Abstract
Background & Aims
Gut microbiota and microbial factors regulate the pathogenesis of nonalcoholic fatty liver disease (NAFLD) in patients with obesity and metabolic abnormalities, but little is known about their roles in nonobese NAFLD. Expansion of Escherichia is associated with NAFLD pathogenesis. We aimed to investigate the pathogenic role of Escherichia fergusonii and its products in the development of nonobese NAFLD.
Methods
We characterized the intestinal microbiome signature in a cohort of NAFLD patients and healthy controls by 16S ribosomal RNA sequencing. The role of E fergusonii was estimated in rats after 16 weeks of administration, and features of NAFLD were assessed. E fergusonii–derived microRNA-sized, small RNAs (msRNAs) were analyzed by deep sequencing.
Results
We detected an expansion of Escherichia_Shigella in NAFLD patients compared with healthy controls, and its increase was associated with disease severity independent of obesity. E fergusonii, a member of the genus Escherichia, induced the development of nonobese NAFLD characterized by hepatic steatosis and hepatocyte ballooning in rats without obesity. It disturbed host lipid metabolism by inhibiting hepatic lipid β-oxidation and promoting de novo lipogenesis. We also showed that E fergusonii caused the development of hepatic inflammation and fibrosis in a sizable fraction of animals at an advanced stage of NAFLD. Mechanistically, E fergusonii–derived msRNA 23487 down-regulated host hepatic peroxisome proliferator-activated receptor α expression, which could contribute to lipid accumulation in the liver.
Conclusions
These results suggest that E fergusonii promotes the pathogenesis of steatohepatitis and fibrosis in nonobese rats by secreting msRNA 23487, and it might be a potential biomarker for predicting steatohepatitis in nonobese NAFLD.
Keywords: Steatosis, Gut Microbiota, Lipid Metabolism, Peroxisome Proliferator-Activated Receptor α
Abbreviations used in this paper: ALT, alanine aminotransferase; AST, aspartate aminotransferase; con, control; HDL-C, high-density lipoprotein cholesterol; HFHC, high-fat, high-cholesterol; LDL-C, low-density lipoprotein cholesterol; msRNA, microRNA-sized, small RNA; NAFL, nonalcoholic fatty liver; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; NC, negative control; PCR, polymerase chain reaction; PPARα, peroxisome proliferator-activated receptor α; rRNA, ribosomal RNA; sRNA, small RNA
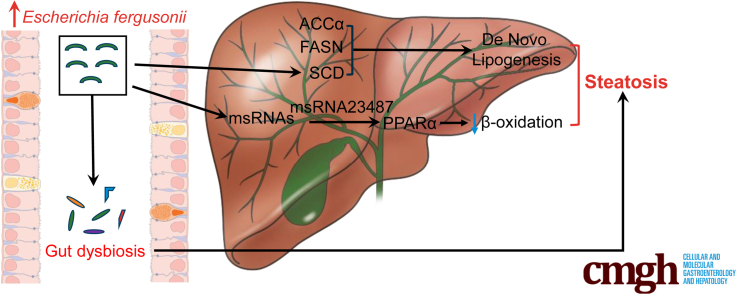
Graphical abstract
Summary.
Our study suggests that gut microbiota dysregulation and expansion of Escherichia_Shigella are associated with the progression of nonobese nonalcoholic fatty liver disease. Escherichia fergusonii promotes the pathogenesis of steatohepatitis and fibrosis in nonobese rats by secreting msRNA 23487.
Nonalcoholic fatty liver disease (NAFLD) is a metabolic stress-induced liver injury that is closely related to genetic susceptibility and insulin resistance in the absence of alcohol consumption. Currently, NAFLD has become the second-leading liver disease among patients awaiting liver transplantation in the United States.1 The spectrum of NAFLD ranges from simple steatosis (or nonalcoholic fatty liver, NAFL) to nonalcoholic steatohepatitis (NASH), with the development of fibrosis, cirrhosis, and, finally, hepatocellular carcinoma.2 NAFLD is seen commonly in patients with metabolic abnormalities associated with obesity, such as dyslipidemia, type II diabetes, and metabolic syndrome.3 Importantly, NAFLD has been reported in nonobese individuals based on liver biopsy, particularly in Asians.4 Studies have suggested that the incidence of NASH and fibrosis does not differ between obese and nonobese NAFLD patients.5 Considering the clinical significance and prevalence of nonobese NAFLD, studies are needed to understand its pathogenesis to identify high-risk patients and manage their metabolic profile.
A growing body of evidence suggests an intricate linkage between the gut microbiome and the pathogenesis of NAFLD.6 The gut microbiome is a diverse microbial community comprising trillions of bacteria, fungi, viruses, archaea, and protists that encode more functional genes than the human genome.6 In the past decade, compositional changes in the gut microbiota and its metabolites, such as short-chain fatty acids, branched-chain amino acids, indoles, and secondary bile acids have been reported in patients and animal studies in NAFLD.6,7 Zhu et al8 compared the gut microbiota of healthy, obese, and NASH patients. Escherichia was the only abundant genus that differed between obese and NASH patients, suggesting a possible role of Escherichia in driving NASH progression independent of obesity. Similar to eukaryotic cells, prokaryotes also can produce microRNA-sized, small RNAs (msRNAs) that play important roles in communicating between bacteria and hosts.9 In the current study, we investigated the role of Escherichia fergusonii and its product msRNA 23487 in the pathogenesis of nonobese NAFLD in rats.
Results
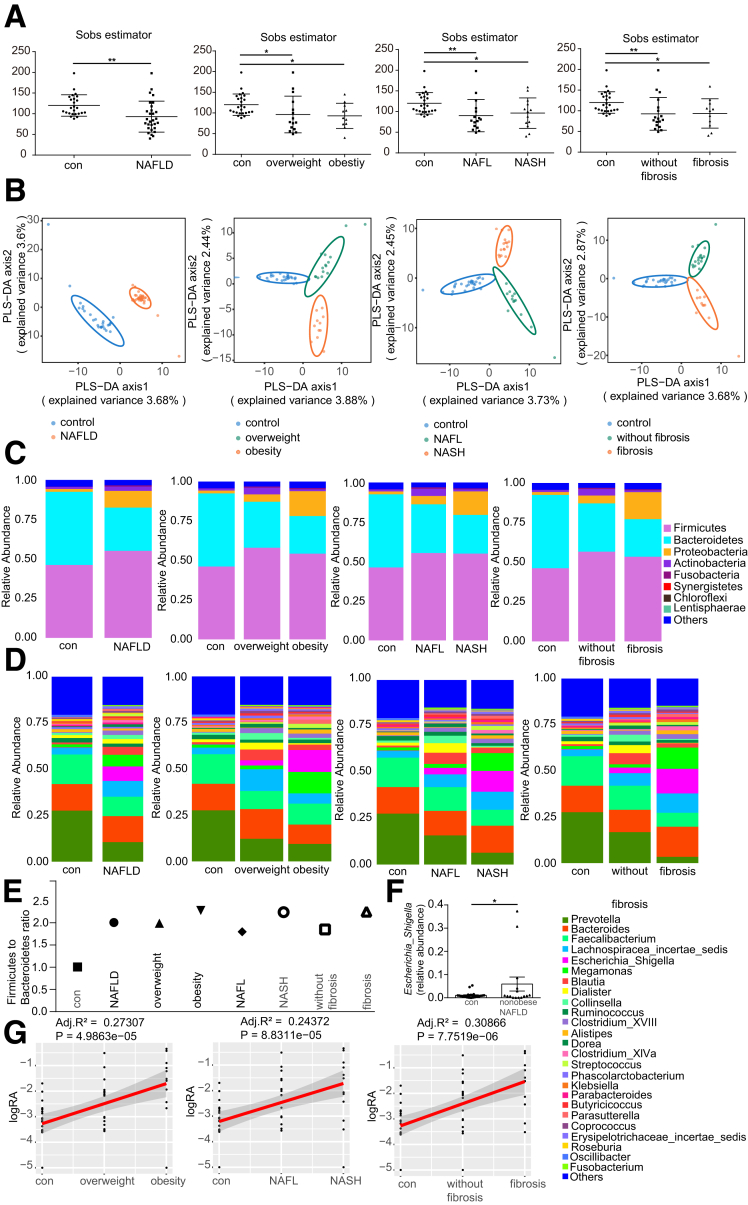
Expansion of Escherichia_Shigella Was Associated With NAFLD Severity
To determine whether NAFLD and NASH are associated with an altered composition of gut microbiota, we performed 16S ribosomal RNA (rRNA) gene sequencing using fecal samples from healthy controls (n = 25) and NAFLD patients (n = 29). Overall, the bacterial richness within the groups based on the Sobs estimator was reduced significantly in NAFLD patients compared with healthy controls. The Sobs estimator in patients with overweight or obesity, with NAFL or NASH, and with or without fibrosis also was decreased significantly compared with healthy controls (Figure 1A). Furthermore, partial least-squares discrimination analysis showed a significant change in the composition of the gut microbiota in NAFLD patients compared with healthy controls (Figure 1B). In different subgroups of NAFLD (patients with overweight or obesity, with NAFL or NASH, with or without fibrosis), we also detected a clustering difference between NAFLD subgroups and controls (Figure 1B). Moreover, the relative abundance at the phylum or genus levels showed alterations in the gut microbiota profile in NAFLD patients. Finally, the relative abundance of Proteobacteria (Figure 1C and D) and the Firmicutes:Bacteroidetes ratio were increased in the NAFLD group compared with healthy controls (2.02 vs 1.00) (Figure 1E).
Figure 1.
Escherichia_Shigella is associated with disease severity in NAFLD patients. (A) Bacterial richness was estimated based on the Sobs estimator in healthy controls (n = 25), NAFLD (n = 29), and NAFLD subgroups. (B) Partial least-squares discrimination analysis (PLS-DA) was used to show β-diversity among the groups. (C and D) Relative abundance of bacteria at the (C) phylum and (D) genus levels in healthy controls, NAFLD patients, and NAFLD subgroups. (E) Firmicutes:Bacteroidetes ratio in healthy controls and NAFLD patients. (F) The relative abundance of Escherichia_Shigella in the healthy controls (n = 25) and nonobese NAFLD patients (n = 15). (G) Spearman correlation between the relative abundance of Escherichia_Shigella and the severity of hepatic inflammation and fibrosis. ∗P < .05, P > .01; ∗∗P < .01, P > .001. Adj., adjusted; RA, relative abundance.
One substantial difference that we observed was an increase in the proportion of Escherichia_Shigella, a member of Proteobacteria, in NAFLD patients (Figure 1D). Moreover, the relative abundance of Escherichia_Shigella was increased significantly in nonobese NAFLD patients compared with healthy controls (Figure 1F), suggesting a possible pathogenic role of Escherichia_Shigella in nonobese NAFLD development. Spearman correlation analysis (Figure 1G) showed that the relative abundance of Escherichia_Shigella was associated with the development of hepatic inflammation (adjusted R2 = 0.244; P < .0001) and fibrosis (adjusted R2 = 0.309; P < .0001).
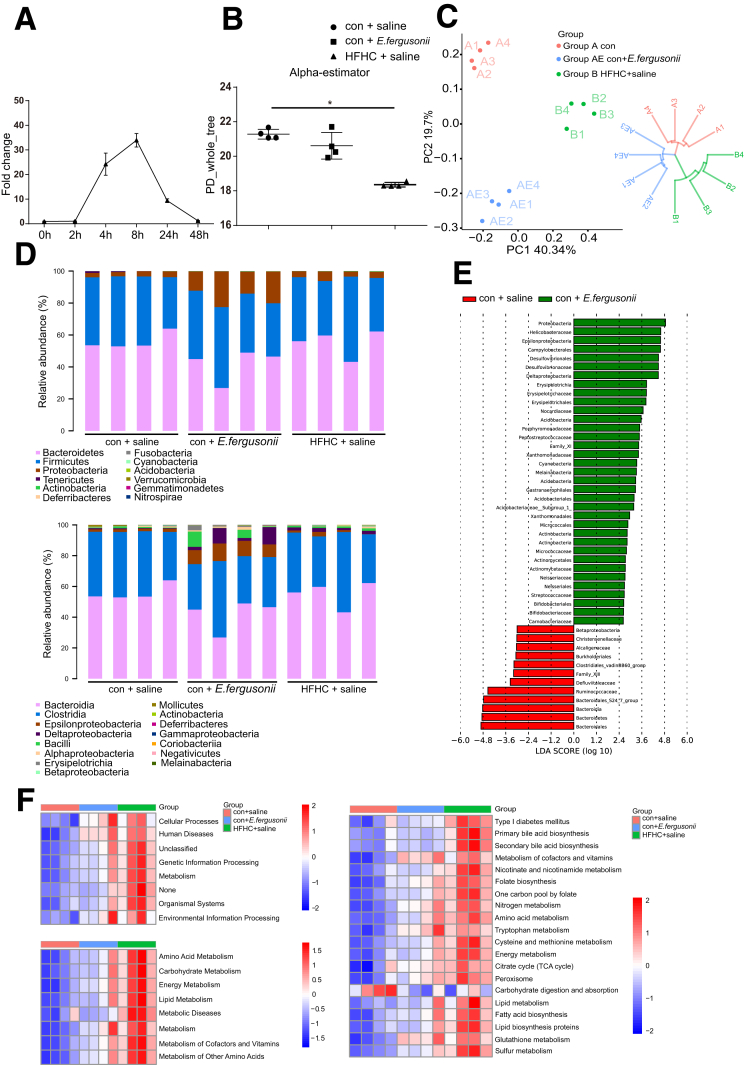
E fergusonii Altered the Gut Microbiome Profile in Rats
To date, 7 species have been identified in the genus Escherichia, among which E fergusonii is a rare pathogen that can infect both human beings and animals.10 To determine the effect of E fergusonii on the gut microbiome profile and biological functions, we treated rats with E fergusonii daily for 16 weeks by oral gavage. Daily administration was necessary because E fergusonii does not colonize rats (Figure 2A). We found that E fergusonii administration caused a shift in gut microbiota and tended to reduce the α-diversity under phylogenetic diversity_whole_tree measurement (Figure 2B). β-diversity based on principal coordinates and the unweighted pair-group method with arithmetic means analysis showed that the E fergusonii and control groups clustered differently (Figure 2C). Interestingly, E fergusonii administration increased the abundance of Proteobacteria (13.35-fold vs the control [con]+saline group) and decreased the abundance of Bacteroidetes (0.75-fold vs con+saline group) at the phylum level (Figure 2D). Linear discriminant effect-size analysis was performed to identify the features most likely to account for differences among groups (Figure 2E). In addition, E fergusonii altered gut microbiota functions based on Phylogenetic Investigation of Communities by Reconstruction of Unobserved States analysis. Pathways associated with metabolism, metabolic diseases, energy, amino acids, carbohydrate metabolism, and lipid metabolism were up-regulated with high-fat, high-cholesterol (HFHC) treatment, and the same trends were observed after E fergusonii administration. Importantly, pathways associated with lipid synthesis and metabolism also were dysregulated by E fergusonii (Figure 2F). These data indicate that E fergusonii disturbs host metabolic homeostasis by altering the gut microbiome profile, which may lead to the progression of NAFLD.
Figure 2.
E fergusonii altered the gut microbiome composition and functions. (A) E fergusonii was detected in feces from rats gavaged with E fergusonii (3∗109 colony-forming units), assessed by quantitative PCR. n = 6 per group. (B) Bacterial richness was estimated based on the Sobs estimator in rats from the con+saline, con+E fergusonii, and HFHC+saline groups. (C) β-diversity in rats from the different groups. (D) Relative abundance of bacteria at the phylum (upper) and class (lower) levels in rats from different groups. (E) Linear discriminant analysis effect size analysis between the con+saline and con+E fergusonii groups. (F) Phylogenetic Investigation of Communities by Reconstruction of Unobserved States analysis predicted gene functions in rats from different groups. ∗P < .05. LDA, linear discriminant analysis; PC, principal coordinate; PD, phylogenic diversity; TCA, tricarboxylic acid cycle.
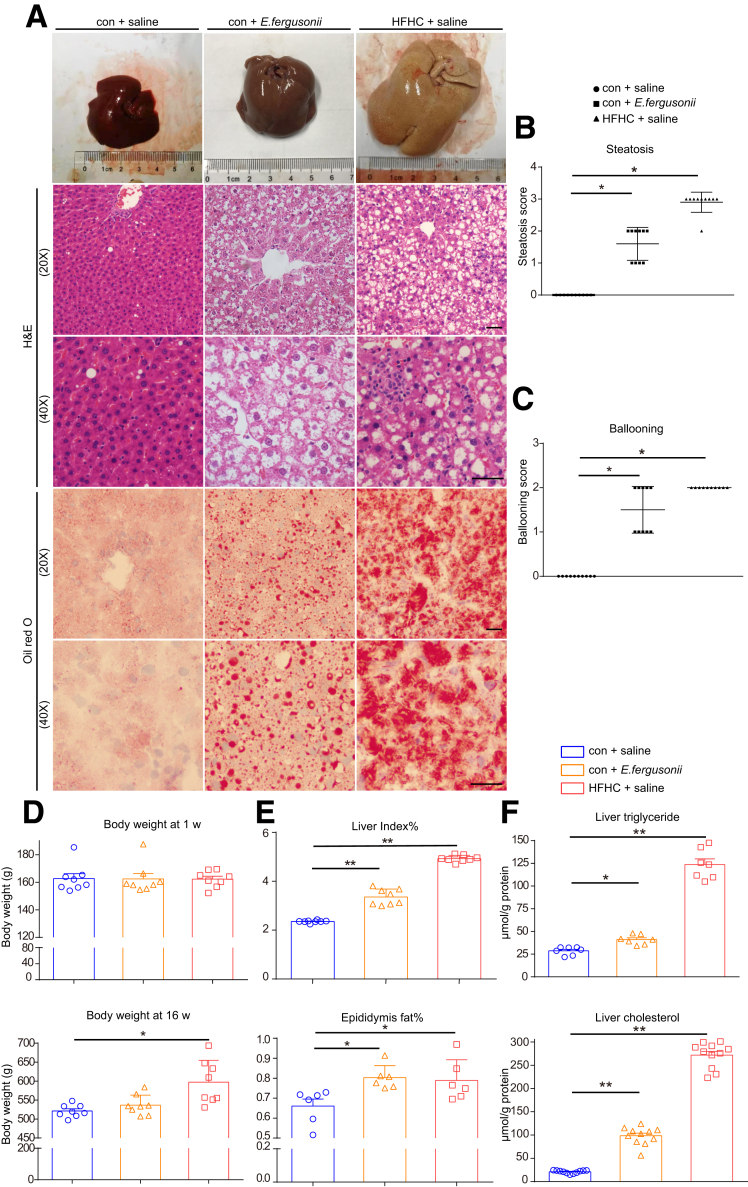
E fergusonii Induced Pathologic Features of Nonobese NAFLD in Rats
Next, we analyzed the effect of E fergusonii on hepatic pathologic features associated with NAFLD. Livers from the E fergusonii treatment group were larger in size and appeared pale in color compared with the con+saline group. We also detected lipid droplet accumulation, hepatomegaly, and hepatocellular ballooning in the E fergusonii group (Figure 3A). Steatosis and the hepatocellular ballooning score were significantly higher in the E fergusonii group than in the con+saline group (Figure 3B and C). Rats fed the HFHC diet showed a dramatic body weight gain after 16 weeks, while E fergusonii strongly aggravated liver lesions with no effect on body weight (Figure 3D). Increased hepatic triglyceride and total cholesterol levels further supported that E fergusonii disturbed host hepatic lipid metabolism (Figure 3F). Finally, the percentages of liver index and epididymal fat were increased significantly in the E fergusonii group (Figure 3E). All of these findings suggested that E fergusonii induced the development of nonobese NAFLD characterized by hepatic steatosis and hepatocellular ballooning without causing obesity in rats.
Figure 3.
E fergusonii induced hepatic steatosis and hepatocellular ballooning. (A) Representative images of liver, H&E, and Oil Red O staining in rats on the con+saline, con+E fergusonii, and HFHC+saline diets. (B) Steatosis score and (C) hepatocyte ballooning score in rats from different groups. (D) Body weight at 1 week and 16 weeks in each group. (E) Liver index (%) and epididymis fat (%) in rats from different groups. (F) Liver triglyceride levels and cholesterol in rats from different groups. ∗P < .05, P > .01; ∗∗P < .01, P > .001.
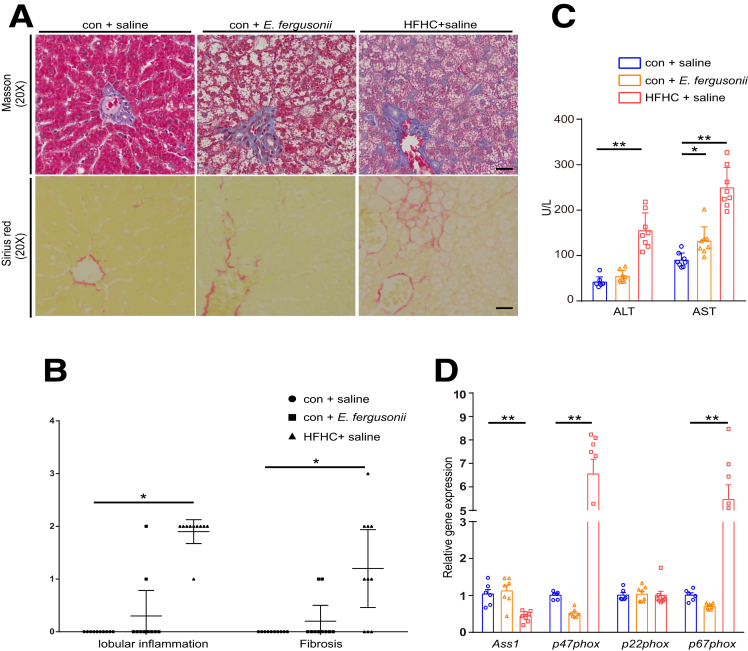
E fergusonii Induced Steatohepatitis in a Proportion of Animals
NASH is the most serious form of NAFLD, characterized by hepatic inflammation and fibrosis as the result of a second hit.11 We therefore explored whether E fergusonii administration induced hepatic inflammation and fibrosis in rats. As we expected, the HFHC group showed marked inflammatory and fibrotic phenotypes by histology (Figure 4A and B). Liver injury assessed by serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) was increased significantly in the HFHC diet group (Figure 4C). Reactive oxygen species–related cytoplasmic subunits, such as p47phox and p67phox, were up-regulated in the HFHC diet group, while the toxic substance binding protein Ass1 was decreased with the HFHC diet (Figure 4D). Furthermore, quantification of Masson trichrome and Sirius red staining showed that the HFHC diet caused fibrosis in 70% of animals that were fed that diet. Specifically, 7 of 10 rats displayed a fibrosis score greater than 1 (Figure 4A and B). Among all animals in the E fergusonii group, 20% (2 of 10) developed hepatic inflammation, and 20% (2 of 10) showed sinus fibrosis (fibrosis score 1) with scattered inflammatory cell infiltration (Figure 4A and B). We confirmed that rats that developed either inflammation or fibrosis were different animals. Therefore, 40% (4 of 10) of rats in the E fergusonii group developed advanced-stage NAFLD. Aside from increased AST levels (Figure 4C), ALT levels tended to increase with E fergusonii administration, although the increase was not statistically significant. Taken together, these data showed that E fergusonii induced steatohepatitis in a sizable fraction of rats.
Figure 4.
Effects of E fergusonii administration on hepatic inflammation and fibrosis. (A) Representative images of liver sections stained with Masson trichrome and Sirius red in rats on the con+saline, con+E fergusonii, and HFHC+saline diets. (B) Lobular inflammation and fibrosis scores in rats from different groups. (C) Serum levels of ALT and AST in each group. (D) Reactive oxygen species–related cytoplasmic subunit expression levels in rats from different groups. ∗P < .05, P > .01; ∗∗P < .01, P > .001.
E fergusonii Dysregulated Host Lipid Metabolism
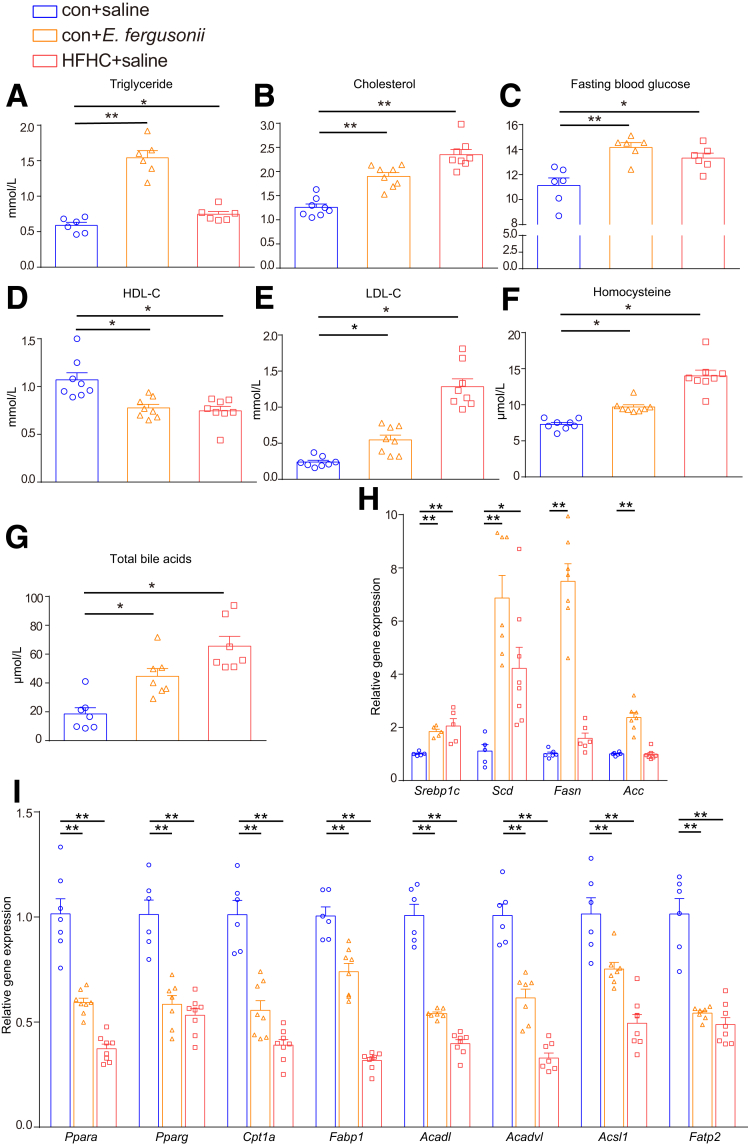
To understand the mechanisms underlying the increased hepatic lipid accumulation, we next measured biochemical parameters critically involved in fatty acid and cholesterol metabolism. The HFHC diet significantly increased the levels of triglycerides (Figure 5A), cholesterol (Figure 5B), fasting blood glucose level (Figure 5C), low-density lipoprotein cholesterol (LDL-C) (Figure 5E), homocysteine (Figure 5F), and total bile acids (Figure 5G) compared with those of the con+saline group, while the high-density lipoprotein cholesterol (HDL-C) level was decreased (Figure 5D). In comparison, E fergusonii increased serum triglyceride, cholesterol, fasting blood glucose, LDL-C, homocysteine, and total bile acid levels, while reducing HDL-C levels (Figure 5A–G).
Figure 5.
E fergusonii disturbed the serum lipid profile and hepatic lipid metabolism. (A–G) Serum triglyceride, cholesterol, fasting blood glucose, HDL-C, LDL-C, homocysteine, and total bile acid levels in the con+saline, con+E fergusonii, and HFHC+saline groups. (H) Expression levels of Srebp1c, Scd, Fasn, and Acc in rats from different groups. (I) Expression levels of Ppara, Pparg, Cpt1α, Fabp1, Acadl, Acadvl, Acsl1, and Fatp2 in rats from different groups. ∗P < .05, P > .01; ∗∗P < .01, P > .001.
Next, we evaluated gene expression related to lipid metabolism in the liver. As shown in Figure 5H, E fergusonii increased the expression of genes associated with de novo lipogenesis, such as Srebp1c, Scd, Fasn, and Acc. Furthermore, E fergusonii significantly reduced ligand-activated transcription factor peroxisome proliferator-activated receptor α (Ppara) expression, a master regulator of fatty acid uptake, β oxidation, ketogenesis, bile acid synthesis, and triglyceride turnover.12 Genes associated with fatty acid oxidation (Cpt1a, Fabp1, Acadl, and Acadvl) and lipid transport (Acsl1 and Fatp2) also were down-regulated with E fergusonii treatment or HFHC diet (Figure 5I). These data suggest that E fergusonii disturbs host hepatic lipid metabolism by inhibiting fatty acid oxidation and enhancing lipid de novo lipogenesis.
msRNA Sequencing Analysis of E fergusonii
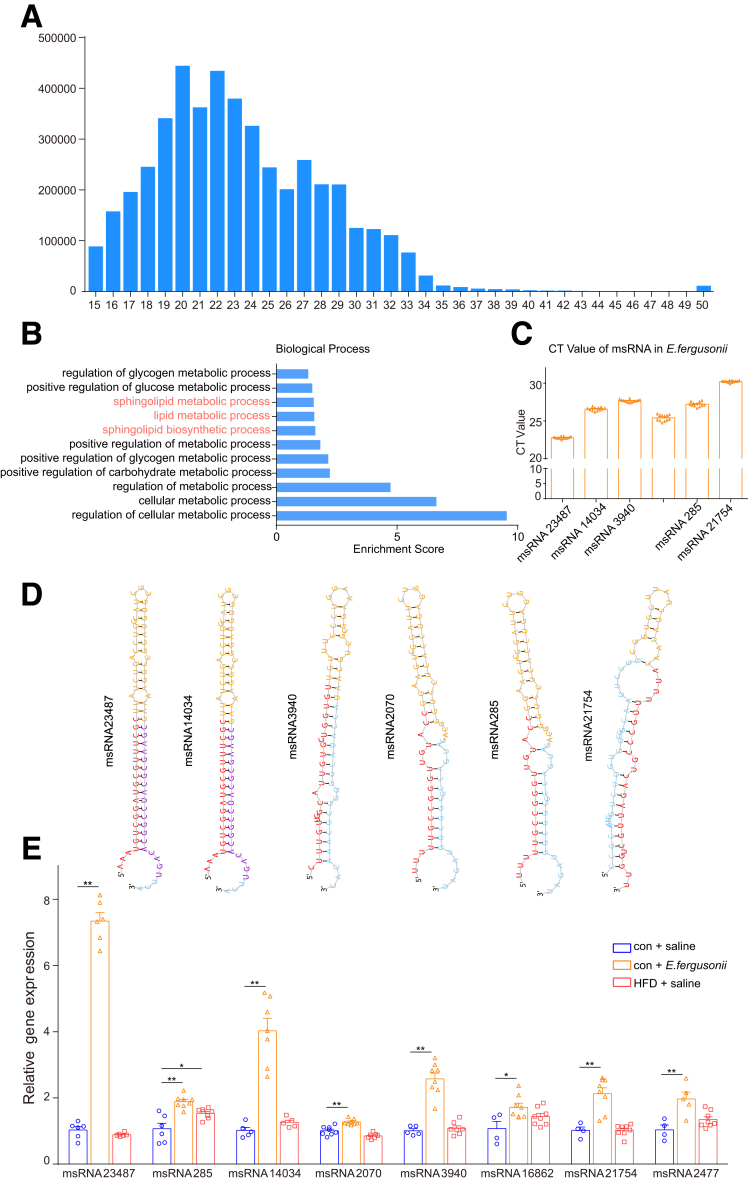
To understand the mechanisms underlying the hepatic lipid accumulation caused by E fergusonii, we next focused on E fergusonii–derived products. Recently, bacteria-derived msRNAs have been reported to regulate host physiological functions.13 To characterize the msRNA profile of E fergusonii and investigate whether it affected host lipid metabolism, we performed deep sequencing analysis on E fergusonii. As shown in Figure 6A, most reads were detected between 15 and 33 nt, which was the typical size of miRNAs. A list of msRNAs with a high miRDeep2 score or cloning copy number reads is summarized in Table 1. Among all msRNAs identified, msRNA 15919 and msRNA 23487 showed the highest miRDeep2 score and cloning copy number. Moreover, enrichment analysis of biological processes showed that msRNAs from E fergusonii had an appreciable impact on host hepatic metabolic processes, including lipid metabolism (Figure 6B). According to the number of reads and the enrichment results, msRNA 23487, msRNA 14034, msRNA 3940, msRNA 2070, msRNA 285, and msRNA 21754 were selected for further analysis. As shown in Figure 6D, we predicted the secondary structures of the precursor hairpin of the msRNAs. Furthermore, msRNA expression levels were verified by quantitative polymerase chain reaction (PCR) (Figure 6C). Among all msRNAs, msRNA 23487 had the highest expression compared with other msRNAs after E fergusonii treatment. Although all the msRNAs mentioned earlier were up-regulated after E fergusonii administration, msRNA 23487 showed the highest increase. To confirm that msRNA 23487 was secreted from E fergusonii, we mapped the whole sequence of msRNA 23487 against the database nucleotide collection (nt) using the web-based NCBI tool Nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). As we expected, msRNA 23487 was aligned to E fergusonii ATCC 35469 with 100% query cover (Table 2). Taken together, E fergusonii had a unique msRNA signature that may function to disturb host lipid metabolism.
Figure 6.
Deep sequencing analysis of E fergusonii. (A) Identification of msRNAs in E fergusonii. (B) Biological process of E fergusonii–derived msRNAs. (C) cycle threshold values of msRNAs in E fergusonii. (D) Secondary structure prediction of E fergusonii–derived msRNAs. (E) Expression levels of E fergusonii–derived msRNAs in livers from the con+saline, con+E fergusonii, and HFHC+saline groups. ∗P < .05, P > .01; ∗∗P < .01, P > .001.
Table 1.
List of msRNAs With High Numbers of Reads or High miRDeep2 Scores
| msRNA name | miRDeep2 score | Reads, n | msRNA sequence | Length | Validation |
|---|---|---|---|---|---|
| msRNA 15919 | 63,000 | 124,422 | AUUUGUGGAGCCCAUCAACC | 20 | qPCR |
| msRNA 23487 | 1100 | 2168 | AAAUGUCGGAUGCGUUUCGC | 20 | qPCR |
| msRNA 17782 | 180 | 364 | GACGACAAAUCGCUGUUUCUUGAC | 24 | |
| msRNA 10285 | 100 | 218 | CUGCGAUCUGUUAUUACUC | 1 | |
| msRNA 14555 | 96 | 194 | UUGUCGCUGCCGUCAGC | 17 | |
| msRNA 9728 | 94 | 189 | AGUUCGUCGUCGUGAAUU | 18 | qPCR |
| msRNA 285 | 1 | 558 | UUUGCCACGGAACGGUCUG | 19 | qPCR |
| msRNA 9506 | 0.6 | 4453 | UUAACGAUGACGGUCUGGUUUUACG | 25 | |
| msRNA 3940 | 0.6 | 184 | CUUUGUUGGCAUUGUGCUGGUGU | 23 | qPCR |
| msRNA 14034 | 0.5 | 439 | CUCGACGAUGCCGGGCAAUGGCCUG | 25 | qPCR |
| msRNA 21754 | 0.4 | 176 | AUUUUUGCCCGUCAGAUUGCUGGUU | 25 | qPCR |
| msRNA 25190 | 0.2 | 2871 | UAUUGCUGCUGUUGGCGCC | 19 |
qPCR, quantitative PCR.
Table 2.
Basic Local Alignment Search Tool Nucleotide (BLASTn) Results for msRNA 23487 Against the Nucleotide Collection (nt) Database With 100% Query Cover
| Description | Maximum score | Total score | Query cover | E value | Percentage identity | Accession |
|---|---|---|---|---|---|---|
| E fergusonii ATCC 35469 chromosome, complete genome | 139 | 139 | 100% | 3.00E-30 | 100.00% | NC_011740.1 |
| E fergusonii strain ATCC 35473 chromosome, complete genome | 134 | 134 | 100% | 1.00E-28 | 98.67% | NZ_CP042942.1 |
| E fergusonii strain ATCC 35470 chromosome, complete genome | 134 | 134 | 100% | 1.00E-28 | 98.67% | NZ_CP042946.1 |
| E coli strain EC42405 chromosome, complete genome | 134 | 134 | 100% | 1.00E-28 | 98.67% | NZ_CP043414.1 |
| E coli strain 2013C-4282 chromosome, complete genome | 134 | 134 | 100% | 1.00E-28 | 98.67% | NZ_CP027579.1 |
| E coli strain EC42405 chromosome, complete genome | 134 | 134 | 100% | 1.00E-28 | 98.67% | CP043414.1 |
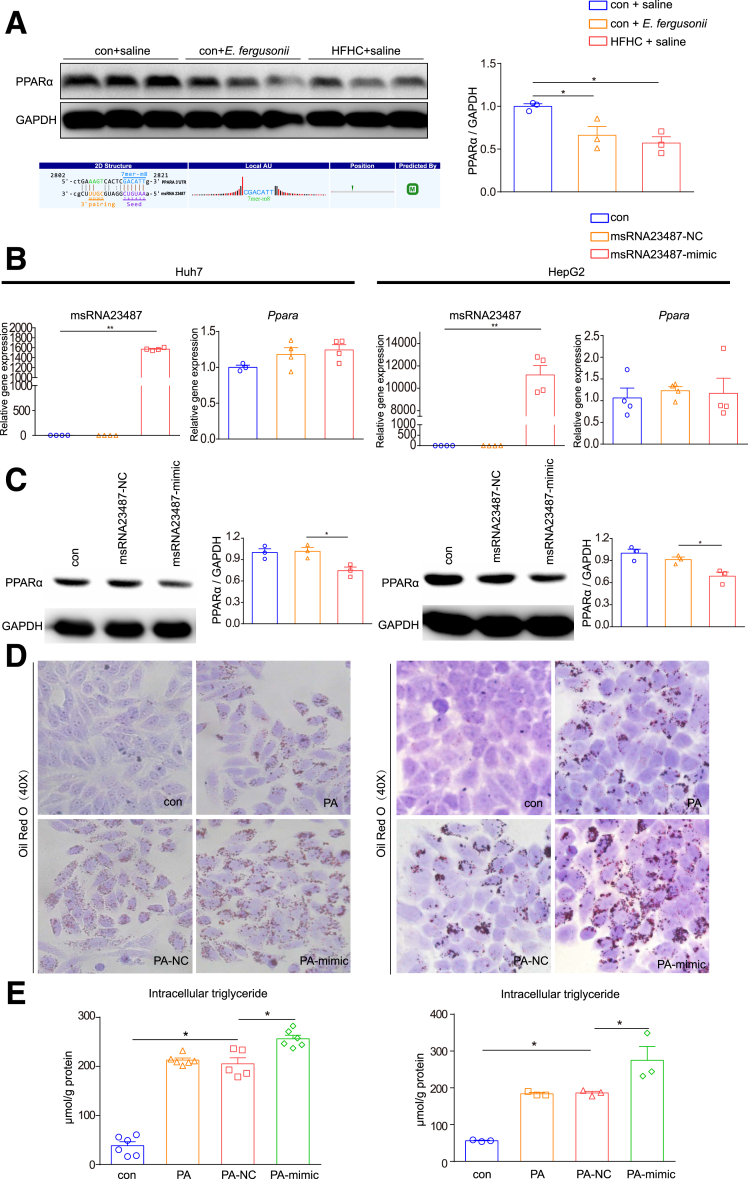
msRNA 23487 From E fergusonii Promoted Lipid Accumulation by Regulating PPARα
To test whether the increased expression of msRNA 23487 is involved in regulating host lipid metabolism, we first examined its target genes predicted by miRanda software.14 Interestingly, PPARα was predicted to be one of msRNA 23487 target genes, which was confirmed by a decreased PPARα protein level in rat livers after E fergusonii treatment (Figure 7A). We next evaluated the effect of msRNA 23487 on PPARα levels by transfecting Huh7 and HepG2 cells with msRNA 23487 mimic or the corresponding negative control (NC). The expression level of msRNA 23487 was increased significantly after transfection with the msRNA 23487 mimic. There was no change in the messenger RNA levels of Ppara, while the protein levels showed a significant reduction when Huh7 and HepG2 cells were transfected with the msRNA 23487 mimic (Figure 7B and C).
Figure 7.
msRNA 23487 from E fergusonii–induced lipid accumulation by interacting with PPARα. (A) Western blot for PPARα protein in liver. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is shown as a loading control for liver protein (upper panel); binding sites of PPARα messenger RNA with msRNA 23487 predicted by miRanda software (lower panel). (B) Expression levels of msRNA 23487 and PPARa after transfection with msRNA 23487 NC or mimic in Huh7 and HepG2 cell lines. (C) PPARα protein levels after transfection with msRNA 23487 NC or mimic in Huh7 and HepG2 cell lines. (D) Oil Red O staining after transfection with msRNA 23487 NC or mimic in Huh7 and HepG2 cell lines treated with palmitic acid (PA) at a concentration of 0.3 mmol/L. (E) Intracellular triglyceride levels after transfection with msRNA 23487 NC or mimic in Huh7 and HepG2 cell lines. ∗P < .05, P > .01; ∗∗P < .01, P > .001.
To further evaluate the function of msRNA 23487 on host hepatic lipid metabolism in vitro, we transfected Huh7 and HepG2 cells with NC or msRNA 23487 mimic co-cultured with 0.3 mmol/L palmitic acid solution. As expected, cells transfected with the msRNA 23487 mimic showed more lipid accumulation than NC cells, as shown by Oil Red O staining and triglyceride assays (Figure 7D and E). Taken together, msRNA 23487 promoted liver lipid accumulation by down-regulating PPARα expression, possibly at the post-transcriptional level.
Discussion
The role of the gut microbiome–liver axis in the pathogenesis of NAFLD has been studied predominantly in patients with metabolic abnormalities. However, the impact and precise mechanisms of gut microbiota in nonobese NAFLD are poorly understood. Herem we provide an example of the pathogenic effect of E fergusonii on inducing the development of nonobese NAFLD in rats. We also proposed a novel mechanism by which E fergusonii regulated host lipid metabolism by producing msRNA 23487, which down-regulated PPARα expression at the protein level.
There is a growing body of evidence suggesting that an overall decrease in microbial diversity and the expansion of selected bacterial families are associated with NAFLD pathogenesis.6 Among them, the most widely known contributors are Firmicutes and Bacteroidetes, the 2 major components of the human gut microbiota.15 Increased Firmicutes levels with decreased Bacteroidetes levels have been detected in a number of metabolic-related diseases including NAFLD.16,17 Consistent with previous findings,7,18 we found an increased Firmicutes:Bacteroidetes ratio and an increased abundance of Proteobacteria in patients with NAFLD, and their levels were higher in patients with obesity, inflammation, and fibrosis. Moreover, we found that Escherichia_Shigella was expanded in NAFLD patients, and its abundance was correlated significantly with NAFLD severity. Consistent with our study, Zhu et al8 also confirmed that Escherichia was the only genus that was significantly different between obese and NASH patients, suggesting a pathogenic role of Escherichia in NASH progression independent of obesity. There are 7 identified species in the genus Escherichia, of which E fergusonii initially was isolated in human blood samples in 1985.19 Because E fergusonii and E coli are congeneric species with 64% similarity in DNA hybridization, they could be difficult to differentiate using PCR methods.20 To solve this problem, we used both PCR and lactose-negative experiments to differentiate E fergusonii from E coli before the animal experiment (data not shown).
Although obesity is a common clinical phenotype associated with NAFLD, a nonobese population without obvious risk factors also can develop NAFLD. In our study, we found that administration of E fergusonii alone could induce the development of nonobese NAFLD, characterized by hepatic steatosis and hepatocellular ballooning without causing body weight gain in rats. In particular, hepatic triglyceride levels were induced significantly by E fergusonii administration, indicating a potential role in disturbing lipid metabolism to promote lipid accumulation, possibly through regulating the PPARα signaling pathway. Furthermore, we detected that a sizable fraction of animals developed advanced NAFLD, characterized by hepatic inflammation and fibrosis. These histopathologic characteristics indicated that 16 weeks of E fergusonii administration to rats induced hepatic pathology indicative of nonobese NAFLD that potentially can progress to NASH. It is possible that a higher dose or longer administration time of E fergusonii may cause a higher proportion of NASH development.
It is widely known that the gut microbiome contributes to NAFLD pathogenesis by producing microbial metabolites, such as bile acids, short-chain fatty acids, branched-chain amino acids, lipopolysaccharides, and indoles.6 In our study, we proposed a novel mechanism by which E fergusonii promotes lipid accumulation by secreting msRNAs. The existence of msRNAs has been shown in E coli and Streptococcus mutans by using a deep sequencing approach.21 To investigate whether E fergusonii exerts its functions via msRNAs, we performed deep sequencing to characterize the msRNA profile of E fergusonii in the context of NAFLD. We identified msRNA 23487 as the most up-regulated msRNA after E fergusonii administration, and msRNA 23487 secreted by E fergusonii regulated host lipid metabolism by down-regulating hepatic PPARα expression.
Our study can be improved by using third-generation sequencing to characterize bacterial signatures in healthy controls and NAFLD patients, which will provide higher resolution of microbial taxonomy compared with our current 454 pyrosequencing technology.22 Studies are needed to confirm the abundance of E fergusonii in nonobese NAFLD patients, and to further investigate whether a longer administration or higher doses will cause an advanced stage of NAFLD in a higher proportion of animals. In addition, studies are needed to validate the effects of E fergusonii in germ-free mice or mice treated with antibiotics to reduce the amount of intestinal bacteria. Last, a recent study from Tu et al23 suggested that mice fed with a HFHC cholate diet developed nonobese NAFLD in 3 weeks. Including the HFHC cholate group in our study would be informative to evaluate the role of E fergusonii in the pathogenesis of nonobese NAFLD.
In summary, we have shown that Escherichia_Shigella was increased in NAFLD patients, and its expansion was associated with disease aggravation independent of obesity. E fergusonii, an important species in the Escherichia genus, induced the development of nonobese NAFLD in rats, which is characterized by hepatic steatosis and hepatocyte ballooning with partial development of fibrosis and inflammation without weight gain. By identifying msRNA 23487 as a regulator of PPARα expression, we proposed a new mechanism by which E fergusonii regulates host lipid metabolism in nonobese NAFLD. Targeting E fergusonii and its product msRNA 23487 might contribute to the clinical management of the disease.
Materials and Methods
Patient Cohorts
A total of 54 subjects were included in this study, of which 29 subjects had biopsy-proven NAFLD and 25 subjects were healthy controls. There was no significant difference in sex or age between the NAFLD group and the healthy controls. In the NAFLD group, 15 subjects were diagnosed with overweight and 11 subjects were diagnosed as obesity according to the Chinese diagnostic criteria for obesity.24 Furthermore, 16 subjects were diagnosed with NAFL and 13 subjects were diagnosed with NASH; 18 subjects had fibrosis, and 11 subjects had no fibrosis based on the steatosis, activity, and fibrosis score.25 All subjects provided informed consent for participation before enrollment. The protocol was approved by the Ethics Committee of Xinhua Hospital affiliated with Shanghai Jiao Tong University School of Medicine.
Animal Experiments
E fergusonii (ATCC 35469) was obtained from Guangdong Culture Collection Center (China). E fergusonii for oral administration was obtained from subcultured bacterial cryopreservation in Luria–Bertani broth and suspended in sterile saline at a concentration of 3∗109 colony-forming units/mL for subsequent animal experiment use.
Specified pathogen-free male Sprague Dawley rats (Sippurbec Laboratory Animal Co, Ltd, Shanghai, China) were kept in a controlled environment with free access to water and a standard chow diet. After adaptive feeding for 7 days, rats were assigned to 3 groups according to the diet type and bacterial intervention (n = 10 per group): chow diet with saline (con+saline), chow diet with E fergusonii (con+E fergusonii), and a HFHC diet with saline (HFHC+saline). The HFHC diet contains 88% standard diet, 10% lard, and 2% cholesterol. Saline (1 mL) containing 3∗109 colony-forming units of E fergusonii was gavaged daily, starting from day 1 of the con+E fergusonii group. The con+saline and HFHC+saline groups were fed 1 mL of sterile saline. After 16 weeks, all rats were fasted overnight and killed to collect serum, liver, intestine, and fecal samples for the following analysis. All animal experiments were approved by the Institutional Animal Care and Use Committee of Xinhua Hospital affiliated with Shanghai Jiao Tong University School of Medicine and were conducted in accordance with the National Research Council Guide for Care and Use of Laboratory Animals.
Biochemical Assays
Serum ALT, AST, alkaline phosphatase, fasting blood glucose, HDL-C, LDL-C, total bile acids, homocysteine, total triglyceride, and total cholesterol were measured by a fully automatic biochemical analyzer (SYSMEX, Kobe, Japan) with 6–8 samples per experimental group according to the manufacturer’s instructions. Hepatic and cellular triglycerides and cholesterol were evaluated using triglyceride and cholesterol assay kits (E1013-105 and E1015-105; Applygen Technologies, Inc, Beijing, China). The final concentrations of triglycerides were corrected according to the protein concentration.
Histologic Analysis of Liver Tissues
Formalin-fixed tissue samples were embedded in paraffin and stained with H&E for morphologic observation. Masson trichrome staining and Sirius Red staining were performed to evaluate fibrosis. The steatosis, activity, and fibrosis score was evaluated based on histologic features. Frozen sections were embedded in optimal cutting temperature compound and stained with Oil Red O to analyze lipid accumulation.
Real-Time Quantitative PCR
Total RNA from liver or cells was extracted using TRIzol (TaKaRa, Tokyo, Japan), and complementary DNA was generated using PrimeScript RT master mix (TaKaRa, Tokyo). Poly-A modification and first-strand complementary DNA synthesis were performed with 1000 ng total RNA using the Mir-X miRNA First-Strand Synthesis Kit (TaKaRa, Tokyo, Japan) according to the manufacturer’s instructions for the reverse-transcription reactions of miRNA. Quantitative PCR was performed on the Applied Biosystems Vii7 platform using SYBR Green Master Mix (Low Rox Plus; YEASEN, Shanghai, China) for messenger RNA or SYBR Premix Ex Taq II (TaKaRa, Tokyo, Japan) for small RNA (sRNA). Primer sequences are summarized in Table 3. All primer specificities were confirmed by dissociation curves using Vii7 system SDS software (Applied Biosystems by Life Technologies, Austin, TX). Ribosomal Protein S18 or U6 was used as the internal control.
Table 3.
Primer Sequences for Quantitative PCR
| Gene name | Forward sequence | Reverse sequence |
|---|---|---|
| Rat | ||
| Ass1 | CACAGCACATCCTTGGACCTCTTC | AAGCGGTTCTCCACGATGTCAATG |
| p47phox | GCCCAAAGATGGCAAGAATA | ATGACCTCAATGGCTTCACC |
| p22phox | GTAGATGCCGCTCGCAATGGCCAG | ATGGGGCAGATCGAGTGGGCCATGT |
| p67phox | AGCAGAAGAGCAGTTAGCATTGG | TGCTTTCCATGGCCTTGTC |
| Tnf | TGCCTCAGCCTCTTCTCATT | GAGCCCATTTGGGAACTTCT |
| Il6 | AGTTGCCTTCTTGGGACTGA | CCTCCGACTTGTGAAGTGGT |
| Il1b | GAAGTCAAGACCAAAGTGG | TGAAGTCAACTATGTCCCG |
| Ccl2 | AGAATCACCAGCAGCAAGTGTCC | TTGCTTGTCCAGGTGGTCCATG |
| Ccr2 | CACCGTATGACTATGATGATG | CAGGAGAGCAGGTCAGAGAT |
| Acta2 | TGTGCTATGTCGCTCTGGAC | CCAATGAAAGATGGCTG GAA |
| Srebp1c | CCAGCCTTTGAGGATAACCA | TGCAGGTCAGACACAGGAAG |
| Scd | AGCTGGTGATGTTCCAGAGG | CAAGAAGGTGCTGACGAACA |
| Fasn | GCCTAACACCTCTGTGCAGT | GGCAATACCCGTTCCCTGAA |
| Acc | GAATATCCAGATGGCCGAGA | CCTTCTGCTCTGGCAAGTTC |
| Ppara | AATGCAATCCGTTTTGGAAG | TTGGCCAGAGATTTGAGGTC |
| Pparg | ACAAGAGCTGACCCAATGGT | GGCTCTTCATGTGGCCTGTT |
| Cpt1α | CCACGAAGCCCTCAAACAGA | CACACCCACCACCACGATAA |
| Fabp1 | GTCTGCCTGAGGACCTCATCCAG | TCATGGTCTCCAGTTCGCACTCC |
| Acadl | ACTCCGCCTCCGCTTCCATG | TACCACCGTAGATCGGCTGAACTC |
| Acadvl | TTGGAGAAGGTGGAGGAGGACAC | CTCTGCCAAGCGAGCGTACTG |
| Acsl1 | ATGCCAGAGCTGATTGACATTCGG | CAAGGACTGCTGATCTTCGGACAC |
| Fatp2 | CACGACAGAGTTGGAGACACCTTC | CCGATGCGACCTTCATGACCTG |
| Human and bacteria | ||
| PPARA | ACGATTCGACTCAAGCTGGT | GTTGTGTGACATCCCGACAG |
| E fergusonii | AGATTCACGTAAGCTGTTACCTT | CGTCTGATGAAAGATTTGGGAAG |
Western Blot Analysis
Protein levels of PPARα in rat liver tissues and cell lines (Huh7 and HepG2) were determined by Western blot analysis. Briefly, proteins were subjected to 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and electroblotted to nitrocellulose membranes. PPARα polyclonal antibody (ABclonal, Wuhan, China) and glyceraldehyde-3-phosphate dehydrogenase monoclonal antibody (Cell Signaling Technology, Boston, MA) were used as primary antibodies. Horseradish-peroxidase–conjugated anti-rabbit IgG (Beyotime, Shanghai China) was used as the secondary antibody. Immune complexes were detected using a Western chemiluminescent horseradish-peroxidase substrate (Millipore Corporation, Billerica, MA). Densitometry of immunoblot analysis was performed using ImageJ software (National Institutes of Health, Bethesda, MD).
sRNA High-Throughput Sequencing of E fergusonii
RNA sequencing, high-throughput sequencing, and subsequent bioinformatics analysis were performed by Cloud-Seq Biotech (Shanghai, China). One microgram of total RNA extracted from E fergusonii was treated with Antarctic phosphatase (New England Biolabs, Ipswich, MA) and polynucleotide kinase (PNK) enzyme (New England Biolabs). The RNA library was prepared according to the manufacturer’s instructions for the TruSeq Small RNA Kit (Illumina, San Diego, CA). The libraries were size-selected for the sRNA fraction (137–170 nt) with 15% Tris-Borate-EDTA (TBE) polyacrylamide gel. Paired-end reads were harvested from an Illumina HiSeq 4000 sequencer and quality controlled by Q30. After 3’ adaptor trimming, low-quality reads were removed with cutadapt software (v1.9.3).26 sRNA candidates are predicted using an sRNA scanner based on transcriptional signals. sRNAs overlapping with known genes were excluded, and the rest were added to the gene annotation of the genome. The clean reads were aligned to the reference genome using Hisat2 software.27 HTSeq was used to count the read numbers mapped to each gene.28 edgeR was used to normalize the read counts for each library.29 Sequence data are available in the NCBI SRA with BioProject PRJNA747160.
Bacterial DNA Extraction and 16S rRNA Sequencing
For healthy controls and NAFLD patients, DNA was extracted from fecal samples. V3/V5 hypervariable regions of the 16S rDNA gene were amplified and sequenced on the 454 GS-FLX platform (Roche, Branford, CT) as previously described.18 For rats, total genomic DNA from fecal samples was extracted using a DNA Extraction Kit according to the manufacturer’s instructions. The V3–V4 region of the 16S rRNA gene was amplified, purified, and sequenced on an Illumina MiSeq Platform (Illumina). Paired-end reads were processed using Trimmomatic software to detect and cut off ambiguous bases.30 After trimming, paired-end reads were assembled using FLASH software.31 Reads with 75% of bases above Q20 were retained, and reads with chimeras were detected and removed using QIIME software (version 1.8.0).32 Clean reads were subjected to primer sequence removal and clustering to generate operational taxonomic units using Vsearch software with a 97% similarity cut-off level.33 The representative read of each operational taxonomic unit was selected using the QIIME package. All representative reads were annotated and blasted against the Silva database using the ribosomal database project (RDP) classifier (confidence threshold, 70%).34 The metabolic function of the gut microbiota was analyzed using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States.35 Sequence data were registered at NCBI under BioProject PRJNA746446 (rats) and PRJNA747151 (human beings).
Cell Culture and sRNA Mimic Transfection
Huh7 and HepG2 cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum (Gibco by Invitrogen, Carlsbad, CA) was used to culture cells under an atmosphere of 5% CO2 at 37°C. Palmitic acid powder (Sigma-Aldrich, St. Louis, MO) was dissolved in 1% fatty acid–free bovine serum albumin at 65°C and filtered through a 0.22-μm filter at a concentration of 5 mmol/L. The final concentration of palmitic acid used for experiments was 0.3 mmol/L. Huh7 or HepG2 cells were transfected with 50 nmol/L msRNA 23487 mimic or NC (RiboBio, Guangzhou, China) by Lipofectamine 3000 in Opti-MEM (Gibco) according to the manufacturer’s instructions. Experiments were performed 24 hours after transfection.
Statistical Analysis
Results are expressed as the means ± SEM. Statistical analysis was performed using analysis of variance and Student–Newman–Keuls multiple-range tests. Differences were considered significant at P < .05. Linear discriminant effect-size analysis was performed to determine the features most likely to account for differences between groups.36
Acknowledgments
The authors want to thank Lian Li and Jia-lin Yu (Oebiotech, China) for the gut microbiome analysis; Zhi-cai Yu, Nan-nan Zheng, and Dai-xi Jiang for their help with molecular biology experiments; as well as Dr Wei Cai for providing an excellent experimental platform and guidance.
CRediT Authorship Contributions
Feng-Zhi Xin (Data curation: Lead; Formal analysis: Lead; Writing – original draft: Lead)
Ze-Hua Zhao (Investigation: Equal; Methodology: Equal)
Xiao-Lin Liu (Investigation: Equal; Methodology: Equal)
Qin Pan (Investigation: Supporting)
Zi-Xuan Wang (Investigation: Supporting)
Lin Zeng (Data curation: Equal)
Qian-Ren Zhang (Methodology: Equal)
Lin Ye (Methodology: Equal)
Meng-Yu Wang (Methodology: Equal)
Rui-Nan Zhang (Investigation: Supporting)
Zi-Zhen Gong (Resources: Equal)
Lei-Jie Huang (Methodology: Equal)
Chao Sun (Resources: Equal)
Feng Shen (Resources: Equal)
Lu Jiang (Conceptualization: Equal; Investigation: Equal; Writing – review & editing: Lead)
Jian-Gao Fan, MD (Conceptualization: Lead; Project administration: Lead; Supervision: Lead; Writing – review & editing: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was supported by the National Key R&D Program of China (2017YFC0908903), and the National Natural Science Foundation of China (81873565, 81470840, and 81700503).
Contributor Information
Lu Jiang, Email: jianglu@xinhuamed.com.cn.
Jian-Gao Fan, Email: fanjiangao@xinhuamed.com.cn.
References
- 1.Wong R.J., Aguilar M., Cheung R., Perumpail R.B., Harrison S.A., Younossi Z.M., Ahmed A. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology. 2015;148:547–555. doi: 10.1053/j.gastro.2014.11.039. [DOI] [PubMed] [Google Scholar]
- 2.Diehl A.M., Day C. Cause, pathogenesis, and treatment of nonalcoholic steatohepatitis. N Engl J Med. 2017;377:2063–2072. doi: 10.1056/NEJMra1503519. [DOI] [PubMed] [Google Scholar]
- 3.Kim D., Kim W.R. Nonobese fatty liver disease. Clin Gastroenterol Hepatol. 2017;15:474–485. doi: 10.1016/j.cgh.2016.08.028. [DOI] [PubMed] [Google Scholar]
- 4.Das K., Das K., Mukherjee P.S., Ghosh A., Ghosh S., Mridha A.R., Dhibar T., Bhattacharya B., Bhattacharya D., Manna B., Dhali G.K., Santra A., Chowdhury A. Nonobese population in a developing country has a high prevalence of nonalcoholic fatty liver and significant liver disease. Hepatology. 2010;51:1593–1602. doi: 10.1002/hep.23567. [DOI] [PubMed] [Google Scholar]
- 5.Cusi K. Nonalcoholic steatohepatitis in nonobese patients: not so different after all. Hepatology. 2017;65:4–7. doi: 10.1002/hep.28839. [DOI] [PubMed] [Google Scholar]
- 6.Sharpton S.R., Schnabl B., Knight R., Loomba R. Current concepts, opportunities, and challenges of gut microbiome-based personalized medicine in nonalcoholic fatty liver disease. Cell Metab. 2021;33:21–32. doi: 10.1016/j.cmet.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou D., Pan Q., Xin F.Z., Zhang R.N., He C.X., Chen G.Y., Liu C., Chen Y.W., Fan J.G. Sodium butyrate attenuates high-fat diet-induced steatohepatitis in mice by improving gut microbiota and gastrointestinal barrier. World J Gastroenterol. 2017;23:60–75. doi: 10.3748/wjg.v23.i1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu L., Baker S.S., Gill C., Liu W., Alkhouri R., Baker R.D., Gill S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 9.Ghosal A., Upadhyaya B.B., Fritz J.V., Heintz-Buschart A., Desai M.S., Yusuf D., Huang D., Baumuratov A., Wang K., Galas D., Wilmes P. The extracellular RNA complement of Escherichia coli. Microbiologyopen. 2015;4:252–266. doi: 10.1002/mbo3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaastra W., Kusters J.G., van Duijkeren E., Lipman L.J. Escherichia fergusonii. Vet Microbiol. 2014;172:7–12. doi: 10.1016/j.vetmic.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 11.Buzzetti E., Pinzani M., Tsochatzis E.A. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD) Metabolism. 2016;65:1038–1048. doi: 10.1016/j.metabol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Liss K.H., Finck B.N. PPARs and nonalcoholic fatty liver disease. Biochimie. 2017;136:65–74. doi: 10.1016/j.biochi.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watkins D., Arya D.P. Regulatory roles of small RNAs in prokaryotes: parallels and contrast with eukaryotic miRNA. Non-coding RNA Investig. 2019;3:28. [Google Scholar]
- 14.Betel D., Koppal A., Agius P., Sander C., Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bäckhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 16.Qian M., Hu H., Zhao D., Wang S., Pan C., Duan X., Gao Y., Liu J., Zhang Y., Yang S., Qi L.-W., Wang L. Coordinated changes of gut microbiome and lipidome differentiates nonalcoholic steatohepatitis (NASH) from isolated steatosis. Liver Int. 2020;40:622–637. doi: 10.1111/liv.14316. [DOI] [PubMed] [Google Scholar]
- 17.Mouzaki M., Comelli E.M., Arendt B.M., Bonengel J., Fung S.K., Fischer S.E., McGilvray I.D., Allard J.P. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120–127. doi: 10.1002/hep.26319. [DOI] [PubMed] [Google Scholar]
- 18.Shen F., Zheng R.-D., Sun X.-Q., Ding W.-J., Wang X.-Y., Fan J.-G. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat Dis Int. 2017;16:375–381. doi: 10.1016/S1499-3872(17)60019-5. [DOI] [PubMed] [Google Scholar]
- 19.Farmer J.J., 3rd, Fanning G.R., Davis B.R., O'Hara C.M., Riddle C., Hickman-Brenner F.W., Asbury M.A., Lowery V.A., 3rd, Brenner D.J. Escherichia fergusonii and Enterobacter taylorae, two new species of Enterobacteriaceae isolated from clinical specimens. J Clin Microbiol. 1985;21:77–81. doi: 10.1128/jcm.21.1.77-81.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence J.G., Ochman H., Hartl D.L. Molecular and evolutionary relationships among enteric bacteria. J Gen Microbiol. 1991;137:1911–1921. doi: 10.1099/00221287-137-8-1911. [DOI] [PubMed] [Google Scholar]
- 21.Kang S.M., Choi J.W., Lee Y., Hong S.H., Lee H.J. Identification of microRNA-size, small RNAs in Escherichia coli. Curr Microbiol. 2013;67:609–613. doi: 10.1007/s00284-013-0411-9. [DOI] [PubMed] [Google Scholar]
- 22.Johnson J.S., Spakowicz D.J., Hong B.Y., Petersen L.M., Demkowicz P., Chen L., Leopold S.R., Hanson B.M., Agresta H.O., Gerstein M., Sodergren E., Weinstock G.M. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10:5029. doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tu L.N., Showalter M.R., Cajka T., Fan S., Pillai V.V., Fiehn O., Selvaraj V. Metabolomic characteristics of cholesterol-induced non-obese nonalcoholic fatty liver disease in mice. Sci Rep. 2017;7:6120. doi: 10.1038/s41598-017-05040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C., Lu F.C. The guidelines for prevention and control of overweight and obesity in Chinese adults. Biomed Environ Sci. 2004;17(Suppl):1–36. [PubMed] [Google Scholar]
- 25.Bedossa P., Poitou C., Veyrie N., Bouillot J.L., Basdevant A., Paradis V., Tordjman J., Clement K. Histopathological algorithm and scoring system for evaluation of liver lesions in morbidly obese patients. Hepatology. 2012;56:1751–1759. doi: 10.1002/hep.25889. [DOI] [PubMed] [Google Scholar]
- 26.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:1–10. [Google Scholar]
- 27.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyon D., Tsai S.Q., Khayter C., Foden J.A., Sander J.D., Joung J.K. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A., Clemente J.C., Burkepile D.E., Vega Thurber R.L., Knight R., Beiko R.G., Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]