Abstract
The incidence of tuberculosis in Norway is one of the lowest in the world, and approximately half of the cases occur in first- and second-generation immigrants. In the present study, the genetic diversity of 92% of all strains of Mycobacterium tuberculosis isolated in Norway in 1994 to 1998 was assessed using restriction fragment length polymorphism (RFLP) analysis, with the insertion sequence IS6110 and the repetitive element DR as probes, to determine the degree of active transmission between patients. The DR probe was used as a secondary molecular marker to support or rule out clustering of strains with fewer than five copies of IS6110. After exclusion of 20 cultures representing laboratory contamination, 573 different IS6110 patterns were found among the 698 strains analyzed. Of these 573 patterns, 542 were observed only once and 31 were shared by 2 to 14 isolates. Among 81 strains (11.5%) carrying fewer than five copies of IS6110, 56 RFLP patterns were found when the results of both the IS6110 and DR methods were combined. Among the 698 strains, 570 were considered to be independent cases. A total of 14.5% of the native Norwegians and 19.7% of the foreign patients were part of a cluster. Thus, the degree of recent transmission of tuberculosis in Norway is low and the great majority of the cases are due to reactivation of previous disease. Transmission between immigrants and native Norwegians is uncommon. Two outbreaks, one among native Norwegians and one mainly among immigrants, have been ongoing for several years, indicating that, even in a low-incidence country such as Norway, with a good national program for tuberculosis surveillance, certain transmission chains are difficult to break.
In recent years, DNA fingerprinting of Mycobacterium tuberculosis based on restriction fragment length polymorphism (RFLP) using IS6110 as a probe has been performed on isolates from various parts of the world (1, 3, 8–11, 13, 15, 21, 25–30, 34–36). The stability and reproducibility of the technique, as well as its usefulness in epidemiological studies, have been convincingly demonstrated (16, 17, 32, 33), and new insight into the nature of tuberculosis transmission has been obtained. Furthermore, RFLP has become an indispensable tool for quality assurance of the processing and culturing of patient samples, since it offers an opportunity to verify suspected cases of cross-contamination (2, 5, 27).
However, isolates of M. tuberculosis that possess few copies of IS6110 do not generate sufficient polymorphism to be readily distinguished by this technique (33); furthermore, a few strains of M. tuberculosis lack IS6110. Therefore, for strains with fewer than five copies of IS6110, other fingerprinting techniques must be employed to ascertain the epidemiological relationships between cases.
Norway has a population of 4.48 million people. Unemployment is low, representing 3.1% of the work force. In the last part of the 1990s, 5.5% of the population were immigrants (defined as persons born in a foreign country) and 0.14% of the population were homeless. Approximately 10,000 people were active injecting drug users and 25,000 individuals were treated for abuse of alcohol and drugs annually (23). The incidence of AIDS is low in Norway: from 1983 until the end of 1998, a total of 639 patients were reported to have developed AIDS. Of these, 509 had died by 31 December 1998 (20). From 1947 to 1995, M. bovis BCG vaccination was obligatory for 12- to 14-year-old children. The incidence of tuberculosis in Norway declined until the middle of the 1980s. Since then, the decline has stopped, mainly due to immigration. While in 1975, 5% of the tuberculosis patients in Norway were of foreign origin, this percentage had increased to 53% in 1998 (13, 19). Despite this increase, the incidence of tuberculosis is still low, with 200 to 250 cases per year (incidence, <5 cases per 100,000 inhabitants). The aims of the present study were to determine the genetic diversity of the population of M. tuberculosis in Norway and to detect the degree of active tuberculosis transmission between patients. In addition, we wanted to assure the quality of the processing and culturing work performed by checking isolates for possible cross-contamination.
MATERIALS AND METHODS
Patient population and bacterial isolates.
The study population comprised 92% of all patients in Norway from whom at least one sample positive for M. tuberculosis by culture was collected from 1994 to 1998. A total of 19 microbiological laboratories, servicing the entire nation, performed the isolation of M. tuberculosis from patient samples. The strains were collected at the National Institute of Public Health in Oslo, which serves as a National Reference Laboratory for tuberculosis. In this period, a total of 816 isolates of M. tuberculosis recovered from 717 different patients were received and analyzed consecutively. Patient information was obtained from the records of the National Health Screening Service, which collects data on tuberculosis patients in Norway.
The species identification of the isolates was based on a 16S rRNA gene hybridization technique (AccuProbe; GenProbe Inc., San Diego, Calif.) and standard microbiological tests.
RFLP analyses.
Chromosomal DNA of the isolates was prepared as described by van Soolingen et al. (31). The RFLP analyses included IS6110 probing for all strains and direct-repeat (DR) probing for strains with fewer than five copies of IS6110 (31). The IS6110 probe is a 245-bp PCR-amplified probe directed against the right arm of IS6110 (32). The DR probe is a 36-bp synthetic oligonucleotide directed against the directly repeated sequences of 36 bp which are clustered in one region of the M. tuberculosis genome and are interspersed with nonrepetitive sequences of 36 to 41 bp (14).
The DNA was digested with the restriction endonuclease PvuII prior to hybridization to the IS6110 probe and with AluI before hybridization to the DR probe. An external 1-kb ladder (Boehringer, Mannheim, Germany) was included in the first, middle, and last lanes of each gel. After separation by electrophoresis, Southern blotting was performed as described by van Soolingen et al. (31). The IS6110 probe was labeled with the digoxigenin-dUTP labeling and detection kit (Boehringer), and the DR probe was labeled with the enhanced chemiluminescence kit (ECL; Amersham International plc, Little Chalfont, United Kingdom). After hybridization, the insertion sequences and DR sequences were visualized following the recommendations of the kit's manufacturer.
The IS6110 fingerprint patterns were compared by visual examination and computer-assisted analyses by use of the GelCompar version 4.1 software (Applied Maths, Kortrijk, Belgium). To facilitate the comparison of the fingerprints, normalization was done using the molecular weight standards on each gel. Band position tolerance was set up to 0.80%, and the optimization was 0.50%. Similarity measures were calculated using the Dice coefficient. Cluster analysis was performed using the unweighted pair-group average method. The patterns obtained with the DR sequence were compared by visual examination. A cluster of isolates was defined as two or more isolates which exhibited 100% identical RFLP patterns.
RESULTS
The number of cases of tuberculosis reported annually in Norway from 1994 to 1998 varied between 205 and 244 (results not shown). Approximately 80% of these were new cases (12, 18, 19). Between 127 and 171 cases were bacteriologically verified for each of these years. A total of 816 strains of M. tuberculosis were isolated from 717 patients, and all strains were fingerprinted and hybridized to the IS6110 probe. The number of IS6110 copies varied from 0 to 19. The isolates without IS6110 copies were verified as M. tuberculosis by sequence analysis of the 16S rRNA gene as described elsewhere (22).
Multiple isolates from individual patients were usually identical, and one strain per patient was further considered for analysis. The only exception was one 64-year-old man of Pakistani origin, who contracted tuberculosis twice during the period. In 1997, a strain of M. tuberculosis that carried a unique RFLP pattern, based on three copies of IS6110 and three copies of the DR sequence, was isolated from this patient. One year later, he was infected with a strain that carried no copies of IS6110. This latter strain was isolated repeatedly on different days and consistently failed to hybridize with the IS6110 probe. The DR pattern of the latter isolates from this patient was also different, thus confirming that the second strain represented exogenous reinfection.
Detection of laboratory cross-contamination.
Of the 718 strains, a total of 20 isolates (2.8%) were suspected to be laboratory cross-contamination. Such contamination was suspected when two or more strains with identical fingerprint patterns were received from the same laboratory within a short period of time and with no epidemiological data connecting the patients. The clinical presentation of the patients and additional laboratory data were considered to determine the most likely actual patient in each case. Laboratory contamination was found in nine different laboratories, and the 20 strains were excluded from further analyses.
Patient origin and age.
After exclusion of the suspected laboratory cross-contaminants, 698 strains from 697 patients remained. A total of 331 (47.5%) of these were native Norwegians, and 366 (52.5%) were first- or second-generation immigrants. The immigrants originated from Somalia (n = 92), Pakistan (n = 51), Vietnam (n = 40), the former Yugoslavia (n = 32), India (n = 17), The Philippines (n = 12), Sri Lanka (n = 11), Ethiopia (n = 9), Thailand (n = 8), China (n = 6), and 35 other countries (n = 53). For 35 patients, the nationality was unknown, but based on their names, they were considered to be immigrants.
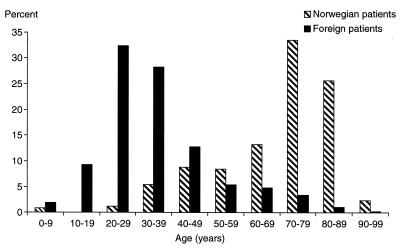
The age distribution of the patients is shown in Fig. 1. About 75% of the patients of Norwegian origin and 10% of the patients of foreign origin were more than 60 years of age. The patients less than 40 years of age represented 72% of the immigrant group and less than 8% of the Norwegians.
FIG. 1.
Age distribution of Norwegian and foreign patients with bacteriologically verified tuberculosis in Norway, 1994 to 1998.
DNA polymorphism of M. tuberculosis strains.
Among the 698 strains, 573 different IS6110 patterns were found. Of these, 542 were observed only once and 31 were shared by two or more isolates (Tables 1 and 2). Two clusters of 27 and 15 strains harbored a single copy of IS6110 on a DNA fragment of the same size. The next most common pattern, characterized by 15 copies of IS6110, included 14 strains. The rate of diversity of the patterns obtained by RFLP analysis with IS6110 for all of the M. tuberculosis strains in this survey was 82.1%. Forty-eight (14.5%) of Norwegian patients and 72 (19.7%) of the foreign patients were infected with an M. tuberculosis strain which was part of a cluster. Overall, 17.2% of the patients were part of a cluster.
TABLE 1.
Clustering of M. tuberculosis isolates in Norway from 1994 to 1998 based on IS6110 and DR fingerprinting
| Clustering using IS6110 probe
|
Clustering using DR probe
|
||||||
|---|---|---|---|---|---|---|---|
| No. of IS6110 copies | No. of isolates | No. of clusters | Cluster sizea | No. of nonclustered isolates | No. of clusters | Cluster sizea | Country of origin of patients in clusters (no. of patients) |
| 0 | 4 | 2 | 1 | 2 | Vietnam (1), Pakistan (1) | ||
| 1 | 54 | 4 | 27 | 15 | 5 | 3 | Somalia (3) |
| 3 | Sri Lanka (2), Pakistan (1) | ||||||
| 2 | Somalia (1), Vietnam (1) | ||||||
| 2 | Vietnam (1), Thailand (1) | ||||||
| 2 | Somalia (1), Ethiopia (1) | ||||||
| 2 | 2 | 0 | |||||
| 15 | 4 | 2 | 9 | Vietnam (9) | |||
| 2 | Somalia (2) | ||||||
| 10 | 2 | 1 | 8 | Ethiopia (1), Norway (4), Somalia (3) | |||
| 2 | 6 | 0 | 6 | 0 | |||
| 3 | 11 | 1 | 2 | 9 | 1 | 2 | Somalia (2) |
| 4 | 6 | 1 | 3 | 6 | 0 | ||
Cluster size is expressed as the number of isolates per cluster.
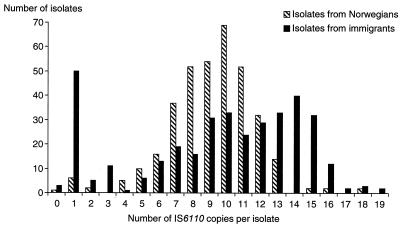
The distribution of IS6110 copies among isolates from patients of foreign and Norwegian backgrounds is shown in Fig. 2. High-copy-number and low-copy-number strains were more commonly isolated from immigrants than from patients of Norwegian background. Of the 81 strains (11.5%) with fewer than five copies of IS6110, 71 were recovered from first- or second-generation immigrants (87.5%).
FIG. 2.
Numbers of IS6110 elements in M. tuberculosis isolates from Norwegian and foreign patients.
The DR-RFLP pattern was used as a secondary molecular marker to support or rule out clustering of the 81 strains with fewer than five copies of IS6110. Among these 81 isolates, 24 IS6110 patterns and 40 DR patterns were found. When the results of the two probes were combined, a total of 56 RFLP patterns were distinguished (Table 1).
Cluster analysis.
Based on the combined IS6110 and DR analyses, a total of 570 isolates (81.7%) had unique fingerprints. These strains were considered to be independent cases of infection. The remaining 128 patients (18.3%) had strains belonging to 1 of 35 clusters, suggesting that they belonged to groups of individuals among whom tuberculosis was recently transmitted. Clusters contained 2 to 14 isolates each (mean, 3.7 patients). Sixteen clusters contained more than two patients.
For the strains with fewer than five copies of IS6110, we considered strains to be part of a cluster when both fingerprinting techniques generated identical results for two or more strains. In some cases, DR fingerprinting gave identical patterns for isolates from patients of the same ethnic origin, even when the IS6110 patterns were different. This was, for example, the case for the isolates from 11 Vietnamese patients who all carried the same four-copy DR pattern. Nine of these strains harbored one identical copy of IS6110, one strain carried a different-sized IS6110 copy, and one isolate carried an additional copy of IS6110.
The four strains that did not hybridize to the IS6110 probe demonstrated three different DR patterns. The two strains with identical DR patterns (Table 1) were recovered from patients in the same hospital, but the sputum specimens had been processed six months apart. One patient was of Vietnamese origin, and the other was the 64-year-old man from Pakistan previously described. Of the two remaining strains, one was recovered from the sputum of another patient of Vietnamese origin and the other was from the feces of a 6-year-old Norwegian girl with long-lasting, diffuse gastrointestinal pain after extensive but inconclusive preceding examinations. The child presented no clinical signs of tuberculosis and was not registered as a tuberculosis patient.
One cluster contained 27 strains that all had one copy of IS6110 on a DNA fragment of the same size. These strains were isolated from patients from eight different countries with ages ranging from 13 to 73 years. When the analysis of DR elements was carried out, this group was subdivided in five clusters and 15 unique strains. The sizes of the clusters and the origins of the patients in the clusters are shown in Table 1.
Another group of 15 isolates carried one IS6110 band of the same size. These strains demonstrated six RFLP patterns when hybridized to the DR probe. They constituted two clusters based on the combined IS6110 and DR results (Table 1). Eight of 10 isolates that clustered together with one identical IS6110 band were confirmed to be identical by the DR probe (Table 1).
The IS6110 and DR fingerprinting results were in agreement for all strains that contained two and three copies of IS6110 (Table 1). Of the strains that had four copies of IS6110, none were found to be identical by DR probing. Three Norwegian patients, living in different parts of the country and with no epidemiological connection between them, were clustered according to their IS6110 RFLP but harbored different DR patterns (Table 1).
The clusters identified among the strains carrying more than four copies of IS6110 are presented in Table 2. Patients who were part of an active chain of transmission were generally of the same ethnic origin. Clustered isolates from native Norwegians often included those from patients with a history of tuberculosis, social difficulties, or abuse of alcohol or drugs.
TABLE 2.
Distribution of RFLP patterns of isolates of M. tuberculosis with more than four copies of IS6110 isolated from patients in Norway from 1994 to 1998a
| No. of IS6110 copies | No. of isolates | No. of clusters | No. of nonclustered isolates | Cluster sizeb | Geographic origin (ages or age range of patients [yr]) and comments about patients in clusters |
|---|---|---|---|---|---|
| 5 | 16 | 2 | 8 | 4 | Norway (74–89), 2 diagnosed with TBc in 1939 and 1950 |
| 4 | Former Yugoslavia (24–46), family | ||||
| 6 | 28 | 1 | 24 | 4 | Somalia (21–33) |
| 7 | 55 | 5 | 40 | 6 | Norway (44–83), family and friends, 1 diagnosed with TB in 1959, social difficulties, alcohol and drug abuse |
| 3 | Unknown (27–34) | ||||
| 2 | Norway (35, 35), sisters | ||||
| 2 | Uganda (36, 52) | ||||
| 2 | Former Yugoslavia (22, 34) | ||||
| 8 | 64 | 2 | 56 | 6 | Norway (33–66), friends, two diagnosed HIV+d, 1 traveled frequently to Thailand |
| 2 | Norway (80, 81) | ||||
| 9 | 83 | 2 | 78 | 3 | Somalia (19–24) |
| 2 | Norway (73, 84) | ||||
| 10 | 97 | 3 | 89 | 4 | Norway (45–79) |
| 2 | Norway (44, 51), one confirmed drug abuser | ||||
| 2 | Somalia (13), unknown (17) | ||||
| 11 | 75 | 5 | 55 | 12 | Norway (36–86), family and friends, social difficulties, alcohol abuse |
| 2 | Norway (62, 84) | ||||
| 2 | Norway (42, 88) | ||||
| 2 | Norway (67, 81), TB diagnosed in 1930s and 1941 | ||||
| 2 | Turkey (20, 24) | ||||
| 12 | 58 | 3 | 50 | 4 | Norway (39–50), social difficulties, alcohol abuse |
| 2 | Norway (82), Somalia (29) | ||||
| 2 | Sri Lanka (43, 54) | ||||
| 13 | 45 | 0 | 45 | ||
| 14 | 39 | 0 | 39 | ||
| 15 | 34 | 2 | 17 | 14 | 11 from Somalia (21–35), 2 from Ethiopia (31, 32), Norway (1) |
| 3 | India (34), Pakistan (24), unknown (3) | ||||
| 16 | 14 | 0 | 14 | ||
| 17 | 2 | 0 | 2 | ||
| 18 | 5 | 0 | 5 | ||
| 19 | 2 | 0 | 2 |
The non-Norwegian patients included first- and second-generation immigrants.
Cluster size is expressed as the number of isolates per cluster.
TB, tuberculosis.
HIV+, human immunodeficiency virus positive.
One cluster included 12 Norwegian patients within a family and group of friends with social and alcohol problems. For older Norwegian patients, reactivation of previous epidemic strains was the most common cause of tuberculosis.
The largest cluster identified during this period included 14 strains isolated from 11 Somali patients, two Ethiopians, and a Norwegian 1-year-old boy. No epidemiological connection was found between the immigrants and the Norwegian patient. This cluster was confirmed by DR fingerprinting. Disturbingly, all of these were resistant to isoniazid, including six multidrug-resistant strains (resistant to at least isoniazid and rifampin).
DISCUSSION
The population of M. tuberculosis analyzed in this study represents 92% of all of the bacteriologically verified cases of tuberculosis in Norway. This includes 63% of all of the tuberculosis patients reported to the National Health Screening Service (12, 13, 18, 19). Thus, the isolates studied are representative of the population of M. tuberculosis present in Norway from 1994 to 1998. RFLP analysis of M. tuberculosis applying the IS6110 element as a probe is an internationally established method well suited to the study of the epidemiology and transmission of tuberculosis (1, 3, 8–11, 13, 15, 17, 21, 25–29, 34–36; van Embden and van Soolingen, Editorial, Int. J. Tuberc. Lung Dis. 4:285–286, 2000). It enabled us to define the magnitude of tuberculosis transmission in Norway during this period and to identify the active chains of transmission. In addition, it was possible to calculate the genetic diversity present in this population of M. tuberculosis and compare it with that of other populations.
The occurrence of false-positive cultures may have serious clinical consequences (5). In a recent review, it was demonstrated that this is a common problem and that the median false-positive rate is 3.1% (interquartile range, 2.2 to 10.5%) (5). In our study, we found that 2.8% of the cases represented laboratory cross-contamination. Of the 20 false-positive patients, 8 (40%) were treated for tuberculosis. In an attempt to eliminate this problem, DNA fingerprinting results are now reported monthly to the National Health Screening Service. We believe that positive cultures are evaluated more critically and that clinical and laboratory staff exercise intensified care to avoid cross-contamination.
No international consensus has yet been reached regarding a secondary method for typing of strains with few copies of IS6110. Thus, isolates that carry no or only a few copies of IS6110 are often excluded from studies (2, 10, 25, 34, 35), since IS6110 is not sufficiently powerful to discriminate among them (6, 7, 24, 32). Previous studies have demonstrated that the DR probe could discriminate among such strains and should corroborate IS6110 results when strains are truly epidemiologically related (17, 28). Nowadays, low-IS6110-copy-number strains are commonly differentiated using a commercial kit for spoligotyping. In this study, 11.5% of the strains carried fewer than five copies of IS6110. By use of DR fingerprinting, it was possible to further subgroup them. When the DR and IS6110 patterns were combined, the results correlated with the epidemiological data.
The present study demonstrated a great diversity of IS6110 and DR fingerprints among isolates of M. tuberculosis in Norway. On this basis, it was clear that there was a low degree of active transmission of tuberculosis between patients. Thus, it was confirmed that tuberculosis in a low-incidence country such as Norway is mostly due to reactivation (13). The largest cluster identified by IS6110 fingerprinting included 14 patients, of whom the first was identified in 1994. This outbreak was still ongoing by the end of the year 2000, and by then it included 20 patients, 11 of whom were infected with multidrug-resistant M. tuberculosis (results not shown).
The high diversity among isolates of M. tuberculosis in Norway is in contrast to the situation in Denmark, a country with a public social security system and standard of living that are comparable to those of Norway. In Denmark, 49% of the population was part of a cluster (3). The difference in diversity between the populations of M. tuberculosis in the two countries was unexpected but may reflect the fact that the population density of Denmark (5.33 million/43,094 km2) is higher than that of Norway (4.48 million/324,220 km2). Also, in the Danish study (3), two large clusters (n = 110 and 90) were evidenced while no outbreak of such a magnitude was observed in Norway. The diversity of M. tuberculosis is also lower in many other countries than that in Norway (8, 10, 11, 28, 29, 34–36). The proportion of tuberculosis patients in Norway that was part of a cluster was similar to the situation in Pisa, Italy (15%) (9), and Zurich, Switzerland (17.5%) (21).
A majority of isolates from patients of foreign origin exhibited unique RFLP patterns, and only 19.7% were part of a cluster. This demonstrated that also among immigrants tuberculosis was due mainly to reactivation of a previous infection. Many immigrants in Norway come from high-incidence countries. A public concern that increased immigration may result in increased transmission of tuberculosis has arisen. We found little evidence of such transmission in the period studied. In Norway, as in Denmark (3), there was a low degree of transmission between natives and foreigners. This was in contrast to a study in The Netherlands, where approximately half of the transmission to native Dutch citizens was from immigrants (4).
A detailed tuberculosis surveillance in Norway is possible due to the small population size and low incidence of disease. The results of the present study support previous findings and the statement of van Soolingen and van Embden that the incidence of active transmission in Norway is extremely low (13; van Embden and van Soolingen, Editorial). Relatively few patients (both native and foreign born) were reported to have had a recent infection. Also, the spread of imported tuberculosis by immigrants plays virtually no role in Norway. It was still difficult to break certain transmission chains, even in a country like Norway, with a good national program of tuberculosis surveillance, effective treatment, and contact tracing.
ACKNOWLEDGMENTS
We thank Berit Gregussen, Anne M. Klem, Elisabet Rønnild, Solveig Undseth, and Ingun Ytterhaug for excellent technical assistance and Nanne Brathås and Vigdis Dahl for help in data collection.
This work was supported by a grant from Laurine Maarschalk's fund to D.A.C.
REFERENCES
- 1.Alland D, Kalkut G E, Moss A R, McAdam R A, Hahn J A, Bosworth W, Drucker E, Bloom B R. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N Engl J Med. 1994;330:1710–1716. doi: 10.1056/NEJM199406163302403. [DOI] [PubMed] [Google Scholar]
- 2.Bauer J, Thomsen V O, Poulsen S, Andersen A B. False-positive results from cultures of Mycobacterium tuberculosis due to laboratory cross-contamination confirmed by restriction fragment length polymorphism. J Clin Microbiol. 1997;35:988–991. doi: 10.1128/jcm.35.4.988-991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer J, Yang Z, Poulsen S, Andersen A B. Results from 5 years of nationwide DNA fingerprinting of Mycobacterium tuberculosis complex isolates in a country with a low incidence of M. tuberculosis infection. J Clin Microbiol. 1998;36:305–308. doi: 10.1128/jcm.36.1.305-308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borgdorff W M, Nagelkerke N, Van Soolingen D, De Haas P E, Veen J, van Embden J D A. Analysis of tuberculosis transmission between nationalities in The Netherlands in the period 1993 to 1995 using DNA fingerprinting. Am J Epidemiol. 1998;147:187–195. doi: 10.1093/oxfordjournals.aje.a009433. [DOI] [PubMed] [Google Scholar]
- 5.Burman W J, Reves R R. Review of false-positive cultures of Mycobacterium tuberculosis and recommendations for avoiding unnecessary treatment. Clin Infect Dis. 2000;31:1390–1395. doi: 10.1086/317504. [DOI] [PubMed] [Google Scholar]
- 6.Burman W J, Reves R R, Hawkes A P, Rietmeijer C A, Yang Z, El Hajj H, Bates J H, Cave M D. DNA fingerprinting with two probes decreases clustering of Mycobacterium tuberculosis. Am J Respir Crit Care Med. 1997;155:1140–1146. doi: 10.1164/ajrccm.155.3.9117000. [DOI] [PubMed] [Google Scholar]
- 7.Chaves F, Yang Z H, El-Hajj H, Alonso M, Burman W J, Eisenach K D, Dronda F, Bates J H, Cave M D. Usefulness of the secondary probe pTBN12 in DNA fingerprinting of Mycobacterium tuberculosis. J Clin Microbiol. 1996;34:1118–1123. doi: 10.1128/jcm.34.5.1118-1123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevrel-Dellagi D, Abderrahman A, Haltiti R, Koubaji H, Gicquel B, Dellagi K. Large-scale DNA fingerprinting of Mycobacterium tuberculosis strains as a tool for epidemiological studies of tuberculosis. J Clin Microbiol. 1993;31:2446–2450. doi: 10.1128/jcm.31.9.2446-2450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garzelli C, Lari N, Nguon B, Pistello M, Falcone G. DNA restriction fragment length polymorphism of Mycobacterium tuberculosis isolates in Pisa, Italy. Eur J Epidemiol. 1997;13:845–851. doi: 10.1023/a:1007337319454. [DOI] [PubMed] [Google Scholar]
- 10.Gutiérrez M C, Vincent V, Aubert D, Bizet J, Gaillot O, Lebrun L, Le Pendeven C, Le Pennec M P, Mathieu D, Offredo C, Pangon B, Pierre-Audigier C. Molecular fingerprinting of Mycobacterium tuberculosis and risk factors for tuberculosis transmission in Paris, France, and surrounding area. J Clin Microbiol. 1998;36:486–492. doi: 10.1128/jcm.36.2.486-492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas W H, Engelmann G, Amthor B, Shyamba S, Mugala F, Felten M, Rabbow M, Leichsenring M, Oosthuizen O J, Bremer H J. Transmission dynamics of tuberculosis in a high-incidence country: prospective analysis by PCR DNA fingerprinting. J Clin Microbiol. 1999;37:3975–3979. doi: 10.1128/jcm.37.12.3975-3979.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heldal E. Tuberculosis in Norway 1995–1996. MSIS-rapport. 1997;25:44. [Google Scholar]
- 13.Heldal E, Döcker H, Caugant D A, Tverdal A. Pulmonary tuberculosis in Norwegian patients. The role of reactivation, reinfection and primary infection assessed by previous mass screening data and restriction fragment length polymorphism analysis. Int J Tuberc Lung Dis. 2000;4:300–307. [PubMed] [Google Scholar]
- 14.Hermans P W M, van Soolingen D, Bik E M, De Haas P E W, Dale J W, van Embden J D A. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermans P W, Messadi F, Guebrexabher H, van Soolingen D, de Haas P E, Heersma H, de Neeling H, Ayoub A, Portaels F, Frommel D, et al. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia, and The Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J Infect Dis. 1995;171:1504–1513. doi: 10.1093/infdis/171.6.1504. [DOI] [PubMed] [Google Scholar]
- 16.Hermans P W, van Soolingen D, Dale J W, Schuitema A R, McAdam R A, Catty D, van Embden J D. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J Clin Microbiol. 1990;28:2051–2058. doi: 10.1128/jcm.28.9.2051-2058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kremer K, van Soolingen D, Frothingham R, Haas W H, Hermans P W M, Martín C, Palittapongarnpim P, Plikaytis B B, Riley L W, Yakrus M A, Musser J M, van Embden J D A. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;37:2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Health Screening Service. Tuberculosis in Norway 1996–1997. Rep Syst Commun Dis. 1998;26:21. [Google Scholar]
- 19.National Health Screening Service. Tuberculosis in Norway 1997–1998. Resistens. 1999;3:5. [Google Scholar]
- 20.Nilsen Ø, Aavitsland P, Lystad A. The AIDS-situation in Norway per December 31st 1998. MSIS-rapport. 1999;27:8. [Google Scholar]
- 21.Pfyffer G E, Strassle A, Rose N, Wirth R, Brandli O, Shang H. Transmission of tuberculosis in the metropolitan area of Zurich: a 3-year survey based on DNA fingerprinting. Eur Respir J. 1998;11:804–808. doi: 10.1183/09031936.98.11040804. [DOI] [PubMed] [Google Scholar]
- 22.Rogall T, Flohr T, Böttger E C. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J Gen Microbiol. 1990;136:1915–1920. doi: 10.1099/00221287-136-9-1915. [DOI] [PubMed] [Google Scholar]
- 23.Rognerud M, Strand B H, Hesselberg Ø. The health of disadvantaged groups in Norway. Norwegian country report for the EU project. Oslo, Norway: National Institute of Public Health; 2000. [Google Scholar]
- 24.Sahadevan R, Narayanan S, Paramasivan C N, Prabhakar R, Narayanan P R. Restriction fragment length polymorphism typing of clinical isolates of Mycobacterium tuberculosis from patients with pulmonary tuberculosis in Madras, India, by use of direct-repeat probe. J Clin Microbiol. 1995;33:3037–3039. doi: 10.1128/jcm.33.11.3037-3039.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samper S, Iglesias M J, Rabanaque M J, Leacano M A, Vitoria L A, Rubio M C, Gómez-Lus R, Gómez L I, Oral I, Martin C. The molecular epidemiology of tuberculosis in Zaragoza, Spain: a retrospective epidemiological study in 1993. Int J Tuberc Lung Dis. 1998;2:281–287. [PubMed] [Google Scholar]
- 26.Small P M, Hopewell P C, Singh S P, Paz A, Parsonnet J, Ruston D C, Schecter G F, Daley C L, Schoolnik G K. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N Engl J Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 27.Small P M, McClenny N B, Singh S P, Schoolnik G K, Tompkins L S, Mickelsen P A. Molecular strain typing of Mycobacterium tuberculosis to confirm cross-contamination in the mycobacteriology laboratory and modification of procedures to minimize occurrence of false-positive cultures. J Clin Microbiol. 1993;31:1677–1682. doi: 10.1128/jcm.31.7.1677-1682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sola C, Horgen L, Goh K S, Rastogi N. Molecular fingerprinting of Mycobacterium tuberculosis on a Caribbean island with IS6110 and DRr probes. J Clin Microbiol. 1997;35:843–846. doi: 10.1128/jcm.35.4.843-846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torrea G, Levee G, Grimont P, Martin C, Chanteau S, Gicquel B. Chromosomal DNA fingerprinting analysis using the insertion sequence IS6110 and the repetitive element DR as strain-specific markers for epidemiological study of tuberculosis in French Polynesia. J Clin Microbiol. 1995;33:1899–1904. doi: 10.1128/jcm.33.7.1899-1904.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Soolingen D, de Haas P E W, Hermans P W M, van Embden J D A. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 1994;235:196–205. doi: 10.1016/0076-6879(94)35141-4. [DOI] [PubMed] [Google Scholar]
- 32.van Soolingen D, de Haas P E W, Hermans P W M, Groenen P M A, van Embden J D A. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Soolingen D, Hermans P W, de Haas P E, Soll D R, van Embden J D. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warren R, Hauman J, Beyers N, Richardson M, Schaaf H S, Donald P, Van Helden P. Unexpectedly high strain diversity of Mycobacterium tuberculosis in a high-incidence community. S Afr Med J. 1996;86:45–49. [PubMed] [Google Scholar]
- 35.Yang Z, Barnes P F, Chaves F, Eisenach K D, Weis S E, Bates J H, Cave M D. Diversity of DNA fingerprints of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol. 1998;36:1003–1007. doi: 10.1128/jcm.36.4.1003-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z H, Mtoni I, Chonde M, Mwasekaga M, Fuursted K, Askgård D S, Bennedsen J, de Haas P E, van Soolingen D, van Embden J D, Andersen A B. DNA fingerprinting and phenotyping of Mycobacterium tuberculosis isolates from human immunodeficiency virus (HIV)-seropositive and HIV-seronegative patients in Tanzania. J Clin Microbiol. 1995;33:1064–1069. doi: 10.1128/jcm.33.5.1064-1069.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]