Abstract
Background
N6-methyladenosine (m6A) mRNA modification is essential for mammalian and plant viability. The U6 m6A methyltransferases in other species regulate S-adenosylmethionine (SAM) homeostasis through installing m6A in pre-mRNAs of SAM synthetases. However, U6 m6A methyltransferase has not been characterized in Arabidopsis and little is known about its role in regulating photomorphogenesis and flowering.
Results
Here we characterize that FIONA1 is an Arabidopsis U6 m6A methyltransferase that installs m6A in U6 snRNA and a small subset of poly(A)+ RNA. Disruption of FIONA1 leads to phytochrome signaling-dependent hypocotyl elongation and photoperiod-independent early flowering. Distinct from mammalian METTL16 and worm METT-10, FIONA1 neither installs m6A in the mRNAs of Arabidopsis SAM synthetases nor affects their transcript expression levels under normal or high SAM conditions. We confirm that FIONA1 can methylate plant mRNA m6A motifs in vitro and in vivo. We further show that FIONA1 installs m6A in several phenotypic related transcripts, thereby affecting downstream mRNA stability and regulating phytochrome signaling and floral transition.
Conclusion
FIONA1 is functional as a U6 m6A methyltransferase in Arabidopsis, distinct from mammalian METTL16 and worm METT-10. Our results demonstrate that FIONA1-mediated m6A post-transcriptional regulation is an autonomous regulator for flowering and phytochrome signaling-dependent photomorphogenesis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13059-022-02612-2.
Keywords: m6A, RNA modification, Methyltransferase, mRNA stability, Photomorphogenesis, Early flowering, Arabidopsis
Background
Light is one of the most important external signals affecting plant physiology and developmental processes. Upon exposure to light, the critical plant developmental processes, like floral transition and photomorphogenesis, are allowed to proceed. During photomorphogenesis, light inhibits hypocotyl growth, promotes cotyledon opening, and activates the expression of light-regulated genes [1, 2]. Plants have evolved an array of photoreceptors: red and far-red light-absorbing phytochromes (phyA and phyB), blue/ultraviolet (UV)-A light-absorbing cryptochromes (CRY1 and CRY2), and UV-B sensing photoreceptor (UVR8), which transduce light signals to modulate the transcriptome and to trigger photomorphogenic growth and development [3–5]. However, little is known about whether RNA modifications regulate photomorphogenesis.
The most abundant internal mRNA modification N6-methyladenosine (m6A) is reversible and vital in many biological processes through post-transcriptional regulation of mRNA processing and metabolism [6–17]. In both mammals and plants, the m6A modification is written by methyltransferase, erased by demethylase, and read by m6A-binding proteins [18–21]. Similar to mammals, several Arabidopsis YTH family proteins (ECT2-4 and CPSF30-L) have been characterized as m6A binding protein that regulate trichome morphology, leaf growth, nitrate signaling, flowering, and abscisic acid (ABA) response [22–27]; two Arabidopsis (ALKBH9B [28] and ALKBH10B [29]) and one tomato AlkB family proteins (SIALKBH2) [30] have been identified as m6A demethylases that regulate viral response, flowering time, and fruit ripening, respectively. Mammals contain two types of mRNA m6A methyltransferase: multi-protein complex and methyltransferase like 16 (METTL16) [31, 32]. Multiple subunits of m6A methyltransferase complex (comprising METTL3, METTL14, and WTAP three key subunits and others) have been identified in both mammal and plants and are responsible for majority mRNA m6A installation [31, 33–39]. Knockout key subunit of m6A methyltransferase complex displays embryo lethal in both mammal and plants [36, 40]. Plant WTAP homolog (Arabidopsis FIP37 and rice OsFIP) affects Arabidopsis shoot stem cell fate and early degeneration of rice microspores [38, 41].
Mammalian METTL16 and worm (Caenorhabditis elegans) METT-10 were found to install m6A on the “UACm6AGAGAA” motif of U6 snRNA [32, 42]. The U6 snRNA is an essential component of the spliceosome, contributing to splicing of nuclear pre-mRNAs and serving as ribozyme catalysts of two consecutive transesterifications to ligate two exons concomitant with removal of an intron [43]. They both also methylate pre-mRNAs of SAM synthetases to affect SAM homeostasis with distinct molecular features. Mammalian METTL16 installs m6A on the UACm6AGAGAA motifs at 3′ UTR of SAM synthetases MAT2A pre-mRNA, which leads to intron retention/decay of the RNA and affects downstream SAM homeostasis [32, 44]. The regulation function of METTL16 in SAM homeostasis is essential for mouse early embryonic development [45]. Although depletion of human METTL16 affects plenty of m6A sites in mRNAs [32], recent finding revealed that human METTL16 only methylates U6 m6A motif UACm6AGAGAA in mRNAs and the increased m6A methylation sites lacking U6 m6A motif in METTL16 deficiency were mediated by the reduced intercellular SAM [46]. Caenorhabditis elegans (C. elegans) METT-10 methylates a variant motif UACm6AGAAAC on the 3′ splice site (AG) of SAM synthetase (SAMS) pre-mRNA. The m6A on the 3′ splice site (AG) hinders the binding of splicing factor U2AF for splicing; therefore, the unspliced SAMS pre-mRNA is subjected to nuclear degradation [42]. Worms use METT-10’s function in SAM homeostasis to respond to change in worm diet [42]. The UACm6AGAGAA and UACm6AGAAAC motifs are only conserved in vertebrate and invertebrate SAM synthetase transcripts, respectively [32, 42]. Although sequence alignment suggests that Arabidopsis FIONA1 protein is the homolog of human METTL16 [32], it has not been validated and characterized. It is unknown that whether plant METTL16 would install m6A on SAM synthetase to affect SAM homeostasis and whether plant METTL16 relies on the U6 m6A motif or a variant U6 m6A motif to install m6A in poly (A)+ RNA.
FIONA1 was identified as a genetic regulator of period length in Arabidopsis circadian clock. The FIONA1 mutant (fio1-1) leads to early flowering and longer hypocotyl in a photoperiod-dependent manner, and lengthens the free-running circadian period of leaf movement [47]. Although FIONA1 was reported to affect the period length of four central oscillator genes’ transcript expression (CCA1, LHY, TOC1, and LUX) and the mRNA expression of key flowering regulatory genes CONSTANS (CO) and FLOWERING LUCUS T (FT) [47], the molecular mechanism has not been fully elucidated. Little is known about whether and how the methylation activity of FIONA1 regulates the phenotypes in the fiona1 mutant.
Here we characterized FIONA1 is an Arabidopsis U6 m6A methyltransferase that installs m6A in U6 snRNA and a small subset of poly(A)+ RNA. We asked whether the methylation activity of FIONA1 is required for the reported phenotypes of photoperiod-dependent hypocotyl growth and flowering. Our two homozygous mutant lines fiona1-1 and fiona1-2 generated by CRISPR-cas9 displayed hypocotyl cell elongation selectively under continuous red (Rc) and far-red light (FRc) conditions and photoperiod-independent early flowering. Complementation experiments in fiona1-1 mutant show that the methylation activity of FIONA1 is required for phytochrome signaling-dependent photomorphogenesis and photoperiod-independent flowering time control. We found that the function and methylation activity features of FIONA1 are distinct from mammalian METTL16 and worm METT-10. FIONA1 can methylate plant m6A motifs in vitro and in vivo but does not install m6A on SAM synthetase transcripts. Then, we asked how the methylation activity of FIONA1 regulates the observed phenotypes. Our results demonstrate that FIONA1-mediated m6A post-transcriptional regulation is an autonomous regulator for floral transition and phytochrome signaling-dependent photomorphogenesis.
Results
FIONA1 installs m6A modification in U6 snRNA and a small subset of poly(A)+ RNA in Arabidopsis
To characterize Arabidopsis U6 m6A methyltransferase, we processed phylogenetic analysis and sequence alignments and found a single, putative homolog of human METTL16 in Arabidopsis, FIONA1 (AT2G21070) (Additional file 1: Fig. S1), which prompted us to test whether FIONA1 deposits m6A modification in plants. We generated fiona1 mutants through CRISPR/Cas9 genome editing system that carried two guiding RNA targeting the coding region of FIONA1 [48] (Additional file 1: Fig. S2a). Two mutant lines fiona1-1 and fiona1-2 were confirmed as homozygous InDel mutants by Sanger sequencing (Additional file 1: Fig. S2b).
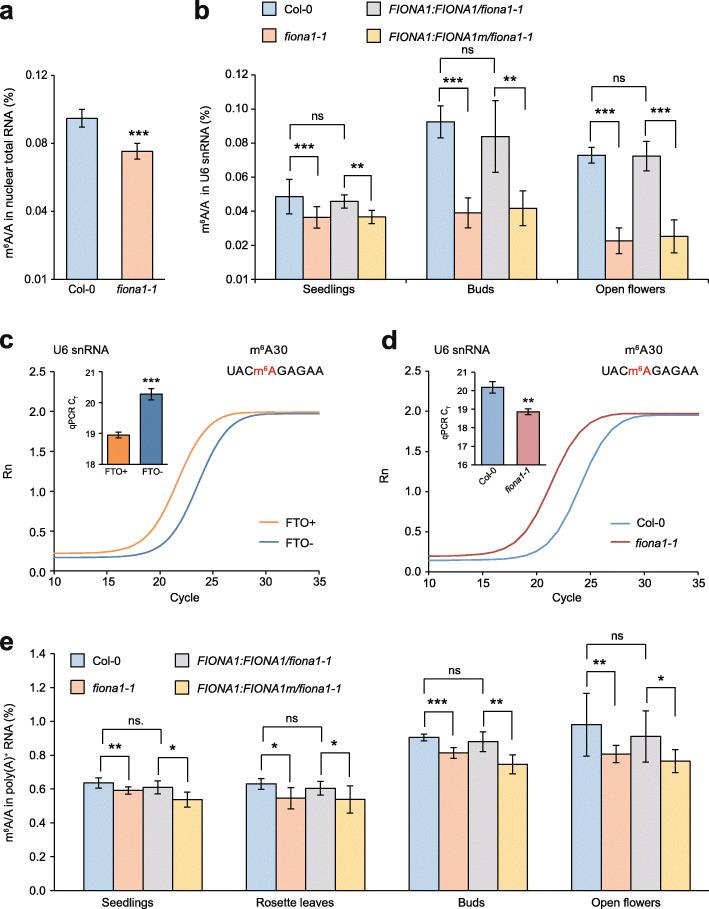
To identify the physiological substrates of FIONA1, we firstly separated the nuclear and cytoplasmic fractions of 12-day-old Col-0 and fiona1-1 seedlings (Additional file 1: Fig. S3a) and isolated their nuclear total RNA for m6A quantification. LC-MS/MS results showed that the m6A level (the ratio of m6A/A) in nuclear total RNA was decreased in fiona1-1 mutant compared with WT (Fig. 1a). As known that mammalian METTL16 is an m6A writer of U6 snRNA [32], we therefore tested whether U6 snRNA is a physiological substrate of FIONA1 in plants. We isolated U6 snRNA from different tissues including 12-day-old seedlings, buds, and flowers (Additional file 1: Fig. S3b) and found that knockout of FIONA1 significantly reduced m6A level in U6 snRNA isolated from these tissues (Fig. 1b). Note that the crystal structure of human METTL16 identified NPPF (residues 184–187) as the catalytic motif for binding substrate adenine base [49] and further biochemical enzymatic assays confirmed METTL16 variants with P185A/P186A or F187G mutations abolish the methyltransferase activity [32, 45]. Based on the sequence alignments among multiple species (Additional file 1: Fig. S1b), we designed an inactive FIONA1 P237A/F239G with mutations of putative catalytic ligand NPPF and named as FIONA1m. To further confirm that U6 snRNA is the physiological substrate of FIONA1, we generated two transgenic complementation lines (FIONA1:FIONA1/fiona1-1 and FIONA1:FIONA1m/fiona1-1) by respectively expressing wild-type FIONA1 and the catalytically inactive mutant FIONA1m in fiona1-1 mutant plants with the native FIONA1 promoter (Additional file 1: Fig. S4). The LC-MS/MS results showed that expression of wild-type FIONA1, but not the catalytically inactive mutant FIONA1m in fiona1-1 mutant, can recover the m6A levels in U6 snRNA (Fig. 1b). These results confirmed that U6 snRNA is FIONA1’s substrate and the NPPF ligands in FIONA1 indeed are the catalytic binding motif for adenine base.
Fig. 1.
FIONA1 installs m6A modification on U6 snRNA and poly(A)+ RNA. a LC-MS/MS quantification of the m6A/A ratio in nuclear total RNA in 12-day-old Col-0 and fiona1-1 seedlings. b LC-MS/MS quantification of the m6A/A ratios of U6 snRNA in the indicated genotypic plant tissues. c-d Real-time fluorescence amplification curves and bar plot of the threshold cycle (CT) of qPCR showing SELECT results for detecting m6A30 site in U6 snRNA with and without FTO demethylation treatment (c) and in Col-0 and fiona1-1 seedlings (d). Rn is the raw fluorescence for the associated well normalized to the fluorescence of the passive reference dye (ROX). e LC-MS/MS quantification of the m6A/A ratios of poly(A)+ RNA in the indicated genotypic plant tissues. Data are means ± SD for 3 biological replicates × 2 technical replicates. * p < 0.05, ** p < 0.01, *** p < 0.001 by t test (two-tailed). ns, no significance
The m6A modification was reported to locate at A43 position of human U6 snRNA with a hairpin structured conserve sequence UACm6AGAGAA [50], but the m6A position in plant U6 has not been mapped. We next conducted our developed SELECT method to map m6A position in Arabidopsis U6 snRNA. SELECT is an elongation- and ligation-based qPCR quantification method to detect m6A locus and fraction in single mRNA/lncRNA at single-base resolution [51]. We run SELECT in two parallel total RNA samples with or without m6A demethylase FTO treatments where more than 95% of m6A modifications in total RNA were removed by FTO (Additional file 1: Fig. S5a). The FTO-assisted SELECT results showed that m6A located at A30 position in Arabidopsis U6 snRNA with the same consensus sequence as human U6 (Fig. 1c; Additional file 1: Fig. S5b). We further performed SELECT in total RNA isolated from 12-day-old Col-0 and fiona1-1 seedlings and confirmed that FIONA1 is an Arabidopsis U6 m6A methyltransferase that is responsible for the m6A installation at UACm6AGAGAA motif of U6 snRNA (Fig. 1d; Additional file 1: Fig. S5c).
We subsequently assessed the in vivo m6A methylation activity of FIONA1 on poly(A)+ RNA. Various tissues including seedlings, rosette leaves, buds, and flowers were collected, and isolated poly(A)+ RNA were analyzed with LC-MS/MS. In all these tissues, the fiona1-1 mutant plants had a significantly reduced extent (around 10~15%) of m6A modification in poly(A)+ RNA compared with Col-0 plants (Fig. 1e). The decreased m6A level in fiona1-1 mutant was restored by expression by wild-type FIONA1, but not by expression of inactive FIONA1m (Fig. 1e). In addition, we measured m6A levels in the isolated 28S, 18S, 5S rRNA, and tRNA and revealed that there were no significant differences of m6A levels in rRNA and tRNA between Col-0 and fiona1-1 (Additional file 1: Fig. S6), which excludes rRNA and tRNA from FIONA1’s substrates.
We further monitored mRNA expression levels of known key m6A methyltransferase subunits (MTA, MTB, FIP37) and demethylase (ALKBH10B) in seedlings, buds, and flowers by RT-qPCR. We detected no significant differences in mRNA expression of these genes between Col-0 and fiona1-1 plants (Additional file 1: Fig. S7). The results exclude the effects of MTA-MTB m6A methyltransferase complex or m6A demethylase on the m6A level of poly(A)+ RNA in fiona1-1. Collectively, we established the finding that FIONA1 is an Arabidopsis U6 m6A methyltransferase that installs m6A modification on U6 snRNA and a small subset of poly(A)+ RNA.
FIONA1 retains methylation activity towards U6 m6A motif and plant mRNA m6A motifs in vitro
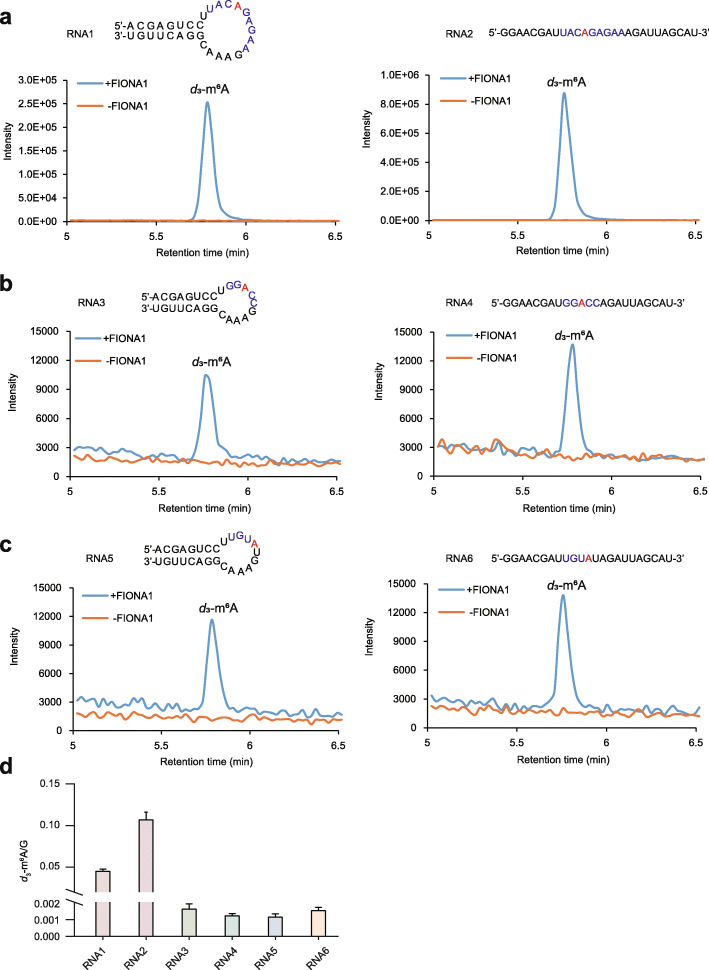
Considering that mammalian METTL16 and worm METT-10 respectively install m6A modifications only on U6 m6A motif UACm6AGAGAA or its variant motif UACm6AGAAAC within a stem-loop structured RNA [32, 42], we next performed the in vitro N6-adenosine methylation activity assay to examine whether FIONA1 consistently exhibits the substrate sequence and structure specificity towards U6 m6A motif (Additional file 1: Fig. S8). We synthesized six unmethylated RNA oligos with single-strand or stem-loop structure incorporating three different sequences: U6 m6A motif UACAGAGAA, and the reported plant mRNA m6A motifs GGACC and UGUAU [24, 52]. S-(5′-Adenosyl)-L-methionine-d3 (d3-SAM) was used as the cofactor in the methylation activity assay and the formation of d3-m6A was used for accurate MS quantification. The peak area ratio of d3-m6A versus one G in the probe (as internal control) was used to evaluate the methylation activities of FIONA1 towards different RNA oligos. The results showed that FIONA1 can transfer methyl group to N6-adenosine in all six RNA oligos (Fig. 2a–c). Clearly, FIONA1 exhibits higher activity for U6 m6A motif over plant mRNA m6A motifs GGACC and UGUAU (Fig. 2d). Furthermore, the methylation activity of FIONA1 towards U6 m6A motif showed a preference for single-stranded over stem-looped structure (Fig. 2d). The results suggest that unlike mammalian METTL16 and worm METT-10, plant FIONA1 can methylate non-U6 m6A motif sequences and its methylation activity does not require RNA secondary structure.
Fig. 2.
In vitro methylation activity of FIONA1. a-c Representative LC-MS/MS chromatograms of d3-m6A formed in RNA probes treated with or without FIONA1. The in vitro RNA N6-adenosine methylation activities of FIONA1 were tested using different RNA probes (numbered 1–6) containing sequence of UACAGAGAA, GGACC and UGUAU in a linear or looped structure in the presence of isotope-labeled cofactor d3-SAM. d The peak area ratio of d3-m6A versus one G in the probe (as internal control) was used to evaluate the methylation activities of FIONA1 towards different RNA oligos. Data are means ± SD for 2 biological replicates × 2 technical replicates
The m6A methylation activity of FIONA1 is required for phytochrome signaling-dependent photomorphogenesis
Consistent with the previous finding that FIONA1 is a nuclear protein [47], our confocal analysis of FIONA1:FIONA1-GFP/fiona1-1 revealed that FIONA1 was indeed localized in the nuclei of root tip (Additional file 1: Fig. S9). RT-qPCR showed that FIONA1 was widely expressed in diverse Arabidopsis tissues, with especially high expression in floral organs including buds and flowers (Additional file 1: Fig. S10).
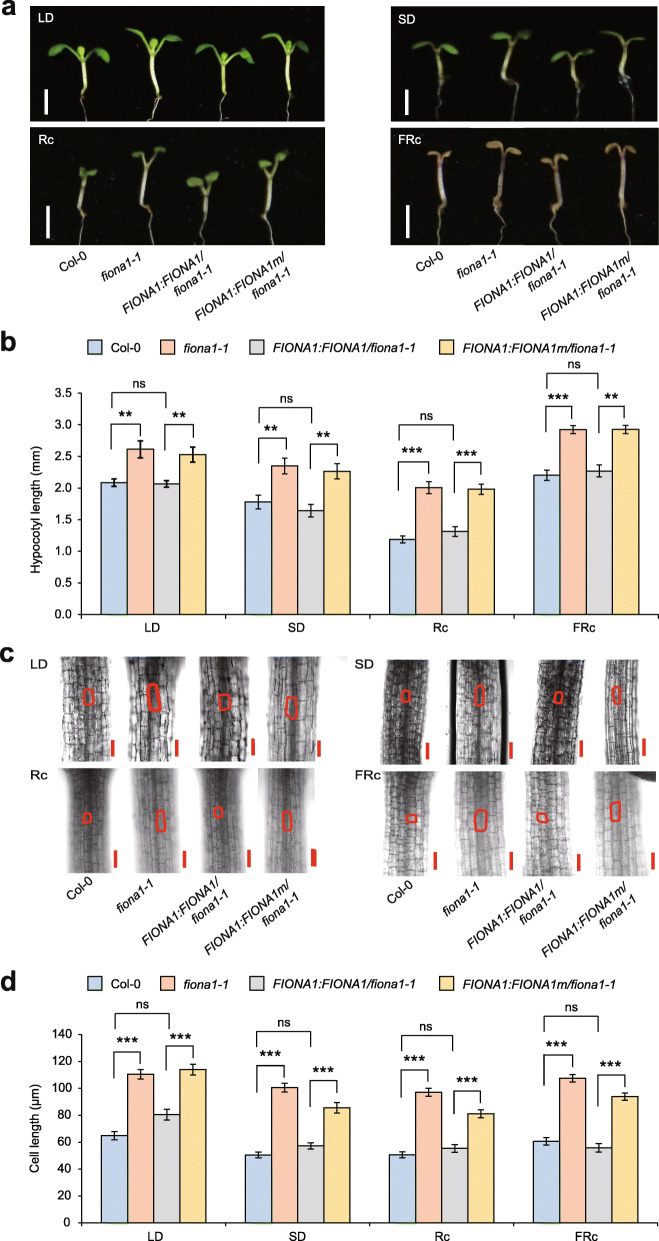
FIONA1 was previously found to affect photoperiodic hypocotyl growth [47]. Our detailed examination of the hypocotyl growth under long-day (LD, 16 h of light/8 h of dark) and short-day (SD, 8 h of light/16 h of dark) conditions revealed that disruption of FIONA1 indeed leads to hypocotyl elongation under both LD and SD conditions (Fig. 3a); however, the hypocotyl length ratio of fiona1-1 versus Col-0 (1.25 for LD and 1.32 for SD) did not show a significant difference between these two photoperiodic conditions (Fig. 3b). In contrast with the previous finding [47], our results suggest the phenotype of hypocotyl elongation in fiona1-1 is not dependent on photoperiod. Although it was reported that FIONA1 does not affect hypocotyl growth under constant light [47], we next investigated the hypocotyl growth in fiona1-1 under continuous light with different wavelength. The results showed that fiona1-1 exhibited elongated hypocotyls only under continuous red (Rc) and far-red light (FRc), but not under continuous blue light (Bc), white light (Wc), and dark (Dk) (Fig. 3a, b; Additional file 1: Fig. S11a, b). Consistently, the homozygous fiona1-2 mutant also led to hypocotyl elongation phenotype selectively under Rc and FRc conditions compared with Col-0 (Additional file 1: Fig. S12). In higher plants, phytochrome A (phyA) and phytochrome B (phyB) are the most abundant members of the photoreceptors family and their deficiencies are most evident under continuous Rc and FRc, respectively. Although phyA and phyB activities occur under different light conditions, the end-point responses (e.g., hypocotyl growth, cotyledon unfolding, flowering) controlled by which are largely the same [53]. Therefore, the selective hyposensitivity to Rc and FRc in FIONA1-defective mutants suggests FIONA1 is a positive regulator of photomorphosis, especially in phyA and phyB signaling transduction in Arabidopsis.
Fig. 3.
The methylation activity of FIONA1 positively regulates the sensitivity of red light and far-red light and cell elongation. a Representative phenotype of Col-0, fiona1-1, FIONA1:FIONA1/fiona1-1, and FIONA1:FIONA1m/fiona1-1 seedlings grown under LD, SD, Rc (30 μmol m−2 s−1), and FRc (11 μmol m−2 s−1) for 7 days. Bar = 2 mm. b Hypocotyl lengths of the seedlings shown in (a). Data are means ± SE (n ≥ 25). c Confocal microscope of hypocotyl epidermal cells of Col-0, fiona1-1, FIONA1:FIONA1/fiona1-1, and FIONA1:FIONA1m/fiona1-1 seedlings grown in LD, SD, Rc, and FRc for 7 days. The cell sizes were marked with red lines. Bar = 100 μm. d The hypocotyl epidermal cell lengths shown in (c). Data are means ± SE (n ≥ 20). ** p < 0.01, *** p < 0.001 by t test (two-tailed). ns, no significance
We subsequently investigated whether hypocotyl photomorphogenesis is dependent on the methylation activity of FIONA1. The rescue experiments showed that the selective hyposensitivity to Rc and FRc in fiona1-1 can be restored by expression of wild-type FIONA1, but not inactive FIONA1m (Fig. 3a, b). Thus, the methylation activity of FIONA1 regulates phytochrome signaling-dependent hypocotyl photomorphogenesis. Furthermore, we sought clue about altered hypocotyl at the histological level and found that the epidermal cells in the upper part of the hypocotyl (the part near the cotyledon) were elongated in fiona1-1 under LD, SD, Rc, and FRc conditions (Fig. 3c, d; Additional file 1: Fig.S11c, d), suggesting the phenotype of hypocotyl elongation in fiona1-1 is due to the cell elongation. Expectedly, the elongation of hypocotyl epidermal cells is dependent on the m6A methylation activity of FIONA1 (Fig. 3c, d; Additional file 1: Fig. S11c, d).
The m6A methylation activity of FIONA1 is required for floral transition
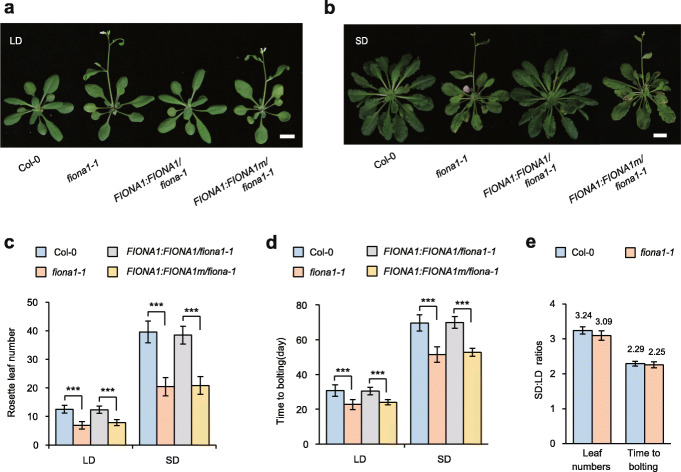
FIONA1 was reported to affect daylength-dependent flowering [47] but its underlying biological function is unknown. We next asked whether the phenotype caused in the fiona1 mutant was dependent on the m6A catalytically activity of FIONA1. Our two CRISPR-cas9 generated homozygous mutant lines fiona1-1 and fiona1-2 both display early flowering phenotypes compared with Col-0 under LD and SD conditions (Fig. 4a–d; Additional file 1: Fig.S13), consistent with the previous finding [47]. However, we did not observe a significant difference in photoperiodic flowering response under LD and SD conditions in our fiona1 homozygous mutant line (Fig. 4e). Expression of wild-type FIONA1 in fiona1-1 (FIONA1:FIONA1 /fiona1-1) can completely recover the early flowering phenotype in fiona1-1 under LD and SD conditions; however, complementation of the inactive mutant FIONA1m with P237A/F239G mutations (FIONA1:FIONA1m/fiona1-1) did not complement the phenotype (Fig. 4a–d). These results demonstrated that the observed flowering phenotype is dependent on the m6A methylation activity of FIONA1.
Fig. 4.
The methylation activity of FIONA1 is required for floral transition. a-b Phenotypes of the floral transition in Col-0, fiona1-1, FIONA1:FIONA1/fiona1-1, and FIONA1:FIONA1m/fiona1-1 plants grown under LD (a, 16 L/8D) and SD (b, 8 L/16D) conditions. Bar = 1 cm. c-d Statistical analysis of total rosette leaf number (c) and days to flowering (d) at the bolting stage in Col-0, fiona1-1, FIONA1:FIONA1/fiona1-1, and FIONA1:FIONA1m/fiona1-1 plants under LD (16 L/8D) and SD (8 L/16D) conditions. Data are means ± SD (n ≥ 20) *** p < 0.001 by t test (two-tailed). e The SD: LD ratios of rosette leaf number and time to bolting at the bolting stage in Col-0 and fiona1-1 plants
Disruption of FIONA1 leads to transcriptomic m6A hypomethylation
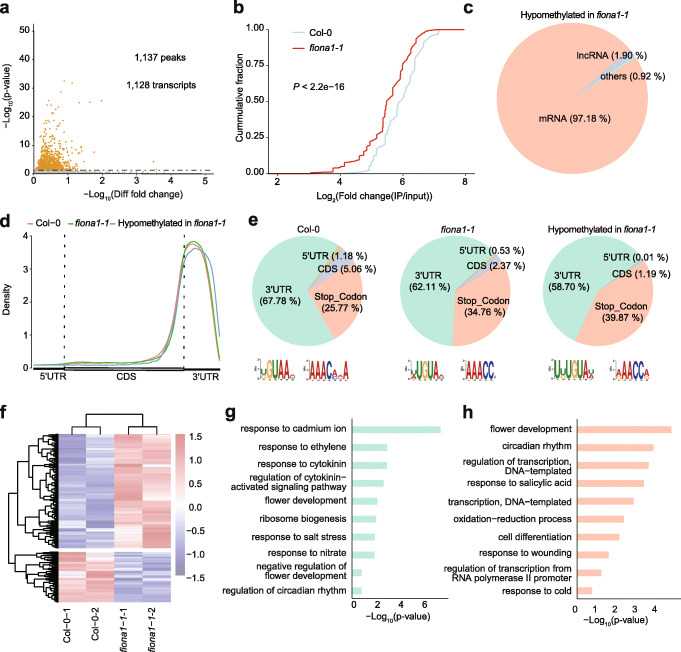
To investigate the global effects of FIONA1-mediated m6A poly(A)+ RNA modification landscape, we performed m6A-seq that combines anti-m6A antibody immunoprecipitation (m6A-IP) and high-throughput sequencing [54–56] in 12-day-old Col-0 and fiona1-1 seedlings (three biological replicates for each genotype). As summarized in Additional file 3: Table S1, around 47–166 million reads per sample were mapped to Arabidopsis TAIR 10 genome. We used MACS2 algorithm with an estimated false discovery rate (FDR) < 0.05 and an enrichment of ≥ 2 to call m6A peaks. The m6A peaks in both replicates were classified as “confident peaks.” We identified 10,840 m6A confident peaks corresponding 12,345 transcripts/genes in Col-0 and 11,238 m6A confident peaks corresponding to 12,461 transcripts/genes in fiona1-1 (Additional file 1: Fig. S14; Additional file 4: Table S2; Additional file 5: Table S3). To seek the potential m6A methylation targets of FIONA1, we conducted ExomePeak algorithm (| fold change | > 1 and FDR < 0.05) to calculate the differential m6A peaks between Col-0 and fiona1-1. We identified 1137 confident hypomethylated m6A peaks corresponding 1128 transcripts/genes in fiona1-1(Fig. 5a, b; Additional file 6: Table S4), which predominantly belong to mRNA (Fig. 5c). To validate our m6A-seq results were reliable and accurate, we randomly selected four m6A hypomethylated genes and conducted m6A-IP-qPCR of fragmented poly(A)+ RNA from Col-0 and fiona1-1. These results showed that the m6A methylation on these four transcripts were significantly reduced in fiona1-1 (Additional file 1: Fig. S15), suggesting the identified confident hypomethylated m6A peaks in fiona1-1 could be the methylation targeted sites of FIONA1.
Fig. 5.
Disruption of FIONA1 leads to transcriptome m6A hypomethylation. a Volcano plot of the hypomethylated m6A peaks identified in fiona1-1 across three biological replicates. b Cumulative fraction of log2 (m6A enrichment fold change) for the hypomethylated m6A peaks in fiona1-1 and Col-0. Pie chart presenting RNA types of the confident hypomethylated m6A peaks identified in fiona1-1. d Metagene profile presenting the distributions of m6A peaks identified in Col-0 and fiona1-1 and the hypomethylated m6A peaks in fiona1-1 across the indicated mRNA segments. e Pie chart presenting the fractions of m6A peaks identified in Col-0 and fiona1-1 and the hypomethylated m6A peaks in fiona1-1 among non-overlapping transcript segments and the MEME-identified motifs for those m6A peaks. f Heatmap showing differentially expressed genes in two biological replicated Col-0 and fiona1-1 seedlings. g GO analysis of the hypomethylated m6A containing genes in fiona1-1. h GO analysis of the differentially expressed genes in fiona1-1 compared with Col-0
We next evaluated the substrates preference of FIONA1. Three groups of m6A peaks—the identified confident hypomethylated m6A peaks in fiona1-1, the confident m6A peaks in Col-0, and the confident m6A peaks in fiona1-1—were used to investigate the m6A distribution across transcripts and the m6A motif. The metagene profiles revealed that the confident hypomethylated m6A peaks in fiona1-1 highly located around stop codon and within 3′ UTR, which was the same as the whole m6A distribution pattern of Col-0 and fiona1-1 (Fig. 5d). We divided transcripts into five non-overlapping segments: 5′ UTRs, Start codon (100-nucleotide window centered on the start codon), coding sequences (CDS), Stop codon (100-nucleotide window centered on the stop codon), and 3′ UTRs. The m6A peaks from the three groups were mainly located in 3′ UTR and stop codon region, but the fraction of the hypomethylated m6A peaks in stop codon portion is slightly higher compared to Col-0 (Fig. 5e). We clustered these three groups of m6A peaks in MEME software package to search motifs. We did not observe U6 m6A motif enriched in the hypomethylated m6A peaks. Instead, we found that the enriched motifs (UKUGUAW (K=U or G; W=U or A) and RAACCR (R = A or G)) in the confident hypomethylated m6A peaks in fiona1-1 were similar as those identified in all m6A peaks in either Col-0 or fiona1-1 (Fig. 5e; Additional file 1: Fig. S16). Note that UGUA is a known plant-unique m6A motif [24] and RAACC resembles m6A consensus motif RRACH [52, 57]. Thus, FIONA1 exhibits no obvious preference in m6A position and motif in comparison to whole m6A peaks in Col-0 and fiona1-1.
We subsequently analyzed our RNA-sequencing performed in 12-day-old Col-0 and fiona1-1 seedlings and identified 267 downregulated genes and 605 upregulated genes in fiona1-1 compared with Col-0 (threshold criteria with | log2(fold change) | ≥ 0.48 and P value < 0.05) (Fig. 5f; Additional file 7: Table S5). To gain functional insights into the role of FIONA1, we performed Gene Ontology (GO) analysis using the DAVID tools on 1128 m6A hypomethylated genes and 872 differential expressed genes in fiona1-1, respectively. GO analysis revealed the m6A hypomethylated genes were positively enriched in response to cadmium ion, ethylene, and cytokinin, and regulation of cytokinin-activated signaling pathway, flower development, and circadian rhythm (Fig. 5g). GO analysis of the differential expressed genes showed that FIONA1 deficiency affects several developmental pathways, including flower development, circadian rhythm, response to salicylic acid, and cell differentiation (Fig. 5h). These gene functions showed clear correlation with the observed phenotypes in fiona1 mutant.
mRNA modification m6A regulates alternative splicing in mammals and mammalian METTL16 affects the splicing of MAT2A mRNA for intron retention [12, 13, 32, 58]. It was predicated that removal of m6A modification from U6 snRNA also could affect splicing [59, 60]. Thus, we investigated whether FIONA1 affects alternative splicing. We analyzed the changes of alternative splicing between Col-0 and fiona1-1 using rMATS tools [39, 61] and found only 43 alternative splicing events (FDR < 0.05) occurred in fiona1-1 compared with Col-0 (Additional file 8: Table S6). Among them, 4 genes contain confident m6A hypomethylation peaks. Therefore, these results showed that disruption of FIONA1 does not affect global alternative splicing.
Differences in methylation sites between FIONA1 and MTA / MTB/FIP37 complex
The MTA/MTB/FIP37 methyltransferase complex is responsible for the majority of mRNA methylation in plants. Disruption of the key subunit of the methyltransferase complex leads to embryonic lethal in Arabidopsis [36, 38]. Conditional expression of FIP37 driven by LEC1 promoter in the fip37 mutant causes more than 80% of m6A decrease in poly(A)+ RNA [38]. However, FIONA1 deficiency has good viability and only reduces 10~15% of m6A level in poly(A)+ RNA, indicating the small subset of m6A sites methylated by FIONA1 would be distinct from the targets of the MTA/MTB/FIP37 methyltransferase complex. To validate it, we termed the hypomethylated m6A in fiona1-1 as “FIONA1-dependent m6A” and the rest m6A peaks in Col-0 excluding FIONA1-dependent m6A as “FIONA1-independent m6A.” We compared these two groups with FIP37-dependent m6A peaks, which were identified from the published m6A-seq results in LEC1:FIP37/fip37-4 and control plants [38]. We found that 84% (3126 out of 3699) of FIP37-dependent m6A peaks were overlapped with FIONA1-independent m6A but only 10% (379 out of 3699) overlapped with FIONA1-dependent m6A (Additional file 1: Fig. S17a). We further calculated the distance between stop codon and m6A sites from the three groups. The results showed that both FIP37-dependent m6A and FIONA1-independent m6A peaks were highly enriched around the stop codon, while FIONA1-dependent m6A were enriched at the downstream of stop codon (Additional file 1: Fig. S17b). Collectively, these results suggest that FIONA1 only methylates a small subset of m6A sites in poly(A)+ RNA and its targeted sites are distinct from the methylation sites of MTA/MTB/FIP37 methyltransferase complex.
FIONA1 does not methylate SAM synthetase transcripts
As U6 m6A methyltransferase, mammalian METTL16 and worm METT-10 install m6A in U6 or U6-like m6A motifs of SAM synthetase to affect its gene expression and SAM homeostasis under high SAM conditions [32, 42]. Considering that FIONA1 is Arabidopsis U6 methyltransferase, we next investigated whether FIONA1 methylates SAM synthetases. Arabidopsis contains four genes encoding SAM synthetase: MAT1 (AT1G02500), MAT2 (AT4G01850), MAT3 (AT2G36880), and MAT4 (AT3G17390). We found none of them contain U6 m6A motif UACm6AGAGAA or U6-like m6A motif UACm6AGAAAC (Additional file 1: Fig. S18). Our m6A-seq results showed that the m6A modifications were localized on the transcripts of SAM synthetases but were not installed by FIONA1 (Additional file 1: Fig. S19a). RNA-seq and RT-qPCR results revealed that FIONA1 deficiency did not affect their expression levels compared with WT under normal growth condition (Additional file 1: Fig. S19b, c). Next, we investigated whether FIONA1 would methylate SAM synthases under high SAM conditions; therefore, we grew Arabidopsis seedlings on the agar plates with different concentrations of SAM or L-methionine (L-Met) for 12 days. We measured and confirmed that the SAM concentration inside seedlings was continuously increased under SAM or L-Met treatments and elevated by 2-fold under 100 mg/L SAM or 50 mg/L L-Met treatment compared non-treatments (Additional file 1: Fig. S20a, b). The RT-qPCR results revealed that no significant differences in pre-mRNA and mRNA expression levels of SAM synthetases MAT1-4 in fiona1-1 compared to WT seedlings grown under high SAM conditions (Additional file 1: Fig. S20c, d). These results suggest that unlike mammalian and worms, Arabidopsis does not use FIONA1’ methylation activity to affect the expression level of SAM synthetase and SAM homeostasis.
FIONA1 installs m6A on non-U6 m6A motifs in photomorphogenic- and flowering-related transcripts in vivo
Excluding the methylation function of FIONA1 on SAM synthetases, we subsequently explored the molecular mechanism by which FIONA1-mediated m6A methylation regulates phytochrome signaling-dependent photomorphogenesis and floral transition. The GO analysis revealed that the m6A hypomethylated genes and the differential expressed genes in fiona1 mutant were enriched in flower development pathway (Fig. 5g, h). The m6A-seq and RNA-seq results showed that m6A at 3′ UTR of CRY2 (CRYPTOCHROMES 2) and FLC (FLOWERING LOCUS C) transcripts were significantly reduced in fiona1-1 (Additional file 1: Fig. S21; Additional file 6: Table S4) and their mRNA expression levels were also significantly changed (Additional file 7: Table S5). CO (CONSTANS) is a key regulator of the photoperiodic flowering pathway [62]. The RNA-seq showed that CO mRNA was significantly upregulated in fiona1-1, consistent with previous report [47]. We did not identify m6A peak on CO transcript in the m6A-seq results, which might be the low expression level of CO at Zeitgeber time (ZT) 13 of LD condition. Thus, we chose three flowering-related genes CRY2, FLC, and CO for further validation. PIF4 (PHYTOCHROME INTERACTING FACTOR4) directly interacts with light-activated phytochromes and is a positive regulator in cell elongation [63]. The hypocotyl elongation of pif4 mutants is specifically defective in responsiveness to red light, the double mutant of PIF4 and its close homolog PIF5 (pif4pif5) show hypersensitivity to far-red light, suggesting that they redundantly control the far-red light responses [64]. In consistent with phenotype of hypocotyl cell elongation selectively under Rc and FRc in fiona1 mutants, RNA-seq revealed upregulation of PIF4 expression level in fiona1-1 (Additional file 7: Table S5). Among three biological replicate m6A-seq, we observed the significant m6A decreases in two biological replicates of fiona1-1 samples (Additional file 1: Fig. S21). Therefore, we selected PIF4 for further validation.
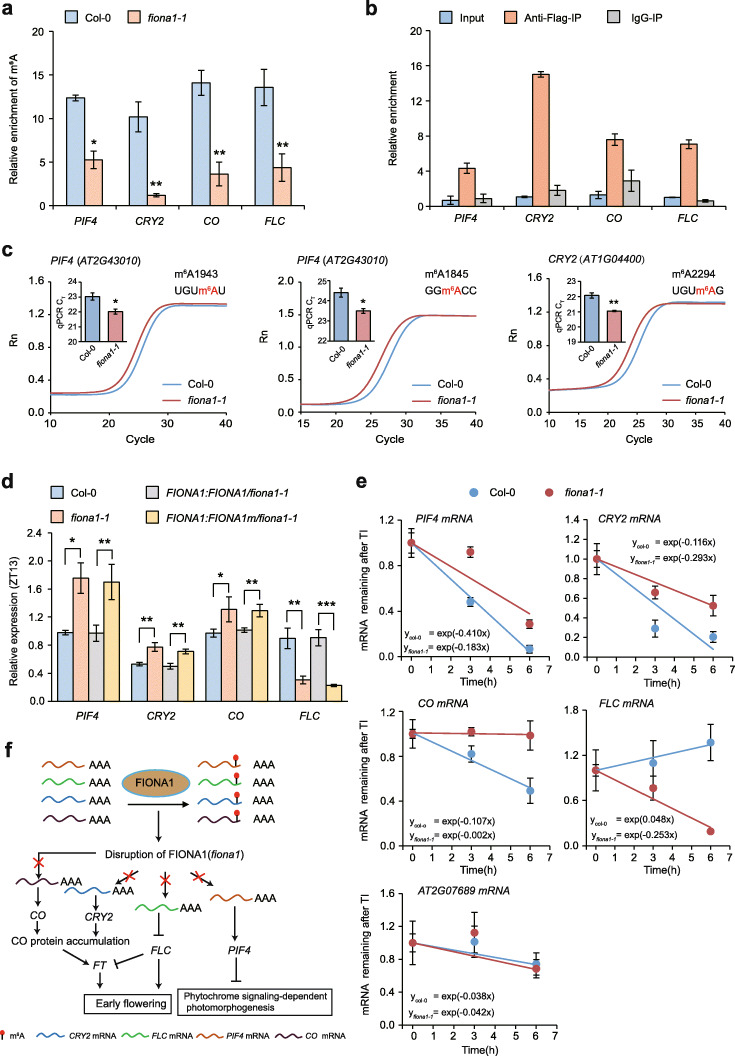
To further verify PIF4, CRY2, CO, and FLC were directly methylation targets of FIONA1, we performed m6A-IP qPCR assays on fragment poly(A)+ RNA isolated from 12-day-old Col-0 and fiona1-1 seedlings at ZT13 of LD condition. We confirmed that the m6A levels (or m6A enrichment) on these identified m6A peaks of PIF4, CRY2, CO, and FLC were significantly reduced in fiona1-1 compared with Col-0 (Fig. 6a). RNA immunoprecipitation (RIP)-qPCR assays using 12-day-old FIONA1:FIONA1-Flag/fiona1-1 seedlings at ZT13 of LD condition showed that FIONA1 protein directly binds PIF4, CRY2, CO, and FLC transcripts (Fig. 6b). Considering FIONA1 is an m6A methyltransferase and does not affect the intercellular SAM level, these results revealed that FIONA1 directly installs m6A on PIF4, CRY2, CO, and FLC transcripts.
Fig. 6.
FIONA1-mediated m6A installation affects the stability of PIF4, CRY2, and FLC transcripts. a m6A-IP-qPCR results showing the relative m6A levels of PIF4, CRY2, and FLC transcripts in 12-day-old Col-0 and fiona1-1 seedlings. Data are means ± SD for 3 biological replicates × 3 technical replicates. * p < 0.05, ** p < 0.01 by t test (two-tailed). b RIP-qPCR assays in FIONA1:FIONA1-FLAG/ fiona1-1 plants showing that FIONA1 directly binds PIF4, CRY2, and FLC transcripts. Data are means ± SD for 3 biological replicates × 2 technical replicates. c Real-time fluorescence amplification curves and bar plot of the threshold cycle (CT) of qPCR showing SELECT results for detecting the FIONA1-targeted m6A sites in PIF4 and CRY2 mRNAs in Col-0 and fiona1-1 seedlings. Rn is the raw fluorescence for the associated well normalized to the fluorescence of the passive reference dye (ROX). Data for bar plot are means ± SD for 3 biological replicates × 2 technical replicates. * p < 0.05, ** p < 0.01 by t test (two-tailed). d The relative expression levels of PIF4, CRY2 and FLC at ZT13 in the indicated genotypic plants under LD conditions. TUB2 was used as the internal control gene. Data are means ± SD for 3 biological replicates × 3 technical replicates. * p < 0.05, ** p < 0.01 by t test (two-tailed). e The mRNA lifetimes of PIF4, CRY2, and FLC in Col-0 and fiona1-1. The AT2G07689 was used as the negative control. TI: transcription inhibition. Data are represented as means ± SD for 2 biological replicates × 3 technical replicates. f Proposed model describing the molecular mechanism through which FIONA1-mediated m6A installation regulates Arabidopsis phytochrome signaling-dependent photomorphogenesis and floral transition
There are no U6 m6A motifs or variant U6 m6A motifs in PIF4, CRY2, CO, and FLC transcripts. Considering our finding that FIONA1 can methylate plant mRNA m6A motifs GGACC and UGUAU in vitro (Fig. 2), we next chose PIF4 and CRY2 to investigate whether FIONA1 can install m6A on non-U6 m6A motif in vivo. The expression levels of PIF4 and CRY2 are highly abundant, which allows us to determine m6A sites using SELECT method, a qPCR method for quantitative detection of m6A locus and fraction in mRNA at single-base resolution [51]. We firstly performed FTO-assisted SELECT on Arabidopsis total RNA to determine m6A positions in the identified FIONA1-targeted m6A peaks (Fig. 6a, b). Two m6A sites in UGUm6A1943U and GGm6A1845CC sequences and one m6A site in UGUm6A2294G sequence were respectively identified within 3′ UTR regions of PIF4 and CRY2 (Additional file 1: Fig. S22). Direct SELECT performed in total RNA isolated from Col-0 and fiona1-1 showed that these three m6A sites identified in PIF4 and CRY2 were methylated by FIONA1 in vivo (Fig. 6c; Additional file 1: Fig. S23). These findings collectively reveal that FIONA1 directly installs m6A on non-U6 m6A motifs in PIF4, CRY2, CO, and FLC transcripts in vivo.
FIONA1-mediated m6A methylation in PIF4, CRY2, CO, and FLC transcripts affects post-transcriptional gene regulation
We next investigated the subsequent effects of the reduced m6A on PIF4, CRY2, CO, and FLC in fiona1 mutant. We measured the transcript expression levels of these four genes in 12-day-old Col-0 and fiona1-1 seedlings under different growth conditions. Consistent with the observed phenotypes in fiona1 mutants, the RT-qPCR results showed that the expression levels of CRY2 and CO transcripts were increased and FLC expression level was decreased in fiona1-1 mutant compared with Col-0 under both LD and SD conditions (Fig. 6d; Additional file 1: Fig. S24). PIF4 transcript expression level was increased in fiona1-1 mutant compared with Col-0 under LD (ZT13), SD (ZT10), and continuous Rc and FRc conditions (Fig. 6d; Additional file 1: Fig. S25a). Consistently, PIF4 protein level was also increased in fiona1-1 mutant plants (Additional file 1: Fig. S25b). Furthermore, the expression levels of these four transcripts in fiona1-1 can be recovered in FIONA1:FIONA1 /fiona1-1 plants but not in FIONA1:FIONA1m/fiona1-1 (Fig. 6d; Additional file 1: Fig. S24e and S25a), revealing that the expression effect of these four transcripts was dependent on the m6A methylation activity of FIONA1.
The m6A modification in human has been confirmed to facility protein translation, mRNA degradation, and mRNA stabilization [10, 18, 65]. The m6A functions in Arabidopsis also have been found to promote mRNA stabilization or mRNA degradation [19, 24, 38], thus, we further performed transcription inhibition assays using actinomycin D to measure the lifetimes of these four transcripts. The results showed that in fiona1-1 mutant, PIF4, CRY2, and CO transcripts were degraded more slowly and FLC transcript was degraded more rapidly compared to Col-0 (Fig. 6e). Thus, our results collectively demonstrate that loss-of-function of FIONA1 reduces m6A in PIF4, CRY2, CO, and FLC transcripts, which promotes stabilization of PIF4, CRY2, and CO mRNA and degradation of FLC mRNA and thereby leads to the observed phenotypes related in phytochrome signaling-dependent photomorphogenesis and floral transition (Fig. 6f).
Additionally, we further showed that the expression level of the downstream florigen FT (FLOWERING LOCUS T) was increased in fiona1-1 mutant under both LD and SD conditions whereas its m6A level was not altered in fiona1-1 mutant compared with Col-0 (Additional file 1: Fig. S26), confirming FT is not the direct m6A methylation target of FIONA1. Thus, the increased FT transcript expression is due to the positively transcription regulation from FIONA1-mediated m6A regulation in its upstream FLC and photoperiodic regulator CRY2 and CO (Fig. 6f).
Discussion
m6A modification is the ubiquitous mRNA modification in eukaryotes [52, 66–68]. Characterization of plant mRNA m6A writer complex, erasers, and readers revealed that mRNA modification m6A regulates developmental timing, leaf morphogenesis, flowering transition, trichome morphology, leaf growth, nitrate signaling, viral response, fruit ripening, ABA response, and salt tolerance [22–30, 69, 70]. However, whether Arabidopsis contains other independent mRNA m6A writers is not fully known. Here, we characterized that FIONA1 as Arabidopsis U6 snRNA m6A methyltransferase methylates m6A on non-U6 m6A motifs in a small subset of poly(A)+ RNA, and demonstrated the molecular mechanism how the m6A methylation activity of FIONA1 regulates phytochrome signaling-dependent photomorphogenesis and floral transition.
As U6 m6A methyltransferase, mammalian METTL16 and worm METT-10 install m6A on U6 or variant U6 m6A motif in pre-mRNAs of SAM synthetases, which affects splicing and subsequent SAM homeostasis [32, 42], the effect of intercellular SAM level by METTL16 or METT-10-mediated m6A methylation in SAM synthetases could tune other DNA/histone/RNA methylation modifications formed by SAM as a donor. It has been confirmed that some mRNA m6A sites written by METTL3/METTL14 complex and some cap m6Am sites written by PCIF1 are affected by disruption of human METTL16 [46]. Here we found several unique features of FIONA1, which are distinct from mammalian METTL16 and worm METT-10. (1) FIONA1 neither installs m6A in the transcripts of Arabidopsis SAM synthetases (MAT1-4) nor affects their transcript expression levels under normal or high SAM conditions (Additional file 1: Fig. S19, 20). (2) FIONA1 can install m6A on plant mRNA m6A motifs in vitro and in vivo. We performed in vitro methylation experiments to reveal that FIONA1 not only methylates U6 m6A motif, but also assembles m6A for RNA containing GGAAC or UGUA motif (Fig. 2). The single-base m6A site validation by SELECT method in fiona1-1 and WT plants further confirmed that FIONA1 can install m6A in UGUAU and GGACC sequences of PIF4 mRNA and UGUAG sequence of CRY2 mRNA in vivo (Fig. 6c). (3) The methylation activity of FIONA1 does not depend on stem-loop structure. We found FIONA1 exhibits higher activity towards U6 m6A motif in a linear RNA over in a stem-loop structured RNA (Fig. 2). Furthermore, we showed that FIONA1 prefers U6 m6A motif over plant mRNA m6A motifs in vitro (Fig. 2); however, we confirmed that FIONA1 indeed installs m6A on plant mRNA m6A motifs in vivo (Fig. 5e and Fig. 6c). Therefore, we speculate there might be some interaction proteins to assist FIONA1’s methylation on mRNA in vivo.
It is well known that the m6A writer complex comprising the key subunits MTA, MTB, and FIP37 is responsible for the majority of poly(A)+ RNA m6A methylation. More than 80% of m6A level in poly(A)+ RNA was reduced in the fip37-4 LEC1:FIP37 plants. We found around 10~15% of m6A level of poly(A)+ RNA was decreased in the fiona1-1 mutant plants compared with Col-0 (Fig. 1e). The m6A sequencing results identified 1137 hypomethylated m6A peaks in fiona1-1 compared with Col-0 (Fig. 5a, b; Additional file 6: Table S4), which could be potential methylation targets of FIONA1. Considering 10,840 m6A confident peaks identified in poly(A)+ RNA of Col-0 (Additional file 1: Fig. S14a), the fraction of the potential FIONA-methylated m6A sites (1,137 hypomethylated m6A peaks) was around 10% of total m6A sites in Col-0 poly(A)+ RNA, in consistent with the 10~15% of m6A level decreased in the fiona1-1 mutant plants. A comparison among the published FIP37 potential methylation targets, FIONA1 potential methylation targets, and non-FIONA1 targets revealed that FIONA1 and FIP37 methylate different m6A sites and FIP37 targets are largely overlapped with non-FIONA targets (Additional file 1: Fig. S17). Collectively, our results suggest that FIONA1 methylates no more than 15% of m6A sites in poly(A)+ RNA; the rest m6A sites in poly(A)+ RNA are installed by the m6A writer complex containing MTA/MTB/FIP37.
The function of m6A in U6 snRNA was predicted to affect splicing [32, 60]. Although mammalian METTL16 and worm METT-10 were discovered as U6 m6A methyltransferase, their regulatory functions on U6 still remain unclear. Here we preliminarily investigated whether m6A in U6 affects splicing. Our analysis showed that disruption of FIONA1 only affects 43 genes’ alternative splicing events and none of them are related with the observed phenotypes (Additional file 8: Table S6), suggesting removal of U6 m6A does not globally affect splicing. Our results rule out the possible effects of loss of U6 m6A methylation on the phenotypes in fiona1-1 mutant. The m6A function on U6 snRNA needs further exploration.
FIONA1 was previously found to affect circadian rhythms and daylength-dependent flowering and hypocotyl growth [47]; however, the underlying molecular mechanisms have not been fully illuminated. Our phenotypic analysis showed that disruption of FIONA1 leads to early flowering under both LD and SD conditions, and its early flowering phenotype is not clearly dependent on daylength/photoperiod (Fig. 4; Additional file 1: Fig. S13). Our two fiona1 homozygous mutant lines displays hypocotyl and its cell elongation phenotypes selectively under continuous Rc and FRc conditions, confirming that FIONA1 regulates phytochrome signaling-dependent photomorphogenesis, not daylength-dependent hypocotyl growth (Fig. 3; Additional file 1: Fig. S11, 12).
In mechanism study, we found that FIONA1 directly binds and methylates PIF4, CRY2, CO, and FLC transcripts and showed FIONA1-mediated m6A post-transcriptional regulation in these transcripts (Fig. 6; Additional file 1: Fig. S24, 25). The FIONA1 transcript level is not affected by diurnal cycle and light [47], suggesting its m6A methylation activity is not affected by diurnal cycle and light. Indeed, our results showed that the FIONA1 regulation on CRY2, CO, and FLC transcripts is independent on LD and SD growth conditions (Fig. 6d; Additional file 1: Fig. S24e). CRY2 and CO are positive regulators and FLC is a negative regulator in flowering time control [62, 71–73]. CRY2 is a blue light receptor [74]; the transcript expression levels of CO and FLC are respectively regulated by photoperiod and vernalization/autonomous pathways [62, 75, 76]. FIONA1-mediated m6A post-transcriptional regulation in these three transcripts explained our phenotypic observation that disruption of FIONA1 leads to early flowering that is independent on photoperiod (Fig. 4, 6; Additional file 1: Fig. S24, 26). PIF4 is phytochrome-interacting bHLH transcription factor that directly interacts with red light-activated phyB and weakly binds to far-red light-activated phyA [3, 63, 77]. PIF4 positively regulates cell elongation [1, 64, 77], and overexpression of PIF4 causes long hypocotyl phenotype both under Rc (30 μmol m−2 s−1) and FRc (10 but not 7 μmol m−2 s−1). We found disruption of FIONA1 reduces m6A on PIF4 transcript and increases its mRNA stability (Fig. 6), in consistent with our phenotype that disruption of FIONA1 led to cell length and long hypocotyl phenotypes selectively under Rc (30 μmol m−2 s−1) and FRc (11 μmol m−2 s−1) conditions (Fig. 3). We should note that other genes/regulators could be the methylation targets of FIONA1 that collectively contribute to the observed phenotypes in flowering and phytochrome signaling. Thus, like histone modifications, FIONA1-mediated m6A post-transcriptional regulation is an autonomous regulator for flowering and phytochrome signaling-dependent photomorphogenesis.
FIONA1 was reported as a genetic regulator of period length in the circadian clock, which affects the free-running circadian period of leaf movement and the period length of four central oscillator genes’ transcript expression (CCA1, LHY, TOC1, and LUX) [47]. Consistently, our GO analysis revealed that the differentially expressed genes in fiona1-1 mutant were enriched in circadian rhythm (Fig. 5h). Our m6A-seq results showed that m6A modification level in CCA1 and LHY transcripts were reduced in fiona1-1 mutant (Additional file 6: Table S4), suggesting CCA1 and LHY could be the methylation targets of FIONA1. It needs to further investigate how FIONA1-mediated m6A post-transcriptional regulation in central oscillator genes. Moreover, GO analysis revealed pathways in hormone responses, especially response to ethylene, cytokinin, and cytokinin-activated signaling pathway (Fig. 5h). PIF4 has been reported to transcriptionally regulate ethylene biosynthesis and to affect their signaling pathways [77]. Whether FIONA1 functions in hormone signaling and the related phenotypes through FIONA1-mediated m6A regulation in PIF4 or other genes remains unknown. In summary, our findings demonstrate that the m6A methylation of FIONA1-mediated post-transcriptional regulation is a new autonomous regulator for flowering and phytochrome signaling-dependent photomorphogenesis.
Conclusions
FIONA1 as Arabidopsis U6 m6A methyltransferase is distinct from mammalian METTL16 and worm METT-10: (1) FIONA1 does not install m6A in pre-mRNAs of SAM synthetases to regulate intercellular SAM level under normal and high SAM conditions; (2) FIONA1 can install m6A on plant mRNA m6A motifs in vitro and in vivo; (3) The methylation activity of FIONA1 does not depend on stem-loop structure. Our results demonstrate that FIONA1-mediated m6A post-transcriptional regulation is an autonomous regulator for flowering and phytochrome signaling-dependent photomorphogenesis.
Methods
Generation of fiona1 mutant by the CRISPR/Cas9 system
The fiona1 mutant was obtained following the published protocol [48]. In brief, two pair’s primers containing two single guide RNAs (sgRNAs) sequences of FIONA1, the pDT1T2 vector as template, were programed PCR amplified. The products were purified and digested with BsaI and ligated into a binary vector pHEE401E. We transformed the PHEE401E-2sgRNA vector into Col-0 plants via Agrobacterium strain GV3101-mediated floral dip method. The seeds were collected from the T0 plants and screened on 1/2MS plates containing 25 mg/L hygromycin. The positive seedlings (T1) obtained were transplanted to soil. To analyze mutations of FIONA1, genomic DNA of rosette leaf from T1 transgenic plants grown in soil were extracted, and the fragments surrounding the target sites amplified by PCR using gene-specific primers fiona1-Mut-F-1/R-1, fiona1-Mut-F-2/R-2, respectively. We submitted purified PCR products for direct Sanger sequencing with the same primers.
Plasmid construction
One microgram of total RNA was extracted using M5 SuperPure Total RNA Extraction Reagent (SuperTRIgent) (Mei5, China) from Col-0 seedlings and reverse transcribed into cDNA using PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Japan). The full-length FIONA1 cDNA was amplified via 2× Phanta Master (Vazyme, China) with specific primers, which were replaced other deoxynucleotide sequences to ensure that the mRNA of FIONA1 would not be cut by Cas9 protein, while the amino acid sequence of FIONA1 protein remained unchanged in vivo. By aligning Arabidopsis FIONA1 proteins with those of other organisms, we found conserved amino acid residues-NPPFF in the methyltransferase domain. According to previous reports, both the METTL16 PP185/186AA and F187G mutants are catalytically inactive in human and mouse [32, 45]. Therefore, two conserved amino acid residues located in the methyltransferase domain were mutated to generate a putative catalytically function-abolished form of FIONA1, FIONA1m (FIONA1P237A/F239G). The cloned native promoter (1.5 kb upstream of FIONA1) and full-length FIONA1 (or FIONA1m) CDS were cloned into a pCAMBIA1305 vector between the EcoRI and SalI sites via ClonExpress MultiS One Step Cloning Kit (Vazyme, China). The pCAMBIA1305 vector was modified before and contains GFP or 3× Flag tag. Thus, the expression vector containing PFIONA1-FIONA1-Flag, PFIONA1-FIONA1m-Flag, and PFIONA1-FIONA1-eGFP and PFIONA1-FIONA1m-eGFP constructs were obtained. Moreover, the clone 35S promotor and FIONA1 fragment were also cloned into pCAMBIA1305 between the EcoRI and SalI sites to generate P35S-FIONA1-eGFP. Schematic representations of the constructs are shown in Additional file 1: Fig. S4a-c. All constructs were confirmed by Sanger sequencing, and the primers used in their generation are shown in Additional file 9: Table S7.
Plant materials and growth conditions
Arabidopsis thaliana genotypes in this study included wild-type Col-0 and the following two representative mutant lines: fiona1-1, fiona1-2. All of the mutant lines were in Col-0 background and obtained via CRISPR/Cas9 editing system. The complementation lines (FIONA1:FIONA1-GFP/fiona1-1, FIONA1:FIONA1m-GFP/fiona1-1, FIONA1:FIONA1-Flag/fiona1-1 and FIONA1:FIONA1m-Flag/fiona1-1) were obtained by transforming plasmids into the fiona1-1. The transgenic lines described are simplified as FIONA1:FIONA1/fiona1-1 and FIONA1:FIONA1m/fiona1-1 in this paper.
Surface-sterilized Arabidopsis seeds were plated on half-strength Murashige–Skoog (1/2 MS) (PhytoTechnology Laboratories, USA) and incubated at 4 °C for 2 days. The seeds then were grown the under different lighting conditions. In terms of flowering phenotype, the seeds were grown at 22 °C under long-day (LD) conditions (16 h light/8 h dark, 80 μmol m−2 s−1) or short-day (SD) conditions (8 h light/16 h dark, 350 μmol m−2 s−1) for 12 days, then the seedlings were transferred into the soil until they bloom under same conditions, respectively. To observe the morphology of hypocotyl, the plants were grown under continuous red light (30 μmol m−2 s−1), far-red light (11 μmol m−2 s−1), blue light (2 μmol m−2 s−1), white light (30 μmol m−2 s−1), or darkness at 22 °C.
Seedlings were collected from 12-day-old plants grown on 1/2MS nutrient agar plates at Zeitgeber time 13 (ZT13) of LD condition and Zeitgeber time 10 (ZT10) of SD condition. The rosette leaves and cauline leaves were harvested at flowering time. Flowers and buds were collected randomly from 5-week-old plants and divided into three biological replicates. The root and juvenile siliques were collected from 2-week-old seedlings and 7-week-old plants respectively. All samples described above were used for LC-MS/MS or RT-qPCR analyses. In addition, 12-day-old seedlings were collected every 3 h during one photoperiod cycles under LD and SD conditions, and subsequently the expression levels of photoperiod - related genes were detected via RT-qPCR for the diurnal course. The primers used for RT-qPCR are shown in Additional file 9: Table S7.
Phenotypic analysis
The epidermal cells of hypocotyl were photographed by laser scanning confocal microscopy (Zeiss LSM 700, Germany). The hypocotyl length and cell length were measured using ImageJ software (http://imagej.nih.gov/ij/).The flowering phenotype under LD and SD conditions and hypocotyl of seedlings grown under various light conditions as indicated were photographed with a digital camera. Flowering time was determined as the number of days before the first flower opened and the number of rosette leaves at flowering.
Extraction of nuclear total RNA, U6 snRNA, and tRNA
According to a published method [78], we collected 12-day-old Col-0 and fiona1-1 seedlings (one biological repeat per 3 g) to perform nuclear-cytoplasmic fractionation. The final cytoplasmic fraction (supernatant) and the pellet (nuclear fraction) were subjected to western blotting respectively to determine whether both of that were completely separated (Additional file 1: Fig.S3a). As a quality control of the fractionation procedure, PEPC (phosphoenolpyruvate carboxylase) and HSP90, as the cytoplasmic markers, were detected using anti-PEPC polyclonal antibody (Huaxingbio, China) and anti-HSP90 polyclonal antibody (Huaxingbio, China), and histone H3 (Sigma-Aldrich, USA) was used as the nuclear marker. An appropriate amount of SuperTRIgent was added to the isolated nucleus to extract the total RNA in the nucleus.
The 8 μg total RNA from 12-day-old Col-0 and fiona1-1 seedlings were analyzed with 10% TBE gel analysis. The U6 or tRNA bands were cut and extracted for LC-MS/MS analysis (Additional file 1: Fig. S3b).
LC-MS/MS for m6A quantification
In total, 100 or 200 ng of RNA was digested with 1 U Nuclease P1 in 40 μL of buffer containing 10% 0.1 M ammonium acetate NH4AC (pH 5.3) at 42 °C for 3 h, followed by the addition of 1 U Shrimp Alkaline Phosphatase (NEB, USA) and 4.5 μl 1 M of 2-(N-morpholino) ethanesulfonic acid (MES). The mixture was incubated at 37 °C for an additional 3 h. The samples were then centrifuged at 15,000 rpm for 30 min, and the aqueous phase was injected into an LC-MS/MS system. Nucleosides were separated using a UPLC pump (Shimadzu, Japan) with a ZORBAX SB-Aq column (Agilent, USA) and analyzed by MS/MS using a Triple QuadTM 5500 (AB SCIEX, USA) mass spectrometer running in positive ion mode and the multiple reaction-monitoring (MRM) feature. MS parameters were optimized for m6A detection. Nucleosides were quantified using the nucleoside-to-base ion mass transitions of m/z 268.0 to 136.0 (A), m/z 282.0 to 150.1 (m6A), m/z 244.0 to 112.0 (C), m/z 284.0 to 152.0 (G), m/z 245.0 to 113.1 (U). Standard curves were generated using a concentration series of pure commercial nucleosides (Sigma-Aldrich, USA) analyzed using the same method. Concentrations of nucleosides and m6A/A ratio in samples were calculated by fitting the signal intensities to the standard curves.
Expression and purification of FIONA1 protein
The MBP tag and coding sequence of FIONA1 were cloned into PET28a vector containing His tag for protein expression and purification. The MBP tag can promote the expression and stability of the recombinant protein. The recombinant plasmid containing MBP-FIONA1-His was transfected into E. coli strain BL-21 Gold competent cells. The E. coli cells were grown at 37 °C to an OD600 of 0.6–0.8, and recombinant protein expression was then induced at 18 °C with 500 nM IPTG for 20 h. Then the pellet from each 2 L culture was collected, resuspended in 30 mL of lysis buffer (10 mM Tris-HCl, pH 8.0, 500 mM NaCl, 1 mM PMSF, 3 mM DTT, 5% glycerol), and sonicated for 10 min. The sample was centrifuged at 13,000 rpm for 30 min, and the supernatant was filtered through a 0.22-μm filter (Millipore), then loaded on a Ni-NTA column (GE Healthcare). After washing in 20 ml Buffer A (10 mM Tris pH 7.9, 150 mM NaCl) and then 20 ml 8% Buffer B (10 mM Tris-HCl pH 7.9, 150 mM NaCl, 500 mM imidazole), protein was eluted by Buffer B. The collected fraction was then purified by a Superdex 75 gel-filtration column (GE Healthcare, 10 mM Tris-HCl pH 7.9, 150 mM NaCl, and 3 mM DTT. Protein was concentrated into 17.8 mg ml− 1 and 20% glycol was added. Aliquots of protein were frozen by liquid N2 then stored in − 80 °C.
Biochemistry assay for m6A methyltransferase activity in vitro
The in vitro methyltransferase activity following a previously published method [31]. In short, the activity assay was performed in a standard 50 μL of reaction mixture containing the following components: 0.15 nmol RNA oligos, 1.5 nmol (for RNA1 and RNA2) or 3 nmol (for RNA3-RNA6) fresh recombinant FIONA1 protein, 0.8 mM d3-SAM, 80 mM KCl, 1.5 mM MgCl2, 0.2 U μL− 1 RNasin, 2 mM DTT, 4% glycerol, and 15 mM HEPES (pH 8.0). Prior to the reaction, the RNA probes were annealed with a program of (i) 90 °C for 3 min and (ii) − 2 °C/cycle for 40 cycles within 30 min. The reaction was incubated at 16 °C for 12 h. The resultant RNA was recovered by phenol/chloroform extraction followed by ethanol precipitation and was digested by nuclease P1 and alkaline phosphatase for QQQ LC/MS/MS analysis. The formation of d3-m6A (the nucleoside-to-base ion mass transitions of 285 to 153) in oligo RNAs through methylation was used for accurate MS quantification. The peak area ratio of d3-m6A versus one G in the probe (as internal control) was used to evaluate the methylation activities of FIONA1 towards different RNA oligos. The RNA oligos (RNA1-RNA6) sequence are listed in Additional file 9: Table S7.
FTO-assisted SELECT method
The FTO-assisted SELECT method is used for determining a putative m6A site which is m6A-modified in mRNAs and lncRNAs from biological samples and the m6A fraction at biological sites [51]. In this method, m6A in RNA is selected twice in a one-tube reaction. In the first of two selection steps, an m6A mark hinders the ability of DNA polymerase to elongate the target sequence by preventing the addition of a thymidine on the “Up Probe” opposite to the m6A site. In the second selection step, m6A marks that are present in the RNA template selectively prohibit DNA-ligase-catalyzed nick ligation between the elongated Up Probe and Down Probe. The final elongated and ligated products are then quantified by qPCR.
Total RNA was treated with FTO following a previously published method [51]. FTO-treated and untreated total RNA were mixed with 800 fmol Up Primer, 800 fmol Down Primer, and 1 pmol dTTP in 1 × CutSmart Buffer. The primers and RNA were annealed by incubating under the following conditions: 90 °C for 1 min, 80 °C for 1 min, 70 °C for 1 min, 60 °C for 1 min, 50 °C for 1 min, and then 48 °C for 6 min hold. Then, 5 μl mixture of 0.001 U Bst 2.0 WarmStart DNA Polymerase in 1× CutSmart Buffer was added in the former mixture. The reaction was incubated at 48 °C for 20 min and kept at 35 °C. Subsequently, a 10 μl mixture containing 0.5 U SplintR ligase and 10 nmol ATP was added to the final volume 20 μl. The reaction mixture was incubated at 35 °C for 15 min, denatured at 95 °C for 5 min, and then kept at 4 °C. RT-qPCR was performed with 2 μl reaction mixture as per the DNA template. Data was analyzed with QuantStudio RT-qPCR Software v.1.3. The detected results from the m6A site were corrected by the neighboring control site. All primers used in SELECT are listed in Additional file 9: Table S7.
Gene expression analysis by RT-qPCR
One microgram of total RNA was extracted from different plant tissues and reverse transcribed into cDNA according to the kit mentioned above. RT-qPCR was carried out using Hieff qPCR SYBR Green Master Mix (Low Rox) (Yeasen, China) on a ViiA 7 Dx instrument (Applied Biosystems, USA). The relative expression levels were determined based on ACTIN2 or TUB2 as the internal control. The 2−ΔΔCT method was used to calculate the gene expression levels. The qPCR primers involving all genes are listed in Additional file 9: Table S7.
M6A-Seq
m6A sequencing was carried out according to previously described m6A-seq method with only slight changes [54]. Briefly, 5 μg of poly(A)+ RNA was enriched from total RNA of 12-day-old Col-0 and fiona1-1 seedlings at the ZT13 stage using a DynabeadsTM mRNA Direct TM kit (Invitrogen, USA). The poly(A)+ RNA samples were then fragmented into ~ 100-nt-nucleotide-long fragments using RNA Fragmentation Reagents (Ambion, USA). Fifty nanograms fragmented poly(A)+ RNA samples were as input for RNA-seq. The left mRNA samples were programmed according to Instruction Manual of N6-Methyladenosine Enrichment Kit (NEB, USA) for enrichment of m6A-containing fragments. Input poly(A)+ RNA and immunoprecipitated poly(A)+ RNA were used to construct libraries with a NEBNext Ultra II RNA Library Prep Kit. Sequencing was performed on the Illumina HiSeq X Ten platform. Read numbers for two biological replicates are summarized in Additional file 3: Table S1.
Analysis of m6A-seq data
For m6A profiling, sequencing reads were trimmed and mapped to the reference genome (TAIR10) by using Cutadapt (v1.18 ) [79], and the length of trimmed reads ≥ 20 nt were retained. Clean reads were mapped to the reference genome (TAIR10) with HISAT2 (v2.1.0) [80]. Picard Toolkit was employed to remove PCR duplication. The m6A-enriched regions in Col-0 and fiona1-1 were identified using the MACS2 [81] peak-calling algorithm based on enrichment criteria (IP/Input) ≥ 5 and FDR < 0.05. FIONA1-dependent m6A peaks were identified by exomePeak [82] based on enrichment criteria of fold change < 1 and FDR < 0.05. We used Bedtools [83] and python scripts (https://github.com/joybio/m6A-seq/tree/main/feature_annotation/) for peak annotation. GO functional annotations (GO enrichment) were performed using DAVID.
Analysis of RNA-seq data
Sequencing reads were trimmed and mapped to the reference genome (TAIR10) by using Cutadapt (v1.18) [79] and HISAT2 (v2.1.0) [80], respectively. The differentially expressed genes between fiona1-1 and Col-0 were screened by R package (edgR) based on a cutoff criterion of | log2(fold change) | > 0.48 and P value < 0.05. We used rMATS [61] to test the effects of fiona1 on global splicing. The Col-0 and fiona1-1 were compared using the --cstat parameter set to 0.1, summary outputs filtered by FDR < 0.01.
Measurement of SAM contents
The 12-day-old seedlings were used for the SAM concentration measurements. In short, 100 mg seedlings from different treatment conditions were ground into powder with in nitrogen. We added 300 μl PBS buffer (pH 7.4) to 100 mg plant powder, and vortexed for 1 min, extracted for 30 min on ice, and then centrifuged at 4 °C for 10 min at 12,000g. The supernatant was filtered and then analyzed via Plant SAM ELISA Kit (Shuangying, China). After the reaction is completed according to the kit, the OD value of samples is measured at 450 nm by enzyme labeling instrument, and the concentration of SAM is calculated according to the formula. The experiments were performed in three independent biological replicates with technical triplicate.
Gene specific of m6A-IP-qPCR
To determine mRNA methylation level of individual genes in different plant lines, we performed an m6A antibody IP-enrichment according to previous reports [54] with minor modification. The 300~400 ng of mRNA (for every biological sample) was extracted from 12-day-old Col-0 and fiona1-1 seedlings at ZT13 seeing above m6A-seq method. Add 5 pg of single strand of oligonucleotide containing m6A as spike in to these RNA samples. Since its m6A level is determined, it can be used as a control for normalization. Specifically, 40 ng mRNA was saved as input sample, the rest mRNA was incubated in 200 μl IP buffer (150 Mm NaCl, 0.1% NP-40, 10 mM Tris, pH 7.4, 100 U RiboLock RNase Inhibitor ( Thermo) at 4 °C for 4 h, which contained complex of m6A antibody (Synaptic Systems, Germany) and Dynabeads® Protein A (Invitrogen, USA). The Dynabeads were washed three times with IP buffer, and then m6A IP portion was eluted by 200 and 150 μL m6A-elute buffer (IP buffer, 6.7 mM m6A, 30 U RNase inhibitor) with incubating and shaking at 4 °C for 1 h, respectively. Finally, m6A IP portion was washed with 50 μl IP buffer once again, the supernatant is preserved. The total 400 μl supernatant was recovered by phenol-chloroform extraction and 70% ethanol precipitation. The resultant RNA concentration was measured with EqualbitTM RNA HS Assay Kit (Vazyme, China). The input mRNA was further analyzed by RT-qPCR along with the m6A-IP mRNA using primers listed in Additional file 9: Table S7. The relative enrichment of m6A in each sample was calculated by normalizing the value of amplification cycle (Cq) of the m6A-IP portion to the Cq of the corresponding input portion.
In vivo RNA immunoprecipitation (RIP)-qPCR
RNA immunoprecipitation was performed according to previously described method [24]. Briefly, 12-day-old FIONA1:FIONA1-Flag/fiona1-1 seedlings were harvested at ZT13, fixed 1% formaldehyde for 15 min under vacuum, and terminated with 150 mM glycine for additional 10 min. Two grams of fixed plant material was ground and homogenized in 2 ml of lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM KCl, 5 mM EGTA, 1 mM PMSF, 0.1 U/μl Ribolock RNase inhibitor, and 1 × Roche Protease inhibitor Cocktail). Take part of lysate was saved as the input sample. The remainder was divided into two equal volumes and subsequently immunoprecipitated with anti-Flag M2 magnetic beads (Sigma-Aldrich, USA) or normal rabbit IgG (Cell Signaling Technology, USA) bound to Dynabeads Protein A, respectively. After washing and ethanol precipitation, the recovered RNA fractions were used for reverse transcribed into cDNA to calculate the relative enrichment fold via qRT–PCR. AT2G07689, which does not have an m6A peak from m6A profiling data, was used as the internal control.
mRNA stability measurements
An mRNA stability measurement assay in vivo was performed as previously described [84] with minor modification. Briefly, 12-day-old Col-0, fiona1-1 Arabidopsis seedlings grown on 1/2 MS medium were transferred to 10-cm Petri dishes containing 1/2 MS liquid medium at ZT13. After 30 min incubation, 0.2 mM actinomycin D was added to the buffer. The tissues were collected at 1 h after the transcription inhibitor was added; these samples are referred to as 0 h samples. The 3 h and 6 h samples were collected and immediately frozen in liquid nitrogen. The total RNA was isolated from these tissues at 3 different time points, and the remaining mRNA levels were quantified by RT-qPCR with gene-specific qPCR primers (Additional file 9: Table S7). 18S RNA was used as the internal control, and AT2G07689 was used as a negative control [24].
Supplementary Information
Additional file 1: Figure S1. Phylogenetic relationships and sequence alignment of METTL16 proteins among different species. Figure S2. The generation of fiona1 mutants by CRISPR/Cas9 genome editing. Figure S3. Separation of nuclear-cytoplasmic fractions, U6 snRNA, and tRNA. Figure S4. The transgenic plants associated with FIONA1. Figure S5. The FTO-assisted SELECT for identification of m6A site in U6 snRNA. Figure S6. LC-MS/MS quantification of the m6A/A ratios in rRNA and tRNA isolated from Col-0 and fiona1-1 seedlings. Figure S7. Relative expression levels of m6A-related regularity genes in Col-0 and fiona1-1 plants. Figure S8. SDS-PAGE gel showing the purified recombinant Arabidopsis FIONA1 proteins for in vitro methylation assays. Figure S9. FIONA1 is a nuclear localized protein in Arabidopsis. Figure S10. FIONA1 is ubiquitously expressed in diverse Arabidopsis tissues. Figure S11. Hypocotyl phenotypes of the indicated genotypic seedlings under continuous blue light, white light, and dark. Figure S12. Disruption of FIONA1 leads to hyposensitivity of fiona1 mutants to red and far-red lights. Figure S13. Disruption of FIONA1 leads to early flowering. Figure S14. Transcriptome m6A profiling in Col-0 and fiona1-1. Figure S15. Representative integrative genomics viewer of hypomethylated m6A peaks in fiona1-1 and verification of m6A-seq results. Figure S16. m6A-binding motif identified by MEME. Figure S17. Differences in methylation sites between FIONA1 and m6A writer complex containing MTA/MTB/FIP37. Figure S18. Homologous sequence alignment between mammalian MAT2A gene and Arabidopsis SAM synthetases MAT1 (AT1G02500), MAT2 (AT4G01850), MAT3 (AT2G36880) and MAT4 (AT3G17390). Figure S19. m6A level and transcriptional expression results of SAM synthetase genes MAT1 (AT1G02500), MAT2 (AT4G01850), MAT3 (AT2G36880) and MAT4 (AT3G17390) in fiona1-1 and Col-0 plants. Figure S20. FIONA1 does not affect the transcript expression levels of Arabidopsis SAM synthetases under normal and high SAM conditions. Figure S21. Genomics viewer showing the m6A-seq results for PIF4, CRY2, and FLC mRNA in Col-0 and fiona1-1. Figure S22. The FTO-assisted SELECT for identification of m6A sites in PIF4 and CRY2 mRNA. Figure S23. SELECT for identification of the FIONA1-targeted m6A sites in PIF4 and CRY2 transcripts. Figure S24. The expression level of CRY2, CO, and FLC in the indicated genotypic plants. Figure S25. The relative expression levels of PIF4 in the indicated genotypic plants under SD, Rc, and FRc. Figure S26. The expression level of FT in the indicated genotypic plants.
Additional file 2. Uncropped western blotting and gel analysis.
Additional file 3: Table S1. Statistics of mapping rate.
Additional file 4: Table S2. Confident m6A peaks identified by m6A-seq in WT.
Additional file 5: Table S3. Confident m6A peaks identified by m6A-seq in fiona1-1.
Additional file 6: Table S4. Hypomethylated m6A peaks identified in fiona1-1.
Additional file 7: Table S5. Differentially expressed genes identified in fiona1-1 compared to WT by RNA-seq.
Additional file 8: Table S6. Summary of splicing events altered in the fiona1-1.
Additional file 9: Table S7. Primers and oligonucleotide probes used in this study.
Acknowledgements
We would like to acknowledge H. Chen for providing illumination incubator.
Review history
The review history is available as Additional file 10.
Peer review information
Wenjing She was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Authors’ contributions
G.J. conceived the project; C.W. performed the experiments with the help of P.S., W.Z., Q.L., and Q.Y.; J.Y. analyzed the sequencing data; G.J. and C.W. designed the experiments, interpreted the results, and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Basic Research Program of China (2019YFA0802201 and 2017YFA0505201), the National Natural Science Foundation of China (nos. 21822702, 21820102008, 92053109, and 21432002), and Beijing Natural Science Foundation (Z200010).
Availability of data and materials
The raw sequencing data reported in this paper have been deposited in the Genome Sequence Archive in the National Genomics Data Center (NGDC), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA004052) which is publicly accessible at https://ngdc.cncb.ac.cn/gsa [85]. The published sequencing data related with FIP37 can be downloaded from the NCBI database (GSE75508) [38]. All the other datasets supporting the conclusions in this study are included in the article and the Additional files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chunling Wang and Junbo Yang contributed equally to this work.
References
- 1.de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451(7177):480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 2.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405(6785):462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 3.Leivar P, Monte E. PIFs: systems integrators in plant development. Plant Cell. 2014;26(1):56–78. doi: 10.1105/tpc.113.120857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galvao VC, Fankhauser C. Sensing the light environment in plants: photoreceptors and early signaling steps. Curr Opin Neurobiol. 2015;34:46–53. doi: 10.1016/j.conb.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Heng Y, Jiang Y, Zhao X, Zhou H, Wang X, Deng X, et al. Correction for Heng et al., BBX4, a phyB-interacting and modulated regulator, directly interacts with PIF3 to fine tune red light-mediated photomorphogenesis. Proc Natl Acad Sci U S A. 2020;117(8):4429–4430. doi: 10.1073/pnas.2001373117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, Okamura H. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155(4):793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 8.Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540(7632):301–304. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, Tienda SM, Chryplewicz A, Zhu AC, Yang Y, Huang JT, Chen SM, Xu ZG, Leng XH, Yu XC, Cao J, Zhang Z, Liu J, Lengyel E, He C. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20(9):1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, Wang X, Ma HL, Huang CM, Yang Y, Huang N, Jiang GB, Wang HL, Zhou Q, Wang XJ, Zhao YL, Yang YG. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61(4):507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 13.Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, Jin KX, Wang X, Huang CM, Fu Y, Ge XM, Song SH, Jeong HS, Yanagisawa H, Niu Y, Jia GF, Wu W, Tong WM, Okamoto A, He C, Danielsen JMR, Wang XJ, Yang YG. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24(12):1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RPG, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15(5):293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 16.Yue YN, Liu JZ, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2020;29(13):1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16(2):191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28(6):616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng H, Li S, Zhang X, Sui N. Functional implications of active N6-methyladenosine in plants. Front Cell Dev Biol. 2020;8:291. doi: 10.3389/fcell.2020.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang Z, Riaz A, Chachar S, Ding Y, Du H, Gu X. Epigenetic modifications of mRNA and DNA in plants. Mol Plant. 2020;13(1):14–30. doi: 10.1016/j.molp.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Arribas-Hernandez L, Brodersen P. Occurrence and functions of m6A and other covalent modifications in plant mRNA. Plant Physiol. 2020;182(1):79–96. doi: 10.1104/pp.19.01156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arribas-Hernandez L, Bressendorff S, Hansen MH, Poulsen C, Erdmann S, Brodersen P. An m6A-YTH module controls developmental timing and morphogenesis in Arabidopsis. Plant Cell. 2018;30(5):952–967. doi: 10.1105/tpc.17.00833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scutenaire J, Deragon J-M, Jean V, Benhamed M, Raynaud C, Favory J-J, Merret R, Bousquet-Antonelli C. The YTH domain protein ECT2 is an m6A reader required for normal trichome branching in Arabidopsis. Plant Cell. 2018;30(5):986–1005. doi: 10.1105/tpc.17.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei LH, Song P, Wang Y, Lu Z, Tang Q, Yu Q, Xiao Y, Zhang X, Duan HC, Jia G. The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell. 2018;30(5):968–985. doi: 10.1105/tpc.17.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song P, Yang J, Wang C, Lu Q, Shi L, Tayier S, Jia G. Arabidopsis N6-methyladenosine reader CPSF30-L recognizes FUE signal to control polyadenylation site choice in liquid-like nuclear body. Mol Plant. 2021;14(4):571–587. doi: 10.1016/j.molp.2021.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Hou Y, Sun J, Wu B, Gao Y, Nie H, Nie Z, Quan S, Wang Y, Cao X, Li S. CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis. Mol Plant. 2021;14(4):688–699. doi: 10.1016/j.molp.2021.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Arribas-Hernández L, Simonini S, Hansen MH, Botterweg Paredes E, Bressendorff S, Dong Y, Østergaard L, Brodersen P. Recurrent requirement for the m6A-ECT2/ECT3/ECT4 axis in the control of cell proliferation during plant organogenesis. Development. 2020;147:dev189134. doi: 10.1242/dev.189134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martínez-Pérez M, Aparicio F, López-Gresa MP, Bellés JM, Sánchez-Navarro JA, Pallás V. Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc Natl Acad Sci U S A. 2017;114(40):10755–10760. doi: 10.1073/pnas.1703139114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duan HC, Wei LH, Zhang C, Wang Y, Chen L, Lu Z, Chen PR, He C, Jia G. ALKBH10B is an RNA N6-methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell. 2017;29(12):2995–3011. doi: 10.1105/tpc.16.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou L, Tian S, Qin G. RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019;20(1):156. doi: 10.1186/s13059-019-1771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, Conrad NK. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169(5):824–835. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell. 2016;63(2):306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, Zou T, Yin P. Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534(7608):575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 35.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, Zhao X, Li A, Yang Y, Dahal U, Lou XM, Liu X, Huang J, Yuan WP, Zhu XF, Cheng T, Zhao YL, Wang X, Danielsen JMR, Liu F, Yang YG. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20(5):1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bodi Z, Zhong S, Mehra S, Song J, Graham N, Li H, May S, Fray RG. Adenosine methylation in Arabidopsis mRNA is associated with the 3' end and reduced levels cause developmental defects. Front Plant Sci. 2012;3:48. doi: 10.3389/fpls.2012.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen L, Liang Z, Gu X, Chen Y, Teo ZW, Hou X, et al. N6-methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev Cell. 2016;38(2):186–200. doi: 10.1016/j.devcel.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruzicka K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, et al. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017;215(1):157–172. doi: 10.1111/nph.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347(6225):1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 41.Zhang F, Zhang YC, Liao JY, Yu Y, Zhou YF, Feng YZ, Yang YW, Lei MQ, Bai M, Wu H, Chen YQ. The subunit of RNA N6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice. PLoS Genet. 2019;15(5):e1008120. doi: 10.1371/journal.pgen.1008120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendel M, Delaney K, Pandey RR, Chen KM, Wenda JM, Vagbo CB, et al. Splice site m6A methylation prevents binding of U2AF35 to inhibit RNA splicing. Cell. 2021;184(12):3125–3142. doi: 10.1016/j.cell.2021.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawa H, Abelson J. Evidence for a base-pairing interaction between U6 small nuclear-RNA and the 5' splice site during the splicing reaction in yeast. Proc Natl Acad Sci U S A. 1992;89(23):11269–11273. doi: 10.1073/pnas.89.23.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shima H, Matsumoto M, Ishigami Y, Ebina M, Muto A, Sato Y, Kumagai S, Ochiai K, Suzuki T, Igarashi K. S-Adenosylmethionine synthesis is regulated by selective N6-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 2017;21(12):3354–3363. doi: 10.1016/j.celrep.2017.11.092. [DOI] [PubMed] [Google Scholar]
- 45.Mendel M, Chen KM, Homolka D, Gos P, Pandey RR, McCarthy AA, et al. Methylation of structured RNA by the m6A writer METTL16 is essential for mouse embryonic development. Mol Cell. 2018;71(6):986–1000. doi: 10.1016/j.molcel.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koh CWQ, Goh YT, Goh WSS. Atlas of quantitative single-base-resolution N6-methyl-adenine methylomes. Nat Commun. 2019;10(1):5636. doi: 10.1038/s41467-019-13561-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J, Kim Y, Yeom M, Kim JH, Nam HG. FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell. 2008;20(2):307–319. doi: 10.1105/tpc.107.055715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015;16(1):144. doi: 10.1186/s13059-015-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doxtader KA, Wang P, Scarborough AM, Seo D, Conrad NK, Nam Y. Structural basis for regulation of METTL16, an S-adenosylmethionine homeostasis factor. Mol Cell. 2018;71(6):1001–1011. doi: 10.1016/j.molcel.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Epstein P, Reddy R, Henning DaB H. The nucleotide sequence of nuclear U6 (4.7 S) RNA. J Biol Chem. 1980;255(18):8901–8906. doi: 10.1016/S0021-9258(18)43587-9. [DOI] [PubMed] [Google Scholar]
- 51.Xiao Y, Wang Y, Tang Q, Wei L, Zhang X, Jia G. An elongation- and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-specific N6-methyladenosine modification. Angew Chem Int Ed Engl. 2018;57(49):15995–16000. doi: 10.1002/anie.201807942. [DOI] [PubMed] [Google Scholar]
- 52.Luo GZ, MacQueen A, Zheng G, Duan H, Dore LC, Lu Z, Liu J, Chen K, Jia G, Bergelson J, He C. Unique features of the m6A methylome in Arabidopsis thaliana. Nat Commun. 2014;5(1):5630. doi: 10.1038/ncomms6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome-A and phytochrome-B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994;104(4):1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8(1):176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- 55.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 56.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xi L, Carroll T, Matos I, Luo JD, Polak L, Pasolli HA, Jaffrey SR, Fuchs E. m6A RNA methylation impacts fate choices during skin morphogenesis. Elife. 2020;9:e56980. doi: 10.7554/eLife.56980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y, Zheng H, Klungland A, Yan W. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3'-UTR mRNAs in male germ cells. Proc Natl Acad Sci U S A. 2018;115(2):E325–E333. doi: 10.1073/pnas.1717794115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84(1):291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan C, Wan R, Shi Y. Molecular mechanisms of pre-mRNA splicing through structural biology of the spliceosome. Cold Spring Harb Perspect Biol. 2019;11(1):a032409. doi: 10.1101/cshperspect.a032409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A. 2014;111(51):E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303(5660):1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 63.Huq E, Quail P. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002;21(10):2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lorrain S, Trevisan M, Pradervand S, Fankhauser C. Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J. 2009;60(3):449–461. doi: 10.1111/j.1365-313X.2009.03971.x. [DOI] [PubMed] [Google Scholar]
- 65.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, Hu YC, Hüttelmaier S, Skibbe JR, Su R, Deng X, Dong L, Sun M, Li C, Nachtergaele S, Wang Y, Hu C, Ferchen K, Greis KD, Jiang X, Wei M, Qu L, Guan JL, He C, Yang J, Chen J. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, Carr SA, Lander ES, Fink GR, Regev A. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155(6):1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li YL, Wang XL, Li CP, Hu SN, Yu J, Song SH. Transcriptome-wide N6-methyladenosine profiling of rice callus and leaf reveals the presence of tissue-specific competitors involved in selective mRNA modification. RNA Biol. 2014;11(9):1180–1188. doi: 10.4161/rna.36281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5' terminus of hela-cell messenger-RNA. Cell. 1975;4(4):379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 69.Zhou L, Tang R, Li X, Tian S, Li B, Qin G. N6-methyladenosine RNA modification regulates strawberry fruit ripening in an ABA-dependent manner. Genome Biol. 2021;22(1):168. doi: 10.1186/s13059-021-02385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu J, Cai J, Park SJ, Lee K, Li Y, Chen Y, Yun JY, Xu T, Kang H. N6-Methyladenosine mRNA methylation is important for salt stress tolerance in Arabidopsis. Plant J. 2021;106(6):1759–1775. doi: 10.1111/tpj.15270. [DOI] [PubMed] [Google Scholar]
- 71.Liu Y, Li X, Ma D, Chen Z, Wang JW, Liu H. CIB1 and CO interact to mediate CRY2-dependent regulation of flowering. EMBO Rep. 2018;19(10):e45762. doi: 10.15252/embr.201845762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11(5):949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, et al. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18(11):2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shalitin D, Yang HY, Mockler TC, Maymon M, Guo HW, Whitelam GC, Lin C. Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature. 2002;417(6890):763–767. doi: 10.1038/nature00815. [DOI] [PubMed] [Google Scholar]
- 75.Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006;46(2):183–192. doi: 10.1111/j.1365-313X.2006.02686.x. [DOI] [PubMed] [Google Scholar]
- 76.Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286(5446):1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- 77.Choi H, Oh E. PIF4 integrates multiple environmental and hormonal signals for plant growth regulation in Arabidopsis. Mol Cells. 2016;39(8):587–593. doi: 10.14348/molcells.2016.0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang W, Ye R, Xin Y, Fang X, Li C, Shi H, Zhou X, Qi Y. An importin beta protein negatively regulates MicroRNA activity in Arabidopsis. Plant Cell. 2011;23(10):3565–3576. doi: 10.1105/tpc.111.091058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 80.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meng J, Cui X, Rao MK, Chen Y, Huang Y. Exome-based analysis for RNA epigenome sequencing data. Bioinformatics. 2013;29(12):1565–1567. doi: 10.1093/bioinformatics/btt171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seo E, Yu J, Ryu KH, Lee MM, Lee I. Werewolf, a regulator of root hair pattern formation, controls flowering time through the regulation of FT mRNA stability. Plant Physiol. 2011;156(4):1867–1877. doi: 10.1104/pp.111.176685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang C, Yang J, Song P, Zhang W, Lu Q, Yu Q, et al. FIONA1 is an RNA N6-methyladenosine methyltransferase affecting Arabidopsis photomorphogenesis and flowering. NGDC. https://ngdc.cncb.ac.cn/gsa/browse/CRA004052. 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Phylogenetic relationships and sequence alignment of METTL16 proteins among different species. Figure S2. The generation of fiona1 mutants by CRISPR/Cas9 genome editing. Figure S3. Separation of nuclear-cytoplasmic fractions, U6 snRNA, and tRNA. Figure S4. The transgenic plants associated with FIONA1. Figure S5. The FTO-assisted SELECT for identification of m6A site in U6 snRNA. Figure S6. LC-MS/MS quantification of the m6A/A ratios in rRNA and tRNA isolated from Col-0 and fiona1-1 seedlings. Figure S7. Relative expression levels of m6A-related regularity genes in Col-0 and fiona1-1 plants. Figure S8. SDS-PAGE gel showing the purified recombinant Arabidopsis FIONA1 proteins for in vitro methylation assays. Figure S9. FIONA1 is a nuclear localized protein in Arabidopsis. Figure S10. FIONA1 is ubiquitously expressed in diverse Arabidopsis tissues. Figure S11. Hypocotyl phenotypes of the indicated genotypic seedlings under continuous blue light, white light, and dark. Figure S12. Disruption of FIONA1 leads to hyposensitivity of fiona1 mutants to red and far-red lights. Figure S13. Disruption of FIONA1 leads to early flowering. Figure S14. Transcriptome m6A profiling in Col-0 and fiona1-1. Figure S15. Representative integrative genomics viewer of hypomethylated m6A peaks in fiona1-1 and verification of m6A-seq results. Figure S16. m6A-binding motif identified by MEME. Figure S17. Differences in methylation sites between FIONA1 and m6A writer complex containing MTA/MTB/FIP37. Figure S18. Homologous sequence alignment between mammalian MAT2A gene and Arabidopsis SAM synthetases MAT1 (AT1G02500), MAT2 (AT4G01850), MAT3 (AT2G36880) and MAT4 (AT3G17390). Figure S19. m6A level and transcriptional expression results of SAM synthetase genes MAT1 (AT1G02500), MAT2 (AT4G01850), MAT3 (AT2G36880) and MAT4 (AT3G17390) in fiona1-1 and Col-0 plants. Figure S20. FIONA1 does not affect the transcript expression levels of Arabidopsis SAM synthetases under normal and high SAM conditions. Figure S21. Genomics viewer showing the m6A-seq results for PIF4, CRY2, and FLC mRNA in Col-0 and fiona1-1. Figure S22. The FTO-assisted SELECT for identification of m6A sites in PIF4 and CRY2 mRNA. Figure S23. SELECT for identification of the FIONA1-targeted m6A sites in PIF4 and CRY2 transcripts. Figure S24. The expression level of CRY2, CO, and FLC in the indicated genotypic plants. Figure S25. The relative expression levels of PIF4 in the indicated genotypic plants under SD, Rc, and FRc. Figure S26. The expression level of FT in the indicated genotypic plants.
Additional file 2. Uncropped western blotting and gel analysis.
Additional file 3: Table S1. Statistics of mapping rate.
Additional file 4: Table S2. Confident m6A peaks identified by m6A-seq in WT.
Additional file 5: Table S3. Confident m6A peaks identified by m6A-seq in fiona1-1.
Additional file 6: Table S4. Hypomethylated m6A peaks identified in fiona1-1.
Additional file 7: Table S5. Differentially expressed genes identified in fiona1-1 compared to WT by RNA-seq.
Additional file 8: Table S6. Summary of splicing events altered in the fiona1-1.
Additional file 9: Table S7. Primers and oligonucleotide probes used in this study.
Data Availability Statement
The raw sequencing data reported in this paper have been deposited in the Genome Sequence Archive in the National Genomics Data Center (NGDC), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA004052) which is publicly accessible at https://ngdc.cncb.ac.cn/gsa [85]. The published sequencing data related with FIP37 can be downloaded from the NCBI database (GSE75508) [38]. All the other datasets supporting the conclusions in this study are included in the article and the Additional files.