Abstract
Innate immune mechanisms are central players in response to the binding of pathogens to pattern-recognition receptors providing a crucial initial block on viral replication. Moreover, innate immune response mobilizes cells of the cellular-mediated immune system, which develop into effector cells that promote viral clearance. Here, we observed circulating leukocyte T cell response in healthy subjects, COVID-19 infected, and in healthy vaccinated subjects. We found a significant CD8+ T cells (p < 0,05) decrease and an augmented CD4+/CD8+ ratio (p < 0,05) in COVID-19 infected group compared with vaccinated subjects. In addition, healthy vaccinated subjects have a significant increased expression of CD8+ T cells, and a reduction of CD4+/CD8+ ratio with respect to subjects previously COVID-19 infected. Central Memory and Terminal Effector Memory cells (TEMRA) increased after vaccine but not among groups.
Keywords: COVID-19 infection, COVID-19 mRNA BNT162b2 (Pfizer-BioNTech) vaccine, T cell immune response, Flow cytometry characterization
1. Introduction
T cells play a relevant role in the cellular-mediated immune response to viral infection (Napoli et al., 2021; Lund et al., 2008; Swain et al., 2012; Liston et al., 2021; Yatim et al., 2021). In particular, viruses can activate CD4+ T cells which are strong effectors of the cellular-mediated immune responses for their ability to help B cell-related antibody production and to interact with CD8+ T cells driving the cytotoxic response able to kill the infected cells (Swain et al., 2012). Indeed, CD8+ T cells directly recognize viral peptides which are presented at the surfaces of infected cells causing apoptosis and preventing the virus from spreading further (Napoli et al., 2021; Swain et al., 2012). When the infection is resolved, the majority of CD4+ T cells die but a smaller population of memory CD4+ T cells persists long-term (Williams et al., 2008). These memory CD4+ T cells respond more rapidly and effectively during viral re-infection and give their potential contribution to immunity when they are induced by either infection or vaccination. In the viral pneumonia context, T cell characterization of patients infected with SARS-CoV (2002/03) demonstrated that CD4+ T cell responses strongly correlated with improved clinical outcomes (Channappanavar et al., 2014; Li et al., 2008). Recent evidence has shown that SARS-CoV-2-specific T cells are present relatively early and increase over time (Chen and John Wherry, 2020; Marfella et al., 2022). Besides, the presence of pre-existing SARS-CoV-2-reactive T cells in a subset of SARS-CoV-2 naïve healthy subjects is of high interest in the prevention of COVID-19; however, larger-scale prospective cohort studies are needed to assess whether their presence may correlate with the protection or severity of COVID-19. First, we evaluated T cell response in subjects who have been infected by SARS-CoV-2 both asymptomatic and/or with middle/severe symptoms and in healthy subjects. Then, we also documented T cell response in a group that received COVID-19 mRNA BNT162b2 (Pfizer-BioNTech) vaccine.
2. Methods
2.1. Subjects
In this study, we selected three groups of subjects who were enrolled in our ongoing DEMETRA clinical trial (NCT04746521). Basic demographic informations are summarized in Supplementary Table S1. In the first group, we included 10 subjects who were never infected with SARS-CoV-2 (Healthy); the second group included 10 subjects who manifested symptoms after infection (Infected); the third group included 10 healthy subjects who received the vaccine (Vaccinated). Our observations are made about 1 month from second dose of vaccine for healthy subjects and after 2 months from negative nasopharyngeal swabs for COVID-19 disease in infected subjects. Since the kits used in the present study are commercially available and routinely employed in the normal clinical practice, the protocol was just approved by the institutional review board and registered in the NHC web control sites as NCT04746521. In any case, we received the written informed consent from each participant and the study has conducted by the Helsinki declaration.
2.2. Chemiluminescent microparticle immunoassay for SARS-CoV-2 IgG and IgM detection
For healthy and infected subjects, qualitative detection of IgG and IgM was performed by chemiluminescent microparticle immunoassay (CMIA) using SARS-CoV-2 IgG (6R86) and IgM (6R87) Reagent Kit (Abbott Laboratories, Diagnostics Division, Abbott Park, IL 60064 USA). In addition, for healthy vaccinated subjects, quantitative measurement of IgG antibodies against the spike receptor-binding domain (RBD) of SARS-CoV-2 was performed by using SARS-CoV-2 IgG II Quant assay (Abbott Laboratories, Diagnostics Division, Abbott Park, IL 60064 USA).
2.3. Fluorescence reagents for flow cytometer staining
The expression of CD3, CD4, CD8, CD19, and CD16-CD56 on cells in peripheral blood samples was detected by using BD Multitest 6-Color TBNK Reagent (BD Biosciences San Jose, CA, USA) a 6-color direct immunofluorescence reagent to identify and determine the percentages and absolute counts of T, B, and natural killer (NK) cells, as well as the CD4 and CD8 subpopulations of T cells in peripheral blood. Cell counts of lymphocytes were calculated using BD Trucount tubes.
In addition, whole blood samples were stained using preconfigured lyophilized reagent tubes (BD Lyotube TM 8-color CD4 and CD8 bundle, (BD Biosciences San Jose, CA, USA including CD4 specific and CD8 specific “Lyotubes”)): CD4 Lyotube: CD95 FITC/ CCR7 PE / CD3 PerCPCy 5.5/ CD25 PE-Cy7/ CD127 Alexa Fluor 647/ CD45APC-H7/ CD45RA V450 / CD4 V500-C; CD8 Lyotube: CD38 FITC/ CCR7 PE / CD3 PerCPCy 5.5/ CD69 PE-Cy7/ CD127 Alexa Fluor 647/ CD45 APC-H7 / CD45RA V450/ CD8 V500-C.
2.4. Blood sampling and staining procedure
Blood was drawn via sterile venipuncture into EDTA vacutainers (Becton Dickinson, BD). Then, 50 uL of blood were incubated for 15 min with 20 uL of BD Multitest 6-Color TBNK Reagent in Trucount tubes. Samples were lysed with 2 mL of lysing buffer 1× (BD PharmLyse) at room temperature in the dark for 10 min and processed immediately. The second Lyotubes staining procedure carried out with rehydration by using 100 uL of blood and incubated for 5 min before mixing. After an additional 30 min of incubation at room temperature, samples were lysed with 2 mL of lysing buffer 1× in the dark. After washing, samples were resuspended with 800 uL, of buffer (FACS Flow, BD), and processed within 1 h.
2.5. Instrument and data analysis details
Sera sample CMIA was performed by using the ARCHITECT i2000SR system (Abbott Laboratories, Diagnostics Division, Abbott Park, IL 60064 USA) (data not shown).
Flow cytometry characterization was performed by using the BD FACSLyric Flow Cytometer and the analysis was made in conjunction with BD FACSuite Clinical software (v. 1.4) for BD Multitest 6-Color TBNK Reagent with BD Trucount Tubes and BD FACSuite software v. 1.4 for BD Lyotube CD4 and CD8 kit (BD Biosciences San Jose, CA, USA).
BD CS&T beads are used for control and setting of flow cytometer (Colacurci et al., 2020). These beads allow the FACSuite software to automatically characterize, track, and report measurements of supported digital flow cytometers. CS&T beads are dyed with fluorochromes which are excited by the cytometer's lasers. In addition, setting by using BD FC beads for all fluorochromes witch in conjunction with FACSuite Clinical software and CS&T beads, is performed to establish standardized fluorescence compensation for the FACSLyric flow cytometer.
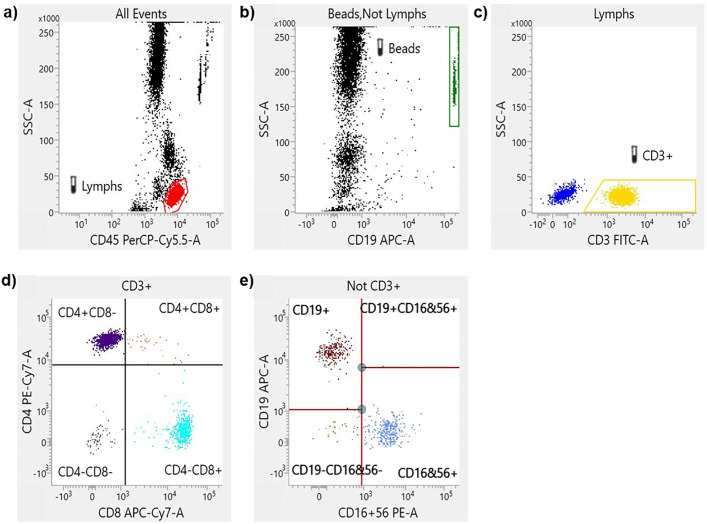
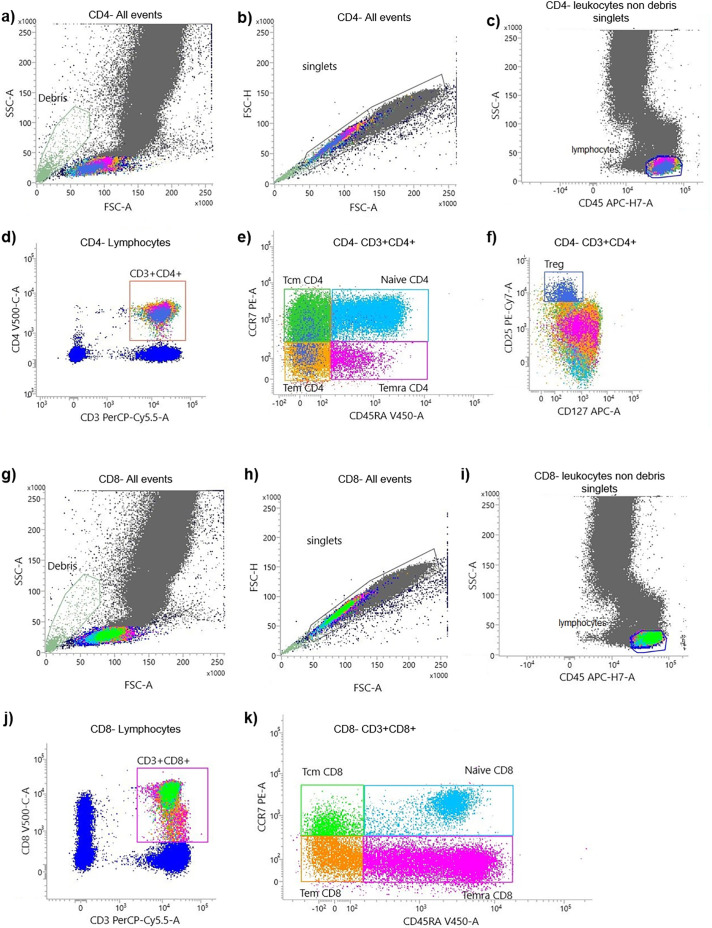
The different flow cytometry gating strategy for each test were described in the Fig. 1, Fig. 2 .
Fig. 1.
Flow cytometry gating strategy of TBNK kit. A representative flow cytometry plot from an infected individual showing: a, gated lymphocytes and monocytes-granulocytes. b, Truecount beads, not Lymphocytes. c, CD3+ T cells, and CD3− lymphocytes. d, T cells (CD3+) are divided basing on CD4+ and CD8+ expression. e, NK cells are CD19− and CD16/CD56+.
Fig. 2.
Flow cytometry gating strategy of maturation stages of T cell by using T CD4 and CD8 lyotubes kit. A representative flow cytometry plot from a vaccinated individual showing: Identification of CD4+ (a, b, c) and CD8+ (g, h, i) leukocytes non debris singlets. After gating on CD3+CD4+ T cells (d) and on CD3+CD8+ T cell (j) we identified CD45RA+CCR7+ naive cells; CD45RA−CCDR7+ central memory T cells (Tcm); CD45RA−CCR7+ effector memory T cells (Tem) and CD45RA+CCR7− terminal differentiated effector memory T cells (Temra) (e). Same gating strategy is applied to T CD8+ cells (k). Regulatory T cells were classified as CD4+CD25+CD127dim (f).
2.6. Statistical analysis
One-way ANOVA analysis of variance was used for the comparison among the three groups (Table 1 and Table 3), and the Bonferroni t-test was used for multiple comparisons among groups (Table 2 ). Statistical significance was set at 5%. The normal distribution of data was evaluated with the Kolmogorov-Smirnov test. P values less than 0.05 were considered statistically significant.
Table 1.
One-way ANOVA variance analysis for TBNK results, p-value ≤ 0.05 is statistically significant
| Factor |
Healthy |
Infected |
Vaccinated |
p-value |
|---|---|---|---|---|
| N | 10 | 10 | 10 | |
| CD4+, mean (SD) | 47.8 (8.5) | 50.7 (7.3) | 43.8 (5.4) | 0.12 |
| CD8+, mean (SD) | 28.9 (7.2) | 20.8 (5.3) | 30.5 (11.5) | 0.035 |
| CD4+/CD8+, mean (SD) | 1.8 (0.6) | 2.7 (1.2) | 1.7 (0.8) | 0.030 |
| NK, mean (SD) | 13.3 (4.5) | 13.5 (6.0) | 14.5 (7.0) | 0.90 |
Table 3.
One-way ANOVA variance analysis for T lyotube results.
| Factor |
Healthy |
Infected |
Vaccinated |
p-value | |
|---|---|---|---|---|---|
| N | 10 | 10 | 10 | ||
| CD4+ | Naive, mean (SD) | 31.5 (12.2) | 39.2 (11.6) | 31.8 (11.7) | 0.28 |
| Tcm, mean (SD) | 37.0 (9.1) | 39.7 (5.7) | 34.3 (10.7) | 0.39 | |
| Tem, mean (SD) | 24.5 (9.8) | 16.9 (7.7) | 24.0 (10.1) | 0.14 | |
| Temra, mean (SD) | 6.3 (7.4) | 2.1 (2.0) | 7.8 (11.3) | 0.26 | |
| Treg, mean (SD) | 3.3 (1.2) | 3.4 (1.0) | 2.4 (0.9) | 0.079 | |
| CD8+ | Naive, mean (SD) | 21.9 (12.1) | 24.9 (10.6) | 19.0 (13.4) | 0.56 |
| Tcm, mean (SD) | 10.8 (5.4) | 11.1 (4.5) | 11.1 (5.5) | 0.99 | |
| Tem, mean (SD) | 24.9 (11.2) | 25.1 (9.4) | 28.9 (12.5) | 0.67 | |
| Temra, mean (SD) | 39.0 (15.2) | 35.6 (14.2) | 39.0 (18.6) | 0.86 | |
Table 2.
Bonferroni t-test for multiple comparisons between groups, p-value ≤ 0.05 is statistically significant
| Factor | (I) soort | (J) soort | Mean Difference (I-J) | Std. Error | P | 95% Confidence Interval |
|
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| CD8+ | Healthy | Infected | 8.1400 | 3.7655 | 0.119 | −1.471 | 17.751 |
| Vaccinated | −1.5500 | 3.7655 | 1.000 | −11.161 | 8.061 | ||
| Infected | Healthy | −8.1400 | 3.7655 | 0.119 | −17.751 | 1.471 | |
| Vaccinated | −9.6900⁎ | 3.7655 | 0.048 | −19.301 | −0.079 | ||
| Vaccinated | Healthy | 1.5500 | 3.7655 | 1.000 | −8.061 | 11.161 | |
| Infected | 9.6900⁎ | 3.7655 | 0.048 | 0.079 | 19.301 | ||
| CD4+/CD8+ | Healthy | Infected | −0.90800 | 0.39017 | 0.083 | −1.9039 | 0.0879 |
| Vaccinated | 0.09400 | 0.39017 | 1.000 | −0.9019 | 1.0899 | ||
| Infected | Healthy | 0.90800 | 0.39017 | 0.083 | −0.0879 | 1.9039 | |
| Vaccinated | 1.00200⁎ | 0.39017 | 0.048 | 0.0061 | 1.9979 | ||
| Vaccinated | Healthy | −0.09400 | 0.39017 | 1.000 | −1.0899 | 0.9019 | |
| Infected | −1.00200⁎ | 0.39017 | 0.048 | −1.9979 | −0.0061 | ||
3. Results
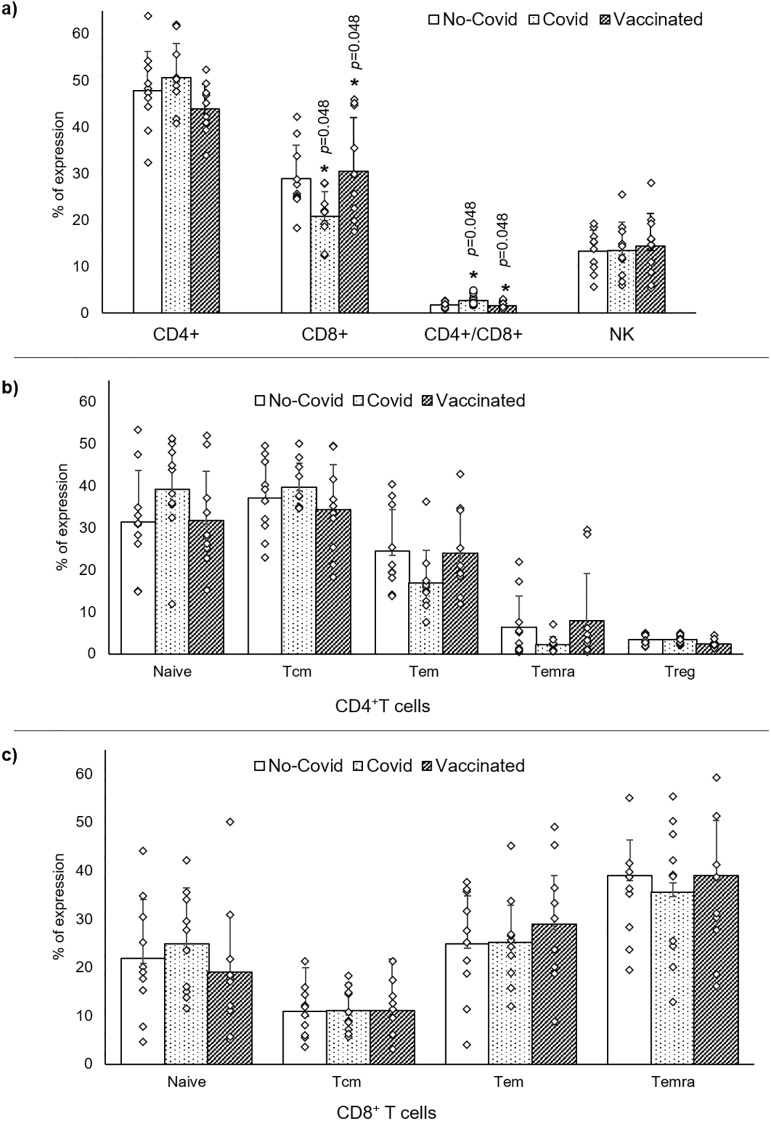
Since data obtained from cell counts and percentages of lymphocyte expression did not show differences, statistical analysis is performed and presented according to the expression percentages of differentiation clusters observed. Statistical variance analysis revealed a significant variation in expression of CD8+ T cells (p = 0.035) and CD4+/CD8+ ratio (p = 0.030) (Table 1). In particular, subjects who received the COVID-19 mRNA BNT162b2 vaccine have shown an increased expression of CD8+ T cells (p = 0.048), and a reduction of CD4+/CD8+ ratio (p = 0.048) compared to subjects previously infected (Table 2 and Fig. 3a). In infected subjects we found a significant reduction of CD8+ T cells (p = 0.048) and a significant increase of CD4+/CD8+ ratio (p = 0.048) respect to vaccinated subjects (Table 2 and Fig. 3a). No significant difference was observed for Naïve, Central Memory, Effector Memory, and Terminal Effector Memory both CD4+ and CD8+ and Treg CD4+ cells comparing groups (Table 3 and Fig. 3b and c).
Fig. 3.
Dot plots summarize data analysis of: a, CD4+, CD8+ and NK cells comparing the three different groups: healthy (no Covid), infected (Covid) and healthy vaccinated (Vaccinated) subjects. b, T CD4+ maturation profile (naïve, central memory T cells (Tcm), effector memory T cells (Tem) and terminal differentiated effector memory T cells (Temra)) including Treg cells and c, T CD8+ maturation profiles.
4. Discussion
The role of T cells and their potential to provide long-term protection from reinfection with SARS-CoV-2 remains debated, yet. Here, we report T cell characterization by using commercial kits which have been widely used in infectious disease studies (Chen and John Wherry, 2020) and recent observations on SARS-CoV-2 infection (Bordoni et al., 2012). Our results are in agreement with recent evidence (Huang et al., 2020; Chen et al., 2020; Wang et al., 2020; De Biasi et al., 2020). We observe that: 1) T cells subset remains within the normal range in all groups, 2) an increased expression of CD8+ T cells and a decreased CD4+/CD8+ ratio in vaccinated respect to infected subjects and 3) a reduction of CD8+ T cells and an increase of CD4+/CD8+ ratio in infected subjects compared to vaccinated group. Although our results revealed different CD4+ and CD8+ T maturation profiles, no significant difference was found comparing groups. Of note, our clinical approach could be improved in the future with additional data on other immune cell types and/or comprehensive data for circulating inflammatory mediators for a larger cohort of subjects with different age (Napoli et al., 2020).
Funding
This study was funded by: PRIN2017F8ZB89 from the Italian Ministry of University and Research (MIUR) (Claudio Napoli) and Ricerca Corrente (RC) 2019 from the Italian Ministry of Health (Claudio Napoli); grants GR-2016-02364785 from the Italian Ministry of Health (Vincenzo Grimaldi and Claudio Napoli).
Declaration of Competing Interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jim.2022.113230.
Appendix A. Supplementary data
Basic demographic information
References
- Bordoni V., et al. A novel 8-color flow cytometry panel to study activation, maturation and senescence of CD4 and CD8 T lymphocytes in HIV-infected individuals at different stages of disease. Int. J. Immunopathol. Pharmacol. 2012;25(2):415–424. doi: 10.1177/039463201202500211. [DOI] [PubMed] [Google Scholar]
- Channappanavar R., Zhao J., Perlman S. T cell-mediated immune response to respiratory coronaviruses. Immunol. Res. 2014;59(1–3):118–128. doi: 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., John Wherry E. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020;20(9):529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., et al. Clinical and immunologic features in severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colacurci N., et al. Flow cytometry characterization of pluripotent transmembrane glycoproteins on resident cervix uteri cells in patients screened for cervical cancer. Cancer Investig. 2020;38(4):228–239. doi: 10.1080/07357907.2020.1742349. [DOI] [PubMed] [Google Scholar]
- De Biasi S., et al. Marked T cell activation, senescence, exhaustion and skewingtowards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020;11(1):3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., et al. Lymphocyte subset counts in COVID-19 patients: a meta-analysis. Cytometry. A. 2020;97(8):772–776. doi: 10.1002/cyto.a.24172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.K., et al. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008;181(8):5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A., et al. Human immune diversity: from evolution to modernity. Nat. Immunol. 2021;22(12):1479–1489. doi: 10.1038/s41590-021-01058-1. [DOI] [PubMed] [Google Scholar]
- Lund J.M., et al. Coordination of early protective immunity to viral infection by regulatory T cells. Science. 2008;320(5880):1220–1224. doi: 10.1126/science.1155209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfella R., et al. Does poor glycaemic control affect the immunogenicity of the COVID-19 vaccination in patients with type 2 diabetes: the CAVEAT study. Diabetes Obes. Metab. 2022;24(1):160–165. doi: 10.1111/dom.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C., et al. Immunosenescence exacerbates the COVID-19. Arch. Gerontol. Geriatr. 2020;90 doi: 10.1016/j.archger.2020.104174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C., et al. Immune reactivity during COVID-19: implications for treatment. Immunol. Lett. 2021;231:28–34. doi: 10.1016/j.imlet.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain S.L., McKinstry K.K., Strutt T.M. Expanding roles for CD4+ T cells in immunity to viruses. Nat. Rev. Immunol. 2012;12(2):136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J. Infect. Dis. 2020;221(11):1762–1769. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M.A., Ravkov E.V., Bevan M.J. Rapid culling of the CD4+ T cell repertoire in the transition from effector to memory. Immunity. 2008;28(4):533–545. doi: 10.1016/j.immuni.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatim N., et al. Immune checkpoint inhibitors increase T cell immunity during SARS-CoV-2 infection. Sci. Adv. 2021;7(34):eabg4081. doi: 10.1126/sciadv.abg4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Basic demographic information