Abstract
The COVID-19 pandemic has had a profound, detrimental effect on economies and societies worldwide. Where the pandemic has been controlled, extremely high rates of diagnostic testing for the SARS-CoV-2 virus have proven critical, enabling isolation of cases and contact tracing. Recently, diagnostic testing has been supplemented with wastewater measures to evaluate the degree to which communities have infections. Whereas much testing has been done through traditional, centralized, clinical, or environmental laboratory methods, point-of-care testing has proven successful in reducing time to result. As the pandemic progresses and becomes more broadly distributed, further decentralization of diagnostic testing will be helpful to mitigate its spread. This will be particularly both challenging and critical in settings with limited resources due to lack of medical infrastructure and expertise as well as requirements to return results quickly. In this article, we validate the tiny isothermal nucleic acid quantification system (TINY) and a novel loop-mediated isothermal amplification (LAMP)–based assay for the point-of-care diagnosis of SARS-CoV-2 infection in humans and also for in-the-field, point-of-collection surveillance of wastewater. The TINY system is portable and designed for use in settings with limited resources. It can be powered by electrical, solar, or thermal energy and is robust against interruptions in services. These applied testing examples demonstrate that this novel detection platform is a simpler procedure than reverse-transcription quantitative polymerase chain reaction, and moreover, this TINY instrument and LAMP assay combination has the potential to effectively provide both point-of-care diagnosis of individuals and point-of-collection environmental surveillance using wastewater.
INTRODUCTION
Following the identification of the SARS-CoV-2 virus in December 2019, this infectious pathogen of COVID-19 disease has spread to more than 200 countries, infecting millions of people and resulting in hundreds of thousands of confirmed deaths.1 In February 2020, COVID-19 was declared a public health emergency of international concern, and it was declared a pandemic on March 11, 2020. SARS-CoV-2 is the seventh coronavirus to be detected in humans, and the third to cause acute respiratory distress syndrome.2,3 Despite the apparent lower mortality rate of COVID-19 compared with previous outbreaks,4–6 its greater reproductive number (Ro) has led to significantly more detrimental economic disruption.7 To reduce the spread of the disease, many countries have issued severe restrictions on social, educational, and economic activity, and these restrictions are likely to have increasingly significant long-term consequences the longer they need to be in place.8–10
In recent years, point-of-care (PoC) diagnostics have proven to be useful in enabling diagnosis of infectious diseases outside of traditional clinical laboratory settings. The main advantage of PoC over regular tests is that they provide more rapid results (on a time scale consistent with the patient visit), require less operator training, and can often be operated in nontraditional remote settings.11–15 Since the beginning of the COVID-19 pandemic, the development of PoC technologies for infection confirmation has been identified as a priority.16 Several policies have been put in place to help commercial manufacturers and laboratories accelerate the development of more rapid and widespread testing. As of June 2020, the US Food and Drug Administration (FDA) has granted approval for emergency use to a number of tests, of which only a small percentage are classified as PoC.17
In addition to clinical diagnostics, measurements of wastewater provide estimates of COVID-19 infection rates from communities contributing towards the wastewater source.18–20 Compared with clinical diagnostics, wastewater surveillance provides information about the entire community with fewer analyses and lower costs. Moreover, the levels of SARS-CoV-2 in wastewater are indicative of infection rates among both symptomatic and asymptomatic populations.21 Going forward, a major challenge with COVID-19 diagnostics is the need to provide enabling technologies where traditional infrastructure may be limited, unreliable, or nonexistent.
There are many factors that underlie this critical need. A primary factor that has been reported22–24 is that there are a significant number of asymptomatic carriers of the virus. In the absence of symptoms, these carriers are unlikely to report for testing and thus could be spreaders without their knowledge. Another factor is that in countries (or regions of countries) with limited access to advanced large-scale, clinical-laboratory–based testing infrastructure, cases are potentially underreported and the spread of the virus is likely broader than has been reported to date.22 In that case, there could be a very large number of carriers, symptomatic or otherwise, who are lacking an appropriate diagnosis and remain within the population, continuing to spread the disease. In situations where human populations are not diagnosed as fully as needed, wastewater-based testing may prove to be effective as both an assessment and predictive tool. In this article, we demonstrate the use of the tiny isothermal nucleic acid quantification system (TINY) device, in conjunction with the New England Biolabs (NEB) loop-mediated isothermal amplification (LAMP) assay, as a viable solution for SARS-CoV-2 PoC screening and for wastewater measurements.
METHODS AND MATERIALS
The TINY System
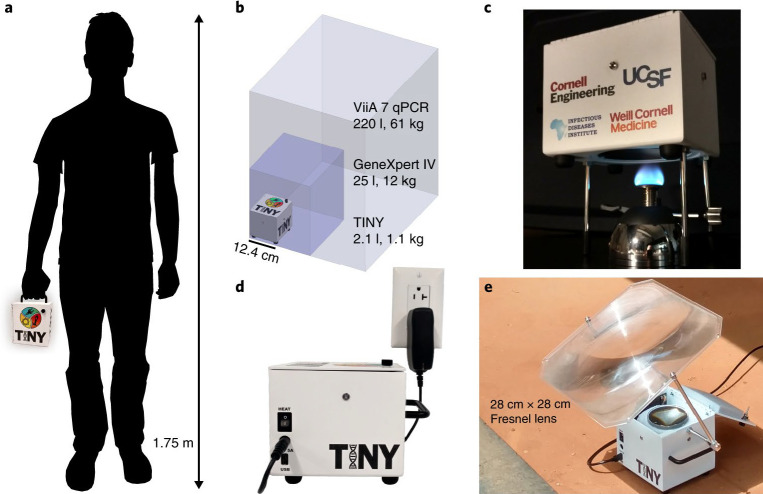
The TINY system (Fig. 1) is a field-portable, PoC, rugged diagnostic tool for nucleic acid-based diagnostics. LAMP nucleic acid detection assays can be used in conjunction with TINY to detect targets in approximately 30 minutes using a constant temperature of 65°C, enabling field testing of wastewater where infrastructure is limited or unreliable for COVID-19 testing.25 Details of the system are available in our recent article.26 As previously demonstrated, a key innovation of the system is the incorporation of phase-change materials, ultimately enabling a much more uniform temperature profile within the system (ensuring equivalent amplification) and robustness against power interruptions, enabling complete tests in the event of unreliable infrastructure. The TINY system's temperature-regulation unit is responsible for heat collection and isothermal stabilization, whereas its measurement unit is responsible for tracking the progress of the nucleic acid amplification through colorimetric, fluorescence, or absorbance means. Extensive analysis of the performance of the TINY system has been performed under multiple operating conditions, in multiple locations, and by multiple user groups.26 This includes laboratory-based analysis in which we showed near identical sensitivities and specificities to quantitative polymerase chain reaction (qPCR), and field testing in which we show near equivalent results independent of the training status of the user, energy source, or location. That work also reported a time to result (for amplification) of approximately 30 minutes, a limit of detection around 20–30 copies per reaction, and roughly equivalent quantification capabilities to qPCR above that limit of detection.
FIGURE 1.
The tiny isothermal nucleic acid quantification system (TINY) is a portable, easy-to-use, point-of-care diagnostic system for nucleic acid diagnostics. TINY can be operated from a Bunsen burner, electricity, or solar energy. a, b, TINY is portable and easily carried in one hand (a), in contrast to other nucleic acid quantification systems such as the GeneXpert IV by Cepheid (footprint outlined by the dark purple box), or the ViiA 7 Real-Time PCR System by Thermo Fisher Scientific (light purple box; b). c, TINY heated by a Bunsen burner through an opening in the bottom of the system. d, TINY heated via electricity, using an integrated cartridge heater. e, TINY heated via concentrated sunlight. Figure taken from https://www.nature.com/articles/s41551-018-0286-y.
TINY-LAMP Combination
The detection of SARS-CoV-2 RNA using the SARS-CoV-2 Rapid Colorimetric LAMP assay kit (No. E2019S; NEB) with the TINY instrument (TINY-LAMP assay) was evaluated using known concentrations of synthetic SARS-CoV-2 RNA Control 2 (No. 102024; Twist). The Twist Synthetic SARS-CoV-2 RNA Control 2 provides coverage for approximately 99.9% of the viral genome, so serial dilutions of the Twist Control 2 allow for specific concentrations (copies/uL) of input to be assessed with TINY. As part of the National Institutes of Health–funded South Florida RADx-radical SARS-CoV-2 wastewater surveillance research project at the University of Miami, 3 wastewater samples that had been previously concentrated, extracted, and quantified by both reverse-transcription (RT)-qPCR and novel Volcano-2G PCR (V2G-PCR)21 were assessed with the TINY system in the Onco-Genomics Shared Resource (OGSR) of the Sylvester Comprehensive Cancer Center at the University of Miami. Replicated experiments were also performed at the OGSR to investigate the limit of detection of the TINY system, and preliminary results (data not shown) indicated that further investigation could be productive.
SalivaDirect
SalivaDirect is a protocol developed at Yale University through the lab of Anne Wiley and Nathan Grubaugh (https://publichealth.yale.edu/salivadirect/). SalivaDirect received an FDA emergency use authorization in August 2020. The protocol is hosted as an open-source document (https://www.protocols.io/view/salivadirect-rna-extraction-free-sars-cov-2-diagno-bkjgkujw). In brief, 0.5 mL of saliva is collected, combined with proteinase K, and vortexed. This mixture is heated for 5 minutes at 95°C and then taken into an RT-PCR reaction.
This method was tested on the TINY at Dr Wiley's lab at Yale. For the proof-of-principle experiment, positive saliva (24.6 cycle threshold [Ct]) was serially diluted into negative saliva. Given that 10× is approximately 3 Ct values, TINY lamp detected up to 33.6 Ct and did not detect the next closest sample at 36.6, which showed excellent sensitivity (Fig. 2).
FIGURE 2.
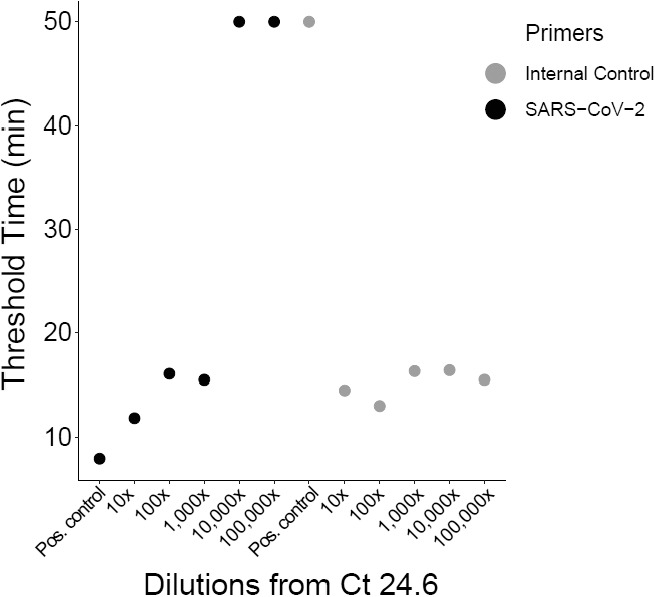
Limit of detection measured on the TINY device. Dilutions of a clinical sample with a cycle threshold value of 24.6 are plotted against the threshold of detection on the TINY device, measured as a change of raw fluorescence units (RFU) over time.
RESULTS
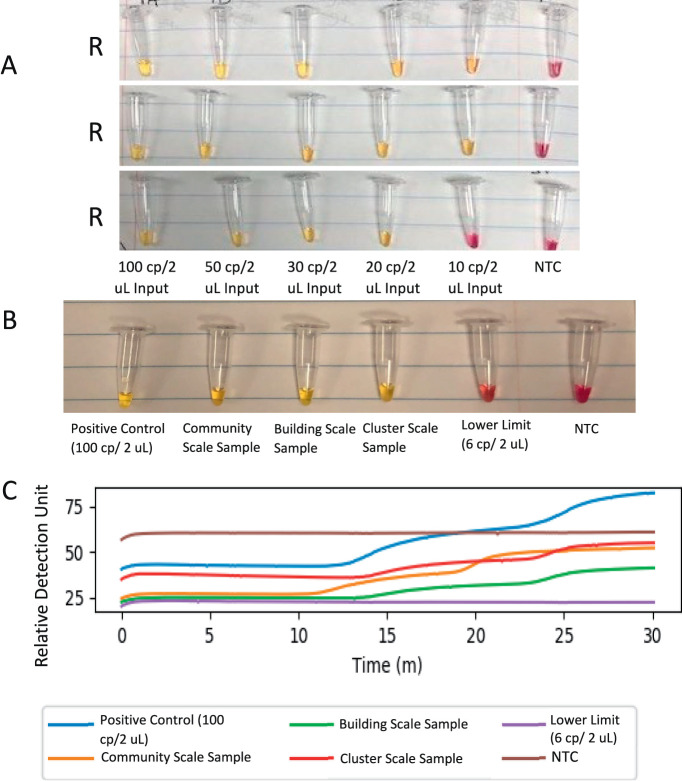
To investigate the dynamic range of TINY, a 1-μg/uL diluted stock of Human T-cell Leukemia (Jurkat) Total RNA (Cat No. AM7858; Ambion) was used in combination with serial dilutions of Twist Synthetic SARS-CoV-2 RNA Control 2 (diluted to 100, 50, 30, 20, and 10 copies/reaction) as input for a TINY-LAMP assay. Colorimetric change from red (negative) to yellow (positive) was seen in every positive replicate down to 10 copies/reaction, which only had 2 of 3 true positive colorimetric changes; no false positives were seen in any replicate of the no-template controls' (NTC's; Fig. 3A). The TINY system's on-board photodiodes recorded the same sensitivity and specificity for the dilution series during its blue LED excitation cycle. These findings are in line with previous estimates of TINY having a general limit of detection of approximately 20–30 copies/reaction for SARS-CoV-2 RNA, and the determined logarithm of odds of novel LAMP assays of 10–100 copies/reaction (25 uL).27–28
FIGURE 3.
TINY-LAMP colorimetric changes after exposure to SARS-CoV-2 template and known positive wastewater. (A) Colorimetric changes in LAMP assay of 3 replicates (R1, R2, and R3) of serial dilutions using Twist synthetic SARS-CoV-2 templates after incubation in TINY. (B) Colorimetric changes in LAMP assay of wastewater samples after incubation in TINY. (C) Relative change of photon detection when excited with blue LED in LAMP assay of wastewater samples during incubation in TINY. Assays performed at the Sylvester OGSR, University of Miami. Abbreviations: LAMP, loop-mediated isothermal amplification; OGSR, Onco-Genomics Shared Resource; TINY, tiny isothermal nucleic acid quantification system (Photographs by Kristina Babler).
A preliminary assessment of the utility of TINY was performed to determine the effectiveness of the system in a field setting for the rapid detection of SARS-COV-2 RNA, using quantified wastewater samples that had been previously evaluated using orthogonal methods (Fig. 3C). Three samples were assessed, each at a different level of wastewater collection, from individual building scale, cluster of buildings scale, and community scale. The samples for building and cluster analysis were collected February 9, 2021, and the sample for community scale analysis was collected February 23, 2021. Each sample assessed was quantified by both V2G-qPCR and RT-qPCR and calculated on a copy per liter basis prior to use in the TINY system. The calculated concentrations of each wastewater sample can be viewed in Table 1. The samples assessed were confirmed to have a positive presence of SARS-CoV-2 RNA by the PCR techniques performed prior to utilization in TINY. The assay investigating the wastewater samples used the 3 wastewater samples individually, a positive control (roughly 100 cp/2 uL), a lower limit of detection (roughly 6 cp/2 uL), and a NTC. The 3 wastewater samples were shown to be positive by their colorimetrically changing from red to yellow in the TINY-LAMP assay (Fig. 3B). These results are in agreement with the orthogonal methods, indicating the feasibility of the TINY-LAMP assay as a wastewater field test.
TABLE 1.
Concentration of 3 wastewater samples quantified by RT-qPCR, and V2G-PCR assaysa
|
Wastewater sample ID & date
|
Sample concentration (gc/L raw sewage) by V2G-PCR
|
Sample concentration (gc/L raw sewage “N” target) by RT-qPCR
|
| WG0P_2/9/21 | 510 975 (= 91 cp/2 uL) | 2 116 027 (= 376 cp/2 uL) |
| WG02_2/9/21 | 614 325 (= 109 cp/2 uL) | 2 400 171 (= 427 cp/2 uL) |
| WC0Dc_2/23/21 | 8640 (= 1.2 cp/2 uL) | 118 411 (= 16 cp/2 uL) |
Abbreviations: RT-qPCR, reverse-transcription quantitative polymerase chain reaction; V2G-PCR, Volcano 2G polymerase chain reaction.
WG0P correlates with building-level wastewater collection, WG02 correlates with cluster-level wastewater collection, and WC0Dc correlates with community-level wastewater collection.
Initial COVID-19 testing on the TINY device was performed using extracted total nucleic acids (TNA) from clinical nasopharyngeal (NP) swabs. Reliable amplification was seen in all 12 clinical positive samples, where Ct values were consistently under 10 minutes, with the earliest being 7.92 and the latest being 10.42 (Fig. 4). Nonamplification was seen in all 12 clinical negative samples. Following successful use of the TINY device for NP sample testing, saliva testing was attempted using the SalivaDirect protocol. SalivaDirect was first validated on a TINY device using a known clinical positive saliva sample (Ct of 24.6). The sample was serially diluted into negative saliva and amplified for 50 minutes. Given that a 10× dilution corresponds to approximately 3 Ct values, amplification reported at 1000× corresponds to a Ct value of 33.6, whereas the next sample at 36.6 did not amplify.
FIGURE 4.
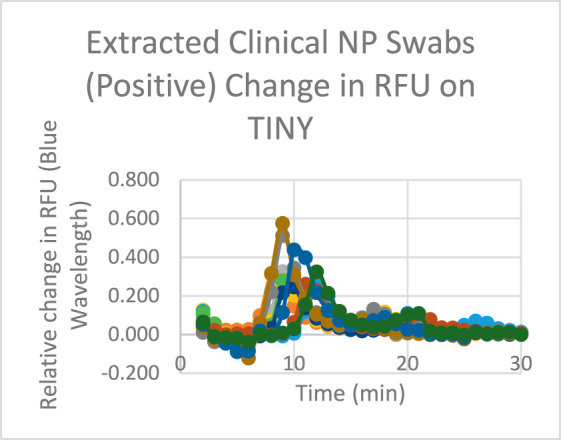
Positive NP swab samples. Relative change in raw fluorescence units (RFU) is plotted against the total time the samples are run on the TINY device. Note the change at around 10 minutes indicating the exponential amplification of the LAMP reaction. Abbreviations: LAMP, loop-mediated isothermal amplification; TINY, tiny isothermal nucleic acid quantification system
With successful use of the SalivaDirect protocol on serially diluted samples, further testing was performed on clinical saliva samples. Positive samples with known qPCR Ct values were tested in triplicate on a TINY device (Fig. 5). Reliable amplification of 3 replicates was seen in 5 of 12 samples, all of which were on the lower end of Ct values tested. Amplification was seen in fewer than 3 replicates in an additional 4 samples. Only 3 samples had no amplification in any replicates. Clinical negative saliva samples were also tested in triplicate with TINY and had no amplification.
FIGURE 5.
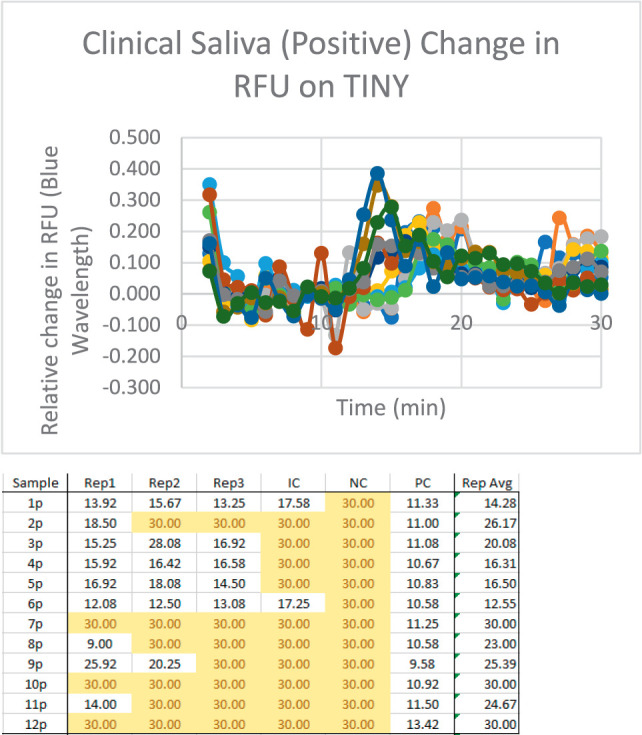
Saliva testing on site for LAMP (a) amplification curves for saliva samples tested at Mirimus (https://www.salivaclear.com). There is a similar jump to photodiode, but this is noisier with saliva. (b) Cycle threshold (Ct) values for each replicate of saliva samples tested at Mirimus. Note many samples are at or above a Ct value of 30, indicating a small amount of virus within the sample. Abbreviation: LAMP, loop-mediated isothermal amplification.
CONCLUSIONS
These data showed that the TINY device worked well with extracted total nucleic acids and is simpler and cheaper than RT-qPCR, which indicates that such tools can help complement testing for organisms at disparate locations, such as across a city,29 and they can also serve to expand the range of other testing methods.30 Of note, the low-Ct, positive clinical samples consistently exhibited detection values under 10 minutes, whereas negative samples showed no amplification during the 30-minute assay. SalivaDirect experiments at Yale showed excellent sensitivity with a Ct value as high as 33.6. However, clinical saliva samples produced less-reliable results, especially at higher Ct values (>30). Clinical saliva samples with lower qPCR-confirmed Ct values exhibited mostly reliable amplification in TINY, whereas higher Ct samples showed unreliable or no amplification. Modifications to the saliva testing protocol to ensure complete removal of contaminants would increase complexity and cost, but likely would provide much more sensitivity in higher-CT, lower-copy number samples.
The TINY system was also found to work well with the NEB LAMP kit for detecting the presence of SARS-CoV-2 RNA. The limits of detection with TINY are still in the process of being determined, including the lower limits of detection. Furthermore, using TINY for in-the-field application of point-of-collection for wastewater analysis is still in the process of being optimized, but known positive wastewater samples (quantified by established, orthogonal PCR assays) were found to be colorimetrically positive with TINY, though more replicate assays need to be run to evaluate robustness and to validate this application. Furthermore, it should be noted that although the proof-of-principle results presented here are promising, the incorporation of TINY in a field setting for detecting the presence of SARS-CoV-2 RNA in wastewater will also require testing and validation of field-compatible methods for sample concentration and nucleic acid extraction. Nonetheless, these results show the utility of these methods for rapid, portable, and easy detection of viruses in a range of settings.
Acknowledgments
Support for this study was received from the University of Miami and the National Institute On Drug Abuse of the National Institutes of Health under award no. U01DA053941. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was performed in part at the Cornell NanoScale Facility, a member of the National Nanotechnology Coordinated Infrastructure, which is supported by the National Science Foundation (grant ECCS-1542081). The authors acknowledge support for this research from the US National Cancer Institute under grant no. UH2 CA202723.
We would like to thank the Epigenomics Core Facility at Weill Cornell Medicine, the Scientific Computing Unit (SCU), XSEDE Supercomputing Resources, the National Center for Advancing Translational Sciences of the National Institutes of Health (UL1TR000457, CTSC), the Starr Cancer Consortium (I13-0052), and funding from the Irma T. Hirschl and Monique Weill-Caulier Charitable Trusts, Bert L. and N. Kuggie Vallee Foundation, the WorldQuant Foundation, The Pershing Square Sohn Cancer Research Alliance, the National Institutes of Health (U01DA053941, R01AI151059, R35GM138152), the Bill and Melinda Gates Foundation (OPP1151054), and the Alfred P. Sloan Foundation (G-2015-13964).
Footnotes
Institutional Review Board Nasopharyngeal samples were collected and processed through the Weill Cornell Medicine Institutional Review Board protocol 19-11021069. All relevant ethical regulations for work with human participants were complied with.
Competing Interests C.E.M is a cofounder and board member for Aevum, Biotia, and Onegevity, as well as an advisor or compensated speaker for Abbvie, Acuamark Diagnostics, ArcBio, BioRad, Bumrungrad, Carverr, DNA Genotek, F!nd, Genialis, Genpro, Homo Deus, Karius, Illumina, MilliporeSigma, Nano-Lit, Nanostring, New England Biolabs, Novartis, Promega, QIAGEN, Resilience Health, Tecan, Tempus, Thermo Fisher, Twist, Whole Biome, and Zymo Research.
REFERENCES
- 1.Verity R, Okell LC, Dorigatti I, et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis . 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol . 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun . 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iuliano AD, Roguski KM, Chang HH, et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet . 2018;391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cumulative Number of Reported Probable Cases of Severe Acute Respiratory Syndrome (SARS) World Health Organization: 2004.
- 6. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) World Health Organization: 2019.
- 7.Liu Y, Gayle AA, Wilder-Smith A, Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med . 2020;27:1–4. doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebrahim SH, Ahmed QA, Gozzer E, Schlagenhauf P, Memish ZA. Covid-19 and community mitigation strategies in a pandemic. BMJ. 2020. 368:m1066. [DOI] [PubMed]
- 9.Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med . 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linton NM, Kobayashi T, Yang Y, et al. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: a statistical analysis of publicly available case data. J Clin Med . 2020;9:538. doi: 10.3390/jcm9020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Geng Z, Fan Z, Liu J, Chen H. Point-of-care testing based on smartphone: the current state-of-the-art (2017–2018) Biosens Bioelectron . 2019;132:17–37. doi: 10.1016/j.bios.2019.01.068. [DOI] [PubMed] [Google Scholar]
- 12.Kozel TR, Burnham-Marusich AR. Point-of-care testing for infectious diseases: past, present, and future. J Clin Microbiol . 2017;55:2313–2320. doi: 10.1128/JCM.00476-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsons LM, Somoskovi A, Gutierrez C, et al. Laboratory diagnosis of tuberculosis in resource-poor countries: challenges and opportunities. Clin Microbiol Rev . 2011;24:314–350. doi: 10.1128/CMR.00059-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yager P, Domingo GJ, Gerdes J. Point-of-care diagnostics for global health. Annu Rev Biomed Eng . 2008;10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- 15.Drain PK, Hyle EP, Noubary F, et al. Diagnostic point-of-care tests in resource-limited settings. Lancet Infect Dis . 2014;14:239–249. doi: 10.1016/S1473-3099(13)70250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.US Food and Drug Administration. Policy for diagnostic tests for coronavirus disease-2019 during the public health emergency: immediately in effect guidance for clinical laboratories, commercial manufacturers, and Food and Drug Administration staff. Published in 2020. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/policy-coronavirus-disease-2019-tests-during-public-health-emergency-revised Accessed January 13, 2022.
- 17.US Food and Drug Administration. Emergency use authorizations. Published 2020. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations Accessed January 13, 2022.
- 18.Ahmed W, Angel N, Edson J, et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020. 728:138764. Accessed January 13, 2022. [DOI] [PMC free article] [PubMed]
- 19.La Rosa G, Iaconelli M, Mancini P, et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci Total Environ . 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ Sci Technol Lett . 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 21.Sharkey ME, Kumar N, Mantero AMA, et al. Lessons learned from SARS-CoV-2 measurements in wastewater. Sci Total Environ . 2021;798:149177. doi: 10.1016/j.scitotenv.2021.149177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizumoto K, Kagaya K, Zarebski A, Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill . 2020;25:2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai Y, Yao L, Wei T, et al. Presumed asymptomatic carrier transmission of COVID-19. JAMA . 2020;323:1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothe C, Schunk M, Sothmann P, et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med . 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu AD, Galatsis K, Zheng J, et al. 2021. Development of a saliva-optimized RT-LAMP assay for SARS-CoV-2. medRxiv Preprint posted online January 2. [DOI]
- 26.Snodgrass R, Gardner A, Semeere A, et al. A portable device for nucleic acid quantification powered by sunlight, flame, or electricity. Nat Biomed Eng . 2018;2:657–665. doi: 10.1038/s41551-018-0286-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butler DJ, Mozsary C, Meydan C, et al. Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. Nat Commun . 2021;12:1660. doi: 10.1038/s41467-021-21361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu R, Wu X, Wan Z, et al. Development of a novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Virol Sin . 2020;35:344–347. doi: 10.1007/s12250-020-00218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenfeld J, Reeves D, Brugler MR, et al. Genome assembly, annotation, and urban phylogenomics of the bedbug (Cimex lectularius) Nat Commun . 2016;7:10164. doi: 10.1038/ncomms10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacKay MJ, Hooker AC, Afshinnekoo E, et al. The COVID-19 XPRIZE and the need for scalable, fast, and widespread testing. Nat Biotechnol . 2020;38:1021–1024. doi: 10.1038/s41587-020-0655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]