of molecular complexity and larger 3D conformational space than typically available commercial collections [88] and possesses the drug-like properties similar to the property space of known drugs [89]. Successful application of TPIMS library collection in preclinical and clinical studies has been extensively reviewed [85, 90].
In our work [91], we screened a collection of TPIMS libraries consisting of more than 30 million compounds designed around 75 molecular scaffolds in order to identify the hits against binding between ICAM-1 (involved in CM) and the corresponding binding domain PF11_0521 DBL2β3 from the 3D7 line PfEMP1 protein [11, 46, 47], with IC50 in a nanomolar range.
Based on the initial screening, dithiazole Positional Scanning Synthetic Combinatorial Library (TPI-2103, 3990 compounds) with the highest inhibitory activity and lowest complexity (two replacement groups) was selected for positional scanning deconvolution of the binding-inhibiting compounds. Individual compounds (104 in total) derived from the deconvolution of TPI-2103 represented all combinations of the selected most active R1 and R2 replacement groups. Their testing revealed two compounds (2648-40 and 2648-33) that inhibited ICAM-1 binding by more than 50% at 1 µM concentration. These two compounds were then characterized at a range of concentrations for the inhibition of ICAM-1 binding (i) in vitro using the bead-immobilized domain and (ii) with live erythrocytes infected by heterologous ICAM-1-binding 3G8 parasite line, as well as for cytotoxicity against two human cell lines.
The two compounds had the IC50 values of 362 nM (2648-40) and 696 nM (2648-33) in vitro, which was about 50 and ~25 times, respectively, better than for the compounds identified in our previous work with the small-size ChemBridge sub-library [47]. Importantly, their inhibitory activity with live IEs was similar (IC50 of 352 nM and 669 nM for 2648-40 and 2648-33, respectively). Thus, the same compound provided stronger inhibition of ICAM-1 binding on the beads and of binding between IEs and surface-immobilized ICAM-1, which was in accordance with our previous observations in [47]. Furthermore, the ICAM-1-binding domains of the PfEMP1 protein expressed on the surface of erythrocytes infected with NF54 line (used in in vitro experiments) and 3G8 line (used in experiments with live IEs) significantly differed in their amino acid sequence (44% identities, 55% positives, and 15% gaps by BLAST). Nevertheless, active compounds identified using a particular genetic background in the in vitro assays were successful in inhibiting the binding of live IEs of very different genetic background, which again corroborated our results obtained using completely different compounds [47]. We believe that our approach might be universally employed for identification of anti-adhesion compounds against various receptor–domain pairs despite a substantial diversity of PfEMP1 domains/proteins.
The cytotoxicity studies demonstrated the selectivity index (SI, ratio of 50% cytotoxic concentration (CC50) toward human cell lines to 50% binding-inhibiting concentration (IC50) between the surface-immobilized receptors and live IE) of ~70 and ~37 for the two compounds, respectively. This is an excellent result, as SI > 10 is considered the minimally acceptable value and SI > 50 is an ideal value for the target candidate profile (TCP-1) [48], because after getting into the bloodstream, common malaria parasite-killing drugs should cross at least 3 membranes (red blood cell membrane, parasitophorous vacuole membrane, and parasite plasma membrane) for their effect. The anti-adhesion drugs are supposed to work immediately at the outer surface of IEs after entering the blood. Therefore, the two identified lead compounds provide an excellent starting point for the development of anti-adhesion drugs against SM and CM, in which IE adhesion to ICAM-1 receptor plays an extremely important pathological role.
As we mentioned above, the binding to ICAM-1 and EPCR is implicated in the development of SM and CM. Many parasites isolated from SM or CM patients demonstrated the presence of a tandem of ICAM-1- and EPCR-binding domains in the expressed PfEMP1 molecules [59, 60, 92-96]. Even if both these domains in a tandem are required for the development of SM and/or CM, this does not mean that the drugs should inhibit the binding of both domains to the corresponding receptors to successfully block IE sequestration. In this case, inhibition of ICAM-1 binding only may significantly reduce the efficacy of IE sequestration, especially in the brain microvasculature, where EPCR is expressed at low levels, while the content of ICAM-1 is high and further increases during brain inflammation (reviewed in [97]), in particular, during CM-associated inflammation induced by TNFα (reviewed in [98]). In this respect, identification of other compounds that will not only prevent but also reverse ICAM-1 sequestration, may have a significant advantage in the anti-adhesion adjunct treatment. As demonstrated in our studies and works of other authors, ICAM-1 binds to the ICAM-1-binding DBL domains with an extremely high avidity (2-4 nM) [46, 59]. Therefore, identification of such compounds might not be a trivial task. Most likely, they will have an allosteric, and not competitive mode of action. The leads identified in [47, 91] did not reverse the established ICAM-1–domain interactions. Hence, molecules with this activity are still to be discovered. Nevertheless, just inhibition of ICAM-1 binding might be sufficient to reduce or prevent the development of pathologies related to the continuous IE sequestration in SM or CM, although this hypothesis should be clarified experimentally.
CHALLENGES IN TESTING ANTI-MALARIAL ANTI-ADHESION DRUGS IN PRE-CLINICAL STUDIES
Since anti-adhesion drugs do not usually kill malaria parasites (if they did, this would be an additional important bonus), it is difficult to justify their trials in human patients with severe forms of malaria. One of the effects these drugs should produce is reduction of inflammation of endothelial cells (ECs) by preventing or reversing IE cytoadhesion. This can be studied in vitro by incubation of ECs with IEs that can adhere to these cells [for example, CSA-binding IEs and human trophoblast-derived BeWo cells expressing CSA, or ICAM-1 binding IEs and TNFα-stimulated human umbilical vascular endothelial cells (HUVEC) that express ICAM-1] in the presence and absence of anti-adhesion compounds, followed by detection of inflammatory molecules secreted by the cells. However, these effects are not easy to demonstrate in vivo in the preclinical studies, as no appropriate animal model for P. falciparum malaria is available. PfEMP1 proteins are specific for P. falciparum only, while malaria parasite species that infect rodents are very different and do not share homologous PfEMP1 proteins. Even humanized mouse malaria model [99], in which immunodeficient laboratory NOD-scid gamma (NSG) mice are injected with human erythrocytes for several days to substitute mouse erythrocytes and then challenged with P. falciparum to establish the infection, will not be appropriate, as mouse endothelial receptors, though homologous, are different from the human receptors interacting with PfEMP1 proteins and may not provide adequate IE cytoadhesion. Therefore, appropriate models should be designed for testing anti-adhesion compounds (beyond their cellular, animal, and human cytotoxicity) before moving to trials in humans. Potentially, some non-human primate models, like P. falciparum infection of Aotus monkeys [100], may be used to test IE sequestration and activity of anti-adhesion drugs.
IMPLICATIONS OF ANTI-ADHESION DRUGS FOR OTHER TARGETS
The two-step approach developed by us for the HT identification of anti-adhesion compounds might be useful in the identification of compounds for treatment of other diseases, for which interactions between two known molecules/molecular structures play an important role in the disease pathology, e.g., infection of cells by viral, parasitic, or bacterial pathogens through specific receptors. Identification of anti-adhesion compounds from large libraries via HT screening in vitro followed by their in vivo or ex vivo testing may significantly speed up the discovery of novel therapeutics for various diseases.
Acknowledgments
I would like to thank all co-authors of our highly collaborative published papers reviewed in this article.
Abbreviations
- CM
cerebral malaria
- DMSO
dimethyl sulfoxide
- HT
high-throughput
- IEs
erythrocytes infected with Plasmodium falciparum
- PM
placental malaria
- SM
severe malaria
Funding
The funding for our work was provided by the Bill & Melinda Gates Foundation (grants 29202 and 1634); NIH (grants 1R21AI064503, 1R56AI083668, and 1R01AI092120), and Florida Atlantic University start-up fund.
Ethics declarations
The author declares no conflicts of interest. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
References
- 1.World Health Organization (2019) World malaria report 2019. URL: https://apps.who.int/iris/handle/10665/330011.
- 2.Hughes K. R., Biagini G. A., Craig A. G. Continued cytoadherence of Plasmodium falciparum infected red blood cells after antimalarial treatment. Mol. Biochem. Parasitol. 2010;169:71–78. doi: 10.1016/j.molbiopara.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duffy, P. E., Acharya, P., Oleinikov. A. V. (2014) Cytoadherence, in: Encyclopedia of Malaria (Hommel, M., and Kremsner, P., eds) Springer, New York, 10.1007/978-1-4614-8757-9_39-1.
- 4.Fried M., Duffy P. E. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science. 1996;272:1502–1504. doi: 10.1126/science.272.5267.1502. [DOI] [PubMed] [Google Scholar]
- 5.Miller L. H., Good M. F., Milon G. Malaria pathogenesis. Science. 1994;264:1878–1883. doi: 10.1126/science.8009217. [DOI] [PubMed] [Google Scholar]
- 6.Newbold C., Warn P., Black G., Berendt A., Craig A., et al. Receptor-Specific Adhesion and Clinical Disease in Plasmodium falciparum. Am. J. Trop. Med. Hyg. 1997;57:389–398. doi: 10.4269/ajtmh.1997.57.389. [DOI] [PubMed] [Google Scholar]
- 7.Turner G. D., Morrison H., Jones M., Davis T. M., Looareesuwan S., et al. An immunohistochemical study of the pathology of fatal malaria. Evidence for widespread endothelial activation and a potential role for intercellular adhesion molecule-1 in cerebral sequestration. Am. J. Pathol. 1994;145:1057–1069. [PMC free article] [PubMed] [Google Scholar]
- 8.Duffy P. E., Fried M. Antibodies that inhibit Plasmodium falciparum adhesion to chondroitin sulfate a are associated with increased birth weight and the gestational age of newborns. Infect. Immun. 2003;71:6620–6623. doi: 10.1128/IAI.71.11.6620-6623.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fried M., Duffy P. Maternal malaria and parasite adhesion. J. Mol. Med. (Berl.) 1998;76:162–171. doi: 10.1007/s001090050205. [DOI] [PubMed] [Google Scholar]
- 10.Fried M., Avril M., Chaturvedi R., Fernandez P., Lograsso J., et al. Multilaboratory approach to preclinical evaluation of vaccine immunogens for placental malaria. Infect. Immun. 2013;81:487–495. doi: 10.1128/IAI.01106-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oleinikov A. V., Amos E., Frye I. T., Rossnagle E., Mutabingwa T. K., et al. High throughput functional assays of the variant antigen PfEMP1 reveal a single domain in the 3D7 Plasmodium falciparum genome that binds ICAM1 with high affinity and is targeted by naturally acquired neutralizing antibodies. PLoS Pathog. 2009;5:e1000386. doi: 10.1371/journal.ppat.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen S., McGregor I., Carrington S. Gamma-globulin and acquired immunity to human malaria. Nature. 1961;192:733–737. doi: 10.1038/192733a0. [DOI] [PubMed] [Google Scholar]
- 13.Harrington W. E., Mutabingwa T. K., Muehlenbachs A., Sorensen B., Bolla M. C., et al. Competitive facilitation of drug-resistant Plasmodium falciparum malaria parasites in pregnant women who receive preventive treatment. Proc. Natl. Acad. Sci. USA. 2009;106:9027–9032. doi: 10.1073/pnas.0901415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh F. L., Turner L., Bolla J. R., Robinson C. V., Lavstsen T., et al. The structural basis for CD36 binding by the malaria parasite. Nat. Commun. 2016;7:12837. doi: 10.1038/ncomms12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fairhurst R. M., Wellems T. E. Modulation of malaria virulence by determinants of Plasmodium falciparum erythrocyte membrane protein-1 display. Curr. Opin. Hematol. 2006;13:124–130. doi: 10.1097/01.moh.0000219655.73162.42. [DOI] [PubMed] [Google Scholar]
- 16.Ch'ng J.‑H., Moll K., Quintana M., Chan S. C., Masters E., et al. Rosette-disrupting effect of an anti-plasmodial compound for the potential treatment of Plasmodium falciparum malaria complications. Sci. Rep. 2016;6:29317. doi: 10.1038/srep29317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnwell J. W., Asch A. S., Nachman R. L., Yamaya M., Aikawa M., et al. A human 88-kD membrane glycoprotein (CD36) functions in vitro as a receptor for a cytoadherence ligand on Plasmodium falciparum-infected erythrocytes. J. Clin. Invest. 1989;84:765–772. doi: 10.1172/JCI114234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berendt A. R., Simmons D. L., Tansey J., Newbold C. I., Marsh K. Intercellular adhesion molecule-1 is an endothelial cell adhesion receptor for Plasmodium falciparum. Nature. 1989;341:57–59. doi: 10.1038/341057a0. [DOI] [PubMed] [Google Scholar]
- 19.Barragan A., Spillmann D., Carlson J., Wahlgren M. Role of glycans in Plasmodium falciparum infection. Biochem. Soc. Trans. 1999;27:487–493. doi: 10.1042/bst0270487. [DOI] [PubMed] [Google Scholar]
- 20.David P. H., Handunnetti S. M., Leech J. H., Gamage P., Mendis K. N. Rosetting: A new cytoadherence property of malaria-infected erythrocytes. Am. J. Trop. Med. Hyg. 1988;38:289–297. doi: 10.4269/ajtmh.1988.38.289. [DOI] [PubMed] [Google Scholar]
- 21.Handunnetti S. M., David P. H., Perera K. L., Mendis K. N. Uninfected erythrocytes form “Rosettes” around Plasmodium Falciparum infected erythrocytes. Am. J. Trop. Med. Hyg. 1989;40:115–118. doi: 10.4269/ajtmh.1989.40.115. [DOI] [PubMed] [Google Scholar]
- 22.Dondorp A. M. Clinical significance of sequestration in adults with severe malaria. Transfus. Clin. Biol. 2008;15:56–57. doi: 10.1016/j.tracli.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Vásquez A. M., Segura C., Blair S. Induction of pro-inflammatory response of the placental trophoblast by Plasmodium falciparum infected erythrocytes and TNF. Malar. J. 2013;12:421. doi: 10.1186/1475-2875-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basilico N., Mondani M., Parapini S., Speciale L., Ferrante P., et al. Plasmodium falciparum parasitized red blood cells modulate the production of endothelin-1 by human endothelial cells. Minerva Med. 2004;95:153–158. [PubMed] [Google Scholar]
- 25.Yipp B. G., Robbins S. M., Resek M. E., Baruch D. I., Looareesuwan S., et al. Src-family kinase signaling modulates the adhesion of Plasmodium falciparum on human microvascular endothelium under flow. Blood. 2003;101:2850–2857. doi: 10.1182/blood-2002-09-2841. [DOI] [PubMed] [Google Scholar]
- 26.Jenkins N., Wu Y., Chakravorty S., Kai O., Marsh K., et al. Plasmodium falciparum Intercellular adhesion molecule-1-based cytoadherence-related signaling in human endothelial cells. J. Infect. Dis. 2007;196:321–327. doi: 10.1086/518795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams S., Brown H., Turner G. Breaking down the blood–brain barrier: signaling a path to cerebral malaria? Trends Parasitol. 2002;18:360–366. doi: 10.1016/S1471-4922(02)02353-X. [DOI] [PubMed] [Google Scholar]
- 28.Cruz L. N., Wu Y., Craig A. G., Garcia C. R. Signal transduction in plasmodium–red blood cells interactions and in cytoadherence. An. Acad. Bras. Cienc. 2012;84:555–572. doi: 10.1590/S0001-37652012005000036. [DOI] [PubMed] [Google Scholar]
- 29.Su X. Z., Heatwole V. M., Wertheimer S. P., Guinet F., Herrfeldt J. A., et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 30.Smith J. D., Chitnis C. E., Craig A. G., Roberts D. J., Hudson-Taylor D. E., et al. Switches in expression of Plasmodiumfalciparumvar genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gardner M. J., Hall N., Fung E., White O., Berriman M., et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biggs B. A., Goozé L., Wycherley K., Wilkinson D., Boyd A. W., et al. Knob-independent cytoadherence of Plasmodium falciparum to the leukocyte differentiation antigen CD36. J. Exp. Med. 1990;171:1883–1892. doi: 10.1084/jem.171.6.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scherf A., Lopez-Rubio J. J., Riviere L. Antigenic variation in Plasmodium falciparum. Annu. Rev. Microbiol. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- 34.Roberts D. J., Craig A. G., Berendt A. R., Pinches R., Nash G., et al. Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature. 1992;357:689–692. doi: 10.1038/357689a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith J. D., Craig A. G., Kriek N., Hudson-Taylor D., Kyes S., et al. Identification of a Plasmodium falciparum intercellular adhesion molecule-1 binding domain: A parasite adhesion trait implicated in cerebral malaria. Proc. Natl. Acad. Sci. USA. 2000;97:1766–1771. doi: 10.1073/pnas.040545897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rask T. S., Hansen D. A., Theander T. G., Gorm Pedersen A., Lavstsen T. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes – divide and conquer. PLoS Comp. Biol. 2010;6:e1000933. doi: 10.1371/journal.pcbi.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McHenry A. M., Adams J. H. The crystal structure of P. knowlesi DBPalpha DBL domain and its implications for immune evasion. Trends Biochem. Sci. 2006;31:487–491. doi: 10.1016/j.tibs.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh S. K., Hora R., Belrhali H., Chitnis C. E., Sharma A. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature. 2006;439:741–744. doi: 10.1038/nature04443. [DOI] [PubMed] [Google Scholar]
- 39.Higgins M. K. The structure of a chondroitin sulfate-binding domain important in placental malaria. J. Biol. Chem. 2008;283:21842–21846. doi: 10.1074/jbc.C800086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh K., Gittis A. G., Nguyen P., Gowda D. C., Miller L. H., et al. Structure of the DBL3x domain of pregnancy-associated malaria protein VAR2CSA complexed with chondroitin sulfate A. Nat. Struct. Mol. Biol. 2008;15:932–938. doi: 10.1038/nsmb.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein M. M., Gittis A. G., Su H. P., Makobongo M. O., Moore J. M., et al. The cysteine-rich interdomain region from the highly variable plasmodium falciparum erythrocyte membrane protein-1 exhibits a conserved structure. PLoS Pathog. 2008;4:e1000147. doi: 10.1371/journal.ppat.1000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howell D. P., Samudrala R., Smith J. D. Disguising itself – insights into Plasmodium falciparum binding and immune evasion from the DBL crystal structure. Mol. Biochem. Parasitol. 2006;148:1–9. doi: 10.1016/j.molbiopara.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 43.Chakravorty S. J., Hughes K. R., Craig A. G. Host response to cytoadherence in Plasmodium falciparum. Biochem. Soc. Trans. 2008;36:221–228. doi: 10.1042/BST0360221. [DOI] [PubMed] [Google Scholar]
- 44.Oleinikov A. V., Rossnagle E., Francis S., Mutabingwa T. K., Fried M., et al. Effects of sex, parity, and sequence variation on seroreactivity to candidate pregnancy malaria vaccine antigens. J. Infect. Dis. 2007;196:155–164. doi: 10.1086/518513. [DOI] [PubMed] [Google Scholar]
- 45.Oleinikov A. V., Voronkova V. V., Frye I. T., Amos E., Morrison R., et al. A plasma survey using 38 PfEMP1 domains reveals frequent recognition of the Plasmodium falciparum antigen VAR2CSA among young tanzanian children. PLoS One. 2012;7:e31011. doi: 10.1371/journal.pone.0031011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gullingsrud J., Saveria T., Amos E., Duffy P. E., Oleinikov A. V. Structure–function–immunogenicity studies of PfEMP1 domain DBL2betaPF11_0521, a malaria parasite ligand for ICAM-1. PLoS One. 2013;8:e61323. doi: 10.1371/journal.pone.0061323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gullingsrud J., Milman N., Saveria T., Chesnokov O., Williamson K., et al. High throughput screening platform identifies small molecules that prevent sequestration of Plasmodium falciparum-infected erythrocytes. J. Infect. Dis. 2015;211:1134–1143. doi: 10.1093/infdis/jiu589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chesnokov O., Merritt J., Tcherniuk S. O., Milman N., Oleinikov A. V. Plasmodium falciparum infected erythrocytes can bind to host receptors integrins alphaVbeta3 and alphaVbeta6 through DBLdelta1_D4 domain of PFL2665c PfEMP1 protein. Sci. Rep. 2018;8:17871. doi: 10.1038/s41598-018-36071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quintana M. D. P., Ecklu-Mensah G., Tcherniuk S. O., Ditlev S. B., Oleinikov A. V., et al. Comprehensive analysis of Fc-mediated IgM binding to the Plasmodium falciparum erythrocyte membrane protein 1 family in three parasite clones. Sci. Rep. 2019;9:6050. doi: 10.1038/s41598-019-42585-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salanti A., Dahlbäck M., Turner L., Nielsen M. A., Barfod L., et al. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J. Exp. Med. 2004;200:1197–1203. doi: 10.1084/jem.20041579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salanti A., Staalsoe T., Lavstsen T., Jensen A. T., Sowa M. P., et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 52.Viebig N. K., Levin E., Dechavanne S., Rogerson S. J., Gysin J., et al. Disruption of Var2csa gene impairs placental malaria associated adhesion phenotype. PLoS One. 2007;2:e910. doi: 10.1371/journal.pone.0000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duffy M. F., Maier A. G., Byrne T. J., Marty A. J., Elliott S. R., et al. VAR2CSA is the principal ligand for chondroitin sulfate A in two allogeneic isolates of Plasmodium falciparum. Mol. Biochem. Parasitol. 2006;148:117–124. doi: 10.1016/j.molbiopara.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 54.Fried M., Nosten F., Brockman A., Brabin B. J., Duffy P. E. Maternal antibodies block malaria. Nature. 1998;395:851–852. doi: 10.1038/27570. [DOI] [PubMed] [Google Scholar]
- 55.Soerli J., Barfod L., Lavstsen T., Bernasconi N. L., Lanzavecchia A., et al. Human monoclonal IgG selection of Plasmodium falciparum for the expression of placental malaria-specific variant surface antigens. Parasite Immunol. 2009;31:341–346. doi: 10.1111/j.1365-3024.2009.01097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ochola L. B., Siddondo B. R., Ocholla H., Nkya S., Kimani E. N., et al. Specific receptor usage in Plasmodium falciparum cytoadherence is associated with disease outcome. PLoS One. 2011;6:e14741. doi: 10.1371/journal.pone.0014741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tuikue Ndam N., Moussiliou A., Lavstsen T., Kamaliddin C., Jensen A. T. R., et al. Parasites causing cerebral falciparum malaria bind multiple endothelial receptors and express EPCR and ICAM-1-binding PfEMP1. J. Infect. Dis. 2017;215:1918–1925. doi: 10.1093/infdis/jix230. [DOI] [PubMed] [Google Scholar]
- 58.Moxon C. A., Wassmer S. C., Milner D. A., Jr., Chisala N. V., Taylor T. E., et al. Loss of endothelial protein C receptors links coagulation and inflammation to parasite sequestration in cerebral malaria in African children. Blood. 2013;122:842–851. doi: 10.1182/blood-2013-03-490219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lennartz F., Adams Y., Bengtsson A., Olsen R. W., Turner L., et al. Structure-guided identification of a family of dual receptor-binding PfEMP1 that is associated with cerebral malaria. Cell Host Microbe. 2017;21:403–414. doi: 10.1016/j.chom.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Avril M., Bernabeu M., Benjamin M., Brazier A. J., Smith J. D. Interaction between endothelial protein C receptor and intercellular adhesion molecule 1 to mediate binding of Plasmodium falciparum-infected erythrocytes to endothelial cells. mBio. 2016;7:e00615–16. doi: 10.1128/mBio.00615-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Howell D. P. G., Levin E. A., Springer A. L., Kraemer S. M., Phippard D. J., et al. Mapping a common interaction site used by Plasmodium falciparum Duffy binding-like domains to bind diverse host receptors. Mol. Microbiol. 2008;67:78–87. doi: 10.1111/j.1365-2958.2007.06019.x. [DOI] [PubMed] [Google Scholar]
- 62.Bengtsson A., Joergensen L., Rask T. S., Olsen R. W., Andersen M. A., et al. A novel domain cassette identifies Plasmodium falciparum PfEMP1 proteins binding ICAM-1 and is a target of cross-reactive, adhesion-inhibitory antibodies. J. Immunol. 2013;190:240–249. doi: 10.4049/jimmunol.1202578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bernabeu M., Smith J. D. EPCR and malaria severity: The center of a perfect storm. Trends Parasitol. 2017;33:295–308. doi: 10.1016/j.pt.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith J. D. The role of PfEMP1 adhesion domain classification in Plasmodium falciparum pathogenesis research. Mol. Biochem. Parasitol. 2014;195:82–87. doi: 10.1016/j.molbiopara.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gonçalves B. P., Huang C.-Y., Morrison R., Holte S., Kabyemela E., et al. Parasite burden and severity of malaria in Tanzanian children. New Engl. J. Med. 2014;370:1799–1808. doi: 10.1056/NEJMoa1303944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Høgh B. Clinical and parasitological studies on immunity to Plasmodium falciparum malaria in children. Scand. J. Infect. Dis. Suppl. 1996;102:1–53. [PubMed] [Google Scholar]
- 67.Jensen A. R., Adams Y., Hviid L. Cerebral Plasmodium falciparum malaria: The role of PfEMP1 in its pathogenesis and immunity, and PfEMP1-based vaccines to prevent it. Immunol. Rev. 2020;293:230–252. doi: 10.1111/imr.12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nishanth G., Schlüter D. Blood–brain barrier in cerebral malaria: Pathogenesis and therapeutic intervention. Trends Parasitol. 2019;35:516–528. doi: 10.1016/j.pt.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 69.Cunnington A. J., Walther M., Riley E. M. Piecing together the puzzle of severe malaria. Sci. Transl. Med. 2013;5:211ps18. doi: 10.1126/scitranslmed.3007432. [DOI] [PubMed] [Google Scholar]
- 70.Francischetti I. M., Seydel K. B., Monteiro R. Q. Blood coagulation, inflammation, and malaria. Microcirculation. 2008;15:81–107. doi: 10.1080/10739680701451516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim H., Higgins S., Liles W. C., Kain K. C. Endothelial activation and dysregulation in malaria: a potential target for novel therapeutics. Curr. Opin. Hematol. 2011;18:177–185. doi: 10.1097/MOH.0b013e328345a4cf. [DOI] [PubMed] [Google Scholar]
- 72.Van der Heyde H. C., Nolan J., Combes V., Gramaglia I., Grau G. E. A unified hypothesis for the genesis of cerebral malaria: sequestration, inflammation and hemostasis leading to microcirculatory dysfunction. Trends Parasitol. 2006;22:503–508. doi: 10.1016/j.pt.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 73.Dormeyer M., Adams Y., Kramer B., Chakravorty S., Tse M. T., et al. Rational design of anticytoadherence inhibitors for Plasmodium falciparum based on the crystal structure of human intercellular adhesion molecule 1. Antimicrob. Agents Chemother. 2006;50:724–730. doi: 10.1128/AAC.50.2.724-730.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Janes J. H., Wang C. P., Levin-Edens E., Vigan-Womas I., Guillotte M., et al. Investigating the host binding signature on the Plasmodium falciparum PfEMP1 protein family. PLoS Pathog. 2011;7:e1002032. doi: 10.1371/journal.ppat.1002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mu J., Awadalla P., Duan J., McGee K. M., Joy D. A., et al. Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol. 2005;3:e335. doi: 10.1371/journal.pbio.0030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Preston M. D., Campino S., Assefa S. A., Echeverry D. F., Ocholla H., et al. A barcode of organellar genome polymorphisms identifies the geographic origin of Plasmodium falciparum strains. Nat. Commun. 2014;5:4052. doi: 10.1038/ncomms5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Padilla A., Dovell S., Chesnokov O., Hoggard M., Oleinikov A. V., et al. Conus venom fractions inhibit the adhesion of Plasmodium falciparum erythrocyte membrane protein 1 domains to the host vascular receptors. J. Proteomics. 2021;234:104083. doi: 10.1016/j.jprot.2020.104083. [DOI] [PubMed] [Google Scholar]
- 78.Jin A. H., Muttenthaler M., Dutertre S., Himaya S. W. A., Kaas Q., et al. Conotoxins: Chemistry and biology. Chem. Rev. 2019;119:11510–11549. doi: 10.1021/acs.chemrev.9b00207. [DOI] [PubMed] [Google Scholar]
- 79.Kaas Q., Yu R., Jin A. H., Dutertre S., Craik D. J. ConoServer: Updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 2012;40:D325–330. doi: 10.1093/nar/gkr886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fu Y., Li C., Dong S., Wu Y., Zhangsun D., et al. Discovery methodology of novel conotoxins from Conus species. Mar. Drugs. 2018;16:417. doi: 10.3390/md16110417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bingham J. P., Andrews E. A., Kiyabu S. M., Cabalteja C. C. Drugs from Slugs. Part II – Conopeptide bioengineering. Chem. Biol. Interact. 2012;200:92–113. doi: 10.1016/j.cbi.2012.09.021. [DOI] [PubMed] [Google Scholar]
- 82.Dhiman V., Pant D. Human health and snails. J. Immunoassay Immunochem. 2021;42:211–235. doi: 10.1080/15321819.2020.1844751. [DOI] [PubMed] [Google Scholar]
- 83.Heddini A., Pettersson F., Kai O., Shafi J., Obiero J., et al. Fresh isolates from children with severe Plasmodium falciparum malaria bind to multiple receptors. Infect. Immun. 2001;69:5849–5856. doi: 10.1128/IAI.69.9.5849-5856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Houghten R. A., Pinilla C., Giulianotti M. A., Appel J. R., Dooley C. T., et al. Strategies for the use of mixture-based synthetic combinatorial libraries: scaffold ranking, direct testing invivo, and enhanced deconvolution by computational methods. J. Comb. Chem. 2008;10:3–19. doi: 10.1021/cc7001205. [DOI] [PubMed] [Google Scholar]
- 85.Pinilla C., Edwards B. S., Appel J. R., Yates-Gibbins T., Giulianotti M. A., et al. Selective agonists and antagonists of formylpeptide receptors: duplex flow cytometry and mixture-based positional scanning libraries. Mol. Pharmacol. 2013;84:314–324. doi: 10.1124/mol.113.086595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reilley K. J., Giulianotti M., Dooley C. T., Nefzi A., McLaughlin J. P., et al. Identification of two novel, potent, low-liability antinociceptive compounds from the direct in vivo screening of a large mixture-based combinatorial library. AAPS J. 2010;12:318–329. doi: 10.1208/s12248-010-9191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pinilla, C., Appel, J. R., Houghten, R. A. (1996) Tea Bag Synthesis of Positional Scanning Synthetic Combinatorial Libraries and Their Use for Mapping Antigenic Determinants, in Epitope Mapping Protocols. Methods in Molecular Biology (Morris, G. E., eds) Vol. 66, Humana Press, 10.1385/0-89603-375-9:171. [DOI] [PubMed]
- 88.López-Vallejo F., Giulianotti M. A., Houghten R. A., Medina-Franco J. L. Expanding the medicinally relevant chemical space with compound libraries. Drug Discov. Today. 2012;17:718–726. doi: 10.1016/j.drudis.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 89.Singh N., Guha R., Giulianotti M. A., Pinilla C., Houghten R. A., et al. Chemoinformatic analysis of combinatorial libraries, drugs, natural products, and molecular libraries small molecule repository. J. Chem. Inf. Model. 2009;49:1010–1024. doi: 10.1021/ci800426u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu J., Zhang Y., Maida L. E., Santos R. G., Welmaker G. S., et al. Scaffold ranking and positional scanning utilized in the discovery of nAChR-selective compounds suitable for optimization studies. J. Med. Chem. 2013;56:10103–10117. doi: 10.1021/jm401543h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chesnokov O., Visitdesotrakul P., Kalani K., Nefzi A., Oleinikov A. V. Small molecule compounds identified from mixture-based library inhibit binding between plasmodium falciparum infected erythrocytes and endothelial receptor ICAM-1. Int. J. Mol. Sci. 2021;22:5659. doi: 10.3390/ijms22115659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turner L., Lavstsen T., Berger S. S., Wang C. W., Petersen J. E., et al. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature. 2013;498:502–505. doi: 10.1038/nature12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kessler A., Dankwa S., Bernabeu M., Harawa V., Danziger S. A., et al. Linking EPCR-binding PfEMP1 to brain swelling in pediatric cerebral malaria. Cell Host Microbe. 2017;22:601–614.e5. doi: 10.1016/j.chom.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Olsen R. W., Ecklu-Mensah G., Bengtsson A., Ofori M. F., Lusingu J. P. A., et al. Natural and vaccine-induced acquisition of cross-reactive IgG-inhibiting ICAM-1-specific binding of a Plasmodium falciparum PfEMP1 subtype associated specifically with cerebral malaria. Infect. Immun. 2018;86:e00622–17. doi: 10.1128/IAI.00622-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bernabeu M., Gunnarsson C., Vishnyakova M., Howard C. C., Nagao R. J., et al. Binding heterogeneity of Plasmodium falciparum to engineered 3D brain microvessels is mediated by EPCR and ICAM-1. mBio. 2019;10:e00420–19. doi: 10.1128/mBio.00420-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Storm J., Jespersen J. S., Seydel K. B., Szestak T., Mbewe M., et al. Cerebral malaria is associated with differential cytoadherence to brain endothelial cells. EMBO Mol. Med. 2019;11:e9164. doi: 10.15252/emmm.201809164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dietrich J. B. The adhesion molecule ICAM-1 and its regulation in relation with the blood-brain barrier. J. Neuroimmunol. 2002;128:58–68. doi: 10.1016/s0165-5728(02)00114-5. [DOI] [PubMed] [Google Scholar]
- 98.Gimenez F., Barraud de Lagerie S., Fernandez C., Pino P., Mazier D. Tumor necrosis factor alpha in the pathogenesis of cerebral malaria. Cell. Mol. Life Sci. 2003;60:1623–1635. doi: 10.1007/s00018-003-2347-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jiménez-Díaz M. B., Mulet T., Viera S., Gómez V., Garuti H., et al. Improved murine model of malaria using Plasmodium falciparum competent strains and non-myelodepleted NOD-scid IL2Rgammanull mice engrafted with human erythrocytes. Antimicrob. Agents Chemother. 2009;53:4533–4536. doi: 10.1128/aac.00519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schmidt L. H. Infections with Plasmodium falciparum and Plasmodium vivax in the owl monkey – model systems for basic biological and chemotherapeutic studies. Trans. R Soc. Trop. Med. Hyg. 1973;67:446–474. doi: 10.1016/0035-9203(73)90077-1. [DOI] [PubMed] [Google Scholar]