Abstract
Basic leucine zipper (bZIP) is a conserved transcription factor (TF) widely present in eukaryotes, and it plays an important role in regulating plant growth and stress responses. To better understand the white pear bZIP gene family, comprehensive bioinformatics analysis of the pear genome was performed. A total of 84 PbbZIP genes were identified, which were divided into 13 subfamilies by phylogenetic analysis. The 84 PbbZIP genes were all located in the nucleus, and 77 of those genes were unevenly distributed across the 17 chromosomes of white pear. The other 7 PbbZIP genes were located on the scaffold. Subsequent expression profile analysis showed that PbbZIP genes in exocarp were significantly upregulated or downregulated in ‘Huangguan’ pear with brown spot (BS) compared with healthy pear and in response to hormonal treatment with gibberellin A3 (GA3). These results provide helpful insights into the characteristics of PbbZIP genes and their responses to BS in ‘Huangguan’ pear.
Keywords: Pear, bZIP gene family, brown spot, hormone, expression profile
Introduction
Basic leucine zippers (bZIPs) play important regulatory roles in many key biological processes and are each comprised of a DNA binding domain and a leucine zipper dimer domain; they are one of the largest gene families and the most widely distributed class of eukaryotic TFs (Glover et al., 1995; Amorim et al., 2017). bZIP family members each have a highly conserved domain comprised of 60~80 amino acids, and they can be divided into 10 subfamilies that exhibit different expression levels and perform different functions in different plants and tissues. bZIP transcription factors (TFs) have been identified in many plants, such as Arabidopsis (Jakoby et al., 2002), Gramineae (Wei et al., 2012; Nijhawan et al., 2008; Wang et al., 2019), olive (Rong et al., 2020), and many horticultural crops (Wang et al., 2017; An et al., 2018; Lu et al., 2020; Jin et al., 2021).
During plant growth and development, bZIP TFs play important roles in seed maturation and germination, cell elongation, flower induction and development, hormone stress and so on (Wang et al., 2011). bZIP TFs are also involved in plant responses to biotic and abiotic stresses. For example, the expression of bZIP related genes can be induced under drought, salt, cold stress, and auxin (IAA) and abscisic acid (ABA) hormone treatment (Hossain et al., 2010). In cucumber, CsbZIP12 and CsbZIP44 genes have been found to be upregulated in the roots after drought stress treatment (Baloglu et al., 2014). In tomato, the bZIP TF SIAREB is involved in the response to water deficiency and salt stress (Hsieh et al., 2010). In ‘Gala’ apple, the expression of MdAREB2 has been shown to increase rapidly after ABA treatment and thereby affect the expression of some stress-resistant genes under high temperature or light stress (Liu et al., 2018). In addition, the expression of MdbZIP26 was found to be significantly upregulated under drought and salt stresses, or exogenous ABA treatment thereby enhancing plant stress resistance through the ABA signaling pathway. This evidence suggests that the bZIP family is involved in plant stress resistance through hormone signaling.
White pear (Pyrus bretschneideri Rehd.) is a deciduous fruit tree of Pyrus genus of Rosaceae (Teng et al., 2004), and its fruit is juicy, sweet and refreshing and is well favored by consumers. Complete genome sequencing of pear has laid a foundation for biological information analysis of the white pear TF family. Many TF families have been characterized, such as WRKY and NAC (Huang et al., 2015; Gong et al., 2019), but there have been no reports on the bZIP TF family in white pear.
In this study, a bioinformatics method was used for genome-wide identification of the bZIP gene family in white pear, and the gene structure, physicochemical properties, conserved motifs, phylogenetic relationships, chromosomal locations, collinearity and cis-acting elements of bZIP family members were analyzed. In our previous study, we found that gibberellin A3 (GA3) has a promoting effect on brown spot (BS) (Wang et al., 2021). In the present study, the expression levels of bZIP genes in exocarp of ‘Huangguan’ pear were evaluated in normal fruit, fruit with BS and fruit treated with GA3. These results enhance our understanding the characteristics of the bZIP gene family in the pear genome and provide insights into how bZIPs participate in regulating BS on the skin of ‘Huangguan’ pear through gibberellin (GA) signaling.
Material and Methods
Plant material
‘Huangguan’ pear was used as a material in this study, and its fruit samples were grown in a 30-year-old horticulture orchard (Dangshan, Anhui, China). GA3 (300 mg/L, Sigma G8040) and water were sprayed on ‘Huangguan’ pear at 10, 20 and 30 days after full bloom (DAFB). After treatment, the fruits were bagged individually. Each treatment had three biological replicates, and each tree had approximately 120 treated fruits. All fruit samples were collected during the harvest season in 2018. The peel was cut into slices manually, immediately frozen in liquid nitrogen, and stored at -80 °C.
Identification of PbbZIP family members
The PF00170 and PF07716 domain model files of the bZIP family members were downloaded from the PFAM website (https://www.pfam.org), and the coding sequence (CDS) nucleotide sequence files, transcript amino acid sequence files and gene annotation files were downloaded from the Genome Database for Rosaceae (GDR) (https://www.rosaceae.org) and the Phytozome Genome Data Resource Library (https://genome.jgi.doe.gov/portal). The first batch of candidate genes containing PF00170 and PF07716 domains was identified (E-value=e-10) using HMMER v.3.2 (Yap et al., 2016).
The AtbZIP genes were obtained by querying the Arabidopsis database (TAIR) (https://www.arabidopsis.org/), and a local BLAST library was constructed using the amino acid sequence files of each species. Then, a BLASTP search was carried out using the local BLAST library (E-value= e-10) with the AtbZIP amino acids, and another batch of candidate genes was obtained.
The two batches of candidate genes were merged and the domains were identified using HMMSCAN (Finn et al., 2015) (E-value=e-10). Low similarity and repetitive sequences were manually removed. Finally, the prephenate dehydratase and ACT domains of the family members were identified using SMART (http://smart.embl- heidelberg.de) and PFAM (https://www.pfam.org), respectively.
Phylogenetic analysis of the PbbZIP family genes
The bZIP protein sequences of white pear and Arabidopsis were extracted and aligned. The InterProScan program was used on all of the candidate protein pairs and confirmed the presence of the diagnostic domain using the Pfam and SMART databases. MAFFT was used with the default parameters to align the sequences of the multiple homologous bZIP genes. A phylogenetic tree was constructed using the maximum likelihood method and IQ-TREE 1.6.9 software (Nguyen et al., 2015). The support values displayed next to the branches were inferred from 1000 replicate trees.
Gene structural and conserved motif analyses of the PbbZIP family genes
Based on the amino acid sequences, the conserved motifs of the PbbZIP family members were analyzed with Motif EM for Motif Elicitation (MEME) v.5.0 software (Bailey et al., 2009). The motif value was set to 10, and the minimum and maximum motif lengths were set to 6 and 50, respectively.
The chromosome position information of the bZIP family members was extracted from the gene annotation files. The major features, including the coding and noncoding regions and the exon-intron pattern, were characterized with Gene Structure Display server (GSDS) v.2.0 software (http://gsds.cbi.pku.edu.cn/index.php). The amino acid sequences of the bZIPs in pear and Arabidopsis were aligned and divided into different domains using Jalview v.2.10 software (Waterhouse et al., 2009).
Analysis of the physical and chemical properties of the PbbZIP familiy genes
The protein physicochemical properties of the bZIP family members were predicted via the ExPASy website (https://www.expasy.org/) (Gasteiger et al., 2003). The signal peptides were analyzed through SignalP (http://www.cbs.dtu.dk/services/SignalP/) (Armenteros et al., 2019). The subcellular locations were calculated using CELLO v.2.5 software (http://cello.life.nctu.edu.tw/) (Yu et al., 2004). The transmembrane structures were predicted by TMHMM Server v.2.0 software (http://www.cbs.dtu.dk/services/TMHMM/) (Krogh et al., 2001).
Gene collinearity relationships of the PbbZIP family genes
The multiple collinearity scan toolkit (MCScanX) (Wang et al., 2012) was used for analysis of collinearity between multiple genomes and the pear genome, and the homologous regions between pears and Arabidopsis were anchored. Gene collinearity relationships were visualized using the Python package circos (https://github.com/Tanghaibao/circos).
Transcriptional profiling of the PbbZIP family genes
An RNA-seq dataset (PRJNA682706) of pear subjected to several different treatments, including NaH2PO4·2H2O (0.2%, Sigma 04269), ABA (100 μM, Sigma A1049), and GA3 (300 mg/L, Sigma G8040), as well as normal fruits and fruits with BS disease, was obtained from our previous work (Wang et al., 2021). Each had three biological replicates. Data from normal fruit, fruit with BS and fruit treated with GA3 was used in this study. The raw data were filtered, and the fragments per kilobase of transcript per million mapped reads (FPKM) values were calculated and investigated for the expression of PbbZIP members. A heatmap of the gene expression profiles of all PbbZIP genes was constructed using TBtools software (Chen et al., 2020).
Coexpression network of PbbZIP genes and GA signaling genes in BS formation
To explore the expression patterns of PbbZIP genes and GA signaling genes during BS formation, RNA sequencing (RNA-seq) data of exocarp of healthy ‘Huangguan’ pear and ‘Huangguan’ pear with BS disease were used to measure the expression similarity between gene pairs as Pearson’s correlation coefficient (PCC) values. The values were then filtered with Excel software (with the parameter set as > 0.4). Visualizations of the data were performed with Cytoscape software (Shannon et al., 2003).
qRT-PCR analysis of PbbZIP genes
To confirm the expression of the PbbZIP genes, total RNA was extracted from each fruit sample with a total RNA purification kit and used to perform reverse transcription with the Prime Script™ RT Reagent Kit. Beacon Designer 7.9 software was used to design the specific primers (Table S1). qRT-PCR was conducted in a 20 µL reaction volume that comprised 10 µL SYBR Premix ExTaq II, 2 µL template cDNA, 0.8 µL each of forward and reverse primer and the remaining volume with nuclease-free water. The PCR conditions were as follows: 94°C for 45 s, followed by 40 cycles of 94°C for 15 s, 60°C for 20 s and 72°C for 20 s; and then holding at 4°C forever. GAPDH in pear was used as the reference gene. The 2-ΔΔCT method was used to calculate the relative expression levels of the PbbZIP genes.
Results
Identification of bZIP TFs in P. bretschneideri
Genome-wide analysis was performed to search for bZIP members in the P. bretschneideri genome (Wu et al., 2013). Eighty-four bZIP genes were identified in pear by Pfam and inter-ProScan confirmation, and several manual checks were performed. Based on their positions on the chromosomes, the bZIP genes in pear were denominated as PbBZIP1-PbBZIP84 (Table S1). Our analysis confirmed that all of the identified PbbZIP proteins each contained conserved bZIP_1 (PF00170) and bZIP_2 (PF07716) domains, which are the specific conserved domains of the PbbZIP gene family.
The lengths of the PbbZIP genes ranged from 369 bp to 2,229 bp, and the average length was 1,021 bp. The PbbZIP proteins contained 123 to 743 amino acids, with an average of 340 amino acids. Molecular weight (MW) ranged from 30,140.67 Da to 183,693.94 Da, with an average of 83,865.54 Da. The predicted isoelectric points of the PbbZIP proteins ranged from 4.91 to 5.23 (Table S2). In addition, all 84 PbbZIP genes were predicted to be located in the nucleus.
Phylogenetic relationships of the PbbZIP proteins
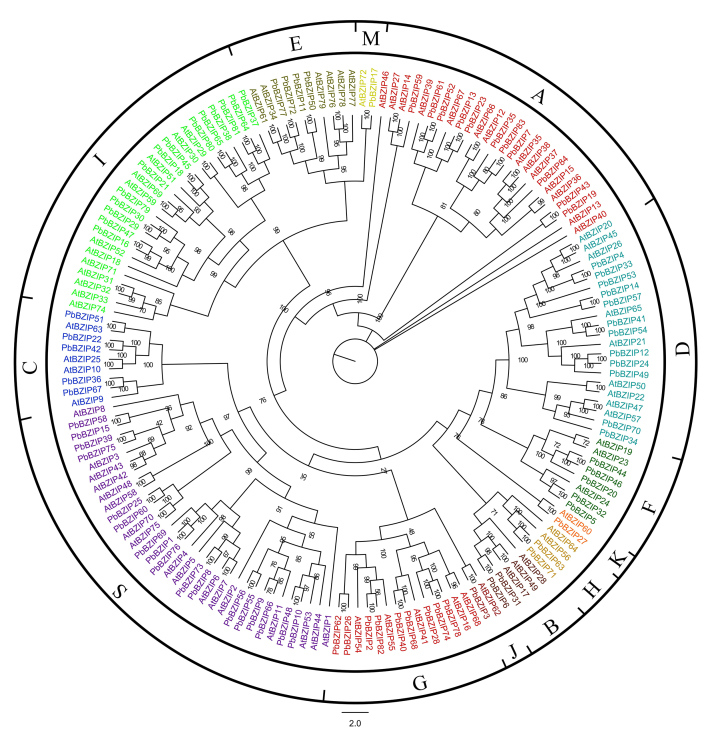
To further investigate the phylogenetic relationships among the bZIP proteins in P. bretschneideri and Arabidopsis, a phylogenetic tree was constructed. The tree showed that the bZIP proteins of Chinese white pear and Arabidopsis could be divided into 13 subfamilies (denoted Groups A, B, C, D, E, F, G, H, I, J, K, S and M). PbbZIP genes in both P. bretschneideri and Arabidopsis contributed to all of the subfamilies A-M (Figure 1). Among these subfamilies, Group S had the largest number of PbbZIP gene members, 17, while Groups J, K and M had the smallest number, 1. These results may indicate special functions of bZIP members in white pear.
Figure 1 -. Phylogenetic analysis of bZIPs from pear and Arabidopsis thaliana. The tree was constructed with IQ-TREE v.1.6 software. The bZIP TFs clustered into 13 distinct clades, marked by curves of different colors.
Conserved structure and intron-exons of the PbbZIP gene family
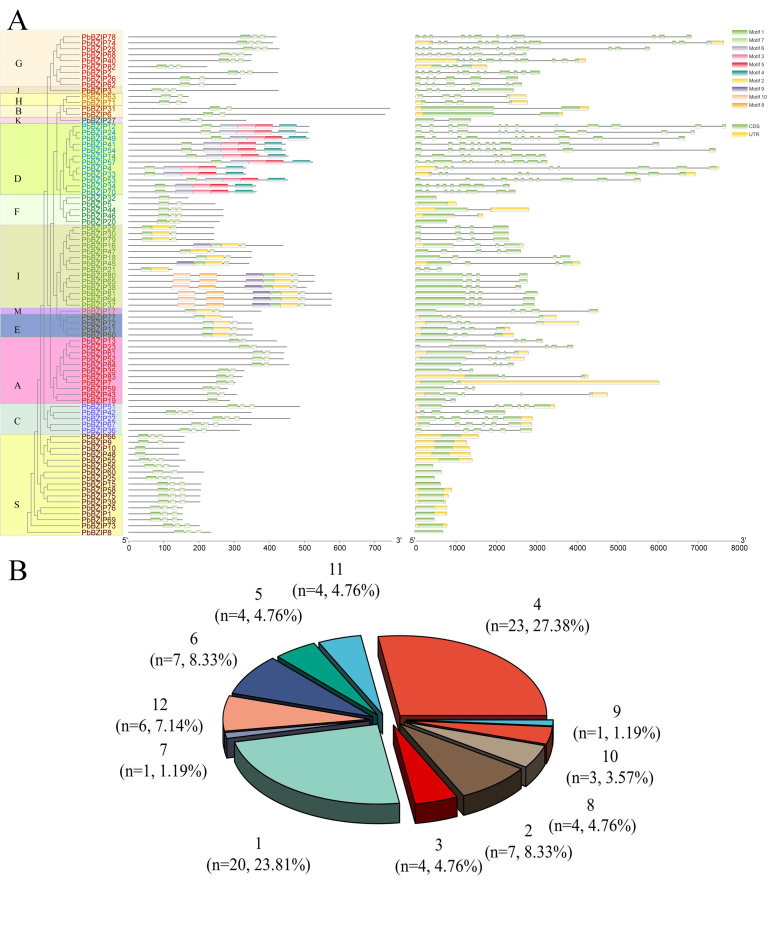
The genomic structures of the 84 PbbZIP genes were quite different from one another. All contained at least one exon, and the maximal number was 12 (Figure 2B, Table S3). Six PbbZIPs contained 12 exons (7.14%), 4 members contained 11 exons (4.76%), 3 contained 10 (3.57%), 1 contained 9 (1.19%), 4 contained 8 (4.76%), 1 contained 7 (1.19%), 7 contained 6 (8.33%), 4 contained 5 (4.76%), 23 contained 4 (27.38%), 4 contained 3 (4.76%), 7 contained 2 (8.33%) and 20 contained 1 (23.81%). In addition, the members within a subfamily had similar gene structures, including similar lengths and numbers of exons, which supported the classification and the identified evolutionary relationships.
Figure 2 -. Phylogenetic relationships, motif compositions and gene structures of PbbZIPs. (A) The phylogenetic tree was produced by MEGA using the neighbor-joining method with 1,000 bootstrap replicates. (B) A schematic representing the conserved motifs of the PbbZIPs identified by MEME. Each motif is indicated by a colored box numbered at the bottom. (C) Exon number distribution of the PbbZIP family TFs in white pear.
In total, 10 motifs were detected from the PbbZIP genes. Within a subfamily, the numbers and types of conserved motifs of PbbZIP were similar, while these were quite diverse among the different subfamilies (Figure 2A). Motif 1 domain is present in all PbbZIP proteins. In addition, some motifs have obvious specificity and belong to specific subgroups. For example, motif 6, motif 3, motif 5, and motif 4 only appear in the D subfamily; motif 8, motif 9 and motif 10 only appear in the I subfamily; and motif 2 only appears in the I and M subgroups. The other subgroups have only the motif 1 domain. Different subgroups with specific motifs may contribute to specific functions.
Chromosomal locations and syntenic relationships of the PbbZIP genes
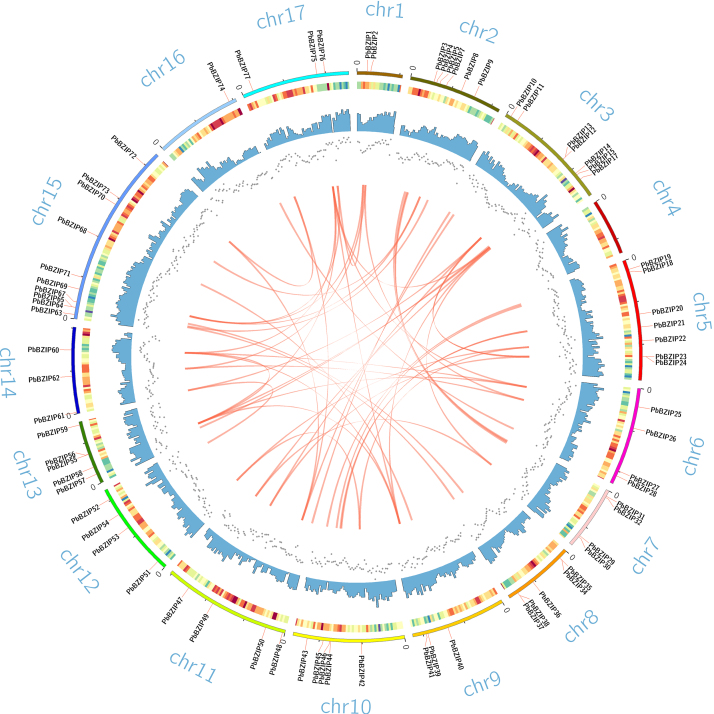
To determine the distribution of the PbbZIP genes on the pear chromosomes, their chromosomal locations were visualized with MCScanX software. The 77 PbbZIP genes were found to be randomly and unevenly distributed on the 17 chromosomes across the white pear genome and the other 7 PbbZIP genes were located on the scaffold (Figure 3, Table S4). The number of PbbZIP genes on each individual chromosome ranged from 0 (Chr4) to 11 (Chr15). The same number of PbbZIP genes (5) were distributed on chromosomes 7, 8, 10 and 13. There were eight PbbZIP genes located on chromosomes 3 and 5. Chromosomes 6, 11 and 12 harbored four PbbZIP genes, and three PbbZIP genes were located on chromosomes 9, 14 and 17.
Figure 3 -. The chromosomal locations of 77 PbbZIPs. The genes were mapped to the pear chromosomes by TBtools. The chromosomes of pear are shown arranged in a circle. The outer circle represents the chromosome. Heatmaps, scatterplots and histograms were used to record the gene density of each chromosome segment. The length of each chromosome segment was set at 500,000 bp.
Gene tandem and fragment repeats are key factors that promote the number of genes in a particular gene family and lead to functional diversity during evolution. In this study, three pairs of PbbZIP genes (PbbZIP34/PbbZIP35, PbbZIP37/PbbZIP38, PbbZIP64/PbbZIP65) were identified as the result of tandem duplication, and 53 pairs were identified as the result of segmental duplication, indicating that segmental duplication was the main force driving the expansion of the PbbZIP family in pear.
Phenotype of ‘Huangguan’ pear and the expression patterns of the PbbZIP genes
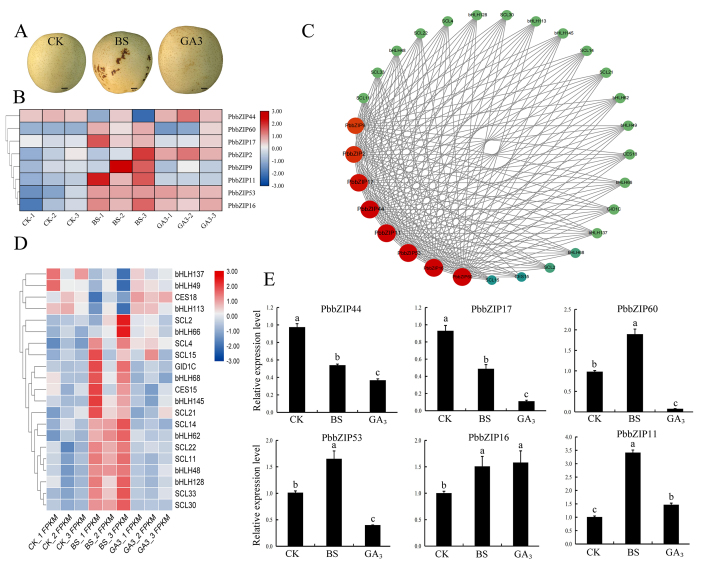
The phenotypes of healthy ‘Huangguan’ pear, ‘Huangguan’ pear with BS and ‘Huangguan’ pear treated with GA3 were observed. The results showed that the fruit size increased significantly after GA3 treatment (Figure 4A). In our previous study, we found that the GA signaling pathway may be the key pathway regulating the occurrence of BS (Wang et al., 2021). Based on the transcriptome analysis, 8 PbbZIP genes were identified, most of which were significantly upregulated in pears with BS compared with healthy pears. Among them, PbbZIP44, PbbZIP2, PbbZIP53 and PbbZIP16 were also significantly upregulated with GA3 treatment (Figure 4B, Table S4). These results suggest that the expression of some PbbZIP genes can be activated by GA3 treatment and promote the occurrence of BS, which demonstrates that PbbZIP family genes play an important role in regulating the formation of BS and are responsive to GA3 hormone.
Figure 4 -. Phenotypic characteristics of BS, expression profiles of PbbZIP genes and a coexpression network and heatmap for PbbZIPs with GA signaling genes in pear. (A) Phenotypes of normal ‘Huangguan’ pear (CK), ‘Huangguan’ pear with BS disease and ‘Huangguan’ pear treated with GA3. (B) The expression patterns of PbbZIP genes in ‘Huangguan’ pear. (C) Interaction network analysis of PbbZIPs with GA signaling genes in pear was performed using the STRING database. (D) Heatmap of GA signaling genes expressed in ‘Huangguan’ and ‘Huangguan’ with BS based on the fold change (log2) in FPKM values. The color scale at the top represents log2-FPKM values. (E) Relative expression profiles of PbbZIP genes in the exocarp of ‘Huangguan’ pear. Error bars represent the SE of three biological replicates. Lowercase letters indicate significant differences between treatments at the P < 0.05 level. CK: normal ‘Huangguan’ pear; BS: ‘Huangguan’ pear with BS; GA3: ‘Huangguan’ pear treated with GA3.
Coexpression network of BS-related genes
Coexpression network analysis was performed to illuminate the collaboration among the PbbZIP genes, analysis of the transcriptome data showed that 8 bZIP genes could be classified into different coexpression clusters with GA signaling genes (Figure 4C, Table S5). These PbbZIP genes had high correlations with 21 GA signaling genes, which were highly expressed in BS (Figure 4D). These results further supported the hypothesis that PbbZIP TFs may regulate the formation of BS through the GA signaling pathway.
qRT-PCR analysis of PbbZIPs in the exocarp of ‘Huangguan’ pear
The expression levels of PbbZIP11, PbbZIP16, PbbZIP53, PbbZIP60, and PbCPRF2 in ‘Huangguan’ pear with BS were significantly higher than those in normal ‘Huangguan’ pear. After GA3 treatment, the expression of the PbbZIP53, PbbZIP60 and PbHY5 genes in ‘Huangguan’ pears decreased and was significantly lower than that in normal ‘Huangguan’, while the expression of PbbZIP11 was significantly higher than that in normal ‘Huangguan’ pear (Figure 4E).
Discussion
The bZIP gene family is a large and complicated family that has many members belonging to different subfamilies. bZIP genes are involved in the responses to abiotic stress and biotic stresses. In a previous study, bZIP genes were identified in many species. For example, 50, 116, 47 and 45 bZIP genes were identified in Malus domestica (apple), Prunus persica (peach), Fragaria vesca (strawberry) and Chinese jujube, respectively (Wang et al., 2017; Zhang et al., 2020). In the present study, a total of 84 bZIP members were identified from white pear, this number is higher than that in Malus domestica, Fragaria vesca and Chinese jujube and lower than that in Prunus persica. These results indicate that bZIP gene loss might occur in some genomes.
Phylogenetic analysis showed that the PbbZIPs could be divided into 13 subfamilies (Figure 2), which is consistent with the results for Arabidopsis thaliana. Notably, some PbbZIP genes originally belonging to subgroup I became isolated from their clusters, which has also been seen in Pyrus communis, Arabidopsis thaliana and Vitis vinifera (Liu et al., 2014). The gene structure and conserved motif results further validated the phylogenetic analysis results (Figure 2). Among the PbbZIPs, approximately 23.8% had no introns, and all of them were classified into the S and F groups, which is consistent with previous reports in maize (Zea mays) (Wei et al., 2012). In addition, we found that Group D belongs to a specific subgroup, and similar findings have been obtained in celery (Apium graveolens).
During evolution, plant genomes have become more complex to adapt to changes in the environment (Fernie and Gutierrez-Marcos, 2019). TFs play important roles in plant environmental adaptation. NAC, bZIP, MYB and WRKY are common TFs in plants that play crucial roles in the regulation of plant hormone-mediated signals for disease and stress resistance. In recent years, increasing evidence has suggested that bZIP genes are involved in plant responses to abiotic and biotic stresses, including phytohormone ABA signaling (An et al., 2018), pathogen defense (Thurow et al., 2005; Kaminaka et al., 2006), drought and high salinity (Huang et al., 2016), cold stresses (Shimizu et al., 2005) and light irradiation (Ulm et al., 2004; Osterlund et al., 2000). For example, the CabZIP1 TF in pepper (Piper nigrum) plays a regulatory role in disease defense and stress responses (Lee et al., 2006). In studies of Arabidopsis thaliana, transcription of AtbZIP44 was responsive to temperature, and under ABA, salt and osmotic stress, transcription of AtbZIP53 was upregulated in seeds, and transcription of AtbZIP2 was downregulated in roots (Kang et al., 2002; Weltmeier et al., 2009). BS is reported to be associated with light irradiation, sudden drops in temperature, calcium deficiency, and various physiological, developmental and hormonal responses (Li et al., 2008; Wang et al., 2011; Wang et al., 2021). These results provide evidence of the potential involvement of bZIP genes in the formation of BS on the surface of ‘Huangguan’ pear.
Functional diversity in bZIP genes has been observed in various plant species; for example, bZIP11 can redundantly repress primary root growth by directly activating IAA3/SHY2 transcription (Weiste et al., 2017), and it participates in regulating the metabolism of trehalose and other minor carbohydrates and amino acid metabolism in Arabidopsis by sucrose signaling (Weltmeier et al., 2009; Ma et al., 2011). These observations potentially reflect some valuable functions of its target genes. In the present study, the expression of PbbZIP11 in ‘Huangguan’ pear with BS was higher than that in CK pear, while the expression of PbbZIP11 in ‘Huangguan’ pear was decreased significantly after GA3 treatment. These results demonstrate that PbbZIP11 could be induced by GA3 and participate in BS formation on the surface of ‘Huangguan’ pear. Our results also provide insights into how bZIPs potentially regulate BS disease through GA signaling.
Acknowledgements
This project was supported by the National Natural Science Foundation of China (31972985), China Agriculture Research System of MOF and MARA and Anhui Province Fruit-Tree Industry Technology System.
Supplementary material.
The following online material is available for this article:
References
- Amorim LLB, Santos RF, Bezerra JP, Neto, Guida-Santos M, Crovella S, Benko-Iseppon AM. Transcription factors involved in plant resistance to pathogens. Curr Protein Pept Sci. 2017;18:335–351. doi: 10.2174/1389203717666160619185308. [DOI] [PubMed] [Google Scholar]
- An JP, Yao JF, Xu RR, You CX, Wang XF, Hao YJ. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid‐promoted anthocyanin accumulation. Plant Cell Environ. 2018;41:2678–2692. doi: 10.1111/pce.13393. [DOI] [PubMed] [Google Scholar]
- Armenteros JJA, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, Heijne GV, Nielsen H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloglu MC, Eldem V, Hajyzaeh M, Unver T. Genome-wide analysis of the bZIP transcription factors in cucumber. PloS One. 2014;9:e96014. doi: 10.1371/journal.pone.0096014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Fernie AR, Gutierrez-Marcos J. From genome to phenome: genome‐wide association studies and other approaches that bridge the genotype to phenotype gap. Plant J. 2019;97:5–7. doi: 10.1111/tpj.14219. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, Bateman A, Eddy SR. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43:W30– W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JNM, Harrison SC. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- Gong X, Zhao L, Song X, Lin Z, Gu B, Yan J, Zhang S, Tao S, Huang X. Genome-wide analyses and expression patterns under abiotic stress of NAC transcription factors in white pear (Pyrus bretschneideri) BMC Plant Biol. 2019;19:161. doi: 10.1186/s12870-019-1760-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Lee Y, Cho JI, Ahn CH, Lee SK, Jeon JS, Kang H, Lee CH, An G, Park PB. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol Biol. 2010;72:557–566. doi: 10.1007/s11103-009-9592-9. [DOI] [PubMed] [Google Scholar]
- Hsieh TH, Li CW, Su RC, Cheng CP, Sanjaya Tsai YC, Chan MT. A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta. 2010;231:1459–1473. doi: 10.1007/s00425-010-1147-4. [DOI] [PubMed] [Google Scholar]
- Huang C, Zhou J, Jie Y, Xing H, Zhong Y, She W, Wei G, Yu W, Ma Y. A ramie (Boehmeria nivea) bZIP transcription factor BnbZIP3 positively regulates drought, salinity and heavy metal tolerance. Mol Breeding. 2016;36:120 [Google Scholar]
- Huang X, Li K, Xu X, Yao Z, Jin C, Zhang S. Genome-wide analysis of WRKY transcription factors in white pear (Pyrus bretschneideri) reveals evolution and patterns under drought stress. BMC Genomics. 2015;16:1104. doi: 10.1186/s12864-015-2233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7 doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Jin M, Gan S, Jiao J, He Y, Liu H, Yin X, Zhu Q, Rao J. Genome-wide analysis of the bZIP gene family and the role of AchnABF1 from postharvest kiwifruit (Actinidia chinensis cv. Hongyang) in osmotic and freezing stress adaptations. Plant Sci. 2021;308:110927. doi: 10.1016/j.plantsci.2021.110927. [DOI] [PubMed] [Google Scholar]
- Kaminaka H, NäKe C, Epple P, Dittgen J, Schütze K, Chaban C, Holtlll BF, Merkle T, Schäfer E, Harter K, et al. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 2006;25:4400–4411. doi: 10.1038/sj.emboj.7601312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell. 2002;14:343–357. doi: 10.1105/tpc.010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lee SC, Choi HW, Hwang IS, Choi DS, Hwang BK. Functional roles of the pepper pathogen-induced bZIP transcription factor, CAbZIP1, in enhanced resistance to pathogen infection and environmental stresses. Planta. 2006;224:1209–1225. doi: 10.1007/s00425-006-0302-4. [DOI] [PubMed] [Google Scholar]
- Li LM, Cheng FM, Guan JF, Ji H, Feng YX, Sun YL, Gong XM. Effects of precooling on the storage quality and peel browning of Huangguan pear. Acta Agric Bor Sin. 2008;23:156–160. [Google Scholar]
- Liu J, Chen N, Chen F, Cai B, Santo SD, Tornielli GB, Pezzotti M, Cheng ZM. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera) BMC Genomics. 2014;15:281. doi: 10.1186/1471-2164-15-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Ma QJ, Lu J, Hao YJ, You CX. Primary analysis of the function of bZIP transcription factor gene MdAREB2 in apple. Acta Hortic Sin. 2018;45:1–10. [Google Scholar]
- Lu B, Wang Y, Zhang G, Feng Y, Yan Z, Wu J, Chen X. Genome-wide identification and expression analysis of the strawberry FvbZIP gene family and the role of key gene FabZIP46 in fruit resistance to gray mold. Plants (Basel) 2020;9:1199. doi: 10.3390/plants9091199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Hanssen M, Lundgren K, Hernández L, Delatte T, Ehlert A, Liu CM, Schluepmann H, Dröge-Laser W, Moritz T, et al. The sucrose-regulated Arabidopsis transcription factor bZIP11 reprograms metabolism and regulates trehalose metabolism. New Phytol. 2011;191:733–745. doi: 10.1111/j.1469-8137.2011.03735.x. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhawan A, Jain M, Tyagi AK, Khurana JP. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008;146:333–350. doi: 10.1104/pp.107.112821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Rong S, Wu Z, Cheng Z, Zhang S, Liu H, Huang Q. Genome-wide identification, evolutionary patterns, and expression analysis of bZIP gene family in olive (Olea europaea L.) Genes (Basel) 2020;11:510. doi: 10.3390/genes11050510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H, Sato K, Berberich T, Miyazaki A, Ozaki R, Imai R, Kusano T. LIP19, a basic region leucine zipper protein, is a Fos-like molecular switch in the cold signaling of rice plants. Plant Cell Physiol. 2005;46:1623–1634. doi: 10.1093/pcp/pci178. [DOI] [PubMed] [Google Scholar]
- Teng YW, Chai ML, Li XG. A historic retrospect and the progress in the taxonomy of the genus Pyrus. J Fruit Sci. 2004;21:252–257. [Google Scholar]
- Thurow C, Schiermeyer A, Krawczyk S, Butterbrodt T, Nickolov K, Gatz C. Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J. 2005;44:100–113. doi: 10.1111/j.1365-313X.2005.02513.x. [DOI] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Máté Z, Ádám É, Oakeley EJ, Schäfer E, Nagy F. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci U S A. 2004;101:1397–1402. doi: 10.1073/pnas.0308044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HB, Guo JY, Tang LZ, Li C. Genome-wide identification and expression analysis of bZIP transcription factor family in Jatropha curcas. Acta Agric Bor Sin. 2019;34:44–58. [Google Scholar]
- Wang J, Zhou J, Zhang B, Vanitha J, Ramachandran S, Jiang SY. Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J Integr Plant Biol. 2011;53:212–231. doi: 10.1111/j.1744-7909.2010.01017.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Wu X, Liu L, Yao D, Li J, Fang J, Chen X, Zhu L, Liu P, Ye Z, et al. Transcriptome and metabolomic analysis to reveal the browning spot formation of ‘Huangguan’ pear. BMC Plant Biol. 2021;21:321. doi: 10.1186/s12870-021-03049-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XL, Chen X, Yang TB, Cheng Q, Cheng ZM. Genome-wide identification of bZIP family genes involved in drought and heat stresses in strawberry (Fragaria vesca) Int J Genomics. 2017;2017:3981031. doi: 10.1155/2017/3981031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K, Chen J, Wang Y, Chen Y, Chen S, Lin Y, Pan S, Zhong X, Xie D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012;19:463–476. doi: 10.1093/dnares/dss026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiste C, Lorenzo P, Selvanayagam J, Muralidhara P, Fröschel C, Novák O, Ljung K, Hanson J, Dröge-Laser W. The Arabidopsis bZIP11 transcription factor links low-energy signalling to auxin-mediated control of primary root growth. PLoS Genet. 2017;13:e1006607. doi: 10.1371/journal.pgen.1006607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltmeier F, Rahmani F, Ehlert A, Dietrich K, Schütze K, Wang X, Chaban C, Hanson J, Teige M, Harter K, et al. Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol Biol. 2009;69:107–119. doi: 10.1007/s11103-008-9410-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S, Khan M.A, Tao S, Korban S, Wang H, et al. The genome of the pear (Pyrus bretschneideri Rehd.) Genome Res. 2013;23:396–408. doi: 10.1101/gr.144311.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap CK, Eisenhaber B, Eisenhaber F, Wong WC. xHMMER3x2: Utilizing HMMER3’s speed and HMMER2’s sensitivity and specificity in the glocal alignment mode for improved large-scale protein domain annotation. Biol Direct. 2016;11:63. doi: 10.1186/s13062-016-0163-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CS, Lin CJ, Hwang JK. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004;13:1402–1406. doi: 10.1110/ps.03479604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao W, Li H, Wang Y, Li D, Xuan C, Liu Z, Liu M, Zhao J. Genome-wide analysis of the bZIP gene family in Chinese jujube (Ziziphus jujuba Mill.) BMC Genomics. 2020;21:483. doi: 10.1186/s12864-020-06890-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.