Abstract
Nucleic acid amplification of clinical specimens with low target concentration has variable sensitivity. We examined whether testing multiple aliquots of extracted DNA increased the sensitivity and reproducibility of Chlamydia pneumoniae detection by PCR. Nested and non-nested C. pneumoniae PCR assays were compared using 10 replicates of 16 serial dilutions of C. pneumoniae ATCC VR-1310. The proportion positive versus the C. pneumoniae concentration was modeled by probit regression analysis. To validate the model, 10 replicates of 26 previously positive patient specimens of peripheral blood mononuclear cells (PBMC), sputum, or nasopharyngeal swabs (NPS) were tested. The proportion of replicates that were positive varied with the concentration of C. pneumoniae in the sample. At concentrations above 5 infection-forming units (IFU)/ml, both nested and non-nested PCR assay sensitivities were 90% or greater. The nested PCR was more sensitive (median detection, 0.35 versus 0.61 IFU/ml; relative median detection, 0.58; 95% confidence interval, 0.31 to 0.99; P = 0.04). In clinical specimens, replicate PCR detected 15 of 26 (nested) versus 1 of 26 (non-nested, P < 0.001). For PBMC specimens, testing 1, 3, or 5 replicates detected 3, 5, or 9 of 10 positive specimens, respectively. Median C. pneumoniae concentrations were estimated at 0.07 IFU/ml for PBMC and at <0.03 IFU/ml for NPS specimens. We conclude that performing 5 or 10 replicates considerably increased the sensitivity and reproducibility of C. pneumoniae PCR and enabled quantitation for clinical specimens. Due to sampling variability, PCR tests done without replication may miss a large proportion of positive specimens, particularly for specimens with small amounts of target C. pneumoniae DNA present.
Controversy surrounds the association of Chlamydia pneumoniae with atherosclerotic heart disease (7, 9, 18), asthma (1, 10), multiple sclerosis (11, 22), and Alzheimer's disease (8, 16, 19), primarily because of the lack of a definitive test for detecting C. pneumoniae. Culture is performed successfully by few laboratories and was much less sensitive than PCR for detection in vascular tissue (13). Serology has been considered the “gold standard” for the diagnosis of infection (12) but did not correlate with the presence of C. pneumoniae DNA or antigen in tissue (5, 13, 17). A superior marker of current or recent infection is required to clarify the clinical importance of C. pneumoniae infection in chronic diseases such as atherosclerosis.
Nucleic acid amplification tests such as PCR enable the detection of low concentrations of organism in clinical specimens. However, great variability of detection has been reported. For atherosclerotic tissue, reports of between 0 and 100% detection have been published, as recently summarized (3, 13). Similarly, the prevalence of C. pneumoniae DNA in peripheral blood mononuclear cells (PBMC) varied between 9% (27) and 59% (4) among patients with proven atherosclerotic heart disease. Some of this discrepancy may be attributable to differences between assays, but sampling variability is an alternative explanation. Whether replicate testing improves sensitivity or reproducibility has not, to our knowledge, been systematically examined for C. pneumoniae nucleic acid amplification tests.
In a previous study, we compared five C. pneumoniae PCRs for both analytical and clinical sensitivity (15), and we noted a major discrepancy between the tests. Despite relatively similar analytical test sensitivities, only the nested PCR based on the ompA gene (23) routinely detected a number of PBMC positives. We hypothesized that sampling variability as well as differences in PCR performance explained the results, and we inferred that clinical specimens had low concentrations of C. pneumoniae DNA.
We test here these hypotheses with probit regression analysis. Specifically, we sought to determine the following. (i) Can replicate C. pneumoniae PCR increase test sensitivity over testing a single time (analytical sensitivity)? (ii) Does replicate testing increase C. pneumoniae detection in clinical specimens (clinical sensitivity)? (iii) Can probit analysis quantitate C. pneumoniae in clinical specimens?
MATERIALS AND METHODS
PCR methods.
A comparison of five PCRs for C. pneumoniae was previously described (15), and two of these PCRs—a nested (23) and a non-nested procedure (6)–were used in this study. Samples (200 μl) of laboratory-cultured strains or clinical specimens (see details below) were extracted using QIAamp DNA Mini-Kits (Qiagen, Mississauga, Ontario, Canada) following a tissue or blood extraction protocol and eluted in 100 μl of buffer. PCR was performed on 2.5-μl purified DNA samples in a total volume of 25 μl. The components of the reaction mixture and thermocycling conditions were previously described by Campbell et al. (6) for the non-nested PCR and by Tong and Sillis (23) for the nested PCR. The non-nested procedure consisted of 40 rounds and amplified a 437-bp cloned PstI fragment. The nested PCR consisted of 40 rounds of amplification of a 333-bp fragment of the ompA gene, followed by 30 rounds of amplification of a 207-bp internal fragment. AmpliTaq Gold (Perkin-Elmer, Branchburg, N.J.) was used for all amplifications. All amplification products were analyzed by 2% (wt/vol) agarose gel electrophoresis followed by ethidium bromide staining. Stringent procedures to minimize or detect contamination included extraction and amplification in separate rooms after changing gloves and lab coats, use of plugged pipette tips and positive displacement pipettors, and insertion of at least one blank every 5 to 10 tubes. Every fifth blank was left open during specimen addition to detect aerosol contamination.
Dilution series of C. pneumoniae.
C. pneumoniae ATCC VR-1310 was cultured in U-937 human mononuclear cells for 40 to 48 h and then spiked into a repeatedly negative volunteer-derived PBMC fraction (CPT tube; BD Vacutainer Systems, Franklin Lakes, N.J.). In PCR replicates of two, serial 10-fold dilutions were tested to establish an upper threshold (all tests positive) and lower thresholds (all tests negative) of test sensitivity as 4 inclusion forming units (IFU)/ml (0.01 IFU/2.5-μl PCR) and 0.04 IFU/ml (0.0001 IFU/2.5-μL PCR), respectively. For each of the non-nested and nested PCRs, we performed 10 replicates of six dilutions between 4 and 0.04 IFU/ml and, at a separate time, 10 replicates of 10 dilutions between 4 and 0.008 IFU/ml. The final probit regression model included data from both of these two dilution series (total of 160 tests for each of the nested and non-nested PCRs).
Clinical validation.
For the clinical validation set, 26 clinical specimens (each from unique patients) were tested in 10 replicates by both PCR assays (total of 520 PCR tests): 10 PBMC specimens from patients undergoing elective coronary angiography, 6 pediatric and 8 adult nasopharyngeal specimens (NPS) from patients with acute respiratory symptoms, and 2 sputum specimens from adults with chronic airway limitation. A patient's specimen was considered positive if one or more of the 10 individual determinations were positive. All specimens were previously C. pneumoniae positive in at least one of three replicates using the nested PCR and were confirmed by Southern blotting and oligonucleotide hybridization with C. pneumoniae-specific probe.
Statistical methods.
The relationship between the proportion positive from each replicate of 10 and the corresponding log concentrations of C. pneumoniae was examined using probit regression analysis (SPSS for Windows 10.0; SPSS, Inc., Chicago, Ill.). Using the probit model, the two tests were compared, the median detection concentration of C. pneumoniae was estimated, and the concentrations corresponding to probits of 0.01 to 0.99 were calculated. Proportions were tested with StatXact version 3.02 (Cytel Software Corp., Cambridge, Mass.). A P value of <0.05 was taken as statistically significant (two tailed). The relationship between replicate number and test sensitivity was calculated in Excel 97 (Microsoft Corp., Redmond, Wash.), using the calculated probit at a given concentration and the following formula: probability of at least one positive in n replicates = 1 − (1 − probit)n. To estimate median C. pneumoniae concentration in clinical specimens, the concentration corresponding to the observed proportion positive of 10 replicates was interpolated from the nested PCR probit analysis table.
RESULTS
Constructing probit regression models.
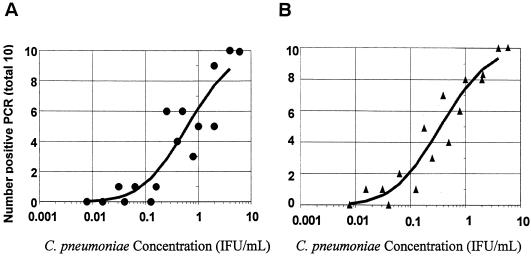
Separate probit regression curves were constructed for the non-nested and nested PCR assays using data from 10 replicates each of 16 dilutions of culture-grown C. pneumoniae ATCC VR-1310. Figure 1 shows the relationships between the number of positives at each dilution and the concentration of C. pneumoniae for the non-nested and nested PCRs. The circles (non-nested, panel A) or triangles (nested, panel B) represent the number of positive results from the 10 replicates at each concentration of C. pneumoniae, and the solid line represents the fitted probit regression line for the assay. At a concentration of 4 IFU/ml, both PCRs detected all 20 replicates as positive. At a concentration of 1 IFU/ml, the non-nested PCR detected 5 of 10 versus 8 of 10 for the nested PCR; and at a concentration of 0.1 IFU/ml, the non-nested PCR detected 0 of 10 versus 1 of 10 for the nested PCR.
FIG. 1.
Number of positives per 10 replicates versus the concentration of C. pneumoniae ATCC VR-1310 for non-nested PCR (A) and nested PCR (B) and a regression curve determined by probit regression analysis (SPSS).
The SPSS statistical program generated the probit (predicted proportion positive) versus the C. pneumoniae concentration with 95% confidence intervals (CI) shown in Table 1. For example, for the nested PCR, a concentration of 0.15 IFU/ml was associated with a probit of 0.30. Thus, repeated enough times, a positive result would be obtained in 30% of replicates. Conversely, at a concentration of 5 IFU/ml or greater, 9 of 10 replicates would be expected to be positive with either PCR, and a single PCR determination would be positive 90 to 95% of the time.
TABLE 1.
Predicted proportion of replicates that were positive versus the C. pneumoniae concentration for non-nested and nested C. pneumoniae PCR tests
| Probita |
C. pneumoniae IFU/ml (95% CI)
|
|
|---|---|---|
| Non-nested PCRb | Nested PCRc | |
| 0.01 | 0.02 (0.01, 0.03) | 0.01 (0.003, 0.02) |
| 0.05 | 0.05 (0.02, 0.08) | 0.03 (0.01, 0.05) |
| 0.10 | 0.08 (0.04, 0.13) | 0.05 (0.02, 0.08) |
| 0.15 | 0.12 (0.07, 0.18) | 0.07 (0.04, 0.11) |
| 0.20 | 0.16 (0.10, 0.24) | 0.09 (0.06, 0.14) |
| 0.30 | 0.27 (0.17, 0.40) | 0.15 (0.10, 0.23) |
| 0.40 | 0.41 (0.28, 0.61) | 0.24 (0.16, 0.35) |
| 0.50 | 0.61 (0.42, 0.92) | 0.35 (0.24, 0.52) |
| 0.60 | 0.92 (0.62, 1.42) | 0.53 (0.36, 0.80) |
| 0.70 | 1.41 (0.94, 2.29) | 0.81 (0.55, 1.28) |
| 0.80 | 2.34 (1.51, 4.08) | 1.34 (0.88, 2.23) |
| 0.90 | 4.69 (2.82, 9.25) | 2.70 (1.66, 5.12) |
| 0.95 | 8.34 (4.68, 18.4) | 4.79 (2.77, 10.2) |
| 0.99 | 24.50 (11.9, 67.8) | 14.1 (7.07, 37.3) |
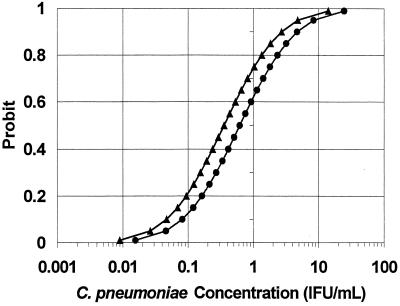
In Table 1 and Fig. 2, the probit models for the non-nested and nested C. pneumoniae PCRs were directly compared. The nested PCR regression line (triangles) is shifted up and to the left of the non-nested PCR line (circles). For any concentration of C. pneumoniae, the probability of detection was greater with the nested PCR, and the nested PCR regression curve was statistically significantly different from the non-nested PCR curve. The predicted median detected concentrations (probit = 0.50) were 0.35 IFU/ml (nested) and 0.61 IFU/ml (non-nested) for a relative median detection of 0.58 (95% CI = 0.31 to 0.99, P = 0.04). Model fit was assessed and adequate [Pearson goodness of fit χ2 (29 df) = 34.7, P = 0.22; parallelism test χ2 (1 df) < 0.001, P = 1.00].
FIG. 2.
Comparison of probit regression curves for a non-nested (●) and nested (▴) C. pneumoniae PCR (SPSS). The probit (predicted proportion of replicates positive) versus the C. pneumoniae ATCC VR-1310 concentration was obtained from 10 replicates of 16 dilutions (see the text).
Relationship between detection and number of replicates.
For a given probit and its corresponding C. pneumoniae concentration, the sensitivity of replicate testing and the number of replicates are related. The number of replicates needed for various probits of 0.01 to 0.99, to achieve an overall test sensitivity of 50, 80, 90, or 95%, were calculated using the nested C. pneumoniae PCR data (Table 2). For example, for a C. pneumoniae concentration of 0.35 IFU/ml (probit of 0.50), a single PCR determination has 50% sensitivity, a three-replicate assay would detect at least one positive with 80% sensitivity, a four-replicate assay with 90% sensitivity, and a five-replicate assay with 95% sensitivity. At lower concentrations, more replicates are required for a given overall test sensitivity. At 0.05 IFU/ml (probit 0.10), a single test achieves 10% sensitivity, compared with 7 replicates (50% sensitivity), 16 replicates (80% sensitivity), or 29 replicates (95% sensitivity). Conversely, at >5 IFU/ml, a single PCR determination is 95% sensitive for detecting C. pneumoniae, and replicates would not increase sensitivity further.
TABLE 2.
Relationship between the predicted number of PCR replicates needed to achieve various test sensitivities and C. pneumoniae concentrations
| Probita | C. pneumoniae concn (IFU/ml)b | No. of replicates needed to achieve sensitivity ofc:
|
|||
|---|---|---|---|---|---|
| 50% | 80% | 90% | 95% | ||
| 0.01 | 0.01 | 69 | 161 | 230 | 299 |
| 0.05 | 0.03 | 14 | 32 | 45 | 59 |
| 0.10 | 0.05 | 7 | 16 | 22 | 29 |
| 0.20 | 0.09 | 4 | 8 | 11 | 14 |
| 0.30 | 0.15 | 2 | 5 | 7 | 9 |
| 0.40 | 0.24 | 2 | 4 | 5 | 6 |
| 0.50 | 0.35 | 1 | 3 | 4 | 5 |
| 0.60 | 0.53 | 1 | 2 | 3 | 4 |
| 0.70 | 0.81 | 1 | 2 | 2 | 3 |
| 0.80 | 1.34 | 1 | 1 | 2 | 2 |
| 0.90 | 2.70 | 1 | 1 | 1 | 2 |
| 0.95 | 4.79 | 1 | 1 | 1 | 1 |
| 0.99 | 14.10 | 1 | 1 | 1 | 1 |
Probit, predicted proportion of replicates positive from probit regression analysis (SPSS), based on 10 replicates of 16 dilutions of C. pneumoniae ATCC VR-1310 (see the text).
C. pneumoniae ATCC VR-1310 concentration as calculated from probit analysis.
Replicates calculated in Microsoft Excel spreadsheet from the following equation: probability (at least 1 positive in n replicates) = 1 − (1 − probit)n.
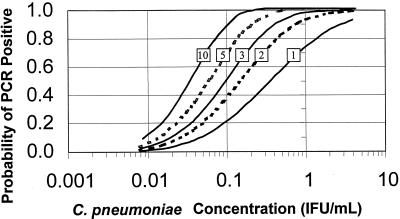
In Fig. 3, the relationship between increasing number of replicates and overall test sensitivity is presented in graphical form, with the curve for the 10 replicate PCR curve on the far left and that for a single PCR on the far right. At higher C. pneumoniae concentrations, repeating the sample increases the test sensitivity very little. At a lower concentration of 0.1 IFU/ml, a single PCR is 20% sensitive, compared with 40% for duplicate PCRs, 50% for replicates of 3, 70% for replicates of 5, and 90% for replicates of 10.
FIG. 3.
Predicted probability of PCR positive test versus the C. pneumoniae concentration for nested PCR by number of replicates. Replicates of 10, 5, 3, or 2 versus single PCR illustrated from left to right. Single PCR curves obtained from probit regression analysis of 10 replicates of 16 dilutions of C. pneumoniae ATCC VR-1310 (see text) are also shown. The remaining four curves were calculated in the spreadsheet program Microsoft Excel using the following formula: probability (at least 1 positive in n replicates) = 1 − (1 − probit)n.
From the probit model, we predicted that the nested PCR would have better sensitivity than the non-nested PCR for the detection of C. pneumoniae in clinical specimens, particularly at lower concentrations of target DNA, and at these concentrations replicates of 3, 5, or 10 would identify increasingly more specimens as positive compared with a single PCR.
Detection of C. pneumoniae in clinical specimens.
We next compared the performance of non-nested and nested PCR assays using clinical specimens. For 10 previously positive PBMC specimens from coronary angiography patients, the non-nested PCR detected a single positive only on the tenth repeat of that specimen (data not shown). In the first one, three, or five replicates, no positives were detected. For the 14 nasopharyngeal and 2 sputum specimens, no positives were detected in any of 10 replicates. In summary, the non-nested PCR identified 1 of 26 patient specimens, and 1 of 260 PCR tests, as positive.
With the nested PCR, all 10 PBMC specimens were identified as positive (Table 3). In the first 1, 3, or 5 replicates, 3, 5, and 9 of the 10 specimens were positive, respectively. In comparison, for a C. pneumoniae concentration of approximately 0.1 IFU/ml, the probit model predicted 2, 5, and 7 positives in 1, 3, or 5 replicates, respectively (Fig. 3). One of the two sputa was positive in 2 of 10 replicates, and the second sputum was negative in all 10 replicates. The nested PCR detected 4 of 8 adult NPS specimens (Table 3, specimens Resp A164, Resp A190, Resp A192, and Resp A269) but did not detect any C. pneumoniae in 6 pediatric NPS specimens. In summary, the nested PCR detected 15 of 26 clinical specimens or 34 of 260 individual PCR determinations and was superior to the non-nested PCR for the detection of C. pneumoniae (15 of 26 versus 1 of 26, P < 0.001). Had only three replicates been done per specimen, and a definition of 1 of 3 or more as C. pneumoniae positive been used, 0 of 26 non-nested and 6 of 26 nested specimens would have been positive (P = 0.03).
TABLE 3.
Nested PCR detection and quantitation of C. pneumoniae in individual clinical specimens by number of replicates
| Identification | Specimen | No. of PCR replicates
|
Proportion positivea | C. pneumoniae IFN/mlb (95% CI) | |||
|---|---|---|---|---|---|---|---|
| 1 | 3 | 5 | 10 | ||||
| Angio 202B | PBMCc | 0 | 0 | 1 | 1 | 0.10 | 0.05 (0.02, 0.08) |
| Angio 208B | PBMC | 1 | 1 | 1 | 2 | 0.20 | 0.09 (0.06, 0.14) |
| Angio 257B | PBMC | 0 | 0 | 0 | 1 | 0.10 | 0.05 (0.02, 0.08) |
| Angio 287A | PBMC | 0 | 0 | 1 | 2 | 0.20 | 0.09 (0.06, 0.14) |
| Angio 244B | PBMC | 1 | 2 | 5 | 9 | 0.90 | 2.70 (1.66, 5.12) |
| Angio 246C | PBMC | 0 | 1 | 2 | 4 | 0.40 | 0.24 (0.16, 0.35) |
| Angio 276A | PBMC | 0 | 0 | 1 | 1 | 0.10 | 0.05 (0.02, 0.08) |
| Angio 291B | PBMC | 0 | 0 | 1 | 1 | 0.10 | 0.05 (0.02, 0.08) |
| Angio 321B | PBMC | 0 | 1 | 1 | 1 | 0.10 | 0.05 (0.02, 0.08) |
| Angio 337C | PBMC | 1 | 3 | 5 | 6 | 0.60 | 0.53 (0.36, 0.80) |
| Bronch 10 | Sputumd | 0 | 1 | 1 | 2 | 0.20 | 0.09 (0.06, 0.14) |
| Resp A164 | NPSe | 1 | 0 | 0 | 0 | 0.10 | 0.05 (0.02, 0.08) |
| Resp A190 | NPS | 0 | 0 | 0 | 1 | 0.10 | 0.05 (0.02, 0.08) |
| Resp A192 | NPS | 0 | 0 | 0 | 1 | 0.10 | 0.05 (0.02, 0.08) |
| Resp A269 | NPS | 0 | 0 | 0 | 1 | 0.10 | 0.05 (0.02, 0.08) |
Proportion positive of 10 PCR replicates. All 26 patient samples previously positive by at least one of three Tong and Sillis (23) PCRs.
C. pneumoniae concentration as interpolated from a probit model of nested PCR of ATCC VR-1310 dilution series (Probit regression analysis, SPSS). For details, see the text.
PBMC were from patients undergoing coronary angiography.
Induced sputum from patients with chronic airflow limitation. One additional sputum was negative in all PCR replicates.
NPS were from an adult with acute respiratory symptoms. An additional four adult and six pediatric NPS were negative in all PCR replicates.
Quantitation of C. pneumoniae in clinical specimens.
By interpolation from the probit regression analysis for the nested PCR (Table 1), estimates of C. pneumoniae in clinical specimens were made (Table 3). For the 10 PBMC specimens, the median probit was estimated at 0.15, for an interpolated concentration of 0.07 IFU/ml (95% CI = 0.04 to 0.11). For specific patient specimens, probits varied between 0.1 and 0.9 or 0.05 to 2.7 IFU/ml. The single positive sputum was positive in 2 of 10 replicates for a probit of 0.2 (0.09 IFU/ml, 95% CI = 0.06 to 0.14). In four of eight adult NPS specimens, 1 of 10 replicates was positive (probit of 0.10) for an interpolated concentration of 0.05 IFU/ml (95% CI = 0.02 to 0.08). In the remaining four adult and all pediatric NPS specimens none of the 10 replicates detected C. pneumoniae (<0.03 IFU/ml). At a probit of <0.05, the median concentration for the nasopharyngeal specimens was <0.03 IFU/ml (95% CI = 0.00 to 0.05).
DISCUSSION
We have demonstrated here that replicate C. pneumoniae PCR markedly increased analytical sensitivity compared with performing a single PCR test. We validated the model by demonstrating that replicate testing increased C. pneumoniae detection in clinical specimens, particularly with the nested PCR, and that the sensitivity levels in analytical and clinical samples were consistent with the probit analysis predictions. We then used probit analysis to quantitate C. pneumoniae in clinical specimens and inferred a higher concentration of C. pneumoniae in PBMC compared with NPS.
The interpretation of replicate testing was facilitated by probit regression analysis, which has been utilized in particular for toxicology studies. In microbiology, probit analysis has been used very rarely: we found only four references in a MEDLINE search of the literature between 1967 and 2000. Vrielink and colleagues used probit analysis to compare the diagnostic sensitivities of enzyme immunoassays for human T-cell leukemia virus types 1 and 2 or hepatitis C virus (24–26), and Saldanha used this regression technique to quantitate the hepatitis C virus genome and compare PCR sensitivities (20).
The sensitivity of a diagnostic test is often considered a constant property, apart from some variation due to laboratory technique or specimen type. However, interpreting the probit as the test sensitivity, we demonstrated that PCR sensitivity varied between 0 and 100%, depending on the C. pneumoniae concentration. The finding that PCR was approximately 100% sensitive above a certain threshold and 0% sensitive below a certain threshold is not surprising. What is surprising is the 100-fold interval of concentrations (between 4 and 0.04 IFU/ml) in which PCR results were intermittently positive. Within this interval, repeat testing and probit modeling could be exploited for detection and quantitation.
Our findings may have important implications for the routine detection of C. pneumoniae in clinical specimens such as blood or respiratory specimens. Conversely, the lack of recognition of these concentrations of intermittently positive values may yield unreliable results.
In an excellent review of the molecular diagnosis of C. pneumoniae, Boman et al. discuss specimen collection, the preparation of nucleic acid from samples, the choice of gene target and primer selection, the optimal amplification conditions, and the detection of the amplification product (3). These authors briefly review sampling variation as a cause of false-negative results and discuss increasing the sample volume as a possible strategy to increase sensitivity, while acknowledging that this strategy may cause an unacceptable increase in the level of PCR inhibitors. We suggest adding the issue of PCR replicates to their list of areas where standardization is required. Readers need to know how many PCR replicates were done by a laboratory and how a positive specimen was operationally defined.
We acknowledge two potentially serious limitations of our study: face validity and feasibility. By face validity, we refer to whether most readers or laboratory directors would have confidence that a single positive in 5 or 10 PCR determinations represented a true positive. At a more stringent requirement of 2 PCR positives per 10 replicates, 0 of 26 clinical specimens were positive by the non-nested PCR, and 6 of 26 specimens were positive by the nested PCR (P = 0.03). A single positive PCR determination may represent contamination, a nonspecific reaction, or a true positive. We demonstrated that lower analytical concentrations were only intermittently PCR positive, and this relationship was predictable from a statistical viewpoint. These results are not likely to be due to contamination, which would not have varied predictably with the concentration. In addition, 0 of 200 negative controls tested with this assay by our laboratory have been positive. To ensure the specificity of the reaction, all first-time positive PCR clinical specimens were confirmed with Southern blotting and oligonucleotide hybridization, and 12 specimens had DNA sequencing of PCR product. The results all confirmed a C. pneumoniae-specific amplification product. If contamination and nonspecificity are ruled out, the results are true positives. Nevertheless, we would not consider a single 1 of 10 samples positive as a “confirmed” C. pneumoniae positive. In our PBMC study, we verified positive specimens by independent re-extraction, followed by PCR in triplicate (21). As more sensitive assays are developed, confirmation by amplifying a different target will be preferable (2).
Regarding feasibility, we acknowledge that a trade-off may be required in determining the optimal number of replicates for different specimen types. For the detection of C. pneumoniae DNA in clinical specimens, replicates of two or three, depending on the specimen type, may be adequate if the laboratory can demonstrate high reproducibility. A larger number of replicates will likely not be feasible, but the laboratory may wish to test a small number of positives in 5 or 10 replicates to examine reproducibility. In a research setting, replicates of up to 10 may be desirable for specimen types in which C. pneumoniae concentration is likely to be low. We currently test blood and respiratory specimens in replicates of three, but we have increased both the concentration (DNA eluted in 50 μl rather than 100 μl) and the sample size (5 μl per 50-μl PCR mixture rather than 2.5 μl per 25-μl PCR mixture) to approximate the same sensitivity as previously achieved with 10 replicates. Nevertheless, we suggest replicate PCR as a “reference standard” only until methods of extraction and detection are improved to the point where single or duplicate PCR will provide comparable sensitivity and reliability. Methods to concentrate target, such as monocyte enrichment using CD14 antibodies (14), or nucleic acid molecule selection using capture probes, may obviate the need for replication altogether.
We conclude that repeat testing of the same specimen markedly increases the sensitivity and reliability of a PCR assay, particularly for clinical specimens with a low C. pneumoniae concentration. Replicate testing may improve the development and comparison of PCRs and provide more precise estimates of organism prevalence in various chronic disease states. Conversely, failure to recognize the low sensitivity of a single PCR determination may cause frustration when positive results cannot be reliably confirmed. Further validation is needed with other C. pneumoniae assays and other clinical specimens.
ACKNOWLEDGMENTS
This study was supported by a grant-in-aide from the Father Sean O'Sullivan Research Centre, St. Joseph's Hospital, Hamilton, Ontario, Canada. M. Smieja is a Research Fellow of the Heart and Stroke Foundation of Canada.
REFERENCES
- 1.Bjornsson E, Hjelm E, Janson C, Fridell E, Boman G. Serology of chlamydia in relation to asthma and bronchial hyperresponsiveness. Scand J Infect Dis. 1996;28:63–69. doi: 10.3109/00365549609027152. [DOI] [PubMed] [Google Scholar]
- 2.Boman J, Gaydos C A. Polymerase chain reaction detection of Chlamydia pneumoniae in circulating white blood cells. J Infect Dis. 2000;181:S452–S454. doi: 10.1086/315609. [DOI] [PubMed] [Google Scholar]
- 3.Boman J, Gaydos C A, Quinn T C. Molecular diagnosis of Chlamydia pneumoniae infection. J Clin Microbiol. 1999;37:3791–3799. doi: 10.1128/jcm.37.12.3791-3799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boman J, Soderberg S, Forsberg J, Birgander L S, Allard A, Persson K, Jidell E, Kumlin U, Juto P, Waldenstrom A, Wadell G. High prevalence of Chlamydia pneumoniae DNA in peripheral blood mononuclear cells in patients with cardiovascular disease and in middle-aged blood donors. J Infect Dis. 1998;178:274–277. doi: 10.1086/517452. [DOI] [PubMed] [Google Scholar]
- 5.Campbell L A, O'Brien E R, Cappuccio A L, Kuo C C, Wang S P, Stewart D, Patton D L, Cummings P K, Grayston J T. Detection of Chlamydia pneumoniae TWAR in human coronary atherectomy tissues. J Infect Dis. 1995;172:585–588. doi: 10.1093/infdis/172.2.585. [DOI] [PubMed] [Google Scholar]
- 6.Campbell L A, Perez M M, Hamilton D J, Kuo C C, Grayston J T. Detection of Chlamydia pneumoniae by polymerase chain reaction. J Clin Microbiol. 1992;30:434–439. doi: 10.1128/jcm.30.2.434-439.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danesh J, Whincup P, Walker M, Lennon L, Thomson A, Appleby P, Wong Y, Bernardes-Silva M, Ward M. Chlamydia pneumoniae IgG titres and coronary heart disease: prospective study and meta-analysis. Br Med J. 2000;321:208–213. doi: 10.1136/bmj.321.7255.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gieffers J, Reusche E, Solbach W, Maass M. Failure to detect Chlamydia pneumoniae in brain sections of Alzheimer's disease patients. J Clin Microbiol. 2000;38:881–882. doi: 10.1128/jcm.38.2.881-882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grayston J T, Campbell L A. The role of Chlamydia pneumoniae in atherosclerosis. Clin Infect Dis. 1999;28:993–994. doi: 10.1086/514764. [DOI] [PubMed] [Google Scholar]
- 10.Hahn D L. Chlamydia pneumoniae, asthma, and COPD: what is the evidence? Ann Allergy Asthma Immunol. 1999;83:271–288. doi: 10.1016/S1081-1206(10)62666-X. , 291. [DOI] [PubMed] [Google Scholar]
- 11.Hammerschlag M R, Ke Z, Lu F, Roblin P, Boman J, Kalman B. Is Chlamydia pneumoniae present in brain lesions of patients with multiple sclerosis? J Clin Microbiol. 2000;38:4274–4276. doi: 10.1128/jcm.38.11.4274-4276.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo C C, Jackson L A, Campbell L A, Grayston J T. Chlamydia pneumoniae (TWAR) Clin Microbiol Rev. 1995;8:451–461. doi: 10.1128/cmr.8.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo C, Campbell L A. Detection of Chlamydia pneumoniae in arterial tissues. J Infect Dis. 2000;181:S432–S436. doi: 10.1086/315615. [DOI] [PubMed] [Google Scholar]
- 14.Maass M, Jahn J, Gieffers J, Dalhoff K, Katus H A, Solbach W. Detection of Chlamydia pneumoniae within peripheral blood monocytes of patients with unstable angina or myocardial infarction. J Infect Dis. 2000;181:S449–S451. doi: 10.1086/315610. [DOI] [PubMed] [Google Scholar]
- 15.Mahony J B, Chong S, Coombes B K, Smieja M, Petrich A. Analytical sensitivity, reproducibility of results, and clinical performance of five PCR assays for detecting Chlamydia pneumoniae DNA in peripheral blood mononuclear cells. J Clin Microbiol. 2000;38:2622–2627. doi: 10.1128/jcm.38.7.2622-2627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nochlin D, Shaw C M, Campbell L A, Kuo C C. Failure to detect Chlamydia pneumoniae in brain tissues of Alzheimer's disease. Neurology. 1999;53:1888. doi: 10.1212/wnl.53.8.1888-a. [DOI] [PubMed] [Google Scholar]
- 17.Puolakkainen M, Kuo C C, Shor A, Wang S P, Grayston J T, Campbell L A. Serological response to Chlamydia pneumoniae in adults with coronary arterial fatty streaks and fibrolipid plaques. J Clin Microbiol. 1993;31:2212–2214. doi: 10.1128/jcm.31.8.2212-2214.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ridker P M, Kundsin R B, Stampfer M J, Poulin S, Hennekens C H. Prospective study of Chlamydia pneumoniae IgG seropositivity and risks of future myocardial infarction. Circulation. 1999;99:1161–1164. doi: 10.1161/01.cir.99.9.1161. [DOI] [PubMed] [Google Scholar]
- 19.Ring R H, Lyons J M. Failure to detect Chlamydia pneumoniae in the late-onset Alzheimer's brain. J Clin Microbiol. 2000;38:2591–2594. doi: 10.1128/jcm.38.7.2591-2594.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saldanha J. Assays for viral sequences and their value in validation of viral elimination. Dev Biol Stand. 1993;81:231–236. [PubMed] [Google Scholar]
- 21.Smieja M, Chong S, Natarajan M, Petrich A, Rainen L, Mahony J B. Circulating nucleic acids of Chlamydia pneumoniae and cytomegalovirus in patients undergoing coronary angiography. J Clin Microbiol. 2001;39:596–600. doi: 10.1128/JCM.39.2.596-600.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sriram S, Mitchell W, Stratton C. Multiple sclerosis associated with Chlamydia pneumoniae infection of the CNS. Neurology. 1998;50:571–572. doi: 10.1212/wnl.50.2.571. [DOI] [PubMed] [Google Scholar]
- 23.Tong C Y, Sillis M. Detection of Chlamydia pneumoniae and Chlamydia psittaci in sputum samples by PCR. J Clin Pathol. 1993;46:313–317. doi: 10.1136/jcp.46.4.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vrielink H, Reesink H, Habibuw M, Schuller M, van der Meer C, Lelie P. Comparison of four HTLV-I and HTLV-I + II ELISAs. Vox Sang. 1999;76:187–191. doi: 10.1159/000031047. [DOI] [PubMed] [Google Scholar]
- 25.Vrielink H, Reesink H W, Zaaijer H L, van der Poel C L, Cuypers H T, Lelie P N. Sensitivity and specificity of four assays to detect human T-lymphotropic virus type I or type I/II antibodies. Transfusion. 1996;36:344–346. doi: 10.1046/j.1537-2995.1996.36496226150.x. [DOI] [PubMed] [Google Scholar]
- 26.Vrielink H, Zaaijer H L, Reesink H W, van der Poel C L, Cuypers H T, Lelie P N. Sensitivity and specificity of three third-generation anti-hepatitis C virus ELISAs. Vox Sang. 1995;69:14–17. doi: 10.1111/j.1423-0410.1995.tb00341.x. [DOI] [PubMed] [Google Scholar]
- 27.Wong Y K, Dawkins K D, Ward M E. Circulating Chlamydia pneumoniae DNA as a predictor of coronary artery disease. J Am Coll Cardiol. 1999;34:1435–1439. doi: 10.1016/s0735-1097(99)00391-5. [DOI] [PubMed] [Google Scholar]