Abstract
As the second year of the COVID-19 pandemic begins, it remains clear that a massive increase in the ability to test for SARS-CoV-2 infections in a myriad of settings is critical to controlling the pandemic and to preparing for future outbreaks. The current gold standard for molecular diagnostics is the polymerase chain reaction (PCR), but the extraordinary and unmet demand for testing in a variety of environments means that both complementary and supplementary testing solutions are still needed. This review highlights the role that loop-mediated isothermal amplification (LAMP) has had in filling this global testing need, providing a faster and easier means of testing, and what it can do for future applications, pathogens, and the preparation for future outbreaks. This review describes the current state of the art for research of LAMP-based SARS-CoV-2 testing, as well as its implications for other pathogens and testing. The authors represent the global LAMP (gLAMP) Consortium, an international research collective, which has regularly met to share their experiences on LAMP deployment and best practices; sections are devoted to all aspects of LAMP testing, including preanalytic sample processing, target amplification, and amplicon detection, then the hardware and software required for deployment are discussed, and finally, a summary of the current regulatory landscape is provided. Included as well are a series of first-person accounts of LAMP method development and deployment. The final discussion section provides the reader with a distillation of the most validated testing methods and their paths to implementation. This review also aims to provide practical information and insight for a range of audiences: for a research audience, to help accelerate research through sharing of best practices; for an implementation audience, to help get testing up and running quickly; and for a public health, clinical, and policy audience, to help convey the breadth of the effect that LAMP methods have to offer.
INTRODUCTION
The need to expand molecular testing options beyond that which polymerase chain reaction (PCR) can cost-effectively deliver has been put into sharp focus by the COVID-19 pandemic. Resource-poor and resource-rich countries alike need to be able to track the virus in real time to mitigate its spread. Here, we show how loop-mediated isothermal amplification (LAMP)-based testing solutions have become a major part of that testing expansion, and we highlight its use in other applications, which includes cost-effective expansion-by-volume (more tests per day), expansion-by-location (more, smaller testing centers to expand the geographic reach of testing), and bringing the test closer to the patient (point-of-care and home testing). The collection of LAMP methods presented here provide a toolbox to construct testing solutions for most real-world applications. Although LAMP is by no means the only testing method to augment PCR, its relative ease of implementation, lower cost, and simple equipment requirements mean it can be broadly, quickly, and cheaply established.
No method is without its flaws; reverse transcription–LAMP (RT-LAMP) is certainly no exception, and valuable scientific work continues to improve specificity, sensitivity, scalability, and usability. However, a tipping point has been reached: progress over the past year has led to the development of many useful variations on a core SARS-CoV-2 RT-LAMP assay; wide-scale LAMP-based testing has moved from being aspirational to being deployed at state and national levels. Multiple tests have been approved by regulators (Table S1), including a colorimetric LAMP test being approved for home use (Lucira Health) and many more under review. Here, we provide a methodologically useful guide for developing and deploying a LAMP-based test for COVID-19 and, more broadly, to bring us closer to a world of democratized diagnostics in which everyone can benefit from advances in modern genomics to address public health challenges.
Tour of the Review
Our emphasis is primarily methodological and although focused on LAMP, many aspects are applicable to other isothermal methods (Piepenburg et al. 2006, Niemz et al. 2011, Li et al. 2018, Chaouch 2021, Eftekhari et al. 2021, Tran et al. 2021, Yu et al. 2021). This review provides readers new to the field with an introduction to SARS-CoV-2 testing and the underlying isothermal technology, the preanalytic sample processing, target amplification and amplicon detection and a consideration of the infrastructure required for deployment in the current regulatory landscape. The Discussion considers the current state of the art and future directions, methodologically and from a public health perspective. The main text is augmented with 4 Topics A– through D that address specialist topics in greater depth.
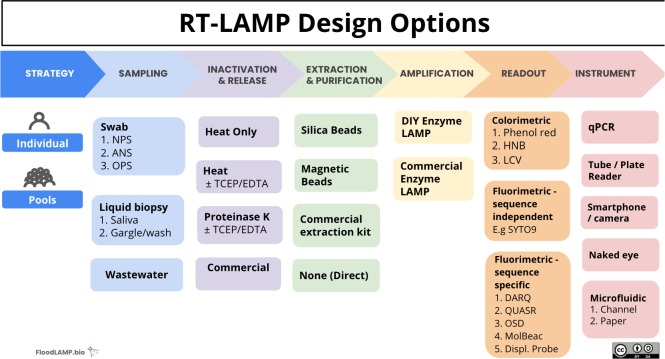
We have highlighted the different lenses and perspectives through which researchers can view LAMP technology, depending on whether they provide 1-to-1 patient care in a remote rural environment or conversely frequent mass testing for surveillance and epidemiology in a large public health laboratory. The series of choices (and corresponding compromises) that one makes to create a test that is fit for a specific purpose is a common theme: regardless of one's setting, budget, or scale of testing, “there is a LAMP for that” (Fig. 1).
FIGURE 1.
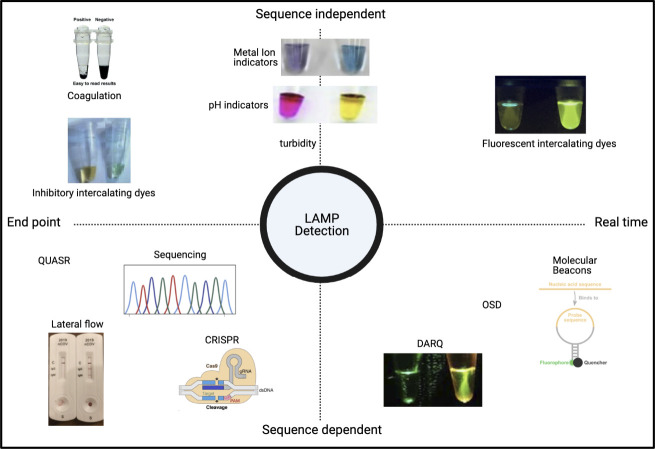
“We have a LAMP for that”: major design choices when developing RT-LAMP tests. At each stage in the design process, a series of decisions affect the final configuration of the test, be that for an individual patient or for surveillance testing with pooled screening. The inherent flexibility and comparative simplicity of LAMP means that, for almost all settings and uses, there is 1 configuration of the LAMP toolbox that is fit for the purpose. The LAMP tests for use in any 2 settings or geographies can be dramatically different and can use NPS, ANS, OPS, TCEP, EDTA, DIY, HNB, LCV, DARQ, QuasR, OSD, or MolBeac. Abbreviations: ANS, anterior nares swab; DIY, do-it yourself; DARQ, dark quenching technique; EDTA, ethylenediamine tetraacetic acid; HNB, hydroxynaphthol blue; LAMP, loop-mediated isothermal amplification; LCV, leuco crystal violet; MolBeac, Molecular Beacons. NPS, nasopharyngeal swabs; OPS, oropharyngeal swab; OSD, oligonucleotide strand displacement; QuasR, quenching of unincorporated amplification signal reporters; RT-LAMP, reverse transcription–LAMP; TCEP, tris(2-carboxyethyl) phosphine.
COVID-19 Testing
The emergence of a cluster of acute respiratory syndrome cases in Wuhan City, China, drew worldwide attention in January 2020, with rapid epidemiologic investigation identifying a novel coronavirus, SARS-CoV-2 as the causative agent of coronavirus disease 2019 (COVID-19). Once the viral genomic RNA sequence was released, the global scientific community began many investigations with a particular focus on diagnostic testing and vaccine development. The potential for global spread was clear, and the ability to rapidly and accurately detect SARS-CoV-2 became paramount to tracing the infection and controlling what quickly became a global pandemic. Researchers from Charité—Universitätsmedizin Berlin reported a quantitative reverse transcription–polymerase chain reaction (RT-qPCR) test for the novel coronavirus in January 2020 with the protocol and primers being endorsed by the World Health Organization (Geneva, Switzerland); the US Centers for Disease Control and Prevention (CDC, Atlanta, GA) followed suit with its own PCR test and began limited distribution in early February 2020 (Abraham et al. 2020).
The rapid production of these tests and the ability to begin testing in diagnostic laboratories was an impressive and valuable achievement. However, with limited access to these tests and a high demand by health care systems overwhelmed by cases of COVID-19, it became clear that relying only on traditional clinical testing infrastructures would be insufficient to track and contain the growing public health threat from SARS-CoV-2. Antigen and antibody test development began in earnest to broaden surveillance capabilities, but the need for alternative, sensitive molecular methods remained. The PCR tests have long been the reference method for molecular diagnostics, but the typical test workflow presents limitations to accessible, wide-scale testing. Specifically, the conventional workflow has significant requirements for nucleic acid extraction from samples, sophisticated and expensive real-time fluorescence thermocyclers, and trained personnel in certified Biosafety Level 2 (BSL2) or BSL3 testing laboratories.
LAMP Testing
Nucleic acid amplification techniques that avoid such constraints and hold promise as a companion method to RT-qPCR testing have been developed. Notable among these is reverse transcription–loop-mediated isothermal amplification (RT-LAMP). First described in 2000 (Notomi et al. 2000), LAMP uses 6 target-specific primers for highly specific and fast amplification (Nagamine et al. 2002). The method is based on a unique primer design that creates a “dumbbell” shaped, looped DNA structure that is self-priming. The DNA polymerase used in LAMP can displace bound strands of DNA as it advances, and thus, there is no need to thermocycle the reaction for the DNA to be amplified. The cycling steps characteristic of qPCR are functionally replaced by enzymatic strand-displacement activity of the Bst polymerase. This means that LAMP reactions operate at a single temperature (usually a single temperature 63°C to 67°C), which greatly simplifies the methodology and required instrumentation. When combined with relatively straightforward optical detection, these properties have enabled LAMP to be used for point-of-care and field diagnostics (e.g., Cook et al. 2015, Calvert et al. 2017, Snodgrass et al. 2018, da Silva Gonçalves et al. 2019).
The detailed mechanism underpinning DNA amplification in LAMP is complex. This arises largely from the multiple sites of polymerase initiation, which is, itself, a consequence of using 6 primers to target multiple locations on the target DNA (rather than 2 as in PCR; Fig. 2). The more complex elongation cycle is described in Supplemental Information S1, whereas the reader may find this figure more intuitive.
FIGURE 2.
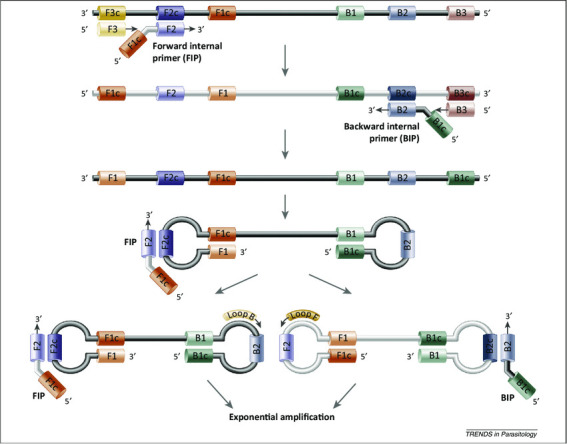
LAMP mechanism. The LAMPs employ 2 sets of primers, forward/backward internal primers (FIP and BIP) and outer primers (F3 and B3) to target 6 distinct regions (F1c, F2c, F3c sites on 1 end, and B1, B2, B3 sites on the other). The reaction is initiated by the binding of FIP to the F2c region on the double-stranded DNA. As the polymerase elongates the DNA from the FIP, the outer primer F3, which is shorter in length and lower in concentration than the FIP, binds onto its complementary region on the DNA and starts to displace the newly synthesized DNA. The replaced strand then forms a loop structure at one end because of the complementarity of F1 and F1c. This results in a single-stranded, double-stem-loop DNA structure (the so-called “dumb-bell” structure) with similar performance for BIP and B3. This dumbbell-structured DNA enters the amplification cycle because it is already self-primed. Elongation by the polymerase can occur from the free 3′-end of the single-stranded DNA (ssDNA) and from binding of the FIP/BIP primers to the single-stranded loop or from the optional accelerating loop primers (see Supplemental Fig. S1, with permission from Alhassan et al. 2015).
The first LAMP assays for SARS-CoV-2 were described in preprints starting in February, March, and April 2020 (Broughton et al. 2020, Butler et al. 2020, Lamb et al. 2020, Yu et al. 2020, Zhang et al. 2020a). As demand for testing increased, laboratories normally devoted to genomics, developmental biology, or plant pathogens shifted focus onto the development of diagnostic tests for SARS-CoV-2.
Specialist Topics: Lenses on LAMP
By design, LAMP is adaptable. In these Topics, authors share various first-hand experiences of development and deployment and of methodologic improvements.
Topic A: developing LAMP tests in and for resource-limited settings
Testing with LAMP is inherently simpler and more portable than conventional RT-qPCR, so interest has been keen by those engaged in distributed (e.g., primary care) and low-resource (e.g., developing nations) settings. We (re)introduce the (RE)ASSURED framework as criteria, such as affordability and robustness to which diagnostics designed for resource-limited settings should aspire. This includes the compromises in analytic sensitivity and specificity that are often necessary to maximize affordability and accessibility of the test to the largest patient population. These pragmatic choices are exemplified with a case study on testing in resource-limited communities in Southeast Asia. Open-source methods, resources, and approaches for affordable production of LAMP enzymes and hardware (see Open Research Infrasturcture) seek to further extend the more-affordable footprint of LAMP across the globe.
Topic B: hardware, reagent, and software considerations for LAMP testing
One of the most attractive features of LAMP is its inherent simplicity, with reactions able to be performed with either a qPCR reader or a water heater and the visual detection of color changes. However, there is a growing “middle ground” for smaller, more-portable equipment that provides most of the functionality at a fraction of the cost and footprint. The first is a LAMP-reader software application that improves the objectivity of colorimetric methods by replacing visual inspection with a dedicated App on a simple smartphone camera. Two contributors describe the adaptation of small, semiportable benchtop incubators and software readers to LAMP testing with COVID: 1, the Axxin Fluorometer, (Fairfield, VIC, Australia) comes from a medical perspective, and the other, the BioRanger (New England BioLabs, Ipswich, MA), was repurposed from it original agricultural use. Finally, we summarize the current status of engineered mutant Bst polymerase enzymes with improved catalytic properties, underscoring the critical enzymatic strand displacement in isothermal LAMP reactions.
Topic C: emerging protocols and methods from the gLAMP Consortium
The focus of this review is the measurement of SARS-CoV2 in oral samples (saliva, nasopharyngeal [NP] swabs, among others). However, here, we describe the application of direct LAMP methods to monitor SARS-CoV-2 shedding via environmental sources, employing municipal wastewater and raw sewage samples as an epidemiologic tool. Monitoring and adapting primer designs to emerging mutations in the SARS-CoV-2 genome detected by sequencing and surveillance studies is similarly highlighted, a topic of considerable interest at the time of writing (December, 2021) because of the spread of the “UK COVID” variant (Pangolin lineage B.1.1.7, Nextstrain clade 20B/501Y.V1, or the Omicron variant). The importance of the preanalytic sample processing, RNA extraction, and/or purification cannot be overstated. Throughout 2020, participants exchanged (mixed) experiences with different preanalytic methods. Two case studies provide examples of 2 very different types of contribution. The first is the building of a laboratory developing a saliva-based LAMP assay “from the ground-up,” using many of the guidelines outlined in this review. Successfully transitioning a laboratory-developed test from bench to bedside (or to schools) is the ultimate objective of any diagnostic development process. We conclude with an account of repeat RT-LAMP surveillance testing of a single cohort of individuals (a K–12 school) with at-home participant saliva collection and onsite sample processing.
Topic D: review highlights and takeaways
The “Review highlights and takeways” section provides some important considerations for building LAMP tests and considers the scale of testing, the intended use, the location or setting, and the budget or level of infrastructure available. It considers a series of questions related to the anticipated use, including:
What is the right test for the given application?
How will the test result be used?
How does the hidden cost of not testing affect utility?
What location will the test be used in?
How will frequency and time-to-results influence the design choice?
What sensitivity and specificity are required, i.e., being “positive” and being contagious are not always synchronous physiologic states?
How does the test fit into the clinical practice and triage?
How will the training and skills-level of the testing personnel influence the design?
It concludes with a summary of some technical features of a “good” LAMP-test methodology upon which the developer can adapt its individual approach.
PREANALYTIC SAMPLE PROCESSING
Preanalytically, SARS-CoV-2 testing begins with a choice of sampling site and then a choice of viral inactivation method. Thereafter, one can choose to employ or omit an RNA “extraction” step (“a direct method”). Generally, methods that employ extraction have better sensitivity and precision, whereas omitting that step is less expensive, faster, and simpler logistically. The unprecedented demand caused by the SARS-CoV-2 pandemic has prompted methodologic refinements to allow the same (or similar) test performance to be achieved faster, cheaper, and simpler.
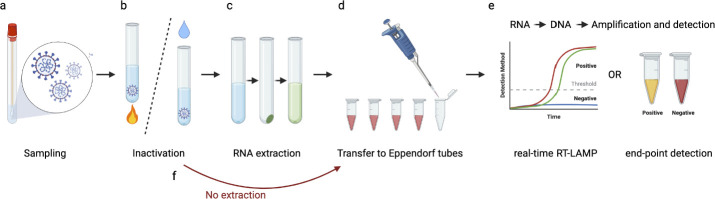
In “Component Methods,” we discuss sampling, viral inactivation, RNA extraction, and direct processing, and conclude with a detailed discussion of the specific issues that arise when pH-based colorimetric LAMP assays are used (Fig. 3). We consider viral inactivation to be the process of preventing the replication of the virus (a safety procedure), whereas RNA extraction involves purposeful release of the viral RNA from the nucleocapsid and, in some protocols, its purification (an analytic procedure). Some methods that involve heating (e.g., Aritzi-Sans et al. 2020) can fully or partially serve both purposes.
FIGURE 3.
A) Biospecimens taken from the patient are inactivated, and the virus is lysed by heating or enzymatic treatment, with or without the addition of chemical agents. B) RNA can be extracted and purified from contaminating proteins and inhibitory contaminants, or C) the step can be omitted (direct methods). F) After transfer of the processed RNA sample into the reverse transcription–loop-mediated isothermal amplification (RT-LAMP) reaction mixture. D) Detection of positive reactions can be achieved through a variety of methods (“Amplicon Detection”), often using real-time fluorescent or visual endpoint readouts E).
COMPONENT METHODS
Biosample Types
Swabs
Because SARS-CoV-2 replicates in the epithelial cells of the respiratory tract (Sungnak et al. 2020), samples for testing are usually from the upper (NP, oropharyngeal [OP], nasal swabs, or saliva) or the lower (sputum, tracheal aspirate, and bronchoalveolar lavage) respiratory tract (Mawaddah et al. 2020). The viral load of SARS-CoV-2 is commonly in the upper respiratory tract, as measured indirectly by viral RNA, because it is elevated during the first week after the onset of symptoms, peaking at 104 to 107 copies/mL after 4 to 6 days (To et al. 2020). The lower respiratory tract is more commonly sampled in symptomatic or severe cases of suspected infection, although the load is highly variable. Swabs have also been used in hospital environments and subways for detecting the presence of the virus (Brune et al. 2021), which also works for other pathogens (Rei et al. 2020).
Washes
Sampling using nasal and throat washes or gargling with a simple 0.9% wt/vol saline or salt solution (Hanks balanced salt solution) is also employed because they are significantly less invasive and have fewer negative effects on the patient (Kellner et al. 2020; see also https://www.maxperutzlabs.ac.at). Encouragingly, respiratory secretions and cells removed from the upper respiratory tract contain comparable or greater viral loads than those obtained from NP or OP swabs (Mawaddah et al. 2020). Similar to saliva (next section), the increased viscosity of throat wash samples can cause problems during sample processing (i.e., samples cannot be pipetted and/or mucous threads pose a cross-contamination risk). This can be easily addressed by incubation of the sample with fresh dithiothreitol (DTT; 5 to 10 mM) for 10 to 15 minutes before further processing to reduce disulfide bonds.
Saliva
Collection of saliva can be done by the patient without the need for the assistance of a health care professional, which reduces the stress on the health care systems and alleviates the need for nasal swabs. The reference NP samples used at the start of the pandemic were progressively augmented with saliva specimens, based on patient saliva collection and drooling (not spitting), including at home collection. Using saliva specimens reduces the risk of exposure to medical workers from viral droplets and reduces the time and cost of the testing procedure compared with that of NP swabs (NPSs).
First applied to RT-qPCR-based detection (Vogels et al. 2020; SalivaDirect, Yale, New Haven, CT), saliva-based LAMP methods have increasingly been developed, although the reported diagnostic sensitivity varies between approximately 70% and 100% (Nagura-Ikeda et al. 2020, To et al. 2020). Comparisons between NPS specimens and saliva concluded that both types of specimen have equivalent sensitivity to detect SARS-CoV-2 (Kellner et al. 2020, Vogels et al. 2020, Zhu et al. 2020). Although collection is more straightforward than NPSs, saliva is a more biologically complex and challenging sample matrix to use, particularly if “minimal” nucleic acid extraction (“Direct”) methods are used. In addition, the composition of saliva varies among individuals, with researchers reporting nonspecific effects of acidic saliva on the assays, an effect that must be mitigated by preneutralization of the sample before LAMP testing when using poorly buffered solutions for the pH-based readout (Nagura-Ikeda et al. 2020). The interplay of competing technical factors in the preanalytic and analytic phases of the test is not uncommon.
Excreta
Clinically, viral RNA is far less commonly isolated from urine (e.g., Kim et al. 2020, Peng et al. 2020), and although virus has been reported in stool samples, its level (approximately 103 to 106 copies/mL) is 10 to 100-fold less than seen in oral or respiratory samples (Wolfel et al. 2020). As elsewhere, detection of the viral RNA does not necessarily correlate with clinical symptoms (Gupta et al. 2020, Peng et al. 2020). More commonly, viruses are detected in excreta in the context of wastewater and sewage surveillance (Wu et al. 2020).
Virus Inactivation Methods
Infectious SARS-CoV-2 can only be worked with in high-containment BSL3 laboratories, whereas inactivation allows handling in an environment with a lower biocontainment level. There are 3 principle methods for inactivating SARS-CoV-2: (1) biological, including antibodies (Martí et al. 2020), (2) physical, including heat (Abraham et al. 2020, Cimolai 2020, Hu et al. 2020, Jureka et al. 2020, Kampf et al. 2020, Pastorino et al. 2020, Yap et al. 2020), cold plasma (Filipić et al. 2021), and ultraviolet light (Buonanno et al. 2020, Martí et al. 2020, Loveday et al. 2021), and (3) chemicals, including detergents, cross-linking agents, oxidizing reagents, chaotropes, alcohols, and other organic solvents (Jureka et al. 2020, Martí et al. 2020, Welch et al. 2020). Frequently, full inactivation is achieved by combining methods, such as heat and a chaotrope (Westhaus et al. 2020).
The effectiveness of inactivation depends on many factors, including viral and reagent concentration, protein content, and treatment time. SARS-CoV-2 inactivation is measured by a reduction in the number of infected tissue culture cells, frequently by plaque assay, or by the nondetection of viral RNA when cell-passaging tests are performed (e.g., Kim et al. 2020, Welch et al. 2020). Inactivating SARS-CoV-2 is relatively easy compared with inactivating nonenveloped viruses, where detergents are frequently employed to disperse or puncture the lipid membrane. However, the virus under physiologic conditions has been demonstrated to maintain infectivity for weeks at room temperature and for months at 4°C (Westhaus et al. 2020).
Although inactivation can be achieved using a wide variety of methods, maintaining RNA integrity for subsequent detection is more difficult. RNA is among the most-fragile biomolecules, with rates of degradation up to a million-fold greater than that of DNA, which can affect downstream profiling (Li et al. 2014). Many inactivation methods, including heat, chaotropes, and cross-linking reagents (such as formaldehyde), lead to loss of RNA integrity, particularly if there is an extended period between collection and molecular analysis. Commercially available reagents to inactivate SARS-CoV-2 include 70% ethanol, 70% isopropanol, 70% acetone, Virkon (Lanxess, Cologne, Germany), NP-40, Triton X-100, 4-M guanidine isothiocyanate with 2% Triton X-100, Primestore MTM (Longhorn Vaccines and Diagnostics, Bethesda, MD), Buffer AVL and RLT with β-mercaptoethanol (Qiagen, Hilden, Germany), virusPHIX (RNAssist, Cambridge, England), VPSS solution (E & O Laboratories, Bonnybridge, Scotland), MagNA Pure Lysis Buffer (F. Hoffmann-La Roche, Basel, Switzerland), and Omnigene Oral DNA (DNA Genotek, Kanata, ON, Canada), as described in Welch et al. 2020; see also Public Health England, https://www.gov.uk/government/organisations/public-health-england).
Many SARS-CoV-2 RNA purification procedures employ an initial lysis step using guanidine or TriZol, which reduces or eliminate viral infectivity without affecting RNA yields or integrity (Batéjat et al. 2021). However, this step increases costs and time, uses commercial products in short supply, and requires several of the constituents (e.g., guanidinium chloride [GnHCl] or guanidinium thiocyanate [GnITC]) that are often incompatible with either the RT step or the LAMP reaction.
As such, physical methods that avoid use of harsh chemicals have become increasingly popular. Although ultraviolet light efficiently inactivates the virus, it also reduces SARS-CoV-2 RNA detection (of note, 1 of the reference control reagents commonly used is γ-irradiated virus NR-52287 from BEI Resources, Manassas, VA). Alternatively, heat can be used for inactivation, although the relatively high temperatures required (ranging from 56°C to 95°C; Batéjat et al. 2021) may also be associated with decreased RNA integrity, particularly if a divalent metal ion chelator is not used to remove the Mg2+ and Mn2+ ions. Extended inactivation (65°C, 30 minutes) provides a more than 105-fold reduction in viral titers (Pastorino et al. 2020), and that is the method used to prepare heat-inactivated virus by BEI Resources (NR-52286). A shorter heating step (56°C, 5 minutes) is not likely sufficient for complete SARS-CoV-2 inactivation (Westhaus et al. 2020).
In summary, although SARS CoV2 inactivation is relatively straightforward, the challenges rest on the delicate balance among clinically complete viral inactivation, maintenance of RNA integrity, use of simple methods, and the compatibility of the viral inactivation method with the downstream analytic RT and LAMP steps.
RNA Extraction
The rationale for RNA extraction
Depending on the application and use, there can be compelling reasons to include an RNA extraction step because it holds certain clear advantages over minimally processed (Direct) samples.
First, patient samples have the potential to be highly heterogeneous, with the degree being dependent on the sample type. The NPSs, and arguably other types of swabs, are less variable among individuals, likely because they are diluted in a common medium, such as Viral Transport Media (VTM), saline, or phosphate-buffered saline (PBS). Saliva, on the other hand, is heavily influenced by factors such as diet, time of day, smoking, and personal behavior. Nucleic acid purification, which aims to remove RNA from the patient specimen and reconstitute it in a common matrix, thus, eliminates confounding factors that may be present in minimally processed specimens. This is particularly important in colorimetric LAMP, which relies upon either pH or free magnesium content as a readout and is, therefore, potentially subject to greater interference from confounding substances.
Second, nucleic acid purification serves to concentrate RNA to improve diagnostic sensitivity by approximately 1 to 2 log10 units, depending on the original specimen, the method of purification, and the downstream assay. Given these 2 advantages, and despite the promise of (and enthusiasm for) Direct methods, for some uses, it may be worthwhile to introduce a nucleic acid purification step. Indeed, of the 10 RT-LAMP-based methods granted Emergency Use Authorization (EUA) as of April 2021, 5 use some form of nucleic acid purification (Table 1). In the following sections, we describe several open-source, low-cost methods for nucleic acid purification used by several of the authors as simple yet effective alternatives to commercial RNA extraction kits.
TABLE 1.
Summary of literature review of different loop-mediated isothermal amplification (LAMP) processing methods
|
Processing method
|
Source,a y
|
Approximate sensitivity or LoD
|
Sample type
|
| 65°C for 30 min | L'Helgouach et al. 2020 | 73% Sensitivity at qPCR Ct < 35. | Saliva |
| 95°C for 15 min | Alekseenko et al. 2021 | Sensitivity comparable to qPCR at Ct = 20–25 | NP swabs |
| 95°C for 5 min | Thi et al. 2020 | Sensitivity comparable to qPCR at Ct = 25–30 | NP swabs |
| Chelex-100, DTT | Howson et al 2020 | Sensitivity comparable to qPCR at Ct = 25–30 | Saliva |
| Chelex-100 | Flynn et al. 2020 | 100 copies/μL | Saliva |
| Proteinase K | Ben-Assa et al. 2020 | Sensitivity comparable to qPCR at Ct = 28 | NP swabs in UTM, saliva |
| Proteinase K | Azmi et al. 2020* | 10 copies/reaction | Saliva |
| Proteinase K (QuickExtract) | Joung et al. 2020* | 100 copies/reaction | NP swab in VTM |
| Proteinase K (QuickExtract) | Nguyen et al. 2020* | 1 copy/μL | NP swabs |
| Semialkaline proteinase | Yamazaki et al. 2021 | 170–230 copies/μL | Saliva |
| TCEP/EDTA (Quick Extract) | Agrawal et al. 2021* | 40 copies/μL | Saliva |
| TCEP/EDTA or DTT/EDTA | Rabe and Cepko 2020 | 50 copies/μL | Saliva, NP swabs |
| TCEP/EDTA | Sherrill-Mix et al. 2021a and 2021b | 100 copies/μL | Saliva |
| TCEP, proteinase K | Yang et al. 2021 | 200 copies/μL | Saliva |
| TCEP, RNASecure, proteinase K | Lalli et al. 2021 | 25 copies/μL | Saliva |
| Thermolabile proteinase K | Wei et al. 2020 | 2.5 copies/μL | NP swabs in VTM |
Abbreviations: Ct, cycle threshold; DTT, dithiothreitol; LAMP, loop-mediated isothermal amplification; LoD, limit of detection; qPCR, quantitative polymerase chain reaction; TCEP, tris(2-carboxyethyl)phosphine; UTM, universal transport medium; VTM, viral transport medium.
Asterisks (*) indicate sensitivity reported using a combination reverse transcription–LAMP/recombinase polymerase amplification (RT-LAMP/RPA) with CRISPR-Cas detection; otherwise, only RT-LAMP was used.
Silica-based methods
The propensity of nucleic acids to bind to silica in alkaline and high-salt conditions was first identified when DNA was demonstrated to bind to glass fibers when solubilized in sodium iodide (NaI). Since then, silica matrices have seen widespread adoption in molecular laboratories, most commonly in the form of silica spin columns. Although effective, the additional cost and centrifugation requirement of spin columns are antithetical toward the development of an accessible, low-cost test. Nevertheless, many efforts have sought to exploit this property of silica in a more-affordable fashion.
Researchers returned to the use of a silica particle suspension (glass milk) in conjunction with NaI for purifying RNA from either NPSs in saline/PBS or saliva (Rabe and Cepko 2020). Rabe and Cepko (2020) found that their glass-milk-NaI purifications achieved sensitivity down to 1 copy/μL in 500 μL of material in a pH-based, colorimetric RT-LAMP assay at a processing cost approximately $0.07/sample. Although the original method demands several centrifugation steps, they found that centrifugation can be substituted by a 5- to 10-minute settling step when NPSs in saline/PBS are used. However, saliva proved too viscous to be amenable toward this settling step.
However, the centrifugation demands of the Rabe and Cepko (2020) protocol are relatively light and, thus, can, theoretically, be fulfilled through low-cost alternatives, such as the Paperfuge (Bhamla et al. 2017). Li et al. (2020) validated the glass-milk protocol with saliva on in their open-source Handyfuge device, which they estimate at under $5 to construct. Intriguingly, Garneret et al. (2021) repurposed the concept of the silica gel membrane found in spin columns for use in a folding-card device. They embed a silica membrane on 1 side of the folding card and freeze-dried fluorescence-based RT-LAMP reactions on the other side. The NPS samples are injected into the silica membrane before being washed and are eluted into the freeze-dried RT-LAMP reactions. Although their article does not describe the chemical parameters of their assay, the principle of the silica membrane likely operates in a fashion similar to that of the Rabe and Cepko (2020) glass-milk purification.
Magnetic bead–based methods
Although glass milk likely represents the least-expensive option for nucleic acid purification, it does not reach scaling as well as other platforms do. If used with centrifugation, glass-milk purification becomes largely incompatible with most forms of liquid-handling automation; whereas if it is used without centrifugation, sample-processing time increases by up to 30 minutes. Magnetic beads operate on the same principle as glass milk—in fact, they usually consist of ferrite cores with a silica or carboxyl coating—but require only a simple magnet for bead separation. Although more expensive when purchased from commercial suppliers (see below), magnetic beads offer facile, centrifugation-free handling and improved automation compatibility.
Multiple groups have found commercial products that are compatible with RT-LAMP. Klein et al. (2020) present a protocol using SiMAG-N-DNA magnetic beads, combined with a homemade GnITC solution to purify nucleic acids from NPSs for use in both pH-based colorimetric and fluorometric RT-LAMP assays. Kellner et al. (2020) demonstrated the capacity of AmpureXP RNAClean to purify RNA from multiple sample types for use in magnesium-sensing colorimetric RT-LAMP. Altogether, they validated NPSs, gargle, and sputum samples mixed with Sputolysin (buffered DTT) solution and estimated their limit of detection from all sample specimens at 10 copies/μL.
An open-source alternative to AmpureXP beads for purification from saliva has been described (Yu et al. 2020), reducing processing costs from approximately $1/sample to approximately $0.20 per sample, reducing the processing time from 20 to 10 minutes, and improving the yield from saliva, relative to the commercial option, achieving a limit of detection of 3.7 copies/μL in pH-based colorimetric RT-LAMP. Interestingly, they found that typical magnetic bead–based purification, which involves removing supernatant from the beads during washing, introduced too much carryover of flocculent matter from saliva, which interfered with the subsequent RT-LAMP reaction. Instead, they used a 3-dimensional (3D)-printed magnetic stick with a disposable tip to remove the beads from the supernatant, which selectively removes the beads over the flocculent matter. Bektaş et al. (2021) also developed a magnetic stick. However, unlike Yu et al. (2020), who found it to be necessary for purification from saliva, Bektaş et al. (2021) were motivated to use a magnetic stick to produce a test that eliminated micropipetting and so was compatible with home testing.
In addition to solid-phase extraction through silica or carboxyl coatings, magnetic beads have also been used to purify nucleic acids through hybridization. Bokelmann et al. (2021) bound biotinylated oligonucleotides complementary to the Orf1b and N genes to streptavidin-coated magnetic beads and used them to purify specific regions of the SARS-CoV-2 genome from gargle samples in sterile water. They estimate their limit of detection to be 5 to 25 copies/μL. Whether this method proves to be substantially advantageous remains to be seen, but unlike solid-phase extraction methods, it does not require high-salt conditions to bind the nucleic acids, thus obviating the need for wash removal of that salt before the input into the RT-LAMP.
Direct Methods
The preceding section makes the case for nucleic acid purification in diagnostic testing to normalize potentially heterogeneous patient samples and also to increase sensitivity through nucleic acid concentration. However, despite widespread use, the necessity of such a cost- and time-intensive procedure may be an unchallenged axiom, rather than an empirically determined need. Given the unprecedented scale of testing demanded by the SARS-CoV-2 pandemic, many have sought to determine whether nucleic acid purification may be eliminated and yet maintain (sufficient) sensitivity and specificity across different patients (“Direct Methods”).
Any direct method must, nevertheless, still fulfill all 4 preanalytic criteria: (1) samples must be rendered noninfectious to ensure the safety of the technician or clinical staff; (2) viral RNA must be released from the viral envelope and made available for assaying; (3) ribonuclease (RNase) present in the patient sample must be inactivated, so that they do not reduce the amount of available RNA before assaying; and (4) the sample must be compatible with the assay in question by minimizing the interfering effect of compounds endogenous to the sample.
Samples processed under these criteria are referred to as minimally processed samples, and we provide an overview of such methods below.
Heat and chemical agents
Perhaps the most intuitive means to fulfill the first 2 criteria above is to denature the undesirable proteins with heat. Indeed, treatments from 60°C to 95°C for various amounts of time have been demonstrated to be effective at eliminating SARS-CoV-2 viral replication (see “Virus Inactivation Methods” above) and releasing RNA. At least 3 studies have found that direct-heat treatment of both saliva and NPS samples is compatible with downstream RT-LAMP, representing the simplest and most-minimal form of pretreatment.
However, RNases are known to resist denaturation at even boiling temperatures and are capable of refolding after denaturation. Therefore, heat treatment of patient samples is often augmented with reducing agents, either DTT or, preferably, the more-stable tris(2-carboxyethyl)phosphine (TCEP). Both RNA preservation agents exert an inhibitory effect on RNases by cleaving disulfide bonds, are compatible with downstream enzymatic assays, and have their activity enhanced at the high temperatures used for heat inactivation. Many studies have used TCEP to great effect, finding it to be an effective reagent in preprocessing saliva and swab specimens alike (Rabe and Cepko. 2020).
Proteinase K and surfactants
An alternative to heat denaturation of proteins is proteolytic digestion, which is, theoretically, effective against even the most heat-stable proteins, and it precludes the possibility of refolding. Like TCEP, proteolytic digestion stands to offer protection against RNases. Many approaches have validated the use of Proteinase K as an effective reagent for processing saliva and swab specimens. In general, this preprocessing step involves digestion for 5 to 15 minutes at 37°C to 65°C, during which, the Proteinase K remains active, followed by protease inactivation at 95°C for 5 to 10 minutes to ensure the Proteinase K does not interfere with downstream reactions.
In the event that the second 95°C incubation step is undesirable, thermolabile Proteinase K, which is active at 37°C and is inactivated at the 65°C used for LAMP assays, can be used. Its use decreases the potential for tubes to “pop” and create cross-contamination during elevated heating and simplifies the protocol, although there is comparatively less experimental validation than for the thermolabile variant (Wei et al. 2020), possibly because of its higher cost. Further validation and increased accessibility of thermolabile Proteinase K may be useful for future diagnostic efforts. In addition, 1 study (Yamazaki et al. 2021) has validated the effectiveness of semi-alkaline proteinase (SAP) on saliva samples as a possible alternative to proteinase K.
Mild surfactants can be used to disrupt cell membranes and release intracellular materials. Nonionic detergents, with uncharged, hydrophilic head groups, such as Triton X-100 and Tween-20, are routinely used to lyse cells (Johnson 2013) and are components of some RT-qPCR (e.g., Ranoa et al. 2020) and RT-LAMP (Azmi et al. 2020, Wei et al. 2020) methods. However, some researchers have observed that the addition of 0.1% Triton X-100 (but not the gentler surfactant Tween-20) during lysis of negative SARS-CoV-2 saliva samples resulted in RT-LAMP false-positive results (Andres Bendesky, July 2021, unpublished data), whereas others have noted the need for removal (i.e., washing) of Triton-X100 in some applications (Ma et al. 2020). Finally, Bendesky and colleagues (unpublished) report that the subsequent RT-LAMP reaction is more sensitive if Tween-20 is combined with “regular” (but not thermolabile) proteinase K during the extraction; presumably, the protease degrades the viral proteins that have become accessible after lysis of the lipid envelope.
Chelating agents
Rather than degrading RNases, RNA may also be protected by denying RNases the cofactors required for their function, such as magnesium. In conjunction with the metal ion chelator EDTA, TCEP exhibits a partial protective effect against some, but not all, RNases. Because Mg2+ is also a required cofactor for RT-LAMP, the allowable carryover of EDTA into the RT-LAMP reaction is limited. Therefore, other researchers (Flynn et al. 2020) added a chelating resin (Chelex-100) to saliva before a 95°C heat treatment, pelleted the resin via centrifugation, and used the supernatant as their RT-LAMP input. Although this benefits from the chelating effect of Chelex-100 without carrying over into the downstream reaction, it adds the requirement for a simple centrifugation step, which may be undesirable in certain cases. Recently, Howson et al. (2021) published encouraging results with a centrifugation-free saliva-processing method involving Sputolysin (buffered DTT), heating, and Chelex-100 resin, which resulted in a notably low false-positive rate (<1:3000) using a fluorescent intercalation detection scheme.
Considerations for pH-based colorimetric RT-LAMP
Of the 2 main colorimetric methods used to monitor LAMP reactions (changes in pH and [Mg2+]free), the pH-based readout is more commonly found in the literature. In this process, the reaction changes color as deoxynucleoside triphosphate (dNTP) incorporation by polymerase releases protons, thereby acidifying a weakly buffered reaction medium containing a pH indicator (see “Amplicon Detection”). As such, the initial pH of the reaction is critically important. When coupling a colorimetric readout with RNA purification, ensuring sample compatibility is achieved simply with a suitable buffer for reconstituting the RNA. However, when working with minimally processed samples, the sample itself must often be chemically adjusted to a suitable pH. To that end, many protocols include sodium hydroxide (NaOH), especially those that use TCEP, which is inherently acidic.
For swab samples, which are typically reconstituted in a diluent or transport medium, the contribution of the medium on the pH can be expected to prevail over the contribution from the respiratory specimen, meaning that one can expect a relatively homogenous spectrum of pH values, making it easy to ensure consistent compatibility with pH-based RT-LAMP methods (Butler et al. 2021). However, when assaying saliva samples directly, the saliva itself becomes the sole determinant of the initial pH. Saliva occupies a wide pH range and is easily influenced by factors such as time of day and diet, which confound efforts at developing a universal solution for ensuring saliva compatibility with colorimetric RT-LAMP. Efforts to test this have suggested final concentrations of NaOH ranging from 1.2 mM to 1.45 mM, but optimization is still required. At high concentrations of NaOH, alkaline samples exhibit reduced color changes, whereas, at low concentrations of NaOH, acidic samples exhibit color changes before incubation. This may be an acceptable compromise in low-throughput environments, in which expert, subjective determination and/or resampling for a compatible specimen are possible. However, minimally processed saliva has limited compatibility with pH-based colorimetric RT-LAMP in a middle- to large-scale diagnostic environment.
Partly for that reason, some laboratories prefer the use of alternative colorimetric dyes that operate on a different principle and in a buffered solution (e.g., hydroxynaphthol blue [HNB] for Mg2+) or use a fluorescence-based readout (see “Amplification Detection”). However, it is worth noting that, although the issue of compatibility is most obvious when developing a protocol for pH-based colorimetric RT-LAMP, it is likely a factor in other assays as well. The full chemical composition and degree of variation in respiratory samples among individuals is not well understood and their interaction with nucleic acid detection technologies also requires research. Such interaction variation in clinical samples suggests that certain sample types may be less amenable than others to detection, with corresponding propensities for false-positive or false-negative results. In the absence of a more-complete understanding of the interdependencies, as well as how that may change with variants of a virus (Alpert et al. 2021) or regional differences (Danko et al. 2021), this spectrum of compatibility tends to be functionally replaced in favor of a binary mode of interpretation: does the process either work or not. The degree to which patient sample heterogeneity affects purification-free testing is currently incompletely understood. Although it is an acceptable, and many would argue, necessary, compromise given the urgency of the pandemic and the need for widespread and rapid testing, future efforts may be well served by a more-systematic characterization of respiratory samples. As elsewhere, it is probable that such research will indicate that there is not a “1-size-fits-all” minimal processing solution.
TARGET AMPLIFICATION
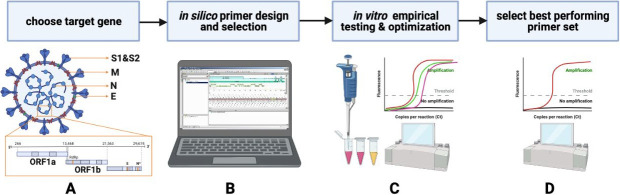
Nucleic acid amplification tests rely on 3 molecular steps: (1) sequence-specific recognition of the target through primer-base pairing, (2) enzyme-catalyzed amplification of the targeted DNA segment(s), and (3) detection of amplified products (amplicons). For SARS-CoV2, because the target is RNA, it must be initially reverse transcribed into complementary DNA before amplification (unless using direct RNA sequencing, as in Liu et al. 2019). The “Primer Selection” section reviews the general design of primers, reaction optimization, possible assay configurations, controls, and validation strategies. Figure 4 depicts a flow chart of the general means to design RT-LAMP primers and to optimize reactions applied to SARS-CoV-2.
FIGURE 4.
A flowchart for SARS-CoV-2 reverse transcription–loop-mediated isothermal amplification (RT-LAMP) primer design and selection. A) Having chosen a preferred viral target genomic sequence based on, for example, abundance or mutagenesis consideration, B) primer sets are designed and selected in silico, considering potential primer-dimers or other undesired interactions, inclusivity across SARS-CoV-2 variants, and exclusivity from other coronaviruses (i.e., Middle East respiratory syndrome [MERS]) or species. C) Having selected promising primer sets in silico, empirical testing D) (time-to-threshold, limit-of-detection, etc.), and reaction optimization in the laboratory identifies the set(s) with the desired empirical properties.
Primer Selection
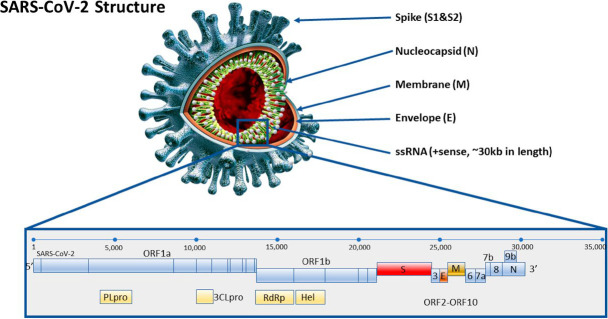
Specific and sensitive target amplification of SARS-CoV-2 by RT-LAMP requires selection of optimal primer sequences (based on the targeted genome sequence) and reaction conditions (often based on literature precedent from other RT-LAMP assays). The SARS-CoV-2 genome comprises approximately 30 000 RNA bases, encoding genes for structural (e.g., nucleocapsid [N] and spike [S]) and nonstructural (e.g., RNA-dependent RNA polymerase [RdRp]) components of the virus (Fig. 5). Early diagnostic efforts (eg, Corman et al. 2020) were based on the published sequence (Wu et al. 2020) and used several potential genes (ORF1a, ORF-1ab, RdRp, S, N, and E), which have become the focus of RT-LAMP and other nucleic acid tests (Table 2). Because the N gene is at the 3′-end of the viral genome, it is contained in all coronaviral, subgenomic RNAs (expressed as a nested set) and is the most-abundant viral RNA in infected cells (Finkel et al. 2021). Consequently, although the N gene is a good target, based on abundance, areas of the viral genome (e.g., the spike gene) harboring low frequency but functionally important mutations (see https://nextstrain.org; please see “Topic C”) might be predicted to be useful targets for examination of SARS-CoV-2 evolution.
FIGURE 5.
Representation of the physical and genomic RNA structure of SARS-CoV-2. The genome of the virus is shown at the bottom, and a rendering of the viral structure is shown on the top.
TABLE 2.
A collection of primers, samples, and further information used in the generation of this review
|
Primers
|
Samples
|
||||
|
Target
|
Typical use
|
Concentration, μM (BIP and FIP/B3 and F3/ LF and LB)
|
Type
|
Extraction
|
Other procedures
|
| Orf1ab | MV | 1.6/0.2/0.4 | Twist 102019 | No | (1) TCEP/EDTA (2) glass milk, chaotropic salts |
| Orf1ab | EUA, MV | 1.6/0.2/0.4 | Spiked, simulated patient samples | No | |
| Orf1ab | 1.6/0.2/0.4 | Patient samples | Yes | ||
| E1 N2 |
EUA, MV | 1.6/0.2/0.4 | Twist 102024; 10 ng/μL Jurkat RNA | ||
| Orf1ab S Orf8 N |
1.17/0.15/0.59 | VTM clinical samples; saliva | No | Heat at 95°C for 1 min. | |
| Orf1ab | RPA 0.48 F3/B3; LAMP 1.6/0.2/0.8 | Synthetic DNA | |||
| Orf1ab Orf1ab Orf1ab E1 N2 |
1.6/0.2/0.4 | Transcribed RNA; Heat inactivated virus | No | (1) Proteinase K, (2) Hudson | |
| Orf3a-A Orf3a-B Orf7a N |
1.6/0.2/0.4 | Patient NP swabs | Heat at 98°C for 15 min. | ||
| Orf8 | 1.6/0.2/0.8 | Patient NP swabs; transcribed RNA | RNA, Swab | Heat | |
| Orf1 (RdRp) | 1.6/0.2/0.4 | NP and throat swabs | Yes | ||
| E N |
EUA, MV | 1.6/0.2/0.8 | Patient NP swabs; transcribed RNA | Yes | |
| S RdRp |
1.6/0.2/0.8 | NP swab samples; synthetic construct; MS2 as control | Yes | ||
| N | 20/5/5 | Transcribed RNA; culture isolate | Yes | ||
| N S Orf1ab |
1.6/0.2/0.4 | Clinical throat swab; synthetic RNA | Yes | ||
| N | 1.6/0.2/0.4 | Throat swab | Yes | ||
| N | 0.8/0.2/0.4 | Synthetic gene | |||
Abbreviations: BIP, backward inner primer; cDNA, complementary DNA; EDTA, ethylenediaminetetraacetic acid; EUA, emergency use authorization, see “Regulatory Landscape”; FAM, fluorescein amidite; FIP, forward internal primer; GeneFinder, OSANG Healthcare (Seoul, South Korea), http://www.osanghc.com; ISO, International Organization for Standardization, Geneva, Switzerland; LAMP, loop-mediated isothermal amplification; LB, loop backward; LCV, leuco crystal violet; LF, loop forward; MV, multiple validation; ND, no data; NEB, New England BioLabs, Ipswich, MA; RPA, recombinase polymerase amplification; Sherlock, specific high-sensitivity enzymatic reporter unlocking; TCEP, tris(2-carboxyethyl)phosphine.
TABLE 2.
(Extended)
|
LAMP Reaction
|
Further Information
|
|||||
|
Amplification, min
|
LoD (copies/reaction)
|
Detection
|
Enzyme Mix
|
Volume/input, μL
|
Method notes
|
Source, y
|
| 60 | 1–50 | Colorimetric (pH); fluorometric | NEB E1700; NEB M0380; NEB M0538; NEB M1800 | 10–25/5 | Used TTTT linker between F1c, F2 regions in FIP and BIP; saliva as matrix | Rabe and Cepko 2020 |
| 30 | Colorimetric (pH); fluorometric | NEB M0372; NEB M0380; NEB M0538 | 25/1 | LoD: 1 fg 8 mM MgSO+. 63°C. | Lamb et al. 2020 | |
| 20 | 10-1000 | Colorimetric (pH); fluorometric | NEB M1800 | 20/1–5 | Used GeneFinder nucleic acid stain (D039); Mineral oil to avoid evaporation | Yu et al. 2020 |
| 20-40 | 10–40 | Colorimetric (pH); fluorometric | NEB M1800 | 20/3 | Combiplex N2, E1 As1e primers. Guanidine chloride to enhance sensitivity | Zhang et al. 2020 |
| 60 | 5–250 | Fluorometric | NEB M0538; NEB M0380 | 16/2 | N gene via 2-step protocol to maximize sensitivity. RT then cDNA amplification. | Ganguli et al 2020 |
| RPA 20; LAMP 40 | 4 | Fluorometric; colorimetric (LCV) | OptiGene ISO-001 | 50/1 | Two-stage amplification. RPA 38°C; LAMP 63°C | El-Tholoth et al 2020 |
| ND | <100 | Colorimetric (pH); fluorometric | NEB M0538; NEB M1800 | 20/1 | Lalli et al 2021 | |
| 40–50 | Dependent on primers | Colorimetric (pH) | NEB M1804 | 20/0.5–1.9 | Superior performance with Orf7a and Orf3a primer sets. Singleplex and multiplex, compared with Sherlock | Schermer et al 2020 |
| 25 | 100 | Fluorometric | NEB E1700; NEB M0380 | 25/5 | Notable for above-average amplification temperature. 67°C | Mautner et al 2020 |
| 60 | 25 | Colorimetric (pH) | NEB M1800 | 25/5 | Nawattanapaiboon et al 2021 | |
| 20–30 | 10 | Fluorometric | NEB M0538; NEB M0380 | 10/2 | RT-LAMP coupled with CRISPR–Cas12-based lateral flow. | Broughton et al 2020 |
| 30 | 25 | Fluorometric | NEB M0538; NEB M0380 | 25/12 | LAMP primers are reconstituted freshly each day; 40 mM guanidine hydrochloride pH 8.0 | Mohon et al 2019 |
| 30 | 100 | Colorimetric (pH) | NEB M0538; NEB M0380 | 10 / 2 | Much greater primer concentrations than typical LAMP | Baek et al 2020 |
| 30 | 80 | Colorimetric (pH) | NEB M0538; NEB M0380 | 25/1 | Used FAM labeled FIP primer for fluorometric detection. | Huang et al 2018 |
| 30–40 | 118 | Colorimetric (pH); fluorometric | NEB M0374; NEB M0380; NEB M0491 | 25/3 | RT-LAMP on mismatch-tolerant amplification | Lu et al 2020 |
| 50 | 6 | Colorimetric (pH); fluorometric | NEB M0374 | 50/3 | Simplified water bath setup for visual RT-LAMP; 55–61°C | Wang et al 2020 |
The original LAMP publications (Notomi et al. 2000, Mori et al. 2001) used 4 primers targeting 6 discrete sites on a sequence of interest (2 of the primers, forward inner primer [FIB] and backward inner primer [BIP], are effectively dual-function primers because they contain 2 discrete binding sequences; see Fig. 2). The addition of 2 “loop” primers (Nagamine et al. 2002) provides additional sites for the Bst polymerase to propagate DNA amplification with the consequence that the time-to-positive for many tests is reduced to well within 30 minutes. Robust reactions with comparatively little sensitivity to matrix interference (Francois et al. 2011, Hu et al. 2017, Kaneko 2007 see also “RNA Extraction” through “Direct Methods”) combined with rapid, supralinear, factorial-amplification kinetics allow target detection with a sensitivity comparable with that of RT-qPCR (Elvira-Gonzalez et al. 2017; Lucigen 2018). Information on previously reported sets is collated in Table 2 and Supplemental Table S1).
In silico primer design and selection
Several open-access software tools have been developed to help design multiple primers, each of which must bind simultaneously, but uniquely, to the target. They predict the potential for undesirable base-pairing interactions among primers and for the unwanted formation of secondary structures, such as stem-loop structures in the primers.
The first steps in such design are to select the target gene and ensure that unique sequences can be targeted (Fig. 4A–C). Basic Local Alignment Search Tool (BLAST, National Center for Biotechnology Information [NCBI], Bethesda, MD) analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to ensure primer sequences are nonrepetitive and unique compared with related pathogens (e.g., Middle East respiratory syndrome [MERS]) or with the human genome's repetitive areas (Rosenfeld et al. 2012) is essential. Several programs exist to design LAMP primer sequences for a target of interest. The more commonly used programs are the (1) PrimerExplorer V5 alone or with the MorphoCatcher (Shirshikov et al. 2019) website plug-in, which can scan individual 2000-bp regions or alignments of such regions; (2) a more-recent alternative from New England BioLabs (Primer Design) and open-source programs; (3) LAVA (Torres et al. 2011); or (4) GLAPD, which is able to accommodate full genomes as the input (Jia et al. 2019). Because of the complexity of the primer design and the many possible primers, the default settings in the chosen primer design software tools are generally a good starting point for many users.
It is important to ensure the target sequences are not predicted to cross-react with, and hence, amplify target nucleic acids from related coronaviruses (e.g., MERS; Shirato et al. 2014) or other species (Pipes et al. 2013), but especially from those that are human. In addition to assessing exclusivity (that primers bind only to the pathogen or target region of interest), it is also necessary to evaluate their inclusivity by comparing them against known variations in the targeted region (Corman et al. 2020, Sapoval et al. 2020). This is particularly (and increasingly) important as new variants of the virus emerge (https://www.gisaid.org/; https://nextstrain.org) to ensure newly arising viral variants are still detectable with these primer sets, including for low-input samples (Parker et al. 2020). From an inclusivity perspective, LAMP is, in general, quite tolerant of small changes to the target sequence, although the converse is that it is more difficult to create specificity for single-nucleotide polymorphisms (SNPs) or variants at the level of primer selection (see “Topic C”).
Empirical testing
Using these programs, several (2–5) basic primer sets (FIP/BIP and F3/B3) are typically designed for each target and then tested experimentally, initially separately (i.e., single-plex) using simpler DNA/RNA controls as the template, screening for amplification speed (shortest time-to-positive), sensitivity (lowest limit of detection [LoD]), and a low frequency of spurious amplification in nontemplate controls (NTCs; e.g., water; Fig. 4D–E). Initial tests ideally use “real-time” amplification conditions, for example, with an intercalating dye (e.g., SYTO9) or other continuous readout (see “Sequence-Independent Detection of RT-LAMP Amplification Products).
After the best of the 4-core LAMP primers are chosen, 2 loop primers, designed to enhance specificity and accelerate the amplification (Nagamine et al. 2002), are typically included. Further in silico tests with a variety of multiple-primer analyzers (e.g., Multiple Primer Analyzer, Thermo Fisher Scientific, Waltham, MA; Oligo Calc, Northwestern University, Evanston, IL; and OligoAnalyzer, Integrated DNA Technologies, Coralville, IA) allow assessment of potential base-pairing interactions between and among chosen primers to rule out undesirable formation of stem-loops or unintended complementary base-pairing, especially at their 3′-ends. These additional checks seek to maximize the specificity of amplification by the primers and to avoid cross-priming (see also Supplemental Information S2).
Despite the undoubted necessity and value of in silico design, the most sensitive and specific set of primers is selected empirically. The best primer sets can detect single-digit copies of their target per reaction within about a half hour; a fit-for-purpose assay for many applications will be able to detect several hundred target copies per reaction. Enhanced sensitivity has been reported by including forward and backward “swarm” primers targeting areas upstream of the FIP/BIP hybridization sites (Martineau et al. 2017), an approach achieving single-digit or reaction sensitivity for SARS-CoV-2 with an HNB/Mg2+ colorimetric readout (Lau et al. 2020).
Troubleshooting
If a complete primer set with acceptable properties cannot be found or if improvements are needed after empirical testing (see below), it is possible to find alternative sets in the region of interest, the adenine-thymine/guanine-cytosine (AT/GC) content can be increased or other individual parameters (such as melting temperature, region length requirements, spacing, among others) can be readjusted. Early studies with LAMP included 4 dTs that separated the 2 approximately 20- to 25-nucleotide component F1c/B1c and F2/B2 regions that make up the longer FIP/BIP primers. This approach is sometimes still used (e.g., the As1e primer set; Rabe and Cepko 2020) and remains an option because the TTTT sequence is predicted to disrupt potential secondary structures. When ordering primers, high-performance liquid chromatography (HPLC) purification is particularly recommended for the longer FIP/BIP primers, which, although more expensive than a simple desalting purchase, eliminates truncated primers formed during synthesis, which can reduce sensitivity and reaction efficiency. Therefore, if the first set of poorly performing primers were not HPLC purified for cost reasons, this is an avenue to explore.
Although most LAMP assays use a relatively standard range for the absolute and relative concentrations of each primer (0.2 μM F3/B3, 0.4 μM Loop F/B, 1.6 μM BIP/FIP), this practice is common but not absolute (e.g., Allgower et al. 2020). For example, reducing the F3/B3 concentration relative to the other 4 primers can sometimes improve efficiency (Sridar Chittur, June 2021, personal communication). Alternatively, doubling the concentration of loop primers can increase sensitivity, whereas, in other cases, somewhat counterintuitively, complete omission of 1 of the loop primers can also enhance assay specificity and/or amplification efficiency (S. Chittur, personal communication). Similar to PCRs, modifications to primer concentrations and altering reaction conditions ([Mg2+]; dNTP; buffer type; and addition of “enhancing” additives, such as betaine, dimethylsulfoxide [DMSO], and notably, GnHCl; Zhang et al. 2020b) can lead to improved assays. However, many researchers prefer the convenience (albeit at increased cost) of using a standard, preprepared master-mix from a commercial supplier of LAMP reagents because those products have already undergone assay or buffer optimization.
If open-source enzymes are used (see “Open-Research Infrastructure”), individual optimization will necessarily be required for each important reaction variable. It is clear that some enzymes and/or their engineered derivatives have altered biochemical properties that can be affected by solution conditions (e.g., salt concentrations; please see “Topic B”). As in many cases with LAMP technology, the researcher can choose to take a more “active” bottom–up building approach to developing a test or primer set or a more “passive” kit-based approach, based on literature precedent, again, depending on his or her expertise, needs, timeframe, and budget.
Multiplexed primer sets
Multiplexing is performed in 1 of 2 ways, depending on the test's objective. If the objective is to enhance genomic coverage, speed, and/or sensitivity of LAMP reactions, one approach is to combine primer sets for multiple genes from the target, for example, genes N, E, and Orf1a from SARS Co-V-2. In this configuration, the particular gene or amplicon being amplified within the combiplex is not sought (Dudley et al. 2020, Zhang et al. 2020b, Butler et al. 2021). In contrast, when multiple targets are to be distinguished in a single reaction (e.g., SARS-CoV2 and influenza or SARS-CoV2 and human internal-extraction control genes; Zhang et al. 2020b), then sequence-specific, multicolor detection must be employed for such bone fide multiplexes (see “Sequence-dependent detection of RT-LAMP amplification products”).
In either case, the design phase necessarily becomes more complex because of the combinatorial potential for base-pairing interactions among primers. Depending on the individual sequences used, combining 2 (12 primers) or 3 (18 primers) targets into a single reaction can be achieved, although the potential for spurious amplification in nontemplate controls should be carefully monitored. Nonspecific amplification curves are typically shallower (reflecting less-efficient amplification), can often be multiphasic, and can yield a melt curve that has a maximum slope (−dF/dT) that is sometimes noticeably (approximately 2°C to 3°C) shifted away from that of bone fide amplicons. Of course, the only true means to distinguish template amplification from nonspecific amplification is ultimately to perform sequencing (“Sequencing Approaches Using RT-LAMP Amplicons”), although that is rarely used in routine practice for most or many laboratories.
Enzymes
Strand-displacing DNA polymerase
The DNA polymerase used for DNA amplification in LAMP reactions must have both DNA template-dependent 5′→3′ polymerase activity but also strand displacement activity at a single elevated temperature (60°C to 74°C, depending on enzyme, usually between 63°C and 65°C). The most commonly used polymerase is Bst DNA polymerase from the thermophilic bacterium Geobacillus stearothermophilus (formerly Bacillus stearothermophilus, hence Bst) (Phang et al. 1995, Kiefer et al. 1997). More specifically, the Bst-LF (large fragment) lacking the smaller N-terminal domain responsible for 5′→3′ exonuclease activity is used because of its ability to perform strand-displacement synthesis in the absence of the nuclease activity (Maranhao et al. 2020).
Engineered enzymes capable of nucleic acid amplification have recently been reviewed (Yasukawa et al. 2020). Thermophilic versions of DNA polymerases, capable of resisting high temperatures (e.g., during sample treatment), have been evolved in vitro via emulsion-based directed evolution (i.e., high-temperature isothermal compartmentalized self-replication in Milligan et al. [2018]) with some being used successfully in RT-LAMP (Alekseenko et al. 2021). Further improvements to increase purification yields include replacing the N-terminal domain of Bst with the small F-actin-binding protein villin, a modification that improved folding and protein solubility (Maranhao et al. 2020). Taking a different approach, Ignatov et al. (2014) developed a derivative of the classic Taq polymerase (SD DNA polymerase, Boca Scientific, Dedham, MA) possessing stronger strand-displacement activity and thus suitable for LAMP. The Lucigen (Middleton, WI, https://www.lucigen.com/) LavaLAMP uses the OmniAmp polymerase (developed from PyroPhage3173 DNA polymerase; Chander et al. 2014), which operates at higher temperatures (68°C to 74°C), potentially leading to improved sensitivity, specificity, and, in some conditions, faster reaction time (Lucigen LAMP Information).
Other commercial developments include in silico design and evolution to improve amplification speed, salt tolerance, thermostability, and yield of LAMP reactions. For instance, New England BioLabs (https://www.neb.com) created 2 Bst variants: (1) Warmstart Bst 2.0 allows preparation and assembly of reactions at room temperature because the polymerase is prepared with a reversibly bound aptamer, which inhibits polymerase activity at temperatures below 45°C; and (2) Bst3.0, designed to have increased RT activity to enable 1-enzyme amplification of RNA targets without addition of reverse transcriptase (see below).
Reverse transcriptases and dual-function polymerases
For SARS CoV-2 detection, the reaction needs to detect the viral positive-sense RNA, and thus, RT of RNA to complementary DNA (cDNA) is required before amplification. By simply altering buffer conditions, some polymerase can act as both a reverse transcriptase (RT, RNA-dependent DNA polymerase) and DNA polymerase (Bhadra et al. 2020a, Bhadra et al. 2020b, Roche Diagnostics 2016). More purposefully, dual function DNA polymerases with RT activity have been developed as protein chimeras (Schönbrunner et al. 2006), and a Bst variant with enhanced RT activity has been developed (Bst 3.0, New England BioLabs).
There is general consensus that diagnostic assays that use a single, dual-function enzyme system are less sensitive than those employing dedicated RT and strand-displacing DNA polymerases in combination. If the 2 enzymes are to be combined and if both RT and DNA amplification steps are to be performed at a single temperature, the RT must retain sufficient activity at 60°C to 65°C. Historically, the avian myeloblastosis virus (AMV) RT was the enzyme of choice for RT-LAMP, and this remains a viable option. Recently, RT from HIV has been demonstrated to be a functional, open-source alternative (Kellner et al. 2020) to AMV/murine leukemia virus (MMLV) and even used to detect RNA modifications (Saletore et al. 2012, Liu et al. 2019), but other options include engineered RTs, such as WarmStart RTx from New England BioLabs (Zhang et al. 2020a, Zhang et al. 2020b) and SuperScript IV RT (Park 2020).
Other considerations
Three other considerations include the following:
Storage: Lyophilized mixes for LAMP reactions were first described in studies to detect the Newcastle disease virus (Pham et al. 2005). The approach, pioneered commercially by Eiken Chemical (Tokyo, Japan) with freeze-dried pellets being placed in the caps of PCR tubes, has been followed by Optigene (Horsham, England; http://www.optigene.co.uk/; http://www.optigene.co.uk/polymerase-selection-guide/).
Two-temperature RT-LAMP: If RT-LAMP reactions are intended to be performed in a laboratory setting with a conventional qPCR thermocycler, a “1-pot-2-temperature” design can sometimes lead to increased sensitivity, whereby the RT reaction is allowed to proceed at a lower temperature (i.e., 10 to 20 minutes at 55°C) than polymerization (approximately 65°C) (Ganguli et al. 2020, Bektaş et al. 2021).
Open-source enzymes: Finally, to both reduce costs and improve the supply chain in the face of high demand, researchers (notably in developing countries) have increasingly begun to explore in-house production of enzymes required for RT-LAMP. These “DIY” (do it yourself) alternatives to commercial enzymes include RTs and DNA polymerases (Bst-LF lysates; Kellner et al. 2020, Sherril-Mix et al. 2021 Bhadra et al. 2020, Alekseenko et al. 2021; see also “Topic B” (Bst enzyme engineering) and “Open-Source Reagents and Biomolecular Tools”).
Controls and Reference Standards
As a matter of routine, positive and negative reaction controls must always be run in parallel (SEQC/MAQC-III Consortium 2014, Foox et al. 2021) with each unknown sample (or each test-batch of unknowns, depending on the scale of testing). If amplicons are not detected in the positive control, the functionality of the reaction mix may be suspect; if amplicons are detected in the negative control, spurious (mispriming) amplification or cross-contamination (with amplicon or spurious DNA) could have occurred.
Negative controls and contamination mitigation
The NTCs most commonly use DNA/RNA-free, nuclease-free distilled, deionized water or Tris-EDTA (TE). Although autoamplification in NTC wells from primer:primer interactions can be observed after prolonged incubation (≳40 to 60 minutes), if amplicons are detected in NTC wells significantly earlier in the reaction time course (≲40 minutes), cross-contamination with previously synthesized amplicon product could also be suspected. Given the exceptionally large amount of multimeric amplicons formed in LAMP (cf. PCR) reactions and their particular stability, the potential for cross-contamination of amplicon from 1 run to the next is very significant. Laboratories should employ strict inventory and storage procedures (e.g., single-use aliquots) and location- or isolation-related standard operating procedures. Thus, isolation of “clean” areas (for aliquoting primers and assembling “mastermix” solutions) from sites in which amplicons could be present is strongly recommended; researchers often employ separate laboratory coats for each area. Whatever procedures are applied, maintaining a heightened awareness of the potential for contamination is essential in all LAMP laboratories.
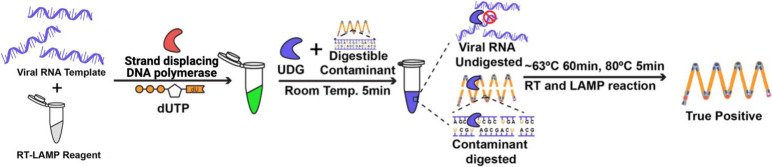
Even great care to avoid cross-contamination may still be insufficient. The deoxyuridine triphosphate (dUTP)/uracil DNA glycosylase (UDG) (or uracil-DNA N-glycosylase [UNG]) system (Kim et al. 2016) incorporates dUTP into LAMP amplicons in “experiment N.” If such labeled amplicons are unintentionally carried over into “experiment N + 1,” then they are cleaved by the UDG enzyme present in the LAMP reaction mixture before being spuriously amplified (Kellner et al. 2020; Fig. 6). The dUTP/UDG system can greatly reduce such risk, with limited effect on the RT-LAMP reactions (a marginal loss in sensitivity), but they cannot be completely removed. Confirmation that the amplicons are, indeed, specific for the target of interest can be accomplished by direct sequencing, but melting-curve analyses can also characterize different amplicon products in a multiplex reaction (e.g., Sherril-Mix 2021), without resorting to the opening of postamplification tubes.
FIGURE 6.
Uracil-DNA-glycosylase (UDG)-supplemented reverse transcription–loop-mediated isothermal amplification (RT-LAMP): the system (Kim et al. 2016) removes carryover DNA contamination from one experiment (N) to subsequent ones (N + 1). In the first experiment, uracil is incorporated into contaminants through the use of approximately one-third of the deoxyuridine triphosphate (dUTP): approximately two-thirds of the deoxythymidine triphosphate (dTTP) is used in the amplification reaction—amplicons so derived contain a mixture of T and U bases. In the subsequent (N + 1) experiment, UDG is added to the input sample before amplification. The UDG specifically cleaves uracil-containing contaminants that were inadvertently carried over from the first (N) experiment at room temperature. Upon elevation of the reaction to approximately 65°C, the UDG is heat inactivated, ensuring that only the target RNA (or 100% thymine-containing DNA) target is amplified.
External positive controls
Beyond target DNA for testing the LAMP primer amplification, the simplest positive control is the isolated nucleic acid itself, such as synthetic viral RNAs (from, e.g., Integrated DNA Technologies, Twist Biosciences [South San Francisco, CA], or from https://www.beiresources.org/) or RNA transcribed in vitro with a T7 promoter in the appropriate primer context (e.g., for the N gene; Zhang et al. 2020a). Such naked RNAs can control for reverse transcriptase activity and subsequent DNA amplification, but, because they are already purified nucleic acid, they do not allow for testing extraction efficiencies and cannot readily mimic methods designed to be used with “direct” or “extraction-free” methods as applied to, for example, saliva. Encapsulated viral particles are available, albeit at higher cost (e.g., Accuplex 0505-0126 (Antech, Exeter, England) or ZeptoMetrix [Buffalo, NY] NATFRC-6C) and with a greater chance of between-lot, and even within-lot, variations. Consequently, some have abandoned their use in favor of internally benchmarked patient samples (by serial dilution; Bendesky, unpublished results; Vogels et al. 2021). Probably the most-common sources of controls are heat-inactivated (NR-52286, BEI Resources) or γ-irradiated (NR-52287, BEI Resources) cell lysates of VeroE6 kidney epithelial cells infected with SARS CoV-2 isolates available twice yearly from.
Extraction-positive control
An essential component of molecular diagnostic tests is an extraction or process-positive control. This serves to ensure not only the proper functioning of the assay components (enzymes, primers, buffers, Mg2+, and dNTP) but also the successful extraction of nucleic acid from the sample of interest. A set of primers targeting DNA or messenger RNA (mRNA) is expected to be in every sample, regardless of infection status; for human diagnostic tests, RNase P (e.g., Color Genomics [EUA], Color Health, Burlingame, CA) or β-actin (e.g., Zhang et al. 2020a) are most commonly used. Designing primers across an exon-exon junction—a feature present only in RNA, not genomic DNA—would additionally confirm activity of the reverse transcriptase for robust RT-LAMP assays and is preferable but not essential.
Incorporating such internal controls provides greater confidence for a true-negative result because that sample material was known to be efficiently extracted, that nucleic acid was transferred to the LAMP reaction, and that amplification occurred. During clinical testing, a failed internal control invalidates the test, such that no result is reported to the sample provider. During basic research or method development, a negative result suggests that 1 or more essential components of the reaction were omitted and/or that they have lost functional activity.
Extraction controls are either external (separate reaction from the clinical sample with SARS-CoV-2 primers, such as MS2) or internal (RNaseP or actin amplification duplexed with SARS-CoV-2 primers). Similar to RT-qPCR tests, the latter is obviously preferable if 2-color fluorescence detection can be achieved with sequence-specific methods (Sequence-Dependent Detection of RT-LAMP Amplification Products”). However, the 2-component–2-reactions should be kinetically balanced so that 1 reaction does not dominate and consume all the limiting reagents (e.g., dNTPs, polymerase). If not optimized, sensitivity for SARS-CoV-2 will be affected, whereas, conversely, a strongly positive sample for SARS-CoV-2 could erroneously appear negative in the internal RNaseP/actin control. The former situation can be readily avoided by reducing the RNaseP/actin primer concentrations or omitting the loop primer(s) to slow that reaction, despite the presence of high DNA/complementary DNA (cDNA) template concentrations.
AMPLICON DETECTION
Much amplicon is generated in LAMP reactions (a consequence of very efficient amplification), which leads to more detection options. The fluorescent methods are akin to those used in RT-qPCR, whereas others reflect changes to the bulk concentrations of reaction components (notably Mg2+, inorganic pyrophosphate [PPi], and H+), leading to a plethora of different detection methods for LAMP (Becherer et al. 2020). We focus on those most commonly used, which generate an optical (rather than, e.g., an electrochemical) signal or which rely on sequencing of amplicons. Two important distinctions among common detection methods include end-point vs. real-time detection (Fig. 7A and B) and sequence-dependent vs. sequence independent methods (Fig. 7C) (Zhang et al. 2014, Becherer et al. 2020). End-point tests are measured at a defined timepoint (e.g., t = 30 minutes), whereas real-time tests follow the progress curves kinetically during the amplification process, yielding the LAMP equivalent of a qPCR cycle threshold (Ct) value. The latter is clearly richer in information and, so, is useful during method development or when semiquantification is sought (LAMP is not generally used for absolute quantification), but it does impose the need for at least a simple fluorescence reader, even if that reader is a smartphone camera combined with a light diode and optical filters (“Hardware Designed for LAMP-Based Assays”).
FIGURE 7.
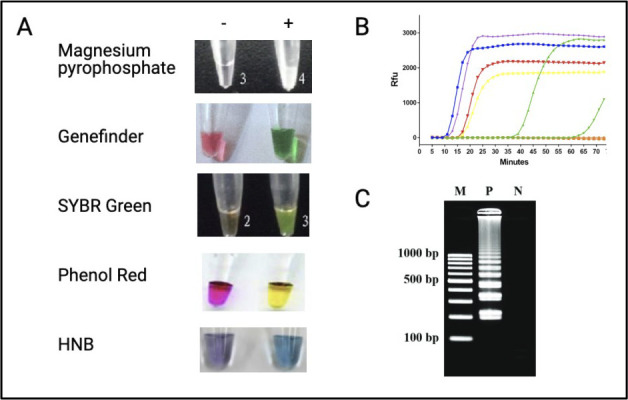
LAMP detection methods overview. A) Visual endpoint readouts use dyes that exhibit simple color or turbidimetric changes upon amplification. B) Similar to qPCR, real-time detection methods use fluorescent dyes to monitor the increase in viral load as the amplification progresses. The fluorescent signal can be sequence-independent (e.g., DNA intercalating; see “Sequence-Independent Detection of RT-LAMP Amplification Products”) or sequence-dependent (hybridization-based; see “Sequence-dependent detection of RT-LAMP amplification products”). C) The loop-mediated isothermal amplification (LAMP) products can, in principle, be verified by agarose gel electrophoresis followed by DNA staining, although that requires postamplification manipulation and the corresponding very real risk of between-experiment cross-contamination.
LAMP amplification products can be detected by running a portion of the finished reaction through an agarose gel along with a DNA-staining dye or fragment analyzer (Agilent Bioanalyzer [Agilent Technologies, Santa Clara, CA], TapeStation [Agilent], or an equivalent). Amplification leads to production of a distinctive ladder-like pattern. However, routine handling of positive LAMP products on the bench is not advisable because of the very considerable quantity of DNA produced (10 to 20 μg vs. 0.1 to 1 μg for PCR; Mori et al. 2001). Consequently, detection methods requiring opening reaction tubes after amplification (e.g., Zhang et al. 2021) are increasingly less common and should be avoided unless extreme measures are taken to avoid contamination (“CRISPR-Cas Cleavage Systems”) and which should almost certainly include the dUTP/UDG system (Kim et al. 2016; “Negative Controls and Contamination Mitigation”). Because many sequence-independent and sequence-dependent “closed tube” detection methods have been developed, we suggest that these should be the default to avoid the very real (and very frustrating) risk of cross-contamination and hence false-positive results.
Sequence-Independent Detection of RT-LAMP Amplification Products
In this section, we consider detection methods that rely on bulk changes to the concentrations of substrates or products, but which are not dependent upon the nucleotide sequence of the amplicon being created (compare with “Sequence-Dependent Detection of RT-LAMP Amplification Products) (Fig. 8).
FIGURE 8.
Categorization of detection methods. Reactions can be monitored using simpler, but less-specific, sequence-independent methods (e.g., pH changes) or the somewhat more complex but equally more-specific sequence-dependent methods. These, in turn, can either be monitored in real time (e.g., detection of amplification by releasing of quenching [DARQ], intercalating dyes) allowing amplicon formation to be monitored kinetically or as an end-point, stopping/recording the result at a defined time (e.g., quantify and annotate short reads in R [QuasR]). DNA sequencing is the ultimate sequence-dependent endpoint method. When sequence-independent methods are used, false-positive results can be an issue. Most sequence-dependent methods also allow for multiplexing multiple targets in the same reaction. Sequencing of amplicons can allow detection of different variants. Variations on these themes have been described—the location of the icon in the 4-box is purely indicative.
Turbidimetry
Historically, LAMP reactions were monitored in real time using turbidimetry arising from the formation of a white magnesium pyrophosphate precipitate that correlated with the amount of DNA generated (Mori et al. 2001). Some disadvantages of this method (Zhang et al. 2014) included a low signal-to-noise ratio (improved by adding calcium; Almasi et al. 2012), the relatively low abundance of real-time turbidimeters, and an incompatibility with minimally processed turbid samples. Turbidimetry was used in an early SARS-CoV-2 article (Yan et al. 2020), along with the fluorescent metal indicator calcein (Tomita et al. 2008), which can also be detected by eye (light orange to light green) or under a handheld ultraviolet (UV) lamp. In general, turbidimetry has been relatively infrequently used for SARS-CoV-2 LAMP assays.
pH-based dyes
Originally developed by New England BioLabs and used in early SARS-CoV2 LAMP articles (Butler et al. 2020, Zhang et al. 2020a, Zhang et al. 2020b), the widely used method of pH-based dyes is based on the color change of the simple pH-sensitive dye phenol red in a minimally buffered reaction as protons are released upon dNTP incorporation. In idealized conditions, the difference between a positive and negative result is visually striking (pink/yellow) but can be harder to determine for actual patient samples by eye because of intermediate or ambiguous color changes (Ben-Assa et al. 2020, Huang et al. 2018, Coehlo et al. 2021). In addition, the time window between the color change in positive reactions and negative controls can be small (Dao Thi et al. 2020, Fowler et al. 2020), so close monitoring of incubation time is required. Because the reaction is minimally buffered, the pH of the input clinical sample can have a measurable effect on the colorimetric signal, independent of (and appearing as) DNA amplification. This is particularly problematic when minimally processed samples are combined with minimally buffered reactions and is seen most acutely with saliva, although the effect has been reported with some viral transport media. Some of these shortcomings can be overcome if reactions are followed in real time using a plate reader and data are analyzed using either derivatives or the difference of measurements in 2 separate channels (Dao Thi et al. 2020), but that can increase the complexity for testing at scale or for point-of-care testing in resource-limited settings.
To address the color discrimination challenges associated with phenol red, Brown et al. (2020) synthesized 2 pH-sensitive dyes—LAMPShade Violet (LSV) and LAMPShade Magenta (LSM)—which, respectively, offer a purple/clear and pink/clear color change to distinguish between negative and positive results. The LSV is reported to have fewer intermediate products and greater contrast, relative to phenol red, in similar minimally buffered reaction conditions (Brown et al. 2020, Yu et al. 2021).
pH-independent colorimetric dyes
Leuco crystal violet (LCV) (Miyamoto et al. 2015) turns from colorless to violet in the presence of double-stranded DNA. Similarly, HNB (or eriochrome Black T) turns from violet to blue when free magnesium is removed because of the formation of Mg·PPi (Goto et al. 2009; there is also a change in fluorescence from 540 nm to 610 nm; Seok et al. 2017). These dyes generate relatively weak color changes but can be amplified with algorithmic image transformations (Martineau et al. 2017, Kellner et al. 2020, see also “Software”). Both were recently used for the colorimetric detection of SARS-CoV-2 (Kellner et al. 2020, Lau et al. 2020, Park et al. 2020).
However, another type of colorimetric detection method was recently developed that involves the use of spermine, silica, and charcoal (Mason and Botella 2019). Spermine destabilizes amplified DNA, which, in turn, causes rapid flocculation of suspended particles of silica and charcoal. This is a low-cost method with results (gray/clear) that can be read via the naked eye but, to date, has only been applied to detecting DNA targets (Mason and Botella, 2020).
Intercalating fluorescence
Classically, fluorescent DNA intercalating dyes have been used for both real-time and endpoint detection. Dyes can differ in respect to their optical properties, signal-to-noise ratio, optimal concentration (which can also change depending on the fluorescence-detection instrument being used), and inhibitory effects on the LAMP reactions (Seyrig et al. 2015, Oscorbin et al. 2016, Quyen et al. 2019). SYTO 9 (Thermo Fisher) for green fluorescence and SYTO 82 (Thermo Fisher) for orange fluorescence were found to be the least inhibiting and to have the best signal-to-noise ratios in multiple studies, but more-common dyes, such as SYBR Safe (Thermo Fisher; Carter et al. 2017), EvaGreen (Biotium [Fremont, CA]; García-Bernalt Diego et al. 2019; García-Bernalt Diego et al. 2021), and even ethidium bromide have been successfully used (Nagamine et al. 2002, Almasi et al. 2012).
Two additional dyes that have also been reported for SARS-CoV-2 detection and which are both colorimetric and fluorescent include the Genefinder dye (Yu et al. 2020) and SYBR Green I (Lamb et al. 2020, L'Helgouach et al. 2020, Garcia-Bernalt Diego et al. 2019). Unfortunately, SYBR Green I is inhibitory to LAMP, so it must be added after the reaction is complete, which can be solved without opening tubes by wax encapsulation (Zhang et al. 2013). In contrast, the GeneFinder dye can be added before the reaction (Alamasi et al. 2012) but is not yet widely available outside of China.
Bioluminescence
The bioluminescence real-time (BART) assay (Kiddle et al. 2012) relies on bioluminescence produced when inorganic pyrophosphate release is coupled enzymatically first to adenosine triphosphate (ATP) production via ATP sulfurylase and then to light formation via the luciferase/luciferin reaction. Although less widely adopted, BART has demonstrated high sensitivity (down to single copies), amenability to field detection (Hardinge et al. 2018), and has recently been demonstrated for SARS-CoV-2 detection (Fei et al. 2021)
Sequence-Dependent Detection of RT-LAMP Amplification Products
In contrast to sequence-independent detection methods outlined above, sequence-dependent methods generate a signal that is exclusively (or heavily) dependent upon the specific nucleotide sequence of the amplicon. With assays based on optical, magnetic, piezoelectric, electrochemical, or magneto-resistive sensing (Becherer et al. 2020), we focus exclusively on those creating a fluorescent signal (Fig. 9).
FIGURE 9.
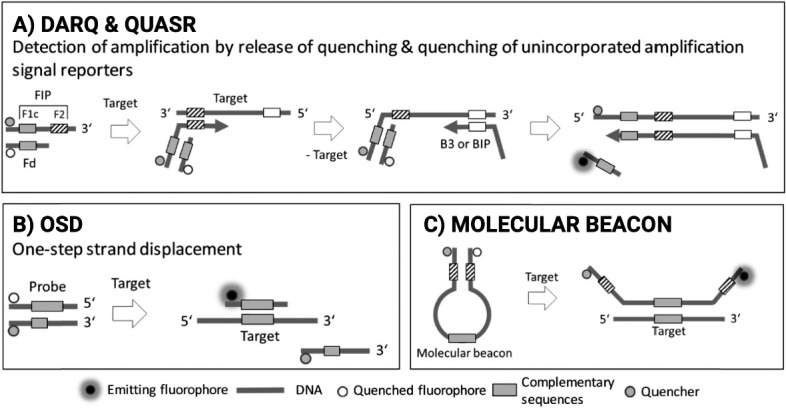
Schematic illustrations of some sequence-dependent fluorescent detection methods. The A) detection of amplification by releasing of quenching (DARQ) and quantify and annotate short reads in R (QuasR), B) one-step strand displacement (OSD), and C) molecular beacon methods can improve specificity of detection and processing for viral and other genome targets.
In all LAMP assays, amplification of nonspecific products, such as primer dimers can be common because of the high primer concentrations needed and the processivity of the polymerases—this is an “Achilles heel” of the methodology. Because sequence-dependent detection methods, in principle, will avoid detection of nonspecific products, these methods are anticipated to be associated with fewer false-positive results (Moehling et al. 2021). Another advantage of sequence-dependent detection is that 2 or more targets can be detected in 1 reaction tube via multiplexing of primer sets with different colored detection probes. This enables internal (within reaction) process controls using primer and probe sets for RNaseP/actin in addition to SARS-CoV2 genes and/or allows detection of viruses causing symptoms similar to SARS-CoV-2, such as influenza (e.g., Zhang et al. 2020b).
Most sequence-specific reporting methods use a variation of a fluorophore-quencher duplex or hemiduplex probe. Signal results from the incorporation of part of the probe into the amplification product, which eliminates fluorescence resonance energy transfer (FRET) between the fluorophore and quencher to produce a fluorescent signal. Six such methods are described below, of which “MB (Molecular Beacons)” and “CRISPR-Cas” reporting require designs of more bioinformatically complex probes or Cas protein/guide RNA complexes. In contrast, QuasR (quantify and annotate short reads in R), DARQ (detection of amplification by releasing of quenching), DP (displaceable probe), and OSD (one-step strand displacement) can be more readily developed based upon existing RT-LAMP primer sets. QuasR reporting generates a bright fluorescent visual signal in comparison with DARQ and potentially has a less-inhibited reaction because of the shorter length of the quencher oligonucleotide. OSD has an added advantage in comparison to QuasR of further discriminating against false-positive results because of the toehold strand displacement of its hemiduplex reporter and the potential for discriminating variants. An important distinction is that, although DARQ, DP, MolBeac (Molecular Beacons), and OSD enable real-time kinetic monitoring, QuasR is inherently an endpoint measurement.
QuasR (quenching of unincorporated amplicon signal reporters)
Ball et al. (2016) reported an endpoint fluorescent method that stands out because it can be readily applied to an already established LAMP primer set for sequence-specific reporting. The modification involves adding a fluorophore on the 5′-end of internal (FIP/BIP) or accelerating (LF/loop backward [LB]) primers. An additional oligonucleotide, complementary to, but shorter than, the fluorescently labeled primer, is added with a quencher on its 3′-end. The melting temperature (Tm) of the fluorophore-quencher oligonucleotide complex is designed to be at least 10°C lower than the LAMP reaction incubation temperature (approximately 65°C), allowing for the incorporation of the fluorophore-tagged primer into the amplicons generated by the reaction (see Supplemental Information S2). At the end of the assay, the reaction is cooled and unincorporated fluorophore-tagged primers anneal to the quencher oligonucleotide resulting, preventing fluorescence. However, if fluorophore primers are incorporated into the amplicons, they are protected from annealing to the quencher oligonucleotide, and positive tubes will fluoresce. Thus far 2 previously published LAMP primer sets, NA (Zhang et al., 2020) and NM (Mammoth Biosciences) have been successfully modified to use QuasR reporting (Aidelberg and Aronoff 2020, Bektaş et al. 2021).
DARQ (detection of amplification by releasing of quenching)
Tanner et al. (2012) describe a method similar to QuasR (see Moehling et al. 2021), in which the locations of the fluorophore and quencher on the primers/quenchers are reversed. The main functional distinction is that the quencher oligonucleotide is released from the FIP/BIP primer by way of the strand-displacement activity intrinsic to Bst and related enzymes, and that enables a continuous real-time readout (cf. with QuasR). DARQ has been employed in detecting SARS-CoV-2 as well as having been shown to be compatible with multiplex detection of influenza A and B and a human RNA control (Zhang and Tanner, 2021). DARQ, similar to OSD, allows for real-time monitoring of amplification but has been suggested to suffer from inhibition because of the full-length complementary quencher sequence, as well as having a less-bright signal in comparison to QuasR (Ball et al. 2016). The OMEGA amplification system from Atila Biosystems (Palo Alto, CA), the first company to receive an EUA for RT-LAMP-based SARS-CoV-2 diagnostics in April 2020, also uses the strand-displacement activity of Bst to produce a sequence-specific fluorescent signal (https://atilabiosystems.com/).
OSD (one-step strand displacement)
Jiang et al. (2015) used a fluorophore/quencher oligonucleotide hemiduplex with a 10 to 11 base toehold on the fluorophore-modified strand. This allows for competitive strand displacement as the amplicon is generated, leading to a continuous fluorescent signal as the hemiduplex is disrupted by thermodynamic competition. Although this presents an added constraint on primer design and the OSD hemiduplex reporter must be prepared in advance, OSD has been shown to be especially robust in SNP detection using LAMP. The OSD has been employed in the detection of SARS-CoV-2 successfully (Bhadra et al. 2020b, Maranhao et al. 2020). Another method, very similar to OSD, and employing the strand displacement activity of Bst, is DP RT-LAMP, which has been used for the detection of SARS-CoV-2 alongside the multiplex detection of an internal control RNaseP target (Yaren et al. 2020; see also a similar methodology described by Jang et al. 2021).
MB (molecular beacons)
Molecular Beacons (MBs) have long been used as probes in real-time PCR reactions (Tyagi and Kramer 1996). They have a hairpin structure in which the loop is complementary to the sequence being probed, whereas the complementary stem sequences have a fluorophore and quencher attached. Upon annealing to the target, the complementary sequences in the stem of the MB are separated, ending the FRET between the fluorophore and the quencher and resulting in fluorescence. The melt curves for specific- and nonspecific amplification using MBs are also very specific to their targets (Sherrill-Mix et al. 2020). Unlike other sequence-specific methods that adapt existing primers, use of MBs necessitates the design of a unique probe, which, although relatively cheap when purchased at scale, can add significant cost, time, and troubleshooting during method development.
Molecular beacons have the key benefit of targeting sequences that are not present in any of the primer sequences, for example, in the loop region of the amplicon and, thus, can be even more resistant to the detection of nonspecific amplification. However, this benefit, shared by OSD, also presents a challenge in finding a suitable sequence that provides stable hybridization at temperatures used for LAMP reactions. Previous studies have employed MBs containing locked nucleic acids to achieve annealing at LAMP temperatures and to allow multiplexed detection of multiple SARS-CoV-2 and human control amplicons (Sherrill-Mix et al. 2020) and, recently, of SARS-CoV2 variants (Sherrill-Mix et al. 2021). The very significant potential for molecular beacons as specific probes for LAMP is clear; the challenge will be to enable their comparatively easy, rapid and cost-effective design.
CRISPR-Cas cleavage systems
Emerging recently, CRISPR-Cas cleavage systems have also been employed as a reporting mechanism for isothermal amplification products arising from LAMP (van Dongen et al. 2021) (Fig. 10). First demonstrated as an endpoint detection platform for recombinase polymerase amplification (RPA), (Sherlock [specific high-sensitivity enzymatic reporter unlocking] Biosciences, Boston, MA; Gootenberg et al. 2017), Cas proteins are coupled with a guide RNA specific to the genomic region being amplified. Molecular recognition of the CRISPR-Cas complex activates collateral single-stranded deoxyribonuclease (ssDNAase) or single-stranded RNase (ssRNase) activity that can be readily measured using either a quenched fluorescence reporter or a biotin-FAM-labeled probe if a lateral flow dipstick method is to be employed. This method, coupled with RT-LAMP rather than RPA, was commercialized by Sherlock Biosciences (Boston, MA) and received an EUA from the US Food and Drug Administration (FDA, Silver Spring, MD) in May 2020. This general schema has been further applied to SARS-CoV-2 detection in what has been dubbed DETECTR (DNA endonuclease-targeted CRISPR trans reporter) (Broughton et al. 2020) with several other systems being developed in parallel (e.g., Guo et al. 2020, Agrawal et al. 2021, Garcia-Venzor et al. 2021; see Fig. 10 and Table 1).
FIGURE 10.
Schematic of the diagnostics with coronavirus enzymatic reporting (DISCoVER) loop-mediated isothermal amplification (LAMP)-CRISPR-Cas (Agrawal et al. 2020). Viral RNA is reverse transcribed and amplified via LAMP then converted back to RNA using T7 polymerase. Cas13 enzymes are programmed with a guide RNA to specifically recognize the desired RNA molecules over non-specifically amplified products. Subsequent activation of Cas13 ribonuclease activity results in cleavage of quenched fluorescence reporter molecules. The CRISPR-Cas provides additional layers of specificity and sensitivity, albeit at increased cost and complexity.
CRISPR-Cas-based methods have undoubted potential and several key advantages, such as increased specificity (via guide RNA [gRNA]) or the potential for robust amplification-free Cas-based diagnostics using only the intrinsic collateral ribonuclease activity of the Cas enzyme (Joung et al. 2020). However, in their most commonly deployed formats, they too suffer from a number of limitations, in addition to the increased complexity and cost per assay. Similar to MB-facilitated readouts, the already stringent primer design necessary for LAMP's particular amplification dynamics is further limited because of the requirement for a suitable protospacer adjacent to the motif sites for the Cas-enzyme–gRNA complex. Furthermore, the Cas-gRNA must be added to the reaction after the RT-LAMP reaction is finalized, so as not to interfere with the LAMP amplification. This can require opening of tubes after amplification to add the Cas reagents and, hence, is associated with risk of contamination.
Two groups have attempted to solve this problem either by designing custom caps for reaction tubes, which hold the Cas/gRNA complex for postamplification cleavage of the ssDNA reporter and keep it separate from the RT-LAMP reaction (Wu et al. 2020) or by simply adding the Cas/gRNA reagent to the cap of a standard tube to be mixed in at the end of amplification (Pang et al. 2020, Wang et al. 2020). Alternatively, a self-contained cartridge from Lotus (Norwich, England), which houses the test strip and amplification tube, makes a functional closed-tube lateral flow system, albeit at increased cost (adds approximately $5/test). In a similar vein, Reboud et al. (2019) used a paper-based microfluidic technology for on-field diagnostic, although they highlight the difficulties in the postamplification flow of products onto the test strip as a major challenge. As with MBs, current results show very promising future potential, but the design and experimental methods for both need to be simplified to enable robust use at the bench-side in a variety of settings and deployments (MacKay et al. 2020).
Sequencing Approaches Using RT-LAMP Amplicons
Because high-throughput multiplexing and sequence verification of samples are particularly important for many applications (Rendeiro et al. 2021), the use of both next-generation high-throughput sequencing (NGS; Illumina, San Diego, CA; Oxford Nanopore Technologies, Oxford, England) and traditional Sanger sequencing are necessary. This combination enables the analysis of thousands of samples in a single analysis run and provides detection by counting and sequencing using short-read and long-read sequencing platforms, such as those of Illumina and Oxford Nanopore Technologies, respectively. Sanger sequencing, in contrast, allows low-throughput sequence verification to confirm the molecular identity of LAMP-positive samples. Although Sanger sequence verification is less commonly used in routine testing (notably it suffers from the need to open a tube with amplified DNA), it may be required for some applications on a case-by-case basis.
Sequencing methods are increasingly important for detection of variants and mutations in the viral genome (“Topic C”) because they can provide a phylogenetic map of the epidemic (Butler et al. 2020), are useful downstream of the LAMP test as a secondary validation of the results, can correct false-positive results, can inform on new strains of the virus, and can aid in contact tracing (Bull et al. 2020). However, sequencing methods rely on more-expensive instrumentation (>$80 000), specialized sample preparations, and computational expertise typically found only in an advanced genomics laboratory. Thus, although many of the methods described in this review aim to meet the spirit of the REASSURED (affordable, sensitive, specific, user-friendly, rapid, equipment-free, delivered plus digital technology and mobile health) criteria (Land et al. 2020) criteria, LAMP linked to NGS largely seeks to fill a distinctly different and complementary niche (Table 3).
TABLE 3.
Qualitative comparison of methods: each method is broadly rated against the REASSURED criteria (see “Topic A”), supplemented with an assessment of the effect of interferences introduced via the sample specimena
Abbreviations: DARQ, quenching of unincorporated amplicon signal reporters; DP, displaceable probes; eBT, eriochrome black T; EvaGreen/SYBR Green/SYTO9, cyanine-dye based (green) fluorescent dyes; HNB, hydroxynaphthol blue; LAMP, loop-mediated isothermal amplification; LCV, leuco crystal violet; LSV/M, lampshade violet/magenta; MolBeac, molecular beacons; OSD, one-step strand displacement; QuasR, quenching of unincorporated amplicon signal reporters; RPA, recombinase polymerase amplification.
We assume that the use of smartphones to capture data and relay results will become increasingly common. For clarity, the qualitative coloring scheme is only intended to be indicative, primarily to provide the reader with a high-level appreciation of the choices and trade-offs that the developer makes when selecting an assay format. Exceptions to these broad categorizations are expected to be numerous. Combination methods of LAMP + CRISPR-Cas (e.g., Broughton et al. 2020) or of RPA + LAMP (El Tholoth et al. 2020) are omitted.
NGS for LAMP
The ability to sequence LAMP amplification products prevents false-positive results from nonspecific amplification because the complete sequence of each amplicon can unambiguously distinguish between the target gene and other mispriming events. Thus, if not for the high cost and long turnaround times incurred, it would be desirable to apply NGS to LAMP amplicons. By analogy with PCR, one method to increase throughput is by multiplexing many samples in 1 sequencing run (Yelagandula et al. 2020). By designing a clinical sample-specific DNA barcode sequence that is either part of each primer set (James et al. 2020, Schmid-Burgk et al. 2020) or ligated to amplicons after amplification (Dao Thi et al. 2020), amplicons from multiple samples can be pooled after being amplified in isolated LAMP reactions. Once the pool is sequenced, the barcode of each amplicon is used to associate it back to its clinical sample of origin, whereas the genomic sequence of the amplicon is used to indicate whether the sample contains the target virus. This multiplexed approach harnesses the rapidly growing scalability of NGS platforms to drive the average cost per sample well below that of the upstream LAMP reaction itself. Nonetheless, the risks and benefits of acquiring the short sequences from these complex concatemeric amplicons is high, and generally, sequence analyses to follow evolving variants are performed by applying the ARTIC protocol for Illumina or Oxford Nanopore sequencing (more below) for at least 10 times the coverage.
Although the sequencing time per sample is low because of the many samples pooled in 1 run, the turnaround time can often be of the order of days. An exception to this is sequencing using nanopores, which can take as little as 1 hour when samples are not multiplexed at high numbers (James et al. 2020). For example, Parker et al. (2020) describe purifying the amplified LAMP DNA using a traditional column-based method and synthesizing a single, rapid nanopore library with RAD004 or a multiplex, rapid library with RBK004. This technique is similar to RT-LAMP sequencing devised by Dao Thi et al. (2020) that multiplexes many RT-LAMP samples after an intermediate PCR step. Because both techniques use rapid tagmentation—a method that combines transposase-assisted fragmentation of the target DNA with adaptor labeling—they benefit from speed and can be sequenced using either a standard Oxford Nanopore flow cell or an Illumina MiSeq.
LamPORE, Oxford Nanopore Technologies sequencing
LamPORE from Oxford Nanopore Technologies combines LAMP with extremely portable and rapid nanopore sequencing (McIntyre et al. 2016, McIntyre et al. 2019) and, thus, has some parallels with the corresponding RT-qPCR-NGS method SARSeq (saliva analysis by RNA sequencing; Yelagandula et al. 2020). This technique involves a simple library preparation that adds unique molecular identifier (UMI) barcodes to each sample and sequencing using a standard Oxford Nanopore Technologies flow cell, allowing for many samples to be multiplexed and analyzed simultaneously (Ptasinska et al. 2020). For example, using 12 different LAMP barcodes combined with 96 rapid nanopore barcodes results in 1152 multiplexed samples. With nanopore sequencing for 4 hours on either a MinION (Oxford Nanopore Technologies) or GridIon (Oxford Nanopore Technologies) sequencer to generate sufficient coverage for each sample (James et al. 2020), it has a theoretical maximum capacity of 15 000 samples per 24 hours. Using the GridIon equipped with 5 flow cells allowed for scalability and high throughput with laboratory information management system (LIMS)-compatible quality control reports. This high-throughput approach has been successfully employed in a large-scale European study involving more than 20 000 samples (Ptasinska et al. 2020). In a 3-week clinical validation study with 1200 participants (3966 swab and 19 461 saliva samples) LamPORE showed a sensitivity of greater than 99.5% using both swab and saliva samples from asymptomatic participants, compared with a reference Conformité Européenne in vitro diagnostic (CE-IVD)-marked RT-qPCR assay. In the symptomatic cohort (incidence, 13.4%), the sensitivity and specificity were 100% (Ptasinska et al. 2020).
Another advantage of LamPORE is the ability to amplify several targets simultaneously, from multiple genomes and/or from multiple regions of the same genome. Because the amplicons are sequenced, a multiplexed LamPORE reaction may detect a broad range of pathogens or strains distinguished by specific variants in a sample in a single sequencing run. Specifically, James et al. (2020) monitored 3 regions of the SARS-CoV-2 genome, in addition to a β-actin internal control, the sequencing counterpart of the multiplexed fluorescence assays described above (“Extraction-Positive Control” and “Sequencing Approaches Using RT-LAMP Amplicons; e.g., Yaren et al. 2020, Zhang et al. 2020a, Bektaş et al. 2021).
Verification of LAMP-positive samples using Sanger and Oxford Nanopore sequencing
Verification of positive RT-LAMP products can be accomplished using traditional molecular techniques, such as the low-throughput, low-cost method of Sanger sequencing (Parker et al. 2020). That technique involves DNA extraction of the LAMP-positive sample using a column-based DNA extract (QIAamp DNA mini kit, Qiagen), quantification using spectrofluorometry (Qubit Instrument Life Technologies, Carlsbad, CA [now Thermo Fisher]), followed by Sanger sequencing. Previous studies have indicated that 6 ng of purified amplification product can be cycle sequenced in the presence of 10% GC Melt (Takara Bio, Kusatsu, Japan) or DMSO using the Loop B or Loop F primer with the resulting sequences used in NCBI BLAST comparisons.
INFRASTRUCTURE: HARDWARE, SOFTWARE, AND BIOWARE
Introduction
A core strength of LAMP is its applicability to multiple settings, many scales of testing, different budgets, and the extent of available infrastructure. It can be readily adapted to existing laboratory equipment because the most basic components are a device that can maintain approximately 65°C for approximately 30 minutes. This inherent simplicity has led to a slew of creative hardware solutions, including a simple sous vide (under vacuum) heater. Thus, isothermal LAMP hardware joins its low-cost PCR counterparts (e.g., the 5-tube PocketPCR, GaudiShop, Almaty, Kazakhstan). Many open-source options described below cost between $400 and $1000 and are able to perform 8 reactions at a time, thus lowering the cost to entry for LAMP.
However, for those with greater resources or a need for greater throughput, midpriced commercial options offer greater technical support and come in more-complete, user-friendly packages using 8- or 96/384-well reaction incubation and plate-reader technology. One (of many) example is the 384-well incubator-reader Neo2 (BioTek Instruments, Winooski, VT), which enables both colorimetric (e.g., pH and Mg2+ sensing) and fluorogenic (DNA intercalation and sequence-specific probe) readouts. In principle, 1536-well screening with a fluorogenic LAMP should be viable in 2- to 10-μL reaction volumes, if evaporation can be controlled.
Budget Breakdowns
A summary of different deployment models for LAMP testing is provided below, distinguished by throughput, cost, and resources needed (Table 4. Two important notes: first, we quote prices as if the items were purchased in the mainland USA (i.e., these are the lowest reference prices); and second, the descriptions and builds below are purely exemplary, being one configuration used by a US laboratory; alternatives can be substituted based on individual budgets and circumstances.
TABLE 4.
Open-science and collaborative organizations; the column headings are intended only to be an approximate reflection of the focus of the organization
|
National/international
|
Bioware/protocols
|
Hardware/software
|
| Global LAMP R&D Consortium (gLAMP) | Various LAMP protocols and methods | https://groups.google.com/g/glamp |
| Access to COVID Tools Accelerator | Saliva direct | https://channels.plos.org/open-source-toolkit |
| UN Technology Access Partnership | https://www.rtlamp.org | http://openhardware.science/ |
| WHO COVID-19 Technology Access Pool | SARS-CoV-2 Genes Collection | GMO Detective Detector |
| https://wellcome.org/coronavirus-covid-19/open-data | ADDGENE | https://channels.plos.org/open-source-toolkit |
| Open COVID Pledge | FreeGenes | OpenTrons OT2 |
| Coronavirus Method Development Community | ReClone Research in Diagnostics Collection | Pocket PCR |
| Coronavirus Standards Working Group | BMRI-ERIC BioBanks | Bomb.bio |
| Just One Giant Laboratory (JOGL) | Open Source Medical Supplies | |
| African Collaborative Initiative to Advance Diagnostics | Helpful Engineering |
Tier 1: “basic LAMP”
Tier 1 is suitable for those interested in piloting the use of LAMP with a minimum investment or with an intended throughput of fewer than 100 tests per day, for example, using individual PCR tubes or 8-well strip tubes. The equipment will be available in most biologic laboratories and many high school science classrooms. The main cost is the LAMP test itself (approximately $8 per test if purchased as a preformulated kit).
Tier 2: moderate-throughput LAMP
Tier 2 expands LAMP capabilities, moves to 96-well plate formats for parallel temperature-controlled incubation, and includes a plate reader that allows for quantitation of the signal beyond reporting results visually as a color change. This system has the capacity for up to approximately1000 tests per day, depending on staffing.
Tier 3: high-throughput RT-LAMP
The tier 3 solution described here expands LAMP capabilities beyond 1000 tests per day by automation of many of the core processes using benchtop (versus integrated)-automation systems. These solutions would be found in, for example, pathology laboratories or centralized testing facilities. Because they likely fall within a regulated realm, testing using such systems also have Medical device, regulatory, and personal data privacy and security consequences. In a research setting, the Mason Laboratory (New Haven, CT) received institutional review board (IRB) approval for the use of a colorimetric LAMP kit (New England BioLabs) with the TINY (tiny isothermal nucleic acid quantification system) testing device (Snodgrass et al. 2018), the results from which were automatically and securely uploaded to a local pathology department for review and potential follow-up with a certified SARS-CoV2 test (“Regulatory Landscape”).
Hardware Designed for LAMP-Based Assays
The most common hardware used for RT-LAMP is a preexisting qPCR reader or conventional plate readers capable of 65°C incubation (e.g., Neo2 [BioTek], SpectraMAX M65C [Molecular Devices, Sunnyvale, CA], or Nivo [PerkinElmer, Waltham, MA]). More simply, a wet/dry heating bath or block can be combined with either visual colorimetric detection (e.g., phenol red) or simple light-emitting diode (LED) fluorescence excitation with smartphone data capture (e.g., SYTO9). In this section, we exemplify—but do not necessarily endorse—some of the now-many midpriced hardware solutions available to LAMP researchers, with references for use with SARS-CoV-2 where available.
The Genie
The Optigene (Horsham, England) Genie II or III performs real-time, fluorescence-based LAMP, providing quantification of the progress curve, in addition to melt curve analysis of the products, as an aid to quality control (template-specific amplicons often have a characteristic melt temperature). Devices cost between $3000 and $15 000 and are capable of processing 8 or 16 reactions at a time. A recent example of its use was for a large, saliva-based clinical study investigating the use of chelex-100 during the preanalytic processing step (Howson et al. 2021)
The Simprova
Eiken Chemical Co. (Tokyo, Japan) has developed an automated cartridge-based system for sample processing/RNA extraction and molecular testing (functionally similar to the Cepheid [Sunnyvale, CA] GeneXpert PCR system). Using a variety of biospecimens, the makers of the Simprova system state it has good or better RNA/DNA extraction performance compared with Qiagen Mini Kits, with a shorter turnaround time (15 minutes vs. 40 minutes). The multiwell testing chip (1 input, 25 test reactions) contains dried LAMP reagents within a closed system to minimize cross-contamination and a fluorophore-guanine quench system for detection (Yonekawa et al. 2020). Although results using Simprova with influenza, respiratory syncytial virus, and metapneumovirus have been published (Takayama et al. 2020), we are unaware of SARS-CoV2-specific publications beyond the Eiken Loopamp kit approved in March 2020.
The ESEQuant
The ESEQuant TS2 (Qiagen) is a small, fluorescence plate reader suitable for fluorescence-based LAMP that uses light-emitting diode and filter technology combined with a touchscreen display, barcodes, radiofrequency identification (RFID) reader and LIMS connectivity. With fluorescence measured in up to 6 channels, it is well suited to multicolor, multiplexed readouts (see ”Amplicon Detection”), whereas melt-curve capabilities provide for quality control, as described previously. Publications citing this reader appear limited at the time of the writing.
The Testing Cube
Designed for use in concert with the FRANKD test strips for SARS-CoV2 (GeneMe, Gdańsk, Poland), the Testing Cube (GeneMe) is a small (2.5 kg), affordable (approximately $250, depending on country and off-the-shelf components), open-source diagnostic device (https://www.geneme.eu). The design specifications have been openly shared on hardware sites, including Team OSV (https://www.teamosv.com), GitLab (https://about.gitlab.com), and Wevolver (https://www.wevolver.com). The Testing Cube consists of a single strip of 8 wells (6 tests, 2 controls) to run the FRANKD (and other isothermal) tests for approximately 30 minutes before delivering the results.
The T8 benchtop fluorometer
The Axxin (Fairvield, VIC, Australia) T8 reader is a small-footprint, lower-cost (retail, $5000), 8-PCR tube, dual-wavelength incubator reader with a 10-second scan-read time. It has the ability to mix viscous liquids if a magnetic ball bearing is included in each tube (this feature was designed for the high-viscosity solutions used for RPA). Natoli et al. (2021) describe methods used to adapt the T8 for use with LAMP at 65°C (vs. RPA at 41°C), particularly the need to add mineral or paraffin oil to limit evaporation because the T8 does not have a heated lid.
The BioRanger
The BioRanger (Digenetix, Manoa, HI) is a handheld device to support LAMP for field-based agricultural diagnostics and its related “Assimilating Probe” sequence-based fluorescence technology (Kubota and Jenkins 2015). The 8-tube, 2-wavelength device incorporates control with a programmed heating cycle. It is intended for low-resource field applications (approximately $3000) and interfaces wirelessly through bluetooth to an Android application with a range of connectivity features. For use with thermally stable polymerases (Chander et al. 2014), the BioRanger can be programmed with a “lysis” step to promote release of nucleic acids before user-defined amplification programming, including an endpoint melting analysis to confirm amplicon purity. A colorimetric version of the instrument has been developed for use with, for example, phenol red/pH changes (Diaz et al. 2021; submitted this issue) with very promising results and times-to-positives that are shorter than when detected visually (N. Tanner, unpublished data).
The Franklin
The thermocycler device (Franklin) developed by Biomeme (Philadelphia, PA) for qPCR can, in principle, also be used for isothermal measurements, although we have found no reports of its use with RT-LAMP. Different models enable either 1, 2, or 3 wavelength detections in 9 tubes simultaneously, when measured on an Android smartphone (cost, $6000 to $10 000).
The SnapDx
The SnapDx (https://www.snapdx.org/), an off-shoot of the Prakash Laboratory at Stanford University, develops numerous, relatively simply built, portable devices for low-resource or home-testing settings using saliva RT-LAMP. The core SnapDx product is currently (April 2021) undergoing trials. Other devices developed include the $5 Handyfuge, an open-source solution for low-speed centrifugation steps in point-of-care RT-LAMP tests, which was successfully applied to the Rabe and Cepko (2020) protocol.
The Pebble
The Pebble incubator-absorbance reader from Biopix-T (Voutes, Greece; https://biopix-t.com) is a compact device for performing real-time colorimetric LAMP with 8 PCR tubes and is operated via an Android application and connecting via bluetooth to a smartphone (Papadakis et al. 2020).
The Genelyzer FIII
The Genelyzer FIII (Canon, Tokyo, Japan) is a portable, battery-operated, fluorescence-based incubator-reader platform for low-infrastructure settings (https://jp.medicalcanon/products/dnachip/genelyzer_F/index). The Genelyzer FIII was demonstrated to have good diagnostic performance with NPS-based, RNA-extracted and, to a lesser extent, extraction-free protocols (LoD, 25 and 1400 copies/reaction) using the Canon 1-enzyme isothermal mastermix and Orf1b primers (Yoshikawa et al. (2020).
SOFTWARE
Primer Design Tools
Given the need to simultaneously design multiple primers, which are predicted to bind specifically and uniquely to the target, the only reasonable path is to employ dedicated, open-access software tools to help guide primer selection (”Primer Selection”). The most commonly used tools are listed below along with their developers:
PrimerExplorer V5 (Fujitsu, Tokyo, Japan/Eiken Chemical)
LAMP Primer Design Tool (New England BioLabs).
OligoAnalyzer (IDT Corp, Newark NJ)
LAVA: Lawrence Livermore National Laboratory (Torres et al. 2011)
GLAPD: Shanghai: (Jia et al. 2019)
Primer Analyzer (Thermo Fisher)
OligoCalc (Northwestern University)
Although many SARS-CoV2 primer sets have been published (Table 2; Supplemental Table S3, see also Janikova et al. 2021), these tools can be used for those interested in deeper exploration, and they can be an important hedge against “standardized” primers becoming less effective or biased as the virus evolves (“Topic C”). Primer design performed well is nontrivial; although these in silico tools are both valuable and necessary, several selected sets should be tested experimentally, especially for cross-reactivity, in multiplexed or comboplexed assays (“Primer Selection”).
Color Interpretation
Colorimetry.net (Mg/HNB-based assays)
The https://colorimetry.net/about/ website is a simple web service for automatic computer-assisted enhancement of images of color changes associated with use of HNB dyes as a measure of LAMP amplification. Its enhancement strategies seek to increase perceptual accuracy when performing a visual readout. The biochemical impetus for the work is the original publication of Goto et al. (2009), as exemplified by Kellner et al. (2020).
Phenol red (pH-based assays)
Colorimetric RT-LAMP assays using weakly buffered solutions and a pH indicator can, at times, lead to subjective and ambiguous results (neither a definitive red nor yellow displayed). In one such assay, the differences in color among the positive and negative samples were identified through color decomposition and analysis in the color CIELab space (Gonzalez-Gonzalez et al. 2020). Yoo and colleagues (2020) have developed a tablet personal computer (PC)-based portable device (FlagMan) using 3 signal categories: DEF (decisive [main color peak], effective [purity of color], and fuzzy [clarity]), which are akin to hue-saturation value (HSV) coordinates (Yoo et al. 2020). McLaughlin et al. (this issue) have similarly developed a software application for color assessment, as described in “Topic B” (LAMPPlatereader.app).
Artificial intelligence LAMP
Rohaim and colleagues (2020) combined automated image acquisition with the use of artificial intelligence (ai) to “engineer a novel hand-held smart diagnostic device,” ai-LAMP, applied to a colorimetric (pH, phenol red) readout. Collected images were converted from RGB (red, green, blue) color space to YUV (luminance [Y], color difference [U and V]) color space for use with a convolutional neural network (CNN), which has been shown effective in many contexts (Haibe-Kains et al. 2020). Using approximately200 suspected COVID-19 patients the platform was shown to be reliable, highly specific, reducing assay run-time and detection subjectivity (Rohaim et al. 2020).
EMERGING TECHNOLOGIES
Most SARS-CoV2 research into RT-LAMP uses comparatively well-established base technology. In this section, we review examples of emerging technologies and methodologies, often based on the adaptation of previous work with other viruses or pathogens. Some are relatively “high tech” (e.g., Qin et al. 2020), whereas others are purposefully “low tech,” being fit for a very different purpose.
Smartphone-Enabled LAMP
Technologic integration of the light and camera features of smartphones or tablets into testing devices has become increasingly popular, creating tube- and microfluidic sensory systems (Farshidfar and Hamedani 2020, Nguyen et al. 2020). Building on the ability to receive and process data from other sensors and devices, their appeal lies in their comparatively low cost, widespread availability, and the ability to connect medical professionals and patients. A second benefit of remote testing is to allow patients exhibiting mild respiratory symptoms to get tested without having to attend, often highly overcrowded, centralized health care facilities.
Several researchers have used smartphones or tablets for tube-based readouts ( Rohaim et al. 2020, Yoo et al. 2020), predominantly for measurement at one wavelength. In contrast, during development of tests for West Nile virus (WNV) and Chikungunya virus (CHKV), Ball et al. (2016) developed the QuasR methodology (see “Sequence-Dependence Detection of RT-LAMP Amplification Products), enabling dual-color duplexing of targets using a flashlight LED, red/green plastic lighting gel sheets, and detection with an dedicated application-enabled iPhone 6.
In addition to tubes or plates, smartphone detection is also used with microfluidic “lab-on-a-chip” approaches (see below). Developed first in a nonclinical model system (Sun et al. 2020), Ganguli et al. (2020) combined a spatially multiplexed microfluidic chip with a cradle to house blue LEDs and a filter or smartphone to detect the green fluorescence and were able to detect SARS-CoV2 RNA in simulated nasal samples.
Microfluidic Platforms
Most devices use classical tube or plate formats. However, for other viruses (HIV, Safavieh et al. 2017; Zika, Roy et al. 2017; Song et al. 2016), some have explored assays performed in microfluidic devices, driven largely by their potential to enable closer to point-of-care testing and the use of predried reagent cartridges with less molecular enzyme and reagent consumption (Zhang et al. 2019; Berkenbrock et al. 2020; Farshidfar and Hamedani 2020; Augustine et al. 2020). Although paper-based devices are formed by a series of hydrophilic cellulose fibers that move reaction liquids by capillary action via absorption, another class of channel-based devices have been engineered through nanofabrication (Basiri et al. 2020, Rodriguez-Manzano et al. 2021).
Paper-based microfluidic devices
Paper-based microfluidic diagnostics emerged largely to enable very low cost, portable, disposable, true point-of-are tests (Gong and Sinton 2017, Carrell et al. 2019). Indeed, Seok et al. (2016) were able to detect meningitis DNA using a LAMP-based 3-layer, stacked-paper microfluidic device. Using dried (not lyophilized) LAMP reagents, the sample is distributed into 3 test zones and 1 control zone via capillary action; amplification is monitored by changes in HNB fluorescence (cf. absorbance) (excitation/emission wavelengths [λex/em] = 540 nm/610 nm) allowing detection of 102 to 105 DNA copies. Similarly, Varsha et al. (2020) describe an even-simpler paper device for Leptospira DNA using commercial Whatman (Maidstone, England) no. 1 paper, again with dried LAMP reagents detected via smartphone. Kaarj et al. (2018) used the wax-printed paper microfluidic as both a means for partial purification of Zika virus RNA (see “Preanalytic Sample Processing”) and also as a substrate for the smartphone-based colorimetric RT-LAMP reaction.
Despite this precedent and potential, only 1 paper-based RT-LAMP diagnostic device (COVIDISC) for SARS-CoV-2 has been described (Garneret et al. 2021). Viral RNA is extracted onto a fiberglass membrane, dried, and then folded onto paper-based “discs” that contain freeze-dried LAMP mixture and primers. The RNA is eluted onto the discs. and the heating of the portable system enables fluorescence readout of the discs using QuasR (“Sequence-Dependent Detection of RT-LAMP Amplification Products”; Ball et al. 2016, Bektaş et al. 2021).
Nanofabricated channel based-microfluidic devices
An early channel-based microfluidic devices for SARS-CoV2 included a disposable microfluidic polymer cartridge containing 2 luer-lock inlet ports (sample and RT-LAMP reagents), a 3D reagent mixing, and an serpentine, heated region. This battery-powered device was able to detect SARS-CoV2 using a fluorescent dye/smartphone combination (Ganguli et al. 2020). Two microfluidic systems based on centrifugal liquid flow have been reported. With the objective of simplifying the sample processing step, Soares et al. (2020) developed an integrated smartphone-enabled, centrifugal, microfluidic platform for a sample-to-result fluorescence-based RT-LAMP test. Using modified agarose beads, their device enhances the signal specificity and mitigates the effect of collection medium on weakly buffered colorimetric LAMP assays (“Considerations for pH-Based Colorimetric RT-LAMP”). de Oliveira et al. (2021) describe a polystyrene-toner (PS-T), centrifugal, microfluidic device manually controlled by a fidget spinner. The amplification (5 μL) was controlled with a thermoblock at 72°C for 10 minutes with automated, on-chip visual detection. Finally, an elegant, but significantly more-complex, solution uses electric field-driven microfluidics (isotachophoresis [ITP]) combined with CRISPR-based diagnostics to enable aspects of RNA purification from NPS, ionic focusing/concentration, and detection in a single microfluidic device (Ramachandran et al. 2020).
The path to commercial and clinical use for Lab-on-a-Chip (Royal Society of Chemistry, London, England) (and similar) devices has been long and has not been without its challenges; the goal has been to go from out of the laboratory to a scaled-up, robustly manufactured product with a simple procedure for routine use by nonexperts. Certainly, that goal is possible—several such commercial systems exist, for example, the Cepheid GeneXpert or the Eiken Simprova—but these systems are currently the exception rather than the rule. We have watched with interest to see whether the COVID pandemic has provided the impetus for microfluidic devices to reach a tipping point at which their potential for low-cost mass production will be realized.
Digital LAMP
Digital amplification methods partition bulk samples into many small or localized compartments so that target viral nucleic acid molecules are isolated in small volumes. Therefore, molecules are amplified independently, generating a localized positive or negative signal that can be considered digital. Each signal is counted in an endpoint measurement, and the sum of all positive-compartment results represents the total number of viral copies detected in the sample.
Although several implementations of a digital LAMP (dLAMP) have been previously reported (Rolando et al. 2020, Yuan et al. 2020), none have, to date, been applied to detect SARS COV-2. Prior work has explored the dynamics and sensitivity of dLAMP using micron-sized chambers to compartmentalize the sample (Khorosheva et al. 2016, Rolando et al. 2019). Others have used droplet microfluidics, in which the LAMP mixture is partitioned into micron-scale drops in oil, and fluorescence from the drops is detected using a simple optical system and camera (Schuler et al. 2016, Hu et al. 2020). Partitioning can also occur within a gel matrix environment, limiting the diffusion of the LAMP-amplified amplicons and generating localized fluorescent spots that can be imaged using a smartphone with no additional optical magnification (Huang et al. 2018). Finally, a dLAMP can operate on a commercial membrane filter (Lin et al. 2019) in which the sample is partitioned into the pores and oil isolates the pores from each other. Although these methods differ in their implementations, they all share the same advantages derived from the isolation of templates: (1) absolute quantification of viral copies compared with real-time qPCR, which requires a standard curve; and (2) template isolation, which limits the effects of false-positive results from contamination or nonspecific amplification in the sample.
SOLUTIONS FOR USE IN LIMITED-RESOURCE SETTINGS
Developing and deploying nucleic acid detection systems that have low capital costs, have little reliance on traditional medical infrastructure, are easy to fix or require no maintenance, can be transported among small rural clinics, and are easy enough to use without extensive training can significantly affect the delivery of diagnostic services in limited-resource settings (Urdea et al. 2006, Yager et al. 2008, Jani and Peter 2013). For example, it has been shown (Cox et al. 2015) that decentralization of diagnostic testing by implementing the GeneXpert PCR system was able to reduce the time for treatment of tuberculosis from 71 days to 8 days in Cape Town, South Africa. The relatively high cost of these established PCR systems and moderate infrastructure requirements, however, tend to limit their distribution beyond midscale facilities and to limit their portability.
The LAMP-based tools have proven a popular method for addressing these challenges, primarily because the single-temperature amplification reduces the required complexity of the instrument, and they maintain high diagnostic accuracy. Successful development and deployment of systems that can address the challenges mentioned above could catalyze the expansion of LAMP, particularly for use in remote, limited-resource settings, or as part of a much-more-distributed testing network.
Intermittent or Zero-Electricity LAMP devices
Liao et al. (2016) created a portable, electronics-free “Smart Cup” LAMP on a microfluidic chip, with heating accomplished by coupling to an exothermic Mg-Fe–alloy hand-warming pouch (approximately $0.15), which enabled a relatively constant temperature of 68°C to be maintained for an hour. With a fluorescence and a smartphone system, they were able to perform 4 reactions in parallel, detecting herpes simplex virus type 2 DNA down to 100 copies/reaction. Snodgrass et al. (2018) describe detection of Kaposi sarcoma herpes virus in human biopsy samples in Uganda with a portable device that can operate based on various live or stored energy sources, including electricity, solar energy, or a flame—performing 6 reactions in parallel. They demonstrated the system was robust against power interruptions and could be used with equivalent efficacy by trained laboratory personnel and local providers. The technology was based on earlier versions by Jiang et al. (2014) and Snodgrass et al. (2016), which used solar energy focused on a traditional microfluidic chip. Labarre et al. (2011), Singleton et al. (2014), and Curtis et al. (2016) all demonstrate innovative, small-scale, typically single-use devices that do not require the use of external electricity to perform detection on a small set of devices. Stedtfeld et al. (2012) demonstrated an early system that uses mobile technology (iPod touch) and microfluidics to perform up to 4 parallel samples.
Battery-operated and other low-cost systems and accessories
Velders et al. (2018) built and programmed a single-tube prototype of a portable, 1-temperature, battery-operated, open-source ArduinoShield for LAMP with a fluorescence readout. With molded polydimethylsiloxane (PDMS) to fit PCR tubes, heating was achieved via a nichrome wire and is designed to be straightforward to assemble. Priye et al. (2017) developed a LAMP box to detect arboviruses using a smartphone, which uses resistors and an aluminum plate as a heat source. That 8-reaction device costs approximately $100 but requires specialized reaction vessels to work optimally. Using 3D printing, Gonzalez-Gonzalez et al. (2020) describe a simple circulating-water holder for standard 0.2-mL PCR tubes when coupled to phenol red detection. Priye et al. (2017) developed a LAMP box to detect arboviruses with resistors and an aluminum plate as a heat source, combining QuasR with chromaticity-based smartphone analysis. Although practical and comparatively cheap (approximately $100), the device requires specialized reaction vessels to work optimally. Some open-hardware devices that could be adapted for LAMP temperature control include PocketPCR from GaudiLabs (pocket-size USB [universal serial bus]-powered PCR ThermoCycler and PocketPCR), NinjaPCR (Tokyo, Japan; for PCR and LAMP), and a 3D-printed RT-qPCR device created for infectious disease diagnostics (Mulberry et al. 2017).
The miniPCR (Cambridge, MA) blueBox S or Pro transilluminators with imaging hood can illuminate plates with a smartphone docking system ($250 to $350), whereas the battery-operated P51 Molecular Fluorescence Viewer (miniPCR) can illuminate 8-tubes ($30), providing a low-cost manufactured solution. Similar readers can be built individually using open-source methods (e.g., for education purposes; https://GMO Detective.com).
Lyophilization and dry storage of RT-LAMP reagents
A reliable supply chain and storage capacity are essential in all settings but are particularly an issue in regions with poorly developed infrastructure, notably those lacking cold chain or storage (2°C to 8°C or −20°C). Dried and/or lyophilized reagents are components of several cartridge-based systems (TwistDx [Abbot Laboratories, Abbott Park, IL] for RPA, Simprova [Eiken] for LAMP, GenExpert [Cepheid] for PCR) and also components of some paper-based microfluidic devices (see above, Seok et al. 2017, Varsha et al. 2020, Garneret et al. 2021). Others have sought to prepare lyophilized (freeze-dried) reagents as standalone reagents with the objective of stabilizing and prolonging room-temperature shelf-life (Pack et al. 2008, Challener 2017, Chen and Ching 2017). Using lyophilized and premixed reagents (e.g., mastermix containing the required primers, detection dye, etc.) necessarily also simplifies and shortens assay preparation (Carter et al. . 2017).
Although individual methods (e.g., temperatures and pressures for lyophilization) mostly contain d-trehalose as a protein stabilizer, avoid the use of hygroscopic glycerol-based stock solutions (Aidelberg and Aronoff 2020, Xu et al. 2020), and contain additional excipients (mannitol, bovine serum albumin, and polyethylene glycol), reagent stability under these various conditions can be measured with an accelerated aging/Q10method (the factor by which the rate of spoilage increases when the temperature is raised by 10°C; Chua et al. 2010).
Dried (not lyophilized) RT-LAMP master mixes for SARS-CoV2 containing primer sets ORF1ab, N5, or N15 and amplification enzymes have been described (Garcia-Bernalt Diego et al. 2021a) with a projected shelf-life (via the Q10 method) of 2 months at room temperature. Similarly, Yaren et al. (2020) first removed glycerol from commercial enzyme preparations before preparing lyophilized master mixes for their RT-LAMP assay.
OPEN-RESEARCH INFRASTRUCTURE
Open Science in Research: For Covid-19 and Beyond
The global urgency to contain the COVID-19 pandemic has led to large-scale global, collaborative efforts among organizations, communities, and individuals from many different backgrounds (e.g., Global LAMP Consortium [gLAMP], Just One Giant Laboratory [JOGL; Paris, France], the access to COVID Tools Accelerator [World Health Organization (WHO), Geneva, Switzerland], Helpful Engineering [Columbus, OH], Open Source Medical Supplies [Washington, DC], WHO COVID-19 Technology Access Pool, and UN Technology Access Partnership [Gebze, Turkey]). To ensure knowledge flows efficiently and advances our understanding of the virus and how best to contain the pandemic, open access to COVID-related publications and data was encouraged (https://wellcome.org/coronavirus-covid-19/open-data). The Open COVID Pledge was established to enable more than 600 000 holders of patents to waive their intellectual property (IP) to help alleviate the effects of the pandemic (Contreras et al. 2020). Throughout the pandemic, scientists around the world have come together to develop and improve RT-LAMP assays for detecting SARS-CoV-2. Scientists in various regions and under distinct regulatory regimes have congregated within and across several consortia and working groups; the gLAMP community is one such group. Here and elsewhere, various research outputs, such as protocols, have been shared via, for example, the Coronavirus Method Development Community (https://www.protocols.io), and multiple websites have been established by researchers to share knowledge and protocols related to their tests (e.g., SalivaDirect, Yale School of Public Health). One such group is the Vienna COVID-19 Detection Initiative (VCDI, Vienna, Italy), informally known as the Vienna group. Their most notable LAMP-related efforts include development of colorimetric LAMP with HNB dyes; development of “bead-LAMP,” featuring purification with magnetic beads; development of a gargle-based sampling (see Kellner et al. 2020); and the development of an in-house preparation of Bst-LF and HIV-RT as lesser cost and that has scalable alternatives to commercial counterparts.
Open-source approaches also have a critical role in addressing the unprecedented demand for reagents and infrastructure. During the pandemic, several projects have developed open hardware devices (Maia Chagas et al. 2020); open genetic resources, such as the Biobanking and BioMolecular Resources Research Infrastructure–European European Research Infrastructure Consortium (BBMRI-ERIC, Graz, Austria) BioBanks, and the African Collaborative Initiative to Advance Diagnostics (AFCAD, Cairo, Egypt; Peeling et al. 2020), which aims to ensure equitable access to biobank samples for diagnostics developers; as well as biologic tools, such as the SARS-CoV-2 Genes Collection (https://www.ncbi.nlm.nih.gov/sars-cov-2/), the Research in Diagnostics Collection (ReClone, https://reclone.org/), and numerous plasmids and materials that were made available for test development and benchmarking. These initiatives are often inspired by the free/libre open-source software movement and provide freedom to use, study, replicate, and distribute instrumentation, protocols, and biomolecular tools with minimal conditions, except those required to acknowledge sources, protect patient privacy, and comply with laws and regulations.
Open Hardware for LAMP
Free and open-source hardware (FOSH) approaches can bring significant cost savings (Pearce 2018 and can allow customization of hardware to meet different experimental and diagnostic needs. For instance, applying FOSH approaches to scientific and medical devices (Pearce 2013, Gibb and Abadie 2014, Baden et al. 2015, Chagas 2018; Gathering for Open Science Hardware [http://openhardware.science/], PLOS Open-Source Toolkit [https://collections.plos.org]) has led to the development of a range of instruments to undertake each step of an isothermal amplification reaction, and some devices now combine steps, such as incubation and real-time readout (see “Battery Operated and Other Low-Cost Systems and Accessories”; e.g., NinjaLAMP, https://github.com/hisashin/NinjaLAMP, Miriam, https://www.miroculus.com/miriam/, GMO Detective Detector, https://gmodetective.com/, and PocketPCR, https://gaudi.ch/PocketPCR/), whereas others use simple consumer hardware, such as sous vide–precision cooking devices as low-cost immersion heaters for water baths (Kellner et al. 2020). Other open-hardware instruments that have been applied to COVID testing and could scale up throughput of LAMP assays include automation tools, such as the OpenTrons OT2 liquid handling robots which have been deployed to accelerate RT-qPCR based testing in the Pandemic Response Lab in NYC (https://pandemicresponselab.com/). Other developments include small devices for specific tasks, such as low-cost centrifuges used to separate reaction inhibitors from inactivated samples before LAMP (Li et al. 2020), salad spinners to spin down 96/384 well plates (Morán and Galindo 2011, Motohashi 2020), and a 3D printed vortex mixer (Patel et al. 2018).
Open reagents and biomolecular tools
Open-source approaches have also spanned to free-to-use molecular biology tools, such as plasmid DNA-expressing enzymes for RT-LAMP and SARS CoV-2 viral genes distributed by Addgene (Cambridge, MA) and Free Genes (BioBricks Foundation, San Francisco, CA) under open material-transfer agreements (Kahl et al. 2018), which enables use of those tools by companies, in addition to academics. Other reagents have been distributed freely for noncommercial use (Bhadra et al. 2020, Maranhao 2020, Sherril-Mix et al. 2021a), Alekseenko et al. 2021. ReClone, a reagents collaboration network, has also emerged to provide access to protocols and off-patent reagents, with the aim of mitigating shortages in the short term and building preparedness in the long term. That network aims to establish an infrastructure to manufacture critical reagents, such as reverse transcriptases (e.g., MMLV and HIV) and Bst LF enzymes used in RT-LAMP. Community biolaboratories have contributed designs for peripheral hardware, such as the simple spectrophotometer and bioreactor developed in the Swiss open-public laboratory Hackuarium Biohackerspace (Écublens, Switzerland).
Developing and deploying diagnostic tests requires more than reagents, it also requires controls and standards, which many researchers have shared, both as data (e.g., DNA sequences) and as materials. The Coronavirus Standards Working Group at Stanford University is an open-membership consortium that is developing validation panels for diagnostics testing and compiling lists of control materials through public meetings, chat channels, and collaboration, similar to other work in genomics standards for laboratory settings (Mason et al. 2017) as well as remote (Nangle et al. 2020) and harsh (Tighe et al. 2017) environments.
The provision of materials by initiatives, such as those described above, has frequently been coupled to openly published protocols for their use, for example, via https://protocols.io (Teytelman et al. 2016), as already mentioned, which in comparison with journal-article methods sections, offers more-detailed insight into experimental knowledge and the ability to comment, discuss, and modify protocols and to retain a version history (see also Table 4). Although analysis of these platforms and their use during COVID-19 is only just beginning, this ability to rapidly publish, adapt, and republish protocols and materials offers great potential for accelerating innovation at times of crisis, when the research is taking place under urgent time pressure.
REGULATORY LANDSCAPE
Regulatory and statutory considerations shape the landscape of clinical diagnostic research, development, and deployment. For those wanting their test to progress into clinical use—from bench to bedside—we provide this overview of the regulatory context, partly as a case study for those new to regulatory affairs. We highlight the changes that may arise from the forthcoming EU in vitro diagnostic regulation (IVDR) in May 2022 (“The Upcoming In Vitro Diagnostic Regulation, 2022”). As that section title implies, regulations are coordinated geographically. We, therefore, consider, first, the USA, and then, the European Union, mindful that we are leaving undiscussed the regulations that affect most of the world's population (e.g., Asia; the Association of Southeast Asian's Medical Device Directive).
Regulatory Perspective in the USA
In the USA, achieving approval from the FDA for a COVID diagnostic test has been a moving target, with significant revisions during the past year after the first major guidance in May 2020 (FDA produced templates for EUA submissions; Supplemental Table S4). With so many new laboratory tests being developed, EUAs were enabled to accelerate the conventional path to medical-device approval, given the immediacy and severity of the emerging COVID pandemic (Hasell et al. 2020; https://www.worldometers.info/coronavirus/). To expedite approval of the growing number of EUA applications, the FDA sought to standardize and clarified a streamlined process with a series of templates being provided over a 6 month period.
Currently, there are 4 primary regulatory pathways for COVID testing involving various degrees of oversight in the USA:
FDA EUA
Clinical Laboratory Improvement Amendments of 1998 (CLIAs) laboratory-developed test (LDT)
Testing as a part of a research study conducted under IRB approval for research on human subjects
Surveillance testing in which the test results are not returned as if those results were from a CLIA laboratory, but rather, patients with a positive result are referred to a CLIA testing facility.
FDA EUA
The EUAs granted by the FDA comprise the (time-bound) regulatory approval for most COVID-19 tests. The EUAs are issued under a more-streamlined process, compared with that of conventional approvals for IVD tests. For example, tests approved with an EUA do not require good manufacturing practices/International Organization for Standardization (GMP/ISO; Geneva, Switzerland)-grade reagents (research-use–only reagents are acceptable), and the size of the clinical validation studies is typically considerably smaller (e.g., 30 positives and 30 negatives for SARS-CoV-2). Individual EUAs for molecular tests for SARS-CoV-2 are classified either as IVDs or LDTs. Although LDTs are limited to use in the CLIA laboratory that developed them, IVDs can be sold and used by other laboratories.
At the time of this writing, 10 LAMP IVD tests have been granted approval under the EUA scheme (Supplemental Table S1). The first (Atila Biosystems) was a direct swab-based LAMP test, which was soon followed by a CRISPR-based assay from Sherlock Biosciences; in both cases, fluorescence was monitored with a qPCR or a plate reader. Color Health (previously Color Genomics) was the first EUA-approved LAMP test to use a 96-well colorimetric readout combined with commercial lysis/inactivation and bead-purification reagents. Finally, Pro-Lab Diagnostics (Round Rock, TX) nasal swab test, with extraction-based or extraction-free (“direct”) protocols. When paired with the Optigene GenieHT fluorescence reader, the test includes a melting-curve analysis of the amplification products, requiring the melting temperature to fall within a specific narrow range (83°C to 85°C). Pro-Lab claims a low LoD of 125 genome equivalents (GEs) per flocked swab (using γ-irradiated cell lysate from BEI Resources).
The LoD has been a key driving metric for SARS-CoV-2 tests but is the source of much confusion and not a little controversy. Viral loads (Wofel et al. 2020) typically rise quickly and spike over 1 or 2 days and decay more slowly thereafter (Larremore et al. 2020). The infectiousness threshold for replication-competent virus is approximately1000 copies/μL (Newman et al. 2020). This corresponds to approximately 2000 to 5000 copies/reaction for most RT-LAMP tests, far above the analytic LoD of the reference RT-PCR test used for purified RNA (single digit copies/μL). Nevertheless, less-sensitive tests can detect most COVID infections that are capable of spreading the virus, especially those used frequently and with approximately 24-hour turnaround times. The question of what constitutes fit-for-purpose analytic sensitivity for clinical practice remains open (see also “Discussion”).
To add further confusion, in September 2020, the FDA released the first round of results of a comparison of LoDs for molecular diagnostic tests that had received EUA approval by May 2020. Although dominated by PCR, rather than LAMP tests, the LoDs varied by 4 orders of magnitude, with the LoDs of many flagship tests being 20- to 50-fold greater than the LoDs for self-reported EUA submission documents (see Supplemental Table S2 and Supplemental Fig. S3; US FDA 2020). Three rounds of reference-panel LoD results have been released; however, we are not aware of any explanation for the large discrepancy in LoDs, but that range also matches what was observed from the EUA submissions themselves (MacKay et al. 2020). Currently, the reference panel materials (comprising 1 concentration of heat-inactivated virus and 4 blinded controls) are only available to EUA holders; wider access of these materials for those making EUA submissions would greatly improve objectivity and might accelerate the validation process.
CLIA and LDTs
According to the CDC, CLIA regulations include federal standards applicable to all US facilities or sites that test human specimens for health assessment or to diagnose, prevent, or treat disease. The FDA's relationship to regulating LDTs, both for nonemergency declaration tests and EUAs, is complex and evolving. Under normal circumstances, CLIA-approved laboratories can develop LDTs (i.e., “home brew” tests) that are limited to use in their specific laboratory and require more limited analytic validation data sets. Once the COVID-19 Emergency Declaration by the Department of Health and Human Services (HHS, Washington, DC) went into effect, CLIA laboratories were required to submit EUA LDTs. Liability protection (under the Public Readiness Emergency Preparedness Act [Pub L No. 109–148, 119 Stat 2818]) and reimbursement (under the Families First Coronavirus Response Act [Public L 116–127, 116 Stat 127]) both require an EUA. (In August 2020, HHS ruled that the FDA did not have regulatory authority to require premarket authorization for LDTs under the scheme, although that was subsequently reversed on November 16, 2020). Although many laboratories purchased commercial tests under an existing EUA, others developed and validated their own tests and were allowed to provide the FDA with their validation data in a defined timeframe as part of an EUA request. With a shift in priorities (December 2, 2020) to tests that increased accessibility (e.g., point-of-care tests) and capacity (high-volume tests), it appears that no further LDT EUAs have since been granted authorization.
State authorization
The FDA is providing flexibility to states who want to authorize laboratories certified to conduct high-complexity tests in that state to develop and perform coronavirus testing. Under this policy, the state or territory takes responsibility for the safety and accuracy of COVID-19 testing by laboratories in its state/territory, and the laboratory does not submit an EUA request to the FDA.
Institutional review boards
In addition, IRBs are formally designated groups that review and monitor clinical research involving human subjects. The purpose of the IRB review is to ensure, both in advance and by periodic review, that appropriate steps are taken to protect the rights and welfare of humans participating as subjects in the research. To accomplish that purpose, IRBs use a group process to review research protocols and related materials (e.g., informed-consent documents) to ensure protection of the rights and welfare of human subjects of research.
When any researcher is planning to gather personal data and samples from research subjects to validate their laboratory test (e.g., against a reference such as RT-PCR), they must submit their planned study protocol and consent forms to an IRB at their own institution (if they are part of a university) or via a commercial IRB (http://www.circare.org/info/commercialirb.htm). Rules governing the function of an IRB reside within 2 government agencies—the Office for Human Research Protections (Rockville, MD) and the FDA—both under the HHS.
Surveillance and asymptomatic testing
Although the regulated pathways of COVID-19 testing have, understandably, received the most attention, a separate pathway has opened that is not subject to the same regulatory controls. “Surveillance testing” is primarily used to gain information at a population, rather than an individual, level. Such testing may involve random sampling of a specific population, for example, to monitor disease prevalence and to determine the population-level effect of community interventions, such as social distancing. Prompted by a case involving the Bill & Melinda Gates Foundation (Seattle, WA) in July and August 2020, the FDA confirmed that, if the result of surveillance testing was purely to suggest to an individual that they go for a confirmatory test in a CLIA-certified laboratory, then the surveillance testing itself is not regulated by the Centers for Medicare Services (Baltimore, MD) or the FDA. In contrast, returning results from tests on asymptomatic people using a CLIA-certified laboratory is regulated by the FDA. Although commonly associated with screening of university staff and students, because surveillance testing is not formally regulated in the USA, it is difficult to estimate the extent of testing performed under this pathway. A subset of the authors of this article are actively engaged in a separate workgroup to explore the potential of expanding this type of RT-LAMP surveillance testing, not only in the USA but also elsewhere.
As the rollout of vaccines continues, US COVID-19 testing is projected to remain elevated for at least much of 2021 and, with new variants appearing, likely for years beyond. Although the role that LAMP tests will have in meeting that demand remains to be seen, continued progress on both the regulatory and commercial fronts will certainly help realize the immense potential of this important and flexible molecular-detection technology.
Regulatory Perspective in the European Union
Current IVD medical device directive
Tests for SARS-CoV-2 are classified as IVDs and must be CE-marked in accordance with the in vitro diagnostic medical devices directive (IVDD) and 98/79/EC, before being placed on the market in Europe. Moreover, CE marking is an administrative marking that indicates conformity with health, safety, and environmental protection standards for products sold within the European Economic Area (EEA) and that it can be sold legally within the EEA, but it is not, however, a quality indicator by itself.
There is no central approval system for IVDD in the European Union. For SARS-CoV-2 diagnostics designed for professional use (general IVDs), manufacturers self-certify that their test meets the requirements of the IVD Directive. Manufacturers are required to provide a certificate of conformity and a technical file that includes the intended use, a risk assessment, and the route by which they claim to be in conformity with the medical device directive. The file may also contain a clinical evidence report with sensitivity and specificity claims, among other reports. This pathway can be complex and Byzantine, given the different regulatory standards and classes of testing devices involved.
The IVDD specifies that devices must be designed and manufactured in such a way that they are suitable for the intended purpose specified by the manufacturer, taking account of the generally acknowledged state of the art. The devices must achieve the relevant performance, in particular, in terms of analytic sensitivity, diagnostic sensitivity, analytic specificity, and diagnostic specificity, accuracy, repeatability, and reproducibility and include control of known relevant interference and LoDs as stated by the manufacturer. The test must be registered with the competent authority in the country in which they are legally based. A competent authority in the context of IVDs is likely to be a given nation's body with authority to ensure that the requirements of the IVDD are fulfilled in that member state. For COVID-19 devices that are designed for use by lay persons (self-tests), the manufacturer must also apply to a third party body, called a notified body, which will do additional verification and issue a certificate.
In-house or LDTs
Somewhat analogous to US CLIA standards, ISO15189 (Medical Laboratories—Requirements for Quality and Competence) is a primary standard to which medical laboratories in Europe must adhere to perform clinical testing. These certifications may be held concurrently in the USA (Schneider et al. 2017). Usually, clinical testing laboratories purchase IVDs from market suppliers. However, when pandemic-driven demand for certain IVD products outstrips market supplies of those products, such health institute laboratories may work to develop their own in-house or LDTs. The IVD tests so developed are not considered to be placed on the market or put into service and are, thus, exempt from the requirements of directive 98/79/EC. Included are in-house exemptions such as the following:
IVDs that are manufactured in house are excluded from the legislation if they are used to test patient samples from the same institution.
If IVDs are manufactured in house and used to test patient samples from outside the manufacturing institution (i.e., from external medical practices or other health institutes), they are excluded from the legislation.
If health institutes use these non-CE–marked tests, the health institute itself must validate the test for their own use.
If these in-house, manufactured IVDs are transferred to another laboratory and that laboratory is outside the health institution's single legal entity, they are then within the scope of the IVD directive and, therefore, must be CE marked. In this scenario, the originating health institution is considered the legal manufacturer of the device (HPRA 2020, https://www.hpra.ie/).
Upcoming in vitro diagnostic regulation for 2022
The regulatory landscape described above is changing with the full implementation of the new IVDR (European Union 2017/746) as of May 26, 2022. Regulations are being tightened noticeably. After that May 2022 date, most “general category IVDs” will be reclassified and can no longer be self-certified, including those already on the market. This will increase the percentage of IVDs requiring CE marks from the current 20% under the IVD directive to 80% under the IVDR (BSI Group, www.bsigroup.com). In addition, all in-house IVDs, such as LDTs, will be regulated, including those that are currently exempt from IVDD. There will be a few limited exemptions if an equivalent device available on the market cannot already meet the clinical needs at the appropriate level of performance, but that scenario will apply to only a very few proportion of LDTs.
In the interest of protection of health, the directive states that a Member State may, in response to a duly justified request, authorize the placing on the market individual devices within its territory for which the applicable conformity-assessment procedures have not yet been performed (e.g., pending the completion of the device's evaluation). In adopting such national derogations, the nationally competent authority of the Member State must carefully consider any risk against the benefit of having the device available for immediate use. The national processes for adopting these derogations vary across Member States (EU Commission 2020, ec.europa.eu). Any SARS-Cov-2 IVD intended to be used in the European Union or to test samples originating in the European Union, even if the laboratory performing the test is based in another country, will be required to comply with IVDR, and therefore, manufacturers and clinical laboratories should start preparing for the relevant regulatory submissions with all new tests.
DISCUSSION
The unprecedented demand for diagnostics caused by the COVID-19 pandemic has demonstrated the glaring weaknesses of the current global, molecular diagnostics infrastructure. Using RT-qPCR as the reference method, some nations fared better than others, but testing needs exposed a strained global supply chain. Delays in turnaround time, limited throughput, the inability to cost-effectively increase the throughput scale in many settings, and restricted geographic and economic accessibility for both hardware and laboratory supplies showcased the operational weaknesses of RT-qPCR, despite its acknowledged value as a reference method.
As established in other fields and, recently, in the COVID-19 pandemic, LAMP technology is an accurate, rapid, easily deployable alternative to RT-qPCR, spanning infectious diseases, agriculture, food safety, and environmental monitoring. Clinically, LAMP has been used within low-resource settings, for example, for tuberculosis (WHO, TB-LAMP) and malaria (Eiken Chemical and Meridian Biosciences Inc [Cincinnati, OH]; Mohon et al. 2019). The COVID-19 pandemic has driven the need for expanded, lower-cost, and simple testing solutions from the bench to the bedside and from the Global South to all corners of the globe. Despite sensitivity that is comparable with RT-qPCR—and importantly, sufficient to detect most clinically important cases—LAMP is also not without its limitations and challenges, notably its propensity for nontemplate amplification, complex primer design, and the potential for laboratory cross-contamination.
What Is Fit-for-Purpose?
Community-wide, school-based, workplace, at-home, primary clinic, and tertiary hospital testing are each unique milieux, each with their own challenges and considerations. Given the variety of design options available (Figs. 1 and 8), developers must first define what combination of features are most suited to the intended use (e.g., total costs, usability, scalability, accessibility and convenience, diagnostic sensitivity and specificity, and effective clinical turnaround time [patient to result]). In resource-limited settings, the chosen combination is paramount because it most clearly articulates the (RE)ASSURED criteria (topic A).
Some choices are relatively obvious. For a portable testing system, ease of use, small footprint, and procedural simplicity are paramount (Newman et al. 2020), whereas those factors are less important for a tertiary hospital. In the community and in schools and colleges, among others, it is possible that the staff running the tests may initially be only minimally familiar with molecular biology techniques. In such cases, training materials should fully describe all steps of the process, even basic instructions, such as “mix” should be expanded to “mix by pipetting,” and instructional videos can also be quite helpful in capturing the nuances and helping staff to troubleshooting. Thus, whether the patient (sample) comes to the test or vice versa is a key first-order consideration, followed closely by the experience and training of those performing the testing.
High-analytic sensitivity is important when clinically meaningful levels of virus are at the LoD of the assay. However, if the intended purpose is to identify patients that pose the largest public health risk because of higher viral loads, then absolute analytic sensitivity is less important. The question of what viral load constitutes contagiousness continues to be debated. Several groups (Dudley et al. 2020, Fowler et al. 2020, Mina et al. 2020, Yaren et al. 2021;) have argued that transmission is primarily associated with an approximately 100-fold higher viral loads (≳100–1000 copies/μL) than the detection limit of many RT-qPCR reference assays (1 to 10 copies/μL). This would correspond to a 6- to 7-Ct “left-shift” relative to the qRT-PCR limit of detection (Ct approximately 40). Recently Coehlo et al. (2021) presented a thorough analysis of the clinical sensitivity of colorimetric RT-LAMP vs. RT-qPCR on 466 patient swab samples. They conclude that their RT-LAMP assay would reliably assign patients with Ct < 30 as positive (yellow), with Ct > 35 as negative (red) and those with 30 < Ct < 35 having an intermediate orange result color (determined by spectrophotometrically). Taking the estimate (from La Scola et al. [2020]) of Ct > 33 to 34 as the threshold for contagiousness, the totality of the data suggest that sensitivities in the 10 to 100 copies/μL range, as observed in LAMP, are well within the range required to identify clinically (and epidemiologically) relevant patients. In several recent studies (Fowler et al. 2020, Howson et al. 2020, Kellner et al. 2020, de Oliveira Coelho 2021, Egerer et al. 2021, Fowler et al. 2021, Natoli 2021, Nawattanapaiboon et al. 2021, Schellenberg et al. 2021), RT-LAMP has generally demonstrated comparable diagnostic performance to RT-qPCR, particularly for samples with the lower, and arguably more clinically relevant, Ct values in RT-PCR, and has also been shown to be close to RNA sequencing (RNA-seq) metrics (Butler et al. 2021).
Arguably, testing with an adequately sensitive method (LAMP) at higher frequency (twice weekly or even daily) to catch infection earlier is preferable to a higher-sensitivity test that cannot be administered as often and/or that has a longer patient-to-result turnaround time (RT-qPCR) (Pilcher et al. 2020, Cevik et al. 2021). In the USA, such testing with appropriate reporting of results is free from regulation by the FDA and has enabled surveillance programs in various US colleges. Regardless of frequency, shortening the time between test and actionable result is important (Larremore et al. 2020). The ability of LAMP to provide rapid (approximately 30 minutes), portable, at or near point-of-care testing at a reasonable (and reducing) cost argues for its use in such relatively high-frequency, community-based, distributed testing programs. More frequent testing is better, but a higher-sensitivity test is not necessarily better. Mining existing clinical and RT-qPCR (Ct value) data to help better address the question of what absolute sensitivity is fit for a clinical purpose appears an important, but outstanding, question, as well as the other biomarkers that can be found to stratify patients as high or low risk (Ng et al. 2021).
In a similar vein, the clinical test is but one aspect of the clinical diagnosis. False-positive results, which undoubtedly occur, can often be mitigated by repeated testing, particularly if the test is quick and comparatively affordable, or by confirmation using a more laborious and expensive RT-qPCR test on the few preliminary positives. False-positive rates should continue to fall as the use of dUTP/UDG and sequence-specific detection methods becomes more commonplace (see also Howson et al. 2021). False-negative results, on the other hand, can be mitigated by more-frequent testing and/or contact tracing in situations in which testing is applied broadly. Some testing regimens include duplicate testing (1 of 2 considered positive, e.g., Dudley et al. 2020, Newman et al. 2021). Fortunately, LAMP has sufficient sensitivity such that false-negative results are comparatively rare at the viral loads generally encountered clinically (>100 copies/μL).
Geographic and Economic Accessibility
Any discussion of test sensitivity should consider the cost of not testing those patients for whom the gold standard is equally inaccessible, geographically or economically. Given the transmissibility of the virus (and particularly of the newer variants), high testing coverage is in everyone's collective best interests. Driving down the cost and clinical turnaround time (patient-to-result) to allow frequent testing becomes increasingly important. The manufacture of in-house LAMP reagents by several laboratories argues well for this trend (“Open-Research Infrastructure”). Nowhere is that more important than in countries for which access to testing for even symptomatic patients is limited. In Manchester, England, the cost of the test is likely to be a secondary consideration; in Manila, cost is often the only consideration. In remote regions, a hospital with an RT-qPCR facility can easily be a day or more away. The clinical sensitivity of a test that is never run is zero.
Attributes of a “Good” RT-LAMP test
We initially sought to answer a seemingly simple question: “For someone starting to develop a LAMP SARS-CoV-2 assay now, what methodology would we recommend?” Given LAMP's inherent flexibility and applicability to very different types of use, making recommendations about “the best method” is necessarily fraught with difficulties. With that caveat, we summarize some of the important technical attributes that a “good” test would be expected to possess, making reference to the decision points outlined in Figure 1.
Collection method. Several methods have been published using anterior nares (AN) swabs (as more convenient alternatives to NPS) and now, particularly, for saliva/drool (Table 1) and gargle (Kellner et al. 2020; https://www.rtlamp.org). These are reasonable starting points for most applications, which are chosen primarily for their procedural simplicity and the ability of personnel with limited training (or the patient themselves) to perform the sampling. The challenge of saliva is its biologic complexity, particularly if used directly without RNA extraction and undiluted (e.g., with buffer or saline), and the tropism of each virus is unique, with some more abundant in saliva than others.
Virus inactivation. Several simple, low-cost, and effective methods exist (“Preanalytic Sample Processing”; Table 1), notably, heat inactivation in the presence of a reducing agent, a metal ion chelator, and/or a protease. Importantly, these simple methods enable scalable inactivation at near-to-the-point collection, requiring only limited infrastructure (e.g., a heating block) to reduce the required biosafety level for subsequent processing.
RNA extraction. It is during RNA extraction that the developer arguably makes their most important design decisions, which affects many upstream (sample selection) and downstream (amplification/detection) steps. The intended use dictates whether to include a dedicated RNA-extraction step. Even so, most LAMP methods do not now use the classic RNA extraction kits used for RT-qPCR testing, partly because of the greater tolerance of LAMP/Bst polymerase to amplification inhibitors and partly because of the low-cost, simplified processes of LAMP methods. In resource-limited environments, the time, cost, and equipment savings of omitting this step far outweigh the loss in sensitivity (“Direct Methods”), whereas if resources are available, the increased sensitivity can be useful for, for example, pooling testing schema. Both magnetic and glass milk extractions are viable and increasingly cost effective (“RNA Extraction”).
Detection chemistry. Colorimetric readouts (ΔpH or ΔMg2+-free) remain mainstays of LAMP testing primarily because of their simplicity. However, they suffer from sample matrix interferences (phenol red) and/or from difficulties of naked-eye color discrimination (HNB). With the increasing advent of low-cost, more-portable fluorescence readers, the detection options have increased correspondingly. Of the 6 main fluorescence-based methods, 5 are sequence specific (Table 3) and, thus, offer the promise of increased matrix robustness, of multiplexing (multiple SARS genes, multiple pathogens, and/or an internal process control), and likely, of mitigation of the real concern of nonspecific amplification (Moehling et al. 2021). These fluorescence approaches provide a “second stage” of specificity, in addition to that from the amplification primers alone (Bhadra et al. 2020, Sherill-Mix et al. 2020, Zhang et al. 2020b, Bektas et al. 2021). Although that increases the complexity of the assay design, we see this category of techniques as the future norm for multiplexed fluorescence-based LAMP detection, just as Taqman is for qPCR.
Hardware/software for quantification. Both colorimetric and fluorigenic LAMP tests can now be objectively quantified with a range of smartphone or tablet applications (“Software”) and many midpriced, dedicated incubator/fluorescence readers, some with melt-curve capabilities. From the simplest water bath to low-cost plate- and tube-based heaters (many being open source) to more-advanced microfluidic platforms, there is no shortage of hardware options for the relatively simple process of maintaining a LAMP reaction at 1 or 2 elevated temperatures (“Hardware Designed for LAMP-Based Assays”). With a reasonably small budget, one can now afford the infrastructure to perform sequence-specific, multicolor, multiplexed, fluorescence-based (kinetic) LAMP assays with 1 or more targets and an internal extraction control. Arguably, if LAMP is to attain the widespread clinical penetration enjoyed by multiplexed qPCR, this type of low-cost experimental setup will need to become the norm.
Future Technical Directions
As vaccinations are becoming increasingly available and accessible, novel variants with increased transmissibility and infectivity remind us that diagnostics remain an integral component to our response to COVID-19 (Alpert et al. 2021) and integration with changes in clinical care (Afshinnekoo et al. 2021). The past year has seen the maturation of LAMP-based diagnostics from the bench to the bedside and from the Global South to worldwide. Isothermal methods have a number of inherent logistical advantages over qPCR; however, LAMP has several remaining challenges to address for more-widespread use in clinical settings. Even relatively low rates of false-positive results caused by mispriming (or cross-contamination) in LAMP assays represents a shortcoming for diseases with relatively low (postpandemic) prevalence (Moehling et al. 2021). Therefore, we highlight the variety of sequence-specific, fluorescence-based methods (“Sequence-Dependent Detection of RT-LAMP Amplification Products”) as a likely future trajectory for clinical LAMP testing.
The intrinsic biochemical sensitivity of LAMP is directly comparable with that of qPCR (single-digit copies per reaction). However, many LAMP developers have consciously addressed a different, near point-of-care use, with minimal sample processing. This contrasts with the highly resource and automation-intensive methods required in central RT-qPCR testing facilities that perform RNA extraction as a matter of clinical normality. Given this fundamental difference in objective, the question regarding what represents a fit-for-that-purpose LAMP test sensitivity remains. Focusing excessively on sensitivity, as but 1 of the 8 REASSURED dimensions, appears unnecessarily self-limiting. The effective sensitivity of a qPCR test that is never deployed because it is geographically or economically inaccessible is far below that of an accessible, but lower sensitivity, LAMP test; it is zero.
High-quality, rapid antigen tests clearly have clinical utility. If advances in LAMP technology, reagents, and supply chains made over the past 12 months continue, we can foresee a time when LAMP molecular tests can compete with all but the cheapest rapid-antigen tests. Because LAMP tests provide the starting material for direct sequencing of positive results, it provides a strong rationale for wider adoption, particularly because the price differential with rapid-Ag tests continues to diminish. This allows wider-scale identification of new COVID-19 variants, emerging pathogens, and a wide variety of otherwise-undetectable public health threats. The ability to sequence directly from LAMP reactions (LAMP-seq and LAMPore) also distinguishes it favorably from qPCR (“Sequencing Approaches Using RT-LAMP Amplicons”).
CONCLUSION
During the past year, we have learned that existing diagnostic systems and implementations globally have failed the enormous challenge posed by SARS-CoV-2 and that a massive increase in rapid-testing methods are needed to mitigate the spread of viruses. Whether testing more-symptomatic patients in resource-limited settings or more-asymptomatic individuals in countries in which testing is more widely available, there needs to be a shift in thinking to include regular, repeated testing of large swaths of the population.
The flexibility to create multiple versions of LAMP suited to different settings is a preeminent concern. Such flexibility does more than create design and supply chain choices, it fundamentally changes the landscape of what is possible for infectious disease. The potential to expand testing opens up public health responses and policies that have, to this point, seemed unrealizable. Parallels to the transition of computing from one of large, centralized mainframe systems to include, and then become dominated by, personal computing are quite clear. New technologies have provided individuals and small groups with access to computational power previously centralized in large companies. As a trend, we embrace a more-distributed future for diagnostics in which local entities can perform work that was previously reserved for large, highly resourced laboratories.
Commensurate with the focus of the Journal of Biomolecular Techniques, we have sought to highlight some of the more relevant and innovative methodologic advances to enable such a transformation. Although much remains to be done, LAMP methods have greatly matured in the past year, with increasing acceptance that it can now take its place squarely in the cast of available clinical technologies in clinical laboratories. With a broad and vibrant open-science movement enabling lower-cost innovations in bioware, software, and hardware, the accessibility of diagnostics by a larger proportion of the globe is an increasingly realistic possibility.
If the future of testing is, indeed, distributed and closer to the point of patient care, the inherent advantages of LAMP mean it can have a significant role, just as PCs (and later laptops and then cell phones) did for information and communication. Whatever the ultimate trajectory of diagnostic testing, lessons learned need to continue to be translated into robust and reliable clinical-testing platforms. Although we will eventually curtail the spread of SARS-CoV-2, the next major infectious agent is surely on the horizon somewhere on our increasingly small and interconnected globe.
Topic D: review highlights and takeaways
Topic D highlights some important considerations for building LAMP tests based on the scale of testing, the intended use, the location or setting, and the budget and level of infrastructure available.
The right test for the right application. “We have a LAMP for that.” Given the variety of design options available, the testing problem dictates the most-appropriate testing configuration. Unlike the qPCR platform, the LAMP testing schema can vary widely in total cost, usability, scalability, accessibility and convenience, diagnostic sensitivity and specificity, and effective clinical turnaround time (patient to result).
How the test result will be used. Will the clinical diagnosis be fore the individual or for epidemiologic surveillance of a population? Will testing be in singlicate or pooled format? These very different uses dictate consciously made decisions about trade-offs among sensitivity, specificity, speed of result, and total cost of testing.
How the hidden cost of not testing affects utility. If your test is geographically or economically inaccessible to “the many,” it is of far less aggregate value than a high-quality test that is accessible by only “the few.” The logistical advantages of LAMP over qPCR have a real clinical value: the sensitivity of a test that is never run is zero.
Location, location, location! The inexorable drive toward distributed testing requires designs that are “As simple as possible… but no simpler.” With hardware becoming smaller, more-portable, and cheaper and with readouts becoming both simple and specific, impediments to testing based on geographic inaccessibility will continue to diminish.
Frequency and time-to-results matter. Once demand for symptomatic testing is met, twice weekly testing in congregate living settings should be an achievable objective of many testing regimes. The viral load rises quickly in a newly infected person, so frequent testing is important for containment. If possible, take the test to the patient, not the other way around, particularly for use in remote or rural locations.
Being positive and being contagious may not be the same thing. Basing clinical and policy decisions on the analytic sensitivity of qPCR might not be the most clinically relevant measure of test utility. If the purpose is to identify individuals with the greatest probability of transmission, then a LAMP test with sufficient test sensitivity but greater population accessibility likely has the more-preferable profile overall.
Think beyond a single test. The diagnostic test, as evaluated in the literature, is only a partial reflection of clinical practice. Repeat or duplicate testing improves clinical diagnosis, either with the same test (LAMP→LAMP) or with an orthogonal test (e.g., LAMP→PCR). If the test is local or distributed, relatively fast, and relatively cheap, confirmation or repeats of positive results are a viable triaging strategy.
Think beyond testing personnel in the laboratory. As testing becomes increasingly distributed (either by choice or out of necessity), the test is likely to be run by health care workers with minimal formal training in molecular biology techniques. Take nothing for granted, ensure user-acceptance testing occurs in the intended settings, and consider the increasing accessibility of video as a communication and training tool.
Attributes of a good SARS-CoV2 RT-LAMP test include the following:
Collection method. Saliva, gargle, or OP/AN swabs are validated biospecimens and are highly preferable to NP swabs because of simplicity and effect on the patient.
Viral inactivation. There are several methods for viral inactivation based on heating, typically in TCEP/EDTA/proteinase K buffers to stabilize viral RNA.
RNA extraction and purification. In resource-limited environments, the savings in cost, equipment, and training far outweigh the loss in sensitivity associated with omitting the extraction step. With greater resources, and if increased sensitivity adds value (e.g., for pooling samples), simplified RNA extraction methods based on magnetic and glass milk or silica beads are available.
Control RNA. RNA heat- and γ-radiation-inactivated viral controls from BEI Resources are a good starting point for preclinical method development.
Target and control genes. Most LAMP assays use 1 or more nucleocapsid (N) primer sets, alone or augmented with envelope (E), RdRp, and sometimes spike (S) sets. RNaseP and β-actin are common positive extraction controls.
Reaction monitoring. If a pH-based method is used, neutralize the input samples to avoid false-positive results; if a colorimetric readout is used, use Smartphone applications to quantify color changes; and if fluorescence is used, newer sequence-specific methods are increasingly preferred over dye intercalation to reduce false-positive results.
Hardware/software. Every day, seemingly, brings new hardware and software, both from commercial suppliers of lower-cost alternatives to qPCR readers and from laboratories taking advantage of the free and open-source hardware movement.
TABLE 5.
Deployment models for LAMP testing
|
|
Itema
|
Example (part no.)
|
Cost, approximate US$
|
| L | Pipette tips, nitrile gloves, biohazard bag, Lo-bind 1.5-mL microfuge tubes; PCR plate or 0.2-mL tubes | 76322-160; 89428-750; 10035-976; 80077-230; 10049-108 | 1000 |
| L | Manual P100 pipette | 76335-742 | 300 |
| L | SARS-CoV-2 LAMP kit; (100–500 RXN) | e.g., NEB E2019S (colorimetric); e.g., OptiGene RT-LAMP KIT-500 | 8/test |
| L | Sous vide water (65°C heat source) | approximately $40 | 40 |
| M | Thermocycler (tube/plate) | 71003-564 | 9000 |
| M | Biosafety cabinet | 89260-050 | 11 000 |
| M | Precision convection oven | PR305220M | 3000 |
| M | Plate reader | e.g., Biotek Neo 2 | 36 000 |
| M | Corning LSE vortex mixer | 6775 | 200 |
| M | Corning LSE mini microfuge | 6770 | 200 |
| H | Refrigerated centrifuge | 5942000245 | 17 000 |
| H | HTP plate reader | e.g., BioTek Neo2S | 43 000 |
| H | PX1 PCR plate sealer | 1814000 | 4500 |
| H | Xpeel plate seal remover | 1150L21 | 37 000 |
| H | BioTek MultiFlo FX MultiMode Dispenser | BTMFXP1 | 18 000 |
| H | Hamilton Starlet liquid handling robot (LHR) | 145 000 | |
| H | Chemagic 360 (automated magnetic-bead based nucleic acid extraction) | 2024-0020 | 130 000 |
The hardware infrastructure required for different scales of testing is indicated by progressive increases in the capital infrastructure needed. In general, testing moves from tubes to plates and from manual pipetting/reading to automated processes. The items (and their approximate prices) are only intended to be exemplary and indicative, as recorded by 1 US-based genomics research laboratory.
Supplementary Material
Acknowledgments
We would like to thank the Epigenomics Core Facility at Weill Cornell Medicine, the Scientific Computing Unit (SCU), XSEDE Supercomputing Resources, the National Center for Advancing Translational Sciences of the National Institutes of Health (UL1TR000457, CTSC), the Saphier Endowment for Translational Research, the Irma T. Hirschl and Monique Weill-Caulier Charitable Trusts, the Bert L. and N. Kuggie Vallee Foundation, the WorldQuant Foundation, the Pershing Square Sohn Cancer Research Alliance, and the Bill and Melinda Gates Foundation (OPP1151054). We also thank Testing for America (501c3), OpenCovidScreen Foundation, the Bert L and N Kuggie Vallee Foundation, Igor Tulchinsky and the WorldQuant Foundation, Bill Ackman and Olivia Flatto and the Pershing Square Foundation, Ken Griffin and Citadel, the US National Institutes of Health (R01AI125416, R21AI129851, R01AI151059, U01DA053941), the Saphier Endowment for Translational Research, and the Alfred P. Sloan Foundation (G-2015-13964). We would like to thank the researchers cited here who have helped further LAMP-based diagnostics, in particular, members of the Global LAMP Consortium not listed. We acknowledge the original ground-breaking work of Eiken Chemical to develop the technique and the contributions of the Vienna Group (VCDI) to methods development, the Artic Network and GISAID as sources of SARS-related sequence information, and BEI Resources, which continues to supply reference controls to the global research community. We thank ABRF and the reviewers and editors of the Journal of Biomolecular Techniques for their patience, support, and encouragement. We are grateful for the Bayan Biomedical undergraduate team at Ateneo for their literature research. K.J.M.M. dedicates his contribution to the memory of Dr. Ian R. Moore (1964–2018), University of Oxford.
REFERENCES
- Abraham JP, Plourde BD, Cheng L. Using heat to kill SARS-CoV-2. Rev Med Virol . 2020;30:e2115. doi: 10.1002/rmv.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshinnekoo E, Bhattacharya C, Burguete-García A et al. MetaSUB Consortium. COVID-19 drug practices risk antimicrobial resistance evolution. Lancet Microbe . 2021;2:e135–e136. doi: 10.1016/S2666-5247(21)00039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S, Fanton A, Chandrasekaran S, et al. Rapid, point-of-care molecular diagnostics with Cas13. medRxiv . 2020. Posted online April 5. and in revised form 2021. [DOI]
- Aidelberg G, Aronoff R. Freeze-drying (lyophilization) of CoronaDetective tubes V.1. 2020. dx.doi.org/10.17504/protocols.io.bk44kyyw Available at: https://www.protocols.io/view/freeze-drying-lyophilization-of-coronadetective-tu-bk44kyyw.html Posted September 8.
- Alekseenko A, Barrett D, Pareja-Sanchez Y, et al. Direct detection of SARS-CoV-2 using non-commercial RT-LAMP reagents on heat-inactivated samples. Sci Rep . 2021;11:1820. doi: 10.1038/s41598-020-80352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allgöwer SM, Hartmann CA, Lipinski C, et al. LAMP-LFD based on isothermal amplification of multicopy gene ORF160b: applicability for highly sensitive low-tech screening of allergenic soybean (Glycine max) in Food. Foods . 2020;9:1741. doi: 10.3390/foods9121741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasi M.A, Moradi A, Nasiri J, et al. Assessment of performance ability of three diagnostic methods for detection of Potato Leafroll Virus (PLRV) using different visualizing systems. Appl Biochem Biotechnol . (2012); 168 :770–784. doi: 10.1007/s12010-012-9818-1. [DOI] [PubMed] [Google Scholar]
- Alpert T, Brito AF, Lasek-Nesselquist E, et al. Early introductions and transmission of SARS-CoV-2 variant B.1.1.7 in the United States. Cell . 2021;184:2595–2605.e13. doi: 10.1016/j.cell.2021.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arizti-Sanz J, Freije CA, Stanton AC, et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat Commun. 2020 Nov 20;11(1):5921. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine R, Hasan A, Das S, et al. Loop-mediated isothermal amplification (LAMP): a rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 Pandemic. Biology (Basel) . 2020;9:182. doi: 10.3390/biology9080182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden T, Chagas AM, Gage GJ. Open Labware: 3-D printing your own lab equipment. PLoS Biol. Mar 20; (2015);13(3):e1002086. doi: 10.1371/journal.pbio.1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball CS, Light YK, Koh CY, et al. Quenching of unincorporated amplification signal reporters in reverse-transcription loop-mediated isothermal amplification enabling bright, single-step, closed-tube, and multiplexed detection of RNA viruses. Anal Chem . 2016;88:3562–3568. doi: 10.1021/acs.analchem.5b04054. [DOI] [PubMed] [Google Scholar]
- Basiri A, Heidari A, Nadi MF, et al. Microfluidic devices for detection of RNA viruses. Rev Med Virol . 2021;31:1–11. doi: 10.1002/rmv.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batéjat C, Grassin Q, Manuguerra JC, et al. Heat inactivation of the severe acute respiratory syndrome coronavirus 2. J Biosaf Biosecur . 2021;3:1–3. doi: 10.1016/j.jobb.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherer L, Borst N, Bakheit M, et al. Loop-mediated isothermal amplification (LAMP) —review and classification of methods for sequence-specific detection. Anal Methods . 2020;12:717–746. [Google Scholar]
- Bektaş A, Covington M, Aidelberg G, et al. Accessible LAMP-Enabled Rapid Test (ALERT) for detecting SARS-CoV-2. medRxiv . 2021. Published online February 20. 10.1101/2021.02.18.21251793. [DOI] [PMC free article] [PubMed]
- Ben-Assa N, Naddaf R, Gefen T, et al. Direct on-the-spot detection of SARS-CoV-2 in patients. Exp Biol Med . 2020;245:1187–1193. doi: 10.1177/1535370220941819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkenbrock JA, Grecco-Machado R, Achenbach S. Arsenal of microfluidic testing devices may combat COVID-19 pandemic. MRS Bull . 2020;45:511–514. doi: 10.1557/mrs.2020.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra S, Maranhao AC, Paik I, et al. One-enzyme reverse transcription qPCR using Taq DNA polymerase. Biochemistry . 2020a;59:4638–4645. doi: 10.1021/acs.biochem.0c00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra S, Riedel T, Lakhotia S, et al. High-surety isothermal amplification and detection of SARS-CoV-2, including with crude enzymes. bioRxiv . 2020b. Posted on May 7. now accepted for publication in mSphere. [DOI] [PMC free article] [PubMed]
- Bhamla M, Benson B, Chai C, et al. Hand-powered ultralow-cost paper centrifuge. Nat Biomed Eng 1, 0009. (2017). [DOI]
- Bokelmann L, Nickel O, Maricic T, et al. Point-of-care bulk testing for SARS-CoV-2 by combining hybridization capture with improved colorimetric LAMP. Nat Commun . 2021;12:1467. doi: 10.1038/s41467-021-21627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton JP, Deng X, Yu G, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol . 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TA. Tsang Arthur, et al., editors. Schaefer, Direct detection of SARS-CoV-2 RNA using high-contrast pH-sensitive dyes. 2020. https://www.medrxiv.org/content/10.1101/2020.12.26.20248878v2 . [DOI] [PMC free article] [PubMed]
- Brune Z, Kuschner CE, Mootz J, et al. Effectiveness of SARS-CoV-2 decontamination and containment in a COVID-19 ICU. Int J Environ Res Public Health . 2021;18:2479. doi: 10.3390/ijerph18052479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull RA, Adikari TN, Ferguson JM, et al. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat Commun . 2020;11:6272. doi: 10.1038/s41467-020-20075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonanno G, Stabile L, Morawska L. Estimation of airborne viral emission: quanta emission rate of SARS-CoV-2 for infection risk assessment. Environ Int . 2020;141:105794. doi: 10.1016/j.envint.2020.105794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler D, Mozsary C, Meydan C, et al. Host, viral, and environmental transcriptome profiles of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). bioRxiv . 2020. Posted online April 21. [DOI]
- Butler D, Mozsary C, Meydan C, et al. Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. Nat Commun . 2021;12:1660. doi: 10.1038/s41467-021-21361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert AE, Biggerstaff BJ, Tanner NA, Lauterbach M, Lanciotti RS. Rapid colorimetric detection of Zika virus from serum and urine specimens by reverse transcription loop-mediated isothermal amplification (RT-LAMP) PLoS One. 2017 Sep 25;12(9):e0185340. doi: 10.1371/journal.pone.0185340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell C, Kava A, Nguyen M, et al. Beyond the lateral flow assay: a review of paper-based microfluidics. Microelectron Eng . 2019;206:45–54. [Google Scholar]
- Carter C, Akrami K, Hall D, et al. Lyophilized visually readable loop-mediated isothermal reverse transcriptase nucleic acid amplification test for detection Ebola Zaire RNA. J Virol Methods . 2017;244:32–38. doi: 10.1016/j.jviromet.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik M, Tate M, Lloyd O, et al. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe . 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagas MA. Haves and have nots must find a better way: The case for open scientific hardware. PLoS Biol. Sep 27; (2018);16(9):e3000014. doi: 10.1371/journal.pbio.3000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challener CA. For lyophilization, excipients really do matter. Biopharm Int . 2017;30:32–35. https://www.biopharminternational.com/view/lyophilization-excipients-really-do-matter . [Google Scholar]
- Chander Y, Koelbl J, Puckett J, et al. A novel thermostable polymerase for RNA and DNA loop-mediated isothermal amplification (LAMP) Front Microbiol . 2014;5:395. doi: 10.3389/fmicb.2014.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouch M. Loop-mediated isothermal amplification (LAMP): An effective molecular point-of-care technique for the rapid diagnosis of coronavirus SARS-CoV-2. Rev Med Virol . 2021;31:e2215. doi: 10.1002/rmv.2215. Posted online ahead of print January 21, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HW, Ching WM. Evaluation of the stability of lyophilized loop-mediated isothermal amplification reagents for the detection of Coxiella burnetii. Heliyon . 2017;3:e00415. doi: 10.1016/j.heliyon.2017.e00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Niu J, Zhang Y, et al. Preparation of His-tagged armored RNA phage particles as a control for real-time reverse transcription-PCR detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol . 2006;44:3557–3561. doi: 10.1128/JCM.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua AL, Elina HT, Lim BH, et al. Development of a dry reagent-based triplex PCR for the detection of toxigenic and non-toxigenic Vibrio cholerae. J Med Microbiol . 2011;60(pt 4):481–485. doi: 10.1099/jmm.0.027433-0. [DOI] [PubMed] [Google Scholar]
- Cimolai N. Potentially repurposing adamantanes for COVID-19. J Med Virol . 2020;92:531–532. doi: 10.1002/jmv.25752. [DOI] [PubMed] [Google Scholar]
- Contreras JL, Eisen M, Ganz A, et al. Pledging intellectual property for COVID-19. Nat Biotechnol . 2020;38:1146–1149. doi: 10.1038/s41587-020-0682-1. [DOI] [PubMed] [Google Scholar]
- Cook J, Aydin-Schmidt B, González IJ, et al. Loop-mediated isothermal amplification (LAMP) for point-of-care detection of asymptomatic low-density malaria parasite carriers in Zanzibar. Malar J . 2015;14:43–43. doi: 10.1186/s12936-015-0573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill . 2021;25:2000045.. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox HS, Daniels JF, Muller O, et al. Impact of decentralized care and the Xpert MTB/RIF test on rifampicin-resistant tuberculosis treatment initiation in Khayelitsha, South Africa. Open Forum Infect Dis. . 2015; 2 :ofv014.. doi: 10.1093/ofid/ofv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko D, Bezdan D, Afshin EE. et al ; International MetaSUB Consortium. A global metagenomic map of urban microbiomes and antimicrobial resistance. Cell . 2021;184:3376–3393.e17. doi: 10.1016/j.cell.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao Thi VL, Herbst K, Boerner K,et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci T ransl Med. . 2020;12:eabc7075.. doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Coelho B, Sanchuki H, Zanette DL, et al. Essential properties and pitfalls of colorimetric reverse transcription loop-mediated isothermal amplification as a point-of-care test for SARS-CoV-2 diagnosis. Mol. M ed . 2021;27:30. doi: 10.1186/s10020-021-00289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Gonçalves D, Hooker DJ, Dong Y, et al. Detecting wMel Wolbachia in field-collected Aedes aegypti mosquitoes using loop-mediated isothermal amplification (LAMP) Parasit Vectors . 2019;12:404. doi: 10.1186/s13071-019-3666-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz L, Johnson B, Jenkins D. Real-time optical analysis of a colorimetric LAMP assay for SARS-CoV-2 in saliva with a handheld instrument improves accuracy compared to endpoint assessment. medRxiv . 2021. Posted online on January 15. [DOI] [PMC free article] [PubMed]
- Dudley DM, Newman CM, Weiler AM, et al. Optimizing direct RT-LAMP to detect transmissible SARS-CoV-2 from primary nasopharyngeal swab samples. PloS One . 2020;15:e0244882. doi: 10.1371/journal.pone.0244882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftekhari A, Alipour M, Chodari L, et al. A comprehensive review of detection methods for SARS-CoV-2. Microorganisms . 2021;9:232. doi: 10.3390/microorganisms9020232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egerer R, Edel B, Löffler B, et al. Performance of the RT-LAMP-based Eazyplex® SARS-CoV-2 as a novel rapid diagnostic test. J Clin Virol . 2021;138:104817. doi: 10.1016/j.jcv.2021.104817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira-González L, Puchades AV, Carpino C, et al. Fast detection of Southern tomato virus by one-step transcription loop-mediated isothermal amplification (RT-LAMP) J Virol Methods . 2017;241:11–14. doi: 10.1016/j.jviromet.2016.12.004. [DOI] [PubMed] [Google Scholar]
- Farshidfar N, Hamedani S. The potential role of smartphone-based microfluidic systems for rapid detection of COVID-19 using saliva specimen. Mol Diagn Ther . 2020;24:371–373. doi: 10.1007/s40291-020-00477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Z, Wei R, Cheng C, et al. A novel approach to the bioluminescent detection of the SARS-CoV-2 ORF1ab gene by coupling isothermal RNA reverse transcription amplification with a digital PCR approach. Int J Mol Sci . 2021;22:1017. doi: 10.3390/ijms22031017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipić A, Dobnik D. Tušek Žnidarič M, et al Inactivation of pepper mild mottle virus in water by cold atmospheric plasma. Front Microbiol . 2021;12:618209. doi: 10.3389/fmicb.2021.618209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel Y, Mizrahi O, Nachshon A, et al. The coding capacity of SARS-CoV-2. Nature . 2021;589:125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- Flynn M, Snitser O, Flynn J, et al. A simple direct RT-LAMP SARS-CoV-2 saliva diagnostic. medRxiv . 2020. Posted online November 22. [DOI]
- Foox J, Tighe SW, Nicolet CM, et al. Performance assessment of DNA sequencing platforms in the ABRF Next-Generation Sequencing Study. Nat Biotechnol . 2021;39:1129–1140. doi: 10.1038/s41587-021-01049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler VL, Armson B, Gonzales JL, et al. A highly effective reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the rapid detection of SARS-CoV-2 infection. J Infect . 2021;92:117–125. doi: 10.1016/j.jinf.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler J, Hill S, Levin R, et al. The effect of stay-at-home orders on COVID-19 cases and fatalities in the United States. medRxiv . 2020. Posted online on April 20. now published in PLoS One. [DOI]
- Francois P, Tangomo M, Hibbs J, et al. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med. Microbiol . 2011;62:41–48. doi: 10.1111/j.1574-695X.2011.00785.x. [DOI] [PubMed] [Google Scholar]
- Ganguli A, Mostafa A, Berger J, et al. Rapid isothermal amplification and portable detection system for SARS-CoV-2. bioRxiv . 2020. Posted online on May 21. now published in Proc Natl Acad Sci USA. 10.1073/pnas.2014739117 https://doi.org/10.1101/2020.05.21.108381. [DOI] [PMC free article] [PubMed]
- García-Bernalt Diego J, Fernández-Soto P, Crego-Vicente B,et al. Progress in loop-mediated isothermal amplification assay for detection of Schistosoma mansoni DNA: towards a ready-to-use test. Sci. Rep. . 2019;9:14744. doi: 10.1038/s41598-019-51342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Bernalt Diego J, Fernández-Soto P, Domínguez-Gil M,et al. A simple, affordable, rapid, stabilized, colorimetric, versatile RT-LAMP assay to detect SARS-CoV-2. Diagnostics (Basel) . 2021a;11:438. doi: 10.3390/diagnostics11030438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Bernalt Diego J, Fernández-Soto P, Febrer-Sendra B,et al. Loop-mediated isothermal amplification in schistosomiasis. J Clin Med. . 2021b;10:511. doi: 10.3390/jcm10030511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Venzor A, Rueda-Zarazua B, Marquez-Garcia E, et al. SARS-CoV-2 direct detection without RNA isolation with loop-mediated isothermal amplification (LAMP) and CRISPR-Cas12. Front Med . 2021;8:627679. doi: 10.3389/fmed.2021.627679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneret P, Coz E, Martin E, et al. Performing point-of-care molecular testing for SARS-CoV-2 with RNA extraction and isothermal amplification. PloS One . 2021;16:e0243712. doi: 10.1371/journal.pone.0243712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong MM, Sinton D. Turning the page: advancing paper-based microfluidics for broad diagnostic application. Chem Rev . 2017;117:8447–8480. doi: 10.1021/acs.chemrev.7b00024. [DOI] [PubMed] [Google Scholar]
- González-González E, Lara-Mayorga I, Rodríguez-Sánchez I, et al. Scaling diagnostics in times of COVID-19: colorimetric loop-mediated isothermal amplification (LAMP) assisted by a 3D-printed incubator for cost-effective and scalable detection of SARS-CoV-2. medRxiv . 2020. Posted online on May 6. 10.1101/2020.04.09.20058651.
- Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science . 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M, Honda E, Ogura A, et al. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Bioechniques . 2009;46:167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- Guo L, Sun X, Wang X, et al. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov . 2020;6:34. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Parker J, Smits S, et al. Persistent viral shedding of SARS-CoV-2 in faeces —a rapid review. Colorectal Dis . 2020;22:611–620. doi: 10.1111/codi.15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haibe-Kains B, Adam GA, Hosny A et al. Massive Analysis Quality Control (MAQC) Society Board of Directors. Transparency and reproducibility in artificial intelligence. Nature . 2020;586:E14–E16. doi: 10.1038/s41586-020-2766-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardinge P, Kiddle G, Tisi L, et al. Optimised LAMP allows single copy detection of 35Sp and NOSt in transgenic maize using bioluminescent assay in real time (BART) Sci Rep . 2018;8:17590. doi: 10.1038/s41598-018-36207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasell J, Mathieu E, Beltekian D, et al. A cross-country database of COVID-19 testing. Sci Data . 2020;7:345. doi: 10.1038/s41597-020-00688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howson E, Kidd SP, Armson B, et al. Preliminary optimisation of a simplified sample preparation method to permit direct detection of SARS-CoV-2 within saliva samples using reverse-transcription loop-mediated isothermal amplification (RT-LAMP) J Virol Methods . 2021;289:114048. doi: 10.1016/j.jviromet.2020.114048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Guo H, Zhou P, et al. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol . 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Kalsi S, Zeimpekis I, et al. Ultra-fast electronic detection of antimicrobial resistance genes using isothermal amplification and thin film transistor sensors. Biosens Bioelectron . 2017;96:281–287. doi: 10.1016/j.bios.2017.05.016. [DOI] [PubMed] [Google Scholar]
- Hu F, Li J, Zhang Z, et al. Smartphone-based droplet digital LAMP device with rapid nucleic acid isolation for highly sensitive point-of-care detection. Anal Chem . 2020;92:2258–2265. doi: 10.1021/acs.analchem.9b04967. [DOI] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 Feb 15;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Lin X, Urmann K, et al. Smartphone-based in-gel loop-mediated isothermal amplification (gLAMP) system enables rapid coliphage MS2 quantification in environmental waters. Environ Sci Technol . 2018;52:6399–6407. doi: 10.1021/acs.est.8b00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatov KB, Barsova EV, Fradkov AF, et al. A strong strand displacement activity of thermostable DNA polymerase markedly improves the results of DNA amplification. Biotechniques . 2014;57:81–87. doi: 10.2144/000114198. [DOI] [PubMed] [Google Scholar]
- James P, Stoddart D, Harrington E, et al. LamPORE: rapid, accurate and highly scalable molecular screening for SARS-CoV-2 infection, based on nanopore sequencing. medRxiv . 2020. Posted online on August 11. [DOI]
- Jang WS, Lim DH, Yoon J, et al. Development of a multiplex Loop-Mediated Isothermal Amplification (LAMP) assay for on-site diagnosis of SARS CoV-2. PLoS One. Mar 3; (2021);16(3):e0248042. doi: 10.1371/journal.pone.0248042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani IV, Peter TF. How point-of-care testing could drive innovation in global health. N Engl J Med . 2013;386:2319–2324. doi: 10.1056/NEJMsb1214197. [DOI] [PubMed] [Google Scholar]
- Janíková M, Hodosy J, Boor P, et al. Loop-mediated isothermal amplification for the detection of SARS-CoV-2 in saliva. Microb Biotechnol . 2021;14:307–316. doi: 10.1111/1751-7915.13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B, Li X, Liu W, et al. GLAPD: whole genome based lamp primer design for a set of target genomes. Front Microbiol . 2019;10:2860. doi: 10.3389/fmicb.2019.02860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Mancuso M, Lu Z, et al. Solar thermal polymerase chain reaction for smartphone-assisted molecular diagnostics. Sci Rep . 2014;4:4137. doi: 10.1038/srep04137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. Detergents: Triton X-100, Tween-20, and more. Mater Methods . 2013;3:163. [Google Scholar]
- Jureka AS, Silvas JA, Basler CF. Propagation, inactivation, and safety testing of SARS-CoV-2. Viruses . 2020;12:622. doi: 10.3390/v12060622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J, Ladha A, Saito M, et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N Engl J Med. 2020 Oct 8;383(15):1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaarj K, Akarapipad P, Yoon JY. Simpler, faster, and sensitive Zika virus assay using smartphone detection of loop-mediated isothermal amplification on paper microfluidic chips. Sci Rep . 2018;8:12438. doi: 10.1038/s41598-018-30797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl L, Molloy J, Patron N, Matthewman C, Haseloff J, Grewal D, Johnson R, Endy D. Opening options for material transfer. Nat Biotechnol. 2018 Oct 11;36(10):923–927. doi: 10.1038/nbt.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf G, Todt D, Pfaender S, et al. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect . 2020;104:246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H, Kawana T, Fukushima E, et al. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J Biochem Biophys Methods . 2007;70:499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Kellner M, Ross J, Schnabl J, et al. A rapid, highly sensitive and open-access SARS-CoV-2 detection assay for laboratory and home testing. bioRxiv . 2020. Posted online on July 23. [DOI] [PMC free article] [PubMed]
- Khorosheva EM, Karymov MA, Selck DA, et al. Lack of correlation between reaction speed and analytical sensitivity in isothermal amplification reveals the value of digital methods for optimization: validation using digital real-time RT-LAMP. Nucleic Acids Res . 2016;44:e10. doi: 10.1093/nar/gkv877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer JR, Mao C, Hansen CJ, et al. Crystal structure of a thermostable Bacillus DNA polymerase I large fragment at 2.1 A resolution. Structure . 1997;5:95–108. doi: 10.1016/s0969-2126(97)00169-x. [DOI] [PubMed] [Google Scholar]
- Kim EM, Jeon HS, Kim JJ, et al. Uracil-DNA glycosylase-treated reverse transcription loop-mediated isothermal amplification for rapid detection of avian influenza virus preventing carry-over contamination. J Vet Sci . 2016;17:421–425. doi: 10.4142/jvs.2016.17.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Kim HM, Lee EJ, et al. Detection and isolation of SARS-CoV-2 in serum, urine, and stool specimens of COVID-19 patients from the Republic of Korea. Osong Public Health Res Perspect . 2020;11:112–117. doi: 10.24171/j.phrp.2020.11.3.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Müller TG, Khalid D, et al. SARS-CoV-2 RNA extraction using magnetic beads for rapid large-scale testing by RT-qPCR and RT-LAMP. Viruses . 2020;12:863. doi: 10.3390/v12080863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota R, Jenkins DM. Real-time duplex applications of loop-mediated AMPlification (LAMP) by assimilating probes. Int J Mol Sci. 2015 Mar 3;16(3):4786–99. doi: 10.3390/ijms16034786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B, Le Bideau M, Andreani J, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis . 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBarre P, Hawkins KR, Gerlach J, et al. A simple, inexpensive device for nucleic acid amplification without electricity-toward instrument-free molecular diagnostics in low-resource settings. PloS One . 2011;6:e19738. doi: 10.1371/journal.pone.0019738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli MA, Langmade JS, Chen X, et al. Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric reverse-transcription loop-mediated isothermal amplification. Clin Chem . 2021;67:415–424. doi: 10.1093/clinchem/hvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb LE, Bartolone SN, Ward E, et al. Rapid detection of novel coronavirus/severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PloS One . 2020;15:e0234682. doi: 10.1371/journal.pone.0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land KJ, Boeras DI, Chen XS, et al. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat Microbiol . 2019;4:46–54. doi: 10.1038/s41564-018-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larremore D, Wilder B, Lester E, et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 surveillance. medRxiv . 2020. Posted online September 8. now published in Sci Adv. doi: [DOI] [PMC free article] [PubMed]
- Lau YL, Ismail I, Mustapa NI, et al. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of SARS-CoV-2. PeerJ . 2020;8:e9278. doi: 10.7717/peerj.9278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Helgouach N, Champigneux P, Schneider F, et al. EasyCOV: LAMP based rapid detection of SARS-CoV-2 in saliva. medRxiv . 2020. Posted online May 30. [DOI]
- Li E, Larson A, Kothari A, et al. Handyfuge-LAMP: low-cost and electricity-free centrifugation for isothermal SARS-CoV-2 detection in saliva. Posted online July 1, 2020. [DOI]
- Li J, Macdonald J, von Stetten F. Review: a comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst . 2018;144:31–67. doi: 10.1039/c8an01621f. [DOI] [PubMed] [Google Scholar]
- Li S, Tighe SW, Nicolet CM, et al. Multi-platform assessment of transcriptome profiling using RNA-Seq in the ABRF Next- Generation Sequencing Study. Nat Biotechnol . 2014;32:915–25. doi: 10.1038/nbt.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao SC, Peng J, Mauk MG, et al. Smart Cup: a minimally-instrumented, smartphone-based point-of-care molecular diagnostic device. Sens Actuators B Chem . 2016;229:232–238. doi: 10.1016/j.snb.2016.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Huang X, Urmann K, et al. Digital loop-mediated isothermal amplification on a commercial membrane. ACS Sens . 2019;4:242–249. doi: 10.1021/acssensors.8b01419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Begik O, Lucas MC, et al. Accurate detection of m6A RNA modifications in native RNA sequences. Nat Commun . 2019;10:4079. doi: 10.1038/s41467-019-11713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveday EK, Hain KS, Kochetkova I, et al. Effect of inactivation methods on SARS-CoV-2 virion protein and structure. Viruses . 2021;13:562. doi: 10.3390/v13040562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Su C, Hu S, et al. The effect of residual Triton X-100 on structural stability and infection activity of adenovirus particles. Mol Ther Methods Clin Dev . 2020;19:35–46. doi: 10.1016/j.omtm.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay MJ, Hooker AC, Afshinnekoo E, et al. The COVID-19 XPRIZE and the need for scalable, fast, and widespread testing. Nat Biotechnol . 2020;38:1021–1024. doi: 10.1038/s41587-020-0655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia Chagas A, Molloy JC, Prieto-Godino LL,et al. Leveraging open hardware to alleviate the burden of COVID-19 on global health systems. PLoS Biol. . 2020;18:e3000730. doi: 10.1371/journal.pbio.3000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardi A, Meidaninikjeh S, Nikfarjam S, et al. Interleukin-1 in COVID-19 Infection: Immunopathogenesis and Possible Therapeutic Perspective. Viral Immunol. 2021 Dec;34(10):679–688. doi: 10.1089/vim.2021.0071. [DOI] [PubMed] [Google Scholar]
- Maranhao A, Bhadra S, Paik I, et al. An improved and readily available version of Bst DNA Polymerase for LAMP, and applications to COVID-19 diagnostics. medRxiv . 2020. Posted online October 5. [DOI]
- Martineau AR, Jolliffe DA, Hooper RL, et al. Vitamin D supplementation to prevent acute respiratory tract infections: systematic review and meta-analysis of individual participant data. BMJ . j2017;356: i6583. [DOI] [PMC free article] [PubMed]
- Mason CE, Afshinnekoo E, Tighe S, et al. International standards for genomes, transcriptomes, and metagenomes. J Biomol Tech . 2017;28:8–18. doi: 10.7171/jbt.17-2801-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Botella JR. A simple, robust and equipment-free DNA amplification readout in less than 30 seconds. RSC Adv . 2019;9:24440–24450. doi: 10.1039/c9ra04725e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Botella JR. Rapid (30-second), equipment-free purification of nucleic acids using easy-to-make dipsticks. Nat Protoc. 2020 Nov;15(11):3663–3677. doi: 10.1038/s41596-020-0392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawaddah A, Gendeh HS, Lum SG, et al. Upper respiratory tract sampling in COVID-19. Malays J Pathol . 2020;42:23–35. [PubMed] [Google Scholar]
- McIntyre ABR, Alexander N, Grigorev K, et al. Single-molecule sequencing detection of N6-methyladenine in microbial reference materials. Nat Commun . 2019;10:579. doi: 10.1038/s41467-019-08289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre ABR, Rizzardi L, Yu AM, et al. Nanopore sequencing in microgravity.”. NPJ Microgravity . 2016;2:16035. doi: 10.1038/npjmgrav.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan JN, Shroff R, Garry DJ, et al. Evolution of a thermophilic strand-displacing polymerase using high-temperature isothermal compartmentalized self-replication. Biochemistry . 2018;57:4607–4619. doi: 10.1021/acs.biochem.8b00200. [DOI] [PubMed] [Google Scholar]
- Mina MJ, Parker R, Larremore DB. Rethinking Covid-19 test sensitivity —a strategy for containment. N Engl J Med . 2020;383:e120. doi: 10.1056/NEJMp2025631. [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Sano S, Takahashi K, et al. Method for colorimetric detection of double-stranded nucleic acid using leuco triphenylmethane dyes. Anal Biochem . 2015;473:28–33. doi: 10.1016/j.ab.2014.12.016. [DOI] [PubMed] [Google Scholar]
- Moehling TJ, Choi G, Dugan LC, et al. LAMP diagnostics at the point-of-care: emerging trends and perspectives for the developer community. Expert Rev Mol Diagn . 2021;21:43–61. doi: 10.1080/14737159.2021.1873769. [DOI] [PubMed] [Google Scholar]
- Mohon AN, Getie S, Jahan N, et al. Ultrasensitive loop mediated isothermal amplification (US-LAMP) to detect malaria for elimination. Malar J . 2019;18:350. doi: 10.1186/s12936-019-2979-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morán P, Galindo J. A practical home-made microcentrifuge for teaching purposes. Biochem Mol Biol Educ. 2011 Jul;39(4):298–9. doi: 10.1002/bmb.20504. [DOI] [PubMed] [Google Scholar]
- Mori Y, Nagamine K, Tomita N, et al. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun . 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- Motohashi K, Simple A. and Fast Manual Centrifuge to Spin Solutions in 96-Well PCR Plates. Methods Protoc. 2020 May 25;3(2):41. doi: 10.3390/mps3020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulberry G, White KA, Vaidya M, et al. 3D printing and milling a real-time PCR device for infectious disease diagnostics. PloS One . 2017;12:e0179133. doi: 10.1371/journal.pone.0179133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes . 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Nagura-Ikeda M, Imai K, Tabata S, et al. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J Clin Microbiol . 2020;58:e01438–20. doi: 10.1128/JCM.01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangle SN, Wolfson MY, Hartsough L, et al. The case for biotechnology on Mars. Nat Biotechnol . 2020;38:401–407. doi: 10.1038/s41587-020-0485-4. [DOI] [PubMed] [Google Scholar]
- Natoli M, Kundrod K, Chang M, et al. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) for point-of-care detection of SARS-CoV-2: a clinical study to evaluate agreement with RT-qPCR. Lancet Glob Health . 2021;9:S3. [Google Scholar]
- Nawattanapaiboon K, Pasomsub E, Prombun P, et al. Colorimetric reverse transcription loop-mediated isothermal amplification (RT-LAMP) as a visual diagnostic platform for the detection of the emerging coronavirus SARS-CoV-2. Analyst . 2021;146:471–477. doi: 10.1039/d0an01775b. [DOI] [PubMed] [Google Scholar]
- Newman C, Ramuta M, McLaughlin M, et al. Initial evaluation of a mobile SARS-CoV-2 RT-LAMP testing strategy. medRxiv . 2021. Posted online February 27. [DOI] [PMC free article] [PubMed]
- Ng DL, Granados AC, Santos YA, et al. A diagnostic host response biosignature for COVID-19 from RNA profiling of nasal swabs and blood. Sci Adv . 2021. 7:eabe5984. [DOI] [PMC free article] [PubMed]
- Nguyen HV, Nguyen VD, Liu F, et al. An integrated smartphone-based genetic analyzer for qualitative and quantitative pathogen detection. ACS Omega . 2020;5:22208–22214. doi: 10.1021/acsomega.0c02317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemz A, Ferguson TM, Boyle DS. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol . 2011;29:240–250. doi: 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res . 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa EM, Mason CE, Mattick JS. Charting the unknown human epitranscriptome. Nat Rev Mol Cell Biol . 2017;18:339–340. doi: 10.1038/nrm.2017.49. [DOI] [PubMed] [Google Scholar]
- Oliveira K, Estrela P, Mendes G, et al. Rapid molecular diagnostics of COVID-19 by RT-LAMP in a centrifugal polystyrene-toner based microdevice with end-point visual detection. Analyst . 2021;146:1178–1187. doi: 10.1039/d0an02066d. [DOI] [PubMed] [Google Scholar]
- Oscorbin IP, Belousova EA, Zakabunin AI, et al. Comparison of fluorescent intercalating dyes for quantitative loop-mediated isothermal amplification (qLAMP) Biotechniques . 2016;61:20–25. doi: 10.2144/000114432. [DOI] [PubMed] [Google Scholar]
- Pack TD, Deng X. inventors; Meridian Bioscience Inc, assignee. Stable reagents and kits useful in loop-mediated isothermal amplification (LAMP). US patent application 20,080,182,312 A1. January 16, Status: abandoned. 2008. https://patents.google.com/patent/US20080182312A1/en .
- Pang B, Xu J, Liu Y, et al. Isothermal Amplification and Ambient Visualization in a Single Tube for the Detection of SARS-CoV-2 Using Loop-Mediated Amplification. Technology CRISPR, editor. Anal Chem. 2020 Dec 15;92(24):16204–16212. doi: 10.1021/acs.analchem.0c04047. [DOI] [PubMed] [Google Scholar]
- Papadakis G, Pantazis A, Fikas N, et al. Real-time colorimetric LAMP methodology for quantitative nucleic acids detection at the point-of-care. bioRxiv . 2020. Posted online July 24,l. [DOI]
- Park GS, Ku K, Baek SH, Kim SJ, Kim SI, Kim BT, Maeng JS. Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) J Mol Diagn. 2020 Jun;22(6):729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CW, Singh N, Tighe S, et al. End-to-end protocol for the detection of SARS-CoV-2 from built environments. mSystems . 2020;5:e00771–20. doi: 10.1128/mSystems.00771-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino B, Touret F, Gilles M, et al. Evaluation of chemical protocols for inactivating SARS-CoV-2 infectious samples. Viruses . 2020;12:624. doi: 10.3390/v12060624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K, Jha A, Patwardhan A Design and Fabrication of Low Cost Vortex Mixer Using Additive Manufacturing. International Journal of Research in Advent Technology, Vol.6, No.12, 6. 3513-3516. (2018).
- Pearce Joshua M. “Sponsored Libre Research Agreements to Create Free and Open Source Software and Hardware”. Inventions . 2018. 3, no. 3: 44. [DOI]
- Peeling RW, Boeras D, Wilder-Smith A, et al. Need for sustainable biobanking networks for COVID-19 and other diseases of epidemic potential. Lancet Infect Dis. Oct; (2020);20(10):e268–e273. doi: 10.1016/S1473-3099(20)30461-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Liu J, Xu W, et al. SARS-CoV-2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J Med Virol . 2020;92:1676–1680. doi: 10.1002/jmv.25936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham HM, Konnai S, Usui T, Chang KS, Murata S, Mase M, Ohashi K, Onuma M. Rapid detection and differentiation of Newcastle disease virus by real-time PCR with melting-curve analysis. Arch Virol. 2005 Dec;150(12):2429–38. doi: 10.1007/s00705-005-0603-0. [DOI] [PubMed] [Google Scholar]
- Phang SM, Teo CY, Lo E, et al. Cloning and complete sequence of the DNA polymerase-encoding gene (BstpolI) and characterisation of the Klenow-like fragment from Bacillus stearothermophilus. Gene . 1995;163:65–68. doi: 10.1016/0378-1119(95)00387-l. [DOI] [PubMed] [Google Scholar]
- Piepenburg O, Williams CH, Stemple DL, et al. DNA detection using recombination proteins. PLoS Biol . 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilcher CD, Westreich D, Hudgens MG. Group testing for severe acute respiratory syndrome –coronavirus 2 to enable rapid scale-up of testing and real-time surveillance of incidence. J Infect Dis . 2020;222:903–909. doi: 10.1093/infdis/jiaa378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipes L, Li S, Bozinoski M, et al. The non-human primate reference transcriptome resource (NHPRTR) for comparative functional genomics. Nucleic Acids Res . 2013. ;41 :D906–D914. [DOI] [PMC free article] [PubMed]
- Priye A, Bird SW, Light YK, et al. A smartphone-based diagnostic platform for rapid detection of Zika, chikungunya, and dengue viruses. Sci Rep . 2017;7:44778. doi: 10.1038/srep44778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptasinska A, Whalley C, Bosworth A, et al. Diagnostic accuracy of Loop mediated isothermal amplification coupled to nanopore sequencing (LamPORE) for the detection of SARS-CoV-2 infection at scale in symptomatic and asymptomatic populations. medRxiv . 2020. Posted online December 16. now published in Clin Microbiol Infect. doi: [DOI] [PMC free article] [PubMed]
- Qin Z, Peng R, Baravik IK, et al. Fighting COVID-19: integrated micro- and nanosystems for viral infection diagnostics. Matter . 2020;3:628–651. doi: 10.1016/j.matt.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quyen TL, Nordentoft S, Vinayaka AC, et al. A sensitive, specific and simple loop mediated isothermal amplification method for rapid detection of Campylobacter spp. in broiler production. Front Microbiol . 2019;10:2443. doi: 10.3389/fmicb.2019.02443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabe BA, Cepko C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc Natl Acad Sci USA . 2020;117:24450–24458. doi: 10.1073/pnas.2011221117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran A, Huyke DA, Sharma E, Sahoo MK, Huang C, Banaei N, Pinsky BA, Santiago JG. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc Natl Acad Sci USA. 2020 Nov 24;117(47):29518–29525. doi: 10.1073/pnas.2010254117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrill-Mix S, Hwang Y, Roche AM, Glascock A, Weiss SR, Li Y, Haddad L, Deraska P, Monahan C, Kromer A, Graham-Wooten J, Taylor LJ, Abella BS, Ganguly A, Collman RG, Van Duyne GD, Bushman FD. Detection of SARS-CoV-2 RNA using RT-LAMP and molecular beacons. Genome Biol. 2021 Jun 3;22(1):169. doi: 10.1186/s13059-021-02387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranoa D, Holland R, Alnaji F, et al. Saliva-based molecular testing for SARS-CoV-2 that bypasses RNA extraction. bioRxiv . 2020. Posted online June 18. [DOI]
- Reboud J, Xu G, Garrett A, et al. Paper-based microfluidics for DNA diagnostics of malaria in low resource underserved rural communities. Proc Natl Acad Sci USA . 2019;116:4834–4842. doi: 10.1073/pnas.1812296116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendeiro AF, Ravichandran H, Bram Y, et al. The spatial landscape of lung pathology during COVID-19 progression. Nature . 2021;593:564–569. doi: 10.1038/s41586-021-03475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rei CK, Li C, Bertrand D, et al. Cartography of opportunistic pathogens and antibiotic resistance genes in a tertiary hospital environment. Nat Med . 2020;26:941–951. doi: 10.1038/s41591-020-0894-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Manzano J, Malpartida-Cardenas K, Moser N, et al. Handheld point-of-care system for rapid detection of SARS-CoV-2 extracted RNA in under 20 min. ACS Cent Sci . 2021;7:307–317. doi: 10.1021/acscentsci.0c01288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohaim MA, Clayton E, Sahin I, et al. Artificial intelligence-assisted loop mediated isothermal amplification (AI-LAMP) for rapid detection of SARS-CoV-2. Viruses . 2020;12:972. doi: 10.3390/v12090972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolando JC, Jue E, Barlow JT, et al. Real-time kinetics and high-resolution melt curves in single-molecule digital LAMP to differentiate and study specific and non-specific amplification. Nucleic Acids Res . 2020;48:e42. doi: 10.1093/nar/gkaa099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolando JC, Jue E, Schoepp NG, et al. Real-time, digital LAMP with commercial microfluidic chips reveals the interplay of efficiency, speed, and background amplification as a function of reaction temperature and time. Anal Chem . 2019;91:1034–1042. doi: 10.1021/acs.analchem.8b04324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld J, Mason CE, Smith T. Limitations of the human genome reference. PLoS One . 2012;7:e40294. doi: 10.1371/journal.pone.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Mohd-Naim NF, Safavieh M, et al. Colorimetric nucleic acid detection on paper microchip using loop mediated isothermal amplification and crystal violet dye. ACS Sens . 2017;2:1713–1720. doi: 10.1021/acssensors.7b00671. [DOI] [PubMed] [Google Scholar]
- Safavieh M, Kaul V, Khetani S, et al. Paper microchip with a graphene-modified silver nano-composite electrode for electrical sensing of microbial pathogens. Nanoscale . 2017;9:1852–1861. doi: 10.1039/c6nr06417e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saletore Y, Meyer K, Korlach J, et al. The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biol . 2012;13:175. doi: 10.1186/gb-2012-13-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapoval N, Mahmoud M, Jochum MD, et al. Hidden genomic diversity of SARS-CoV-2: implications for qRT-PCR diagnostics and transmission. bioRxiv . 2020. Posted online July 2. [DOI] [PMC free article] [PubMed]
- Schellenberg JJ, Ormond M, Keynan Y. Extraction-free RT-LAMP to detect SARS-CoV-2 is less sensitive but highly specific compared to standard RT-PCR in 101 samples. J Clin Virol . 2021;136:104764. doi: 10.1016/j.jcv.2021.104764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk J, Schmithausen R, Li D, et al. LAMp-seq: population-scale COVID-19 diagnostics using combinatorial barcoding. bioRxiv . 2020. Posted online June 8. [DOI]
- Schneider F, Maurer C, Friedberg RC. International Organization for Standardization (ISO) 15189. Ann Lab Med. 2017 Sep;37(5):365–370. doi: 10.3343/alm.2017.37.5.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönbrunner NJ, Fiss EH, Budker O, et al. Chimeric thermostable DNA polymerases with reverse transcriptase and attenuated 3′-5′ exonuclease activity. Biochemistry . 2006;45:12786–12795. doi: 10.1021/bi0609117. [DOI] [PubMed] [Google Scholar]
- Schuler F, Siber C, Hin S, et al. Digital droplet LAMP as a microfluidic app on standard laboratory devices. Anal Methods . 2016;8:2750–2755. [Google Scholar]
- Seok Y, Joung HA, Byun JY, et al. A paper-based device for performing loop-mediated isothermal amplification with real-time simultaneous detection of multiple DNA targets. Theranostics . 2017;7:2220–2230. doi: 10.7150/thno.18675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [SEQC/MAQC-III Consortium] Sequencing Quality Control/MicroArray Quality Control Consortium. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequence Quality Control Consortium. Nat Biotechnol . 2014;32:903–14. doi: 10.1038/nbt.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyrig G, Stedtfeld RD, Tourlousse DM, et al. Selection of fluorescent DNA dyes for real-time LAMP with portable and simple optics. J Microbiol Methods . 2015;119:223–227. doi: 10.1016/j.mimet.2015.11.004. [DOI] [PubMed] [Google Scholar]
- Sherrill-Mix S, Duyne G, Bushman F. Molecular beacons allow specific RT-LAMP detection of B.1.1.7 variant SARS-CoV-2. medRxiv . 2021. Posted online March 26. [DOI] [PMC free article] [PubMed]
- Sherrill-Mix S, Hwang Y, Roche A, et al. LAMP-BEAC: detection of SARS-CoV-2 RNA Using RT-LAMP and Molecular Beacons. medRxiv . 2020. Posted online November 25. now published in Genome Biol. doi: [DOI] [PMC free article] [PubMed]
- Shirato K, Yano T, Senba S, et al. Detection of Middle East respiratory syndrome coronavirus using reverse transcription loop-mediated isothermal amplification (RT-LAMP) Virol J . 2014;11:139. doi: 10.1186/1743-422X-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirshikov FV, Pekov YA, Miroshnikov KA. MorphoCatcher: a multiple-alignment based web tool for target selection and designing taxon-specific primers in the loop-mediated isothermal amplification method. PeerJ. 2019 Apr 26;7:e6801. doi: 10.7717/peerj.6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AAMD, Lima-Neto LG, Azevedo CMPES, et al. Population-based seroprevalence of SARS-CoV-2 and the herd immunity threshold in Maranhão. Rev Saude Publica . 2020;54:131. doi: 10.11606/s1518-8787.2020054003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton J, Osborn JL, Lillis L, et al. Electricity-free amplification and detection for molecular point-of-care diagnosis of HIV-1. PloS One . 2014;9:e113693. doi: 10.1371/journal.pone.0113693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snodgrass R, Gardner A, Jiang L. KS-Detect —validation of solar thermal PCR for the diagnosis of Kaposi's sarcoma using pseudo-biopsy samples. PloS One . 2016;11:e0147636. doi: 10.1371/journal.pone.0147636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snodgrass R, Gardner A, Semeere A, et al. A portable device for nucleic acid quantification powered by sunlight, a flame or electricity. Nat Biomed Eng . 2018;2:657–665. doi: 10.1038/s41551-018-0286-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares R, Akhtar A, Pinto I, et al. Point-of-care detection of SARS-CoV-2 in nasopharyngeal swab samples using an integrated smartphone-based centrifugal microfluidic platform. medRxiv . 2020. Posted online November 5. 10.1101/2020.11.04.20225888 now published in Lab Chip doi: 10.1039/D1LC00266J.
- Song J, Mauk MG, Hackett BA, et al. Instrument-free point-of-care molecular detection of Zika virus. Anal Chem . 2016;88:7289–7294. doi: 10.1021/acs.analchem.6b01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedtfeld RD, Tourlousse DM, Seyrig G, et al. Gene-Z: a device for point of care genetic testing using a smartphone. Lab Chip . 2012;12:1454–1462. doi: 10.1039/c2lc21226a. [DOI] [PubMed] [Google Scholar]
- Sun F, Ganguli A, Nguyen J, et al. Smartphone-based multiplex 30-minute nucleic acid test of live virus from nasal swab extract. Lab Chip . 2020;20:1621–1627. doi: 10.1039/d0lc00304b. [DOI] [PubMed] [Google Scholar]
- Sungnak W, Huang N, Bécavin C, et al. HCA Lung Biological Network. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. (2020);26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama I, Semba S, Yokono K, et al. Clinical evaluation of fully automated molecular diagnostic system “Simprova” for influenza virus, respiratory syncytial virus, and human metapneumovirus. Sci Rep . 2020;10:13496. doi: 10.1038/s41598-020-70090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner NA, Zhang Y, Evans TC., Jr Simultaneous multiple target detection in real-time loop-mediated isothermal amplification. Biotechniques . 2012;53:81–89. doi: 10.2144/0000113902. [DOI] [PubMed] [Google Scholar]
- Teytelman L, Stoliartchouk A, Kindler L, et al. Protocols.io: Virtual Communities for Protocol Development and Discussion. PLoS Biol. Aug 22; (2016);14(8):e1002538. doi: 10.1371/journal.pbio.1002538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe S, Afshinnekoo E, Green SJ, et al. Genomic methods and microbiological technologies for profiling novel and extreme environments for the Extreme Microbiome Project (XMP). J Biomol Tech . 2017;28:31–39. doi: 10.7171/jbt.17-2801-004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KK, Tsang OT, Yip CC, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis . 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3(5):877–82. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Torres C, Vitalis EA, Baker BR, et al. LAVA: an open-source approach to designing LAMP (loop-mediated isothermal amplification) DNA signatures. BMC Bioinformatics . 2011;12:240. doi: 10.1186/1471-2105-12-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran D, Cuong H, Tran H, et al. A comparative study of isothermal nucleic acid amplification methods for SARS-CoV-2 detection at point-of-care. bioRxiv . 2021. Posted online February 5. [DOI]
- Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol . 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- Urdea M, Penny LA, Olmsted SS, et al. Requirements for high impact diagnostics in the developing world. Nature . 2006;444(suppl 1):73–79. doi: 10.1038/nature05448. [DOI] [PubMed] [Google Scholar]
- van Dongen JE, Berendsen J, Steenbergen R, et al. Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities. Biosens B ioelectron . 2020;166:112445. doi: 10.1016/j.bios.2020.112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varsha V, Aishwarya S, Murchana S, et al. Correction pen based paper fluidic device for the detection of multiple gene targets of Leptospira using loop mediated isothermal amplification. J Microbiol Methods . 2020;174:105962. doi: 10.1016/j.mimet.2020.105962. [DOI] [PubMed] [Google Scholar]
- Velders AH, Schoen C, Saggiomo V. Loop-mediated isothermal amplification (LAMP) shield for Arduino DNA detection. BMC Res N otes . 2018;11:93. doi: 10.1186/s13104-018-3197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels CBF, Brito AF, Wyllie AL, Fauver JR, et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat Microbiol. 2020 Oct;5(10):1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels CBF, Breban MI, Ott IM, et al. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol . 2021a;19:e3001236. doi: 10.1371/journal.pbio.3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels CBF, Watkins AE, Harden CA. SalivaDirect: a simplified and flexible platform to enhance SARS-CoV-2 testing capacity. Med (NY) . 2021b;2:263–280.e6. doi: 10.1016/j.medj.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhong M, Liu Y, Ma P, Dang L, Meng Q, Wan W, Ma X, Liu J, Yang G, Yang Z, Huang X, Liu M. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout. [DOI] [PMC free article] [PubMed]
- Wei S, Kohl E, Djandji A, et al. Direct diagnostic testing of SARS-CoV-2 without the need for prior RNA extraction. medRxiv . 2020. Posted online June 20. now published in Sci Rep. doi: [DOI] [PMC free article] [PubMed]
- Westhaus S, Widera M, Rabenau H, et al. Evaluation of stability and inactivation methods of SARS-CoV-2 in context of laboratory settings. bioRxiv . 2020. Posted online December 10. now published in Med Microbiol Immunol (with first author Widera). [DOI] [PMC free article] [PubMed]
- Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature . 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature . 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang J, Zhong Z, et al. Room-temperature-storable PCR mixes for SARS-CoV-2 detection. Clin Biochem . 2020;84:73–78. doi: 10.1016/j.clinbiochem.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager P, Domingo GJ, Gerdes J. Point-of-care diagnostics for global health. Annu Rev Biomed Eng . 2008;10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- Yan C, Cui J, Huang L, et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect . 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Meyerson N, Clark S, et al. Saliva TwoStep for rapid detection of asymptomatic SARS-CoV-2 carriers. medRxiv . 2021. Posted online February 16. [DOI] [PMC free article] [PubMed]
- Yaren O, McCarter J, Phadke N, et al. Ultra-rapid detection of SARS-CoV-2 in public workspace environments. PloS One . 2021;16:e0240524. doi: 10.1371/journal.pone.0240524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa K, Yanagihara I, Fujiwara S. Alteration of enzymes and their application to nucleic acid amplification (review) Int J Mol Med . 2020;46:1633–1643. doi: 10.3892/ijmm.2020.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelagandula R, Bykov A, Vogt A, et al. SARSeq, a robust and highly multiplexed NGS assay for parallel detection of SARS-CoV2 and other respiratory infections. medRxiv . 2020. Posted online November 3. [DOI]
- Yoo W, Han H, Kim J, et al. Development of a tablet PC-based portable device for colorimetric determination of assays including COVID-19 and other pathogenic microorganisms. RSC Adv . 2020;10:32946–32952. doi: 10.1039/d0ra05866a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa R, Abe H, Igasaki Y, et al. Development and evaluation of a rapid and simple diagnostic assay for COVID-19 based on loop-mediated isothermal amplification. PLoS Negl Trop Dis. Nov 4; (2020);14(11):e0008855. doi: 10.1371/journal.pntd.0008855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Chan KG, Yean CY, et al. Nucleic acid-based diagnostic tests for the detection SARS-CoV-2: an update. Diagnostics (Basel) . 2021;11:53. doi: 10.3390/diagnostics11010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Wu S, Hao X, et al. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin Chem . 2020;66:975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Chao Y. Shum HC. droplet and microchamber-based digital loop-mediated isothermal amplification (dLAMP) Small . 2020;16:e1904469. doi: 10.1002/smll.201904469. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zheng T, Wang H, et al. Rapid one-pot detection of SARS-CoV-2 based on a lateral flow assay in clinical samples. Anal Chem . 2021;93:3325–3330. doi: 10.1021/acs.analchem.0c05059. [DOI] [PubMed] [Google Scholar]
- Zhang H, Xu Y, Fohlerova Z, et al. LAMP-on-a-chip: revising microfluidic platforms for loop-mediated DNA amplification. Trends Analyt Chem . 2019;113:44–53. doi: 10.1016/j.trac.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Tanner N. Development of multiplexed reverse-transcription loop-mediated isothermal amplification for detection of SARS-CoV-2 and influenza viral RNA. BioTechniques. (2021);70(3) doi: 10.2144/btn-2020-0157. Vol. No. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Liu YN, Chen LL, et al. One simple DNA extraction device and its combination with modified visual loop mediated isothermal amplification for rapid on field detection of genetically modified organisms. Anal. Chem . (2013). 85 75–82. [DOI] [PubMed]
- Zhang XO, Wang HB, Zhang Y. Complementary sequence-mediated exon circularization. Cell . 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Odiwuor N, Xiong J, et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv . 2020. Posted online February 29. a. doi: [DOI]
- Zhang Y, Ren G, Buss J, et al. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. Biotechniques . 2020b;69:178–185. doi: 10.2144/btn-2020-0078. [DOI] [PubMed] [Google Scholar]
- Zhu J, Guo J, Xu Y, et al. Viral dynamics of SARS-CoV-2 in saliva from infected patients. J Infect . 2020;81:e48–e50. doi: 10.1016/j.jinf.2020.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.