Abstract
Background
Pathology reports serve as an auditable trial of a patient’s clinical narrative, containing text pertaining to diagnosis, prognosis, and specimen processing. Recent works have utilized natural language processing (NLP) pipelines, which include rule-based or machine-learning analytics, to uncover textual patterns that inform clinical endpoints and biomarker information. Although deep learning methods have come to the forefront of NLP, there have been limited comparisons with the performance of other machine-learning methods in extracting key insights for the prediction of medical procedure information, which is used to inform reimbursement for pathology departments. In addition, the utility of combining and ranking information from multiple report subfields as compared with exclusively using the diagnostic field for the prediction of Current Procedural Terminology (CPT) codes and signing pathologists remains unclear.
Methods
After preprocessing pathology reports, we utilized advanced topic modeling to identify topics that characterize a cohort of 93,039 pathology reports at the Dartmouth-Hitchcock Department of Pathology and Laboratory Medicine (DPLM). We separately compared XGBoost, SVM, and BERT (Bidirectional Encoder Representation from Transformers) methodologies for the prediction of primary CPT codes (CPT 88302, 88304, 88305, 88307, 88309) as well as 38 ancillary CPT codes, using both the diagnostic text alone and text from all subfields. We performed similar analyses for characterizing text from a group of the 20 pathologists with the most pathology report sign-outs. Finally, we uncovered important report subcomponents by using model explanation techniques.
Results
We identified 20 topics that pertained to diagnostic and procedural information. Operating on diagnostic text alone, BERT outperformed XGBoost for the prediction of primary CPT codes. When utilizing all report subfields, XGBoost outperformed BERT for the prediction of primary CPT codes. Utilizing additional subfields of the pathology report increased prediction accuracy across ancillary CPT codes, and performance gains for using additional report subfields were high for the XGBoost model for primary CPT codes. Misclassifications of CPT codes were between codes of a similar complexity, and misclassifications between pathologists were subspecialty related.
Conclusions
Our approach generated CPT code predictions with an accuracy that was higher than previously reported. Although diagnostic text is an important source of information, additional insights may be extracted from other report subfields. Although BERT approaches performed comparably to the XGBoost approaches, they may lend valuable information to pipelines that combine image, text, and -omics information. Future resource-saving opportunities exist to help hospitals detect mis-billing, standardize report text, and estimate productivity metrics that pertain to pathologist compensation (RVUs).
Keywords: BERT, Current procedural terminology, Deep learning, Machine learning, Pathology reports, XGBoost
1. Background and significance
Electronic Health Records (EHR)1 refer to both the structured and unstructured components of patients’ health records/information (PHI), synthesized from a myriad of data sources and modalities. Such data, particularly clinical text reports, are increasingly relevant to “Big Data” in the biomedical domain. Structured components of EHR, such as clinical procedural and diagnostic codes, are able to effectively store the patient’s history,2, 3, 4 whereas unstructured clinical notes reflect an amalgamation of more nuanced clinical narratives. Such documentation may serve to refresh the clinician on the patient’s history, highlight key aspects of the patient’s health, and facilitate patient handoff among providers. Further, analysis of clinical free text may reveal physician bias or inform an audit trail of the patient’s clinical outcomes for purposes of quality improvement. As such, utilizing sophisticated algorithmic techniques to assess text data in pathology reports may improve decision making and hospital processes/efficiency, possibly saving hospital resources while prioritizing patient health.
NLP3,5, 6, 7, 8 is an analytic technique that is used to extract semantic and syntactic information from textual data. Traditionally, rule-based approaches cross-reference and tabulate domain-specific key words or phrases with large biomedical ontologies and standardized vocabularies, such as the Unified Medical Language System (UMLS).9,10 However, although these approaches provide an accurate means of assessing a narrow range of specified patterns, they are neither flexible nor generalizable since they require extensive annotation and development from a specialist. Machine-learning approaches (e.g. support vector machine (SVM), random forest)11,12 employ a set of computational heuristics to circumvent manual specification of search criteria to reveal patterns and trends in the data. Bag-of-word approaches13,14 study the frequency counts of words (unigrams) and phrases (bigrams, etc.) to compare the content of multiple documents for recurrent themes, whereas deep learning approaches15, 16, 17 simultaneously capture syntax and semantics with artificial neural network (ANN) techniques. Recent deep learning NLP approaches have demonstrated the ability to capture meaningful nuances that are lost in frequency-based approaches; for instance, these approaches can effectively contextualize short- and long-range dependencies between words.18,19 Despite potential advantages conferred from less structured approaches, the analysis of text across any domain usually necessitates balancing domain-specific customization (e.g. a medical term/abbreviation corpora) with generalized NLP techniques.
The analysis of pathology reports using NLP has been particularly impactful in recent years, particularly in the areas of information extraction, summarization, and categorization. Noteworthy developments include information extraction pipelines that utilize regular expressions (regex), to highlight key report findings (e.g., extraction of molecular test results),20, 21, 22, 23 as well as topic modeling approaches that summarize a document corpus by common themes and wording.24 In addition to extraction methods, machine-learning techniques have been applied to classify pathologist reports25; notable examples include the prediction of ICD-O morphological diagnostic codes26,27 and the prediction of CPT codes based only on diagnostic text.28,29 Widespread misspelling of words and jargon specific to individual physicians have made it difficult to reliably utilize the rule-based and even machine-learning approaches for report prediction in a clinical workflow. In addition, hedging and uncertainty in text reports may further obfuscate findings.30
The CPT codes are assigned to report reimbursable medical procedures for diagnosis, surgery, and ordering of additional ancillary tests.31,32 Assignments of CPT codes are informed by guidelines and are typically integrated into the Pathology Information System. As such, the degree to which new technologies and practices are implemented and disseminated are often informed by their impact on CPT coding practices. Reimbursements from CPT codes can represent tens to hundreds of millions of dollars of revenue at mid-sized medical centers, and thus systematic underbilling of codes could lead to lost hospital revenue, whereas overbilling patterns may lead to the identification of areas of redundant or unnecessary testing (e.g., duplication of codes, ordering of unnecessary tests, or assignment of codes representing more complex cases, etc.).
Ancillary CPT codes represent procedural codes that are automatically assigned when ancillary tests are ordered (e.g., immunohistochemical stains; e.g., CPT 88341, 88342, 88313, 88360, etc.). In contrast, primary CPT codes (e.g., CPT 88300, 88302, 88304, 88305, 88307, and 88309) are assigned based on the pathologist examination of the specimen, where CPT 88300 represents an examination without requiring the use of a microscope (gross examination), whereas CPT 88302-88309 include gross and microscopic examination of the specimen and are ordered by the case’s complexity level (as specified by the CPT codebook; an ordinal outcome; e.g., CPT 88305: Pathology examination of tissue using a microscope, intermediate complexity), which determines reimbursement. The assignment of such codes is not devoid of controversy. Although it is expected that raters will not report a specimen with a higher/lower code level, some may argue that such measures may not reflect the degree of difficulty for a particular case or there may not be a specific language that denotes primary CPT code placement of the phenomena (i.e., unlisted specimen, where it is at the pathologist’s discretion to determine placement). For these codes, case complexity may ultimately be traced back to the clinical narrative reported in the pathology report text.33
Since the assignment of case complexity is sometimes unclear to the practicing pathologist as guidelines evolve, the prediction of these CPT codes from the diagnostic text using NLP algorithms can be used to inform whether a code was assigned that matches the case complexity. Recently developed approaches to predict CPT codes demonstrate remarkable performance; however, they only rely on the first 100 words from the report text, do not compare across multiple state-of-the-art NLP prediction algorithms, and do not consider report text outside of the diagnosis section.28 Further, report lexicon is hardly standardized, as it may be littered with language and jargon that is specific to the sign-out pathologist and may vary widely in length for the same diagnosis, which can make it difficult to build an objective understanding of the report text.
Comparisons of different algorithmic techniques and relevant reporting text to use for the prediction of primary CPT codes are essential to further understand their utility for curbing under/overbilling issues. In addition, contextualizing primary code findings by ancillary findings and building a greater understanding of how pathologists differ in their lexical patterns may provide further motivation for the standardization of reporting practices and how report text can optimize the ordering of ancillary tests.34
1.1. Objective
The primary objective of this study is to compare the capacity to delineate primary CPT procedural codes (CPT 88302, 88304, 88305, 88307, 88309) corresponding to case complexity across state-of-the-art machine-learning models over a large corpus of more than 93,039 pathology reports from the Dartmouth-Hitchcock Department of Pathology and Laboratory Medicine (DPLM), a midsized academic medical center. Using XGBoost, SVM, and BERT techniques, we hope to gain a better understanding of which algorithms are useful for predicting primary CPT codes representing case complexity, which will prove helpful for the detection of under/overbilling.
1.2. Secondary objectives
We have formulated various secondary objectives that are focused on capturing additional components of reporting variation:
1. Expanded reporting subfields: Exploration of methods that incorporate other document subfields outside of the diagnostic text into the modeling approaches, which may contain additional information.
2. Ancillary Testing Codes: Predicting the assignment of 38 different CPT procedure codes, largely comprising secondary CPT codes, under the hypothesis that nondiagnostic text provides additional predictive accuracy as compared with primary CPT codes, which may rely more heavily on the diagnostic text. Although the prediction of whether an ancillary test was ordered via secondary CPT codes has limited potential for incorporation into the Pathology Information System, as these codes are automatically assigned after test ordering, prediction of the ancillary tests can provide an additional context for the prediction of primary codes.
3. Pathologist-Specific Language: Investigate whether the sign-out pathologist can be predicted based on word choice. Although the sign-out pathologist can be found through an SQL query in the Pathology Information System, we are interested in translating sign-outs to a unified language that is consistent across sign-outs (i.e., a similar lexicon across pathologists, given diagnosis, code assignments, and subspecialty). As an example, some pathologists may more verbosely describe a phenomenon that could be succinctly summarized to match a colleague’s description, though this could be difficult to disentangle without a quantitative understanding of lexical differences. To do this, we need to identify several components of variation (i.e., within a subspecialty, where reports from pathologists may vary widely); we want to further understand this heterogeneity to standardize communications within our department.
Although the final two objectives (ancillary testing and pathologist prediction) can be resolved by using an SQL query, we emphasize that these secondary objectives were selected to better identify the potential sources of reporting inconsistency with the aim of informing optimal reporting standards rather than imputing information that can be readily queried through the Pathology Information System.
1.3. Approach and procedure
1.3.1. Data acquisition
We obtained Institutional Review Board approval and accessed more than 96,418 pathologist reports from DPLM, collected between June 2015 and June 2020. We removed a total of 3,379 reports that did not contain any diagnostic text associated with CPT codes, retaining 93,039 reports (Supplementary Table 1). Each report was appended with metadata, including corresponding EPIC (EPIC systems, Verona, WI),35 Charge Description Master (CDM), and CPT procedural codes, the sign-out pathologist, the amount of time to sign out the document, and other details. Fuzzy string matching using the fuzzywuzzy package was used to identify whether any pathologists’ names were misspelled (or resolve potential last name changes) between documents.36 First, all unique pathologist names were identified. Then, for each pair of names, the token sort ratio was calculated, thresholded by whether the ratio exceeded 0.7 to establish a unipartite graph of pathologist names connected to their candidate duplicates. Finally, clusters of similar names were identified by using connected component analysis. In most cases, unique names were assigned to each cluster of names, though in select cases, names were kept separate.37 The documents were deidentified by stripping all PHI-containing fields and numerals from the text and replacing with holder characters (e.g. 87560 becomes #####). As a final check, we used regular expressions (regex) to remove mentions of patient names in the report text. This was accomplished by first compiling and storing several publicly available databases of 552,428 first and last names (Supplementary Materials, section “Additional Information on Deidentification Approach”). Then, using regex, we searched for the presence of each first and last name in the report subsections and replaced names at matched positions with white spaces. However, we did not remove mention of the physicians and consulting pathologist. The information on the physicians and consulting pathologist were identified in the “ordered by,” “reports to,” and “verified by” fields of the pathology report using known personal identifiers. The deidentification protocol was approved by the Institutional Review Board, Office of Research Operations and Data Governance. A total of 17,744 first and last names were stripped from the in-house data.
1.3.2. Preprocessing
We used regular expressions (regex) to remove punctuation from the text, and the text was preprocessed by using the Spacy package,38 to tokenize the text. We utilized Spacy’s en_core_web_sm processing pipeline (https://spacy.io/models/en#en_core_web_sm) to remove English stop words and words shorter than three characters. Out of concern for removing pathologist lexicon germane to pathologist sign-out, for this preliminary assessment, we did not attempt to prune additional words from the corpus outside of the methods used to generate word frequencies for the bag of words approaches. We also split up each pathology report into their structured sections: Diagnosis, Clinical Information, Specimen Processing, Discussion, Additional Studies, Results, and Interpretation. This allowed for an equal comparison between the machine-learning algorithms. The deep learning algorithm BERT can only operate on 512 words at a time due to computational constraints (See the “Limitations” and Supplementary Materials section “Additional Information on BERT Pretraining”). Sometimes, the pathology reports exceeded this length when considering the entire document (1.77% exceeded 512 words) and as such these reports were limited to the diagnosis section (0.02% exceeded 512 words) when training a new BERT model (Supplementary Table 1; Supplementary Fig. 1). We removed all pathology reports that did not contain a diagnosis section.
1.3.3. Characterization of the text corpus
After preprocessing, we encoded each report tabulating the occurrence of all contiguous one- to two-word sequences (unigram and bigrams) to form sparse count matrices, where each column represents a word or phrase and each row represents the document, and the value is the frequency of occurrence in the document. Although the term “frequency” may be representative of the distribution of words/phrases in a corpus, high-frequency words that are featured across most of the document corpus are less likely to yield an informative lexicon that is specific to a subset of the documents. To account for less important but ubiquitous words, we transformed raw word frequencies to term frequency inverse document frequency (tf-idf) values, which up-weights the importance of the word based on its occurrence within a specific document (term frequency), but down-weights the importance if the word is featured across the corpus (inverse document frequency) (see the Supplementary Material section “Additional Description of Topic Modeling and Report Characterization Techniques”). We summed the tf-idf value of each word across the documents to capture the word’s overall importance across the reports and utilized a word cloud algorithm to display the relative importance of the top words.
After constructing count matrices, we sought to characterize and cluster pathology documents as they relate to each other and ascribe themes to the clusters. Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP)39 dimensionality reduction was used to project the higher dimensional word frequency data into lower dimensions while preserving important functional relationships. Each document could then be represented by a 3D point in the Cartesian coordinate system; these points were clustered by using a density-based clustering algorithm called HDBSCAN40 to simultaneously estimate characteristic groupings of documents while filtering out noisy documents that did not explicitly fit in these larger clusters. To understand which topics were generally present in each cluster, we deployed Latent Dirichlet Allocation (LDA),13 which identifies topics characterized by a set of words, and then derives the distribution of topics over all clusters. This is accomplished via a generative model that attempts to recapitulate the original count matrix, which is further outlined in greater detail in the Supplementary Material section “Additional Description of Topic Modeling and Report Characterization Techniques.” The individual topics estimated using LDA may be conceptualized as a Dirichlet/multinomial distribution (“weight” per each word/phrase) over all unigrams and bigrams, where a higher weight indicates membership in the topic. The characteristic words pertaining to each topic were visualized by using a word cloud algorithm. Finally, we correlated the CPT codes with clusters, topics, and select pathologists by using Point-Biserial and Spearman correlation measures41 to further characterize the overall cohort.
1.3.4. Machine learning models
We implemented the following three machine-learning algorithms in our study as a basis for our text classification pipeline [Fig. 1]:
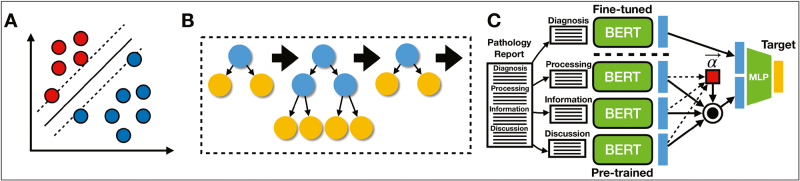
Fig. 1.
Model Descriptions: Graphics depicting: (A) SVM, where hyperplane linearly separates pathology reports, which are represented by individual datapoints; (B) XGBoost, which sequentially fits decision trees based on residuals from sum of conditional means of previous trees and outcomes; (C) All-Fields BERT model, where a diagnosis-specific neural network extracts relevant features from the diagnostic field, whereas a neural network trained on a separate clinical corpus extracts features for the remaining subfields; subfields are weighted and summed via the attention mechanism, indicated in red; subfields are combined with diagnostic features and fine-tuned with a multilayer perceptron for the final prediction.
1.3.4.1. SVM
We trained an SVM42,43 to make predictions by using the UMAP embeddings formed from the tf-idf matrix. The SVM operates by learning a hyperplane that obtains maximal distance (margin) to datapoints of a particular class [Fig. 1A]. However, because datapoints/texts from different classes may not be separable in the original embedding space, the SVM model projects data to a higher dimensional space where data can be linearly separated. We utilized GPU resources via the ThunderSVM package44 to train the model in reasonable compute time.
1.3.4.2. Bag of words with XGBoost
XGBoost algorithms45 operate on the entire word by report count matrix and ensemble or average predictions across individual Classification and Regression Tree (CART) models.46 Individual CART models devise splitting rules that partition instances of the pathology notes based on whether the count of a particular word or phrase in a pathology note exceeds an algorithmically derived threshold. Important words and thresholds (i.e. partition rules) are selected from the corpus based on their ability to partition the data, based on the purity of a decision leaf through the calculation of an entropy measure. Each successive splitting rule serves to further minimize the entropy or maximize the information gained. Random Forest models47 bootstrap which subsets of predictors/words and samples are selected for a given splitting rule of individual trees and aggregate the predictions from many such trees; Extreme Gradient Boosting Trees (XGBoost) fit trees (structure and the conditional means of the terminal nodes) sequentially based on the residual (in the binary classification setting, misclassification is estimated using a Bernoulli likelihood) between the outcome and the sum of both the conditional means of the previous trees (which are set) and the conditional means of the current tree (which is optimized). This gradient-based optimization technique prioritizes samples with a large residual/gradient from the previous model fit to account for the previous “weak learners” [Fig. 1B]. In both scenarios, random forest (a bagging technique) and XGBoost (a boosting technique), individual trees may exhibit bias but together cover a larger predictor space. Our XGBoost classifier models were trained by using the XGBoost library, which utilizes GPUs to speed up calculation.
1.3.4.3. BERT
ANN48 are a class of algorithms that use highly interconnected computational nodes to capture relationships between predictors in complex data. The information is passed from the nodes of an input layer to the individual nodes of subsequent layers that capture additional interactions and nonlinearities between predictors while forming abstractions of the data in the form of intermediate embeddings. The BERT18 model first maps each word in a sentence to its own embedding and positional vectors, which captures key semantic/syntactic and contextual information that is largely absent from the bag of words approaches. These word-level embeddings are passed to a series of self-attention layers (the Transformer component of the BERT model), which contextualizes the information of a single word in a sentence based on short- and long-term dependencies between all words from the sentence. The individual word embeddings are combined with the positional/contextual information, obtained via the self-attention mechanism, to create embeddings that represent the totality of a sentence. Finally, this information is passed to a series of fully connected layers that produce the final classification. With BERT, we are also able to analyze the relative importance and dependency between words in a document by extracting “attention matrices.” We are also able to retrieve sentence-level embeddings encoded by the network by extracting vectors from the intermediate layers before they pass for the final classification.
We trained the BERT models by using the HuggingFace Transformers package,49 which utilizes GPU resources through the PyTorch framework. We used a collection of models that have already been pretrained on a large medical corpus50 in order to both improve the predictive accuracy of our model and significantly reduce the computational load compared with training a model from scratch. Because significant compute resources are still required to train the model, most BERT models limit the document characterization length to 512 words. To address this, we split pathology reports into document subsections when training BERT models.
In training a BERT model, we updated the word embeddings through fine-tuning a pretrained model on our diagnostic corpus. This model, which had been trained solely on diagnostic text, could be used to predict the target of interest (Dx Model). However, we then used this fine-tuned model to extracted embeddings that were specific to the diagnosis subfield to serve as input for a model that could utilize text from other document subfields. We separately utilized the original pretrained model to extract embeddings from the other report subfields that are less biased by diagnostic codes and thus more likely to provide contextual information (All Fields Model). We developed a global/gating attention mechanism procedure that serves to dynamically prune unimportant, missing, or low-quality document subsections for classification [Fig. 1C]. Predictions may be obtained when some/all report subfields are supplied via the following method:
Where represents the embeddings extracted from the pretrained and fine-tuned BERT embeddings on respective report subsections, and is a vector of attention scores between 0 and 1 that dictates the importance of particular subsections. These attention scores are determined by using a separate gating neural network, fgate , which maps , a 768-dimensional vector to a scalar for each document subsection through two projection matrices: W1 a 768-dimension (dimensionality of BERT embeddings) by 100-dimensional matrix, and W2 a 100-dimension (dimensionality of BERT embeddings) by 1-dimensional matrix that generates the attention scores. A softmax transformation is used to normalize the scores between zero and one across the subsections. Finally, fMLP are a set of fully connected layers that operate on the concatenation between the BERT embeddings that were fine-tuned on the diagnosis-specific section and those extracted by using the pre-trained BERT model on the other document subfields, as weighted by using the gated attention mechanism (Supplementary Section “Additional Description of Explanation Techniques”). To train this model, we experimented with an ordinal loss function,51 based off of the proportional odds cumulative link model specification, which respects the ordering of the primary CPT codes by case complexity, though ultimately, we opted for using a Cross-Entropy loss since ordinal loss functions are not currently configured for the other machine-learning methods (e.g., XGBoost).
1.4. Prediction of primary current procedural terminology codes
We developed machine-learning pipelines to delineate primary CPT codes requiring examination with a microscope (CPT 88302, 88304, 88305, 88307, 88309) using BERT, XGBoost, and SVM, with reports selected based on whether they contained only one of the five codes (where the primary codes were present in the following proportions: CPT 88302:0.67%, 88304:6.59%, 88305:85.97%, 88307:6.32%, and 88309:0.44%). The prevalence of most of the five codes did not change over time (Supplementary Fig. 2; Supplementary Table 2). Given the characterization of the aforementioned deep learning framework, we utilized a BERT model that was pretrained first on a large corpus of biomedical research articles from PubMed, and then pretrained by using a medical corpus of free text notes from an intensive care unit (MIMIC3 database; Bio-ClinicalBERT; Supplementary Materials section “Additional Information on BERT Pretraining”).50,52,53 Finally, the model was fine-tuned on our DHMC pathology report corpus (to capture institution-specific idiosyncrasies) for the task of classifying particular CPT codes from diagnostic text. XGBoost was trained on the original count matrix, whereas SVM was trained on a 6-dimensional UMAP projection; a UMAP projection was utilized for computational considerations. The models were evaluated by using five-fold cross-validation as a means to compare the model performances. Internal to each fold is a validation set used for identifying optimal hyperparameters (supplementary section “Additional Information on Hyperparameter Scans”) through performance statistics and a held-out test set. For each approach, we separately fit a model considering only the Diagnosis text (Dx Models) and all of the text (All Fields Models) to provide additional contextual information. We calculated the Area Under the Receiver Operating Curve (AUC-Score; considers sensitivity/specificity of the model at a variety of probability cutoffs; anything above a 0.5 AUC is better than random), F1-Score (which considers the tradeoff between sensitivity and specificity) and macro-averaged these scores across the five CPT codes, which gives greater importance to rare codes. Since codes are also ordered by complexity (ordinal variable), we also report a confusion matrix, which tabulates the real versus predicted codes for each approach and measures both a spearman correlation coefficient and linear-weighted kappa between predicted and real CPT codes as a means to communicate how the model preserves the relative ordering of codes (i.e., if the model is incorrect, better to predict a code of a similar complexity).
1.5. Ancillary testing current procedural terminology codes and pathologist prediction tasks
To contextualize findings for primary codes, these machine-learning techniques were employed to predict each of 38 different CPT codes (38 codes remained after removing codes that occurred less than 150 times across all sign-outs) (e.g., if the prediction of primary codes relies on the diagnostic section, do secondary codes rely on other document sections more?). The primary code model predicted a categorical outcome, whereas ancillary testing models were configured in the multitarget setting, where each code represents a binary outcome. We compared cross-validated AUC statistics between and across the 38 codes to further explore the reasons that some codes yielded lower scores than others. We also compared different algorithms via the sensitivity/specificity reported via their Youden’s index (the optimal tradeoff possible between sensitivity and specificity from the receiver operating curve), averaged across validation folds.
We similarly trained all models to recognize the texts of the 20 pathologists with the most sign-outs to see whether the models could reveal pathologist-specific text to inform future efforts to standardize text lexicon. We retained reports from the 20 pathologists with the most sign-outs, reducing our document corpus from 93,039 documents to 64,583 documents, and we utilized all three classification techniques to predict each sign-out pathologist simultaneously. The selected pathologists represented a variety of specialties. Choosing only the most prolific pathologists removed the potential for biased associations by a rare outcome in the multiclass setting.
1.6. Model interpretations
Finally, we used shapley additive explanations (SHAP; a model interpretation technique that estimates the contributions of predictors to the prediction through credit allocation)54 to estimate which words were important for the classification of each of these codes, visualized by using a word cloud. For the BERT model, we utilized the Captum55 framework to visualize backpropagation from the outcome to predictors/words via IntegratedGradients56 and attention matrices. Additional extraction of attention weights also revealed not only which words and their relationships contributed to the prediction of the CPT code (i.e. self-attention denotes word-to-word relationships), but also which document subfields other than the diagnosis field were important for assignment of the procedure code (i.e. global/gating attention prunes document subfields by learning to ignore irrelevant information; the degree of pruning can be extracted during inference). Further description of these model interpretability techniques (SHAP, Integrated Gradients, Self-Attention/“word-to-word”, Attention) may be found in the supplementary material (section “Additional Description of Explanation Techniques: SHAP, Integrated Gradients, Self-Attention, Attention Over Pathology Report Subfields”). Pathologist-specific word choice was extracted by using SHAP/Captum from the resulting model fit and visualized by using word clouds and attention matrices.
2. Results
2.1. Corpus preprocessing and Uniform Manifold Approximation and Projection for Dimension Reduction results
After initial filtering, we amassed a total of 93,039 pathology reports, which were broken into the following subsections: Diagnosis, Clinical Information, Specimen Processing, Discussion, Additional Studies, Results, and Interpretation. The median word length per document was 119 words (Interquartile Range; IQR=90). Very few reports contained subfields that exceeded the length acceptable by the BERT algorithm (2% of reports containing a Results section exceeded this threshold; Supplementary Table 1; Supplementary Fig. 1).
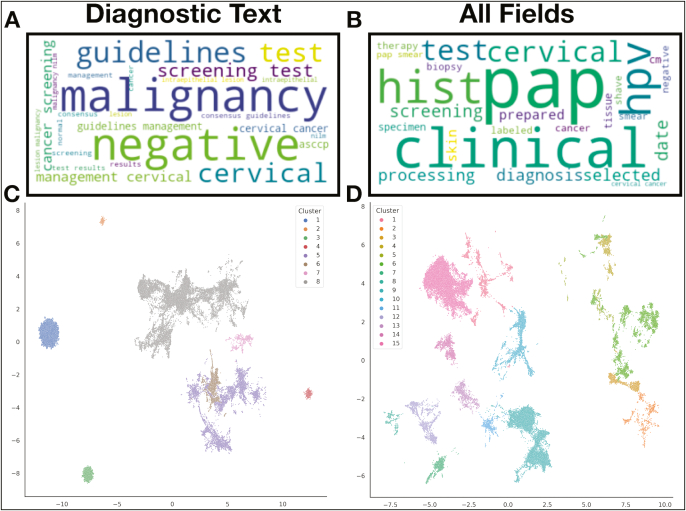
Displayed first are word clouds of the top 25 words in only the diagnostic document subsection [Fig. 2A] and across all document subsections [Fig. 2B], with their size reflecting their tf-idf scores [Fig. 2A and B]. As expected, the diagnostic-field cloud contains words that are pertinent to the main diagnosis, whereas the all-field cloud contains words that are more procedural, suggesting that other pathology document subfields yield distinct and specific clinical information that may lend complementary information versus analysis solely on diagnostic fields. We clustered and visualized the diagnostic subsection and also all document subsections after running UMAP, which yielded 8 and 15 distinct clusters, respectively [Fig. 2C and D]. The number of words per report correlated poorly with the number of total procedural codes assigned (Spearman r=0.066,p<0.01 ). However, when these correlations were assessed within the HDBSCAN report clusters (subset to reports within a particular cluster for cluster-specific trends), 33% of the all-fields report clusters reported moderate correlations (Supplementary Table 3). Interestingly, one of the eight report clusters from the diagnostic fields experienced a moderate negative correlation with the number of codes assigned.
Fig. 2.
Pathology report corpus characterization: (A and B) Word cloud depicting words with the highest aggregated tf-idf scores across the corpus of: (A) diagnostic text only, (B) all report subfields (all-fields); important words across the corpus indicated by relative size of the word in the word cloud; (C and D) UMAP projection of the tf-idf matrix, clustered and noise removal via HDBSCAN for: (C) diagnostic texts only, and (D) all report subfields (all-fields).
2.2. Topic modeling with Latent Dirichlet Allocation and additional topic associations
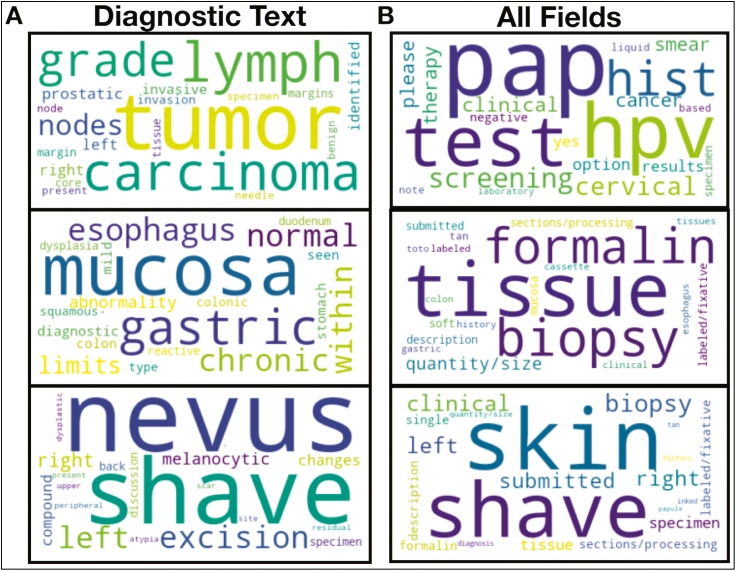
From our LDA analysis on all document subsections, we discovered 10 topics [Fig. 3; Supplementary Table 4]. Correlations between these topics with clusters, pathologists, and CPT codes are displayed in the supplementary material (Supplementary Figs 3-6). We discovered additional associations between CPT codes, clusters, and pathologists (Supplementary Fig. 7A), suggesting a specialty bias in document characterization. We clustered pathologists using co-occurrence of procedural code assignments in order to establish “subspecialties” (e.g., pathologist who signs out multiple specialties) that could be used to help interpret sources of bias in an evaluation of downstream modeling approaches.
Fig. 3.
LDA Topic Words: Important words found for three select LDA Topics from: (A) diagnostic text only and (B) all report subfields (all-fields); important words across the corpus indicated by relative size of the word in the word cloud.
2.3. Primary current procedural terminology code classification results
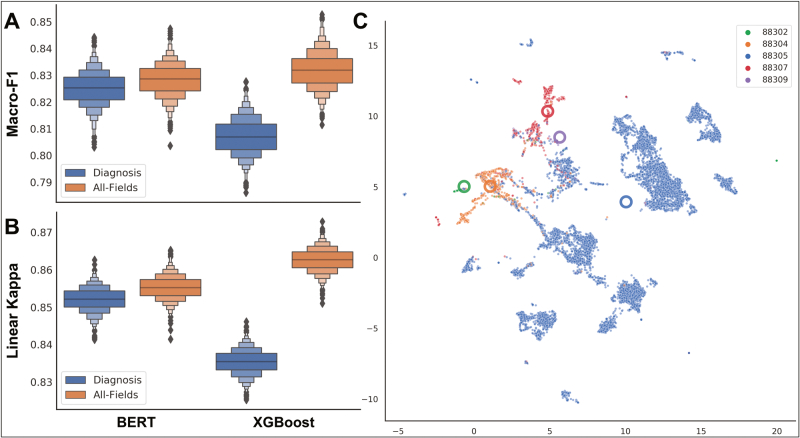
The XGBoost and BERT models significantly outperformed the SVM model for the prediction of primary CPT codes [Table 1; Fig. 4A and B; Supplementary Table 5]. The BERT model made more effective use of the diagnostic text (macro-f1=0.825; κ = 0.852) as compared with the XGBoost model (macro-f1=0.807; κ = 0.835). Incorporating the text from other report subfields provided only a marginal performance gain for BERT (macro-f1=0.829; κ = 0.855) and both a large and significant performance gain for XGBoost (macro-f1=0.831; κ = 0.863) [Fig. 4A and B]. Across the BERT and XGBoost models, codes were likely to be misclassified if they were of a similar complexity [Table 1; Supplementary Table 5]. Plots of low-dimensional text embeddings extracted from the BERT All-Fields model demonstrated clustering by code complexity and relative preservation of the ordering of code complexity (i.e., reports pertaining to codes of lower/higher complexity clustered together) [Fig. 4C].
Table 1.
Predictive performances for primary CPT code algorithms
| Approach | Type | Macro-F1 ± SE | ± κ se | AUC ± SE | Spearman |
|---|---|---|---|---|---|
| BERT | Diagnosis All fields |
0.825 ± 0.0064 0.828 ± 0.0062 |
0.852 ± 0.0033 0.855 ± 0.0032 |
0.99 ± 0.0008 0.99 ± 0.0006 |
0.84 ± 0.0044 0.843 ± 0.0044 |
| XGBoost | Diagnosis All fields |
0.807 ± 0.0069 0.832 ± 0.0069 |
0.835 ± 0.0034 0.863 ± 0.0032 |
0.99 ± 0.0007 0.994 ± 0.0004 |
0.824 ± 0.0045 0.855 ± 0.0042 |
| SVM | Diagnosis All fields |
0.497 ± 0.0047 0.518 ± 0.0048 |
0.644 ± 0.0043 0.668 ± 0.0044 |
0.554 ± 0.0021 0.554 ± 0.0014 |
0.637 ± 0.0056 0.652 ± 0.0058 |
Macro-Fl and AUC measures are agnostic to the ordering of the CPT code complexity; whereas Linear Kappa (κ) and Spearman correlation coefficients respect the CPT code ordering (88302, 88304, 88305, 88307, and 88309).
Fig. 4.
Primary CPT Code Model Performance: (A and B) Grouped boxenplots demonstrating the performance of machine-learning models (BERT, XGBoost) for the prediction of primary CPT codes (bootstrapped performance statistics; A) macro-averaged F1-Score, (B) Linear-Weighted Kappa for performance across different levels of complexity, which takes into account the ordinal nature of the outcome; reported across five CPT code), given analysis of either the diagnostic text (blue) or all report subfields (orange); (C) UMAP projection of All-Fields BERT embedding vectors after applying the attention mechanism across report subfields; each point is reported with information aggregated from all report subfields; individual points represent reports, colored by the CPT code; large thick circles represent the report centroids for each CPT code; note how codes CPT 88302 and CPT 88304 cluster together and separately CPT 88307 and CPT 88309 cluster together, whereas CPT 88305 sits in between clustered reports of low and high complexity.
2.4. Ancillary current procedural terminology code and pathologist classification results
We were able to accurately assign ancillary CPT codes to each document, regardless of which machine learning algorithm was utilized (Supplementary Fig. 8; Supplementary Table 6). Across all ancillary codes, we found that XGBoost (median AUC=0.985) performed comparably to BERT (median AUC=0.990; P = 0.64) when predicting CPT codes based on the diagnostic subfield alone, whereas SVM performed worse (median AUC=0.966) than both approaches, per cross-validated AUC statistics (Supplementary Tables 6-10; Supplementary Fig. 9). In contrast to results obtained for the primary codes, we discovered that classifying by including all of the report subelements (All Fields) performed better than just classifying based on the diagnostic subsection (P < 0.001 for both BERT and XGBoost approaches; Supplementary Tables 6, 8-10; Supplementary Figs 9 and 10), suggesting that these other more procedural/descriptive elements contribute meaningful contextual information for the assignment of ancillary CPT codes (Supplementary Materials section “Supplementary Ancillary CPT Code Prediction Results”). We also report that the sign-out pathologist can also be accurately identified from the report text, with comparable performance between the BERT (macro-f1=0.72) and XGBoost (macro-f1=0.71) models, and optimal performance when all report subfields are used (macro-f1=0.77 and 0.78, respectively) (Supplementary Materials section “Supplementary Pathologist Prediction Results”; Supplementary Table 11; Supplementary Figure 11).
2.5. Model interpretation results
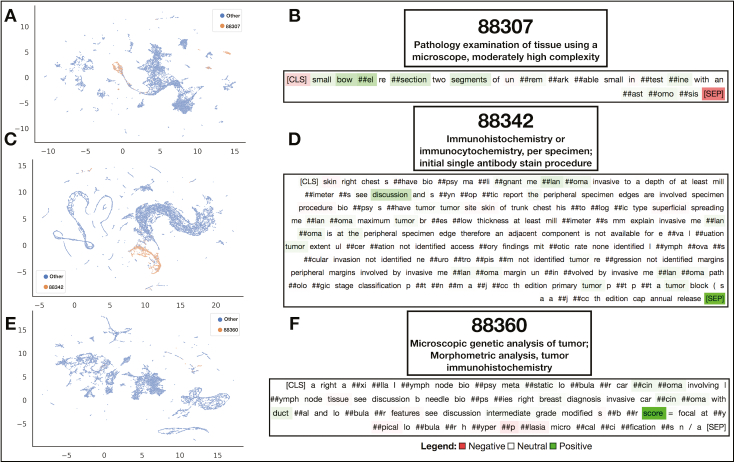
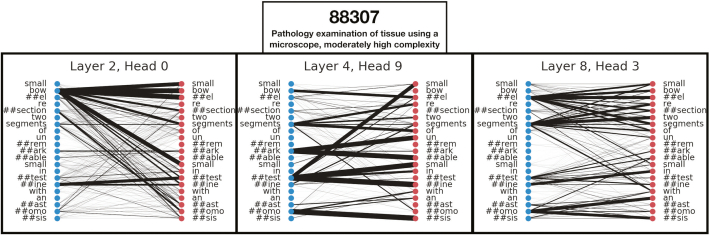
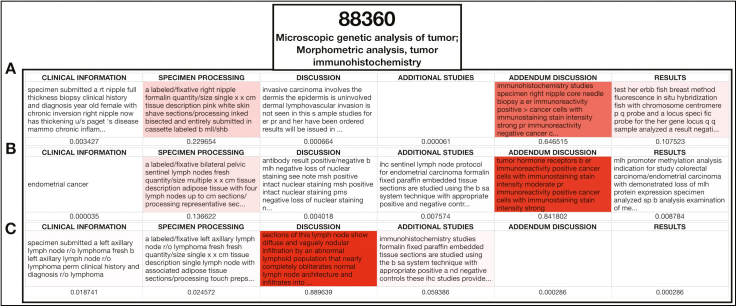
We also visualized which words were found to be important for a subsample of primary and ancillary procedural codes by using the XGBoost algorithm [Fig. 5; Supplementary Figure 12]. In the Supplementary Materials, we have also included a table that denotes the relevance of the top 30 words for the XGBoost All Fields model for the prediction of specific primary CPT codes, as assessed through SHAP (Supplementary Table 12). Reports that were assigned the same ancillary CPT code clustered together in select low-dimensional representations learned by some of the All Fields BERT models [Fig. 6A, C, and E]. Model-based interpretations of a few sample sentences for CPT codes using the Diagnosis BERT approach revealed important phrases that aligned with assignment of the respective CPT code [Fig. 6C, D, and F]. Finally, we included a few examples of the attention mechanism used in the BERT approach, which highlights some of the many semantic/syntactic dependencies that the model finds within text subsections [Fig. 7]. These attention matrices were plotted along with importance assigned to subsections of pathology reports using the All-Fields model [Fig. 8], all with their respective textual content. Additional interpretation of reports for pathologists may be found in the Supplementary Materials (Supplementary Figures 13 and 14).
Fig. 5.
SHAP interpretation of XGBoost predictions: Word clouds demonstrating words found to be important using the XGBoost algorithm (All-Fields) for the prediction of primary CPT codes, found via shapley attribution; important words pertinent to each CPT code indicated by the relative size of the word in the word cloud; word clouds visualized for word importance (A) across all five primary CPT codes and (B-F) for the following CPT codes: (B) CPT code 88302; (C) CPT code 88304; (D) CPT code 88305; (E) CPT code 88307; and (F) CPT code 88309; note that the size of the word considers strength but not directionality of the relationship with the code, which may be negatively associated in some cases.
Fig. 6.
Embedding and Interpretation of BERT Predictions: (A, C, and E) UMAP projection of All-Fields BERT embedding vectors after applying the attention mechanism across report subfields; each point is reported with information aggregated from all report subfields; (B, D, and F) Select diagnostic text from individual reports interpreted by Integrated Gradients to elucidate words positively and negatively associated with calling the CPT code; Integrated Gradients was performed on the diagnostic text BERT models; Utilized CPT codes: (A and B) CPT code 88307, (C and D) CPT code 88342, and (E and F) CPT code 88360.
Fig. 7.
BERT Diagnostic Model Self-Attention: Output of self-attention maps for select self-attention heads/layers from the BERT diagnostic text model visualizes various layers of complex word-to-word relationships for the assessment of a select pathology report that was found to report CPT code 88307.
Fig. 8.
BERT All-Fields Model Interpretation: Visualization of importance scores assigned to pathology report subfields outside of the diagnostic section for three separate pathology reports (A-C) that were assigned by raters CPT code 88360; information from report subfields that appear more red was utilized more by the model for the final prediction of the code; attention scores listed below the text from the subfields and title of each subfield supplied.
3. Discussion
In this study, we characterized a large corpus of almost 100,000 pathology reports at a mid-sized academic medical center. Our studies indicate that the XGBoost and BERT methodologies produce highly accurate predictions of both primary and ancillary CPT codes, which has the potential to save operating costs by first suggesting codes prior to manual inspection and flagging potential manual coding errors for review. Further, both the BERT and XGBoost models preserved the ordering of the code/case complexity, where most of the misclassifications were made between codes of a similar complexity. The model interpretations via SHAP suggest a terminology that is consistent with code complexity. For instance, “vulva,” “uterus,” and “adenocarcinoma” were associated with CPT code 88309. We noted associations between “endometrium diagnosis” and “esophagus” and CPT code 88305. “Biopsy” was associated with CPT codes 88305 and 88307, while “myocyte” was associated with CPT code 88307 (myocardium). In addition, we noticed a positive association between “products of conception” and lower complexity codes (CPT code 88304) and a negative association with higher complexity codes. The aforementioned associations uncovered using SHAP are consistent with reporting standards for histological examination.31,32,57
Previous studies predicting CPT codes have largely been unable to characterize the importance of different subsections of a pathology report. Using the BERT and XGBoost methods, we were also able to show that significant diagnostic/coding information is contained in nondiagnostic subsections of the pathology report, particularly the Clinical Information and Specimen Processing sections. Such information was more pertinent when predicting ancillary CPT codes, as nondiagnostic subfields are more likely to contain test ordering information, though performance gains were observed for primary codes when employing the XGBoost model over an entire pathology report. This is expected, as many of the CPT codes are based on procedure type/specimen complexity and ancillary CPT codes are expected to contain more informative text in the nondiagnostic sections. Potentially, the variable presence/absence of different reporting subfields may have made predicting primary codes using the BERT model more difficult, as the extraction of information different subsections was not optimized for aside from how much weight to apply to each section.
Although our prediction accuracy is comparable to previous reports of CPT prediction using machine-learning methods, our work covers a wider range of codes than previously reported, compares the different algorithms through rigorous cross-validation, reports a significantly higher sensitivity and specificity, and demonstrates the importance of utilizing other parts of the pathology report for procedural code prediction. Further, previous works had only considered the first 100 words of the diagnostic section and had failed to properly account for class-balancing, potentially leading to inflated performance statistics; however, our study carefully considers the ordinality of the response and reports macro-averaged measures that take into account infrequently assigned codes.
We also demonstrated that the pathology report subfields contained pertinent diagnostic and procedural information that could adequately separate our text corpus based on ancillary CPT codes and the signing pathologist. With regard to ancillary testing, it was interesting to note how some of the clinical codes for acquisition and quantification of markers on specialized stains (CPT 88341, 88342, 88344, 88360) performed the worst overall, which may potentially suggest inconsistent reporting patterns for the ordering of specialized stains.34 The revision of CPT codes 88342 and 88360, and the addition of CPT codes 88341 and 88344 in 2015 lay just outside of the range of the data collection period, which was from June 2015 to June 2020.58 Evolving coding/billing guidelines will always present challenges when developing NLP guidelines for clinical tests, though our models’ optimal performance and the fact that major coding changes occurred outside of the data collection period suggest that temporal changes in coding patterns did not likely impact the ability to predict CPT codes. We did not find significant changes in the assignment of most of the primary codes over the study period. Since major improvements were obtained through incorporating the other report subfields for the codes, nondiagnostic text may be more important for records of specialized stain processing and should be utilized as such.
4. Limitations
There are a few limitations to our work. For instance, due to computational constraints, most BERT models can only take as input 512 words at a time (Supplementary Section “Additional Information on BERT Pretraining”). We utilized a pretrained BERT model that inherited knowledge from large existing biomedical data repositories at the expense of flexibility in sequence length size (i.e. we could not modify the word limit while utilizing this pretrained model). We noticed that in our text corpus, less than 2% of reports were longer than this limitation and thus had to be truncated when input into the deep learning model, which may impact results. Potentially, longer pathology reports describe more complicated cases, which may utilize additional procedures. From our cluster analysis, we demonstrated that this appeared to be the case for a subset of report clusters, though for one cluster, the opposite was true. However, a vast majority of pathology reports fell within the BERT word limits, so we considered any word length-based association with CPT code complexity to have negligible impact on the model results. The XGBoost model, alternatively, is able to operate on the entire report text. Thus, XGBoost may more directly capture interactions between words spanning across document subsections pertaining to complex cases, which may serve as one plausible explanation of its apparent performance increase with respect to the BERT approaches. Although we attempted to take into account the ordinality of case complexity for the assignment of primary CPT codes, such work should be revisited as ordinal loss functions for both deep learning and tree-based models become more readily available. There were also cases where multiple primary codes were assigned; whereas the ancillary codes were predicted by using a multitarget objective, and the primary code prediction can be configured similarly though this was outside the scope of the study.32 Although we conducted coarse hyperparameter scans, we note that generally such methods are deemed both practical and acceptable. Although other advanced hyperparameter scanning techniques exist (e.g., Bayesian optimization or genetic algorithm), in many cases, these methods obtain performance similar to randomized hyperparameter searches and may be far more resource intensive.59
5. Future directions
Given the secondary objectives of our study (e.g., prediction of ancillary codes, studying sources of variation in text, i.e. pathologist), we were able to identify additional areas for follow-up.
First, we were able to assess nuanced pathologist-specific language, which was largely determined by specialty (e.g. subspecialties such as cytology use highly regimented language, making it more difficult to separate practitioners). There is also potentially useful information to be gained by working to identify text that can distinguish pathologists within subspecialties (found as a flag in the Pathology Information System) and conditional on code assignment rather than identify pathologists across subspecialties. This information can be useful in helping to create more standardized lexicons/diagnostic rubrics (for instance, The Paris System for Urine Cytopathology60). Research into creating a standard lexicon for particular specialties or converting raw free text into a standardized report could be very fruitful, especially for the positive impact it would have in allowing nonpathologist physicians to more easily interpret pathology reports and make clinical decisions. As an example of how nonstandardized text lexicon can impact reporting, it has long been suspected that outlier text can serve as a marker of uncertainty or ambiguity about the diagnosis. For instance, if there is a text content outlier in a body of reports with the same CPT code, then we can hypothesize that such text may be more prone to ambiguous phrases or hedging, from which pathologists may articulate their uncertainty for a definitive diagnosis. As such, we would also like to assess the impact of hedging in the assignment of procedural codes, and further its subsequent impact on patient care. As another example, excessive ordering of different specialized stains and pathology consults may suggest indecisiveness, as reflected in the pathology report. To ameliorate these differences in reporting patterns, generative deep learning methods can be employed to summarize the text through the generation of a standard lexicon.
Other excellent applications of BERT-based text models include the prediction of relative value units (RVU’s) via report complexity for pathologist compensation calculations (which is related to primary code assignment) and the detection of cases that may have been mis-billed (e.g., a code of lower complexity was assigned), which can potentially save the hospital resources.61 We are currently developing a web application that will both interface with the Pathology Information System and can be used to estimate the fiscal impact of underbilling by auditing reports with false positive findings. Tools such as Inspirata can also provide additional structuring for our pathology reports outside of existing schemas.62
Although much of the patient’s narrative may be told separately through text, imaging, and omics modalities,63 there is tremendous potential to integrate semantic information contained in pathologist notes with imaging and omics modalities to capture a more holistic perspective of the patient’s health and integrate potentially useful information that could otherwise be overlooked. For instance, the semantic information contained in a report may highlight specific morphological and macro-architectural features in the correspondent biopsy specimen that an image-based deep learning model might struggle to identify without additional information. Although XGBoost demonstrated equivalent performance with the deep learning methods used for CPT prediction, its usefulness in a multimodal model is limited because these machine-learning approaches rely heavily on the feature extraction approach, where feature generation mechanisms using deep learning can be tweaked during optimization to complement the other modalities. Alternatively, the semantic information contained within the word embedding layers of the BERT model can be fine-tuned when used in conjunction with or directly predicting on imaging data, allowing for more seamless integration of multimodal information. Integrating such information, in addition to structured text extraction systems (i.e., named entity recognition) that can recognize and correct the mention of such information in the text, may provide a unique search functionality that can benefit experiment planning.34
Although comparisons between different machine-learning models may inform the optimal selection of tools that integrate with the Pathology Information System, we acknowledge that such comparisons can benefit from updating as new machine-learning architectures are developed. As such, we plan to incorporate newer deep learning architectures, such as the Reformer or Albert, which do not suffer from the word length limitations of BERT, though training all possible language models was outside of the scope of our study since pretrained medical word embeddings were not readily available at the time of modeling.
6. Conclusion
In this study, we compare three cutting-edge machine learning techniques for the prediction of CPT codes from pathology text. Our results provide additional evidence for the utility of machine-learning models to predict CPT codes in a large corpus of pathology reports acquired from a mid-sized academic medical center. Further, we demonstrated that utilizing text from parts of the document other than the diagnostic section aids in the extraction of procedural information. Although both the XGBoost and BERT methodologies yielded comparable results, either method can be used to improve the speed and accuracy of coding by the suggestion of relevant CPT codes to coders, though deep learning approaches present the most viable methodology for incorporating text data with other pathology modalities.
Financial support and sponsorship
This work was supported by NIH grants R01CA216265, R01CA253976, and P20GM104416 to BC, Dartmouth College Neukom Institute for Computational Science CompX awards to BC and LV, and Norris Cotton Cancer Center, DPLM Clinical Genomics and Advanced Technologies EDIT program. JL is supported through the Burroughs Wellcome Fund Big Data in the Life Sciences at Dartmouth. The funding bodies above did not have any role in the study design, data collection, analysis and interpretation, or writing of the manuscript.
Authors’ contributions
The conception and design of the study were contributed by JL and LV. Initial analyses were conducted by JL and NV. All authors contributed to writing and editing of the manuscript and all authors read and approved the final manuscript.
Declaration of Competing Interest
There are no conflicts of interest.
Acknowledgments
We would like to thank Matthew LeBoeuf for thoughtful discussion.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.4103/jpi.jpi_52_21.
Appendix A. Supplementary data
Supplementary material
References
- 1.Mantas J., Hasman A. IOS Press; Amsterdam: 2013. Informatics, Management and Technology in Healthcare. [Google Scholar]
- 2.Wilson R.A., Chapman W.W., Defries S.J., Becich M.J., Chapman B.E. Automated ancillary cancer history classification for mesothelioma patients from free-text clinical reports. J Pathol Inform. 2010;1:24. doi: 10.4103/2153-3539.71065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheikhalishahi S., Miotto R., Dudley J.T., Lavelli A., Rinaldi F., Osmani V. Natural language processing of clinical notes on chronic diseases: Systematic review. JMIR Med Inform. 2019;7 doi: 10.2196/12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Assale M., Dui L.G., Cina A., Seveso A., Cabitza F. The revival of the notes field: Leveraging the unstructured content in electronic health records. Front Med (Lausanne) 2019;6:66. doi: 10.3389/fmed.2019.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spasic I., Nenadic G. Clinical text data in machine learning: systematic review. JMIR Med Inform. 2020:8. doi: 10.2196/17984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horsky J., Drucker E.A., Ramelson H.Z. Accuracy and completeness of clinical coding using ICD-10 for ambulatory visits. AMIA Annu Symp Proc. 2017;2017:912–920. [PMC free article] [PubMed] [Google Scholar]
- 7.Senders J.T., Cote D.J., Mehrtash A., Wiemann R., Gormley W.B., Smith T.R. Deep learning for natural language processing of free-text pathology reports: A comparison of learning curves. BMJ Innovations. 2020;6:192–198. [Google Scholar]
- 8.Savova G.K., Danciu I., Alamudun F., et al. Use of natural language processing to extract clinical cancer phenotypes from electronic medical records. Cancer Res. 2019;79:5463–5470. doi: 10.1158/0008-5472.CAN-19-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alawad M., Hasan S.M.S., Christian J.B., Tourassi G. 2018 IEEE International Conference on Big Data (Big Data) 2018. Retrofitting word embeddings with the UMLS metathesaurus for clinical information extraction; pp. 2838–2846. [Google Scholar]
- 10.Levis M., Leonard Westgate C., Gui J., Watts B.V., Shiner B. Natural language processing of clinical mental health notes may add predictive value to existing suicide risk models. Psychol Med. 2020;51:1–10. doi: 10.1017/S0033291720000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghassemi M., Naumann T., Schulam P., Beam A.L., Chen I.Y., Ranganath R. A review of challenges and opportunities in machine learning for health. AMIA Jt Summits Transl Sci Proc. 2020;2020:191–200. [PMC free article] [PubMed] [Google Scholar]
- 12.Weng W.H., Wagholikar K.B., McCray A.T., Szolovits P., Chueh H.C. Medical subdomain classification of clinical notes using a machine learning-based natural language processing approach. BMC Med Inform Decis Mak. 2017;17:155. doi: 10.1186/s12911-017-0556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blei D.M., Ng A.Y., Jordan M.I. Latent Dirichlet Allocation. J Mach Learn Res. 2003;3:993–1022. [Google Scholar]
- 14.Ramos J. Proceedings of the first Instructional Conference on Machine Learning, Association for Computing Machinery, New York, NY. Vol. 242. 2003. Using TF-IDF to determine word relevance in document queries; pp. 133–142. [Google Scholar]
- 15.Nagendran M., Chen Y., Lovejoy C.A., et al. Artificial intelligence versus clinicians: Systematic review of design, reporting standards, and claims of deep learning studies. BMJ. 2020;368 doi: 10.1136/bmj.m689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajkomar A., Oren E., Chen K., Dai A.M., Hajaj N., Hardt M. Scalable and accurate deep learning with electronic health records. NPJ Digital Medicine. 2018;1:1–10. doi: 10.1038/s41746-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shickel B., Tighe P.J., Bihorac A., Rashidi P. Deep EHR: A survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. IEEE J Biomed Health Inform. 2018;22:1589–1604. doi: 10.1109/JBHI.2017.2767063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devlin J., Chang M.-W., Lee K., Toutanova K. Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers) Association for Computational Linguistics; Minneapolis, MN: 2019. BERT: Pre-training of deep bidirectional transformers for language understanding; pp. 4171–4186. [Google Scholar]
- 19.Vaswani A., Shazeer N., Parmar N., et al. Proceedings of the 31st International Conference on Neural Information Processing Systems. NIPS’17. Red Hook, NY: Curran Associates. 2017. Attention is all you need. 6000-10. [Google Scholar]
- 20.Qiu J., Yoon H.-J., Oak R.N.L., Fearn P.A., Tourassi G.D. Deep learning for automated extraction of primary sites from cancer pathology reports. IEEE J Biomed Health Inform. 2017;22:244–251. doi: 10.1109/JBHI.2017.2700722. [DOI] [PubMed] [Google Scholar]
- 21.Gao S., Young M.T., Qiu J.X., et al. Hierarchical attention networks for information extraction from cancer pathology reports. J Am Med Inform Assoc. 2018;25:321–330. doi: 10.1093/jamia/ocx131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buckley J.M., Coopey S.B., Sharko J., et al. The feasibility of using natural language processing to extract clinical information from breast pathology reports. J Pathol Inform. 2012;3:23. doi: 10.4103/2153-3539.97788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliwa T., Maron S.B., Chase L.M., et al. Obtaining knowledge in pathology reports through a natural language processing approach with classification, named-entity recognition, and relation-extraction heuristics. JCO Clin Cancer Inform. 2019;3:1–8. doi: 10.1200/CCI.19.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnold C.W., El-Saden S.M., Bui A.A., Taira R. Clinical case-based retrieval using latent topic analysis. AMIA Annu Symp Proc. 2010;2010:26–30. [PMC free article] [PubMed] [Google Scholar]
- 25.Kalra S., Li L., Tizhoosh H.R. arXiv:190307406 [cs, stat] March 2019. Automatic classification of pathology reports using TF-IDF features. [Google Scholar]
- 26.Xu K., Lam M., Pang J., et al. Machine Learning for Healthcare Conference. PMLR; 2019. Multimodal machine learning for automated ICD coding; pp. 197–215. [Google Scholar]
- 27.Saib W., Chiwewe T., Singh E. arXiv:200900542 [cs] August 2020. Hierarchical deep learning classification of unstructured pathology reports to automate ICD-O morphology grading. [Google Scholar]
- 28.Ye J.J. Construction and utilization of a neural network model to predict current procedural terminology codes from pathology report texts. J Pathol Inform. 2019;10:13. doi: 10.4103/jpi.jpi_3_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dotson P. CPT® codes: What are they, why are they necessary, and how are they developed? Adv Wound Care (New Rochelle) 2013;2:583–587. doi: 10.1089/wound.2013.0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanauer D.A., Liu Y., Mei Q., Manion F.J., Balis U.J., Zheng K. Hedging their mets: The use of uncertainty terms in clinical documents and its potential implications when sharing the documents with patients. AMIA Annu Symp Proc. 2012;2012:321–330. [PMC free article] [PubMed] [Google Scholar]
- 31.Deeken-Draisey A., Ritchie A., Yang G.Y., et al. Current procedural terminology coding for surgical pathology: A review and one academic center’s experience with pathologist-verified coding. Arch Pathol Lab Med. 2018;142:1524–1532. doi: 10.5858/arpa.2017-0190-RA. [DOI] [PubMed] [Google Scholar]
- 32.Dimenstein I.B. Principles and controversies in CPT coding in surgical pathology. Lab Med. 2011;42:242–249. [Google Scholar]
- 33.Joo H., Burns M., Kalidaikurichi Lakshmanan S.S., Hu Y., Vydiswaran V.G.V. Neural machine translation-based automated current procedural terminology classification system using procedure text: Development and validation study. JMIR Form Res. 2021;5 doi: 10.2196/22461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye J.J. Using an R program to monitor pathology reports for omissions in reporting ancillary tests and errors in test names. Arch Pathol Lab Med. 2020;144:917–918. [Google Scholar]
- 35.Milinovich A., Kattan M.W. Extracting and utilizing electronic health data from epic for research. Ann Transl Med. 2018;6:42. doi: 10.21037/atm.2018.01.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bosker H.R. Using fuzzy string matching for automated assessment of listener transcripts in speech intelligibility studies. Behav Res Methods. 2021;53:1945–1953. doi: 10.3758/s13428-021-01542-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neamatullah I., Douglass M.M., Lehman L.W., et al. Automated de-identification of free-text medical records. BMC Med Inform Decis Mak. 2008;8:32. doi: 10.1186/1472-6947-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montani I., Honnibal M., Honnibal M., et al. Zenodo; 2021. SpaCy: Industrial-Strength Natural Language Processing in Python. [Google Scholar]
- 39.McInnes L., Healy J., Saul N., Groẞberger L. UMAP: Uniform manifold approximation and projection. J Open Source Softw. 2018;3:861. [Google Scholar]
- 40.McInnes L., Healy J., Astels S. HDBSCAN: Hierarchical density based clustering. J Open Source Softw. 2017;2:205. [Google Scholar]
- 41.Bonett D.G. Point-biserial correlation: Interval estimation, hypothesis testing, meta-analysis, and sample size determination. Br J Math Stat Psychol. 2020;73:113–144. doi: 10.1111/bmsp.12189. [DOI] [PubMed] [Google Scholar]
- 42.Platt J.C. In: Advances in Large Margin Classifiers. Smola A.J., Bartlett P., Schölkopf B., Schuurmans D., editors. MIT Press; Cambridge, MA: 1999. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods; pp. 61–74. [Google Scholar]
- 43.Hearst M., Dumais S.T., Osman E., Platt J., Scholkopf B. Support vector machines. IEEE Intel Syst Appl. 1998;13:18–28. [Google Scholar]
- 44.Wen Z., Shi J., Li Q., He B., Chen J. ThunderSVM: A fast SVM library on GPUs and CPUs. J Mach Learn Res. 2018;19:1–5. [Google Scholar]
- 45.Chen T., Guestrin C. Proceedings of the 22Nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. KDD ’16. ACM; New York, NY: 2016. XGBoost: A scalable tree boosting system; pp. 785–794. [Google Scholar]
- 46.Loh W.-Y. Classification and regression trees. Wiley Interdiscip Rev Data Min Knowl Discov. 2011;1:14–23. doi: 10.1002/widm.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 48.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 49.Wolf T., Debut L., Sanh V., et al. Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations. Association for Computational Linguistics; Online: 2020. Transformers: State-of-the-Art natural language processing; pp. 38–45. [Google Scholar]
- 50.Alsentzer E., Murphy J., Boag W., et al. Proceedings of the 2nd Clinical Natural Language Processing Workshop. Association for Computational Linguistics; Minneapolis, MN: 2019. Publicly available clinical BERT embeddings; pp. 72–78. [Google Scholar]
- 51.McCullagh P. Wiley StatsRef: Statistics Reference Online. American Cancer Society; Hoboken, NJ: 2014. Proportional odds model: Theoretical background. [Google Scholar]
- 52.Khattak F.K., Jeblee S., Pou-Prom C., Abdalla M., Meaney C., Rudzicz F. A survey of word embeddings for clinical text. J Biomed Inform. 2019;100S doi: 10.1016/j.yjbinx.2019.100057. [DOI] [PubMed] [Google Scholar]
- 53.Johnson A.E., Pollard T.J., Shen L., et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3 doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lundberg S.M., Erion G., Chen H., et al. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell. 2020;2:56–67. doi: 10.1038/s42256-019-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kokhlikyan N., Miglani V., Martin M., et al. arXiv:200907896 [cs, stat] September 2020. Captum: A unified and generic model interpretability library for PyTorch. [Google Scholar]
- 56.Sundararajan M., Taly A., Yan Q. International Conference on Machine Learning. PMLR. Toulon, France. Vol. 5. 2017. Axiomatic attribution for deep networks; pp. 3319–3328. [Google Scholar]
- 57.Bonert M., Zafar U., Maung R., et al. Evolution of anatomic pathology workload from 2011 to 2019 assessed in a regional hospital laboratory via 574,093 pathology reports. PLoS One. 2021;16 doi: 10.1371/journal.pone.0253876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Look A. Ahead: Pathology CPT Changes for 2015 | APS Medical Billing. https://apsmedbill.com/whitepapers/look-ahead-pathology-cpt-changes-2015 Available from: [Last accessed on 2021 Feb 11]
- 59.Mayhew M.B., Tran E., Choi K., et al. Optimization of genomic classifiers for clinical deployment: Evaluation of Bayesian optimization to select predictive models of acute infection and in-hospital mortality. Pac Symp Biocomput. 2021;26:208–219. [PubMed] [Google Scholar]
- 60.Vaickus L.J., Suriawinata A.A., Wei J.W., Liu X. Automating the Paris system for urine cytopathology-A hybrid deeplearning and morphometric approach. Cancer Cytopathol. 2019;127:98–115. doi: 10.1002/cncy.22099. [DOI] [PubMed] [Google Scholar]
- 61.Kim Y., Lee J.H., Choi S., et al. Validation of deep learning natural language processing algorithm for keyword extraction from pathology reports in electronic health records. Sci Rep. 2020;10:20265. doi: 10.1038/s41598-020-77258-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cernile G., Heritage T., Sebire N.J., et al. Network graph representation of COVID-19 scientific publications to aid knowledge discovery. BMJ Health Care Inform. 2021;28 doi: 10.1136/bmjhci-2020-100254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frolov S., Hinz T., Raue F., Hees J., Dengel A. Adversarial text-to-image synthesis: A review. Neural Netw. 2021;144:187–209. doi: 10.1016/j.neunet.2021.07.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material