Abstract
In this study, we compared serum hepatitis C virus (HCV) RNA concentrations with HCV RNA concentrations in whole blood collection tubes, including two different types of EDTA tubes and nucleic acid stabilization tubes (NASTs). We also investigated the impact of a processing delay on HCV RNA concentration in these tubes. In NASTs, the mean HCV RNA concentration was comparable to the mean serum HCV RNA concentration at “date zero.” In EDTA tubes, mean baseline HCV RNA concentrations were higher. Storage at room temperature up to 96 h did not result in a decline of HCV RNA concentration in any of the whole blood collection tubes. In NASTs, HCV RNA concentrations remained stable during the whole study period, whereas a significant increase of HCV RNA was observed in both types of EDTA tubes at 96 h compared to date zero. We concluded that HCV RNA remains stable in NASTs at room temperature for at least 96 h, allowing greater flexibility in sample collection and transport.
Hepatitis C virus (HCV) is the most important agent of posttransfusion and community-acquired non-A, non-B hepatitis (5, 9). HCV has been associated with liver cirrhosis and hepatocellular carcinoma (6, 19). Furthermore, various extrahepatic disorders have been described in a subgroup of HCV-infected patients (2, 18).
Molecular techniques based on amplification of viral nucleic acids by PCR have been shown to be effective tools for direct detection of HCV. To meet the routine needs of the diagnostic laboratory, PCR amplification and detection of amplified products have recently been automated with the COBAS AMPLICOR system (3, 11, 13, 14). For detection of serum HCV RNA, both qualitative and quantitative tests are now available.
The success of molecular methods in the clinical diagnostic laboratory depends largely on the quality of the nucleic acid purified from the clinical specimen, which is directly related to how the specimen is stored and transported to the laboratory after it has been collected. In clinical practice and in multicenter trials, specimens may be in transit for several days. To ensure accurate results, it is important to define optimum specimen handling and shipping conditions. Results from studies of the stability of HCV RNA in whole blood specimens have been controversial. When plasma HCV RNA levels are measured in whole blood samples collected and stored in EDTA tubes, significant declines as well as stable levels are reported (7, 8, 10, 20). Less automated molecular assays have usually been used for those studies, and intra-assay variabilities have been inadequately reported or not reported at all.
Previous studies have shown that HCV can infect peripheral blood mononuclear cells (PBMCs) in patients with chronic hepatitis C (4, 22, 23). The presence of HCV in PBMCs may have implications in the patient's response to antiviral therapy (16, 17, 21). It has been demonstrated recently that detection of HCV RNA in PBMCs may be an additional tool to demonstrate the persistence of HCV RNA. Reappearance of HCV RNA is detected earlier in whole blood samples collected in EDTA tubes than in serum samples (15). Therefore, recovery of total HCV RNA, i.e., intracellular RNA as well as plasma RNA, appears to be of major importance to detect low-level viremia.
This study compared serum HCV RNA concentrations with HCV RNA concentrations in different types of whole blood collection tubes and examined the impact of a processing delay on the HCV RNA concentrations in several types of whole blood collection tubes.
MATERIALS AND METHODS
Blood samples were collected from 18 patients (female/male ratio, 8:10; age range, 25 to 62 years) who had previously been found to be serum HCV RNA positive. The local ethics committee approved the study protocol, and all patients gave informed consent.
For quantitation of serum HCV RNA, blood from all patients was collected in 9-ml tubes (BD Vacutainer Systems, Franklin Lakes, N.J.). In addition, whole blood was collected in the following types of collection tubes: 3.0-ml Vacuette EDTA tubes (greiner bio-one GmbH, Kremsmünster, Austria), 3.5-ml Vacuette nucleic acid stabilization tubes (NASTs) (greiner bio-one GmbH), which contain a liquid nucleic acid stabilizer, and 3.0-ml Vacutainer EDTA tubes (BD Vacutainer Systems). First, intra-assay variability was determined. Blood from two patients was collected in each of the three types of whole blood collection tubes mentioned above. Intra-assay variability was defined by testing seven replicates of each of the three whole blood samples. Then HCV RNA stability was tested. Blood was collected from 16 patients. For each of these patients, three of each type of collection tube were used, totaling nine whole blood collection tubes per patient.
Within 2 h of drawing the blood, 9-ml tubes were centrifuged at 1,500 × g for 20 min at room temperature. After centrifugation, aliquots were prepared and immediately frozen at −70°C until tested. Simultaneously, three whole blood collection tubes (one of each type) were labeled “time zero” and frozen at −70°C. For investigation of HCV RNA stability, whole blood collection tubes were stored at room temperature (minimum, 23.5°C; maximum, 26.5°C). At 48 h after time zero, three whole blood collection tubes (one of each type) from the room temperature group were frozen at −70°C until tested, and at 96 h, another three-tube set from the room temperature group was also frozen as above.
Individual-donor sample sets were processed together to minimize the effect of inter-assay variability. For quantitation of serum HCV RNA, the COBAS AMPLICOR HCV monitor test, version 2.0 (Roche Diagnostic Systems, Pleasanton, Calif.) was used according to the manufacturer's package insert. The COBAS AMPLICOR instrument automatically determines the HCV RNA concentrations for samples and controls. The HCV RNA concentration is expressed in international units per milliliter.
For isolation of HCV RNA from whole blood, the High Pure Viral Nucleic Acid kit, version 3 (Roche Molecular Biochemicals, Mannheim, Germany) was used. When EDTA tubes were used, 6 μl of internal quantitation standard (QS), taken from the COBAS AMPLICOR HCV monitor test, was added to 200 μl of whole blood from EDTA tubes, 200 μl of working solution [binding buffer supplemented with poly(A) carrier RNA], and 40 μl of proteinase K. Then HCV RNA was isolated according to the manufacturer's package insert. When NASTs were used, flakes were resuspended by gentle shaking, and 3 μl of QS was added to 275 μl of NAST blood (corresponding to 100 μl of whole blood) and 162.5 μl of preincubation solution. After incubating at room temperature for 15 min and vortexing, 85 μl of isopropanol was added. Next, HCV RNA was extracted according to the High Pure Viral Nucleic Acid kit protocol. Immediately after HCV RNA isolation, amplification and hybridization of amplification products followed by detection of hybridization products were all accomplished by using the COBAS AMPLICOR HCV monitor test, version 2.0, on the COBAS AMPLICOR instrument.
Data were analyzed with standard descriptive statistics. To demonstrate the effects of storage time, data were standardized according to serum HCV RNA concentrations. To compare the values for whole blood samples with those for serum values, paired sample tests were used. The effects of storage time were analyzed by repeated measurements using a generalized linear model. These effects were also confirmed by nonparametric analysis. Multiple comparisons were made when appropriate with the Dunnett test for the generalized linear model, to adjust the significance level, and with Bonferroni-adjusted Wilcoxon tests in the nonparametric analyses. For computerized statistical analysis, the packages SPSS (SPSS Inc., Chicago, Ill.) and S-plus (MathSoft Inc., Searrle, Wash.) software were used.
RESULTS AND DISCUSSION
First, the intra-assay variability of the quantitative molecular assay designed to measure recovery of HCV RNA was determined for each of the whole blood collection tubes (Table 1). With EDTA tubes, the coefficient of variation (CV) ranged between 10 and 16%; with NASTs, the CV ranged from 23 to 34%.
TABLE 1.
Intra-assay variability of whole blood collection tubesa
| Tube type | HCV RNA quantitation results for patient no.:
|
|||||
|---|---|---|---|---|---|---|
| Mean HCV RNA (IU/ml)
|
SD
|
CV (%)
|
||||
| 1 | 2 | 1 | 2 | 1 | 2 | |
| Vacuette EDTA | 196,000 | 611,000 | 20,300 | 78,600 | 10 | 13 |
| Vacuette NAST | 163,000 | 348,000 | 54,800 | 80,700 | 34 | 23 |
| Vacutainer EDTA | 217,000 | 594,000 | 22,700 | 93,400 | 10 | 16 |
Data were obtained by testing seven replicates.
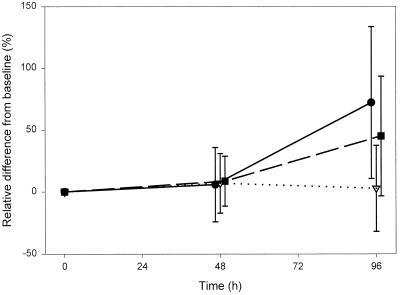
Then HCV RNA stability was tested. At time zero, the mean serum HCV RNA concentration was 4.2 × 105 IU/ml. The corresponding values for whole blood from EDTA tubes were 9.1 × 105 IU/ml and 7.2 × 105 IU/ml. In NASTs, the mean HCV RNA concentration was 4.7 × 105 IU/ml. At 48 h, HCV RNA levels were comparable to those measured at time zero in all whole blood collection tubes studied (9.2 × 105 IU/ml, 4.5 × 105 IU/ml, and 7.9 × 105 IU/ml) (Fig. 1). At 96 h, HCV RNA levels were comparable to those measured at time zero in NASTs (4.8 × 105 IU/ml). However, in EDTA collection tubes a significant increase of HCV RNA levels was found (1.1 × 106 IU/ml in Vacuette EDTA tubes [P < 0.001] and 1.6 × 106 IU/ml in Vacutainer EDTA tubes [P < 0.007]) (Fig. 1).
FIG. 1.
Effect of storage on different blood collection tubes. Symbols: ●, Vacuette EDTA; ▿, Vacuette NAST; ▪, Vacutainer EDTA.
To obtain reliable quantitation results, a molecular system that reveals reasonable intra-assay variability must be chosen. With the COBAS AMPLICOR HCV monitor test, version 2.0, for quantitation of serum HCV RNA, the CV ranges between 7 and 51% according to the manufacturer's package insert. Recently, the intra-assay CV was even reported to range between 0.55 and 2.95% (1). Such results can be achieved only when maximum automated systems are used, and because the COBAS AMPLICOR HCV monitor test is not designed for application on whole blood samples, a novel molecular assay had to be created for this study. HCV RNA was extracted with a commercially available assay for viral nucleic isolation from whole blood. Extraction was achieved by incubation of the sample in a special lysis/binding buffer in the presence of proteinase K and subsequent specific binding of RNA to the surface of glass fibers in the presence of a chaotropic salt. To make quantitation possible, a defined amount of internal QS, taken from the COBAS AMPLICOR HCV monitor test, was added. After HCV RNA isolation, reverse transcription, amplification, hybridization, and detection were done on the automated COBAS AMPLICOR instrument. With this molecular assay, reliable quantitation can be achieved.
Compared to the mean serum HCV RNA concentration, the mean whole blood HCV RNA concentrations were higher in both of the tested EDTA tubes. This might be explained by the presence of HCV RNA in PBMCs and recovery of intracellular viral RNA in addition to plasma RNA. Release of reverse transcription (RT)-PCR inhibitors and RNAses from the cells, which could affect quantitation results, can be ruled out. The mean HCV RNA concentration in NASTs was comparable to the mean serum HCV RNA concentration.
Storage of whole blood samples at room temperature did not result in a decrease of HCV RNA concentrations. Inhibitors of RT or PCR did not build up over time. Surprisingly, an increase of the mean HCV RNA concentration was observed in EDTA tubes at 96 h after time zero. The reason might be an insufficient recovery of intracellular viral RNA by the nucleic acid isolation assay used in this study and a “natural” release of intracellular viral RNA by the ongoing cellular decay. With the NASTs, HCV RNA concentrations were comparable throughout the whole study. NASTs contained a liquid nucleic acid stabilizer, which was capable of stabilization of nucleic acids within the blood samples at room temperature for at least 96 h.
Others have recently reported similar stability of HCV RNA in whole blood collection tubes for storage of samples at temperatures less than 37°C. One study showed that HCV RNA was stable in EDTA tubes for up to 72 h at room temperature, and the authors estimated that a 25% reduction of HCV RNA concentration would require more than 200 h of storage at room temperature (20). Another study found that more than 75% of HCV RNA was retained after 5 days of storage at room temperature in EDTA tubes (10). One recent storage study showed an insignificant decline of HCV RNA concentration in EDTA tubes when stored for 72 h at 25°C (12).
In summary, with the molecular assay described in this study, reliable results were achieved for quantitation of HCV RNA. In NASTs, HCV RNA remained stable for considerable periods of time. We concluded that if processing of samples is guaranteed within 96 h, tubes could be stored at room temperature without sample degradation. These findings will allow greater flexibility in sample collection and transport, and they have considerable implications for control of the costs incurred in handling blood samples for quantitation of HCV RNA.
ACKNOWLEDGMENTS
We gratefully acknowledge Anja Herrmann for stimulating discussions.
This project was supported in part by a grant from greiner bio-one GmbH.
REFERENCES
- 1.Afonso A R M, Didier J, Plouvier E, Falissard B, Ferey M P, Bogard M, Dussaix E. Performance of an automated system for quantification of hepatitis C virus RNA. J Virol Methods. 2000;86:55–60. doi: 10.1016/s0166-0934(99)00179-2. [DOI] [PubMed] [Google Scholar]
- 2.Agnello V, Chung R T, Kaplan L M. A role for hepatitis C virus infection in type II cryoglobulinemia. N Engl J Med. 1992;327:1490–1495. doi: 10.1056/NEJM199211193272104. [DOI] [PubMed] [Google Scholar]
- 3.Albadalejo J, Alonso R, Antinozzi R, Bogard M, Bourgault A M, Colucci G, Fenner T, Petersen H, Sala E, Vincelette J, Young C. Multicenter evaluation of the COBAS AMPLICOR HCV assay, an integrated PCR system for rapid detection of hepatitis C virus RNA in the diagnostic laboratory. J Clin Microbiol. 1998;36:862–865. doi: 10.1128/jcm.36.4.862-865.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouffard P, Hayashi P H, Acevedo R, Levy N, Zeldis J B. Hepatitis C virus is detected in a monocyte/macrophage subpopulation of peripheral blood mononuclear cells of infected patients. J Infect Dis. 1992;166:1276–1280. doi: 10.1093/infdis/166.6.1276. [DOI] [PubMed] [Google Scholar]
- 5.Choo Q L, Weiner A J, Overby L R, Kuo G, Houghton M. Hepatitis C virus: the major causative agent of viral non-A, non-B hepatitis. Br Med Bull. 1990;46:423–441. doi: 10.1093/oxfordjournals.bmb.a072408. [DOI] [PubMed] [Google Scholar]
- 6.Colombo M, Kuo G, Choo Q L, Donato M F, Del Ninno E, Tommasini M A, Houghton M. Prevalence of antibodies of hepatitis C virus in Italian patients with hepatocellular carcinoma. Lancet. 1989;ii:1006–1008. doi: 10.1016/s0140-6736(89)91016-7. [DOI] [PubMed] [Google Scholar]
- 7.Cuypers H T M, Bresters D, Winkel I N, Reesink H W, Weiner A J, Houghton M C, van der Poel L, Lelie P N. Storage conditions of blood samples and primer selection affect the yield of cDNA polymerase chain reaction products of hepatitis C virus. J Clin Microbiol. 1992;30:3220–3224. doi: 10.1128/jcm.30.12.3220-3224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damen M, Sillekens P, Sjerps M, Melsert R, Frantzen I, Reesink H W, Lelie P N, Cuypers H T M. Stability of hepatitis C virus RNA during specimen handling and storage prior to NASBA amplification. J Virol Methods. 1998;72:175–184. doi: 10.1016/s0166-0934(98)00024-x. [DOI] [PubMed] [Google Scholar]
- 9.Dana F, Becherer P R, Bacon B R. Hepatitis C virus. What recent studies can tell us. Postgrad Med. 1994;95:125–130. [PubMed] [Google Scholar]
- 10.Dockter J, Gaichetti C, McDonough S, Mimms L, Schoening V, Kuramoto K. Stability of hepatitis C virus RNA in blood specimens as measured by the Gen-Probe assay. Transfusion. 1998;38(Suppl.):S246. [Google Scholar]
- 11.Doglio A, Laffont C, Caroli-Bosc F X, Rochet P, Lefebvre J C. Second generation of the automated COBAS AMPLICOR HCV assay improves sensitivity of hepatitis C virus RNA detection and yields results that are more clinically relevant. J Clin Microbiol. 1999;37:1567–1569. doi: 10.1128/jcm.37.5.1567-1569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant P R, Kitchen A, Barbara J A J, Hewitt P, Sims C M, Garson J A, Tedder R S. Effects of handling and storage of blood on the stability of hepatitis C virus RNA: implications for NAT testing in transfusion practice. Vox Sang. 2000;78:137–142. doi: 10.1159/000031171. [DOI] [PubMed] [Google Scholar]
- 13.Jungkind D, DiRenzo S, Beavis K G, Silverman N S. Evaluation of an automated COBAS AMPLICOR PCR system for detection of several infectious agents and the impact on laboratory management. J Clin Microbiol. 1996;34:2778–2783. doi: 10.1128/jcm.34.11.2778-2783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kessler H H, Dragon E A, Pierer K, Santner B I, Liao Y, Stünzner D, Stelzl E, Marth E. Performance of the automated COBAS AMPLICOR system for the detection of hepatitis C virus RNA. Clin Diagn Virol. 1997;7:139–145. doi: 10.1016/s0928-0197(96)00263-2. [DOI] [PubMed] [Google Scholar]
- 15.Kessler H H, Pierer K, Santner B I, Vellimedu S K, Stelzl E, Marth E, Fickert P, Stauber R E. Evaluation of molecular parameters for routine assessment of viremia in patients with chronic hepatitis C who are undergoing antiviral therapy. J Hum Virol. 1998;1:314–319. [PubMed] [Google Scholar]
- 16.Kusaka S, Okusa T, Araki A, Fujiki K, Takashimizu I, Okayasu I, Yamamoto N, Sato C. Prediction of relapses after interferon-alpha therapy by hepatitis C virus RNA in peripheral blood mononuclear cells. J Med Virol. 1995;46:265–268. doi: 10.1002/jmv.1890460317. [DOI] [PubMed] [Google Scholar]
- 17.Ounanian A, Gueddah N, Rolachon A, Thelu M A, Zarski J P, Seigneurin J M. Hepatitis C virus RNA in plasma and blood mononuclear cells in patients with chronic hepatitis C treatment with alpha-interferon. J Med Virol. 1995;45:141–145. doi: 10.1002/jmv.1890450205. [DOI] [PubMed] [Google Scholar]
- 18.Pascual M, Perrin L, Giostra E, Shifferli J A. HCV in patients with cryoglobulinemia type II. J Infect Dis. 1990;162:569–570. doi: 10.1093/infdis/162.2.569. [DOI] [PubMed] [Google Scholar]
- 19.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuci S, Watanabe Y, Koi S, Onji M, Ohta Y. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:2059–2061. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stramer S L, Porter R A, Brodsky J P, Waldman K J, Pollack S R, Dodd R Y, Smith R I F, Billyard B, Rainen L. Stability of HCV RNA for pooled genome amplification testing (GAT) Transfusion. 1998;38(Suppl.):S263. [Google Scholar]
- 21.Taliani G, Badolato C, Lecce R, Poliandri G, Bozza A, Duca F, Pasquazzi C, Clementi C, Furlan C, de Bac C. Hepatitis C virus RNA in peripheral blood mononuclear cells: relation with response to interferon treatment. J Med Virol. 1995;47:16–22. doi: 10.1002/jmv.1890470105. [DOI] [PubMed] [Google Scholar]
- 22.Wang J T, Sheu J C, Lin J T, Wang T H, Chen D S. Detection of replicative form of hepatitis C virus RNA in peripheral blood mononuclear cells. J Infect Dis. 1992;166:1167–1169. doi: 10.1093/infdis/166.5.1167. [DOI] [PubMed] [Google Scholar]
- 23.Zignego A L, Macchia D, Monti M, Thiers V, Mazzetti M, Foschi M, Maggi E, Romagnani S, Gentilini P, Brechot C. Infection of peripheral mononuclear blood cells by hepatitis C virus. J Hepatol. 1992;15:382–386. doi: 10.1016/0168-8278(92)90073-x. [DOI] [PubMed] [Google Scholar]