Abstract
Computational biology is one of many scientific disciplines ripe for innovation and acceleration with the advent of high-performance computing (HPC). In recent years, the field of machine learning has also seen significant benefits from adopting HPC practices. In this work, we present a novel HPC pipeline that incorporates various machine-learning approaches for structure-based functional annotation of proteins on the scale of whole genomes. Our pipeline makes extensive use of deep learning and provides computational insights into best practices for training advanced deep-learning models for high-throughput data such as proteomics data. We showcase methodologies our pipeline currently supports and detail future tasks for our pipeline to envelop, including large-scale sequence comparison using SAdLSA and prediction of protein tertiary structures using AlphaFold2.
Keywords: high-performance computing, computational biology, machine learning, deep learning, protein sequence alignment, protein structure prediction
I. Introduction
Correctly inferring a gene’s function from its sequence, i.e., gene functional annotation, is of utmost importance to the biological sciences. Dramatic advances in next-generation sequencing technologies have driven exponential growth in the number of sequenced genomes, leading to a growing bottleneck in accurate gene annotation [10], [64]. Indeed, the need to extract information about gene function from massive amounts of sequence data is approaching the scale of data generated in particle and astrophysics [62]. Computational approaches can take advantage of this massive amount of data and play a key role in eliminating the gene-annotation bottleneck. An efficient and accurate predictive computational mapping of large genomic datasets to information about organism fitness, metabolism, and response to its environment will help the understanding of domains of unknown function, thus facilitating major advances in genomic sciences research. Because the function of a protein fundamentally relies on its three-dimensional structure, and the same protein structure can result from different protein amino acid sequences due to redundancy in the physical properties of different amino acids, the inclusion of structure into functional analysis can provide essential information about protein function that purely sequence-based methods may not detect [57].
A main challenge in tackling the growing gene-sequence annotation bottleneck is the need to scale up sequence analysis and other important bioinformatics tasks such as protein structure prediction, detection of homologs, and prediction and modeling of protein-protein interactions. The use of high performance computing (HPC) resources could, in principle, assist genome annotation at large-scale tremendously. However, bioinformatics and computational biology applications have often not made use of HPC or even code acceleration with general-purpose graphics processing units (GPUs), both of which were adopted by other computational fields with large computing needs, such as astrophysics. Barriers to the use of HPC and GPUs have included the heterogeneous sizes of biological datasets, their storage formats in many small text files due to the community compendia that are sequence databases, and the many different steps required for each part of a lengthy processing and analysis pipeline, each of which requires specialized programs often developed by third-parties. The latter is a direct result of the difficult and messy nature of the biological sciences: the variability in genome structure across the taxa, the multi-scale and multi-disciplinary scientific fields that are involved (from quantum-mechanical considerations of catalysis to ecological-scale considerations of population genetics), and the complexity of gene expression and regulatory networks in living organisms.
The convergence of HPC and artificial intelligence (AI), especially deep learning (DL), promises to open the door to the use of HPC resources for gene functional annotation. Indeed, the emergence of GPU accelerators has helped to spark the “resurrection” of neural networks in the past decade and has led to increases in their accuracy, generalizability and predictive power [63]. In addition, the use of DL and other AI methods in the biosciences has recently flourished [49]. Here, we describe our efforts to make the necessary connections between bioinformatics, deep-learning, and HPC to develop next-generation predictive tools for inferring gene function from sequence, using structural information both explicitly and implicitly. Designed to bridge the genome-annotation technology gap, we focus on using leadership computing resources to tackle the gene annotation problem for proteins on the scale of full genomes. We have developed, deployed, and tested a number of different methods to perform genome-scale analyses, from DL-based sequence alignments, to protein structure prediction and error analysis, to the prediction of protein-protein interactions. Our toolbox, which is continually developing and growing, can be used in many different types of pipelines and workflows designed to crack the code that translates protein sequence to protein function across thousands of protein sequences in organisms’ proteomes.
II. Programs and methods in the toolbox
Our HPC toolbox consists of several deep-learning-based modules, for protein sequence alignment, structure prediction (including the assessment of predicted structure quality), domain assignment, and prediction of the interactions between proteins. In order to take advantage of HPC resources, we have been developing and optimizing these tools. In this section we first describe the different methods from a theoretical perspective, and then detail their algorithmic components.
A. SAdLSA alignment
In order to annotate the function of a new gene, a vital step is to search computationally for its homologs that have been experimentally studied. Since all species are related by evolution, one can infer the functions of new gene sequences from previously characterized ones provided that they share an appropriate level of sequence similarity. This step of sequence comparison, or alignment, has become a central focus of bioinformatics. Indeed, the classic sequence alignment algorithm BLAST, was once nearly synonymous with bioinformatics. Sequence alignment is not merely a direct comparison between the characters that represent the sequences being compared, but involves advanced statistical methods and substitution matrices that take into consideration common substitutions found in nature and similarities between physical properties of different amino acids that make up the sequence alphabet, as well as each sequence element’s neighbors. While the classic BLAST algorithm and the improved, iterative sequence-profile method PSI-BLAST [1] are efficient heuristic approaches designed to rapidly search a large sequence database and accurately return related sequences that may have only 40-50% direct sequence similarity, they often fail to detect subtle, yet significant, similarity for sequences in the “twilight zone” of sequence identity [52]. Here, only about 20 to 35% of aligned residues are identical, and more than 95% of sequence pairs within this zone are from structurally dissimilar proteins; however, many highly similar protein structures which are also evolutionarily related may have such a low sequence identity. Their performance becomes much worse in the “midnight zone” at < 20% pairwise sequence identity [43], [52]. To address this issue, sequence comparison methods based on Hidden Markov Models have been proposed, e.g., the widely adopted HMMER [15] and HHsearch approaches [60]. These are routinely used to classify protein families such as Pfam [18], or search for potential structural homologs within the Protein Data Bank (PDB) [30]. Although these methods are more sensitive, the difficulty of identifying sequence and structural similarity in the twilight zone remains. In a typical bacterial proteome, we estimate that about 20 to 30% of protein sequences have an unknown or a low-confidence family classification. Even for those sequences with a reliable protein family assignment, this does not necessarily mean that they are functionally or structurally characterized; such knowledge for most of these protein families is not provided in existing databases such as Pfam [18] or UniProt [10]. SAdLSA was developed to address the “twilight zone” issue. It is a DL-based approach for protein sequence alignment that incorporates structural information [25]. It is trained to learn the structural relationship encoded by two protein sequences subjected to comparison. As such, it can sense the structural fold of input sequences and obtain more sensitive alignment not possible with classic sequence alignment approach [26]. SAdLSA uses a deep residual convolutional neural network trained on hundreds of thousands of structural alignments computed using three-dimensional alignment tools applied to experimental protein structures [25]. This allows the program to detect remote relationships useful for genome annotation or functional predictions. The advantage of SAdLSA was demonstrated in benchmark tests against HMM-based HHsearch [60]. For challenging cases, SAdLSA is ~150% more accurate at generating pairwise alignments and ~50% more accurate at identifying the proteins with the best alignments in a sequence library [25].
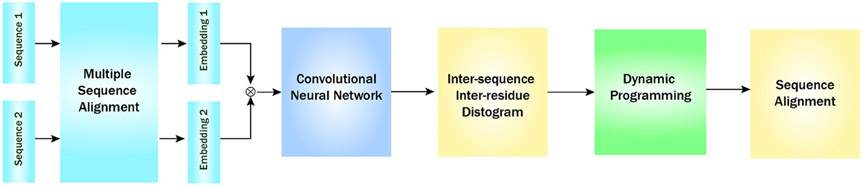
A flowchart of SAdLSA is presented in figure 1. The two input sequences are first compared against a large sequence library, such as a UniProt reference library [10] using a fast sequence alignment tool such as HHblits [50], yielding multiple sequence alignments (MSAs). The MSAs are then employed to generate two position-specific sequence profiles, or embeddings, each of dimension Nk × 20, where Nk is the length of the kth sequence (k = 1,2), and the 20 columns represent the 20 different amino acids at each residue position. Note that these two MSAs contains close homologs to the input sequences, and remote homologs are typically absent in these alignments. The outer product of these two 1D sequence features yields a 2D matrix of features, where at position (i, j) of the matrix the elements are a concatenation of the 20 columns formed from the i-th residue of sequence 1 and the j-th residue of sequence 2. Subsequently, these 2D features are fed into a fully convolutional neural network consisting of residual convolutional blocks [28]. The main objective of this neural network model is to predict residue-residue distances between the two input sequences that recapitulates their optimal structural alignment, using observed structural alignments as the training ground truth. The training distance labels are created from structural alignments by APoc [22], which takes advantage of a global alignment provided by TM-align [75]. The inter-sequence, inter-residue distogram is then converted into a scoring table for subsequent dynamic programming, which generates an alignment for the two input sequences. For practical applications, such as comparing an input sequence against a library of sequences, SAdLSA is designed to run efficiently in a multiple node, multiple GPU mode, using pre-processed sequence embeddings.
Fig. 1.
Overview of SAdLSA, a deep-learning algorithm for protein sequence alignment. The symbol ⊗ denotes outer concatenation of two embedding vectors
SAdLSA’s deep-learning neural network is composed of multiple residual blocks, either conventional [28] or dilated [72] in slightly different design variants. The residual block design is a key to train a deep neural network model. Within a residual block, each convolutional layer is composed of about 64 filters with a kernel size of 3 × 3 in dilated residual blocks or 5 × 5 in regular residual blocks. After the residual blocks, the last 2D convolutional layer outputs 22 channels, representing 21 distance bins (1 to 20 at 1 Å each, and >20 Å) and channel 0 which is reserved for ignored pairs (e.g., gap residues missing in a structure, or large distances >30 Å). Finally, a softmax layer calculates the probability scores for each distance bin. For this study, we used the consensus (mean) scores from two ensembles of DL models. The first set of DL models (RN14) consists of six models with 14 residual blocks and 64 5 × 5 kernels in each convolutional layer. The second set consists of three from the first set, and three dilated models (DRN34) with 34 residual blocks (alternating 1,2,8,16,32 dilate rates) and 50 to 75 3 × 3 kernels. The regular RN14 and dilated DRN34 models have 2.9 and 2.4 million parameters, respectively. Empirically, we found that the mixture of these two types of models delivers better performance than a single uniform set of DL models. The outputs from the DL model are the probabilities of distance bins forming inter-protein residue-residue distance matrices. To build an alignment using dynamic programming (DP), we convert this probability matrix into a mean distance matrix D, whose element where i, j are target/template sequence positions, is the probability for bin k at position (i, j), bk are distance labels. D is subsequently adapted as the scoring matrix to obtain the optimal alignment using a Smith-Waterman-like DP algorithm [59]. The distance matrix D is also used to calculate an estimated TM-score [73] for ranking the significance of an alignment. The constant c is set at 1 such that a perfect alignment gives an estimated TM-score of 1. In practical applications, one typically needs to run a large number of sequence comparisons. Since these comparisons are independent, one can easily distribute these calculations into different GPUs if they are available. For this purpose, we employed the multiprocess Python module to implement a queue that distributes multiple sequence alignment runs across multiple GPUs. This enables efficient runs for applications.
B. Protein structure prediction
Another important method in translating protein sequence to function is the analysis of its three-dimensional structure. Experimentally, protein structures are obtained via x-ray crystallography, nuclear magnetic resonance (NMR) techniques, and cryo-Electron Microscopy (cryo-EM), which are very time consuming and costly. As an example, only about 17% of the human proteome has an experimentally determined three-dimensional structure [36]. Protein structure prediction via computational approaches can facilitate the structural characterization of protein sequences [74]. One critical challenge in structure prediction is to correctly model long-range, many-body effects from scratch, that are traditionally best dealt with by template-based methods [58]. Over the past several years, exciting breakthroughs have been made to include these long-range residue-residue interactions in predictions using deep residual convolutional neural networks [68]. Significant success has been demonstrated in protein structure prediction by taking advantage of deep-learning based contact predictions, as demonstrated in the Critical Assessment of protein Structure Prediction (CASP) contests; CASP is a blind biannual protein structure prediction competition. In CASP13, all four top-ranked groups in the most challenging, free-modeling category used residue-residue contacts or distance matrices predicted via deep-learning [23], [34], [54]. Very recently, Google DeepMind’s AlphaFold2 achieved impressive accuracy in CASP14 [36]. For 38 free-modeling domain targets, it achieved a mean GDT-score ~0.85. When applied to the human proteome, it yielded a coverage of 58% by amino acids (over the total length of all sequences), in comparison to the 17% by experiments [65].
It is important to point out that a protein’s structure by itself does not immediately yield functional annotation. Further computational steps such as structural comparisons [32], [75], structure analysis such as ligand-binding pocket analysis [22] or protein-protein interface analysis [24], molecular docking and simulation [40], [67], and prediction of interactions and interaction interfaces of the protein on a three-dimensional level, can be performed to further elucidate relationships and refine predictions. Ultimately, it will be important to match predictions to validating experiments, which, with the help of high-throughput techniques, are also providing larger datasets.
1). Computational quality assessment and estimation of model accuracy for predicted structures using deep learning:
Estimation of model accuracy (EMA) or Quality Assessment (QA) plays a core role not only in protein structure prediction, but also for confident use of the predicted tertiary structure in subsequent steps of the functional annotation pipelines. Quality assessment of predicted protein structure models plays a key role in improving the quality of the final prediction: if accurate information about the similarity or discrepancy between the predicted models and the native structure along all residues can be acquired, it is possible to considerably reduce the search space for further refinement algorithms and improve the overall model quality [31]. Another challenge for EMA is to identify the most confident structural models out of a set of predicted structures [9]; this becomes even more important for genome-scale applications where thousands of predicted structures may be generated. We developed two different methods: a deep learning-based graph method (GEN) [5] that estimates both per-residue accuracy and correct coordinates which can be used to select the predicted models that potentially give the highest quality structure, using a set of geometric features, and a vision transformer-based EMA model (VTM) which uses only the single raw protein residue pairwise distance map as the input for predicting a decoy’s quality score.
For the QA using the GEN method, we used datasets from CASP8-14 [9], [38] with targets in CASP13 as the validation set and CASP14 as the test set. Models are first filtered by removing those with incomplete or inconsistent residues compared to the corresponding reference structure. As a result, there are 477, 82, and 70 targets used for training, validation, and final testing, respectively, and of all 109318 structure models from the corresponding targets, 12118 and 10435 were held out for validation and testing, respectively. For a given protein structure model presented as a file in Protein Data Bank (PDB) format, the following 3 types of output are generated: per-residue local distance difference test (LDDT) scores, distributions of error of per-residue-pair carbon alpha error using native PDBs as reference structure, and finally, the updated alpha-carbon coordinates for the input model. The input features consist of protein sequences, pairwise orientation information, coordinates of the alpha carbon atoms in the models. We included model representations from DeepAccNet [31] as atom-level information and DeepDist [27] as co-evolution information. After processing all features, we represent them as L × c or L × L × c data structures (where L is the sequence length) for input to the deep learning model.
For EMA using the VTM, we were inspired by the outstanding performance of transformer-based in computer vision (CV) tasks, and we developed a vision transformer-based single EMA model. Since 2017, the self-attention mechanism has become popular [4], [48]. In recent years, researchers tried to apply self-attention or transformer architecture to CV. The Vision Transformer (ViT) [13] was the first method to utilize the transformer model in CV tasks. Recently, increasing numbers of efforts [12], [70] combined the CNN and transformer for reducing the computational requirements and for getting better performance. Protein inter-chain distance information is a key source of information for predicting protein’s three-dimensional structure and many methods which output inter-chain distance information can contribute data to the EMA [7], [9]. We applied the attention mechanism to predict protein residue-residue contact [6]. In a recent study, the DISTEMA method [8] used only the single raw distance map as the input for predicting a decoy structure’s quality score. Building on our this work, we inserted self-attention modules into the CNN to catch the global attention information by using the depth-wise separable convolution layer to generate the query key value matrices for self-attention. Comparing to the original transformer architecture, CNN with self-attention could reduce the model’s parameters. However, training a vision-transformer model demands significant computational power. To train the model, we used CASP8-13 as our training dataset, and we generated a difference map [8] for each decoy as the input feature.
2). Reconstructing protein structures from CryoEM density maps using deep learning:
Near-atomic resolution images of large macromolecules and macromolecular assemblies can be obtained using cryogenic electron microscopy (cryo-EM) [11]. However, inferring the structures of proteins from the cryo-EM density map data is still a challenging task. Deep learning recently emerged a useful technique to tackle the problem [39], [56]. We used a deep learning architecture to predict the alpha carbon and backbone atoms of the protein structures. The images generated by cryoEM are stored in a 3D-grid structure in a file. Each voxel (pixel in 3D) represents the probability of the presence of atoms based on the scattering electrons detected by the cryoEM technique. Based on this data and metadata about the protein density map, we create labeled data and train our model to predict the voxel of alpha carbon and backbone atoms in the density map. The predicted result is used to determine the final 3D protein structure.
C. Boundary prediction for protein structure domains
Protein domains are regions of a single protein chain that can fold into a stable tertiary structure and/or have some distinct function [16], [33]. Domains can be functional building blocks, and may be recombined over the course of molecular evolution in different arrangements to create proteins with different functions. The study and prediction of protein domains is therefore important for understanding the function of proteins [16], [33], [35]. Annotation of the domains of proteins is an important effort, both for the classification of proteins with known structures in order to find common patterns in structure-function relationships [57] and for use sequence-based analysis that can infer structural domains to perform domain annotation [51]. In addition, protein domains can be a useful feature to assist protein structure prediction [16], [19]. However, considering the large amount of sequence data, it is practically impossible for human experts to manually annotate protein domains. To mitigate this intensive process, computational methods are necessary to facilitate the automatic prediction of protein domain boundaries. Here we used a DL model for domain boundary prediction from protein distance maps [71]. The output is the probability of a residue being in the domain boundary.
D. Protein-protein interactions
Proteins are frequently found in complexes called oligomers which can be functionally obligatory and which may involve multiple copies of the same protein, or several different proteins. A dimer is a complex of two proteins; a homodimer contains two copies of the same protein, and a heterodimer contains two different proteins. Prediction of protein-protein interactions (PPIs) is an important part of efforts to understand protein function [76]. Here, we focus on predicting the location where two proteins will interface, or protein interface prediction (PIP), which is an important computational effort [69], relevant to drug discovery and to protein design and engineering [44], [47].
1). Prediction of protein-protein interaction using structure:
For our toolkit, we adapted two state-of-the-art (SOTA) graph neural network architectures for PIP [42], Neighborhood Average (NeiA) and Neighborhood Weighted Average (NeiWA), from Liu et al. [41]. The input for our method is the tertiary structures of two protein chains that form a dimer. We represent each chain’s tertiary structure as a 3D graph, defining the nodes of each graph as a chain’s alpha-carbon atoms and its edges between nodes as their 20-nearest neighbors in Euclidean space. The output of our network is a two-dimensional (2D) binary-valued tensor of shape M × N for each pair of chains, where M and N are the numbers of alpha-carbon atoms in the first and second chain, respectively. To prevent these models from overfitting, we added dropout layers and a cosine annealing optimizer to their training procedure. Inspired by the high-order pairwise interaction (HOPI) module in Liu et al. [41], we then constructed a multi-layer convolutional neural network module for dense prediction of inter-chain interacting residue pairs, using the ResNet architecture [28] as a baseline for its design. The flow of input data in our models is such that, once the NeiA or NeiWA layers have finished updating their initial input node and edge features for both graphs using shared weights in their graph convolutions, the updated node representations from both graphs are then interleaved to form a 3D tensor. This 3D tensor then serves as input to our deep residual convolution module for dense prediction of inter-chain residue-residue interactions, after which the model is scored using a class-weighted binary cross-entropy loss function to see how well it was able to distinguish interacting residue pairs from those not in interaction.
2). Prediction of dimerization from sequence:
Accurately predicted inter-chain contacts are important for PIP and PPI prediction [46]. If the Euclidean distance between the heavy atoms of two residues in different chains of a protein complex is ≤ 6 Å, we say that the two residues are in inter-chain contact [46]. If we consider a multimeric (oligomeric) complex to be composed of individual dimers, predicting the inter-chain contacts of all the possible dimers can allow us to reassemble the structure of the entire oligomer. Here, we train a deep neural network using co-evolution features that are derived from multiple sequence alignments or MSA, focusing on homodimers. For this case, since both the monomers are the same, the MSA of the monomer may contain both the co-evolutionary information of intra-chain as well as inter-chain interactions [46], [66]. Our input consists of the protein sequence of the monomer which is then used to generate MSA, from which, a co-evolutionary, secondary structural, etc. feature matrix is derived as the input for training our deep neural network. The model outputs a 2D contact map of size L × L, where L is the sequence length. This contact map contains probabilities of residues being in inter-chain contact with each other.
III. Deployment on HPC systems
Here we describe development and deployment of our methods on HPC systems, including the Summit supercomputer at the Oak Ridge Leadership Computing Facility (OLCF). Scaling to large portions of the supercomputer is the key to genome-scale inference, and scaling to multiple GPUs is important for training efficiency. The latter is accomplished both with data-parallel and model-parallel methods, as described in the following sections. Large-scale inference is performed by parallelizing over the databases for each sequence alignment with SAdLSA [25], and over proteins in a genome for structure prediction.
A. Systems used
The Summit supercomputer is a 200 petaFLOP IBM AC922 system consisting of 4608 nodes each with six NVIDIA Volta V100 GPUs and two POWER9 CPU sockets providing 42 usable cores per node. The Andes cluster at the OLCF is used for preprocessing and data analysis, and is a 704-compute node commodity-type Linux cluster with AMD EPYC 7302 16-core processors. The ORNL Raptor workstations contain two POWER9 CPU sockets and a single NVIDIA Volta V100 GPU. The initial deployment and testing of genome-scale structure prediction with AlphaFold was carried out using computational resources supported by the Partnership for an Advanced Computing Environment (PACE) at the Georgia Institute of Technology: CPU nodes used were dual Intel Xeon Gold 6226 CPUs each with 12 cores, and 40 NVIDIA RTX6000 GPUs were used for model predictions.
B. Deploying SAdLSA on a genomic scale on the OLCF Summit supercomputer
Despite the exciting development of deep-learning based computational tools for annotating protein sequences, it is not straightforward to apply them at the genome-scale, e.g., hundreds to thousands of sequences from an organism. The largest data load in the SAdLSA workflow for microbial organisms is usually the database against which each sequence in a proteome is compared, as these can contain tens of thousands of sequences, while a microbial genome typically contains less than five thousand. Here we focus on a microbial genome for our SAdLSA deployment, namely the Desulfovibrio vulagris, strain Hildenborough, bacteria, a model sulfur-reducing organism [29] which is important to the Department of Energy’s Office of Biological and Environmental Research. For initial deployment we focus on the 559 proteins labeled as “hypothetical” in the March 2020 GenBank annotated genome file found on the National Center for Biotechnology Information of the Desulfovibrio vulgaris Hildenborough reference genome.
Figure 2 illustrates the pipeline for deployment of SAdLSA with parallelization over the alignment database and illustrating the portions that make use of the GPUs and the non-volatile memory (NVM) on Summit’s compute nodes and the NVM Express (NVME) specification. Each protein sequence is aligned using many nodes, each working on a portion of the database, which is usually tens of thousands of sequences, but may be as large as millions to billions. The proteins in the genome are worked on by SAdLSA in a serial manner. SAdLSA performs DL inference on the GPUs, and a final Smith-Waterman (SW) dynamic programming procedure on the CPU. While each portion of the database is worked on in a completely parallel manner, the initial task-list creation, the final reduction, and the SW procedure is performed in serial. Aligning the entire set of 559 hypothetical proteins to the Pfam database, which contains 19,179 entries (sequence profiles from hidden Markov models) representing protein families, takes less than 15 minutes using 1000 Summit nodes. Performance tests are shown in Figure 3 using a reduced set of 120 sequences so that jobs using lower node counts can finish within queue limits. Time for an alignment to Pfam varies nearly linearly with query sequence length, and time to solution is reduced by ~1000× when scaling to 1000 nodes from 1 node. Average time per sequence alignment to Pfam using 1000 nodes is 1.3 seconds, averaged over the 120 sequences. Aligning these 120 sequences to the PDB70, which contains 88,284 entries takes on average 16 seconds per sequence using 1000 nodes. Figure 4 displays the weak scaling of the PDB70 from 96 nodes to 1000 nodes using 12 and 120 sequences, and the times per sequence inference over the set. The set of 12 was chosen to have similar sequence length variance as the the set of 120.
Fig. 2.
Scheme for SAdLSA deployment at scale on Summit
Fig. 3.
Performance of SAdLSA on large sets of proteins and scaling across Summit. A: Runtime, in seconds, against the Pfam database for each sequence using one node. B: Runtime, in seconds, against the Pfam database for each sequence using one thousand nodes. Parallelization was over the Pfam database for each sequence input for inference calculation.
Fig. 4.
Performance of SAdLSA on the PDB70 database on Summit. Top: Weak scaling– total time in minutes for a given number of sequences to be aligned to the PDB70 using 12 sequences, 96 nodes, and 120 sequences, 1000 nodes. The set of 12 was chosen to have a similar length distribution ad the set of 120. Bottom: Runtime, in seconds, against the PDB70 database for each sequence in the 120 sequence set used in Figure 3 above, using one thousand nodes. Parallelization was over the PDB70 database for each sequence input for inference calculation.
C. Deploying AlphaFold for genome-scale structure prediction
On July 15, 2021, DeepMind released a fully open-source version of their AlphaFold (version 2) program which had performed exceptionally well at the CASP14 competition [36]. The accuracy of this method’s predictions is due in part to the size of the neural network as well as innovations in the approach and network architecture. The use of GPUs to both train and perform inference is important for its high performance, and essential for genome-scale applications. Deployment of AlphaFold at scale to predict the structure of an organism’s full genome is a natural desire, and has been pioneered by AlphaFold together with the European Bioinformatics Institute (EMBL-EBI) for 21 organisms, including human and pathogenic microbes [65]. The computing power of the Summit supercomputer is an excellent resource for this type of efforts, and our aim is to supplement the AlphaFold/EMBL-EBI database with organisms of interest to the Department of Energy, and to produce full-genome structural predictions for organisms central to our research. Challenges to deploying AlphaFold on Summit include the Power9 CPU architecture that prevents the use of package managers to download pre-compiled binaries, combined with the use of build systems non-standard in the HPC setting. Here we describe our current progress in testing and deploying AlphaFold on our compute resources and preparation for full-scale deployments on Summit. AlphaFold provides a Docker1 recipe for building all of its dependencies and packaging, but that recipe is not directly usable on Summit or on other systems which do not support Docker.
1). Prediction of the structures of all uncharacterized proteins in the Desulfovibrio vulagris Hildenborough Proteome:
As an initial test of genome-scale protein structure prediction with AlphaFold, we predicted the structures of the hypothetical proteins presented in Section III-B which were also subjected to alignment with SAdLSA. Due to the need to build from source on Summit for the Power9 architecture, and incompatibility of the Google Bazel build system with HPC toolchains (described in Section III-C2 below), we first tested this initial deployment method using the PACE computational resources at the Georgia Institute of Technology to validate our re-orchestrated pipeline and to obtain estimates of the computational cost of these types of deployments: the program in its released form could not be deployed on academic clusters that do not support Docker containers, and thus the AlphaFold workflow was extracted from the Docker recipe and deployed with Python scripts. Furthermore, for deployment on Summit, reconfiguration of the pipeline is also needed so that CPU-based feature generation steps could be calculated on resources other than Summit in order not to waste expensive GPU node hours. We included all steps in the AlphaFold Docker-based workflow and utilized the reduced BFD dataset which was provided by DeepMind (to reduce the size of the databases from 2.2 TB to 410 GB without reduction in accuracy). Results from tests on the CASP13 and CASP14 protein targets indicated that our reconstructed pipeline was consistent with its expected performance. For the 559 hypothetical proteins, we used 30 of the Intel Xeon Gold CPU nodes for deriving input features, and 40 of the NVIDIA RTX6000 GPUs for model predictions on PACE. The total walltime for predicting structures of this full set was about 4.7 hours for feature generation, 8 hours for the DL inference, and about 1.5 hours for the structure relaxation portion, for a total of about 14 hours for the 559 proteins. For Eukaryotic organisms, the total number of proteins in a genome is usually around 30-40 thousand, which would require about 3 months using an academic system such as PACE for a single proteome (not including queue time), illustrating the need for large leadership resources for genome-scale structure prediction campaigns.
2). Towards deploying AlphaFold at genomic scale on the Summit supercomputer:
The primary challenge in preparing AlphaFold for Summit was the preparation of software dependencies, within AlphaFold, to run on Summit’s Power9 architecture. The AlphaFold Docker recipe depends on prebuilt x86 binaries for JAX, Tensorflow, and some of their dependencies. The core dependency that needed to be enabled on Power9 was JAX2, a high performance machine learning tool that uses the XLA (Accelerated Linear Algebra) compiler3. By using XLA, which is a domain-specific compiler for linear algebra, JAX is able to just-in-time compile NumPy code on accelerators. Most of our time in preparing AlphaFold for Summit was spent on getting JAX built and functioning for Power9 with NVIDIA GPUs. We initially tried to build JAX outside of a container, directly on Summit, in an attempt to deploy the pipeline in a similar way as on the PACE resources. However, we quickly ran into issues as the required build system for JAX, Google’s Bazel4, does not operate well in an HPC environment. Ultimately, we were unable to pass a compiler from a non-standard location (i.e. outside of /usr/bin) to all the build steps within Bazel. In HPC, it is common to use compilers outside the standard location, but this was not operable with Bazel. This is where gaining access to a Power9 system in which we could build containers became essential.
In the end, we used a Raptor external POWER9 system with compilers in the standard /usr/bin locations, and built a Singularity5 container that could run on Summit. This Raptor workstation had support for Podman [21] as the container build mechanism. We altered the AlphaFold Dockerfile to build JAX and other components from source, and to remove feature generation steps which did not require GPUs and thus could be pre-computed on our Andes cluster. Podman enabled us to build a container image from our altered AlphaFold Docker recipe, that builds JAX and other components from source. The resulting image built with Podman was around 38 GB, since Podman builds images in layers, but the final image was 9.8 GB after converting to Singularity’s format which strips the unneeded layer information. From there, we were able to successfully transfer and run the resulting AlphaFold container image on Summit with Singularity. Summit does not yet support container builds, but it does provide a functioning container runtime with Singularity. Tests against the CASP benchmarks indicated that our deployment was providing correct results, and that this container was ready to be used at scale across Summit nodes at genome-scale– work that is currently in progress.
D. Training QA and EMA models for structure assessment on Summit
For the GEN method, training we used 8 nodes of Summit and the PyTorch [45] as distributed deep learning training framework with the Horovod backend [55]. The batch size is set to 1 for each GPU, which is equivalent to an effective batch size of 48. We used SGD as optimizer with learning rate 1e-6, momentum 0.9 and decay 5e-5. For predicted LDDT scores and coordinates, we used mean square error (MSE) as the loss function. For distributions of error of pairwise alpha-carbon distance, we used categorical cross-entropy as the loss function. We compared our results trained using different coordinate-superposing methods (TMAlign [75] or Kabsch least-squares superposition [37]) and different labels for coordinate loss (using real-value distances or coordinates directly) and DeepAccNet [31]. Future work can explore how to make use of more nodes of Summit to achieve even higher accuracy for prediction. For the VTM, we tested training the model on the Summit computer with a batch size of 1 per GPU core, and using all 6 GPUs on a node. In data parallel mode, each batch approximately requires 40 minutes of wall time. The model usually converges around 100 epochs. Compared to training with only one V100 GPU, using multiple GPUs in data-parallel mode speeds up training by a factor of 4. Future work will scale this parallel training to larger datasets which can make use of multiple nodes on Summit.
E. Deep learning training on Summit for cryoEM reconstruction
To train our model, we utilized distributed data parallel (DDP) and distributed data parallel sharded (DDP-sharded) techniques on multiple GPUs. We used the multi-GPU training capabilities in PyTorch Lightning [17], by specifying the accelerator training flag ddp for DDP and using the DDP-sharded plugin with the ddp_sharded flag. DDP in PyTorch works by initiating a process and replicating the model into each GPU, splitting the batch data across GPUs, training individual models on these data splits, and then resolving the multiple models into a single version after a set of steps by synchronizing the gradients and buffers through communication between GPUs6. This allows us to increase our batch size by the number of GPUs available, effective batch size = original batch size × number of GPUs available. We tested this method with our application on 6 GPUs of a Summit node, which means each GPU processed a single protein density map in parallel with the others. The model trains the forward and backward pass, and gradients are synced across the GPUs. Finally, the gradients are averaged and all the optimizers in each individual GPU updates its weights. Alternately, to lower the memory overhead per GPU, the sharded technique shards the optimizer and gradients across the GPUs. The integration of sharded training is provided by FairScale [2], which is built to be PyTorch-compatible. Using sharded training, we are able to train the model faster with efficient memory utilization on the high memory V100 GPUs (32 GB) on Summit. In a small test example, using the DDP-sharded training increased the model training speed by 6% in comparison to the DDP enabled training, where the model’s trainable parameters, number of epochs, and batch size were kept constant. Using 10 cores on Summit’s CPUs, we initialize 10 data-loader workers, which helps to speed up the training process as well. The input to the model is a 3D cryoEM density map and the output is also the 3D map with only alpha carbon or backbone atoms present on the 3D grid. After the preprocessing steps of normalizing the high intensity and low intensity data, resampling the grid to 1 Å, and generating the labels, we train our model. We then track the training of the model in real time with the cloud sync feature of Weights & Biases [3].
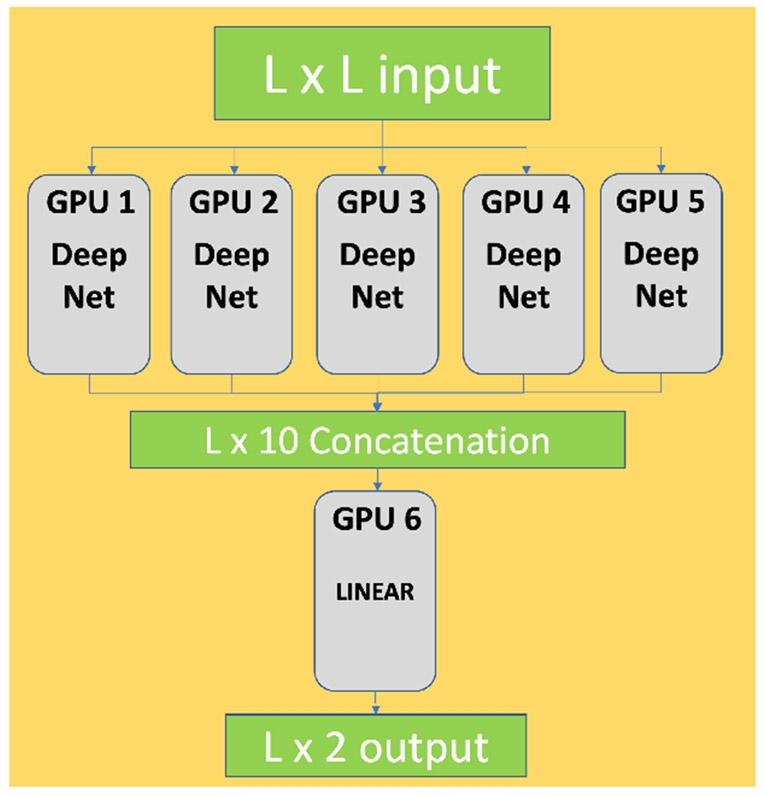
F. Protein domain boundary prediction using model parallelization on Summit
As more complex models are developed in order to solve problems with higher precision and more generalization to real-world data, models may be too large to fit on the memory of one GPU. Model parallelization is a technique used to alleviate such complications. A model is split into several parts by layer, and each of those parts is distributed into several GPUs. For example, if a model contains m layers, and there are n GPUs available, each GPU will have m/n layers of the model in a parallel, or ensemble, approach. We developed a model parallelization strategy, using the six GPUs on a Summit node for protein domain boundary prediction using an ensemble of neural network models. Each model in the ensemble produces an output; these are then all concatenated together to produce the final domain boundary prediction. The model ensembles are distributed on five GPUs. On the sixth GPU, the concatenated output from the five GPUs is processed through a linear neural network layer, producing the final output and prediction of the domain boundary. The model layout and the GPU allocation is shown in Figure 5. The training is done with a batch size of 4 and cross-entropy loss is used as the loss function. Each model uses 12.665 GB out of the available 16.130 GB of GPU memory, and the linear layer uses 1.453 GB, illustrating the need for a parallelization technique for proper deployment. In this same manner, the model can be parallelized across nodes as well, and more complex execution schemes can be developed to enable efficient use of all GPUs.
Fig. 5.
Scheme of the model parallelization method on a Summit node for domain boundary prediction
G. Creating protein interface prediction datasets
Success in PIP, as in most machine learning efforts, rests in large part on the quality and quantity of training data. Towards this end, using OLCF resources, we developed a curated dataset named DIPS-Plus [42], large enough and with enough features to develop computational models that can reliably predict the residues that will form the interface between two given proteins. In addition, accompanying DIPS-Plus, we provide a set of benchmarks in Table I describing the contribution DIPS-Plus makes to existing SOTA methods for PIP (i.e., NeiA and NeiWA). We observe that we can increase the latest SOTA performance for PIP by training NeiA and NeiWA on DIPS-Plus and testing them on a standardized test dataset for PIP methods, DB5-Plus [42].
TABLE I.
DIPS-Plus’ effect on the MedAUROC of SOTA algorithms for protein interface prediction.1
| # GNN Layers | ||||
|---|---|---|---|---|
| Method | 1 | 2 | 3 | 4 |
| NGF [14] | 0.865 (0.007) | 0.871 (0.013) | 0.873 (0.017) | 0.869 (0.017) |
| DTNN [53] | 0.867 (0.007) | 0.880 (0.007) | 0.882 (0.008) | 0.873 (0.012) |
| Node and Edge Average [20] | 0.876 (0.005) | 0.898 (0.005) | 0.895 (0.006) | 0.889 (0.007) |
| NeiA+HOPI [41] | 0.902 (0.012) | 0.919 (0.015) | 0.921 (0.009) | 0.915 (0.009) |
| NeiWA+HOPI [41] | 0.908 (0.019) | 0.930 (0.016) | 0.924 (0.011) | 0.914 (0.013) |
| NeiA+HOPI+DIPS-Plus [42] | 0.9426 (0.001) | 0.9438 (0.001) | 0.9432 (0.005) | 0.9426 (0.007) |
| NeiWA+HOPI+DIPS-Plus [42] | 0.9065 (0.029) | 0.9342 (0.01) | 0.9335 (0.006) | 0.9384 (0.004) |
MedAUROC: median area under the receiver operating characteristic curve. SOTA: state-of-the-art.
We trained both NeiA and NeiWA for 10 epochs on our standardized 80%-20% cross-validation split of DIPS-Plus’ complexes to observe both models’ behavior on DB5-Plus’s test complexes thereafter. We ran each of our experiments ten times, with each experiment using a random seed and between one and four GNN layers of NeiA or NeiWA, respectively. The results for each experiment’s entry in Table I were derived by computing the sample mean and standard deviation (in parentheses) of the ten runs’ median area under the receiver operating characteristic curve (MedAUROC) scores for the experiment. For the experiments, we used the following architecture and hyperparameters: (1) 1-4 NeiA/NeiWA GNN layers; (2) 3 residual CNN blocks, each employing a 2D convolution module, ReLU activation function, another 2D convolution module, followed by adding the block input’s identity map back to the output of the block; (3) an intermediate channel dimensionality of 212 for each residual CNN block; (4) a learning rate of 1e-5; (5) a batch size of 32; (6) a weight decay of 1e-7; and (7) a dropout (forget) probability of 0.3. DIPS-Plus’ curation required the use of 16 data processing nodes on Andes running in parallel for multiple sequence alignment (MSA) generation using HH-suite3 [61]. Moreover, running several grid search experiments over DIPS-Plus for deep learning benchmarks in a short time span is only possible with the use of large GPU nodes such as those available on Summit. For our benchmark experiments, we trained a total of 80 separate deep learning models, each using a GPU on Summit. We used Python 3.8, PyTorch 1.7.1, and PyTorch Lightning 1.3.8 to run our deep learning benchmarks. PyTorch Lightning was used to facilitate model checkpointing, metric reporting, and, in some earlier experiments, data parallelism across six GPUs.
H. Training models for prediction of homodimer interactions on Summit
We developed a data-parallel method to train DL models on Summit to find protein homodimer interactions. This model was trained utilizing a node with high memory V100 GPUs (32 GB). Because of the large number of trainable parameters, one high memory GPU could fit only a batch size of 2. To increase training speed, we used distributed deep learning based on PyTorch using the Horovod [55]. The effective batch size using the distributed approach becomes 12. The feature size of 600 × 600 × 592 is memory intensive. To help reduce latency, the 42 CPU cores and 2TB RAM on Summit’s high-memory nodes were also used by the data loader to load the data into the memory, enabling overlap of data transfer and compute and reducing total time to solution. This substantially sped up the training process, with each training epoch taking on average 1 hour and 40 minutes versus approximately 9 hours using only one GPU and no data loader optimizations. The size of the training data for this project was 5,975 protein monomers, and the whole training process was executed with only 300 node hours which is approximately 5.5 times faster than with only 1 GPU and no optimizations for data loading.
IV. Example pipeline for genome-scale functional annotation
There are numerous ways to use the methods and tools in our HPC toolbox for genome annotation studies. Currently we are developing a pipeline that uses a consensus between several methods to arrive at a prediction of protein function. The functions of known proteins found by SAdLSA alignment to crystallographic and sequence-based protein databases such as Pfam, and by three-dimensional geometric alignment between predicted structures, ranked by QA and EMA methods, and those found on non-redundant structural databases are compared for similarities. Regions of the sequences with the highest numbers of conserved residues are tabulated. Predicted structures are stored in databases for in-depth analysis and for structure-based modeling and simulation. Protein interface prediction is currently being incorporated into the pipeline, as are various analyses of the predicted structures, for which protein domain boundary prediction will be important. In the future, cryoEM density refinement in addition to structure and PPI prediction modules can discover information about large complexes. Figure 6 shows possible pipelines that can be deployed for genome-scale functional annotation using our structure-based HPC toolbox. Currently we are focused on genome-scale prediction for several microbial organisms, and plant species. Our hope is that through a combination of genome-scale computational efforts and experimental methods, we can not only characterize and annotate the functions of unknown proteins in the proteomes of these organisms, which can help to engineer new strains that can be more resilient, or in the development of new energy solutions, but also to discover new information about proteins whose function has been partially characterized, through the collection of predicted functions, structures, and complex modeling.
Fig. 6.
Simplified scheme for annotation pipeline with consensus using our toolbox.
V. Conclusions and future work
We have presented our new HPC toolbox for protein functional annotation using structure-based deep-learning methods. We have shown both the deployment of inference at large-scale using the SAdLSA DL-based alignment method, and the development of distributed training methods that utilized multiple GPUs and Summit nodes, and will next be scaled up further accommodate even larger training datasets. We also report the re-organization and deployment of the AlphaFold structure prediction program both on Summit using Singularity containers and with a prototype small genome-scale test case on the PACE resources. Our toolbox, which contains multiple methods useful for structure-based functional annotation, will be used in pipelines to generate such annotations for large sets of proteins with unknown function or low-confidence annotations, or even to assist in validation of proteins of known function and the prediction of their structural properties to provide more detailed information about catalytic mechanisms and metabolic pathways that these proteins may be involved in. In future work, we hope to build on top of our toolbox to support new emerging tasks in bioinformatics, including large-scale prediction of protein tertiary and quaternary structures, and the development of new pipelines using the various tools to provide high-confidence hypotheses for informing and guiding bench-top experiments.
Acknowledgment
This research was partly sponsored by Office of Biological and Environmental Research’s Genomic Science program within the US Department of Energy Office of Science, under award number ERKP917, the Laboratory Directed Research and Development Program at Oak Ridge National Laboratory (ORNL), and used resources of the Oak Ridge Leadership Computing Facility, which is a DOE Office of Science User Facility supported under Contract DE-AC05-00OR22725, granted in part by the Advanced Scientific Computing Research (ASCR) Leadership Computing Challenge (ALCC) program. The development of the deep learning tools for protein domain prediction, protein model quality assessment, protein interaction prediction, and cryo-EM data analysis was supported by the National Science Foundation (DBI1759934 and IIS1763246), National Institutes of Health (R01GM093123), Department of Energy, USA (DE-AR0001213, DE-SC0020400 and DE-SC0021303), and the Thompson Missouri Distinguished Professorship. The development of SAdLSA was supported in part by the National Institute Health (NIH R35GM118039) and used resources supported by the Partnership for an Advanced Computing Environment (PACE) at Georgia Tech.
Footnotes
References
- [1].Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, and Lipman DJ. Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Research, 25(17):3389–3402, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Baines Mandeep, Bhosale Shruti, Caggiano Vittorio, Goyal Naman, Goyal Siddharth, Ott Myle, Lefaudeux Benjamin, Liptchinsky Vitaliy, Rabbat Mike, Sheiffer Sam, Sridhar Anjali, and Xu Min. Fairscale: A general purpose modular pytorch library for high performance and large scale training. [Google Scholar]
- [3].Biewald Lukas. Experiment tracking with weights and biases, 2020. Software available from wandb.com. [Google Scholar]
- [4].Brown Tom B, Mann Benjamin, Ryder Nick, Subbiah Melanie, Kaplan Jared, Dhariwal Prafulla, Neelakantan Arvind, Shyam Pranav, Sastry Girish, Askell Amanda, et al. Language models are few-shot learners. arXiv preprint arXiv:2005.14165, 2020. [Google Scholar]
- [5].Chen Chen, Chen Xiao, Wu Tianqi, Alex Morehead, and Cheng Jianlin. Improved protein structure accuracy estimation with graph-based equivariant networks. In preparation, 2021. [Google Scholar]
- [6].Chen Chen, Wu Tianqi, Guo Zhiye, and Cheng Jianlin. Combination of deep neural network with attention mechanism enhances the explain-ability of protein contact prediction. Proteins: Structure, Function, and Bioinformatics, 89(6):697–707, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen Xiao, Akhter Nasrin, Guo Zhiye, Wu Tianqi, Hou Jie, Shehu Amarda, and Cheng Jianlin. Deep ranking in template-free protein structure prediction. In Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, pages 1–10, 2020. [Google Scholar]
- [8].Chen Xiao and Cheng Jianlin. Distema: distance map-based estimation of single protein model accuracy with attentive 2d convolutional neural network. bioRxiv, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen Xiao, Liu Jian, Guo Zhiye, Wu Tianqi, Hou Jie, and Cheng Jianlin. Protein model accuracy estimation empowered by deep learning and inter-residue distance prediction in casp14. Scientific Reports, 11(1):1–12, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].The UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Research, 47(D1):D506–D515, 11 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cressey Daniel and Callaway Ewen. Cryo-electron microscopy wins chemistry nobel. Nature News, 550(7675): 167, 2017. [DOI] [PubMed] [Google Scholar]
- [12].Dai Zihang, Liu Hanxiao, Le Quoc V, and Tan Mingxing. Coatnet: Marrying convolution and attention for all data sizes. arXiv preprint arXiv:2106.04803, 2021. [Google Scholar]
- [13].Dosovitskiy Alexey, Beyer Lucas, Kolesnikov Alexander, Weissenborn Dirk, Zhai Xiaohua, Unterthiner Thomas, Dehghani Mostafa, Minderer Matthias, Heigold Georg, Gelly Sylvain, et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929, 2020. [Google Scholar]
- [14].Duvenaud David, Maclaurin Dougal, Aguilera-Iparraguirre Jorge, Gómez-Bombarelli Rafael, Hirzel Timothy, Aspuru-Guzik Alán, and Adams Ryan P. Convolutional networks on graphs for learning molecular fingerprints. arXiv preprint arXiv:1509.09292, 2015. [Google Scholar]
- [15].Eddy Sean R.. Accelerated profile hmm searches. PLOS Computational Biology, 7(10):e1002195, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Eickholt Jesse, Deng Xin, and Cheng Jianlin. Dobo: Protein domain boundary prediction by integrating evolutionary signals and machine learning. BMC bioinformatics, 12(1):1–8, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].et al. Falcon WA. Pytorch lightning. GitHub. Note: https://github.com/PyTorchLightning/pytorch-lightning, 3, 2019. [Google Scholar]
- [18].Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, and Bateman A. The pfam protein families database. Nucleic Acids Research, 36:D281–D288, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Folkers GE, van Buuren BNM, and Kaptein R. Expression screening, protein purification and nmr analysis of human protein domains for structural genomics. Journal of structural and functional genomics, 5(1):119–131, 2004. [DOI] [PubMed] [Google Scholar]
- [20].Fout Alex, Byrd Jonathon, Shariat Basir, and Ben-Hur Asa. Protein interface prediction using graph convolutional networks. In Guyon I, Luxburg UV, Bengio S, Wallach H, Fergus R, Vishwanathan S, and Garnett R, editors, Advances in Neural Information Processing Systems, volume 30, pages 6530–6539. Curran Associates, Inc., 2017. [Google Scholar]
- [21].Gantikow Holger, Walter Steffen, and Reich Christoph. Rootless containers with podman for hpc. In International Conference on High Performance Computing, pages 343–354. Springer, 2020. [Google Scholar]
- [22].Gao M and Skolnick J. Apoc: large-scale identification of similar protein pockets. Bioinformatics, 29(5):597–604, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gao M, Zhou H, and Skolnick J. Destini: A deep-learning approach to contact-driven protein structure prediction. Sci Rep, 9(1):3514, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gao Mu and Skolnick Jeffrey. iAlign: a method for the structural comparison of protein–protein interfaces. Bioinformatics, 26(181:2259–2265, 07 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gao Mu and Skolnick Jeffrey. A novel sequence alignment algorithm based on deep learning of the protein folding code. Bioinformatics, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gao Mu and Skolnick Jeffrey. A general framework to learn tertiary structure for protein sequence characterization. Frontiers in Bioinformatics, 1(14), 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Guo Zhiye, Wu Tianqi, Liu Jian, Hou Jie, and Cheng Jianlin. Improving deep learning-based protein distance prediction in casp14. bioRxiv, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].He Kaiming, Zhang Xiangyu, Ren Shaoqing, and Sun Jian. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 770–778, 2016. [Google Scholar]
- [29].Heidelberg John F, Seshadri Rekha, Haveman Shelley A, Hemme Christopher L, Paulsen Ian T, Kolonay James F, Eisen Jonathan A, Ward Naomi, Methe Barbara, Brinkac Lauren M, et al. The genome sequence of the anaerobic, sulfate-reducing bacterium desulfovibrio vulgaris hilden-borough. Nature biotechnology, 22(5):554–559, 2004. [DOI] [PubMed] [Google Scholar]
- [30].Hildebrand A, Remmert M, Biegert A, and Söding J. Fast and accurate automatic structure prediction with hhpred. Proteins, 77(Suppl 9): 128–32, 2009. [DOI] [PubMed] [Google Scholar]
- [31].Hiranuma Naozumi, Park Hahnbeom, Baek Minkyung, Anishchenko Ivan, Dauparas Justas, and Baker David. Improved protein structure refinement guided by deep learning based accuracy estimation. Nature communications, 12(1): 1–11, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Holm L and Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol, 233(1):123–38, 1993. [DOI] [PubMed] [Google Scholar]
- [33].Hong Seung Hwan, Joo Keehyoung, and Lee Jooyoung. Condo: protein domain boundary prediction using coevolutionary information. Bioinformatics, 35(14):2411–2417, 2019. [DOI] [PubMed] [Google Scholar]
- [34].Hou Jie, Wu Tianqi, Cao Renzhi, and Cheng Jianlin. Protein tertiary structure modeling driven by deep learning and contact distance prediction in casp13. Proteins: Structure, Function, and Bioinformatics, 87(12): 1165–1178, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jiang Yuexu, Wang Duolin, and Xu Dong. Deepdom: Predicting protein domain boundary from sequence alone using stacked bidirectional lstm. In BIOCOMPUTING 2019: Proceedings of the Pacific Symposium, pages 66–75. World Scientific, 2018. [PMC free article] [PubMed] [Google Scholar]
- [36].Jumper John, Evans Richard, Pritzel Alexander, Green Tim, Figurnov Michael, Ronneberger Olaf, Tunyasuvunakool Kathryn, Bates Russ, Žídek Augustin, Potapenko Anna, et al. Highly accurate protein structure prediction with alphafold. Nature, page 1, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kabsch Wolfgang. A discussion of the solution for the best rotation to relate two sets of vectors. Acta Crystallographica Section A: Crystal Physics, Diffraction, Theoretical and General Crystallography, 34(5):827–828, 1978. [Google Scholar]
- [38].Kwon Sohee, Won Jonghun, Kryshtafovych Andriy, and Seok Chaok. Assessment of protein model structure accuracy estimation in casp14: Old and new challenges. Proteins: Structure, Function, and Bioinformatics, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lawson Catherine L, Kryshtafovych Andriy, Adams Paul D, Afonine Pavel V, Baker Matthew L, Barad Benjamin A, Bond Paul, Burnley Tom, Cao Renzhi, Cheng Jianlin, et al. Cryo-em model validation recommendations based on outcomes of the 2019 emdataresource challenge. Nature methods, 18(2):156–164, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].LeGrand Scott, Scheinberg Aaron, Tillack Andreas F, Thavappiragasam Mathialakan, Vermaas Josh V, Agarwal Rupesh, Larkin Jeff, Poole Duncan, Santos-Martins Diogo, Solis-Vasquez Leonardo, Koch Andreas, Forli Stefano, Hernandez Oscar, Smith Jeremy C., and Sedova Ada. GPU-accelerated drug discovery with docking on the summit supercomputer: porting, optimization, and application to covid-19 research. In Proceedings of the 11th ACM international conference on bioinformatics, computational biology and health informatics, pages 1–10, 2020. [Google Scholar]
- [41].Liu Yi, Yuan Hao, Cai Lei, and Ji Shuiwang. Deep learning of high-order interactions for protein interface prediction. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, pages 679–687, 2020. [Google Scholar]
- [42].Morehead Alex, Chen Chen, Sedova Ada, and Cheng Jianlin. Dips-plus: The enhanced database of interacting protein structures for interface prediction, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Muller A, MacCallum RM, and Sternberg MJE. Benchmarking psiblast in genome annotation. Journal of Molecular Biology, 293(5):1257–1271, 1999. [DOI] [PubMed] [Google Scholar]
- [44].Murakami Yoichi, Tripathi Lokesh P, Prathipati Philip, and Mizuguchi Kenji. Network analysis and in silico prediction of protein–protein interactions with applications in drug discovery. Current opinion in structural biology, 44:134–142, 2017. [DOI] [PubMed] [Google Scholar]
- [45].Paszke Adam, Gross Sam, Massa Francisco, Lerer Adam, Bradbury James, Chanan Gregory, Killeen Trevor, Lin Zeming, Gimelshein Natalia, Antiga Luca, Desmaison Alban, Kopf Andreas, Yang Edward, DeVito Zachary, Raison Martin, Tejani Alykhan, Chilamkurthy Sasank, Steiner Benoit, Fang Lu, Bai Junjie, and Chintala Soumith. Pytorch: An imperative style, high-performance deep learning library. In Wallach H, Larochelle H, A. Beygelzimer, d'Alché-Buc F, Fox E, and Garnett R, editors, Advances in Neural Information Processing Systems 32, pages 8024–8035. Curran Associates, Inc., 2019. [Google Scholar]
- [46].Quadir Farhan, Roy Raj S, Halfmann Randal, and Cheng Jianlin. Dncon2_inter: predicting interchain contacts for homodimeric and homomultimeric protein complexes using multiple sequence alignments of monomers and deep learning. Scientific reports, 11(1):1–10, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Quadir Farhan, Roy Raj S., Soltanikazemi Elham, and Cheng Jianlin. Deepcomplex: A web server of predicting protein complex structures by deep learning inter-chain contact prediction and distance-based modelling. Frontiers in Molecular Biosciences, 8:827, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Radford Alec, Wu Jeffrey, Child Rewon, Euan David, Amodei Dario, Sutskever Ilya, et al. Language models are unsupervised multitask learners. OpenAI blog, 1(8):9, 2019. [Google Scholar]
- [49].Ramsundar Bharath, Eastman Peter, Walters Patrick, and Pande Vijay. Deep learning for the life sciences: applying deep learning to genomics, microscopy, drug discovery, and more. ” O’Reilly Media, Inc.”, 2019. [Google Scholar]
- [50].Remmert M, Biegert A, Hauser A, and Soding J. Hhblits: lightning-fast iterative protein sequence searching by hmm-hmm alignment. Nat Methods, 9(2):173–5, 2011. [DOI] [PubMed] [Google Scholar]
- [51].Rentzsch Robert and Orengo Christine A. Protein function prediction using domain families. In BMC bioinformatics, volume 14, pages 1–14. BioMed Central, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Rost B. Twilight zone of protein sequence alignments. Protein Eng, 12(2): 85–94, 1999. [DOI] [PubMed] [Google Scholar]
- [53].Schütt Kristof T, Arbabzadah Farhad, Chmiela Stefan, Müller Klaus R, and Tkatchenko Alexandre. Quantum-chemical insights from deep tensor neural networks. Nature communications, 8(1):1–8, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Senior Andrew W., Evans Richard, Jumper John, Kirkpatrick James, Sifre Faurent, Green Tim, Qin Chongli, Žídek Augustin, Nelson Alexander W. R., Bridgland Alex, Penedones Hugo, Petersen Stig, Simonyan Karen, Crossan Steve, Kohli Pushmeet, Jones David T., Silver David, Kavukcuoglu Koray, and Hassabis Demis. Improved protein structure prediction using potentials from deep learning. Nature, 577(7792):706–710, 2020. [DOI] [PubMed] [Google Scholar]
- [55].Sergeev Alexander and Del Balso Mike. Horovod: fast and easy distributed deep learning in tensorflow. arXiv preprint arXiv:1802.05799, 2018. [Google Scholar]
- [56].Si Dong, Moritz Spencer A, Pfab Jonas, Hou Jie, Cao Renzhi, Wang Figuo, Wu Tianqi, and Cheng Jianlin. Deep learning to predict protein backbone structure from high-resolution cryo-em density maps. Scientific reports, 10(1): 1–22, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sillitoe Ian, Fewis Tony E., Cuff Alison, Das Sayoni, Ashford Paul, Dawson Natalie F., Furnham Nicholas, Faskowski Roman A., Fee David, Fees Jonathan G., Fehtinen Sonja, Studer Romain A., Thornton Janet, and Orengo Christine A.. CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Research, 43(D1):D376–D381, 10 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Skolnick Jeffrey and Gao Mu. The role of local versus nonlocal physicochemical restraints in determining protein native structure. Current Opinion in Structural Biology, 68:1–8, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Smith TF and Waterman MS. Identification of common molecular subsequences. J Mol Biol, 147(1):195–7, 1981. [DOI] [PubMed] [Google Scholar]
- [60].Soding J. Protein homology detection by hmm-hmm comparison. Bioinformatics, 21(7):951–60, 2005. [DOI] [PubMed] [Google Scholar]
- [61].Steinegger Martin, Meier Markus, Mirdita Milot, Vöhringer Harald, Haunsberger Stephan J, and Söding Johannes. Hh-suite3 for fast remote homology detection and deep protein annotation. BMC bioinformatics, 20(1):1–15, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Stephens Zachary D, Fee Skylar Y, Faghri Faraz, Campbell Roy H, Zhai Chengxiang, Efron Miles J, Iyer Ravishankar, Schatz Michael C, Sinha Saurabh, and Robinson Gene E. Big data: astronomical or genomical? PLoS biology, 13(7):e1002195, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Tan Kar-Han and Fim Boon Pang. The artificial intelligence renaissance: deep learning and the road to human-level machine intelligence. APSIPA Transactions on Signal and Information Processing, 7, 2018. [Google Scholar]
- [64].Tatusova Tatiana, DiCuccio Michael, Badretdin Azat, Chetvernin Vyacheslav, Nawrocki Eric P., Zaslavsky Feonid, Fomsadze Alexandre, Pruitt Kim D., Borodovsky Mark, and Ostell James. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Research, 44(14):6614–6624, 06 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Tunyasuvunakool Kathryn, Adler Jonas, Wu Zachary, Green Tim, Zielinski Michal, Žídek Augustin, Bridgland Alex, Cowie Andrew, Meyer Clemens, Faydon Agata, Velankar Sameer, Kleywegt Gerard J., Bateman Alex, Evans Richard, Pritzel Alexander, Figurnov Michael, Ronneberger Olaf, Bates Russ, Kohl Simon A. A., Potapenko Anna, Ballard Andrew J., Romera-Paredes Bernardino, Nikolov Stanislav, Jain Rishub, Clancy Ellen, Reiman David, Petersen Stig, Senior Andrew W., Kavukcuoglu Koray, Birney Ewan, Kohli Pushmeet, Jumper John, and Hassabis Demis. Highly accurate protein structure prediction for the human proteome. Nature, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Uguzzoni Guido, Fovis Shalini John, Oteri Francesco, Schug Alexander, Szurmant Hendrik, and Weigt Martin. Large-scale identification of coevolution signals across homo-oligomeric protein interfaces by direct coupling analysis. Proceedings of the National Academy of Sciences, 114(13):E2662–E2671, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Vermaas Josh Vincent, Sedova Ada, Baker Matthew B, Boehm Swen, Rogers David M, Farkin Jeff, Glaser Jens, Smith Micholas D, Hernandez Oscar, and Smith Jeremy C. Supercomputing pipelines search for therapeutics against covid-19. Computing in Science & Engineering, 23(1):7–16, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Wang Sheng, Sun Siqi, Li Zhen, Zhang Renyu, and Xu Jinbo. Accurate de novo prediction of protein contact map by ultra-deep learning model. PLOS Comput Biol., 13(1):e1005324, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wells James A and McClendon Christopher F. Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature, 450(7172):1001–1009, 2007. [DOI] [PubMed] [Google Scholar]
- [70].Wu Haiping, Xiao Bin, Codella Noel, Liu Mengchen, Dai Xiyang, Yuan Lu, and Zhang Lei. Cvt: Introducing convolutions to vision transformers. arXiv preprint arXiv:2103.15808, 2021. [Google Scholar]
- [71].Wu Tianqi, Guo Zhiye, Hou Jie, and Cheng Jianlin. Deepdist: real-value inter-residue distance prediction with deep residual convolutional network. BMC bioinformatics, 22(1):1–17, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Yu Fisher, Koltun Vladlen, and Funkhouser Thomas. Dilated residual networks, 2017. [Google Scholar]
- [73].Zhang Y and Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins-Structure Function and Bioinformatics, 57(4):702–710, 2004. [DOI] [PubMed] [Google Scholar]
- [74].Zhang Yang. Progress and challenges in protein structure prediction. Current Opinion in Structural Biology, 18(3):342–348, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Zhang Yang and Skolnick Jeffrey. Tm-align: a protein structure alignment algorithm based on the tm-score. Nucleic acids research, 33(7):2302–2309, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zhao Bihai, Hu Sai, Li Xueyong, Zhang Fan, Tian Qinglong, and Ni Wenyin. An efficient method for protein function annotation based on multilayer protein networks. Human genomics, 10(1):1–15, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]