Abstract
The K+-Cl– cotransporter KCC2, encoded by the Slc12a5 gene, is a neuron-specific chloride extruder that tunes the strength and polarity of GABAA receptor-mediated transmission. In addition to its canonical ion transport function, KCC2 also regulates spinogenesis and excitatory synaptic function through interaction with a variety of molecular partners. KCC2 is enriched in the vicinity of both glutamatergic and GABAergic synapses, the activity of which in turn regulates its membrane stability and function. KCC2 interaction with the submembrane actin cytoskeleton via 4.1N is known to control its anchoring near glutamatergic synapses on dendritic spines. However, the molecular determinants of KCC2 clustering near GABAergic synapses remain unknown. Here, we used proteomics to identify novel KCC2 interacting proteins in the adult rat neocortex. We identified both known and novel candidate KCC2 partners, including some involved in neuronal development and synaptic transmission. These include gephyrin, the main scaffolding molecule at GABAergic synapses. Gephyrin interaction with endogenous KCC2 was confirmed by immunoprecipitation from rat neocortical extracts. We showed that gephyrin stabilizes plasmalemmal KCC2 and promotes its clustering in hippocampal neurons, mostly but not exclusively near GABAergic synapses, thereby controlling KCC2-mediated chloride extrusion. This study identifies gephyrin as a novel KCC2 anchoring molecule that regulates its membrane expression and function in cortical neurons.
SIGNIFICANCE STATEMENT Fast synaptic inhibition in the brain is mediated by chloride-permeable GABAA receptors (GABAARs) and therefore relies on transmembrane chloride gradients. In neurons, these gradients are primarily maintained by the K/Cl cotransporter KCC2. Therefore, understanding the mechanisms controlling KCC2 expression and function is crucial to understand its physiological regulation and rescue its function in the pathology. KCC2 function depends on its membrane expression and clustering, but the underlying mechanisms remain unknown. We describe the interaction between KCC2 and gephyrin, the main scaffolding protein at inhibitory synapses. We show that gephyrin controls plasmalemmal KCC2 clustering and that loss of gephyrin compromises KCC2 function. Our data suggest functional units comprising GABAARs, gephyrin, and KCC2 act to regulate synaptic GABA signaling.
Keywords: cerebral cortex, chloride, GABA transmission, KCC2, proteomics, synaptic transmission
Introduction
Fast inhibitory synaptic transmission in the brain is mediated by chloride-permeable GABAA and glycine receptors. To maintain a hyperpolarizing driving force for transmembrane chloride fluxes through these receptors, neurons are equipped with a unique chloride extrusion capacity mediated by the K/Cl cotransporter KCC2, a member of the Slc12 cation-coupled chloride transporters (Chamma et al., 2012; Kaila et al., 2014a). The delayed expression onset of KCC2 is responsible for the progressive hyperpolarizing shift in the polarity of GABA signaling in the developing brain (Rivera et al., 1999). Conversely, downregulation of KCC2 expression in a variety of neurologic and neuropsychiatric conditions is associated with paradoxical, depolarizing actions of GABA signaling that may underlie pathologic network activities (Cohen et al., 2002; Coull et al., 2003; Huberfeld et al., 2007; Kahle et al., 2008; Boulenguez et al., 2010; Pallud et al., 2014; Tyzio et al., 2014; Kaila et al., 2014a; Banerjee et al., 2016).
In addition to its canonical role in regulating transmembrane chloride gradients, KCC2 is involved in numerous biological processes, independently of its ion transport function (Chamma et al., 2012; Blaesse and Schmidt, 2015; Chevy et al., 2020). Thus, KCC2 expression is required for spinogenesis during neuronal maturation (Li et al., 2007; Fiumelli et al., 2013; Awad et al., 2018), controls the confinement of postsynaptic glutamate receptors within dendritic spines (Gauvain et al., 2011), and gates long-term potentiation at glutamatergic synapses (Chevy et al., 2015). These effects involve the interaction of the large C-terminal domain (CTD) of KCC2 with actin-related proteins, such as the FERM domain, spectrin/actin-binding protein 4.1N (Li et al., 2007), and the Rac1-specific guanine nucleotide exchange factor βPIX (Chevy et al., 2015; Llano et al., 2015), which controls actin dynamics through regulation of cofilin activity. In addition, KCC2 regulates neuronal excitability through interaction with the two-pore leak potassium channel Task-3 (Goutierre et al., 2019) and is involved in molecular interactions that regulate its traffic and membrane expression (Ivakine et al., 2013; Mahadevan et al., 2017; Pressey et al., 2017).
The multiple functions of KCC2 imply that its membrane expression is tightly regulated at the subcellular level. Although KCC2 shows diffuse expression throughout the somatodendritic plasma membrane (Gulyás et al., 2001; Baldí et al., 2010; Gauvain et al., 2011; Chamma et al., 2013), it also forms perisynaptic but not postsynaptic aggregates near both GABAergic (Gulyás et al., 2001; Chamma et al., 2013) and glutamatergic (Gulyás et al., 2001; Gauvain et al., 2011) synapses. Such clustering may be directly related to KCC2 function, as its specific disruption with no change in total membrane expression results in a loss of KCC2 transport activity in hippocampal neurons (Watanabe et al., 2009). The interaction of KCC2-CTD with 4.1N near glutamatergic synapses hinders KCC2 membrane diffusion and likely participates in its clustering. Blocking KCC2-4.1N interaction by overexpressing KCC2-CTD, knocking down 4.1N expression, or pharmacologically disrupting actin polymerization results in reduced dwell time of KCC2 in the vicinity of glutamatergic synapses (Chamma et al., 2013). However, these manipulations do not affect KCC2 dwell time near GABAergic synapses, suggesting the existence of synapse-specific molecular interactions contributing to the formation of perisynaptic KCC2 clusters. Although KCC2 was shown to interact with both GABAA (Huang et al., 2012; Wan et al., 2020) and GABAB (Wright et al., 2017) receptors, the molecular mechanisms of KCC2 clustering near GABAergic synapses remain to be identified. Here, we addressed this issue using proteomic analysis of the KCC2 interactome in cortical principal neurons. Of the 271 candidate KCC2 partners identified, we focused on the scaffolding protein gephyrin, which is known to stabilize GABAA and glycine receptors at inhibitory synapses (Feng et al., 1998; Kneussel et al., 1999; Lévi et al., 2004; Jacob et al., 2005; Yu et al., 2007). We confirmed gephyrin interaction with native KCC2 in cortical neurons and showed that gephyrin tightly regulates KCC2 membrane expression and clustering, notably but not exclusively near GABAergic synapses. Furthermore, gephyrin knockdown was associated with a significant reduction in transmembrane chloride extrusion. These results identify gephyrin as a major KCC2 interacting protein that regulates its membrane expression and function in cortical neurons.
Materials and Methods
Animals.
Adult male Sprague Dawley rats were obtained from Janvier Labs. All procedures conformed to the International Guidelines on the ethical use of animals and to the French Agriculture and Forestry Ministry guidelines for handling animals (decree 87 849, license A 75–05-22), and were approved by the Charles Darwin Ethics Committee (APAFIS2015111011588776v3). All efforts were made to minimize animal suffering and to reduce the number of animals used.
Virus and stereotaxic viral injection.
Rat KCC2-Flag sequence (Chamma et al., 2013) was introduced in a pAAV vector under CaMKII promoter (pAAV-CW3SL-EGFP; plasmid #61463, Addgene) and used to produce purified adenoviral particles (Atlantic Gene Therapy).
The 4- to 5-week-old rats were anesthetized by intraperitoneal injection of ketamine/xylazine (75/10 mg/kg) and subcutaneous injection of buprenorphine (0.05 mg/kg) for analgesia. Animals were then head fixed in a stereotaxic apparatus. One microliter of adeno-associated virus (AAV) suspension (concentrated at 7.3 × 1012 transduction units/ml) was injected unilaterally in the cortex at four injection sites [(1) anteroposterior (AP), +2 mm; mediolateral (ML), ±2.5 mm; dorsoventral (DV), −2 mm; (2) AP, −0.5 mm; ML, ±2.5 mm; DV, −1.8 mm; (3) AP, −3 mm; ML, ±2.5 mm; DV, −1.2 mm; (4) AP, −5.5 mm; ML, ±2.5 mm; DV, −1.2 mm] at a rate of 125 nl/min.
Immunohistochemistry.
KCC2-Flag expression was verified at least 2 weeks following viral injections. Rats were anesthetized by intraperitoneal ketamine/xylazine injection (110/15 mg/kg) and perfused transcardially with ice-cold choline-based solution containing the following (in mm): 110 choline Cl, 25 glucose, 25 NaHCO3, 11.6 ascorbic acid, 3.1 pyruvic acid, 1.25 NaH2PO4, 2.5 KCl, 0.5 CaCl2, and 7 MgCl2 saturated with 95% O2/5% CO2. Brains were removed, postfixed in 4% paraformaldehyde for 48 h and stored at 4°C in 30% sucrose PBS. Parasagittal sections (40 µm thick) were obtained using a cryotome. After washes, brain slices were incubated for 3 h with 0.3% Triton X-100 and 10% goat serum in PBS, and then for 48 h at 4°C with primary antibodies: KCC2 (rabbit, 2.5 µg/ml; catalog #07–432, Merck) and Flag (mouse, 2.5 µg/ml; M2 clone, catalog #F3165, Merck). After washing in PBS, slices were incubated for 3 h with secondary antibodies (Cy3-coupled goat anti-mouse and Alexa Fluor 647-coupled goat anti-rabbit, 3 µg/ml; Jackson ImmunoResearch), washed with PBS and mounted with Mowiol/Dabco (25 mg/ml). Images were acquired with a DM6000-2 upright epifluorescence microscope with a 12 bit cooled CCD camera (Micromax, Roper Scientific) run with MetaMorph software (Roper Scientific).
Ex vivo KCC2 coimmunoprecipitation.
Rat cortices were dissected and homogenized by sonication in coimmunoprecipitation buffer (1 ml/100 mg tissue) containing (in mm): 50 Tris-HCl pH 7.4, 150 NaCl, 1 EDTA, as well as 1% Triton X-100 and protease inhibitors, and solubilized by rotation for 4–6 h at 4°C. After centrifugation at 20,000 × g for 40 min at 4°C, the protein content of the supernatant was measured by BCA Protein Assay (Thermo Fisher Scientific). Solubilized proteins (10 mg) were incubated overnight at 4°C with 50 µl of anti-Flag magnetic beads (Sigma-Aldrich). After two 30 min washes at 4°C with coimmunoprecipitation buffer containing 1% Triton X-100, beads were washed twice with detergent-free coimmunoprecipitation buffer, first for 30 min at 4°C and then overnight at 4°C. Immunoprecipitated proteins were gently eluted by rotating beads for 1 h at room temperature in 57 µl of elution buffer containing 250 mm Tris-HCl, pH 6.8, 1% SDS, 20% glycerol, and bromophenol blue, to limit the amount of bait in the eluate (EL), reduce the concomitant elution of IgGs and avoid interference with subsequent identification of partners by MS. Eluates were transferred in a new tube where SDS and β-mercaptoethanol were added for a 1 h denaturation at 37°C. A control Western blot was performed with 50 µg of lysate as input (IN), 10% of the EL, 5% of the total flow-through (FT) fraction, and all material remaining bound to the beads after mild elution. The latter was obtained by denaturing solubilization for 1 h at 37°C in Laemmli buffer.
Mass spectrometry analysis.
Proteins were separated by SDS-PAGE (4–15% polyacrylamide; Mini-PROTEAN TGX Precast Gels, BIO-RAD) and stained with Protein Staining Solution (Euromedex). Gel lanes were cut into five gel pieces and destained with 50 mm triethylammonium bicarbonate/50% acetonitrile and three washes in 100% acetonitrile. Proteins were digested In-Gel using trypsin (1 µg/band; Gold, Promega) as described previously (Thouvenot et al., 2008).
The generated peptides were analyzed online by nano-flow liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) using a Quadrupole-Orbitrap mass spectrometer (Q-Exactive +, Thermo Fisher Scientific) coupled to an Ultimate RSLC (Thermo Fisher Scientific). Peptides were introduced onto the column in buffer A (0.1% formic acid) and separated with a 103 min gradient of 5–40% of buffer B (80% acetonitrile, 0.1% formic acid), both at a flow rate of 300 nl/min. Eluted peptides were electrosprayed online at a voltage of 1.8 kV into the Q-Exactive+ programmed in data-dependent acquisition mode. Full scans [375–1500 mass/charge ratio (m/z)] were acquired in the Orbitrap mass analyzer with a resolution of 70,000 at 200 m/z. For the full scans, 3E6 ions were accumulated within a maximum injection time of 60 ms and detected in the Orbitrap analyzer. The 12 most intense ions with charge states ≥2 were sequentially isolated to a target value of 1e5 with a maximum injection time of 45 ms and fragmented by HCD in the collision cell (normalized collision energy, 28%) and detected in the Orbitrap analyzer at 17,500 resolution.
Raw MS spectra were processed using the MaxQuant environment (version 1.5.5.1; Cox and Mann, 2008) and Andromeda for database search (Cox et al., 2011). The MS/MS spectra were matched against the UniProt Reference Proteome (Proteome ID UP000002494) of Rattus norvegicus (release 2017_12; http://www.uniprot.org) and 250 frequently observed contaminants, as well as reversed sequences of all entries. The following settings were applied: spectra were searched with a mass tolerance of 7 ppm (MS) and 0.5 m/z (MS/MS). Enzyme specificity was set to trypsin/P, and the search included oxidation of methionine and acetylation (protein N-term) as variable modifications. Up to two missed cleavages were allowed for protease digestion. The false discovery rate was set at 0.01 for peptides and proteins, and the minimal peptide length was set at 7.
The representative protein ID in each protein group was automatically selected using the in-house developed Leading tool (version 3.2; Raynaud et al., 2018). Experiments were repeated three times to assess biological reproducibility. Only proteins with a greater than twofold enrichment (based on the MS/MS ratio) and one unique peptide per protein were considered as potential partners of KCC2 (Table 1).
Table 1.
KCC2 Protein partners identified by MS
| Gene name | UniProt ID | Protein name | TM | Ratio KCC2/sham MS-MS count |
Mean ratio KCC2/sham. MS-MS count | ||
|---|---|---|---|---|---|---|---|
| N1 | N2 | N3 | |||||
| * Slc12a5 | Q63633 | Solute Carrier Family 12 Member 5 | x | 214.0 | 11.7 | 77.4 | 101.0 |
| * Canx | P35565 | Calnexin | x | 4.4 | 1.9 | 27.0 | 11.1 |
| Npepps | F1M9V7 | Aminopeptidase Puromycin Sensitive | x | 3.0 | 1.7 | 28.0 | 10.9 |
| Dpysl5 | Q9JHU0 | Dihydropyrimidinase-Like 5 | 2.0 | 1.5 | 21.0 | 8.2 | |
| * Jagn1 | Q4KM64 | Jagunal Homolog 1 | x | 5.0 | 17.0 | 2.0 | 8.0 |
| Tkt | G3V826 | Transketolase | 1.7 | 2.1 | 20.0 | 7.9 | |
| Opa1 | Q2TA68 | Opa1, Mitochondrial Dynamin Like Gtpase | x | 2.5 | 1.0 | 20.0 | 7.8 |
| * Sfpq | A0A0G2K8K0 | Splicing Factor Proline and Glutamine Rich | 4.0 | 1.1 | 18.0 | 7.7 | |
| Immt | A0A0G2JVH4 | Inner Membrane Mitochondrial Protein | x | 2.0 | 1.0 | 20.0 | 7.7 |
| Slc3a2 | A0A0H2UHQ0 | Solute Carrier Family 3 Member 2 | 2.7 | 1.3 | 19.0 | 7.6 | |
| * Cpsf6 | D3ZPL1 | Cleavage and Polyadenylation Specific factor 6 | 4.0 | 1.6 | 15.0 | 6.9 | |
| Hsp90b1 | Q66HD0 | Heat Shock Protein 90 β Family Member 1 | 5.0 | 1.1 | 14.0 | 6.7 | |
| Eef2 | P05197 | Eukaryotic Translation Elongation factor 2 | 3.5 | 1.6 | 14.0 | 6.4 | |
| Qars | Q66H61 | Glutaminyl-Trna Synthetase | 2.0 | 1.0 | 16.0 | 6.3 | |
| Ptprs | Q64605 | Protein Tyrosine Phosphatase, Receptor Type, S | x | 1.6 | 3.3 | 14.0 | 6.3 |
| Jup | Q6P0K8 | Junction Plakoglobin | 3.0 | 0.7 | 15.0 | 6.2 | |
| Slc25a12 | F1LX07 | Solute Carrier Family 25 Member 12 | x | 2.5 | 0.8 | 15.0 | 6.1 |
| Adam22 | F1M542 | Adam Metallopeptidase Domain 22 | 3.0 | 1.0 | 14.0 | 6.0 | |
| Uggt1 | Q9JLA3 | Udp-Glucose Glycoprotein Glucosyltransferase 1 | 3.0 | 1.0 | 13.0 | 5.7 | |
| Hspa4l | F7F2F3 | Heat Shock Protein 4-Like | 3.5 | 1.4 | 12.0 | 5.6 | |
| Acsl6 | P33124 | Acyl-Coa Synthetase Long-Chain Family Member 6 | x | 3.0 | 1.3 | 12.0 | 5.4 |
| Gucy1b3 | P20595 | Guanylate Cyclase 1 Soluble Subunit β 3 | 4.0 | 1.2 | 11.0 | 5.4 | |
| Gys1 | A2RRU1 | Glycogen Synthase 1 | 4.0 | 0.9 | 11.0 | 5.3 | |
| Dhx36 | D4A2Z8 | Deah-Box Helicase 36 | 6.0 | 1.4 | 8.0 | 5.1 | |
| * Slc8a2 | P48768 | Solute Carrier Family 8 Member A2 | x | 7.0 | 1.3 | 7.0 | 5.1 |
| Slc27a1 | Q6GMM8 | Solute Carrier Family 27 Member 1 | 3.0 | 1.3 | 11.0 | 5.1 | |
| Letm1 | Q5XIN6 | Leucine Zipper and Ef-Hand Containing Transmembrane Protein 1 | x | 1.0 | 2.1 | 12.0 | 5.0 |
| Aldh1l1 | P28037 | Aldehyde Dehydrogenase 1 Family, Member L1 | 5.0 | 1.1 | 9.0 | 5.0 | |
| Mars | D3Z941 | Methionyl-Trna Synthetase | 2.0 | 0.9 | 12.0 | 5.0 | |
| * Sec23a | A0A0G2JZM2 | Sec23 Homolog A, Coat Complex II Component | 2.0 | 0.9 | 12.0 | 5.0 | |
| Rph3a | F1LPB9 | Rabphilin 3A | 3.7 | 0.9 | 10.0 | 4.9 | |
| Sirpa | P97710 | Signal-Regulatory Protein α | x | 2.0 | 3.8 | 8.0 | 4.6 |
| Trim28 | O08629 | Tripartite Motif-Containing 28 | 1.0 | 2.5 | 10.0 | 4.5 | |
| Kpnb1 | F2Z3Q8 | Karyopherin Subunit β 1 | 1.3 | 2.0 | 10.0 | 4.4 | |
| Ago1 | D4AC38 | Argonaute 1, Risc Catalytic Component | 3.3 | 0.8 | 9.0 | 4.4 | |
| Elob | P62870 | Transcription Elongation factor B Subunit 2 | 10.0 | 2.3 | 0.7 | 4.3 | |
| Slitrk5 | M0R9U3 | Slit and Ntrk-Like Family, Member 5 | 3.0 | 0.9 | 9.0 | 4.3 | |
| Exoc3 | Q62825 | Exocyst Complex Component 3 | 3.0 | 0.8 | 9.0 | 4.3 | |
| Syn3 | O70441 | Synapsin III | 2.0 | 0.8 | 10.0 | 4.3 | |
| Sf3b1 | G3V7T6 | Splicing factor 3B, Subunit 1 | 4.0 | 0.7 | 8.0 | 4.2 | |
| Scyl1 | Q5M9F8 | Scy1 Like Pseudokinase 1 | 2.7 | 0.9 | 9.0 | 4.2 | |
| Srp72 | D4A7R0 | Signal Recognition Particle 72 | 2.0 | 1.5 | 9.0 | 4.2 | |
| * Gls | P13264 | Glutaminase | 4.0 | 1.4 | 7.0 | 4.1 | |
| Pspc1 | Q4KLH4 | Paraspeckle Component 1 | 1.0 | 2.0 | 9.0 | 4.0 | |
| Mta2 | B2GV01 | Metastasis Associated 1 Family, Member 2 | 4.0 | 0.9 | 7.0 | 4.0 | |
| Dlg4 | P31016 | Discs Large Maguk Scaffold Protein 4 | 2.0 | 0.9 | 9.0 | 4.0 | |
| Rpn2 | P25235 | Ribophorin II | x | 4.0 | 1.4 | 6.0 | 3.8 |
| N/A | A0A0G2JY03 | Uncharacterized protein | 2.0 | 1.2 | 8.0 | 3.7 | |
| FbxO21 | F1M5Q6 | F-Box Protein 21 | 4.5 | 0.7 | 6.0 | 3.7 | |
| Trim32 | Q66H79 | Tripartite Motif-Containing 32 | 2.0 | 1.2 | 8.0 | 3.7 | |
| Cadm2 | Q1WIM2 | Cell Adhesion Molecule 2 | x | 4.0 | 3.0 | 4.0 | 3.7 |
| Copg1 | Q4AEF8 | Coatomer Protein Complex, Subunit γ 1 | 2.0 | 1.0 | 8.0 | 3.7 | |
| Pygm | G3V8V3 | Phosphorylase, Glycogen, Muscle | 2.0 | 0.9 | 8.0 | 3.6 | |
| Hnrnpl | F1LQ48 | Heterogeneous Nuclear Ribonucleoprotein L | 4.0 | 0.7 | 6.0 | 3.6 | |
| Rap1gap | F1LV89 | Rap1 Gtpase-Activating Protein | 2.7 | 0.8 | 7.0 | 3.5 | |
| Cttn | Q66HL2 | Cortactin | 3.5 | 0.9 | 6.0 | 3.5 | |
| Plaa | P54319 | Phospholipase A2, Activating Protein | 6.0 | 1.4 | 3.0 | 3.5 | |
| Vps33a | Q63615 | Vps33A Corvet/Hops Core Subunit | 2.3 | 0.9 | 7.0 | 3.4 | |
| Khsrp | Q99PF5 | Kh-type Splicing Regulatory Protein | 2.0 | 1.1 | 7.0 | 3.4 | |
| Upf1 | A0A0U1RS25 | Upf1, Rna Helicase and Atpase | 2.0 | 1.1 | 7.0 | 3.4 | |
| Ndufs2 | Q641Y2 | Nadh Dehydrogenase Ubiquinone Fe-S Protein 2 | 6.0 | 2.0 | 2.0 | 3.3 | |
| Hyou1 | Q63617 | Hypoxia Up-Regulated 1 | 1.0 | 2.0 | 7.0 | 3.3 | |
| Iars | A0A0G2JVL8 | Isoleucyl-Trna Synthetase | 2.0 | 1.0 | 7.0 | 3.3 | |
| Mut | D3ZKG1 | Methylmalonyl Coa Mutase | 2.0 | 1.0 | 7.0 | 3.3 | |
| Faah | P97612 | Fatty Acid Amide Hydrolase | x | 1.0 | 3.0 | 6.0 | 3.3 |
| * Adgrl2 | O88923 | Adhesion G Protein-Coupled Receptor L2 | x | 1.0 | 3.0 | 6.0 | 3.3 |
| Eif2ak3 | Q9Z1Z1 | Eukaryotic Translation Initiation factor 2 Alpha Kinase 3 | x | 4.0 | 0.9 | 5.0 | 3.3 |
| Rangap1 | F1MAA5 | Ran Gtpase Activating Protein 1 | 2.0 | 0.7 | 7.0 | 3.2 | |
| * Prrt2 | D3ZFB6 | Proline-Rich Transmembrane Protein 2 | x | 2.0 | 2.6 | 5.0 | 3.2 |
| Aak1 | P0C1X8 | Ap2 Associated Kinase 1 | 2.0 | 1.8 | 5.8 | 3.2 | |
| Palm | Q920Q0 | Paralemmin | 3.0 | 1.1 | 5.5 | 3.2 | |
| Apoe | P02650 | Apolipoprotein E | 3.0 | 6.0 | 0.3 | 3.1 | |
| Ivns1abp | A0A0G2K427 | Influenza Virus Ns1A Binding Protein | 2.4 | 0.8 | 6.0 | 3.1 | |
| Slain2 | G3V6A2 | Slain Motif Family, Member 2 | 2.0 | 3.0 | 4.0 | 3.0 | |
| Dnm3 | Q08877 | Dynamin 3 | 1.0 | 2.0 | 6.0 | 3.0 | |
| Sdha | Q920L2 | Succinate Dehydrogenase Complex Flavoprotein Subunit A | 3.0 | 0.9 | 5.0 | 3.0 | |
| Slc6a11 | P31647 | Solute Carrier Family 6 Member 11 | x | 1.0 | 3.7 | 4.0 | 2.9 |
| Madd | O08873 | Map-Kinase Activating Death Domain | 1.0 | 2.6 | 5.0 | 2.9 | |
| Ptpn11 | P41499 | Protein Tyrosine Phosphatase, Non-Receptor type 11 | 4.0 | 1.6 | 3.0 | 2.9 | |
| Camk2b | P08413 | Calcium/Calmodulin-Dependent Protein Kinase II Beta | 5.3 | 1.2 | 2.0 | 2.8 | |
| * Actn4 | Q9QXQ0 | Actinin α 4 | 2.5 | 1.0 | 5.0 | 2.8 | |
| Mccc1 | Q5I0C3 | Methylcrotonoyl-Coa Carboxylase 1 | 2.0 | 0.4 | 6.0 | 2.8 | |
| Gps1 | P97834 | G Protein Pathway Suppressor 1 | 4.0 | 1.3 | 3.0 | 2.8 | |
| Vps45 | O08700 | Vacuolar Protein Sorting 45 | 2.0 | 1.3 | 5.0 | 2.8 | |
| Dlg1 | Q62696 | Discs Large Maguk Scaffold Protein 1 | 3.0 | 1.3 | 4.0 | 2.8 | |
| Sort1 | O54861 | Sortilin 1 | x | 3.0 | 1.3 | 4.0 | 2.8 |
| Ubxn6 | A0A0G2K012 | Ubx Domain Protein 6 | 4.0 | 1.2 | 3.0 | 2.7 | |
| Edf1 | P69736 | Endothelial Differentiation-Related factor 1 | 4.0 | 2.8 | 1.4 | 2.7 | |
| * Ndufs1 | Q66HF1 | Nadh Dehydrogenase Ubiquinone Fe-S Protein 1 | 2.0 | 1.1 | 5.0 | 2.7 | |
| Soga3 | D4A0A1 | Soga Family Member 3 | 2.3 | 0.8 | 5.0 | 2.7 | |
| Cops3 | Q68FW9 | Cop9 Signalosome Subunit 3 | 5.0 | 2.3 | 0.8 | 2.7 | |
| Cds2 | Q91XU8 | Cdp-Diacylglycerol Synthase 2 | 3.0 | 2.0 | 3.0 | 2.7 | |
| Syt2 | P29101 | Synaptotagmin 2 | x | 2.0 | 2.0 | 4.0 | 2.7 |
| Hint1 | P62959 | Histidine Triad Nucleotide Binding Protein 1 | 2.0 | 5.0 | 1.0 | 2.7 | |
| Psmb6 | P28073 | Proteasome Subunit β 6 | 1.0 | 3.0 | 4.0 | 2.7 | |
| * Dhx9 | D4A9D6 | Dexh-Box Helicase 9 | 5.0 | 1.0 | 2.0 | 2.7 | |
| Lmna | G3V8L3 | Lamin A/C | 2.0 | 1.0 | 5.0 | 2.7 | |
| Rpl12 | P23358 | Ribosomal Protein L12 | 4.0 | 1.0 | 3.0 | 2.7 | |
| Pank4 | Q923S8 | Pantothenate Kinase 4 | 2.0 | 1.0 | 5.0 | 2.7 | |
| Cul1 | B1WBY1 | Cullin 1 | 3.0 | 1.0 | 4.0 | 2.7 | |
| Psma3 | P18422 | Proteasome Subunit Alpha 3 | 3.0 | 3.5 | 1.5 | 2.7 | |
| Epb41l2 | D3ZAY7 | Erythrocyte Membrane Protein Band 4.1-Like 2 | 2.0 | 1.7 | 4.3 | 2.7 | |
| Mpp2 | D3ZAA9 | Membrane Palmitoylated Protein 2 | 2.3 | 1.5 | 4.2 | 2.7 | |
| Napb | F8WFM2 | Nsf Attachment Protein β | 5.0 | 2.3 | 0.4 | 2.6 | |
| Cntn1 | Q63198 | Contactin 1 | 1.5 | 2.7 | 3.5 | 2.6 | |
| Ehd1 | Q641Z6 | Eh-Domain Containing 1 | 2.0 | 2.7 | 3.0 | 2.6 | |
| Add1 | Q63028 | Adducin 1 | 2.0 | 1.0 | 4.6 | 2.5 | |
| Usp15 | A0A0A0MY07 | Ubiquitin Specific Peptidase 15 | 2.3 | 0.8 | 4.5 | 2.5 | |
| Synj1 | Q62910 | Synaptojanin 1 | 3.0 | 1.7 | 2.8 | 2.5 | |
| Rbmx | Q4V898 | Rna Binding Motif Protein, X-Linked | 5.0 | 2.0 | 0.5 | 2.5 | |
| Slc24a2 | O54701-2 | Sodium/potassium/calcium exchanger 2 | x | 1.0 | 4.5 | 2.0 | 2.5 |
| Hepacam | D3ZEI4 | Hepatic and Glial Cell Adhesion Molecule | 2.0 | 1.5 | 4.0 | 2.5 | |
| Brsk2 | D3ZML2-2 | Serine/threonine-protein kinase BRSK2 | 2.0 | 1.4 | 4.0 | 2.5 | |
| Epb41l3 | G3V874 | Erythrocyte Membrane Protein Band 4.1-Like 3 | 2.0 | 1.3 | 4.0 | 2.4 | |
| Klhl22 | D3ZZC3 | Kelch-Like Family Member 22 | 2.0 | 1.3 | 4.0 | 2.4 | |
| Map3k7 | P0C8E4 | Mitogen Activated Protein Kinase Kinase Kinase 7 | 2.0 | 1.3 | 4.0 | 2.4 | |
| Wdr48 | D3Z8C7 | Wd Repeat Domain 48 | 3.0 | 1.3 | 3.0 | 2.4 | |
| Acadvl | P45953 | Acyl-Coa Dehydrogenase, Very Long Chain | 2.0 | 1.3 | 4.0 | 2.4 | |
| Gpd1 | O35077 | Glycerol-3-Phosphate Dehydrogenase 1 | 3.0 | 3.3 | 0.9 | 2.4 | |
| Krt77 | Q6IG01 | Keratin 77 | 5.0 | 2.0 | 0.2 | 2.4 | |
| Gphn | Q03555 | Gephyrin | 2.0 | 1.2 | 4.0 | 2.4 | |
| Ppm1e | Q80Z30 | Protein Phosphatase, Mg2+/Mn2+ Dependent, 1E | 2.0 | 1.1 | 4.0 | 2.4 | |
| Cadm3 | Q1WIM3 | Cell Adhesion Molecule 3 | x | 2.0 | 3.0 | 2.0 | 2.3 |
| Acyp2 | P35745 | Acylphosphatase 2 | 2.0 | 3.0 | 2.0 | 2.3 | |
| Aplp1 | F1LRS5 | Amyloid Beta Precursor Like Protein 1 | x | 2.0 | 3.0 | 2.0 | 2.3 |
| Poldip2 | E9PT51 | Dna Polymerase δ Interacting Protein 2 | 2.0 | 3.0 | 2.0 | 2.3 | |
| Brcc3 | B2RYM5 | Brca1/Brca2-Containing Complex, Subunit 3 | 3.0 | 2.0 | 2.0 | 2.3 | |
| Gria2 | P19491 | Glutamate Ionotropic Receptor Ampa type Subunit 2 | x | 1.0 | 2.0 | 4.0 | 2.3 |
| Ggt7 | Q99MZ4 | γ-Glutamyltransferase 7 | x | 1.0 | 2.0 | 4.0 | 2.3 |
| Cpne7 | H1UBN0 | Copine 7 | 1.0 | 3.0 | 3.0 | 2.3 | |
| Mtmr9 | Q5XIN4 | Myotubularin Related Protein 9 | 3.0 | 1.0 | 3.0 | 2.3 | |
| Gsta3 | P04904 | Glutathione S-Transferase β 1 | 1.0 | 2.0 | 4.0 | 2.3 | |
| Lrpprc | Q5SGE0 | Leucine-Rich Pentatricopeptide Repeat Containing | 4.0 | 0.7 | 2.3 | 2.3 | |
| Phgdh | O08651 | Phosphoglycerate Dehydrogenase | 2.0 | 3.7 | 1.2 | 2.3 | |
| Plec | Q6S399 | Plectin | 4.0 | 0.8 | 2.0 | 2.3 | |
| Eif5 | Q07205 | Eukaryotic Translation Initiation factor 5 | 4.0 | 2.0 | 0.8 | 2.3 | |
| Gap43 | P07936 | Growth Associated Protein 43 | 3.2 | 1.3 | 2.3 | 2.3 | |
| Pde1a | A0A0G2JZX9 | Phosphodiesterase 1A | 3.0 | 2.5 | 1.3 | 2.3 | |
| Sel1l | Q80Z70 | Sel1L Erad E3 Ligase Adaptor Subunit | x | 2.0 | 1.7 | 3.0 | 2.2 |
| Rab3d | Q63942 | Rab3D, Member Ras Oncogene Family | 3.0 | 2.0 | 1.7 | 2.2 | |
| Ndrg2 | Q8VBU2-2 | Protein NDRG2 | 4.0 | 2.3 | 0.3 | 2.2 | |
| Prag1 | D3ZMK9 | Pragma Of Rnd2 | 4.0 | 0.5 | 2.0 | 2.2 | |
| Kcnj10 | P49655 | Potassium Voltage-Gated Channel Subfamily J Member 10 | x | 1.5 | 2.0 | 3.0 | 2.2 |
| Ngef | Q5BKC9 | Neuronal Guanine Nucleotide Exchange Factor | 2.5 | 1.0 | 3.0 | 2.2 | |
| Rps5 | P24050 | Ribosomal Protein S5 | 3.0 | 3.0 | 0.4 | 2.1 | |
| Wasf3 | M0R7F3 | Was Protein Family, Member 3 | 3.0 | 0.4 | 3.0 | 2.1 | |
| Tnrc6b | A0A0G2K6R0 | Trinucleotide Repeat Containing 6B | 4.0 | 0.3 | 2.0 | 2.1 | |
| Dnajb12 | Q5FVC4 | Dnaj Heat Shock Protein Family Hsp40 Member B12 | x | 3.0 | 2.0 | 1.3 | 2.1 |
| Dnajc5 | P60905 | Dnaj Heat Shock Protein Family (Hsp40) Member C5 | 3.0 | 2.7 | 0.6 | 2.1 | |
| Gdap1 | D4A5X7 | Ganglioside-Induced Differentiation-Associated-Protein 1 | 2.0 | 3.5 | 0.7 | 2.1 | |
| Aldh2 | P11884 | Aldehyde Dehydrogenase 2 Family Mitochondrial | 3.0 | 2.5 | 0.7 | 2.1 | |
| Copg2 | D4ABY2 | Coatomer Protein Complex, Subunit γ 2 | 2.0 | 1.2 | 3.0 | 2.1 | |
| Phactr1 | P62024 | Phosphatase and Actin Regulator 1 | 2.5 | 1.0 | 2.7 | 2.1 | |
| Nipsnap2 | Q5RK08 | Glioblastoma Amplified Sequence | 3.0 | 1.1 | 2.0 | 2.0 | |
| Lsm4 | D4A2C6 | Lsm4 Homolog, U6 Small Nuclear Rna and Mrna Degradation Associated | 2.0 | 2.0 | 2.0 | 2.0 | |
| Dnajb14 | A0A0G2JTM9 | Dnaj Heat Shock Protein Family Hsp40 Member B14 | x | 2.0 | 2.0 | 2.0 | 2.0 |
| Lin7b | Q9Z252 | Lin-7 Homolog B, Crumbs Cell Polarity Complex Component | 1.0 | 3.0 | 2.0 | 2.0 | |
| Ndrg3 | Q6AYR2 | Ndrg Family Member 3 | 1.0 | 3.0 | 2.0 | 2.0 | |
| Hspa2 | P14659 | Heat Shock Protein Family A Member 2 | 2.0 | 3.0 | 1.0 | 2.0 | |
| Zwint | Q8VIL3 | Zw10 Interacting Kinetochore Protein | 3.0 | 2.0 | 1.0 | 2.0 | |
| Synm | A0A096MK54 | Synemin | 2.0 | 3.0 | 1.0 | 2.0 | |
| Gad2 | Q05683 | Glutamate Decarboxylase 2 | 1.0 | 2.0 | 3.0 | 2.0 | |
| * Fasn | P12785 | Fatty Acid Synthase | 2.0 | 0.8 | 3.0 | 2.0 | |
| Vars | Q04462 | Valyl-Trna Synthetase | 2.0 | 1.6 | 2.3 | 1.9 | |
| Aspscr1 | A0A0G2JWF4 | Aspscr1, Ubx Domain Containing Tether For Slc2A4 | 2.0 | 0.8 | 3.0 | 1.9 | |
| * Lgi3 | D3ZN61 | Leucine-Rich Repeat Lgi Family, Member 3 | 2.0 | 3.0 | 0.8 | 1.9 | |
| Rtn1 | Q64548 | Reticulon 1 | x | 2.1 | 1.2 | 2.4 | 1.9 |
| Pitpna | P16446 | Phosphatidylinositol Transfer Protein, α | 2.0 | 3.3 | 0.4 | 1.9 | |
| Dync1i2 | Q62871 | Dynein cytoplasmic 1 intermediate chain 2 | 2.0 | 0.7 | 3.0 | 1.9 | |
| Magi2 | A0A0G2JXR9 | Membrane Associated Guanylate Kinase, Ww AND Pdz Domain Containing 2 | 2.5 | 0.2 | 3.0 | 1.9 | |
| Ubxn2b | P0C627 | Ubx Domain Protein 2B | 2.0 | 1.7 | 2.0 | 1.9 | |
| Vps16 | Q642A9 | Vps16 Corvet/Hops Core Subunit | 2.0 | 0.6 | 3.0 | 1.9 | |
| Vsnl1 | P62762 | Visinin-Like 1 | 3.0 | 2.1 | 0.4 | 1.8 | |
| Adgrl1 | O88917 | Adhesion G Protein-Coupled Receptor L1 | x | 2.0 | 0.9 | 2.6 | 1.8 |
| Gatad2b | Q4V8E1 | Gata Zinc Finger Domain Containing 2B | 2.0 | 0.5 | 3.0 | 1.8 | |
| Fbxl16 | Q5MJ12 | F-Box and Leucine-Rich Repeat Protein 16 | 3.0 | 2.0 | 0.5 | 1.8 | |
| * Ddx17 | E9PT29 | Dead-Box Helicase 17 | 2.0 | 0.5 | 3.0 | 1.8 | |
| Mgp | P08494 | Matrix Gla Protein | 2.0 | 1.0 | 2.5 | 1.8 | |
| Abi1 | Q9QZM5 | Abl-Interactor 1 | 2.0 | 1.5 | 2.0 | 1.8 | |
| * Cct5 | Q68FQ0 | Chaperonin Containing Tcp1 Subunit 5 | 2.2 | 1.1 | 2.1 | 1.8 | |
| Elavl1 | B5DF91 | Elav Like Rna Binding Protein 1 | 3.0 | 2.0 | 0.4 | 1.8 | |
| Sec22b | Q4KM74 | Sec22 Homolog B, Vesicle Trafficking Protein | x | 2.0 | 1.3 | 2.0 | 1.8 |
| Rap1gds1 | A0A0G2K1D2 | Rap1 Gtpase-Gdp Dissociation Stimulator 1 | 2.3 | 2.2 | 0.8 | 1.8 | |
| Serpinh1 | Q5RJR9 | Serpin Family H Member 1 | 2.0 | 2.7 | 0.5 | 1.7 | |
| Tmx1 | Q52KJ9 | Thioredoxin-Related Transmembrane Protein 1 | 1.0 | 2.0 | 2.0 | 1.7 | |
| LOC367746 | F1LPW6 | Similar To Spindlin-Like Protein 2, Spin-2 | 2.0 | 1.0 | 2.0 | 1.7 | |
| Fn3k | D3ZZU8 | Fructosamine 3 Kinase | 2.0 | 2.0 | 1.0 | 1.7 | |
| Purb | Q68A21 | Purine Rich Element Binding Protein B | 2.0 | 1.0 | 2.0 | 1.7 | |
| Gstm5 | Q9Z1B2 | Glutathione S-Transferase, Mu 5 | 2.0 | 0.8 | 2.0 | 1.6 | |
| Napg | A0A0G2K350 | Nsf Attachment Protein γ | 2.0 | 2.2 | 0.5 | 1.6 | |
| Rab10 | Q5RKJ9 | Rab10, member Ras oncogene family | 2.0 | 2.0 | 0.6 | 1.5 | |
| Naca | B2RYX0 | Nascent Polypeptide-Associated Complex α Subunit | 2.0 | 2.0 | 0.6 | 1.5 | |
| * Ina | G3V8Q2 | Internexin Neuronal Intermediate Filament Protein, α | 2.0 | 0.5 | 2.0 | 1.5 | |
| Cpsf7 | Q5XI29 | Cleavage and Polyadenylation Specific Factor 7 | 2.0 | 0.5 | 2.0 | 1.5 | |
| Habp4 | A1L1K8 | Hyaluronan Binding Protein 4 | 2.0 | 0.3 | 2.2 | 1.5 | |
| Vps26b | B1WBS4 | Vps26 Retromer Complex Component B | 2.0 | 2.0 | 0.4 | 1.5 | |
| Hapln4 | D3Z9H2 | Hyaluronan and Proteoglycan Link Protein 4 | 2.0 | 2.0 | 0.3 | 1.4 | |
| Tst | P24329 | Thiosulfate Sulfurtransferase | 2.0 | 2.0 | 0.3 | 1.4 | |
| Acsm3 | Q6SKG1 | Acyl-Coa Synthetase Medium-Chain Family Member 3 | 0.0 | 12.0 | 5.0 | 5.7 | |
| Acly | P16638 | Atp Citrate Lyase | 0.0 | 2.5 | 10.0 | 4.2 | |
| Ezr | P31977 | Ezrin | 0.0 | 5.0 | 7.0 | 4.0 | |
| Acad9 | B1WC61 | Acyl-Coa Dehydrogenase Family, Member 9 | 0.0 | 2.0 | 9.0 | 3.7 | |
| Fip1l1 | Q5U317 | Factor Interacting With Papola and Cpsf1 | 0.0 | 3.0 | 8.0 | 3.7 | |
| Ehd4 | Q8R3Z7 | Eh-Domain Containing 4 | 0.0 | 6.0 | 4.0 | 3.3 | |
| Dpp10 | Q6Q629 | Dipeptidylpeptidase 10 | x | 0.0 | 2.0 | 8.0 | 3.3 |
| Pdia3 | P11598 | Protein Disulfide Isomerase Family A, Member 3 | 0.0 | 2.8 | 7.0 | 3.3 | |
| Uba1 | Q5U300 | Ubiquitin-Like Modifier Activating Enzyme 1 | 0.0 | 2.8 | 7.0 | 3.3 | |
| Tf | A0A0G2QC06 | Transferrin | 0.0 | 2.5 | 7.0 | 3.2 | |
| Plxnb1 | D3ZDX5 | Plexin B1 | x | 0.0 | 3.0 | 6.0 | 3.0 |
| Nomo1 | D3ZSA9 | Nodal Modulator 1 | 0.0 | 5.0 | 4.0 | 3.0 | |
| Stxbp5 | Q9WU70 | Syntaxin Binding Protein 5 | 0.0 | 2.0 | 7.0 | 3.0 | |
| Ogt | P56558 | O-Linked N-Acetylglucosamine Glcnac Transferase | 0.0 | 2.3 | 6.0 | 2.8 | |
| Cspg5 | Q9ERQ6-2 | Chondroitin Sulfate Proteoglycan 5 | x | 0.0 | 2.3 | 6.0 | 2.8 |
| Efr3b | F1LTW9 | Efr3 Homolog B | 0.0 | 5.0 | 3.0 | 2.7 | |
| Kif21a | D4A1V5 | Kinesin Family Member 21A | 0.0 | 2.0 | 6.0 | 2.7 | |
| Tpp2 | Q64560 | Tripeptidyl Peptidase 2 | 0.0 | 3.7 | 4.0 | 2.6 | |
| Alcam | O35112 | Activated Leukocyte Cell Adhesion Molecule | x | 0.0 | 2.3 | 5.0 | 2.4 |
| Lingo1 | G3V881 | Leucine Rich Repeat and Ig Domain Containing 1 | 0.0 | 4.0 | 3.0 | 2.3 | |
| ATP8 | P11608 | Atpase Subunit 8 | x | 0.0 | 3.0 | 4.0 | 2.3 |
| Itfg1 | Q5U355 | Integrin Alpha Fg-Gap Repeat Containing 1 | x | 0.0 | 2.0 | 5.0 | 2.3 |
| Pik3r4 | P0C0R5 | Phosphoinositide-3-Kinase, Regulatory Subunit 4 | 0.0 | 3.0 | 4.0 | 2.3 | |
| Scamp5 | Q9JKE3 | Secretory Carrier Membrane Protein 5 | x | 0.0 | 3.0 | 4.0 | 2.3 |
| Mogs | O88941 | Glucosidase 1 | x | 0.0 | 2.0 | 5.0 | 2.3 |
| Cd200 | A0A5D0 | Cd200 Molecule | 0.0 | 5.0 | 2.0 | 2.3 | |
| Gpm6b | Q9JJK1 | Glycoprotein M6B | x | 0.0 | 4.0 | 2.5 | 2.2 |
| Pithd1 | D4ABS5 | Pith Domain Containing 1 | 0.0 | 2.5 | 4.0 | 2.2 | |
| S100b | P04631 | S100 Calcium Binding Protein B | 0.0 | 4.0 | 2.5 | 2.2 | |
| Pak1 | P35465 | P21 Activated Kinase 1 | 0.0 | 3.0 | 3.0 | 2.0 | |
| Abcb7 | Q704E8 | Atp Binding Cassette Subfamily B Member 7 | x | 0.0 | 4.0 | 2.0 | 2.0 |
| * Hk1 | M0RAQ6 | Hexokinase 1 | 0.0 | 4.0 | 2.0 | 2.0 | |
| Arl3 | P37996 | Adp Ribosylation factor Like Gtpase 3 | 0.0 | 4.0 | 2.0 | 2.0 | |
| Pfkl | P30835 | Phosphofructokinase, Liver Type | 0.0 | 2.0 | 4.0 | 2.0 | |
| Pex5l | Q925N3 | Peroxisomal Biogenesis factor 5-Like | 0.0 | 2.0 | 4.0 | 2.0 | |
| Llgl1 | Q8K4K5 | Llgl1, Scribble Cell Polarity Complex Component | 0.0 | 4.0 | 2.0 | 2.0 | |
| RGD1307235 | D3ZJ01 | Similar to Riken Cdna 2310035C23 | 0.0 | 2.5 | 3.5 | 2.0 | |
| Ppp2r5d | F1MAA3 | Serine/Threonine-Protein Phosphatase 2A 56 kDa Regulatory Subunit δ Isoform-Like | 0.0 | 2.0 | 4.0 | 2.0 | |
| Hgs | Q9JJ50 | Hepatocyte Growth Factor-Regulated Tyrosine Kinase Substrate | 0.0 | 4.0 | 2.0 | 2.0 | |
| Tgm3 | D4A5U3 | Transglutaminase 3 | 4.0 | 2.0 | 0.0 | 2.0 | |
| Cadps | Q62717 | Calcium Dependent Secretion Activator | 0.0 | 2.0 | 3.3 | 1.8 | |
| Cd9 | P40241 | Cd9 Molecule | x | 0.0 | 3.0 | 2.0 | 1.7 |
| Map2k6 | Q925D6 | Mitogen-Activated Protein Kinase Kinase 6 | 0.0 | 3.0 | 2.0 | 1.7 | |
| Vps33b | Q63616 | Vps33B, Late Endosome and Lysosome Associated | 0.0 | 2.0 | 3.0 | 1.7 | |
| Ccdc47 | Q5U2X6 | Coiled-Coil Domain Containing 47 | 0.0 | 2.0 | 3.0 | 1.7 | |
| Acyp1 | D4A6X4 | Acylphosphatase 1 | 0.0 | 2.0 | 3.0 | 1.7 | |
| Trappc8 | F1M9W9 | Trafficking Protein Particle Complex 8 | 0.0 | 2.0 | 3.0 | 1.7 | |
| Nup85 | Q4QQS8 | Nucleoporin 85 | 3.0 | 2.0 | 0.0 | 1.7 | |
| Stxbp5l | D3ZU84 | Syntaxin Binding Protein 5-Like | 0.0 | 3.0 | 2.0 | 1.7 | |
| Hectd3 | F1LVZ9 | Hect Domain E3 Ubiquitin Protein Ligase 3 | 0.0 | 3.0 | 2.0 | 1.7 | |
| Rtca | Q68FS8 | Rna Terminal Phosphate Cyclase Domain 1 | 0.0 | 3.0 | 2.0 | 1.7 | |
| Slc12a2 | E9PTX9 | Solute Carrier Family 12 Member 2 | x | 0.0 | 3.0 | 2.0 | 1.7 |
| Hbs1l | Q6AXM7 | Hbs1-Like Translational Gtpase | 0.0 | 2.0 | 3.0 | 1.7 | |
| Pi4k2a | Q99M64 | Phosphatidylinositol 4-Kinase type 2 α | 2.0 | 3.0 | 0.0 | 1.7 | |
| Padi2 | P20717 | Peptidyl Arginine Deiminase 2 | 0.0 | 2.0 | 3.0 | 1.7 | |
| Lypla1 | P70470 | Lysophospholipase I | 0.0 | 2.0 | 2.5 | 1.5 | |
| Gnpnat1 | B1H249 | Glucosamine-Phosphate N-Acetyltransferase 1 | 0.0 | 2.0 | 2.0 | 1.3 | |
| Nub1 | F7ES73 | Negative Regulator of Ubiquitin-Like Proteins 1 | 0.0 | 2.0 | 2.0 | 1.3 | |
| Nolc1 | P41777 | Nucleolar and Coiled-Body Phosphoprotein 1 | 2.0 | 2.0 | 0.0 | 1.3 | |
| Pkp1 | D3ZY51 | Plakophilin 1 | 2.0 | 0.0 | 2.0 | 1.3 | |
| Ddt | P80254 | d-Dopachrome Tautomerase | 0.0 | 2.0 | 2.0 | 1.3 | |
| Ptpn9 | Q641Z2 | Protein Tyrosine Phosphatase, Non-Receptor Type 9 | 0.0 | 2.0 | 2.0 | 1.3 | |
| Exoc8 | O54924 | Exocyst Complex Component 8 | 0.0 | 2.0 | 2.0 | 1.3 | |
| Carm1 | Q4AE70 | Coactivator-Associated Arginine Methyltransferase 1 | 0.0 | 2.0 | 2.0 | 1.3 | |
| Rapgef2 | F1M386 | Rap Guanine Nucleotide Exchange Factor 2 | 0.0 | 2.0 | 2.0 | 1.3 | |
| * Acat1 | P17764 | Acetyl-Coa Acetyltransferase 1 | 0.0 | 2.0 | 2.0 | 1.3 | |
| Dnajc19 | M0R6L8 | Dnaj Heat Shock Protein Family Hsp40 Member C19 | 0.0 | 2.0 | 2.0 | 1.3 | |
| Shoc2 | Q6AYI5 | Shoc2 Leucine-Rich Repeat Scaffold Protein | 0.0 | 2.0 | 2.0 | 1.3 | |
| Nrbp2 | A0A0G2JT23 | Nuclear Receptor Binding Protein 2 | 0.0 | 2.0 | 2.0 | 1.3 | |
| Cul4a | B2RYJ3 | Cullin 4A | 0.0 | 2.0 | 2.0 | 1.3 | |
| Anxa6 | P48037 | Annexin A6 | 0.0 | 2.0 | 2.0 | 1.3 | |
| Fabp3 | P07483 | Fatty Acid Binding Protein 3 | 0.0 | 2.0 | 2.0 | 1.3 | |
| Wasl | O08816 | Wiskott-Aldrich Syndrome-Like | 0.0 | 2.0 | 2.0 | 1.3 | |
Gene name, corresponding UniProt ID, and protein name are indicated for each candidate. TM, Single or multiple predicted transmembrane domains, based on UniProt (04/2020). Only candidate protein partners with MS/MS ratio KCC2/sham >2 in at least two of the three replicates are shown, ranked first by the number of replicates in which they were identified and then by the descending mean MS-MS count in the three replicates.
*Candidates identified in the previous KCC2 interactome (Mahadevan et al., 2017).
Venn diagrams were generated using the Venny tool (http://bioinfogp.cnb.csic.es/tools/venny/). Gene ontology enrichment in the KCC2 interactome (271 candidate partners) was analyzed using the Cytoscape (version 3.6.1) ClueGO plugin (version 2.5.0; Shannon et al., 2003; Bindea et al., 2009), and the 2464 proteins identified in the three biological replicates as a reference dataset. The GO domains (GO Biological Processes, GO Cellular Components, and GO Molecular Functions) from EBIQuickGO-GOA (updated on April 4, 2018) were used. The UniProt identifiers were mapped to the selected annotations (GO evidence without inference from electronic annotations) with default ClueGO options in addition to the following selection criteria: 3–8 GO tree interval, a minimum of 10 genes from the list of 271 KCC2 candidate partners found to be associated to a term and that these genes represent at least 13% from the total number of associated genes. The statistical test used was an enrichment right-sided hypergeometric test with the Benjamini–Hochberg correction method.
Coimmunoprecipitation from rat cortical homogenates.
Cortices were dissected from adult Sprague Dawley male rats and homogenized by sonication in coimmunoprecipitation buffer (1 ml/100 mg tissue) containing the following (in mm): 50 Tris-HCl, pH 7.4, 150 NaCl, 1 EDTA, 1% Triton X-100, and protease inhibitors, and solubilized by rotation for 4–6 h at 4°C. After centrifugation at 20,000 × g for 40 min at 4°C, the protein content of the supernatant was measured using the BCA Protein Assay (Thermo Fisher Scientific). Solubilized proteins (5 mg/assay) were incubated with 5 μg of primary antibody [mouse anti-KCC2 (Merck); or mouse anti-gephyrin (Synaptic System)] or nonimmune IgG from the same species (Jackson ImmunoResearch) overnight at 4°C under rotation and immune complexes were precipitated with 20 µl of protein-G magnetic beads (Thermo Fisher Scientific) for 2 h at 4°C. Beads were then washed twice with 1 ml coimmunoprecipitation buffer and once with Triton-free coimmunoprecipitation buffer. Immunoprecipitated proteins were eluted in Novex NuPAGE LDS (4×) sample buffer for 1 h at 37°C separated by SDS-PAGE and analyzed by Western blotting.
Transfection of Neuro2a cells.
Neuro2a cells were grown in DMEM GlutaMAX (Thermo Fisher Scientific) supplemented with 1 g/L glucose and 10% fetal bovine serum. For biochemical assays, cells (60–70% confluent) were cotransfected using a ratio of 1:3 DNA and polyethyleneimine (PEI), respectively (5 µg of DNA and 15 µg of PEI; Polysciences), with plasmids encoding full-length, Flag-tagged rat KCC2b or KCC2–CTD (Chamma et al., 2013; Chevy et al., 2015) and EGFP-tagged rat gephyrin (1:1 ratio; Battaglia et al., 2018) or EGFP as control (4:1 ratio). For immunocytochemistry, cells at 60–70% confluence were cotransfected using transfectin (BIO-RAD) according to manufacturer instructions with plasmids encoding full-length Flag-tagged rat KCC2 and either EGFP-tagged rat gephyrin (1:1 ratio) or EGFP (3:1 ratio). Coimmunoprecipitation and immuncytochemistry experiments were performed 2 d post-transfection.
Coimmunoprecipitation from Neuro2a cells.
Forty-eight hours after transfection, cells were homogenized by brief sonication in coimmunoprecipitation buffer containing the following (in mm): 50 Tris-HCl, pH 7.4, 150 NaCl, 1 EDTA, 0.5% Triton X-100, and protease inhibitors. Cells were then solubilized by rotation for 2 h at 4°C and centrifuged at 20,000 × g for 30 min at 4°C. Solubilized proteins were incubated with 20 µl of prewashed anti-GFP-coupled beads (Chromoteck) overnight at 4°C under rotation. Beads were then washed twice with 1 ml of coimmunoprecipitation buffer and once with Triton-free coimmunoprecipitation buffer. Immunoprecipitated proteins were eluted in Novex NuPAGE LDS (4×) sample buffer for 1 h at 37°C, separated by SDS-PAGE and analyzed by Western blotting.
Western blotting.
Proteins were separated on a 4–12% SDS polyacrylamide gradient gel (Invitrogen) and transferred onto a nitrocellulose membrane (GE Healthcare). For coimmunoprecipitation assays from rat cortical homogenates, blots were probed with antibodies against KCC2 (rabbit, 1 µg/ml; catalog #MABN88, Merck) and gephyrin (mouse, 2 µg/ml; clone 3B11, Synaptic Systems). For coimmunoprecipitation assays from Neuro2a cells, blots were probed with GFP (mouse, 1 µg/ml; 4B10 clone, Cell Signaling Technology) and KCC2 (rabbit, 1 µg/ml; Merck) antibodies. Primary antibodies were detected with fluorescent secondary antibodies (0.3–1 µg/ml; DyLight 700 or 800, Rockland; or 0.07 µg/ml; IR700 or 800, LI-COR) using an Odyssey infrared imaging system (LI-COR). All biochemical assays were repeated at least three times on independent cortical extracts or cultures.
Immunocytochemistry.
Immunocytochemistry was used to determine the surface/total KCC2 ratio. To stain the surface pool of KCC2, Neuro2a cells were incubated with anti-Flag antibody (mouse, 1 µg/ml; Merck) at 4°C for 30 min in HEPES-based medium, fixed with paraformaldehyde (4% w/v) supplemented with 4% (w/v) sucrose in PBS and incubated for 30 min in blocking buffer (10% normal goat serum in PBS). Then cells were incubated with goat anti-mouse Cy3 for 1 h at room temperature (0.75 µg/ml; Jackson ImmunoResearch) in blocking buffer. To reveal total (surface + intracellular) KCC2 pools, cells were then permeabilized for 4 min with 0.25% (v/v) Triton X-100 in PBS. After washes in PBS, cells were incubated for 30 min in blocking buffer and then with the anti-Flag antibody (mouse, 1 µg/ml; Merck) for 1 h at room temperature. After washes in PBS, cells were incubated with secondary antibodies (goat anti-mouse Alexa Fluor 647; 1.5 µg/ml; Jackson ImmunoResearch) for 1 h at room temperature. Coverslips were washed in PBS and mounted in Mowiol/Dabco (25 mg/ml) solution. Quantification of surface (Cy3) to total (A647) KCC2 integrated immunofluorescence ratio was computed after background subtraction using ImageJ.
Primary hippocampal cultures.
Neurons were prepared as described previously (Gauvain et al., 2011). Briefly, hippocampi were dissected from embryonic day 18–19 Sprague Dawley rats of both sexes. Tissue was then trypsinized (0.25% v/v), and mechanically dissociated in 1× HBSS (Thermo Fisher Scientific) containing 10 mm HEPES (Invitrogen). Neurons were plated at a density of 120 × 103 cells/ml onto 18-mm-diameter glass coverslips (Assistent) precoated with 55 µg/ml poly-d,l-ornithine (Sigma-Aldrich) in plating medium composed of Minimum Essential Medium (Sigma-Aldrich) supplemented with horse serum (10% v/v; Thermo Fisher Scientific), l-glutamine (2 mm), and Na-pyruvate (1 mm; Thermo Fisher Scientific). After attachment to coverslips for 3–4 h, cells were incubated in culture medium composed of Neurobasal Medium supplemented with B27 (1×), l-glutamine (2 mm), and antibiotics (penicillin, 200 U/ml; streptomycin, 200 µg/ml; Thermo Fisher Scientific) for up to 4 weeks at 37°C in a 5% CO2 humidified incubator. Each week, one-third of the culture medium was renewed. For pharmacological silencing (see Figs. 3, 4), tetrodotoxin (TTX, 1 μm; Latoxan), (RS)-α-methyl-4-carboxyphenylglycine (MCPG; 500 μm), 2,3-dihydroxy-6-nitro-7-sulfonyl-benzo[f]quinoxaline (NBQX; 10 μm), and d-2-amino-7-phosphonovalerate (d-APV; 50 μm; all from HelloBio) were directly added to the culture medium, and neurons were used 12–16 h later for immunocytochemistry or electrophysiology.
Figure 3.
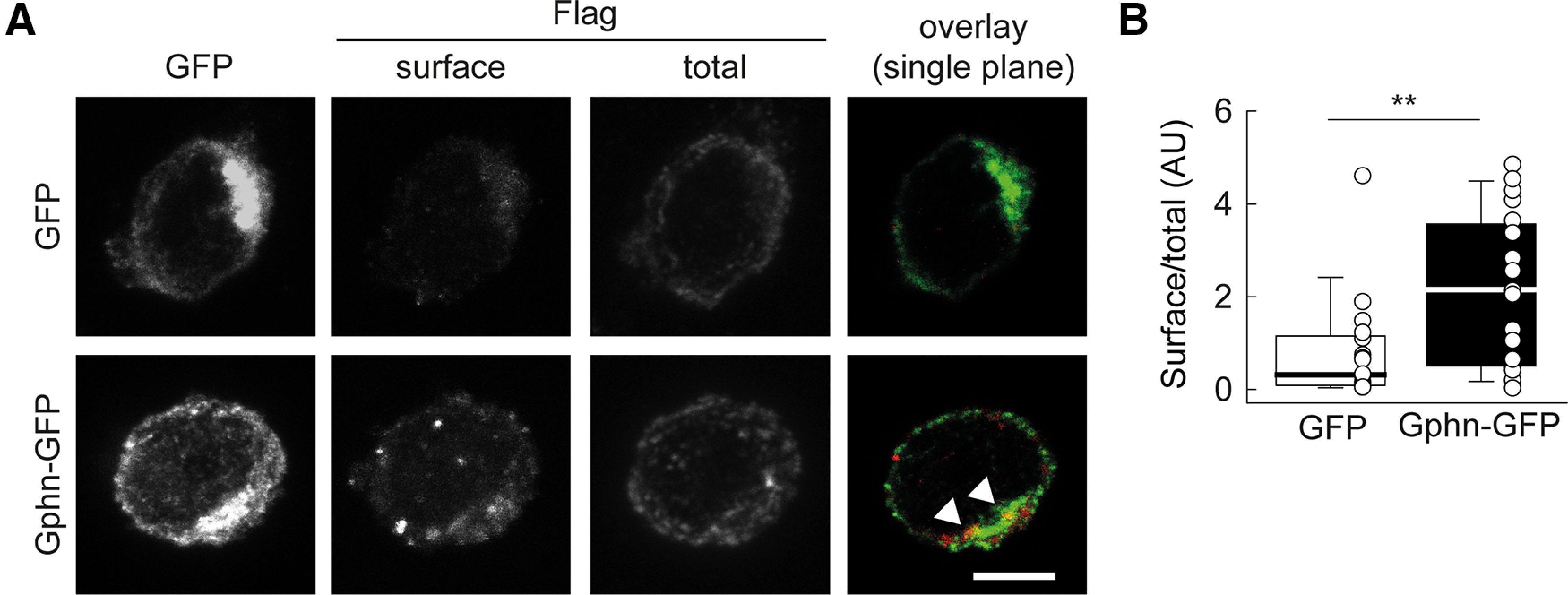
Gephyrin regulates KCC2 membrane expression and clustering in Neuro2a cells. A, Representative confocal maximum projection images of Neuro2a cells cotransfected with plasmids encoding either GFP or GFP-tagged gephyrin (Gphn-GFP) and a plasmid encoding Flag-tagged KCC2. Cells were immunostained for surface and total KCC2 using anti-Flag antibodies. Right, Single confocal image showing overlay of surface Flag (red) and GFP (green) immunostainings. Note the colocalization of surface Flag-KCC2 and GFP-gephyrin immunofluorescent clusters (arrowheads). Scale bar, 5 µm. B, Summary boxplots showing normalized surface/total Flag immunofluorescence intensity ratios (GFP, n = 18; GFP-gephyrin, n = 20 cells; three independent experiments). **p < 0.01, Mann–Whitney rank-sum test.
Figure 4.
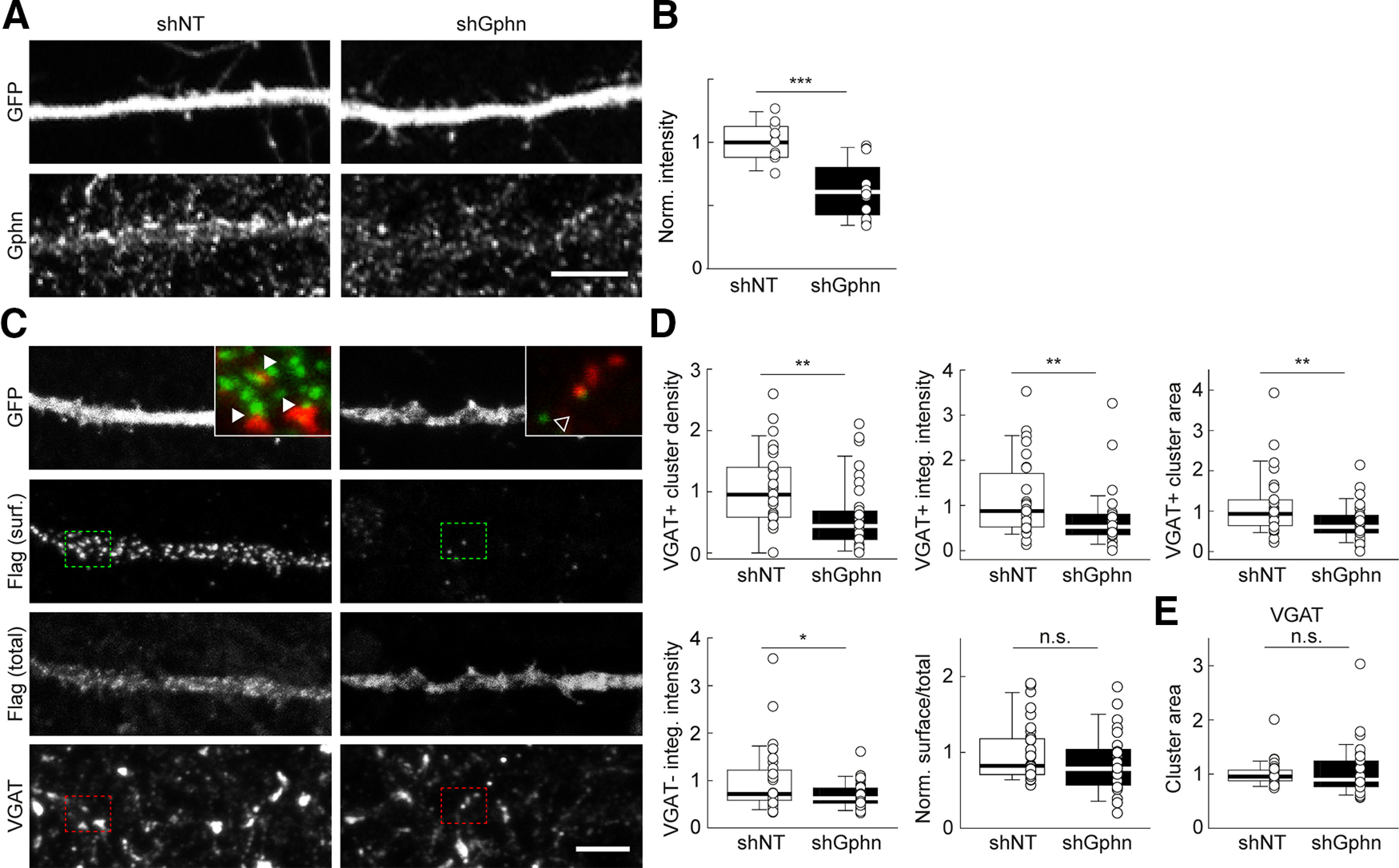
Gephyrin regulates plasmalemmal KCC2 clustering in hippocampal neurons. A, Representative confocal maximum projection images of dendritic sections of hippocampal neurons transfected with a plasmid encoding GFP and either shNT or shGphn, and immunostained for GFP and gephyrin. Note that dendrites of neighboring, nontransfected neurons display gephyrin immunostaining overlapping with that of transfected neurons. Scale bar, 10 µm. B, Summary boxplots showing the intensity of gephyrin immunofluorescence of neurons transfected with shNT and shGphn, normalized to that of shNT-expressing neurons. ***p < 0.001, Mann–Whitney rank-sum test. C, Representative confocal maximum projection images of hippocampal neurons as in A, after live immunostaining for Flag to probe plasmalemmal recombinant KCC2 [Flag (surf.)] and postfixation immunostaining for Flag [Flag (total)] and VGAT. Insets, overlays of live Flag-KCC2 (red) and postfixation VGAT (green) immunostaining from areas boxed in full-scale images. Note the apposition of surface Flag-KCC2 and VGAT immunofluorescent clusters (arrowheads) in control but not gephyrin knock-down neurons (open arrowhead). Scale bar, 5 µm. D, Summary boxplots showing the properties of Flag-KCC2-immunopositive clusters colocalizing (VGAT+, top three plots) or not (VGAT–, lower left boxplot) with VGAT-immunopositive clusters, all normalized to control (shNT). Gephyrin knock-down neurons showed reduced Flag-KCC2 VGAT+ and VGAT– mean cluster density, intensity, and area. This effect was not accompanied by significant changes in surface/total Flag-KCC2 immunofluorescence (bottom middle boxplot). E, No change in the mean area of VGAT clusters was observed on gephyrin knockdown. shNT, n = 32 cells; shGphn, n = 35 cells; from three independent experiments for cluster analysis (n = 39 and n = 25 cells from three independent experiments for surface/total Flag-KCC2). **p < 0.01, *p < 0.05; Mann–Whitney rank-sum tests.
Transfection of primary hippocampal cultures.
Transfections were conducted at day in vitro (DIV_ 13–14 using transfectin, according to the manufacturer instructions (DNA/transfectin ratio, 1 µg:3 µl), with 1 µg of plasmid DNA per 20 mm well. Plasmids encoding Flag-KCC2 (Chamma et al., 2013), EGFP, and shRNA sequences (either nontarget or gephyrin specific; Battaglia et al., 2018) were cotransfected in a ratio of 0.4:0.2:0.4 μg. Immunocytochemistry experiments were performed 8 d post-transfection. For chloride imaging, neurons were cotransfected with a vector expressing cyan fluorescent protein (CFP)-yellow fluorescent protein (YFP)-based chloride sensor (a gift from P. Bregestovski, Institut de Neurosciences des Systèmes, Marseille, France; Markova et al., 2008; Chamma et al., 2013) and shRNA plasmids as above, at a ratio of 1:3 for Cl-sensor and shRNA vectors, respectively.
Immunocytochemistry of KCC2 surface clusters in neurons.
For KCC2 surface immunostaining, neurons were incubated with anti-Flag antibody (mouse; 1 µg/ml; Merck) at 13°C for 20 min in conditioned culture medium (Friedel et al., 2015), rinsed with HEPES-based buffer, fixed with paraformaldehyde (4% w/v) supplemented with 4% (w/v) sucrose in PBS, and incubated for 30 min in blocking buffer (10% v/v normal goat serum in PBS). Then cells were incubated with goat anti-mouse Cy3 for 1 h at room temperature (0.75 µg/ml; Jackson ImmunoResearch) in blocking buffer and permeabilized with 0.25% (v/v) Triton X-100 in PBS. After washing in PBS, cells were incubated for 30 min in blocking buffer and then with anti-Flag antibody (mouse; 1 µg/ml; Merck), anti-VGAT antibody (rabbit; 2 µg/ml; catalog #131002, Synaptic Systems), and anti-GFP antibody (chicken; 2 µg/ml; catalog #AB16901, Merck) at 4°C overnight. After washing in PBS, cells were incubated with secondary antibodies [donkey anti-mouse 7-amino-4-methylcoumarin-3-acetate (AMCA), goat anti-rabbit Alexa Fluor 647, and donkey anti-chicken Alexa Fluor 488, all at 1.5 µg/ml; Jackson ImmunoResearch] for 1 h at room temperature. Coverslips were washed in PBS and mounted in Mowiol-DABCO (25 mg/ml) solution.
Fluorescence image acquisition and analysis.
Image acquisition was performed using an upright epifluorescence microscope (model DM6000 or DM6000-2, Leica) equipped with a 63× objective [numerical aperture (NA), 1.32] and a 12 bit cooled CCD camera (Micromax, Roper Scientific) run by MetaMorph software (Roper Scientific). For analysis, image exposure time was set on bright cells to maximize fluorescence to noise ratio and avoid saturation. All cells from the same culture were then imaged using these acquisition parameters. For illustration, stacks (z-step: 0.2 µm) of immunofluorescence images were acquired with an upright confocal microscope (Leica TCS SP5), using a 63× 1.40–N.A (zoom, 5–6×) objective and lasers set at 405, 488, 561 and 633 nm for excitation of DAPI or AMCA, EGFP, Cy3 and Alexa 647, respectively.
Analysis of VGAT and KCC2 clusters was performed using MetaMorph software (Roper Technologies). Image files were first randomized before analysis. For each neuron, a well focused dendrite was chosen and a region of interest (ROI) surrounding the dendrite was manually selected. Only primary dendrites were considered. The region of interest started after the cell body and, when possible, the entire dendrite (∼20–130 µm) was considered so that no cluster was excluded from the analysis. Images were then flattened, background filtered (core size, 3 × 3 × 2) to enhance cluster contours, and a user-defined intensity threshold was applied to select clusters and avoid their coalescence. A VGAT cluster was only considered if it exceeded a size of three pixels. This excluded most of the mobile, nonsynaptic VGAT clusters or background. Clusters were then delineated, and the corresponding regions were transferred to the raw images to determine the average number of clusters, and their area and fluorescence intensity. For quantification of perisynaptic KCC2 clusters, only clusters comprising at least three pixels and colocalized over at least 1 pixel with clusters of VGAT were considered. Then, the ROI area was measured to determine the number of clusters per unit area. For each culture, we analyzed ∼10 cells per experimental condition. Because of variability in synapse density between cultures, cluster density in each culture was normalized to control values, allowing for comparisons between cultures. The density, area, and integrated intensity of all clusters in the ROI was then computed. For surface/total KCC2 ratios, 2–5 dendritic sections per neuron were selected using ImageJ based on GFP fluorescence of transfected neurons. These ROIs were then copied to Cy3 (surface KCC2) or Cy5 (total KCC2) images to extract mean fluorescence intensities that were then corrected for background.
Chloride imaging.
Transfected neurons were imaged 7–10 d post-transfection in extracellular medium containing the following (in mm): NaCl 120, d-glucose 20, HEPES 10, MgCl2 3, KCl 2, CaCl2 2, pH 7.4, in a temperature-controlled open chamber maintained at 31°C, mounted onto an Olympus IX71 inverted microscope equipped with a 60× objective (1.42 NA; Olympus). CFP and YFP were detected using a Lambda DG-4 monochromator (Sutter Instruments) coupled to the microscope through an optic fiber with appropriate filters (excitation, D436/10× and HQ485/15×; dichroic, 505DCXR; emission, HQ510lp; CFP and YFP filters; Chroma Technology). Images were acquired using an ImagEM EMCCD camera (Hamamatsu Photonics) and MetaFluor software (Roper Scientific). The mean fluorescence intensity was measured in the somatic area, and a dendritic section on the last 10 consecutive frames. Mean background fluorescence was subtracted and the ratio F440/F480 was computed using ImageJ software.
Cellular electrophysiology.
Neurons were maintained at 33°C in an extracellular medium containing the following (in mm): NaCl 120, d-glucose 20, HEPES 10, MgCl2 3, KCl 2, CaCl2 2, pH 7.4, d-APV 0.05 (HelloBio), NBQX 0.01 (HelloBio), CGP54626 0.1 (Tocris Bioscience), MCPG 0.5 (HelloBio), and TTX 0.001 (Latoxan) in a recording chamber (Luigs & Neumann) equipping an upright microscope (model BX51WI, Olympus). Neurons were patch clamped in whole-cell configuration, with a borosilicate glass pipette containing (in mm): K-gluconate 104, KCl 25.4, HEPES 10, EGTA 10, MgATP 2, Na3GTP 0.4, and MgCl2 1.8, pH 7.4, and held at −65 mV. GABAergic currents were induced at the somatic or dendritic level (∼50–100 µm from the soma), by local photolysis 4 ms/10–50 mW on the soma, 8 ms/60–90 mW on the distal dendrites of caged GABA (30 μm; RuBi-GABA, Abcam). The photolysis was performed using the beam (3–5 µm) of a laser diode digitally modulated at 405 nm (Deepstar, Omicron; Photon Lines) and focused through the lens using a photolysis module (Prairie Technologies). Since the low concentration of RuBi-GABA is unlikely to activate synaptic receptors, we did not attempt to specifically target GABAergic synapses. Neurons were voltage clamped from −85 to −5 mV with 3.5 s step increments of 10 mV. The current–voltage relation (I–V) of somatic and dendritic currents was then calculated from the peak amplitude of GABAergic currents recorded at each potential, corrected for the liquid junction potential (−15.2 mV in our conditions) and voltage drop across the access resistance.
Statistics.
Data were compared using the nonparametric Mann–Whitney test using SigmaPlot13 (SPSS). If normality (Shapiro-Wilk test) and equal variance tests passed, means were instead compared using a t test (two tailed). Significance was determined as p < 0.05.
Results
KCC2 interactome in rat neocortex
We used an affinity-purification coupled to MS proteomic strategy to identify KCC2 protein partners in rat neocortical neurons. As antibodies raised against intracellular KCC2 domains may prevent interaction with multiple known partners (Inoue et al., 2004; Li et al., 2007; Mahadevan et al., 2017), we infected rat neocortex with an AAV vector expressing a recombinant, extracellularly Flag-tagged KCC2b construct previously validated for normal membrane traffic and expression in hippocampal neurons in vitro (Fig. 1A,B; Chamma et al., 2013). Expression was restricted to principal neurons by the use of a CaMKII promoter. Two weeks after infection, infected rat cortices showed robust expression of recombinant KCC2, as detected by Western blotting using a Flag antibody (Fig. 1C). Immunoprecipitation with Flag antibody yielded concentrated recombinant KCC2, in both monomeric (150 kDa) and oligomeric (>300 kDa) forms (Fig. 1D).
Figure 1.
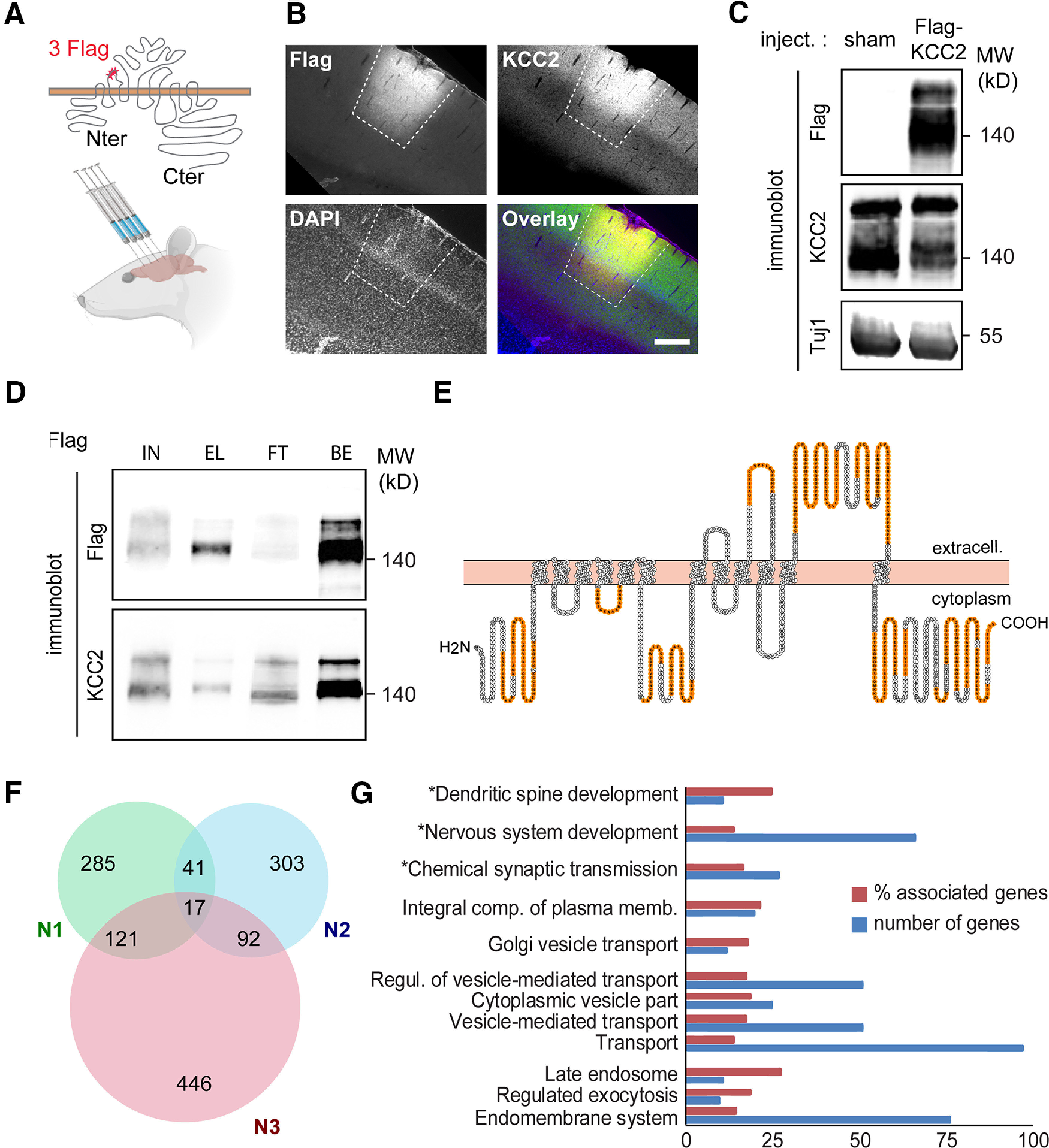
Recombinant Flag-tagged KCC2 ex vivo interactome. A, Experimental paradigm. An AAV2.1 vector expressing an extracellularly Flag-tagged, full-length rat KCC2b construct was stereotaxically injected in the cortex of young adult rats (four injection sites per animal). Sham animals received carrier (PBS) injection. B, Confocal fluorescent micrographs (and overlay, with DAPI staining, blue) of cortical slices of an infected rat, immunostained with Flag (red) and KCC2 (green) antibodies, showing widespread expression of recombinant Flag-KCC2 at one virus injection site (dashed lines). Scale bar, 250 µm. C, Western blots of protein extracts from cortices of rats infected with Flag-KCC2 expressing AAV (Flag-KCC2) or sham animals (sham), using Flag and KCC2 antibodies, showing massive expression of recombinant KCC2 in the former. D, Western blots of protein extracts from cortices of rats infected with Flag-KCC2-expressing AAV (KCC2), immunoprecipitated with Flag antibody, and probed with Flag (top) and KCC2 (bottom) antibodies, showing specific enrichment of recombinant KCC2 in the EL used for LC-MS/MS analysis. Most of solubilized Flag-KCC2 present in the total lysate (IN, here 50 µg proteins) was immunoprecipitated (EL), as evidenced by the very low Flag signal in the FT fraction. BS, Flag-KCC2 (bait) fraction bound to the beads on elution. E, KCC2 amino acid sequence (from Protter; Omasits et al., 2014) with peptides identified by LC-MS/MS in orange. F, Venn diagram showing the total number of Flag-KCC2 interacting proteins in the three biological replicates (N1, N2, and N3). Proteins identified in at least two of three replicates (n = 271) were considered for further analysis. G, Gene ontology enrichment analysis of the 271-protein list using all identified proteins as a reference list performed with ClueGO. Number of genes, Number of genes from our list that were associated with the GO term; % associated genes, number of genes from our list associated with the GO term divided by the total number of genes associated with the term. Only GO terms with a significant enrichment (right-sided hypergeometric test, p < 0.05) are shown. GO terms comprising KCC2 are marked with an asterisk.
The immunoprecipitated fraction was then processed for LC-MS/MS, and detected peptides were matched to the protein database (see Materials and Methods). We observed peptides corresponding to both KCC2 isoforms a and b with a mean MS/MS ratio >100 in extracts from Flag-KCC2-expressing cortices compared with sham rat cortices (Fig. 1E, Table 1). A total of 2464 proteins were identified with at least one unique peptide from the three biological replicates. Of these, 271 proteins showed at least a twofold enrichment, assessed by spectral count, in protein extracts from Flag-KCC2-expressing cortices, compared with sham in at least two of three replicates, and 17 proteins were enriched in KCC2 immunoprecipitates in the three replicates (Fig. 1F, Table 1). The latter included the amyloid precursor protein Aplp1 recently identified as a regulator of KCC2 expression (Doshina et al., 2017) as well as the chaperone protein Calnexin (Canx) and the endoplasmic reticulum transmembrane protein Jagunal Homolog 1 (Jagn1), both identified in a KCC2 interactome study under native conditions using a multiepitope strategy (Mahadevan et al., 2017), thus validating the experimental strategy used in the current study.
Gene ontology analysis of the KCC2 interactome shows enrichment in “integral component of plasma membrane” GO annotation and, more interestingly, in “synaptic transmission” and “dendritic spine development” annotations (Fig. 1G), two major known functions of KCC2 in neurons (Chamma et al., 2012; Blaesse and Schmidt, 2015). In particular, several members of the 4.1 family known to interact with KCC2 (Li et al., 2007) were identified in the latter. We also identified many proteins enriched in biological processes related to vesicular trafficking, such as “Golgi vesicle transport,” “late endosome,”and “regulated exocytosis.” These include exocyst complex component 8 (Exoc8), actinin α 4 (Actn4), and Ras-associated binding protein 10 (Rab10), as well as proteins involved in intracellular microtubule-dependent transport such as dynein cytoplasmic 1 intermediate chain 2 (Dync1i2), and kinesin family member 21A (Kif21a). Those may be of great interest for dissecting molecular and cellular mechanisms of KCC2 trafficking.
KCC2 interacts with gephyrin via its cytoplasmic C-terminal domain
Candidate KCC2 interacting proteins also included gephyrin (Table 1), the main scaffolding protein of GABAA and glycine receptors at inhibitory synapses (Tyagarajan and Fritschy, 2014). We therefore evaluated KCC2–gephyrin interaction in coimmunoprecipitation assays of native proteins using protein lysates from adult rat cortices. We showed that an anti-KCC2 antibody pulled down gephyrin (Fig. 2A), indicating an interaction between the endogenous proteins in cortical neurons. However, just like gephyrin interaction with GABAA receptors (GABAARs; Kasaragod and Schindelin, 2018), interaction with KCC2 may be relatively weak, and we did not always observe KCC2 immunoprecipitation using gephyrin antibody (Fig. 2A). However, to further characterize KCC2–gephyrin interaction, we also performed coimmunoprecipitation assays from proteins extracts from Neuro2a cells expressing recombinant Flag-tagged KCC2 and EGFP-tagged gephyrin (Fig. 2B). In lysates from cotransfected cells, the anti-GFP nanobodies coupled to agarose (GFP-Trap) pulled down Flag-KCC2 in EGFP-gephyrin-expressing cells but not in cells expressing EGFP only.
Figure 2.
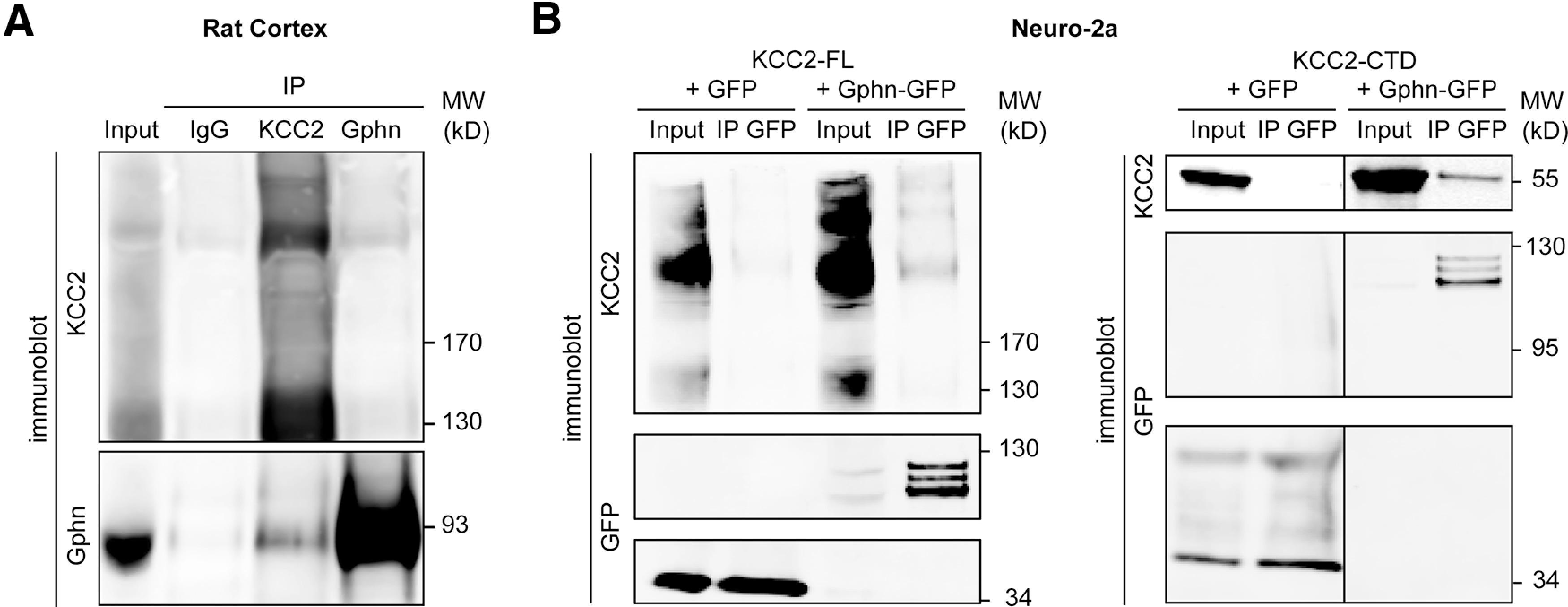
KCC2 interacts with gephyrin via its C-terminal domain. A, Immunoblot of coimmunoprecipitation samples prepared from adult rat cortex. Solubilized membrane homogenates (input) were immunoprecipitated with antibodies raised against gephyrin (Gphn), KCC2 or with control IgG. Input and immunoprecipitated proteins were probed with both KCC2 (top) and gephyrin (bottom) antibodies. B, Immunoblots of immunoprecipitates from Neuro2a cell homogenates coexpressing either Flag-tagged full-length KCC2 (KCC2-FL, left) or KCC2-CTD (right) and either GFP-tagged gephyrin (Gphn-GFP) or GFP. Input and immunoprecipitated proteins were probed with both KCC2 (top) and GFP (bottom) antibodies. Immunoprecipitation with GFP antibody pulled down full-length KCC2 and KCC2-CTD in cells expressing Gphn-GFP but not GFP only. Representative blots from four and three independent experiments are shown in A and B, respectively.
KCC2 interaction with the FERM-domain, spectrin/actin binding protein 4.1N and brain-type creatine kinase has been shown to involve the long KCC2-CTD (Inoue et al., 2004; Li et al., 2007). We then tested whether this cytoplasmic domain (KCC2-CTD) was also sufficient to recruit gephyrin. Reminiscent of the results obtained in cells expressing full-length KCC2, EGFP immunoprecipitation pulled down KCC2-CTD in protein extracts from cells expressing EGFP-gephyrin but not cytosolic EGFP (Fig. 2B). This result demonstrates that KCC2 interacts with gephyrin at least in part via its C-terminal cytoplasmic domain.
Gephyrin regulates KCC2 surface expression and clustering near GABAergic synapses
Whereas the mechanisms of KCC2 clustering near glutamatergic synapses have been partly identified (Chamma et al., 2013), those involved near GABAergic synapses remain unknown. We therefore asked whether interaction with gephyrin may contribute to KCC2 aggregation near GABAergic synapses.
We first overexpressed recombinant, Flag-tagged KCC2 together with either recombinant EGFP-tagged gephyrin or cytosolic EGFP alone in Neuro2a cells. We performed dual immunostaining of both plasmalemmal and total Flag-KCC2 (see Materials and Methods). Quantification of plasmalemmal and total Flag-KCC2 immunofluorescent signals showed that surface/total KCC2 immunostaining was twice as elevated in Neuro2a cells coexpressing EGFP-gephyrin compared with cells expressing EGFP (Fig. 3A,B; p < 0.01). In addition, KCC2 and gephyrin clusters were observed and often colocalized in cells coexpressing recombinant EGFP-gephyrin and Flag-KCC2 (Fig. 3A, arrowheads). These results suggest that gephyrin interaction with KCC2 may act to promote KCC2 membrane expression in heterologous cells.
To further characterize the role of gephyrin on KCC2 plasma membrane localization and clustering, we then explored the impact of chronic gephyrin knockdown on KCC2 plasmalemmal expression in hippocampal neurons. Primary hippocampal cultures were transfected with nontarget shRNA (shNT) or a previously validated gephyrin-directed shRNA (shGphn; Yu et al., 2007; Battaglia et al., 2018) together with EGFP. Gephyrin immunostaining revealed an ∼40% reduction in immunofluorescence in gephyrin knock-down neurons compared with controls (Fig. 4A,B; p < 0.001). However, this may underestimate the actual reduction in gephyrin expression, since the dendrites of transfected neurons were often overlapping with those of nontransfected neurons expressing gephyrin. Since gephyrin knockdown may reduce the inhibitory synaptic drive onto hippocampal neurons (Lévi et al., 2004) and promote hyperexcitability, transfected cultures were kept for 24 h in the presence of blockers of glutamatergic transmission and TTX (see Materials and Methods). In these conditions, neurons expressing nontarget shRNA showed surface KCC2 immunostaining typical of mature hippocampal neurons, with numerous clusters in apposition to the vesicular transporters VGAT (Fig. 4C, inset). The mean area of KCC2 clusters facing VGAT-immunopositive terminals in primary dendrites of hippocampal neurons was 0.57 ± 0.14 µm2 (n = 28 neurons). In gephyrin knock-down neurons, the density of KCC2 clusters apposed to VGAT-immunopositive terminals was reduced by ∼45% (p = 0.002), suggesting that KCC2 clustering near GABAergic synapses depends on gephyrin. In addition, mean cluster intensity and area were reduced by ∼44% and 40%, respectively (p = 0.004 and 0.006, respectively; Fig. 4D). These changes were not associated with a significant change in the area of VGAT-immunopositive terminals (p = 0.62; Fig. 4E) and were also observed, yet to a lesser extent, for KCC2 clusters not facing VGAT-positive terminals. Thus, the mean intensity of recombinant KCC2 clusters not colocalized with VGAT-immunopositive clusters was also reduced by ∼30% (p = 0.048; Fig. 4D). Finally, the ratio of plasmalemmal over total recombinant KCC2 ratio was not affected in gephyrin knock-down neurons compared with controls, suggesting that the lack of gephyrin may prevent clustering of plasmalemmal KCC2 without reducing its total membrane expression. These results demonstrate that KCC2 clustering in the plasma membrane, both near and outside GABAergic synapses, is strongly regulated by gephyrin expression in hippocampal neurons.
Gephyrin expression controls chloride extrusion in hippocampal neurons
KCC2 plasma membrane expression and clustering are tightly correlated with its ion transport function (Watanabe et al., 2009; Chamma et al., 2013; Heubl et al., 2017). We, therefore, examined whether gephyrin expression also regulates KCC2-dependent chloride extrusion in hippocampal neurons. This was first tested by measuring steady-state intracellular chloride concentrations using a ratiometric CFP/YFP-based chloride sensor (Markova et al., 2008; Chamma et al., 2013). In gephyrin knock-down neurons, F440/F480 ratio (which we previously showed positively correlates with intracellular chloride concentration in the 5–100 mm range; Chamma et al., 2013), increased by ∼30% (p = 0.016; Fig. 5A,B), suggesting that gephyrin knockdown led to increased intracellular chloride compared with control neurons.
Figure 5.
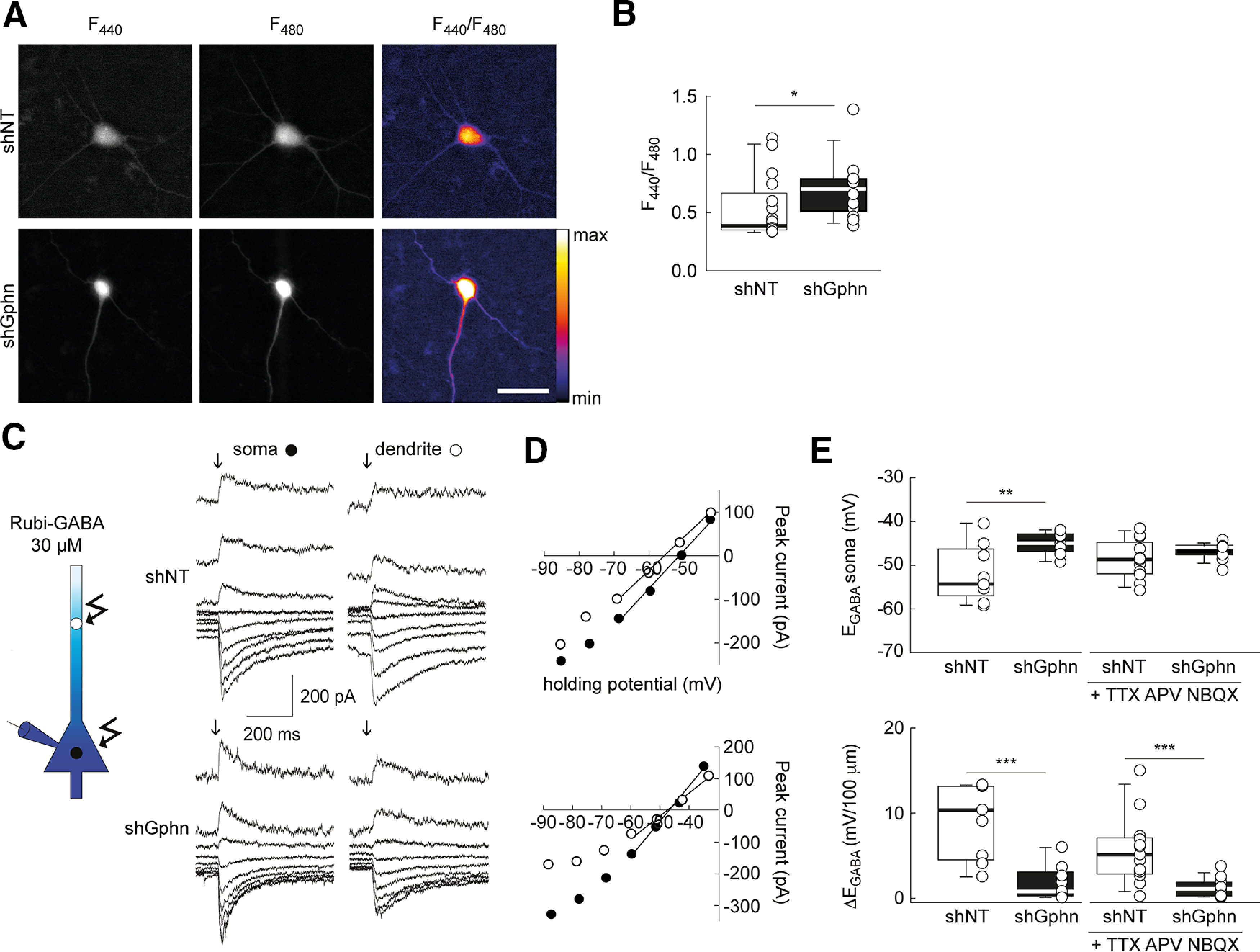
Gephyrin expression is essential for chloride extrusion in hippocampal neurons. A, Representative widefield images of Cl-sensor fluorescence of hippocampal neurons expressing Cl-sensor and either shNT or shGphn, on excitation at 440 and 480 nm. Right, Ratio of the two images at 440 and 480 nm illumination. B, Summary boxplots showing F440/F480 ratios from 17 (shNT) and 14 (shGphn) neurons from two independent experiments. *p < 0.05, Mann–Whitney rank-sum test. C, Left, Representation of experimental setting. RuBi-GABA (30 μm) was locally photolyzed onto the soma or dendrite of transfected neurons whole-cell patch clamped with a high-chloride intracellular solution. Right, Representative recordings of GABA-evoked currents evoked at varying potentials by RuBi-GABA photolysis on the soma or dendrite of hippocampal neurons (DIV 21–24) expressing either shNT or shGphn. D, I–V relations corresponding to traces shown in C. E, Summary graphs showing reversal potential (EGABA, top) and somatodendritic EGABA gradient (ΔEGABA, bottom) in shNT- and shGphn-expressing neurons either in basal culture conditions (n = 9 neurons each, two independent cultures) or on 24 h treatment with TTX and glutamate receptor antagonists (d-APV, NBQX, and MCPG; see Materials and Methods; n = 15 neurons each, n = 2 independent cultures). **p < 0.01, ***p < 0.001; Mann–Whitney rank-sum tests.
However, such steady-state chloride measurements do not provide a direct evaluation of the efficacy of KCC2-mediated chloride extrusion (as discussed in the review by Virtanen et al., 2021). We thus tested transmembrane chloride export by comparing the reversal potential of GABAAR-mediated currents (EGABA) evoked by local photolysis of RuBi-GABA onto the soma and dendrite of hippocampal neurons whole-cell recorded with high-chloride (29 mm) intracellular solution. We and others had shown that the somatodendritic gradient of EGABA measured in these conditions provides a reliable estimate of KCC2 function as it is nearly abolished by a specific KCC2 antagonist (Khirug et al., 2008; Gauvain et al., 2011; Otsu et al., 2020; Fig. 5C). Under these recording conditions, somatic EGABA in neurons transfected with nontarget shRNA was more hyperpolarized than estimated ECl derived from the Nernst equation (approximately −40 mV; Fig. 5E), as previously observed (Otsu et al., 2020). Dendritic EGABA was likewise consistently more hyperpolarized than somatic EGABA by ∼5 mV/100 µm (Fig. 5D). In neurons expressing the shRNA against gephyrin, somatic EGABA was depolarized by ∼7 mV (p < 0.01) compared with neurons expressing nontarget shRNA, while the somatodendritic EGABA gradient (ΔEGABA) was significantly reduced and nearly abolished (p < 0.001; Fig. 5E). These observations were independent of whether cultures were chronically silenced with TTX and glutamate receptor antagonists, suggesting that they do not reflect changes in synaptic or intrinsic activity induced by gephyrin knockdown. Together, these results demonstrate that gephyrin expression strongly regulates membrane KCC2 expression and aggregation as well as its ion transport function in hippocampal neurons.
Discussion
Our data provide a novel KCC2 interactome confirming previously demonstrated protein partners and identifying new ones. They demonstrate for the first time the critical role of gephyrin in regulating KCC2 membrane expression and function in hippocampal neurons. We show that endogenous KCC2 interacts with gephyrin ex vivo and that recombinant tagged gephyrin and KCC2 interact in heterologous cells. This interaction regulates KCC2 plasma membrane localization in both Neuro2a and hippocampal neurons in vitro. In the latter, gephyrin is not strictly required for the formation of plasmalemmal KCC2 clusters but strongly regulates their size and content. Finally, we show that gephyrin expression critically regulates KCC2-mediated chloride extrusion in mature hippocampal neurons, revealing gephyrin as a novel key KCC2 regulator.
A novel KCC2 interactome
KCC2 forms macromolecular complexes with protein partners that control both its membrane expression and function, and, reciprocally, that are regulated through their interaction with KCC2. Thus, identifying KCC2 protein partners has shed light on novel modalities of KCC2 regulation as well as unsuspected roles for KCC2 in neurons. Most interactions have been identified using yeast two-hybrid screens and coimmunoprecipitation assays. For example, the clathrin-binding AP-2 (adaptor protein-2; Zhao et al., 2008), Rab11b (Ras-associated binding protein 11b; Roussa et al., 2016), and the actin-associated protein 4.1N (Li et al., 2007) were shown to interact with KCC2 and control its endocytosis, recycling, and clustering, respectively. Conversely, KCC2 interaction with the guanylyl exchange factor βPIX controls Rac1 and cofilin activity (Chevy et al., 2015; Llano et al., 2015), while its interaction with the two-pore leak potassium channel Task-3 regulates its trafficking (Goutierre et al., 2019).
A first native KCC2 interactome recently identified several known partners such as ATPase subunits, as well as unsuspected ones like the endocytic adaptor protein PACSIN1 (Mahadevan et al., 2017). Twenty putative partners identified in this study were also identified in our analysis (Table 1). These include the chaperone Canx, the transmembrane protein Jagn1, Prrt2 (proline-rich transmembrane protein 2), Slc8a2 (Na+-Ca2+ exchanger 2), and Actn4. In addition, we found several partners identified in previous studies that were not recovered in the proteomic analysis of Mahadevan et al. (2017), like members of the 4.1 (Li et al., 2007) and amyloid precursor protein families (Chen et al., 2017). Finally, our study identified several novel putative interactors, including the scaffolding molecule gephyrin and the NKCC1 transporter. Although we did not further examine this interaction in the present study, the occurrence of both NKCC1 and Na/K-ATPase in our interactome supports the notion that cation chloride cotransporters and ATPases may be part of a functional macromolecular complex, or metabolon (Kaila et al., 2014b).
Apparent discrepancies between our new KCC2 interactome and the previously reported one (Mahadevan et al., 2017) likely reflect differences in experimental approaches. First, Mahadevan et al. (2017) used the nonionic detergent C12E9 and nondenaturing conditions that may better preserve native, higher-order multiprotein complexes than the Triton X-100 used in our study. Although C12E9 is usually preferred to stabilize transmembrane proteins, it is, however, remarkable that extraction with Triton X-100 yielded a comparable transmembrane/total protein ratio (17% and 22% in this study and in the study by Mahadevan et al., 2017, respectively). In addition, the previous study used endogenous KCC2 as bait and antibodies against various domains of the protein to immunoprecipitate KCC2. Instead, we used recombinant Flag-tagged KCC2 as bait to promote antibody access to critical interaction regions, such as the C-terminal domain involved in KCC2 dimerization (Agez et al., 2017); and interaction with 4.1N (Li et al., 2007), βPIX (Chevy et al., 2015; Llano et al., 2015), and gephyrin (this study; Fig. 2). This may explain why 4.1N and gephyrin were not identified in the previously described interactome. Our approach also allowed us to restrict our interactome to the KCC2b isoform, the major isoform in the adult brain (Uvarov et al., 2009), whereas KCC2 antibody-based pull-down indistinctly recovered KCC2a and KCC2b interacting proteins. It also let us restrict our analysis to principal cortical neurons (using CaMKII promoter to drive Flag-KCC2b expression in the isocortex), whereas whole-brain extracts were previously used. Since KCC2 is expressed in various neuron subtypes and brain regions where its noncanonical functions, which rely on molecular interactions, substantially differ (e.g., spinogenesis; Li et al., 2007; Seja et al., 2012; Fiumelli et al., 2013; Awad et al., 2018), interactomics studies performed from whole-brain extracts may reveal interactions that do not occur in cortical principal neurons.
Gephyrin interacts with KCC2 and regulates its membrane clustering and function
KCC2 has been shown to form clusters near both glutamatergic and GABAergic synapses (Gulyás et al., 2001; Gauvain et al., 2011; Chamma et al., 2013), and these clusters were suggested to represent the ion transport active pool of KCC2 in the plasma membrane (Watanabe et al., 2009). By analogy to postsynaptic receptors, plasmalemmal KCC2 clustering is likely to reflect a diffusion-capture behavior (Choquet and Triller, 2013), with free diffusion in the membrane and local molecular constraints promoting clustering in the vicinity of synaptic zones (Chamma et al., 2013; Côme et al., 2019). The nature of these constraints is not fully elucidated. Whereas KCC2 interaction with 4.1N was shown to hinder KCC2 membrane diffusion near glutamatergic synapses in dendritic spines, it does not seem to fulfill a similar role near GABAergic synapses. KCC2 has been reported to interact with GABAA (Huang et al., 2012) and GABAB (Wright et al., 2017) receptor subunits, but these interactions were not found in KCC2 interactomics studies (Mahadevan et al., 2017; this study), and no evidence supports the idea that KCC2 interaction with these receptors controls its membrane expression.
Gephyrin acts as the main scaffolding protein that anchors GABAA receptors to the submembrane cytoskeleton (Tyagarajan and Fritschy, 2014) and was therefore considered a promising partner for KCC2 clustering near GABAergic synapses. KCC2 interaction with gephyrin was confirmed in coimmunoprecipitation assays on native proteins. As reported for 4.1N binding with KCC2 (Li et al., 2007), KCC2–gephyrin interaction involved the KCC2 C-terminal domain since coimmunoprecipitation was preserved in cells expressing only this intracellular domain. Gephyrin subdomains involved in this interaction remain to be identified. Whereas the N-terminal G domain is primarily involved in the trimerization of gephyrin, the central C domain and C-terminal E domain were shown to mediate most interactions with GABAA receptor subunits and other protein partners (Fritschy et al., 2008) and are candidates for mediating interaction with KCC2.
Our results demonstrate that gephyrin promotes plasmalemmal KCC2 clustering in hippocampal neurons. In this context, it is remarkable that gephyrin expression is maximal around the onset of KCC2 expression in rodent isocortex (Clayton et al., 1998; Pinto et al., 2013). The density, size, and intensity of KCC2-immunopositive clusters facing VGAT-immunopositive terminals were all reduced on gephyrin knockdown, supporting the idea that KCC2–gephyrin interaction strongly promotes KCC2 aggregation near GABAergic synapses. However, as described previously, KCC2 clusters are mostly formed at the periphery of both GABAergic and glutamatergic synapses (Chamma et al., 2013). Exclusion of KCC2 from the postsynapse may reflect a higher affinity of gephyrin for α1/2-containing synaptic GABAA receptors, steric hindrance (Kokolaki et al., 2020) or stronger interaction with other extrasynaptic partners. Thus, α5-containing GABAA receptors are excluded from the postsynapse in part because of stronger interaction with radixin (Hausrat et al., 2015). Super-resolution microscopy may help resolve the nanoscopic organization of gephyrin, GABAAR, and KCC2 clusters at GABAergic synapses.
Interestingly, gephyrin knockdown resulted in reduced surface KCC2 cluster intensity not only near but also, to a lesser extent, outside GABAergic synapses (Fig. 3). This observation suggests that gephyrin promotes KCC2 clustering not only near GABAergic synapses but also throughout the dendritic shaft membrane. This effect likely reflects the presence of gephyrin and gephyrin clusters outside GABAergic synapses, as recently revealed by PALM (photoactivated localization microscopy) imaging (Pennacchietti et al., 2017). Finally, although plasmalemmal/total KCC2 expression was increased in N2a cells on coexpression of gephyrin, no significant change in this ratio was observed in hippocampal neurons on gephyrin knockdown. This observation suggests that KCC2 membrane trafficking is not compromised by gephyrin knockdown in hippocampal neurons and that the lack of gephyrin may instead promote a redistribution of plasmalemmal KCC2, from a clustered to a more diffuse state. This effect is somewhat reminiscent of that observed on dephosphorylation of tyrosine residues of the C-terminal domain of KCC2 (Watanabe et al., 2009), which was associated with reduced ion transport.
Indeed, reduced plasmalemmal KCC2 clustering on gephyrin knockdown was associated with a significant increase in intracellular chloride concentration, as detected using Cl-sensor imaging. More importantly, a striking reduction in transmembrane chloride extrusion capacity, as demonstrated in our electrophysiological assay, was observed on gephyrin knockdown. In this assay, neurons are loaded through the patch pipette with a high and fixed concentration of chloride such that the reversal potential of GABAAR-mediated responses at dendritic sites distant from the soma directly reflects chloride cytosolic diffusion and transmembrane extrusion (Khirug et al., 2008; Gauvain et al., 2011). Suppressing KCC2 function or expression has been shown to largely compromise somatodendritic EGABA gradient in such an experimental setting (Gauvain et al., 2011). Our observation that gephyrin knockdown also strongly reduces the EGABA gradient, therefore, suggests that even a partial reduction of KCC2 dendritic clustering is sufficient to profoundly compromise chloride extrusion in cortical neurons. Interestingly, preventing KCC2 interaction with 4.1N perturbed KCC2 clustering near glutamatergic synapses by promoting its lateral diffusion (Chamma et al., 2013). This manipulation, however, had no detectable effect on the somatodendritic EGABA gradient (Gauvain et al., 2011). Together, these results suggest that dendritic, KCC2-mediated chloride export in cortical neurons primarily relies on gephyrin-dependent, but not on 4.1N-dependent, plasmalemmal KCC2 clustering. Whether KCC2 clustering in dendritic spines specifically controls the efficacy of GABAergic synapses they harbor (Kubota et al., 2007; Virtanen et al., 2018) remains to be explored.
Finally, KCC2 clustering and function are tightly controlled by neuronal and synaptic activity (Woodin et al., 2003; Rivera et al., 2004; Lee et al., 2011; Chamma et al., 2012, 2013; Medina et al., 2014; Heubl et al., 2017). In particular, glutamatergic (Chamma et al., 2013) and GABAergic (Heubl et al., 2017) synaptic transmission exert opposing control over KCC2 by regulating its lateral diffusion and clustering, resulting in a reduced or enhanced KCC2-mediated chloride export, respectively. This regulation relies on signaling cascades involving, at least in part, phosphorylation/dephosphorylation of specific serine and threonine residues in the KCC2 C-terminal domain (Kahle et al., 2005; Lee et al., 2011; Chamma et al., 2013; de Los Heros et al., 2014; Friedel et al., 2015; Heubl et al., 2017). It seems improbable, however, that reduced synaptic GABAA receptor function induced by gephyrin knockdown may significantly contribute to reduce KCC2 membrane clustering. Thus, complete genetic ablation of gephyrin resulted in only 20% reduction in the amplitude of miniature IPSCs with no change in frequency (Lévi et al., 2004). In contrast, complete blockade of GABAA receptors by gabazine reduced KCC2 cluster intensity by only 20% (compared with >40% in our experiments; Heubl et al., 2017). Instead, by analogy with activity-dependent regulation of postsynaptic receptor aggregation (Choquet and Triller, 2013), it is then tempting to speculate that changes in the phosphorylation state of KCC2 and/or its scaffolding partners including gephyrin (Battaglia et al., 2018) and 4.1N may induce conformational modifications that may act to perturb KCC2–scaffold interactions and reduce clustering and/or membrane anchoring of the transporter.
In conclusion, our study identified gephyrin as a novel key KCC2 partner that regulates its membrane clustering and function. Our results further illustrate the importance of functional macromolecular complexes regulating KCC2 membrane clustering and function. KCC2 clustering near glutamatergic synapses involves anchoring to the submembrane actin cytoskeleton via 4.1N (Li et al., 2007; Chamma et al., 2013), which in turn binds to the AMPAR GluA1 subunit (Lin et al., 2009). Similarly, near GABAergic synapses, coclustering of GABAA receptors and KCC2 with gephyrin may involve interaction with subsynaptic microtubule cytoskeleton (Kirsch et al., 1991) and promote the formation of a stable, functional macromolecular complex acting to regulate GABA signaling in cortical neurons (Heubl et al., 2017; Côme et al., 2020).
Footnotes
This work was supported in part by Fondation pour la Recherche Médicale (Grant DEQ20140329539 to J.C.P.), ERANET-Neuron (funded by Agence Nationale de la Recherche; to J.C.P.), and the Fondation Française pour la Recherche sur l'Epilepsie–Fédération pour la Recherche sur le Cerveau (to J.C.P.). F.D. and M.G. were recipients of fellowships from Sorbonne University. The Poncer-Lévi laboratory is affiliated with the Sorbonne University i-Bio Initiative. We thank Jean-Antoine Girault for advice on protein extraction, and Kai Kaila for discussions while preparing the manuscript. We also thank the Proteomics Platform of Biocampus Montpellier and the Photonic Imaging Platform of Institut du Fer à Moulin, where mass spectrometry experiments and image acquisition, respectively, were carried out.
The authors declare no competing financial interests.
References
- Agez M, Schultz P, Medina I, Baker DJ, Burnham MP, Cardarelli RA, Conway LC, Garnier K, Geschwindner S, Gunnarsson A, McCall EJ, Frechard A, Audebert S, Deeb TZ, Moss SJ, Brandon NJ, Wang Q, Dekker N, Jawhari A (2017) Molecular architecture of potassium chloride co-transporter KCC2. Sci Rep 7:16452. 10.1038/s41598-017-15739-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad PN, Amegandjin CA, Szczurkowska J, Carrico JN, Fernandes do Nascimento AS, Baho E, Chattopadhyaya B, Cancedda L, Carmant L, Di Cristo G (2018) KCC2 regulates dendritic spine formation in a brain-region specific and BDNF dependent manner. Cereb Cortex 28:4049–4062. 10.1093/cercor/bhy198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldí R, Varga C, Tamás G (2010) Differential distribution of KCC2 along the axo-somato-dendritic axis of hippocampal principal cells. Eur J Neurosci 32:1319–1325. 10.1111/j.1460-9568.2010.07361.x [DOI] [PubMed] [Google Scholar]
- Banerjee A, Rikhye RV, Breton-Provencher V, Tang X, Li C, Li K, Runyan CA, Fu Z, Jaenisch R, Sur M (2016) Jointly reduced inhibition and excitation underlies circuit-wide changes in cortical processing in Rett syndrome. Proc Natl Acad Sci U S A 113:E7287–E7296. 10.1073/pnas.1615330113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia S, Renner M, Russeau M, Côme E, Tyagarajan SK, Lévi S (2018) Activity-dependent inhibitory synapse scaling is determined by gephyrin phosphorylation and subsequent regulation of GABAA receptor diffusion. eNeuro 5:ENEURO.0203-17.2017. 10.1523/ENEURO.0203-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25:1091–1093. 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaesse P, Schmidt T (2015) K-Cl cotransporter KCC2–a moonlighting protein in excitatory and inhibitory synapse development and function. Pflugers Arch 467:615–624. 10.1007/s00424-014-1547-6 [DOI] [PubMed] [Google Scholar]
- Boulenguez P, Liabeuf S, Bos R, Bras H, Jean-Xavier C, Brocard C, Stil A, Darbon P, Cattaert D, Delpire E, Marsala M, Vinay L (2010) Down-regulation of the potassium-chloride cotransporter KCC2 contributes to spasticity after spinal cord injury. Nat Med 16:302–307. 10.1038/nm.2107 [DOI] [PubMed] [Google Scholar]
- Chamma I, Chevy Q, Poncer JC, Lévi S (2012) Role of the neuronal K-Cl co-transporter KCC2 in inhibitory and excitatory neurotransmission. Front Cell Neurosci 6:5. 10.3389/fncel.2012.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamma I, Heubl M, Chevy Q, Renner M, Moutkine I, Eugène E, Poncer JC, Lévi S (2013) Activity-dependent regulation of the K/Cl transporter KCC2 membrane diffusion, clustering, and function in hippocampal neurons. J Neurosci 33:15488–15503. 10.1523/JNEUROSCI.5889-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Wang J, Jiang J, Zheng X, Justice NJ, Wang K, Ran X, Li Y, Huo Q, Zhang J, Li H, Lu N, Wang Y, Zheng H, Long C, Yang L (2017) APP modulates KCC2 expression and function in hippocampal GABAergic inhibition. eLife 6:e20142. 10.7554/eLife.20142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevy Q, Heubl M, Goutierre M, Backer S, Moutkine I, Eugène E, Bloch-Gallego E, Lévi S, Poncer JC (2015) KCC2 gates activity-driven AMPA receptor traffic through cofilin phosphorylation. J Neurosci 35:15772–15786. 10.1523/JNEUROSCI.1735-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevy Q, Simonnet C, Al Awabdh S, Lévi S, Poncer JC (2020) Transport-dependent and independent functions of KCC2 at excitatory synapses. In: Neuronal chloride transporters in health and disease (Tang X, ed), pp 33–158. Amsterdam: Academic. [Google Scholar]
- Choquet D, Triller A (2013) The dynamic synapse. Neuron 80:691–703. 10.1016/j.neuron.2013.10.013 [DOI] [PubMed] [Google Scholar]
- Clayton GH, Owens GC, Wolff JS, Smith RL (1998) Ontogeny of cation-Cl- cotransporter expression in rat neocortex. Brain Res Dev Brain Res 109:281–292. 10.1016/s0165-3806(98)00078-9 [DOI] [PubMed] [Google Scholar]
- Cohen I, Navarro V, Clemenceau S, Baulac M, Miles R (2002) On the origin of interictal activity in human temporal lobe epilepsy in vitro. Science 298:1418–1421. 10.1126/science.1076510 [DOI] [PubMed] [Google Scholar]
- Côme E, Heubl M, Schwartz EJ, Poncer JC, Lévi S (2019) Reciprocal regulation of KCC2 trafficking and synaptic activity. Front Cell Neurosci 13:48. 10.3389/fncel.2019.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côme E, Marques X, Poncer JC, Lévi S (2020) KCC2 membrane diffusion tunes neuronal chloride homeostasis. Neuropharmacology 169:107571. 10.1016/j.neuropharm.2019.03.014 [DOI] [PubMed] [Google Scholar]
- Coull JA, Boudreau D, Bachand K, Prescott SA, Nault F, Sík A, De Koninck P, De Koninck Y (2003) Trans-synaptic shift in anion gradient in spinal lamina I neurons as a mechanism of neuropathic pain. Nature 424:938–942. 10.1038/nature01868 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- de Los Heros P, Alessi DR, Gourlay R, Campbell DG, Deak M, Macartney TJ, Kahle KT, Zhang J (2014) The WNK-regulated SPAK/OSR1 kinases directly phosphorylate and inhibit the K+-Cl- co-transporters. Biochem J 458:559–573. 10.1042/BJ20131478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshina A, Gourgue F, Onizuka M, Opsomer R, Wang P, Ando K, Tasiaux B, Dewachter I, Kienlen-Campard P, Brion JP, Gailly P, Octave JN, Pierrot N (2017) Cortical cells reveal APP as a new player in the regulation of GABAergic neurotransmission. Sci Rep 7:370. 10.1038/s41598-017-00325-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Tintrup H, Kirsch J, Nichol MC, Kuhse J, Betz H, Sanes JR (1998) Dual requirement for gephyrin in glycine receptor clustering and molybdoenzyme activity. Science 282:1321–1324. 10.1126/science.282.5392.1321 [DOI] [PubMed] [Google Scholar]
- Fiumelli H, Briner A, Puskarjov M, Blaesse P, Belem BJ, Dayer AG, Kaila K, Martin JL, Vutskits L (2013) An ion transport-independent role for the cation-chloride cotransporter KCC2 in dendritic spinogenesis in vivo. Cereb Cortex 23:378–388. 10.1093/cercor/bhs027 [DOI] [PubMed] [Google Scholar]
- Friedel P, Kahle KT, Zhang J, Hertz N, Pisella LI, Buhler E, Schaller F, Duan J, Khanna AR, Bishop PN, Shokat KM, Medina I (2015) WNK1-regulated inhibitory phosphorylation of the KCC2 cotransporter maintains the depolarizing action of GABA in immature neurons. Sci Signal 8:ra65. 10.1126/scisignal.aaa0354 [DOI] [PubMed] [Google Scholar]
- Fritschy JM, Harvey RJ, Schwarz G (2008) Gephyrin: where do we stand, where do we go? Trends Neurosci 31:257–264. 10.1016/j.tins.2008.02.006 [DOI] [PubMed] [Google Scholar]
- Gauvain G, Chamma I, Chevy Q, Cabezas C, Irinopoulou T, Bodrug N, Carnaud M, Lévi S, Poncer JC (2011) The neuronal K-Cl cotransporter KCC2 influences postsynaptic AMPA receptor content and lateral diffusion in dendritic spines. Proc Natl Acad Sci U S A 108:15474–15479. 10.1073/pnas.1107893108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutierre M, Al Awabdh S, Donneger F, François E, Gomez-Dominguez D, Irinopoulou T, Menendez de la Prida L, Poncer JC (2019) KCC2 regulates neuronal excitability and hippocampal activity via interaction with Task-3 channels. Cell Rep 28:91–103.e7. 10.1016/j.celrep.2019.06.001 [DOI] [PubMed] [Google Scholar]
- Gulyás AI, Sík A, Payne JA, Kaila K, Freund TF (2001) The KCl cotransporter, KCC2, is highly expressed in the vicinity of excitatory synapses in the rat hippocampus. Eur J Neurosci 13:2205–2217. 10.1046/j.0953-816x.2001.01600.x [DOI] [PubMed] [Google Scholar]
- Hausrat TJ, Muhia M, Gerrow K, Thomas P, Hirdes W, Tsukita S, Heisler FF, Herich L, Dubroqua S, Breiden P, Feldon J, Schwarz JR, Yee BK, Smart TG, Triller A, Kneussel M (2015) Radixin regulates synaptic GABAA receptor density and is essential for reversal learning and short-term memory. Nat Commun 6:6872. 10.1038/ncomms7872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heubl M, Zhang J, Pressey JC, Al Awabdh S, Renner M, Gomez-Castro F, Moutkine I, Eugène E, Russeau M, Kahle KT, Poncer JC, Lévi S (2017) GABAA receptor dependent synaptic inhibition rapidly tunes KCC2 activity via the Cl−-sensitive WNK1 kinase. Nat Commun 8:1776. 10.1038/s41467-017-01749-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Ko H, Cheung ZH, Yung KK, Yao T, Wang JJ, Morozov A, Ke Y, Ip NY, Yung WH (2012) Dual actions of brain-derived neurotrophic factor on GABAergic transmission in cerebellar Purkinje neurons. Exp Neurol 233:791–798. 10.1016/j.expneurol.2011.11.043 [DOI] [PubMed] [Google Scholar]
- Huberfeld G, Wittner L, Clemenceau S, Baulac M, Kaila K, Miles R, Rivera C (2007) Perturbed chloride homeostasis and GABAergic signaling in human temporal lobe epilepsy. J Neurosci 27:9866–9873. 10.1523/JNEUROSCI.2761-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Ueno S, Fukuda A (2004) Interaction of neuron-specific K+-Cl– cotransporter, KCC2, with brain-type creatine kinase. FEBS Lett 564:131–135. 10.1016/S0014-5793(04)00328-X [DOI] [PubMed] [Google Scholar]
- Ivakine EA, Acton BA, Mahadevan V, Ormond J, Tang M, Pressey JC, Huang MY, Ng D, Delpire E, Salter MW, Woodin MA, McInnes RR (2013) Neto2 is a KCC2 interacting protein required for neuronal Cl- regulation in hippocampal neurons. Proc Natl Acad Sci U S A 110:3561–3566. 10.1073/pnas.1212907110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob TC, Bogdanov YD, Magnus C, Saliba RS, Kittler JT, Haydon PG, Moss SJ (2005) Gephyrin regulates the cell surface dynamics of synaptic GABAA receptors. J Neurosci 25:10469–10478. 10.1523/JNEUROSCI.2267-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle KT, Rinehart J, de Los Heros P, Louvi A, Meade P, Vazquez N, Hebert SC, Gamba G, Gimenez I, Lifton RP (2005) WNK3 modulates transport of Cl- in and out of cells: implications for control of cell volume and neuronal excitability. Proc Natl Acad Sci U S A 102:16783–16788. 10.1073/pnas.0508307102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle KT, Staley KJ, Nahed BV, Gamba G, Hebert SC, Lifton RP, Mount DB (2008) Roles of the cation-chloride cotransporters in neurological disease. Nat Clin Pract Neurol 4:490–503. 10.1038/ncpneuro0883 [DOI] [PubMed] [Google Scholar]
- Kaila K, Price TJ, Payne JA, Puskarjov M, Voipio J (2014a) Cation-chloride cotransporters in neuronal development, plasticity and disease. Nat Rev Neurosci 15:637–654. 10.1038/nrn3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaila K, Ruusuvuori E, Seja P, Voipio J, Puskarjov M (2014b) GABA actions and ionic plasticity in epilepsy. Curr Opin Neurobiol 26:34–41. 10.1016/j.conb.2013.11.004 [DOI] [PubMed] [Google Scholar]
- Kasaragod VB, Schindelin H (2018) Structure-function relationships of glycine and GABA(A) receptors and their interplay with the scaffolding protein gephyrin. Front Mol Neurosci 11:317. 10.3389/fnmol.2018.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khirug S, Yamada J, Afzalov R, Voipio J, Khiroug L, Kaila K (2008) GABAergic depolarization of the axon initial segment in cortical principal neurons is caused by the Na-K-2Cl cotransporter NKCC1. J Neurosci 28:4635–4639. 10.1523/JNEUROSCI.0908-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch J, Langosch D, Prior P, Littauer UZ, Schmitt B, Betz H (1991) The 93-kDa glycine receptor-associated protein binds to tubulin. J Biol Chem 266:22242–22245. [PubMed] [Google Scholar]
- Kneussel M, Brandstätter JH, Laube B, Stahl S, Müller U, Betz H (1999) Loss of postsynaptic GABAA receptor clustering in gephyrin-deficient mice. J Neurosci 19:9289–9297. 10.1523/JNEUROSCI.19-21-09289.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokolaki ML, Fauquier A, Renner M (2020) Molecular crowding and diffusion-capture in synapses. iScience 23:101382. 10.1016/j.isci.2020.101382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Hatada S, Kondo S, Karube F, Kawaguchi Y (2007) Neocortical inhibitory terminals innervate dendritic spines targeted by thalamocortical afferents. J Neurosci 27:1139–1150. 10.1523/JNEUROSCI.3846-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Deeb TZ, Walker JA, Davies PA, Moss SJ (2011) NMDA receptor activity downregulates KCC2 resulting in depolarizing GABAA receptor-mediated currents. Nat Neurosci 14:736–743. 10.1038/nn.2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévi S, Logan SM, Tovar KR, Craig AM (2004) Gephyrin is critical for glycine receptor clustering but not for the formation of functional GABAergic synapses in hippocampal neurons. J Neurosci 24:207–217. 10.1523/JNEUROSCI.1661-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Khirug S, Cai C, Ludwig A, Blaesse P, Kolikova J, Afzalov R, Coleman SK, Lauri S, Airaksinen MS, Keinänen K, Khiroug L, Saarma M, Kaila K, Rivera C (2007) KCC2 interacts with the dendritic cytoskeleton to promote spine development. Neuron 56:1019–1033. 10.1016/j.neuron.2007.10.039 [DOI] [PubMed] [Google Scholar]
- Lin DT, Makino Y, Sharma K, Hayashi T, Neve R, Takamiya K, Huganir RL (2009) Regulation of AMPA receptor extrasynaptic insertion by 4.1N, phosphorylation and palmitoylation. Nat Neurosci 12:879–887. 10.1038/nn.2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano O, Smirnov S, Soni S, Golubtsov A, Guillemin I, Hotulainen P, Medina I, Nothwang HG, Rivera C, Ludwig A (2015) KCC2 regulates actin dynamics in dendritic spines via interaction with β-PIX. J Cell Biol 209:671–686. 10.1083/jcb.201411008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan V, Khademullah CS, Dargaei Z, Chevrier J, Uvarov P, Kwan J, Bagshaw RD, Pawson T, Emili A, De Koninck Y, Anggono V, Airaksinen M, Woodin MA (2017) Native KCC2 interactome reveals PACSIN1 as a critical regulator of synaptic inhibition. eLife 6:e28270. 10.7554/eLife.28270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markova O, Mukhtarov M, Real E, Jacob Y, Bregestovski P (2008) Genetically encoded chloride indicator with improved sensitivity. J Neurosci Methods 170:67–76. 10.1016/j.jneumeth.2007.12.016 [DOI] [PubMed] [Google Scholar]
- Medina I, Friedel P, Rivera C, Kahle KT, Kourdougli N, Uvarov P, Pellegrino C (2014) Current view on the functional regulation of the neuronal K(+)-Cl(-) cotransporter KCC2. Front Cell Neurosci 8:27. 10.3389/fncel.2014.00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U, Ahrens CH, Müller S, Wollscheid B (2014) Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30:884–886. 10.1093/bioinformatics/btt607 [DOI] [PubMed] [Google Scholar]
- Otsu Y, Donneger F, Schwartz EJ, Poncer JC (2020) Cation-chloride cotransporters and the polarity of GABA signalling in mouse hippocampal parvalbumin interneurons. J Physiol 598:1865–1880. 10.1113/JP279221 [DOI] [PubMed] [Google Scholar]
- Pallud J, Le Van Quyen M, Bielle F, Pellegrino C, Varlet P, Labussiere M, Cresto N, Dieme MJ, Baulac M, Duyckaerts C, Kourdougli N, Chazal G, Devaux B, Rivera C, Miles R, Capelle L, Huberfeld G (2014) Cortical GABAergic excitation contributes to epileptic activities around human glioma. Sci Transl Med 6:244ra289. 10.1126/scitranslmed.3008065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchietti F, Vascon S, Nieus T, Rosillo C, Das S, Tyagarajan SK, Diaspro A, Del Bue A, Petrini EM, Barberis A, Cella Zanacchi F (2017) Nanoscale molecular reorganization of the inhibitory postsynaptic density is a determinant of GABAergic synaptic potentiation. J Neurosci 37:1747–1756. 10.1523/JNEUROSCI.0514-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto JG, Jones DG, Murphy KM (2013) Comparing development of synaptic proteins in rat visual, somatosensory, and frontal cortex. Front Neural Circuits 7:97. 10.3389/fncir.2013.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressey JC, Mahadevan V, Khademullah CS, Dargaei Z, Chevrier J, Ye W, Huang M, Chauhan AK, Meas SJ, Uvarov P, Airaksinen MS, Woodin MA (2017) A kainate receptor subunit promotes the recycling of the neuron-specific K(+)-Cl(-) co-transporter KCC2 in hippocampal neurons. J Biol Chem 292:6190–6201. 10.1074/jbc.M116.767236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud F, Homburger V, Seveno M, Vigy O, Moutin E, Fagni L, Perroy J (2018) SNAP23-Kif5 complex controls mGlu1 receptor trafficking. J Mol Cell Biol 10:423–436. 10.1093/jmcb/mjy031 [DOI] [PubMed] [Google Scholar]
- Rivera C, Voipio J, Payne JA, Ruusuvuori E, Lahtinen H, Lamsa K, Pirvola U, Saarma M, Kaila K (1999) The K+/Cl- co-transporter KCC2 renders GABA hyperpolarizing during neuronal maturation. Nature 397:251–255. 10.1038/16697 [DOI] [PubMed] [Google Scholar]
- Rivera C, Voipio J, Thomas-Crusells J, Li H, Emri Z, Sipilä S, Payne JA, Minichiello L, Saarma M, Kaila K (2004) Mechanism of activity-dependent downregulation of the neuron-specific K-Cl cotransporter KCC2. J Neurosci 24:4683–4691. 10.1523/JNEUROSCI.5265-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussa E, Speer JM, Chudotvorova I, Khakipoor S, Smirnov S, Rivera C, Krieglstein K (2016) The membrane trafficking and functionality of the K+-Cl- co-transporter KCC2 is regulated by TGF-beta2. J Cell Sci 129:3485–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seja P, Schonewille M, Spitzmaul G, Badura A, Klein I, Rudhard Y, Wisden W, Hübner CA, De Zeeuw CI, Jentsch TJ (2012) Raising cytosolic Cl- in cerebellar granule cells affects their excitability and vestibulo-ocular learning. EMBO J 31:1217–1230. 10.1038/emboj.2011.488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thouvenot E, Urbach S, Dantec C, Poncet J, Séveno M, Demettre E, Jouin P, Touchon J, Bockaert J, Marin P (2008) Enhanced detection of CNS cell secretome in plasma protein-depleted cerebrospinal fluid. J Proteome Res 7:4409–4421. 10.1021/pr8003858 [DOI] [PubMed] [Google Scholar]
- Tyagarajan SK, Fritschy JM (2014) Gephyrin: a master regulator of neuronal function? Nat Rev Neurosci 15:141–156. 10.1038/nrn3670 [DOI] [PubMed] [Google Scholar]
- Tyzio R, Nardou R, Ferrari DC, Tsintsadze T, Shahrokhi A, Eftekhari S, Khalilov I, Tsintsadze V, Brouchoud C, Chazal G, Lemonnier E, Lozovaya N, Burnashev N, Ben-Ari Y (2014) Oxytocin-mediated GABA inhibition during delivery attenuates autism pathogenesis in rodent offspring. Science 343:675–679. 10.1126/science.1247190 [DOI] [PubMed] [Google Scholar]
- Uvarov P, Ludwig A, Markkanen M, Soni S, Hübner CA, Rivera C, Airaksinen MS (2009) Coexpression and heteromerization of two neuronal K-Cl cotransporter isoforms in neonatal brain. J Biol Chem 284:13696–13704. 10.1074/jbc.M807366200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen MA, Lacoh CM, Fiumelli H, Kosel M, Tyagarajan S, de Roo M, Vutskits L (2018) Development of inhibitory synaptic inputs on layer 2/3 pyramidal neurons in the rat medial prefrontal cortex. Brain Struct Funct 223:1999–2012. 10.1007/s00429-017-1602-0 [DOI] [PubMed] [Google Scholar]
- Virtanen MA, Uvarov P, Mavrovic M, Poncer JC, Kaila K (2021) The multifaceted roles of KCC2 in cortical development. Trends Neurosci 44:378–392. 10.1016/j.tins.2021.01.004 [DOI] [PubMed] [Google Scholar]
- Wan L, Chen L, Yu J, Wang G, Wu Z, Qian B, Liu X, Wang Y (2020) Coordinated downregulation of KCC2 and GABA(A) receptor contributes to inhibitory dysfunction during seizure induction. Biochem Biophys Res Commun 532:489–495. 10.1016/j.bbrc.2020.08.082 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Wake H, Moorhouse AJ, Nabekura J (2009) Clustering of neuronal K+-Cl- cotransporters in lipid rafts by tyrosine phosphorylation. J Biol Chem 284:27980–27988. 10.1074/jbc.M109.043620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodin MA, Ganguly K, Poo M-m (2003) Coincident pre- and postsynaptic activity modifies GABAergic synapses by postsynaptic changes in Cl− transporter activity. Neuron 39:807–820. 10.1016/s0896-6273(03)00507-5 [DOI] [PubMed] [Google Scholar]
- Wright R, Newey SE, Ilie A, Wefelmeyer W, Raimondo JV, Ginham R, McLlhinney RAJ, Akerman CJ (2017) Neuronal chloride regulation via KCC2 is modulated through a GABAB receptor protein complex. J Neurosci 37:5447–5462. 10.1523/JNEUROSCI.2164-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Jiang M, Miralles CP, Li RW, Chen G, de Blas AL (2007) Gephyrin clustering is required for the stability of GABAergic synapses. Mol Cell Neurosci 36:484–500. 10.1016/j.mcn.2007.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wong AY, Murshid A, Bowie D, Presley JF, Bedford FK (2008) Identification of a novel di-leucine motif mediating K(+)/Cl(-) cotransporter KCC2 constitutive endocytosis. Cell Signal 20:1769–1779. 10.1016/j.cellsig.2008.06.011 [DOI] [PubMed] [Google Scholar]


